Abstract
This study aimed to investigate the association of miRNA-21 with mutant p53 expression, prognosis, interaction, and clinicopathological features of non-small cell lung cancer (NSCLC). Tissue specimens from 200 NSCLC patients were collected for qRT-PCR analysis of miR-21 and p53 expression, and p53 mutations were analyzed by Sanger sequencing. NSCLC cell lines were used to determine the effects of miR-21 knockdown on cell viability, cell cycle distribution, and p53 expression. We found that miR-21 expression was upregulated in NSCLC tissues, which was associated with an increase in p53 mRNA levels and with advanced tumor-node-metastasis (TNM) stages and lymph node metastasis. The most common mutant sites of p53 in NSCLC were R175H and R248Q. Moreover, elevated miR-21 and p53 expression levels were associated with shorter overall survival. Knockdown of miR-21 reduced NSCLC cell viability, arrested NSCLC cells at the G0-to-G1 phase of the cell cycle, and downregulated mutant p53 mRNA levels and phosphorylated p53 protein expression in A549 and H1650 cells compared to control cells. miR-21 is associated p53 at mutant sites R175H and R248Q, which seems not to be oncogenic, as it is being reported, since in a normal cell, without a mutated p53, it will probably have a protective role.
Keywords: non-small cell lung cancer, miR-21, p53, knockdown, prognosis, interaction
Graphical Abstract
miR-21 is associated with p53 at mutant sites R175H and R248Q, which seems not to be oncogenic, as it is being reported, since in a normal cell, without a mutated p53, it will probably have a protective role.
Introduction
Lung cancer accounts for most cancer incidences and mortality in most parts of the world,1 although effective tobacco control and smoking cessation have reduced lung cancer risk globally in recent years.2,3 Progress in lung cancer treatment, including improved surgical resection, chemoradiotherapy, and/or molecular and targeting therapy, has improved long-term survival of lung cancer patients.4 However, the current challenges facing clinicians are early detection, effective treatment options to control tumor progression, and means to predict survival and/or treatment responses. Lung cancer patients often suffer from tumor recurrence and metastasis, leading to poor overall survival (OS).5 Thus, better understandings of lung cancer pathogenesis, molecular alterations, and novel therapeutics could aid in early lung cancer diagnosis, effective treatment or control of tumor lesions and progression, and predict treatment responses and survival of patients.
MicroRNAs (miRNAs or miRs) are a group of small non-coding RNAs that are approximately 25 nt in length and regulate expression of protein-coding genes by binding to the 3′ untranslated region of the targeted mRNA. Thus, miRNAs can post-translationally regulate protein expression, which adds another level of gene regulation in cells.6 Previous studies have demonstrated the role of miRNAs in lung cancer development, progression, and control.7, 8, 9 For example, miR-21 was upregulated in lung cancer and used as a novel biomarker for screening non-small cell lung cancer (NSCLC),10 as well as in many types of solid tumors.11, 12, 13 Another study found abnormal miR-21 levels in cancer stem-like cells, which were associated with lung cancer cell resistance to radiation.14 Taken together, miR-21 could be considered an oncogene or at least possess oncogenic effects in human cancers. Furthermore, p53, a transcription factor, can regulate expression of various human genes that are related to cell cycle progression and apoptosis,15 and p53 expression is induced in response to DNA damage or cell stress.16 A previous study demonstrated that tobacco carcinogens induce p53 mutations in lung cancer cells.17 p53 executes most of these cellular processes as a transcription factor, by binding to DNA regulatory modules and regulating gene expression. Until now, p53 has the closest co-relationship with human tumors, which occupied the core position among the tumor molecular network. Wild-type (WT) p53 protein has a shorter half-life in vitro, leading to difficulty in p53 detection, whereas the mutant p53 (mtp53) protein has a longer half-life, which increases the ability to be detected.18, 19, 20 p53 is frequently mutated and highly expressed in tumor tissue, with more than 80% of mutations occurring in the DNA-binding domains and exons.5, 6, 7, 8 Mutation types mainly include insertions, frameshifts, missense mutations, and such, and missense mutations dominantly account for about 75%.18,19 In addition, several miRNAs have been shown to be involved in the regulation of p53 and p53-related pathways.21, 22, 23 For example, a previous study reported that downregulating miR-21 expression in glioblastoma cells decreased WT p53 expression and inhibited p53 activity and cycle arrest.24
Thus, in this study, we first analyzed expression of miR-21 and p53 in NSCLC tissue samples to determine their association with NSCLC prognosis and clinicopathological NSCLC features. We also confirmed the mutant sites of p53 and investigated the effects of miR-21 knockdown on regulation of NSCLC cell viability, cell cycle distribution, and p53 expression. Our findings provide a better understanding of the role of miR-21 and p53 at different sites in NSCLC development and progression.
Results
Patient Characteristics
Among the 200 NSCLC patients, there were 116 males and 84 females, with 109 cases >60 years of age. There were 113 patients who were classified as ever tobacco smokers and 87 as never smokers. 142 patients were classified as having adenocarcinoma of the lung and 58 as having squamous cell carcinoma of the lung. 64 patients were at stage I, 51 at stage II, 44 at stage III, and 39 at stage IV (Table 1). The 5-year OS rate of these NSCLC patients was 65.5%, and 69 patients died during the follow-up, among whom 29 were stage IV (5-year OS, 25.6%), 24 were stage III (5-year OS, 45.4%), 11 were stage II (5-year OS, 78.4%), and 5 were stage I (5-year OS, 92.2%).
Table 1.
