Abstract
Objective
Solute carrier family 38 (SLC38s) transporters play important roles in amino acid transportation and signaling transduction. However, their genetic alterations and biological roles in tumors are still largely unclear. This study aimed to elucidate the genetic signatures of SLC38s transporters and their implications in esophageal squamous cell carcinoma (ESCC).
Methods
Analyses on somatic mutation and copy number alterations (CNAs) of SLC38A3 were performed as described. Immunohistochemistry (IHC) assay and Western blot assay were used to detect the protein expression level. MTS assay, colony formation assay, transwell assay and wound healing assay were used to explore the malignant phenotypes of ESCC cells. Immunofluorescence assay was used to verify the colocalization of two indicated proteins and immunopreciptation assay was performed to confirm the interaction of proteins.
Results
Our findings revealed that SLC38s family was significantly disrupted in ESCC, with high frequent CNAs and few somatic mutations. SLC38A3 was the most frequent loss gene among them and was linked to poor survival and lymph node metastasis. The expression of SLC38A3 was lower in tumor tissues compared to that in normal tissues, which was also significantly associated with worse clinical outcome. Further experiments revealed that depletion of SLC38A3 could promote EMT in ESCC cell lines, and the interaction of SLC38A3 and SETDB1 might lead to the reduced transcription of Snail. Pharmacogenomic analyses demonstrated that fifteen inhibitors were showed significantly correlated with SLC38A3 expression.
Conclusions
Our investigations have provided insights that SLC38A3 could act as a suppressor in EMT pathway and serve as a prognostic factor and predictor of differential drug sensitivities in ESCC.
Keywords: Amino acid transporter, esophageal squamous cell carcinoma, epithelial-mesenchymal transition, genomic alterations, SLC38A3, SETDB1, Snail
Introduction
Esophageal cancer is one of the top ten malignant tumors and the sixth leading cause of cancer-related death in the world (1). More than half of global esophageal cancer cases occur in China, with 477.9 thousand new cases diagnosed in 2015 (2,3). As a histological subtype of esophageal cancer, esophageal squamous cell carcinoma (ESCC) is the major type in Asian populations (e g., 90% of Chinese patients) (4). Although the diagnosis, staging and treatment of ESCC have been greatly improved in the last decade, the five-year survival rate is still far from satisfying (5). Recently, genomic studies have provided comprehensive insights into the genetic variations of ESCC and highlighted a panel of driver somatic alterations involved in ESCC development (5-11). However, there are still no effective therapeutic targets existing in ESCC clinical practice. Therefore, it is extremely urgent to have a deeper understanding of the molecular mechanism underlying ESCC tumorigenesis and thus to identify new therapeutic targets.
Solute carrier family 38 (SLC38s) are ubiquitously expressed in all kinds of human cells, which mediate Na+-dependent net uptake and efflux of small neutral amino acids (12). This family comprises 11 members, which are also referred to as sodium neutral amino acid transporters, or system N/A transporters (13), and functional studies have been carried out on the first six members (12). Unlike those transporters that have been extensively studied in cancer, such as SLC1s, SLC3s, SLC6s and SLC7s (14), the genomic alterations and molecular mechanisms of SLC38s in cancer remain unclear. However, their biological functions have been studied in mammalian brain, kidney, liver and Xenopus oocytes in the physiological status (12). All SLC38s transporters mediate glutamine transportation via a Na+-amino acid co-transport manner (13). It is well recognized that glutamine is a key onco-metabolite and glutamine metabolism significantly affects tumor growth and progression (15), indicating that the disruption of these glutamine transporters may have a great impact on tumor initiation and propagation. Overexpression of SLC38A1 in breast cancer has been reported to correlate with tumor size, nodal metastasis and advanced tumor stage (16). Inhibiting SLC38A1 could decrease the growth and migration of pancreatic cancer cells (17). However, a study showed that SLC38A3 was undetectable in 5 renal cell carcinoma cell lines (KH39, CAKI-1, CAKI-2, KRC/Y and TK-10), cervical carcinoma line HeLa and small cell lung carcinoma (SCLC) line U2020, while was expressed in normal human kidney (NHK) cells (18). The transcript of SLC38A3 was not found or was greatly reduced in mouse fibrosarcoma xenografts (18). Nevertheless, a recent study confirmed that SLC38A3 was up-regulated in metastatic NSCLC cells and this genetic alteration promoted metastasis of non-small cell lung cancer cell A549 through activating PDK1/AKT pathway (19). These contradictory alteration patterns imply that amino acid transporters were sophisticatedly orchestrated by tumor cells and they might play other key roles in neoplasia rather than simply uptake several amino acids for biosynthesis. However, the genomic alterations of SLC38s in ESCC and their relevance to clinical features remain to be defined.
Epithelial-mesenchymal transition (EMT) is a developmental regulatory program that transiently places epithelial cells into mesenchymal-like cell states (20-23). EMT modifies the adhesion molecules expressed by the cell, which leads to the detachment of epithelial cells from each other, allowing it to adopt a migratory and invasive behavior and a new transcriptional program is activated to promote the mesenchymal fate (24). EMT confers on cancer cells increased potential of metastasis and even greater resistance to anti-tumor therapies. A set of transcriptional factors, including Snail, Slug, Twist, and Zeb1/2, which are regarded as EMT “master” transcription factors, are vital to orchestrate the EMT and relate migratory processes (24). Typically, Snail represses the E-cadherin-encoding gene, thereby depriving neoplastic epithelial cells of this key suppressor of motility and invasiveness (25).
SETDB1 plays an important role in methylation and gene silencing. Recent studies indicate that SETDB1 is abnormally expressed in various human cancer conditions and contributes to enhanced tumor growth and metastasis (26). Du et al. reported that the SETDB1 repressed EMT by Smad3 (27). However, Yang et al. indicated that SETDB1 acted as an EMT inducer by direct binding to the promoter of Snail (28). Zhang et al. also reported that SETDB1 promoted cell proliferation and migration in hepatocellular carcinoma (29). Thus, the role of SETDB1 in EMT seems ambiguous.
In the present study, we sought to depict the genomic alterations of SLC38s (SLC38A1-7) in ESCC, and elucidate their molecular mechanism in tumorigenesis and clinical implications. Through integration of our previous genomic studies and other groups’ sequencing data, we provided the first insight into the genetic variations of SLC38s, which showed frequent copy number alterations (CNAs) and few somatic mutations. Intriguingly, SLC38A3 CNA loss was the most frequent alterations among this family and was significantly associated with poor prognostic outcome. SLC38A3 was frequently lowly expressed in ESCC tissue samples and significantly correlated with poor survival. Functional experiments revealed that defect of SLC38A3 benefited cancer cells. Mechanistically, SLC38A3 physically interacted with SETDB1 that was involved in the transcription of Snail. Knockdown of SLC38A3 promoted the EMT. Moreover, we also showed that SLC38A3 might be a useful predictor for differential drug sensitivity.
Materials and methods
Genome data collection and processing
This study integrated the individual genomic data and clinical data of subjects from seven independent whole genome sequencing (WGS) and whole-exome sequencing (WES) of ESCC (integrated ESCC cohort) (5-11). Somatic mutation and CNA analyses were performed as described (30). Somatic CNA detection was based on the quantification of deviations from the log-ratio of sequence coverage depth within a tumor-normal pair (31). We used an empirical threshold of log ratio (log2R) >0.3 for genomic gain and a log ratio <−0.3 for genomic loss. The genetic alterations of SLC38s were visualized by the online tool Oncoprinter ( http://www.cbioportal.org/oncoprinter) (32,33).
Immunohistochemistry (IHC)
IHC analysis was performed and diagnosed by two pathologists blindly on the 105 paraffin-embedded ESCC tissue sections [tissue microarray (TMA), including 75 paired adjacent normal tissue samples and ESCC samples]. TMA of ESCC specimens were obtained from Shanghai Outdo Biotech Co., Ltd. (SOBC), with the approval of the Institutional Review Board of Taizhou Hospital, Zhejiang Province. The IHC assay was performed as previous described (34). Tissue sections were incubated with rabbit anti-SLC38A3 (1:100; Proteintech, Wuhan, China), overnight at 4 °C. After washing, the tissue sections were treated with goat anti-mouse/rabbit IgG HRP-polymer (ZSGB-BIO, Beijing, China) for 30 min. 3, 3’-Diaminobenzidine was used as the chromogen. IHC scores were determined by combining the intensity of staining and the proportion of positively stained tumor cells. First, the intensity was graded as follows: 0, negative; 1, weak; 2, moderate; 3, strong. Second, the proportion of positive tumor cells was graded: 0, <5%; 1, 5%−25%; 2, 26%−50%; 3, 51%−75%; 4, >75%. A final score was derived by multiplication of these two primary scores. Final scores of 0−4 were defined as low “expression” (−); scores of 6−12 as “high expression” (+).
Cell lines and cell culture
The human ESCC cell lines YES2, KYSE30, KYSE70, KYSE140, KYSE150, KYSE180, KYSE410, KYSE450, KYSE510, NE2 and NE3 were kindly provided by Dr. Y. Shimada (Kyoto University). These cells were cultured in RPMI 1640 (Lonza, Switzerland) supplemented with 10% fetal bovine serum (FBS) (Gibco, America) and 1% penicillin/streptomycin (Gibco, America). Both types of ESCC cells were maintained at 37 °C and 5% CO2. We have recently authenticated the source of cell lines and tested for mycoplasma contamination, and did not find cross-contaminated cell lines and mycoplasma contamination.
Antibodies
The antibodies used included SLC38A3 (Proteintech, 14315-1-AP), E-cadherin (Cell Signaling Technology, Rabbit mAb#3195), Vimentin (Cell Signaling Technology, Rabbit mAb#5741), Snail (Cell Signaling Technology, Rabbit mAb#3879), Twist (Cell Signaling Technology, #46702), SETDB1 (Proteintech, 11231-1-AP), β-actin (Proteintech, 66009-1-Ig), and GAPDH (Proteintech, 60004-1-Ig).
