Abstract
Ribosomally synthesized and post-translationally modified peptides (RiPPs) compose a large structurally and functionally diverse family of natural products. The biosynthesis system of RiPPs typically involves a precursor peptide comprising of a leader and core motif and nearby processing enzymes that recognize the leader and act on the core for producing modified peptides. Interest in RiPPs has increased substantially in recent years as improvements in genome mining techniques have dramatically improved access to these peptides and biochemical and engineering studies have supported their applications. A less understood, intriguing feature in the RiPPs biosynthesis is the precursor peptides of multiple RiPPs families produced by bacteria, fungi and plants carrying multiple core motifs, which we term “multicore”. Herein, we present the prevalence of the multicore systems, their biosynthesis and engineering for applications.
Keywords: RiPPs, Natural Products, Multicore, Biosynthesis, Engineering
Natural products have long inspired drug discovery and development due to their intrinsic bioactivity. A major type of natural products, peptides possess diverse biological roles and have received tremendous attention in basic and translational research [43,86]. One specific utility for peptides is to target “undruggable” space, e.g., protein-protein interactions, which is less accessible with other types of small molecules since peptidic compounds demonstrate increased specificity and less off-target effects [115,43,86]. Ribosomally synthesized and post-translationally modified peptides (RiPPs) are one category of peptidic natural peptides that are produced by all major domains of life and contain a wide variety of structural diversity [2]. These peptides demonstrate a wide array of biological activities, e.g., inhibiting nucleotides/proteins synthesis, chelating metal ions, and permeabilizing cell membranes [5,21,41,70,127]. RiPPs also play important roles in internal regulation and host-bacteria interactions [5,70,127]. Of note, the RiPP nisin has been used for food preservation since the late 1960s. As new RiPPs are rapidly discovered, their applications in drug development and other areas will continue to expand.
So far, over 22 RiPP families have been discovered with distinct structural features, e.g., lanthipeptides, thiopeptides and lasso peptides, while their biosynthesis follows a common logic (Fig. 1A) [5]. The RiPP biosynthetic gene clusters (BGCs) minimally consist of one precursor peptide and one or more pathway-specific modification enzymes. The precursor peptide generally includes an N-terminal leader peptide (LP), which often dictates the binding of modification enzymes, and a C-terminal core peptide (CP), which is enzymatically modified and then released to form the advanced biosynthetic intermediate or final product (Fig. 1A). A long list of peptide modification enzymes promotes a wide variety of modifications on the CP, e.g., methylation, oxidation/reduction, dehydration, crosslinks between amino acid residues, and macrocyclization; the chemical diversity of RiPPs rivals that of nonribosomal peptides, another major category of peptidic natural products [3,5,23,87,114,127]. Importantly, capable bioinformatics tools have been developed to predict and discover new RiPP analogs and even novel families from organism genomes [11,20,55–58,64,106–108,113]. The known and newly mined RiPPs gene clusters can be further categorized into three groups based on the status of their precursor peptides. The gene clusters in the first group encode only a single precursor peptide that contains a single CP, while those in the second group carry multiple precursor peptides (Fig. 1B). The single CP of these multiple precursors can range from identical sequences to very low similarity. The RiPP gene clusters in the last group have one or more precursor peptides, each of which consists of multiple CPs after the single LP (Fig. 1B). Compared with the first two RiPP groups, the biosynthetic understanding of the multicore RiPPs remains underexplored. On the other hand, the multicore system holds the promising potential to expand peptide chemical diversity with an economical genetic cost, aiding improved target selectivity and potency in peptide drug research [8,25,44,97,101,125,126]. Herein, we review current knowledge of all known multicore RiPPs systems in bacteria, fungi and plants, highlighting their prevalence, distribution, bioactivity, and biosynthetic logic compared to their respective single-core systems. We will further highlight engineering efforts in developing these RiPPs for biomedical applications.
Fig. 1.
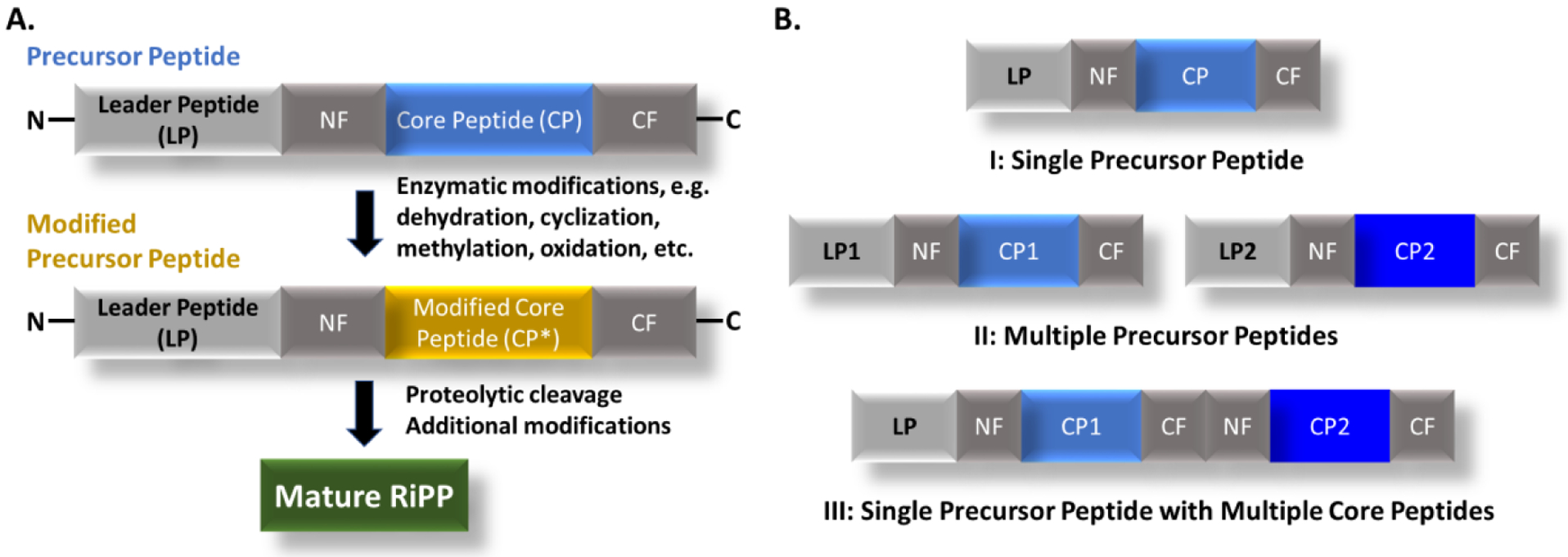
A. General scheme of RiPPs biosynthesis. B. Three groups of precursor peptides found in the RiPP gene clusters. The core peptide of the precursor peptide sometimes carries flanking sequences at its N- and C-termini, named as N-flanking (NF) and C-flanking (CF), respectively.
1. Bacterial multicore RiPPs
The vast majority of RiPPs are of bacterial origins but only a few demonstrate apparent multicore features in the biosynthesis, with repeated CP units within a single precursor peptide, including cyanobactins and microviridins as well as some Ω-ester peptides. We will review each of these RiPPs families along with putative multicore RiPPs discovered through genome mining efforts.
1.1. Cyanobactins:
Cyanobactins were first discovered in 1989 as natural products of marine ascadians, but were later shown to be biosynthesized by the symbiotic cyanobacterium Prochloron didemni [40,47,90,101]. To date, dozens of cyanobactins have been isolated from multiple cyanobacterial species (Fig. 2). Importantly, over 20% of cyanobacterial genomes are predicted to contain at least one cyanobactin cluster [66,105], while the similar clusters have been discovered in other bacterial phyla, such as Actinomycetes [38,53]. The native functions of cyanobactins remain unclear, but these peptides have been shown to execute diverse biological activities, including cancer cytotoxicity, growth and protease inhibition, metal chelation, and reducing antibiotic drug resistance [45,105].
Fig. 2.
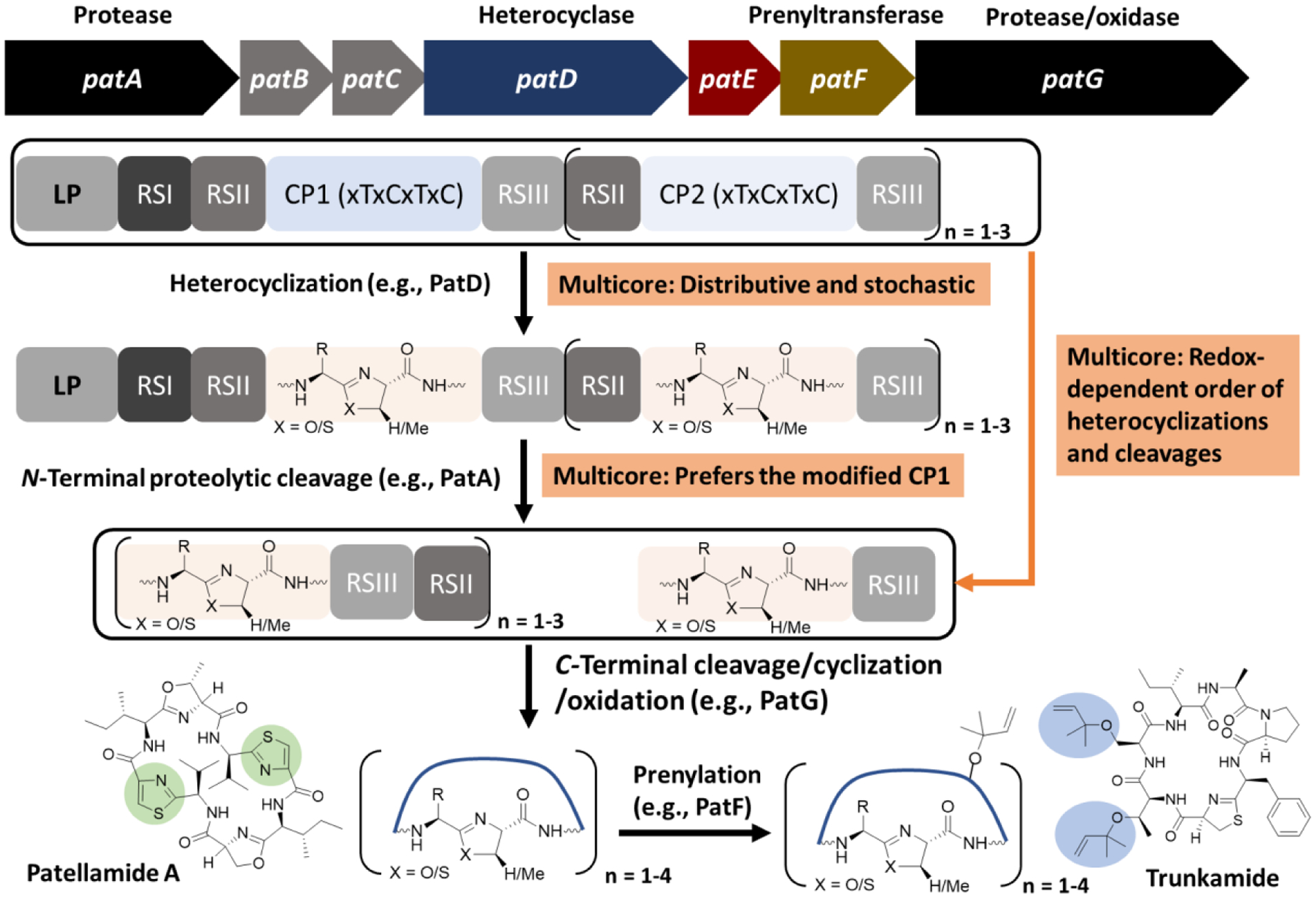
Biosynthetic scheme of cyanobactins. The representative gene cluster (pat) of patellamide is shown at the top. Pathway-specific enzymes demonstrate different features toward precursor peptides with a single CP and multiple CPs. Chemical structures of representative cyanobactins, patellamide A and trunkamide, are shown and their thiazole and prenyl moieties are shadowed in green and light blue, respectively. RS: recognition site.
