Abstract
Parasites are strictly associated with their hosts and present a great diversity of life histories, often resulting in different diversity patterns than those observed in free-living species. However, ecological approaches have detected that, in some cases, mammal-associated helminths respond similarly to non-parasitic species in terms of diversity patterns. Using 2200 recorded interactions, we analysed the diversity patterns of helminths (Acanthocephala, Nematoda and Platyhelminthes) harbored by humans, wild and domestic mammals of Mexico, depending on latitude, host body mass and trophic guild (carnivore, herbivore, insectivore, omnivore), considering helminth richness and average taxonomic distinctness, and host phylogenetic diversity and phylogenetic clustering. Latitude was positively correlated with the average taxonomic distinctness encompassing the three parasite phyla and nematodes. Northern latitudes had less taxonomically related parasite assemblages. Host body mass had a significant negative relationship with the average taxonomic distinctness of acanthocephalans and the richness of helminths associated to wild hosts. The omnivore hosts had greater parasite richness, while insectivores had a less taxonomically related parasite assemblage and herbivores had a more heterogeneous parasite assemblage. Our results highlight the importance of incorporating different dimensions of diversity, such as average taxonomic distinctness and to consider the composition of parasite assemblages to better understand their diversity patterns.
Keywords: Helminth, Mammals, Richness, Phylogenetic diversity, Taxonomic distinctness
Graphical abstract
Highlights
-
•
Four diversity measures were used to describe diversity patterns of helminths.
-
•
Latitude was positively correlated with helminth average taxonomic distinctness.
-
•
Host body mass was negatively related with the helminth richness of wildlife hosts.
-
•
Helminth sets of omnivore hosts were richer in parasite species.
-
•
Helminth sets of insectivore hosts had a wider taxonomic breadth.
1. Introduction
Parasites are not distributed homogeneously in space, in time, or in their hosts and despite representing a significant portion of Earth's biodiversity, key determinants influencing parasite diversity are still not well understood. Macroecological approximations have been used to attempt to explain how patterns of parasite diversity are related to ecological, evolutionary, historical, and geographical processes (Dallas et al., 2018, 2019; Kamiya et al., 2014a; Huang et al., 2015), and in some cases these studies have revealed that parasite diversity follows patterns similar to those found in non-parasitic wild species (Morand, 2015; Stephens et al., 2016).
Particularly in mammal-helminth associations, the geographical range size and population density of mammals have been positively related to the richness of parasites (Torres et al., 2006). At the same time, the latitudinal diversity gradient (LDG), a well-documented pattern of decreasing diversity with increasing latitude in several taxonomic groups, seems to be reversed (Lindenfors et al., 2007; Kamiya et al., 2014b) or nonexistent (Nunn et al., 2005; Bordes et al., 2010) when investigated in mammal-associated helminths. Host body mass also seems to influence parasite richness. Larger hosts are expected to harbor more parasites because they represent a larger “patch” of habitat, with which various parasites can be associated, and tend to have a greater probability of parasite exposure due to a higher resource consumption and a more extensive habitat range (Poulin, 1995; Ezenwa et al., 2006; Huang et al., 2015). Several authors have found a positive relationship between mammal body sizes and helminth richness (Vitone et al., 2004; Ezenwa et al., 2006; Lindefors et al., 2007; Kamiya et al., 2014b; Huang et al., 2015), although others have not (Morand and Poulin, 1998; Nunn et al., 2003; Dallas et al., 2018; Preisser, 2019).
Host diet also could play a key role in shaping helminth diversity and communities (Monello and Gompper, 2011; Locke et al., 2014; Gutiérrez et al., 2017). This is expected since many helminths require trophic transmission, and are acquired through the ingestion of infective larval stages, either directly or incidentally from the environment or from infected prey serving as intermediate or paratenic hosts (Lafferty, 1999; Poulin and Morand, 2004). Furthermore, species with a broader diet breadth are expected to harbor more parasites than hosts with a more specialized diet (Gutiérrez et al., 2019; Leung and Koprivnikar, 2019). In primate hosts diet has been found to be positively related with helminth and nematode richness (Vitone et al., 2004). This trait has also been associated to greater richness of cestodes in cricetid rodents (Preisser, 2019). However, Poulin (1995) and Nunn et al. (2003) reported no significant relationship between host diet and helminth richness in mammals.
Aspects of study design could obscure or modify the influence of predictors like latitude, host body mass and diet on parasite diversity. The geographical scale of the study (regional, continental, global) can influence the effect the predictors have on parasite richness (Kamiya et al., 2014b),similarly, lack of control in sampling effort and host phylogenetic relationships, might generate inflated estimates or mask these effects (Morand and Poulin, 1998; Nunn et al., 2003; Vitone et al., 2004; Krasnov et al., 2008b; Garrido-Olvera et al., 2012). Parasites are subject to environmental restrictions, the variation and availability of definitive or intermediate hosts across sites, which could extend or restrict their spatial distribution (Bozick and Real, 2015). Futhermore, other host traits like longevity, host density and social group behavior can also be determinants in the variation of parasite richness among hosts and act as covariables; making difficult to discern the true effect on the relationships between these predictors and parasite richness, as well as, producing different diversity patterns than those of non-parasitic organisms (Vitone et al., 2004; Nunn et al., 2003; Cooper et al., 2012; Violante-González et al., 2012).
A further potential limitation in studies of parasite diversity is the fact that studies on species diversity usually use species richness (S) as a measure. This measure is limiting because it considers all species to be equally distinct, and does not allow the incorporation of data about their ecological and evolutionary identity, or attempt to understand the structure of species assemblages more broadly (Purvis and Hector, 2000; Marrugan, 2004; Poulin, 2004; Gómez-Ortiz et al., 2017). For these reasons, the use of measures such as average taxonomic distinctness, phylogenetic and functional diversity, is currently increasing, providing information about processes or patterns that may have been overlooked (Krasnov et al., 2008a, 2019; Poulin and Leung, 2011; Rasmussen and Randhawa, 2018; Fecchio et al., 2020).
Taxonomic distinctness of parasites has been used as a complementary measure to parasite richness in studies about determinants of parasite diversity and have been shown to provide insight on how parasite assemblages are formed (Luque et al., 2004; Randhawa and Poulin, 2010; Rasmussen and Randhawa, 2018). A narrow taxonomic range in a parasite assemblage could be associated with intra-host speciation or colonization by a limited taxonomic spectrum of parasites. In contrast, when parasites in an assemblage are completely unrelated, the assemblage was likely formed by repeated colonisations by a broad taxonomic range of parasites. This allows the identification of features influencing the total number of parasites harboured by a host species, and the variety of parasite taxa a host will has; thus providing different explanations of the determinants affecting parasite biodiversity (Luque et al., 2004; Quiroz-Martínez and Salgado-Maldonado, 2013; Rasmussen and Randhawa, 2018).
Dimensions of diversity like phylogenetic or functional diversity have been used to understand the ecological mechanisms behind the correlation between host and parasite species richness. The positive correlation between these variables is frequently assumed to emerge because communities with higher host diversity offer additional niches for different parasites. Halliday et al. (2019) proposed that this relationship depends on how host communities assemble, and that shifts in host phylogenetic diversity during community assemblage can alter the relationship between host and parasite richness. More distantly related hosts are less likely to share parasites and communities with higher host phylogenetic diversity may harbor more parasite species. Nevertheless, a higher host phylogenetic diversity can also impede parasite transmission reducing parasite richness and weaken the relationship between host and parasite richness (Halliday et al., 2019, 2020).
In Mexico, 564 species of mammals have been recorded (Gómez-Ortiz et al., 2017), but less than 25% of these mammal species have been examined for helminths (García-Prieto et al., 2012). This percentage reflects the lack of knowledge on helminth richness associated with different groups of mammals. In addition, the close contact between humans, wildlife and domestic species generated by the change in land use, the movement of hosts for livestock activities, and the introduction of non-native species provide opportunities for parasitic helminths to associate and interact with new hosts. (Clark et al., 2018; Wells et al., 2019). This set of factors could alter the biological patterns and processes of associations between parasite and host (Kutz et al., 2005; Kennedy, 2006; Wells et al., 2018; Fernandez, 2019). In this study, we aim to describe the richness, average taxonomic distinctness, host phylogenetic diversity and host phylogenetic clustering of helminths associated with humans, wild and domestic mammals considering latitude, host body mass and trophic guild.
2. Material and methods
2.1. Database construction
A database was constructed using the taxonomic checklist of helminths associated with wild mammals in Mexico published by García-Prieto et al. (2012). This database was updated by gathering records published up until 2017 using search tools including the ISI Web of Science, CAB Abstracts, Biological Abstracts and Google Scholar with the key words: “mammal*“, “helminth” and “Mexico”. Information on records of helminths in humans and domestic animals was obtained from a bibliographic review (Lamothe-Argumedo and García-Prieto, 1988; Quiroz Romero, 1987) and electronic resources generated by the Parasitology Department of the School of Veterinary Medicine at UNAM and other departments at the UNAM (Alcalá et al., 2008; https://datosabiertosunam.mx/; http://oreon.dgbiblio.unam.mx/). Complementary to this review, we searched the ISI Web of Science (https://apps.webofknowledge.com) using the keywords “dog”, “cat”, “cattle”, “goat”, “sheep”, “rabbit”, “horse”, and “donkey” combined with the words “helminth” and “Mexico".
