Abstract
With the explosion of normal mode analyses (NMAs) based on elastic network models (ENMs) in the last decade, and the proven precision of MD simulations for visualizing interactions at atomic scale, many hybrid methods have been proposed in recent years. These aim at exploiting the best of both worlds: the atomic precision of MD that often fall short of exploring time and length scales of biological interest, and the capability of ENM-NMA to predict the cooperative and often functional rearrangements of large structures and assemblies, albeit at low resolution. We present an overview of recent progress in the field with examples of successful applications highlighting the utility of such hybrid methods and pointing to emerging future directions guided by advances in experimental characterization of biomolecular systems structure and dynamics.
Keywords: Hybrid simulation, Molecular dynamics, normal mode analysis, elastic network model, protein-protein complex, integrating experiments, cryo-EM, NMR, SAXS, conformational landscape
Introduction
Recent years have seen an explosion in the number of studies that have used either coarse-grained (CG) or atomic models for normal mode analyses (NMAs) of complex biomolecular structures. Among CG models, elastic network models (ENMs), and especially the anisotropic network model (ANM), have found wide utility, due to their simplicity and computational efficiency as well as proven robustness and relevance of predictions to functional mechanisms. These studies present the advantage of predicting a unique solution for the spectrum of modes of motion accessible to each structure, each mode representing a collective coordinate for ‘escaping’ the original energy minimum in the multidimensional conformational energy landscape (see Box 1). More importantly, numerous comparisons with experimental data demonstrated that the most easily accessible modes (that entail the least ascent for a given size departure from the energy minimum) are those naturally selected and deployed to accommodate functional interactions, hence the increased focus on NMA and the emergence of new concepts such as the evolution of structures to favor functional dynamics [1]. The fact that mutants exhibit structural changes along these ‘soft’ modes further suggests that these modes often provide a path to alleviate the effect of perturbations.
While such studies have shown considerable success, it also became clear that they could benefit from combined use with classical molecular dynamics (MD) simulations, and vice versa. For example, commonly used ENMs are agnostic to residue type and are purely based on geometry. The predicted modes represent entropically driven fluctuations. Such mode evaluations could benefit from refinements/corrections by full atomic or forcefield-based (FF-based) simulations. MD simulations, on the other hand, are usually limited by computing time, entailing a trade-off between the system size and the duration of simulations. Small proteins’ dynamics can be simulated up to milliseconds [2], but large systems typically characterized by cryo-EM cannot be simulated for long enough to explore events of biological interest, even with specialized hardware. Such systems can be analyzed using NMA, albeit at low resolution [3,4].
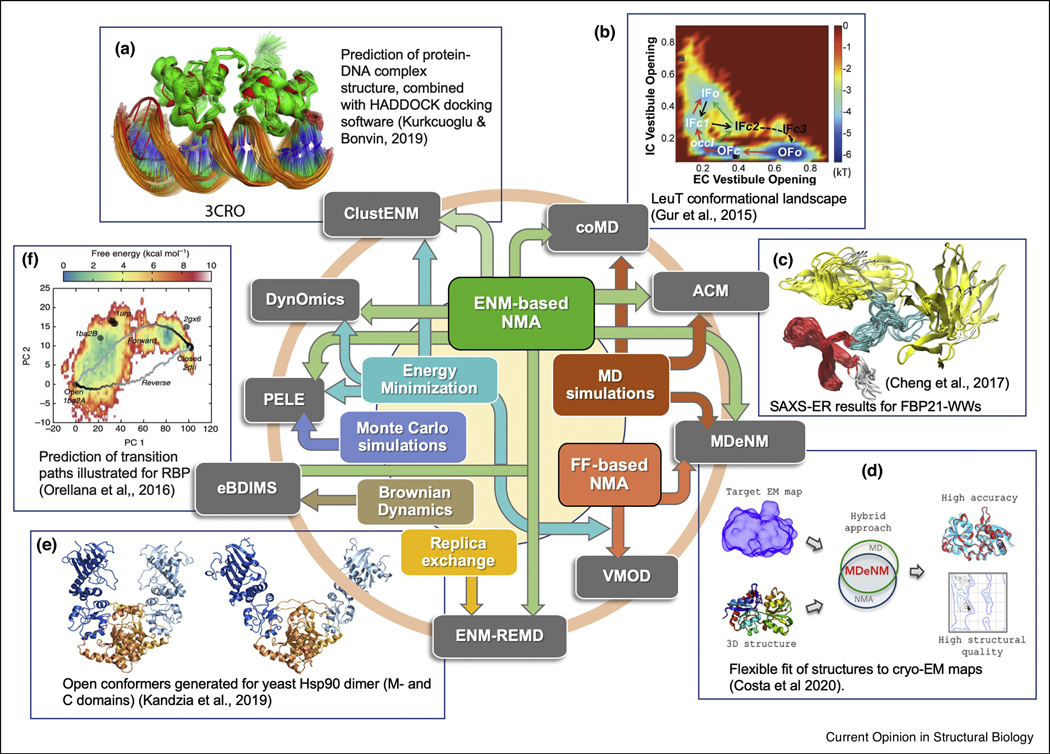
