Abstract
Purpose
Astaxanthin (Ast) has been reported to reduce oxidative stress induced by diabetes mellitus (DM). The aim of this research was to give a systematic overview of the biological basis for this process.
Methods
Ast-targeted proteins were collected from the BATMAN database, Comparative Toxicogenomics Database, and STITCH database. Putative DM-related protein targets were collected from the GeneCards database. A DM-rat model was then built with streptozotocin (STZ) combined with a high-sugar, high-fat diet for 30 days. Total cholesterol (TC), triglycerides (TGs), and insulin levels were examined using whole tail-vein blood from overnight-fasted rats. SOD, GSH, and MDA activy was detected in liver tissue (p<0.05). In addition, we used RNA-sequencing analysis to detect gene-transcription level in liver tissue of rats and GO biological process analysis to show all the log2FC≥2 genes in the Ast-fed DM rats compared with the DM group using the STRING database. Ast-intersecting targets were collected with Venn analysis. Docking analysis between Ast and targeted proteins was down with the SwissDock server. Ast targets–pathway networks were built using Cytoscape 3.7.2 software.
Results
A total of 120 Ast-targeted proteins and 13,784 DM-related targets were collected. Ast functioned in reducing TC, TG, and MDA levels, promoting SOD activity and GSH expression, and alleviating islet-cell injury in Ast-fed DM rats compared with DM control rats. Furthermore, genes involved in MAPK, TNF, AMPK, and FOXO signaling pathways were differently expressed in Ast-treated DM rats compared with DM rats. The differentially expressed genes were enriched in euchromatin, thyroid cancer, and metaphase-plate congression. Three Ast-intersecting targets — Col5A1, Nqo1, and Notch2 — were then identified. We found possible binding patterns of Ast with Nqo1 and Notch2, respectively. Ast targets–pathway networks were finally built to show a systematic overview of how Ast works in multiple pathways to reduce oxidative stress. Taken together, Ast is predicted to target Col5A1, Nqo1, and Notch2 to form a network of systemic pharmacological effects to: 1) promote insulin-releasing balance and relieve insulin resistance, 2) reduce testicular cell apoptosis, and 3) maintain normal size in marginal-zone B cells and inhibit autoimmune DM, all of which contribute to the balance of lipid metabolism and reduction of oxidative stress in DM patients.
Conclusion
Ast functions in reducing oxidative stress in DM rats by regulating a variety of targets to form a comprehensive antioxidative network.
Keywords: astaxanthin, diabetes mellitus, oxidative stress, RNA-seq analysis, network pharmacy
Introduction
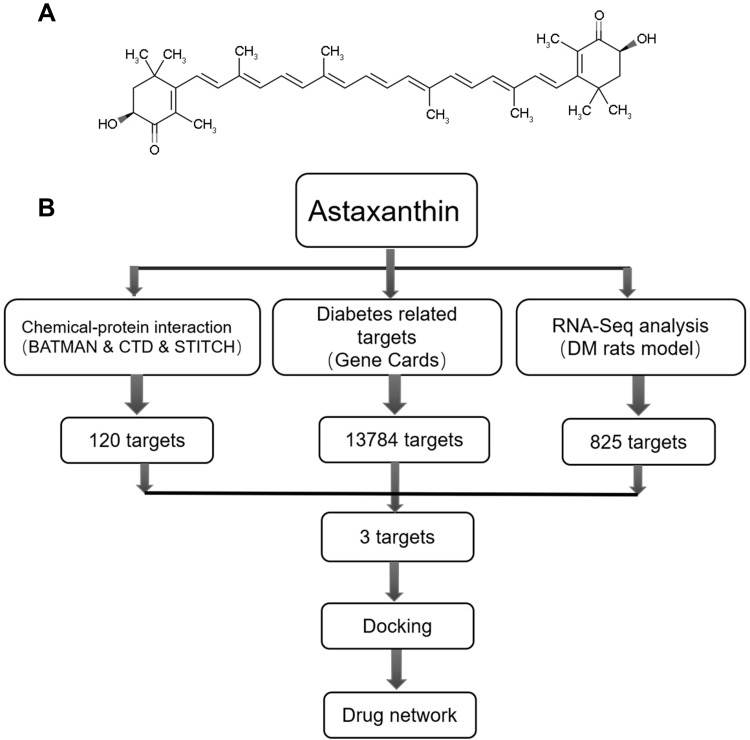
Astaxanthin (Ast; 3,3′-dihydroxy-β,β′-carotene-4,4′-dione) is a type of red carotenoid that can be extracted from various microorganisms and marine animals, eg, algae, salmon, and shrimp.1,2 Ast is red due to conjugated double bonds at the center of the compound (Figure 1A). These bonds are considered a strong antioxidant through donating electrons and reacting with free radicals to stabilize the latter and terminate free-radical chain reactions in huge amounts of living organisms.3,4 Furthermore, with its special combined ketone–hydroxyl structure, Ast possesses better biological activity than other antioxidants, as it is long enough to link with cell membranes from inside to outside.
Figure 1.
