Abstract
miR-205 plays important roles in the physiology of epithelia by regulating a variety of pathways that govern differentiation and morphogenesis. Its aberrant expression is frequently found in human cancers, where it was reported to act either as tumor-suppressor or oncogene depending on the specific tumor context and target genes. miR-205 expression and function in different cell types or processes are the result of the complex balance among transcription, processing and stability of the microRNA. In this review, we summarize the principal mechanisms that regulate miR-205 expression at the transcriptional and post-transcriptional level, with particular focus on the transcriptional relationship with its host gene. Elucidating the mechanisms and factors regulating miR-205 expression in different biological contexts represents a fundamental step for a better understanding of the contribution of such pivotal microRNA to epithelial cell function in physiology and disease, and for the development of modulation strategies for future application in cancer therapy.
Subject terms: Cancer epigenetics, miRNAs
Facts
miR-205 contributes to the development of epithelia.
miR-205 is up- or down-modulated in different epithelial cancers.
miR-205 expression is regulated at the transcriptional and post-transcriptional level.
miR-205 biogenesis is intimately linked to the processing of its host gene (MIR205HG).
miR-205 and MIR205HG-derived long noncoding RNA play independent but complementary functions in epithelial cells.
Open questions
Which is the exact role of the sequences upstream or downstream of miR-205/MIR205HG locus?
Do specific regions account for a MIR205HG-independent miR-205 biogenesis?
Which mechanisms or factors modify the reciprocal production of miR-205 and MIR205HG long noncoding RNA?
Introduction
MicroRNAs (miRNAs) are small, noncoding RNAs that play a critical role in a wide range of physiologic and pathologic processes, mainly by acting as negative post-transcriptional regulators of their target genes1. A tight control of miRNA expression is essential for tissue homeostasis and development. Indeed, several studies showed that dysregulated miRNA expression is functionally related to the onset of different human diseases, including cancer2.
Intergenic miRNAs are transcribed by RNA polymerase II or III, in the nucleus, as primary miRNA transcripts (pri-miRNA), which are then processed by Drosha-DGCR8 microprocessor complex into 70–90 bases long hairpin precursor miRNAs (pre-miRNA). Once exported to the cytoplasm, the pre-miRNA is cleaved by the RNAse III, Dicer, to produce the miRNA duplex, of which one strand is incorporated into the Argonaute complex to be guided to the target mRNA3. Alternative miRNA biogenesis pathways may occur for non-intergenic miRNAs, such as intronic ones, which are transcribed together with their “host genes” and then directly processed by the splicing machinery4,5. Therefore, the location of the miRNA transcription unit dictates different mechanisms of biogenesis.
MicroRNA-205 (miR-205) is a highly conserved miRNA expressed in epithelial tissues of different species6,7. In the human genome, the pre-miR-205 sequence is located in the last intron/exon junction of a gene annotated as miR-205 Host gene (MIR205HG), on chromosome 1q32.2. To date, miR-205 has been widely characterized for its functions in normal development and in cancer, where it was reported to be aberrantly expressed (either up- or downregulated) and exert pro- or anti-tumorigenic roles depending on the cellular context and target genes. Elucidating the mechanisms and factors regulating miR-205 expression in different biological contexts represents a fundamental step for a better understanding of the contribution of such pivotal miRNA in epithelial cell function and cancer.
This review highlights the recent understanding of miR-205 transcriptional and post-transcriptional regulation in normal and cancer cells and reveals new insights into the miRNA/host gene architecture. Knowledge in such field may potentially uncover new opportunities to manipulate miR-205 expression for therapeutic purposes in cancer.
miR-205 expression and functions
miR-205 in normal physiology
miR-205 is highly expressed in human epithelial tissues of breast, prostate, skin, eye, and thymus, where it plays a critical role in tissutal morphogenesis and homeostasis. In general, it sustains the epithelial phenotype through the direct targeting of zinc finger E-box-binding homeobox 1 (ZEB1) and ZEB2, two transcription factors (TFs) that repress E-cadherin and other polarity genes8.
In the early stages of embryogenesis, miR-205 is expressed in trophoblasts, where it regulates the placental development through the suppression of Mediator of RNA polymerase II transcription subunit 1 (MED1)9. During the embryonic development, miR-205 regulates the extraembryonic endoderm differentiation and spermatogenesis by targeting genes governing cell migration and adhesion10.
In the mammary gland, miR-205 is highly expressed in the basal stem cells. Overexpression of miR-205 was shown to induce the expansion of the progenitor cell population, reduce cell size and increase cellular proliferation. These effects are mediated by the repression of phosphatase and tensin homolog (PTEN) tumor-suppressor11. In addition, miR-205 expression is enriched during gestation and late involution stages, suggesting a dynamic regulation in developmental processes12. Similarly, in the human prostate, it is highly expressed in the basal cells, which are presumed as the progenitors of the epithelium. In this context, it regulates the production of the basement membrane protein complex laminin-332 and its receptor integrin-β4, thus ensuring proper tissue polarity and morphogenesis13. A miR-205 involvement in the stemness program has been reported in the skin epidermis and in the stratified epithelia of esophagues and tongue. Here, it plays a pivotal role in the expansion of stem cell population via regulation of PI(3)-kinase signaling14. Moreover, by acting on the same signaling pathways, miR-205 increases the migration of human epidermal and corneal epithelial keratinocytes, playing an essential role in the wound healing and in the corneal development15. Consistent with the aforementioned functions, miR-205 knock-out mice perinatally die due to severe skin defects and compromised proliferation of multiple stratified epithelia tissues14,16. In addition, in mice, miR-205 plays a critical role in the early lacrimal gland development by modulating fibroblast growth factor 10 (FGF10) signaling17. Finally, miR-205 is highly expressed in thymic epithelial cells, where it helps to preserve T-cell maturation in response to strong inflammatory insults, such as infections, radiation exposure, and steroids. This happens through regulation of chemokine/chemokine receptor pathways, antigen processing components, and Wnt signaling system, as a consequence of miR-205-mediated repression of Forkhead Box N1 (Foxn1) TF18.
