Abstract
Current developments in protein docking aim at improvement of applicability, accuracy and utility of modeling macromolecular complexes. The challenges include the need for greater emphasis on protein docking to molecules of different types, proper accounting for conformational flexibility upon binding, new promising methodologies based on residue co-evolution and deep learning, affinity prediction, and further development of fully automated docking servers. Importantly, new developments increasingly focus on realistic modeling of protein interactions in vivo, including crowded environment inside a cell, which involves multiple transient encounters, and propagating the system in time. This opinion paper offers the author’s perspective on these challenges in structural modeling of protein interactions and the future of protein docking.
Keywords: molecular recognition, structure prediction, macromolecular modeling
Introduction
Protein docking has come a long way since its early days in the last century when first consequential approaches laid the foundation for the field [1]. Reflecting on the past, it is appropriate to think about the future of the protein docking, and to talk about its challenges - the ones within our immediate reach, for which current research is rapidly progressing, and the more distant ones, for which it is important to formulate basic paradigms and outline strategic directions.
The current developments and the future directions can be roughly put in two major categories: structural modeling of (a) static and (b) dynamic protein interaction systems. The “static” interactions do not involve propagation of the system in time (except molecular dynamics-driven simulations of individual complexes, which by design involve the time coordinate, but aim primarily at prediction/refinement of the equilibrium state, possibly including binding pathways [2]). Such modeling deals with improvement of the applicability, accuracy and utility of the equilibrium state prediction of a complex of two or several proteins or, more generally, proteins and other molecules. Here, we intentionally leave aside protein complexes with small compounds, which is a separate field of study due to important differences in the systems. The protein-small ligand docking, typically, involves predefined binding site on the protein receptor and thus has no requirement for the global docking search. Instead, it emphasizes determination of the precise atomic details of the interaction. Thus, it employs different techniques and often has different goals (e.g. drug design). The static interactions of proteins are modeled in dilute environment, with no regards to the presence of other molecules, except water for docking of soluble proteins and lipids for docking of integral membrane ones. Predictions can focus on a single complex, several complexes, or the entire interactome (all protein complexes in an organism, or in multiple organisms [3–5]).
The dynamic interactions inherently involve propagating the system in time. They deal with realistic modeling of protein interactions in vivo, including crowded environment inside a cell, which involves multiple transient interactions [6]. Such modeling would allow simulation of structure-based molecular diffusion and the multiplicity of binding/unbinding events describing molecular mechanisms in living systems. The modeling could scale up from individual pathways to whole cells [7–9] at different levels of approximation, from extreme coarse-graining [10] to potentially involving conformational flexibility of the molecules.
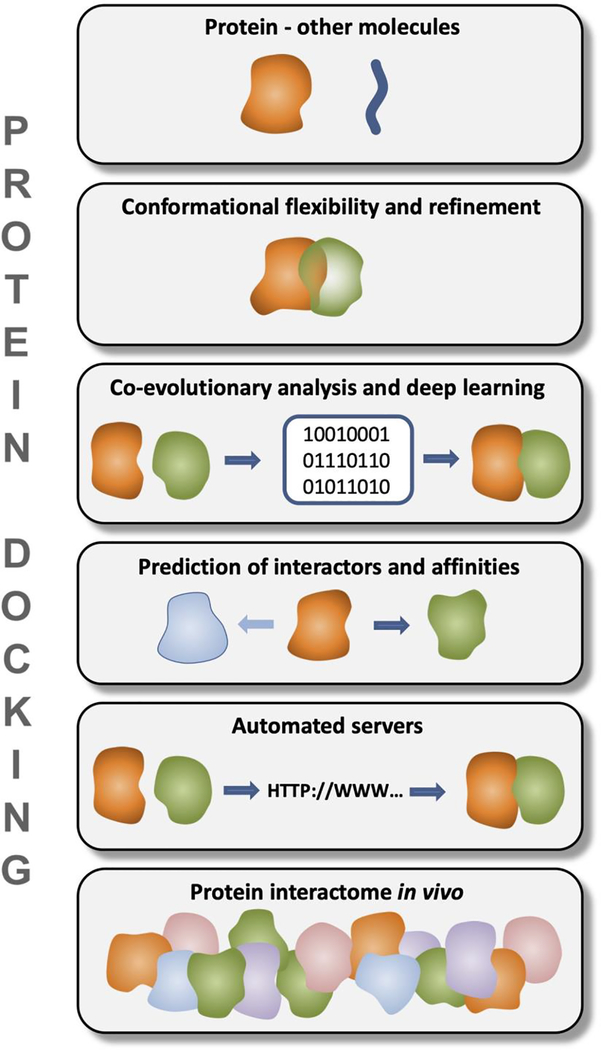
This opinion paper offers the author’s perspective on these challenges in structural modeling of protein interactions (Figure 1) and the future of protein docking.