miR-21 and p53 mRNA Levels in NSCLC Tissue Samples
| Variables | n | miR-21 Level (2−ΔΔCt Value) | p Value | p53 Level (2−ΔΔCt Value) | p Value |
|---|---|---|---|---|---|
| Tumor versus Normal | |||||
| NSCLC | 200 | 18.04 ± 2.09 | 0.0001 |
5.81 ± 1.05 | 0.02 |
| Normal tissues | 200 | 1.23 ± 0.16 | 1.14 ± 0.12 | ||
| Histology | |||||
| Adenocarcinoma | 142 | 19.15 ± 1.62 | 0.03 |
7.76 ± 1.14 | 0.032 |
| SCC | 58 | 14.93 ± 1.21 | 4.92 ± 0.99 | ||
| Sex | |||||
| Male | 116 | 16.02 ± 2.01 | 0.58 |
5.61 ± 1.03 | 0.63 |
| Female | 84 | 19.25 ± 2.59 | 7.89 ± 1.25 | ||
| History of Smoking | |||||
| Ever | 113 | 19.36 ± 2.51 | 0.65 |
7.42 ± 1.12 | 0.021 |
| Never | 87 | 17.99 ± 2.07 | 4.98 ± 1.05 | ||
| Tumor Size (cm) | |||||
| T1 (>3) | 79 | 23.36 ± 2.61 | 0.04 |
8.12 ± 1.52 | 0.031 |
| T1 (0–3) | 121 | 16.46 ± 2.01 | 3.92 ± 1.07 | ||
| Distant Metastasis | |||||
| Yes | 46 | 24.38 ± 2.63 | 0.01 |
10.52 ± 1.52 | 0.013 |
| No | 154 | 13.89 ± 1.81 | 2.89 ± 0.98 | ||
| Stage | |||||
| I | 64 | 11.02 ± 1.89 | 2.98 ± 0.54 | ||
| II | 51 | 16.25 ± 2.32 | 4.77 ± 0.78 | ||
| III | 44 | 21.28 ± 2.67 | 6.59 ± 1.03 | ||
| IV | 39 | 24.65 ± 2.98 | 8.43 ± 1.12 | ||
| TNM Stage | |||||
| I and II | 0.001 |
0.001 |
|||
| III and IV | |||||
| Tumor (T) | |||||
| T1 | 53 | 10.99 ± 1.56 | 3.18 ± 0.59 | ||
| T2 | 52 | 17.02 ± 2.42 | 4.76 ± 0.62 | ||
| T3 | 49 | 21.98 ± 2.81 | 7.59 ± 1.16 | ||
| T4 | 46 | 25.85 ± 3.12 | 8.43 ± 1.23 | ||
| T Stage | |||||
| T1 and T2 | 0.003 |
0.02 |
|||
| T3 and T4 | |||||
| Lymph Node Metastasis (N) | |||||
| N0 | 72 | 11.29 ± 1.66 | 2.88 ± 0.55 | ||
| N1 | 49 | 18.12 ± 2.62 | 3.75 ± 0.63 | ||
| N2 | 45 | 21.18 ± 2.51 | 6.52 ± 0.73 | ||
| N3 | 34 | 24.77 ± 3.02 | 8.46 ± 1.52 | ||
| N Stage | |||||
| N0 and N1 | 0.01 | 0.02 | |||
| N2 and N3 | |||||
SCC, squamous cell carcinoma.
High Expression of miR-21 and p53 in NSCLC
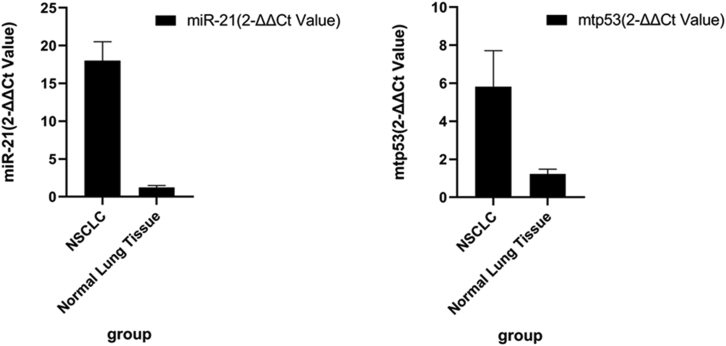
We first measured miR-21 and p53 mRNA levels in the 200 matched NSCLC and adjacent normal lung tissues using qRT-PCR. We found that miR-21 and p53 mRNA levels were significantly higher in NSCLC specimens compared to the corresponding normal lung tissues (18.04 ± 2.09 versus 1.23 ± 0.16, p = 0.002; 5.81 ± 1.05 versus 1.14 ± 0.12, p = 0.03, respectively; Figure 1). Furthermore, we determined the association of aberrant miR-21 levels with p53 mRNA expression using the cutoff values of high versus low expression of miR-21 and p53 in NSCLC tissues (72/85 versus 4/115, p = 0.003 when we used the cutoff value of miR-21 at 10 and the cutoff value of p53 at 3). High miR-21 levels were associated with increases in p53 mRNA in NSCLC tissues samples (Figure 1, 72/85 versus 4/115, p = 0.003).
Figure 1.
Upregulation of miR-21 and p53 mRNA in NSCLC Tissue Samples
A total of 200 NSCLC and matched non-tumor tissues were collected and analyzed using qRT-PCR. miR-21 and p53 levels in NSCLC tissues were compared to the paired adjacent noncancerous tissues and normal lung tissues (18.04 ± 2.09 versus 1.23 ± 0.16, p = 0.002; 5.81 ± 1.05 versus 1.14 ± 0.12, p = 0.03, respectively).
Association of miR-21 and p53 Expression with Clinicopathological Features and Prognosis of NSCLC Patients
We next determined the association of miR-21 and p53 mRNA levels using the above stated cutoff values, with clinicopathological data. We found that miR-21 and p53 mRNA levels were associated with advanced tumor-node-metastasis (TNM) stages (p = 0.01 and p = 0.02 for miR-21 and p53, respectively) and lymph node metastasis; however, there were no statistically significant differences between miR-21 and p53 levels and other clinicopathological features (such as patient sex and tumor location, differentiation, or pathology [adenocarcinoma or squamous cell carcinoma]). p53 expression in patients who had a smoking history was higher than those without a history of smoking, indicating that tobacco smoking can induce p53 mutations (Table 2).
Table 2.
Association of miR-21 and p53 mRNA with Clinicopathological Data from NSCLC Patients
| Variable/Group | High miR-21 Group | Low miR-21 Group | High p53 Group | Low p53 Group |
|---|---|---|---|---|
| Histological Classification | ||||
| AD | 70 | 45 | 68 | 47 |
| SCC | 51 | 29 | 46 | 34 |
| Sex | ||||
| Male | 77 | 59 | 73 | 63 |
| Female | 35 | 29 | 30 | 34 |
| History of Smoking | ||||
| Ever | 120 | 26 | 125 | 21 |
| Never | 12 | 43 | 9 | 46 |
| Stage | ||||
| I | 4 | 60 | 7 | 57 |
| II | 13 | 38 | 8 | 43 |
| III | 33 | 14 | 27 | 17 |
| IV | 35 | 4 | 34 | 5 |
| Tumor (T) | ||||
| T1 | 6 | 47 | 4 | 49 |
| T2 | 15 | 37 | 12 | 40 |
| T3 | 32 | 17 | 36 | 13 |
| T4 | 39 | 7 | 41 | 5 |
| Lymph Node Metastasis (N) | ||||
| N0 | 10 | 62 | 12 | 60 |
| N1 | 19 | 30 | 17 | 32 |
| N2 | 31 | 14 | 34 | 11 |
| N3 | 30 | 4 | 29 | 5 |
AD, adenocarcinoma; SCC, squamous cell carcinoma.