Each record in the database represented the detection of a parasite in a mammal species and contained information on the hosts (species, family) and parasites (species, family, class, order, phylum), as well as their geographical location (state, locality, geographic coordinates), and their bibliographic reference. In the records where the latitude and longitude were missing, we used the geographic coordinates provided by Google maps after entering the name of the locality and the state described for that record.
Helminths were classified into three categories: associated with wildlife mammals (W), associated with domestic mammals (D), and associated with both (D-W), based on the type of host in which the helminth species has been recorded in the literature. Since humans and domestic animals live in close proximity of each other, inhabit almost all terrestrial environments and the subsets of parasites they share increase the longer the animal species has been domesticated (Morand et al., 2014; Wells et al., 2018), we included Homo sapiens in the domestic animals category. Synanthropic rodents were also classified within this group, because despite not going through a process of direct domestication their frequent contact and long-time commensal relationships with humans, has selected traits that differentiate them from wild rodents, and have favored their adaptation to anthropogenic environments (Weissbrod et al., 2017; Geiger et al., 2018). The domestic mammals included were: cow (Bos taurus), dog (Canis familiaris), goat (Capra hircus), donkey (Equus asinus), horse (Equus caballus), cat (Felis catus), human (Homo sapiens), mouse (Mus musculus), rabbit (Oryctolagus cuniculus), sheep (Ovis aries), brown rat (Rattus norvegicus), black rat (Rattus rattus), and pig (Sus scrofa).
2.2. Correcting sampling bias
The inventory of mammal-associated helminths distributed in the Mexican territory is far from complete; the sampling effort has been spotty and heterogeneous among wildlife hosts and geographic areas. In general, more intensely studied host species like domestic animals would be expected to have more parasite species recorded in the literature simply due to sampling effort, generating potential bias in helminth records and interactions. To address this problem, following the values of average helminth richness per mammal species presented by Dobson et al. (2008), we selected mammals with a minimum of 5 species of helminths recorded. Then, to account for sampling bias, we followed the methodology of several authors (Poulin, 1995; Morand and Poulin, 1998; Vitone et al., 2004; Rasmussen and Randhawa, 2018) to adjust diversity measure values (i.e. richness, average taxonomic distinctness) by sampling effort (i.e. number of publications). We did a linear or simple quadratic regression of the diversity measure as a function of the sampling effort (Rasmussen and Randhawa, 2018) and if significant (P < 0.05), used the residuals of the regression in place of the original diversity measure to correct for the influence of sampling effort in further analysis. The regressions were calculated using the lm() function implemented in the stats package in R version 3.5.0 (R Core Team, 2018). The results of these regressions are shown in Table 1 of the Supplementary Material.
Table 1.
Correlations between the latitude, parasite richness (S), host phylogenetic diversity (PD), host phylogenetic clustering (PC) and parasite average taxonomic distinctness (Δ+) (dependent variables were controlled for number of publications, before calculating the correlations).
| Measure | Correlation | p-value | |
|---|---|---|---|
| Total helminth | S | r = −0.29 | 0.21 |
| PD | r = −0.47 | 0.04* | |
| PC | r = −0.20 | 0.41 | |
| Δ+ | r = 0.52 | 0.02* | |
| Acanthocephala | S | r = −0.07 | 0.85 |
| PD | r = −0.44 | 0.27 | |
| PC | r = 0.28 | 0.49 | |
| Δ+ | r = 0.6 | 0.35 | |
| Nematoda | S | r = −0.31 | 0.18 |
| PD | r = −0.45 | 0.05 | |
| PC | r = −0.38 | 0.10 | |
| Δ+ | r = 0.50 | 0.02* | |
| Platyhelminthes | S | r = −0.15 | 0.55 |
| PD | r = −0.36 | 0.14 | |
| PC | rs = −0.11 | 0.64 | |
| Δ+ | r = −0.26 | 0.37 | |
| Associated with domestic animals | S | r = −0.09 | 0.83 |
| PD | r = −0.42 | 0.33 | |
| PC | r = 0.41 | 0.35 | |
| Δ+ | r = 0.90 | 0.09 | |
| Associated with both domestic and wildlife hosts (D–W) | S | rs = −0.18 | 0.45 |
| PD | r = −0.32 | 0.17 | |
| PC | r = −0.31 | 0.19 | |
| Δ+ | r = −0.02 | 0.92 | |
| Associated with wildlife hosts (W) | S | rs = −0.16 | 0.61 |
| PD | r = −0.18 | 0.56 | |
| PC | r = 0.52 | 0.08 | |
| Δ+ | r = 0.07 | 0.81 |
2.3. Diversity measures
Four main dependent variables related to patterns of helminth and host diversity were analysed with respect to latitude, host body mass, and trophic guild: richness (S), host phylogenetic diversity (PD), host phylogenetic clustering (PC), and average taxonomic distinctness (Δ+). For latitude, using the geographic coordinates of the records we created a new column in the database containing the degree of latitude value without considering the decimals (i.e. 14⁰, 15⁰) as a classificatory variable. From the Phylacine and MOM v4.1 databases (Faurby et al., 2018; Smith et al., 2003) we compiled data on body mass (g), and consumed percentage of plants, vertebrates and invertebrates for each mammal host.
According to Faurvy et al. (2018) the values on host body mass were obtained in several ways. Values obtained from Smith et al. (2003) where species body mass was compiled taking into account these considerations: a) For gender-specific estimates obtained by averaging male or female values reported from across different geographic localities, these were then averaged to obtain an overall species value, b) When no geographic information was provided but gender-specific values were available, these were averaged for an overall species body mass estimate, c) If both male and female masses were not available, male mass was used preferentially, averaging over geographic localities if such information was available, d) If only female body mass was available, it was used, averaging over geographic localities as available. Values obtained from Faurby and Svenning (2016) were compiled from separate data sources. Lastly, for species for which body mass was not recorded, the assumption of isometries for related similar-size species and phylogenetic imputation were used to assign the value (Faurby et al., 2018; Faurby and Svenning, 2016).
Likewise the percentage on the diet categories, were obtained from other data sources and phylogenetic imputation (Faurby et al., 2018). These percentages were used to create a column in which we classified mammal-hosts into four trophic guilds: herbivores (species consuming 100% of plants), carnivores (species consuming 100% of vertebrates), insectivores (species consuming 100% of invertebrates) and omnivores (species consuming two or more diet categories).
Parasite richness (S) was measured as the total number of parasite species. Host phylogenetic diversity (PD) was the total sum of the branch lengths between host species present within a grouping variable (i.e. parasite phylum, type of host with which the helminth is associated) (Faith, 1992), and was measured using the pd() function implemented in the picante package (Kembel et al., 2010) in R (Team, 2018). The function ses.pd() or standardized effect size of phylogenetic diversity (SES.PD) from the picante package (Kembel et al., 2010) was used to calculate the phylogenetic clustering (PC). We generated 999 null model randomizations using the null model “taxa.labels”, where taxa labels of all taxa in the phylogeny are repeatedly shuffled in order to assess whether phylogenetic diversity is high or low for a group of hosts (Poulin et al., 2011). Negative SES. PD values and low quantiles (P < 0.05) indicate that host species within the group are more related than expected by chance, while positive values and high quantiles (P > 0.95) indicate overdispersion or the co-occurrence of distantly related hosts within the group (Poulin et al., 2011; Walker et al., 2017). The calculation of host phylogenetic diversity and phylogenetic clustering were based on the phylogeny of mammals published by Faurvy et al. (2018).
The average taxonomic distinctness (Δ+), was calculated exclusively for helminths using the taxondive() function implemented in the vegan package in R (Oksanen et al., 2016). The average taxonomic distinctness (Δ+) arranges helminth species in a taxonomic hierarchy and calculates the average number of steps that two species within a group require to reach the same level in that hierarchy (Poulin and Mouillot, 2004). Step lengths are standardized so parasite species connected at the highest taxonomic level (Phylum) is set to be equal to 100, with six levels require to reach species, such that each steps accumulates a value equal to 16.66 (Poulin and Mouillot, 2004). The helminth hierarchy was obtained from the classifications published by Amin (2013) for acanthocephalans, Jones et al. (2005) for trematodes, Caira and Jensen (2017) for cestodes and Anderson et al. (1974–1985), Gibbons (2010) and Hodda (2011) for nematodes. The taxonomic categories recorded were: Species, Genus, Family, Order, Class and Phylum. The taxonomic distinctness is associated with a variance (Λ+), which measures the heterogeneity in the sets of parasites found in a host, since, it is possible to have two groups of hosts with the same Δ+ value but which differ in their taxonomic range (Poulin and Mouillot 2004; Quiroz-Martínez and Salgado-Maldonado, 2013).