Figure 1.
Challenges in protein docking.
Protein - other molecules
Protein-protein interactions are a key component of life processes at the molecular level. Thus, protein-protein docking has been arguably the most popular branch of docking (again, leaving aside docking of small ligands - see Introduction). Whereas the basic motivation for that is the centrality of the protein-protein interactions in molecular mechanisms, an important pragmatic consideration for this focus is that proteins in general, due to the uniqueness or limited variety of their global fold (at least for the non-intrinsically disordered protein types) are well-suited for the rigid body docking approximation. Such approximation is extremely important for the docking protocols because it allows one to exclude the enormous multitude of the internal degrees of freedom of the interacting molecules, reducing the global search space to only six coordinates of a two rigid-bodies system. Another important consideration is that the protein shape, reflecting the protein fold, to a large extent determines the docking solution, even at coarse-grained representation [11]. The rise of the comparative modeling of protein-protein complexes (template-based docking), based on the rapid expansion of PDB, makes the availability of templates for docking procedures routine [12,13]. Thus, an argument can be made that the protein-protein docking field is fairly advanced towards “solving” the protein-protein docking problem, at least at the coarse-grained resolution, for the “traditional” equilibrium (e.g. co-crystallizable) complexes of soluble globular proteins.
While other types of molecules interacting with proteins, as well as protein-protein interactions in membranes, have been studied extensively through the years [14–18], the progress there might be less obvious compared to the soluble globular protein-protein case. Beyond the arguably lesser focus of the docking community on such targets (empirically assessed by the number of research groups working on the subject, reflected in publications), an important “natural” obstacle is the global conformational flexibility of the non-protein component, in cases of peptides, RNA and lipids. Such flexibility limits the applicability of the powerful structure-alignment-based comparative docking [19] and significantly increases the dimensionality of the docking search space by adding the multiplicity of the internal degrees of freedom of the non-protein component. In cases where the conformational flexibility is less of a problem, such as protein-protein complexes in membranes and protein-DNA complexes, the dimensionality of the docking space may actually be less than that for the soluble protein-protein complexes. Protein-protein translational degrees of freedom in membranes are largely constrained in two dimensions by the membrane, and movement of proteins interacting with DNA may be modeled by sliding them along the one-dimensional DNA chain until they recognize the intended patch of nucleotides. However, the problem in these cases is that the recognition factors are smaller in scale than in the soluble protein-protein complexes (where coarse-grained representation determined by the global fold often suffices for a meaningful prediction) and require atomic-level prediction accuracy.
Conformational flexibility and refinement
Proteins, as well as other macromolecules, change conformation upon binding. While the global flexibility (multiplicity of conformations of the overall structure), may not be a problem for a number of protein docking cases (see above), in general, it is an important consideration for the docking approaches [20]. In a long-standing debate between the proponents of the “induced-fit” and the “conformational selection” binding mechanisms [21–23], the evidence pointing to the latter allows one to precompute the conformational ensemble of the interacting molecule(s) and dock the separate conformers with limited conformational search [24,25]. That, in principle, should solve the problem in a number of cases. However, often the determination of the adequate conformational ensemble is complicated (e.g. for inherently flexible molecules, like peptides, lipids, etc.) and/or the binding involves a significant contribution of the induced fit mechanisms.
