Abstract
This study aimed to evaluate the prevalence, multidrug-resistance traits, PCR-detection of virulence, and antibiotic-resistance genes of E. coli isolated from secondary infections following FMD-outbreak in cattle. A total of 160 random samples were gathered from private dairy farms in Damietta Province, Egypt. The specimens were subjected to bacteriological examination, serotyping, congo-red binding assay, antibiogram-testing, and PCR-monitoring of virulence-determinant genes (tsh, phoA, hly, eaeA, sta, and lt) as well as the antibiotic-resistance genes (blaTEM, blaKPC, and blaCTX). The prevalence of E. coli was 30% (n = 48) distributed in 8 serogroups (40/48, 83.3%), while 8 isolates (8/48, 16.6%) were untypable. Besides, 83.3% of the examined isolates were positive for CR-binding. The tested strains harbored the virulence genes phoA, hly, tsh, eaeA, sta, and lt with a prevalence of 100% and 50%, 45.8%, 25%, 8.4%, and 6.2%, respectively. Furthermore, 50% of the recovered strains were multidrug-resistant (MDR) to penicillins, cephalosporins, and carbapenems, and are harboring the blaTEM, blaCTX, and blaKPC genes. Moreover, 25% of the examined strains are resistant to penicillins, and cephalosporins, and are harboring the blaTEM and blaCTX genes. To the best of our knowledge, this is the first report concerning the E. coli secondary bacterial infections following the FMD-outbreak. The emergence of MDR strains is considered a public health threat and indicates complicated treatment and bad prognosis of infections caused by such strains. Colistin sulfate and levofloxacin have a promising in vitro activity against MDR-E. coli.
Subject terms: Bacteriology, Microbiology, Infectious-disease diagnostics
Introduction
Foot and mouth disease (FMD) is a primary contagious disease of a significant threat to ruminants1, 2. Globally, it causes severe financial loss in the veterinary sector owing to the high cost of treatment, vaccination, and production losses3, 4. Recently, three common strains, A, O and SAT 2 are endemic in Egypt. Despite the routine application of vaccination programs in Egypt, a high prevalence of FMD-outbreaks was recorded5. The FMD-Vaccination is usually accompanied by immunosuppression that may lead to secondary bacterial infections in the vaccinated animals3. Escherichia coli (E. coli) is an opportunistic microorganism that is usually inhabitants of the intestinal tract of both humans and animals. E. coli represents a common bacterial pathogen which incriminated in various secondary infections6. The pathogenicity of E. coli is governed by several virulence factors such as; hemolysins, enterotoxins, Shiga-toxins, intimin, fimbria-mannose binding type1-H adhesion, alkaline phosphatase, and Temperature Sensitive Haemagglutinin (Tsh-protein) which are encoded by the specific virulence genes: hly, lt, sta, stx1, stx2, eaeA, fimH, phoA, and tsh, respectively7, 8.
Concerning the site of infection, E. coli is categorized into (1)-intestinal pathogenic E. coli, and (2)-extra-intestinal pathotype. Moreover, Virulent E. coli strains, which usually affect both animals and humans, are categorized in various pathotypes according to the mechanism of disease occurrence, including; Enterotoxigenic, Enteropathogenic, Enteroinvasive, Enteroaggregative, and Shiga-toxigenic pathotypes9. Enterotoxigenic E. coli is the main pathotype that incriminated in white scour in calves, both enterotoxins (heat-labile and heat-stable) and fimbrial-adhesions govern the pathogenesis of the disease. Although Enteropathogenic E. coli doesn't produce enterotoxins, it causes severe watery diarrhea in cattle by other mechanisms. Briefly, the bacteria do intimate-adhesion (non-fimbrial adhesion called intimin) with the enterocyte apical cell membrane resulting in the demolition of the intestinal brush border. Furthermore, the Enteroinvasive E. coli can invade the epithelial cells of the large intestine, causing ulcerations and inflammation. The invasion process is controlled by a specific plasmid (140 MDa) encoding for the release of various outer membrane proteins involved in the disease pathogenesis10, 11.
Globally, the β-lactam antibiotics (cephalosporins, carbapenems, and penicillins) represent about 60% of the used antimicrobial agents12–14. The emerging multidrug-resistant E. coli is considered a public health threat. The antimicrobial resistance in E. coli is mainly attributed to the Extended-Spectrum Beta-Lactamases (ESBLs); which could destroy various β-lactam antimicrobial agents as penicillins, various generations of cephalosporins, and carbapenems15. ESBLs are encoded by specific ESBL-genes such as; blaTEM (encoded for penicillins-resistance), blaKPC (encoded for carbapenems-resistance), and blaCTX (encoded for cephalosporins-resistance). The emergence of multidrug-resistant virulent E. coli has been described by previous studies16–18.
This study was performed to inspect the prevalence, antibiogram, PCR detection of virulence genes (tsh, phoA, hly, eaeA, sta, and lt) as well as the antibiotic-resistance genes (blaTEM, blaKPC, and blaCTX) of E. coli isolated from secondary bacterial infections following FMD-outbreak in cattle.
Methods
Animal ethics
All methods performed consistent with relevant guidelines and regulations. Well-trained experts conducted the handling of animals and experimental procedures. Handling of animals and all protocols were approved by the Animal Ethics Review Committee of Suez Canal University (AERC-SCU), Egypt.
Sampling and clinical examination
One hundred and sixty specimens; milk (n = 40), blood (n = 40), fecal swabs (n = 40), and nasal swabs (n = 40) were randomly collected under complete aseptic conditions from two private cattle farms (native breeds cows of both sexes with average two years old age and with a history of FMD-outbreak) at Damietta Province, Egypt (From March 2019 to August 2019). The sampling was carried out after FMD-outbreak. The examined farms are very close to each other and sharing the same management practices, nutrition, and water supply. The sampling was performed according to the clinical signs. Blood specimens were gathered from animals suffering from fever, milk specimens were collected from clinically mastitic animals, fecal swabs were collected from diarrheic animals, and nasal swabs were collected from animals that exhibited respiratory manifestations. The examined animals were previously treated with trimethoprim and amoxicillin without improvement. The obtained specimens were processed as soon as possible at the same day of collection and were collected on tryptic soy broth (Oxoid, Hampshire, UK).
Isolation and identification of E. coli and other pathogens
For isolation of E. coli, swabs from the obtained specimens were inoculated in McConkey's broth (Oxoid, Hampshire, UK), followed by incubation for 24 h at 37 °C. A loopful of broth-culture was streaked onto MacConkey's agar, and eosin methylene blue agar (Oxoid, Hampshire, UK). The suspected colonies were identified according to their colonial characters, hemolytic activity, microscopical examination using Gram's staining, motility test, hemolytic activity on blood agar, and biochemical reactions (oxidase, catalase, indole, lactose fermentation, methyl-red, citrate-utilization, H2S, Voges-Proskauer, and urease tests) as described by Quinn19.
For isolation of other bacterial pathogens, swabs from the processed specimens were inoculated on nutrient agar, blood agar, mannitol salt agar, cetrimide agar, and MacConkey's agar (Oxoid, Hampshire, UK), then the inoculated plates were incubated for 24–48 h at 37 °C. The obtained pure colonies were identified according to their colonial characters, morphological characters, and biochemically as described by Quinn19.
E. coli serotyping
The retrieved isolates were serotyped for somatic antigen (O-antigen) by the aid of slide agglutination test using standard polyvalent and monovalent commercial E. coli antisera (Denka Seiken-Co., Ltd., Tokyo, Japan) at the Animal-Health Research-Institute, Dokki, Egypt as described by Starr20.
Congo-red binding
To emphasize the pathogenicity and the invasiveness of the isolated strains, the assessment of congo-red binding was performed on trypticase agar (containing 0.03% CR dye) (Oxoid, UK). The tested strains were inoculated on trypticase agar and then incubated at 37 °C for 24 h. Then plates were preserved at room temperature (for 48 h). The positive result is indicated by the appearance of red colonies as previously reported by Panigrahy and Yushen21.
