Abstract
Computational protein-protein docking is one of the most intensively studied topics in structural bioinformatics. The field has made substantial progress through over three decades of development. The development began with methods for rigid-body docking of two proteins, which have now been extended in different directions to cover the various macromolecular interactions observed in a cell. Here, we overview the recent developments of the variations of docking methods, including multiple protein docking, peptide-protein docking, and disordered protein docking methods.
Keywords: protein docking, protein structure prediction, macromolecular complexes, structural bioinformatics
Introduction
Protein-protein interactions (PPIs) are involved in many essential biological processes in a cell. To understand the mechanisms of PPIs’ functions, tremendous efforts have been made to determine the tertiary structures of protein complexes using experimental methods. To supplement experimental approaches, computational methods have been developed that build predicted structures of protein-protein complexes. Computational prediction models do not always have an atomic-detailed accuracy; however, models with a lower accuracy are still able to provide useful information such as interface residues and overall docking orientation depending on the application. Furthermore, as we describe later, computational modeling and experiments can be combined.
Conventionally, the main development of protein docking methods is for modeling pairwise interaction of proteins that have more or less rigid structures. Although the focus of this article is to overview computational protein docking approaches that are outside of the conventional type, it would be appropriate to provide a quick summary of the conventional protein docking methods. Computational protein-protein docking methods can be roughly categorized into two types, template-based and ab initio (template-free) docking. Template-based docking approaches are increasingly applicable as experimentally determined complex structures have been rapidly accumulated in the Protein Data Bank (PDB) [1]. The algorithms range from those which use global templates [2–4] to those which use local interaction patterns of complexes [5–8].
In cases where no appropriate template is available, docking models can be constructed by an ab initio method, which constructs a pool of docking models by essentially exhaustively exploring candidate poses with different mutual rotation angles and translations. Then, a scoring function is used to select the most plausible models out of the pool. Various protein structure representations [9,10] and docking conformation search algorithms [11–13] were employed in ab initio methods. Scoring functions developed include those which are based on the statistics of atom interactions in known protein structures (statistical knowledge-based potentials), which give favorable scores to structure models that follow the distribution of atom pair distances and angles of native protein structures [14,15], scoring functions based on numerical optimization, which optimize parameters (energy values) so that near-native models can be selected [16], and re-parameterization of molecular mechanics force field [17]. There are also attempts to compute the binding free energy of protein docking models using molecular dynamics (MD) simulations [18,19]. Recently, 3D deep learning was applied for docking model selection [20]. A task that remains difficult in pairwise protein docking is to consider conformational changes of proteins that may occur upon docking. Methods used for generating docking conformations include Monte-Carlo (MC) approaches [21], elastic network models (ENMs) that consider proteins as mass-and-spring networks to simulate flexibility [13,22,23], and MD simulations [24,25].
The progress of docking methods has been periodically monitored by a community-wide protein docking experiment, the Critical Assessment of PRedicted Interactions (CAPRI). In the recent 7th edition of the community-wide CAPRI assessment [26], targets were not limited to pairwise interactions, but have included a variety of interesting modeling targets, including large multimeric protein complexes, protein-peptide complexes, artificial protein complexes, and protein-oligosaccharide interactions. Modeling a complex is easy if a known protein complex structure can be used as a template. Oppositely, the level of difficulty increases if an available template has significant difference with the target complex, or a suitable template for a complex is not available. Another general lesson from CAPRI is that biological information, such as known interface residues, can drastically reduce the possible docking poses and lead to correct modeling. In the 7th CAPRI assessment, there were eight protein-protein targets, among which five were dimers and three were multimers. Models of acceptable quality or better, were obtained for a total of six protein-protein complexes, three dimer and all three multimer targets. Participants were also able to model all three peptide targets and all five polysaccharide targets. Readers are referred to the recent assessment reports from the CAPRI organizers [26,27]. For more detailed information about recent protein docking development, refer to review articles [28,29].
Multiple protein docking
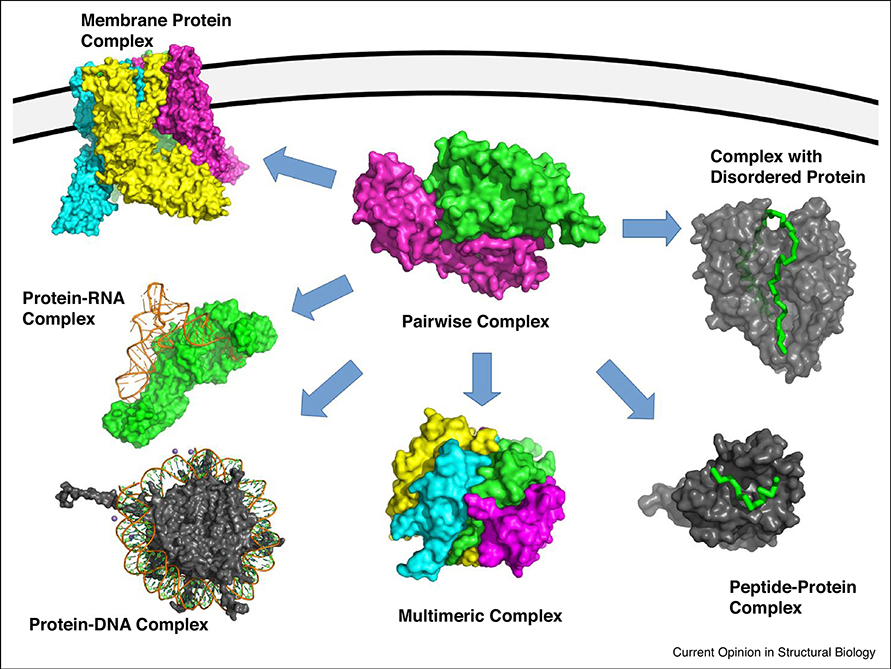
Starting from pairwise protein docking, docking methods have been expanded into several directions to reflect different types of interactions observed in a cell (Figure 1). One obvious need for an extension is methods for docking multiple subunits into a complex.