In addition to the global flexibility, the local conformational adjustment (refinement) remains a challenge. Although for many biological applications, the approximate docking prediction from the global search may suffice (e.g. prediction of the binding interfaces for functional assessment, etc.), in a number of cases the atomic resolution of the predicted match is required (e.g. inhibition of interaction, affinity assessment, etc.). The local conformational search involves less degrees of freedom than the global one, and as such is more tractable. However, despite significant progress in this direction [26–29], the problem remains a challenge, especially in cases of conformational changes in the backbone.
Co-evolutionary analysis and deep learning
Co-evolution of residues is correlated with their position in protein structure. Thus, the co-evolution data can be useful for structure prediction algorithms. In recent years, there has been major progress in utilizing the residue co-evolution information for predicting structures of individual proteins, based on the rapid growth of experimental data on proteins and new ideas on how to use it, combined with spectacular advances in computer science (deep learning) [30–33]. A similar advancement in structural modeling of protein assemblies has not occurred yet. The co-evolution of residues provides structural information from sequence data by inferring distances between co-evolving residues, propagating beyond the first layer of the immediate neighbors to subsequent layers of indirect, weaker but more numerous co-evolutionary relationships. The current obstacle for applying this approach to docking is that it is not clear how to distinguish the ínter-molecular co-evolutionary information, needed for docking, from the intra-molecular one, which is not directly relevant to docking. The other problem is a perceived lack of sufficient amount of sequence data on protein-protein (and/or protein-other molecules) interfaces needed for the deep learning. More sophisticated utilization of current and future data on protein interaction should provide a path towards solving these problems.
Prediction of interactors and affinities
Knowledge of the strength of protein interaction is essential for understanding and characterizing biomolecular mechanisms. Computational determination of this key characteristic of protein association is highly non-trivial. In its simplest, but still very useful formulation, one can think of it as the ability to distinguish interacting from non-interacting proteins. Prediction of protein interaction, as a term, has dual meaning. One is predicting that two proteins, or a protein and another molecule, interact - e.g. predicting the fact of interaction, or predicting interactors, often in the context of reconstructing networks of protein interactions. The other is predicting the mode of interaction, given the fact of interaction obtained by other means, experimental or computational. Docking traditionally addresses only the second problem. Docking algorithms have not been specifically designed for or capable of distinguishing interacting and non-interacting proteins. Assuming that the fact of interaction is determined by (a) co-localization and (b) the strength of interaction, docking in principle can address the second aspect, including related problems of predicting binding affinities [34–39], discrimination of non-biological interfaces [40], and binding specificity [41–44]. However, although typical docking scores are correlated with the energy of interaction (otherwise they would not be able to make correct predictions of the mode of interaction), the correlation is too loose to distinguish weakly interacting (non-interacting) from strongly interacting molecular pairs, based on the absolute values of the docking scores. Still, the correlation of the docking scores with the actual energy of association is strong enough to reflect the intermolecular energy landscape. Since these landscapes should be different for strongly and weakly interacting proteins, studies have shown that exploration of the intermolecular energy landscapes based on the docking output is helpful in distinguishing interacting and non-interacting proteins [45,46]. Combining these methodologies with alternative approaches (based on protein co-localization, sequence analysis and such) will improve our ability to predict protein interactions.
Automated servers: Docking without human intervention
Fully automated (queryable) modeling servers are extremely important for biology. Firstly, they are the most convenient tools for the broader biological community of researchers who lack expertise in modeling and would appreciate a user-friendly hands-off utility to answer their biological question. Secondly, the lack of human intervention provides the purest test of the computational methodology, which is important for the objective assessment of its value in relation to competing approaches. Such servers are common in prediction of individual protein structures [31]. However, in that regard, the docking community still lags behind. The issue with some leading publicly available docking servers is that an important part of the prediction protocols is based on data which serves as constraints for docking and is supposed to be supplied by the user. Automated hands-off generation of such data is often non-trivial. Such automated procedures, like text mining of publicly available online publications [47] face copyright restrictions and are hindered by still limited amount of open-access publications. The growth of popularity of the open access publishing will increase the utility of such approaches.