Such considerations led to the development of a plethora of hybrid models in recent years in the interest of taking advantage of the collective/global nature of ‘soft’ modes and the atomic-scale accuracy of MD simulations. These methods, broadly described as normal mode (NM)-driven simulations, use FF- or ENM-based NMA for enhancing the efficiency of atomic simulations such as MD, accelerated MD, Brownian dynamics, or replica exchange MD (REMD). Conversely, a second group of studies focuses on conformational changes driven by CG NMs, and refines them by MD or energy minimization to reconstruct their atomic counterparts. These may be classified as simulation-refined ENM predictions. Finally, a third group resorts to the incorporation or consolidation of experimental data, shortly referred to as experimentally-guided ENM-NMA. While the separation between these three groups is not clear-cut, we present in the next three sections a few examples which predominantly belong to these respective groups, some illustrated in Figure 1.
Figure 1: Examples of hybrid methods combining elastic network model (ENM)- or force-field (FF)-based NMA with various kinds of simulations.
The colored arrows connect the core methods of the same color to the hybrid methods on the outside, and panels A–E show example applications. (A) Combination of ClustENM with the HADDOCK docking software enables generation of more realistic protein-protein and protein-DNA complexes, such as the example shown. (B) CoMD was used to fill in missing regions of the conformational landscape of the leucine transporter (LeuT), enabling the transport cycle to be elucidated. (C) The ACM method was integrated with SAXS experiments for ensemble refinement (SAXS-ER), yielding an ensemble of structures compatible with the SAXS data. (D) MDeNM-EMfit is a version of MDeNM where modes are selected that increase fitting to an electron density map. This method yields high quality structures with good fits to EM density maps as shown. (E) ENMREMD enhances sampling of various conformations along normal modes as shown for the Hsp90 dimer. (F) The eBDIMs method enables efficient transition path sampling as illustrated.
One major success of NM-driven simulations has been their ability to efficiently sample conformational space
NMs can be introduced into MD in many ways. One approach that has found utility in recent years [5,6] is MD with excited normal modes (MDeNM; see Figure 1D) [7]. The idea therein is to enhance the atomic velocities in the direction of a set of linear combinations of either FF- or ENM-based NMs, effectively exciting the associated degrees of freedom to a higher temperature, followed by relaxation of the structure via local rearrangements over a few picoseconds, within a multi-replica strategy. This excitation may, for example, mimic the modes being deployed in response to a perturbation, such as ligand binding.
The strategy of iteratively directing the conformational changes along NMs (by targeted MD) followed by short MD runs to relax the target conformation is also the underlying idea of collective MD (CoMD; Figure 1B) [8]. Yet, CoMD differs from MDeNM as the combination of soft modes is generated by taking tens to thousands of steps along them to iteratively create intermediate conformers at each cycle. CoMD samples conformational transitions by using a Metropolis Monte Carlo scheme that selectively biases the trajectory toward the endpoint; and it may alternatively perform unbiased sampling starting from one or more structures. CoMD has proven useful in assessing the conformational landscape of proteins [9]. A similar approach has been employed in the Perturb-Scan-Pull approach [10] where steered MD is used to drive conformational sampling along directions defined by the ENM-based perturbation response scanning (PRS) method [11,12].
Distance-dependent biasing potentials based on single ENM modes have also been used in a Hamiltonian replica exchange framework [13–15], shortly designated as ENM-REMD, which successfully characterized yeast Hsp90 dimer motions [13] (Figure 1E).