Figure 1.
Extending protein docking from pairwise to diverse categories. Methods for pairwise docking include LZerD ClusPro, SwarmDock, PatchDock, HADDOCK, and ZDOCK. For multiple docking, tools such as CombDock, Multi-LZerD, M-ZDOCK, SymmDock, and SAM are available. Membrane protein docking tools include Memdock and MPDock. Peptide-protein docking tools include PIPER-FlexPepDock CABS-Dock, HPepDock, AutoDock CrankPep, MDockpep, and pepATTRACT. For docking with a disordered protein, IDP-LZerD is available. Methods for docking a protein with DNA/RNA include FoldX, RnaX, HDOCK, and NPDock.
CombDock is a pioneering method that performs multiple protein docking [30]. The method first predicts pairwise interactions between subunits of a target protein complex using PatchDock [31]. Then, it performs a hierarchical combinatorial assembly of sub-complexes, starting from building sub-complex models of three subunits, then four subunits, until models of the full-subunit complex are build, by each time combining models of smaller sub-complexes. At each stage, top-scoring models are selected to reduce the number of possible combinations. Another multiple protein docking method, Multi-LZerD [32], also starts from pre-computed pairwise models of all possible subunit pairs using a pairwise docking method, LZerD [10]. In Multi-LZerD, these pairwise docking models are assembled into full-subunit models using a genetic algorithm (GA). The GA constructs a population of different models at each stage (called a generation), which are modified to produce a new set of models in the next generation. At each generation, a population of models (~200) is selected by a scoring function, and the procedure is iterated until convergence. The two methods are applicable to complexes with up to ~8 subunits but would face difficulty for larger complexes because of a combinatorially growing docking conformational space. Also, the strategy of using pairwise models as starting may not be appropriate for larger complexes where many subunit pairs do not tightly interact. For assembling larger proteins, extra information of interacting and non-interacting subunit pairs or interface residues from experiments would be helpful. Multi-LZerD was later extended to predict the assembly pathway of multimeric complexes (Path-LZerD) [33]. The complex building and optimization process by the GA in Multi-LZerD can be considered as a simulation of the assembly process, from which the assembly order can be observed.
CombDock and Multi-LZerD do not require additional constraints for modeling and are able to model asymmetric hetero complexes. On the other hand, model construction is significantly easier for homo-symmetric complexes. M-ZDOCK [34], SymmDock [35], and SAM [36] perform an exhaustive docking conformation search restricted to specified point group symmetries. The first two methods are for complexes with a symmetric symmetry (Cn), whereas SAM can also handle higher-order symmetries, dihedral (Dn), tetrahedral (T), octahedral (O), and icosahedral (I) groups.
Membrane protein docking
Application to membrane proteins is an important sub-category of docking as a relatively lower fraction of structures are available in PDB despite their biological importance. Membrane proteins are also important targets for drug discovery [37]. Docking of membrane proteins typically starts by placing two membrane proteins in the correct topology relative to the membrane. Then docking conformations are sampled under the constraints that interacting residues from two proteins are at a similar height relative to the membrane and that the two proteins keep more or less perpendicular orientation to the membrane. Memdock [38] takes this approach. Membrane proteins are initially oriented using the Orientations of Proteins in Membranes (OPM) database and the PPM server [39], which determines the spatial position of a protein in the membrane by optimizing the transfer free energy from water to the lipid bilayer [40]. Another method, MPDock [41], combines an MC algorithm implemented in RosettaDock [17] with a membrane energy function and conformational sampling that account for the membrane environment. A benchmark dataset for membrane protein docking consisting of 37 membrane proteins was collected by the Bonvin group [42], which encourages future developments of novel docking algorithms for membrane proteins.
Peptide & intrinsic disordered protein docking
Protein-peptide docking, another class of docking, is important since protein-peptide interactions are part of various signaling pathways, and also due to the emergence of peptide therapeutics [43]. Likely reflecting their importance in therapeutics application, several notable new developments have been recently observed. In this problem setting, flexibility cannot be readily neglected since detailed atomic interactions are usually of interest in peptide docking. In this category, we also consider assembly involving intrinsically disordered proteins (IDPs), since their flexibility needs to be explicitly modelled.