Dynamic and realistic representation of protein interactome in vivo
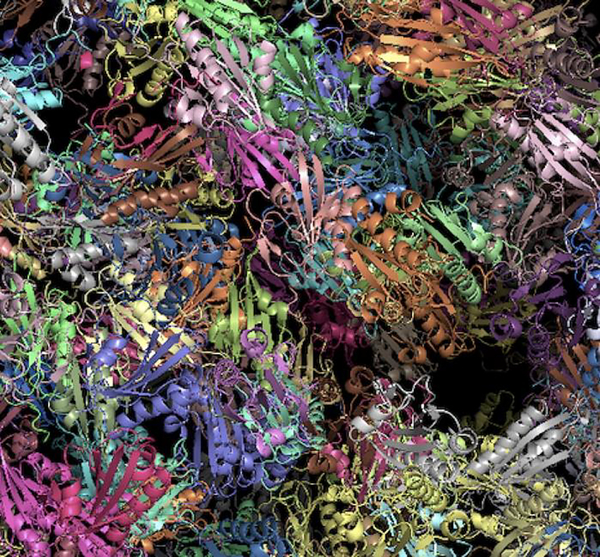
Structure-based modeling of dynamic and realistic interactome in vivo will allow deeper insights into molecular mechanisms of life processes. It will also lead to our ability to model whole cells at molecular/atomic resolution [8,9,48–52], complementing alternative approaches to cell modeling that are based on differential equations, imaging data, and other integrative techniques [53]. In our opinion, such modeling is the ultimate strategic goal of protein docking, along with a number of other modeling techniques (such as rapidly progressing molecular dynamics [2,54,55]). Arguably, many modeling techniques needed for a first-pass approximation (e.g. coarse-graining [56–58]) already exist, requiring “just” proper scaling-up and integration into a self-consistent system. The whole-cell modeling, effectively simulating “life in silico,” will be a true milestone in life sciences, providing unprecedented opportunities for biology and medicine. To the question why such modeling is needed, the simple answer is: to interpolate and extrapolate the existing data on the cell function. The experimentally and computationally determined atomic resolution data currently provide snapshots of the molecules in the cellular environment. The whole-cell modeling, will propagate the system in time, using these snapshots as data points for fitting and validating the trajectory. From the docking perspective, such trajectory would involve a dynamic protocol incorporating the multiplicity of the docking encounters in the crowded cellular environment (Figure 2) involving molecules of different types, conformational flexibility and other phenomena. Obviously, this direction due to its extreme scale and complexity, is an open-ended long-term proposition. However, first steps on this long path are already being made by the community.
Figure 2. Modeling representation of protein packing in cytosol.
The extremely tight packing of the proteins, which is close to physiological, illustrates the exceptional challenge of atomistic modeling of protein interactions in the crowded environment of the cell.
Highlights.
Docking developments aim at improvement of modeling macromolecular complexes
Challenges include proper accounting for conformational flexibility
Docking methods benefit from new data on protein interactions
New developments focus on realistic modeling of protein interactions in vivo
Acknowledgments
This study was supported by grants R01GM074255 from the NIH and DBI1565107 from the NSF. The author thanks Petras Kundrotas for helpful discussions and Taras Dauzhenka for assistance with illustrations.
Footnotes
Disclosure
The author declares no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vakser IA: Protein-protein docking: From interaction to interactome. Biophys. J. 2014, 107:1785–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan AC, Jacobson D, Yatsenko K, Sritharan D, Weinreich TM, Shaw DE: Atomic-level characterization of protein-protein association. Proc. Nati. Acad. Sci. USA 2019, 116:4244–4249.• The paper describes molecular dynamics-based simulation of protein association. The molecular dynamics protocols, so far, have been unsuccessful in solving the general docking problem, due to the complexity of the system’s energy landscape. The paper appears to be the first successful application of the molecular dynamics-based simulation to protein docking, predicting the binding modes of several complexes, at the same time allowing exploration of the binding pathways.