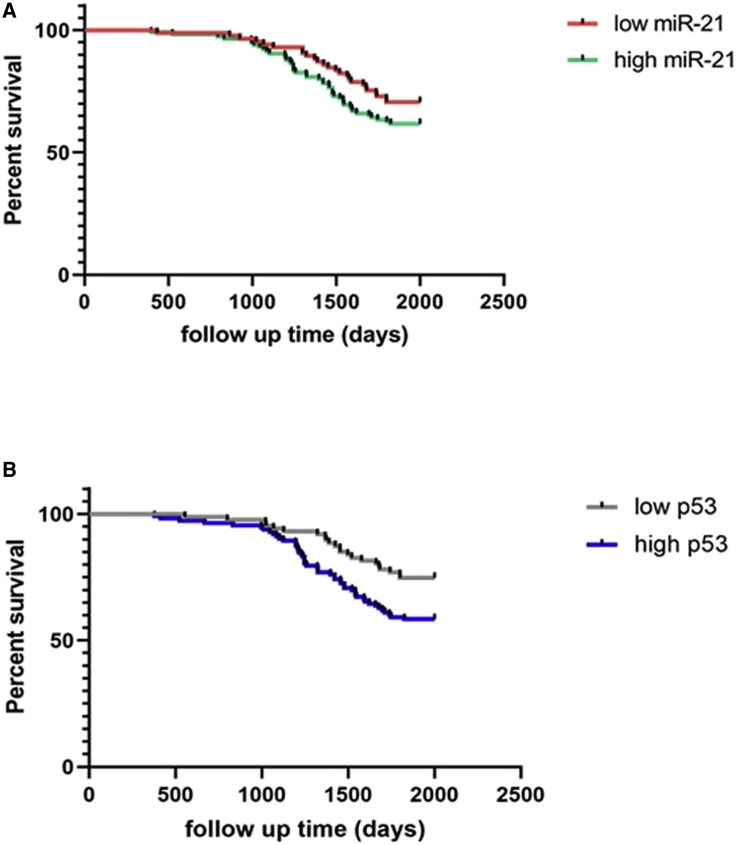
Furthermore, our data showed that NSCLC patients with high miR-21 and p53 mRNA levels in tumor lesions had shorter survival times compared to those with low miR-21 and/or p53 levels (11.6 ± 1.66 versus 2.13 ± 0.84, p = 0.04; 6.4 ± 1.15 versus 1.5 ± 0.14, p = 0.01, respectively). The Kaplan-Meier curve analysis showed that high miR-21 and p53 levels were associated with shorter OS in NSCLC patients compared to those with low miR-21 and p53-expressed NSCLCs (Figure 2).
Figure 2.
Kaplan-Meier Curve Analysis of OS of NSCLC Patients Stratified by miR-21 and p53 Levels
(A and B) miR-21 (A) and p53 (B) levels. The cutoff values of miR-21 were 2−ΔΔCt values <10 for low expression and >10 for high expression; the cutoff values of p53 were <3 for low expression and >3 for high expression.
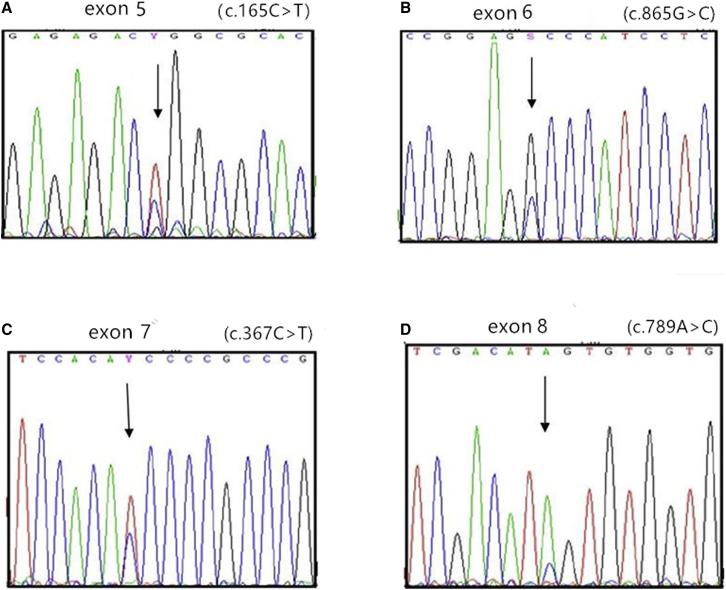
Mutation Sites of R175H and R248Q
A total of 93 mutation sites were found in 100 samples. Missense mutations occurred in 78 sites, accounting for 83.8% of the total mutations (78/93). Nine sites had suffered frameshift mutations, accounting for 9.7% (9/93), and nonsense mutations occurred in six sites, accounting for 6.5% (6/93) of the total mutations. Splicing mutations were not found in the samples. Exon 5 had the highest mutation frequency, which is 44.1% (41/93). In this study, we observed that 15 mutations occurred in codon 175, 10 mutations occurred in codon 248, 7 mutations occurred in codon 249, 5 mutations occurred in codon 273, 4 mutations occurred in codon 282, and 2 mutations occurred in codon 245 (Table 3; Figure 3). The mutant sites of R175H and R248Q is what we wanted to study.
Table 3.
