Abstract
As biosensing research is rapidly advancing due to significant developments in materials, chemistry, and electronics, researchers strive to build cutting-edge biomedical devices capable of detecting health-monitoring biomarkers with high sensitivity and specificity. Biosensors using nanomaterials are highly promising because of the wide detection range, fast response time, system miniaturization, and enhanced sensitivity. In the recent development of biosensors and electronics, graphene has rapidly gained popularity due to its superior electrical, biochemical, and mechanical properties. For biomarker detection, human saliva offers easy access with a large variety of analytes, making it a promising candidate for its use in point-of-care (POC) devices. Here, we report a comprehensive review that summarizes the most recent graphene-based nanobiosensors and oral bioelectronics for salivary biomarker detection. We discuss the details of structural designs of graphene electronics, use cases of salivary biomarkers, the performance of existing sensors, and applications in health monitoring. This review also describes current challenges in materials and systems and future directions of the graphene bioelectronics for clinical POC applications. Collectively, the main contribution of this paper is to deliver an extensive review of the graphene-enabled biosensors and oral electronics and their successful applications in human salivary biomarker detection.
Keywords: Graphene, Salivary biomarkers, Nanobiosensors, Bioelectronics, Wearables
1. Introduction
Early disease diagnosis significantly reduces the mortality rates while accelerating patients’ recovery time with a timely, customized treatment (Siegel et al. 2017). Recent data shows that the US’s cancer death rate has fallen 27% between 1991 and 2016 because of the enabled early detection (Siegel et al. 2019). As a result, it is of utmost importance to find viable solutions that can provide rapid and affordable diagnosis for treating these diseases. There has been increasing momentum in the search for such solutions. Recent advances in materials science, electronics, and medical technology are paving the way to develop novel disease diagnostic devices (Botta et al. 2016; Dai et al. 2015). In this context, wearable sensors that can monitor the patient’s condition are gaining increasing interest due to their non-invasive nature, ease of remote operation, portability, and real-time monitoring capability. Recent articles summarize the unique advantages, essential materials, and critical functions of wearable biosensors and bioelectronics (Bandodkar et al. 2016; Kim et al. 2019; Lim et al. 2020; Ray et al. 2019). To enhance biosensors’ performance, many different types of nanomaterials have been used (Herbert et al. 2018; Mishra et al. 2016; Park et al. 2020). Among them, graphene has attracted significant interest due to its superior mechanical and electrical properties. The antimicrobial properties of graphene have been reported in many research articles (Akhavan and Ghaderi 2010; Jastrzębska et al. 2012) and its biocompatibility in wearable applications (Kwon et al. 2020). The versatility of graphene has made it a perfect candidate for electrochemical sensors (Parate et al. 2020; Torrente-Rodríguez et al. 2020) or electronic sensors (Gao et al. 2020; Selvarajan et al. 2020).
Our body continuously provides natural fluids that can be used to detect specific analytes in different locations, including serum, whole blood, saliva, tears, sweat, and urine. The exploitation of graphene-based biosensors for body fluid analysis has already produced exciting results for screening disease-related biomarkers and monitoring human health (Albishri and Abd El-Hady 2019; Janyasupab et al. 2019; Torrente-Rodríguez et al. 2020; Zou et al. 2020). The saliva represents a highly promising choice among the body fluids due to its easy accessibility, non-invasiveness of its collection, and its analyte-rich contents (Eftekhari et al. 2019; Gug et al. 2019). Moreover, it has been proved that out of about 2,000 proteins contained in saliva, 26% of those are also shared with blood, demonstrating their close correlation (Goswami et al. 2015; Javaid et al. 2016). However, while saliva possesses many advantages over other biofluids, it is still susceptible to intra-subject variability in composition and the lower concentration of the target analytes than whole blood (Franco-Martínez and Castillo-Felipe 2020).
Although many published articles study graphene as biosensors, none of the existing papers summarizes the capability of graphene nanobiosensors for the detection of various types of human salivary biomarkers.
Here, this review reports a comprehensive summary of graphene-based biosensors and bioelectronics that have been utilized for the investigation of human saliva samples. We introduce the essential properties of graphene and its derivatives and the well-characterized production of methods of those materials. The second section is entirely dedicated to the summary of saliva biomarkers for rapid disease diagnosis. Afterward, we review various types of graphene-based biosensors to detect salivary biomarkers, including viruses, bacteria, cancer, drugs, and health-indicating factors. Figure 1 captures the overview of the advantages of the graphene biosensors and their applications for targeting multiple biomarkers. We also discuss the existing challenges that need to be overcome to translate this technology into low-cost, miniaturized wearable POC devices.
Figure 1. Overview of graphene-based nanobiosensors for salivary biomarker detection.
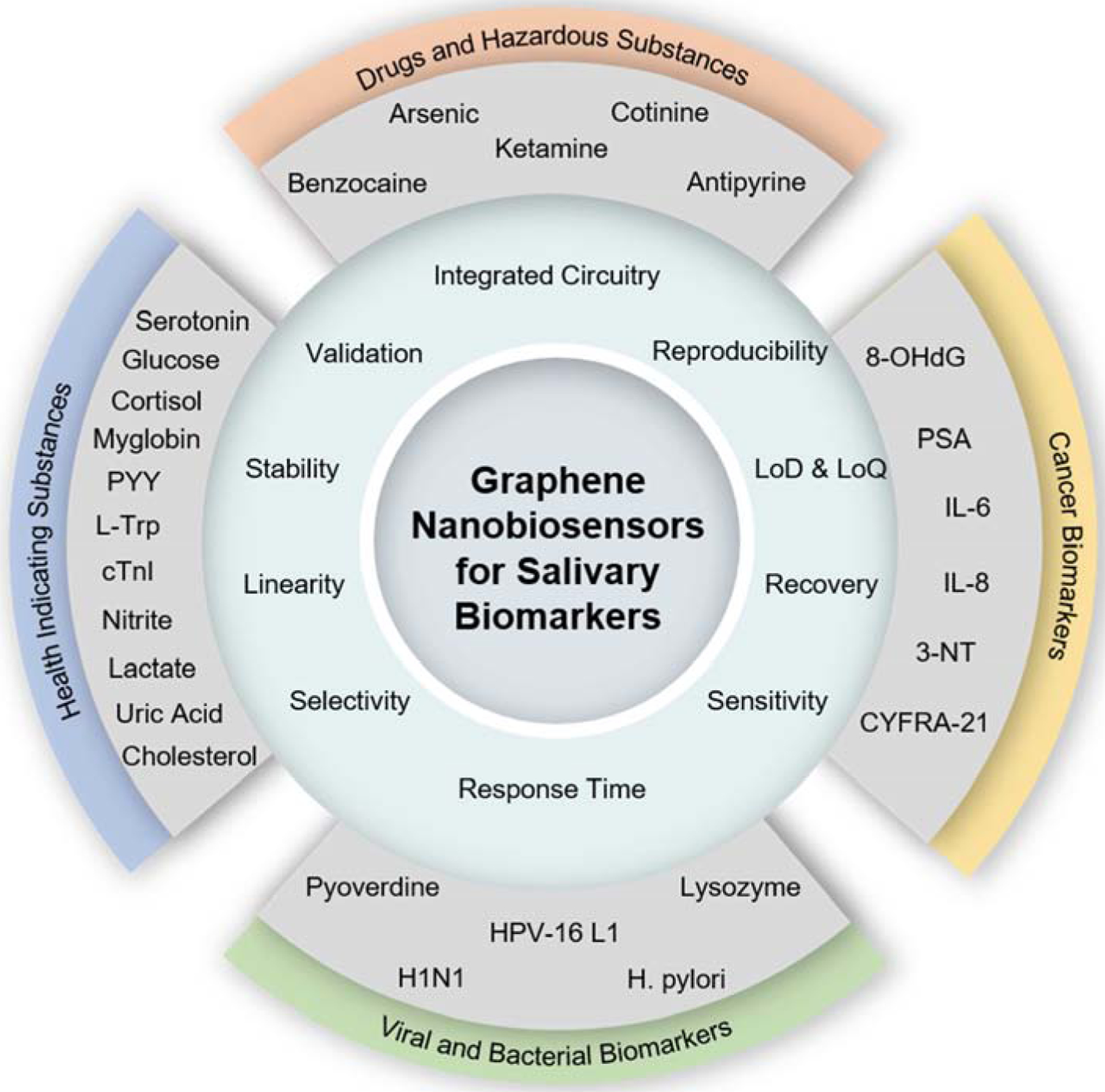
This chart summarizes the key properties and performance indicators of biosensors and targeted salivary biomarkers in four different categories.
2. Graphene-based biosensors
2.1. Key properties of graphene and graphene derivatives
Graphene is one of the most attractive 2D nanomaterials for the development of biosensors and bioelectronics due to its superior physical, chemical, and structural properties (Ferrari et al. 2015; Liu et al. 2014). Table 1 summarizes the key properties of graphene and its derivatives for the development of high-performance biosensors. Graphene consists of a layer of sp2 hybridized carbon atoms arranged in a hexagonal network (Burnett et al. 2012; Geim and Novoselov 2007). The thickness of each graphene layer is around 0.35 nm, and the length of the covalent bonds between carbon atoms is 1.42 Å (Castro Neto et al. 2009). Graphite is made up of several graphene layers held together by Van der Waal forces (Chauhan et al. 2017). Graphene possesses unique electronic properties due to its peculiar band structure. The valence and conduction band of graphene overlap at the Dirac point, consequently displaying both metal and semiconductor features. As a result of its crystal lattice and band structure, graphene has the longest mean free path of among any known nanomaterial, meaning that an electron can travel without scattering, generating minimal heat due to the negligible resistance encountered (Avouris 2010). Consequently, it has been reported that graphene has a very high charge carrier mobility of 15,000 cm2/V∙s at room temperature and too high intrinsic mobility in a suspended form (Bolotin et al. 2008; Du et al. 2008; Morozov et al. 2008). However, the electronic properties of graphene change depending on the number of layers. Since there is no specific standard in these materials’ terminology, a single monolayer of carbon atoms is called graphene, while graphene sheets can contain up to 10 layers. When the number of layers exceeds 10, the resulting material can be referred to as graphite thin films (Pumera 2013).
Table 1.
Key properties of graphene and graphene derivatives
| Property | Graphene (GR) | Graphene Oxide (GO) | Reduced Graphene Oxide (rGO) |
|---|---|---|---|
| Electrical (carrier mobility) | 200,000 cm2 V−1 s−1 (Bolotin et al. 2008) | 0.1 ~ 10 cm2 V−1 s−1 (Eda et al. 2011) | 372 cm2 V−1 s−1 (Taufique et al. 2017) |
| Thermal (thermal conductivity) | 5 × 103 W m−1 K−1 (Balandin et al. 2008) | 18 W m−1 K−1 (Mahanta and Abramson 2012) | 1,390 W m−1 K−1 (Kumar et al. 2015a) |
| Structural (specific surface area) | 2,630 m2 g−1 (Stoller et al. 2008) | 736.6 m2 g−1 (Montes-Navajas et al. 2013) | 758 m2 g−1 (Zhang et al. 2011) |
| Structural (Young modulus) | 1 × 1012 Pa (Lee et al. 2008) | 207.6 × 103 Pa (Benchirouf et al. 2016) | 6.3 × 109 Pa (Kumar et al. 2015a) |
| Optical (light adsorption) | 2.3 % (Nair et al. 2008) | 25 % (Goumri et al. 2016) | 20% (Goumri et al. 2016) |
| Biological (dispersibility in water) | Not dispersible (Liu et al. 2011b) | High (Konkena and Vasudevan 2012) | Low (Konkena and Vasudevan 2012) |
Among the electronic properties of graphene, its high conductivity when biased with voltage (Novoselov et al. 2004) and its strong ambipolar electric field effect is quite remarkable for its applications in advanced electronic devices (Geim and Novoselov 2007; Mao et al. 2013; Schedin et al. 2007). From a mechanical perspective, graphene has an extremely high specific surface area of 2,630 m2/g (Lin et al. 2011), a relative tensile strength that is 10-fold that of steel (Lee et al. 2014), and high elasticity. Graphene also has excellent thermal conductivity, high optical transparency (Kim et al. 2017a; Lee et al. 2015), and can be made biocompatible when required (Akhavan and Ghaderi 2010; Jastrzębska et al. 2012). Moreover, these properties can be modified through chemical doping (Liu et al. 2011a). One of the most critical features of graphene is its hydrophobic nature (Georgakilas et al. 2016) because it allows for an easy functionalization, that ultimately affects the electronic properties of the material (Georgakilas et al. 2012). The functionalization by means of chemical reactions can be covalent, resulting in a stable and specific structure with significant defects (Park et al. 2016), or non-covalent. Chemical modifications can be carried out using strong oxidants, as in the case of graphene oxide (GO). GO is prepared through two different oxidations, one with carboxylic acids at the graphene edges, the other with epoxy and hydroxyl groups on the basal plane. The resulting material has increased dispersibility compared to pristine graphene (Chua and Pumera 2013; Georgakilas et al. 2012). Various approaches to modify the molecular structure of GO and restore its electrical conductivity have been tested. Reduced graphene oxide (rGO) is the product of a reduction process operated on graphene oxide through chemical, thermal or electrochemical processes (Lightcap et al. 2010). A large share of the oxygenated groups is eliminated, with an increase in electrical conductivity that is highly dependent upon the reduction technique being used (Gilje et al. 2007; Khan et al. 2017c; Yang et al. 2009). Graphene, GO, and rGO can also undergo non-covalent functionalization via van der Waals or p-p type of interactions. This kind of functionalization allows for a combination of a graphene derivative and a polymer or metal nanoparticle, generating a hybrid compound with new interesting properties (Mandal et al. 2015; Wang et al. 2011).
2.2. Production methods of graphene and graphene derivatives
The first example of pristine monolayer graphene production is the micromechanical exfoliation of graphite (Novoselov et al. 2004). Since then, many advanced techniques have been developed to obtain high-quality graphene. However, the main limitation in the large-scale graphene production resides in its low throughput and high cost. High yield and low-cost graphene production methods typically come at the expense of a larger degree of defects in the final product.
Figure 2 summarizes the representative examples of bottom-up and top-down methods for the production of graphene and graphene derivatives. Methods of producing graphene can be grouped into top-down and bottom-up approaches (Edwards and Coleman 2013). In a top-down process, monolayer or few-layer graphene sheets are obtained from bulk graphite, which involves breaking the van der Walls bonds between individual graphene layers. In a bottom-up approach, on the other hand, graphene is created from a carbon source on a dedicated substrate (usually SiC or Si). Each method has its advantages and drawbacks; thus, it is important to carefully select the best process for a specific application, depending on the requirements of quality or cost. Among the top-down approaches to obtain graphene, chemical reduction of graphene oxide, derived from graphite oxide, is the very first example of the production of graphene presented by Boehm et al. in 1962. The fabrication of rGO has seen many attempts of scaling to large productions. However, a large number of defects and impurities have always been regarded as a limiting factor for many applications. Micromechanical exfoliation is operated by peeling off layers of graphite by using common scotch tape. Even if this technique presents evident limitations for scaling, it is still a common choice for many research groups at a proof-of-concept stage. The unzipping of carbon nanotubes, as it is seen for the fabrication of rGO, can also be achieved via the use of strong oxidants to produce graphene nanoribbons. The properties of the obtained nanoribbons mainly depend on the direction where the nanotube structure has been opened (zigzag or armchair) (Dhakate et al. 2011). This technique can give a high yield at a moderate cost and presents good potential for scalability.
Figure 2. Production methods of graphene and graphene derivatives.