Therefore, tight regulation of miR-205 expression is required for the development of different epithelial tissues. On the other hand, alteration of its normal expression is observed during the initiation and progression of different epithelial cancers.
miR-205 in cancer: ups and downs
miR-205 was found to be either up- or downregulated in several cancers according to the subtype, cell of origin or stage of tumor progression (Table 1). In specific cell types, miR-205 facilitates tumor initiation and proliferation acting as an oncogene; in others, it inhibits cell proliferation, invasion and epithelial-mesenchimal transition (EMT), thus playing a tumor-suppressive role. Inasmuch as miR-205 is a marker of epithelial phenotype, its expression was shown to decrease during EMT8, a key step in the promotion of tumor invasion and metastasis11,19. In accordance to its involvement in this process, the downregulation of miR-205 has been found in invasive and mesenchymal tumors when comparing to normal tissues8. In prostate cancer, miR-205 was demonstrated to act as a tumor-suppressor by repressing the expression of factors (N-chimerin, E2F1, E2F5, ZEB2, and protein kinase Cε) involved in EMT, cell motility and invasion20. Moreover, it was hypothesized to counteract tumor initiation and progression by preserving basement membrane or by repressing the activity of mitogen-activated protein kinase (MAPK) and androgen receptor (AR)13,21. Likewise, miR-205 is downregulated in triple-negative breast cancer (TNBC) and in metastatic breast cancer, indicating a tumor-suppressive role also in these cancer subtypes. Specifically, by targeting human epidermal growth factor receptor 3 (HER3), miR-205 inhibits cell proliferation and migration and, by repressing ZEB1 and ZEB2, it limits tumor invasion8,22,23. Decreased levels of miR-205 have been reported also in melanoma specimens and in renal cancer, where it exerts anti-proliferative and pro-apoptotic functions by repressing E2F1 and Src-family-genes, respectively24,25. On the other hand, miR-205 is over-expressed in endometrial cancer, where it inhibits apoptosis and promotes cell proliferation through the inhibition of the tumor-suppressors PTEN and estrogen-related receptor-γ (ESRRG)26,27. Moreover, in this tumor type, its enhanced levels are related to advanced stage and poor overall survival, suggesting a possible use as prognostic marker28,29. Likewise, increased expression of miR-205 in nasopharyngeal carcinoma is associated with PTEN reduction, followed by tumor promotion and increased resistance to radiotherapy in patients with higher clinical stages30,31.
Table 1.
Summary of the expression, regulation, and functions of miR-205 in different cancers.
| Cancer type | miR-205 Expression | Regulatory mechanisms | Functions | Target genes | References |
|---|---|---|---|---|---|
| Prostate cancer | Downregulated | TFs (p63/ΔNp63α, HIF-1α), hypermetilation, deacetylation | Tumor-suppressor | N-chimerin, E2F1, E2F5, ZEB2, protein kinase Cε, MED1, MAPK, AR | 13,20,21,37,39,40,45,48 |
| Breast cancer | Downregulated (TNBC) | TF (p53), hypermetilation, deacetylation, lncRNA (linc-ROR) | Tumor-suppressor, oncogene | ZEB1, ZEB2, PTEN, HER3, VEGFA, FGF2 | 8,11,22,23,38,65 |
| Lung cancer | Downregulated (adenocarcinoma) or upregulated (squamous cell carcinoma) | Hypermethylation, deacetylation | Tumor-suppressor or oncogene; biomarker | PTEN, PHLPP2, RUNX1 | 35,41,51 |
| Renal cancer | Downregulated | lncRNA (LINC00152) | Tumor-suppressor | Src-family members, Ras/Raf/ERK1/2 | 25,66 |
| Head and neck cancer | Upregulated | TF (p53) | Oncogene, prognostic marker | PTEN | 30 |
| Melanoma | Downregulated | TFs (p73/ΔNp73) | Tumor-suppressor | E2F1, BCL2, VEGF | 24 |
| Bladder cancer | Downregulated invasive bladder cancer) or upregulated (non-invasive bladder cancer) | TF (ΔNp63α, Twist1), hypermetilation, lncRNA (HOTAIR) | Tumor-suppressor or oncogene | ZEB1, ZEB2 | 36,47,67 |
| Endometrial cancer | Upregulated | lncRNAs (RP11–395G23.3 and LA16c-313D11.11) | Oncogene, prognostic marker | PTEN, ESRRG | 26,27 |
In addition, miR-205 levels were significantly increased in the serum of ovarian cancer patients and high expression of the circulating miR-205 was associated with angiogenesis and ovarian cancer progression32,33. In this context, the reduction of the miR-205 PTEN target leads to a persistent activation of AKT signaling, which results in uncontrolled proliferation and neoplastic angiogenesis33.
Accumulating evidence showed heterogeneity of miR-205 expression within the same tumor type, as reported in non-small cell lung carcinoma and esophageal cancer. In these contexts, miR-205 overexpression is indicative of squamous cell carcinoma, while its down-expression is characteristic of adenocarcinoma, thus suggesting its possible application as a diagnostic marker34,35. Taken together, these studies provide evidence for the dual role of miR-205 in different cancers and arise important questions about upstream factors regulating its expression.
Levels of miR-205 regulation
Transcriptional regulation: the players and the molecular landscape
Chromatin changes
Epigenetic modifications, especially Cytosine (phosphodiester bond) Guanine (CpG) DNA methylation and histone re-modeling, have important roles in gene expression regulation and can also affect miRNA transcription. The recruitment of DNA methyltransferase (DNMT) and histone deacetylases (HDACs) leads to chromatin inactivation and the consequent repression of transcription. CpG sites were identified in a region immediately upstream of the first exon of MIR205HG and in the miR-205 locus22,36–38. For example, DNA methylation in the 300 bases preceding MIR205HG Transcription Start Site (TSS) and deacetylation of lysine 9 of histone 3 (H3K9) in the genomic region coding for pre-miR-205 were shown to contribute to miR-205 downregulation in prostate cancer cells37,39. Other evidence for the miR-205 repressive epigenetic status in prostate cancer cells has been reported by Ke and colleagues, who detected a gain of lysine 27 trimethylation of histone 3 (H3K27me3) mark and a loss of lysine 4 trimethylation of histone 3 (H3K4me3) in miR-205 locus40. Similarly, aberrant DNA hypermethylation, H3K9 deacetylation and H3K27me3 in the pre-miR-205 locus are related to miR-205 silencing in muscle-invasive bladder cancer and in transformed lung epithelial cells36,41. However, treatment with de-methylating agents and histone deacetylase (HDAC) inhibitors demonstrated that these chromatin modifications are not the sole regulators of miR-205 and that other factors may contribute to its transcriptional repression36. In another aggressive subtype of breast cancer, defined as HER2-positive cancer, miR-205 expression is modulated by HER2 signaling via Ras/Raf/MEK/ERK. The activation of this pathway induces the overexpression of DNMT proteins and the consequent hypermethylation of the regions upstream of MIR205HG and of miR-205 locus38. On the contrary, DNA hypomethylation was responsible for the induced miR-205 expression in oral squamous cell and ovarian carcinomas32,42. Indirectly, the epigenetic status can regulate the expression of miR-205 by the modulation of TFs. As reported in TNBC, the restoration of the normal DNA methylation status induces p53 TF up-regulation and the consequent rebalancing of miR-205 levels22.
