Abstract
Three-dimensional microelectrode arrays (3D MEAs) have emerged as promising tools to detect electrical activities of tissues or organs in vitro and in vivo, but challenges in achieving fast, accurate, and versatile monitoring have consistently hampered further advances in analyzing cell or tissue behaviors. In this review, we discuss emerging 3D MEA technologies for in vitro recording of cardiac and neural cellular electrophysiology, as well as in vivo applications for heart and brain health diagnosis and therapeutics. We first review various forms of recent 3D MEAs for in vitro studies in context of their geometry, materials, and fabrication processes as well as recent demonstrations of 3D MEAs to monitor electromechanical behaviors of cardiomyocytes and neurons. We then present recent advances in 3D MEAs for in vivo applications to the heart and the brain for monitoring of health conditions and stimulation for therapy. A brief overview of the current challenges and future directions of 3D MEAs are provided to conclude the review.
Keywords: 3D MEAs, Cardiac Interface, Neuronal Interfaces, Cellular electrophysiology
1. Introduction
The functions of human cells, tissues, and organs have been a major area of pursuit for study in medicine and biological sciences to understand the physiology of the human body. Electrophysiology, in particular, has become intriguing towards this goal because it enables accurate, real-time collection of information which characterizes the inception of extracellular field potentials, as well as for potential propagation from extracellular field potential recordings (Gomes et al., 2016). However, the interpretation of these extracellular properties remains a constant challenge due to several confounding factors and technical limitations (Herreras, 2016). The development of microelectrode arrays (MEAs) have not only demonstrated prospects for in vitro recording of extracellular field potentials of cardiac (Connolly et al., 1990; Dai et al., 2016; Thomasjr et al., 1972) and neural cell behaviors (Borkholder et al., 1997; Gross et al., 1982; Regehr et al., 1989), but also in vivo studies by implantation in biological tissues for real-time mapping, monitoring of organ activity, and electrically/optically/thermally stimulated therapy (Chen et al., 2019; Choi et al., 2019; Lee and Kim, 2019). Conventionally, extracellular potential measurement systems are conducted with the 2D planar electrodes underneath a tissue or slice (Jones et al., 2011; Stett et al., 2003). This system vitally consists of the tissue, and the interface between the tissue and the electrodes, and the substrate embedding microelectrodes (Spira and Hai, 2013).
A major limitation in developing MEAs has been finding appropriate ways to improve their performances with high sensitivity and higher signal to noise ratio (SNR) during electrical recording and stimulation. Thus far, much effort has been devoted to addressing sensitivity and SNR drawbacks for enhanced MEA performances. Adjustments include changing electrode material composition for reduced impedance of the electrode (Kireev et al., 2017), electrode geometry to make effective contact interfaces between the cells and electrodes (Santoro et al., 2014; Weidlich et al., 2017), and electrode topography to increase interfacial surface area (Choi et al., 2020; Smith et al., 2020). Investigation in the aforementioned aspects has recently led the transition from traditional two-dimensional (2D) electrodes to 3D electrodes. This is attributed to greater surface area of the 3D electrodes, which decreases the electrode impedance and enhances signal-to-noise ratio relative to planar electrodes. The ongoing advances in 3D MEAs have brought about promising solutions in versatile electrophysiological assays based on alterations of electrical activity measurement for in vitro and in vivo studies of complex tissue or organs.
In this review, we will discuss emerging trends of in vitro and in vivo 3D MEA systems designed for cardiac and neural monitoring and therapy. A variety of designs, materials, and fabrication methods of electrodes will first be introduced, followed by how these parameters can be controlled or utilized for intracellular/extracellular recordings for in vitro analysis of cardiac or neural cells. Next, we describe recent development in in vivo 3D MEAs, which are attached to the heart or brain for diagnosis or clinical therapy. Future investigations in the material and design of 3D MEAs, along with innovative approaches to comply with the size, shape, and geometry of cells or tissues are needed for extensive analysis of their electrochemistry, mechanics, and genetics. Overcoming current challenges in 3D MEAs may potentially result in playing a key role in in vitro high-throughput screening assays for disease modeling, drug screening, or in vivo personalized healthcare monitoring systems.
2. 3D Microelectrode Array for in vitro Applications
In recent years, studies of electrical activities within cardiac and neural cells have yielded novel designs of 3D MEAs. Sensing changes in the action potential provides better understanding of heart or brain tissue behavior, as well as in vitro differences between diseased and malfunctioning organ tissues. One strategy to reduce the impedance of electrodes, achieve higher signal-to-noise ratio and selectivity has been manipulating the geometry of 3D electrodes and materials defining the electrode architecture. Here, we discuss recent methods that have been reported in order to design 3D MEAs for in vitro analysis of electrical activity of cardiomyocytes and neurons.
2.1. Material, Design, and Fabrication of in vitro 3D MEAs
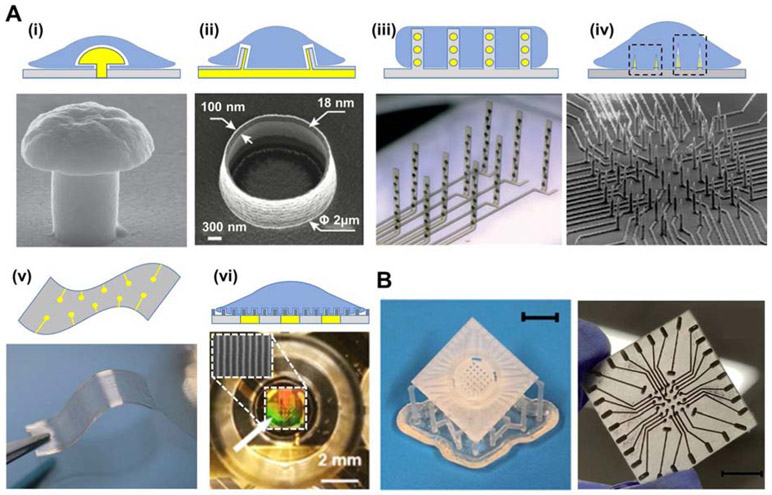
To date, numerous 3D MEAs have been presented with variances in their materials, design, and fabrication methods. The surface modification, material composition, and processing parameters of electrodes are widely adopted approaches to enhance the MEAs. Figure 1A illustrates the representative forms of 3D electrode/tissue interfaces. As shown in Figure 1A(i), mushroom-shaped electrodes are able to enhance the electrical coupling between the neurons and electrodes (Santoro et al., 2014; Weidlich et al., 2017). The void between the extracellular membrane and substrate may result in relatively weak electrical coupling, which interferes with action potential propagation along with reduced signal-to-noise ratio. To yield such architectures, silicon or glass substrates are coated with a Cr/Au layer deposited via sputter deposition technique. This layer is photolithographically defined for traces, bond pads and circular microelectrode sites and these features are etched into the wafer. Further, another spin-coated layer of a sacrificial photoresist (PR) layer is defined on the wafer. Photolithography is subsequently performed on this sacrificial PR layer for the formation of microscale circular arrays. Thick film processes such as electroplating are used to define a thick gold layer on top of the sputter coated circular microelectrode sites on the wafer surface, and finally the sacrificial photoresist layer is removed (Fendyur and Spira, 2012; Hai et al., 2009). Moreover, volcano-shaped electrodes surrounded by cellular membrane have been demonstrated to acquire higher action potential signals compared to flat electrodes (see Figure 1A(ii) (Cools et al., 2017; Desbiolles et al., 2019). This particular structure provides access to intracellular electrophysiology of neonatal cardiomyocyte. The architecture of these 3D electrodes is produced based on local redeposition during ion beam etching to pattern cavities onto the PR layer, followed by photolithography for multilayering of walls for the volcano structure (Desbiolles et al., 2019). Figure 1 A(iii) depicts flexible 3D microelectrode probe arrays which provide interfacing with the neural network in vitro. The hinge regions in the probe may deform and allow the electrode probes to stand upright during electromechanical monitoring. Unlike well-established 2D cell