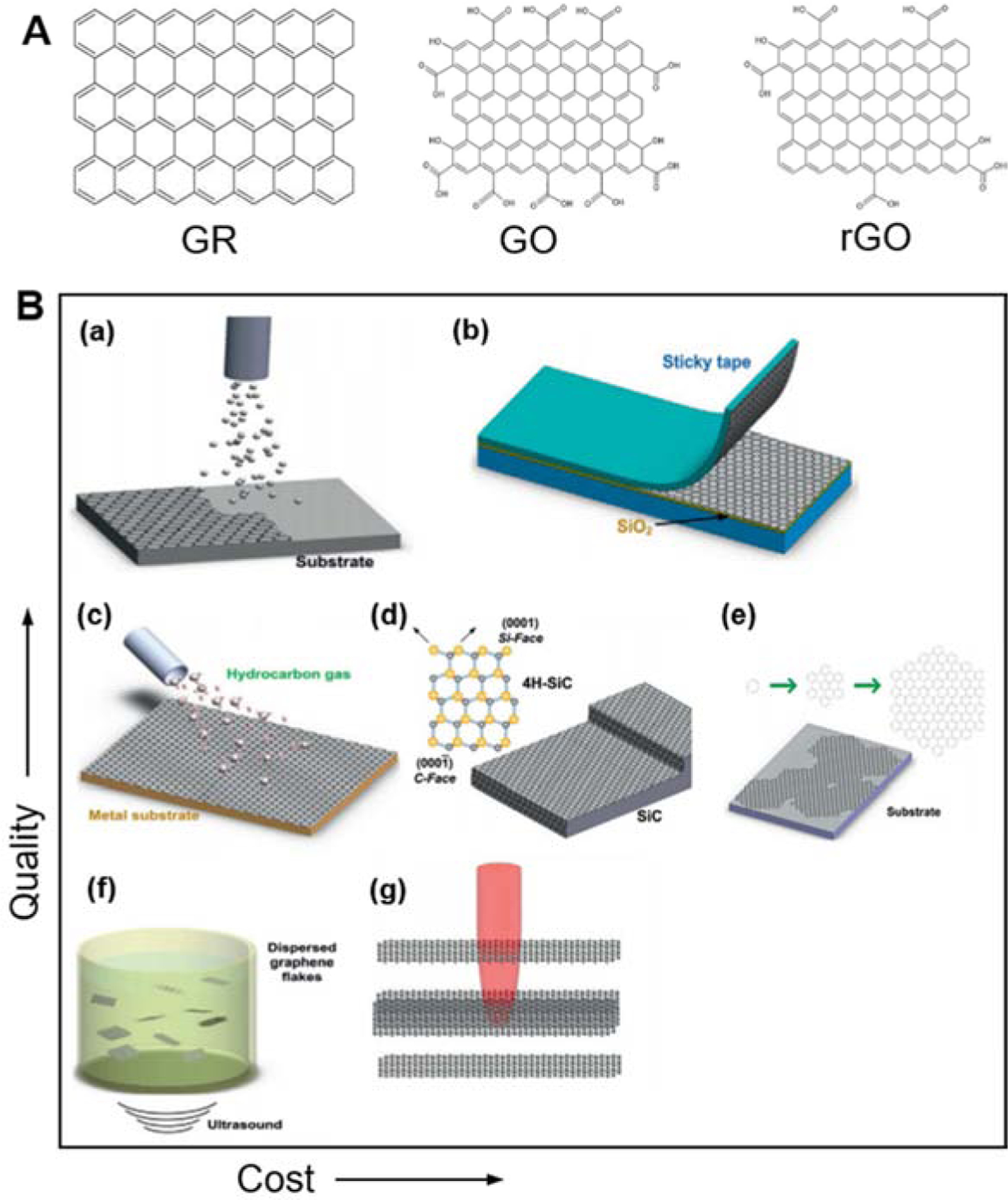
A) Molecular structure of graphene (GR), graphene oxide (GO), and reduced graphene oxide (rGO). B) Comparison of various graphene production techniques according to quality and cost: (a) molecular beam epitaxy, (b) micromechanical exfoliation, (c) chemical vapor deposition, (d) epitaxial growth on SiC, (e) molecular assembly, (f) liquid-phase exfoliation, and (g) photoexfoliation. Adapted with permission from (Bonaccorso et al. 2012)
Bottom-up approaches for the production of graphene include epitaxial growth, chemical vapor deposition, and self-assembly. A graphene thin film with high quality can be obtained by epitaxial growth in ultra-high vacuum (UHV) on SiC at very high temperatures (Camara et al. 2008; Shivaraman et al. 2009). One downside of this technique is the difficulty in precisely controlling the growth process. Chemical vapor deposition (CVD) on metal substrates is one of the most commonly employed methods to produce graphene on a large scale (Dong et al. 2011; Gao et al. 2010). This technique, however, is still considered only moderately scalable for large productions. Self-assembly of chemical modified graphenes (CMGs) is a solution-based technique that uses the pi-pi interactions and van der Waals forces acting between graphene layers. Layer by layer and Langmuir-Blodgett deposition are among the most used self-assembly techniques to produce graphene. Despite the advances in this area, finely tuning the shape and surface chemistry of the obtained material remains a grand challenge, and affordable production strategies are still needed to scale up this technology (Yuan et al. 2018).
2.3. Graphene-based biosensor architecture and performance metrics
In 1977, the term “biosensor” was coined by Cammann (Cammann 1977), which was officially recognized by the IUPAC in 1999 as an analytical device that can translate a biological event into a meaningful electrical signal (Thevenot et al. 1999). Figure 3 shows a schematic illustration of a typical biosensor composed of a transducer, biorecognition elements, and targeted molecules. Biosensors can use different transduction modalities (optical, electrochemical, electronic, etc.), depending on target analytes. Among various biosensors, graphene-based biosensor utilizes graphene, graphene derivatives, or graphene-based nanomaterials as the transducing element. A bioreceptor is immobilized on the surface of the transducing layer via a chemical or physical functionalization process. Examples of widely used biorecognition elements include antibodies (North 1985), DNA single strands (Wang 1998), and enzymes (Wilson and Hu 2000).
Figure 3. Architecture of a graphene-based biosensor.
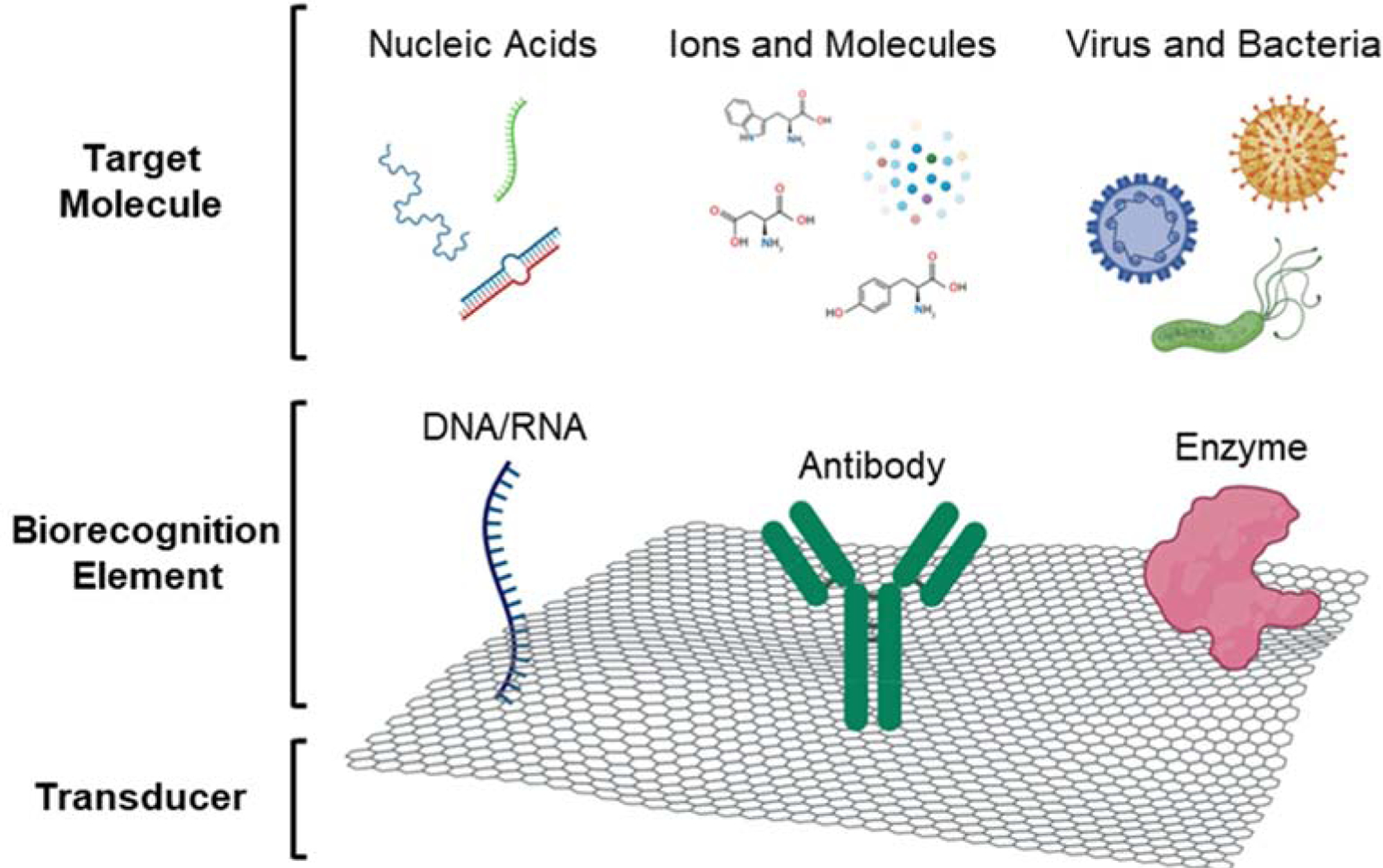
The biorecognition element is immobilized on the surface of the graphene layer, acting as the transducer. The interaction between the target molecule and the bioreceptor generate a biological signal that is translated into a meaningful electrical signal.
Upon developing graphene biosensors, many design considerations should be taken into account since some of the properties of graphene can largely affect the device’s performance. First of all, sensor performance metrics have been introduced to offer the evaluation standard of biosensors. The limit of detection (LOD) of an analytical measurement is defined as the lowest amount of analyte in a given sample that can be detected. While many approaches for calculating the LOD exist, one of the most widely used procedures is to analyze a calibration plot that shows the biosensor response against the analyte concentration. In this case, the LOD is defined as:
where S is the standard deviation of the response and b is the slope of the calibration curve. The slope of the calibration curve b represents the device sensitivity. While most studies calculate the LOD from linear regression using an S/b ratio of 3, some authors can also arbitrarily choose to employ a value of S/b = 2 (Martínez-García et al. 2016). Another critical parameter in evaluating the sensor performance is the sensing linearity, which can be defined as the range of analyte concentration for which the biosensor produces a linear response. In addition, the device stability needs to be evaluated to determine how long the biosensor can maintain the performance. Stability is typically calculated as the ratio of the response to a given analyte concentration, measured before and after the dry storage period. Alternatively, a percent loss can be simply calculated as:
Furthermore, the selectivity of a biosensor is critical to offer reliable and accurate detection of target analytes. Such selectivity is investigated by adding interferent species in a saliva sample while exclusively measuring the sensor response on the designated target. Although there is no specific calculation method on the selectivity, researchers calculate the signal-to-noise ratio where the signal is for the target, and noise is for unwanted other analytes. Another useful evaluation metric is the sensor recovery to detect a target analyte in a complex sample matrix (i.e., whole saliva). The matrix is spiked with a known concentration of the analyte of interest, then a practical measurement using the biosensor is carried out. The percentage ratio between the theoretical (known) and practical (measured) concentration of the analyte provides the recovery value. Lastly, both the repeatability and reproducibility of a sensor are two metrics that reveal the sensor’s reliable use. Typically, the relative standard deviation (RSD) is calculated to relate the standard deviation of measurement to the measurement’s mean. When the RSD is computed on different tests performed on the same biosensor, it is called intra-assay RSD. Conversely, inter-assay RSD is calculated based on measurements carried out on distinct devices.
3. Biomarkers in human saliva
Human saliva belongs to the family of body fluids that also include serum (Albishri and Abd El-Hady 2019), sweat (Torrente-Rodríguez et al. 2020), urine (Janyasupab et al. 2019), and tears (Kim et al. 2017a). Blood tests are still considered the gold standard for examinations, but other fluids that can be less invasively collected are widely used in advanced biosensors and bioelectronics (Ma et al. 2020). As summarized in Table 2, the abundance and continuous renovation, the ease of its collection, and its demonstrated richness in biomarkers (Malon et al. 2014) is fostering saliva as a promising tool for remote diagnostics (Goswami et al. 2015; Javaid et al. 2016). Saliva is produced by the salivary glands in the oral cavity, one of the connecting points between our organism and the outside environment (Figure 4) (Wang et al. 2020). The oral cavity is also a location where many human systems converge. Food processing is carried out by the digestive and muscular system (Breslin 2013). Simultaneously, the assumption of substances by matter exchange has a direct relationship with our health status, reflected in the circulatory, respiratory and nervous system (Haeckel and Hanecke 1996; Ocak et al. 2015; Plummer et al. 2015). The immune system shields our body from several pathogens that could accidentally enter the oral cavity, through the use of various glands controlled by the excretory system (Warinner et al. 2014; Wu et al. 2014).
Table 2.
List of biomarkers in human saliva samples (Gug et al. 2019)
Figure 4. Overview of the human oral cavity.
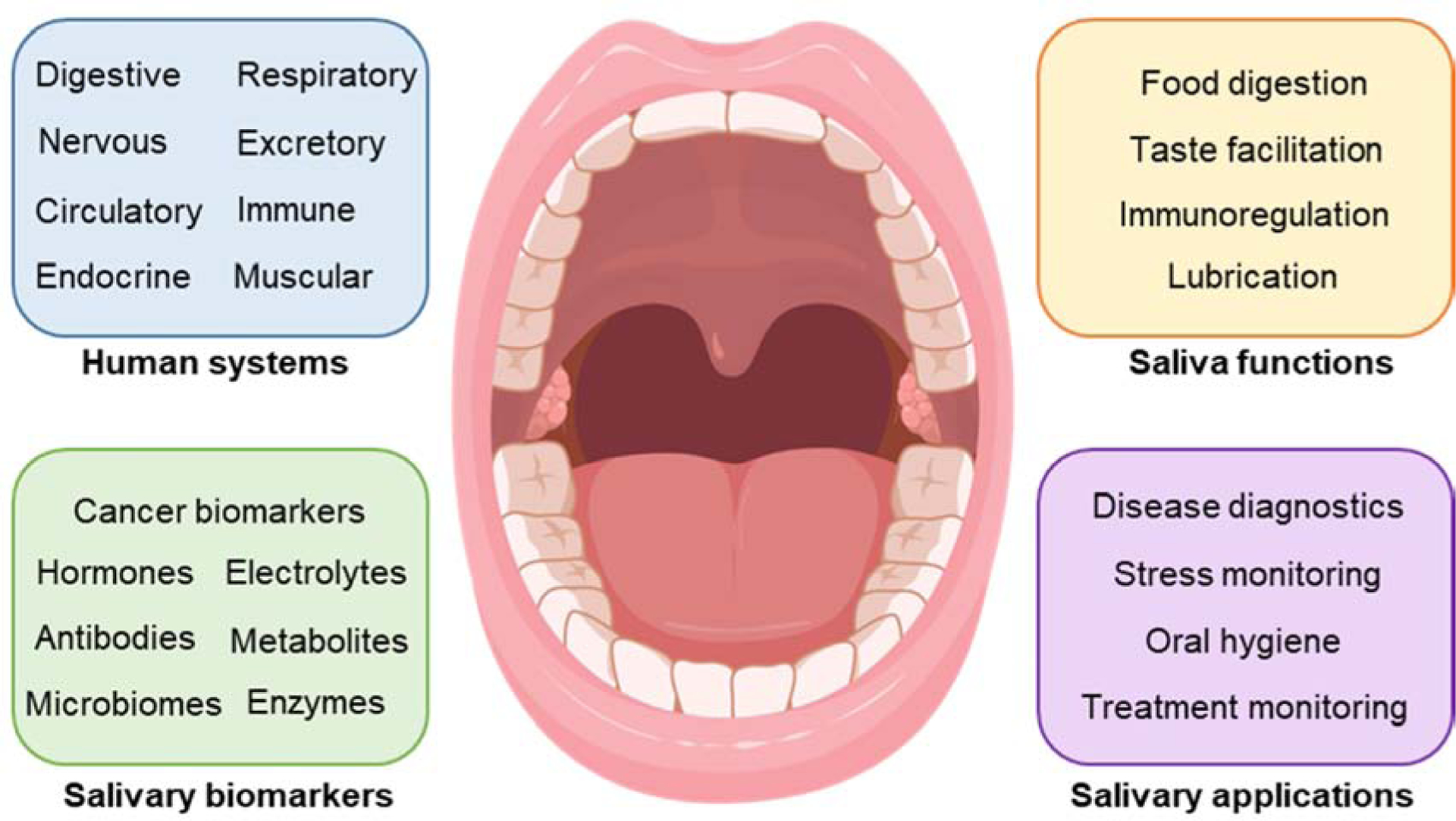
Saliva is produced by the salivary glands contained in the oral cavity, one of the connecting points between our organism and the outside environment. The oral cavity is also a location where many human systems converge, and many types of biomarkers exist.