Oligonucleotide transfection
Specific small interfering RNA (siRNA) targeting SLC38A3 and SETDB1 were custom designed and provided by Guangzhou RiboBio Co., Ltd. (RiboBio). Three siRNAs targeting SLC38A3 include SLC38A3_1, SLC38A3_2 and SLC38A3_3. Cells were seeded on 60-mm culture plates and transfected with oligonucleotides using lipofectamine 2000 (Invitrogen) according to manufacturer’s instructions. Transfected cells were incubated at 37 °C with 5% CO2 for 48 h.
Transwell migration/invasion assays
Migration and invasion assays were performed as described. In brief, migration of cells was assayed in Transwell cell culture chambers with 6.5-mm diameter polycarbonate membrane filters containing 8.0 mm pore size (Costar, 3422). Briefly, 100,000 cells in 100 μL of serum-free medium were added to the upper chamber of the device, and the lower chamber was filled with 600 μL culture medium with 20% FBS. After 10 h of incubation at 37 °C, the non-migration cells were removed from the upper surface of the membrane with a cotton swab. The filters were then fixed in methanol for 10 min, stained with crystal violet solution for 1 h, and counted. Five random micro-scopic fields (×100) were counted per well and the mean was determined. For the Transwell invasion assay, the membrane of the upper chamber was pre-coated with 50 μL of a 2.5 mg/mL solution of Matrigel (Falcon BD).
Colony formation assay
A total of 2,000 transfected cells were seeded into 100-mm culture plates and incubated at 37 °C with 5% CO2 for 10 d. Culture plates were performed in duplicates. After a wash with pre-cooled phosphate buffer saline (PBS), cultures were fixed with pre-cooled methanol for 20 min and stained with crystal violet for 15 min. Colonies were examined and calculated.
Wound healing assay
Transfected cells were plated in a six-well plate at a density of 1×105 cells per well. Cells were grown to 80% confluence and several wound lines were scratched vertically to the bottom with a 200 μL pipette tip. After being washed with PBS three times, cells were incubated in growth medium containing 1.5% serum. The wound width was determined after 24 h at 100× under a Leica microscope. Values were expressed as the percentage of wound healing.
Western blot assay
Cells were harvested in RIPA buffer (Beyotime, China). A total of 20 μg of cellular protein was subjected to 8%−15% SDS-polyacrylamide gel electrophoresis and transferred onto a polyvinylidene difluoride membrane. Incubation with antibodies was performed as described previously (35). The chemiluminescence signals were detected with an Amersham Imager 600 (GE, America).
Immunoprecipitation
Cells were treated with IP buffer (20 mmol/L Tris/HCl, pH=7.6, 100 mmol/L NaCl, 20 mmol/L KCl, and 1.5 mmol/L MgCl2, 0.5% NP-40) adding protease inhibitor cocktail. The whole-cell lysates were incubated with protein A/G-Sepharose beads (MCE), pretreated with anti-SLC38A3 (Proteintech, 14315-1-AP for IP, Western blot and IHC analyses), anti-SETDB1 (Proteintech, 11231-1-AP) antibody at 4 °C overnight. The beads were washed with cell lysis buffer and the immunoprecipitated samples were analyzed by Western blot.
Immunofluorescent staining
Cells were placed on Glass Bottom Cell Culture Dish (Nest, 801002). Twenty-four hours later, the cells were fixed in 4% paraformaldehyde for 15 min at room temperature, blocked with 2% bovine serum albumin (BSA) and then incubated with 0.1% Triton X-100 for 5 min. The cells were incubated with the indicated antibody at 4 °C overnight. The slides were subsequently incubated with an Alexa Fluor 488-labelled (ZSGB-BIO, ZF-0512) or Alexa Fluor 543-labelled secondary antibody (ZSGB-BIO, ZF-0516) in the dark for 2 h at room temperature. Next, the nuclei were detected by staining with 0.5 μg/mL DAPI (4’,6-diamidino-2-phenylindole). Images were captured and visualized by a confocal microscope (Leica ST2, Leica, Germany).
Quantitative real-time polymerase chain reaction (qRT-PCR) assay
The qRT-PCR was performed using the Premix Ex Taqkit (Takara) and a 7500 fast real-time PCR system (Life Technologies) according to manufacturer’s instructions. The mRNA expression level of SNAIL was normalized to the endogenous expression of GAPDH. Primers were provided by Beijing Tianyi Huiyuan Bioscience & Technology Inc. as described below: SNAIL forward, 5’- GAGGACAGTGGGAAAGGCTC-3’; SNAIL reverse, 5’- TGGCTTCGGATGTGCATCTT-3’. GAPDH forward, 5’-TCTCTGCTCCTCCTGTTC-3’; GAPDH reverse, 5’- GTTGACTCCGACCTTCAC-3’.
Proliferation assay
Proliferation ability of different tumor cells was detected by MTS assays (Promega) according to the manufacturer’s instructions. The data were analyzed with GraphPad Prism 5.0 software (GraphPad Software, San Diego, USA) and were presented as the relative cell viability normalized to the control.
Correlation analysis of SLC38A3 and drug sensitivity
Gene expression, CNA (loss of heterozygosity, LOH) and drug sensitivity data of 26 ESCC cell lines were obtained from Genomics of Drug Sensitivity in Cancer (GDSC) (36,37). Pearson or Spearman Correlation Coefficient test was used to determine association between drug sensitivity and SLC38A3 expression or LOH for each cell line.
Statistical analysis
Statistical analysis was carried out using IBM SPSS Statistics (Version 20; IBM Corp., New York, USA) or GraphPad Prism 5.0 for Windows. Pearson correlation coefficient test was used to analyze the association between SLC38A3 CNA and mRNA level in The Cancer Genome Atlas (TCGA) cohort. The association between SLC38A3 mRNA level and drug sensitivity was also analyzed by Pearson correlation coefficient test. The association between SLC38A3 LOH and drug sensitivity was also analyzed by Spearman correlation coefficient test. Two-tailed Student’s t-test and paired two-tailed Student’s t-test were used to analyze the difference of IHC scores between normal tissue samples and ESCC samples and the paired samples respectively. The two-tailed Pearson χ2 test was used to analyze the association between SLC38A3 CNA loss/SLC38A3 expression and clinicpathological parameters. The survival curves were plotted by Kaplan-Meier analysis and compared by log-rank test. Survival data were evaluated by multivariate Cox regression analysis. Differences were considered significant when the P value was less than 0.05.
Results
Genomic alterations of SLC38s family in ESCC
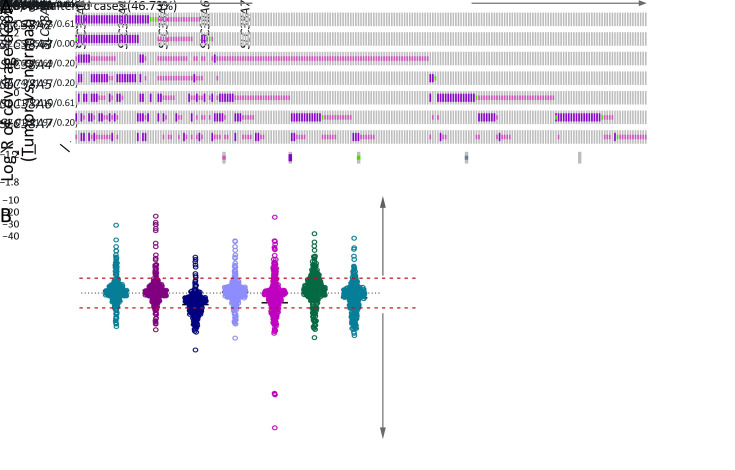
We first investigated their genomic alterations in paired normal-ESCC tumor samples. Our previous sequencing data were pooled with additional data from other groups (5-11) to analyze somatic mutations (n=490) and somatic CNAs (n=314), which generated comprehensive and precise genomic atlas of SLC38s family genes. Interestingly, all genes of SLC38s were disrupted in our integrated ESCC cohort, with prominent CNAs and few somatic mutations (Figure 1A , Supplementary Table S1 ). However, the genetic variation types were different from each other among SLC38s family genes. Intriguingly, system A transporters (SLC38A1, SLC38A2 and SLC38A4) were mainly CNA gain, while system N transporters (SLC38A3 and SLC38A5) were dominated by CNA loss (Figure 1B ). Except for SLC38A3, other SLC38s family genes harbored a few somatic mutations, and more than half of these missense mutations were predicted to affect their protein functions by Polyphen-2b (Supplementary Table S2 ) (38). Nonetheless, SLC38A3 was the most frequent disrupted gene by CNA loss, with 112 perturbed samples among 314 samples (35.67%; Figure 1A ). Additionally, another system N transporter SLC38A5 showed several remarkable losses in ESCC samples (Figure 1B ). Taken together, these observations indicated that SLC38s family genes were significantly disrupted in ESCC, raising the possibility that Na+-dependent net uptake and efflux of small neutral amino acid pathways mediated by SLC38s family might be significant for ESCC progression. In addition, different expression patterns of members in SLC38s family imply that they can play different roles.
1.
Genetic alterations of SLC38s family in integrated ESCC cohort. Somatic mutations of SLC38s family were analyzed in 490 paired ESCC samples and CNAs were analyzed in 314 paired ESCC samples. (A) Genomic landscape of SLC38s family; (B) Scatter plot showed relative copy number variations of SLC38s family. SLC38s, solute carrier family 38; ESCC, esophageal squamous cell carcinoma; CNA, copy number alteration.