culture, each probe consists of eight integrated electrodes which may non-invasively cover the 3D brain tissue for effective interaction (Soscia et al., 2020). As shown in Figure 1A(iv), vertically standing nanowire electrodes have become a widely adopted configuration for extra- or intracellular recordings, independent electrical addressability, high-throughput measurements, and high signal to noise ratio (Abbott et al., 2017; Charvet et al., 2010; Dipalo et al., 2017; Lin et al., 2017; Liu et al., 2017; Sohee Kim et al., 2007; Xie et al., 2012). Other applications for these nanowire electrodes include extracellular recording upon contact with outer regions of the cell membrane (see Figure 1A(iv), left dotted square) and intracellular recording upon penetration of the 3D electrodes into the cell membrane (see Figure 1A(iv), right dotted square). A flexible and porous substrate consisting of electrode arrays and a 3D scaffold has also been proposed to record extracellular electrical signals of cardiac cells (see Figure 1A(v)) (Feiner et al., 2016). The porous constructs are designed to have minimal contact with the tissues, allowing nutrients to circulate and promote cell maturation as well as tissue growth. The 3D MEAs having mechanical properties similar to those of cellular environment, may be rolled or folded with the cell sheets in order to replicate the structure of biological tissues. Furthermore, MEAs with ion conductive, ion permeable nanotopographic surfaces have been applied not only to increase interactive surface area, which guide mass ion transfers generated from the cells to electrodes, but also to provide maturation, organization, and alignment of the cardiac or neuron cells (see Figure 1A (vi) (Choi et al., 2020; Smith et al., 2020). They confirmed that higher ion conductive materials show effective ion transfers to the electrodes, allowing better extracellular recordings without signal loss during the electrical coupling between cells to electrodes (Choi et al., 2020). Such an electrode platform is fabricated through multiple steps of photolithography, metal deposition, and etching. Recently, numerous studies have attempted to utilize 3D printing to develop MEAs with hopes to replace conventional fabrication process. Kundu et. al. (2019) exhibited a combined process which involves 3D printing to produce non-planar MEA designs (Figure 1B). This approach consists of simple steps of 3D printing, casting of conductive ink, electroplating, laminated/spun cast insulation and laser micromachining. However, processes which involve 3D printing, relative to traditional photolithographic methods, are relatively fast, customizable, facile, cost effective, and do not require complex procedures for packaging (Kundu et al., 2019, 2018). In fact, in these techniques instead of being a separate methodology, packaging can be co-designed and cofabricated with the device due to the enormous flexibility in 3D printing. Recently, Kundu et al. (2020) have even demonstrated a proof of concept 6-well and 12-well 3D MEAs with a selfinsulated approach depicting possibilities for rapidly integrating 3D MEAs in multiwell plate formats for integration with standard robotic and high-throughput electrophysiology instruments. Didier et. al. (2020) investigated stretchable and flexible MEA designs with 3D micro-serpentine architectures fabricated via micro-stereolithographic 3D printing. In addition, Azim et. al. (2019) demonstrated 3D microscale towers fabricated by sequential 3D printing, metal deposition, and lamination to insulate the traces. In comparison to most lithographic approaches involving complicated steps, 3D printing enables simplicity and reduced processes and time for the microfabrication and packaging of custom 3D microelectrodes though long-term performance and technological comparisons of these devices with traditional lithography-based devices are at an early stage of evaluation.
Figure 1. Recent advances in 3D microelectrode arrays interface configuration and fabrication.
(A) Representative forms of the 3D electrode/tissue interface. Top panel depict schematic illustrations of the 3D electrodes, while bottom panels exhibit the real images of the 3D electrodes. The 3D electrode/tissue interfaces include (i) mushroom-shaped (Reproduced with permission from Nanotechnology (2017) 28, 095302. Copyright 2017 Nanotechnology) (Weidlich et al., 2017), (ii) volcano-shaped (Reproduced with permission from Nano letter (2019) 19, 6173. Copyright 2019 Nano letter) (Desbiolles et al., 2019), and (iii) flexible standing substrate embedding electrodes. (Reproduced with permission from Lab Chip (2020) 20, 901. Copyright 2020 Lab Chip) (Soscia et al., 2020). (iv) Needle type electrodes with two distinct recording types, namely extracellular (left dotted square) and intracellular (right dotted square). (Reproduced with permission from Nano Lett. (2017) 17, 2757–2764. Copyright 2017 Nano Lett.) (Liu et al., 2017). (v) Freestanding, flexible substrate consisting of a porous network and gold electrodes. (Reproduced with permission from Nature Materials (2016) 15, 679. Copyright 2016 Nature Materials) (Feiner et al., 2016). (vi) SEM image of a nanotopographic substrate (top) applied to a single well of MEA plate (bottom) (Smith et al., 2020). (B) Photographic images of 3D printed MEA (left), which undergo ink casting, pulsed electroplating, lamination, and laser micromachining to produce the final 3D electrode design (right). Scale bar for left image corresponds to 10 mm. (Kundu et al., 2019).
2.2. in vitro Applications of 3D MEAs
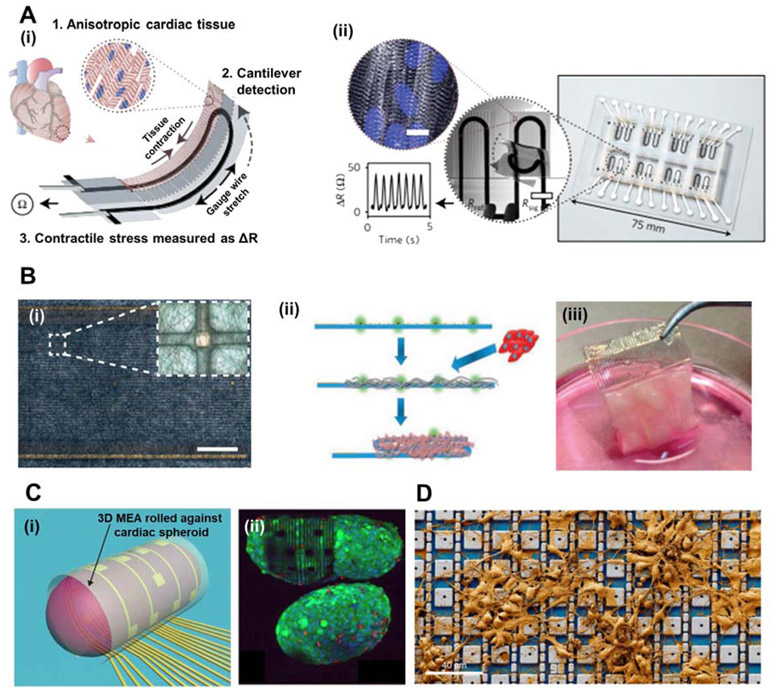
With recent advances in 3D MEA designs, a plethora of studies have demonstrated novel strategies of detecting cardiac/neuronal electrophysiological signals (Athanasiadis et al., 2020; Heuschkel et al., 2018; Manuel et al., 2016; Musick et al., 2009; Tomaskovic-Crook et al., 2019). Figure 2 exhibits 3D MEAs for in vitro applications to understand the electromechanical behaviors of cardiac and neuronal cells and tissues. Conventional planar 2D MEAs are composed of rigid surfaces, which are not appropriate to conform and measure repeated cellular dynamics of contraction and relaxation. Such movements of cardiac/neural cells and tissues may result in the loss of electrical coupling between the cell membrane and electrode surface. Lind et. al. (2017) utilized a 3D printing approach to develop a soft cantilever substrate array which contains flexible multilayers of a base substrate, a strain gauge sensor, and a tissue-guiding layer (see Figure 2A(i)). Throughout repeated contractions of the laminar cardiac tissue on the cantilevers, the strain gauge sensors rolled up simultaneously and responsively to such movements. The tissue-guiding layer not only aids in attaching the cardiac tissues to the substrate, but also recapitulates the aligned cardiac architectures in vitro. As shown in Figure 2A(ii), the device presents a proportional relation between the resistance of strain sensors and the contractile stress of the laminar tissue; this is contrary to many former works which analyze field potential duration for measurement of cardiac contraction period. Figure 2B depicts flexible and freestanding cardiac patches which include arrays of electrodes integrated with polycaprolactone–gelatin nanofibers to sense the electrical activity of cardiac tissues (Feiner et al., 2016). Depicted in Figure 2B(i) these biocompatible nanofibers are designed to cover the electrode arrays and provide in vivo-like microenvironments for cardiac tissue growth and structural organization. Meanwhile, the electrode array is periodically embedded throughout the flexible substrate, enabling signal propagation measurements of the engineered cardiac tissue. The scaffold on the surface of the MEA provides a substrate for cardiac cell plating to create a thick, foldable 3D cardiac patch (see Figure 2B(ii)). Photographic image of the device verifies such phenomena, displaying the thick, folded 3D cardiac patch after 7 days of growth (see Figure 2B(iii)).