A general protocol for peptide-docking is to generate an ensemble of peptide conformations, dock them to a receptor protein, and finally refine selected docked models. Thus, methods can be characterized by examining the techniques used in each step. To generate the initial ensemble of peptide conformers, pepATTRACT [44] and HADDOCK [45] use three simple ideal conformations of peptides while MDockPep [46] uses a homology modeling method, MODELLER [47], to model conformations. HPepDock [48], PIPER-FlexPepDock [49], and IDP-LZerD sample peptide conformations from a fragment library. For the docking step, a common choice is to use rigid-body docking, as adopted by HPepDock, HADDOCK, pepATTRACT, PIPER-FlexDock, and IDP-LZerD [50]. Some methods perform the first two steps, peptide conformation generation and docking, simultaneously. MDockPep uses Autodock [51], a popular protein-small chemical molecule docking program, for flexible peptide docking. AutoDock CrankPep [52] is a recent addition to the AutoDock program series that is specifically designed for peptide docking. Rosetta FlexPepDock [53] starts from an extended peptide conformation placed at a known binding site and folds the peptide on the receptor protein surface using a library of 3- to 5-mer peptide fragments. Anchordock starts from a folded peptide conformation that is placed at a known binding residue on the receptor surface and then optimizes the docked conformation using MD [54]. CABS-dock [55] performs MC simulation of peptide conformation sampling and docks using a coarse-grained protein representation. Most of the methods perform a structure refinement step at the last stage in the protocol.
The peptide docking methods described here have been shown to be able to dock sequences at most 16 residues long ab initio. The exception is IDP-LZerD [50], which has been shown to successfully dock IDPs of up to 69 residues long. In IDP-LZerD, 30 peptide conformations are generated for each of the 9-residue long fragment windows slid along the sequence of an input IDP, which are then independently docked to a receptor. Then, docked peptides are selected on the receptor surface so that they can be connected to the full-length IDP.
DNA/RNA protein docking
As in the case of protein-protein docking, a template-based approach works well as long as an appropriate template structure of a protein-nucleic acid complex exists in PDB [56]. Other existing methods include the application of rigid-body docking programs [57–60] and scoring functions that specifically target protein-DNA/RNA complexes [57,61–63]. For nucleotide docking, modeling flexibility is very important as nucleotides tend to be more flexible than proteins. HADDOCK was combined with ENM-based conformer generation to quickly search the conformational space [64], although the degree of the conformational changes considered by ENM was small, 1 to 2 Å root mean square deviation (RMSD) to the initial structures.
Regarding the nucleotide flexibility, FoldX [65] and RnaX [66] are of particular note. These two methods use fragment pair libraries of double-stranded (ds) DNA and RNA, respectively, bound to protein fragments to generate plausible traces of the nucleic acid bound to the target protein. In FoldX, peptide-DNA fragments collected from PDB have a peptide length of 6–12 amino acids and a dsDNA length of 4–8 base pairs. To model a protein-DNA complex, peptide-DNA fragments are selected and connected along the protein by considering the sequence similarity and evaluating with a force field and geometrical constraints. Similarly, in RnaX, a library of protein-RNA fragments with 6 amino acids and RNA fragments of 4–8 nucleotides were used. Thus, in principle, the methods are able to model protein-nucleotide complexes in the absence of a clear template. FoldX and RnaX have been tested on complexes with fairly long dsDNAs and RNAs, which are over 190 bps and 150 bases, respectively.
Data-assisted protein docking
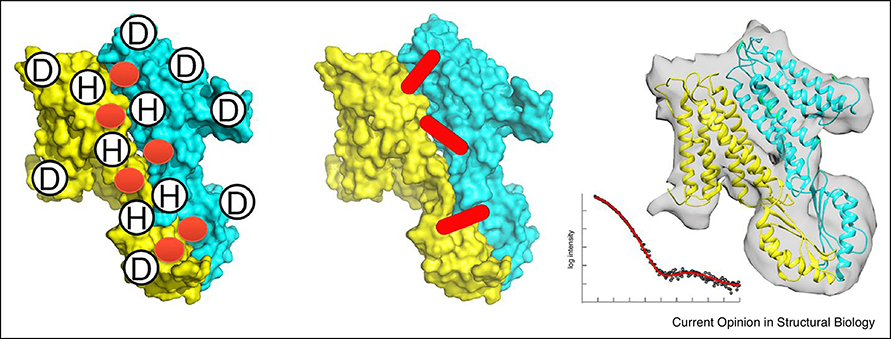
In the last section, we briefly overview methods that use additional information to guide docking. For more details of methods in this category, readers are referred to a recent review [67]. Roughly, types of experimental data can be classified into three categories, those which provides interface and surface residue information, residue and atom distance data, and data that indicate overall shape (Figure 2). Mutagenesis [68] and hydrogen/deuterium (H/D) exchange [69] are experiments that can provide information on binding site residues. On the other hand, XL-MS and FRET provide distance constraints by identifying cross-linked residues (XL-MS) [70] or a donor and an acceptor pair that transfer the energy (FRET) [71,72] between interacting proteins. NMR provides restraints of interface residues with Chemical Shift Perturbations (CSP), orientation with Residual Dipolar Couplings (RDCs), and distance information with Nuclear Overhauser Effects (NOEs) [73]. Interface residues and residue distances information can be readily input into protein docking web servers, such as ClusPro [12], HADDOCK [74], HDOCK [75], LZerD [76], PatchDock [31], and PyDock [77] servers as constraints.