S1. Frequencies of SLC38 family genetic alterations in integrated ESCC cohort in our data.
| Gene name | CNA frequency | Non-silent mutation frequency | |||||||
| Total | CNA gain | CNA loss | Frequency gain (%) | Frequency loss (%) | Total | Mutation | Frequency non-silent
mutation (%) |
||
| SLC38, solute carrier family 38; ESCC, esophageal squamous cell carcinoma; CNA, copy number alteration. | |||||||||
| SLC38A1 | 314 | 29 | 17 | 9.24 | 5.41 | 490 | 3 | 0.61 | |
| SLC38A2 | 314 | 27 | 15 | 8.60 | 4.78 | 490 | 3 | 0.61 | |
| SLC38A3 | 314 | 8 | 112 | 2.55 | 35.67 | 490 | 0 | 0 | |
| SLC38A4 | 314 | 21 | 21 | 6.69 | 6.69 | 490 | 1 | 0.20 | |
| SLC38A5 | 314 | 40 | 69 | 12.74 | 21.97 | 490 | 1 | 0.20 | |
| SLC38A6 | 314 | 60 | 38 | 19.11 | 12.10 | 490 | 3 | 0.61 | |
| SLC38A7 | 314 | 22 | 69 | 7.01 | 21.97 | 490 | 1 | 0.20 | |
S2. Prediction of functional effects of SLC38s family with somatic alterations in integrated ESCC cohort.
| Sample ID | Gene ID | Alteration* | Type** | Polyphen-2 |
| SLC38, solute carrier family 38; ESCC, esophageal squamous cell carcinoma; MISSENSE, missense mutation; TRUNC, truncating mutation; *, somatic genomic alteration; **, alteration type. | ||||
| ESCC-0082 | SLC38A1 | p.K213N | MISSENSE | POSSIBLY DAMAGING |
| ESCC-0432 | SLC38A1 | p.I327T | MISSENSE | BENIGN |
| ESCC-0467 | SLC38A1 | p.L433V | MISSENSE | POSSIBLY DAMAGING |
| ESCC-0171 | SLC38A2 | p.I365V | MISSENSE | BENIGN |
| ESCC-0008 | SLC38A2 | p.A343T | MISSENSE | POSSIBLY DAMAGING |
| ESCC-0304 | SLC38A2 | p.SN55fs | TRUNC | POSSIBLY DAMAGING |
| ESCC-0428 | SLC38A4 | p.R292H | MISSENSE | BENIGN |
| ESCC-0275 | SLC38A5 | p.F302L | MISSENSE | BENIGN |
| ESCC-0089 | SLC38A6 | p.D452H | MISSENSE | BENIGN |
| ESCC-0249 | SLC38A6 | p.D326H | MISSENSE | POSSIBLY DAMAGING |
| ESCC-0421 | SLC38A6 | p.L440F | MISSENSE | POSSIBLY DAMAGING |
| ESCC-0308 | SLC38A7 | p.V218I | MISSENSE | POSSIBLY DAMAGING |
SLC38A3 CNA loss was significantly correlated with clinicopathological characteristics
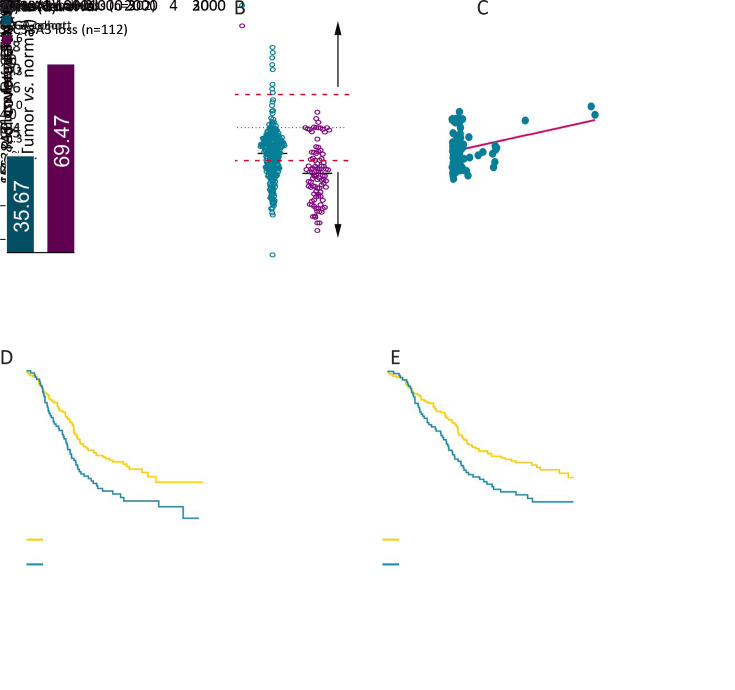
We were particularly intrigued by SLC38A3, the most frequent loss SLC38s family gene in ESCC, whose role in cancer development still remains controversial. To further confirm the CNA loss in ESCC, we evaluated the CNA types and frequency of SLC38A3 in the TCGA cohort. Consistently but unexpectedly, the CNA loss frequency of SLC38A3 in the TCGA cohort was almost twice as much as in our integrated cohort (69.47%, Figure 2A ). And both sets of cohorts showed more CNA loss, compared with CNA gain (Figure 2B ). CNAs often result in mRNA expression changes, thus we examined the correlation between SLC38A CNA and mRNA level in the TCGA cohort, and the result showed a weakly positive correlation (Pearson r=0.2909, P=0.004, Figure 2C ). Next, we further explored the association between SLC38A3 CNA loss and different clinicopathological features of patients with ESCC in our integrated cohort. Statistical analyses revealed that SLC38A3 CNA loss was significantly correlated with lymph node metastasis (LNM, P=0.026), drinking (P=0.002), advanced pathological stage (P=0.022), patient overall survival (OS) (P=0.016) and 5-year survival (P=0.029, Table 1 ).
2.
CNA and clinical association of SLC38A3 in ESCC integrated cohort and TCGA cohort. (A) CNA loss frequency of SLC38A3 in ESCC integrated cohort and TCGA cohort; (B) Scatter plot showed relative copy number variations of SLC38A3 in ESCC integrated cohort and TCGA cohort; (C) Correlation between SLC38A3 CNA and mRNA level in TCGA cohort (r=0.2909, P=0.004, Pearson Correlation Coefficient test); (D,E) Kaplan-Meier survival analysis of ESCC integrated cohort stratified by SLC38A3 CNA loss (n=314; P=0.011 for OS and P=0.014 for 5-year survival, log-rank test). CNA, copy number alteration; SLC38, solute carrier family 38; ESCC, esophageal squamous cell carcinoma; TCGA, The Cancer Genome Atlas; OS, overall survival.
1. Association of SLC38A3 CNV with clinicopathological features in integrated ESCC cohort.
| Clinicopathological features | Total [n (%)] | SLC38A3 CNV status [n (%)] | P | |
| Loss | Non-loss | |||
| SLC38s, solute carrier family 38; CNV, copy number variation; ESCC, esophageal squamous cell carcinoma; LNM, lymph node metastasis. | ||||
| Gender | 0.485 | |||
| Male | 42 (13.4) | 17 (40.5) | 25 (59.5) | |
| Female | 272 (86.6) | 95 (34.9) | 177 (65.1) | |
| Age (year) | 0.357 | |||
| ≤59 | 158 (50.8) | 52 (32.9) | 106 (67.1) | |
| >59 | 153 (49.2) | 58 (37.9) | 95 (62.1) | |
| LNM | 0.026 | |||
| Yes | 153 (48.7) | 64 (41.8) | 89 (58.2) | |
| No | 161 (51.3) | 48 (29.8) | 113 (70.2) | |
| N stage | 0.462 | |||
| N0+N1 | 79 (77.5) | 48 (60.8) | 31 (39.2) | |
| N2 | 23 (22.5) | 12 (52.2) | 11 (47.8) | |
| Grade | 0.225 | |||
| 1 | 100 (34.0) | 29 (29.0) | 71 (71.0) | |
| 2 | 146 (49.7) | 55 (37.7) | 91 (62.3) | |
| 3 | 48 (16.3) | 20 (41.7) | 28 (58.3) | |
| T stage | 0.603 | |||
| T1+T2 | 76 (24.2) | 29 (38.2) | 47 (61.8) | |
| T3+T4 | 238 (75.8) | 83 (34.9) | 155 (66.1) | |
| Smoking | 0.575 | |||
| Yes | 224 (75.9) | 77 (34.4) | 147 (65.6) | |
| No | 71 (24.1) | 27 (38.0) | 44 (62.0) | |
| Drinking | 0.002 | |||
| Yes | 83 (43.5) | 45 (54.2) | 38 (45.8) | |
| No | 108 (56.5) | 35 (32.4) | 73 (67.6) | |
| Pathological stage | 0.022 | |||
| I | 62 (21.0) | 13 (21.0) | 49 (79.0) | |
| II | 93 (31.5) | 35 (37.6) | 58 (62.4) | |
| III+IV | 140 (47.5) | 56 (40.0) | 84 (60.0) | |
| Overall survival | 0.016 | |||
| Dead | 141 (52.0) | 60 (42.6) | 81 (57.4) | |
| Alive | 130 (48.0) | 37 (28.5) | 93 (71.5) | |
| 5-year survival | 0.029 | |||
| Dead | 138 (50.9) | 58 (42.0) | 80 (58.0) | |
| Alive | 133 (49.1) | 39 (29.3) | 94 (70.7) | |
Importantly, SLC38A3 CNA loss was strongly associated with poor OS and 5-year survival of ESCC patients (P=0.011 and 0.014 respectively, Kaplan-Meier survival analysis and log-rank test, Figure 2D ,E ). The median survivals were substantially lower in SLC38A3 CNA loss group than in SLC38A3 CNA non-loss group (795.00 d vs. 1,773.00 d for OS), and the 5-year survival rate of SLC38A3 CNA loss group was also apparently lower compared to SLC38A3 CNA non-loss group (34.82% vs. 46.53%). Collectively, these findings indicate that SLC38A3 CNA loss might contribute to the progression of ESCC and could serve as a prognostic factor.
