Abstract
Objective
Although T-cell immunoglobulin and mucin-domain containing molecule-3 (Tim-3) has been recognized as a promising target for cancer immunotherapy, its exact role in breast cancer has not been fully elucidated.
Methods
Tim-3 gene expression in breast cancer and its prognostic significance were analyzed. Associated mechanisms were then explored in vitro by establishing Tim-3-overexpressing breast cancer cells.
Results
In a pooled analysis of The Cancer Genome Atlas (TCGA) database, Tim-3 gene expression levels were significantly higher (P<0.001) in breast cancer tissue, compared with normal tissues. Tim-3 was a prognosis indicator in breast cancer patients [relapse-free survival (RFS), P=0.004; overall survival (OS), P=0.099]. Tim-3 overexpression in Tim-3low breast cancer cells promoted aggressiveness of breast cancer cells, as evidenced by enhanced proliferation, migration, invasion, tight junction deterioration and tumor-associated tubal formation. Tim-3 also enhanced cellular resistance to paclitaxel. Furthermore, Tim-3 exerted its function by activating the NF-κB/STAT3 signalling pathway and by regulating gene expression [cyclin D1 (CCND1), C-Myc, matrix metalloproteinase-1(MMP1), TWIST, vascular endothelial growth factor (VEGF) upregulation, concomitant with E-cadherin downregulation). Lastly, Tim-3 downregulated tight junction-associated molecules zona occludens (ZO)-2, ZO-1 and occludin, which may further facilitate tumor progression.
Conclusions
Tim-3 plays an oncogenic role in breast cancer and may represent a potential target for antitumor therapy.
Keywords: Breast neoplasm, hepatitis A virus cellular receptor 2, tight junction, aggression, chemoresistance
Introduction
Breast cancer is the most frequently diagnosed malignancy and the main cause of cancer-associated mortality in women (1). Although comprehensive treatments are clinically available, the response of individual breast cancer patients greatly varies, partly due to different antitumor immune responses (2). Dysregulation of immune checkpoints can play an important role in tumor immune evasion, especially through tumor-reactive T-cell exhaustion (3).
T-cell immunoglobulin and mucin-domain containing molecule-3 (Tim-3), also known as hepatitis A virus cellular receptor 2 (HAVCR2), is a negative immune checkpoint molecule expressed on a variety of immune cells including T-cells (4), dendritic cells (5) and macrophages (6). Tim-3 can reduce cell proliferation, decrease the production of effective cytokines and increase apoptosis of effector T-cells, through interaction with its ligands including galectin-9, high mobility group protein B1 (HMGB1), carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM-1) and phosphatidylserine (7,8). Tim-3 is considered a critical mediator in cancer progression and a potential therapeutic target. Tim-3 blockade has demonstrated promising results in multiple preclinical cancer models (9). Evidence suggests that resistance to anti-cytotoxic T-lymphocyte-associated antigen 4 or anti-programmed death-1 (PD-1)/PD-ligand 1 (PD-L1) inhibitors is compensated by upregulation of additional immune checkpoints, including Tim-3 (10). Moreover, PD-1 and Tim-3 co-blockade resulted in a significant survival advantage in a murine lung cancer model (11). These findings support the view that Tim-3 may be a potential target for tumor therapy.
Tim-3 is overexpressed in many types of malignant tumors. Ectopic expression of Tim-3 in tumor cells was correlated with more advanced pathologic T classification in non-small-cell lung carcinoma (12), lymph-vascular invasion in gastric cancer (13), lung metastasis in clear cell renal cell carcinoma (14), and lymphatic metastasis in colon cancer (8). A meta-analysis also suggested that high expression of Tim-3 in solid tumors led to significantly shorter overall survival (OS) (15). Therefore, Tim-3 has been described as a prognostic indicator for cancer patients. However, Tim-3 downregulation promotes invasion and metastasis of colorectal cancer cells (16). Low Tim-3 expression levels in tumor tissues is associated with poor prognosis for metastatic prostate cancer (17). Tim-3 expression in renal cell carcinoma is associated with longer progression-free survival (PFS) and OS (18). These seemingly contradictory results suggest that the role of Tim-3 might be dependent on tumor type. Tim-3 is reported to be overexpressed in breast cancer tissues (15,19), and high Tim-3 levels are associated with poor prognosis (19). Downregulation or overexpression of Tim-3 suppresses or enhances the proliferation, migration and invasion of breast cancer cells, respectively (19).
Previous studies demonstrated that Tim-3 was associated with resistance to anti-angiogenic drug sunitinib and to mTOR inhibitor rapamycin in a renal cell carcinoma cell line (20). Additionally, Tim-3 promoted resistance to adriamycin and carboplatin in lymphoma ATN-1 cells (21). These findings implied a possible role in tumor angiogenesis and chemoresistance. Therefore, the aim of the present study was to evaluate the clinical significance of Tim-3 using large-scale genomic data analysis and to determine the mechanisms underlying the effects of Tim-3 in breast cancer cell lines. The role of Tim-3 in tumor-associated angiogenesis and chemoresistance was also examined.
Materials and methods
Cell lines and culture
Human breast cancer cell lines MDA-MB-231 and MCF7 and human umbilical vein endothelial cells (HUVECs) were purchased from American Type Culture Collection (Middlesex, UK). These cells were maintained in a humidified incubator at 37 °C with 5% CO2.
Stable cell lines overexpressing Tim-3
To establish Tim-3-overexpressing breast cancer cell lines, lentiviral vectors containing full-length Tim-3 (PLV[Exp]-EGFP: T2A: Puro-CMV> hHAVCR2 [NM_032782.4]) or Scramble (Scr) negative control (PLV[Exp]-EGFP: T2A: Puro-CMV>stuffer_300 bp) were transduced into MDA-MB-231 and MCF7 cells (Vector builder, USA) according to the manufacturer’s instructions.
Quantitative real-time polymerase chain reaction (qPCR)
Total RNA from cultured cells was extracted using TRIzol reagent (Sigma-Aldrich, Dorset, UK) according to the manufacturer’s instructions. RNA was then reverse transcribed into cDNA using the GoScriptTM Reverse Transcription System kit (Promega, Madison, USA). Subsequently, qPCR was carried out using an iCycler iQTM (Bio-Rad Laboratories, Hemel Hempstead, UK). The primer sequences used in this study are listed in Table 1 .