Transcription factors
miR-205 expression was shown to be regulated by several TFs, among which the members of p53 family, Specificity protein 1 (Sp1), Hypoxia-Inducible Factor alpha (HIF-1α), Transforming Growth Factor β (TGF-β), and Twist-related protein 1 (Twist1)13,22,36,43–48.
P53 plays multiple roles, including regulation of the cell cycle, apoptosis and genomic stability, thus exerting a tumor-suppressive function49,50. Findings showed that p53 stimulates the expression of miR-205 by interacting with putative p53 responsive elements (p53REs) in a region upstream of miR-205 sequence, in different cellular models22 (Fig. 1). Notably, a decrease or complete loss of miR-205 is frequently found in breast cancers. In this regard, it was reported that inactivating mutations of p53 that result in its defective binding to p53REs are responsible for decreased level of miR-205 in TNBC22. Conversely, p53 gain of function mutants (GOF-mutp53) induce the up-regulation of miR-205, which promotes proliferation in head and neck squamous cell carcinoma. Interestingly, GOF-mutp53 does not recognize the wild type-p53RE in the pre-miR-205 genomic sequence but binds to a region upstream of MIR205HG51. p53 mutants may impinge on miR-205 expression also through different mechanisms. For example, in the prostate cancer context, p53 mutants reduce p63 stability and activity, with a consequent reduction of miR-205 transcriptional rate48. In fact, p63 and p73 proteins, by exhibiting a remarkable sequence and structural homology to p53, can bind to p53 DNA target sites and similarly regulate miR-205 expression24,52. In prostate and bladder epithelia, p63 and its isoform ΔNp63α, lacking the transactivation domain, bind to p53REs upstream of miR-205 sequence and induce its expression13,47,48. As consequence, the loss of p63 observed in these epithelia upon transformation results in a decrease of miR-205 levels and in a reciprocal effect on EMT process47,48. On the other hand, p73 TF, by binding to miR-205 upstream sites, induces miR-205 expression in non-metastatic melanoma cells. Interestingly, the truncated form of p73, ΔNp73, highly expressed in malignant melanoma cells, interferes with p73 activity, thus being responsible for the reduced expression of miR-20524.
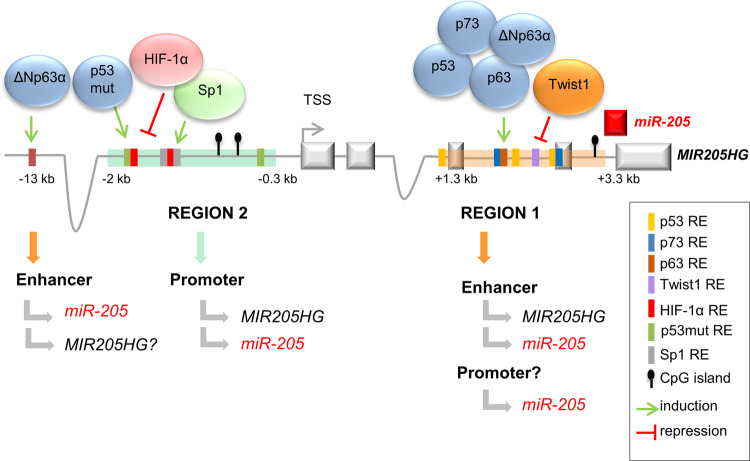
Fig. 1. Human miR-205 host gene (MIR205HG) transcription unit.
The schematic representation shows the transcription factor responsive elements (REs) and methylation sites (CpG islands) localized upstream of MIR205HG and of pre-miR-205 sequence. The miR-205 and MIR205HG regulatory regions are depicted in orange (region 1) and in aquamarine (region 2).
Another DNA-binding factor that regulates miR-205 expression is Sp1. It was shown that induction of Sp1 upon DNA damage activates the expression of miR-205 and its host gene in esophageal squamous cell carcinoma by binding to sites immediately upstream of MIR205HG. Interestingly, the Sp1-mediated transcriptional activation of miR-205 promotes radioresistence and an aggressive phenotype through PTEN-PI3K/AKT pathway46. Conversely, pro-inflammatory cytokines, cell or environmental stresses converge onto the downregulation of miR-205 in EMT contexts. A decreased expression of miR-205 was reported in a canine kidney (MDCK) and in a glioma cell lines upon treatment with TGF-β18,44. Moreover, TGF-β1, co-secreted with IL-6 by aggressive prostate cancer cells, converts stromal cells into cancer-associated fibroblasts, which in turn stabilize HIF-1α. This redox sensitive TF, directly interacting with REs in proximity of MIR205HG, represses miR-205 expression. In this context, downregulation of miR-205 leads to de-repression of ZEB1/2 and PKCε, allowing EMT of prostate cancer cells45. Again, miR-205 expression was shown to be controlled by the EMT-inducing transcription Twist1. Indeed, Twist1 was proven to directly repress miR-205 transcription in invasive bladder cancer, through direct binding to a pre-miR-205 regulatory region36.