Figure 2.
Three categories of experimental data that can assist protein docking. From left, data that indicate residues at the interface or other surface, which include data from mutagenesis, H/D exchange. Middle, residue/atom distance data, which can be provided by FRET, XL-MS, and NMR. Right, complex shape information, which can be provided by SAXS and EM.
Notably, the same types of information, interface residues and contacting residue pairs, can also be predicted from the protein sequence and structure and have been successfully used for guiding docking. Protein docking interface residues can be identified by mutation patterns [78], common interaction sites of ligand proteins [79], or combinations of residue features [80]. PI-LZerD [81] uses predicted interface residue information with an assumption that prediction may not be entirely correct. There is a rapid development of prediction methods for contacting residues and distances between residues in protein structures [82]. Such methods have also been applied for predicting contacts across two proteins for guiding protein docking [83–85].
SAXS and cryo-electron microscopy (cryo-EM) provide low-resolution shape information of macromolecular complexes. A SAXS profile of the complex is used as a filter to select docking models that agree with it [86–89]. In cryo-EM, although near-atomic resolution density maps are more highlighted recently, there are still many maps determined at a lower resolution. These low-resolution maps are still useful for guiding docking [90–92].
Docking methods, including HADDOCK [74] and IMP [67], integrate different sources of experimental information to improve the docking process.
Future direction
We have discussed how the development of protein docking, which started from pairwise protein docking, expanded into various directions to be able to construct and predict major types of molecular interactions that occur in a cell. An interesting future direction would be to model and simulate macromolecular interactions in large scale biomolecular systems in a crowded environment of a cell [93]. Recently, substantial efforts have been made in the dynamics simulation of proteins in a crowded environment [94]. Work has also begun on protein docking methods that explicitly consider interaction in a crowded environment [95]. Uniting structure modeling and dynamics simulations of such macromolecular interactions integrated with large-scale experiments [96,97] could yield a clearer and more realistic picture for understanding the behavior of molecules in a living cell.
Acknowledgements
This work was partly supported by the National Institutes of Health (R01GM123055), the National Science Foundation (DMS1614777, CMMI1825941, and MCB1925643) and the Purdue Institute of Drug Discovery. C.C. is supported by National Institute of General Medical Sciences-funded predoctoral fellowship to C.C. (T32 GM132024).
Footnotes
Conflict of Interest
The authors declare that there is no conflict of interest regarding the publication of this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE: The Protein Data Bank. Nucleic Acids Res 2000, 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen H, Skolnick J: M-TASSER: an algorithm for protein quaternary structure prediction. Biophys J 2008, 94:918–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mukherjee S, Zhang Y: Protein-protein complex structure predictions by multimeric threading and template recombination. Structure 2011, 19:955–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peterson LX, Shin WH, Kim H, Kihara D: Improved performance in CAPRI round 37 using LZerD docking and template-based modeling with combined scoring functions. Proteins 2018, 86 Suppl 1:311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aytuna AS, Gursoy A, Keskin O: Prediction of protein-protein interactions by combining structure and sequence conservation in protein interfaces. Bioinformatics 2005, 21:2850–2855. [DOI] [PubMed] [Google Scholar]
- 6.Gunther S, May P, Hoppe A, Frommel C, Preissner R: Docking without docking: ISEARCH--prediction of interactions using known interfaces. Proteins 2007, 69:839–844. [DOI] [PubMed] [Google Scholar]
- 7.Tuncbag N, Gursoy A, Nussinov R, Keskin O: Predicting protein-protein interactions on a proteome scale by matching evolutionary and structural similarities at interfaces using PRISM. Nat Protoc 2011, 6:1341–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kundrotas PJ, Vakser IA: Accuracy of protein-protein binding sites in high-throughput template-based modeling. PLoS Comput Biol 2010, 6:e1000727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaudhury S, Berrondo M, Weitzner BD, Muthu P, Bergman H, Gray JJ: Benchmarking and analysis of protein docking performance in Rosetta v3.2. PLoS One 2011, 6:e22477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venkatraman V, Yang YD, Sael L, Kihara D: Protein-protein docking using region-based 3D Zernike descriptors. BMC Bioinformatics 2009, 10:407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katchalski-Katzir E, Shariv I, Eisenstein M, Friesem AA, Aflalo C, Vakser IA: Molecular surface recognition: determination of geometric fit between proteins and their ligands by correlation techniques. Proceedings of the National Academy of Sciences 1992, 89:2195–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C, Beglov D, Vajda S: The ClusPro web server for protein-protein docking. Nat Protoc 2017, 12:255–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moal IH, Bates PA: SwarmDock and the use of normal modes in protein-protein docking. Int J Mol Sci 2010, 11:3623–3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang C, Liu S, Zhu Q, Zhou Y: A knowledge-based energy function for protein-ligand, protein-protein, and protein-DNA complexes. J Med Chem 2005, 48:2325–2335. [DOI] [PubMed] [Google Scholar]
- 15.Lu H, Lu L, Skolnick J: Development of unified statistical potentials describing protein-protein interactions. Biophys J 2003, 84:1895–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang SY, Zou X: An iterative knowledge-based scoring function for protein-protein recognition. Proteins 2008, 72:557–579. [DOI] [PubMed] [Google Scholar]
- 17.Gray JJ, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, Rohl CA, Baker D: Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J Mol Biol 2003, 331:281–299. [DOI] [PubMed] [Google Scholar]
- 18.Shinobu A, Takemura K, Matubayasi N, Kitao A: Refining evERdock: Improved selection of good protein-protein complex models achieved by MD optimization and use of multiple conformations. J Chem Phys 2018, 149:195101.* The method uses MD simulation of docking models in explicit water to evaluate the binding free energy as the sum of the conformational energy and the solvation free energy.