2.4. Data analysis
The diversity patterns of parasites and hosts were explored using the previously mentioned measures, considering overall helminths, separating the groups of helminths at the taxonomic level of phylum (Acanthocephala, Nematoda and Platyhelminthes) and at the categories of the type of host with which the parasite was commonly associated (Domestic, Wildlife, Domestic-Wildlife). For latitude, we calculated the value of the dependent variables (parasite richness, host phylogenetic diversity, host phylogenetic clustering and average taxonomic distinctness) considering the subsets of species in the before mentioned groups of phyla and type of host with which the helminth associates at each latitude degree. The association between latitude and the dependent variables was measured using a Pearson or Spearman correlation (when the values of the dependent variable did not have a normal distribution) with the stats package (R Core Team, 2018).
For host body mass, we log-transformed the values to correct the skewed distribution of this variable (Garamszegi, 2014). Then, as phylogenetically close species tend to share common traits and are not independent observations (Gay et al., 2014; Poulin and Mouillot, 2004; Vitone et al., 2004). We computed a Phylogenetic Generalized Least Squares model (PGLS) with the caper package in R (Orme et al., 2018); where host body mass was the predictor for parasite richness and parasite average taxonomic distinctness. The phylogenetic information was included from the Phylacine mammal phylogenetic tree (Faurby et al., 2018). PGLS is an extension of general linear model which allows the incorporation of phylogenetic dependence in the error term. This term was conformed by a matrix of expected trait covariances calculated using the phylogeny and the maximum likelihood (ML) estimate of Pagel's lambda λ (Cooper et al., 2012; Garamszegi, 2014). Finally, to assess whether trophic guild of the host had a relationship with parasite richness and average taxonomic distinctness, we generated a PGLS using trophic guild as predictor variable and parasite richness and average taxonomic distinctness as response variables. As described in section 2.2, whenever there was a significant relationship between sampling effort and some diversity measures, we used residuals from that regression rather than raw diversity measures to account for sampling bias (Supplementary Material Table S2).
3. Results
The database contained 2403 records, consisting of 118 host species, 346 helminth species, ten acanthocephalans, 84 platyhelminths, and 252 nematodes. Of the 118 nominal host species, 41 complied with the minimum of five helminth species, leaving 2200 records with 250 helminths, ten acanthocephalans, 210 nematodes, and 71 platyhelminths. The mammal order Carnivora had the greatest richness of helminths, whereas Cingulata had the lowest parasite richness (Supplementary Material Table S1). The average number of parasites per host was 6.09, with the horse (Equus caballus) having the greatest parasite richness (40). Seventeen helminths were associated with domestic animals, 158 were associated with both domestic and wild mammals, and 116 were associated with wildlife only.
Host phylogenetic diversity was higher for nematodes (1844.03, 41 host species) than for platyhelminths (1634.34, 35 host species) or acanthocephalans (770.50, 10 host species). None of the three phyla had significant phylogenetic clustering (ses.pd = 0.85, p = 0.80 for nematodes; ses.pd = 0.30 for platyhelminths, p = 0.60 and ses.pd = 0.19, p = 0.59 for acanthocephalans). Acanthocephalans had a higher average taxonomic distinctness and variance (Δ+ = 86.16, Λ+ = 472.91) than nematodes (Δ+ = 78.53, Λ+ = 459.23) and platyhelminths (Δ+ = 77.91, Λ+ = 444.21). Helminths associated with domestic and wild mammals presented a larger host phylogenetic diversity (1665.65, 35 host species), than those associated with domestic (572.73, 8 host species) and wild (1520.37, 27 host species) mammals. Helminths associated with wild mammals had a higher average taxonomic distinctness (Δ+ = 80.72) than those associated with both domestic and wild mammals (Δ+ = 77.36) and domestic (Δ+ = 76.89) mammals. Helminths associated with both domestic and wild mammals had higher variance (Λ+ = 474.25) than helminths associated with domestic (Λ+ = 404.89) and wild (Λ+ = 431.72) mammals. None of the groups had significant phylogenetic clustering (Domestic-Wildlife: ses.pd = 0.56, p = 0.70, Wildlife: ses.pd = 1.24, p = 0.90, Domestic: ses.pd = −0.87, p = 0.19).
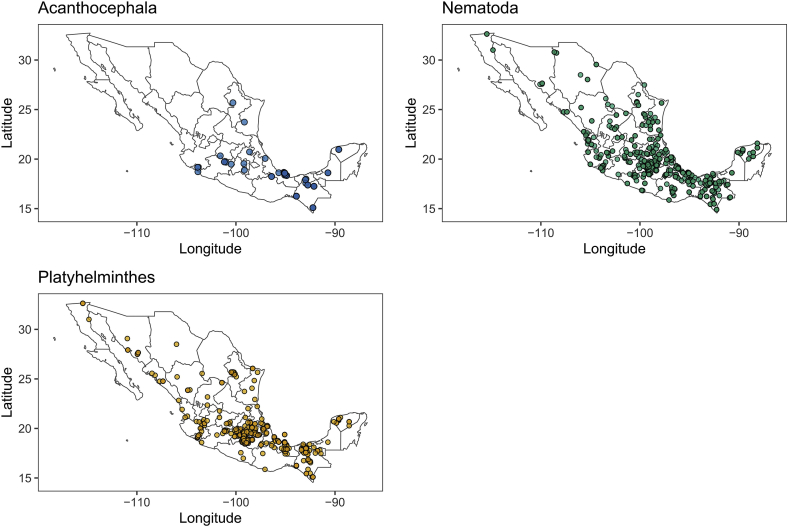
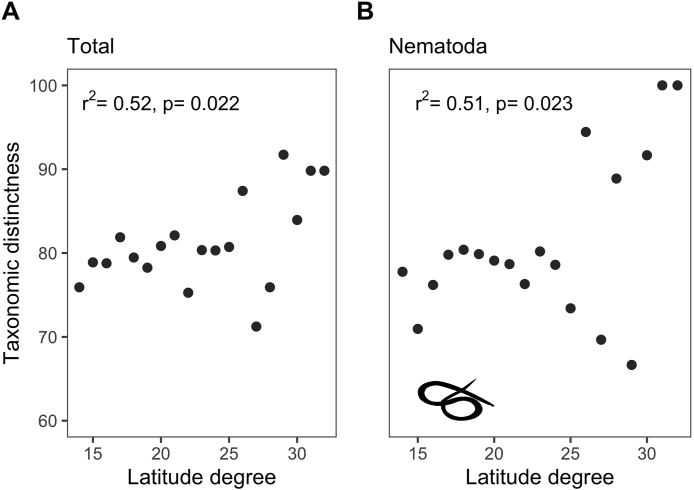
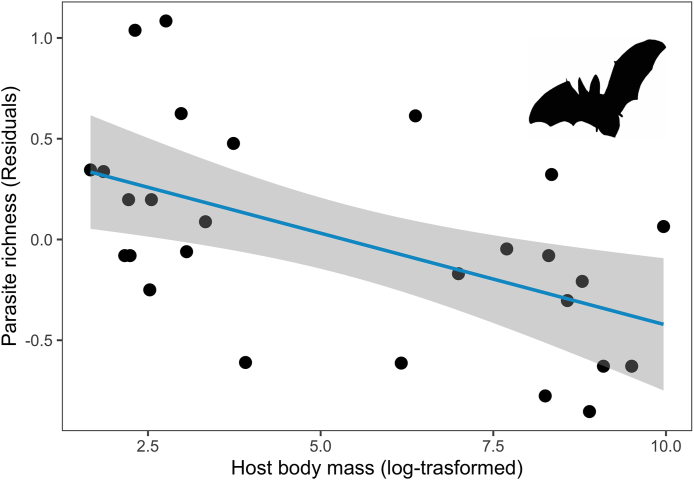
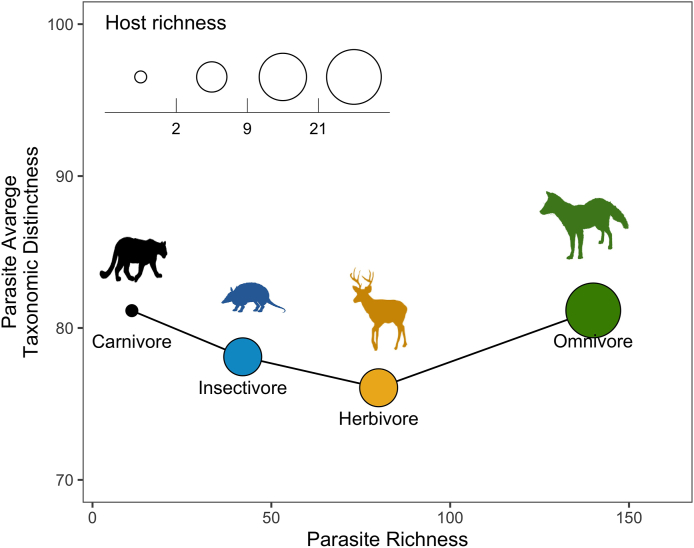
Only 1861 records of the 2200 filtered records reported geolocation coordinates (Fig. 1). Latitude was positively correlated with parasite and nematode average taxonomic distinctness (r = 0.52, p = 0.02; r = 0.51, p = 0.02 respectively) (Table 1) (Fig. 2A and B). Host body mass had a significant negative relationship with acanthocephalan average taxonomic distinctness (Table 2) and helminth associated to wildlife mammal richness (Fig. 3). Omnivore hosts had the greatest parasite richness among the trophic guilds (Fig. 4), but insectivores the higher average taxonomic distinctness (Δ+ = 79.73) and herbivores had the largest variance in average taxonomic distinctness (Λ+ = 486.12) (Table 3). There were no significant relationships between host trophic guild, parasite richness and average taxonomic distinctness (Table 4).
