Abstract
We used a quantitative microbial risk assessment approach to relate log10 disinfection reductions of SARS-CoV-2 bioburden to COVID-19 infection risks. Under low viral bioburden, minimal log10 reductions may be needed to reduce infection risks for a single hand-to-fomite touch to levels lower than 1:1,000,000, as a risk comparison point. For higher viral bioburden conditions, log10 reductions of more than 2 may be needed to achieve median infection risks of less than 1:1,000,000.
Key Words: SARS-CoV-2, Fomite, QMRA, Disinfection
While droplet and bioaerosol transmission are considered the main contributors to COVID-19 transmission,1 SARS-CoV-2 detection on surfaces2 indicates the potential for fomite-mediated transmission and the need for surface disinfection in multibarrier mitigation approaches. Data indicate that surfaces most likely to facilitate coronavirus transmission are surfaces which are frequently touched by many people (eg, door and tap handles) and that disinfection practices should be targeted at these surfaces.3 , 4
Disinfectant efficacy on surfaces contaminated with coronavirus has been evaluated,5 , 6 but the log10 reductions obtained have not been quantitatively linked to infection risk reduction. This challenges health authorities in specifying disinfectant dilutions and contact times required to reduce viral bioburden to safety target levels, but risk assessment bridges the divide between environmental virus quantification and implementation of health targets. Here we use a quantitative microbial risk assessment (QMRA) approach to estimate and compare COVID-19 infection risks after single hand-to-fomite-to-mucosal membrane contacts for high and low levels of viral bioburden and variable disinfection efficacy.
METHODS
We estimated infection risks for a single hand-to-fomite and hand-to-facial mucosal membrane (mouth, eyes, and nose) contact scenario, where reduction efficacy of the virus was varied between 1 and 5 log10. We then compared estimated infection risks to 1:10,000 and 1:1,000,000 risks. This approach has been used in previous QMRAs for relating surface disinfection efficacies against bacteria and viruses to estimated health outcomes.7 , 8
A Monte Carlo approach was used to account for variability and uncertainty in the following: transfer efficiencies, fractions of the hand used for surface and face contacts, viral bioburden, disinfection log10 reductions, and surface areas of the hand and of fomites available for contact (Supplementary Table S1). Ten-thousand iterations were used.
Currently, data are lacking describing infective virus bioburdens on fomites in part due to detection limits for current culture assays being higher than viral concentrations on surfaces.9 Therefore, we assumed a range of viral bioburden (0.1 to 10,000 genome copies (gc)/cm2) to evaluate the effect of variable viral bioburden on infection risk reductions offered by various log10 viral bioburden reductions and used 1 gc/cm2, an assumed limit of detection, to compare low versus high viral bioburden conditions. To account for variations in the level of infectivity of viral genome copies, bioburdens were adjusted to assume either 1% or 10% of gc/cm2 were infective.
As shown in the supplementary notes, the viral concentration on hands for each scenario was estimated from the viral bioburden, which was adjusted for the total surface area of fomites available for contact. For every estimated viral concentration on hands, a dose was then calculated for the subsequent hand-to-facial mucous membrane contact. These doses were then inputted into an exact beta-Poisson dose-response curve for relating estimated doses to probability of infection. Spearman correlation coefficients were used to evaluate the strength of monotonic relationships between model inputs and infection risk. Model equations and sensitivity analysis results are provided in Supplementary Materials.