- 19.Kingsley LJ, Esquivel-Rodriguez J, Yang Y, Kihara D, Lill MA: Ranking protein-protein docking results using steered molecular dynamics and potential of mean force calculations. J Comput Chem 2016, 37:1861–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, Terashi G, Christoffer CW, Zhu M, Kihara D: Protein docking model evaluation by 3D deep convolutional neural networks. Bioinformatics 2020, 36:2113–2118.** This article describes 3D Deep Convolutional Neural Networks (3DCNN) for selecting protein docking models. Positions of atoms and interaction energy distribution at docking interface are considered as input structural features for the 3DCNN.
- 21.Marze NA, Roy Burman SS, Sheffler W, Gray JJ: Efficient flexible backbone protein-protein docking for challenging targets. Bioinformatics 2018, 34:3461–3469.* A recent development of RosettaDock. It uses three conformer generation methods in the Rosetta package to consider protein flexibilty in docking.
- 22.Roel-Touris J, Don CG, R VH, Rodrigues J, Bonvin A: Less Is More: Coarse-Grained Integrative Modeling of Large Biomolecular Assemblies with HADDOCK. J Chem Theory Comput 2019, 15:6358–6367.* A recent development of HADDOCK. It uses the MARTINI coarse-grained model to sample chain conformations in docking. The results of docking at the coarse-grained level is converted back to all-atom resolution using distance restraints.
- 23.Glashagen G, de Vries S, Uciechowska-Kaczmarzyk U, Samsonov SA, Murail S, Tuffery P,Zacharias M: Coarse-grained and atomic resolution biomolecular docking with the ATTRACT approach. Proteins 2019, 10 10.02/prot.25860. [DOI] [PubMed] [Google Scholar]
- 24.Plattner N, Doerr S, De Fabritiis G, Noe F: Complete protein-protein association kinetics in atomic detail revealed by molecular dynamics simulations and Markov modelling. Nat Chem 2017, 9:1005–1011. [DOI] [PubMed] [Google Scholar]
- 25.Pan AC, Jacobson D, Yatsenko K, Sritharan D, Weinreich TM, Shaw DE: Atomic-level characterization of protein-protein association. Proc Natl Acad Sci U S A 2019, 116:4244–4249.** Long-timescale (microseconds) molecular dynamics simulation coupled with enhanced sampling were performed on five protein complexes, starting from bound experimentally determined structures. Binding free energy and association-dissociation kinetics between monomeric chains of the protein complex were estimated.
- 26.Lensink MF, Nadzirin N, Velankar S, Wodak SJ: Modeling protein-protein, protein-peptide, and protein-oligosaccharide complexes: CAPRI 7th edition. Proteins 2019, 10 10.02/prot.25870.** This is the 7th CAPRI evaluation report. There were over 36,000 models of protein complexes submitted for evaluation by 57 groups, out of which 8 were protein-protein, 3 were protein-peptide, and 5 were protein-oligosaccharide targets.
- 27.Lensink MF, Brysbaert G, Nadzirin N, Velankar S, Chaleil RAG, Gerguri T, Bates PA, Laine E, Carbone A, Grudinin S, et al. : Blind prediction of homo- and hetero-protein complexes: The CASP13-CAPRI experiment. Proteins 2019, 87:1200–1221.** This is the evaluation report for the CASP13-CAPRI held in summer 2018. There were 20 targets in these round, out of which 14 were homo-oligomers and 6 were heterocomplexes. There are abundance of template and ab-initio targets.