Figure 2. 3D MEAs for in vitro applications in cardiac and neuron tissue engineered constructs.
(A) (i) Schematic illustration of a soft, flexible cantilever substrate array which contains multilayers of a base substrate, a strain gauge sensor, and a tissue-guiding layer. (ii) Deflection of the cantilever substrate upon contraction of the anisotropic engineered cardiac tissue, generating resistance change proportional to the contractile stress of the tissue. (Reproduced with permission from Nature Materials (2017) 16, 303. Copyright 2017 Nature Materials) (Lind et al., 2017). (B) (i) Optical microscopy (OM) image of nanocomposite fibres of polycaprolactone-gelatin weaved and deposited onto an electronic mesh. Scale bar, 1.5 mm. Inset shows a single sensing/stimulating electrode covered with the nanofibre scaffold. (ii) Schematic illustration outlining the maturation of cardiac cells within the electrospun fibres that are deposited onto the electronic mesh. (iii) Photographic image of the folded microECP after 7 days of cultivation with cardiac cells. (Reproduced with permission from Nature Materials (2016) 15, 679. Copyright 2016 Nature Materials) (Feiner et al., 2016). (C) (i) Cardiac spheroid encapsulated in a 3D rolling MEA, allowing electrical mapping and signal propagation in 3D. (ii) Fluorescent staining image depicting the live/dead assay of the cardiac spheroid encapsulated by the MEA. (Reproduced with permission from Science Advances (2019) 5, eaax0729. Copyright 2019 Science Advances) (Kalmykov et al., 2019). (D) A CMOS-activated PtB electrode array with cultured neurons. (Reproduced with permission from Nature Biomedical Engineering (2020) 4, 232–24. Copyright 2020 from Nature Biomedical Engineering) (Abbott et al., 2020).
Figure 2C(i) illustrates a 3D self-rollable biosensor array which encapsulates 3D cardiac spheroids for 3D mapping and propagation of electrical signals (Kalmykov et al., 2019). 3D cardiac spheroids have been viewed favorably over 2D reconstitutions because they can better recapitulate the structure and microenvironment of native, in vivo human tissues and cellular organizations. As shown in fluorescence image in Figure 2C(ii) the self-rolled electrode array provides improved contact interface between the cardiac spheroid surface and the 2D electrodes. Multisite, simultaneous measurements of electrophysiological signals from this 3D multicellular system is also demonstrated. This MEA system may potentially enable testing of drug effects on spheroids, as well as comparison between diseased and healthy phenotypes.
Figure 2D represents a complementary metal-oxide semiconductor (CMOS)-activated electrode array which connects to cultured neuronal networks and records intracellular electrical signals (Abbott et al., 2020, 2017). The platinum-black (PtB) electrode array consists of 4,096 platinum-black electrodes with nanoscale roughness. Nanotopography of the PtB electrode design provides increases in surface area and lowering of impedance that enhances the SNR of each of the 4,096 electrodes. This yields not only an enhancement of cell-to-amplifier signal transfer for higher recording sensitivity, but also a decrease in the possible gas bubble generation during the electrode current injection. Prevention of bubble formation is critical because they may disrupt the membrane-electrode interface and electrical coupling. Another advantage of the PtB nanoscale roughness, beside reduced impedance, is its physical role in tightening and stabilizing its seal with the cell membrane (Dipalo et al., 2017; Hai et al., 2010; Lin et al., 2014; Liu et al., 2017).
3. 3D Microelectrode Arrays for in vivo Applications
Recent developments of 3D MEA systems have also opened up opportunities for these devices to be applied to the heart and brain, which are recognized as two of the most vital components of the human body (Hong and Lieber, 2019; Whyte et al., 2018). Malignances between the two organs have recently drawn much attention, as they make up most of the illnesses that occur among the human population (Khodagholy et al., 2013; Mendis et al., 2011). Providing amplified interfacial contact area, sensitivity, and adaptability against the physical complexities of the heart and the brain, 3D MEAs have become attractive technologies to provide stable, accurate, and real-time propagation of signals which provide vital information regarding the electrophysiology of organs (Fattahi et al., 2014; Kim et al., 2016). From here, this review will cover recent advances in 3D MEAs which can monitor health conditions of the heart and the brain in vivo.
3.1. 3D MEAs for in vivo heart applications
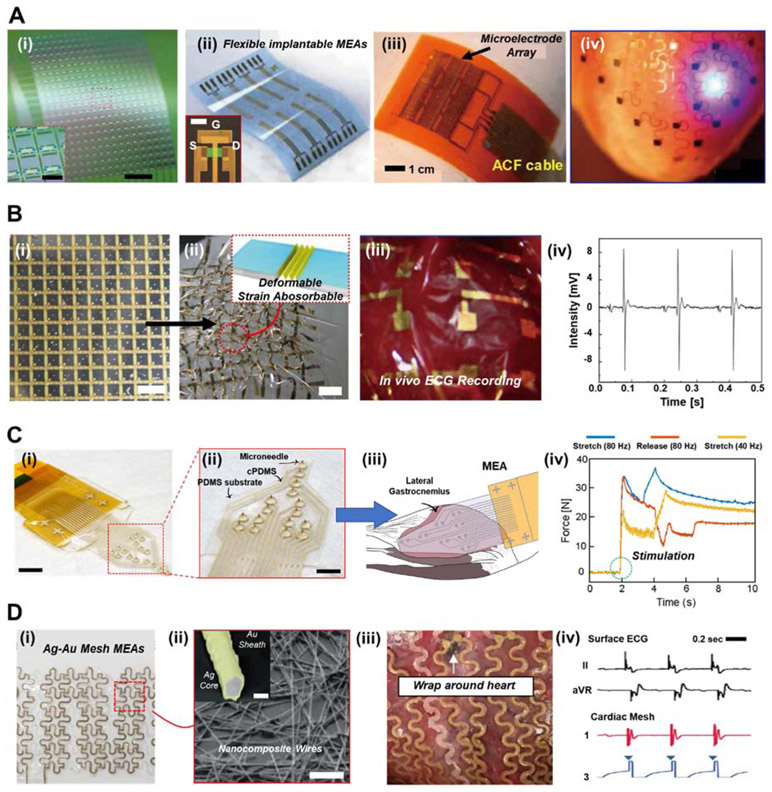
Cardiovascular diseases are a leading cause of morbidity and mortality around the globe, which accounts for a total death toll as high as ~ 40% (Hong et al., 2019; Nie et al., 2018; Wu et al., 2017). In order to alleviate the severity of cardiovascular diseases, the demand for developing healthcare technologies which could continuously monitor heart conditions has progressively increased. This is because cardiovascular diseases are easier to treat at earlier stages, and are easily interpreted using patterns of electrocardiograms (ECGs), blood pressure, and blood oxygen saturation (Khan et al., 2016). In recent years, soft implantable MEA devices have become attractive to provide real-time monitoring of heart conditions, as well as stimulation for therapeutic purposes. Figure 3A illustrates a variety of MEA systems that have been presented for in vivo heart applications. As shown in Figure 3A(i-iii), many of these MEAs are fabricated on 2D, flexible planar substrates (Fang et al., 2017), which enable the attachment of the device against the curvatures of the organ. Photolithographic processes are then utilized in order to deposit layers of electrodes, insulation, sensors, and/or field-effect transistors (FETs) onto these substrates. Figure 3A(iv) depicts a mesh-type MEA which wraps around a heart for in vivo clinical therapy (Xu et al., 2014). This device exhibits one of the earliest demonstrations of bioelectronics which enabled bulk metallic electrodes to be assembled into fabric-like devices, providing relative stretchability and organ conformability. Despite remarkable performance of these developments, the heart is a complex organ with irregular surface morphologies and ultrasoft material properties (Ghista et al., 1975; Le Ven et al., 2016). Drawbacks such as limited coverage of 2D MEA interfaces, as well as detection inaccuracy, instability, etc. hamper their utilization in future healthcare applications (Hong et al., 2019).