Antimicrobial susceptibility testing
The recovered E. coli strains were assessed for their antimicrobial resistance using the disc diffusion method on Mueller–Hinton agar (Oxoid, UK). The following antimicrobial agents were involved; ampicillin (AMP) (10 μg), meropenem (MEM) (10 μg), amikacin (AK) (30 μg), trimethoprim–sulfamethoxazole (SXT) (19:1 μg), imipenem (IMP) (10 μg), amoxicillin–clavulanic acid (AMC) (30 μg), ceftazidime (CAZ) (30 μg), cefotaxime (30 μg) (CTX), levofloxacin (LEV) (5 μg), amoxicillin (AMX) (10 μg), and colistin sulfate (CT) (10 μg) (Oxoid, Basingstoke, UK). The E. coli-ATCC 25922 was used as a reference strain. Zone-diameters were interpreted according to CLSI22. The tested antibiotics are the most commonly used antimicrobial agents in Egypt in both veterinary and health sectors. The tested strains are classified into MDR, XDR and PDR as previously described by Magiorakos15.
Molecular typing of virulence-determinant genes and antibiotic-resistance genes
PCR-monitoring of virulence-determinant genes (tsh, phoA, hly, eaeA, sta, and lt) and the antibiotic-resistance genes (blaTEM, blaKPC, and blaCTX) was carried out. The selection of these antibiotic-resistance genes was based upon the results of the antimicrobial susceptibility testing, moreover, the selection of the current virulence genes is based upon their significant role in the pathogenesis of the disease as described in previous studies9, 10, 18. Genomic DNA of the examined strains was extracted regarding the manufacturer's guidelines of the QIAamp DNA Mini Kit (Qiagen, GmbH, Germany/Catalogue No.51304). The reaction volume was adjusted at 25-μl (3 μl of genomic-DNA, 5 μl of 5 × Master Mix, and 20 pmol of each prime, the reaction volume was completed by adding distilled H2O). Positive controls (provided by A.H.R.I, Egypt) and negative controls (DNA-free) were used in all reactions. The sequences of the used primers (Metabion International AG, Germany) and the PCR-cycling conditions are illustrated in Table 1. Finally, the separation of the obtained products was performed using the agar gel electrophoresis (1.5% agarose stained with ethidium bromide 0.5 μg/ml), and the gel was photographed.
Table 1.
Oligonucleotides sequences, target genes, specific amplicon size, and PCR re-cycling conditions.
| Target gene | Primers sequences | Amplicon size (bp) | Amplification (35 cycles) | References | ||
|---|---|---|---|---|---|---|
| Denaturation | Annealing | Extension | ||||
| lt | GGTTTCTGCGTTAGGTGGAA | 605 |
94 °C 30 s |
57 °C 45 s |
72 °C 45 s |
23 |
| GGGACTTCGACCTGAAATGT | ||||||
| sta | GAAACAACATGACGGGAGGT | 299 |
94 °C 30 s |
57 °C 30 s |
72 °C 30 s |
23 |
| GCACAGGCAGGATTACAACA | ||||||
| eaeA | ATGCTTAGTGCTGGTTTAGG | 248 |
94 °C 30 s |
51 °C 30 s |
72 °C 30 s |
24 |
| GCCTTCATCATTTCGCTTTC | ||||||
| tsh | GGT GGT GCA CTG GAG TGG | 620 |
94 °C 30 s |
54 °C 40 s |
72 °C 45 s |
25 |
| AGT CCA GCG TGA TAG TGG | ||||||
| phoA | CGATTCTGGAAATGGCAAAAC | 720 |
94 °C 30 s |
60 °C 40 s |
72 °C 1 min |
26 |
| CGTGATCAGCCCTGACTATGAC | ||||||
| hly | AACAAGGATAAGCACTGTTCTGGCT | 1177 |
94 °C 30 s |
54 °C 40 s |
72 °C 45 s |
27 |
| ACCATATAAGCGGTCATTCCCGTCA | ||||||
| blaKPC | ATGTCACTGTATCGCCGTCT | 882 |
94 °C 1 min |
55 °C 1 min |
72 °C 1 min |
28 |
| TTACTGCCCGTTGACGCCC | ||||||
| blaCTX | ATG TGC AGY ACC AGT AAR GTK ATG GC | 593 |
94 °C 30 s |
54 °C 40 s |
72 °C 45 s |
29 |
| TGG GTR AAR TAR GTS ACC AGA AYC AGC GG | ||||||
| blaTEM | ATCAGCAATAAACCAGC | 516 |
94 °C 30 s |
54 °C 40 s |
72 °C 45 s |
30 |
| CCCCGAAGAACGTTTTC | ||||||
Statistical analyses
The Chi-square test was performed to analyse the obtained results (SAS software, version 9.4, SAS Institute, Cary, NC, USA) (significance level; P < 0.05). Furthermore, the correlation analysis was conducted using R software (version 4.0.2; https://www.r-project.org/), it was calculated using the “cor” function and visualization using the “corrplot” functions from the “corrplot” package.
Results
Prevalence of E. coli and other bacterial pathogens in the examined animals
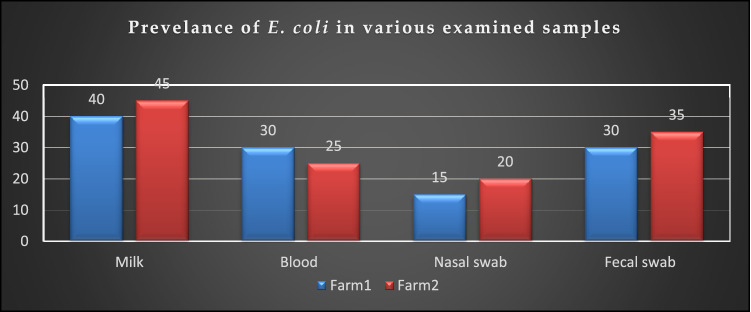
Regarding the phenotypic characteristics of the retrieved E. coli; isolates were identified as E. coli based on their morphology and biochemical characteristics. Microscopically, the bacteria appeared as Gram-negative moderate size, motile, and non-sporulated rods. The bacteria grew well on MacConkeys's agar and gave characteristic pink colonies due to lactose fermentation. On blood agar, the colonies are hemolytic, moreover on EMB; the bacteria gave characteristic metallic sheen colonies. Biochemically, all isolates were positive for catalase, lactose fermentation, indole, and methyl-red, tests. Simultaneously, they were negative for cytochrome oxidase, Voges-Proskauer, citrate-utilization, H2S production, and urease tests. The bacteriological inspection proved that the total prevalence of E. coli was 30% (48/160); the prevalence of E. coli was 28.75% (23/80) in the farm (1), while it was 31.25% (25/80) in the farm (2) as described in Table 2. Concerning the types of the tested samples, the prevalence of E. coli was 42.5%, 27.5%, 17.5%, and 32.5% in the examined milk samples, blood specimens, nasal, and fecal swabs, respectively (Table 2, Fig. 1). Statistically, there is no significant difference in the prevalence of E. coli between the examined farms (P > 0.05).
Table 2.
Prevalence of E. coli in various types of samples obtained from diseased cattle.
| Examined samples | Number and percentage of E. coli | |||||
|---|---|---|---|---|---|---|
| Farm 1 n = 80 |
Farm 2 n = 80 |
Total | ||||
| N | % | N | % | N | % | |
| Milk | 8/20 | 40 | 9/20 | 45 | 17/40 | 42.5 |
| Blood | 6/20 | 30 | 5/20 | 25 | 11/40 | 27.5 |
| Nasal swabs | 3/20 | 15 | 4 /20 | 20 | 7/40 | 17.5 |
| Fecal swabs | 6/20 | 30 | 7/20 | 35 | 13/40 | 32.5 |
| Total | 23/80 | 28.75 | 25/80 | 31.25 | 48/160 | 30 |
Chi-square value = 3.112, P value = 0.375.