p53 Mutation Type, Site, and Specific Distribution in 100 NSCLC Patients
| No. | Exon | Base Variation | Amino Acid Variation |
|---|---|---|---|
| 1 | 8 | c.797G>A | p.G266E |
| 2 | 8 | c.907dupT | p.P301fs |
| 3 | 5 | c.527G>T | p.R273H |
| 4 | 7 | c.746A>T | p.R249W |
| 6 | 5 | c.657G>A | p.R175H |
| 7 | 8 | c.817C>T | p.R273C |
| 8 | 5 | c.564G>C | p.R175H |
| 9 | 6 | c.577C>T | p.H193Y |
| 10 | 5 | c.848G>T | p.G245S |
| 11 | 6 | c.524G>A | p.R175H |
| 14 | 5 | c.382C>A | p.P128T |
| 15 | 7 | c.607C>G | p.R282W |
| 16 | 8 | c.844C>T | p.R248Q |
| 17 | 6 | c.584T>C | p.I195T |
| 18 | 5 | c.700T>C | p.Y234H |
| 19 | 6 | c.637C>T | p.R213X |
| 20 | 7 | c.743C>T | p.R248Q |
| 21 | 6 | c.549A>T | p.R282W |
| 22 | 5 | c.455dupCGC | p.G154fs |
| 23 | 5 | c.379C>T | p.R248Q |
| 24 | 5 | c.765G>C | p.R175H |
| 25 | 6 | c.103dupC | p.R202fs |
| 26 | 8 | c.503G>T | p.R249S |
| 27 | 5 | c.476C>T | p.R273H |
| 28 | 6 | c.583dupA | p.I195fs |
| 29 | 6 | c.586C>T | p.R282W |
| 30 | 5 | c.148G>C | p.R175H |
| 31 | 5 | c.337C>T | p.R248Q |
| 32 | 7 | c.709A>G | p.Y234C |
| 33 | 5 | c.487A>T | p.R175H |
| 34 | 5 | c.517G>T | p.V173L |
| 36 | 8 | c.839dupG | p.R273H |
| 37 | 5 | c.360_406del34 | p.L130fs |
| 39 | 5 | c.226_229delG | p.V143fs |
| 40 | 5 | c.767C>G | p.R282W |
| 41 | 6 | c.592G>T | p.R249S |
| 42 | 5 | c.451C>T | p.P151S |
| 43 | 7 | c.907T>A | p.R175H |
| 44 | 5 | c.493dupC | p.Q165fs |
| 45 | 6 | c.567_610dup44 | p.E204fs |
| 46 | 5 | c.469G>T | p.V157F |
| 47 | 5 | c.366A>T | p.R175H |
| 48 | 8 | c.818G>A | p.R273H |
| 49 | 5 | c.233G>C | p.R213W |
| 50 | 8 | c.797G>A | p.G249E |
| 52 | 7 | c.733G>A | p.G245S |
| 53 | 7 | c.680dupC | p.S227fs |
| 54 | 5 | c.234C>G | p.R175H |
| 55 | 6 | c.737C>T | p.G266E |
| 56 | 6 | c.537C>T | p.R249S |
| 57 | 5 | c.212C>G | p.R248Q |
| 58 | 6 | c.346_378delGA | p.Q192del |
| 59 | 7 | c.1037A>T | p.E192C |
| 60 | 8 | c.211C>G | p.R175H |
| 61 | 5 | c.537C>T | p.R356W |
| 63 | 6 | c.637G>T | p.R248Q |
| 64 | 5 | c.119A>G | p.R175H |
| 65 | 8 | c.785delG | p.G262fs |
| 66 | 5 | c.490G>T | p.R248Q |
| 67 | 8 | c.101delC | p.R213Q |
| 68 | 5 | c.568G>C | p.R175H |
| 69 | 6 | c.243G>T | p.R356W |
| 71 | 7 | c.769C>G | p.L257V |
| 72 | 7 | c.713G>A | p.C238Y |
| 73 | 6 | c.1106G>T | p.R175H |
| 75 | 5 | c.451C>T | p.P151S |
| 76 | 5 | c.428T>G | p.V143G |
| 77 | 8 | c.853_857delGAGG | p.E285fs |
| 79 | 5 | c.547G>C | p.R249S |
| 80 | 6 | c.208A>T | p.R238W |
| 81 | 5 | c.524G>T | p.R175H |
| 82 | 8 | c.670G>C | p.W238T |
| 83 | 5 | c.527G>T | p.C176F |
| 84 | 5 | c.536A>G | p.H179R |
| 85 | 5 | c.404G>T | p.C135F |
| 86 | 7 | c.743C>G | p.R248Q |
| 87 | 6 | c.574G>C | p.R175H |
| 88 | 5 | c.464C>A | p.T155N |
| 89 | 5 | c.512A>G | p.E171G |
| 90 | 5 | c.337A>T | p.G245S |
| 91 | 7 | c.748G>C | p.R249S |
| 92 | 6 | c.570C>T | p.R248Q |
| 93 | 5 | c.524G>A | p.R175H |
| 94 | 6 | c.599delA | p.N200fs |
| 95 | 7 | c.790delC | p.L287K |
| 96 | 5 | c.535C>T | p.H179Y |
| 97 | 7 | c.743C>G | p.R248Q |
| 99 | 6 | c.643A>C | p.S215R |
| 100 | 7 | c.730G>T | p.G244C |
Figure 3.
Peak of Missense Mutations at Different Loci by Sanger Sequencing
(A) Fifth exon: c.165C>T (the mutant peak is higher than the normal peak). (B) Sixth exon: c.865G>C (the mutant peak is lower than the normal peak). (C) Seventh exon: c.367C>T (the mutant peak is higher than the normal peak). (D) Eighth exon: c.789A>C (the mutant peak is higher than the normal peak).
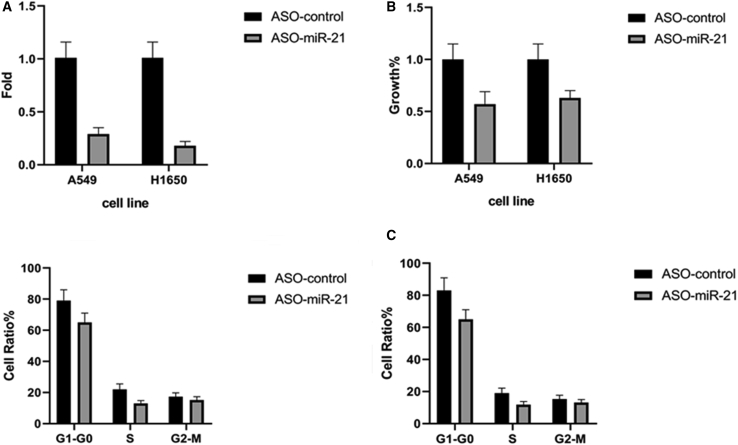
Reduction of NSCLC Cell Viability after Knockdown of miR-21 Expression
We next assessed whether knockdown of miR-21 expression could suppress NSCLC cell growth and alter gene expression. Our first step was to transfect mir-21 antisense oligonucleotides (ASOs) into A549 and H1650 cells. Our data showed that miR-21 levels were knocked down by more than 70% compared to cells transfected with the negative control ASO (Figure 4A). Knockdown of miR-21 expression reduced NSCLC cell viability (Figure 4B). Moreover, our cell cycle distribution analysis revealed that knockdown of miR-21 arrested NSCLC cells at the G0-to-G1 phase of the cell cycle and reduced the number of cells in the S phase compared to the control ASO-transfected A549 and H1650 cell lines (p = 0.02; Figure 4C). This finding indicates that knockdown of miR-21 levels reduces NSCLC cell proliferation, which further confirms the oncogenic role of miR-21 in NSCLC (Table 1). Moreover, in order to exclude the contribution of mutant p53 to the ability of knocking down miR-21 to suppress NSCLC cells, we first silenced p53 in A549 and H1650 cells with mutant p53 and then assessed the ability of silencing miR-21 to further suppress the cells and induce cell death or cell cycle arrest. We found that the result was similar to purely silencing miR-21.