Figure 3. 3D MEA systems for in vivo cardiac applications.
(A) (i) Photograph of nanomembrane transistors for high-resolution electrophysiological mapping. Scale bar, 2 mm. Inset shows magnified image of the nodes. Inset scale bar, 500 mm. (Reproduced with permission from Nature Biomedical Engineering (2017) 1, 0038. Copyright 2017 Nature Biomedical Engineering) (Fang et al., 2017). (ii) Ultra-thin flexible MEA system. Inset shows OM image of a single transistor structure. (Reproduced with permission from Advanced Functional Materials (2018) 28, 1702284. Copyright 2018 Advanced Functional Materials.) (Song et al., 2018) (iii) Integrated patch-type MEA device for in vivo heart diagnosis. (Reproduced with permission from Proc. Natl. Acad. Sci. (2014) 111, 1927-1932. Copyright 2014 Proc. Natl. Acad. Sci.) (Dagdeviren et al., 2014) (iv) Photograph of Multifunctional mesh-type bioelectronics attached to a rabbit epicardial surface. (Reproduced with permission from Nature Communications (2014) 5, 3329. Copyright 2014 Nature Communications) (Xu et al., 2014). (B) (i) Photograph of the adhesive gel MEAs with 3D strain-absorbing designs within an active matrix. Scale bar, 1 cm. (ii) Photograph of the MEA under severe compressive strain (100%). Inset shows structure of the MEAs during deformation for stress absorbance. Scale bar, 1 cm. (iii) Photograph of the adhesive gel MEAs attached to a rodent heart for in vivo monitoring of electrocardiogram (ECG) signals. (iv) Representative measurement of ECG signals of rodent heart in vivo. (Reproduced with permission from Nature Communications (2014) 5, 5898. Copyright 2014 Nature Communications) (Lee et al., 2014). (C) (i) Photograph of the MEA with 3D arrowhead-shaped microneedles. Scale bar, 8 mm. (ii) Magnified photograph of the MEA with microneedles, conductive PDMS (cPDMS), and PDMS substrate. Scale bar, 5 mm. (iii) Schematic illustration of electrical stimulation and measurement of EMG signals with the microneedle MEA against feline’s lateral gastrocnemius muscle. (iv) Real-time in vivo EMG measurements of the microneedle MEA with varying stimulation frequencies or stretch/release displacements. (Reproduced with permission from IEEE Transactions on Neural Systems and Rehabilitation Engineering (2017) 25, 1440–1452. Copyright 2017 IEEE Transactions on Neural Systems and Rehabilitation Engineering) (Guvanasen et al., 2017). (D) (i) Photograph of the mesh-type MEA with Ag-Au nanocomposites. (ii) Scanning electron microscopy (SEM) image of the Ag-Au nanocomposites wires. Scale bar, 2mm. Inset shows backscattered electron (BSE) image of the Ag core and Au sheath of a single wire. Inset scale bar, 100 nm. (iii) Representative photograph of the 3D MEA wrapping around the pig heart. (iv) In vivo ECG measurements of the mesh-type MEA applied to pig heart during cardiac pacing. (Reproduced with permission from Nature Nanotechnology (2019) 28, 095302. Copyright 2019 Nature Nanotechnology) (Choi et al., 2018)
In order to overcome such limitations, 3D MEA systems have been highly promising. For instance, Lee et al. developed an ultra-flexible adhesive gel MEA with 3D strain resistant structures for in vivo monitoring of the heart (Lee et al., 2014). As shown in Figure 3B(i), the MEA is composed of an active matrix of 12 × 12 organic transistors coupled with adhesive gel electrodes composed of poly(vinyl alcohol) (PVA) dispersed in a polyrotaxane matrix. Under severe compressive strain (~ 100%), the 3D wrinkled structures of the MEA are able to absorb the strain (see Figure. 3B(ii)), which provide stable electrical measurements even under large deformations. The adhesive gel-coated MEA was able to maintain conformal contact upon implantation against a rodent heart, as shown in Figure 3B(iii). Ultra-flexible, electromechanically stable properties of the MEA enabled real-time detection of electrical signals from the ventricle surface, exhibiting all the P, Q, R, S, T and U waves necessary to understand cardiac health (see Figure. 3B(iv)).
3D MEAs with more direct interfacing with the heart tissue surface have shown to enable accurate detection of their subtle biosignals. Guvanasen et al. developed a stretchable MEA with 3D microneedles that can to stimulate and measure the electrical activity of muscle across multiple sites (see Figure 3C(i)) (Guvanasen et al., 2017). Fabricated through laser micromachining and photochemically milling, the microneedles are in the form of 3D arrowhead shapes which facilitate effective adherence of the electrode to the muscle tissue. As shown in Figure 3C(ii), the microneedle electrode array was assembled with traces of conductive PDMS (cPDMS) onto a PDMS substrate to provide stretchability up to ~ 63% strain. As Figure 3C(iii) depicts, the MEA is designed to cover the entire posterior surface of a feline’s lateral gastrocnemius muscle, to enable stimulation and measurement of that muscle’s electromyographic (EMG) activity. During in vivo applications, the device was able to maintain stable connection with the moving muscle, and provide real-time detection of EMG signals for varying frequencies of electrical stimulation and stretch/release displacements, as shown in Figure 3C(iv).
Various soft cardiac devices, particularly in the form of meshes and sleeves, have become attractive, because they enable MEAs to be deformable and conform with organ morphology, while providing vital information about the electrophysiology of the heart (Park et al., 2016; Roche et al., 2017). Choi et al. developed multifunctional bioelectronics composed of Ag-Au MEAs that could wrap around the epicardial surface for 3D diagnostics of a pig heart (see Fig. 3C(i)) (Choi et al., 2018). Shown in Fig. 3C(ii), the Ag-Au composite nanowires are dispersed throughout the MEA, fabricated via galvanic-free deposition of Au onto the Ag nanowires. Through such a process, Au cations selectively bind onto the Ag surface, consequently lowering the reduction potential of Au. The cardiac mesh-type MEA conformably wraps around the surface of the pig heart (see Figure 3C(iii)), and effectively measures ECG waveforms throughout with continuous electrical stimulation. The bioelectronic device demonstrates simultaneous cardiac monitoring and acts as a pacemaker, demonstrating next-generation 3D MEAs with both diagnostic and therapeutic functionalities.