The presence of various biomarkers that are contained in saliva can be originated by either inflammatory cells proper of chronic diseases (Prasad et al. 2016; Rathnayake et al. 2017) or molecules that are produced by the immune system (Choromańska et al. 2017; Khurshid et al. 2016; Tartaglia et al. 2017). Saliva can also be used to monitor the pharmacokinetics of a specific drug during a treatment (Idkaidek and Arafat 2012; Kaczor-Urbanowicz et al. 2017). Moreover, being it continuously renovated (Jokerst et al. 2009), saliva is the perfect candidate for the continuous monitoring of electrolytes and metabolites in the oral cavity through POC devices (Herr et al. 2007; Tan et al. 2008). Compared to the blood draw, the use of saliva requires easier collection and processing procedures, more appropriate for less-skilled personnel (Campuzano et al. 2017). This becomes even more important when the patient is an infant or an older adult, that usually suffers from pain and anxiety associated with the blood sampling procedure (Malon et al. 2014). As compared to blood, saliva also offers additional advantages related to sampling and handling. Saliva does not clot, and its analytes are stable over time, while blood could present issues of this kind. Moreover, saliva presents almost no risk of cross-contamination between patients or exposure to pathogens, which is quite more frequent with blood samples (Malon et al. 2014). Despite these advantages, the saliva collection and processing workflow still lack a widely accepted and standardized set of rules (Malon et al. 2014). As a result, it becomes difficult to precisely compare data coming from distinct research groups or different acquisition techniques.
Saliva is usually centrifuged to remove big unwanted particles and food residues that are not of interest for the detection. The supernatant is then typically diluted, but the dilution process is again not standardized. However, according to recent studies, a dilution of 1/10 for human saliva samples seems to be the most commonly operated procedure (Liu et al. 2017). In some cases, human saliva samples are then spiked with a specified amount of the analytes of interest. Storage of saliva samples is also another factor that could potentially affect the analysis and should be taken into account when reporting the results. Established laboratory techniques for the analysis of saliva already exist and are highly reliable, even in terms of standardization of the procedure. However, since these processes are only performed in centralized laboratories, they suffer from slow and expensive operations, including sample preparation, data acquisition, and shipment (Rusling et al. 2010). Standard techniques for the detection of biomarkers in saliva include immunoassays (radioimmunoassay (Walker et al. 1978), chemiluminescence immunoassay (De Boever et al. 1990), electrophoretic immunoassay (Koutny et al. 1996)), DNA-based techniques (Garbieri et al. 2017), salivary proteomics (Delaleu et al. 2015; Hu et al. 2008) and the most widely employed Enzyme-Linked Immunosorbent Assay (ELISA) (Holmström et al. 1990; Oba et al. 2000; Pinho et al. 1999). These techniques are notably slow, expensive, and require highly trained personnel (Choudhary et al. 2013).
Considering the limitations posed by conventional methods, there has been an increasing effort in the scientific community to develop biosensors that would allow rapid and affordable diagnostic tools for salivary analysis in either clinical or non-clinical settings. The development of biosensors that are accurate, reliable, and reproducible is still very challenging. One of these challenges is the target recognition layer; the enzyme-based biosensor can be degraded by the changes of environmental factors such as pH and temperature. Another challenge is the concern of biofouling on the sensor surface since it reduces the detection accuracy of the device. Many researchers are studying various materials and surface coating methods for surface anti-fouling. In addition, wearable biosensors have challenges in developing reusable biosensors that are hindered by the strong binding between the receptor and the target molecule. While there are remaining challenges, the advancements in nanotechnology and new materials have helped to design accurate POC devices. Among many examples, a group of investigators reported how engineering the fractal dimension of the biosensing surface by combining different nanomaterials could enhance the device sensitivity (Sadana and Sadana 2008).
The electrochemical techniques that can be utilized to detect the analytes and measure their concentration are various and mainly dependent upon the design constraints imposed by the application of the biosensor. The most common techniques use electronic transducers, such as field-effect transistors, produce current-voltage data that contain a richer amount of information when compared to conventional electrochemical techniques. As reported by a recent article (Schroeder et al. 2019), interactions between the analyte and the sensing element leading to doping, Schottky Barrier modulation and change in mobility can be identified by characteristic traits of the current-voltage plot. However, they need a semiconductor parameter analyzer to interpret the data. Chemiresistive biosensors offer a very intuitive detection method that is limited in information but does not require complicated readout circuitry to interpret the data. A similar advantage can be found from chronoamperometry and other similar potentiometric techniques. The suitability of a detection technique is primarily given by the specific application of the biosensor and its design constraints.
4. Detection of various types of salivary biomarkers by using graphene biosensors
To overcome the existing limitation of salivary biomarker detection, recent works have focused on developing nanoscale biosensors that could be integrated with readout circuitry for a fast and accurate diagnosis. However, only a few of them have seen the successful realization of a reliable device for salivary analysis. Figure 5 summarizes the available examples that use wearable devices to detect specific analytes from human saliva. The device, realized by Mannoor et al. (Mannoor et al. 2012) in 2012 (Figure 5a), is the first example of a graphene-based biosensor with an integrated readout circuitry. Such biosensor, however, was not capable of allowing for continuous real-time analysis due to the limitation of a near field communication (NFC) interrogating device. Moreover, this is the only biosensor that employs graphene as sensing material and possesses a feature of wearability and wireless data communication. Kim and coworkers prepared a mouthguard biosensor (Figure 5b) with a three-electrode configuration directly screen printed on PET (Kim et al. 2014). While the direct integration of screen-printed electrodes (SPEs) on the mouthguard makes the sensor fully wearable, this device still lacked the integrated electronics needed to continuously perform its monitoring function without being necessarily connected to an external electrochemical analyzer. The second version of this device was realized shortly later to remedy these shortcomings (Kim et al. 2015) (Figure 5c). The updated version of the mouthguard biosensor included the same SPEs configuration, with an optimized design and a specific enzyme functionalization, for the selective detection of uric acid in saliva. The biosensor includes miniaturized instrumentation electronics featuring Bluetooth communication, all embedded on the plastic mouthguard for continuous real-time monitoring.
Figure 5. Wearable biosensors for salivary biomarker detection.
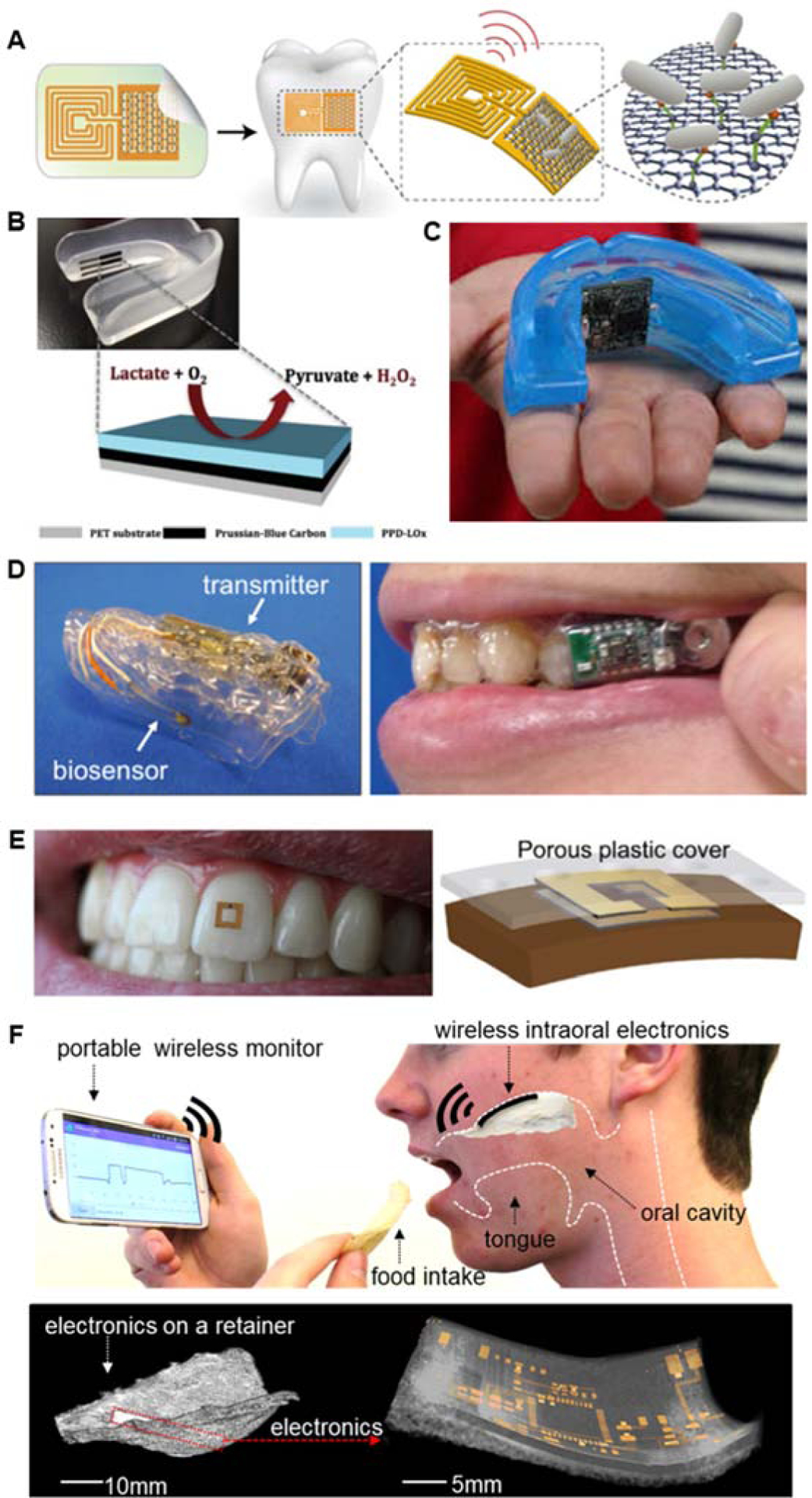
A) Only example of a graphene-based biosensor for the wireless bacteria detection. Mannoor et al. 2012. B) Mouthguard biosensor for monitoring of metabolites in saliva Kim et al. 2014. C) Mouthguard biosensor with integrated wireless electronics for the detection of uric acid in saliva Kim et al. 2015. D) Mouthguard biosensor with integrated electronics for selective monitoring of glucose in saliva Arakawa et al. 2016, Mitsubayashi et al. 2016. E) Wearable miniaturized RF sensor with wireless capability for monitoring of food consumption Tseng et al. 2018. F) Wireless intraoral biosensor for monitoring of sodium intake on a soft retainer Lee et al. 2018
Another example of a mouthguard integrated biosensor can be found in Arakawa’s work (Figure 5d) (Arakawa et al. 2016; Mitsubayashi and Arakawa 2016). The mouthguard integrates both the sensing electrodes and the on-board electronics with BLE communication for real-time analysis of saliva samples. The printed circuit board (PCB) is clearly visible on one side of the transparent PETG mouthguard while the mouthguard featured the electronics on the inside of the mouthguard (Kim et al. 2015). Figure 5e shows the RF sensor can be used for monitoring of food consumption (Tseng et al. 2018). The device, consisting of two open ring resonators, has an ultra-small footprint that allows for seamless attachment to a human tooth. The RF capability allows for data transfer when an interrogating device is placed near to the resonating coils. Recently, Lee et al. presented a wireless intraoral electronic system placed on the upper palate (Figure 5f) (Lee et al. 2018a). The hybrid biosensing platform with integrated wireless soft electronics monitored the sodium intake continuously, showcasing a non-invasive practical POC solution for patients suffering from hypertension.
4.1. Graphene biosensors for the detection of viral and bacterial markers in saliva
Viral and bacterial infections affect millions of people every year while killing thousands of them. The possibility to perform screening of such diseases in saliva with POC devices would represent great advantage in terms of both response time and costs compared to the conventional blood draws. Viral and bacterial infections can be recognized by the presence of just a single pathogen, as compared to systemic diseases where only the assessment of multiple biomarkers can certify an accurate diagnosis (Corstjens et al. 2012).
Table 3 reports an up-to-date summary of graphene-based biosensors for salivary analysis, where the main performance parameters are specified. Great relevance is given to those studies that have been validated with a gold standard (i.e., ELISA) since that provides a direct comparison between a highly precise conventional method (usually immunological or separation-type) and a novel electrochemical approach. Among the biosensors for the screening of viral or bacterial markers reported herein, none has been validated with a gold standard. Mannoor et al. developed the first graphene-based biosensor with an integrated readout coil for NFC data communication (Mannoor et al. 2012). Graphene is deposited on a bioresorbable silk substrate using transfer printing, followed by e-beam evaporation of gold to create the interdigitated electrodes structure and the associated readout coil for NFC data transmission. The device showed an average response time of 10 min, but a very small sampling volume (1 μL) was needed for the analysis. The sensor was able to detect as low as 100 cells of the bacteria H. pylori and maintaining a linear response in the range between 102 and 106 cells. The simple chemo-resistive detection mechanism, where a change in the output resistance indicates the detection of bacterial cells, is particularly suitable for fast and easy integration with the readout circuitry since it does not require complicated electrochemical analyzers. Moreover, the absence of active components and power sources makes this platform particularly promising for wearable applications where real-time monitoring is not a primary interest.
Table 3.
Graphene biosensors for the detection of viral and bacterial markers in saliva
| Biomarker | Sensing Platform | Detection Method | LODa | Detection Range | Recoveryb | Assay Time | Sample Volume | Preparation | Reference |
|---|---|---|---|---|---|---|---|---|---|
| H. Pylori | Graphene on Au/Silk | Chemi-resistive | 100 cells | 102 – 106 cells | N/A | 10 min | 1 μL | Whole saliva | (Mannoor et al. 2012) |
| HPV-16 L1 | rGO/MoS2/GCE | DPV | 0.1 ng/mL (1.75 pM) | 0.2 – 2 ng/mL (5.3 – 35.3 pM) | 97.1–105.3% | N/A | N/A | Whole saliva | (Chekin et al. 2018a) |
| H1N1 | Shellac-derived TrGO/ITO/glass | EIS | 33.11 PFU/mLc 26.04 PFU/mLd |
0 – 10000 PFU/mL | N/A | N/A | N/A | 80% v/v dilution with 1 × PBS | (Joshi et al. 2020) |
| Lysozyme | LBA-GR/GCE | EIS | 6 fmol/L | 0.01 – 0.5 pmol/L | N/A | N/A | N/A | Diluted | (Xiao et al. 2013) |
| Lysozyme | FAM-aptamers/ssDNA/rGO assembly | Fluorescence | 1.4 μL (21.8 pM)c 6.837 pMd |
1.6 – 14.4 μLc 25 – 175 pMd |
85% | 20 min | 1 mL | 10-fold dilution | (Liu et al. 2017) |
| Pyoverdine | AuNPs/Ppy-COOH/GR modified SPE | DPV | 0.33 μM | 1 – 100 μM | 97.8 – 104.93% | N/A | N/A | 1: 10 dilution with PBS | (Cernat et al. 2018) |
| Pyoverdine | AuNPs/Ppy-COOH/GR modified SPE | DPV | 333.33 nmol/L | 1 – 100 μmol/L | 102.12% | N/A | N/A | 1:100 dilution with PBS | (Gandouzi et al. 2018) |
S/N = 3, otherwise differently specified
Values obtained in real saliva samples, otherwise differently specified
in saliva
in buffer
The Checkin group reported the fabrication of an rGO/MoS2 modified glassy carbon electrode (GCE) platform for the detection of human papillomavirus type 16 (HPV-16 L1), which is among the indicators of genital cancer (Chekin et al. 2018a). A low limit of detection of 0.1 ng∙mL−1 allows for accurate screening of saliva samples with great precision. The recovery values when the biosensor is tested on human saliva are in the 97–105% range, which is considered good for electrochemical biosensors. The sensor has been tested for stability, showing only a 5% loss in performance after 1-month dry storage. The inter-assay reproducibility has also been tested, resulting in an RSD of 9.3% that the highest among the analyzed biosensors. This shows poor reproducibility during the fabrication process, which can lead to inconsistent results and an imprecise diagnosis. The selectivity of the device has been proven in the presence of two disturbing species, ovalbumin (OVA) and human serum albumin (HSA), which did not affect the detection of HPV-16 L1. Another biosensor for the detection of viral agents has been developed by Yoshi et al. (Joshi et al. 2020) that used shellac, a natural biopolymer, for the production of thermally reduced graphene oxide (TrGO) as a sensing element. The limit of detection was quantified both in real saliva samples and in buffer solution, showing comparable results. The short-term stability of the sensor has been tested, with the sensor retaining 90% of its performance after two weeks. The device has also been tested for selectivity against adenovirus, influenza B virus, and MS2 bacteriophage, which caused a negligible response in the electrochemical impedance spectroscopy (EIS) analysis. The inter-assay RSD value of 2.06% proves a substantially better reproducibility when compared to the previously analyzed device for HPV-16 L1 (Chekin et al. 2018a).