Frequently down regulation of SLC38A3 in human ESCC tissues
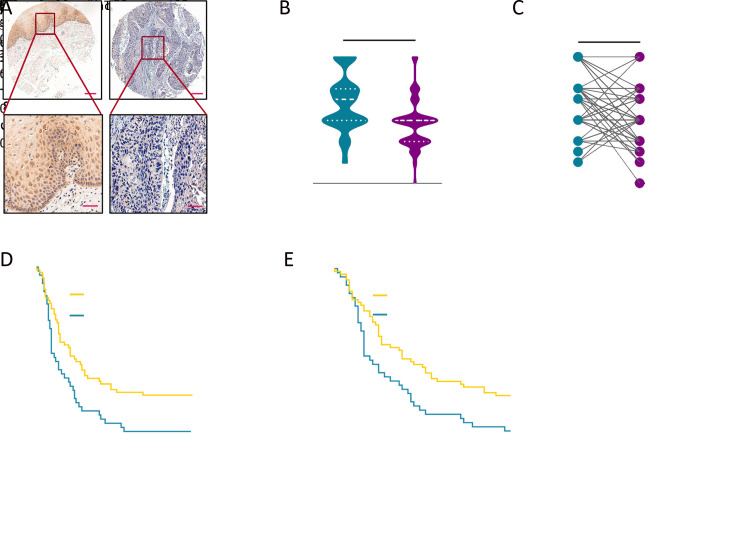
We sought to determine whether the protein level of SLC38A3 was also down regulated and related to clinicopathological characteristics in patients with ESCC. We first examined the expression level of SLC38A3 in TMA, including 75 paired adjacent normal tissues and ESCC tissues and 30 additional ESCC tissues via IHC assay. As expected, the results showed that SLC38A3 was significantly down regulated in ESCC tissues compared to normal tissues (Figure 3A −C ). Among the paired group, there were 57.33% (43/75) of patients showed a lower SLC38A3 level in ESCC tissues.
3.
Clinical implication of SLC38A3 in ESCC tumor samples. (A) Representative IHC photos of SLC38A3 expression in ESCC sample and normal tissue sample. Scale bar: 200 μm (upper) and 50 μm (bottom); (B) Quantitative IHC scores of SLC38A3 in normal tissue samples (n=75) and ESCC samples (n=105, Student’s t-test); (C) Quantitative IHC scores of SLC38A3 in 75 paired normal tissue samples and ESCC samples (paired Student’s t-test); (D,E) Kaplan-Meier survival analysis of ESCC tissue microarray stratified by SLC38A3 CNA loss (n=104; P=0.002 for OS and P=0.004 for 5-year survival, log-rank test). ***, P<0.001. SLC38s, solute carrier family 38; ESCC, esophageal squamous cell carcinoma; IHC, immunohistochemistry; OS, overall survival.
We also performed statistical analysis to explore the correlation between SLC38A3 expression level and different clinicopathological features of patients with ESCC. Consistently, the low expression level of SLC38A3 was significantly correlated with LNM (P=0.038), advanced American Joint Committee on Cancer (AJCC) stage (P=0.003), OS (P=0.004) and 5-year survival (P=0.007, Table 2 ). Notably, SLC38A3 low expression was strongly associated with poor OS and 5-year survival of ESCC patients (P=0.002 and P=0.004, respectively, Kaplan-Meier survival analysis and log-rank test, Figure 3 D ,E ). The median OS and 5-year survival of SLC38A3 low expression group were both 10 [95% confidence interval (95% CI), 8.19−13.81] months, while SLC38A3 high expression group was both 23 (95% CI, 9.50−36.50) months. The 5-year survival rate in SLC38A3 low expression group (7.14%, 3/42) was substantially lower than that of SLC38A3 high expression group (28.57%, 18/63).
2. Association of SLC38A3 expression with clinicopathological features in ESCC TMA cohort.
| Clinicopathological features | Total [n (%)] | SLC38A3 level [n (%)] | P | |
| High | Low | |||
| SLC38s, solute carrier family 38; ESCC, esophageal squamous cell carcinoma; TMA, tissue microarray; LNM, lymph node metastasis; AJCC, American Joint Committee on Cancer; OS, overall survival. | ||||
| Gender | 0.589 | |||
| Male | 77 (73.3) | 45 (58.4) | 32 (41.6) | |
| Female | 28 (26.7) | 18 (64.3) | 10 (35.7) | |
| Age (year) | 0.339 | |||
| ≤65 | 54 (51.4) | 30 (55.6) | 24 (44.4) | |
| >65 | 51 (48.6) | 33 (64.7) | 18 (35.3) | |
| LNM | 0.038 | |||
| Yes | 56 (54.4) | 28 (50.0) | 28 (50.0) | |
| No | 47 (45.6) | 33 (70.2) | 14 (29.8) | |
| N stage | 0.462 | |||
| N0+N1 | 79 (77.5) | 48 (60.8) | 31 (39.2) | |
| N2 | 23 (22.5) | 12 (52.2) | 11 (47.8) | |
| Grade | 0.328 | |||
| 1 | 36 (34.3) | 25 (69.4) | 11 (30.6) | |
| 2 | 45 (42.9) | 21 (46.7) | 24 (53.3) | |
| 3 | 24 (22.9) | 10 (41.7) | 14 (58.3) | |
| T stage | 0.235 | |||
| T1+T2 | 16 (15.7) | 12 (75.0) | 4 (25.0) | |
| T3+T4 | 86 (84.3) | 51 (59.3) | 35 (40.7) | |
| AJCC stage | 0.003 | |||
| 1 | 6 (6.0) | 6 (100) | 0 (0) | |
| 2 | 42 (42.0) | 30 (71.4) | 12 (28.6) | |
| 3 | 52 (52.0) | 25 (48.1) | 27 (51.9) | |
| OS | 0.004 | |||
| Dead | 86 (81.9) | 46 (53.5) | 40 (46.5) | |
| Alive | 19 (18.1) | 17 (89.5) | 2 (10.5) | |
| 5-year survival | 0.007 | |||
| Dead | 84 (80.0) | 45 (53.6) | 39 (46.4) | |
| Alive | 21 (20.0) | 18 (85.7) | 3 (14.3) | |
Interestingly, multivariate Cox regression survival analysis adjusting for SLC38A3 protein level, gender, age, T stage, LNM and pathological grade consistently showed strong correlation between SLC38A3 low expression and shorter OS (HR=1.799, 95% CI, 1.126−2.875, P=0.014) and 5-year survival (HR=1.796, 95% CI, 1.123−2.874, P=0.015,Table 3 ), indicating that SLC38A3 expression level was an independent prognostic factor for outcomes in ESCC. In fact, the stratification by SLC38A3 level displayed similar prognostic significance than the widely employed LNM (P=0.019, HR=1.789, 95% CI, 1.101−2.907,Table 3 ). Taken together, our results supported the notion that SLC38A3 was greatly involved in the ESCC progression and raised the possibility that SLC38A3 could be used as an independent prognostic factor in clinical practice.
3. Multivariate Cox regression analysis of risk factors associated with OS and 5-year survival.
| Risk factor | OS | 5-year survival | |||
| Adjusted HR (95% CI) | P | Adjusted HR (95% CI) | P | ||
| OS, overall survival; LNM, lymph node metastasis; SLC38, solute carrier family 38; HR, hazard ratio; 95% CI, 95% confidence interval. | |||||
| Gender | 1.245 (0.712−2.179) | 0.442 | 1.198 (0.684−2.096) | 0.528 | |
| Age | 1.182 (0.732−1.906) | 0.494 | 1.154 (0.712−1.868) | 0.561 | |
| LNM | 1.768 (1.093−2.859) | 0.020 | 1.789 (1.101−2.907) | 0.019 | |
| T stage (3/4 vs. 1/2) | 2.381 (1.157−4.899) | 0.018 | 2.621 (1.228−5.591) | 0.013 | |
| Grade (2/3 vs. 1) | 0.754 (0.439−1.297) | 0.308 | 0.758 (0.440−1.305) | 0.317 | |
| SLC38A3 low expression | 1.799 (1.126−2.875) | 0.014 | 1.796 (1.123−2.874) | 0.015 | |
Knockdown of SLC38A3 promotes malignant phenotypes of ESCC
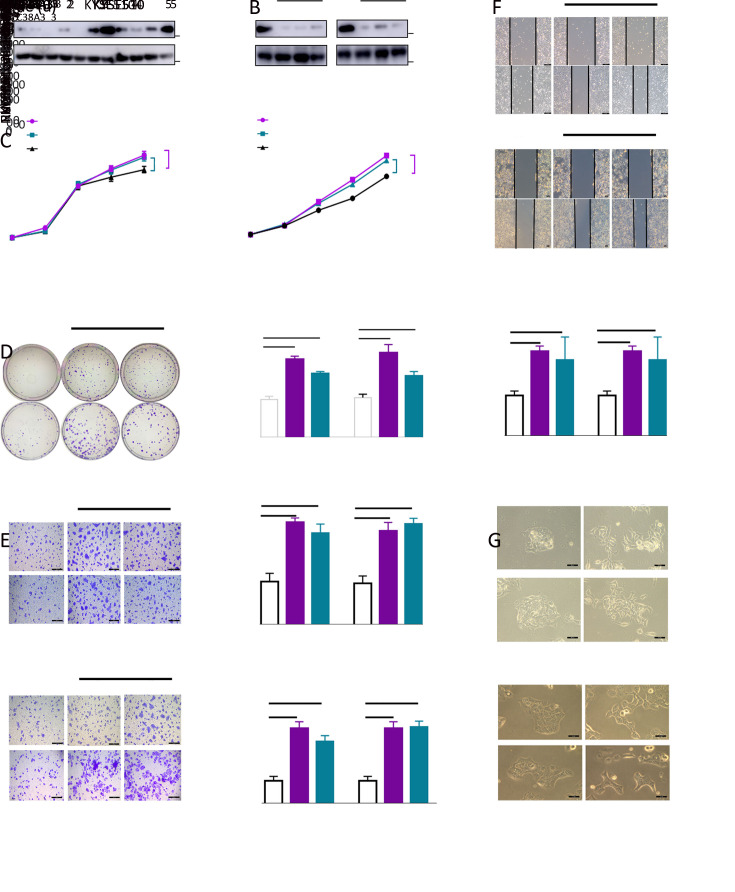
To elucidate the biological functions of SLC38A3 loss in ESCC, we firstly assessed SLC38A3 expression in nine ESCC cell lines and two immortalized normal esophageal cell lines NE2 and NE3. We found that SLC38A3 was lowly expressed in nearly all ESCC cell lines compared with normal controls, which was consistent with the clinical data (Figure 4A ). Then a set of phenotypes assays associated with tumor progression were conducted to determine the role of SLC38A3 in ESCC. We knocked down SLC38A3 through siRNA in KYSE410 and KYSE510 cells, which express relatively higher levels of SLC38A3 (Figure 4B ). Then we found that depletion of SLC38A3 augmented cellular malignant phenotypes including cell proliferation, colony formation, migration, invasion and wound healing (Figure 4C-F ). To further confirm whether the loss of SLC38A3 could induce EMT, we scrutinized the cell morphology with or without transforming growth factor beta (TGF-β) after knocking down SLC38A3. Consistently, a change of epithelium-like morphology of KYSE410 and KYSE510 cells to a mesenchyme-like morphology was observed after the depletion of SLC38A3 (Figure 4G ). These observations suggested that low expression of SLC38A3 played an important role in promoting migration and invasive potential of ESCC and it might function as a tumor suppressor gene.