RESULTS
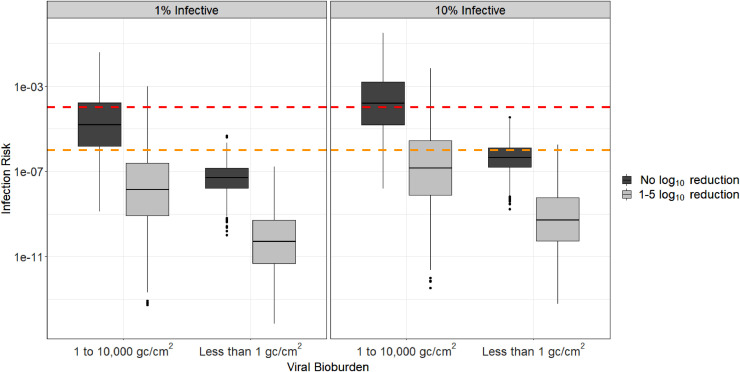
Under low viral bioburden conditions (<1 genome copies (gc)/cm2), the median infection risks were below 1:1,000,000, regardless of whether there was log10 reduction or whether 1% or 10% of the genome copies were assumed to be infectious (Fig 1 ). For the scenarios where there were high viral bioburdens (1-10,000 gc/cm2) and there was no log10 reduction in viral bioburden, few infection risks were below 1:1,000,000 regardless of whether 1% or 10% of genome copies/cm2 were infective. Under these same high viral bioburden conditions, median infection risks were below 1:1,000,000 when viral bioburden was reduced by 1 to 5 log10 (Fig 1).
Fig 1.
Infection risk distributions for low and high surface bioburdens, associated with either no log10 reduction or a 1-5 log10 reduction of bioburden on surfaces, and assuming either 1% or 10% of detected viral genome copies were infectious*. *Red and orange dashed lines represent 1/10,000 and 1/1,000,000 risk targets, respectively
For low viral bioburdens and 1% infectivity of detected RNA, all infection risks where disinfection with a 1-5 log10 reduction was used were below 1:1,000,000 (Fig 2 ). Note that infection risks with no log10 reduction for this scenario were nearly all below 1:1,000,000 as well. When 10% of gc/cm2 were assumed to be infective and concentrations were <1 gc/cm2, nearly all infection risks estimated for disinfection scenarios were below 1:1,000,000 whereas 1:1,000,000 was in the interquartile range of infection risks for the no log10 reduction scenario (Fig 2).
Fig 2.
Infection risk distributions for low and high surface bioburdens associated with no log10 reduction or a range of log10 reductions achieved by use of disinfectant* assuming either 1% or 10% of detected viral genome copies were infectious. *Log10 reduction ranges include no reduction (0 log10), greater than or equal to 1 and less than or equal to 2 log10, greater than 2 and less than or equal to 3 log10, greater than 3 and less than or equal to 4 log10, and greater than 4 and less than or equal to 5 log10 reduction. Red and orange dashed lines represent 1/10,000 and 1/1,000,000 risk targets, respectively.
For higher viral bioburden scenarios (1-10,000 gc/cm2), median infection risks for surfaces treated with disinfectant to produce 1-5 log10 disinfection reductions were below 1:1,000,000 when it was assumed 1% of gc/cm2 were infective. When 10% of gc/cm2 was assumed infective, 2-5 log10 reductions were required to reduce median infection risk to less than 1:1,000,000.
DISCUSSION
These simulations indicate that under low viral bioburden conditions, minimal log10 reductions may be needed to achieve risks less than 1:1,000,000. For higher viral bioburden conditions, log10 reductions of more than 2 may be needed to achieve median risks of less than 1:1,000,000, especially when assuming 10% of gc/cm2 represent infective virus (Fig 2).
The CDC recommends a 1000 ppm bleach dilution for those isolated in home care for nonporous surface disinfection, where appropriate.10 Sattar (1989) quantified a >3 log10 reduction of human coronavirus 229E with a 1000 ppm and a 5000 ppm bleach dilution.5 Other biocidal agents, such as 70% ethanol, have demonstrated similar log10 reductions in carrier tests with coronaviruses.6 Our model demonstrates that a 2-3 log10 reduction would, in most cases, result in risks less than 1:1,000,000 for high-viral bioburden scenarios if 1% of gc/cm2 is assumed to be infective. However, this reduction range would be less adequate in achieving risks below 1:1,000,000 when a higher fraction of infective virus is expected (Fig 2). More data are needed describing the relationship between molecularly detected virus and infectious virus concentrations, as this affects infection risk estimates and required log10 reductions needed to protect health at specific risk-informed levels. While 1:1,000,000 was used as a conservative point of comparison for estimated risks, this is a de minimis risk level. Improvements to risk comparisons in future work include comparing infection risks estimates to rates of increased number of illness cases. To better inform scenario-specific targeted surface hygiene, data are needed for (1) SARS-CoV-2 bioburden on different environment-specific (home or health care) fomites and (2) fomite-specific touch frequencies.