Another hybrid method, implemented in the server eBDIMS[16], generates transition paths between, or excursions in the vicinity of, known structures in the conformational landscape. eBDIMS combines ENM modes with Brownian Dynamics (BD) Importance Sampling [17] (Figure 1F). There are several other NM-based methods/servers for sampling transition pathways, including IMODS[18], aANM[19], NMSim[20], NOMAD[21], ANMPathway[22] although the majority do not include MD.
ENM-NMA predictions assist in selecting or refining biomolecular complexes and assemblies
In the second group of methods (ENM-NMAs refined by MD or energy minimization), we distinguish three areas of application: generation of conformers for complexes, cryo-EM structure refinement/fitting, and studies at the structural-proteome level.
ClustENM performs unbiased sampling of the conformational space in conformity with experimental and MD data by iteratively deforming the structures along combinations of soft modes, clustering the generated conformers, and applying energy minimization to cluster representatives in implicit solvent before starting a new cycle [23]. A notable extension [24] is the generation of conformers for protein-protein and protein-DNA complexes, combined with docking software HADDOCK [25], illustrated in Figure 1A. The recently introduced iterANM-IaMD protocol also generates ClustENM conformers followed by exploration of energy landscape by enhanced sampling MD [26]. The BP-Dock algorithm [27] uses a similar approach for ENM-based conformer generation for docking but the directions are based on PRS.
A similar approach is used for individual modes in the DynOmics webserver [28] where bond regularization and energy minimization generate physically realistic full-atomic conformers. A major utility is to generate conformers not only for individual proteins but also biological assemblies. Due to its computational efficiency, DynOmics is rapidly becoming a broadly used tool with applicability to the structural proteome.
A recent extension of MDeNM, called MDeNM-EMfit, demonstrates the utility of hybrid methods in meeting the needs emerging by the cryo-EM revolution [5] (Figure 1D). MDeNM-EMfit generates conformers that best fit the target density map by a biased selection of NMs. This is an advance over flexible fittings of high resolution structures to EM maps [29] using either NMs (usually ENM-based) or MD. This example presents a smooth transition to methods that advantageously utilize experimental data.
Integrating Experimental Data Improves Elucidation of Conformational Ensembles
Recent studies have shown the utility of incorporating experimental data in developing or refining computational methods for complex systems/processes [29–32]. Experimental data not only enhance conformational sampling but also alleviate limitations intrinsic to MD or lack of resolution inherent to CG models [30,31].
Experiments-guided methods are especially useful for interpreting ensemble-averaged data from NMR and SAXS. The amplified collective motions (ACM) method [33], one of the earliest hybrid methods using ANM and atomic simulations, recently proved useful in SAXS-oriented ensemble-refinement (SAXS-ER) [34,35] (Figure 1C). Averaging restraints across multiple replica runs recovers the structural and dynamic features captured by NMR, shown for a number of proteins [30].
In cryo-EM, where single particles in different conformations can be captured but the signal-to-noise ratio is poor, averaging within classes is essential to reconstructing the structure of the complexes under study [36], and the multi-replica approach has proven useful in ensemble fitting to cryo-EM class averages [37]. Hybrid EM NMA (HEMNMA) [38] is another recent method developed for interpreting cryo-EM data, where structures deformed along NMs are converted to density maps that are directly compared against the raw 2D projection images from cryo-EM. This allows continuous classification of the 2D images, which could provide better interpretations of functional mechanisms.
Ensemble analysis can also be performed for multiple structures resolved for a given protein under different experimental conditions. Such comparisons [39] were indeed at the heart of ProDy development [40,41], which found extensive use in recent years. Further examination of CG free energy landscapes in the light of knowledge-based potentials and ENM-based entropies provides new insights into the molecular basis of functional transitions [42]. Such ensemble analyses provide useful information on consensus normal modes or signature dynamics of protein families [43,44], and can be combined into NM-driven sampling as shown for HIV protease [45]. The new toolkit SignDy facilitates a systematic analysis of such signature dynamics, including the classification of protein families into subfamilies based on their dynamics, and incorporation of sequence evolution analyses, providing a better understanding of protein mechanisms and their evolution [43]. Such comparative analyses of sequence conservation and structural dynamics lead to a better understanding of the interplay between evolution and function [1,40,43,46,47]. Integration of network models with sequence evolutionary analysis emerges as a useful tool for elucidating mechanisms of action [48].