4.
Loss of SLC38A3 promotes cell proliferation, migration and invasion of ESCC. (A) Western blotting analysis of SLC38A3 in nine ESCC cell lines and two immortalized normal esophageal cell lines NE2 and NE3; (B) Western blotting analysis of SLC38A3 expression after introducing small interfering RNA (siRNA) of SLC38A3 in KYSE410 and KYSE510. Cell lysates were collected 48 h after transfection; (C) Cell proliferation assays were performed after introducing siRNA of SLC38A3 in KYSE410 and KYSE510, and cell viability was quantified by MTS assay. Data represent
 , n=3 independent experiments. Unpaired t test was performed to calculate P values; (D) Colony formation assays with siCtrl, siSLC38A3_1, siSLC38A3_3 transfected cells for 10 d. Data represent
, n=3 independent experiments. Unpaired t test was performed to calculate P values; (D) Colony formation assays with siCtrl, siSLC38A3_1, siSLC38A3_3 transfected cells for 10 d. Data represent
 , n=3 independent experiments. Unpaired t test was performed to calculate P values; (E) Transwell-based migration and invasion assay with siCtrl, siSLC38A3_1, siSLC38A3_3 transfected cells for 12 h. Scale bars: 50 µm. Data represent
, n=3 independent experiments. Unpaired t test was performed to calculate P values; (E) Transwell-based migration and invasion assay with siCtrl, siSLC38A3_1, siSLC38A3_3 transfected cells for 12 h. Scale bars: 50 µm. Data represent
 , n=3 independent experiments. Unpaired t test was performed to calculate P values; (F) Wound healing assay of siCtrl, siSLC38A3_1, siSLC38A3_3 transfected cells for 24 h. Representative images were taken with phase contrast microscopy. Scale bars: 50 µm; (G) Representative images of siCtrl, siSLC38A3_1 transfected cells with or without TGF-β (10 ng/mL) for 48 h. Scale bars: 50 µm. **, P<0.01; ***, P<0.001; ****, P<0.0001 for three independent experiments analyzed by Student’st test. SLC38s, solute carrier family 38; ESCC, esophageal squamous cell carcinoma; TGF-β, transforming growth factor beta.
, n=3 independent experiments. Unpaired t test was performed to calculate P values; (F) Wound healing assay of siCtrl, siSLC38A3_1, siSLC38A3_3 transfected cells for 24 h. Representative images were taken with phase contrast microscopy. Scale bars: 50 µm; (G) Representative images of siCtrl, siSLC38A3_1 transfected cells with or without TGF-β (10 ng/mL) for 48 h. Scale bars: 50 µm. **, P<0.01; ***, P<0.001; ****, P<0.0001 for three independent experiments analyzed by Student’st test. SLC38s, solute carrier family 38; ESCC, esophageal squamous cell carcinoma; TGF-β, transforming growth factor beta.
SLC38A3 suppresses Snail transcription associated with SETDB1
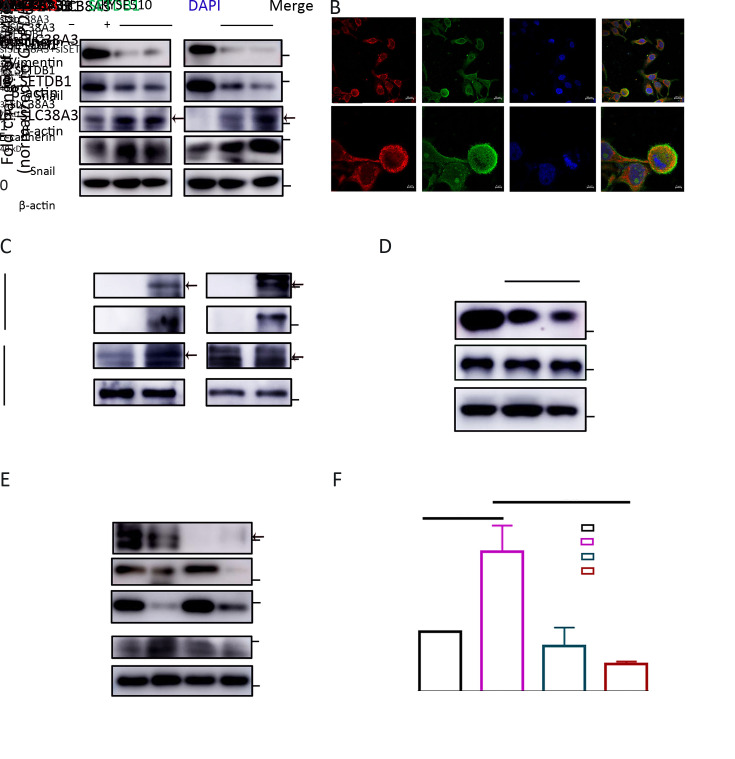
As our data showed that SLC38A3 could serve as a potential tumor suppressor gene in ESCC and it was involved in migration and invasion, which seemed independent of its canonical function of amino acid transporting. The depletion of SLC38A3 led to a remarkable change in EMT signaling pathway. We also noticed that among several master EMT transcription factors, Snail was elevated evidently (Figure 5A ), although others like Twist remained relatively unchanged (Data not shown). It is reasonable to assume that SLC38A3 could directly take part in the signaling pathway associated with EMT, and this effect may occur by regulating the expression of Snail. To explore the molecular mechanism of this implicit association between SLC38A3 and EMT, online database UniHI7 ( http://www.unihi.org/) and HitPredict ( http://www.hitpredict.org/htp_int.php?Value=9225) were employed to predict the direct physical interactors of SLC38A3 (39,40). As a result, four proteins were predicted to directly interact with SCL38A3 and among these protein candidates, a Histone-lysine N-methyltransferase SETDB1 showed high confidence among these interactions (Supplementary Table S3 ). Interestingly, Yang et al. reported that SETDB1 could act as an EMT inducer by binding directly to the promoter of the transcription factor Snail and promoted it’s expression (28). Therefore, we sought to determine whether SLC38A3 could interact with SETDB1 in ESCC cells. We found that SLC38A3 was co-localized with SETDB1 in the cytoplasm by immunofluorescence (Figure 5B ). Furthermore, the interaction between SLC38A3 and SETDB1 was also confirmed by immunoprecipitation (IP) (Figure 5C ). We detected the protein level of SETDB1 after interfering the expression of SLC38A3, and it seemed that their interaction did not influence the stability of SETDB1 (Figure 5D ), which implied that the interaction between SCL38A3 and SETDB1 might lead to subcellular location or modification of SETDBI, so as to regulate the transcription of SNAIL. Next, we sought to determine whether the regulation of Snail by SETDB1 was also affected by SLC38A3. Interestingly, both the protein level and the mRNA level of Snail increased after the depletion of SLC38A3. But this effect was rescued when depleting SLC38A3 and SETDB1 at the same time (Figure 5E ,F ). Therefore, our data indicated that SLC38A3 could interact with SETDB1 directly and suppressed the transcription of EMT-related gene SNAIL.
5.
SLC38A3 suppresses transcription of SNAIL by interacting with SETDB1. (A) Western blotting analyses of expression of indicated proteins in siCtrl; (B) Immunofluorescence images of SLC38A3, SETDB1 in KYSE510 cells. Scale bars: 5 µm; (C) Western blotting analyses of immunoprecipitation (IP) products and whole cell lysates derived from KYSE410; (D) Western blotting analyses of expression of total SETDB1 in siCtrl, siSLC38A3_1, siSLC38A3_3 transfected cells derived from KYSE510; (E) Western blotting analyses of expression of indicated proteins in siCtrl, siSLC38A3, siSETDB1 and siSLC38A3+siSETDB1 transfected cells derived from KYSE410; (F) Quantitative RT-PCR analyses of SNAIL levels in siCtrl, siSLC38A3, siSETDB1 and siSLC38A3+siSETDB1 transfected cells derived from KYSE410. SLC38s, solute carrier family 38; IgG, immunoglobulin G; RT-PCR, real-time polymerase chain reaction.