Figure 1.
Prevalence of E. coli in various examined samples. The prevalence of E. coli was 42.5%, 27.5%, 17.5%, and 32.5% in the examined milk samples, blood specimens, nasal, and fecal swabs, respectively.
Besides, 70% of the examined diseased animals (n = 112) are infected with other bacterial pathogens including; in mastitis: Streptococcus uberis (10/40, 25%), Streptococcus bovis (8/40, 20%) and Enterococcus faecalis (5/40, 12.5%), in fever: Pseudomonas aeruginosa (10/40, 25%), and Mannheimia hemolytica (6/40, 15%), in respiratory manifestations: Pasturella multocida (10/40, 25%), Mannheimia hemolytica (4/40, 10%), and Pseudomonas aeruginosa (4/40, 10%), and in diarrhea; Proteus mirabilis (17/40, 42.5% ) and Enterococcus faecalis (10/40, 25%).
Serotyping of the recovered E. coli isolates
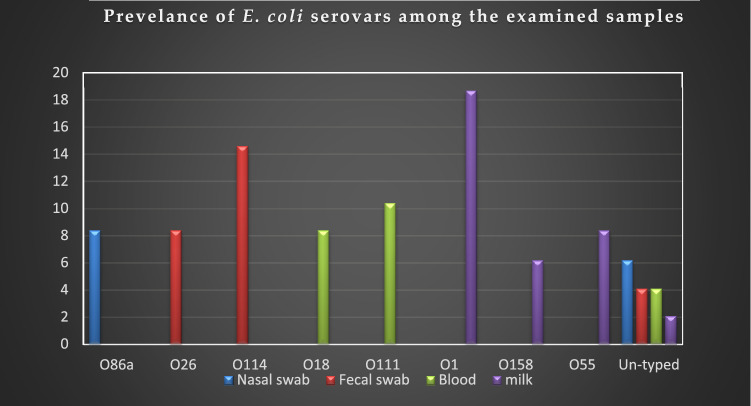
The serotyping of the retrieved isolates showed that 40 isolates belonged to 8 O-serogroups and were distributed as the following: O1 (9/48, 18.7%), O114 (7/48, 14.6%), O111 (5/48, 10.4%), O18 (4/48, 8.4%), O26 (4/48, 8.4%), O55 (4/48, 8.4%), O86a (4/48, 8.4%), and O158 (3/48, 6.2%). Furthermore, the remaining isolates (8/48, 16.6%) were untypable (Table 3; Fig. 2). Regarding the type of the examined samples, the E. coli serovars scattered as the following; nasal swabs: O86a (4/48) and untyped strains (3/48), fecal swabs: O114 (7/48), O26 (4/48), and untyped strains (2/48), blood samples: O111 (5/48), O18 (4/48), and untyped strains (2/48), milk samples: O1 (9/48), O55 (4/48), O158 (3/48), and untyped strains (1/48). Statistically, there is a significant difference in the prevalence of different serovars retrieved from various types of samples (P < 0.05).
Table 3.
Prevalence of E. coli serovars isolated from the examined diseased cattle.
| Sample-types | Serovars | Number | % |
|---|---|---|---|
| Nasal swabs | 086a | 4 | 8.4 |
| Untyped | 3 | 6.2 | |
| Fecal swabs | O114 | 7 | 14.6 |
| O26 | 4 | 8.4 | |
| Untyped | 2 | 4.1 | |
| Blood-samples | O111 | 5 | 10.4 |
| O18 | 4 | 8.4 | |
| Untyped | 2 | 4.1 | |
| Milk samples | O1 | 9 | 18.7 |
| O55 | 4 | 8.4 | |
| O158 | 3 | 6.2 | |
| Untyped | 1 | 2.1 | |
| Total | 48 | 100 |
Chi-square value = 116.588, P < 0.0001.
Figure 2.
The distribution of E. coli serovars among various examined samples. The most prevalent E. coli serovar accompanied the respiratory infection was 086a, diarrhea: O114, fever: O111, and mastitis: O1.
Congo-red binding assay
In the present study, 83.3% of the examined isolates (40/48) were positive for CR-binding assay. All the tested serovars were positive, including; O1 (9/48), O114 (7/48), O111 (5/48), O18 (4/48), O26 (4/48), O55 (4/48), O86a (4/48), and O158 (3/48), while the untyped strains were negative (8/48).
Antimicrobial-resistance traits of the recovered isolates
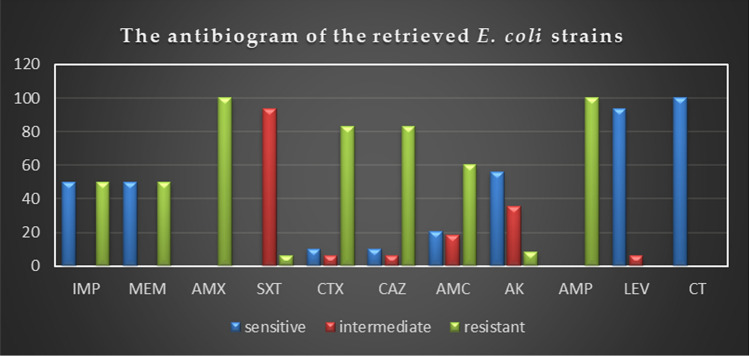
The in-vitro antimicrobial susceptibility testing revealed that the retrieved isolates displayed high resistance pattern to penicillins: ampicillin and amoxicillin (100%), and amoxicillin-clavulanic acid (60.4%), cephalosporins: cefotaxime and ceftazidime (83.3%), and carbapenems: imipenem and meropenem (50%), while showed intermediate resistance to trimethoprim-sulfamethoxazole (93.8%). Besides, the examined strains were highly susceptible to colistin sulfate (100%), followed by levofloxacin (93.8%) and amikacin (56.2%) as described in Table 4 and Fig. 3. Statistically, there is a significant difference in the resistance of the retrieved isolates to various tested antimicrobial agents (P < 0.05). The correlation analysis among the tested antimicrobial-agents was conducted. Our results revealed strong positive correlations (r = 0.5–0.86) between: AK and LEV (r = 0.83); AK and CT (r = 0.86); IMP, MEM, and CT (r = 0.5); IMP, MEM, and AMX (r = 0.5); IMP, MEM, and AMP (r = 0.5); IMP, MEM, and AMC (r = 0.54); IMP, MEM, and CAZ (r = 0.54); IMP, MEM, and CTX (r = 0.54). Furthermore, a moderate positive correlation was noticed between IMP and MEM (r = 0.45) (Fig. 4).
Table 4.
Antimicrobial resistance pattern of the retrieved E. coli strains (n = 48).
| Antibiotic classes | Specific tested antibiotic | Interpretation | |||||
|---|---|---|---|---|---|---|---|
| Sensitive | Intermediate | Resistance | |||||
| N | % | N | % | N | % | ||
| Penicillins | Amoxicillin | – | – | – | – | 48 | 100 |
| Ampicillin | – | – | – | – | 48 | 100 | |
| Amoxicillin-Clavulanic acid | 10 | 20.8 | 9 | 18.7 | 29 | 60.4 | |
| Cephalosporins | Cefotaxime | 5 | 10.4 | 3 | 6.2 | 40 | 83.3 |
| Ceftazidime | 5 | 10.4 | 3 | 6.2 | 40 | 83.3 | |
| Carbapenems | Imipenem | 24 | 50 | – | – | 24 | 50 |
| Meropenem | 24 | 50 | – | – | 24 | 50 | |
| Aminoglycosides | Amikacin | 27 | 56.2 | 17 | 35.4 | 4 | 8.4 |
| Fluoroquinolones | Levofloxacin | 45 | 93.8 | 3 | 6.2 | – | – |
| Polymyxins | Colistin sulfate | 48 | 100 | – | – | – | – |
| Sulfonamides | Trimethoprim-sulfamethoxazole | – | – | 45 | 93.8 | 3 | 6.2 |
| P value | P < 0.0001 | P < 0.0001 | P < 0.0001 | ||||
Figure 3.