Footnotes
A.M. Wilson was supported by the University of Arizona Foundation and the Hispanic Women's Corporation/Zuckerman Family Foundation Student Scholarship Award through the Mel and Enid Zuckerman College of Public Health, University of Arizona. Code is available under a Creative Commons License at: https://github.com/awilson12/QMRA_bleach
Conflicts of interest: A.M. Wilson reports grants from Allied BioScience, Inc.; ZoonoUSA; Gojo Industries, Inc.; and Ecolab outside the submitted work. M.H. Weir reports no conflicts of interest. SF Bloomfield reports no conflicts of interest. E.A. Scott reports no conflicts of interest. K.A. Reynolds reports grants from GOJO Industries, Inc. and Ecolab outside the submitted work.
Supplementary material associated with this article can be found in the online version at https://doi.org/10.1016/j.ajic.2020.11.013.
Appendix. SUPPLEMENTARY MATERIALS
References
- 1.Jones RM. Relative contributions of transmission routes for COVID-19 among healthcare personnel providing patient care. J Occup Environ Hyg. 2020;17:408–415. doi: 10.1080/15459624.2020.1784427. http://www.ncbi.nlm.nih.gov/pubmed/32643585 Available at: [DOI] [PubMed] [Google Scholar]
- 2.Ong SWX, Tan YK, Chia PY, et al. Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from a symptomatic patient. JAMA. 2020;323:1610–1612. doi: 10.1001/jama.2020.3227. https://jamanetwork.com/journals/jama/fullarticle/2762692 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weir MH, Shibata T, Masago Y, Cologgi DL, Rose JB. Effect of surface sampling and recovery of viruses and non-spore-forming bacteria on a quantitative microbial risk assessment model for fomites. Environ Sci Technol. 2016;50:5945–5952. doi: 10.1021/acs.est.5b06275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maillard J-Y, Bloomfield SF, Courvalin P, et al. Reducing antibiotic prescribing and addressing the global problem of antibiotic resistance by targeted hygiene in the home and everyday life settings: a position paper. Am J Infect Control. 2020;48:1090–1099. doi: 10.1016/j.ajic.2020.04.011. https://linkinghub.elsevier.com/retrieve/pii/S0196655320302091 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sattar SA, Springthorpe VS, Karim Y, Loro P. Chemical disinfection of non-porous inanimate surfaces experimentally contaminated with four human pathogenic viruses. Epidemiol Infect. 1989;102:493–505. doi: 10.1017/s0950268800030211. https://www.cambridge.org/core/product/identifier/S0950268800030211/type/journal_article Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kampf G, Todt D, Pfaender S, Steinmann E. Persistence of coronaviruses on inanimate surfaces and its inactivation with biocidal agents. J Hosp Infect. 2020;104:246–251. doi: 10.1016/j.jhin.2020.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ryan MO, Haas CN, Gurian PL, Gerba CP, Panzl BM, Rose JB. Application of quantitative microbial risk assessment for selection of microbial reduction targets for hard surface disinfectants. Am J Infect Control. 2014;42:1165–1172. doi: 10.1016/j.ajic.2014.07.024. [DOI] [PubMed] [Google Scholar]
- 8.Wilson AM, Reynolds KA, Sexton JD, Canales RA. Modeling surface disinfection needs to meet microbial risk reduction targets. Appl Environ Microbiol. 2018;84 doi: 10.1128/AEM.00709-18. e00709–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou J, Otter JA, Price JR, et al. Investigating SARS-CoV-2 surface and air contamination in an acute healthcare setting during the peak of the COVID-19 pandemic in London [e-pub ahead of print] Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa905. Accessed November 23, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. Coronavirus disease 2019 detailed disinfection guidance. 2020. Available at: https://www.cdc.gov/coronavirus/2019-ncov/prevent-getting-sick/cleaning-disinfection.html. Accessed May 28, 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.