S3. List of potential interactors of SLC38A3*.
| Interaction | Interactor | Name | Experiments | Category | Method score | Annotation score | Interaction score | Confidence |
| SLC38, solute carrier family 38; *, Generated from HitPredict database (04 Feb 2019). | ||||||||
| 706501 | Q9NY72 | SCN3B | 1 | High-throughput | 0.38 | 1.00 | 0.618 | High |
| 573103 | Q15047 | SETB1 | 1 | High-throughput | 0.49 | 0.55 | 0.518 | High |
| 435188 | P46934 | NEDD4 | 1 | Small-scale | 0.38 | 0.55 | 0.458 | High |
| 186279 | P05114 | HMGN1 | 1 | High-throughput | 0.49 | 0.16 | 0.282 | High |
| 266349 | P12110 | CO6A2 | 1 | High-throughput | 0.49 | 0.16 | 0.282 | High |
Defect of SLC38A3 might confer ESCC cells with differential drug sensitivity
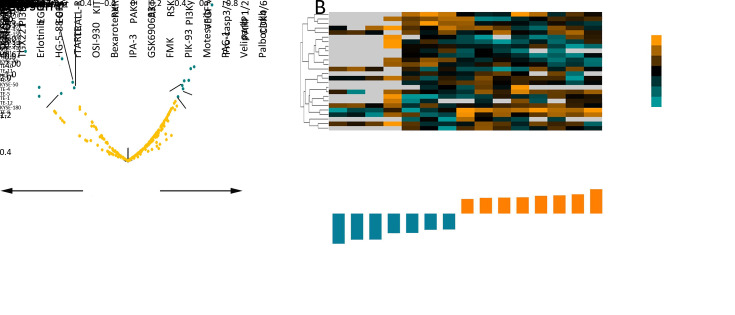
Dysregulation of amino acid transporters has been shown to significantly inference the signaling pathway activity (14). Thus, we tested whether SLC38A3 expression or CNA was associated with susceptibility to targeted pathway inhibitors using data from the Genomics of Drug Sensitivity in Cancer database (GDSC) (36,37). We obtained response data of 251 inhibitors tested on 26 ESCC cell lines and performed Pearson and Spearman correlation coefficient test to calculate the association between reported ln (IC50) values and SLC38A3 mRNA level and CNAs, respectively. Interestingly, fifteen inhibitors were showed significantly correlated with SLC38A3 expression (P<0.05, r>0.4 or r<−0.4,Figure 6A ). It seemed that SLC38A3 high expression cell lines were more resistant to inhibitors of CDK4/6, PARP1/2, Procaspase-3/7, VEGFR, PI3Kγ, RSK, AKT and PAK1, while more sensitive to RXR agonist, KIT inhibitor, TRAIL receptor agonist, EGFR inhibitors, PI3Kβ inhibitor and IKK inhibitor (Figure 6B , Supplementary Table S4 ). Consistently, loss of heterozygosity (LOH) of SLC38A3 was confirmed to be more resistant to VEGFR inhibitor and more sensitive to TRAIL receptor agonist (Supplementary Table S5 ). Palbociclib, an inhibitor of CDK4/6, was most significantly positively correlated with SLC38A3 mRNA level, while GSK319347A, an inhibitor of IKK, was most significantly negative correlated with SLC38A3 mRNA level (Figure 6A ), indicating that SLC38A3 was very likely involved in cell cycle regulation and IKK-NF-κB signaling. Additionally, the heatmap of drug sensitivity among cell lines showed substantial heterogeneity of cell-drug response, suggesting different drug combinations might exert a more prominent anticancer effect (Figure 6B ). Together, the expression and alterations of SLC38A3 might make ESCC show different sensitivity to drugs.
6.
SLC38A3 expression level correlated with several target therapeutic drugs. (A) Volcano plot displaying association between SLC38A3-loss and 251 pharmacologic inhibitors from GDSC study. X axis, Pearson correlation coefficient r; Y axis, significance −log 10 P (P<0.05, r>0.4 or r<−0.4); (B) Heatmap showed IC50 of SLC38A3 significantly correlated drugs in different ESCC cell lines. The ln (IC50) data were used for z-score calculation. SLC38s, solute carrier family 38; GDSC, Genomics of Drug Sensitivity in Cancer database; IC50, half inhibitory concentration; ESCC, esophageal squamous cell carcinoma.
S4. Sensitivities of significant SLC38A3 mRNA level associated drugs in ESCC cell lines.
| Cell
line name |
GSK319347A | TGX221 | Erlotinib | HG-5-
88-01 |
rTRAIL | OSI-930 | Bexarotene | IPA-3 | GSK690693 | FMK | PIK-93 | Motesanib | PAC-1 | Veliparib | Palbociclib |
| SLC38, solute carrier family 38; ESCC, esophageal squamous cell carcinoma; na, not available. | |||||||||||||||
| COLO-680N | na | na | na | na | 0.88 | 5.65 | 4.59 | 3.84 | 3.06 | 4.82 | 6.23 | 3.67 | 1.51 | 4.42 | 1.74 |
| EC-GI-10 | 4.07 | na | 2.72 | 3.91 | 0.19 | 4.39 | 4.60 | 5.49 | 5.04 | 5.71 | 2.73 | 2.29 | 3.66 | 4.30 | 1.01 |
| HCE-4 | na | na | na | na | na | 3.99 | 3.10 | 2.44 | 4.50 | 4.95 | 6.23 | na | na | na | na |
| KYSE-140 | na | na | na | 3.70 | na | 4.37 | 3.01 | 2.59 | 0.54 | na | 2.73 | 2.14 | 0.59 | 4.53 | 1.98 |
| KYSE-150 | na | na | na | 4.81 | −0.26 | 4.43 | 3.34 | 3.43 | 3.23 | 4.58 | 3.08 | 3.26 | 1.06 | 2.46 | −0.01 |
| KYSE-180 | na | na | na | na | −1.42 | 4.40 | 0.78 | 5.88 | 3.64 | 4.67 | 2.44 | na | 1.17 | na | na |
| KYSE-220 | na | na | na | 2.87 | 0.32 | 5.07 | 4.44 | 5.88 | 1.66 | 3.83 | 3.01 | 2.96 | 2.99 | 4.20 | 2.25 |
| KYSE-270 | na | na | na | na | −0.29 | 4.08 | 3.74 | 2.32 | 3.64 | 4.46 | 1.61 | na | 0.09 | na | na |
| KYSE-410 | na | na | na | 2.62 | −0.15 | 5.10 | 3.97 | 3.53 | 2.94 | 5.22 | 3.34 | 2.59 | 0.17 | 3.42 | −0.14 |
| KYSE-450 | na | na | na | 4.61 | −0.67 | 4.72 | 2.32 | 2.58 | 2.26 | 3.03 | 2.23 | 2.79 | 0.36 | 3.37 | 2.07 |
| KYSE-50 | na | na | na | 3.77 | −2.02 | 2.80 | 2.32 | 2.25 | 2.72 | 5.37 | 3.01 | 2.49 | 3.29 | 4.00 | 1.86 |
| KYSE-510 | na | na | na | na | −0.33 | 4.31 | 2.31 | 2.46 | 3.61 | 4.95 | 2.84 | 2.12 | 0.57 | 2.77 | 1.41 |
| KYSE-520 | na | na | na | na | 0.45 | 5.77 | 4.21 | 5.62 | 3.33 | 5.22 | 3.43 | 2.63 | 2.49 | 3.64 | 0.69 |
| KYSE-70 | na | na | na | na | −0.56 | 4.63 | 2.97 | 3.51 | 4.27 | 5.79 | 3.62 | na | 3.45 | 2.95 | na |
| OE21 | na | na | na | 2.50 | 0.42 | 5.25 | 2.86 | 3.30 | 3.84 | 5.66 | 3.57 | 3.03 | 0.81 | 3.51 | −0.08 |
| TE-1 | 3.10 | 3.07 | −1.40 | 1.07 | −2.45 | 3.90 | 3.19 | 6.74 | 5.56 | 6.17 | 5.49 | 3.96 | 4.18 | 4.87 | 4.66 |
| TE-10 | 4.05 | 3.75 | 1.25 | 3.05 | 0.05 | 3.61 | 3.42 | 3.17 | 4.78 | 5.24 | 4.25 | 3.24 | 3.01 | na | 1.75 |
| TE-11 | na | na | na | na | −0.81 | 4.17 | 3.56 | 4.94 | 4.58 | 5.15 | 4.22 | 3.05 | 2.11 | 3.24 | 1.48 |
| TE-12 | 3.82 | 3.63 | −2.62 | 2.57 | 0.30 | 3.50 | 2.22 | 6.60 | 3.85 | 6.11 | 3.68 | 3.34 | 4.05 | 4.60 | 3.33 |
| TE-15 | 4.48 | 5.11 | 0.96 | na | −0.01 | 3.54 | 3.30 | 2.46 | 2.49 | 3.51 | 1.99 | 3.45 | 1.70 | 4.58 | 1.43 |
| TE-4 | na | na | na | 3.19 | −1.80 | 4.01 | na | na | 3.04 | na | 4.88 | 2.42 | na | 4.19 | 1.92 |
| TE-5 | 4.53 | 4.90 | 0.55 | 2.60 | −0.90 | 4.91 | 2.53 | 4.19 | 3.37 | 5.51 | 4.42 | 3.34 | 3.81 | 4.23 | 1.48 |
| TE-6 | 4.34 | 4.54 | 0.18 | na | 0.41 | 4.18 | 4.03 | 3.38 | 3.54 | 6.25 | 3.70 | 2.81 | 3.50 | 4.86 | 1.93 |
| TE-8 | 4.40 | 4.19 | 1.81 | 4.91 | 0.24 | 3.96 | 2.54 | 3.56 | 4.84 | 5.67 | 3.07 | 3.42 | 2.86 | 4.39 | 0.06 |
| TE-9 | 4.30 | 2.30 | na | 2.64 | 0.00 | 4.37 | 2.73 | 2.94 | 4.94 | 5.44 | 5.79 | 3.41 | 3.31 | 4.41 | 3.01 |
| T-T | na | na | na | na | 0.16 | 4.33 | 1.20 | 4.72 | 4.89 | 5.53 | 2.54 | 2.81 | 3.10 | 3.92 | 1.03 |
| Pearson r | −0.84 | −0.73 | −0.73 | −0.55 | −0.54 | −0.45 | −0.44 | 0.41 | 0.44 | 0.45 | 0.46 | 0.50 | 0.51 | 0.54 | 0.68 |
| P | 0.00 | 0.03 | 0.04 | 0.03 | 0.01 | 0.02 | 0.03 | 0.04 | 0.02 | 0.03 | 0.02 | 0.02 | 0.01 | 0.01 | 0.00 |
S5. Sensitivities of significant SLC38A3 LOH associated drugs in ESCC cell lines.