Fig. 1.
Maps showing the geographic locations of the records, classified by phylum of the subsetted database.
Fig. 2.
Relationships between latitude and the average taxonomic distinctness of overall helminths (A) and nematodes (B).
Table 2.
Phylogenetic generalized least squares model (PGLS) for host body mass (predictor), parasite richness (S) and parasite average taxonomic distinctness (Δ+) (response variables were controlled for number of publications and the predictor variable was log-transformed before calculating PGLS).
| Response | Estimate | λ | t | p-value | |
|---|---|---|---|---|---|
| Total helminth | S | −0.02 | 0.00 | −1.19 | 0.24 |
| Δ+ | 0.001 | 0.00 | 0.34 | 0.73 | |
| Acanthocephala | S | −0.06 | 1.00 | −1.10 | 0.38 |
| Δ+ | −0.05 | 1.00 | −5.57 | 0.03* | |
| Nematoda | S | 0.04 | 1.00 | 1.39 | 0.17 |
| Δ+ | −1.01 | 1.00 | −1.27 | 0.20 | |
| Platyhelminthes | S | 0.006 | 0.90 | 0.39 | 0.70 |
| Δ+ | 0.28 | 0.00 | 0.75 | 0.45 | |
| Associated with domestic hosts (D) | S | −0.007 | 0.00 | −0.07 | 0.94 |
| Δ+ | 0.09 | 0.00 | 0.78 | 0.47 | |
| Associated with both domestic and wildlife hosts (D–W) | S | 0.01 | 0.74 | 0.35 | 0.72 |
| Δ+ | 0.03 | 0.00 | −0.57 | 0.38 | |
| Associated with wildlife hosts (W) | S | −0.07 | 0.00 | −2.45 | 0.02* |
| Δ+ | −0.005 | 0.00 | −0.50 | 0.61 |
Fig. 3.
Phylogenetic generalized least squares (PGLS) regression of host body mass (values were log-transformed) with richness of helminths associated to wildlife hosts (values were corrected for sampling effort).
Fig. 4.
Parasite richness and average taxonomic distinctness by host trophic guild (the size of circle represents the number of hosts belonging to each trophic guild).
Table 3.
Host richness (SM), parasite richness (Sp), parasite average taxonomic distinctness (Δ+) and variance (Λ+) by mammal trophic guild.
| Trophic guild | Host richness (SM) | Parasite richness (SP) | Average taxonomic distinctness (Δ+) | Variance of average taxonomic distinctness (Λ+) |
|---|---|---|---|---|
| Herbivores | 9 | 103 | 76.17 | 486.12 |
| Carnivores | 3 | 21 | 77.04 | 375.76 |
| Insectivore | 9 | 43 | 79.73 | 449.45 |
| Omnivores | 21 | 155 | 79.64 | 437.52 |
Table 4.
Phylogenetic generalized least squares model (PGLS) for trophic guild (predictor), parasite richness (S) and parasite average taxonomic distinctness (Δ+) (response variables were controlled for number of publications before calculating PGLS).
| Response | Estimate | λ | t | p-value | |
|---|---|---|---|---|---|
| Carnivore-Herbivore | S | 0.17 | 1.00 | 0.14 | 0.88 |
| Δ+ | 3.11 | 1.00 | 0.26 | 0.78 | |
| Carnivore-Insectivore | S | 0.29 | 1.00 | 0.23 | 0.81 |
| Δ+ | 5.96 | 1.00 | 0.50 | 0.61 | |
| Carnivore-Omnivore | S | 0.11 | 1.00 | 0.09 | 0.92 |
| Δ+ | 3.10 | 1.00 | 0.28 | 0.77 | |
| Herbivore-Insectivore | S | 0.12 | 1.00 | 0.16 | 0.86 |
| Δ+ | 2.85 | 1.00 | 0.41 | 0.67 | |
| Herbivore-Omnivore | S | −0.06 | 1.00 | −0.12 | 0.90 |
| Δ+ | −0.00 | 1.00 | −0.00 | 0.99 | |
| Omnivore-Insectivore | S | 0.18 | 1.00 | 0.30 | 0.76 |
| Δ+ | 2.85 | 1.00 | 0.49 | 0.62 |
4. Discussion
The description of the factors involved in parasite diversity patterns is of paramount importance in explaining how their diversity is distributed and improves our knowledge of parasite-host interactions. Besides understanding how parasite diversity is related to different features, it is fundamental to comprehend why some host species harbor more parasite richness than others (Luque et al., 2004; Locke et al., 2014). Due to differential sampling effort directed towards them for their importance in public health and livestock activities, hosts orders containing domestic species like Carnivora and Cetartiodactyla had more parasite richness. Our analysis also showed that nematodes and platyhelminths had higher parasite richness and host phylogenetic diversity than acanthocephalans, understanding as phylogenetic diversity, the sum of branches length of each group host associated with these two phylas. Both platyhelminths and nematodes are abundant and speciose among vertebrate hosts; they are diverse in their biology and life histories and they have evolved parasitism independently multiple times (Viney, 2018; Weinstein and Kuris, 2016). Acanthocephala, on the other hand, is a small phylum that has undergone multiple radiations from aquatic to terrestrial habitats, presents conservatism of their intermediate host and is mainly harbored by fishes and birds (Kennedy, 2006; Near, 2002).
A low value of average taxonomic distinctness (Δ+) means that on average the parasites in a group (i.e. Phylum) are closely taxonomically related (Quiroz-Martínez and Salgado-Maldonado, 2013). Nematodes of the same clade frequently parasitize different hosts, and the presence of congeneric nematode species is common in mammals (i.e., flock species) (Kennedy and Bush, 1992; Poulin, 2004). Several platyhelminths families also show diversification within mammal groups (e.g Hymenolepididae, Anoplocephalidae) (Morand et al., 2005), which could result in the observed values of Δ+. Despite having the lowest parasite richness, the phylum Acanthocephala presented the largest value of average taxonomic distinctness and their variance (Λ+). High values of Λ+ are associated with different families giving origin to single species, resulting in more heterogeneous assemblages. Since all acanthocephalans require an intermediate host (potentially including paratenic hosts) (Kennedy, 2006), the presence and consumption of different intermediate hosts such as insects or amphipods could result in the heterogenous assemblages we see in this phylum (Kennedy, 2006; Spickett et al., 2017; Prati and Henriksen, 2020).
When considering host categories, helminths associated with wildlife (W) and both domestic and wild (D-W) hosts had a greater richness and host phylogenetic diversity than those that are associated only with domestic mammals (D). Helminths associated with wildlife included marsupials, bats and armadillos among their hosts. The inclusion of these mammal orders increases the branch length measured in host phylogenetic diversity, as these organisms are more distantly related that the other mammals in this group. As for the value of average taxonomic distinctness the incorporation of families of acanthocephalans like Plagiorynchidae and Oligacanthorhynchidae harbored in marsupials, primates and carnivorans, as well as the adding of trematode species from families like Anenterotrematidae and Phaneropsolidae for bats, Rhopalidae, and the cestode order Onchoproteocephalidea for marsupials increased the average taxonomic distinctness, generating a wider taxonomic range for the helminths associated to wildlife. Within the helminths associated with both domestic and wild mammals, although most of the wildlife hosts were phylogenetically close to some domestic mammals, we also included bats, marsupials and the armadillo (Dasypus novemcinctus) which increased their host phylogenetic diversity. Parasite recorded in these hosts like Ancylostoma braziliensis, Paragonimus mexicanus and Plagiorchis verpertilionis besides being found in domestic animals have also been found in humans (Lamothe-Argumedo and García-Prieto, 1988; Guk et al., 2007; Guillot et al., 2012).
The average taxonomic distinctness in the D-W category showed taxonomic relatedness among the parasites, with one parasite order (Rhabditida) comprising almost 50% of the parasite species in that category and parasite families like Strongylidae and Trichostrongylidae contributing several species. Both families contained a variety of mammalian nematodes that live in the gastrointestinal system of artiodactyl and perissodactyl hosts (Chilton et al., 2006). Finally, helminths associated with domestic hosts (D) had the lowest host phylogenetic diversity and average taxonomic distinctness, of the seventeen helminth species classified in this group almost 50% were comprising within two orders (Cyclophyllidea and Spirurida). The cestodes comprised within Cyclophyllidea were associated to human and rodents, similarly Spirurida was represented by nematodes harbored by human, rodents and ruminants. Furthermore, parasites in both groups (Domestic, Domestic and Wildlife) are of human, veterinary and economic importance. (Smout et al., 2017; Wells et al., 2018).