1. Primer sequences.
| Gene | Forward Primers (5’→3’) | Reverse Primers (5’→3’) |
| CCND1, cyclin D1; IL-6, interleukin 6; MMP-1, matrix metalloproteinase-1; Tim-3, T-cell immunoglobulin and mucin-domain containing molecule-3; VEGF, vascular endothelial growth factor; ZO, zona occludens. The Z Sequence is highlighted in bold. | ||
| CCND1 | CGGTGTCCTACTTCAAATGT | ACTGAACCTGACCGTACAGAAGCGGTCCAGGTAGTTC |
| C-Myc | TGCTCCATGAGGAGACAC | ACTGAACCTGACCGTACATGATCCAGACTCTGACCTTT |
| E-cadherin | CACACGGGCTTGGATTT | ACTGAACCTGACCGTACAGACCTCAAAAGGTACCACAT |
| GAPDH | CTGAGTACGTCGTGGAGTC | ACTGAACCTGACCGTACACAGAGATGATGACCCTTTTG |
| IL-6 | TCATCACTGGTCTTTTGGAG | ACTGAACCTGACCGTACACAGGGGTGGTTATTGCATC |
| MMP1 | CTTTTGTCAGGGGAGATCAT | ACTGAACCTGACCGTACAGGTCCACCTTTCATCTTCAT |
| NF-κB | ACAGAGAGGATTTCGTTTCC | ACTGAACCTGACCGTACAGTTGCAGATTTTGACCTGAG |
| Occludin | GAATTCAAACCGAATCATTG | ACTGAACCTGACCGTACATGAAGAATTTCATCTTCTGG |
| STAT3 | CATGGAAGAATCCAACAACG | ACTGAACCTGACCGTACAAATCAGGGAAGCATCACAAT |
| Tim-3 | GCTCCATGTTTTCACATCTT | ACTGAACCTGACCGTACAATTCCACTTCTGAGGACCTT |
| TWIST | AGCAACAGCGAGGAAGAG | ACTGAACCTGACCGTACAGAGGACCTGGTAGAGGAAGT |
| VEGFA | GAGCCGGAGAGGGAG | ACTGAACCTGACCGTACACTGGGACCACTTGGCAT |
| VEGFD | TCCACATTGGAACGATCTGA | ACTGAACCTGACCGTACACTCCACAGCTTCCAGTCCTC |
| ZO-1 | TGACACACATGGTAGACTCA | ACTGAACCTGACCGTACAGTAACTGCGTGAATATTGCT |
| ZO-2 | CAAAAGAGGATTTGGAATTG | ACTGAACCTGACCGTACAGAGCACATCAGAAATGACAA |
| β-catenin | AAAGGCTACTGTTGGATTGA | ACTGAACCTGACCGTACACTGAACTAGTCGTGGAATGG |
Western blotting
Western blot analysis was carried out on whole cell lysates. Antibodies against Tim-3 (Ab241332), NF-κB (p65) (Ab16502), p-NF-κB (p-p65) (Ab194726) and vascular endothelial growth factor (VEGF) A (Ab9570) were purchased from (Abcam Cambridge, UK). GAPDH (sc-47724), p-β-catenin (sc-16743-R), cyclin D1 (CCND1) (sc-8396), C-Myc (sc-70465), matrix metalloproteinase-1 (MMP1) (sc-21731), TWIST (sc-6269), zona occludens (ZO)-1 (sc-10804), ZO-2 (sc-11448), occludin (sc-133256) and VEGFB (sc-13083) antibodies were obtained from Santa Cruz (Insight Biotechnology Limited, Middlesex UK). E-Cadherin (AF748) and VEGFD (MAB286) antibodies were purchased from R&D Systems (Abingdon, Oxfordshire, UK). STAT3 (S5933), p-STAT3 (SAB4504541), interleukin (IL)-6 (17901) and β-catenin (C2206) antibodies were purchased from Sigma-Aldrich (Gillingham, Dorset, UK). Anti-mouse (A5278), anti-rabbit (A0545) and anti-goat (A8919) secondary antibodies were obtained from Sigma-Aldrich (Gillingham, Dorset, UK).
Immunohistochemistry
Cultured cells were fixed using 95% ethanol for approximately 1 h, then treated with a Tim-3 primary antibody (1:500, Ab241332, Abcam, Cambridge, UK) overnight at 4 °C. For the negative control, the primary antibody was replaced with phosphate buffer saline (PBS). Samples were then treated with an anti-rabbit HRP-conjugated secondary antibody (Abcam, UK) for 30 min. The colour was developed by using 0.025% 3, 3’-diaminobenzidine (DAB) solution. Nuclei were counterstained with hematoxylin. Excess staining was removed using hydrochloric acid and alcohol solution. Ammonia was used to restore staining colour.
Cell proliferation and cytotoxicity assay
Cell proliferation was assessed using an AlamarBlue assay (Serotec Ltd., Oxford, UK) according to the manufacturer’s instructions. The percentage of growth during the incubation period was calculated against the fluorescence values at d 0.
For cytotoxicity assays, 8×103 cells/ well were seeded into a 96-well plate with medium containing 1% FBS. After cells were starved overnight, medium was replaced with different concentrations of paclitaxel (Sigma-Aldrich, UK), paclitaxel + Stattic (MedChemExpress, USA) or paclitaxel + SC75741 (MedChemExpress, USA) and incubated for 48 h. Cell viability was then assessed by AlamarBlue assay.
Wound healing assay
Cells were seeded into a 24-well plate at a density of 2×105 cells/well and grown to confluence. The cell monolayer was then scratched using a 1 mL pipette tip. Migration of cells to the wounding gap was monitored using an EVOS® FL imaging system (Life Technologies, Carlsbad, CA, USA) with a 4× objective every 2 h for 24−48 h.
Matrigel invasion assay
A Transwell Matrigel assay was used to assess cell invasion in vitroas described previously (22).
Cell-matrix adhesion assay
The cell-matrix adhesion was evaluated using a 96-well plate coated with 10 μg/well Matrigel (23).
Tube formation assay
Cancer cells stably transfected with Tim-3 overexpression (OE) or Scr control vectors were cultured to reach 70%−80% confluence in complete medium. The cells were then washed twice with PBS and re-suspended in serum-free medium. The supernatant (conditioned medium) was then collected after a 24-h incubation, filtered with a 0.22-μm filter (Millipore) and stored at −80 °C for further use. The tube formation ability of HUVECs mimicking angiogenesis was then measured in response to the tumor-conditioned medium.
Enzyme-linked immunosorbent assay (ELISA)
Cultured cells were concentrated using the Amicon Ultra-4 centrifugal filter units (Sigma-Aldrich; Merck KGaA) and the medium was subsequently used for ELISA. ELISA was performed using the human VEGFC (cat. no. E-EL-H1600; Elabscience) and vascular endothelial growth factor receptor 2 (VEGFR2) ELISA kit (cat. no. E-EL-H1603; Elabscience), according to the manufacturer’s protocol.
Electric cell-substrate impedance sensing (ECIS) assay
ECIS was used to assess cell migration as described previously (24). Briefly, an ECIS Zθ system with a 96W1E+ array plate (Applied Biophysics, Inc., Troy, NY, USA) was used for the measurement of cell function, including initial attachment, spreading and barrier function. The 96W1E+ array plate was stabilized using normal medium 2 h in advance. A total of 5×104 cells per well were then seeded and cultured for 24 h. Each group was set up at least six repetitions. The resistance across the array was recorded at multiple frequencies.