Lysozyme is an antimicrobial enzyme that is usually produced by our immune system when it gets attacked by pulmonary pathogens (Smith et al. 2019). Xiao et al. realized a biosensing platform based on graphene-modified GCEs for the selective detection of lysozyme in saliva, achieving an ultra-low limit of detection in the femtomolar range (Xiao et al. 2013). The selectivity of the biosensor is demonstrated by a marked response to lysozyme in the presence of thrombin and bovine serum albumin (BSA) as disturbing species. Liu and coworkers developed an aptameric biosensing device for lysozyme recognition, using a modified rGO assembly and fluorescence as a detection technique (Liu et al. 2017). The biosensor showed a LOD in the picomolar range when tested in both 10-fold diluted saliva samples and in standard buffer solution, which is higher compared to that achieved by the prior work (Xiao et al. 2013). The values of recovery when tested in real samples are acceptable, and the device proved to be highly selective for lysozyme, producing an 8-fold higher relative fluorescence peak compared to that originated by six control proteins: BSA, interferon-gamma (IFN-γ), myoglobin (Mb), cytochrome C (Cyt C) and nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB).
Pyoverdine is a siderophore (iron-chelating compound) that is secreted by the bacterium Pseudomonas aeruginosa, which is related to potential chronic lung infections (Kang et al. 2018; Peek et al. 2012). Cernat and coworkers developed a biosensing platform based on a combination of several nanomaterials, to take advantage of their combined properties (Cernat et al. 2018). Polypyrrole-3-carboxilic acid (Ppy-COOH) is deposited on graphene-modified SPEs to stabilize the composite structure. Then the platform is decorated with gold nanoparticles (Au NPs) to enhance the device’s sensitivity. The resulting biosensor has a LOD in the micromolar range and showed excellent recovery when tested in both buffer solution and real saliva samples. The selectivity has been confirmed against ascorbic acid, acetylsalicylic acid, uric acid, NADH, dopamine, and glucose. However, the sensor failed to demonstrate satisfactory long-term stability, producing 41.42% higher results after only one week and 61.12% higher results after two weeks. The same group later realized another version of the sensor based on the same nanocomposite platform to overcome the stability, as mentioned earlier limitations (Gandouzi et al. 2018). The already satisfactory recovery, obtained in the previous study, was improved to 102.12%, and the long-term stability substantially increased, with the sensor retaining 91.43% of its initial performance after 30 days in dry storage. However, this device demonstrated poor intra-assay reproducibility after the 5th consecutive test, showing the nanocomposite platform’s limitations in terms of regeneration and consecutive uses. The inter-assay RSD of 5.6% is instead among normal values for biosensors.
4.2. Graphene biosensors for the detection of cancer markers in saliva
The early diagnosis of disease plays a major role, especially for those patients affected by cancer, reducing the risk of mortality and producing a timely response that can substantially affect the outcome of the therapy (Wu and Qu 2015). As a result, tremendous efforts are being devoted to the development of technologies that can accurately detect any trace of carcinogenic molecules (Goossens et al. 2015; Sawyers 2008). A biomarker can be referred to as a biological molecule whose presence in body fluid can be related to the appearance of a disease (Henry and Hayes 2012). In some cases, as for viral and bacterial infections (Corstjens et al. 2012), one molecule is sufficient to unquestionably certify the infection, while in other cases, such as for cancer biomarkers, the contemporary assessment of the presence of specific molecules is a strong sign of the insurgence of the disease.
Table 4 summarizes the latest efforts in developing graphene-based nanobiosensors for the detection of cancer biomarkers in human saliva samples. As already specified in the previous section, greater relevance and importance is given to those studies that have been validated against a conventional screening method. In this case, less than half of the analyzed studies provided a direct comparison with a gold standard (ELISA). CYFRA-21–1 is a cancer marker of new conception, found to be particularly useful in the detection of non-small cell lung cancer (Stieber et al. 1993; Wieskopf et al. 1995). Kumar and coworkers realized an electrochemical biosensor for the detection of CYFRA 21–1 using DPV (Kumar et al. 2016). The nanocomposite platform exploits an rGO/ITO biosensing platform decorated with zirconia nanoparticles to enhance sensitivity, and a further functionalization with 3-aminopropyl triethoxy saline (APTES) and anti-CYFRA-21–1. BSA is then added to the immune electrode to impede non-specific binding on the active surface. The resulting device showed a remarkably low LOD of 0.122 ng mL−1 and a wide linear range. The sensitivity of the device, defined as the ratio of the change in the output signal to the correspondent change in the analyte concentration, was calculated to be 0.756 μA mL∙ng−1. The device was tested for its long-term stability and successfully retained 94% of its performance after a period of 8 weeks in dry storage. The selectivity of the biosensing platform was tested against other biomarkers, such as carcinoembryonic antigen (CEA) and cardiac troponin I (cTnI), as well as other disturbing molecules (NaCM, glucose, endotheline 1, KCl, NaCl, and CaCl2). The interference of these molecules was quantified to be negligible, with a mean RSD of 0.32% having no significant effect on the major peak corresponding to CYFRA-21–1. The study was also validated against ELISA, being the gold standard for the detection of the biomarker, and the device exhibited a high correlation, with an RSD in the range of 0-2-3.35%. The same group (Kumar et al. 2018) later developed a similar biosensor with a slightly modified sensing platform. The novel device employed nanostructured hafnium oxide (nHfO2) as a surface modification for the reduced graphene oxide layer deposited on ITO glass. The results reported in Table 4 proved comparable with their previous study, with a marginally higher LOD and wider linear range. The sensor exhibited lower stability (90% after 27 days of storage) but a substantially higher sensitivity of 18.24 μA mL∙ng−1. The selectivity in detecting CYFRA-21–1 was tested on similar interfering molecules with the addition of NaHCO3, ascorbic acid and lactic acid. The biosensor was also validated against ELISA, showing, however, a lower correlation compared to the previous sensing platform, with an RSD in the 2.2–8% range.
Table 4.
Graphene biosensors for the detection of cancer markers in saliva
| Biomarker | Sensing Platform | Detection Method | LODa | Detection Range | Recoveryb | Validation | Assay Time | Sample Volume | Preparation | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| CYFRA-21–1 | BSA/anti-CYFRA-21–1/APTES/ZrO2-rGO/ITO/glass | DPV | 0.122 ng mL−1 | 2 – 22 ng mL−1 | N/A | ELISA RSD: 0.2– 3.35% | 16 min | N/A | Whole saliva | (Kumar et al. 2016) |
| CYFRA-21–1 | BSA/anti-CYFRA-21–1/APTES/nHfO2@RGO/ITO/glass | DPV | 0.16 ng mL−1 | 0–30 ng mL−1 | N/A | ELISARSD: 2.2–8% | 15 mn | N/A | N/A | (Kumar et al. 2018) |
| Interleukin-6 (IL-6) | Aptameric GFET | Potentio-metric | 12.2 Pm | 0.05 – 0.8 nM | N/A | N/A | 400s | 30 μL | 1:4 dilution with PBS | (Hao et al. 2019) |
| Interleukin-8 (IL-8) | AuNPs-rGO/ITO/glass | DPV | 73 pg mL−1 | 500 fg mL−1 – 4 ng mL−1 | 94.15% | N/A | 9 min | N/A | Whole saliva | (Verma et al. 2017) |
| Interleukin-8 (IL-8) | Anti-IL-8/ZnO-rGO/ITO/glass | DPV | 51.53 pg mL−1 | 100 fg/mL – 5 ng mL−1 | 95 – 100% | N/A | 10 min | N/A | 1:10 dilution with 1 × PBS | (Verma and Singh 2019) |
| Prostate specific antigen (PSA) | Graphene-polymer-PS67-b-PAA27-Au | Chemi-resistive | 40 fg mL−1 | 100 fg/mL – 100 ng mL−1 | 94 – 114% | Self-designed ELISA protocol: 94.02% linearity | 1–3 min | 10 μL | Whole saliva | (Khan et al. 2018) |
| Prostate specific antigen (PSA) | MWCNT/His-rGO/thionine/Ab/GCE | DPV | 2.8 fg mL−1 | 10 fg mL−1 – 20 ng mL−1 | 96.4 – 101.9% | ELISA 97.6% accuracy | 25 min | N/A | 1:10 dilution with PBS | (Farzin et al. 2019) |
| 3-Nitro-l-tyrosine (3-NT) | ZrO2@rGONCs/GCE | Ampero-metric | 0.009 μM | 0.025 – 855.2 μM | 94.61 – 96.24% | N/A | N/A | N/A | Whole saliva | (Maheshwaran et al. 2020) |
| 8-OHdG | Graphene modified SPE | CV | 1.8 nM | 6×10−6 – 6×10−4 M | 95 – 106% | N/A | N/A | N/A | Whole saliva | (Varodi et al. 2019) |
S/N = 3, otherwise differently specified
values obtained in real saliva samples, otherwise differently specified
Interleukins are a family of cytokines secreted by our immune system during an immune response and can be attributed to local inflammations and cancer progression (Bryant and Fitzgerald 2009). The overexpression of Interleukin-6 (IL-6), for instance, has been signaled in all types of reported tumors, therefore becoming an important cancer biomarker (Kumari et al. 2016). Similarly, the presence of interleukin-8 (IL-8) in body fluids is directly linked to tumor progression in a multitude of cancers, including oral cancer (Waugh and Wilson 2008; Xie 2001). Zao et al. (Hao et al. 2019) recently developed a biosensing device with integrated readout circuitry and wireless capability for real-time detection of IL-6 in human saliva samples. The device is based on a graphene field-effect transistor (GFET) that operates as a transducer and produces a characteristic I-V curve as signal output, which contains a richer amount of information compared to standard electrochemical techniques. The limit of detection in the picomolar range makes it suitable for the detection of IL-6 in very small concentrations. The selectivity of the GFET is tested against the epidermal growth factor (EGF) and human growth hormone (GH) that produced a voltage variation only seven times smaller than the IL-6 main peak. The reproducibility of the device has been assessed, registering maximum fluctuations of around 7%, and a coefficient of variation of ~ 0.12. The key features of this biosensor that makes it particularly promising for POC applications are the compact integrated electronic module featuring Wi-Fi communication and extremely fast response of 400 seconds after the addition of 30 μL of saliva. Verma and coworkers developed a biosensor based on rGO-modified ITO glass, decorated with Au NPs, for the detection of IL-8 in saliva samples (Verma et al. 2017). The sensor proved promising for the detection in saliva with recovery values around 94% and selectivity against spiked MAGE-A11, MAGE A-2, CD59, and h-TERT in real samples. Moreover, the device showed excellent inter-assay reproducibility with an RSD of 2.7% and promising reusability. The biosensor exhibited a maximum deflection of 1.4% after three regenerations and a slight increase of 2.9% after the fourth cycle. Remarkable is the data concerning the long-term stability, with a 94.3% performance retainment after three months and a 91.8% stability after four months of dry storage. The same research group recently developed a modified version of the biosensor with a zinc-oxide-reduced graphene oxide sensing platform and a further functionalization with IL-8 antibodies (Verma and Singh 2019). The device exhibited a lower LOD when tested in the same buffer solution, as well as a wider linear range and recovery values in the range of 95–100%. Moreover, data on the sensitivity of the device in real saliva samples (11.57 μA mL∙ng−1) were added, compared to the previous study. The stability of the biosensor was only evaluated for 70 days, without providing quantification of the performance solidity. Excellent reproducibility values were again reported, with an intra assay RSD of 3.2%.
Prostate-specific antigen is a glycoprotein enzyme that is always present in men serum in a physiological concentration that should not exceed 4 ng/mL (Prestigiacomo and Stamey 1996). Whenever PSA values overcome this threshold, men are suggested to undergo examinations to exclude the insurgence of prostate cancer, being PSA one of the most common biomarkers for the disease (Catalona et al. 1991; Stamey et al. 1987). Khan and coworkers (Khan et al. 2018) developed a fully integrated chemiresistive graphene-based biosensor using Whatman filter paper as a substrate, aiming to provide a low-cost solution for rapid PSA screening using saliva samples. The LOD of 40 fg∙mL−1 is sufficient for the detection of PSA below the clinical threshold of relevance, and the linear correlation is maintained for a very wide range. The biosensor responds to the detection of PSA with a sensitivity of 0.875 Ω∙mL∙fg−1. The stability of the device is verified up to a period of 7 weeks in dry storage, and the selectivity is tested against beta 2-microglobulin (B2 M), lactoferrin (LF), and ascorbic acid (AA). The sensing platform is directly interfaced to a miniaturized handheld PCB that makes the developed sensor promising for POC testing. A reproducibility of 88.89% is calculated among 72 identically fabricated sensor chips. The device has also been validated against a self-designed ELISA protocol with a remarkable 94% correlation with the gold standard. The integration of an electronic readout circuitry, the validation against the ELISA test, and an extremely fast response time between 1 and 3 minutes make this device a model for future development on graphene-based biosensors for POC applications. Farzin et al. (Farzin et al. 2019) also developed a nanocomposite sensor for the detection of PSA in saliva. The biosensor features a combination of 1D and 2D nanomaterials deposited on functionalized GCEs. They achieved a substantially lower LOD of 2.8 fg/mL and better values of recovery when compared to Khan et al. (Khan et al. 2018) work. However, the biosensor lacks integrated readout circuitry and consequently needs to be connected to an electrochemical analyzer. The long-term stability of the biosensor has been assessed after 2 and 4 weeks, yielding a 97% and 94.7% stability, respectively. The selectivity of the sensing platform has been verified against the disturbing agents HSA, IgG, and VEGF165. The reproducibility of the biosensor has been tested both inter and intra-assay with RSD values of 2.9% and 5.7%, respectively. Validation with the ELISA gold standard confirms the applicability of the biosensor for PSA screening in saliva, with a 97.6% accuracy with respect to the above-mentioned benchmark.
Oxidative stress induced by free radicals can significantly damage the organism, and as such, it is among the parameters that should always be kept under constant monitoring (Liguori et al. 2018; Sies et al. 2017). 8-hydroxy-2′-deoxyguanosine (8-OHdG) and 3-nitro-L-tyrosine (3-NT) has been reported, among others, as important biomarkers for DNA oxidative damage and potential risk factors for diseases, including cancer (Govindasamy et al. 2018; Kondo et al. 1999; Mohd Azmi et al. 2014; Ng et al. 2017; Valavanidis et al. 2009; Wu et al. 2004). Varodi et al. recently realized a biosensor based on graphene-modified SPEs for the detection of 8-OHdG in human saliva samples (Varodi et al. 2019). The sensor exhibited a sensitivity of 0.018 A∙M−1 and an intra-assay reproducibility of 3%, in alignment with many reported data. When tested in real saliva samples, the recovery values were satisfactory in the range of 95–106%. A recent work (Maheshwaran et al. 2020) shows an amperometric biosensor for the detection of 3-NT in human saliva, exploiting a graphene-based nanocomposite platform built on GCEs. The sensor showed high selectivity toward the detection of 3-NT when tested in the presence of glucose, H202, dopamine, UA, NaDH, NaSO4, K+, 4-nitrophenol, and 2,4-nitrobenzene.
4.3. Graphene biosensors for the detection of drugs and hazardous substances in saliva
Saliva is recently receiving increasing interest as a body fluid for rapidly screening drugs and hazardous substances or therapeutic drug monitoring (Hutchinson et al. 2018; Michalke et al. 2015; Rose et al. 1999). This is mainly due to the blood collection procedure’s invasiveness, still considered the gold standard for most of the screening tests. The use of wearable or even disposable devices can be particularly helpful for specific categories of the population. As an example, people that work in close contact with hazardous substances could be routinely screened for the presence of dangerous molecules. On the other hand, the detection of recreational drugs could also potentially be eased, reducing times and costs. The development of research devoted to wearable and portable biosensors specifically designed to detect these analytes has been significant. However, only a few research groups focused on devices based on graphene, whose studies are reported in Table 5. Moreover, none of these studies have been validated against an established conventional gold standard, as previously seen with viral and bacterial markers.
Table 5.