Biomedical Applications: shedding lights into the mechanisms of diseases, allostery, inhibition, and impact of mutations
As described above, hybrid methods permit us to examine systems/processes beyond the reach of individual methods used in isolation, hence their use in biomedical applications. Here, we highlight a few recent examples.
By combining ENMs with multiple MD trajectories, Orozco and coworkers demonstrated that glioblastoma mutations at the ectodomain of EGFR can favor untethering to a compact intermediate state, removing a steric constraint on kinase activation [49]. Another recent study of the dynamics of a mechanosensitive channel, NOMPC, using a combination of ANM-NMA with MD and continuum mechanics, showed that ankyrins behave as biological springs to open the NOMPC channel [50]. In another, MDeNM was used to study structural transition of prion conformations from the cellular natural (PrPC) to the scrapie (PrPSc), obtaining several intermediate and transition states involved in aggregation [6].
VMOD [45,51,52], based on adding constraints to the FF to bias conformational changes, has elucidated the exposure of binding sites, such as the putative binding regions for human Trx1, upon global conformational changes [53], corroborated by docking simulations and CD spectrometry [54]. VMOD also helped elucidate the exposure of the binding sites on DPP-4 [55], CDK2 and DHFR for ligand binding and virtual screening [56]. Not surprisingly, several tools/servers have been developed for modeling small ligand- and peptide-binding to highly flexible proteins [57], as well as the complexation of trigger-factor with ribosome [58]. Combination of ANM-based computations with suppressor sequence analysis recently showed the capacity of cohesin to ‘hold and release’ DNA [59]. Combined use of ENM/NMA-based linear response theory and REMD helped provide a spatiotemporal description of the mechanism of programmed ribosomal frameshifting [60].
Another area where ENM predictions made an important impact is the evaluation of missense mutations. A newly developed machine learning methodology implemented in the server Rhapsody [61] combines sequence information with ANM predictions to accurately assess the effect of missense mutations on biological function [62]. This study, validated for more than 20,000 single amino acid variants, highlights the significance of collective dynamics (in addition to sequence evolution and structure) in determining the impact of mutations. DynaMut [63], on the other hand, integrates graph-based signatures, structural features and NMs (using an ENM-based contact model [64]), but not sequence data, to estimate the impact of a mutation on protein stability.
Summary and future directions: integrative approaches for structural systems biology
Hybrid approaches combining NMA (using either ENMs or atomic models for representing biomolecular structures) and MD, plus experimental data originating from biophysical techniques such as X-ray crystallography, NMR, SAXS, EM, and FRET, will increasingly be necessary to establish the link between the complex structural dynamics of biomolecular assemblies or supramolecular machines and their mechanisms of function. Although NMA is performed under vacuum conditions, it captures in a robust way the large-scale collective directions of motions that span the subspace of potentially functional changes in conformation. Use of this information in hybrid approaches through energy minimization and/or MD that make use of precise atomistic physical FFs, including if necessary experimental restraining potentials, will lead to powerful possibilities for large systems. Among the main challenges, the determination of the different preferential structures that these systems can adopt, the pathways linking them, and the quantitative evaluation of corresponding time scales remain important aspects to be improved. The use of experimental restraints in hybrid methods will certainly offer more targeted and reliable explorations. In this regard, cryo-EM opened up new outstanding possibilities to study the dynamics of very large systems. As cryo-EM allows for the characterization of a broad spectrum of conformations, advances in hybrid simulations accompanying those in cryo-EM technology will make it possible to thoroughly study the space of conformations and obtain an overall view of functional dynamics.
From a methodological point of view, the combination of NMA with MD allows a powerful way to realize the coupling between slow and fast motions. Such methods will also help treat ordered and disordered regions of a biological system and reveal their mutual influence. They are also essential to characterizing stable or transient structures and interdomain/intersubunit correlations typical of allosteric proteins and designing new strategies for molecular interventions. From an energetic point of view, the integration of MD and NMA offers the possibilities to include the effect of the environment, like the solvent, ions, and membrane, which can influence the structural dynamics, while also allowing for exploring time and length scales well beyond those conventionally examined by full atomic MD. The use of more realistic FFs, such as those that include a polarization contribution, would undoubtedly lead to a better estimation of the relative stabilities of conformational substates. Finally, ever-increasing computational power will allow the entropic contributions to be considered more routinely in these estimations in the near future.