- 28.Porter KA, Desta I, Kozakov D, Vajda S: What method to use for protein-protein docking? Curr Opin Struct Biol 2019, 55:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ritchie DW: Recent progress and future directions in protein-protein docking. Curr Protein Pept Sci 2008, 9:1–15. [DOI] [PubMed] [Google Scholar]
- 30.Inbar Y, Benyamini H, Nussinov R, Wolfson HJ: Prediction of multimolecular assemblies by multiple docking. J Mol Biol 2005, 349:435–447. [DOI] [PubMed] [Google Scholar]
- 31.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ: PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res 2005, 33:W363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esquivel-Rodriguez J, Yang YD, Kihara D: Multi-LZerD: Multiple protein docking for asymmetric complexes. Proteins 2012, 80:1818–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peterson LX, Togawa Y, Esquivel-Rodriguez J, Terashi G, Christoffer C, Roy A, Shin WH,Kihara D: Modeling the assembly order of multimeric heteroprotein complexes. PLoS Comput Biol 2018, 14:e1005937.** This article reports Path-LZerD, the first method for predicting the assemble order of multimeric protein complexes from a set of individual structures of component subunits. The method constructs complex models from subunit structures and the assembly order is predicted by comparing binding energies of subunit pairs in the complex.
- 34.Pierce B, Tong W, Weng Z: M-ZDOCK: a grid-based approach for Cn symmetric multimer docking. Bioinformatics 2005, 21:1472–1478. [DOI] [PubMed] [Google Scholar]
- 35.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ: Geometry-based flexible and symmetric protein docking. Proteins 2005, 60:224–231. [DOI] [PubMed] [Google Scholar]
- 36.Ritchie DW, Grudinin S: Spherical polar Fourier assembly of protein complexes with arbitrary point group symmetry. J Appl Crystallogr 2016, 49:158–167. [Google Scholar]
- 37.Rask-Andersen M, Masuram S, Schioth HB: The druggable genome: Evaluation of drug targets in clinical trials suggests major shifts in molecular class and indication. Annu Rev Pharmacol Toxicol 2014, 54:9–26. [DOI] [PubMed] [Google Scholar]
- 38.Hurwitz N, Schneidman-Duhovny D, Wolfson HJ: Memdock: an alpha-helical membrane protein docking algorithm. Bioinformatics 2016, 32:2444–2450. [DOI] [PubMed] [Google Scholar]
- 39.Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL: OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res 2012, 40:D370–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lomize AL, Pogozheva ID, Mosberg HI: Anisotropic solvent model of the lipid bilayer. 2. Energetics of insertion of small molecules, peptides, and proteins in membranes. J Chem Inf Model 2011, 51:930–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alford RF, Koehler Leman J, Weitzner BD, Duran AM, Tilley DC, Elazar A, Gray JJ: An Integrated Framework Advancing Membrane Protein Modeling and Design. PLoS Comput Biol 2015, 11:e1004398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koukos PI, Faro I, van Noort CW, Bonvin A: A Membrane Protein Complex Docking Benchmark. J Mol Biol 2018, 430:5246–5256.* This paper provides a dataset for membrane protein docking with modeling results with HADDOCK.
- 43.Lau JL, Dunn MK: Therapeutic peptides: Historical perspectives, current development trends, and future directions. Bioorg Med Chem 2018, 26:2700–2707. [DOI] [PubMed] [Google Scholar]
- 44.Schindler CEM, de Vries SJ, Zacharias M: Fully Blind Peptide-Protein Docking with pepATTRACT. Structure 2015, 23:1507–1515. [DOI] [PubMed] [Google Scholar]
- 45.Trellet M, Melquiond AS, Bonvin AM: A unified conformational selection and induced fit approach to protein-peptide docking. PLoS One 2013, 8:e58769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan C, Xu X, Zou X: Fully Blind Docking at the Atomic Level for Protein-Peptide Complex Structure Prediction. Structure 2016, 24:1842–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Webb B, Sali A: Protein Structure Modeling with MODELLER. Methods Mol Biol 2017, 1654:39–54. [DOI] [PubMed] [Google Scholar]
- 48.Zhou P, Jin B, Li H, Huang SY: HPEPDOCK: a web server for blind peptide-protein docking based on a hierarchical algorithm. Nucleic Acids Res 2018, 46:W443–W450.* A new web server by the Huang group who is recently active in developing docking methods.
- 49.Alam N, Goldstein O, Xia B, Porter KA, Kozakov D, Schueler-Furman O: High-resolution global peptide-protein docking using fragments-based PIPER-FlexPepDock. PLoS Comput Biol 2017, 13:e1005905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peterson LX, Roy A, Christoffer C, Terashi G, Kihara D: Modeling disordered protein interactions from biophysical principles. PLoS Comput Biol 2017, 13:e1005485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trott O, Olson AJ: AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010, 31:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y, Sanner MF: AutoDock CrankPep: combining folding and docking to predict protein-peptide complexes. Bioinformatics 2019, 35:5121–5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khramushin A, Marcu O, Alam N, Shimony O, Padhorny D, Brini E, Dill KA, Vajda S,Kozakov D, Schueler-Furman O: Modeling beta-sheet peptide-protein interactions: Rosetta FlexPepDock in CAPRI rounds 38–45. Proteins 2019, 10 10.02/prot.25871.* This article discusses the Rosetta FlexPepDock pipeline and its performance in the recentCAPRI rounds 38–45. In particular, the authors achieved the top modeling performance on the peptide targets, and note the role of beta sheet continuation.