Transepithelial resistance (TER) and paracellular permeability (PCP)
TER is used to assess the integrity of tight junction (TJ) dynamics in cell culture models of epithelial monolayers as a widely accepted quantitative technique. An EVOM voltohmmeter (World Precision Instruments, Aston, Herts, UK), equipped with STX2 chopstick electrodes (World Precision Instruments, Inc., Sarasota, FL, USA) was used to measure TER, and PCP was assessed. The medium in the upper chamber was replaced with medium containing 0.2 mg/mL fluorescein isothiocyanate (FITC)-dextran 10 kDa. Then, 50 μL of medium from outside the insert was transferred into a black 96-well cell culture microplate (Greiner Bio-One) in duplicate every 2 h for 10 h. Basolateral dextran passage was analyzed with a GloMax®-Multi Microplate Multimode Reader (Promega UK Ltd., Southampton, UK) at 490-nm excitation and 510−570 nm emission. Measurement of dextran-indicated PCP was then normalized to the 0 h time point.
Statistical analysis
Data are presented as the
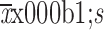 unless stated otherwise. GraphPad Prism (Version 7.0; GraphPad Software, San Diego, CA, USA) or R language (Version 3.6.3; https://www.R-project.org) were used for statistical analysis. The normality of the data was determined using the Shapiro-Wilk normality test. Two-group comparisons were analyzed using a two-sided t-test when data were normally distributed and a Mann-Whitney U test otherwise. Differences between multiple groups were analyzed using Analysis of Variance (ANOVA) followed by Tukey’s post hoc test for pairwise comparison if the data were normal. For non-parametric data, the Kruskal-Wallis test was used instead. For cytotoxicity assay, two-way ANOVA was used to evaluate the significance by considering two factors, paclitaxel doses and cell lines. Pairwise comparison between two cell lines was performed using Holm’spost hoc test. Differences were considered statistically significant if P<0.05. Experiments were repeated 2−4 times unless otherwise stated. The statistical significance in the figures is shown as*, P<0.05 or**, P<0.01 or***, P<0.001.
unless stated otherwise. GraphPad Prism (Version 7.0; GraphPad Software, San Diego, CA, USA) or R language (Version 3.6.3; https://www.R-project.org) were used for statistical analysis. The normality of the data was determined using the Shapiro-Wilk normality test. Two-group comparisons were analyzed using a two-sided t-test when data were normally distributed and a Mann-Whitney U test otherwise. Differences between multiple groups were analyzed using Analysis of Variance (ANOVA) followed by Tukey’s post hoc test for pairwise comparison if the data were normal. For non-parametric data, the Kruskal-Wallis test was used instead. For cytotoxicity assay, two-way ANOVA was used to evaluate the significance by considering two factors, paclitaxel doses and cell lines. Pairwise comparison between two cell lines was performed using Holm’spost hoc test. Differences were considered statistically significant if P<0.05. Experiments were repeated 2−4 times unless otherwise stated. The statistical significance in the figures is shown as*, P<0.05 or**, P<0.01 or***, P<0.001.
Results
Tim-3 is upregulated in breast cancer tissues
Gene expression levels of Tim-3 were analyzed in breast cancer (n=1,097) and normal tissue (n=114) from The Cancer Genome Atlas (TCGA) breast cancer datasets. Although Tim-3 was overexpressed in breast cancer compared with normal tissues (P<0.001) (Figure 1A ), there was no statistical difference among different breast cancer subtypes (P=0.074) (Figure 1B ). The effect of Tim-3 on patient survival was evaluated using the KM-plotter database (http://kmplot.com/analysis/) by entering “Gene symbol = HAVCR2 (235458_at)”, selecting ‘JetSet best probe set’, and dividing patients using “Auto select best cut-off”. Patients with high Tim-3 expression had a significantly worse relapse-free survival (RFS) when over a 20-year follow-up period (P=0.004) (Figure 1C ). OS displayed a similar trend but this was not statistically significant (P=0.099) (Figure 1E ).
1.
Tim-3 gene expression in breast cancer and association with patient survival. (A) Tim-3 mRNA levels in primary tumor vs. normal tissue (P<0.001); (B) Tim-3 mRNA levels among breast cancer subgroups (P=0.074); (C) Association ofTim-3 gene expression with RFS in breast cancer patients [HR, 1.29 (1.08−1.54); log-rank P=0.004]; (D) Association of Tim-3 gene expression levels with RFS in breast cancer subtypes; (E) Association of Tim-3 gene expression with OS in breast cancer patients (P=0.099); (F) Association of Tim-3 gene expression levels with OS in breast cancer subtypes. Tim-3, T-cell immunoglobulin and mucin-domain containing molecule-3; RFS, relapse-free survival; HR, hazard ratio; HER2, human epidermal growth factor receptor 2; OS, overall survival.
In subgroup analysis, high Tim-3 expression was associated with worse RFS in luminal A (P<0.001) and luminal B (P=0.039) subtypes, but improved RFS in basal breast cancer (P<0.001) (Figure 1D ). The human epidermal growth factor receptor 2 (HER2)-positive subtype showed a similar trend with basal subtype, without statistical significance (P>0.05) (Figure 1D ). For OS, high Tim-3 levels were associated with worse prognosis in luminal A subtype (P=0.019). In basal type, patients with high levels of Tim-3 had a better prognosis (P<0.001) (Figure 1F ).
Stable cell lines overexpressing Tim-3
To evaluate the role of Tim-3 in breast cancer in vitro, Tim-3low MDA-MB-231 and MCF7 cell lines (initial assessment shown in Supplementary Figure S1 ) were stably transfected with Tim-3 OE plasmid or Scr, respectively. Tim-3 levels increased in Tim-3 OE cells, compared with the Scr and wildtype (WT) controls both at mRNA and protein levels, confirming that Tim-3 was successfully transfected (Figure 2A ). Tim-3 was expressed in both the cytoplasm and membrane in Tim-3 OE cells (Supplementary Figure S2 ).
S1.
Endogenous gene expression levels of Tim-3 in ten breast cancer, normal human mammary epithelial cells and two vascular endothelial cells accessed by qPCR. Tim-3, T-cell immunoglobulin and mucin-domain containing molecule-3; qPCR, quantitative real-time polymerase chain reaction.
2.
Effect of Tim-3 OE on proliferation, adhesion and signalling in breast cancer cells. (A) Validation of Tim-3 OE in MDA-MB-231 and MCF7 cells by qPCR and western blotting; (B) Effect of Tim-3 OE on proliferation of MDA-MB-231 cells (6 d, P<0.001); (C) Effect of Tim-3 OE on proliferation of MCF7 cells (6 d, P<0.001); (D) Tim-3 OE reduces adhesion in MDA-MB-231 cells (P=0.006); (E) Tim-3 OE affects adhesion of MCF7 cells (P<0.001); (F) Protein levels of STAT3, phosphorylated STAT3, NF-κB, phosphorylated NF-κB, IL-6 and proliferation markers; (G) Quantitative densitometric analysis of western blots (n=3); (H) Gene expression profile of MDA-MB-231 cell line; (I) Gene expression profile of MCF7 cell line (n=3). OE, overexpression; Tim-3, T-cell immunoglobulin and mucin-domain containing molecule-3; qPCR, quantitative real-time polymerase chain reaction; IL-6, interleukin 6.*, P<0.05;**, P<0.01;***, P<0.001.