Figure 4.
Effects of miR-21 Knockdown on NSCLC Cell Viability and Cell Cycle Distribution
(A) qRT-PCR. A549 and H1650 cells were grown and transiently transfected with miR-21 ASOs or negative control ASOs for 24 h and subjected to qRT-PCR analysis of miR-21 expression. miR-21 levels in A549 and H1650 cells were lower than those in the control group (1.01 ± 0.12 versus 0.29 ± 0.06, p = 0.001; 1.01 ± 0.12 versus 0.18 ± 0.04, p = 0.001, respectively). (B) Cell viability MTT assay. A549 and H1650 cells were grown and transiently transfected with miR-21 ASOs or negative control ASOs and subjected to an MTT assay. The growth in A549 and H1650 cells was lower than that in the control group (1 ± 0.12 versus 0.57 ± 0.06, p = 0.02; 1 ± 0.12 versus 0.63 ± 0.07, p = 0.02, respectively). (C) Flow cytometric assay. A549 and H1650 cells were grown and transiently transfected with miR-21 ASOs or negative control ASOs for 24 h and subjected to a flow cytometric cell cycle distribution assay. The percentages in the G1-to-G0 phase were higher than those in the control group (79.05 ± 7.01 versus 65.01 ± 5.97, p = 0.03; 83.07 ± 7.85 versus 65.01 ± 5.97, p = 0.02, respectively).
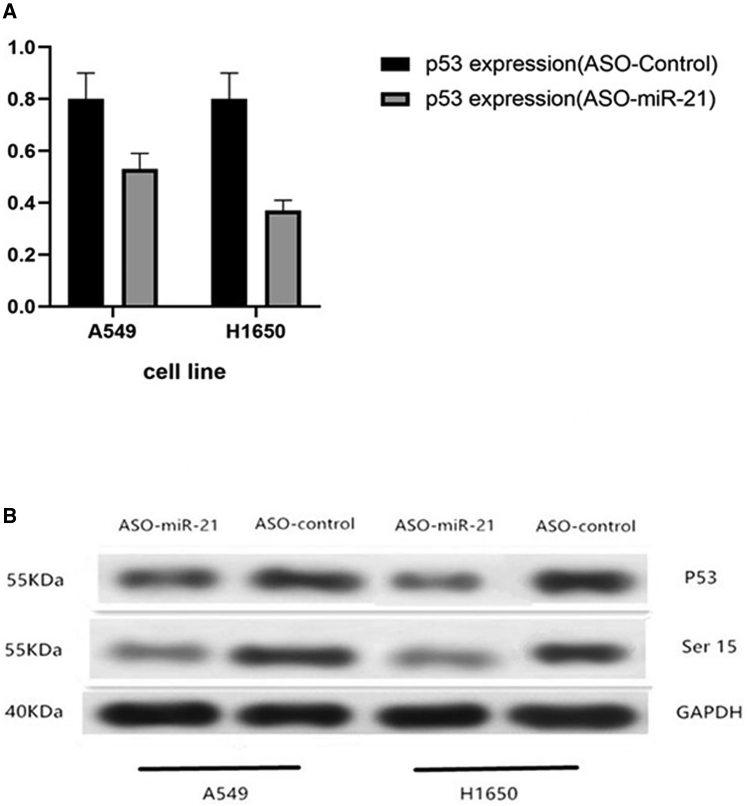
Decrease in p53 and Phosphorylated p53 at Ser15 after Knockdown of miR-21 Expression
Our data also showed that knockdown of miR-21 expression downregulated p53 mRNA levels and phosphorylated (Ser15) p53 protein expression in A549 and H1650 cells compared to the control cells (Figure 5).
Figure 5.
Effects of miR-21 Knockdown on NSCLC p53 Expression
(A) qRT-PCR. A549 and H1650 cells were grown and transiently transfected with miR-21 ASOs or negative control ASOs for 24 h and subjected to qRT-PCR analysis of p53 expression. The expression of p53 in the miR-21 ASOs was higher than that in the control group in A549 and H1650 cells (1.75 ± 0.08 versus 1.01 ± 0.02, p = 0.01; 1.54 ± 0.06 versus 1.01 ± 0.02, p = 0.03, respectively). (B) Western blot. A549 and H1650 cells were grown and transiently transfected with miR-21 ASOs or negative control ASOs for 24 h and subjected to western blot analysis of p53 and phosphorylated p53 protein at Ser15.
Discussion
In our current study, we observed that both miR-21 and p53 levels were upregulated in NSCLC compared to normal lung tissues, which further supported our previous findings of the association of miR-21 with NSCLC.13 Our current study revealed that the most regular mutant sites of p53 in NSCLC were R175H and R248Q, which are the most significant sites. Moreover, miR-21 expression is associated with increased p53 levels. To the best of our knowledge, this is the first reported association of miR-21 and p53 expression of different mutant sites specifically in NSCLC. Furthermore, our in vitro data support the oncogenic role of miR-21 in NSCLC, such that knockdown of miR-21 expression arrested NSCLC cells at the G0-to-G1 phase of the cell cycle and downregulated p53 expression and phosphorylation, indicating that miR-21 and p53 mRNA levels could be used as biomarkers to predict NSCLC prognosis.
Indeed, previous in vitro studies showed that miR-21 promoted lung cancer cell resistance to gefitinib,25 and that miR-21 expression enhanced lung cancer cell migration, invasion, and epithelial-mesenchymal transition of lung adenocarcinoma cancer cells.26 In contrast, solasodine-mediated downregulation of miR-21 expression inhibited NSCLC cell invasion capacity.27 Moreover, hypoxia-inducible factor (HIF)-1α and miR-21 could enhance the effects of cigarette smoke extract on malignant transformation of human bronchial epithelial (HBE) cells.28 miR-21 expression also enhanced NSCLC cell uptake of glucose and increased lactate generation and decreased oxygen consumption in NSCLC cells.29 Taken together, miRNA-21 could be a novel therapeutic target to control lung cancer.30 In addition, another study revealed that miR-145, miR-20a, miR-21, and miR-223 levels were increased in the early stage of NSCLC compared to control samples, indicating that these miRNAs could be biomarkers for lung cancer diagnosis,31 and, more specifically, upregulated miR-21 expression could predict NSCLC metastasis to the brain.32 Our current study also demonstrated that increased miR-21 expression in NSCLC tissues is associated with poor patient OS, which further confirmed the previous studies on the role of miR-21 in NSCLC.