The biosynthesis of cyanobactins obeys the general logic of RiPPs (Fig. 1A) [24,67,73], and demonstrates obvious biosynthetic plasticity in generating chemical diversity. Some cyanobactin gene clusters contain up to 10 precursor peptides, each of which carries a single CP, while others encode a single precursor peptide with up to 4 CP motifs [99]. Our current understanding of cyanobactin biosynthesis mainly comes from the precursor peptides with 2 to 3 CPs, e.g., PatE from the patellamide biosynthetic pathway, while the engineered single-core precursors have often been used for easing enzymatic and engineering studies [33,39]. The cyanobactin CP generally consists of 6 to 8 amino acids with cysteine, serine, threonine residues separated by one nonconserved residue, e.g., xTxCxSxC. Multiple enzymes install modifications on each CP, including heterocyclase (e.g., PatD), N-terminal protease (e.g., PatA), C-terminal protease/cyclase (e.g., PatG), oxidase (e.g., ThcOx), and prenyltransferase (e.g., PatF) (Fig. 2). Some modification enzymes contain a RiPP precursor peptide recognition element (RRE) that is a PqqD domain for recognizing and binding a helix-turn-helix motif typically found in the LP [7]. In addition, some cyanobactin biosynthetic enzymes bind to another type of conserved motifs in the precursor peptide, termed recognition sites (RSs) (Fig. 2). Recognition site 1 (RSI) is N-terminal to the CP and typically has a conserved sequence of LAELSEE. RSI guides the heterocyclization of the conserved cysteine, serine, and threonine in the CP. The cyanobactin heterocyclases contain an ATP-dependent YcaO domain [9]. The heterocyclization within the cyanobactin CP occurs in a C to N direction, with cysteines modified before threonine or serine residues. The majority of cyanobactin CPs contain a C-terminal heterocycle, typically thiazol(in)e [38,67,105]. On the other hand, some cyanobactin BGCs have no heterocyclase and their precursor peptides carry no RSI but often have a C-terminal proline in the CP, e.g., xTxCxSxP [99]. Following RSI, RSII with a conserved sequence of Gx(E/D)xS flanks the CP on the N-terminus, while RSIII as the motif of (A/S)YD is C-terminal to the CP (Fig. 2). RSII interacts with the N-terminal subtilisin-like protease (PatA) to release the heterocyclized CP C-terminal to the conserved Ser of the RSII motif. RSIII in the released modified CP then guides the C-terminal subtilisin-like protease (e.g., PatP) to hydrolyze the amide bond N-terminal to its conserved Ala/Ser residue. In addition, the latter protease often catalyzes head to tail cyclization of the released cyanobactins through a transamidation, but linear cyanobactins have also been found [1,60,61,65,67,76].
The modified CP cleaved from the cyanobactin precursor peptide can undergo additional modifications, commonly oxidation and prenylation, to generate the final product, e.g., patellamides and trunkamides (Fig. 2) [46,77]. The FMN-dependent oxidase can be fused with the C-terminal protease (e.g., PatG) or as a stand-alone enzyme (e.g., ThcOx) [32]. Oxidation on the azolines (e.g., thiazolines) proceeds in an N to C direction on both linear and macrocyclic peptides [32,46]. The oxidase domain contains an RRE that is important to enzyme stability but shows no clear binding to the cyanobactin precursor peptide [32]. The prenylation is often the last possible modification on cyanobactins [75,97]. The prenyltransferase LynF modifies the –OH group of tyrosine residues, while TruF acts on the –OH group of serine and threonine residues. After the O-prenylation, a spontaneous Claisen rearrangement can occur to produce an ortho-C-prenylated cyanobactin. On the other hand, the prenyltransferase is fused with a methyltransferase as a single enzyme AgeMTPT for the biosynthesis of linear cyanobactins aeruginosamides [97]. In this case, after heterocyclization and N-terminal cleavage, the exposed N-terminus is prenylated to prevent the transamidation reaction by the C-terminal protease AgeG that cleaves at RSIII. The AgeMTPT further protects the C-terminus by the methylation [97].
Multicore cyanobactin precursors are often found in the patellamide/trunkamide-like BGCs and contain up to 4 CP motifs. Each CP is flanked by an RSII and RSIII with only one RSI site before the first CP, if heterocyclization enzymes are present (Fig. 2) [39,99]. The modifications on these CPs demonstrate distinct features, although the overall modifications on each CP are the same as the single-core system (Fig. 2) [98]. In contrast to the engineered single-core precursor peptide, the heterocyclase iteratively binds to, releases, and rebinds to the parental substrate and modified intermediates during the reaction, and displays no preferred order in modifying the multiple CPs. Indeed, cyanobactin intermediates have been detected with cyclization on either CP [39]. The N-terminal protease prefers the cleavage at the first CP under non-reductive conditions, but distributive catalysis still occurs with initial cleavage at the second CP as a minor product (Fig. 2). Interestingly, the order of cyclization and cleavage is redox-state dependent [39]. Overall, in the multicore processing, the majority of cyanobactin modification enzymes are distributive and tolerant to core sequence variations, leading to the generation of multiple intermediates depending on the processing order [39].
The inherent biosynthetic plasticity of cyanobactins can be leveraged to produce unnatural analogs. Several libraries of unnatural cyanobactins have been synthesized, with most proteinogenic residues being accepted at several positions within the CP, without affecting enzyme activity [95]. Furthermore, cyanobactin biosynthetic enzymes successfully processed the precursor peptides carrying non-proteinogenic residues in the CP [46,100,112]. For example, the macrocyclase PatG has been shown to cyclize compounds with no natural residues or the presence of heterocycles [87]. Remarkably, the use of modification enzymes from different cyanobactin pathways further expanded the chemical diversity, generating hundreds of new compounds [46,95,98]. Importantly, the engineering of the multicore system, in conjunction with combinatorial biosynthesis, is expected to lead to new or improved activities for a wide range of applications.
1.2. Microviridins:
Microviridins were discovered in 1990 from an extraction of the cyanobacterium Microcystis viridis [49]. Since then, tens of microviridins have been isolated from a variety of cyanobacterial strains [2,128]. These tricyclic peptides possess potent and selective serine protease inhibitory activity (Fig. 3A) [80,93,104,128]. In addition, microviridin J has demonstrated the toxicity to the water flea Daphnia pulicaria that is the predator of the producing cyanobacterium [93,94].
Fig. 3.
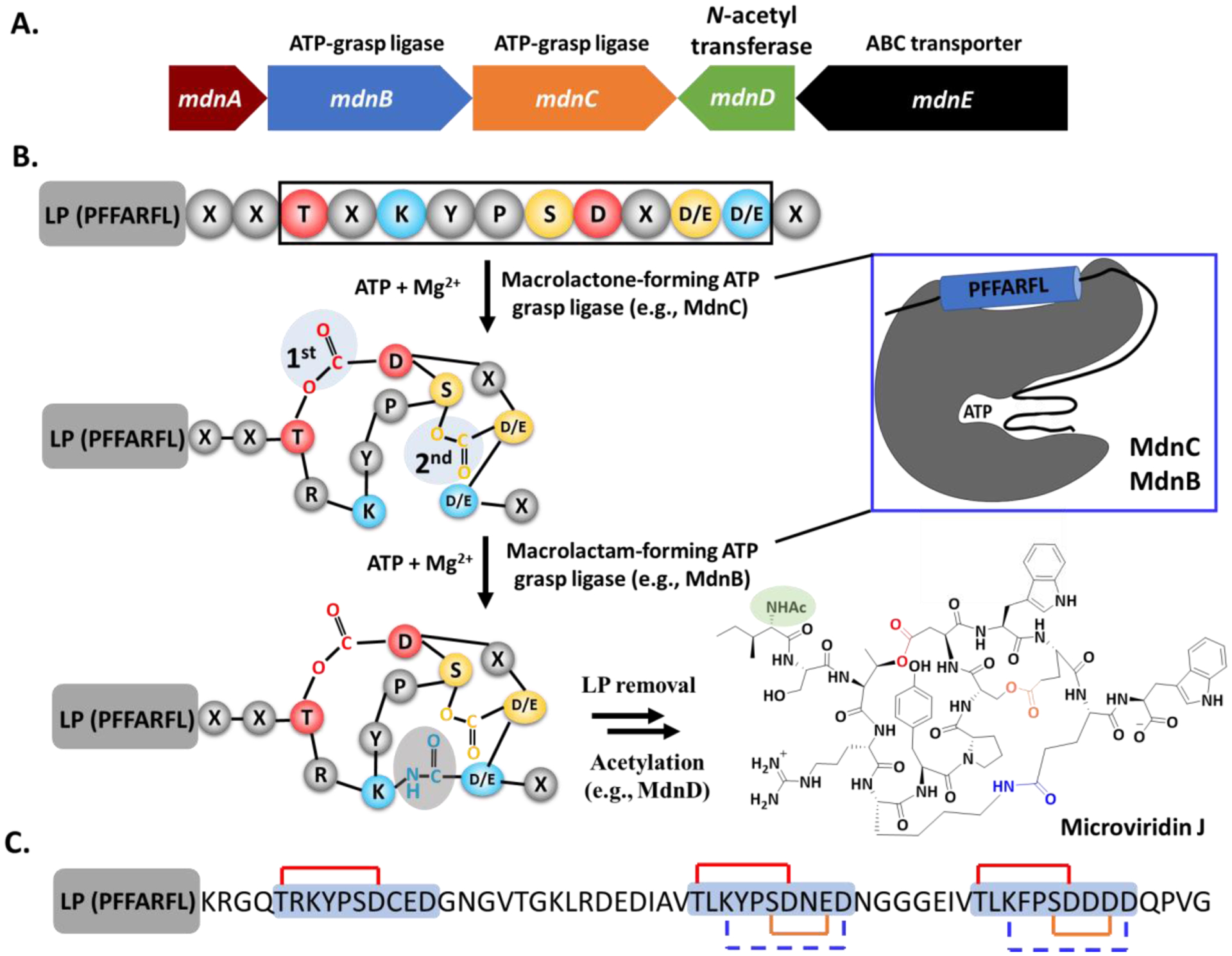
A. A representative gene cluster (mdn) of microviridin. The annotated functions of biosynthetic proteins are shown at the top. MdnE encodes a precursor peptide. B. General scheme of microviridin biosynthesis. The interaction of LP/enzyme is shown on the top right. C. Structure of the most abundant modified AMdnA in the reaction of AMdnC. Each CP in AMdnA is shadowed in light blue and the first and second macrolactones are shown as red and brown lines, respectively. The blue dashed lines indicate the predicted macrolactams.
Microviridin BGCs have a minimal gene set of a precursor peptide and two ATP-grasp ligases but can contain additional genes, including an acyltransferase and an ABC transporter. The ATP-grasp ligases contain an atypical ATP-binding site, named the ATP-grasp fold, and typically catalyze the intermolecular formation of a peptide or ester bond. The microviridin precursor peptide (e.g., MdnA) carries an N-terminal conserved leader motif of PFFARFL and a conserved 10-amino acid core motif of 1-TxKYPSDx(D/E)(D/E)-10 (Fig. 3A) [88]. The two ATP-grasp ligases (e.g., MdnB and MdnC) interact with the conserved leader motif and install three macrocycles on the CP. The first ATP-grasp ligase MdnC lactonizes the CP between the side chains of its Thr1 and Asp7. Upon the formation of the first macrolactone, MdnC further forms the second macrolactone between the side chains of Ser6 and Asp/Glu9. The bicyclic microviridin CP is then modified by the second ATP-grasp ligase MdnB to install one macrolactam between the side chains of Lys3 and Asp/Glu10 (Fig. 3A) [69,88,121]. The order of these three cyclizations is strict [88], while natural bicyclic microviridin analogs have been isolated when their core Lys3 is mutated to Arg [109,111]. Interestingly, recent structural studies revealed that neither MdnC nor MdnB carries the RRE domain [69]. Instead, MdnC and MdnB share the standard ATP-grasp ligase structural motifs comprising of N-domain, central domain and C-domain, and their central domain interacts with an α-helical element formed from the conserved leader motif (Fig. 3A) [69], representing a new strategy of substrate/enzyme recognition in the RiPPs biosynthesis. The LP/enzyme interaction opens the catalytic pocket formed by residues from the N- and C-domains for cyclizing the CP [69]. Furthermore, the leader motif can activate the ATP-grasp ligases for the cyclizations when it is fused to the enzymes or added in trans [2,69,91,121]. The remaining biosynthetic steps of microviridin include the proteolytic cleavage of the LP and the acetylation of N-terminus of the released CP to produce mature microviridins that may be subjected to a pathway-specific ABC transporter [2,121,128].
Recent bioinformatics studies identified hundreds of microviridin BGCs from the genomes of cyanobacteria, as well as those of proteobacteria, bacteriodetes, and acidobacteria phyla [2]. These mined clusters demonstrate tremendous diversity as some encode up to 10 precursor peptides, while the precursor peptides of other clusters contain up to five CPs [2,127]. Of note, the microviridin multicore precursors appear in the clusters found in all four bacterial phyla [2], presumably reflecting the biological significance of the multicore design. The first understanding of the enzymatic processing of the microviridin multicore systems comes from the research of the cluster in the filamentous cyanobacterium Anabaena sp. PCC 7120 [127]. In this cluster, the precursor peptide AMdnA has three predicted CPs and the lactone-forming ATP-grasp ligase AMdnC cyclizes these cores with a favored N to C directionality (Fig. 3B) [127]. On the other hand, the first CP of the most abundant product is macrocyclized only once, while two macrolactones are installed on each of the remaining CPs (Fig. 3B). The order of two cyclization reactions on the two CPs of AMdnA is the same as the single-core containing MdnA. These results together suggest that the modification status on one CP of AMdnA has no, to minimal, influence on the enzymatic macrocyclization of others, highlighting the enzyme’s substrate promiscuity. The broad substrate scope of AMdnC is further supported by its reactions on engineered AMdnA analogs with one to four CPs and the noncognate substrate MdnA. Similar to the biosynthetic enzymes of cyanobactins, AMdnC is distributive in cyclizing CPs within AMdnA [39,127]. The distributive catalysis may be one hallmark of the processing of multicore precursors in the RiPPs biosynthesis. A conserved GG motif is often available between two microviridin CPs that can be cleaved by a C39 protease [2]. However, individual microviridins have not been isolated from any multicore systems yet [2,127].