With increasing volume of structural data, including those determined by cryo-EM and advanced structural modeling tools, as well as other experimental data serving as restraints, a new field, integrative structural biology [32] or structural systems biology [65], is emerging in recent years. It is anticipated that CG models, including ENMs, and associated hybrid methods, will be increasingly important for navigating the space of conformations accessible to supramolecular assemblies, or the network of interactions at the structural proteome level. As recently noted [25], ‘less is more’, i.e. performing operations at a CG level (followed by full atomic reconstruction) is a viable approach for reducing the complexity of conformational searches upon smoothing out the energy landscape. Such efforts for ‘adding structural details’ to protein networks (e.g. Interactome3D [66]) significantly accelerated in the last decade [67,68], and the importance of adding not only structure but also dynamic features is now being recognized [69].
Supplementary Material
Highlights.
Hybrid methods of three kinds combine normal mode analysis and MD simulations
Normal mode (NM)-driven simulations use NMs to enhance energy landscape sampling
MD simulations or energy minimization are also used to refine predictions from NMs
Integrating experiments into computations creates better hybrid methods
These hybrid methods are especially useful for large complexes from cryo-EM
Acknowledgment.
This study was supported by the NIH awards P41GM103712 and P30DA035778 (to I.B.), and FAPESP award 2017/19077-4 (ALS).
Footnotes
Conflict of Interest and Authorship Conformation Form
Please check the following as appropriate:
✓ All authors have participated in (a) conception and design, or analysis and interpretation of the data; (b) drafting the article or revising it critically for important intellectual content; and (c) approval of the final version.
✓ This manuscript has not been submitted to, nor is under review at, another journal or other publishing venue.
✓ The authors have no affiliation with any organization with a direct or indirect financial interest in the subject matter discussed in the manuscript
○ The following authors have affiliations with organizations with direct or indirect financial interest in the subject matter discussed in the manuscript:
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhang Y, Doruker P, Kaynak B, Zhang S, Krieger J, Li H, Bahar I: Intrinsic dynamics is evolutionarily optimized to enable allosteric behavior. Curr Opin Struct Biol 2019, 62:14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hollingsworth SA, Dror RO: Molecular Dynamics Simulation for All. Neuron 2018, 99:1129–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sorzano COS, Jimenez A, Mota J, Vilas JL, Maluenda D, Martinez M, Ramirez-Aportela E, Majtner T, Segura J, Sanchez-Garcia R, et al. : Survey of the analysis of continuous conformational variability of biological macromolecules by electron microscopy. Acta Crystallogr F Struct Biol Commun 2019, 75:19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atilgan C: Computational Methods for Efficient Sampling of Protein Landscapes and Disclosing Allosteric Regions. Adv Protein Chem Struct Biol 2018, 113:33–63. [DOI] [PubMed] [Google Scholar]
- 5.Costa MGS, Fagnen C, Venien-Bryan C, Perahia D: A New Strategy for Atomic Flexible Fitting in Cryo-EM Maps by Molecular Dynamics with Excited Normal Modes (MDeNM-EMfit). J Chem Inf Model 2020, 10.1021/acs.jcim.9b01148. [DOI] [PubMed] [Google Scholar]
- 6.Lima AN, de Oliveira RJ, Braz ASK, de Souza Costa MG, Perahia D, Scott LPB: Effects of pH and aggregation in the human prion conversion into scrapie form: a study using molecular dynamics with excited normal modes. Eur Biophys J 2018, 47:583–590. [DOI] [PubMed] [Google Scholar]
- 7.Costa MG, Batista PR, Bisch PM, Perahia D: Exploring free energy landscapes of large conformational changes: molecular dynamics with excited normal modes. J Chem Theory Comput 2015, 11:2755–2767. [DOI] [PubMed] [Google Scholar]
- 8.Gur M, Madura JD, Bahar I: Global transitions of proteins explored by a multiscale hybrid methodology: application to adenylate kinase. Biophys J 2013, 105:1643–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gur M, Zomot E, Cheng MH, Bahar I: Energy landscape of LeuT from molecular simulations. J Chem Phys 2015, 143:243134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jalalypour F, Sensoy O, Atilgan C: Perturb-Scan-Pull: A Novel Method Facilitating Conformational Transitions in Proteins. J Chem Theory Comput 2020, 10.1021/acs.jctc.9b01222. [DOI] [PubMed] [Google Scholar]