The tumor suppressor protein p53 plays a crucial role in maintaining genomic stability and controlling cell cycle progression, apoptosis, and cell senescence.33 In NSCLC, p53 is frequently mutated and correlates with lung cancer development and progression. Different mutations can have different consequences. Clinically, p53 protein accumulation can be detected using immunohistochemistry because the half-life of mutated p53 protein is longer.34,35 In our current study, we found that p53 mRNA levels were upregulated in NSCLC tissues, mainly represented by the R175H and R248Q mutants. Furthermore, we found that knockdown of miR-21 expression reduced levels of p53 mRNA and phosphorylation of p53 at Ser15 in lung cancer cell lines. Previous studies showed that phosphorylated p53 at Ser15 promoted expression of proapoptotic genes in several types of cancers.36,37 Phosphorylation of p53 at Ser15 activates p53 in response to DNA damage.38 Thus, we speculate that miR-21 interaction with p53 could result in NSCLC development and progression, whereas knockdown of miR-21 expression reduces expression of p53. However, our current association study failed to explain how miR-21 regulates p53 levels in NSCLC and whether upregulated p53 is functional in these NSCLC tissue samples. Importantly, note that p53 protein accumulation in NSCLC has been used an indicator of p53 mutations in previous studies.34,35 In addition, future in vitro studies are needed to confirm whether p53 is functional in A549 and H1650 cells.
In summary, our study demonstrated that miR-21 and p53 expression levels are increased in NSCLC, and that these increases are associated with clinicopathological data and survival of NSCLC patients. We also confirmed the mutant sites of R175H and R248Q in NSCLC. Our in vitro data further support the role of miR-21 and the one-way interaction with p53 in NSCLC. Importantly, according to our study, miR-21 probably is not oncogenic, as has been suggested before, but it is a tumor suppressor as p53. It may play an oncogenic role in NSCLC only because the p53 is mutated, but if it were a normal p53 protein, miR-21 would increase p53 expression and activate several proapoptotic genes. As for how to regulate downstream genes, this will be the next step to be confirmed. Also, future studies will verify whether miR-21 could be as therapeutic target of NSCLC and the bidirectional interaction between miR-21 and p53.
Materials and Methods
NSCLC Patients and Tissue Samples
We retrospectively collected samples from 200 NSCLC patients who underwent surgical resection of NSCLC lesions at the Shanghai Ninth People’s Hospital, Shanghai Jiaotong University (Shanghai, China). Patients were consecutively selected, and any patients who received preoperative chemotherapy and/or radiotherapy were excluded from this study. NSCLC diagnosis and staging were assessed based on Lung Cancer Surgery and Pathology (eighth edition).39 Both NSCLC and adjacent normal lung tissues were collected from each participant during surgery or tumor fine-needle punctures and then snap-frozen in liquid nitrogen and stored at −80°C until use. This study was approved by the Research Ethics Committee of Shanghai Jiaotong University, and written informed consent was obtained from each patient before inclusion in this study. Clinicopathological data were gathered from medical records. All patients were followed up regularly, and the last follow-up was conducted on December 12, 2017.
RNA Isolation and qRT-PCR
Total RNA was isolated from the frozen tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and reverse transcribed into cDNA using a reverse transcription kit (Invitrogen) according to the manufacturers’ instructions. qPCR was amplified using a standard TaqMan PCR protocol in the ABI 7500 HT system (Applied Biosystems, Foster City, CA, USA). The primers used wereas follows: miR-21, 5′-TAGCTTATCAGACTGATGTTGA-3′ and 5′-TGCGTGTCGTGGAGTC-3′; p53, 5′-ACCTATGGAAACTACTTCCTGAAA-3′ and 5′-CTGGCATTCTGGGAGCTTCA-3′; and U6, 5′-CAAATTCGTGAAGCGTTCCATAT-3′. The relative levels of pre-miR-21 and p53 mRNA were determined using the cycle threshold (2−ΔΔCt) method compared to U6 RNA, which was used as an internal control. For example, the amount of the target mRNA = 2−ΔΔCt, where ΔΔCt = (CtmiR-21 − CtU6) tumor − (CtmiR-21 − CtU6) matched non-tumor tissues or ΔΔCt = (Ctp53 − CtU6) tumor − (Ctp53 – CtU6) matched non-tumor tissues. The 2-ΔΔCt cutoff values for miR-21 were <10 for low expression and >10 for high expression; the cutoff values for p53 were <3 for low expression and >3 for high expression.
Sanger Sequencing
We randomly selected 100 out of 200 samples to prepare for sequencing of p53 mutant sites. Each sample was weighed as 100 mg before DNA extraction to ensure enough tissue to be processed. DNA was isolated by a Thermo Fisher Scientific tissue extraction kit recommendations. Samples were re-suspended in buffer GB for further purification and PCR. The purified reagent was added in the proportion of 5 μL of PCR product plus 1.5 μL of ExoSAP-IT reagent (Amersham Biosciences, USA). The purified enzyme could be inactivated at 37°C for 30 min and heated to 80°C for 15 min.
After DNA purity and concentration were determined, template DNA, primer sequences, and Extender PCR-to-gel master mix (2×) (Amresco, USA) were mixed together for the PCR cycle. PCR products were then analyzed by gel electrophoresis, purified, and then sent for Sanger sequencing by an ABI 3730 DNA analyzer (Applied Biosystems, USA). Sequencing results were divided into base sequence data and a base peak map. Data of the base sequence were analyzed by NCBI BLAST database software (https://blast.ncbi.nlm.nih.gov/Blast.cgi), and the base peak map was analyzed with Chromas 2.2.6 software, which finally synthesized results into specific conclusions.
Cell Lines and Culture
Human NSCLC A549 and H1650 cell lines were originally obtained from The Institute of Cell Biology, Shanghai Jiaotong University, and cultured in Roswell Park Memorial Institute Medium (RPMI) 1640 supplemented with 10% fetal bovine serum (FBS; Invitrogen), 100 U/mL penicillin, and 100 mg/mL streptomycin (Invitrogen) in a humidified incubator with 5% CO2 at 37°C.
miR-21 Inhibitor and Cell Transfection
miR-21 ASOs (5′-UCAACAUCAGUCUGAUAAGCUA-3′) and the negative control ASOs (5′-CAUUAAUGUCGGACAACUCAAU-3′) were designed using online tools (Primer 5.0) and synthesized by Sangon (Shanghai, China). For cell transfection, NSCLC cells were grown in a six-well plate overnight and then transfected with 100 pmol per well of miR-21 antisense or the negative control oligonucleotides using Lipofectamine 2000 (Invitrogen) for 24 h. Thereafter, the cells were used for different assays (see below).