The plasticity of microviridin biosynthetic systems has allowed engineering efforts to expand the chemical diversity of these peptides for the discovery of potent and selective serine protease inhibitors [2,128,122]. Almost all proteinogenic residues are tolerant in the nonconserved sites of the CP to generate diverse peptide libraries [91,122]. It is expected that the multicore systems can further facilitate the synthesis of new analogs that can be used to screen for inhibitors of serine proteases essential to all forms of life.
1.3. Ω-Ester peptides:
In the continued effort to uncover microviridin-like peptides, the Kim laboratory predicted a total of 12 putative peptide groups whose precursor peptides demonstrate distinct features but contain multiple potential pairs of Thr/Ser/Lys and Asp/Glu residues for the ATP-grasp ligase mediated macrocyclizations [63]. Remarkably, similar to microviridins (Group 1), some precursor peptides from Groups 2 to 6 also carry multiple CP repeats [63]. Among them, the biosynthesis of plesiocin (Group 2) and thuringinins (Group 3) have been studied in good detail, as discussed below.
The plesiocin gene cluster was first mined from the genome of a marine myxobacterium Plesiocystis pacifica in 2017, and its putative clusters were later found to be widely distributed in actinobacteria, cyanobacteria, firmicutes, proteobacteria, acidobacteria, euryarchaeota, and plantomycetes [63,64]. The understanding of the plesiocin biosynthesis came from the in vitro biochemical characterization of the plesiocin precursor PsnA2 processed by the pathway specific ATP-grasp ligase PsnB (Fig. 4A). PsnA2 carries four repeats of 1-TTLxxGEE-8 separated by 2 to 4 residues, while its homologs can have up to 15 repeats. Recombinant PsnB prepared from E. coli successfully processed all four repeats of PsnA2 by forming two ester linkages between the side chains of Thr1 and Glu8 and of Thr2 and Glu7 in each repeat, yielding four hairpin-like bicyclic units. PsnB was distributive in modifying these four repeats but demonstrated low activity toward engineered repeats with more than four residues between the inner pair of Thr and Glu [62]. Furthermore, the plesiocin precursor does not share the same conserved leader motif as microviridins, suggesting a new way for the interactions of leader/ATP-grasp ligase in the peptide macrocyclization. The minimal leader peptide for the effective processing was determined with serial truncated PsnA2 analogs (Fig. 4A). Although it is still unclear if one or more repeats of plesiocin are proteolytically released in the native producers, both plesiocin and its engineered single repeat inhibit serine proteases elastase and chymotrypsin, but not trypsin, and plesiocin is 10 to 20-fold more potent. On the other hand, the replacement of Leu3 with Arg introduced potent trypsin activity. In this regard, plesiocins can be engineered to target one or multiple proteases at once potently and selectively.
Fig. 4.

The processing of multiple CP repeats within the plesiocins (A) and thuringinin (B) precursor peptides PsnA2 and TgnA by their cognate ATP-grasp ligases PsnB and TgnB, respectively. Conserved key residues in each repeat that is shadowed in light blue are highlighted in bold, while the minimal leader peptides potentially for the interactions with ATP-grasp ligases are shadowed in light grey. The ester bonds formed on each repeat are shown with red lines. In the thuringinin biosynthesis, the inner macrolactone is formed first and the outer cycles are shown with brown lines.
Similar to plesiocins, the thuringinin (Tgn) gene cluster was first mined from the genome of one Bacillus species in 2019 and appeared in multiple species of Firmicutes and Proteobacteria [92]. All clusters consist of at least one precursor peptide (e.g., TgnA), a transmembrane protein (e.g., TgnC), and an ATP-grasp ligase (TgnB) (Fig. 4B). The thuringinin precursor peptides have an N-terminal leader and 2 to 7 repeats of the conserved motif of 1-TxxTxxxExxD-11 at the C-terminus. TgnA from the Bacillus species carries three repeats and the co-expression with its cognate TgnB in E. coli generated the recombinant TgnA that receives six macrolactones, two on each repeat (Fig. 4B) [92]. The first ester linkage is formed between the side chains of Thr4 and Glu8 and the other from Thr1 and Asp11. Remarkably, the conserved leader motif of TgnA for the binding of TgnB is discovered by substrate engineering and distinct from those of microviridin and plesiocin precursor peptides (Fig. 4B), indicating that ATP-grasp ligases can evolve multiple modes of protein/protein interactions in the RiPPs biosynthesis. Processed thuringinins with the attached leader showed no inhibitory activity against several serine proteases. Furthermore, it is unclear if any repeats of thuringinins are proteolytically released.
2. Fungal multicore RiPPs
So far, only several families of fungal RiPPs have been isolated, e.g., amatoxins and phallotoxins [4,5,72,123]. On the other hand, multicore precursor peptides are increasingly being discovered due to improvements of genome mining approaches. The biosynthetic understanding of multicore fungal RiPPs has been elucidated, in part, for dikaritin-type peptides.
Dikaritins are a family of cyclic peptides produced by fungal species of the Dikarya subkingdom [28]. Within this family are currently ustiloxins, phomopsins, and asperipins (Fig. 5) [117]. The characteristic cycle of the dikaritin family is formed between the aryl hydroxyl group of Tyr and ß-carbon of another residue, e.g., Ile for ustiloxins and phomopsins. Ustiloxins were first isolated from parasitic false smut balls growing on rice plants and were shown to inhibit microtubule assembly and mitosis of human tumor cell lines in the low μM range and are extremely toxic against mice [89,90]. Similarly, phomopsins are a group of hexapeptide mycotoxins that are the major cause of lupinosis disease and inhibit microtubule assembly [19,117]. In contrast, asperipins were recently identified from Aspergillus flavus primarily driven by searching biosynthetic enzyme homologs of ustiloxins from fungal genomes [82].
Fig. 5.
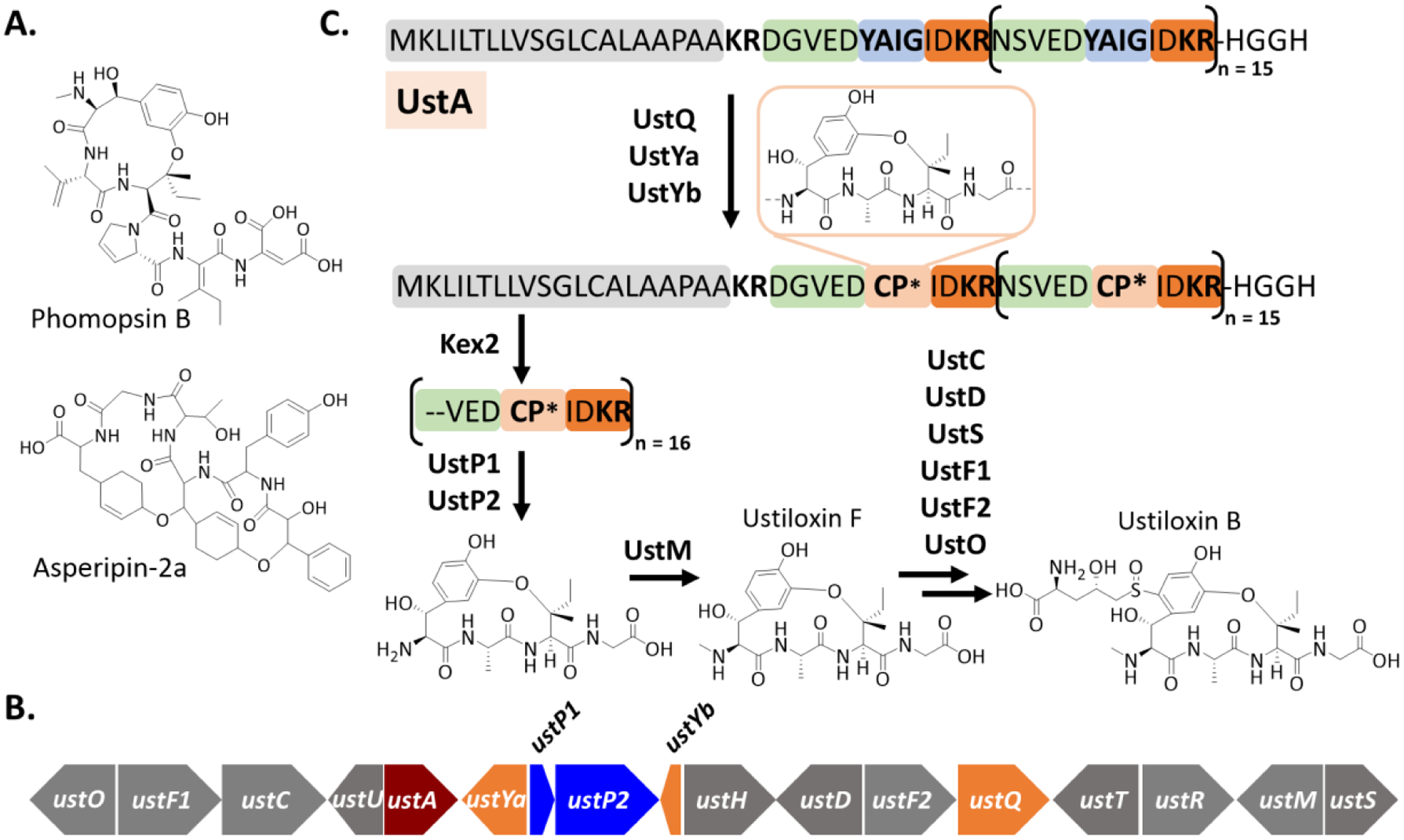
A. Structures of representative phomopsins and asperipins. B. A schematic representation of the ustiloxin B (ust) gene cluster. The precursor peptide gene is shown in dark red, while the ustQ, ustYa and ustYB genes are in orange. Two peptidase genes ustP1 and ustP2 are shown in blue. C. General scheme of ustiloxin biosynthesis. The leader peptide region is in grey. The tetrapeptide core in light blue is flanked with conserved N and C-terminal sequences in green and orange, respectively. CP* represents the modified CP. The details of the majority of biosynthetic steps remain unavailable. Ustiloxin F and B are used as representative examples.
The biosynthesis of dikaritins has been characterized to a certain degree in the biochemical and genetic studies of ustiloxins and phomopsins. The ustiloxin BGCs contain 11 to 17 genes, encoding a precursor peptide (e.g., UstA), a copper-dependent tyrosinase (e.g., UstQ), and a variety of other tailoring enzymes, e.g., flavin-dependent monooxygenases (e.g., UstF1-F2), P450 (e.g., UstC), methyltransferase (e.g., UstM), and pyridoxamine 5′-phosphate (PLP) oxidase (e.g., UstO) [82,116,125]. The precursor peptide UstA from Aspergillus flavus contains 16 repeats with a highly conserved sequence motif of NSVEDYAIGIDKR, of which the tetrapeptide YAIG fragment appears in the final structures of ustiloxins (Fig. 5B). A combination of functional prediction, gene knockouts, and biochemical characterization suggests key modifications on the conserved repeats, but the details of these modifications (e.g., order and distributive catalysis) are still unknown. Briefly, the processing requires multiple proteases to release the conserved repeats and the modified or unmodified tetrapeptide. A subtilisin-like endoprotease Kex2 is expected to release the repeat fragments at the C-terminus of the conserved KR motif but this protease is not pathway-specific. The ustiloxin BGC encodes two S41 family peptidases UstP1 and UstP2 that may remove the N and C-terminal sequences of the tetrapeptide core in the released repeat fragments. The conversion of the tetrapeptide YAIG into ustiloxin F requires a series of reactions, including β-hydroxylation and 3-hydroxylation on the phenol of Tyr, crosslinking between 3-OH of Tyr and β-carbon of Ile and N-methylation of the primary amine of the core motif [82,116,125]. The tyrosinase UstQ is likely responsible for the phenol hydroxylation, while UstM catalyzes the N-methylation (Fig. 5B). Two essential uncharacterized proteins UstYa and UstYb may catalyze the β-hydroxylation of Tyr and the oxidative cyclization. Of note, except UstM, the order and other details of the proteolysis and tailoring reactions in processing the multicore UstA are completely unknown. Furthermore, it is unclear if and how the processing enzymes interact with the LP in the modifications. The synthesis of other ustiloxin analogs requires additional modifications. For example, a norvaline residue is added through a sulfur linkage at the tyrosine benzyl group of ustiloxin F to generate ustiloxin B, which likely requires a cytochrome P450 UstC, a cysteine desulfurase UstD, a glutathione S-transferase UstS, two flavin-dependent enzymes UstF1 and UstF2, and a PLP-dependent oxidase UstO (Fig. 5B) [116,125]. The functions of UstF1, UstF2 and UstD in the biosynthesis of ustiloxin B have been confirmed in both genetic and biochemical studies [116,125]. The biosynthesis of phomopsins is highly similar to ustiloxins [117]. The precursor peptides of phomopsins carry multiple repeats with a highly conserved sequence motif of VEDY(V/A/F)I(G/P)(I/V/A)DKR. The central hexapeptide fragment appears in the final structures of phomopsins. The proteolytic cleavages of the native or modified repeats are presumably mediated by Kex2 and one pathway-specific S41 family peptidease PhomP1, while the modifications on the hexapeptide can be catalyzed by the tyrosinase PhomQ1, one or more of five uncharacterized enzymes PhomYa-e, a hydroxylase PhomE, an oxidoreductase PhomF, and a methyltransferase PhomM [117]. Again, the order and details of these potential processing steps in the phomopsin biosynthesis have not been studied yet. Collectively, the known dikaritin precursor peptides contain multiple repeats of conserved sequence motifs and the biosynthesis requires multiple proteolysis steps and enzymatic modifications, particularly aromatic hydroxylation and oxidative crosslinking, which await further characterization. The similar biosynthetic logic can be employed in the biosynthesis of epichloëcyclins that are produced by Epichloë fungi associate with grasses in mutualistic to parasitic manners [54,55]. Furthermore, the relatively conserved modifications and the variations on several positions of the dikaritin CPs indicate great potential in the creation of diverse libraries and combinatorial biosynthesis [22,82,116,125].