3.2. 3D MEAs for in vivo brain applications
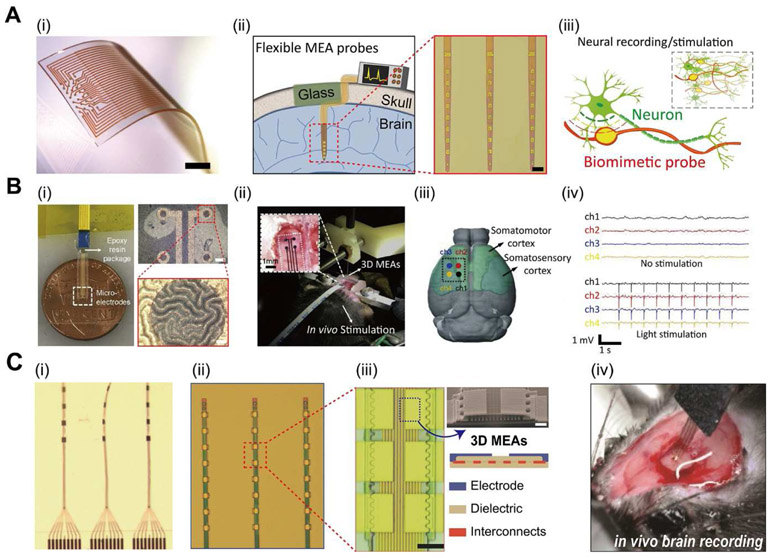
For many years, researchers have been intrigued in developing neuromodulatory techniques which may effectively treat patients suffering from a broad range of neurodegenerative diseases, including Parkinson’s disease, Alzheimer’s disease, and stroke (Acarón Ledesma et al., 2019; Chen et al., 2017; Won et al., 2020). Capabilities for modulating such types of neural functions, especially chemical and electrical signal propagation in neurons, axons, and dendrites have exhibited potential solutions to address neurological health conditions (Koo et al., 2018; Lee et al., 2019). To date, a plethora of in vivo MEA technologies which could monitor disordered neural activities have been produced. As shown in Figure 3A, stretchable MEA membranes with 3D electrodes composed of Pt-coated Au-TiO2 nanowires (i) (Tybrandt et al., 2018), flexible probes for brain implantation (ii) (Luan et al., 2017), and bioelectronics with neuron-like structural or mechanical features (Yang et al., 2019), have all become promising tools for 3D mapping and electrophysiological studies of the neural network.
MEAs on planar substrates, soft patches or membranes, may be laminated adaptably onto the surface of the cerebral cortex, enabling measurement of electrocorticography (ECoG) waveforms (Buzsáki et al., 2012). To demonstrate such capabilities, Ji et al. produced flexible bioelectrodes with 3D wrinkled MEA structures for in vivo brain signal recordings and neuromodulation (Ji et al., 2019). The 3D wrinkled MEAs were fabricated via oil extraction from an elastic substrate, followed by deposition of the Cr/Au electrode layer. Then, poly (3,4ethylenedioxythiophene): polystyrene sulfonate (PEDOT:PSS) and platinum black (Pt-black) are electroplated onto the wrinkled MEA sites. As shown in Figure 4C(i), microscale wrinkles are populated on the surface of the MEAs, which enhance active interfacial area for superior electrochemical properties relative to flat, 2D MEAs. The 3D MEA was then applied to a rodent brain in vivo to exhibit simultaneous functions of optical stimulation and ECoG measurements (see Figure 4C(ii)). Herein, the device was placed specifically onto the surface of somatomotor and somatosensory cortex to compare changes in ECoG signals with/without stimulation (see Figure 4C(iii)). Figure 4C(iv) depicts the advantages of the 3D structures, as heightened cortexdevice interactions achieved clear collection of low-noise ECoG signals as well as the lightsensitive spikes.
Figure 4. 3D MEAs for in vivo neuronal applications.
(A) (i) Photograph of a stretchable MEA device on a PDMS substrate with 3D electrodes composed of Pt-coated Au-TiO2 nanowires. Scale bar, 1mm. (Reproduced with permission from Advanced Materials (2018) 30, 1706520. Copyright 2018 Advanced Materials) (Tybrandt et al., 2018) (ii) Ultraflexible probes for electrical recording and imaging during brain implantation. Inset shows OM image of the probes with microelectrodes. Inset scale bar, 100 mm. (Reproduced with permission from Science Advances (2017) 3, e1601966. Copyright 2017 Science Advances) (Luan et al., 2017) (iii) Schematic illustration of neuron-like electronics for three-dimensional in vivo mapping and electrophysiology studies. (Reproduced with permission from Nature Materials (2019) 18, 510–517. Copyright 2019 Nature Materials) (Yang et al., 2019). (B) (i) Flexible MEAs with PEDOT:PSS 3D wrinkle microstructures. Inset shows magnified OM images of the wrinkles. (Top right) Inset scale bar, 100mm. (Bottom right) Inset scale bar, 10mm. (ii) Photograph of the MEA connected to a rodent brain for in vivo optical stimulation. Inset shows attachment of the MEA to the rodent brain. (iii) Schematic illustration of the electrode position (black line; dashed) with four channels across the somatomotor and somatosensory cortices of the brain. (iv) ECoG signals recorded by the 3D MEA before and after light stimulation. (Reproduced with permission from Bionsensors and Bioelectronics (2019) 135, 181–191. Copyright 2019 Bionsensors and Bioelectronics) (Ji et al., 2019). (C) (i) Optical microscope (OM) image of the nanostructured ultraflexible MEAs. (ii) Magnified image of the linear electrode arrays. (iii) Close-up images of the microelectrode arrays. Scale bar, 10 mm. (Top right) SEM image of the 3D electrode structure. Inset scale bar, 2 mm. (Bottom right) Schematic illustration of the electrode nanostructure. (iv) Photographic image of the implanted MEA for intracortical recordings. (Reproduced with permission from Advanced Science (2018) 5, 1700625. Copyright 2018 Advanced Science) (Wei et al., 2018).
Moreover, MEAs deposited on neural probes may be implanted deep into the brain, which could detect local field potential of single neurons and/or neuron clusters (Kozai et al., 2015; Kringelbach et al., 2007). For example, Wei et al. presented implantable neural probes with electrode arrays composed of 3D microarchitectures for high-density intracortical recording (Wei et al., 2018). As shown in Figure 4B(i), the probes are ultra-flexible to conform with deep brain tissue, and fabricated via photolithography of the flexible probe substrate and electron beam deposition of metallic electrodes. Figure 4B(ii) displays a magnified view of the device, which contains linear array of microelectrodes layered in 3D architectures for improved detection and sorting fidelity of single-unit action potentials (see Figure 4B(iii). Shown in Figure 4B(iv), the ultra-flexible 3D MEA probes were then implanted deep beneath the brain surface, which provides real-time electrophysiological recordings and neuronal circuitry mapping.
4. Perspective and Future Directions
Despite the vast progress in 3D MEAs in vitro or in vivo purposes, their capabilities to effectively monitor the electrophysiology of 3D engineered tissue or anatomical complexity of tissues and organs are still highly limited. The 3D MEAs covered throughout this paper are mostly 2D+ or 2.5D based electrodes. This is because current MEAs are built upwards starting from a flat substrate using photolithography (see Table 1). Moreover, most of the findings throughout this review are based on planar sheets of cells, but in vivo cellular structures and microenvironments are much more complex than such in vitro representations. An emerging trend in tissue engineering is the development of tissue organoids and spheroids, which can mimic the 3D, non-flat morphologies and structures of organ-specific tissues (Cakir et al., 2019; Nugraha et al., 2018; Richards et al., 2020; Voges et al., 2017). However, these 3D cellular and tissue models do not have consistent sizes, shapes, or structures of cells for extended analysis of their biochemistry, genetics, and electrophysiology. MEA systems and devices which can measure vital signals in these 3D cell compositions also remain underdeveloped. An early demonstration of 3D electrical mapping of cardiac spheroids was recently presented, but such organ-on-a-chip cannot provide high-throughput assay to understand alterations of diseased cells and their responses to drugs (Kalmykov et al., 2019). Hence, 3D MEAs capable of stable, accurate, and high-throughput diagnosis of cardiac and/or neural cells for in vitro studies are desperately needed. Recently, one of the authors groups at the University of Central Florida has taken steps to utilize 3D printing and 3D metal-based approaches which have the potential to be truly 3D architectures for in vitro applications and in collaboration with the other authors at Johns Hopkins University are currently characterizing these devices (Azim et al., 2019; Didier et al., 2020; Kundu et al., 2020).
Table 1.