S2.
Tim-3 was expressed in both cytoplasm and membrane in Tim-3 OE MDA-MB-231 cells detected by immunohistochemistry. (A) MDA-MB-231 Scr; (B) MDA-MB-231 Tim-3 OE. Tim-3, T-cell immunoglobulin and mucin-domain containing molecule-3; OE, overexpression.
Effect of Tim-3 in cell proliferation and adhesion in vitro
Effects of Tim-3 on cell proliferation and adhesion were evaluated in vitro. Proliferation was enhanced in MDA-MA-231 Tim-3 OE and MCF7 Tim-3 OE cells, compared with their Scr controls, especially after 6 d (P<0.001 for both) (Figure 2B ,C ), indicating that Tim-3 could promote breast cancer cell proliferation.
Cell-matrix adhesion assay was used to investigate the role of Tim-3 in cell adhesion. The adhesive ability of cells was increased when Tim-3 was overexpressed in the MCF7 cell line (P<0.001) (Figure 2E ). However, Tim-3 OE reduced adhesion of MDA-MB-231 cell line (P=0.006) (Figure 2D ). Thus, the effect of Tim-3 may be dependent on cell type.
The signalling pathways at play in Tim-3 OE breast cancer cells were examined. NF-κB (p65), STAT3 and IL-6 were all upregulated both at gene and protein levels in MDA-MB-231 Tim-3 OE cells. Additionally, p-STAT3 levels were increased in MDA-MB-231 Tim-3 OE cells (Figure 2F ,G ). NF-κB (p65), p-NF-κB (p-p65), STAT3, p-STAT3 and IL-6 were also upregulated in MCF7 Tim-3 OE cells at the protein level (Figure 2F ,G ). Moreover, cell proliferation markers β-catenin, p-β-catenin, CCND1 and C-Myc were upregulated in MDA-MB-231 Tim-3 OE cells (Figure 2G ,H ). β-catenin, p-β-catenin and C-Myc protein levels were also increased in MCF7 Tim-3 OE cells (Figure 2G ), but not at the mRNA level (Figure 2I ). Thus, the NF-κB/STAT3 pathway was involved in breast cancer progression following Tim-3 upregulation.
Tim-3 increases cell invasion and migration in vitro
Effects of Tim-3 on cell invasion and migration were evaluated in Transwell Matrigel invasion assays and wound healing assays. Tim-3 OE cells were significantly more invasive than Scr controls, both in MDA-MB-231 (P=0.030) (Figure 3A ) and MCF7 (P<0.001) (Figure 3B ) cells. Cell migration was also enhanced in MDA-MB-231 (P<0.001 at 24h) (Figure 3C ) and in MCF7 Tim-3 OE cells (P=0.007 at 48 h) (Figure 3D ).
3.
Effect of Tim-3 OE on invasion and migration of breast cancer cells. (A) MDA-MB-231 cell invasion following Tim-3 OE; (B) MCF7 cell invasion following Tim-3 OE; (C) Migration of MDA-MB-231 Tim-3-overexpressing cells; (D) Migration of MCF7 Tim-3-overexpressing cells; (E) Protein levels of MMP1, TWIST and E-cadherin in breast cancer cells (left) and quantitative densitometric analysis (n=3, right); (F) Gene expression levels of MMP1, TWIST and E-cadherin (n=3). OE, overexpression; Tim-3, T-cell immunoglobulin and mucin-domain containing molecule-3; MMP-1, matrix metalloproteinase-1. *, P<0.05;**, P<0.01;***, P<0.001.
As epithelial-mesenchymal transition (EMT) is a key step in metastasis, EMT-associated molecules were also measured. Tim-3 downregulated the epithelial marker E-cadherin, whilst upregulating TWIST in both MDA-MB-231 and MCF7 cells (Figure 3E ) at the protein level. The downstream target of TWIST MMP1 was also upregulated in MDA-MB-231 Tim-3 OE cells at the protein level (Figure 3E ). Upregulation of TWIST and downregulation of E-cadherin in MCF7 Tim-3 OE cells were also observed at the mRNA level (Figure 3F ). Therefore, EMT could promote the invasion of Tim-3 breast cancer overexpressing cells.
Tim-3 disrupts TJ integrity
TJ is a complex structure that cancer cells need to destroy in order to metastasize. We evaluated the role of Tim-3 in TJ function of breast cancer cells. ECIS was used to evaluate the resistance at 1 kHz, as the current at this frequency is mainly flowing outside the cell and therefore is representative of cellular interactions. Tim-3 OE cells displayed lower resistance relative to Scr cells during initial attachment and spreading in MDA-MB-231 and MCF 7 cells (Figure 4A ), suggesting Tim-3 might inhibit TJ function.
4.
Role of Tim-3 in tight junction function in breast cancer cells. (A) Decelerated cell attachment and spreading in MDA-MB-231 and MCF7 cell lines after Tim-3 OE; (B) Effect of Tim-3 OE on TER in both MDA-MB-231 and MCF7 cell lines; (C) Effect of Tim-3 OE on permeability between cells monolayers in MDA-MB-231 and MCF7 cell lines; (D) Protein levels of ZO-2, ZO-1 and occludin following Tim-3 OE; (E) Quantitative densitometric analysis; (F) mRNA expression levels of ZO-2, ZO-1 and occludin. OE, overexpression; Tim-3, T-cell immunoglobulin and mucin-domain containing molecule-3; TER, transepithelial resistance; ZO, zona occludens.*, P<0.05;**, P<0.01;***, P<0.001.
To confirm these findings, we performed TER and PCP assays to examine the effect of Tim-3 on TJ barrier function. TER values in Tim-3 OE cells were reduced, compared with Scr controls both in MDA-MB-231 (P<0.001) and MCF7 (P<0.001) cells (Figure 4B ). PCP was also assessed using FITC-dextran 10 kDa as a tracer. Higher PCP fluorescence signals were detected in Tim-3 OE MDA-MB-231 (10 h, P=0.002) and MCF7 (10 h, P=0.023) cell lines (Figure 4C ), compared with Scr. Thus, Tim-3 OE cells had a looser connection of cell-cell junctions.
There is evidence that the expression or distribution of TJ proteins is usually altered in cancer. While most cell-cell adhesion proteins are downregulated, others may be overexpressed or delocalized. We therefore examined changes in key TJ molecules in cancer cells following Tim-3 OE. ZO-2 was reduced in both MDA-MB-231 and MCF7 Tim-3 OE cells at the protein level (Figure 4D ,E ). The mRNA levels of ZO-2 were also downregulated in MDA-MB-231 Tim-3 OE cells (Figure 4F ). ZO-1 protein was downregulated in MDA-MB-231 Tim-3 OE cells. In addition, occludin was downregulated in MCF Tim-3 OE cells. Therefore, Tim-3 disrupts TJ integrity by regulating the expression of TJ-associated proteins.