3. Plant multicore RiPPs
Plant RiPPs are stable cyclic peptides of significant pharmacological interests. These peptides are very diverse in the size and residue components. Although they can be produced from the precursor peptides containing a single CP, recent genome mining studies revealed the prevalence of multicore precursor peptides. Here we will review the biosynthesis and engineering of three known families, cyclotides, orbitides and lyciumins.
3.1. Cyclotides:
Cyclotides were first discovered from Oldelandia affinis DC, a plant used by African women to accelerate contractions and childbirth [35,36,96], and hundreds of analogs have been identified from many other plants [15,18,79,119]. These compounds have a common characteristic structural feature, a cysteine knot in which a ring formed by two disulfide bonds is threaded by a third disulfide linkage (Fig. 6A). This structural fold is further divided into three subfamilies, bracelet-like in which the backbone is not twisted (e.g., Cycloviolacin O2), Möbius where the peptide is cyclized through a backbone twist (e.g., Kalata B12), resembling a Möbius strip, and the trypsin inhibitor (e.g., MCoTI-II), which has a longer sequence resulting in a looser cysteine knot (Fig. 6A) [34,77]. The bracelet family is the most common, accounting for approximately two-thirds of known cyclotides, followed by the Möbius family [120]. On the other hand, while many cyclotides are also head to tail cyclized, there are known linear cyclotides [18,48,96]. Cyclotides often contain 28 to 37 amino acid residues, including six conserved cysteines at specific positions for forming the characteristic fold. Furthermore, the first and last residues of cyclotides are generally A/G and N/D, respectively [17,52], while the rest are highly variable. The sequence diversity may be related to their potential biological role as a host defense factor as some plants produce large quantities of these compounds (e.g., 1g compound/kg leaf extract) [18], while cyclotides also demonstrate diverse other activities, including antiviral, protease inhibition, insecticidal, and cytotoxic activities.
Fig. 6.
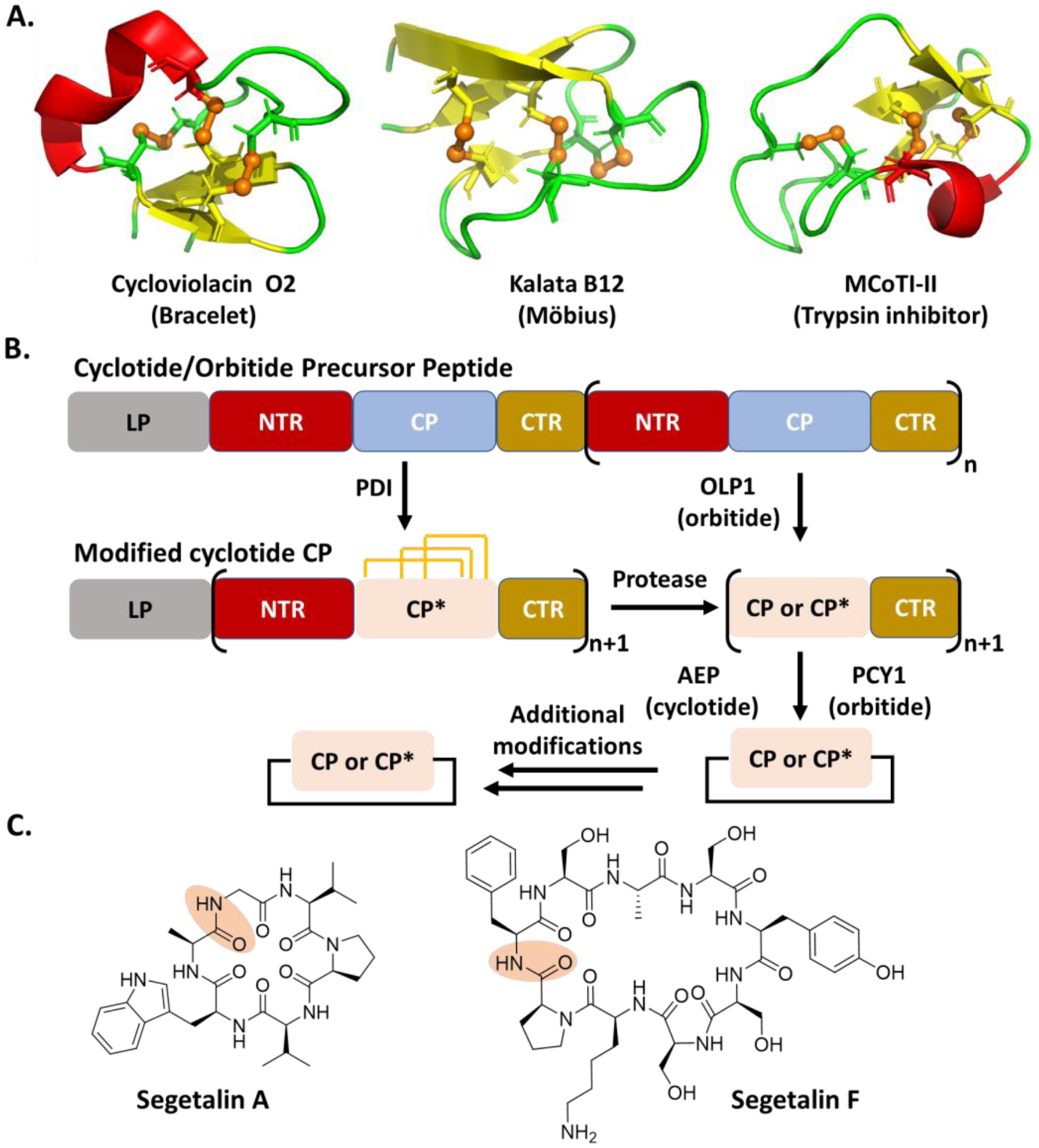
A. Representative cyclotides showing structural variations. The PDB IDs of Cycloviolacin O2, Kalata 12 and MCoTI-II are 2KCG, 2KVX, and 1HA9, respectively. The figures were created by Pymol. The disulfide bond is shown in brown and the brown sphere represents sulfur atom. B. General scheme of cyclotide and orbitide biosynthesis. The orange line represents a disulfide bond. C. Representative orbitide structures. The peptide bond for the cyclization is shadowed in orange.
Cyclotide BGCs have been uncovered with up to 10 precursor peptides, while each precursor peptide can contain up to eight CP motifs. In the multicore systems, these CPs can encode both identical and unique cyclotides, i.e., the cyclotide precursor peptide from Cucurbitacaeae can produce four distinct products from six CP units [17,18,52,81]. Each CP of the cyclotide precursor peptide is flanked by two conserved motifs, termed terminal repeats (NTR/CTR) (Fig. 6B) [41]. On the other hand, the NTR/CTR sequences of the precursor peptides from different plants are highly variable. Furthermore, the cyclotide CPs have been found to be embedded in the albumin gene, which normally encode hormone-like peptides with insecticidal properties [37,89]. The albumin BGC can generate the active cyclotide, without clear cyclotide NTR/CTR [90]. The same “hijacking” mechanism has also been found in the biosynthesis of cyclic plant peptides PawS and PawL [27,30,31,50].
The cyclotide precursor peptide is ribosomally translated and targeted to endoplasmic reticulum (ER) by its N-terminal ER signal peptide, where the signal peptide is removed (Fig. 6B). The details of the remaining steps, particularly the order, in the cyclotide biosynthesis have not been clearly defined. Briefly, a protein-disulfide isomerase (PDI) is expected to catalyze the formation of the three disulfide bonds on each CP in the ER [42]. It is unclear if and how the leader peptide interacts with PDI and other processing enzymes in the cyclotide biosynthesis. The modified precursor peptide is then transported to the vacuole, where a yet-identified enzyme first cleaves the NTR of each modified CP. The exposed amino group of A/G of each modified CP then acts as a nucleophile for intramolecular transpeptidation mediated by an asparaginyl endopeptidase (AEP) to produce cyclic cyclotides (Fig. 6B) [14,17,83,84]. On the other hand, the head-to-tail cyclization mediated by AEP is independent of the presence of disulfide bonds and the enzyme is promiscuous. Additional modifications to the cyclic cyclotides are present, e.g., reductions and alkylations [102]. Furthermore, the current understanding of the cyclotide biosynthesis has mainly relied on the use of natural or engineered single-core precursor peptides, and the processing details of multicore systems remains unknown.
Given the availability of multiple nonconserved sites, cyclotides have been used for molecular grafting, in which epitopes are engineered into cyclotides, for improving the stability and efficacy of the epitope and introducing new bioactivities for the recombinant cyclotides [16,118]. Unfortunately, some epitopes prevent enzymatic cyclization. Recently, an enzymatic method for improved cyclization of epitope-linked cyclotides has been reported [108]. In this case, an alternative cyclization site was introduced, allowing for efficient ligation and efficacy of the bioactive epitope, without affecting normal cyclization of the peptide. This improved method opens new opportunities to create large cyclotide libraries in a rapid and low-cost manner.
3.2. Orbitides:
Orbitides are another family of plant cyclic peptides that contain 5–16 residues (Fig. 6C) [6,28]. Over 130 orbitides have so far been discovered and some possess anti-inflammatory, antiplatelet, antimalarial, immunosuppressive, and cytotoxic activities [12,13,79,85,110,119,124]. Compared with cyclotides, orbitides are also head-to-tail cyclized through the peptide backbone, but have no disulfide bridges. On the other hand, the biosynthesis of cyclotides and orbitides share multiple common features (Fig. 6B). The orbitide precursors contain up to nine CP motifs whose sequence components and sizes can be varied [29,86,103]. Each orbitide CP is flanked by conserved NTR/CTR repeats whose sequences often differ among plants. Similar to cyclotides, the orbitide CPs have been found in the albumin gene cluster [30,50]. The processing of the CP is believed to start by the removal of NTR by a serine protease OLP1, followed by the cleavage of CTR and the intramolecular transpeptidation by a peptide cyclase of the S9A protease family, PCY1 (Fig. 6B) [6,10,13,71]. Additional modifications on the cyclized orbitides have been observed [68,109]. Similar to AEP in the biosynthesis of cyclotides, PCY1 is promiscuous in modifying a variety of non-native substrates [106], suggesting its biotechnological applications for the synthesis of the orbitide libraries. Similar to the cyclotide biosynthesis, the details of the enzymatic processing of the orbitide multicore system, including the LP/protein interactions, have yet been deduced.
3.3. Lyciumins:
Lyciumins are a family of branched cyclic peptides produced by Lyceum barbarum (Fig. 7) and are potent inhibitors of proteases, angiotensin-converting enzymes and renin. The biosynthetic logic of lyciumins was uncovered in 2018 through genome-mining efforts [59]. The first precursor gene discovered, LbaLycA, contains 12 CP motifs coding for four different compounds, while the precursor peptides mined from other plants can carry 1 to 18 CPs [59]. In addition to the CPs, the precursor peptides have an N-terminal ER signal and a C-terminal BURP domain that is specific in terrestrial plants [42,59]. The CPs can reside within the BURP domain or be N-terminal to it. The 12 CPs in the LbaLycA show a conserved motif of 1-QP(W/Y)GVG(I/S)W-8, with a kexin-like cleavage site N-terminal to each core. However, the LP portion of LbaLycA and its role in the CP processing have not been defined. Biochemical studies of the lyciumin biosynthesis have not been available, while the functional prediction and heterologous expression suggested that the processing is started by cyclizing the CP by a C-N bond between the α-carbon Gly4 and the indole nitrogen of Trp8, likely through a radical mechanism (Fig. 7). Subsequently, the N-terminus of each modified CP is exposed after the cleavage by yet-identified endopeptidases. The Gln1 is then converted into a pyroglutamate moiety by a pathway-specific glutamine cyclotransferase. The final products are then generated by proteolytic cleavage C-terminal to each modified CP (Fig. 7). The processing enzymes of lyciumins are tolerant alanine replacement of the CP at every position other than the residues involved in cyclization [59]. Furthermore, the CP with a Gly4Thr mutant was converted into a cyclized product, Lyciumin I, as well as a dehydrothreonine derivative. On the other hand, the details in processing the lyciumin multicore system have not been studied. Collectively, the enzymes involved in the lyciumin biosynthesis show different biotransformations with those of cyclotides and orbitide (Fig. 6).