- 3.Garzon JI, Deng L, Murray D, Shapira S, Petrey D, Honig B: A computational interactome and functional annotation for the human proteome. eLife 2016, 5:e18715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kundrotas PJ, Zhu Z, Vakser IA: GWIDD: A comprehensive resource for genome-wide structural modeling of protein-protein interactions. Human Genomics 2012, 6:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mosca R, Ceol A, Aloy P: Interactome3d: Adding structural details to protein networks. Nature Methods 2013, 10:47–53 [DOI] [PubMed] [Google Scholar]
- 6.Kozakov D, Li K, Hall DR, Beglov D, Zheng J, Vakili P, Schueler-Furman O, Paschalidis IC, Clore GM, Vajda S: Encounter complexes and dimensionality reduction in protein-protein association. eLife 2014, 3:e01370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Im W, Liang J, Olson A, Zhou HX, Vajda S, Vakser IA: Challenges in structural approaches to cell modeling. J. Mol. Biol. 2016, 428:2943–2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vakser IA, Deeds EJ: Computational approaches to macromolecular interactions in the cell. Curr. Opin. Struct. Biol. 2019, 55:59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feig M, Sugita Y: Whole-cell models and simulations in molecular detail. Annu. Rev. Cell Dev. Biol. 2019, 35:191–211.• The paper reviews near- and long-term challenges in building whole-cell models based on physical principles. Such models can connect molecular structure with biological function and are enabled by rapidly expanding experimental data on the composition and structure of cellular components.
- 10.Frembgen-Kesner T, Elcock AH: Computer simulations of the bacterial cytoplasm. Biophys. Rev. 2013, 5:109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vakser IA: Low-resolution structural modeling of protein interactome. Curr. Opin. Struct. Biol. 2013, 23:198–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kundrotas PJ, Zhu Z, Janin J, Vakser IA: Templates are available to model nearly all complexes of structurally characterized proteins. Proc. Natl. Acad. Sci. USA 2012, 109:9438–9441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lensink MF, Brysbaert G, Nadzirin N, Velankar S, Chaleil R, Gerguri T, Bates P, Laine E, Carbone A, Grudinin S, et al. : Blind prediction of homo- and hetero-protein complexes: The CASP13-CAPRI experiment. Proteins 2019, 87:1200–1221.• •The paper presents the results of the most recent community-wide protein assembly prediction conducted jointly by the protein structure prediction (CASP) and prediction of protein interactions (CAPRI) challenges. The paper summarizes comparative evaluation of the results by a broad spectrum of participating groups, and as such reflects the state-of-the-art in today’s protein docking.
- 14.Nithin C, Ghosh P, Bujnicki JM: Bioinformatics tools and benchmarks for computational docking and 3D structure prediction of RNA-protein complexes. Genes 2018, 9:432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Si J, Zhao R, Wu R: An overview of the prediction of protein DNA-binding sites. Int. J. Mol. Sci. 2015, 16:5194–5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hurwitz N, Schneidman-Duhovny D, Wolfson HJ: Memdock: An alpha-helical membrane protein docking algorithm. Bioinformatics 2016, 32:2444–2450. [DOI] [PubMed] [Google Scholar]
- 17.Koukos PI, Faro I, van Noort CW, Bonvin AMJJ: A membrane protein complex docking benchmark. J. Mol. Biol. 2018, 430:5246–5256. [DOI] [PubMed] [Google Scholar]
- 18.Ciemny M, Kurcinski M, Kamel K, Kolinski A, Alam N, Schueler-Furman O, Kmiecik S: Protein-peptide docking: Opportunities and challenges. Drug. Discov. Today 2018, 23:1530–1537. [DOI] [PubMed] [Google Scholar]
- 19.Chakravarty D, McElfresh GW, Kundrotas PJ, Vakser IA: How to choose templates for modeling of protein complexes: Insights from benchmarking template-based docking. Proteins 2020, doi: 10.1002/prot.25875.• Docking utilizing structure similarity of target protein monomers to template protein-protein complexes significantly expands structural coverage of the interactome. The paper presents a comprehensive comparative benchmarking study of the template-based docking utilizing different template detectiont methods.