- 54.Slutzki M, Ben-Shimon A, Niv MY: AnchorDock for Blind Flexible Docking of Peptides to Proteins. Methods Mol Biol 2017, 1561:95–108. [DOI] [PubMed] [Google Scholar]
- 55.Kurcinski M, Jamroz M, Blaszczyk M, Kolinski A, Kmiecik S: CABS-dock web server for the flexible docking of peptides to proteins without prior knowledge of the binding site. Nucleic Acids Res 2015, 43:W419–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng J, Kundrotas PJ, Vakser IA, Liu S: Template-Based Modeling of Protein-RNA Interactions. PLoS Comput Biol 2016, 12:e1005120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perez-Cano L, Romero-Durana M, Fernandez-Recio J: Structural and energy determinants in protein-RNA docking. Methods 2017, 118–119: 163–170. [DOI] [PubMed] [Google Scholar]
- 58.Roberts VA, Pique ME, Ten Eyck LF, Li S: Predicting protein-DNA interactions by full search computational docking. Proteins 2013, 81:2106–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yan Y, Wen Z, Wang X, Huang SY: Addressing recent docking challenges: A hybrid strategy to integrate template-based and free protein-protein docking. Proteins 2017, 85:497–512. [DOI] [PubMed] [Google Scholar]
- 60.Tuszynska I, Magnus M, Jonak K, Dawson W, Bujnicki JM: NPDock: a web server for protein-nucleic acid docking. Nucleic Acids Res 2015, 43:W425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang SY, Zou X: A knowledge-based scoring function for protein-RNA interactions derived from a statistical mechanics-based iterative method. Nucleic Acids Res 2014, 42:e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tuszynska I, Bujnicki JM: DARS-RNP and QUASI-RNP: new statistical potentials for protein-RNA docking. BMC Bioinformatics 2011, 12:348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Setny P, Bahadur RP, Zacharias M: Protein-DNA docking with a coarse-grained force field. BMC Bioinformatics 2012, 13:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kurkcuoglu Z, Bonvin A: Pre- and post-docking sampling of conformational changes using ClustENM and HADDOCK for protein-protein and protein-DNA systems. Proteins 2020, 88:292–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blanco JD, Radusky L, Climente-Gonzalez H, Serrano L: FoldX accurate structural protein-DNA binding prediction using PADA1 (Protein Assisted DNA Assembly 1). Nucleic Acids Res 2018, 46:3852–3863.* This article describes FoldX, a method for modeling protein-DNA interaction byconsidering protein-DNA fragment pairs. In particular, based on a large library of fragment pairs collected from structures solved by experiment, FoldX predicts DNA-binding regions and interacting DNA sequences.
- 66.Delgado Blanco J, Radusky LG, Cianferoni D, Serrano L: Protein-assisted RNA fragment docking (RnaX) for modeling RNA-protein interactions using ModelX. Proc Natl Acad Sci U S A 2019, 116:24568–24573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koukos PI, Bonvin A: Integrative Modelling of Biomolecular Complexes. J Mol Biol 2019, 10.1016/j.jmb.2019.11.009.** A comprehensive review on integrative and data-driven computational approaches for bimolecular structure modeling. It covers types of data used for modeling as well as advancements in the modeling field.
- 68.Chelliah V, Blundell TL, Fernandez-Recio J: Efficient restraints for protein-protein docking by comparison of observed amino acid substitution patterns with those predicted from local environment. J Mol Biol 2006, 357:1669–1682. [DOI] [PubMed] [Google Scholar]
- 69.Zhang MM, Beno BR, Huang RY, Adhikari J, Deyanova EG, Li J, Chen G, Gross ML: An Integrated Approach for Determining a Protein-Protein Binding Interface in Solution and an Evaluation of Hydrogen-Deuterium Exchange Kinetics for Adjudicating Candidate Docking Models. Anal Chem 2019, 91:15709–15717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mintseris J, Gygi SP: High-density chemical cross-linking for modeling protein interactions. Proc Natl Acad Sci U S A 2020, 117:93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brunger AT, Strop P, Vrljic M, Chu S, Weninger KR: Three-dimensional molecular modeling with single molecule FRET. J Struct Biol 2011, 173:497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bonomi M, Pellarin R, Kim SJ, Russel D, Sundin BA, Riffle M, Jaschob D, Ramsden R, Davis TN, Muller EG, et al. : Determining protein complex structures based on a Bayesian model of in vivo Forster resonance energy transfer (FRET) data. Mol Cell Proteomics 2014, 13:2812–2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van Dijk AD, Fushman D, Bonvin AM: Various strategies of using residual dipolar couplings in NMR-driven protein docking: application to Lys48-linked di-ubiquitin and validation against 15N-relaxation data. Proteins 2005, 60:367–381. [DOI] [PubMed] [Google Scholar]
- 74.de Vries SJ, van Dijk M, Bonvin AM: The HADDOCK web server for data-driven biomolecular docking. Nat Protoc 2010, 5:883–897. [DOI] [PubMed] [Google Scholar]