Fig. 7.
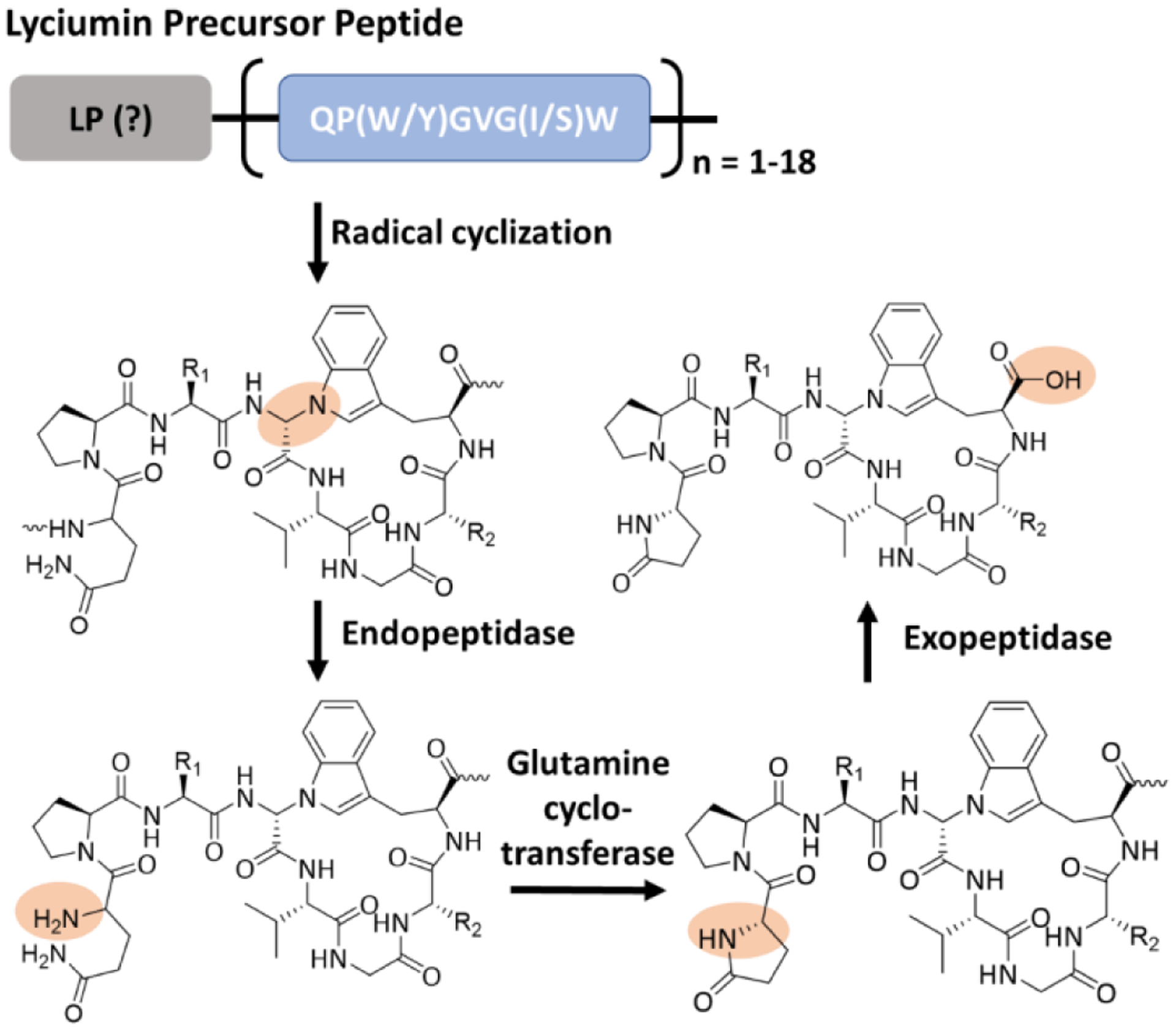
General scheme of lyciumin biosynthesis. Key chemical transformations are shadowed in orange. R1 and R2 represent the side chains of Trp/Tyr and Ile/Ser, respectively.
4. Conclusion and future perspectives
The wide-spread presence of multiple core motifs within the precursors of different RiPPs families is a recently discovered phenomenon. The multicore biosynthesis allows for increased peptide production and diversity, without the added cost of translating multiple precursors or enzymes. This review highlights multicore systems in both eukaryotic and prokaryotic organisms, outlining the current biosynthetic understanding of these systems and differences between multicore and single-core, as well as the engineering. As a bonus for pharmacological research, enzymes in the multicore systems are naturally promiscuous to account for changes in size or residues within the various CPs. This inherent promiscuity enables the incorporation of canonical and nonproteinogenic residues, as well as medically relevant epitopes, to enhance the potency and selectivity. Indeed, the multicore system has been utilized to generate diversity in cyanobactin libraries, clearly demonstrating the potential of these systems for improving peptide production efficiency. However, a major limitation to broad utilization of these multicore systems is our limited understanding of how the enzymes act on multiple core-containing substrates. Indeed, the biosynthetic logic of the multicore systems has often been characterized with engineered single-core substrates that significantly ease the monitoring of CP modifications [47]. However, the simplified substrates lead to the missing of critical biochemical and mechanistic insights into the enzymatic processing of multicore systems, e.g., the timing and order of different modifications on the multiple CPs and the potential cooperation of different biosynthetic enzymes in the processing [39, 127]. In this regard, the actual promiscuity and combinatorial biosynthesis potential of the enzymes of the multicore systems have not been assessed in detail. On the other hand, as the understanding of the multicore biosynthetic logic improves with increased research efforts, we believe that there is tremendous potential for the generation of large non-natural peptide libraries with demonstrated biological activity. In addition, different sets of enzymes used in the biosynthesis of the same RiPP family members and proven enzyme promiscuity enable combinatorial biosynthesis, further diversifying these peptides. While many single-core RiPPs have potent bioactivities, their multicore counterparts can demonstrate increased potency and specificity [64]. Importantly, these multicore peptides can execute different bioactivities by engineering individual CPs [62]. We are optimistic that through their natural diversity-generating pathways in conjunction with substrate and enzyme engineering, multicore RiPPs systems can greatly improve basic and translational studies and expand industrial applications.
Acknowledgements:
This work was financially supported by a start-up funding from the University of Florida and NIH 1R35GM128742 (Y.D.) and the University of Florida Graduate School Preeminence Award (G.M.R.). We thank the current and past members of the Ding group for the critical discussions and the support from Prof. Steven D. Bruner at the University of Florida.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Conflict of interest:
The authors declare that they have no conflict of interest.
References:
- 1.Agarwal Vinayak, Pierce Elizabeth, John McIntosh Eric W. Schmidt, and Nair Satish K.. 2012. “Structures of Cyanobactin Maturation Enzymes Define a Family of Transamidating Proteases.” Chemistry & Biology 19 (11): 1411–22. 10.1016/j.chembiol.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed Muhammad N., Emmanuel Reyna-González Bianca Schmid, Wiebach Vincent, Süssmuth Roderich D., Dittmann Elke, and Fewer David P. 2017. “Phylogenomic Analysis of the Microviridin Biosynthetic Pathway Coupled with Targeted Chemo-Enzymatic Synthesis Yields Potent Protease Inhibitors.” ACS Chemical Biology 12 (6): 1538–46. 10.1021/acschembio.7b00124. [DOI] [PubMed] [Google Scholar]
- 3.Akondi Kalyana B., Muttenthaler Markus, Dutertre Sébastien, Kaas Quentin, Craik David J., Lewis Richard J., Alewood Paul F., and Akondi KB. 2014. “Discovery, Synthesis, and Structure-Activity Relationships of Conotoxins.” Chemical Reviews 114 (11): 5815–47. 10.1021/cr400401e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderl Jan, Echner Hartmut, and Faulstich Heinz. 2012. “Chemical Modification Allows Phallotoxins and Amatoxins to Be Used as Tools in Cell Biology.” Beilstein Journal of Organic Chemistry 8 (November): 2072–84. 10.3762/bjoc.8.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnison Paul G., Bibb Mervyn J., Bierbaum Gabriele, Bowers Albert A., Bugni Tim S., Bulaj Grzegorz, Camarero Julio A., et al. 2013. “Ribosomally Synthesized and Post-Translationally Modified Peptide Natural Products: Overview and Recommendations for a Universal Nomenclature.” Natural Product Reports 30 (1): 108–60. 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barber Carla J.S., Pujara Pareshkumar T., Reed Darwin W., Chiwocha Shiela, Zhang Haixia, and Covello Patrick S.. 2013. “The Two-Step Biosynthesis of Cyclic Peptides from Linear Precursors in a Member of the Plant Family Caryophyllaceae Involves Cyclization by a Serine Protease-like Enzyme.” Journal of Biological Chemistry 288 (18): 12500–510. 10.1074/jbc.M112.437947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burkhart Brandon J., Hudson Graham A., Dunbar Kyle L., and Mitchell Douglas A.. 2015. “A Prevalent Peptide-Binding Domain Guides Ribosomal Natural Product Biosynthesis.” Nature Chemical Biology 11 (8): 564–70. 10.1038/nchembio.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burkhart Brandon J., Kakkar Nidhi, Hudson Graham A., Van Der Donk Wilfred A., and Mitchell Douglas A.. 2017. “Chimeric Leader Peptides for the Generation of Non-Natural Hybrid RiPP Products.” ACS Central Science 3 (6): 629–38. 10.1021/acscentsci.7b00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burkhart Brandon J., Schwalen Christopher J., Mann Greg, Naismith James H., and Mitchell Douglas A.. 2017. “YcaO-Dependent Posttranslational Amide Activation: Biosynthesis, Structure, and Function.” Chemical Reviews. 10.1021/acs.chemrev.6b00623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chekan Jonathan R., Estrada Paola, Covello Patrick S., and Nair Satish K.. 2017. “Characterization of the Macrocyclase Involved in the Biosynthesis of RiPP Cyclic Peptides in Plants.” Proceedings of the National Academy of Sciences of the United States of America 114 (25): 6551–56. 10.1073/pnas.1620499114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung-Lee Wai Ling, and Link A. James. 2019. “Genome Mining for Lasso Peptides: Past, Present, and Future.” Journal of Industrial Microbiology and Biotechnology 46 (9–10): 1371–79. 10.1007/s10295-019-02197-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chuang Pei Hsuan, Pei Wen Hsieh Yu Liang Yang, Kuo Feng Hua Fang Rong Chang, Shiea Jentaie, Hsiung Wu Shih, and Chang Wu Yang. 2008. “Cyclopeptides with Anti-Inflammatory Activity from Seeds of Annona Montana.” Journal of Natural Products 71 (8): 1365–70. 10.1021/np8001282. [DOI] [PubMed] [Google Scholar]
- 13.Condie Janet A., Nowak Goska, Reed Darwin W., Balsevich J. John, Reaney Martin J.T. T., Arnison Paul G., and Covello Patrick S.. 2011. “The Biosynthesis of Caryophyllaceae-like Cyclic Peptides in Saponaria Vaccaria L. from DNA-Encoded Precursors” The Plant Journal 67 (4): 682–90. 10.1111/j.1365-313X.2011.04626.x. [DOI] [PubMed] [Google Scholar]
- 14.Conlan Brendon F., Gillon Amanda D., Barbeta Barbara L., and Anderson Marilyn A.. 2011. “Subcellular Targeting and Biosynthesis of Cyclotides in Plant Cells.” American Journal of Botany 98 (12): 2018–26. 10.3732/ajb.1100382. [DOI] [PubMed] [Google Scholar]
- 15.Craik David J. 2009. “Circling the Enemy: Cyclic Proteins in Plant Defense.” Trends in Plant Science. 14 (6): 328–35. 10.1016/j.tplants.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Craik David J., and Du Junqiao. 2017. “Cyclotides as Drug Design Scaffolds.” Current Opinion in Chemical Biology. 38: 8–16. 10.1016/j.cbpa.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 17.Craik David J., and Malik Uru. 2013. “Cyclotide Biosynthesis.” Current Opinion in Chemical Biology. 17 (4): 546–54 10.1016/j.cbpa.2013.05.033. [DOI] [PubMed] [Google Scholar]
- 18.Craik David J., Daly Norelle L., Bond Trudy, and Waine Clement. 1999. “Plant Cyclotides: A Unique Family of Cyclic and Knotted Proteins That Defines the Cyclic Cystine Knot Structural Motif.” Journal of Molecular Biology 294 (5): 1327–36. 10.1006/jmbi.1999.3383. [DOI] [PubMed] [Google Scholar]
- 19.Culvenor Claude C.J., Edgar John A., Mackay Maureen F., Gorst-Allman Charles P., Marasas Walter F.O., Steyn Pieter S., Vleggaar Robert, and Wessels Philippus L.. 1989. “Structure Elucidation and Absolute Configuration of Phomopsin a, a Hexapeptide Mycotoxin Produced by Phomopsis Leptostromiformis.” Tetrahedron 45 (8): 2351–72. 10.1016/S0040-4020(01)83436-0. [DOI] [Google Scholar]
- 20.de los Santos Emmanuel L.C. 2019. “NeuRiPP: Neural Network Identification of RiPP Precursor Peptides.” Scientific Reports 9 (1): 1–9. 10.1038/s41598-019-49764-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delves-Broughton J 1996. “Applications of the Bacteriocin, Nisin.” Antonie van Leeuwenhoek. 69: 193–202. 10.1007/BF00399424. [DOI] [PubMed] [Google Scholar]
- 22.Ding Wei, Wan Qiu Liu Youli Jia, Li Yongzhen, Van Der Donk Wilfred A., and Zhang Qi. 2016. “Biosynthetic Investigation of Phomopsins Reveals a Widespread Pathway for Ribosomal Natural Products in Ascomycetes.” Proceedings of the National Academy of Sciences of the United States of America 113 (13): 3521–26. 10.1073/pnas.1522907113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong Shi Hui, Liu Andi, Mahanta Nilkamal, Mitchell Douglas A., and Nair Satish K.. 2019. “Mechanistic Basis for Ribosomal Peptide Backbone Modifications.” ACS Central Science 5 (5): 842–51. 10.1021/acscentsci.9b00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donia Mohamed S., and Schmidt Eric W.. 2011. “Linking Chemistry and Genetics in the Growing Cyanobactin Natural Products Family.” Chemistry & Biology 18 (4): 508–19. 10.1016/j.chembiol.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donia Mohamed S., Hathaway Brian J., Sudek Sebastian, Haygood Margo G., Rosovitz MJ, Ravel Jacques, and Schmidt Eric W.. 2006. “Natural Combinatorial Peptide Libraries in Cyanobacterial Symbionts of Marine Ascidians.” Nature Chemical Biology 2 (12): 729–35. 10.1038/nchembio829. [DOI] [PubMed] [Google Scholar]