Graphene biosensors for the detection of drugs and hazardous substances in saliva
| Biomarker | Sensing Platform | Detection Method | LODa | Detection Range | Recoveryb | Assay Time | Sample Volume | Preparation | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Antipyrine (ANT) | TiO2-GO modified CPE | Square wave voltammetry (SWV) | 3.00 × 10−9 M | 1.2 × 10−8 – 8 × 10−5 M | 99.62% | N/A | N/A | Dilution with 5 mL BRB | (Mohamed et al. 2017) |
| Arsenic | MnO2POT/rGO/GCE | DPV | 0.042 ppb | 0.01 – 0.9 ppb | 99.8–101.8% | N/A | N/A | 3-fold dilution with nitric acid | (Ponnaiah et al. 2019) |
| Benzocaine (BEN) | TiO2-GO modified CPE | SWV | 2.48 × 10−7 M | 1 × 10−6 – 1 × 10−4 M | 99.62% | N/A | N/A | Dilution with 5 mL BRB | (Mohamed et al. 2017) |
| Cotinine | PtNP-graphene modified SPCE | CV | 0.33 nMc | 1–100 nMc | N/A | 12 min | N/A | Whole saliva | (Parate et al. 2019) |
| Ketamine | KT-MIM/MOFs@G/SPE | DPV | 4.0×10−11 mol L−1 | 1.0×10−10 – 4.0×10−5 mol L−1 | 98 – 116% | 5 min | N/A | 1:10 dilution with DI water | (Fu et al. 2019) |
S/N = 3, otherwise differently specified
values obtained in real saliva samples, otherwise differently specified
in saliva
Mohamed et al. developed a sensing platform for the simultaneous sensing of two therapeutic drugs: the analgesic antipyrine (ANT) and the anesthetic benzocaine (BEN) (Mohamed et al. 2017). The carbon paste electrode (CPE) platform exploits the decoration of immobilized GO sheets with metal nanoparticles to increase the sensitivity, which has been calculated to be 0.193 μA∙μM−1 and 0.353 μA∙μM−1 for benzocaine and antipyrine, respectively. LODs for both the analytes are below the micromolar range, and the linearity of the response spans across three orders of magnitude. The obtained recovery values are extremely close to 100% (99.62%), as it shows the 30-days stability (98.8 – 99.1%). Reproducibility and repeatability have been tested for the detection of each analyte, producing an intra-assay RSD of 1.9% (BEN) and 2.2% (ANT), and an inter-assay RSD of 2.5% (BEN) and 1.8% (ANT). The selectivity of the biosensor has been examined for the detection of one analyte in the presence of other disturbing species. Ponnaiah et al. realized a biosensor for the detection of arsenic, a toxic metalloid, in real samples of human saliva (Ponnaiah et al. 2019). The sensing platform is based on GCEs modified with an rGO/metal nanocomposite. The values of recovery are exceptionally good, as seen in the previous study, and the 30-days stability is also promising with only a 4% variation after one month. The biosensor proved to be sensitive (0.00163 μA∙ppb−1) and highly specific for the detection of arsenic, when tested in the presence of the disturbing species Cd2+, Pb2+, Hg2+, F−, Cl−, NO3−, uric acid (UA), hydroquinone (Hq), and catechol (Ct). Very low intra assay (0.53%) and inter assay (2.71%) RSD values confirmed the reproducibility and repeatability of the platform.
Parate et al. fabricated an electrochemical biosensor to assess the exposure to smoke and tobacco byproducts (Parate et al. 2019). Specifically, analytes that are cotinine, tobacco alkaloid, and metabolite of nicotine were used as biomarkers in smokers. Screen-printed carbon electrodes (SPCEs) are modified with graphene, decorated with platinum nanoparticles (PtNPs) and biofunctionalized with an electrodeposited molecularly imprinted polymer (MIP). A LOD of 0.33 nM, a linear range of 1–100 nM, and a sensitivity of 1.89 μA/decade have been calculated directly on real saliva samples. Selectivity is tested against similar species, nicotine, and myosmine. Ketamine, a therapeutic anesthetic drug, is increasingly being used as a recreational drug (Morgan et al. 2012). Fu and coworkers realized a biosensor based on graphene-modified SPEs for the detection of ketamine in saliva samples (Fu et al. 2019). The ultra-low LOD close to the picomolar range and the ultra-wide linear range spanning five orders of magnitude assures the sensitive detection of the analyte. The values of recovery in real saliva samples, however, are poorer when compared to the previously analyzed studies. The stability of the sensor is measured to be 90% after a two-months period, and selectivity is tested detecting ketamine in the presence of NKT, MDMA, MA, dopamine (DA), and ascorbic acid (AA). Inter-assay and intra-assay RSD values of reproducibility are below 3.5% in both cases.
4.4. Graphene biosensors for the detection of health-indicating analytes in saliva
The monitoring of salivary biomarkers is a fundamental tool for assessing the oral health status of an individual, which is directly correlated to the overall health status (Celecová et al. 2013; Khanna 2008; Lawrence 2002; Mojon et al. 1999). The absence or presence of specific biomarkers of the oral cavity can be related to bad nutrition (Mojon et al. 1999), the insurgence of systemic diseases (Lawrence 2002), oral carcinogenesis (Khanna 2008; Markopoulos et al. 2010; Nagler et al. 2006), and oxidative stress (Celecová et al. 2013; Kamodyová et al. 2015), to cite a few. As a result, most studies regarding the development of graphene-based biosensors have focused on the detection of such health status markers, which are summarized in Table 6. Only half of the reported biosensors have been validated against a conventional test (ELFA or ELISA), demonstrating once again the need for systematic data reporting to uniformly compare new findings and verify their suitability for clinical applications.
Table 6.
Graphene biosensors for the detection of health-indicating analytes in saliva
| Biomarker | Sensing Platform | Detection Method | LODa | Detection Range | Recoveryb | Validation | Assay Time | Sample Volume | Preparation | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| Cardiac troponin (cTnI) | N-prGO-(py-PEG/Py-COOH=20:1)/GCE | DPV | 1 pg mL−1 | 0.001 – 100 ng mL−1 | N/A | ELFA | N/A | N/A | Whole saliva | (Chekin et al. 2018b) |
| Cortisol | GNPs and PS67-b-PAA27 on Whatman filter paper | EIS | 1 pg mL−1 | 1 pg mL−1 – 10 ng mL−1 | N/A | ELISA RE62011 kit R2 = 0.9386 |
10 min | 20 μL | 1:10 dilution | (Khan et al. 2017a) |
| Cortisol | GNPs and PS67-b-PAA27 on Whatman filter paper | Chemi-resistive | 3 pg mL−1c | 0.018 – 4.552 ng mL−1c 3 pg mL−1 – 10 μg mL−1d |
N/A | ELISA RE62011 kit R2 = 0.9951 |
12 min | 5 μL | Whole saliva | (Khan et al. 2017b) |
| Cortisol | c-Mab conjugated rGO sheet/ITO/glass | Chemi-resistive | 10 pg mL−1 (27.6 pM) | 1 – 10 ng mL−1 | N/A | ELISA | N/A | N/A | 1:1 dilution with buffer solution | (Kim et al. 2017c) |
| Cortisol | anti-cortisol antibody/d-BSA/rGO/quartz | EIS | 10 pM | 10 pM – 100 nM | 92 – 103% | N/A | 30 min | 1 mL | Whole saliva | (Kim et al. 2017b) |
| Cortisol | BSA/anti-Cab/SAM/CBμE/PS_b_PAA/G RP/Paper | EIS | 0.87 pg mL−1 | 1 pg mL−1 – 10 ng mL−1 | 91 – 113% | ELISA 96.3% accuracy | 12 min | 5 μL | Whole saliva | (Khan et al. 2019) |
| Ghrelin (GHRL) | anti-GHRL-Phe-rGO/SPCE | DPV | 1 pg mL−1e | 10−3 – 100 ng/mL | 97% | ELISA LOD = 161 pg/mL | N/A | 3 μL | N/A | (Martínez-García et al. 2016) |
| Glucose | CuO/rGO/CNF/GCE | Chronoamperometric | 0.1 μM | 1 μM – 5.3 mM | 99.7 – 101.3% | N/A | 3 s | N/A | 1:10 dilution | (Ye et al. 2013) |
| L-tryptophan (Try) | AuNPs/rGO/SPE | DPV | 0.39 μmol/L | 0.5 – 500 μmol/L | 94.6 – 107.6% | N/A | N/A | N/A | Whole saliva | (Nazarpour et al. 2020) |
| Myoglobin (Mb) | anti-Mb-Ab/d-BSA/r-HGO/quartz | EIS | 2.37 pM | 5 pM – 10 nM | 90.9–111.3% | N/A | N/A | N/A | Whole saliva | (Yoo et al. 2020) |
| Nitrite (NIT) | 3D printed polylactic acid containing graphene (G-PLA) electrodes | DPV BIA-MPA |
BIA-MPA: 0.03 μmol/L DPV: 30 μmol/L |
BIA-MPA: 0.5–250 μmol/L DPV: 50–1300 μmol/L |
100 – 110% | N/A | N/A | N/A | Dilution | (Cardoso et al. 2020) |
| Peptide YY (PYY) | anti-PYY-Phe-rGO/SPCE | DPV | 0.02 pg/mLe | 10−4 – 10 ng/mL | 99% | ELISA LOD = 2.84 pg/mL | N/A | 3 μL | N/A | (Martínez-García et al. 2016) |
| Serotonin | GO-chitosan modified GSPE | DPV | 3.2 Nm | 0.01 – 100 μM | 104.20% | N/A | 20 min | 10 μL | N/A | (Adumiträchioaie et al. 2019) |
| Uric acid (UA) | 3D printed polylactic acid containing graphene (G-PLA) electrodes | DPV BIA-MPA |
BIA-MPA: 0.02 μmol/L DPV: 0.5 μmol/L |
BIA-MPA: 0.5–250 μmol/L DPV: 10–70 μmol/L |
120% | N/A | N/A | N/A | Dilution | (Cardoso et al. 2020) |
S/N = 3, otherwise differently specified
values obtained in real saliva samples, otherwise differently specified
in saliva
in buffer
S/N = 2
Cardiac troponin I (cTnI) is a cardiac regulatory protein and one of the most accurate biomarkers for detecting acute myocardial infarction and consequent cardiac muscle damage (Mishra et al. 2018; Peacock IV et al. 2008; Sharma et al. 2004). Chekin et al. have developed a biosensor for the detection of cTnI in human serum and saliva samples based on graphene-modified GCEs (Chekin et al. 2018b). The sensor has been validated against the conventional ELFA test in serum, but no data on the correlation are provided. The stability of the sensor has been verified over a one-month period, resulting in only an 8% loss of performance. The sensitivity is calculated as usual as the slope of the current-concentration calibration curve and has been reported to be 41 μA cm−2/decade. Selective detection of cTnI has been certified in the presence of BNP, BSA, and lysozyme as disturbing agents. Another biomarker of acute myocardial infarction is the protein myoglobin, which has been intensely studied for years now (Hendgen-Cotta et al. 2010; Ishii et al. 1997). Yoo et al. (Yoo et al. 2020) realized a sensing platform based on reduced graphene oxide. They achieved a low LOD in the picomolar range and acceptable values of recovery when tested in real saliva samples (91–111%). The selectivity of the biosensor has been tested in the presence of hemoglobin (Hb) and cytochrome C (Cyt-C).
Cortisol is a lipophilic steroid hormone commonly related to psychological stress (Holsboer and Ising 2010). It has been proven that the levels of cortisol in saliva are highly correlated with those found in serum, opening an exciting opportunity for non-invasive monitoring of cortisol through saliva collection and analysis (Aardal and Holm 1995; Teruhisa et al. 1981). Therefore, many research groups started developing biosensors for the detection of cortisol, and Khan and coworkers (Khan et al. 2017a) were the first to realize a graphene-based portable biosensor with integrated handheld EIS analyzer (AD5933 chip). The sensing platform is validated against an ELISA cortisol luminescence immunoassay (RE62011) kit showing a correlation coefficient R2 = 0.9386. The same research group used an identical sensing platform based on Whatman filter paper to design another biosensor, based on a chemiresistive detection mechanism (Khan et al. 2017b). The LOD and linear range of this sensor are determined directly on saliva samples, and a sensitivity of 50 Ω∙pg∙mL−1 is reported. The sensing platform features the same miniaturized handheld PCB seen in their previous version of the device. The sensor is also again compared to the ELISA RE62011 kit, showing a much-improved correlation coefficient R2 = 0.9951. The device performance has also been certified as stable after six weeks of the dry storage period. Khan et al. (Khan et al. 2019) recently developed another biosensor based on a different platform but still having graphene as a sensing element. Recovery values are reported for the first time and are in the acceptable range but far worse than in many other cases analyzed in this review. Once again, the biosensor is validated against ELISA, exhibiting an accuracy of 96.3% with respect to the gold standard. The same stability of 6 weeks as in the previous case, has been reported, and selectivity has also been tested in the presence of ascorbic acid (AA). Kim et al. also developed a cortisol sensing device based on chemiresistive detection. The sensor has been validated against the ELISA gold standard, but no correlation coefficient or accuracy has been provided by the authors (Kim et al. 2017c). The biosensor exhibited high stability, being able to retain its performance for several months. The selectivity has been proved in a neuro-cell culture matrix where different neuroendocrine, neurotransmitter, and protein were present. Kim and coworkers developed another rGO-based sensing platform for cortisol detection in saliva (Kim et al. 2017b). No gold standard validation was performed; however, the sensor was tested for recovery in human saliva samples showing values in the 92–103% range, the best among the two reported herein for cortisol. When cortisol monitoring was performed in the presence of aldosterone and progesterone, two interfering molecules, the resistance increment relative to the cortisol detection was 8-fold higher, showing excellent selectivity for the analyte.
Ghrelin (GHRL) and peptide YY (PYY) are two other hormones that are instead associated with hunger; they have opposite functions, with ghrelin increasing appetite and peptide YY decreasing it (Karamanakos et al. 2008). These hormones are indicators of appetite disorders. A prior study (Maier et al. 2008) shows how deregulated food intake in obese subjects can be monitored by observing specific levels of GHRL and PYY. Martínez-García and others realized an rGO-based biosensor with two different biofunctionalizations to detect each hormone (Martínez-García et al. 2016). The results were then compared with ELISA, and in both cases (GHRL and PPY) the biosensor demonstrated lower limits of detection as reported in Table 6. The sensitivity of the platform is different for GHRL and PPY, with the former being 246 nA/decade and the latter being 422 nA/decade. The stability of the fabricated sensor is limited to only 10 days. However, the sensor proved to be highly selective for both species, in the presence of the interfering molecules adiponectin (APN), insulin (INS), GH, deacylated GHRL (DA-G), and follicle stimulant hormone (FSH). The biosensor also showed high inter-assay reproducibility, both intraday (2.9% and 2.4% for GHRL and PYY respectively) and inter-days (2.8% and 2.9% for GHRL and PYY respectively). The recovery values in human saliva samples are among the best reported herein, 97% for ghrelin, and 99% for peptide YY. Glucose is among the most common investigated analytes and health biomarkers, being the best indicator for the insurgence or control of diabetes mellitus (Satish et al. 2014; Shanbhag and Prasad 2016; Ship 2003). The effort in the scientific community for the realization of glucose devices has been massive in the last decade. However, only a few used saliva as sampling fluid, with the majority of them performing analysis on whole blood or serum. Among those focusing on salivary analysis, Ye and coworkers (Ye et al. 2013) realized a graphene-based platform for the detection of glucose though chronoamperometry. The fabricated sensor reported exceptional recovery values when tested on real saliva samples, as well as a high sensitivity of 912.7 μA∙mM−1∙cm−2. The biosensor is characterized by an ultrafast detection time of less than 3s after the addition of saliva, the lowest among all the reported studies. The stability of 92% has been assessed after 4 weeks of dry storage. The selectivity of the sensor has been tested in the presence of UA, AA, DA, galactose (Gal), mannose (Man), lactose (Lac), acetaminophen (Ace), epinephrine (Ep) and KCl.
The oral cavity is also a host of many biomarkers indicating the mental health status of an individual. L-tryptophan is an amino acid, the mediator of serotonin, and consequently, a good indicator of possible mental health disorders (Murphy et al. 1974), while serotonin itself has already been demonstrated as being closely related to mood changes or depression (Barchas 2012; Blier and El Mansari 2013). Nazarpour et al. have recently developed an electrochemical biosensor for the detection of L-tryptophan (Try) in human saliva samples (Nazarpour et al. 2020). The recovery values in real saliva were acceptable, in the 95–108% range, and the measured reproducibility was high both intra-assay (3.69%) and inter-assay (4.36%). They classified as interference a species causing a current change higher than 10%, and the sensor proved to be selective for the detection of Try only in the presence of AA, arginine, urea, glutamine, vitamin B1, creatine, valine, leucine, and isoleucine. Adumitrăchioaie et al. (Adumitrăchioaie et al. 2019) recently fabricated an immunosensor based on GO-chitosan modified GSPEs with a sensitivity of 0.05 μA∙μM−1. The sensor exhibited recovery of 104.2% when tested in real saliva samples and retained 91.3% of its performance after 4 weeks of dry storage. As in the previous case of the detection of Try (Nazarpour et al. 2020), the sensor was tested against many interfering molecules, namely adrenaline (Adr), noradrenaline (NA), DA, paracetamol (P), aspirin, UA, AA, and glucose. Cardoso et al. (Cardoso et al. 2020) recently reported the first example of 3D-printed graphene-based biosensors for the detection of analytes in real body fluids, measuring the concentration of nitrite (NIT) and uric acid (UA) in human saliva. Nitrites are easily incorporated through food consumption (James et al. 2015) and play an important role in the physiological activity of the body (Ma et al. 2018) while uric acid, produced by the breakdown of purines during metabolism, can be associated to cell death and have a potential pathogenic effect in neurological and infectious diseases (El Ridi and Tallima 2017; Liu et al. 2019). The biosensor consists of electrodes that are printed using graphene-modified polylactic acid (G-PLA). The study was carried out using two different electrochemical detection mechanism, differential pulse voltammetry (DPV) and multiple-pulse amperometry combined with batch-injection analysis (BIA-MPA). Different values of LOD and linearity were obtained with the two techniques (details in Table 6), demonstrating how the detection mechanism influences the sensitivity of the device. The repeatability also varied depending on the detection technique, with an RSD value of 2.1% (UA) and 1.1% (NIT) when using BIA-MPA, and an RSD value of 8.2% (UA) and 7.2% (NIT) when using DPV. When tested in real saliva samples, the recovery performance of the biosensor is acceptable (100–110%) for NIT, but poor for UA (120%).