Various Types of 3D MEAs for in vitro and in vivo applications
| Type of 3D MEA | Fabrication Method | Materials | Geometry / Layout / Features | Ref | |
|---|---|---|---|---|---|
| In vitro | Cardiac |
|
|
|
(Desbiol les et al., 2019) |
|
|
|
(Feiner et al. 2016) | ||
|
|
|
(Lind et al., 2017) | ||
| Neural |
|
|
|
(Soscia et al., 2020) | |
|
|
|
(Liu et al., 2017) | ||
|
|
|
(Abbott et al., 2020) | ||
| In vivo | Heart |
|
|
|
(Lee et al., 2014) |
|
|
|
(Guvana sen et al., 2017) | ||
|
|
|
(Choi et al., 2018) | ||
| Brain |
|
|
|
(Tybran dt et al. 2018) | |
|
|
|
(Ji et al., 2019) | ||
|
|
|
(Luan et al., 2017) | ||
|
|
|
(Wei et al., 2018) | ||
|
|
|
(Yang et al., 2019) | ||
3D MEA systems for in vivo applications are also hampered by poor interaction with the applied biological tissues. This is because human organs, particularly the heart and brain, are very soft, rough, irregularly shaped, and move constantly in rhythmic motions. These properties could interfere with the device conformability to the bio-surface, and interfere with signal propagation for stable, real-time monitoring of health conditions (Rogers et al., 2020; Song et al., 2019). Conventional adhesives composed of cyanoacrylate chemicals have been widely used for stable attachment to engaged tissues. Yet, cytotoxicity, surface contamination, and brittleness upon hardening by contact with water make such adhesives ineffective (Li et al., 2017). In order to enhance interfacing of the MEA device to the organ, novel designs of biocompatible glues such as mussel-inspired catechol, proteins, and other biomaterials have been highly favorable due to their robustness and surface adaptability (Lee et al., 2007; Li and Mooney, 2016; Pagel and Beck-Sickinger, 2017). Their integration to the electrode array matrices, composed of soft, elastic, biocompatible/biodegradable, and or hybrid materials remains as the next step to further developments of in vivo 3D MEAs (Han et al., 2018; Yang and Suo, 2018; Yuk et al., 2019).
To develop such complex 3D MEA systems for both in vitro and in vivo studies, there has been increasing demand in finding novel fabrication methods, which involve top-down, bottomup, or synergetic techniques which provide both tissue-compliant and electromechanically stable properties. Advanced 3D printing of MEAs has become a promising tool to realize this technology, attributed to facile, effective production as well as printing capabilities of a variety of materials (Chortos et al., 2020; Skylar-Scott et al., 2019; Tetsuka and Shin, 2020). However, low resolution of inkjet printing in 3D MEAs, difficulty in mass production, and challenges in layering different materials are major limitations that need to be addressed in this field but offer promising opportunities for materials and device designers. With rapid improvements in 3D printing techniques and apparatus, 3D printing of MEAs may eventually be established as a fundamental platform for in vitro and in vivo applications (Kim et al., 2019; Lin et al., 2019; Valentine et al., 2017).
5. Conclusion
3D MEAs have become a versatile technology to understand human physiology at both the cellular and the organ level. We have described recent advances in 3D MEA devices which can be used in vitro to understand the behaviors of cardiomyocytes or neurons, as well as implanted in vivo on the heart or the brain to diagnose their health conditions and provide therapeutic benefits. A variety of designs, materials, and fabrication methods of electrodes are introduced throughout this review, followed by how these parameters can be controlled or utilized for intracellular/extracellular recordings for in vitro analysis of cardiac or neural cells. Moreover, recent development in in vivo 3D MEAs was also demonstrated, which are attached to the heart or brain for diagnosis or clinical therapy. With further advances in 3D fabrication methods, materials, and structural design, 3D MEAs could open more opportunities for high-throughput screening assays in drug development or disease modeling, as well as real-time health monitoring for physiological complexity of tissues and organs for personalized medicine and clinical therapy.
Highlights.
This manuscript reviews the state-of-the-art technologies of three-dimensional microelectrode arrays for in vitro analyses of cardiac and neural cell electrophysiology
Structural, material properties and fabrication of 3D MEAs for in vitro applications are detailed.
Recent development in in vivo 3D MEAs for the heart or brain diagnosis or clinical therapy are discussed
Recent developments and demonstrations of 3D MEAs for in vitro cardiac/neural cell monitoring are reviewed.
Recent advances in 3D MEAs for in vivo applications for signal detection and stimulation are delineated.
Acknowledgments
This work was supported by National Institutes of Health grants R01 HL135143 and R44 HL131169 (to D.-H.K.). This research was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health and Welfare, Republic of Korea (grant number HI19C0642). (to D.-H.K.). This research was financially supported by the Ministry of Trade, Industry and Energy (MOTIE) and Korea Institute for Advancement of Technology (KIAT) through the International Cooperative R&D program. (Project No. P0004638)
Footnotes
Conflict of Interest
The authors declare the following competing financial interest(s): Deok-Ho Kim is a scientific founder and equity holder of Curi Bio. Swaminathan Rajaraman is a co-founder and equity holder in Axion BioSystems Inc. and an equity holder in BioCircuit Technologies.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Abbott J, Ye T, Krenek K, Gertner RS, Ban S, Kim Y, Qin L, Wu W, Park H, Ham D, 2020. Nat. Biomed. Eng 4, 232–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott J, Ye T, Qin L, Jorgolli M, Gertner RS, Ham D, Park H, 2017. Nat. Nanotechnol 12, 460–466. [DOI] [PubMed] [Google Scholar]
- Acarón Ledesma H, Li X, Carvalho-de-Souza JL, Wei W, Bezanilla F, Tian B, 2019. Nat. Nanotechnol 14, 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiadis M, Afanasenkau D, Derks W, Tondera C, Murganti F, Busskamp V, Bergmann O, Minev IR, 2020. npj Flex. Electron 4, 16. [Google Scholar]
- Azim N, Kundu A, Royse M, Li Sip YY, Young M, Santra S, Zhai L, Rajaraman S, 2019. J. Microelectromechanical Syst 28, 606–618. [Google Scholar]
- Borkholder D., Bao J, Maluf N, Perl E, Kovacs GT, 1997. J. Neurosci. Methods 77, 61–66. [DOI] [PubMed] [Google Scholar]