- 26.Dutton Julie L., Renda Rosemary F., Waine Clement, Clark Richard J., Daly Norelle L., Jennings Cameron V., Anderson Marilyn A., and Craik David J.. 2004. “Conserved Structural and Sequence Elements Implicated in the Processing of Gene-Encoded Circular Protein.” Journal of Biological Chemistry 279 (45): 46858–67. 10.1074/jbc.M407421200. [DOI] [PubMed] [Google Scholar]
- 27.Elliott Alysha G., Delay Christina, Liu Huanle, Phua Zaiyang, Rosengren K. Johan, Benfield Aurélie H., Panero Jose L., et al. 2014. “Evolutionary Origins of a Bioactive Peptide Buried within Preproalbumin.” Plant Cell 26 (3): 981–95. 10.1105/tpc.114.123620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fisher Mark F., Payne Colton D., Rosengren K. Johan, and Mylne Joshua S.. 2019. “An Orbitide from Ratibida Columnifera Seed Containing 16 Amino Acid Residues.” Journal of Natural Products 82 (8): 2152–58. 10.1021/acs.jnatprod.9b00111. [DOI] [PubMed] [Google Scholar]
- 29.Fisher Mark F., Zhang Jingjing, Berkowitz Oliver, Whelan James, and Mylne Joshua S.. 2020. “Novel Cyclic Peptides in Seed of Annona Muricata Are Ribosomally Synthesized.” BioRxiv, February, 647552 10.1101/647552. [DOI] [PubMed] [Google Scholar]
- 30.Fisher Mark F., Zhang Jingjing, Taylor Nicolas L., Howard Mark J., Berkowitz Oliver, Debowski Aleksandra W., Behsaz Bahar, Whelan James, Pevzner Pavel A., and Mylne Joshua S.. 2018. “A Family of Small, Cyclic Peptides Buried in Preproalbumin since the Eocene Epoch.” Plant Direct 2 (2). 10.1002/pld3.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franke B, Mylne JS, and Rosengren KJ. 2018. “Buried Treasure: Biosynthesis, Structures and Applications of Cyclic Peptides Hidden in Seed Storage Albumins.” Natural Product Reports 35 (2): 137–46. 10.1039/c7np00066a. [DOI] [PubMed] [Google Scholar]
- 32.Gao Sisi, Ge Ying, Bent Andrew F., Schwarz-Linek Ulrich, and Naismith James H.. 2018. “Oxidation of the Cyanobactin Precursor Peptide Is Independent of the Leader Peptide and Operates in a Defined Order.” Biochemistry 57 (41): 5996–6002. 10.1021/acs.biochem.8b00835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goto Yuki, Ito Yumi, Kato Yasuharu, Tsunoda Shotaro, and Suga Hiroaki. 2014. “One-Pot Synthesis of Azoline-Containing Peptides in a Cell-Free Translation System Integrated with a Posttranslational Cyclodehydratase.” Chemistry & Biology 21 (6): 766–74. 10.1016/j.chembiol.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 34.Gould Andrew, Ji Yanbin, Aboye Teshome L., and Camarero Julio A.. 2012. “Cyclotides, a Novel Ultrastable Polypeptide Scaffold for Drug Discovery.” Current Pharmaceutical Design 17 (38): 4294–4307. 10.2174/138161211798999438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gran Lorents. 2009. “On the Effect of a Polypeptide Isolated from ‘Kalata-Kalata’ (Oldenlandia Affinis DC) on the Oestrogen Dominated Uterus.” Acta Pharmacologica et Toxicologica 33 (5–6): 400–408. 10.1111/j.1600-0773.1973.tb01541.x. [DOI] [PubMed] [Google Scholar]
- 36.Gran Lorents. 1973. “Oxytocic Principles of Oldenlandia Affinis.” Lloydia 36 (2): 174–78. [PubMed] [Google Scholar]
- 37.Gressent Frédéric, Pedro de Silva Vanessa Eyraud, Karaki Lamis, and Royer Corinne. 2011. “Pea Albumin 1 Subunit b (PA1b), a Promising Bioinsecticide of Plant Origin.” Toxins 3 (12):1502–17. 10.3390/toxins3121502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gu Wenjia, Shi Hui Dong Snigdha Sarkar, Nair Satish K., and Schmidt Eric W.. 2018. “The Biochemistry and Structural Biology of Cyanobactin Pathways: Enabling Combinatorial Biosynthesis” In Methods in Enzymology, 604:113–63. Academic Press Inc. 10.1016/bs.mie.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu Wenjia, Sardar Debosmita, Pierce Elizabeth, and Schmidt Eric W.. 2018. “Roads to Rome: Role of Multiple Cassettes in Cyanobactin RiPP Biosynthesis.” Journal of the American Chemical Society 140 (47): 16213–21. 10.1021/jacs.8b09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hambley Trevor W., Hawkins Clifford J., Lavin Martin F., van den Brenk Anna, and Watters Diane J.. 1992. “Cycloxazoline: A Cytotoxic Cyclic Hexapeptide from the Ascidian Lissoclinum Bistratum.” Tetrahedron 48 (2): 341–48. 10.1016/S0040-4020(01)88146-1. [DOI] [Google Scholar]
- 41.Hansen J. Norman. 1994. “Nisin as a Model Food Preservative.” Critical Reviews in Food Science and Nutrition 34 (1): 69–93. 10.1080/10408399409527650. [DOI] [PubMed] [Google Scholar]
- 42.Hattori J, Boutilier KA, Van Lookeren Campagne MM, and Miki BL. 1998. “A Conserved BURP Domain Defines a Novel Group of Plant Proteins with Unusual Primary Structures.” Molecular and General Genetics 259 (4): 424–28. 10.1007/s004380050832. [DOI] [PubMed] [Google Scholar]
- 43.Hennemann Hanjo, Wirths Sabine, and Carl Claudia. 2015. “Cell-Based Peptide Screening to Access the Undruggable Target Space.” European Journal of Medicinal Chemistry 94 (April): 489–96. 10.1016/j.ejmech.2014.10.038. [DOI] [PubMed] [Google Scholar]
- 44.Hetrick Kenton J., Walker Mark C., and Van Der Donk Wilfred A.. 2018. “Development and Application of Yeast and Phage Display of Diverse Lanthipeptides.” ACS Central Science 4 (4): 458–67. 10.1021/acscentsci.7b00581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Houssen Wael E., and Jaspars Marcel. 2010. “Azole-Based Cyclic Peptides from the Sea Squirt Lissoclinum Patella: Old Scaffolds, New Avenues.” ChemBioChem 11 (13): 1803–15. 10.1002/cbic.201000230. [DOI] [PubMed] [Google Scholar]
- 46.Houssen Wael E., Bent Andrew F., McEwan Andrew R., Pieiller Nathalie, Tabudravu Jioji, Koehnke Jesko, Mann Greg, et al. 2014. “An Efficient Method for the in Vitro Production of Azol(in)e-Based Cyclic Peptides.” Angewandte Chemie International Edition 53 (51): 14171–74. 10.1002/anie.201408082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ireland Chris, and Scheuer Paul J.. 1980. “Ulicyclamide and Ulithiacyclámide, Two New Small Peptides from a Marine Tunicate.” Journal of the American Chemical Society. 102 (17): 5688–91. 10.1021/ja00537a053. [DOI] [Google Scholar]
- 48.Ireland David C., Colgrave Michelle L., Nguyencong Philip, Daly Norelle L., and Craik David J.. 2006. “Discovery and Characterization of a Linear Cyclotide from Viola Odorata: Implications for the Processing of Circular Proteins.” Journal of Molecular Biology 357 (5): 1522–35. 10.1016/j.jmb.2006.01.051. [DOI] [PubMed] [Google Scholar]
- 49.Ishitsuka Midori O., Kusumi Takenori, Kakisawa Hiroshi, Kaya Kunimitsu, and Watanabe Makoto M.. 1990. “Microviridin: A Novel Tricyclic Depsipeptide from the Toxic Cyanobacterium Microcystis viridis.” Journal of the American Chemical Society 112 (22): 8180–82. 10.1021/ja00178a060. [DOI] [Google Scholar]
- 50.Jayasena Achala S., Fisher Mark F., Panero Jose L., Secco David, Kalia Bernath-Levin Oliver Berkowitz, Taylor Nicolas L., Schilling Edward E., Whelan James, and Mylne Joshua S.. 2017. “Stepwise Evolution of a Buried Inhibitor Peptide over 45 My.” Molecular Biology and Evolution 34 (6): 1505–16. 10.1093/molbev/msx104. [DOI] [PubMed] [Google Scholar]
- 51.Jennings Cameron V., Rosengren K. Johan, Daly Norelle L., Plan Manuel, Stevens Jackie, Scanlon Martin J., Waine Clement, Norman David G., Anderson Marilyn A., and Craik David J.. 2005. “Isolation, Solution Structure, and Insecticidal Activity of Kalata B2, a Circular Protein with a Twist: Do Möbius Strips Exist in Nature?” Biochemistry 44 (3): 851–60. 10.1021/bi047837h. [DOI] [PubMed] [Google Scholar]
- 52.Jennings Cameron V., West Jenny, Waine Clement, Craik David J., and Anderson Marilyn A.. 2001. “Biosynthesis and Insecticidal Properties of Plant Cyclotides: The Cyclic Knotted Proteins from Oldenlandia Affinis.” Proceedings of the National Academy of Sciences of the United States of America 98 (19): 10614–19. 10.1073/pnas.191366898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jian Xiao Hong, Hai Xue Pan Ting Ting Ning, Yuan Yuan Shi Yong Sheng Chen, Li Yan, Xiao Wei Zeng Jian Xu, and Tang Gong Li. 2012. “Analysis of YM-216391 Biosynthetic Gene Cluster and Improvement of the Cyclopeptide Production in a Heterologous Host.” ACS Chemical Biology 7 (4): 646–51. 10.1021/cb200479f. [DOI] [PubMed] [Google Scholar]
- 54.Johnson Linda J., Johnson Richard D., Schardl Christopher L., and Panaccione Daniel G.. 2003. “Identification of Differentially Expressed Genes in the Mutualistic Association of Tall Fescue with Neotyphodium Coenophialum.” Physiological and Molecular Plant Pathology 63 (6): 305–17. 10.1016/j.pmpp.2004.04.001. [DOI] [Google Scholar]
- 55.Johnson Richard D., Lane Geoffrey A., Koulman Albert, Cao Mingshu, Fraser Karl, Fleetwood Damien J., Voisey Christine R., et al. 2015. “A Novel Family of Cyclic Oligopeptides Derived from Ribosomal Peptide Synthesis of an in Planta-Induced Gene, GigA, in Epichloë Endophytes of Grasses.” Fungal Genetics and Biology 85 (December): 14–24. 10.1016/j.fgb.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 56.Kaas Quentin, Westermann Jan Christoph, and Craik David J.. 2010. “Conopeptide Characterization and Classifications: An Analysis Using ConoServer.” Toxicon 55 (8): 1491–1509. 10.1016/j.toxicon.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 57.Kaas Quentin, Westermann Jan-C, Halai Reena, Wang Conan K L, and Craik David J. 2008. “ConoServer, a Database for Conopeptide Sequences and Structures.” Bioinformatics 24 (3): 445–46. 10.1093/bioinformatics/btm596. [DOI] [PubMed] [Google Scholar]