miR-205 transcription unit: insights into the regulatory regions
Pre-miR-205 sequence is located within MIR205HG, precisely in the connecting region between its last intron and exon. This peculiar location stimulated interest in investigating the potential transcriptional relationship between the two genes and in identifying the relevant regulatory regions. As mentioned before, TF motifs, DNA methylation and DNAseI-sensitive sites were found in proximity of both pre-miR-205 (region 1, Fig. 1) and MIR205HG (region 2, Fig. 1), suggesting a possible regulatory role for these regions. Region 1 is located within 2 kb upstream of the pre-miR-205 sequence and contains p53 family REs, Twist1-binding sites and methylation sites13,22,24,47,48. Region 2 is upstream of the first exon of MIR205HG and includes binding sites for GOF-mutp53, HIF-1α, Sp1, and chromatin modification sites37–39,43,45,46. In addition, a sequence at –13 kb with respect to the MIR205HG TSS, enriched in p63 sites, is functional in regulating miR-205 expression13. A number of research groups investigated the regulatory role of these regions through in silico and experimental analyses. Region 1 showed to have promoter activity in two different works, when tested by reporter assay upon co-transfection with p63 and p7324,48. However, the same region tested by a different reporter construct showed only enhancer activity upon p53 transfection in HEK-293 cells22. In addition, Chromatin immunoprecipitation (ChIP) experiments demonstrated that the functional binding of the p63 isoform ΔNp63 to responsive elements in region 1 induced a strong enrichment of RNA Polymerase II onto region 2, thus resulting in increased expression of both miR-205 and MIR205HG transcripts47. Consistent with this, region 2 was validated to have promoter activity in reporter assays by different authors22,45,46. The presence of CpG in region 2 again supports its promoter function, as methylation sites in human genes are mostly related to promoter regions37–39,53. A recent work showed that the binding of the mutant p53 to region 2 increases the expression of both MIR205HG and miR-20543, indicating that this region may act as the promoter for both RNAs. Consistent with this, analysis of RNA-seq data from The Cancer Genome Atlas (TCGA, https://www.cancer.gov/tcga) showed marked correlation between MIR205HG and miR-205 expression across tissues54, again supporting co-transcriptional regulation. Recent data from our lab showed that genomic deletion of the sequence spanning from exon 1 to 3 of MIR205HG (including TSS) in prostate basal cells or antisense oligonucleotide-mediated targeting of introns of MIR205HG primary sequence invariably resulted in the abrogation of both MIR205HG and miR-205 expression54, letting to hypothesize that a “promiscuous” MIR205HG/miR-205 primary transcript might exist (see section “Post-transcriptional regulation” for further details).
Altogether, the evidence collected so far suggests two main possible models for miR-205 transcription: the first one accounts for an independent miR-205 transcription driven by pre-miR-205 proximal promoter (region 1, Fig. 1); the latter supports a MIR205HG-dependent expression dictated by the distal promoter (region 2, Fig. 1), which would transcribe for a common MIR205HG/miR-205 primary transcript, with region 1 acting as enhancer.
Post-transcriptional regulation
miR-205 processing: implications for miR-205 and MIR205HG expression
The peculiar pre-miR-205 location in the last intron-exon junction of MIR205HG arises important questions about miR-205 biogenesis from the putative MIR205HG/miR-205 primary transcript. In fact, use of this splice site would be incompatible with the excision of pre-miR-205. Interestingly, Chang and co-workers55, by sequencing pri-miRNA structures using a dominant-negative Drosha mutant, were able to map lowly expressed alternatively spliced MIR205HG transcripts that, by utilizing a distinct 3’ terminal exon (exon 5.2, Fig. 2), make the pre-miR-205 sequence fully intronic, a configuration permissive to miRNA processing. Accordingly, reannotation of all possible MIR205HG transcripts made starting from recently acquired long read sequencing data56 suggested the existence of two locus configurations, one that acts as source of miR-205 (miR-205 compatible transcripts) and the other that gives rise to MIR205HG transcripts only (miR-205 incompatible transcripts)54. These data suggest that alternative splicing may dictate the switch between the two locus configurations.
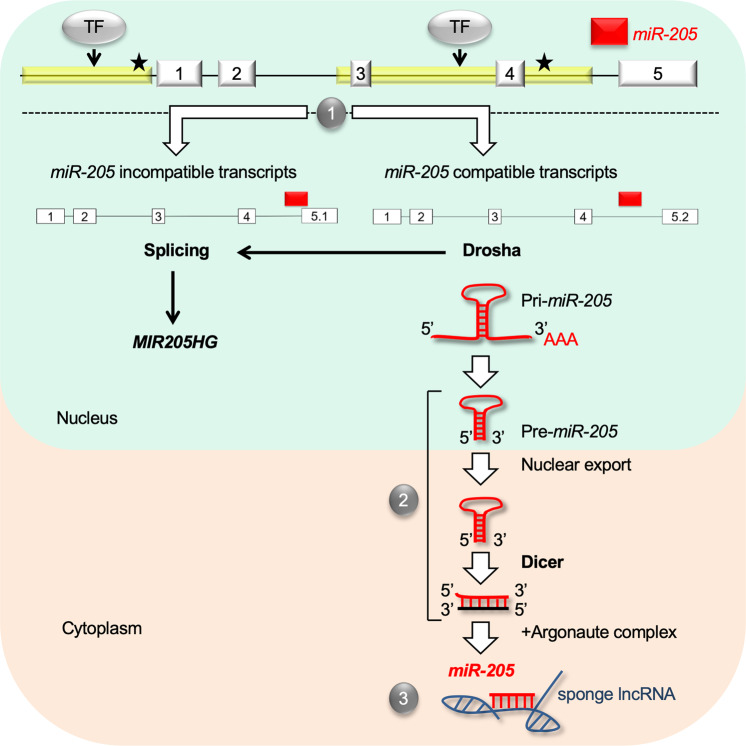
Fig. 2. Scheme of miR-205 regulation.
At the transcriptional level, miR-205 is regulated by transcription factors (TF) or epigenetic modifications (indicated by stars). At the post-transcriptional level, alternative splicing of MIR205HG primary transcript gives origin to miR-205 compatible and non-compatible transcripts (1). Drosha masks the splicing site between MIR205HG exons 4 and 5.1, favoring the use of an alternative exon (exon 5.2) and the excision of pre-miR-205, which is then processed by Dicer into miRNA duplex (2). miR-205 bioavailability is regulated by several cytoplasmic lncRNAs acting as miRNA sponges (3).