Our study registered different patterns of helminth diversity across a geographical and biological predictors. The latitudinal diversity gradient (LDG) has been described for different species of mammals, insects, fishes, and bacteria (Willig et al., 2003), and a similar pattern of diversity could be expected in parasites, since they depend on their hosts for their survival; however, this is not always the case (Kennedy, 2006; Kamiya et al., 2014b; Morand, 2015; Preisser, 2019). We did find a negative correlation between latitude and host phylogenetic diversity (rs = −0.47, p = 0.04), but we did not find a significant correlation between parasite richness and latitude. According to several authors, factors like temperature, precipitation and the capacity of some parasites to disperse across their host ranges could be involved in latitudinal patterns of helminth diversity rather than latitude per se, since these variables influence parasite stages occurring in the environment, the transmission to their hosts, the diversity and availability of intermediate hosts, resulting in different diversity patterns than those observed in their hosts (Lindenfors et al., 2007; Morand, 2015; Dallas et al., 2018; Preisser, 2019).
We did, however found a positive correlation between latitude and average taxonomic distinctness of helminths as a group and nematodes. Although assemblages at lower latitudes were richer in helminth species, they were congregated into families within the same order, while at higher latitudes the richness was lower, but parasites were more widely distributed among families and orders. The variation observed in the Δ+ of the parasite subsets may also be a result of the host species associated with them (Krasnov et al., 2008a); at lower latitudes hosts were mainly cetartiodactyls whereas, at higher latitudes species such as carnivorans and rodents were more frequent. However, is important to consider that parasites do not distribute completely or evenly across the geographic range of their hosts distribution (Cumming, 1999; Shenbrot et al., 2010); factors related to environmental conditions, competition among parasites, presence and distribution of the intermediate hosts could influence parasite species composition (Cumming, 2002; Shenbrot et al., 2010; Krasnov et al., 2010; Warburton et al., 2016). Furthermore, even though we consider sampling effort bias, most of records we used came from latitudes of 18⁰, 19⁰ and 20⁰, future studies should revisit these relationships when more data of higher latitudes have been collected.
Host body mass had a significant negative relationship with average taxonomic distinctness of acanthocephalans; meaning that as host body mass increases, parasite subsets for this phylum become more similar. Our corrected database contained ten acanthocephalan species distributed among eleven hosts, but only for four hosts it was possible to estimate the average taxonomic distinctness, since the remaining hosts had only one species of acanthocephalan (Clarke and Warwick, 2001); this relationship should be re-analised in future studies when more information on acanthocephalans in mammals is available, as small sample sizes could generate unreliable results (Garamszegi, 2014).
Contrary to the works of Ezenwa et al. (2006); Lindefors et al. (2007) and Kamiya et al. (2014b), we found a significant negative relationship between host body mass and the richness of helminth associated to wildlife mammals. Within this group more than a half of the hosts were rodents and bats. A study made by Dáttilo et al., in 2020 found a similar relationship between small-bodied mammals and parasite richness.The authors mentioned that differences in sampling effort, could lead to increase richness values, as small mammals like rodents and bats tended to be more well-sampled in comparison to other mammals. Host body mass has been noted as an important predictor of parasite richness. However, the strength of its effect may vary among taxonomic groups of hosts; for example, according to Kamiya et al. (2014b), the association between host body size and metazoan richness was significantly stronger in fishes than in birds and mammals. In addition, body size variation within host species can be linked to various processes that also influence parasite richness and their opportunity to find a host, such as host density, habitat use, and host life-history traits (e.g., longevity, terrestriality), such that host body size is not a universal covariate of parasite diversity (Dallas et al., 2018; Morand and Poulin, 1998).
The omnivore trophic guild presented the highest parasite richness, while parasites associated with insectivore hosts were less taxonomically related than the parasites in other trophic guilds, and herbivore hosts had the most heterogenous parasite species assemblages. According to Luque et al. (2004), certain host traits may favor the acquisition of a wider taxonomic spectrum of parasites, whereas the presence of other traits may result in the same number of parasite species but with narrower taxonomic spectrum. Due to the variability of their diets, omnivores have a higher probability of encountering infecting stages of parasites than species whose diets only include a few elements (Miller et al., 2009; Albuquerque et al., 2016; Leung and Koprivnikar, 2019). However, the specificity of the intermediate host that some helminths present (Locke et al., 2010; Kennedy, 2006; Near, 2002) and the consumption of several of these type of hosts as occurs in insectivore hosts could result in less taxonomically related assemblages. As for the variance of taxonomic distinctness, the high value presented in herbivores hosts suggests that one main branch in the taxonomic tree contributes proportionally more species than the others and provides information on particular taxa are over- or under-represented (Korallo et al., 2007; Milošević et al., 2012; Rasmussen and Randhawa, 2018). In the case of this trophic guild, two families (Trichostrongylidae, Strongylidae) accounted for nearly half of the parasites within this trophic guild. On average, herbivore hosts had higher parasite richness than insectivore hosts, but their taxonomic distinctness did not reflect this. This could happen because the route by which a host acquires the parasite could be limited to a type of resource (e.g. grass, leaves) and to several parasite species, but with a narrow taxonomic breadth (e.g. strongylids in horses, oxyurids in primates) (Vitone et al., 2004; Morariu et al., 2016). As for the rest of the guilds, in some cases trophically transmitted parasite families contributed several species (e.g. Phaneropsolidae for insectivores, Oligacanthorhynchidae for omnivores), while other families like (e.g. Ascaridae for carnivores or Molineidae for omnivores) also contributed various parasites but are not likely acquired through the diet. In the same way that other factors could be confounded with latitude and host body mass, future analysis should also consider other host traits like habitat use or lifestyle in addition to trophic guild, since they could have an important role in determining the composition of parasite assemblages (Morand et al., 2005; Leung and Koprivnikar, 2016, 2019).
Lastly, parasite richness among our hosts was highly variable, ranging from five to 40 species, whereas average taxonomic distinctness values were much less variable, even for species with many parasites like domestic mammals (Supplementary Material Fig. 1). In addition, this measure did not seem to be affected by sampling effort (Korallo et al., 2007; Poulin and Mouillot, 2004; Rasmussen and Randhawa, 2018). These characteristics make average taxonomic distinctness an interesting complementary measure to consider in future works looking to detect diversity patterns that cannot be observed when we only consider the raw number of species (Quiroz-Martínez and Salgado-Maldonado, 2013; Rasmussen and Randhawa, 2018).
4.1. Conclusions
Although it was possible to describe some patterns already reported in the literature, the results of these approaches should be taken with caution. In many wild mammals, only one helminth species was recorded, and there are still many mammal species that have not been sampled at all. Among our records, taxonomic orders containing domestic animals had more recorded helminths than other host species. Domestic animals are more frequently studied because parasites affect human and animal health and the economics of livestock activities (Walker and Morgan, 2014). Host species richness and sampling effort are often associated with parasite richness (Morand, 2015). These information biases could be obscuring patterns produced by factors measured in this study.
It is therefore vital to continue sampling and identifying parasites associated with mammals, allowing a description of the multiple interactions that these organisms have with their hosts. As parasites, helminths are an essential and abundant component of natural communities. According to Dobson et al. (2008), there are approximately 11,631 helminth species associated with the 4637 mammal species on the planet. As inventories of helminth species become more complete, it will be possible to more accurately identify the factors involved in the diversity patterns of these parasites.
In general, our analysis was enriched by the calculation of the average taxonomic distinctness, since this allowed us to explore the taxonomic composition of the parasite assemblages, facilitating the understanding of the diversity patterns we found in this work.
We consider that it is only through macroecological studies that address multiple dimensions of diversity that we can more accurately describe diversity patterns, the ecological traits involved in the composition of parasite assemblages in their hosts, and how current changes could alter them.
Declaration of competing interest
We warrant that none of the authors has any conflict of interest in regard to this manuscript.
Acknowledgments
We thank the Parasitology department of the UNAM School of Veterinary Medicine for their help and facilitation the review of their material and Dr. Cintli Martínez Ortiz de Montellano for her assistance and for sharing her data on helminths associated with domestic animals. Thanks to the Posgrado en Ciencias de la Producción de la Salud Animal programme of UNAM and to the Consejo Nacional de Ciencia y Tecnología (CONACyT) for the Master's Scholarship (Grant number: 482793)
The Project was funded by PAPIIT IN218020.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijppaw.2020.10.010.
Contributor Information
María del Carmen Villalobos-Segura, Email: macavs324@gmail.com.
Luis García-Prieto, Email: luis.garcia@ib.unam.mx.