- 75.Yan Y, Zhang D, Zhou P, Li B, Huang SY: HDOCK: a web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res 2017, 45:W365–W373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Esquivel-Rodriguez J, Filos-Gonzalez V, Li B, Kihara D: Pairwise and multimeric protein-protein docking using the LZerD program suite. Methods Mol Biol 2014, 1137:209–234. [DOI] [PubMed] [Google Scholar]
- 77.Cheng TMK, Blundell TL, Fernandez-Recio J: pyDock: Electrostatics and desolvation for effective scoring of rigid body protein–protein docking. Proteins: Structure, Function, and Bioinformatics 2007, 68:503–515. [DOI] [PubMed] [Google Scholar]
- 78.La D, Kihara D: A novel method for protein-protein interaction site prediction using phylogenetic substitution models. Proteins 2012, 80:126–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Viswanathan R, Fajardo E, Steinberg G, Haller M, Fiser A: Protein-protein binding supersites. PLoS Comput Biol 2019, 15:e1006704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Qin S, Zhou HX: meta-PPISP: a meta web server for protein-protein interaction site prediction. Bioinformatics 2007, 23:3386–3387. [DOI] [PubMed] [Google Scholar]
- 81.Li B, Kihara D: Protein docking prediction using predicted protein-protein interface. BMC Bioinformatics 2012, 13:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shrestha R, Fajardo E, Gil N, Fidelis K, Kryshtafovych A, Monastyrskyy B, Fiser A: Assessing the accuracy of contact predictions in CASP13. Proteins 2019, 10.1002/prot.25819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ovchinnikov S, Kamisetty H, Baker D: Robust and accurate prediction of residue-residue interactions across protein interfaces using evolutionary information. Elife 2014, 3:e02030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zeng H, Wang S, Zhou T, Zhao F, Li X, Wu Q, Xu J: ComplexContact: a web server for inter-protein contact prediction using deep learning. Nucleic Acids Res 2018, 46:W432–W437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hopf TA, Scharfe CP, Rodrigues JP, Green AG, Kohlbacher O, Sander C, Bonvin AM, MarksDS: Sequence co-evolution gives 3D contacts and structures of protein complexes. Elife 2014, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ignatov M, Kazennov A, Kozakov D: ClusPro FMFT-SAXS: Ultra-fast Filtering Using Small-Angle X-ray Scattering Data in Protein Docking. J Mol Biol 2018, 430:2249–2255. [DOI] [PubMed] [Google Scholar]
- 87.Schindler CEM, de Vries SJ, Sasse A, Zacharias M: SAXS Data Alone can Generate High-Quality Models of Protein-Protein Complexes. Structure 2016, 24:1387–1397. [DOI] [PubMed] [Google Scholar]
- 88.Pons C, D’Abramo M, Svergun DI, Orozco M, Bernado P, Fernandez-Recio J: Structural characterization of protein-protein complexes by integrating computational docking with small-angle scattering data. J Mol Biol 2010, 403:217–230. [DOI] [PubMed] [Google Scholar]
- 89.Schneidman-Duhovny D, Hammel M, Sali A: Macromolecular docking restrained by a small angle X-ray scattering profile. J Struct Biol 2011, 173:461–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pandurangan AP, Vasishtan D, Alber F, Topf M: gamma-TEMPy: Simultaneous Fitting of Components in 3D-EM Maps of Their Assembly Using a Genetic Algorithm. Structure 2015, 23:2365–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Esquivel-Rodriguez J, Kihara D: Fitting Multimeric Protein Complexes into Electron Microscopy Maps Using 3D Zernike Descriptors. J Phys Chem B 2012, 116:6854–6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lasker K, Sali A, Wolfson HJ: Determining macromolecular assembly structures by molecular docking and fitting into an electron density map. Proteins 2010, 78:3205–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vakser IA, Deeds EJ: Computational approaches to macromolecular interactions in the cell. Curr Opin Struct Biol 2019, 55:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nawrocki G, Karaboga A, Sugita Y, Feig M: Effect of protein-protein interactions and solvent viscosity on the rotational diffusion of proteins in crowded environments. Phys Chem Chem Phys 2019, 21:876–883.* This article presents a methodology to determine diffusive properties of proteins incrowded environments using molecular dynamics simulation. The crowding environment model is represented as a concentrated villin headpiece solution.
- 95.Li X, Moal IH, Bates PA: Detection and refinement of encounter complexes for protein-protein docking: taking account of macromolecular crowding. Proteins 2010, 78:3189–3196. [DOI] [PubMed] [Google Scholar]
- 96.Luchinat E, Banci L: In-cell NMR: a topical review. IUCrJ 2017, 4:108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sustarsic M, Kapanidis AN: Taking the ruler to the jungle: single-molecule FRET for understanding biomolecular structure and dynamics in live cells. Curr Opin Struct Biol 2015, 34:52–59. [DOI] [PubMed] [Google Scholar]