Notably, the process of miR-205 production from miR-205 compatible transcripts was found to be dependent on Drosha activity. Specifically, Drosha is not only responsible for miRNA processing, rather it is able to physically mask miR-205-incompatible splicing site, thus favoring production of miR-205-compatible MIR205HG isoforms54. Consistent with this, defects in biogenesis machinery were observed in prostate cancer cells under hypoxia where the decreased levels of Drosha and Dicer resulted in a down-modulation of miR-20545,57. Taken together, these findings illustrate an additional level of miR-205 regulation where competition between the spliceosome and the microprocessor Drosha may direct the processing of MIR205HG primary transcript into miR-205 and/or MIR205HG RNAs. In this regard, it is still to explore whether a functional splicing machinery is requested or dispensable for miR-205 biogenesis.
It is worth mentioning that, though processed from a common primary transcript, mature MIR205HG and miR-205 have been shown to act independently. In fact, recent studies on MIR205HG support its function as long noncoding RNA (lncRNA) in different cell contexts (Table 2)43,54,58–62. LncRNAs are functional non-protein coding transcripts longer than 200 nucleotides63. In the last years, lncRNAs have been increasingly recognized as regulators of pivotal biological processes, with their aberrant activity being implicated in tumorigenesis64.
Table 2.
Summary of the reported functions and mechanisms of action of MIR205HG lncRNA in different tissue/tumor types.
| Species | Tissue/cancer type | Biological role | Mechanism of action | References |
|---|---|---|---|---|
| Human | LUSC | Cell proliferation, migration, EMT | ceRNA for miR-299–3p | 62 |
| Human | LUSC | Cell proliferation, migration, apoptosis | Bcl-2 and Bax regulation | 61 |
| Human | Cervical cancer | Cell proliferation, migration, apoptosis | SRSF1/KRT17 axis regulation | 60 |
| Human | Cervical cancer | Cell proliferation, migration, apoptosis | ceRNA for miR-122-5p | 59 |
| Human | HNSCC | Cell proliferation, migration | ceRNA for miR-590-3p | 43 |
| Human | Prostate basal cells | Basal-luminal differentiation | transcriptional repression of interferon genes | 54 |
| Mouse | Pituitary | Growth hormone and prolactin production | Pit1 regulation | 58 |
LUSC lung squamous cell carcinoma, HNSCC head and neck squamous cell carcinoma.
In head and neck squamous cell carcinoma, where both MIR205HG and miR-205 are over-expressed, MIR205HG lncRNA was shown to enhance cancer cell proliferation and motility by sequestering miR-590–3p and preventing interaction with its target mRNAs, thus acting as “molecular sponge”43. Similar protumorigenic effects were reported for MIR205HG also in cervical cancer59,60 and in lung squamous cell carcinoma61,62, though with different mechanisms of action (Table 2). This oncogenic role is remindful, though independent, to that of miR-205 in squamous cell carcinomas, where the miRNAs was shown to exert protumorigenic function through repression of tumor-suppressor genes46. MIR205HG and miR-205 were reported to play non redundant independent functions also in prostate basal cells, where MIR205HG maintains basal identity by regulating differentiation and miR-205 regulates the production of basement membrane13,54. Notably, also in the mouse context, MIR205HG lncRNA was shown to work independently of miR-205 in regulating growth hormone and prolactin production in the anterior pituitary58.
Reduction of miR-205 availability: role of long noncoding RNAs
One of the numerous mechanisms by which lncRNAs can regulate gene expression is by sponging (sequestering) miRNAs. Owing to their frequent aberrant expression in cancer, they are in part responsible for the dysregulated miRNA expression in such conditions65,66. Growing evidence reported a negative control relationship between miR-205 and lncRNAs in cancer. The long Intergenic Non-Protein Coding 00673 (LINC00673), enriched in hepatocellular carcinoma, by adsorbing miR-205 reduces its availability and consequently prevents miR-205 tumor-suppressor functions67. Small Nucleolar RNA Host Gene 5 (SNHG5) is another tumor-enriched lncRNA that binds and sequesters miR-205, de-repressing the target ATP-binding cassette sub-family C member 2 (ABCC2) and promoting imatinib resistance in chronic myeloid leukemia68. On the other hand, reduced levels of the lncRNA growth arrest specific 5 (GAS5) in human cervical cancer dictate the up-regulation of miR-205 and its oncogenic role in promoting the proliferation and migration of cervical cells69. A crosstalk between lncRNAs and miR-205 has been also observed in renal carcinoma, where a reduction of miR-205 is related to an overexpression of LINC0015270. RP11–395G23.3 and LA16c-313D11.11 are two lncRNAs associated to the pathogenesis of endometrial cancer. They act as endogenous competing RNAs for miR-205-PTEN network and have been shown to inhibit the expression and the activity of miR-205 at the post-transcriptional level through highly conserved miRNA responsive elements71. Again, a recent work reported a link between decreased levels of miR-205 and high expression of lncRNA small nucleolar RNA host gene 16 (SNHG16) in osteosarcoma tissues. These studies revealed that SNHG16, by acting as endogenous sponge of miR-205, upregulates ZEB1 and enhances proliferation of osteosarcoma cells72. Similarly, SNHG16 was found to act as miR-205 sponge in the cardiovascular context, where reduced levels of miR-205 are related to increased proliferation and migration of aortic smooth muscle cells, suggesting a link with atherosclerosis73. In ovarian cancer, an increase of miR-205 levels are related to a reduction of its lncRNA sponge LINC01133. This unbalance results in a reduction of the miR-205 target Leucine-rich repeat kinase 2 (LRRK2) and enhanced proliferatory, migratory and invasive ovarian cancer cell ability74.
The LINC regulator of reprogramming (LINC-ROR) is important for the maintenance of induced pluripotent and embryonic stem cells. Specifically, LINC-ROR, by acting as a molecular sponge for miR-205, prevents the degradation of miR-205 targets (e.g., ZEB1 and ZEB2) and promotes EMT in breast cancer75.
Interestingly, lncRNAs can also indirectly regulate miRNAs. As reported by Sun and co-workers76, the lncRNA HOX transcript antisense RNA (HOTAIR) participates to miR-205 silencing in bladder cancer by breaking the balance between the positive (H3K4me3) and negative (H3K27me3) chromatin marks on miR-205 promoter.