- 58.Kaas Quentin, Yu Rilei, Ai Hua Jin Sébastien Dutertre, and Craik David J.. 2012. “ConoServer: Updated Content, Knowledge, and Discovery Tools in the Conopeptide Database.” Nucleic Acids Research 40 (D1). 10.1093/nar/gkr886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kersten Roland D., and Weng Jing Ke. 2018. “Gene-Guided Discovery and Engineering of Branched Cyclic Peptides in Plants.” Proceedings of the National Academy of Sciences of the United States of America 115 (46): E10961–69. 10.1073/pnas.1813993115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koehnke Jesko, Bent Andrew, Houssen Wael E., Zollman David, Morawitz Falk, Shirran Sally, Vendome Jeremie, et al. 2012. “The Mechanism of Patellamide Macrocyclization Revealed by the Characterization of the PatG Macrocyclase Domain.” Nature Structural and Molecular Biology 19 (8): 767–72. 10.1038/nsmb.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lawton Linda A., Morris Linda A., and Jaspars Marcel. 1999. “A Bioactive Modified Peptide, Aeruginosamide, Isolated from the Cyanobacterium Microcystis Aeruginosa.” Journal of Organic Chemistry 64 (14): 5329–32. 10.1021/jo990247i. [DOI] [PubMed] [Google Scholar]
- 62.Lee Chanwoo, Lee Hyunbin, Park Jung Un, and Kim Seokhee. 2020. “Introduction of Bifunctionality into the Multidomain Architecture of the ω-Ester-Containing Peptide Plesiocin.” Biochemistry 59 (3): 285–89. 10.1021/acs.biochem.9b00803. [DOI] [PubMed] [Google Scholar]
- 63.Lee Hyunbin, Choi Mingyu, Jung Un Park Heejin Roh, and Kim Seokhee. 2020. “Genome Mining Reveals High Topological Diversity of ω-Ester-Containing Peptides and Divergent Evolution of ATP-Grasp Macrocyclases.” Journal of the American Chemical Society 142 (6): 3013–23. 10.1021/jacs.9b12076. [DOI] [PubMed] [Google Scholar]
- 64.Lee Hyunbin, Park Youngseon, and Kim Seokhee. 2017. “Enzymatic Cross-Linking of Side Chains Generates a Modified Peptide with Four Hairpin-like Bicyclic Repeats.” Biochemistry 56 (37): 4927–30. 10.1021/acs.biochem.7b00808. [DOI] [PubMed] [Google Scholar]
- 65.Lee Jaeheon, Mcintosh John, Hathaway Brian J., and Schmidt Eric W.. 2009. “Using Marine Natural Products to Discover a Protease That Catalyzes Peptide Macrocyclization of Diverse Substrates.” Journal of the American Chemical Society 131 (6): 2122–24. 10.1021/ja8092168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leikoski Niina, Fewer David P., and Sivonen Kaarina. 2009. “Widespread Occurrence and Lateral Transfer of the Cyanobactin Biosynthesis Gene Cluster in Cyanobacteria.” Applied and Environmental Microbiology 75 (3): 853–57. 10.1128/AEM.02134-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leikoski Niina, Liu Liwei, Jokela Jouni, Wahlsten Matti, Gugger Muriel, Calteau Alexandra, Permi Perttu, Kerfeld Cheryl A., Sivonen Kaarina, and Fewer David P.. 2013. “Genome Mining Expands the Chemical Diversity of the Cyanobactin Family to Include Highly Modified Linear Peptides.” Chemistry & Biology 20 (8): 1033–43. 10.1016/j.chembiol.2013.06.015. [DOI] [PubMed] [Google Scholar]
- 68.Li Chao Ming, Ning Hua Tan Hui Lan Zheng, Mu Qing, Xiao Jiang Hao Yi Neng He, and Zhou Jun. 1999. “Cyclopeptides from the Seeds of Annona Glabra.” Phytochemistry 50 (6): 1047–52. 10.1016/S0031-9422(98)00631-1. [DOI] [Google Scholar]
- 69.Li Kunhua, Condurso Heather L., Li Gengnan, Ding Yousong, and Bruner Steven D.. 2016. “Structural Basis for Precursor Protein-Directed Ribosomal Peptide Macrocyclization.” Nature Chemical Biology 12 (11): 973–79. 10.1038/nchembio.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lovelace Erica S., Armishaw Christopher J., Colgrave Michelle L., Wahlstrom Maria E., Alewood Paul F., Daly Norelle E., and Craik David J.. 2006. “Cyclic MrIA: A Stable and Potent Cyclic Conotoxin with a Novel Topological Fold That Targets the Norepinephrine Transporter.” Journal of Medicinal Chemistry 49 (22): 6561–68. 10.1021/jm060299h. [DOI] [PubMed] [Google Scholar]
- 71.Ludewig Hannes, Czekster Clarissa M., Oueis Emilia, Munday Elizabeth S., Arshad Mohammed, Synowsky Silvia A., Bent Andrew F., and Naismith James H.. 2018. “Characterization of the Fast and Promiscuous Macrocyclase from Plant PCY1 Enables the Use of Simple Substrates.” ACS Chemical Biology 13 (3): 801–11. 10.1021/acschembio.8b00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luo Shangwen, and Shi Hui Dong. 2019. “Recent Advances in the Discovery and Biosynthetic Study of Eukaryotic RIPP Natural Products.” Molecules. 24 (8): pii: E1541 10.3390/molecules24081541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martins Joana, and Vasconcelos Vitor. 2015. “Cyanobactins from Cyanobacteria: Current Genetic and Chemical State of Knowledge.” Marine Drugs 13 (11): 6910–46. 10.3390/md13116910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Matsumoto T, A Shishido H Morita H Itokawa, and Takeya K. 2001. “Cyclolinopeptides F-I, Cyclic Peptides from Linseed.” Phytochemistry 57 (2): 251–60. 10.1016/s0031-9422(00)00442-8. [DOI] [PubMed] [Google Scholar]
- 75.McIntosh John A., Donia Mohamed S., Nair Satish K., and Schmidt Eric W.. 2011. “Enzymatic Basis of Ribosomal Peptide Prenylation in Cyanobacteria.” Journal of the American Chemical Society 133 (34): 13698–705. 10.1021/ja205458h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McIntosh John A., Robertson Charles R., Agarwal Vinayak, Nair Satish K., Bulaj Grzegorz W., and Schmidt Eric W.. 2010. “Circular Logic: Nonribosomal Peptide-like Macrocyclization with a Ribosomal Peptide Catalyst.” Journal of the American Chemical Society 132 (44): 15499–501. 10.1021/ja1067806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McKeever Benedict, and Pattenden Gerald. 2003. “Total Synthesis of Trunkamide A, a Novel Thiazoline-Based Prenylated Cyclopeptide Metabolite from Lissoclinum Sp.” Tetrahedron 59 (15): 2713–27. 10.1016/S0040-4020(03)00294-1. [DOI] [Google Scholar]
- 78.Merwin Nishanth J., Mousa Walaa K., Dejong Chris A., Skinnider Michael A., Cannon Michael J., Li Haoxin, Dial Keshav, Gunabalasingam Mathusan, Johnston Chad, and Magarvey Nathan A.. 2020. “DeepRiPP Integrates Multiomics Data to Automate Discovery of Novel Ribosomally Synthesized Natural Products.” Proceedings of the National Academy of Sciences of the United States of America 117 (1): 371–80. 10.1073/pnas.1901493116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mulvenna Jason P, Conan Wang, and Craik David J. 2006. “CyBase: A Database of Cyclic Protein Sequence and Structure.” Nucleic Acids Research 34 (Database issue): D192–4. 10.1093/nar/gkj005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Murakami Masahiro, Sun Qi, Ishida Keishi, Matsuda Hisashi, Okino Tatsufumi, and Yamaguchi Katsumi. 1997. “Microviridins, Elastase Inhibitors from the Cyanobacterium Nostoc Minutum (NIES-26).” Phytochemistry 45 (6): 1197–1202. 10.1016/S0031-9422(97)00131-3. [DOI] [Google Scholar]
- 81.Mylne Joshua S., Lai Yue Chan Aurelie H. Chanson, Daly Norelle L., Schaefer Hanno, Bailey Timothy L., Nguyencong Philip, Cascales Laura, and Craik David J.. 2012. “Cyclic Peptides Arising by Evolutionary Parallelism via Asparaginyl-Endopeptidase-Mediated Biosynthesis.” Plant Cell 24 (7): 2765–78. 10.1105/tpc.112.099085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nagano Nozomi, Umemura Myco, Izumikawa Miho, Kawano Jin, Ishii Tomoko, Kikuchi Moto, Tomii Kentaro, et al. 2016. “Class of Cyclic Ribosomal Peptide Synthetic Genes in Filamentous Fungi.” Fungal Genetics and Biology 86 (January): 58–70. 10.1016/j.fgb.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 83.Nguyen Giang K.T., Shujing Wang, Yibo Qiu, Xinya Hemu, Yilong Lian, and Tam James P.. 2014. “Butelase 1 Is an Asx-Specific Ligase Enabling Peptide Macrocyclization and Synthesis.” Nature Chemical Biology 10 (9): 732–38. 10.1038/nchembio.1586. [DOI] [PubMed] [Google Scholar]
- 84.Nguyen Giang K.T., Antony Kam, Shining Loo, Jansson Anna E., Pan Lucy X., and Tam James P.. 2015. “Butelase 1: A Versatile Ligase for Peptide and Protein Macrocyclization.” Journal of the American Chemical Society 137 (49): 15398–401. 10.1021/jacs.5b11014. [DOI] [PubMed] [Google Scholar]
- 85.Northfield Susan E., Wang Conan K., Schroeder Christina I., Durek Thomas, Meng Wei Kan Joakim E. Swedberg, and Craik David J.. 2014. “Disulfide-Rich Macrocyclic Peptides as Templates in Drug Design.” European Journal of Medicinal Chemistry 77: 248–57. 10.1016/j.ejmech.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 86.Okinyo-Owiti Denis P., Young Lester, Burnett Peta-Gaye G., and Reaney Martin J. T.. 2014. “New Flaxseed Orbitides: Detection, Sequencing, and 15N Incorporation.” Biopolymers 102 (2): 168–75. 10.1002/bip.22459. [DOI] [PubMed] [Google Scholar]
- 87.Oueis E, Stevenson H, Jaspars M, Westwood NJ, and Naismith JH. 2017. “Bypassing the Proline/Thiazoline Requirement of the Macrocyclase PatG.” Chemical Communications 53 (91): 12274–77. 10.1039/c7cc06550g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Philmus Benjamin, Guerrette Joshua P., and Hemscheidt Thomas K.. 2009. “Substrate Specificity and Scope of MvdD, a GRASP-like Ligase from the Microviridin Biosynthetic Gene Cluster.” ACS Chemical Biology 4 (6): 429–34. 10.1021/cb900088r. [DOI] [PubMed] [Google Scholar]
- 89.Poth Aaron G., Colgrave Michelle L., Lyons Russell E., Dalya Norelle L., and Craik David J.. 2011. “Discovery of an Unusual Biosynthetic Origin for Circular Proteins in Legumes.” Proceedings of the National Academy of Sciences of the United States of America 108 (25): 10127–32. 10.1073/pnas.1103660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Prinsep Michele R., Moore Richard E., Levine Ira A., and Patterson Gregory M.L.. 1992. “Westiellamide, a Bistratamide-Related Cyclic Peptide from the Blue-Green Alga Westiellopsis Prolifica.” Journal of Natural Products 55 (1): 140–42. 10.1021/np50079a022. [DOI] [PubMed] [Google Scholar]
- 91.Reyna-González Emmanuel, Schmid Bianca, Petras Daniel, Süssmuth Roderich D., and Dittmann Elke. 2016. “Leader Peptide-Free In Vitro Reconstitution of Microviridin Biosynthesis Enables Design of Synthetic Protease-Targeted Libraries.” Angewandte Chemie International Edition 55 (32): 9398–9401. 10.1002/anie.201604345. [DOI] [PubMed] [Google Scholar]
- 92.Roh Heejin, Han Yeji, Lee Hyunbin, and Kim Seokhee. 2019. “A Topologically Distinct Modified Peptide with Multiple Bicyclic Core Motifs Expands the Diversity of Microviridin‐Like Peptides.” ChemBioChem 20 (8): 1051–59. 10.1002/cbic.201800678. [DOI] [PubMed] [Google Scholar]
- 93.Rohrlack Thomas, Christoffersen Kirsten, Poul Erik Hansen Wei Zhang, Czarnecki Olaf, Henning Manfred, Fastner Jutta, Erhard Marcel, Neilan Brett A., and Kaebernick Melanie. 2003. “Isolation, Characterization, and Quantitative Analysis of Microviridin J, a New Microcystis Metabolite Toxic to Daphnia.” Journal of Chemical Ecology 29 (8): 1757–70. 10.1023/A:1024889925732. [DOI] [PubMed] [Google Scholar]