Tim-3 promotes tube formation of endothelial cells
To explore whether Tim-3 plays a role in mediating tumor-associated angiogenesis, tube formation assay was performed using endothelial cells subjected to conditioned medium from stable cells with Tim-3 OE. Tube formation ability of HUVECs cultured in medium from MDA-MB-231 Tim-3 OE cells was significantly increased when cultured for 8 h (P=0.014) (Figure 5A ), a similar phenomenon was also observed in medium from MCF7 Tim-3 OE cells after 16 h (P=0.016) (Figure 5B ), indicating that Tim-3 OE promoted tumor-associated angiogenesis.
5.
Effect of Tim-3 OE on tube formation of endothelial cells. (A) Tube formation ability of HUVECs cultured in conditioned medium from MDA-MB-231 Tim-3-overexpressing cells (P=0.014 vs. Scr); (B) Tube formation ability of HUVECs cultured in conditioned medium from MCF7 Tim-3-overexpressing cells (P=0.016 vs. Scr); (C) Protein levels of VEGFA, VEGFB and VEGFD following Tim-3 OE (left) and quantitative densitometric analysis (right); (D) mRNA expression of VEGFA and VEGFD genes in breast cancer cells. OE, overexpression; Tim-3, T-cell immunoglobulin and mucin-domain containing molecule-3; HUVEC, human umbilical vein endothelial cell; VEGF, vascular endothelial growth factor.*, P<0.05;**, P<0.01;***, P<0.001.
We next determined whether the role of Tim-3 in angiogenesis was VEGF-dependent. The protein levels of VEGFA, VEGFB and VEGFD increased in MDA-MB-231 Tim-3 OE cells, while VEGFA was upregulated in MCF7 Tim-3 OE cells (Figure 5C ). However, gene expression analysis indicated that VEGFD expression increased in MCF7 Tim-3 OE cells (Figure 5D ). We also evaluated the extracellular levels of VEGFC and VEGFR2 proteins. VEGFC expression was significantly increased in MDA-MB-231 Tim-3 OE cells, compared with Scr control (P<0.01) (Supplementary Figure S3 ). The overall levels of VEGFC in MCF7 cells were extremely low, compared with MDA-MB-231 (P<0.05). VEGFR2 expression was increased in MCF7 Tim-3 OE cells, compared with Scr control (P<0.05). Thus, Tim-3-mediated angiogenesis is VEGF-dependent.
S3.
VEGFC and VEGFR2 protein levels in conditioned media of stable cell lines indicated by ELISA. (A) VEGFC; (B) VEGFR2. VEGF, vascular endothelial growth factor; ELISA, enzyme-linked immunosorbent assay.
Tim-3 enhances breast cancer cell resistance to paclitaxel
In order to evaluate the role of Tim-3 in chemo-drug sensitivity in breast cancer, MDA-MB-231 and MCF7 cells were treated with different concentrations of paclitaxel, and their viability was assessed. MDA-MB-231 Tim-3 OE cells were more resistant to paclitaxel than Scr cells at concentrations of 10 (P=0.049), 20 (P=0.003) and 40 nmol/L (P<0.001). MCF7 Tim-3 OE cells were also more resistant to paclitaxel at concentrations of 2.5 nmol/L (P=0.043) and 5 nmol/L (P=0.002), compared with Scr (Figure 6A ,C ).
6.
Effect of Tim-3 OE on paclitaxel resistance in MDA-MB-231 and MCF7 cells. (A) Paclitaxel resistance in MDA-MB-231 cells; (B,E) Protein levels of STAT3, NF-κB, p-NF-κB and CCND1 in MDA-MB-231 cells treated with 10 nmol/L paclitaxel (B) and quantitative densitometry analysis (E); (C) Paclitaxel resistance in MCF7 cells; (D,F) Protein levels of STAT3, NF-κB, p-NF-κB and CCND1 in MCF7 cells treated with 5 nmol/L paclitaxel (D) and quantitative densitometry analysis accordingly (F); (G) Cytotoxicity assay of MDA-MB-231 stable cell lines in response to paclitaxel in absence or presence of NF-κB inhibitor (SC75741) or STAT3 inhibitor (Stattic). OE, overexpression; Tim-3, T-cell immunoglobulin and mucin-domain containing molecule-3; CCND1, cyclin D1; *, P<0.05;**, P<0.01;***, P<0.001.
The levels of STAT3, NF-κB, p-NF-κB, and CCND1 proteins were also altered following treatment with single-dose paclitaxel (10 nmol/L for MDA-MB-231 and 5 nmol/L for MCF7) for 6 h and 24 h. Total NF-κB protein levels significantly increased in Tim-3 OE cells when cultured for 6 h, and p-NF-κB was significantly higher in Tim-3 OE cells when cultured for 24 h in both cell lines, compared with the Scr controls (Figure 6B -F ). After 24-h paclitaxel treatment, protein levels of STAT3 were significantly higher in Tim-3 OE cells than in Scr cells in both cell lines. CCND1 was upregulated in MCF7 Tim-3 OE cells after 6 h, compared with Scr controls (Figure 6B -F ). To validate the functional involvement of NF-κB and STAT3 in the Tim-3 mediated paclitaxel resistance, we performed cytotoxicity assays using the NF-κB inhibitor SC75741 or the STAT3 inhibitor Stattic. In the presence of either SC75741 or Stattic, the paclitaxel resistance induced by Tim-3 was abolished (Figure 6G ). Thus, NF-κB and STAT3 activities were involved in Tim-3 mediated paclitaxel resistance.
Discussion
In recent years, immune checkpoint inhibition in breast cancer, especially in triple-negative breast cancer (TNBC), has attracted accumulating interest to improve the efficacy of targeted therapies. Previous reports have described functions of Tim-3 in the regulation of immune response during cancer progression. As a negative immune regulator, Tim-3 has also been proposed as a prognostic indicator in several types of solid tumor (15).
Previous small-scale studies suggest that Tim-3 is overexpressed in breast cancer. Positive Tim-3 staining is significantly higher in breast cancer than in adjacent tissues (19). Tim-3 expression is also significantly in invasive ductal breast carcinoma cells higher than in normal breast tissues (15). Tim-3 levels are also higher in breast tumor tissues (25). Moreover, high Tim-3 expression is associated with advanced clinical stage, lymph node metastasis, higher Ki67 and a poorer 5-year patient survival rate (15,19). Consistent with previous studies, our pooled analysis of the TCGA and KM-plotter databases further confirms that Tim-3 is upregulated in breast cancer and is associated with poor OS.