Antimicrobial resistance pattern of the retrieved E. coli strains (n = 48). The retrieved isolates displayed high resistance to ampicillin and amoxicillin (100%), cefotaxime and ceftazidime (83.3%), amoxicillin-clavulanic acid (60.4%), and imipenem and meropenem (50%). Besides, the examined strains were highly susceptible to colistin sulfate (100%), and levofloxacin (93.8%).
Figure 4.
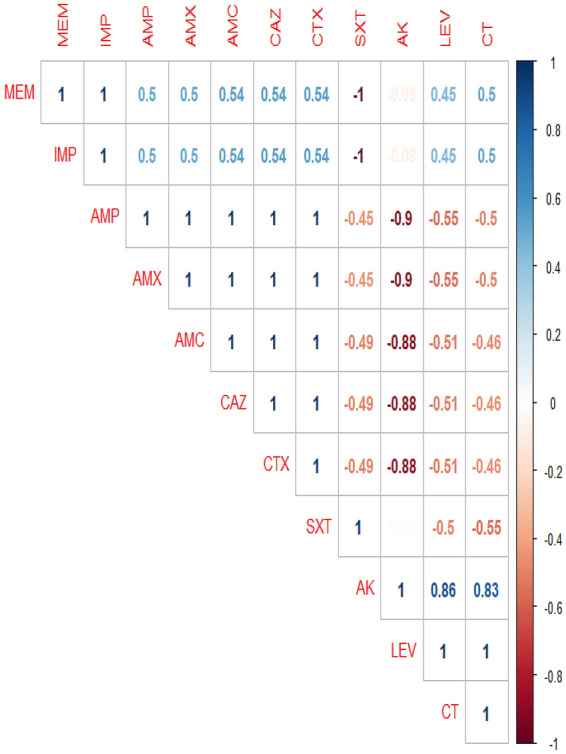
The correlation between the tested antimicrobial agents. The intensity of colors indicates the numerical value of the correlation coefficient (r), red, and blue color refers to the negative and positive correlations, respectively.
The frequency of the virulence-determinant and antibiotic-resistance genes among the recovered strains (n = 48)
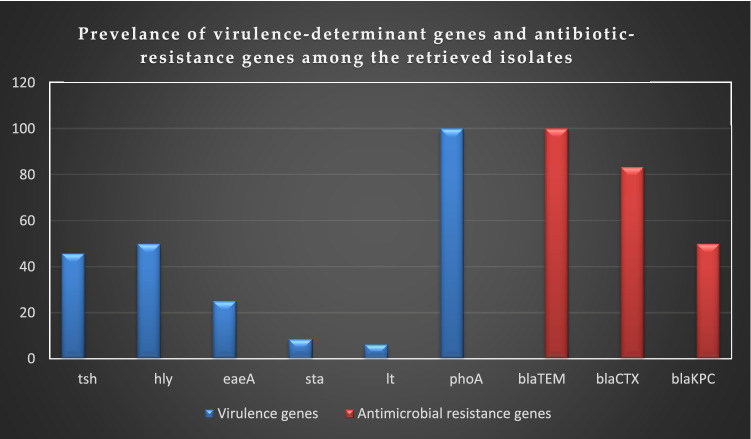
Regarding the virulence-determinant genes, the PCR proved that the tested strains harbored the virulence genes phoA, hly, tsh, eaeA, sta, and lt with a prevalence of 100% and 50%, 45.8%, 25%, 8.4%, and 6.2%, respectively. Concerning the antibiotic-resistance genes, the examined strains were positive for blaTEM, blaCTX, and blaKPC resistance-genes with a prevalence of 100%, 83.3%, and 50%, respectively, as presented in Table 5. The frequency of the virulence-determinant and antibiotic-resistance genes in the retrieved serovars is illustrated in Tables 5 and 6, and Fig. 5. Statistically, there is a significant difference in the prevalence of the virulence-determinant genes and the antibiotic-resistant genes among the tested strains (P < 0.05).
Table 5.
PCR-based screening of virulence and antibiotic resistance genes among the recovered strains (n = 48).
| Target genes | N | % | P value | |
|---|---|---|---|---|
| Virulence-determinant genes | phoA | 48 | 100 | P < 0.0001 |
| hly | 24 | 50 | ||
| tsh | 22 | 45.8 | ||
| eaeA | 12 | 25 | ||
| sta | 4 | 8.4 | ||
| lt | 3 | 6.2 | ||
| Antibiotic-resistance genes | blaTEM | 48 | 100 | P < 0.0001 |
| blaCTX | 40 | 83.3 | ||
| blaKPC | 24 | 50 | ||
Table 6.
Prevalence of virulence genes and antibiotic resistance genes among the retrieved serovars (n = 48).
| Samples | Serovars | N | tsh gene | phoA gene | hly gene | eaeA gene | sta gene | lt gene | blaCTX gene | blaKPC gene | blaTEM gene |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Nasal swabs | O86a | 4 | – | 4 | 4 | – | – | – | 4 | 4 | 4 |
| Untyped | 3 | – | 3 | – | – | – | – | 2 | 1 | 3 | |
| Fecal swabs | O114 | 7 | 7 | 7 | 1 | 3 | 4 | – | 5 | 5 | 7 |
| O26 | 4 | – | 4 | 4 | 1 | – | 3 | 4 | 4 | 4 | |
| Untyped | 2 | – | 2 | – | 2 | – | – | 2 | – | 2 | |
| Blood samples | O111 | 5 | 5 | 5 | 5 | – | – | – | 4 | 4 | 5 |
| O18 | 4 | – | 4 | 2 | – | – | – | 4 | – | 4 | |
| Untyped | 2 | 2 | 2 | 1 | – | – | – | 1 | – | 2 | |
| Milk samples | O1 | 9 | 2 | 9 | 3 | 5 | – | – | 7 | 2 | 9 |
| O55 | 4 | 3 | 4 | 2 | – | – | – | 3 | 4 | 4 | |
| O158 | 3 | 2 | 3 | 2 | – | – | – | 3 | – | 3 | |
| Untyped | 1 | 1 | 1 | – | 1 | – | – | 1 | – | 1 | |
| Total | 48 | 22 | 48 | 24 | 12 | 4 | 3 | 40 | 24 | 48 | |
Figure 5.
The distribution of virulence-determinant and antibiotic-resistance genes among the recovered strains. The tested strains harbored the virulence-determinant genes phoA, hly, tsh, eaeA, sta, and lt with a prevalence of 100%, 50%, 45.8%, 25%, 8.4%, and 6.2%, respectively. Besides, they harbored the blaTEM, blaCTX, and blaKPC resistance genes with a prevalence of 100%, 83.3%, and 50%, respectively.