- 94.Rohrlack Thomas, Christoffersen Kirsten, Kaebernick Melanie, and Neilan Brett A.. 2004. “Cyanobacterial Protease Inhibitor Microviridin J Causes a Lethal Molting Disruption in Daphnia Pulicaria.” Applied and Environmental Microbiology 70 (8): 5047–50. 10.1128/AEM.70.8.5047-5050.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ruffner Duane E., Schmidt Eric W., and Heemstra John R.. 2015. “Assessing the Combinatorial Potential of the RiPP Cyanobactin Tru Pathway.” ACS Synthetic Biology 4 (4): 482–92. 10.1021/sb500267d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Saether Olav, Craik David J., Campbell Iain D., Sletten Knut, Juul Jessie, and Norman David G.. 1995. “Elucidation of the Primary and Three-Dimensional Structure of the Uterotonic Polypeptide Kalata B1.” Biochemistry 34 (13): 4147–58. 10.1021/bi00013a002. [DOI] [PubMed] [Google Scholar]
- 97.Sardar Debosmita, Hao Yue, Lin Zhenjian, Morita Maho, Nair Satish K., and Schmidt Eric W.. 2017. “Enzymatic N- and C-Protection in Cyanobactin RiPP Natural Products.” Journal of the American Chemical Society 139 (8): 2884–87. 10.1021/jacs.6b12872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sardar Debosmita, Lin Zhenjian, and Schmidt Eric W.. 2015. “Modularity of RiPP Enzymes Enables Designed Synthesis of Decorated Peptides.” Chemistry & Biology 22 (7): 907–16. 10.1016/j.chembiol.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sardar Debosmita, Pierce Elizabeth, McIntosh John A., and Schmidt Eric W.. 2015. “Recognition Sequences and Substrate Evolution in Cyanobactin Biosynthesis.” ACS Synthetic Biology 4 (2): 167–76. 10.1021/sb500019b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sarkar Snigdha, Gu Wenjia, and Schmidt Eric W. 2020. “Expanding the Chemical Space of Synthetic Cyclic Peptides Using a Promiscuous Macrocyclase from Prenylagaramide Biosynthesis.” BioRxiv, March, 2020.03.17.996157. 10.1101/2020.03.17.996157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schmidt Eric W., Nelson James T., Rasko David A., Sudek Sebastian, Eisen Jonathan A., Haygood Margo G., and Ravel Jacques. 2005. “Patellamide A and C Biosynthesis by a Microcin-like Pathway in Prochloron Didemni, the Cyanobacterial Symbiont of Lissoclinum Patella.” Proceedings of the National Academy of Sciences of the United States of America 102 (20): 7315–20. 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Serra Aida, Hemu Xinya, Nguyen Giang K.T., Nguyen Ngan T.K., Sze Siu Kwan, and Tam James P.. 2016. “A High-Throughput Peptidomic Strategy to Decipher the Molecular Diversity of Cyclic Cysteine-Rich Peptides.” Scientific Reports 6 (1): 1–13. 10.1038/srep23005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shim Youn Young, Young Lester W., Arnison Paul G., Gilding Edward, and Reaney Martin J.T.. 2015. “Proposed Systematic Nomenclature for Orbitides.” Journal of Natural Products 78 (4): 645–52. 10.1021/np500802p. [DOI] [PubMed] [Google Scholar]
- 104.Sieber Simon, Grendelmeier Simone M., Harris Lonnie A., Mitchell Douglas A., and Gademann Karl. 2020. “Microviridin 1777: A Toxic Chymotrypsin Inhibitor Discovered by a Metabologenomic Approach.” Journal of Natural Products 83 (2): 438–46. 10.1021/acs.jnatprod.9b00986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sivonen Kaarina, Leikoski Niina, Fewer David P., and Jokela Jouni. 2010. “Cyanobactins-Ribosomal Cyclic Peptides Produced by Cyanobacteria.” Applied Microbiology and Biotechnology 86 (5): 1213–25. 10.1007/s00253-010-2482-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Skinnider Michael A, Dejong Chris A, Rees Philip N, Johnston Chad W, Li Haoxin, Webster Andrew L H, Wyatt Morgan A, and Magarvey Nathan A. 2015. “Genomes to Natural Products PRediction Informatics for Secondary Metabolomes (PRISM).” Nucleic Acids Research 43 (20): 9645–62. 10.1093/nar/gkv1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Skinnider Michael A, Merwin Nishanth J, Johnston Chad W, and Magarvey Nathan A. 2017. “PRISM 3: Expanded Prediction of Natural Product Chemical Structures from Microbial Genomes.” Nucleic Acids Research 45 (W1): W49–54. 10.1093/nar/gkx320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Smithies Bronwyn J., Yen Hua Huang Mark A. Jackson, Yap Kuok, Gilding Edward K., Harris Karen S., Anderson Marilyn A., and Craik David J.. 2020. “Circular Permutation of the Native Enzyme-Mediated Cyclization Position in Cyclotides.” ACS Chemical Biology 15 (4): 962–69. 10.1021/acschembio.9b00996. [DOI] [PubMed] [Google Scholar]
- 109.Taichi Misako, Yamazaki Toshimasa, Kimura Terutoshi, and Nishiuchi Yuji. 2009. “Total Synthesis of Marinostatin, a Serine Protease Inhibitor Isolated from the Marine Bacterium Pseudoallteromonas Sagamiensis.” Tetrahedron Letters 50 (20): 2377–80. 10.1016/j.tetlet.2009.02.213. [DOI] [Google Scholar]
- 110.Tan Ning Hua, and Zhou Jun. 2006. “Plant Cyclopeptides.” Chemical Reviews 106 (3): 840–95. 10.1021/cr040699h. [DOI] [PubMed] [Google Scholar]
- 111.Taniguchi M, Kamei K, Kanaori K, Koyama T, Yasui T, Takano R, Harada S, Tajima K, Imada C, and Hara S. 2005. “Relationship between Temporary Inhibition and Structure of Disulfide-Linkage Analogs of Marinostatin, a Natural Ester-Linked Protein Protease Inhibitor.” Journal of Peptide Research 66 (2): 49–58. 10.1111/j.1399-3011.2005.00271.x. [DOI] [PubMed] [Google Scholar]
- 112.Tianero Ma Diarey B., Donia Mohamed S., Young Travis S., Schultz Peter G., and Schmidt Eric W.. 2012. “Ribosomal Route to Small-Molecule Diversity.” Journal of the American Chemical Society 134 (1): 418–25. 10.1021/ja208278k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Truman Andrew W. 2016. “Cyclisation Mechanisms in the Biosynthesis of Ribosomally Synthesised and Post-Translationally Modified Peptides.” Beilstein Journal of Organic Chemistry 12 (1): 1250–68. 10.3762/bjoc.12.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tsomaia Natia. 2015. “Peptide Therapeutics: Targeting the Undruggable Space.” European Journal of Medicinal Chemistry 94 (April): 459–70. 10.1016/j.ejmech.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 115.Tsukui Takahiro, Nagano Nozomi, Umemura Myco, Kumagai Toshitaka, Terai Goro, Machida Masayuki, and Asai Kiyoshi. 2015. “Ustiloxins, Fungal Cyclic Peptides, Are Ribosomally Synthesized in Ustilaginoidea Virens.” Bioinformatics 31 (7): 981–85. 10.1093/bioinformatics/btu753. [DOI] [PubMed] [Google Scholar]
- 116.Umemura Myco, Nagano Nozomi, Koike Hideaki, Kawano Jin, Ishii Tomoko, Miyamura Yuki, Kikuchi Moto, et al. 2014. “Characterization of the Biosynthetic Gene Cluster for the Ribosomally Synthesized Cyclic Peptide Ustiloxin B in Aspergillus Flavus” 68: 23–30. 10.1016/j.fgb.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 117.Vogt Eva, and Markus Künzler. 2019. “Discovery of Novel Fungal RiPP Biosynthetic Pathways and Their Application for the Development of Peptide Therapeutics.” Applied Microbiology and Biotechnology 103 (14): 5567–5581. 10.1007/s00253-019-09893-x. [DOI] [PubMed] [Google Scholar]
- 118.Wang Conan K., and Craik David J.. 2018. “Designing Macrocyclic Disulfide-Rich Peptides for Biotechnological Applications Perspective.” Nature Chemical Biology. 14 (5): 417–427. 10.1038/s41589-018-0039-y. [DOI] [PubMed] [Google Scholar]
- 119.Wang Conan K L, Quentin Kaas, Laurent Chiche, and Craik David J. 2008. “CyBase: A Database of Cyclic Protein Sequences and Structures, with Applications in Protein Discovery and Engineering.” Nucleic Acids Research 36 (Database issue): D206–10. 10.1093/nar/gkm953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Weidmann Joachim, and Craik David J. 2016. “Discovery, Structure, Function, and Applications of Cyclotides: Circular Proteins from Plants.” Journal of Experimental Botany 67 (16): 4801–12. 10.1093/jxb/erw210. [DOI] [PubMed] [Google Scholar]
- 121.Weiz Annika R., Ishida Keishi, Makower Katharina, Ziemert Nadine, Hertweck Christian, and Dittmann Elke. 2011. “Leader Peptide and a Membrane Protein Scaffold Guide the Biosynthesis of the Tricyclic Peptide Microviridin.” Chemistry and Biology 18 (11): 1413–21. 10.1016/j.chembiol.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 122.Weiz Annika R., Ishida Keishi, Quitterer Felix, Meyer Sabine, Jan Christoph Kehr Kristian M. Müller, Groll Michael, Hertweck Christian, and Dittmann Elke. 2014. “Harnessing the Evolvability of Tricyclic Microviridins to Dissect Protease-Inhibitor Interactions.” Angewandte Chemie International Edition 53 (14): 3735–38. 10.1002/anie.201309721. [DOI] [PubMed] [Google Scholar]
- 123.Wieland Theodor, Faulstich Heinz, and Fiume Luigi. 1978. “Amatoxins, Phallotoxins, Phallolysin, and Antamanide: The Biologically Active Components of Poisonous Amanita Mushroom.” Critical Reviews in Biochemistry and Molecular Biology 5 (3): 185–260. 10.3109/10409237809149870. [DOI] [PubMed] [Google Scholar]
- 124.Yang Yu Liang, Kuo Feng Hua Pei Hsuan Chuang, Shih Hsiung Wu Kuen Yuh Wu, Fang Rong Chang, and Yang Chang Wu. 2008. “New Cyclic Peptides from the Seeds of Annona Squamosa L. and Their Anti-Inflammatory Activities.” Journal of Agricultural and Food Chemistry 56 (2): 386–92. 10.1021/jf072594w. [DOI] [PubMed] [Google Scholar]
- 125.Ye Ying, Minami Atsushi, Igarashi Yuya, Izumikawa Miho, Umemura Myco, Nagano Nozomi, Machida Masayuki, et al. 2016. “Unveiling the Biosynthetic Pathway of the Ribosomally Synthesized and Post-Translationally Modified Peptide Ustiloxin B in Filamentous Fungi.” Angewandte Chemie International Edition 55 (28): 8072–75. 10.1002/anie.201602611. [DOI] [PubMed] [Google Scholar]
- 126.Zhang Yi, Chen Manyun, Bruner Steven D., and Ding Yousong. 2018. “Heterologous Production of Microbial Ribosomally Synthesized and Post-Translationally Modified Peptides.” Frontiers in Microbiology. 9: 1801 10.3389/fmicb.2018.01801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang Yi, Li Kunhua, Yang Guang, McBride Joshua L., Bruner Steven D., and Ding Yousong. 2018. “A Distributive Peptide Cyclase Processes Multiple Microviridin Core Peptides within a Single Polypeptide Substrate.” Nature Communications 9 (1): 1–10. 10.1038/s41467-018-04154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ziemert Nadine, Ishida Keishi, Weiz Annika, Hertweck Christian, and Dittmann Elke. 2010. “Exploiting the Natural Diversity of Microviridin Gene Clusters for Discovery of Novel Tricyclic Depsipeptides.” Applied and Environmental Microbiology 76 (11): 3568–74. 10.1128/AEM.02858-09. [DOI] [PMC free article] [PubMed] [Google Scholar]