Tim-3 signalling is associated with the downstream effector NF-κB in negative regulation of T cell function or liver cancer (26-28). NF-κB plays a key role in targeting the IL-6/STAT3 axis, which is associated with pro-tumor activity (29). We therefore examined Tim-3-associated molecules, including NF-κB, STAT3 and IL-6 following Tim-3 OE. NF-κB, STAT3 and IL-6 were upregulated following Tim-3 OE, suggesting that these molecules were involved in Tim-3-mediated function in breast cancer cells. Similarly, a recent study in liver cancer also demonstrated that Tim-3 OE enhanced tumor cell growth by activating the NF-κB/IL-6/STAT3 pathway, while Tim-3 inhibition resulted in suppressed tumor growth, both in vitro and in Tim-3 knockout mice (30). Tim-3 knockdown also suppresses proliferation and invasion of clear cell renal carcinoma cell lines (14).
Phosphorylated STAT3 binds to DNA in response to IL-6 and epidermal growth factor (31). STAT3 plays a critical role in breast cancer and STAT3 inhibitors show efficacy in inhibiting TNBC tumor growth and metastasis (32). Besides regulating downstream gene expression in its phosphorylated state, STAT3 may also be involved in transcriptional regulation by forming complexes with NF-κB in its unphosphorylated form (33). The cooperation of STAT3 and NF-κB has also been reported in fascin expression, which accelerates the migration of breast cancer cells (34). STAT3 signalling promotes breast tumor progression by regulating downstream molecules that control cell proliferation (CCND1, C-Myc, Bcl-2, Bcl-xL and survivin), angiogenesis (HIF1α and VEGF) and epithelial-mesenchymal transition (TWIST, Vimentin, MMP9 and MMP7) (35). In the present study, CCND1 and C-Myc were upregulated in Tim-3 overexpressing cells, which facilitated cell proliferation. A positive correlation between STAT3 and CCND1 in both primary breast tumors and breast cancer cell lines has been suggested (36). CCND1 assembles with the cyclin-dependent kinases 4/6 (CDK4/6), phosphorylates substrates such as retinoblastoma protein, releases E2F transcription factor and promotes entry of cells to the S-phase (36). C-Myc is a proto-oncogene associated with high grade and advanced stage of TNBC, and C-Myc expression correlates with poor prognosis (37). Based on the evidence as above, we propose that Tim-3 upregulates CCND1 and C-Myc by activating STAT3, which promotes cell proliferation in breast cancer.
In our study, Tim-3 promoted cell invasion and migration, implying a potential role in cancer metastasis. EMT is a key process during cancer invasion and metastasis, which confers an aggressive phenotype to tumor cells. Our study shows that Tim-3 OE influences EMT-associated molecules. It has been reported that there is a positive correlation between phosphorylated STAT3 and TWIST in primary breast carcinoma (38). Therefore, we hypothesise that Tim-3 promotes breast cancer invasion by regulating STAT3 and downstream EMT-associated molecules, consistent with a previous study (39).
Loss of intercellular adhesion molecules also facilitates tumor cells detachment from primary tumors, ultimately initiating metastasis. In this study, Tim-3 disrupted TJ integrity by downregulating TJ molecules including ZO-2, ZO-1 and occludin. ZO-2 and ZO-1 belong to the membrane-associated guanylate kinase protein family and interact with numerous molecules, including cell-cell adhesion proteins, cytoskeletal components and nuclear factors (40). STAT3 activation induced by IL-6 increases retinal endothelial permeability and vascular leakage through the reduction of ZO-1 and occludin expression (41). STAT3 also suppresses CLDN1 transcription by direct binding to its promoter (42,43). VEGF can induce phosphorylation and downregulation of ZO-1 and ZO-2 in endothelial cells (44). Moreover, VEGF promotes motility and reduces the expression of ZO-2 in pancreatic cancer cells (45). Thus, we hypothesise that Tim-3 promotes invasion and migration by disrupting TJ through downregulation of ZO-2, ZO1 and occludin, which may be STAT-3 and VEGF dependent.
Tim-3 promotes resistance to the anti-angiogenesis drug sunitinib and mTOR inhibitor rapamycin in renal cell carcinoma cell lines (20). Interestingly, our data indicate that conditioned medium from Tim-3 overexpressing cancer cells accelerates tube formation, compared with the controls in both breast cancer cell lines. We also evaluated the expression levels of VEGF family members in Tim-3 overexpressing cell lines. Levels of VEGFA, VEGFB and VEGFD were increased in MDA-MB-231 Tim-3-overexpressing cells, while VEGFA was upregulated in MCF7 Tim-3-overexpressing cells. STAT3 plays an important role in angiogenesis. For instance, STAT3 and HIF1α cooperatively activate VEGF and haptoglobin genes during hypoxia in breast cancer cell lines (46), which may also partially contribute to the role of Tim-3 in angiogenesis.
Chemoresistance is a major obstacle for the treatment of breast cancer. Previous studies suggest a role of Tim-3 in chemotherapeutic resistance in cancer (21,47,48). In our study, Tim-3 OE in breast cancer cells induces resistance to paclitaxel, possibly due to the upregulation of STAT3 and NF-κB. Numerous studies confirmed the role of STAT3 in cancer chemoresistance. For example, paclitaxel induces apoptosis in human esophageal squamous cell carcinoma cell lines through the reduction of STAT3 expression and phosphorylation (49). Tumor cell sensitivity to paclitaxel can also be improved by targeting STAT3 using microRNAs (50,51). Therefore, Tim-3 may enhance paclitaxel resistance by upregulating STAT3 in breast cancer (Figure 7 ).
7.
Schematic illustration of role of Tim-3 in breast cancer. Upregulation of Tim-3 not only promotes cell proliferation, migration and invasion, but also disrupts cell-cell tight junction, increases angiogenesis of endothelial cells and paclitaxel-resistance. Tim-3 functions in breast cancer cells by activating NF-κB/STAT3 pathway and downstream target genes. Tim-3, T-cell immunoglobulin and mucin-domain containing molecule-3; IL-6, interleukin 6; ZO, zona occludens; VEGF, vascular endothelial growth factor; CCND1, cyclin D1; MMP-1, matrix metalloproteinase-1.
In this study, although Tim-3 was associated with poor prognosis in total breast cancer patients, the prognostic significance varied with different subtypes. High expression of Tim-3 was associated with poor RFS in luminal A and luminal B subtypes, but better RFS in basal breast cancer. Both MCF7 and MDA-MB-231 cells exhibited a more malignant phenotype after Tim-3 OE, which might be associated with poor prognosis in vivo. Recent studies, however, suggest that Tim-3+ tumor-infiltrating lymphocytes (TILs) are associated with better disease-free survival and OS (52). Thus, we hypothesise that the distribution of Tim-3 on tumor or immune cells might have different prognostic significance. The location of Tim-3 in certain types of cells in cancer might determine prognostic outcomes, although further studies including in vivo models would be required to confirm this hypothesis.