Conclusions
miR-205 is one of the most investigated miRNAs due to its involvement in multiple physiologic, oncogenic and tumor-suppressor pathways. Several mechanisms and factors, depending on cell and tumor types, regulate miR-205 expression and contribute to its complex function. In cancer, epigenetic modifications, mutated or alternatively spliced p53 family proteins and components of tumor microenvironment (hypoxia, inflammatory cytokines) mostly contribute to miR-205 dysregulation at the transcriptional level. Post-transcriptionally, lncRNAs are in part responsible for changes in miR-205 availability in tumor cells.
miR-205 regulation is made further complicated by its special genomic location, which rises issues regarding its biogenesis in relationship to that of its host gene. Despite the binding of transcription factors upstream of the precursor miR-205 sequence could account for a MIR205HG-independent transcription, experimental data supporting promoter activity of this region are weak. The most accredited function for this regulatory region could be to serve as enhancer for transcription of a common MIR205HG/miR-205 primary transcript, from which both miR-205 and MIR205HG-processed transcripts are produced. However, additional investigations in the sequences upstream or downstream of the pre-miR-205 could evidence regions involved in specific miR-205 regulation.
The observation that miR-205 can originate from alternatively spliced MIR205HG transcripts with the intervention of Drosha lets to speculate that a diverse commitment between miR-205 compatible and incompatible transcripts and/or a competition between Drosha and spliceosome may represent a further checkpoint in miR-205 regulation to modify miR-205/MIR205HG ratio. The fine tuning between miR-205 and MIR205HG level may have important implications on biological functions, in dependence on the cell type and physio-pathological state. In this regard, additional investigation would be required to identify the context and factors that may modify reciprocal miR-205/MIR205HG biogenesis.
Overall, the reviewed studies provide a further understanding of miR-205 gene organization and regulation, including the recently acquired knowledge about its host gene. Elucidating the complex, multilevel regulation of miR-205 expression during the normal development of epithelia is crucial to determine which factors maintain the balance and contribute to the physiologic state. Subsequently, unraveling the causes responsible for miR-205 dysregulation in cancer is of utmost interest for the future development of miR-205 modulating strategies applicable in therapy.
Acknowledgements
This work in the authors’ laboratory was supported by grants from Italian Ministry of Health (GR-2013-02355625 to P.G.) and CARIPLO Foundation (2015–0866 to P.G.). We thank all the people of the laboratory who contributed, over the years, to shed light on miR-205 and MIR205HG function.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by A. Rufini
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bartel DP. MicroRNA target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee YS, Dutta A. MicroRNAs in cancer. Annu. Rev. Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Westholm JO, Lai EC. Mirtrons: microRNA biogenesis via splicing. Biochimie. 2011;93:1897–1904. doi: 10.1016/j.biochi.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 7.Ryan DG, Oliveira-Fernandes M, Lavker RM. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Mol. Vis. 2006;12:1175–1184. [PubMed] [Google Scholar]
- 8.Gregory PA, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 9.Mouillet JF, Chu T, Nelson DM, Mishima T, Sadovsky Y. MiR-205 silences MED1 in hypoxic primary human trophoblasts. FASEB J. 2010;24:2030–2039. doi: 10.1096/fj.09-149724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li C, Finkelstein D, Sherr CJ. Arf tumor suppressor and miR-205 regulate cell adhesion and formation of extraembryonic endoderm from pluripotent stem cells. Proc. Natl Acad. Sci. USA. 2013;110:E1112–E1121. doi: 10.1073/pnas.1302184110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greene SB, Gunaratne PH, Hammond SM, Rosen JM. A putative role for microRNA-205 in mammary epithelial cell progenitors. J. Cell Sci. 2010;123:606–618. doi: 10.1242/jcs.056812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Avril-Sassen S, et al. Characterisation of microRNA expression in post-natal mouse mammary gland development. BMC Genomics. 2009;10:548. doi: 10.1186/1471-2164-10-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gandellini P, et al. miR-205 regulates basement membrane deposition in human prostate: implications for cancer development. Cell Death Differ. 2012;19:1750–1760. doi: 10.1038/cdd.2012.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang D, et al. MicroRNA-205 controls neonatal expansion of skin stem cells by modulating the PI(3)K pathway. Nat. Cell Biol. 2013;15:1153–1163. doi: 10.1038/ncb2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu J, et al. MicroRNA-205 promotes keratinocyte migration via the lipid phosphatase SHIP2. FASEB J. 2010;24:3950–3959. doi: 10.1096/fj.10-157404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farmer DT, et al. Partially penetrant postnatal lethality of an epithelial specific MicroRNA in a mouse knockout. PLoS ONE. 2013;8:e76634. doi: 10.1371/journal.pone.0076634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farmer DT, et al. miR-205 is a critical regulator of lacrimal gland development. Dev. Biol. 2017;427:12–20. doi: 10.1016/j.ydbio.2017.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoover AR, et al. MicroRNA-205 maintains T cell development following stress by regulating forkhead box N1 and selected chemokines. J. Biol. Chem. 2016;291:23237–23247. doi: 10.1074/jbc.M116.744508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darnell DK, et al. MicroRNA expression during chick embryo development. Dev. Dyn. 2006;235:3156–3165. doi: 10.1002/dvdy.20956. [DOI] [PubMed] [Google Scholar]
- 20.Gandellini P, et al. miR-205 Exerts tumor-suppressive functions in human prostate through down-regulation of protein kinase Cepsilon. Cancer Res. 2009;69:2287–2295. doi: 10.1158/0008-5472.CAN-08-2894. [DOI] [PubMed] [Google Scholar]