- 11.Atilgan C, Atilgan AR: Perturbation-response scanning reveals ligand entry-exit mechanisms of ferric binding protein. PLoS Comput Biol 2009, 5:e1000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atilgan C, Gerek ZN, Ozkan SB, Atilgan AR: Manipulation of conformational change in proteins by single-residue perturbations. Biophys J 2010, 99:933–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kandzia F, Ostermeir K, Zacharias M: Global Dynamics of Yeast Hsp90 Middle and C-Terminal Dimer Studied by Advanced Sampling Simulations. Front Mol Biosci 2019, 6:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ostermeir K, Zacharias M: Hamiltonian replica exchange combined with elastic network analysis to enhance global domain motions in atomistic molecular dynamics simulations. Proteins 2014, 82:3410–3419. [DOI] [PubMed] [Google Scholar]
- 15.Zacharias M: Combining Elastic Network Analysis and Molecular Dynamics Simulations by Hamiltonian Replica Exchange. J Chem Theory Comput 2008, 4:477–487. [DOI] [PubMed] [Google Scholar]
- 16.Orellana L, Gustavsson J, Bergh C, Yoluk O, Lindahl E: eBDIMS server: protein transition pathways with ensemble analysis in 2D-motion spaces. Bioinformatics 2019, 35:3505–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orellana L, Yoluk O, Carrillo O, Orozco M, Lindahl E: Prediction and validation of protein intermediate states from structurally rich ensembles and coarse-grained simulations. Nat Commun 2016, 7:12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez-Blanco JR, Aliaga JI, Quintana-Orti ES, Chacon P: iMODS: internal coordinates normal mode analysis server. Nucleic Acids Res 2014, 42:W271–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Z, Majek P, Bahar I: Allosteric transitions of supramolecular systems explored by network models: application to chaperonin GroEL. PLoS Comput Biol 2009, 5:e1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krüger DM, Ahmed A, Gohlke H: NMSim web server: integrated approach for normal mode-based geometric simulations of biologically relevant conformational transitions in proteins. Nucleic Acids Res 2012, 40:W310–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindahl E, Azuara C, Koehl P, Delarue M: NOMAD-Ref: visualization, deformation and refinement of macromolecular structures based on all-atom normal mode analysis. Nucleic Acids Res 2006, 34:W52–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das A, Gur M, Cheng MH, Jo S, Bahar I, Roux B: Exploring the conformational transitions of biomolecular systems using a simple two-state anisotropic network model. PLoS Comput Biol 2014, 10:e1003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurkcuoglu Z, Bahar I, Doruker P: ClustENM: ENM-Based Sampling of Essential Conformational Space at Full Atomic Resolution. J Chem Theory Comput 2016, 12:4549–4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurkcuogl Z, Bonvi A: Pre- and post-docking sampling of conformational changes using ClustENM and HADDOCK for protein-protein and protein-DNA systems. Proteins 2020, 88:292–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roel-Touris J, Don CG, R VH, Rodrigues J, Bonvin A: Less Is More: Coarse-Grained Integrative Modeling of Large Biomolecular Assemblies with HADDOCK. J Chem Theory Comput 2019, 15:6358–6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang A, Zhang D, Li Y, Zhang Z, Li G: Large-Scale Biomolecular Conformational Transitions Explored by a Combined Elastic Network Model and Enhanced Sampling Molecular Dynamics. J Phys Chem Lett 2020, 11:325–332. [DOI] [PubMed] [Google Scholar]
- 27.Bolia A, Gerek ZN, Ozkan SB: BP-Dock: a flexible docking scheme for exploring protein-ligand interactions based on unbound structures. J Chem Inf Model 2014, 54:913–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Chang YY, Lee JY, Bahar I, Yang LW: DynOmics: dynamics of structural proteome and beyond. Nucleic Acids Res 2017, 45:W374–W380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyashita O, Tama F: Hybrid Methods for Macromolecular Modeling by Molecular Mechanics Simulations with Experimental Data. Adv Exp Med Biol 2018, 1105:199–217. [DOI] [PubMed] [Google Scholar]