5. Conclusions and future perspectives
This review reports a comprehensive summary of graphene-based biosensors for salivary biomarker detection. We have covered the advantages of graphene and its derivatives to build highly sensitive biosensors and bioelectronics. Human saliva has been introduced for biomarker detection due to easy access, noninvasive sample collection, and the source of an abundance of various analytes. Many graphene-based nanobiosensors have been developed to target multiple salivary biomarkers categorized for viral/bacterial biomarkers, cancer biomarkers, drugs/hazardous substances, and health-indicating substances. We have discussed the details of materials, structural designs, analyte detection methods, and performance of graphene bioelectronics, along with system miniaturization for oral-wearable applications.
A schematic illustration in Figure 6 delivers our perspectives of wearable graphene oral biosensors for future clinical applications with enhanced data acquisition, remote health monitoring, and personalized medicine. A patient will become the owner of personal data collected by the wearable POC oral biosensors at home. It will be the wearer’s responsibility to decide when and how to share the data with clinicians. A clinician could then offer the opportunity to store such data on a secure decentralized cloud, aiming to build a patient-specific database containing health information. This process would extremely simplify the workflow for both patients and clinicians since no in-person and frequent hospital visit is required. In addition, both parties will save costs in travel, labor, and system management, while clinicians can offer a timely diagnosis and follow-up treatment based on pre-collected data during daily activities. Overall, rapid and more accurate personalized treatment is possible for patients. In particular, the wearable devices and home healthcare will provide benefits to infants and older adults who always require caregivers for hospital visits.
Figure 6. Wearable oral graphene biosensors for clinical applications.
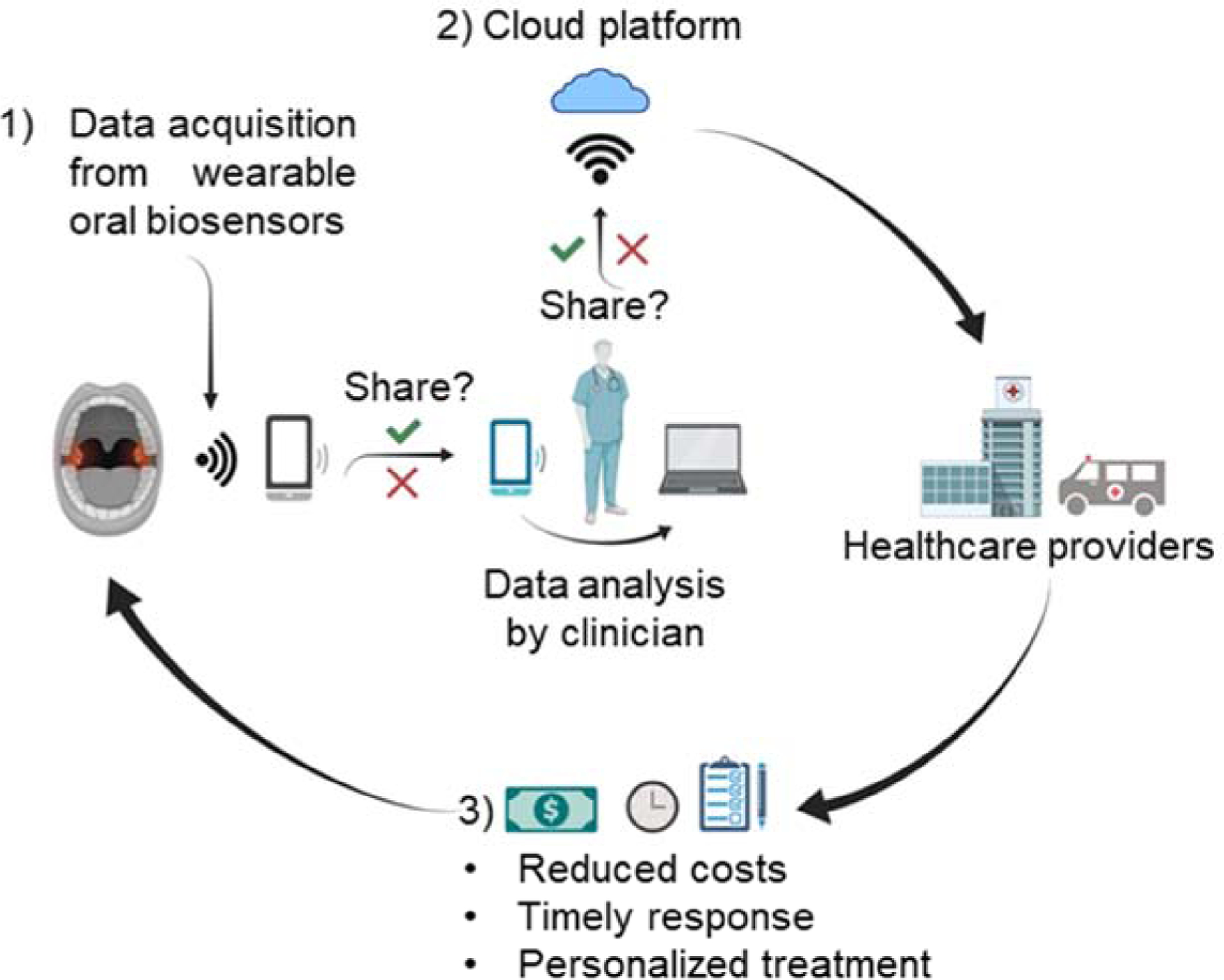
This schematic illustration includes a possible implementation of wearable oral graphene biosensors for future health monitoring and therapeutics: 1) Data from wearable oral biosensors are collected and wirelessly sent to the clinician for data analysis, 2) Data can be stored on the cloud platform, upon permission, to build a customized database of information for each patient, and 3) Healthcare providers suggest a personalized treatment based on the condition and clinical history of the patient.
For the next-generation diagnosis and treatment, we still need to improve the quality of oral-wearable biosensors and bioelectronics. In this regard, we expect graphene and graphene-derivatives to play a pivotal role in developing next-generation oral biosensors. The excellent mechanical and electrical properties of graphene, along with its biocompatibility and antimicrobial traits, make it an up-and-coming candidate for its use in clinical applications. The versatility and ductility of graphene allow it to be easily customized through a tailored functionalization to meet specific needs in terms of analyte detection. Moreover, the increasing efforts in making graphene production a low-cost, reliable, large-scale manufacturing process will foster its use in mass production of high-quality oral-wearable biosystems. Further miniaturization of electronic circuits, wireless communication units, and powering systems will provide additional improvements for ergonomic, easy-to-use devices. In summary, we believe that considering the existing challenges in materials, electronics, and manufacturing will provide new opportunities to utilize the graphene-based biosensors for more practical, home healthcare applications in daily life.
Highlights.
Human saliva offers easy access to a large variety of analytes, making it a promising candidate for its use in point-of-care devices.
This review summarizes the most recent graphene-based nanobiosensors and oral bioelectronics for salivary biomarker detection.
We discuss the details of structural designs of graphene electronics, use cases of salivary biomarkers, the performance of existing sensors, and applications in health monitoring.
This review describes current challenges in materials and systems and future directions of the graphene bioelectronics for clinical point-of-care applications.
Acknowledgments
W.-H.Y. acknowledges the support of the Alzheimer’s Association (2019-AARGD-NTF-643460) and the National Institutes of Health (NIH R21AG064309). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This work was supported by the Center Seed Grant from the Georgia Tech Institute for Electronics and Nanotechnology, a member of the National Nanotechnology Coordinated Infrastructure (NNCI), which is supported by the National Science Foundation (ECCS-2025462).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Conflict of interests
We confirm that there are no known conflicts of interest associated with this publication. There has been no significant financial support for this work that could have influenced its outcome.
References
- Aardal E, Holm AC, 1995. Eur. J. Clin. Chem. Clin. Biochem 33(12), 927–932. [DOI] [PubMed] [Google Scholar]
- Adumitrăchioaie A, Tertiș M, Suciu M, Graur F, Cristea C, 2019. Electrochim. Acta 311, 50–61. [Google Scholar]
- Akhavan O, Ghaderi E, 2010. ACS Nano 4(10), 5731–5736. [DOI] [PubMed] [Google Scholar]
- Albishri HM, Abd El-Hady D, 2019. Talanta 200, 107–114. [DOI] [PubMed] [Google Scholar]
- Arakawa T, Kuroki Y, Nitta H, Chouhan P, Toma K, Sawada S. i., Takeuchi S, Sekita T, Akiyoshi K, Minakuchi S, 2016. Biosens. Bioelectron 84, 106–111. [DOI] [PubMed] [Google Scholar]
- Arellano-Garcia M, Hu S, Wang J, Henson B, Zhou H, Chia D, Wong D, 2008. Oral Dis. 14(8), 705–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avouris P, 2010. Nano Lett. 10(11), 4285–4294. [DOI] [PubMed] [Google Scholar]
- Balandin AA, Ghosh S, Bao W, Calizo I, Teweldebrhan D, Miao F, Lau CN, 2008. Nano Lett. 8(3), 902–907. [DOI] [PubMed] [Google Scholar]
- Bandodkar AJ, Jeerapan I, Wang J, 2016. ACS Sens. 1(5), 464–482. [Google Scholar]
- Barchas J, 2012. Serotonin and Behavior, Elsevier. [Google Scholar]
- Benchirouf A, Müller C, Kanoun O, 2016. Nanoscale Res. Lett 11(1), 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blier P, El Mansari M, 2013. Philos. Trans. R. Soc. B 368(1615), 20120536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin KI, Sikes KJ, Jiang Z, Klima M, Fudenberg G, Hone J, Kim P, Stormer HL, 2008. Solid State Commun. 146(9), 351–355. [Google Scholar]
- Bonaccorso F, Lombardo A, Hasan T, Sun Z, Colombo L, Ferrari AC, 2012. Mater. Today 15(12), 564–589. [Google Scholar]
- Botta A, de Donato W, Persico V, Pescapé A, 2016. Future Gener. Comput. Syst 56, 684–700. [Google Scholar]
- Breslin PA, 2013. Curr. Biol 23(9), R409–R418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant C, Fitzgerald KA, 2009. Trends Cell Biol. 19(9), 455–464. [DOI] [PubMed] [Google Scholar]
- Burnett TL, Yakimova R, Kazakova O, 2012. J. Appl. Phys 112(5), 054308. [Google Scholar]
- Camara N, Rius G, Huntzinger J-R, Tiberj A, Mestres N, Godignon P, Camassel J, 2008. Appl. Phys. Lett 93(12), 123503. [Google Scholar]
- Cammann K, 1977. Fresenius Z. Anal. Chem 287(1), 1–9. [Google Scholar]
- Campuzano S, Yánez-Sedeño P, Pingarrón JM, 2017. Trends Anal. Chem 86, 14–24. [Google Scholar]
- Cardoso RM, Silva PR, Lima AP, Rocha DP, Oliveira TC, do Prado TM, Fava EL, Fatibello-Filho O, Richter EM, Muñoz RA, 2020. Sens. Actuators B Chem 307, 127621. [Google Scholar]
- Castro Neto AH, Guinea F, Peres NMR, Novoselov KS, Geim AK, 2009. Rev. Mod. Phys 81(1), 109–162. [Google Scholar]
- Catalona WJ, Smith DS, Ratliff TL, Dodds KM, Coplen DE, Yuan JJJ, Petros JA, Andriole GL, 1991. N. Engl. J. Med 324(17), 1156–1161. [DOI] [PubMed] [Google Scholar]
- Celecová V, Kamodyová N, Tóthová Ľ, Kúdela M, Celec P, 2013. J. Oral Pathol. Med 42(3), 263–266. [DOI] [PubMed] [Google Scholar]
- Cernat A, Tertis M, Gandouzi I, Bakhrouf A, Suciu M, Cristea C, 2018. Electrochem. Commun 88, 5–9. [Google Scholar]
- Chauhan N, Maekawa T, Kumar DNS, 2017. J. Mater. Res 32(15), 2860–2882. [Google Scholar]
- Chekin F, Bagga K, Subramanian P, Jijie R, Singh SK, Kurungot S, Boukherroub R, Szunerits S, 2018a. Sens. Actuators B Chem 262, 991–1000. [Google Scholar]
- Chekin F, Vasilescu A, Jijie R, Singh SK, Kurungot S, Iancu M, Badea G, Boukherroub R, Szunerits S, 2018b. Sens. Actuators B Chem 262, 180–187. [Google Scholar]
- Choromańska M, Klimiuk A, Kostecka-Sochoń P, Wilczyńska K, Kwiatkowski M, Okuniewska N, Waszkiewicz N, Zalewska A, Maciejczyk M, 2017. Int. J. Mol. Sci 18(10), 2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary M, Kumar V, Singh A, Singh MP, Kaur S, Reddy G, Pasricha R, Singh S, Arora K, 2013. J. Biosens. Bioelectron 4(4), 1–9. [Google Scholar]
- Chua CK, Pumera M, 2013. Chem. Soc. Rev 42(8), 3222–3233. [DOI] [PubMed] [Google Scholar]
- Ciui B, Tertis M, Feurdean CN, Ilea A, Sandulescu R, Wang J, Cristea C, 2019. Sens. Actuators B Chem 281, 399–407. [Google Scholar]
- Corstjens PLAM, Abrams WR, Malamud D, 2012. J. Am. Dent. Assoc 143(10 Suppl), 12S–18S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, Wang B, Yuan Y, Han S, C I, Wang Z, 2015. IEEE Commun. Mag 53(9), 74–81. [Google Scholar]
- De Boever J, Kohen F, Bouve J, Leyseele D, Vandekerckhove D, 1990. Clin. Chem 36(12), 2036–2041. [PubMed] [Google Scholar]
- Delaleu N, Mydel P, Kwee I, Brun JG, Jonsson MV, Jonsson R, 2015. Arthritis Rheumatol. 67(4), 1084–1095. [DOI] [PubMed] [Google Scholar]
- Dhakate SR, Chauhan N, Sharma S, Mathur RB, 2011. Carbon 49(13), 4170–4178. [Google Scholar]
- Dong X, Wang P, Fang W, Su C-Y, Chen Y-H, Li L-J, Huang W, Chen P, 2011. Carbon 49(11), 3672–3678. [Google Scholar]
- Du X, Skachko I, Barker A, Andrei EY, 2008. Nat. Nanotechnol 3(8), 491–495. [DOI] [PubMed] [Google Scholar]
- Eda G, Ball J, Mattevi C, Acik M, Artiglia L, Granozzi G, Chabal Y, Anthopoulos TD, Chhowalla M, 2011. J. Mater. Chem 21(30), 11217–11223. [Google Scholar]
- Edwards RS, Coleman KS, 2013. Nanoscale 5(1), 38–51. [DOI] [PubMed] [Google Scholar]
- Eftekhari A, Hasanzadeh M, Sharifi S, Dizaj SM, Khalilov R, Ahmadian E, 2019. Int. J. Biol. Macromol 124, 1246–1255. [DOI] [PubMed] [Google Scholar]
- El Ridi R, Tallima H, 2017. J. Adv. Res 8(5), 487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzin L, Sadjadi S, Shamsipur M, Sheibani S, 2019. J. Pharm. Biomed 172, 259–267. [DOI] [PubMed] [Google Scholar]
- Ferrari AC, Bonaccorso F, Fal’ko V, Novoselov KS, Roche S, Bøggild P, Borini S, Koppens FHL, Palermo V, Pugno N, Garrido JA, Sordan R, Bianco A, Ballerini L, Prato M, Lidorikis E, Kivioja J, Marinelli C, Ryhänen T, Morpurgo A, Coleman JN, Nicolosi V, Colombo L, Fert A, Garcia-Hernandez M, Bachtold A, Schneider GF, Guinea F, Dekker C, Barbone M, Sun Z, Galiotis C, Grigorenko AN, Konstantatos G, Kis A, Katsnelson M, Vandersypen L, Loiseau A, Morandi V, Neumaier D, Treossi E, Pellegrini V, Polini M, Tredicucci A, Williams GM, Hee Hong B, Ahn J-H, Min Kim J, Zirath H, van Wees BJ, van der Zant H, Occhipinti L, Di Matteo A, Kinloch IA, Seyller T, Quesnel E, Feng X, Teo K, Rupesinghe N, Hakonen P, Neil SRT, Tannock Q, Löfwander T, Kinaret J, 2015. Nanoscale 7(11), 4598–4810. [DOI] [PubMed] [Google Scholar]
- Franco-Martínez L, Castillo-Felipe C, 2020. In: Tvarijonaviciute A, Martínez-Subiela S, López-Jornet P, Lamy E (Eds.), Saliva in Health and Disease: The Present and Future of a Unique Sample for Diagnosis, pp. 49–65. Springer International Publishing, Cham. [Google Scholar]
- Fu K, Zhang R, He J, Bai H, Zhang G, 2019. Biosens. Bioelectron 143, 111636. [DOI] [PubMed] [Google Scholar]