Conclusions
Tim-3 OE in breast cancer promotes tumor cell proliferation, migration and invasion while disrupting TJ function, increasing tumor-associated tube formation and paclitaxel resistance. These effects are achieved by activating the NF-κB/STAT3 signalling pathway and altering gene expression of CCND1, C-Myc, MMP1, TWIST, VEGF and E-cadherin. Moreover, Tim-3 modulates TJ dynamics by downregulating ZO-2, ZO-1 and occludin, which may, in turn, facilitate tumor invasion and migration. Thus, Tim-3 may serve as a prognostic predictor and have therapeutic potential for breast cancer treatment.
Acknowledgements
This study was supported by the key project of research and development plan of Shandong province (No. 2018GSF118125) and Yantai city (No. 2017YD007).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Contributor Information
Guangdong Qiao, Email: qiaogddxy@163.com.
Wenguo Jiang, Email: jiangw@cardiff.ac.uk.
Zhigang Yu, Email: yuzhigang@sdu.edu.cn.
References
- 1.Sun D, Cao M, Li H, et al Cancer burden and trends in China: A review and comparison with Japan and South Korea. Chin J Cancer Res. 2020;32:129–39. doi: 10.21147/j.issn.1000-9604.2020.02.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de la Cruz-Merino L, Chiesa M, Caballero R, et al Breast cancer immunology and immunotherapy: Current status and future perspectives. Int Rev Cell Mol Biol. 2017;331:1–53. doi: 10.1016/bs.ircmb.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 3.Fuertes Marraco SA, Neubert NJ, Verdeil G, et al Inhibitory receptors beyond t cell exhaustion. Front Immunol. 2015;6:310. doi: 10.3389/fimmu.2015.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monney L, Sabatos CA, Gaglia JL, et al Th1-specific cell surface protein Tim-3 regulates macrophage activation and severity of an autoimmune disease. Nature. 2002;415:536–41. doi: 10.1038/415536a. [DOI] [PubMed] [Google Scholar]
- 5.Patel J, Bozeman EN, Selvaraj P Taming dendritic cells with TIM-3: another immunosuppressive strategy used by tumors. Immunotherapy. 2012;4:1795–8. doi: 10.2217/imt.12.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan W, Liu X, Ma H, et al Tim-3 fosters HCC development by enhancing TGF-beta-mediated alternative activation of macrophages. Gut. 2015;64:1593–604. doi: 10.1136/gutjnl-2014-307671. [DOI] [PubMed] [Google Scholar]
- 7.Huang YH, Zhu C, Kondo Y, et al CEACAM1 regulates TIM-3-mediated tolerance and exhaustion. Nature. 2015;517:386–90. doi: 10.1038/nature13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou E, Huang Q, Wang J, et al Up-regulation of Tim-3 is associated with poor prognosis of patients with colon cancer. Int J Clin Exp Pathol. 2015;8:8018–27. [PMC free article] [PubMed] [Google Scholar]
- 9.Ngiow SF, von Scheidt B, Akiba H, et al Anti-TIM3 antibody promotes T cell IFN-gamma-mediated antitumor immunity and suppresses established tumors. Cancer Res. 2011;71:3540–51. doi: 10.1158/0008-5472.CAN-11-0096. [DOI] [PubMed] [Google Scholar]
- 10.Andrews LP, Yano H, Vignali DA Inhibitory receptors and ligands beyond PD-1, PD-L1 and CTLA-4: breakthroughs or backups. Nat Immunol. 2019;20:1425–34. doi: 10.1038/s41590-019-0512-0. [DOI] [PubMed] [Google Scholar]
- 11.Koyama S, Akbay EA, Li YY, et al Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat Commun. 2016;7:10501. doi: 10.1038/ncomms10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhuang X, Zhang X, Xia X, et al Ectopic expression of TIM-3 in lung cancers: a potential independent prognostic factor for patients with NSCLC. Am J Clin Pathol. 2012;137:978–85. doi: 10.1309/AJCP9Q6OVLVSHTMY. [DOI] [PubMed] [Google Scholar]
- 13.Jiang J, Jin MS, Kong F, et al Decreased galectin-9 and increased Tim-3 expression are related to poor prognosis in gastric cancer. PLoS One. 2013;8:e81799. doi: 10.1371/journal.pone.0081799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan J, Jiang B, Zhao H, et al Prognostic implication of TIM-3 in clear cell renal cell carcinoma. Neoplasma. 2014;61:35–40. doi: 10.4149/neo_2014_006. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H, Xiang R, Wu B, et al T-cell immunoglobulin mucin-3 expression in invasive ductal breast carcinoma: Clinicopathological correlations and association with tumor infiltration by cytotoxic lymphocytes. Mol Clin Oncol. 2017;7:557–63. doi: 10.3892/mco.2017.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun QY, Qu CH, Liu JQ, et al Down-regulated expression of Tim-3 promotes invasion and metastasis of colorectal cancer cells. Neoplasma. 2017;64:101–7. doi: 10.4149/neo_2017_112. [DOI] [PubMed] [Google Scholar]
- 17.Wu J, Lin G, Zhu Y, et al Low TIM3 expression indicates poor prognosis of metastatic prostate cancer and acts as an independent predictor of castration resistant status. Sci Rep. 2017;7:8869. doi: 10.1038/s41598-017-09484-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, Yin X, Zhang H, et al Differential expression of TIM-3 between primary and metastatic sites in renal cell carcinoma. BMC Cancer. 2019;19:49. doi: 10.1186/s12885-019-5273-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng SQ, Han FY, Xu YQ, et al Expression of Tim-3 in breast cancer tissue promotes tumor progression. Int J Clin Exp Pathol. 2018;11:1157–66. [PMC free article] [PubMed] [Google Scholar]
- 20.Komohara Y, Morita T, Annan DA, et al The coordinated actions of TIM-3 on cancer and myeloid cells in the regulation of tumorigenicity and clinical prognosis in clear cell renal cell carcinomas. Cancer Immunol Res. 2015;3:999–1007. doi: 10.1158/2326-6066.CIR-14-0156. [DOI] [PubMed] [Google Scholar]
- 21.Horlad H, Ohnishi K, Ma C, et al TIM-3 expression in lymphoma cells predicts chemoresistance in patients with adult T-cell leukemia/lymphoma. Oncol Lett. 2016;12:1519–24. doi: 10.3892/ol.2016.4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marshall J Transwell(®) invasion assays. Methods Mol Biol. 2011;769:97–110. doi: 10.1007/978-1-61779-207-6_8. [DOI] [PubMed] [Google Scholar]
- 23.Zaman MH, Trapani LM, Sieminski AL, et al Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc Natl Acad Sci U S A. 2006;103:10889–94. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin TA, Jordan N, Davies EL, et al Metastasis to bone in human cancer is associated with loss of occludin expression. Anticancer Res. 2016;36:1287–93. [PubMed] [Google Scholar]