Oscar Rico-Chávez, Email: orich@unam.mx.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
References
- Albuquerque A.C.A., Moraes M., Silva A.C., Lapera I.M., Tebaldi J.H., Lux Hoppe E.G. Helminth fauna of chiropterans in Amazonia: biological interactions between parasite and host. Parasitol. Res. 2016;115:3229–3237. doi: 10.1007/s00436-016-5085-3. [DOI] [PubMed] [Google Scholar]
- Alcalá J., Figueroa J.A., Alberti A.B., Posadas E., Cuéllar J.A. Facultad de Medicina Veterinaria y Zootecnia Universidad Nacional Autónoma de México; 2008. Base de datos sobre registros de parásitos de rumiantes en México.https://parasitosderumiantes.com.mx/ [Google Scholar]
- Amin O.A. Classification of the Acanthocephala. Folia Parasitol. 2013;60:273–305. doi: 10.14411/fp.2013.031. [DOI] [PubMed] [Google Scholar]
- Anderson R.C., Chabaud A.G., Willmott S. CAB International; Wallingford (UK): 2009. CIH Keys to the Nematode Parasites of Vertebrates. Archival Volume. [Google Scholar]
- Bordes F., Morand S., Krasnov B.R., Poulin R. The Biogeography of Host-Parasite Interactions. Oxford University Press; 2010. Parasite diversity and latitudinal gradients in terrestrial mammals; pp. 89–98. [Google Scholar]
- Bozick B.A., Real L.A. Integrating parasites and pathogens into the study of geographic range limits. Q. Rev. Biol. 2015;90:361–380. doi: 10.1086/683698. [DOI] [PubMed] [Google Scholar]
- Caira J.N., Jensen K., editors. Planetary Biodiversity Inventory (2008–2017): Tapeworms from Vertebrate Bowels of the Earth. University of Kansas, Natural History Museum, Special Publication No. 25, Lawrence, KS, USA; 2017. [Google Scholar]
- Chilton N.B., Huby-Chilton F., Gasser R.B., Beveridge I. The evolutionary origins of nematodes within the order Strongylida are related to predilection sites within hosts. Mol. Phylogenet. Evol. 2006;40:118–128. doi: 10.1016/j.ympev.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Clark N.J., Seddon J.M., Šlapeta J., Wells K. Parasite spread at the domestic animal - wildlife interface: anthropogenic habitat use, phylogeny and body mass drive risk of cat and dog flea (Ctenocephalides spp.) infestation in wild mammals. Parasites Vectors. 2018;11:8. doi: 10.1186/s13071-017-2564-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke K.R., Warwick R.M. A further biodiversity index applicable to species lists: variation in taxonomic distinctness. Mar. Ecol. Prog. Ser. 2001;216:265–278. doi: 10.3354/meps216265. [DOI] [Google Scholar]
- Cooper N., Kamilar J.M., Nunn C.L. Host longevity and parasite species richness in mammals. PLoS One. 2012;7 doi: 10.1371/journal.pone.0042190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumming G.S. Host distributions do not limit the species ranges of most African ticks (Acari: Ixodida) Bull. Entomol. 1999;89:303–327. doi: 10.1017/S0007485399000450. [DOI] [Google Scholar]
- Cumming G.S. Comparing climate and vegetation as limiting factors for species ranges of African ticks. Ecology. 2002;83:255–268. [Google Scholar]
- Dallas T.A., Aguirre A.A., Budischak S., Carlson C., Ezenwa V., Han B., Huang S., Stephens P.R. Gauging support for macroecological patterns in helminth parasites. Global Ecol. Biogeogr. 2018;27:1437–1447. doi: 10.1111/geb.12819. [DOI] [Google Scholar]
- Dallas T.A., Gehman A.l.L.M., Aguirre A.A., Budischak S.A., Drake J.M., Farrell M.J., Ghai R., Huang S., Morales-Castilla I. Contrasting latitudinal gradients of body size in helminth parasites and their hosts. Global Ecol. Biogeogr. 2019;28:804–813. doi: 10.1111/geb.12894. [DOI] [Google Scholar]
- Dáttilo W., Barrozo-Chávez N., Lira-Noriega A., Guevara R., Villalobos F., Santiago-Alarcon D., Neves F.S., Izzo T., Ribeiro S.P. Species-level drivers of mammalian ectoparasite faunas. J. Anim. Ecol. 2020;89:1754–1765. doi: 10.1111/1365-2656.13216. [DOI] [PubMed] [Google Scholar]
- Dobson A., Lafferty K.D., Kuris A.M., Hechinger R.F., Walter J. hHomage to Linnaeus: How many parasites? How many hosts? In: Avise J.C., Hubbell S.P., Ayala F.J., editors. In the Light of Evolution, Volume II: Biodiversity and Extinction. National Academies Press; Washington D.C, USA: 2008. p. 432. [PubMed] [Google Scholar]
- Ezenwa V.O., Price S.A., Altizer S., Vitone N.D., Cook K.C. Host traits and parasite species richness in even and odd-toed hoofed mammals, Artiodactyla and Perissodactyla. Oikos. 2006;115:526–536. [Google Scholar]
- Faith D.P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 1992;61:1–10. doi: 10.1016/0006-3207(92)91201-3. [DOI] [Google Scholar]
- Faurby S., Davis M., Pedersen R., Schowanek S., Antonelli A., Svenning J. Phylacine 1.2: the phylogenetic atlas of mammal macroecology. Ecology. 2018;99 doi: 10.1002/ecy.2443. 2626–2626. [DOI] [PubMed] [Google Scholar]
- Faurby S., Svenning J.C. Resurrection of the island rule: human-driven extinctions have obscured a basic evolutionary pattern. Am. Nat. 2016;187:812–820. doi: 10.1086/686268. [DOI] [PubMed] [Google Scholar]
- Fecchio A., Bell J.A., Bosholn M., Vaughan J.A., Tkach V.V., Lutz H.L., Cueto V.R., Gorosito C.A., González-Acuña D., Stromlund C., Kvasager D., Comiche K.J.M., Kirchgatter K., Pinho J.B., Berv J., Anciães M., Fontana C.S., Zyskowski K., Sampaio S., Dispoto J.H., Galen S.C., Weckstein J.D., Clark N.J. An inverse latitudinal gradient in infection probability and phylogenetic diversity for Leucocytozoon blood parasites in New World birds. J. Anim. Ecol. 2020;89:423–435. doi: 10.1111/1365-2656.13117. [DOI] [PubMed] [Google Scholar]
- Fernandez J.A. The holistic specimen and parasites of mammals. THERYA. 2019;10:65–67. [Google Scholar]
- Garamszegi Z., editor. Modern Phylogenetic Comparative Methods and Their Application in Evolutionary Biology Concepts and Practice. Springer; Berlin, Germany: 2014. [Google Scholar]
- García-Prieto L., Falcón-Ordaz J., Guzmán-Cornejo C. Helminth parasites of wild Mexican mammals: list of species, hosts and geographical distribution. Zootaxa. 2012;3290:1–92. [Google Scholar]
- Garrido-Olvera L., Arita H.T., Pérez-Ponce De León G. The influence of host ecology and biogeography on the helminth species richness of freshwater fishes in Mexico. Parasitology. 2012;139:1652–1665. doi: 10.1017/S003118201200100X. [DOI] [PubMed] [Google Scholar]
- Gay N., Olival K., Bumrungsri S., Siriaroonrat B., Bourgarel M., Morand S. Parasite and viral species richness of Southeast Asian bats: fragmentation of area distribution matters. Int. J. Parasitol. Parasites Wildl. 2014;3:161–170. doi: 10.1016/j.ijppaw.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger M., Sánchez-Villagra M.R., Lindholm A.K. A longitudinal study of phenotypic changes in early domestication of house mice. R. Soc. Open Sci. 2018;5 doi: 10.1098/rsos.172099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons L. CAB International; Wallinford, England: 2010. Keys to the Nematode Parasites of Vertebrates. [Google Scholar]
- Gómez-Ortiz Y., Dominguez-Vega H., Moreno C.E. Spatial variation of mammal richness, functional and phylogenetic diversity in the Mexican Transition Zone. Community Ecol. 2017;18:121–127. doi: 10.1556/168.2017.18.2.1. [DOI] [Google Scholar]
- Guillot J., Caumes E., Joncour A., Le Lacour S.A., Lecso G. Case Report : molecular characterization of Ancylostoma braziliense larvae in a patient with hookworm-related cutaneous larva migrans. Am. J. Trop. Med. Hyg. 2012;86:843–845. doi: 10.4269/ajtmh.2012.11-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guk A.S., Kim J., Park J., Chai J., Liesenfeld O., Ramı B.E. A human case of Plagiorchis vespertilionis (Digenea : Plagiorchiidae) infection in the Republic of Korea. J. Parasitol. 2007;93:1225–1227. doi: 10.1645/GE-1098R.1. [DOI] [PubMed] [Google Scholar]
- Gutiérrez J.S., Piersma T., Thieltges D.W. Micro- and macroparasite species richness in birds: the role of host life history and ecology. J. Anim. Ecol. 2019;88:1226–1239. doi: 10.1111/1365-2656.12998. [DOI] [PubMed] [Google Scholar]