| Type | Drug name | Target | Estimate rho | P |
| SLC38, solute carrier family 38; LOH, loss of heterozygosity; ESCC, esophageal squamous cell carcinoma. | ||||
| SLC38A3_LOH | LN_IC50: KIN001-244 | PDK1 (PDPK1) | −0.513974 | 0.0072 |
| SLC38A3_LOH | LN_IC50: YM201636 | PYKFYVE | −0.474938 | 0.0142 |
| SLC38A3_LOH | LN_IC50: Alectinib | ALK | −0.448914 | 0.0214 |
| SLC38A3_LOH | LN_IC50: NG-25 | TAK1, MAP4K2 | −0.448914 | 0.0214 |
| SLC38A3_LOH | LN_IC50: Cabozantinib | VEGFR, MET, RET, KIT, FLT1, FLT3, FLT4, TIE2, AXL | −0.448914 | 0.0214 |
| SLC38A3_LOH | LN_IC50: rTRAIL | TRAIL receptor agonist | 0.452059 | 0.0266 |
| SLC38A3_LOH | LN_IC50: MG-132 | Proteasome, CAPN1 | 0.724569 | 0.0272 |
| SLC38A3_LOH | LN_IC50: Pyrimethamine | Dihydrofolate reductase (DHFR) | 0.724569 | 0.0272 |
| SLC38A3_LOH | LN_IC50: VX-11e | ERK2 | −0.409878 | 0.0376 |
| SLC38A3_LOH | LN_IC50: PI-103 | PI3Kalpha, DAPK3, CLK4, PIM3, HIPK2 | −0.416025 | 0.0386 |
| SLC38A3_LOH | LN_IC50: KIN001-266 | MAP3K8 | −0.396866 | 0.0447 |
| SLC38A3_LOH | LN_IC50: KIN001-260 | IKKB | −0.396866 | 0.0447 |
Discussion
Amino acids transporters are critical mediators for tumor cell amino acid homeostasis, which have been shown dysregulation during carcinogenesis (14). However, their genomic signatures and biological functions in ESCC initiation and progression are less investigated yet. In this study, we provided the first evidence of genomic alterations and clinical implications of SLC38s in ESCC and highlighted the significant prognostic value of SLC38A3 in clinical practice. In our integrated ESCC whole-genome sequencing and whole-exome sequencing cohorts, we observed notable genetic disruptions of SLC38s family gene locus, with prominent CNAs and few somatic mutations. SLC38A3 was found to be the member with the most frequently CNA loss among this family, which was significantly correlated with poor survival outcomes of patients with ESCC. CNA loss and low expression of SLC38A3 were associated with LNM, advanced pathological stage and drinking. Subsequent experiments revealed that SLC38A3 physically interacted with SETDB1 and this interaction might lead to impairment of the transcription of SNAIL. Intriguingly, we also found that the mRNA level of SLC38A3 could serve as a predictor for differential drug sensitivity. The expression of SLC38A3 was significantly negatively correlated with CDK4/6 inhibitor Palbociclib, while it was positively correlated with EGFR inhibitor Erlotinib, indicating potential target therapy strategies for clinical practice by stratifying patients with SLC38A3 expression status.
Interestingly, it seemed that the system A type transporters (SLC38A1/2) tended to be affected by CNA gain, while system N transporters (SLC38A3/5/7) were prone to suffer from CNA loss. Additionally, Na+-amino acid co-transport by SLC38s transporters could elicit depolarization of the cell plasma membrane due to excessive Na+ accumulation (41), which might induce apoptosis (42). Probably due to the different transport mechanisms and depolarization risk, tumor cells might have evolved sophisticated manners for orchestrating the sonata of SLC38s family gene expression. Unlike other housekeeping amino acid transporters, which are thought to be expressed constitutively, SLC38s family transporters are dynamically regulated by stimuli such as amino acid shortage, hormones, osmolarity and cell growth (43). There was very little knowledge about these family genes in tumors, except for several papers that have revealed limited biological functions and clinical implications of SLC38A1/2/3/5 in breast cancer, hepatocellular carcinoma, non-small squamous lung cancer, cervical cancer, osteosarcoma and prostate cancer (16,19,44-47). However, the physiological functions of SLC38s family have been comprehensive studied (12). SLC38A2 has been reported to modulate the mTOR signaling pathway indirectly by modulating intracellular amino acids concentration (48-50). Moreover, it could also modulate mTOR signaling pathways and transcriptional signaling more directly by acting as a transceptor (51,52). Conversely, the mTOR signaling has been suggested to play a role in maintaining surface localization of SLC38A2 (53). Recently, SLC38A2 was reported to undergo ubiquitination-dependent degradation mediated by ubiquitin ligase RNF5 in response to endoplasmic reticulum (ER) stress-induced paclitaxel and subsequently reduced the glutamine uptake and inhibited the mTOR signaling in breast cancer (47). Since amino acids homeostasis, mTOR signaling pathways, and ER stress are important for tumorigenesis, it is logical to suspect that SLC38A2 CNA gain may contribute to the progression of ESCC.
SLC38A3 was the most affected gene of SLC38s family members in our integrate ESCC cohort and TCGA cohort, and has significant clinical implications, indicating that SLC38A3 was very likely to play a role in ESCC development. Of the system N transporters, SLC38A3 has been investigated in detail. It has been reported that during chronic metabolic acidosis, SLC38A3 mRNA in the kidney cortex was up-regulated 100-fold, thereby counteracting the reduced activity of the transporter (54). Cancer cells have a slightly elevated cytosolic pH (55), which could allow faster glutamine uptake. Accordingly, it is confusing to interpret the CNA loss of SLC38A3 in ESCC. Since SLC38A3 can bi-directionally co-transport the neutral substrate with Na+ and in antiport with H+, its biological functions and regulations were more complicated. Despite the fact that SLC38A3 was generally identified as a membrane-anchored protein, our data showed it mostly localized in cytoplasm (Figure 5B ), which was consistent with the results of The Human Protein Atlas ( https://www.proteinatlas.org/ENSG00000188338-SLC38A3/cell). On the basis of genomic alterations and clinical associations, our findings support the notion that SLC38A3 may act as a tumor suppressor in ESCC, which was contradictory with other overexpressed amino acids transporters in cancers (18). Therefore, it is logically assumed that SLC38A3 may function as an important signal molecule independent of its canonical role in amino acids transporting. Since many proteins have been identified for more vital functions totally different from their own classical functions, we consider it is reasonable for an amino acid transporter to play a role as a cytosol signaling mediator. As we know, GSK-3β, the last enzyme in glycogen biosynthesis, was identified initially to have important roles in metabolism (56). But later GSK-3β was found to function in a wide range of cellular processes and this pivotal kinase interacts with multiple signaling pathways such as: PI3K/PTEN/Akt/mTORC1, Ras/Raf/MEK/ERK, Wnt/beta-catenin, Hedgehog, Notch and others (57). Recently, Atsushi Hoshino and colleagues revealed that adenine nucleotide translocator (ANT) promotes mitophagy independently of its nucleotide translocase catalytic activity. Instead, the ANT complex is required for inhibition of the presequence translocase TIM23, which leads to stabilization of PINK1, in response to bioenergetic collapse (58). So we wonder if SLC38A3 could take part in other signaling pathways instead of simply amino acids transporting. Based on online protein-protein interaction predictions, we confirmed the SETDB1 interaction of SETDB1 with SLC38A3 by a set of experiments. Further studies illustrated that SLC38A3 could suppress EMT by inhibiting Snail transcription via interaction with SETDB1, indicating that SLC38A3 was able to function as a signal molecule to suppress tumorigenesis independent its canonical role of amino acids transporting. However, the mechanism how SETDB1 was changed to regulate the expression of Snail by SLC38A3 remains to be further investigated.
It is still challenging to identify therapeutic strategies for subsets of tumors with tumor suppressor genes loss. Given that more than 35% of SLC38A3 CNA loss and low expression were seen in our integrated ESCC cohort, TCGA cohort and ESCC TMA cohort, differential drug sensitivity predicted by SLC38A3 level may provide a new avenue for therapeutic strategies. Indeed, by analyzing the association between the mRNA level of SLC38A3 and the target drugs activities, we figured out 15 significant correlated drugs, including several inhibitors approved by Food and Drug Administration, such as Erlotinib, Bexarotene, Veliparib and Palbociclib. These findings raised the possibility that stratified by the expression of SLC38A3, patients with ESCC may benefit from these inhibitors. The gene-drug sensitivity analysis also revealed the potential molecular mechanisms of SLC38A3 in regulating carcinogenesis.
Conclusions
Our results demonstrated for the first time that SLC38s family genes were significantly disrupted in ESCC and implicated with clinical outcomes, highlighting that SLC38A3 could inhibit EMT of ESCC by interacting with SETDB1. The genomic signature of SLC38A3 suggests it can be a promising prognostic factor and predictor for differential drug sensitivity.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81830086, 81988101, 81802780), Beijing Municipal Administration of Hospital’s Mission Plan (No. SML20181101), Beijing Nova Program (No. Z191100001119038), Beijing Hospitals Authority Youth Programme (No. QML20191104).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Contributor Information
Weimin Zhang, Email: wmzhang411@163.com.