- Buzsáki G, Anastassiou CA, Koch C, 2012. Nat. Rev. Neurosci 13, 407–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakir B, Xiang Y, Tanaka Y, Kural MH, Parent M, Kang Y-J, Chapeton K, Patterson B, Yuan Y, He C-S, Raredon MSB, Dengelegi J, Kim K-Y, Sun P, Zhong M, Lee S, Patra P, Hyder F, Niklason LE, Lee S-H, Yoon Y-S, Park I-H, 2019. Nat. Methods 16, 1169–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvet G, Rousseau L, Billoint O, Gharbi S, Rostaing J-P, Joucla S, Trevisiol M, Bourgerette A, Chauvet P, Moulin C, 2010. Biosens. Bioelectron 25, 1889–1896. [DOI] [PubMed] [Google Scholar]
- Chen R, Canales A, Anikeeva P, 2017. Nat. Rev. Mater 2, 16093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Rogers JA, Lacour SP, Hu W, Kim D-H, 2019. Chem. Soc. Rev 48, 1431–1433. [DOI] [PubMed] [Google Scholar]
- Choi C, Lee Y, Cho KW, Koo JH, Kim D-H, 2019. Acc. Chem. Res 52, 73–81. [DOI] [PubMed] [Google Scholar]
- Choi JS, Smith AST, Williams NP, Matsubara T, Choi M, Kim J, Kim HJ, Choi S, Kim D, 2020. Adv. Funct. Mater 1910660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Han SI, Jung D, Hwang HJ, Lim C, Bae S, Park OK, Tschabrunn CM, Lee M, Bae SY, Yu JW, Ryu JH, Lee S-W, Park K, Kang PM, Lee WB, Nezafat R, Hyeon T, Kim D-H, 2018. Nat. Nanotechnol 13, 1048–1056. [DOI] [PubMed] [Google Scholar]
- Chortos A, Hajiesmaili E, Morales J, Clarke DR, Lewis JA, 2020. Adv. Funct. Mater 30,1907375. [Google Scholar]
- Connolly P, Clark P, Curtis ASG, Dow JAT, Wilkinson CDW, 1990. Biosens. Bioelectron 5, 223–234. [DOI] [PubMed] [Google Scholar]
- Cools J, Copic D, Luo Z, Callewaert G, Braeken D, De Volder M, 2017. Adv. Funct. Mater 27, 1701083. [Google Scholar]
- Dagdeviren C, Yang BD, Su Y, Tran PL, Joe P, Anderson E, Xia J, Doraiswamy V, Dehdashti B, Feng X, Lu B, Poston R, Khalpey Z, Ghaffari R, Huang Y, Slepian MJ, Rogers JA, 2014. Proc. Natl. Acad. Sci 111, 1927–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Zhou W, Gao T, Liu J, Lieber CM, 2016. Nat. Nanotechnol 11, 776–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbiolles BXE, de Coulon E, Bertsch A, Rohr S, Renaud P, 2019. Nano Lett. 19, 6173–6181. [DOI] [PubMed] [Google Scholar]
- Didier C, Kundu A, Rajaraman S, 2020. Microsystems Nanoeng. 6, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dipalo M, Amin H, Lovato L, Moia F, Caprettini V, Messina GC, Tantussi F,Berdondini L, De Angelis F, 2017. Nano Lett. 17, 3932–3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H, Yu KJ, Gloschat C, Yang Z, Song E, Chiang C-H, Zhao J, Won SM, Xu S, Trumpis M, Zhong Y, Han SW, Xue Y, Xu D, Choi SW, Cauwenberghs G, Kay M, Huang Y, Viventi J, Efimov IR, Rogers JA, 2017. Nat. Biomed. Eng 1, 0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattahi P, Yang G, Kim G, Abidian MR, 2014. Adv. Mater 26, 1846–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiner R, Engel L, Fleischer S, Malki M, Gal I, Shapira A, Shacham-Diamand Y, Dvir T, 2016. Nat. Mater 15, 679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendyur A, Spira ME, 2012. Front. Neuroeng 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghista DN, Vayo WH, Sandler H, 1975. Med. Biol. Eng 13, 151–161. [DOI] [PubMed] [Google Scholar]
- Gomes J-M, Bédard C, Valtcheva S, Nelson M, Khokhlova V, Pouget P, Venance L, Bal T, Destexhe A, 2016. Biophys. J 110, 234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross GW, Williams AN, Lucas JH, 1982. J. Neurosci. Methods 5, 13–22. [DOI] [PubMed] [Google Scholar]
- Guvanasen GS, Guo L, Aguilar RJ, Cheek AL, Shafor CS, Rajaraman S, Nichols TR, DeWeerth SP, 2017. IEEE Trans. Neural Syst. Rehabil. Eng 25, 1440–1452. [DOI] [PubMed] [Google Scholar]
- Hai A, Dormann A, Shappir J, Yitzchaik S, Bartic C, Borghs G, Langedijk JPM, Spira ME, 2009. J. R. Soc. Interface 6, 1153–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai A, Shappir J, Spira ME, 2010. Nat. Methods 7, 200–202. [DOI] [PubMed] [Google Scholar]
- Han L, Liu K, Wang M, Wang K, Fang L, Chen H, Zhou J, Lu X, 2018. Adv. Funct. Mater 28, 1704195. [Google Scholar]
- Herreras O, 2016. Front. Neural Circuits 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuschkel M, Mor F, Stoppini L, 2018. Front. Cell. Neurosci 12. [Google Scholar]
- Hong G, Lieber CM, 2019. Nat. Rev. Neurosci 20, 330–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong YJ, Jeong H, Cho KW, Lu N, Kim D, 2019. Adv. Funct. Mater 29, 1808247. [Google Scholar]
- Ji B, Wang M, Ge C, Xie Z, Guo Z, Hong W, Gu X, Wang L, Yi Z, Jiang C, Yang B, Wang X, Li X, Li C, Liu J, 2019. Biosens. Bioelectron 135, 181–191. [DOI] [PubMed] [Google Scholar]
- Jones IL, Livi P, Lewandowska MK, Fiscella M, Roscic B, Hierlemann A, 2011. Anal. Bioanal. Chem 399, 2313–2329. [DOI] [PubMed] [Google Scholar]
- Kalmykov A, Huang C, Bliley J, Shiwarski D, Tashman J, Abdullah A, Rastogi SK, Shukla S, Mataev E, Feinberg AW, Hsia KJ, Cohen-Karni T, 2019. Sci. Adv 5, eaax0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan Y, Ostfeld AE, Lochner CM, Pierre A, Arias AC, 2016. Adv. Mater 28, 4373–4395. [DOI] [PubMed] [Google Scholar]
- Khodagholy D, Doublet T, Quilichini P, Gurfinkel M, Leleux P, Ghestem A, Ismailova E, Hervé T, Sanaur S, Bernard C, Malliaras GG, 2013. Nat. Commun 4, 1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HN, Habbit NL, Su C, Choi N, Ahn EH, Lipke EA, Kim D, 2019. Adv. Funct. Mater 29, 1807553. [Google Scholar]
- Kim SJ, Cho KW, Cho HR, Wang L, Park SY, Lee SE, Hyeon T, Lu N, Choi SH, Kim D-H, 2016. Adv. Funct. Mater 26, 3207–3217. [Google Scholar]
- Kireev D, Seyock S, Lewen J, Maybeck V, Wolfrum B, Offenhäusser A, 2017. Adv. Healthc. Mater 6, 1601433. [DOI] [PubMed] [Google Scholar]
- Koo J, MacEwan MR, Kang S-K, Won SM, Stephen M, Gamble P, Xie Z, Yan Y, Chen Y-Y, Shin J, Birenbaum N, Chung S, Kim SB, Khalifeh J, Harburg DV, Bean K, Paskett M, Kim J, Zohny ZS, Lee SM, Zhang R, Luo K, Ji B, Banks A, Lee HM, Huang Y, Ray WZ, Rogers JA, 2018. Nat. Med 24, 1830–1836. [DOI] [PubMed] [Google Scholar]
- Kozai TDY, Catt K, Li X, Gugel ZV, Olafsson VT, Vazquez AL, Cui XT, 2015. Biomaterials 37, 25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kringelbach ML, Jenkinson N, Owen SLF, Aziz TZ, 2007. Nat. Rev. Neurosci 8, 623–635. [DOI] [PubMed] [Google Scholar]
- Kundu A, Ausaf T, Rajaraman S, 2018. Micromachines 9, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu A, Nattoo C, Fremgen S, Springer S, Ausaf T, Rajaraman S, 2019. RSC Adv. 9, 8949–8963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu A, Rozman A, Rajaraman S, 2020. J. Microelectromechanical Syst 1–3. [Google Scholar]
- Le Ven F, Bibeau K, De Larochellière É, Tizón-Marcos H, Deneault-Bissonnette S, Pibarot P, Deschepper CF, Larose É, 2016. Eur. Hear. J. – Cardiovasc. Imaging 17, 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Dellatore SM, Miller WM, Messersmith PB, 2007. Science (80-. ). 318, 426–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Shim HJ, Choi C, Kim D-H, 2019. Nano Lett. 19, 2741–2749. [DOI] [PubMed] [Google Scholar]