- 21.Hagman Z, et al. miR-205 negatively regulates the androgen receptor and is associated with adverse outcome of prostate cancer patients. Br. J. Cancer. 2013;108:1668–1676. doi: 10.1038/bjc.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piovan C, et al. Oncosuppressive role of p53-induced miR-205 in triple negative breast cancer. Mol. Oncol. 2012;6:458–472. doi: 10.1016/j.molonc.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iorio MV, et al. microRNA-205 regulates HER3 in human breast cancer. Cancer Res. 2009;69:2195–2200. doi: 10.1158/0008-5472.CAN-08-2920. [DOI] [PubMed] [Google Scholar]
- 24.Alla V, et al. E2F1 confers anticancer drug resistance by targeting ABC transporter family members and Bcl-2 via the p73/DNp73-miR-205 circuitry. Cell Cycle. 2012;11:3067–3078. doi: 10.4161/cc.21476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Majid S, et al. MicroRNA-205 inhibits Src-mediated oncogenic pathways in renal cancer. Cancer Res. 2011;71:2611–2621. doi: 10.1158/0008-5472.CAN-10-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su N, et al. miR-205 promotes tumor proliferation and invasion through targeting ESRRG in endometrial carcinoma. Oncol. Rep. 2013;29:2297–2302. doi: 10.3892/or.2013.2400. [DOI] [PubMed] [Google Scholar]
- 27.Zhang G, Hou X, Li Y, Zhao M. MiR-205 inhibits cell apoptosis by targeting phosphatase and tensin homolog deleted on chromosome ten in endometrial cancer Ishikawa cells. BMC Cancer. 2014;14:440. doi: 10.1186/1471-2407-14-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chung TK, et al. Dysregulated microRNAs and their predicted targets associated with endometrioid endometrial adenocarcinoma in Hong Kong women. Int. J. Cancer. 2009;124:1358–1365. doi: 10.1002/ijc.24071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karaayvaz M, Zhang C, Liang S, Shroyer KR, Ju J. Prognostic significance of miR-205 in endometrial cancer. PLoS ONE. 2012;7:e35158. doi: 10.1371/journal.pone.0035158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qu C, et al. MiR-205 determines the radioresistance of human nasopharyngeal carcinoma by directly targeting PTEN. Cell Cycle. 2012;11:785–796. doi: 10.4161/cc.11.4.19228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mao Y, Wu S, Zhao R, Deng Q. MiR-205 promotes proliferation, migration and invasion of nasopharyngeal carcinoma cells by activation of AKT signalling. J. Int. Med. Res. 2016;44:231–240. doi: 10.1177/0300060515576556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iorio MV, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 33.He L, et al. Ovarian cancer cell-secreted exosomal miR-205 promotes metastasis by inducing angiogenesis. Theranostics. 2019;9:8206–8220. doi: 10.7150/thno.37455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hezova R, et al. MiR-205 functions as a tumor suppressor in adenocarcinoma and an oncogene in squamous cell carcinoma of esophagus. Tumour Biol. 2016;37:8007–8018. doi: 10.1007/s13277-015-4656-8. [DOI] [PubMed] [Google Scholar]
- 35.Charkiewicz R, et al. Validation for histology-driven diagnosis in non-small cell lung cancer using hsa-miR-205 and hsa-miR-21 expression by two different normalization strategies. Int. J. Cancer. 2016;138:689–697. doi: 10.1002/ijc.29816. [DOI] [PubMed] [Google Scholar]
- 36.Wiklund ED, et al. Coordinated epigenetic repression of the miR-200 family and miR-205 in invasive bladder cancer. Int. J. Cancer. 2011;128:1327–1334. doi: 10.1002/ijc.25461. [DOI] [PubMed] [Google Scholar]
- 37.Hulf T, et al. Epigenetic-induced repression of microRNA-205 is associated with MED1 activation and a poorer prognosis in localized prostate cancer. Oncogene. 2013;32:2891–2899. doi: 10.1038/onc.2012.300. [DOI] [PubMed] [Google Scholar]
- 38.Hasegawa T, et al. ErbB2 signaling epigenetically suppresses microRNA-205 transcription via Ras/Raf/MEK/ERK pathway in breast cancer. FEBS Open Bio. 2017;7:1154–1165. doi: 10.1002/2211-5463.12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhatnagar N, et al. Downregulation of miR-205 and miR-31 confers resistance to chemotherapy-induced apoptosis in prostate cancer cells. Cell Death Dis. 2010;1:e105. doi: 10.1038/cddis.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ke XS, et al. Genome-wide profiling of histone h3 lysine 4 and lysine 27 trimethylation reveals an epigenetic signature in prostate carcinogenesis. PLoS ONE. 2009;4:e4687. doi: 10.1371/journal.pone.0004687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tellez CS, et al. EMT and stem cell-like properties associated with miR-205 and miR-200 epigenetic silencing are early manifestations during carcinogen-induced transformation of human lung epithelial cells. Cancer Res. 2011;71:3087–3097. doi: 10.1158/0008-5472.CAN-10-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiklund ED, et al. MicroRNA alterations and associated aberrant DNA methylation patterns across multiple sample types in oral squamous cell carcinoma. PLoS ONE. 2011;6:e27840. doi: 10.1371/journal.pone.0027840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Agostino S, et al. Long non-coding MIR205HG depletes Hsa-miR-590-3p leading to unrestrained proliferation in head and neck squamous cell carcinoma. Theranostics. 2018;8:1850–1868. doi: 10.7150/thno.22167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duan Y, Chen Q. TGF-β1 regulating miR-205/miR-195 expression affects the TGF-β signal pathway by respectively targeting SMAD2/SMAD7. Oncol. Rep. 2016;36:1837–1844. doi: 10.3892/or.2016.5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gandellini P, et al. miR-205 hinders the malignant interplay between prostate cancer cells and associated fibroblasts. Antioxid. Redox Signal. 2014;20:1045–1059. doi: 10.1089/ars.2013.5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pan F, et al. Sp1-mediated transcriptional activation of miR-205 promotes radioresistance in esophageal squamous cell carcinoma. Oncotarget. 2017;8:5735–5752. doi: 10.18632/oncotarget.13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tran MN, et al. The p63 protein isoform DeltaNp63alpha inhibits epithelial-mesenchymal transition in human bladder cancer cells: role of MIR-205. J. Biol. Chem. 2013;288:3275–3288. doi: 10.1074/jbc.M112.408104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tucci P, et al. Loss of p63 and its microRNA-205 target results in enhanced cell migration and metastasis in prostate cancer. Proc. Natl Acad. Sci. USA. 2012;109:15312–15317. doi: 10.1073/pnas.1110977109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams AB, Schumacher B. p53 in the DNA-damage-repair process. Cold Spring Harb. Perspect. Med. 2016;6:a026070. doi: 10.1101/cshperspect.a026070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shaw PH. The role of p53 in cell cycle regulation. Pathol. Res. Pract. 1996;192:669–675. doi: 10.1016/S0344-0338(96)80088-4. [DOI] [PubMed] [Google Scholar]