- 30.Bonomi M, Heller GT, Camilloni C, Vendruscolo M: Principles of protein structural ensemble determination. Curr Opin Struct Biol 2017, 42:106–116. [DOI] [PubMed] [Google Scholar]
- 31.Allison JR: Using simulation to interpret experimental data in terms of protein conformational ensembles. Curr Opin Struct Biol 2017, 43:79–87. [DOI] [PubMed] [Google Scholar]
- 32.Rout MP, Sali A: Principles for Integrative Structural Biology Studies. Cell 2019, 177:1384–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z, Shi Y, Liu H: Molecular dynamics simulations of peptides and proteins with amplified collective motions. Biophys J 2003, 84:3583–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wen B, Peng J, Zuo X, Gong Q, Zhang Z: Characterization of protein flexibility using small-angle xray scattering and amplified collective motion simulations. Biophys J 2014, 107:956–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng P, Peng J, Zhang Z: SAXS-Oriented Ensemble Refinement of Flexible Biomolecules. Biophys J 2017, 112:1295–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vilas JL, Tabassum N, Mota J, Maluenda D, Jimenez-Moreno A, Majtner T, Carazo JM, Acton ST, Sorzano COS: Advances in image processing for single-particle analysis by electron cryomicroscopy and challenges ahead. Curr Opin Struct Biol 2018, 52:127–145. [DOI] [PubMed] [Google Scholar]
- 37.Bonomi M, Pellarin R, Vendruscolo M: Simultaneous Determination of Protein Structure and Dynamics Using Cryo-Electron Microscopy. Biophys J 2018, 114:1604–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harastani M, Sorzano COS, Jonic S: Hybrid Electron Microscopy Normal Mode Analysis with Scipion. Protein Sci 2020, 29:223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bakan A, Bahar I: The intrinsic dynamics of enzymes plays a dominant role in determining the structural changes induced upon inhibitor binding. Proc Natl Acad Sci U S A 2009, 106:14349–14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bakan A, Dutta A, Mao W, Liu Y, Chennubhotla C, Lezon TR, Bahar I: Evol and ProDy for bridging protein sequence evolution and structural dynamics. Bioinformatics 2014, 30:2681–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bakan A, Meireles LM, Bahar I: ProDy: Protein Dynamics Inferred from Theory and Experiments. Bioinformatics 2011, 27:1575–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sankar K, Liu J, Wang Y, Jernigan RL: Distributions of experimental protein structures on coarse-grained free energy landscapes. J Chem Phys 2015, 143:243153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang S, Li H, Krieger JM, Bahar I: Shared Signature Dynamics Tempered by Local Fluctuations Enables Fold Adaptability and Specificity. Mol Biol Evol 2019, 36:2053–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Batista PR, Robert CH, Marechal JD, Hamida-Rebai MB, Pascutti PG, Bisch PM, Perahia D: Consensus modes, a robust description of protein collective motions from multiple-minima normal mode analysis--application to the HIV-1 protease. Phys Chem Chem Phys 2010, 12:2850–2859. [DOI] [PubMed] [Google Scholar]
- 45.Batista PR, Pandey G, Pascutti PG, Bisch PM, Perahia D, Robert CH: Free Energy Profiles along Consensus Normal Modes Provide Insight into HIV-1 Protease Flap Opening. J Chem Theory Comput 2011, 7:2348–2352. [DOI] [PubMed] [Google Scholar]
- 46.Liu Y, Bahar I: Sequence evolution correlates with structural dynamics. Mol Biol Evol 2012, 29:2253–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haliloglu T, Bahar I: Adaptability of protein structures to enable functional interactions and evolutionary implications. Curr Opin Struct Biol 2015, 35:17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang Z, Verkhivker GM, Hu G: Integration of network models and evolutionary analysis into high-throughput modeling of protein dynamics and allosteric regulation: theory, tools and applications. Brief Bioinform 2019, 10.1093/bib/bbz029. [DOI] [PubMed] [Google Scholar]