- 20.Ozdemir ES, Nussinov R, Gursoy A, Keskin O: Developments in integrative modeling with dynamical interfaces. Curr. Opin. Struct. Biol. 2019, 56:11–17. [DOI] [PubMed] [Google Scholar]
- 21.Ruvinsky AM, Kirys T, Tuzikov AV, Vakser IA: Ensemble-based characterization of unbound and bound states on protein energy landscape. Protein Sci. 2013, 22:734–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Csermely P, Palotai R, Nussinov R: Induced fit, conformational selection and independent dynamic segments: An extended view of binding events. Trends Biochem. Sci. 2010, 35:539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greives N, Zhou HX: Both protein dynamics and ligand concentration can shift the binding mechanism between conformational selection and induced fit. Proc. Natl. Acad. Sci. USA 2014, 111:10197–10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marze NA, Burman SSR, Sheffler W, Gray JJ: Efficient flexible backbone protein-protein docking for challenging targets. Bioinformatics 2018, 34:3461–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurkcuoglu Z, Bonvin AMJJ: Pre- and post-docking sampling of conformational changes using ClustENM and HADDOCK for protein-protein and protein-DNA systems. Proteins 2020, 88:292–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dauzhenka T, Kundrotas PJ, Vakser IA: Computational feasibility of an exhaustive search of side-chain conformations in protein-protein docking. J. Comput. Chem. 2018, 39:2012–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schindler CEM, de Beauchene IC, de Vries SJ, Zacharias M: Protein-protein and peptide-protein docking and refinement using ATTRACT in CAPRI. Proteins 2017, 85:391–398. [DOI] [PubMed] [Google Scholar]
- 28.Roy Burman SS, Yovanno RA, Gray JJ: Flexible backbone assembly and refinement of symmetrical homomeric complexes. Structure 2019, 27:1041–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfeiffenberger E, Bates PA: Refinement of protein-protein complexes in contact map space with metadynamics simulations. Proteins 2019, 87:12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Senior AW, Evans R, Jumper J, Kirkpatrick J, Sifre L, Green T, Qin C, Zídek A, Nelson AWR, Bridgland A, et al. : Improved protein structure prediction using potentials from deep learning. Nature 2020, 577:706–710.• • The paper presents a deep-learning system that provides accurate predictions of the inter-residue distances and can be used to construct a protein-specific potential that represents the protein structure. The method was used for protein structure prediction, showing superior performance in the community-wide assessment of predictive methodologies.
- 31.Kryshtafovych A, Schwede T, Topf M, Fidelis K, Moult J: Critical assessment of methods of protein structure prediction (CASP)-Round XIII. Proteins 2019, 87:1011–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anishchenko I, Ovchinnikov S, Kamisetty H, Baker D: Origins of coevolution between residues distant in protein 3D structures. Proc. Natl. Acad. Sci. USA 2017, 114:9122–9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ovchinnikov S, Park H, Varghese N, Huang PS, Pavlopoulos GA, Kim DE, Kamisetty H, Kyrpides NC, Baker D: Protein structure determination using metagenome sequence data. Science 2017, 355:294–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suh D, Jo S, Jiang W, Chipot C, Roux B: String method for protein-protein binding free-energy calculations. J. Chem. Theory Comput. 2019, 15:5829–5844. [DOI] [PubMed] [Google Scholar]
- 35.Siebenmorgen T, Zacharias M: Evaluation of predicted protein-protein complexes by binding free energy simulations. J. Chem. Theory Comput. 2019, 15:2071–2086. [DOI] [PubMed] [Google Scholar]
- 36.Marin-Lopez M, Planas-Iglesias J, Aguirre-Plans J, Bonet J, Garcia-Garcia J, Fernandez-Fuentes N, Oliva B: On the mechanisms of protein interactions: Predicting their affinity from unbound tertiary structures. Bioinformatics 2018, 34:592–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barlow KA, Conchuir SO, Thompson S, Suresh P, Lucas JE, Heinonen M, Kortemme T: Flex ddG: Rosetta ensemble-based estimation of changes in protein-protein binding affinity upon mutation. J. Phys. Chem. B 2018, 122:5389–5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jankauskaite J, Jimenez-Garcia B, Dapkunas J, Fernandez-Recio J, Moal IH: SKEMPI 2.0: An updated benchmark of changes in protein-protein binding energy, kinetics and thermodynamics upon mutation. Bioinformatics 2019, 35:462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raucci R, Laine E, Carbone A: Local interaction signal analysis predicts protein-protein binding affinity. Structure 2018, 26:905–915. [DOI] [PubMed] [Google Scholar]