- 51.Arenberg DA, et al. The role of CXC chemokines in the regulation of angiogenesis in non-small cell lung cancer. J. Leukoc. Biol. 1997;62:554–562. doi: 10.1002/jlb.62.5.554. [DOI] [PubMed] [Google Scholar]
- 52.Lin C, et al. The microRNA feedback regulation of p63 in cancer progression. Oncotarget. 2015;6:8434–8453. doi: 10.18632/oncotarget.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strichman-Almashanu LZ, et al. A genome-wide screen for normally methylated human CpG islands that can identify novel imprinted genes. Genome Res. 2002;12:543–554. doi: 10.1101/gr.224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Profumo V, et al. LEADeR role of miR-205 host gene as long noncoding RNA in prostate basal cell differentiation. Nat. Commun. 2019;10:307. doi: 10.1038/s41467-018-08153-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang TC, Pertea M, Lee S, Salzberg SL, Mendell JT. Genome-wide annotation of microRNA primary transcript structures reveals novel regulatory mechanisms. Genome Res. 2015;25:1401–1409. doi: 10.1101/gr.193607.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lagarde J, et al. High-throughput annotation of full-length long noncoding RNAs with capture long-read sequencing. Nat. Genet. 2017;49:1731–1740. doi: 10.1038/ng.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu CG, Yang MF, Fan JX, Wang W. MiR-30a and miR-205 are downregulated in hypoxia and modulate radiosensitivity of prostate cancer cells by inhibiting autophagy via TP53INP1. Eur. Rev. Med. Pharmacol. Sci. 2016;20:1501–1508. [PubMed] [Google Scholar]
- 58.Du Q, et al. MIR205HG is a long noncoding RNA that regulates growth hormone and prolactin production in the anterior pituitary. Dev. Cell. 2019;20:618–631. doi: 10.1016/j.devcel.2019.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Y, Wang H, Huang H. Long non-coding RNA MIR205HG function as a ceRNA to accelerate tumor growth and progression via sponging miR-122-5p in cervical cancer. Biochem. Biophys. Res. Commun. 2019;514:78–85. doi: 10.1016/j.bbrc.2019.04.102. [DOI] [PubMed] [Google Scholar]
- 60.Dong M, Dong Z, Zhu X, Zhang Y, Song L. Long non-coding RNA MIR205HG regulates KRT17 and tumor processes in cervical cancer via interaction with SRSF1. Exp. Mol. Pathol. 2019;111:104322. doi: 10.1016/j.yexmp.2019.104322. [DOI] [PubMed] [Google Scholar]
- 61.Chang Y, et al. MIR205HG facilitates carcinogenesis of lung squamous cell carcinoma in vitro revealed by long noncoding RNA profiling. Acta Biochim. Biophys. Sin. 2020;52:371–381. doi: 10.1093/abbs/gmaa006. [DOI] [PubMed] [Google Scholar]
- 62.Liu L, et al. MIR205HG acts as a ceRNA to expedite cell proliferation and progression in lung squamous cell carcinoma via targeting miR-299-3p/MAP3K2 axis. BMC Pulm. Med. 2020;20:163. doi: 10.1186/s12890-020-1174-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 64.Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genom. Proteom. Bioinf. 2016;14:42–54. doi: 10.1016/j.gpb.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang J, Liu L, Li J, Le TD. LncmiRSRN: identification and analysis of long non-coding RNA related miRNA sponge regulatory network in human cancer. Bioinformatics. 2018;34:4232–4240. doi: 10.1093/bioinformatics/bty525. [DOI] [PubMed] [Google Scholar]
- 66.Chen L, Dzakah EE, Shan G. Targetable long non-coding RNAs in cancer treatments. Cancer Lett. 2018;418:119–124. doi: 10.1016/j.canlet.2018.01.042. [DOI] [PubMed] [Google Scholar]
- 67.Zhang LG, Zhou XK, Zhou RJ, Lv HZ, Li WP. Long non-coding RNA LINC00673 promotes hepatocellular carcinoma progression and metastasis through negatively regulating miR-205. Am. J. Cancer Res. 2017;7:2536–2544. [PMC free article] [PubMed] [Google Scholar]
- 68.He B, Bai Y, Kang W, Zhang X, Jiang X. LncRNA SNHG5 regulates imatinib resistance in chronic myeloid leukemia via acting as a CeRNA against MiR-205-5p. Am. J. Cancer Res. 2017;7:1704–1713. [PMC free article] [PubMed] [Google Scholar]
- 69.Yang W, et al. LncRNA GAS5 suppresses the tumorigenesis of cervical cancer by downregulating miR-196a and miR-205. Tumour Biol. 2017;39:1010428317711315. doi: 10.1177/1010428317711315. [DOI] [PubMed] [Google Scholar]
- 70.Wang Y, et al. Long intergenic non-coding RNA 00152 promotes renal cell carcinoma progression by epigenetically suppressing P16 and negatively regulates miR-205. Am. J. Cancer Res. 2017;7:312–322. [PMC free article] [PubMed] [Google Scholar]
- 71.Xin W, et al. Long non-coding RNA derived miR-205-5p modulates human endometrial cancer by targeting PTEN. Am. J. Transl. Res. 2015;7:2433–2441. [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu C, Cheng D, Qiu X, Zhuang M, Liu Z. Long noncoding RNA SNHG16 promotes cell proliferation by sponging microRNA-205 and upregulating ZEB1 expression in osteosarcoma. Cell Physiol. Biochem. 2018;51:429–440. doi: 10.1159/000495239. [DOI] [PubMed] [Google Scholar]
- 73.Lin Y, et al. Long non-coding RNA SNHG16 regulates human aortic smooth muscle cell proliferation and migration via sponging miR-205 and modulating Smad2. J. Cell Mol. Med. 2019;23:6919–6929. doi: 10.1111/jcmm.14576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu M, Shen C, Wang C. Long noncoding RNA LINC01133 confers tumor-suppressive functions in ovarian cancer by regulating leucine-rich repeat kinase 2 as an miR-205 sponge. Am. J. Pathol. 2019;189:2323–2339. doi: 10.1016/j.ajpath.2019.07.020. [DOI] [PubMed] [Google Scholar]
- 75.Hou P, et al. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014;5:e1287. doi: 10.1038/cddis.2014.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun X, et al. Long non-coding RNA HOTAIR regulates cyclin J via inhibition of microRNA-205 expression in bladder cancer. Cell Death Dis. 2015;6:e1907. doi: 10.1038/cddis.2015.269. [DOI] [PMC free article] [PubMed] [Google Scholar]