- 49.Orellana L, Thorne AH, Lema R, Gustavsson J, Parisian AD, Hospital A, Cordeiro TN, Bernado P, Scott AM, Brun-Heath I, et al. : Oncogenic mutations at the EGFR ectodomain structurally converge to remove a steric hindrance on a kinase-coupled cryptic epitope. Proc Natl Acad Sci U S A 2019, 116:10009–10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Argudo D, Capponi S, Bethel NP, Grabe M: A multiscale model of mechanotransduction by the ankyrin chains of the NOMPC channel. J Gen Physiol 2019, 151:316–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Floquet N, Durand P, Maigret B, Badet B, Badet-Denisot MA, Perahia D: Collective motions in glucosamine-6-phosphate synthase: influence of ligand binding and role in ammonia channelling and opening of the fructose-6-phosphate binding site. J Mol Biol 2009, 385:653–664. [DOI] [PubMed] [Google Scholar]
- 52.Guilbert C, Perahia D, Mouawad L: A method to explore transition paths in macromolecules. Applications to hemoglobin and phosphoglycerate kinase. Computer Physics Communications 1995, 91:263–273. [Google Scholar]
- 53.Philot EA, Perahia D, Braz AS, Costa MG, Scott LP: Binding sites and hydrophobic pockets in Human Thioredoxin 1 determined by normal mode analysis. J Struct Biol 2013, 184:293–300. [DOI] [PubMed] [Google Scholar]
- 54.Philot EA, da Mata Lopes D, de Souza AT, Braz AS, Nantes IL, Rodrigues T, Perahia D, Miteva MA, Scott LP: Binding of phenothiazines into allosteric hydrophobic pocket of human thioredoxin 1. Eur Biophys J 2016, 45:279–286. [DOI] [PubMed] [Google Scholar]
- 55.Pantaleao SQ, Philot EA, de Resende-Lara PT, Lima AN, Perahia D, Miteva MA, Scott AL, Honorio KM: Structural Dynamics of DPP-4 and Its Influence on the Projection of Bioactive Ligands. Molecules 2018, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moroy G, Sperandio O, Rielland S, Khemka S, Druart K, Goyal D, Perahia D, Miteva MA: Sampling of conformational ensemble for virtual screening using molecular dynamics simulations and normal mode analysis. Future Med Chem 2015, 7:2317–2331. [DOI] [PubMed] [Google Scholar]
- 57.Kurkcuoglu Z, Doruker P: Ligand Docking to Intermediate and Close-To-Bound Conformers Generated by an Elastic Network Model Based Algorithm for Highly Flexible Proteins. PLoS One 2016, 11:e0158063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Can MT, Kurkcuoglu Z, Ezeroglu G, Uyar A, Kurkcuoglu O, Doruker P: Conformational dynamics of bacterial trigger factor in apo and ribosome-bound states. PLoS One 2017, 12:e0176262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu X, Kanai R, Nakazawa N, Wang L, Toyoshima C, Yanagida M: Suppressor mutation analysis combined with 3D modeling explains cohesin’s capacity to hold and release DNA. Proc Natl Acad Sci U S A 2018, 115:E4833–E4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chang KC, Salawu EO, Chang YY, Wen JD, Yang LW: Resolution-exchanged structural modeling and simulations jointly unravel that subunit rolling underlies the mechanism of programmed ribosomal frameshifting. Bioinformatics 2019, 35:945–952. [DOI] [PubMed] [Google Scholar]
- 61.Ponzoni L, Penaherrera DA, Oltvai ZN, Bahar I: Rhapsody: predicting the pathogenicity of human missense variants. Bioinformatics 2020, 36:3084–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ponzoni L, Bahar I: Structural dynamics is a determinant of the functional significance of missense variants. Proc Natl Acad Sci U S A 2018, 115:4164–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rodrigues CH, Pires DE, Ascher DB: DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res 2018, 46:W350–W355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Frappier V, Najmanovich RJ: A coarse-grained elastic network atom contact model and its use in the simulation of protein dynamics and the prediction of the effect of mutations. PLoS Comput Biol 2014, 10:e1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mih N, Brunk E, Chen K, Catoiu E, Sastry A, Kavvas E, Monk JM, Zhang Z, Palsson BO: ssbio: a Python framework for structural systems biology. Bioinformatics 2018, 34:2155–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mosca R, Ceol A, Aloy P: Interactome3D: adding structural details to protein networks. Nat Methods 2013, 10:47–53. [DOI] [PubMed] [Google Scholar]
- 67.Bertoni M, Aloy P: DynBench3D, a Web-Resource to Dynamically Generate Benchmark Sets of Large Heteromeric Protein Complexes. J Mol Biol 2018, 430:4431–4438. [DOI] [PubMed] [Google Scholar]
- 68.Teyra J, Kim PM: Interpreting protein networks with three-dimensional structures. Nat Methods 2013, 10:43–44. [DOI] [PubMed] [Google Scholar]
- 69.Ozdemir ES, Nussinov R, Gursoy A, Keskin O: Developments in integrative modeling with dynamical interfaces. Curr Opin Struct Biol 2019, 56:11–17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.