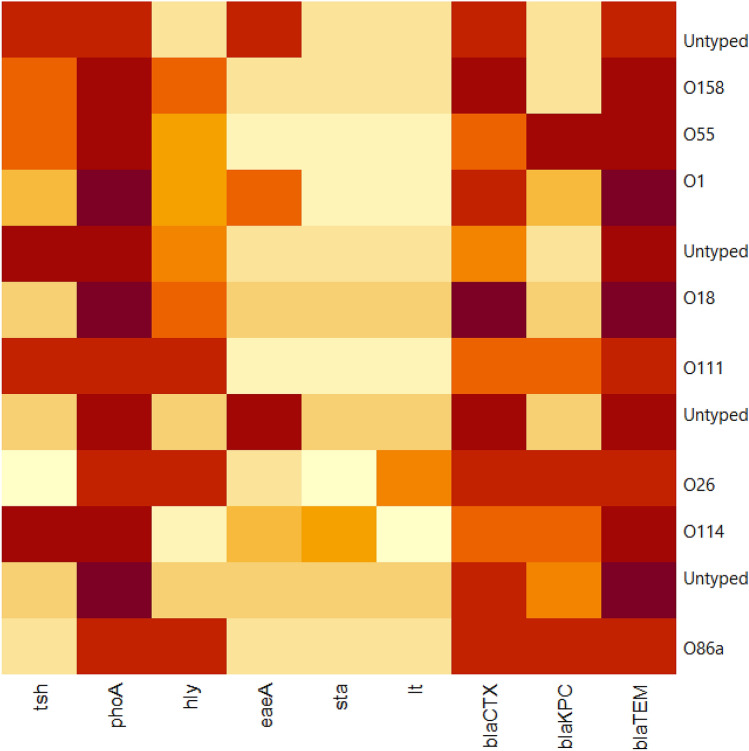
The correlation analysis was determined between various virulence genes and antibiotic-resistance genes. The obtained results revealed strong positive correlations (r = 0.53–0.95) between: blaCTX, blaTEM, and phoA (r = 0.95); tsh and sta (r = 0.72); eaeA, blaTEM, and phoA (r = 0.69); eaeA and blaCTX (r = 0.63), hly and blaKPC (r = 0.6); hly and blaCTX (r = 0.59); blaKPC, blaTEM, and phoA (r = 0.56); blaKPC and blaCTX (r = 0.54); blaKPC and tsh (r = 0.53). Moreover, moderate positive correlation (r = 0.3–0.49) was observed between: tsh, blaTEM, and phoA (r = 0.49); hly, blaTEM, and phoA (r = 0.46); blaKPC and sta (r = 0.46); sta, blaTEM, and phoA (r = 0.43); eaeA and sta (r = 0.39); lt and hly (r = 0.37); blaCTX and tsh (r = 0.32); lt and blaKPC (r = 0.31); blaCTX and sta (r = 0.3). Besides, low positive correlation was noticed between eaeA and tsh (r = 0.25) (Fig. 6). Furthermore, the heat-map illustrates the distribution of virulence genes and the antibiotic-resistance genes among the recovered E. coli serovars. The intensity of colors indicates the numerical value of the distribution (Fig. 7).
Figure 6.
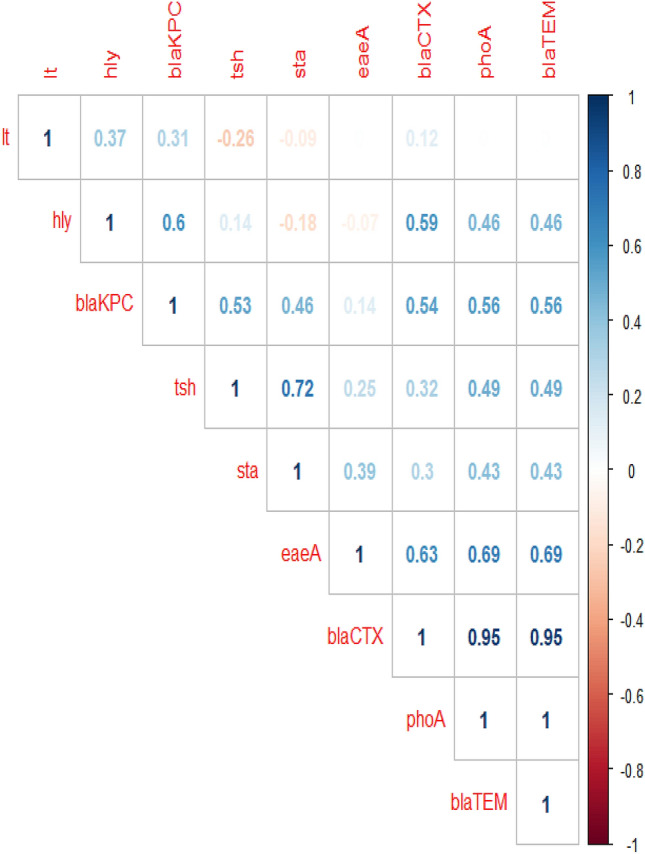
The correlation between virulence genes and the antibiotic-resistance genes. The intensity of colors indicates the numerical value of the correlation coefficient (r), red, and blue color refers to the negative and positive correlations, respectively.
Figure 7.
The heat-map illustrates the distribution of virulence genes and the antibiotic-resistance genes among the recovered E. coli serovars. The intensity of colors indicates the numerical value of the distribution.
The in-vitro multidrug-resistance patterns and the distribution of antibiotic-resistance genes
Concerning the occurrence of multidrug-resistance phenomena, in the present study, 50% of the recovered strains are multidrug-resistant (MDR) (MDR: non-susceptible to ≥ one agent in ≥ three antimicrobial classes); to penicillins: ampicillin, amoxicillin, and amoxicillin–clavulanic acid; cephalosporins: ceftazidime and cefotaxime; carbapenems: meropenem and imipenem, and are harboring the blaTEM, blaCTX, and blaKPC genes. Moreover, 25% of the examined strains are resistant to penicillins: ampicillin, amoxicillin, and cephalosporins: ceftazidime, and cefotaxime, and are harboring the blaTEM and blaCTX genes. Furthermore, 8.3% of the recovered strains were multidrug-resistant (MDR) to penicillins: ampicillin, and amoxicillin; cephalosprins: ceftazidime, cefotaxime, and aminoglycosides: amikacin, and possessed the blaTEM and blaCTX resistance genes (Table 7). The correlation analysis performed between various phenotypic multidrug-resistance patterns and the antibiotic-resistance genes. The obtained results revealed strong positive correlations between: blaCTX gene, CAZ, and CTX (r = 0.99); blaTEM gene, AMX, AMP, and AMC (r = 1); blaKPC gene, MEM, and IMP (r = 1) (Fig. 8).
Table 7.
The frequency of the phenotypic multidrug-resistance and the antibiotic-resistance genes among the retrieved strains (n = 48).
| No. of strains | % | Type of resistance | Phenotypic multidrug resistance | The antibiotic -resistance genes |
|---|---|---|---|---|
| 24 | 50 | MDR | Penicillins: ampicillin, amoxicillin, amoxicillin–clavulanic acid | blaTEM, blaCTX, and blaKPC |
| Cephalosporins: cetazidime, cefotaxime | ||||
| Carbapenems: meropenem and imipenem | ||||
| 12 | 25 | Resistant | Penicillins: ampicillin and amoxicillin | blaTEM and blaCTX |
| Cephalosporins: cetazidime and cefotaxime | ||||
| 4 | 8.3 | MDR | Penicillins: ampicillin and amoxicillin | blaTEM and blaCTX |
| Cephalosporins: cetazidime and cefotaxime | ||||
| Aminoglycosides: amikacin | ||||
| 3 | 6.3 | Resistant | Penicillins: ampicillin, amoxicillin, amoxicillin–clavulanic acid | blaTEM |
| Sulfonamides: trimethoprim–sulfamethoxazole | ||||
| 3 | 6.3 | Resistant | Penicillins: ampicillin, and amoxicillin | blaTEM |
| 2 | 4.1 | Resistant | Penicillins: ampicillin, amoxicillin, and amoxicillin–clavulanic acid | blaTEM |
Characteristics of multidrug resistance (MDR), extensively drug-resistance (XDR), and pandrug-resistance (PDR) in E. coli: PDR non-susceptible to all antimicrobial agents listed, XDR non-susceptible to ≥ one agent in all but ≤ two antimicrobial classes, MDR non-susceptible to ≥ one agent in ≥ three antimicrobial classes.
Figure 8.