- 25.Sasidharan Nair V, El Salhat H, Taha RZ, et al DNA methylation and repressive H3K9 and H3K27 trimethylation in the promoter regions of PD-1, CTLA-4, TIM-3, LAG-3, TIGIT, and PD-L1 genes in human primary breast cancer. Clin Epigenetics. 2018;10:78. doi: 10.1186/s13148-018-0512-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomkowicz B, Walsh E, Cotty A, et al TIM-3 suppresses anti-CD3/CD28-induced TCR activation and IL-2 expression through the NFAT signaling pathway. PLoS One. 2015;10:e0140694. doi: 10.1371/journal.pone.0140694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J, Su EW, Zhu C, et al Phosphotyrosine-dependent coupling of Tim-3 to T-cell receptor signaling pathways. Mol Cell Biol. 2011;31:3963–74. doi: 10.1128/MCB.05297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu F, Liu Y, Chen Z Tim-3 expression and its role in hepatocellular carcinoma. J Hematol Oncol. 2018;11:126. doi: 10.1186/s13045-018-0667-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He G, Karin M NF-κB and STAT3-key players in liver inflammation and cancer. Cel Res. 2011;21:159–68. doi: 10.1038/cr.2010.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H, Song Y, Yang H, et al Tumor cell-intrinsic Tim-3 promotes liver cancer via NF-κB/IL-6/STAT3 axis. Oncogene. 2018;37:2456–68. doi: 10.1038/s41388-018-0140-4. [DOI] [PubMed] [Google Scholar]
- 31.Zhong Z, Wen Z, Darnell JE Jr Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–8. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 32.Kettner NM, Vijayaraghavan S, Durak MG, et al Combined inhibition of STAT3 and DNA repair in palbociclib-resistant ER-positive breast cancer. Clin Cancer Res. 2019;25:3996–4013. doi: 10.1158/1078-0432.CCR-18-3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J, Liao X, Agarwal MK, et al Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFkappaB. Genes Dev. 2007;21:1396–408. doi: 10.1101/gad.1553707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snyder M, Huang J, Huang XY, et al A signal transducer and activator of transcription 3.Nuclear Factor kappaB (Stat3.NFκB) complex is necessary for the expression of fascin in metastatic breast cancer cells in response to interleukin (IL)-6 and tumor necrosis factor (TNF)-α. J Biol Chem. 2014;289:30082–9. doi: 10.1074/jbc.M114.591719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banerjee K, Resat H Constitutive activation of STAT3 in breast cancer cells: A review. Int J Cancer. 2016;138:2570–8. doi: 10.1002/ijc.29923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leslie K, Lang C, Devgan G, et al Cyclin D1 is transcriptionally regulated by and required for transformation by activated signal transducer and activator of transcription 3. Cancer Res. 2006;66:2544–52. doi: 10.1158/0008-5472.CAN-05-2203. [DOI] [PubMed] [Google Scholar]
- 37.Nedeljković M, Tanić N, Dramićanin T, et al Importance of copy number alterations of FGFR1 and C-MYC Genes in triple negative breast cancer. J Med Biochem. 2019;38:63–70. doi: 10.2478/jomb-2018-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lo HW, Hsu SC, Xia W, et al Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of TWIST gene expression. Cancer Res. 2007;67:9066–76. doi: 10.1158/0008-5472.CAN-07-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shang Y, Li Z, Li H, et al TIM-3 expression in human osteosarcoma: Correlation with the expression of epithelial-mesenchymal transition-specific biomarkers. Oncol Lett. 2013;6:490–4. doi: 10.3892/ol.2013.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.González-Mariscal L, Miranda J, Raya-Sandino A, et al ZO-2, a tight junction protein involved in gene expression, proliferation, apoptosis, and cell size regulation. Ann N Y Acad Sci. 2017;1397:35–53. doi: 10.1111/nyas.13334. [DOI] [PubMed] [Google Scholar]
- 41.Yun JH, Park SW, Kim KJ, et al Endothelial STAT3 activation increases vascular leakage through downregulating tight junction proteins: implications for diabetic retinopathy. J Cell Physiol. 2017;232:1123–34. doi: 10.1002/jcp.25575. [DOI] [PubMed] [Google Scholar]
- 42.Ryu WI, Lee H, Bae HC, et al IL-33 down-regulates CLDN1 expression through the ERK/STAT3 pathway in keratinocytes. J Dermatol Sci. 2018;90:313–22. doi: 10.1016/j.jdermsci.2018.02.017. [DOI] [PubMed] [Google Scholar]
- 43.Chen YJ, You ML, Chong QY, et al Autocrine human growth hormone promotes invasive and cancer stem cell-like behavior of hepatocellular carcinoma cells by STAT3 dependent inhibition of CLAUDIN-1 expression. Int J Mol Sci. 2017;18:1274. doi: 10.3390/ijms18061274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chidiac R, Zhang Y, Tessier S, et al Comparative phosphoproteomics analysis of VEGF and angiopoietin-1 signaling reveals ZO-1 as a critical regulator of endothelial cell proliferation. Mol Cell Proteomics. 2016;15:1511–25. doi: 10.1074/mcp.M115.053298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doi Y, Yashiro M, Yamada N, et al VEGF-A/VEGFR-2 signaling plays an important role for the motility of pancreas cancer cells. Ann Surg Oncol. 2012;19:2733–43. doi: 10.1245/s10434-011-2181-6. [DOI] [PubMed] [Google Scholar]
- 46.Pawlus MR, Wang L, Hu CJ STAT3 and HIF1α cooperatively activate HIF1 target genes in MDA-MB-231 and RCC4 cells. Oncogene. 2014;33:1670–9. doi: 10.1038/onc.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu JF, Ma SR, Mao L, et al T-cell immunoglobulin mucin 3 blockade drives an antitumor immune response in head and neck cancer. Mol Oncol. 2017;11:235–47. doi: 10.1002/1878-0261.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Mingo Pulido Á, Gardner A, Hiebler S, et al TIM-3 regulates CD103+ dendritic cell function and response to chemotherapy in breast cancer . Cancer Cell. 2018;33:60–74. doi: 10.1016/j.ccell.2017.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang X, Wu X, Zhang F, et al Paclitaxel induces apoptosis of esophageal squamous cell carcinoma cells by downregulating STAT3 phosphorylation at Ser727. Oncol Rep. 2017;37:2237–44. doi: 10.3892/or.2017.5503. [DOI] [PubMed] [Google Scholar]
- 50.Gao J, Shao Z, Yan M, et al Targeted regulation of STAT3 by miR-29a in mediating Taxol resistance of nasopharyngeal carcinoma cell line CNE-1. Cancer Biomark. 2018;22:641–8. doi: 10.3233/CBM-170964. [DOI] [PubMed] [Google Scholar]
- 51.Fan Z, Cui H, Yu H, et al MiR-125a promotes paclitaxel sensitivity in cervical cancer through altering STAT3 expression. Oncogenesis. 2016;5:e197. doi: 10.1038/oncsis.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Byun KD, Hwang HJ, Park KJ, et al T-cell immunoglobulin mucin 3 expression on tumor infiltrating lymphocytes as a positive prognosticator in triple-negative breast cancer. J Breast Cancer. 2018;21:406–14. doi: 10.4048/jbc.2018.21.e61. [DOI] [PMC free article] [PubMed] [Google Scholar]