- Gutiérrez J.S., Rakhimberdiev E., Piersma T., Thieltges D.W. Migration and parasitism: habitat use, not migration distance, influences helminth species richness in Charadriiform birds. J. Biogeogr. 2017;44:1137–1147. doi: 10.1111/jbi.12956. [DOI] [Google Scholar]
- Halliday F.L., Heckman R., Wilfarht P.A., Mitchell C.E. bioRxiv 857151; 2019. Host Community Assembly Modifies the Relationship between Host and Parasite Richness. [Google Scholar]
- Halliday F.L., Heckman R.W., Wilfahrt P.A., Mitchell C.E. Eutrophication, biodiversity loss, and species invasions modify the relationship between host and parasite richness during host community assembly. Global Change Biol. 2020;26:4854–4867. doi: 10.1111/gcb.15165. [DOI] [PubMed] [Google Scholar]
- Hodda M. Animal biodiversity: an outline of higher-level classification and survey of taxonomic richness. Zootaxa. 2011;3164:63–95. doi: 10.11646/zootaxa.3703.1.1. [DOI] [PubMed] [Google Scholar]
- Huang S., Drake J.M., Gittleman J.L., Altizer S. Parasite diversity declines with host evolutionary distinctiveness: a global analysis of carnivores. Evolution. 2015;69:621–630. doi: 10.1111/evo.12611. [DOI] [PubMed] [Google Scholar]
- Jones A., Bray R., Gibson D., editors. vol. 2. The Natural History Museum; London, Wallinford, England: 2005. (Keys to the Trematoda). [Google Scholar]
- Kamiya Tsukushi, Dwyer K.O., Nakagawa S., Poulin R. Host diversity drives parasite diversity: meta-analytical insights into patterns and causal mechanisms. Ecography. 2014;37:689–697. doi: 10.1111/j.1600-0587.2013.00571.x. [DOI] [Google Scholar]
- Kamiya T., O’Dwyer K., Nakagawa S., Poulin R. What determines species richness of parasitic organisms? A meta-analysis across animal, plant and fungal hosts. Biol. Rev. 2014;89:123–134. doi: 10.1111/brv.12046. [DOI] [PubMed] [Google Scholar]
- Kembel S.W., Cowan P.D., Helmus M.R., Cornwell W.K., Morlon H., Ackerly D.D., Blomberg S.P., Webb C. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. 2010;26:1463–1464. doi: 10.1093/bioinformatics/btq166. [DOI] [PubMed] [Google Scholar]
- Kennedy C.R. Cambridge University Press; New York, NY, USA: 2006. Ecology of the Acanthocephala. [Google Scholar]
- Kennedy C.R., Bush A.O. Species richness in helminth communities : the importance of multiple congeners. Parasitology. 1992;104:189–197. doi: 10.1017/s0031182000060935. [DOI] [PubMed] [Google Scholar]
- Korallo N.P., Vinarski M.V., Krasnov B.R., Shenbrot G.I., Mouillot D., Poulin R. Are there general rules governing parasite diversity? Small mammalian hosts and gamasid mite assemblages. Divers. Distrib. 2007;13:353–360. doi: 10.1111/j.1472-4642.2007.00332.x. [DOI] [Google Scholar]
- Krasnov B.R., Korallo-Vinarskaya N.P., Vinarski M.V., Shenbrot G.I., Mouillot D., Poulin R. Searching for general patterns in parasite ecology: host identity versus environmental influence on gamasid mite assemblages in small mammals. Parasitology. 2008;135:229–242. doi: 10.1017/S003118200700368X. [DOI] [PubMed] [Google Scholar]
- Krasnov B.R., Mouillot D., Shenbrot G.I., Khokhlova I.S., Poulin R. Deconstructing spatial patterns in species composition of ectoparasite communities : the relative contribution of host composition, environmental variables and geography. Glob. Ecol. Biogeogr. 2010:515–526. doi: 10.1111/j.1466-8238.2010.00529.x. [DOI] [Google Scholar]
- Krasnov B.R., Shenbrot G.I., Khokhlova I.S., Mouillot D., Poulin R. Latitudinal gradients in niche breadth: empirical evidence from haematophagous ectoparasites. J. Biogeogr. 2008;35:592–601. doi: 10.1111/j.1365-2699.2007.01800.x. [DOI] [Google Scholar]
- Krasnov B.R., Shenbrot G.S., van der Mescht L., Warburton E.M., Khokhlova I.S. Phylogenetic and compositional diversity are governed by different rules: a study of fleas parasitic on small mammals in four biogeographic realms. Ecography. 2019;42:1000–1011. doi: 10.1111/ecog.04224. [DOI] [Google Scholar]
- Kutz S.J., Hoberg E.P., Polley L., Jenkins E.J. Global warming is changing the dynamics of Arctic host-parasite systems. Proc. R. Soc. B Biol. Sci. 2005;272:2571–2576. doi: 10.1098/rspb.2005.3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafferty K.D. The evolution of trophic transmission. Parasitol. Today. 1999;15:111–115. doi: 10.1016/s0169-4758(99)01397-6. [DOI] [PubMed] [Google Scholar]
- Leung T.L.F., Koprivnikar J. Your infections are what you eat: how host ecology shapes the helminth parasite communities of lizards. J. Anim. Ecol. 2019;88:416–426. doi: 10.1111/1365-2656.12934. [DOI] [PubMed] [Google Scholar]
- Lamothe-Argumedo R., García-Prieto L. A.G.T Editor S.A; Ciudad de México: 1988. Helmintiasis del hombre en México: Tratamiento y profilaxis. [Google Scholar]
- Leung T.L.F., Koprivnikar J. Nematode parasite diversity in birds: the role of host ecology, life history and migration. J. Anim. Ecol. 2016;85:1471–1480. doi: 10.1111/1365-2656.12581. [DOI] [PubMed] [Google Scholar]
- Lindenfors P., Nunn C.L., Jones K.E., Cunningham A.A., Sechrest W., Gittleman J.L. Parasite species richness in carnivores: effects of host body mass, latitude, geographical range and population density. Glob. Ecol. Biogeogr. 2007;16:496–509. doi: 10.1111/j.1466-8238.2006.00301.x. [DOI] [Google Scholar]
- Locke S., McLaughlin J., Marcogliese D. DNA barcodes show cryptic diversity and a potential physiological basis for host specificity among Diplostomoidea (Platyhelminthes: Digenea) parasitizing freshwater fishes in the St. Lawrence River, Canada. Mol. Ecol. 2010;19:2813–2827. doi: 10.1111/j.1365-294X.2010.04713.x. [DOI] [PubMed] [Google Scholar]
- Locke S.A., Marcogliese D.J., Tellervo Valtonen E. Vulnerability and diet breadth predict larval and adult parasite diversity in fish of the Bothnian Bay. Oecologia. 2014;174:253–262. doi: 10.1007/s00442-013-2757-x. [DOI] [PubMed] [Google Scholar]
- Luque J.L., Mouillot D., Poulin R. Parasite biodiversity and its determinants in coastal marine teleost fishes of Brazil. Parasitology. 2004;128:671–682. doi: 10.1017/s0031182004005050. [DOI] [PubMed] [Google Scholar]
- Marrugan A. Blackwell Publishing; Oxford, Oxford, USA: 2004. Measuring Biological Diversity. [Google Scholar]
- Miller D.L., Schrecengost J., Merrill A., Kilgo J., Ray H.S., Miller K.V., Baldwin C. a. Hematology, parasitology, and serology of free-ranging coyotes (Canis latrans) from South Carolina. J. Wildl. Dis. 2009;45:863–869. doi: 10.7589/0090-3558-45.3.863. [DOI] [PubMed] [Google Scholar]
- Milošević D, Simić V, Stojković M, Živić I. Chironomid faunal composition represented by taxonomic distinctness index reveals environmental change in a lotic system over three decades. Hydrobiologia. 2012;683:69–82. doi: 10.1007/s10750-011-0941-8. [DOI] [Google Scholar]
- Monello R.J., Gompper M.E. Effects of resource availability and social aggregation on the species richness of raccoon endoparasite infracommunities. Oikos. 2011;120:1427–1433. doi: 10.1111/j.1600-0706.2011.19260.x. [DOI] [Google Scholar]
- Morand S. (macro-) Evolutionary ecology of parasite diversity: from determinants of parasite species richness to host diversification. Int. J. Parasitol. Parasites Wildl. 2015;4:80–87. doi: 10.1016/j.ijppaw.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morand S., Krasnov B., Poulin R., editors. Micro Mammals and Macroparasites: from Evolutionary Ecology to Management. Springer-Verlag; 2005. Cestodes of small mammals: taxonomy and life cycles; pp. 29–62. [Google Scholar]
- Morand S., McIntyre K., Baylis M. Domesticated animals and human infectious diseases of zoonotic origins: domestication time matters. Infect. Genet. Evol. 2014;24:76–81. doi: 10.1016/j.meegid.2014.02.013. [DOI] [PubMed] [Google Scholar]
- Morand S., Poulin R. Density, body mass and parasite species richness of terrestrial mammals. Evol. Ecol. 1998;12:717–727. doi: 10.1023/A:1006537600093. [DOI] [Google Scholar]
- Morariu S., Mederle N., Badea C., Gheorghe D., Genchi C. The prevalence , abundance and distribution of cyathostomins (small stongyles) in horses from Western Romania. Vet. Parasitol. 2016;223:205–209. doi: 10.1016/j.vetpar.2016.04.021. [DOI] [PubMed] [Google Scholar]