Qimin Zhan, Email: zhanqimin@bjmu.edu.cn.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Sun K, Zheng R, et al Cancer incidence and mortality in China, 2014. Chin J Cancer Res. 2018;30:1–12. doi: 10.21147/j.issn.1000-9604.2018.01.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Torre LA, Bray F, Siegel RL, et al Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 5.Lin DC, Hao JJ, Nagata Y, et al Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat Genet. 2014;46:467–73. doi: 10.1038/ng.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song Y, Li L, Ou Y, et al Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509:91–5. doi: 10.1038/nature13176. [DOI] [PubMed] [Google Scholar]
- 7.Gao YB, Chen ZL, Li JG, et al Genetic landscape of esophageal squamous cell carcinoma. Nat Genet. 2014;46:1097–102. doi: 10.1038/ng.3076. [DOI] [PubMed] [Google Scholar]
- 8.Zhang L, Zhou Y, Cheng C, et al Genomic analyses reveal mutational signatures and frequently altered genes in esophageal squamous cell carcinoma. Am J Hum Genet. 2015;96:597–611. doi: 10.1016/j.ajhg.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin HD, Liao XY, Chen YB, et al Genomic characterization of esophageal squamous cell carcinoma reveals critical genes underlying tumorigenesis and poor prognosis. Am J Hum Genet. 2016;98:709–27. doi: 10.1016/j.ajhg.2016.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agrawal N, Jiao Y, Bettegowda C, et al Comparative genomic analysis of esophageal adenocarcinoma and squamous cell carcinoma. Cancer Discov. 2012;2:899–905. doi: 10.1158/2159-8290.CD-12-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The Cancer Genome Atlas Research Network Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541:169–75. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bröer S The SLC38 family of sodium-amino acid co-transporters. Pflugers Arch. 2014;466:155–72. doi: 10.1007/s00424-013-1393-y. [DOI] [PubMed] [Google Scholar]
- 13.Mackenzie B, Erickson JD Sodium-coupled neutral amino acid (System N/A) transporters of the SLC38 gene family. Pflugers Arch. 2004;447:784–95. doi: 10.1007/s00424-003-1117-9. [DOI] [PubMed] [Google Scholar]
- 14.Bhutia YD, Babu E, Ramachandran S, et al Amino acid transporters in cancer and their relevance to “glutamine addiction”: novel targets for the design of a new class of anticancer drugs. Cancer Res. 2015;75:1782–8. doi: 10.1158/0008-5472.CAN-14-3745. [DOI] [PubMed] [Google Scholar]
- 15.Hensley CT, Wasti AT, DeBerardinis RJ Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013;123:3678–84. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang K, Cao F, Fang W, et al Activation of SNAT1/SLC38A1 in human breast cancer: correlation with p-Akt overexpression. BMC Cancer. 2013;13:343. doi: 10.1186/1471-2407-13-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie J, Chen Z, Liu L, et al shRNA-mediated Slc38a1 silencing inhibits migration, but not invasiveness of human pancreatic cancer cells. Chin J Cancer Res. 2013;25:514–9. doi: 10.3978/j.issn.1000-9604.2013.09.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kholodnyuk ID, Kozireva S, Kost-Alimova M, et al Down regulation of 3p genes, LTF, SLC38A3 and DRR1, upon growth of human chromosome 3-mouse fibrosarcoma hybrids in severe combined immunodeficiency mice. Int J Cancer. 2006;119:99–107. doi: 10.1002/ijc.21794. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Fu L, Cui M, et al Amino acid transporter SLC38A3 promotes metastasis of non-small cell lung cancer cells by activating PDK1. Cancer Lett. 2017;393:8–15. doi: 10.1016/j.canlet.2017.01.036. [DOI] [PubMed] [Google Scholar]
- 20.Nieto MA, Huang RY, Jackson RA, et al Emt: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 21.Thiery JP, Acloque H, Huang RYJ, et al Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 22.Nieto MA Epithelial-mesenchymal transitions in development and disease: old views and new perspectives. Int J Dev Biol. 2009;53:1541–7. doi: 10.1387/ijdb.072410mn. [DOI] [PubMed] [Google Scholar]
- 23.Kalluri R, Weinberg RA The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dongre A, Weinberg RA New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20:69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 25.Cano A, Pérez-Moreno MA, Rodrigo I, et al The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 26.Karanth AV, Maniswami RR, Prashanth S, et al Emerging role of SETDB1 as a therapeutic target. Expert Opin Ther Targets. 2017;21:319–31. doi: 10.1080/14728222.2017.1279604. [DOI] [PubMed] [Google Scholar]
- 27.Du D, Katsuno Y, Meyer D, et al Smad3-mediated recruitment of the methyltransferase SETDB1/ESET controls expression and epithelial-mesenchymal transition. EMBO Rep. 2018;19:135–55. doi: 10.15252/embr.201744250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang W, Su Y, Hou C, et al SETDB1 induces epithelialmesenchymal transition in breast carcinoma by directly binding with Snail promoter. Oncol Rep. 2019;41:1284–92. doi: 10.3892/or.2018.6871. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Huang J, Li Q, et al Histone methyltransferase SETDB1 promotes cells proliferation and migration by interacting withTiam1 in hepatocellular carcinoma. BMC Cancer. 2018;18:539. doi: 10.1186/s12885-018-4464-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du P, Huang P, Huang X, et al Comprehensive genomic analysis of oesophageal squamous cell carcinoma reveals clinical relevance. Sci Rep. 2017;7:15324. doi: 10.1038/s41598-017-14909-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koboldt DC, Zhang Q, Larson DE, et al VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–76. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao J, Aksoy BA, Dogrusoz U, et al Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cerami E, Gao J, Dogrusoz U, et al The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang W, Hong R, Xue L, et al Piccolo mediates EGFR signaling and acts as a prognostic biomarker in esophageal squamous cell carcinoma. Oncogene. 2017;36:3890–902. doi: 10.1038/onc.2017.15. [DOI] [PubMed] [Google Scholar]
- 35.Wu M, Wang Y, Yang D, et al A PLK1 kinase inhibitor enhances the chemosensitivity of cisplatin by inducing pyroptosis in oesophageal squamous cell carcinoma. EBioMedicine. 2019;41:244–55. doi: 10.1016/j.ebiom.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iorio F, Knijnenburg TA, Vis DJ, et al A landscape of pharmacogenomic interactions in cancer. Cell. 2016;166:740–54. doi: 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garnett MJ, Edelman EJ, Heidorn SJ, et al Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–5. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet 2013;Chapter 7: Unit7.20.
- 39.Kalathur RK, Pinto JP, Hernández-Prieto MA, et al UniHI 7: an enhanced database for retrieval and interactive analysis of human molecular interaction networks. Nucleic Acids Res. 2014;42:D408–14. doi: 10.1093/nar/gkt1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.López Y, Nakai K, Patil A HitPredict version 4: comprehensive reliability scoring of physical protein-protein interactions from more than 100 species. Database (Oxford) 2015;2015:bav117. doi: 10.1093/database/bav117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reimann F, Williams L, da Silva Xavier G, et al Glutamine potently stimulates glucagon-like peptide-1 secretion from GLUTag cells. Diabetologia. 2004;47:1592–601. doi: 10.1007/s00125-004-1498-0. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki-Karasaki Y, Suzuki-Karasaki M, Uchida M, et al Depolarization controls TRAIL-sensitization and tumor-selective killing of cancer cells: Crosstalk with ROS. Front Oncol. 2014;4:128. doi: 10.3389/fonc.2014.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGivan JD, Pastor-Anglada M Regulatory and molecular aspects of mammalian amino acid transport. Biochem J. 1994;299:321–34. doi: 10.1042/bj2990321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kondoh N, Imazeki N, Arai M, et al Activation of a system A amino acid transporter, ATA1/SLC38A1, in human hepatocellular carcinoma and preneoplastic liver tissues. Int J Oncol. 2007;31:81–7. [PubMed] [Google Scholar]
- 45.Bröer A, Rahimi F, Bröer S Deletion of amino acid transporter ASCT2 (SLC1A5) reveals an essential role for transporters SNAT1 (SLC38A1) and SNAT2 (SLC38A2) to sustain glutaminolysis in cancer cells. J Biol Chem. 2016;291:13194–205. doi: 10.1074/jbc.M115.700534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okudaira H, Shikano N, Nishii R, et al Putative transport mechanism and intracellular fate of trans-1-amino-3-18F-fluorocyclobutanecarboxylic acid in human prostate cancer. J Nucl Med. 2011;52:822–9. doi: 10.2967/jnumed.110.086074. [DOI] [PubMed] [Google Scholar]
- 47.Jeon YJ, Khelifa S, Ratnikov B, et al Regulation of glutamine carrier proteins by RNF5 determines breast cancer response to ER stress-inducing chemotherapies. Cancer Cell. 2015;27:354–69. doi: 10.1016/j.ccell.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baird FE, Bett KJ, MacLean C, et al Tertiary active transport of amino acids reconstituted by coexpression of System A and L transporters in Xenopus oocytes. Am J Physiol Endocrinol Metab. 2009;297:E822–9. doi: 10.1152/ajpendo.00330.2009. [DOI] [PubMed] [Google Scholar]
- 49.Evans K, Nasim Z, Brown J, et al Acidosis-sensing glutamine pump SNAT2 determines amino acid levels and mammalian target of rapamycin signalling to protein synthesis in L6 muscle cells. J Am Soc Nephrol. 2007;18:1426–36. doi: 10.1681/ASN.2006091014. [DOI] [PubMed] [Google Scholar]
- 50.Evans K, Nasim Z, Brown J, et al Inhibition of SNAT2 by metabolic acidosis enhances proteolysis in skeletal muscle. J Am Soc Nephrol. 2008;19:2119–29. doi: 10.1681/ASN.2007101108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pinilla J, Aledo JC, Cwiklinski E, et al SNAT2 transceptor signalling via mTOR: a role in cell growth and proliferation? Front Biosci (Elite Ed) 2011;3:1289–99. doi: 10.2741/e332. [DOI] [PubMed] [Google Scholar]
- 52.Hundal HS, Taylor PM Amino acid transceptors: gate keepers of nutrient exchange and regulators of nutrient signaling. Am J Physiol Endocrinol Metab. 2009;296:E603–13. doi: 10.1152/ajpendo.91002.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosario FJ, Kanai Y, Powell TL, et al Mammalian target of rapamycin signalling modulates amino acid uptake by regulating transporter cell surface abundance in primary human trophoblast cells. J Physiol. 2013;591:609–25. doi: 10.1113/jphysiol.2012.238014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karinch AM, Lin CM, Wolfgang CL, et al Regulation of expression of the SN1 transporter during renal adaptation to chronic metabolic acidosis in rats. Am J Physiol Renal Physiol. 2002;283:F1011–9. doi: 10.1152/ajprenal.00106.2002. [DOI] [PubMed] [Google Scholar]
- 55.White KA, Grillo-Hill BK, Barber DL Cancer cell behaviors mediated by dysregulated pH dynamics at a glance. J Cell Sci. 2017;130:663–9. doi: 10.1242/jcs.195297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Woodgett JR Molecular cloning and expression of glycogen synthase kinase-3/factor A. EMBO J. 1990;9:2431–8. doi: 10.1002/j.1460-2075.1990.tb07419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCubrey JA, Steelman LS, Bertrand FE, et al GSK-3 as potential target for therapeutic intervention in cancer. Oncotarget. 2014;5:2881–911. doi: 10.18632/oncotarget.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoshino A, Wang WJ, Wada S, et al The ADP/ATP translocase drives mitophagy independent of nucleotide exchange. Nature. 2019;575:375–9. doi: 10.1038/s41586-019-1667-4. [DOI] [PMC free article] [PubMed] [Google Scholar]