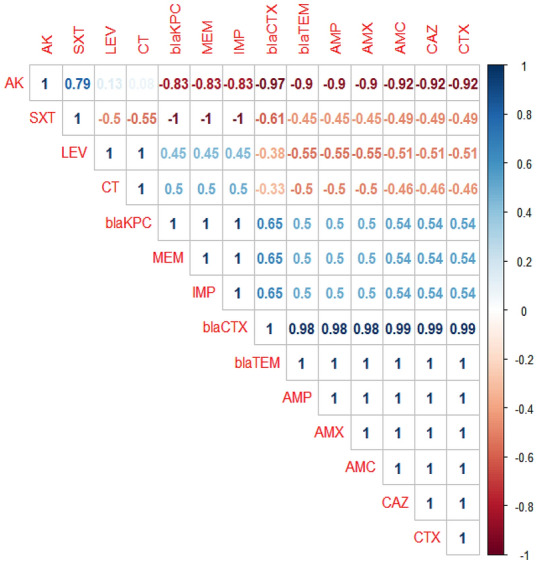
The correlation between various phenotypic multidrug-resistance patterns and the antibiotic-resistance genes. The intensity of colors indicates the numerical value of the correlation coefficient (r), red, and blue color refers to the negative and positive correlations, respectively.
Discussion
Globally, cattle are representing the main supply of high-quality meat and milk. However, few reports explained the role of pathogenic E. coli as a secondary bacterial pathogen following the FMD-outbreaks. The current study was conducted to inspect the prevalence, antibiogram, PCR detection of virulence-determinant genes (tsh, phoA, hly, eaeA, sta, and lt) and the antibiotic-resistance genes (blaTEM, blaKPC, and blaCTX) of E. coli isolated from secondary bacterial infections following FMD-outbreak in cattle.
The bacteriological assay proved that E. coli was detected in 30% of the examined samples. Besides, other bacterial pathogens were isolated from 112 (70%) examined diseased animals. There is no significant difference in the prevalence of E. coli between the surveyed farms (P > 0.05), as the inspected farms are very close to each other and sharing the same management practices, nutrition, and water supply. E. coli is a common opportunistic microorganism that incriminated in several infections, especially diarrhea, mastitis, septicemia, and respiratory manifestations11, 18, 23. In Nigeria, S. uberis and S. bovis clinical mastitis are also reported by Amosun31. In China, the emergence of P. mirabilis as a causative agent of diarrhea was reported by Gong32. Moreover, in Nepal, E. faecalis diarrhea was recorded in immune-compromised persons by Sah33. El-Seedy34 reported that P. multocida and M. hemolytica are major pathogens of calf pneumonia in Egypt, while Algammal11 categorized P. aeruginosa as a common pathogen of pneumonia in calves. In Egypt, although the available FMD-vaccine is efficient to minimize the mortality rate, the vaccination-failure may happen that results in the occurrence of FMD-outbreak and the emergence of secondary bacterial infections due to the immunosuppression35. A previous study in Cambodia reported the occurrence of FMD-vaccination failure in more than 50% of the vaccinated animals. The vaccination failure is mainly attributed to improper technique, insufficient dose, immunological factors, and vaccine cold-chain miscarriage36. Several causes are implicated in the existence of E. coli secondary infection, including; bad sanitation, intensive-breeding management, bad environmental conditions, stress, and weak animal immunity18.
Concerning the E. coli serovars, the most prevalent E. coli serovar accompanied the respiratory infection was 086a (n = 4), diarrhea: O114 (n = 7), fever: O111 (n = 5), mastitis: O1 (n = 9). The investigation of E. coli O-serogroups has a major public health concern. The recovered serovars are analogous to those reported by previous studies, which concerned the E. coli infections37–39. In the present study, the CR-binding assay proved that 83.3% of the examined isolates (40/48) were CR-binding positive. All the tested serovars were positive. Moreover, the untypable strains were negative (8/48). The current results agreed with Algammal18, who reported that 89.8% of the tested strains are invasive by congo-red binding assay, which confirms the pathogenicity of these isolates.
Regarding the in-vitro antimicrobial susceptibility testing, the retrieved strains exhibited a remarkable resistance to penicillins, cephalosporins, and carbapenems which gave a public health alarm. The current findings nearly agreed with those reported by Shahrani40, Gupta41, and Touwendsida42. The uncontrolled widespread use of antibiotics in veterinary and health sectors as well as the bacterial antibiotic-resistant genes are incriminated in the development of such multidrug-resistant strains43, 44. Regrettably, E. coli is capable to resist various antibiotic-classes due to possessing resistant genes and/ or R-plasmids45.
In the current study, the PCR proved that the recovered E. coli strains were found to posse 2–5 virulence genes. The most prevalent virulence genes accompanied the respiratory infections are phoA and hly genes, in diarrhea: phoA, sta, lt, eaeA, and hly genes, in fever and mastitis: phoA, tsh, and hly genes. These findings agreed with those obtained by previously reported by Algammal18, Andrade46 , and Whitelegge47. The pathogenesis of virulent E. coli is controlled by multiple virulence determinants that vary among different pathotypes. The most common virulence determinants that accompanied the E. coli-pathotypes are enterotoxins, hemolysins, siderophores, intimin, fimbria-mannose binding type1-H adhesion, alkaline phosphatase, and temperature-sensitive haemagglutinin (Tsh-protein). Furthermore, the production of these virulence-determinants is regulated by the expression of specific virulence genes48, 49.
In the present study, 50% of the recovered strains are MDR to penicillins, cephalosporins, and carbapenems, and are harboring the blaTEM, blaCTX, and blaKPC genes. Furthermore, 25% of the examined strains are resistant to penicillins and cephalosporins, and are harboring the blaTEM and blaCTX genes. The Extended Spectrum β-lactamases (ESBLs) produced by E. coli incriminated in the β-lactam-antibiotic resistance. The heavy use of penicillin, cephalosporins, and carbapenems-antibiotics in medications is resulting in the evolution of multidrug-resistant strains. The resistance to the β-lactam-antibiotics is mainly mediated by the ESBL-genes; blaTEM, blaCTX, and blaKPC which are encoded for penicillin, cephalosporins, and carbapenem-resistance, respectively50–53. Different mechanisms explain the emergence of MDR-E. coli strains include: 1-Shared resistance mechanisms; occur especially for the antimicrobial agents in the same category due to penicillin-binding protein mutations as well as the β-lactamases. Furthermore, it could happen for different antibiotics in various classes due to the efflux pumps acting on numerous drugs in different species. 2-Linkage among the antibiotic resistance genes, this mechanism plays a significant role in association links between various resistances and to differentiate between resistance mechanisms (either the resistance arise due to alterations in the target protein of the antibiotic or due to a resistance gene encoded for an enzyme that destroys the antibiotic). 3-Correlated drug exposure of the host, it mainly occurs due to routine use of combination therapy and the repeated treatment failure54–59.
Limitations and future recommendations: Future work is recommended to perform phylogenetic analysis either by MLST or PFGE to understand the clonal relatedness of the obtained strains.
In conclusion, to the best of our knowledge, this is the first report concerning the E. coli secondary bacterial infections following the FMD-outbreak. The immunosuppression due to the FMD increases the animal susceptibility to E. coli secondary infections. The most prevalent E. coli serovar associated the respiratory infections was O86a, in diarrhea: O114, in fever: O111, and in mastitis: O1. Furthermore, the most predominant virulence-determinant genes accompanied the E. coli respiratory infections were phoA and hly genes, diarrhea: phoA, sta, lt, eaeA, and hly genes, fever, and mastitis: phoA, tsh, and hly genes. A high percentage of the isolated E. coli strains were multidrug resistant (MDR) to penicillins: ampicillin, amoxicillin, and amoxicillin-clavulanic acid; cephalosporins: ceftazidime and cefotaxime; carbapenems: meropenem and imipenem, and are harboring the blaTEM, blaCTX, and blaKPC genes. In-vitro, colistin sulfate and levofloxacin have promising activity against MDR-E. coli. The emergence of highly pathogenic MDR-E. coli strains constitutes a significant threat to the cattle health resulting in multiple severe infections and huge economic losses in the livestock production. Furthermore, the evolution of penicillins, cephalosporins, and carbapenems-resistant strains is reflecting a public health alarm and specifies the convoluted treatment of the infections caused by these strains. Moreover, it recommends the proper use of antimicrobial agents in the veterinary and health sectors as well as the routine application of the antimicrobial susceptibility testing.