- Gandouzi I, Tertis M, Cernat A, Bakhrouf A, Coros M, Pruneanu S, Cristea C, 2018. Bioelectrochemistry 120, 94–103. [DOI] [PubMed] [Google Scholar]
- Gao J, Gao Y, Han Y, Pang J, Wang C, Wang Y, Liu H, Zhang Y, Han L, 2020. ACS Appl. Electron. Mater 2(4), 1090–1098. [Google Scholar]
- Gao L, Ren W, Zhao J, Ma L-P, Chen Z, Cheng H-M, 2010. Appl. Phys. Lett 97(18), 183109. [Google Scholar]
- Garbieri TF, Brozoski DT, Dionísio TJ, Santos CF, Neves L.T.d., 2017. J. Appl. Oral Sci 25(2), 147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia PT, Dias AA, Souza JAC, Coltro WKT, 2018. Anal. Chim. Acta 1041, 50–57. [DOI] [PubMed] [Google Scholar]
- Geim AK, Novoselov KS, 2007. Nature Mater. 6(3), 183–191. [DOI] [PubMed] [Google Scholar]
- Georgakilas V, Otyepka M, Bourlinos AB, Chandra V, Kim N, Kemp KC, Hobza P, Zboril R, Kim KS, 2012. Chem. Rev 112(11), 6156–6214. [DOI] [PubMed] [Google Scholar]
- Georgakilas V, Tiwari JN, Kemp KC, Perman JA, Bourlinos AB, Kim KS, Zboril R, 2016. Chem. Rev 116(9), 5464–5519. [DOI] [PubMed] [Google Scholar]
- Gilje S, Han S, Wang M, Wang KL, Kaner RB, 2007. Nano Lett. 7(11), 3394–3398. [DOI] [PubMed] [Google Scholar]
- Goossens N, Nakagawa S, Sun X, Hoshida Y, 2015. Transl. Cancer Res 4(3), 256–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami Y, Mishra R, Agrawal AP, Agrawal LA, 2015. IOSR J. Dental Med. Sci 14(3), 80–87. [Google Scholar]
- Goumri M, Lucas B, Ratier B, Baitoul M, 2016. Opt. Mater 60, 105–113. [Google Scholar]
- Govindasamy M, Manavalan S, Chen S-M, Umamaheswari R, Chen T-W, 2018. Sens. Actuators B Chem 272, 274–281. [Google Scholar]
- Gug IT, Tertis M, Hosu O, Cristea C, 2019. Trends Anal. Chem 113, 301–316. [Google Scholar]
- Gupta A, Tripathi A, Patil R, Kumar V, Khanna V, Singh V, 2019. J. Oral Biol. Craniofac. Res 9(1), 19–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeckel R, Hanecke P, 1996. Eur. J. Clin. Chem. Clin. Biochem 34(3), 171–192. [PubMed] [Google Scholar]
- Hamad AWR, Gaphor SM, Shawagfeh MT, Al-Talabani NG, 2011. Acad. J. Cancer Res 4(2), 47–55. [Google Scholar]
- Hao Z, Pan Y, Shao W, Lin Q, Zhao X, 2019. Biosens. Bioelectron 134, 16–23. [DOI] [PubMed] [Google Scholar]
- Hendgen-Cotta U, Flögel U, Kelm M, Rassaf T, 2010. J. Exp. Biol 213(16), 2734–2740. [DOI] [PubMed] [Google Scholar]
- Henry NL, Hayes DF, 2012. Mol. Oncol 6(2), 140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert R, Kim J-H, Kim YS, Lee HM, Yeo W-H, 2018. Materials 11(2), 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr AE, Hatch AV, Throckmorton DJ, Tran HM, Brennan JS, Giannobile WV, Singh AK, 2007. Proc. Natl. Acad. Sci. U.S.A 104(13), 5268–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmström P, Syrjänen S, Laine P, Valle SL, Suni J, 1990. J. Med. Virol 30(4), 245–248. [DOI] [PubMed] [Google Scholar]
- Holsboer F, Ising M, 2010. Annu. Rev. Psychol 61(1), 81–109. [DOI] [PubMed] [Google Scholar]
- Hu S, Arellano M, Boontheung P, Wang J, Zhou H, Jiang J, Elashoff D, Wei R, Loo JA, Wong DT, 2008. Clin. Cancer Res 14(19), 6246–6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson L, Sinclair M, Reid B, Burnett K, Callan B, 2018. Br. J. Clin. Pharmacol 84(6), 1089–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idkaidek N, Arafat T, 2012. Mol. Pharm 9(8), 2358–2363. [DOI] [PubMed] [Google Scholar]
- Ishii J, Wang J. h., Naruse H, Taga S, Kinoshita M, Kurokawa H, Iwase M, Kondo T, Nomura M, Nagamura Y, 1997. Clin. Chem 43(8), 1372–1378. [PubMed] [Google Scholar]
- Izawa S, Miki K, Liu X, Ogawa N, 2013. Brain Behav. Immun 27, 38–41. [DOI] [PubMed] [Google Scholar]
- James PE, Willis GR, Allen JD, Winyard PG, Jones AM, 2015. Nitric Oxide 48, 44–50. [DOI] [PubMed] [Google Scholar]
- Janyasupab M, Liu C-W, Chanlek N, Chio-Srichan S, Promptmas C, Surareungchai W, 2019. Sens. Actuators B Chem 286, 550–563. [Google Scholar]
- Jastrzębska AM, Kurtycz P, Olszyna AR, 2012. J. Nanopart. Res 14(12), 1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javaid MA, Ahmed AS, Durand R, Tran SD, 2016. J. Oral Biol. Craniofac. Res 6(1), 67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokerst JV, Raamanathan A, Christodoulides N, Floriano PN, Pollard AA, Simmons GW, Wong J, Gage C, Furmaga WB, Redding SW, McDevitt JT, 2009. Biosens. Bioelectron 24(12), 3622–3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi SR, Sharma A, Kim G-H, Jang J, 2020. Mater. Sci. Eng. C 108, 110465. [DOI] [PubMed] [Google Scholar]
- Jou Y-J, Hua C-H, Lin C-D, Lai C-H, Huang S-H, Tsai M-H, Kao J-Y, Lin C-W, 2014. Clin. Chim. Acta 436, 121–129. [DOI] [PubMed] [Google Scholar]
- Kaczor-Urbanowicz KE, Martin Carreras-Presas C, Aro K, Tu M, Garcia-Godoy F, Wong DT, 2017. Exp. Biol. Med 242(5), 459–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamodyová N, Červenka T, Celec P, 2015. Front. Cell. Infect. Microbiol 5, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D, Kirienko DR, Webster P, Fisher AL, Kirienko NV, 2018. Virulence 9(1), 804–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamanakos SN, Vagenas K, Kalfarentzos F, Alexandrides TK, 2008. Ann. Surg 247(3), 401–407. [DOI] [PubMed] [Google Scholar]
- Khan M, Dighe K, Wang Z, Srivastava I, Daza E, Schwartz-Dual A, Ghannam J, Misra S, Pan D, 2018. Analyst 143(5), 1094–1103. [DOI] [PubMed] [Google Scholar]
- Khan MS, Dighe K, Wang Z, Schwartz-Duval AS, Misra SK, Pan D, 2017a. 2017 IEEE Healthcare Innovations and Point of Care Technologies (HI-POCT), pp. 184–187. IEEE. [Google Scholar]
- Khan MS, Dighe K, Wang Z, Srivastava I, Schwartz-Duval AS, Misra SK, Pan D, 2019. Analyst 144(4), 1448–1457. [DOI] [PubMed] [Google Scholar]
- Khan MS, Misra SK, Wang Z, Daza E, Schwartz-Duval AS, Kus JM, Pan D, Pan D, 2017b. Anal. Chem 89(3), 2107–2115. [DOI] [PubMed] [Google Scholar]
- Khan QA, Shaur A, Khan TA, Joya YF, Awan M, 2017c. Cogent Chem. 3(1), 1298980. [Google Scholar]
- Khanna S, 2008. Int. J. Environ. Res. Public Health 5(5), 418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurshid Z, Naseem M, Sheikh Z, Najeeb S, Shahab S, Zafar MS, 2016. Saudi Pharm. J 24(5), 515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Campbell AS, de Ávila BE-F, Wang J, 2019. Nature Biotechnol. 37(4), 389–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Imani S, de Araujo WR, Warchall J, Valdés-Ramírez G, Paixão TRLC, Mercier PP, Wang J, 2015. Biosens. Bioelectron 74, 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kim M, Lee M-S, Kim K, Ji S, Kim Y-T, Park J, Na K, Bae K-H, Kyun Kim H, Bien F, Young Lee C, Park J-U, 2017a. Nature Commun. 8(1), 14997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Valdés-Ramírez G, Bandodkar AJ, Jia W, Martinez AG, Ramírez J, Mercier P, Wang J, 2014. Analyst 139(7), 1632–1636. [DOI] [PubMed] [Google Scholar]
- Kim KS, Lim SR, Kim S-E, Lee JY, Chung C-H, Choe W-S, Yoo PJ, 2017b. Sens. Actuators B Chem 242, 1121–1128. [Google Scholar]
- Kim Y-H, Lee K, Jung H, Kang HK, Jo J, Park I-K, Lee HH, 2017c. Biosens. Bioelectron 98, 473–477. [DOI] [PubMed] [Google Scholar]
- Kondo S, Toyokuni S, Iwasa Y, Tanaka T, Onodera H, Hiai H, Imamura M, 1999. Free Radical Biology and Medicine 27(3–4), 401–410. [DOI] [PubMed] [Google Scholar]
- Konkena B, Vasudevan S, 2012. J. Phys. Chem. Lett 3(7), 867–872. [DOI] [PubMed] [Google Scholar]
- Koutny LB, Schmalzing D, Taylor TA, Fuchs M, 1996. Anal. Chem 68(1), 18–22. [DOI] [PubMed] [Google Scholar]
- Kumar P, Shahzad F, Yu S, Hong SM, Kim Y-H, Koo CM, 2015a. Carbon 94, 494–500. [Google Scholar]
- Kumar S, Kumar S, Augustine S, Yadav S, Yadav BK, Chauhan RP, Dewan AK, Malhotra BD, 2018. Biosens. Bioelectron 102, 247–255. [DOI] [PubMed] [Google Scholar]
- Kumar S, Kumar S, Tiwari S, Srivastava S, Srivastava M, Yadav BK, Kumar S, Tran TT, Dewan AK, Mulchandani A, Sharma JG, Maji S, Malhotra BD, 2015b. Adv. Sci 2(8), 1500048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Sharma JG, Maji S, Malhotra BD, 2016. Biosens. Bioelectron 78, 497–504. [DOI] [PubMed] [Google Scholar]
- Kumari N, Dwarakanath BS, Das A, Bhatt AN, 2016. Tumor Biol. 37(9), 11553–11572. [DOI] [PubMed] [Google Scholar]
- Kwon Y-T, Kim Y-S, Kwon S, Mahmood M, Lim H-R, Park S-W, Kang S-O, Choi JJ, Herbert R, Jang YC, 2020. Nature communications 11(1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence HP, 2002. J. Can. Dent. Assoc 68(3), 170–175. [PubMed] [Google Scholar]
- Lazzeri C, Valente S, Chiostri M, Gensini GF, 2015. World J. Cardiol 7(8), 483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Wei X, Kysar JW, Hone J, 2008. Science 321(5887), 385–388. [DOI] [PubMed] [Google Scholar]
- Lee H, Lee Y, Song C, Cho HR, Ghaffari R, Choi TK, Kim KH, Lee YB, Ling D, Lee H, Yu SJ, Choi SH, Hyeon T, Kim D-H, 2015. Nature Commun. 6(1), 10059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-H, Loya PE, Lou J, Thomas EL, 2014. Science 346(6213), 1092–1096. [DOI] [PubMed] [Google Scholar]
- Lee Y, Howe C, Mishra S, Lee DS, Mahmood M, Piper M, Kim Y, Tieu K, Byun H-S, Coffey JP, 2018a. Proc. Natl. Acad. Sci 115(21), 5377–5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Eom KS, Shin K-S, Kang JY, Lee SH, 2018b. Sens. Actuators B Chem 271, 73–81. [Google Scholar]
- Lightcap IV, Kosel TH, Kamat PV, 2010. Nano Lett. 10(2), 577–583. [DOI] [PubMed] [Google Scholar]
- Liguori I, Russo G, Curcio F, Bulli G, Aran L, Della-Morte D, Gargiulo G, Testa G, Cacciatore F, Bonaduce D, 2018. Clin. Interv. Aging 13, 757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HR, Kim HS, Qazi R, Kwon YT, Jeong JW, Yeo WH, 2020. Adv. Mater 32(15), 1901924. [DOI] [PubMed] [Google Scholar]
- Lin Y-M, Valdes-Garcia A, Han S-J, Farmer DB, Meric I, Sun Y, Wu Y, Dimitrakopoulos C, Grill A, Avouris P, Jenkins KA, 2011. Science 332(6035), 1294–1297. [DOI] [PubMed] [Google Scholar]
- Liu D, Yun Y, Yang D, Hu X, Dong X, Zhang N, Zhang L, Yin H, Duan W, 2019. Dis. Markers 2019, 4081962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Liu Y, Zhu D, 2011a. J. Mater. Chem 21(10), 3335–3345. [Google Scholar]
- Liu Q, Shi J, Sun J, Wang T, Zeng L, Jiang G, 2011b. Angew. Chem. Int 50(26), 5913–5917. [DOI] [PubMed] [Google Scholar]
- Liu W-W, Chai S-P, Mohamed AR, Hashim U, 2014. J. Ind. Eng. Chem 20(4), 1171–1185. [Google Scholar]
- Liu X, Li Q, Gu P, Su S, Huang Y, Feng X, Fan Q, Huang W, 2017. IEEE Sens. J 17(17), 5431–5436. [Google Scholar]
- Ma L, Hu L, Feng X, Wang S, 2018. Aging Dis. 9(5), 938–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Zhang Y, Cai S, Han Z, Liu X, Wang F, Cao Y, Wang Z, Li H, Chen Y, 2020. Adv. Mater 32(15), 1902062. [DOI] [PubMed] [Google Scholar]
- Mahanta NK, Abramson AR, 2012. 13th InterSociety Conference on Thermal and Thermomechanical Phenomena in Electronic Systems, pp. 1–6. IEEE. [Google Scholar]
- Maheshwaran S, Akilarasan M, Chen S-M, Chen T-W, Tamilalagan E, Tzu CY, Lou B-S, 2020. J. Electrochem. Soc 167(6), 066517. [Google Scholar]
- Maier C, Riedl M, Vila G, Nowotny P, Wolzt M, Clodi M, Ludvik B, Luger A, 2008. Diabetes 57(9), 2332–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malon RSP, Sadir S, Balakrishnan M, Córcoles EP, 2014. BioMed Res. Int 2014, 962903–962903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Maitra A, Das T, Das CK, 2015. Graphene Materials: Fundamentals and Emerging Applications, pp. 1–23. [Google Scholar]
- Mannoor MS, Tao H, Clayton JD, Sengupta A, Kaplan DL, Naik RR, Verma N, Omenetto FG, McAlpine MC, 2012. Nature Commun. 3, 763. [DOI] [PubMed] [Google Scholar]
- Mao HY, Laurent S, Chen W, Akhavan O, Imani M, Ashkarran AA, Mahmoudi M, 2013. Chem. Rev 113(5), 3407–3424. [DOI] [PubMed] [Google Scholar]
- Markopoulos AK, Michailidou EZ, Tzimagiorgis G, 2010. Open Dent. J 4, 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García G, Agüí L, Yáñez-Sedeño P, Pingarrón J, 2016. Electrochim. Acta 202, 209–215. [Google Scholar]
- Michalke B, Rossbach B, Göen T, Schäferhenrich A, Scherer G, 2015. Int. Arch. of Occ. Env. Hea 88(1), 1–44. [DOI] [PubMed] [Google Scholar]