Cell Viability Assay
NSCLC cell lines were grown and transfected with 100 pmol per well of either the miR-21 ASOs or the control ASOs for 48 h and cells were re-seeded into a 96-well plate at a density of 1 × 104 cells/well and grown for 48 h. We then added 20 μL of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; 5 mg/mL; Sigma-Aldrich, St. Louis, MO, USA) into each well, and the plate was incubated for an additional 4 h. At the end of each experiment, the cell culture medium was replaced with 150 μL of dimethyl sulfoxide (DMSO) per well and then thoroughly mixed. The absorbance rate of the cells was measured at 490 nm using a spectrophotometer (Mapada, Shanghai, China). Experiments were conducted in triplicate and repeated at least three times.
Flow Cytometry Assay
Cells were grown and transfected with miR-21 ASOs or control ASOs for 48 h. After that, the cells were washed twice with ice-cold phosphate-buffered saline (PBS) and fixed overnight with 70% ethanol at 4°C. On the next day, the cells were digested with RNase and then stained with a propidium iodide (PI) (Biotechnology, Shanghai, USA) solution, and cell cycle distribution was analyzed with the BD LSR II flow cytometry system (BD Biosciences, Franklin Lakes, NJ, USA). Experiments were conducted in duplicate and repeated at least twice.
Western Blot Assay
Cells were grown and transfected with miR-21 ASOs or negative control ASOs for 48 h and then collected and lysed in radioimmunoprecipitation assay (RIPA) buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% Nonidet P-40 (NP-40), 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, and the proteinase and phosphatase inhibitor cocktails (Sigma-Aldrich). After quantification, 20 μg of the proteins samples as loaded onto 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels for separation and then electronically transferred onto a polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA, USA). For western blot analysis, the membranes were blocked in 5% skim milk solution for 2 h at room temperature and then incubated with a primary antibody at 4°C overnight. The primary antibodies were rabbit anti-p53 (catalog no. SAB4503018-100UG), mouse anti-phosphorylated p53 at Ser15 (catalog no. 4030S; Cell Signaling Technology, Shanghai, China), and mouse anti-GAPDH (Bioss, Woburn, MA, USA) at a dilution of 1:100. After that, the membranes were washed with Tris-buffered saline (TBS) and then incubated with a horseradish peroxidase (HRP)-conjugated secondary antibody (Tissue Engineering Laboratory, Shanghai,China) at room temperature for 1 h. After three brief washes with TBS with Tween 20, the blots were incubated briefly with an enhanced chemiluminescence substrate (PerkinElmer, MA, USA) and exposed to X-ray films to detect positive protein signals.
Statistical Analysis
We performed a Student’s t test and χ2 test to assess statistical significance for association of miR-21 or p53 levels with clinicopathological data and Kaplan-Meier curves for association of miR-21 or p53 levels with OS. All tests were two-sided, and a p value less than or equal to 0.05 was set as statistical significance. All statistical analyses were performed using SPSS 19.0 software (IBM, Armonk, NY, USA).
Author Contributions
Y. Zhou designed the experiments and reviewed and edited the paper; Y. Zhou and D.G. conducted the experiments; Y. Zhou wrote the original draft; and Y. Zhou and Y. Zhang performed the data analysis.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81902310).
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA Cancer J. Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Clancy L. Reducing lung cancer and other tobacco-related cancers in Europe: smoking cessation is the key. Oncologist. 2014;19:16–20. doi: 10.1634/theoncologist.2013-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vachani A., Sequist L.V., Spira A. AJRCCM: 100-year anniversary. The shifting landscape for lung cancer: past, present, and future. Am. J. Respir. Crit. Care Med. 2017;195:1150–1160. doi: 10.1164/rccm.201702-0433CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 5.Ha D., Choi H., Chevalier C., Zell K., Wang X.F., Mazzone P.J. Survival in patients with metachronous second primary lung cancer. Ann. Am. Thorac. Soc. 2015;12:79–84. doi: 10.1513/AnnalsATS.201406-261OC. [DOI] [PubMed] [Google Scholar]
- 6.Ameres S.L., Zamore P.D. Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Biol. 2013;14:475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- 7.Takamizawa J., Konishi H., Yanagisawa K., Tomida S., Osada H., Endoh H., Harano T., Yatabe Y., Nagino M., Nimura Y. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 8.Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 9.Yanaihara N., Caplen N., Bowman E., Seike M., Kumamoto K., Yi M., Stephens R.M., Okamoto A., Yokota J., Tanaka T. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 10.Yang J.S., Li B.J., Lu H.W., Chen Y., Lu C., Zhu R.X., Liu S.H., Yi Q.T., Li J., Song C.H. Serum miR-152, miR-148a, miR-148b, and miR-21 as novel biomarkers in non-small cell lung cancer screening. Tumour Biol. 2015;36:3035–3042. doi: 10.1007/s13277-014-2938-1. [DOI] [PubMed] [Google Scholar]
- 11.Yaodong Zhou., Bo Sheng. Association of microRNA-21 with biological features and prognosis of neuroblastoma. Cancer Control. 2016;23:78–84. doi: 10.1177/107327481602300113. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y., Gao X., Wei F., Zhang X., Yu J., Zhao H., Sun Q., Yan F., Yan C., Li H., Ren X. Diagnostic and prognostic value of circulating miR-21 for cancer: a systematic review and meta-analysis. Gene. 2014;533:389–397. doi: 10.1016/j.gene.2013.09.038. [DOI] [PubMed] [Google Scholar]
- 13.Zhou Y., Sheng B., Xia Q., Guan X., Zhang Y. Association of long non-coding RNA H19 and microRNA-21 expression with the biological features and prognosis of non-small cell lung cancer. Cancer Gene Ther. 2017;24:317–324. doi: 10.1038/cgt.2017.20. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J., Zhang C., Hu L., He Y., Shi Z., Tang S., Chen Y. Abnormal expression of miR-21 and miR-95 in cancer stem-like cells is associated with radioresistance of lung cancer. Cancer Invest. 2015;33:165–171. doi: 10.3109/07357907.2015.1019676. [DOI] [PubMed] [Google Scholar]