Acknowledgements
The authors are grateful to the Deanship of Scientific Research, King Saud University for funding through Vice Deanship of Scientific Research Chairs.
Author contributions
A.M.A and R.M.E Conceptualization; A.M.A and R.M.E, H.F.H, H.R.H, W.N.H, G.E.B., and W.M.E conducted the experiments. A.M.A and R.M.E drafted the manuscript. A.M.A, R.M.E, H.F.H, H.R.H, W.N.H, G.E.B., W.M.E and A.M.T did the statistical analysis, investigation, data validation and accuracy, and supervision. A.M.A, H.F.H, and R.M.E wrote and revised the manuscript. All authors have revised and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Abdelazeem M. Algammal, Email: abdelazeem.algammal@gmail.com
Reham M. El-Tarabili, Email: rehameltrabely@gmail.com
References
- 1.Verma AK, et al. Studies of the outbreaks of foot and mouth disease in Uttar Pradesh, India, between 2000 and 2006. Asian J. Epidemiol. 2011;3:141–147. doi: 10.3923/aje.2010.141.147. [DOI] [Google Scholar]
- 2.Rodriguez LL, Gay CG. Development of vaccines toward the global control and eradication of foot-and-mouth disease. Expert Rev. Vaccines. 2011;10:377–387. doi: 10.1586/erv.11.4. [DOI] [PubMed] [Google Scholar]
- 3.Verma AK, Kumar A, Rahal A, Bist B. Multi drug resistant pseudomonas aeruginosa: a secondary invader. Glob. J. Med. Res. 2014;14:6–9. [Google Scholar]
- 4.Yao-Zhong D, et al. An overview of control strategy and diagnostic technology for foot-and-mouth disease in China. Virol. J. 2013;10:1–6. doi: 10.1186/1743-422X-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elbayoumy, M. K. et al.Molecular Characterization of Foot-and-Mouth Disease Virus Collected from Al-Fayoum and Beni-Suef Governorates in Egypt (2014).
- 6.Dobrindt U. (Patho-)Genomics of Escherichia coli. Int. J. Med. Microbiol. 2005;295:357–371. doi: 10.1016/j.ijmm.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 7.Ulett GC, et al. Uropathogenic Escherichia coli virulence and innate immune responses during urinary tract infection. Curr. Opin. Microbiol. 2013;16:100–107. doi: 10.1016/j.mib.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Eid HI, Algammal AM, Nasef SA, Elfeil WK, Mansour GH. Genetic variation among avian pathogenic E. coli strains isolated from broiler chickens. Asian J. Anim. Vet. Adv. 2016;11:350–356. doi: 10.3923/ajava.2016.350.356. [DOI] [Google Scholar]
- 9.Smith JL, Fratamico PM, Gunther NW. Extraintestinal pathogenic Escherichia coli. Foodborne Pathogens Dis. 2007;4:134–163. doi: 10.1089/fpd.2007.0087. [DOI] [PubMed] [Google Scholar]
- 10.Clarke SC, Haigh RD, Freestone PPE, Williams PH. Virulence of enteropathogenic Escherichia coli, a global pathogen. Clin. Microbiol. Rev. 2003;16:365–378. doi: 10.1128/CMR.16.3.365-378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Algammal AM, et al. Prevalence, the antibiogram and the frequency of virulence genes of the most predominant bacterial pathogens incriminated in calf pneumonia. AMB Express. 2020;10:1–8. doi: 10.1186/s13568-020-01037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abolghait SK, Fathi AG, Youssef FM, Algammal AM. Methicillin-resistant Staphylococcus aureus (MRSA) isolated from chicken meat and giblets often produces staphylococcal enterotoxin B (SEB) in non-refrigerated raw chicken livers. Int. J. Food Microbiol. 2020;328:108669. doi: 10.1016/j.ijfoodmicro.2020.108669. [DOI] [PubMed] [Google Scholar]
- 13.Algammal AM, Enany ME, El-Tarabili RM, Ghobashy MOI, Helmy YA. Prevalence, antimicrobial resistance profiles, virulence and enterotoxin-determinant genes of MRSA isolated from subclinical bovine mastitis samples in Egypt. Pathogens. 2020;9:1–11. doi: 10.3390/pathogens9050362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Algammal AM, et al. Molecular typing, antibiogram and PCR-RFLP based detection of Aeromonas hydrophila complex isolated from Oreochromis niloticus. Pathogens. 2020;9:238. doi: 10.3390/pathogens9030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magiorakos AP, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infection. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 16.Enany ME, et al. The occurrence of the multidrug resistance (MDR) and the prevalence of virulence genes and QACs resistance genes in E. coli isolated from environmental and avian sources. AMB Express. 2019;9:192. doi: 10.1186/s13568-019-0920-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eid HM, et al. Prevalence, molecular typing, and antimicrobial resistance of bacterial pathogens isolated from ducks. Vet. World. 2019;12:677–683. doi: 10.14202/vetworld.2019.677-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Algammal AM, et al. Genes encoding the virulence and the antimicrobial resistance in enterotoxigenic and Shiga-toxigenic E. coli isolated from diarrheic calves. Toxins (Basel) 2020;12:383. doi: 10.3390/toxins12060383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quinn PJ, Markey BK, Leonard FC, Hartigan P, Fanning S, Fitzpatrick E. Veterinary Microbiology and Microbial Disease. 2. Hoboken: Wiley; 2011. pp. 1–928. [Google Scholar]
- 20.Starr, M. P. Edwards and Ewing's Identification of Enterobacteriaceae.: Fourth Edition. By William H. Ewing. Elsevier Science Publishing Co., Inc., New York. 1986. 536 pages. $65.00. ISBN 0-444-00981-7. Int. J. Syst. Bacteriol.36, 581–582 (1986).
- 21.Panigrahy B, Yushen L. Differentiation of pathogenic and nonpathogenic Escherichia coli isolated from poultry. Avian Dis. 1990;34:941–943. doi: 10.2307/1591387. [DOI] [PubMed] [Google Scholar]
- 22.CLSI, Clinical and Laboratory Standards Institute: Performance standards for antimicrobial susceptibility testing : 27th ed 424 informational supplement. CLSI Doc. M100-S20 (2017).