- Mishra S, Saadat D, Kwon O, Lee Y, Choi W-S, Kim J-H, Yeo W-H, 2016. Biosensors and Bioelectronics 81, 181–197. [DOI] [PubMed] [Google Scholar]
- Mishra V, Patil R, Khanna V, Tripathi A, Singh V, Pandey S, Chaurasia A, 2018. J. Clin. Diagnostic Res 12(7), ZC44–ZC47. [Google Scholar]
- Mitsubayashi K, Arakawa T, 2016. Electroanalysis 28(6), 1170–1187. [Google Scholar]
- Mohamed MA, Atty SA, Merey HA, Fattah TA, Foster CW, Banks CE, 2017. Analyst 142(19), 3674–3679. [DOI] [PubMed] [Google Scholar]
- Mohd Azmi MA, Tehrani Z, Lewis RP, Walker KAD, Jones DR, Daniels DR, Doak SH, Guy OJ, 2014. Biosens. Bioelectron 52, 216–224. [DOI] [PubMed] [Google Scholar]
- Mojon P, Budtz-Jørgensen E, Rapin CH, 1999. Age Ageing 28(5), 463–468. [DOI] [PubMed] [Google Scholar]
- Montes-Navajas P, Asenjo NG, Santamaría R, Menéndez R, Corma A, García H, 2013. Langmuir 29(44), 13443–13448. [DOI] [PubMed] [Google Scholar]
- Morgan CJ, Curran HV, Drugs I.S.C.o., 2012. Addiction 107(1), 27–38. [DOI] [PubMed] [Google Scholar]
- Morozov SV, Novoselov KS, Katsnelson MI, Schedin F, Elias DC, Jaszczak JA, Geim AK, 2008. Phys. Rev. Lett 100(1), 016602. [DOI] [PubMed] [Google Scholar]
- Murphy DL, Baker M, Goodwin FK, Miller H, Kotin J, Bunney WE, 1974. Psychopharmacology 34(1), 11–20. [DOI] [PubMed] [Google Scholar]
- Nagler R, Bahar G, Shpitzer T, Feinmesser R, 2006. Clin. Cancer Res 12(13), 3979–3984. [DOI] [PubMed] [Google Scholar]
- Nair RR, Blake P, Grigorenko AN, Novoselov KS, Booth TJ, Stauber T, Peres NMR, Geim AK, 2008. Science 320(5881), 1308–1308. [DOI] [PubMed] [Google Scholar]
- Napoleon Waszkiewicz AUBZ-SAUAZAUMWAUSDSAUBRAU, 2012. Folia Histochem. Cytobiol 50(2), 248–254. [DOI] [PubMed] [Google Scholar]
- Nazarpour S, Hajian R, Sabzvari MH, 2020. Microchem. J, 104634. [Google Scholar]
- Ng SP, Qiu G, Ding N, Lu X, Wu C-ML, 2017. Biosens. Bioelectron 89, 468–476. [DOI] [PubMed] [Google Scholar]
- Ngamchuea K, Batchelor-McAuley C, Compton RG, 2018. Sens. Actuators B Chem 262, 404–410. [Google Scholar]
- North JR, 1985. Trends Biotechnol. 3(7), 180–186. [Google Scholar]
- Novoselov KS, Geim AK, Morozov SV, Jiang D, Zhang Y, Dubonos SV, Grigorieva IV, Firsov AA, 2004. Science 306(5696), 666–669. [DOI] [PubMed] [Google Scholar]
- Oba IT, Spina AMM, Saraceni CP, Lemos MF, Senhoras R.d.C.F.A., Moreira RC, Granato CFH, 2000. Rev. Inst. Med. Trop. Sao Paulo 42(4), 197–200. [DOI] [PubMed] [Google Scholar]
- Ocak E, Kubat G, Yorulmaz İ, 2015. Balk. Med. J 32(1), 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parate K, Karunakaran C, Claussen JC, 2019. Sens. Actuators B Chem 287, 165–172. [Google Scholar]
- Parate K, Rangnekar SV, Jing D, Mendivelso-Perez DL, Ding S, Secor EB, Smith EA, Hostetter JM, Hersam MC, Claussen JC, 2020. ACS Appl. Mater. Interfaces 12(7), 8592–8603. [DOI] [PubMed] [Google Scholar]
- Park CS, Yoon H, Kwon OS, 2016. J. Ind. Eng. Chem 38, 13–22. [Google Scholar]
- Park S, Kim H, Kim J-H, Yeo W-H, 2020. Materials 13(16), 3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S, Metgud R, 2015. J. Cancer Res. Ther 11(1), 119–123. [DOI] [PubMed] [Google Scholar]
- Peacock IV WF, De Marco T, Fonarow GC, Diercks D, Wynne J, Apple FS, Wu AH, 2008. N. Engl. J. Med 358(20), 2117–2126. [DOI] [PubMed] [Google Scholar]
- Peek ME, Bhatnagar A, McCarty NA, Zughaier SM, 2012. Interdiscip. Perspect. Infect. Dis 2012, 843509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho R, Pedrosa R, Costa-Martins P, Castello-Branco L, 1999. Acta Trop. 72(1), 31–38. [DOI] [PubMed] [Google Scholar]
- Plummer M, Franceschi S, Vignat J, Forman D, de Martel C, 2015. Int. J. Cancer 136(2), 487–490. [DOI] [PubMed] [Google Scholar]
- Ponnaiah SK, Periakaruppan P, Selvam M, Muthupandian S, Jeyaprabha B, Selvanathan R, 2019. J. Clust. Sci, 1–9. [Google Scholar]
- Prasad S, Tyagi AK, Aggarwal BB, 2016. Exp. Biol. Med 241(8), 783–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestigiacomo AF, Stamey TA, 1996. J. Urol 155(6), 1977–1980. [PubMed] [Google Scholar]
- Pumera M, 2013. Electrochem. Commun 36, 14–18. [Google Scholar]
- Rahim MAA, Rahim ZHA, Ahmad WAW, Hashim OH, 2015. Int. J. Med. Sci 12(4), 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathnayake N, Gieselmann D-R, Heikkinen AM, Tervahartiala T, Sorsa T, 2017. Diagnostics 7(1), 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray TR, Choi J, Bandodkar AJ, Krishnan S, Gutruf P, Tian L, Ghaffari R, Rogers JA, 2019. Chem. Rev 119(8), 5461–5533. [DOI] [PubMed] [Google Scholar]
- Rose DM, Muttray A, Mayer-Popken O, Jung D, Konietzko J, 1999. Eur. J. Med. Res 4(12), 529–532. [PubMed] [Google Scholar]
- Rusling JF, Kumar CV, Gutkind JS, Patel V, 2010. Analyst 135(10), 2496–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbagh MN, Shi J, Lee M, Arnold L, Al-Hasan Y, Heim J, McGeer P, 2018. BMC Neurol. 18(1), 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadana A, Sadana N, 2008. In: Sadana A, Sadana N (Eds.), Fractal Analysis of the Binding and Dissociation Kinetics for Different Analytes on Biosensor Surfaces, pp. 19–53. Elsevier, Amsterdam. [Google Scholar]
- Sánchez-Tirado E, González-Cortés A, Yáñez-Sedeño P, Pingarrón JM, 2018. Biosens. Bioelectron 113, 88–94. [DOI] [PubMed] [Google Scholar]
- Satish B, Srikala P, Maharudrappa B, Awanti SM, Kumar P, Hugar D, 2014. J. Int. Oral Health 6(2), 114. [PMC free article] [PubMed] [Google Scholar]
- Sawyers CL, 2008. Nature 452(7187), 548–552. [DOI] [PubMed] [Google Scholar]
- Schedin F, Geim AK, Morozov SV, Hill EW, Blake P, Katsnelson MI, Novoselov KS, 2007. Nature Mater. 6(9), 652–655. [DOI] [PubMed] [Google Scholar]
- Schroeder V, Savagatrup S, He M, Lin S, Swager TM, 2019. Chem. Rev 119(1), 599–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura R, Javierre C, Ventura JL, Lizarraga MA, Campos B, Garrido E, 1996. Br. J. Sports Med 30(4), 305–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvarajan RS, Rahim RA, Majlis BY, Gopinath SC, Hamzah AA, 2020. Sensors 20(9), 2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag VKL, Prasad K, 2016. Anal. Methods 8(33), 6255–6259. [Google Scholar]
- Sharma S, Jackson PG, Makan J, 2004. J. Clin. Pathol 57(10), 1025–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ship JA, 2003. J. Am. Dent. Assoc 134, 4S–10S. [DOI] [PubMed] [Google Scholar]
- Shivaraman S, Barton RA, Yu X, Alden J, Herman L, Chandrashekhar MVS, Park J, McEuen PL, Parpia JM, Craighead HG, Spencer MG, 2009. Nano Lett. 9(9), 3100–3105. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A, 2017. CA Cancer J. Clin 67(1), 7–30. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A, 2019. CA Cancer J. Clin 69(1), 7–34. [DOI] [PubMed] [Google Scholar]
- Sies H, Berndt C, Jones DP, 2017. Annu. Rev. Biochem 86, 715–748. [DOI] [PubMed] [Google Scholar]
- Smith KG, Kamdar AA, Stark JM, 2019. In: Wilmott RW, Deterding R, Li A, Ratjen F, Sly P, Zar HJ, Bush A (Eds.), Kendig’s Disorders of the Respiratory Tract in Children, pp. 120–133.e122, ninth ed ed. Content Repository Only!, Philadelphia. [Google Scholar]
- Stamey TA, Yang N, Hay AR, McNeal JE, Freiha FS, Redwine E, 1987. N. Engl. J. Med 317(15), 909–916. [DOI] [PubMed] [Google Scholar]
- Stieber P, Hasholzner U, Bodenmüller H, Nagel D, Sunder-Plassmann L, Dienemann H, Meier W, Fateh-Moghadam A, 1993. Cancer 72(3), 707–713. [DOI] [PubMed] [Google Scholar]
- Stoller MD, Park S, Zhu Y, An J, Ruoff RS, 2008. Nano Lett. 8(10), 3498–3502. [DOI] [PubMed] [Google Scholar]
- Taba M, Kinney J, Kim AS, Giannobile WV, 2005. Dental Clinics 49(3), 551–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W, Sabet L, Li Y, Yu T, Klokkevold PR, Wong DT, Ho C-M, 2008. Biosens. Bioelectron 24(2), 266–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia GM, Gagliano N, Zarbin L, Tolomeo G, Sforza C, 2017. Arch. Oral Biol 78, 34–38. [DOI] [PubMed] [Google Scholar]
- Taufique MFN, Bhaumik A, Haque A, Karnati P, 2017. J. Mater. Sci. Eng 06. [Google Scholar]
- Teruhisa U, Ryoji H, Taisuke I, Tatsuya S, Fumihiro M, Tatsuo S, 1981. Clin. Chim. Acta 110(2), 245–253.6261989 [Google Scholar]
- Thevenot DR, Toth K, Durst RA, Wilson GS, 1999. Pure Appl. Chem 71(12), 2333–2348. [Google Scholar]
- Torrente-Rodríguez RM, Campuzano S, Ruiz-Valdepeñas Montiel V, Gamella M, Pingarrón JM, 2016. Biosens. Bioelectron 77, 543–548. [DOI] [PubMed] [Google Scholar]
- Torrente-Rodríguez RM, Tu J, Yang Y, Min J, Wang M, Song Y, Yu Y, Xu C, Ye C, IsHak WW, Gao W, 2020. Matter 2(4), 921–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng P, Napier B, Garbarini L, Kaplan DL, Omenetto FG, 2018. Adv. Mater 30(18), 1703257. [DOI] [PubMed] [Google Scholar]
- Valavanidis A, Vlachogianni T, Fiotakis C, 2009. J. Environ. Sci. Health C 27(2), 120–139. [DOI] [PubMed] [Google Scholar]
- Varodi C, Pogacean F, Coros M, Rosu M-C, Stefan-van Staden R-I, Gal E, Tudoran L-B, Pruneanu S, Mirel S, 2019. Sensors 19(19), 4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S, Singh A, Shukla A, Kaswan J, Arora K, Ramirez-Vick J, Singh P, Singh SP, 2017. ACS Appl. Mater. Interfaces 9(33), 27462–27474. [DOI] [PubMed] [Google Scholar]
- Verma S, Singh SP, 2019. MRS Commun. 9(4), 1227–1234. [Google Scholar]
- Walker RF, Riad-Fahmy D, Read G, 1978. Clin. Chem 24(9), 1460–1463. [PubMed] [Google Scholar]
- Wang J, 1998. Biosens. Bioelectron 13(7–8), 757–762. [DOI] [PubMed] [Google Scholar]
- Wang J, Yu J, Wang T, Li C, Wei Y, Deng X, Chen X, 2020. J. Mater. Chem. B 8(16), 3341–3356. [DOI] [PubMed] [Google Scholar]
- Wang Q, Gao P, Cheng F, Wang X, Duan Y, 2014. Talanta 119, 299–305. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li Z, Wang J, Li J, Lin Y, 2011. Trends Biotechnol. 29(5), 205–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warinner C, Rodrigues JFM, Vyas R, Trachsel C, Shved N, Grossmann J, Radini A, Hancock Y, Tito RY, Fiddyment S, 2014. Nature Genet. 46(4), 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waugh DJ, Wilson C, 2008. Clin. Cancer Res 14(21), 6735–6741. [DOI] [PubMed] [Google Scholar]
- Wieskopf B, Demangeat C, Purohit A, Stenger R, Gries P, Kreisman H, Quoix E, 1995. Chest 108(1), 163–169. [DOI] [PubMed] [Google Scholar]
- Wilson GS, Hu Y, 2000. Chem. Rev 100(7), 2693–2704. [DOI] [PubMed] [Google Scholar]
- Wu L, Qu X, 2015. Chem. Soc. Rev 44(10), 2963–2997. [DOI] [PubMed] [Google Scholar]
- Wu LL, Chiou C-C, Chang P-Y, Wu JT, 2004. Clin. Chim. Acta 339(1–2), 1–9. [DOI] [PubMed] [Google Scholar]
- Wu R-Q, Zhang D-F, Tu E, Chen Q-M, Chen W, 2014. Int. J. Oral Sci 6(3), 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Wang Y, Wu M, Ma X, Yang X, 2013. J. Electroanal. Chem 702, 49–55. [Google Scholar]
- Xie K, 2001. Cytokine Growth F. R 12(4), 375–391. [DOI] [PubMed] [Google Scholar]
- Yang D, Velamakanni A, Bozoklu G, Park S, Stoller M, Piner RD, Stankovich S, Jung I, Field DA, Ventrice CA, Ruoff RS, 2009. Carbon 47(1), 145–152. [Google Scholar]
- Ye D, Liang G, Li H, Luo J, Zhang S, Chen H, Kong J, 2013. Talanta 116, 223–230. [DOI] [PubMed] [Google Scholar]
- Yoo SS, Kim SY, Kim KS, Hong S, Oh MJ, Nam MG, Kim W-J, Park J, Chung C-H, Choe W-S, 2020. Sens. Actuators B Chem 305, 127477. [Google Scholar]
- Yuan Z, Xiao X, Li J, Zhao Z, Yu D, Li Q, 2018. Adv. Sci 5(2), 1700626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H-B, Wang J-W, Yan Q, Zheng W-G, Chen C, Yu Z-Z, 2011. J. Mater. Chem 21(14), 5392–5397. [Google Scholar]
- Zhang W, Du Y, Wang ML, 2015. Sens. Biosensing Res 4, 23–29. [Google Scholar]
- Zheng J, Sun L, Yuan W, Xu J, Yu X, Wang F, Sun L, Zeng Y, 2018. J. Oral Pathol. Med 47(9), 830–835. [DOI] [PubMed] [Google Scholar]
- Zhong L, Cheng F, Lu X, Duan Y, Wang X, 2016. Talanta 158, 351–360. [DOI] [PubMed] [Google Scholar]
- Zou R, Shan S, Huang L, Chen Z, Lawson T, Lin M, Yan L, Liu Y, 2020. ACS Biomater. Sci. Eng 6(1), 673–679. [DOI] [PubMed] [Google Scholar]