- 15.Kotler E., Shani O., Goldfeld G., Lotan-Pompan M., Tarcic O., Gershoni A., Hopf T.A., Marks D.S., Oren M., Segal E. A systematic p53 mutation library links differential functional impact to cancer mutation pattern and evolutionary conservation. Mol. Cell. 2018;71:178–190.e8. doi: 10.1016/j.molcel.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 16.Román-Rosales A.A., García-Villa E., Herrera L.A., Gariglio P., Díaz-Chávez J. Mutant p53 gain of function induces HER2 over-expression in cancer cells. BMC Cancer. 2018;18:709. doi: 10.1186/s12885-018-4613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denissenko M.F., Pao A., Tang M., Pfeifer G.P. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science. 1996;274:430–432. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]
- 18.Fortunato O., Boeri M., Moro M., Verri C., Mensah M., Conte D., Caleca L., Roz L., Pastorino U., Sozzi G. Mir-660 is downregulated in lung cancer patients and its replacement inhibits lung tumorigenesis by targeting MDM2-p53 interaction. Cell Death Dis. 2014;5:e1564. doi: 10.1038/cddis.2014.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krasnov G.S., Puzanov G.A., Kudryavtseva A.V., Dmitriev A.A., Beniaminov A.D., Kondratieva T.T., Senchenko V.N. [Differential expression of an ensemble of the key genes involved in cell-cycle regulation in lung cancer] Mol. Biol. (Mosk.) 2017;51:849–856. doi: 10.7868/S0026898417050135. [DOI] [PubMed] [Google Scholar]
- 20.Hollstein M., Sidransky D., Vogelstein B., Harris C.C. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 21.Afanasyeva E.A., Mestdagh P., Kumps C., Vandesompele J., Ehemann V., Theissen J., Fischer M., Zapatka M., Brors B., Savelyeva L. MicroRNA miR-885-5p targets CDK2 and MCM5, activates p53 and inhibits proliferation and survival. Cell Death Differ. 2011;18:974–984. doi: 10.1038/cdd.2010.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar M., Lu Z., Takwi A.A., Chen W., Callander N.S., Ramos K.S., Young K.H., Li Y. Negative regulation of the tumor suppressor p53 gene by microRNAs. Oncogene. 2011;30:843–853. doi: 10.1038/onc.2010.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He L., He X., Lim L.P., de Stanchina E., Xuan Z., Liang Y., Xue W., Zender L., Magnus J., Ridzon D. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papagiannakopoulos T., Shapiro A., Kosik K.S. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68:8164–8172. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 25.Jing C., Cao H., Qin X., Yu S., Wu J., Wang Z., Ma R., Feng J. Exosome-mediated gefitinib resistance in lung cancer HCC827 cells via delivery of miR-21. Oncol. Lett. 2018;15:9811–9817. doi: 10.3892/ol.2018.8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su C., Cheng X., Li Y., Han Y., Song X., Yu D., Cao X., Liu Z. miR-21 improves invasion and migration of drug-resistant lung adenocarcinoma cancer cell and transformation of EMT through targeting HBP1. Cancer Med. 2018;7:2485–2503. doi: 10.1002/cam4.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen K.H., Hung J.H., Chang C.W., Weng Y.T., Wu M.J., Chen P.S. Solasodine inhibits invasion of human lung cancer cell through downregulation of miR-21 and MMPs expression. Chem. Biol. Interact. 2017;268:129–135. doi: 10.1016/j.cbi.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 28.Lu L., Xu H., Yang P., Xue J., Chen C., Sun Q., Yang Q., Lu J., Shi A., Liu Q. Involvement of HIF-1α-regulated miR-21, acting via the Akt/NF-κB pathway, in malignant transformation of HBE cells induced by cigarette smoke extract. Toxicol. Lett. 2018;289:14–21. doi: 10.1016/j.toxlet.2018.02.027. [DOI] [PubMed] [Google Scholar]
- 29.Dai Q., Li N., Zhou X. Increased miR-21a provides metabolic advantages through suppression of FBP1 expression in non-small cell lung cancer cells. Am. J. Cancer Res. 2017;7:2121–2130. [PMC free article] [PubMed] [Google Scholar]
- 30.Markou A., Zavridou M., Lianidou E.S. miRNA-21 as a novel therapeutic target in lung cancer. Lung Cancer (Auckl.) 2016;7:19–27. doi: 10.2147/LCTT.S60341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H., Mao F., Shen T., Luo Q., Ding Z., Qian L., Huang J. Plasma miR-145, miR-20a, miR-21 and miR-223 as novel biomarkers for screening early-stage non-small cell lung cancer. Oncol. Lett. 2017;13:669–676. doi: 10.3892/ol.2016.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong J., Zhang Z., Gu T., Xu S.F., Dong L.X., Li X., Fu B.H., Fu Z.Z. The role of microRNA-21 in predicting brain metastases from non-small cell lung cancer. OncoTargets Ther. 2016;10:185–194. doi: 10.2147/OTT.S116619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rufini A., Tucci P., Celardo I., Melino G. Senescence and aging: the critical roles of p53. Oncogene. 2013;32:5129–5143. doi: 10.1038/onc.2012.640. [DOI] [PubMed] [Google Scholar]
- 34.Shipman R., Schraml P., Colombi M., Raefle G., Dalquen P., Ludwig C. Frequent TP53 gene alterations (mutation, allelic loss, nuclear accumulation) in primary non-small cell lung cancer. Eur. J. Cancer. 1996;32A:335–341. doi: 10.1016/0959-8049(95)00535-8. [DOI] [PubMed] [Google Scholar]
- 35.Bosari S., Viale G., Bossi P., Maggioni M., Coggi G., Murray J.J., Lee A.K. Cytoplasmic accumulation of p53 protein: an independent prognostic indicator in colorectal adenocarcinomas. J. Natl. Cancer Inst. 1994;86:681–687. doi: 10.1093/jnci/86.9.681. [DOI] [PubMed] [Google Scholar]
- 36.Dashzeveg N., Taira N., Lu Z.-G., Kimura J., Yoshida K. Palmdelphin, a novel target of p53 with Ser46 phosphorylation, controls cell death in response to DNA damage. Cell Death Dis. 2014;5:e1221. doi: 10.1038/cddis.2014.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamińska I., Bar J.K. The association between p53 protein phosphorylation at serine 15, serine 20 and sensitivity of cells isolated from patients with ovarian cancer and cell lines to chemotherapy in in vitro study. Pharmacol. Rep. 2018;70:570–576. doi: 10.1016/j.pharep.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 38.Loughery J., Cox M., Smith L.M., Meek D.W. Critical role for p53-serine 15 phosphorylation in stimulating transactivation at p53-responsive promoters. Nucleic Acids Res. 2014;42:7666–7680. doi: 10.1093/nar/gku501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rami-Porta R., Bolejack V., Crowley J., Ball D., Kim J., Lyons G., Rice T., Suzuki K., Thomas C.F., Jr., Travis W.D. The IASLC lung cancer staging project: proposals for the revisions of the T descriptors in the forthcoming eighth edition of the TNM classification for lung cancer. J. Thorac. Oncol. 2015;10:990–1003. doi: 10.1097/JTO.0000000000000559. [DOI] [PubMed] [Google Scholar]