- 40.Xu Q, Dunbrack RL: Principies and characteristics of biological assemblies in experimentally determined protein structures. Curr. Opin. Struct. Biol. 2019, 55:34–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Viswanathan R, Fajardo E, Steinberg G, Haller M, Fiser A: Protein-protein binding supersites. PLoS Comp. Biol. 2019, 15:e1006704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dequeker C, Laine E, Carbone A: Decrypting protein surfaces by combining evolution, geometry, and molecular docking. Proteins 2019, 87:952–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lagarde N, Carbone A, Sacquin-Mora S: Hidden partners: Using cross-docking calculations to predict binding sites for proteins with multiple interactions. Proteins 2018, 86:723–737. [DOI] [PubMed] [Google Scholar]
- 44.Mudgal R, Srinivasan N, Chandra N: Resolving protein structure-function-binding site relationships from a binding site similarity network perspective. Proteins 2017, 85:1319–1335. [DOI] [PubMed] [Google Scholar]
- 45.Yueh C, Hall DR, Xia B, Padhorny D, Kozakov D, Vajda S: ClusPro-DC: Dimer classification by the Cluspro server for protein-protein docking. J. Mol. Biol. 2017, 429:372–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vakser IA: Low-resolution docking: Prediction of complexes for underdetermined structures. Biopolymers 1996, 39:455–464. [DOI] [PubMed] [Google Scholar]
- 47.Badal VD, Kundrotas PJ, Vakser IA: Natural language processing in text mining for structural modeling of protein complexes. BMC Bioinformatics 2018, 19:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goodsell DS, Franzen MA, Herman T: From atoms to cells: Using mesoscale landscapes to construct visual narratives. J. Mol. Biol. 2018, 430:3954–3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goodsell DS, Autin L, Olson AJ: Lattice models of bacterial nucleoids. J. Phys. Chem. B 2018, 122:5441–5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skolnick J: Perspective: On the importance of hydrodynamic interactions in the subcellular dynamics of macromolecules. J. Chem. Phys. 2016, 145:100901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rickard MM, Zhang Y, Gruebele M, Pogorelov TV: In-cell protein-protein contacts: Transient interactions in the crowd. J. Phys. Chem. Lett. 2019, 10:5667–5673. [DOI] [PubMed] [Google Scholar]
- 52.Bortot LO, Bashardanesh Z, van der Spoel D: Making soup: Preparing and validating models of the bacterial cytoplasm for molecular simulation. J. Chem. Inf. Model. 2020, 60:322–331. [DOI] [PubMed] [Google Scholar]
- 53.Szigeti B, Roth YD, Sekar JAP, Goldberg AP, Pochiraju SC, Karr JR: A blueprint for human whole-cell modeling. Curr. Opin. Syst. Biol. 2018, 7:8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jung J, Nishima W, Daniels M, Bascom G, Kobayashi C, Adedoyin A, Wall M, Lappala A, Phillips D, Fischer W, et al. : Scaling molecular dynamics beyond 100,000 processor cores for large-scale biophysical simulations. J. Comput. Chem. 2019, 40:1919–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nawrocki G, Im W, Sugita Y, Feig M: Clustering and dynamics of crowded proteins near membranes and their influence on membrane bending. Proc. Natl. Acad. Sci. USA 2019, 116:24562–24567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vakser IA, Matar OG, Lam CF: A systematic study of low-resolution recognition in protein-protein complexes. Proc. Natl. Acad. Sci. USA 1999, 96:8477–8482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pak AJ, Voth GA: Advances in coarse-grained modeling of macromolecular complexes. Curr. Opin. Struct. Biol. 2018, 52:119–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hafner AE, Krausser J, Saric A: Minimal coarse-grained models for molecular self-organisation in biology. Curr. Opin. Struct. Biol. 2019, 58:43–52. [DOI] [PubMed] [Google Scholar]