- 23.Lee SI, Kang SG, Kang ML, Yoo HS. Development of multiplex polymerase chain reaction assays for detecting enterotoxigenic Escherichia coli and their application to field isolates from piglets with diarrhea. J. Vet. Diagn. Invest. 2008;20:492–496. doi: 10.1177/104063870802000413. [DOI] [PubMed] [Google Scholar]
- 24.Bisi-Johnson MA, Obi CL, Vasaikar SD, Baba KA, Hattori T. Molecular basis of virulence in clinical isolates of Escherichia coli and Salmonella species from a tertiary hospital in the Eastern Cape. South Africa. Gut Pathog. 2011;3:9. doi: 10.1186/1757-4749-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delicato ER, De Brito BG, Gaziri LCJ, Vidotto MC. Virulence-associated genes in Escherichia coli isolates from poultry with colibacillosis. Vet. Microbiol. 2003;94:97–103. doi: 10.1016/S0378-1135(03)00076-2. [DOI] [PubMed] [Google Scholar]
- 26.Hu Q, et al. Development of multiplex PCR assay for rapid detection of Riemerella anatipestifer, Escherichia coli, and Salmonella enterica simultaneously from ducks. J. Microbiol. Methods. 2011;87:64–69. doi: 10.1016/j.mimet.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 27.Piva IC, et al. Virulence markers of enteroaggregative Escherichia coli isolated from children and adults with diarrhea in Brasília, Brazil. J. Clin. Microbiol. 2003;41:1827–1832. doi: 10.1128/JCM.41.5.1827-1832.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pillai DR, et al. Klebsiella pneumoniae Carbapenemase Canada. Emerg. Infectious Dis. 2009;15:827–829. doi: 10.3201/eid1505.081536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Archambault M, et al. Molecular characterization and occurrence of extended-spectrum β-lactamase resistance genes among Salmonella enterica serovar corvallis from Thailand, Bulgaria, and Denmark. Microb. Drug Resist. 2006;12:192–198. doi: 10.1089/mdr.2006.12.192. [DOI] [PubMed] [Google Scholar]
- 30.Colom K, et al. Simple and reliable multiplex PCR assay for detection of blaTEM, blaSHV and blaOXA-1 genes in Enterobacteriaceae. FEMS Microbiol. Lett. 2003;223:147–151. doi: 10.1016/S0378-1097(03)00306-9. [DOI] [PubMed] [Google Scholar]
- 31.Amosun EA, Ajuwape AT, Adetosoye AI. Bovine streptococcal mastitis in Southwest and Northern states of Nigeria. Afr. J. Biomed. Res. 2010;13:33–37. [Google Scholar]
- 32.Gong Z, et al. Characterization of a novel diarrheagenic strain of Proteus mirabilis associated with food poisoning in China. Front. Microbiol. 2019;10:2810. doi: 10.3389/fmicb.2019.02810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sah, R. et al. Vancomycin resistant Enterococcus faecalis causing diarrhea in renal transplant patient. Int. Educ. Appl. Sci. Res. J. 2(9), 1-3 (2017).
- 34.El-Seedy FR, et al. Respiratory affections in calves in upper and middle Egypt: bacteriologic, immunologic and epidemiologic studies. Adv. Anim. Vet. Sci. 2020;8:558–569. [Google Scholar]
- 35.Diab E, et al. Foot-and-mouth disease outbreaks in Egypt during 2013–2014: molecular characterization of serotypes A, O, and SAT2. Vet. World. 2019;12:190–197. doi: 10.14202/vetworld.2019.190-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sieng S, Walkden-Brown SW, Kerr J. Effect of vaccine storage temperatures and dose rate on antibody responses to foot and mouth disease vaccination in Cambodia. Vet. Med. Sci. 2018;4:35–44. doi: 10.1002/vms3.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang JR, et al. Comparison between O serotyping method and multiplex real-time PCR to identify diarrheagenic Escherichia coli in Taiwan. J. Clin. Microbiol. 2007;45:3620–3625. doi: 10.1128/JCM.00596-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blum S, et al. Identification of a bovine mastitis Escherichia coli subset. Vet. Microbiol. 2008;132:135–148. doi: 10.1016/j.vetmic.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 39.Shabana II. Escherichia coli pathotypes associated with diarrhea in human and domestic animals. Am. J. Anim. Vet. Sci. 2014;9:155–161. doi: 10.3844/ajavsp.2014.155.161. [DOI] [Google Scholar]
- 40.Shahrani M, Dehkordi FS, Momtaz H. Characterization of Escherichia coli virulence genes, pathotypes and antibiotic resistance properties in diarrheic calves in Iran. Biol. Res. 2014;47:28. doi: 10.1186/0717-6287-47-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gupta MD, Islam M, Sen A, Sarker MS, Das A. Prevalence and antibiotic susceptibility pattern of Escherichia coli in cattle on Bathan and intensive rearing system. Microb. Health. 2017;6:1–4. doi: 10.3329/mh.v6i1.34062. [DOI] [Google Scholar]
- 42.Touwendsida SB, et al. Antibiotic susceptibility of Escherichia coli and Salmonella strains isolated from raw and curds milk consumed in Ouagadougou and Ziniar, Burkina Faso. Afr. J. Microbiol. Res. 2014;8:1012–1016. doi: 10.5897/AJMR2014.6632. [DOI] [Google Scholar]
- 43.Du X, Shen Z, Wu B, Xia S, Shen J. Characterization of class 1 integrons-mediated antibiotic resistance among calf pathogenic Escherichia coli. FEMS Microbiol. Lett. 2005;245:295–298. doi: 10.1016/j.femsle.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 44.Algammal, A.M. et al. Emerging MDR-Pseudomonas aeruginosa in fish commonly harbor oprL and toxA virulence genes and blaTEM, blaCTX-M, and tetA antibiotic-resistance genes. Scientific Reports10, 15961 (2020). 10.1038/s41598-020-72264-4. [DOI] [PMC free article] [PubMed]
- 45.Perelle S, Dilasser F, Grout J, Fach P. Detection by 5′-nuclease PCR of Shiga-toxin producing Escherichia coli O26, O55, O91, O103, O111, O113, O145 and O157:H7, associated with the world's most frequent clinical cases. Mol. Cell. Probes. 2004;18:185–192. doi: 10.1016/j.mcp.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 46.Andrade GI, et al. Identification of virulence factors by multiplex PCR in Escherichia coli isolated from calves in Minas Gerais, Brazil. Trop. Anim. Health Prod. 2012;44:1783–1790. doi: 10.1007/s11250-012-0139-8. [DOI] [PubMed] [Google Scholar]
- 47.Whitelegge J. Gas-phase structure of the E. coli OmpA dimer. Structure. 2014;22:666–667. doi: 10.1016/j.str.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaipainen T, et al. Virulence factors of Escherichia coli isolated from bovine clinical mastitis. Vet. Microbiol. 2002;85:37–46. doi: 10.1016/S0378-1135(01)00483-7. [DOI] [PubMed] [Google Scholar]
- 49.Pearce MC, et al. Prevalence and virulence factors of Escherichia coli serogroups O26, O103, O111 and O145 shed by cattle in Scotland. Appl. Environ. Microbiol. 2006;72:653–659. doi: 10.1128/AEM.72.1.653-659.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bradford PA. Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 2001;14:933–951. doi: 10.1128/CMR.14.4.933-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bonnet R. Growing group of extended-spectrum β-Lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 2004;48:1–14. doi: 10.1128/AAC.48.1.1-14.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nordmann P, Dortet L, Poirel L. Carbapenem resistance in Enterobacteriaceae: Here is the storm! Trends Mol. Med. 2012;18:263–272. doi: 10.1016/j.molmed.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 53.Humphries RM, et al. Carbapenem-resistant enterobacteriaceae detection practices in california: what are we missing? Clin. Infect. Dis. 2018;66:1061–1067. doi: 10.1093/cid/cix942. [DOI] [PubMed] [Google Scholar]
- 54.Giske CG, Monnet DL, Cars O, Carmeli Y. Clinical and economic impact of common multidrug-resistant gram-negative bacilli. Antimicrob. Agents Chemother. 2008;52:813–821. doi: 10.1128/aac.01169-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Batiha G-S, et al. The pharmacological activity, biochemical properties, and pharmacokinetics of the major natural polyphenolic flavonoid: quercetin. Foods. 2020;9:374. doi: 10.3390/foods9030374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Falagas ME, Bliziotis IA. Pandrug-resistant Gram-negative bacteria: the dawn of the post-antibiotic era? Int. J. Antimicrob. Agents. 2007;29:630–636. doi: 10.1016/j.ijantimicag.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 57.El-Sayed M, et al. Pathogenicity, genetic typing, and antibiotic sensitivity of Vibrio alginolyticus isolated from Oreochromis niloticus and Tilapia zillii. Rev. Méd. Vét. J. 2019;70:80–86. [Google Scholar]
- 58.Piddock LJ. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 2006;19:382–402. doi: 10.1128/cmr.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Enany ME, et al. Molecular typing and evaluation of Sidr honey inhibitory effect on virulence genes of MRSA strains isolated from catfish in Egypt. Pak. J. Pharma. Sci. 2018;31:1865–1870. [PubMed] [Google Scholar]