- Near T. Acanthocephalan phylogeny and the evolution of parasitism. Integr. Comp. Biol. 2002;42:668–677. doi: 10.1093/icb/42.3.668. [DOI] [PubMed] [Google Scholar]
- Nunn C., Altizer S., Jones K., Sechrest W. Comparative tests of parasite species richness in primates. Am. Nat. 2003;162:597–614. doi: 10.1086/378721. [DOI] [PubMed] [Google Scholar]
- Nunn C.L., Altizer S.M., Sechrest W., Cunningham A.A. Latitudinal gradients of parasite species richness in primates. Divers. Distrib. 2005;11:249–256. doi: 10.1111/j.1366-9516.2005.00160.x. [DOI] [Google Scholar]
- Oksanen J., Blanchet F., Friendly M., Kindt R., Legendre P., Mcglinn D., Minchin P.R., Hara R.B.O., Simpson G.L., Solymos P., Stevens M.H.H., Szoecs E. Version 2.4-0; 2016. Package “Vegan”. [Google Scholar]
- Orme D., Freckleton R., Thomas G., Petzoldt T., Fritz S., Isaac N., Pearse W. 2018. Caper: Comparative Analyses of Phylogenetics and Evolution in R. R Package Version 1.0.1. [Google Scholar]
- Poulin R. Macroecological patterns of species richness in parasite assemblages. Basic Appl. Ecol. 2004;5:423–434. doi: 10.1016/j.baae.2004.08.003. [DOI] [Google Scholar]
- Poulin R. Phylogeny, ecology, and the richness of parasite communities in vertebrates. Ecol. Monogr. 1995;65:283–302. [Google Scholar]
- Poulin R., Krasnov B.R., Mouillot D. Host specificity in phylogenetic and geographic space. Trends Parasitol. 2011;27:355–361. doi: 10.1016/j.pt.2011.05.003. [DOI] [PubMed] [Google Scholar]
- Poulin R., Leung T.L.F. Latitudinal gradient in the taxonomic composition of parasite communities. J. Helminthol. 2011;85:228–233. doi: 10.1017/S0022149X10000696. [DOI] [PubMed] [Google Scholar]
- Poulin R., Morand S. Smithsonian Institution Books; Washington D.C, USA: 2004. Parasite Biodiversity. [Google Scholar]
- Poulin R., Mouillot D. The evolution of taxonomic diversity in helminth assemblages of mammalian hosts. Evol. Ecol. 2004;18:231–247. doi: 10.1023/B:EVEC.0000035029.55952.68. [DOI] [Google Scholar]
- Prati S., Henriksen E.H., Knudsen R., Amundsen P.A. Seasonal dietary shifts enhance parasite transmission to lake salmonids during ice cover. Ecol. Evol. 2020;10:4031–4043. doi: 10.1002/ece3.6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preisser W. Latitudinal gradients of parasite richness: a review and new insights from helminths of cricetid rodents. Ecography. 2019;42:1–16. doi: 10.1111/ecog.04254. [DOI] [Google Scholar]
- Purvis A., Hector A. Getting the measure of biodiversity. Nature. 2000;405:212–219. doi: 10.1038/35012221. [DOI] [PubMed] [Google Scholar]
- Quiroz-Martínez B., Salgado-Maldonado G. Taxonomic distinctness and richness of helminth parasite assemblages of freshwater fishes in Mexican hydrological basins. PLoS One. 2013;8 doi: 10.1371/journal.pone.0074419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiroz Romero H. Limusa; Ciudad de México: 1987. Parasitología y enfermedades parasitarias de animales domésticos. [Google Scholar]
- Randhawa H.S., Poulin R. Determinants of tapeworm species richness in elasmobranch fishes: untangling environmental and phylogenetic influences. Ecography. 2010;33:866–877. [Google Scholar]
- Rasmussen T.K., Randhawa H.S. Host diet influences parasite diversity: a case study looking at tapeworm diversity among sharks. Mar. Ecol. Prog. Ser. 2018;605:1–16. [Google Scholar]
- Shenbrot G., Krasnov B.R., Lu L. Geographical range size and host specificity in ectoparasites : a case study with Amphipsylla fleas and rodent hosts. J. Biogeogr. 2010;34:1679–1690. doi: 10.1111/j.1365-2699.2007.01736.x. [DOI] [Google Scholar]
- Smith F.A., Lyons S.K., Ernest S.K.M., Jones K.E., Kaufman D.M., Dayan T., Marquet P.A., Brown J.H., Haskell J.P. Body mass of late Quaternary mammals. Ecology. 2003;84 doi: 10.1890/02-9003. 3403–3403. [DOI] [Google Scholar]
- Smout F.A., Skerratt L.F., Butler J.R.A., Johnson C.N., Congdon B.C., Thompson R.C.A. The hookworm Ancylostoma ceylanicum: an emerging public health risk in Australian tropical rainforests and indigenous communities. One Heal. 2017;3:66–69. doi: 10.1016/j.onehlt.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spickett A., Junker K., Krasnov B.R., Haukisalmi V. Helminth parasitism in two closely related South African rodents: abundance, prevalence, species richness and impinging factors. Parasitol. Res. 2017;116:1395–1409. doi: 10.1007/s00436-017-5419-9. [DOI] [PubMed] [Google Scholar]
- Stephens P.R., Altizer S., Smith K.F., Alonso Aguirre A., Brown J.H., Budischak S.A., Byers J.E., Dallas T., Jonathan Davies T., Drake J.M., Ezenwa V.O., Farrell M.J., Gittleman J.L., Han B.A., Huang S., Hutchinson R.A., Johnson P., Nunn C.L., Onstad D., Park A., Vazquez-Prokopec G.M., Schmidt J.P., Poulin R. The macroecology of infectious diseases: a new perspective on global-scale drivers of pathogen distributions and impacts. Ecol. Lett. 2016;19:1159–1171. doi: 10.1111/ele.12644. [DOI] [PubMed] [Google Scholar]
- Team R.C. 2018. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Torres J., Miquel J., Casanova J., Ribas A., Feliu C., Morand S. Endoparasite species richness of Iberian carnivores: influences of host density and range distribution. Biodivers. Conserv. 2006;15 [Google Scholar]
- Viney M. The genomic basis of nematode parasitism. Brief. Funct. Genomics. 2018;17:8–14. doi: 10.1093/bfgp/elx010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Violante-González J., Monks S., Gil-Guerrero S., Rojas-Herrera A.A., Flores-Rodríguez P. Helminth communities of two species of piscivorous birds, Ardea alba (Linnaeus) and Nyctanassa violacea (Gmelin) (Ciconiiformes: Ardeidae), in two coastal lagoons from Guerrero state, Mexico. Parasitol. Res. 2012;111:309–315. doi: 10.1007/s00436-012-2840-y. [DOI] [PubMed] [Google Scholar]
- Vitone N.D., Altizer S., Nunn C.L. Body size, diet and sociality influence the species richness of parasitic worms in anthropoid primates. Evol. Ecol. Res. 2004;6:183–199. [Google Scholar]
- Walker J.G., Hurford A., Cable J., Ellison A.R., Price S.J., Cressler C.E. Host allometry influences the evolution of parasite host-generalism: theory and meta-analysis. Philos. Trans. R. Soc. B Biol. Sci. 2017;372:20160089. doi: 10.1098/rstb.2016.0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J.G., Morgan E.R. Generalists at the interface: nematode transmission between wild and domestic ungulates. Int. J. Parasitol. Parasites Wildl. 2014;3:242–250. doi: 10.1016/j.ijppaw.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton E.M., Kohler S.L., Vonhof M.L. Patterns of parasite community dissimilarity: the significant role of land use and lack of distance-decay in a bat–helminth system. Oikos. 2016;215:374–385. doi: 10.1111/oik.02313. [DOI] [Google Scholar]
- Weinstein S.B., Kuris A.M. Independent origins of parasitism in Animalia. Biol. Lett. 2016;12:1–5. doi: 10.1098/rsbl.2016.0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissbrod L., Marshall F.B., Valla F.R., Khalaily H., Bar-Oz G., Auffray J.C., Vigne J.D., Cucchi T. Origins of house mice in ecological niches created by settled hunter-gatherers in the Levant 15,000 y ago. Proc. Natl. Acad. Sci. U. S. A. 2017;114:4099–4104. doi: 10.1073/pnas.1619137114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells K., Gibson D.I., Clark N.J. Global patterns in helminth host specificity: phylogenetic and functional diversity of regional host species pools matter. Ecography. 2019;42:416–427. doi: 10.1111/ecog.03886. [DOI] [Google Scholar]
- Wells K., Morand S., Gibson D.I., Clark N.J., Mccallum H.I., Ribas A. Global spread of helminth parasites at the human – domestic animal – wildlife interface. Global Change Biol. 2018;24:3254–3265. doi: 10.1111/gcb.14064. [DOI] [PubMed] [Google Scholar]
- Willig A.M.R., Kaufman D.M., Stevens R.D. Latitudinal gradients of Biodiversity: Pattern, Process , Scale, and Synthesis. Annu. Rev. Ecol. Evol. Syst. 2003;34:273–309. doi: 10.1146/annurev.ecolsys.34.012103.144032. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.