- Lee S, Inoue Y, Kim D, Reuveny A, Kuribara K, Yokota T, Reeder J, Sekino M, Sekitani T, Abe Y, Someya T, 2014. Nat. Commun 5, 5898. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim D-H, 2019. Nat. Biomed. Eng 3, 5–6. [DOI] [PubMed] [Google Scholar]
- Li J, Celiz AD, Yang J, Yang Q, Wamala I, Whyte W, Seo BR, Vasilyev NV, Vlassak JJ, Suo Z, Mooney DJ, 2017. Science (80-. ). 357, 378–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Mooney DJ, 2016. Nat. Rev. Mater 1, 16071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin NYC, Homan KA, Robinson SS, Kolesky DB, Duarte N, Moisan A, Lewis JA, 2019. Proc. Natl. Acad. Sci 116, 5399–5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin ZC, McGuire AF, Burridge PW, Matsa E, Lou H-Y, Wu JC, Cui B, 2017. Microsystems Nanoeng. 3, 16080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin ZC, Xie C, Osakada Y, Cui Y, Cui B, 2014. Nat. Commun 5, 3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind JU, Busbee TA, Valentine AD, Pasqualini FS, Yuan H, Yadid M, Park S-J, Kotikian A, Nesmith AP, Campbell PH, Vlassak JJ, Lewis JA, Parker KK, 2017. Nat. Mater 16, 303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Chen R, Elthakeb AT, Lee SH, Hinckley S, Khraiche ML, Scott J, Pre D, Hwang Y, Tanaka A, Ro YG, Matsushita AK, Dai X, Soci C, Biesmans S, James A, Nogan J, Jungjohann KL, Pete DV, Webb DB, Zou Y, Bang AG, Dayeh SA, 2017. Nano Lett. 17, 2757–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan L, Wei X, Zhao Z, Siegel JJ, Potnis O, Tuppen CA, Lin S, Kazmi S, Fowler RA, Holloway S, Dunn AK, Chitwood RA, Xie C, 2017. Sci. Adv 3, e1601966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuel M, Stefan K, Alfred S, Thoralf H, Gunther Z, Claus B, 2016. Front. Neurosci 10. [Google Scholar]
- Mendis S, Puska P, Norrving B, 2011. World Health Organization. [Google Scholar]
- Musick K, Khatami D, Wheeler BC, 2009. Lab Chip 9, 2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie J-J, Qiao B, Duan S, Xu C, Chen B, Hao W, Yu B, Li Y, Du J, Xu F-J, 2018. Adv. Mater 30, 1801570. [DOI] [PubMed] [Google Scholar]
- Nugraha B, Buono MF, Emmert MY, 2018. Eur. Heart J 39, 4234–4237. [DOI] [PubMed] [Google Scholar]
- Pagel M, Beck-Sickinger AG, 2017. Biol. Chem 398, 3–22. [DOI] [PubMed] [Google Scholar]
- Park J, Choi S, Janardhan AH, Lee S-Y, Raut S, Soares J, Shin K, Yang S, Lee C, Kang K-W, Cho HR, Kim SJ, Seo P, Hyun W, Jung S, Lee H-J, Lee N, Choi SH, Sacks M, Lu N, Josephson ME, Hyeon T, Kim D-H, Hwang HJ, 2016. Sci. Transl. Med 8, 344ra86–344ra86. [DOI] [PubMed] [Google Scholar]
- Regehr WG, Pine J, Cohan CS, Mischke MD, Tank DW, 1989. J. Neurosci. Methods 30, 91–106. [DOI] [PubMed] [Google Scholar]
- Richards DJ, Li Y, Kerr CM, Yao J, Beeson GC, Coyle RC, Chen X, Jia J, Damon B, Wilson R, Starr Hazard E, Hardiman G, Menick DR, Beeson CC, Yao H, Ye T, Mei Y, 2020. Nat. Biomed. Eng 4, 446–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche ET, Horvath MA, Wamala I, Alazmani A, Song S-E, Whyte W, Machaidze Z, Payne CJ, Weaver JC, Fishbein G, Kuebler J, Vasilyev NV, Mooney DJ, Pigula FA, Walsh CJ, 2017. Sci. Transl. Med 9, eaaf3925. [DOI] [PubMed] [Google Scholar]
- Rogers JA, Chen X, Feng X, 2020. Adv. Mater 32, 1905590. [DOI] [PubMed] [Google Scholar]
- Santoro F, Dasgupta S, Schnitker J, Auth T, Neumann E, Panaitov G, Gompper G, Offenhäusser A, 2014. ACS Nano 8, 6713–6723. [DOI] [PubMed] [Google Scholar]
- Skylar-Scott MA, Mueller J, Visser CW, Lewis JA, 2019. Nature 575, 330–335. [DOI] [PubMed] [Google Scholar]
- Smith AST, Choi E, Gray K, Macadangdang J, Ahn EH, Clark EC, Laflamme MA, Wu JC, Murry CE, Tung L, Kim D-H, 2020. Nano Lett. 20, 1561–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Sohee, Tathireddy P, Normann RA, Solzbacher F, 2007. IEEE Trans. Neural Syst. Rehabil. Eng 15, 493–501. [DOI] [PubMed] [Google Scholar]
- Song E, Lee YK, Li R, Li J, Jin X, Yu KJ, Xie Z, Fang H, Zhong Y, Du H, Zhang J, Fang G, Kim Y, Yoon Y, Alam MA, Mei Y, Huang Y, Rogers JA, 2018. Adv. Funct. Mater 28, 1702284. [Google Scholar]
- Song Y, Min J, Gao W, 2019. ACS Nano 13, 12280–12286. [DOI] [PubMed] [Google Scholar]
- Soscia DA, Lam D, Tooker AC, Enright HA, Triplett M, Karande P, Peters SKG, Sales AP, Wheeler EK, Fischer NO, 2020. Lab Chip 20, 901–911. [DOI] [PubMed] [Google Scholar]
- Spira ME, Hai A, 2013. Nat. Nanotechnol 8, 83–94. [DOI] [PubMed] [Google Scholar]
- Stett A, Egert U, Guenther E, Hofmann F, Meyer T, Nisch W, Haemmerle H, 2003. Anal. Bioanal. Chem 377, 486–495. [DOI] [PubMed] [Google Scholar]
- Tetsuka H, Shin SR, 2020. J. Mater. Chem. B 8, 2930–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomasjr C, Springer P, Loeb G, Berwaldnetter Y, Okun L, 1972. Exp. Cell Res 74, 61–66. [DOI] [PubMed] [Google Scholar]
- Tomaskovic-Crook E, Zhang P, Ahtiainen A, Kaisvuo H, Lee C, Beirne S, Aqrawe Z, Svirskis D, Hyttinen J, Wallace GG, Travas-Sejdic J, Crook JM, 2019. Adv. Healthc. Mater 8, 1900425. [DOI] [PubMed] [Google Scholar]
- Tybrandt K, Khodagholy D, Dielacher B, Stauffer F, Renz AF, Buzsáki G, Vörös J, 2018. Adv. Mater. 30, 1706520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine AD, Busbee TA, Boley JW, Raney JR, Chortos A, Kotikian A, Berrigan JD, Durstock MF, Lewis JA, 2017. Adv. Mater 29, 1703817. [DOI] [PubMed] [Google Scholar]
- Voges HK, Mills RJ, Elliott DA, Parton RG, Porrello ER, Hudson JE, 2017. Development 144, 1118–1127. [DOI] [PubMed] [Google Scholar]
- Wei X, Luan L, Zhao Z, Li X, Zhu H, Potnis O, Xie C, 2018. Adv. Sci 5, 1700625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidlich S, Krause KJ, Schnitker J, Wolfrum B, Offenhäusser A, 2017. Nanotechnology 28, 095302. [DOI] [PubMed] [Google Scholar]
- Whyte W, Roche ET, Varela CE, Mendez K, Islam S, O’Neill H, Weafer F, Shirazi RN, Weaver JC, Vasilyev NV, McHugh PE, Murphy B, Duffy GP, Walsh CJ, Mooney DJ, 2018. Nat. Biomed. Eng 2, 416–428. [DOI] [PubMed] [Google Scholar]
- Won SM, Song E, Reeder JT, Rogers JA, 2020. Cell 181, 115–135. [DOI] [PubMed] [Google Scholar]
- Wu J, Dong M, Santos S, Rigatto C, Liu Y, Lin F, 2017. Sensors 17, 2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Lin Z, Hanson L, Cui Y, Cui B, 2012. Nat. Nanotechnol 7, 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Gutbrod SR, Bonifas AP, Su Y, Sulkin MS, Lu N, Chung H-J, Jang K-I, Liu Z, Ying M, Lu C, Webb RC, Kim J-S, Laughner JI, Cheng H, Liu Y, Ameen A, Jeong J-W, Kim G-T, Huang Y, Efimov IR, Rogers JA, 2014. Nat. Commun 5, 3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Suo Z, 2018Nat. Rev. Mater 3, 125–142. [Google Scholar]
- Yang X, Zhou T, Zwang TJ, Hong G, Zhao Y, Viveros RD, Fu T-M, Gao T, Lieber CM, 2019. Nat. Mater 18, 510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuk H, Lu B, Zhao X, 2019. Chem. Soc. Rev 48, 1642–1667. [DOI] [PubMed] [Google Scholar]