(A) Chemical structure of Ast; (B) pipeline for identification of both predicted and detected Ast targets that integrate compound proteins, disease proteins, RNA-seq analysis, GO analysis, docking, and network construction.
Diabetes mellitus (DM) puts a huge burden on health-care systems all over the world.5 There are several complications caused by DM, such as disorders of lipid metabolism, fatty liver, and inflammation.6,7 It has been reported that oxidative stress is induced by hyperglycemia from dysfunction of pancreatic β cells, leading to tissue damage and complications in DM patients.8 The oxidative stress is created by imbalance between reactive oxygen species (ROS) production and antioxidant defense. With increased electrons, ROS are reduced to a highly reactive, radical format in the cells, including O2–, O2−2, H2O2, OH, and OH–.9,10 All these ROS have different actions and kinetics in cellular metabolism. Therefore, mitigating the symptoms of DM by reducing oxidative stress appears to be a feasible approach.
Ast has shown great potential in alleviating oxidative stress induced by DM. Ast reduces oxidative stress caused by hyperglycemia in pancreatic β cells and regulates glucose and serum-insulin levels.11 Oxidative stress in streptozotocin (STZ)-induced DM rats is inhibited by combination treatment of Ast with α-tocopherol.12 Recently, Ast treatment has been reported to be involved in stimulating mitochondrial biogenesis and significantly ameliorating insulin resistance through activation of the AMPK pathway in skeletal muscle.13 Ast inhibits glycation and glycated protein–induced cytotoxicity in human umbilical vein endothelial cells by preventing lipid/protein oxidation.14 Ast also prevents DM nephropathy by reduction of oxidative stress and renal cell damage.15–17 However, the biological basis of Ast in reducing oxidative stress induced by DM is not clear.
Network pharmacy is a new approach for drug discovery. It is a useful and comprehensive tool to elucidate complicated relationships among drugs, targets, and diseases through building a network.18 In this work, we combined network-pharmacy methods and RNA sequencing (RNA-seq) to analyze the interacting network among Ast, target proteins, and DM. We collected the targets of Ast and DM from public databases. Then, differential gene-expression patterns were analyzed using liver tissue of DM rats fed with Ast using RNA-seq. Intersecting targets were identified and docking analysis between Ast and target proteins performed. A compound target–pathway network was finally built to illustrate how Ast targets multiple signaling pathways to reduce oxidative stress caused by DM. The overall procedures for Ast target prediction are shown in Figure 1B.
Methods
Animals
The research was conducted in accordance with international principles for laboratory animal use and care and guidelines of the Animal Care Committee at the Faculty of Medicine, Tianjin University (China). Sprague Dawley rats aged 56 weekswithout specific pathogens and weighing 180–200 g were purchased from Vital River (Beijing, China) and maintained under conditions of controlled temperature (22°C±1°C), 40% humidity, and a daily 12-hour photoperiod.
Experimental Procedure
The rats were randomly divided into two groups: ten in the negative-control group and 40 rats in the DM group. The negative-control group was fed a normal diet, while the model group was fed a high-sugar, high-fat diet for 30 days. All rats were fed standard pellets and kept in standard cages. After 30 days, rats in the model group were given STZ (Sigma-Aldrich) 30 mg/kg intraperitoneally after fasting for 12 hours. Five days after the STZ injection, tail-vein blood glucose of rats injected with STZ was measured. The DM model was successfully established when the blood glucose of rats injected with STZ was >200 mg/dL.
The 40 DM rats were randomly divided into four groups, with ten in each group. Ast was dissolved in olive oil. The low-dose group was treated with 200 mg/kg Ast, the high-dose group 300 mg/kg Ast, and the metformin (Met) group 200 mg/kg Met. No special treatment was given to rats in the DM-model group. All 40 rats were fed the high-sugar, high-fat diet for 30 days. Rats given low-dose Ast (200 mg/kg) were finally used for H&E staining and RNA-seq analysis.
Quantitative Analysis of Triglyceride, Total Cholesterol, and Insulin Levels in Rats
About 50 μL tail-vein blood from overnight-fasted rats was collected in 1.5 mL Eppendorf tubes. Total cholesterol (TC), triglyceride (TG), and insulin levels were then tested using whole tail-vein blood with an ELISA kit produced by DG Biotech, China (DG20220D, DG20167D, and DG20757D) in accordance with the manufacturer’s instructions.
Quantitative Analysis of SOD, GSH, and MDA
SOD activity was measured kinetically as per Sun et al.19 Lipid peroxide levels, expressed in terms of MDA, were determined as per Buege and Aust.20 GSH was measured as per Ellman.21 For SOD and GSH detection, liver homogenates of aliquots of 0.5 mL were used. SOD activity was detected using a SOD-assay kit (WST1) produced by Nanjing Jiancheng, China (A001-3-2). Spectrophotometry (absorbance) at 450 nm was recorded. Hepatic GSH was estimated with a reduced glutathione (GSH)–assay kit produced by Nanjing Jiancheng, China (A006-1-1) at absorbance of 420 nm. For MDA testing, 0.2 mL liver homogenates of aliquots were used. Malondialdehyde (MDA) levels were tested with an MDA-assay kit produced by Nanjing Jiancheng (A003-1-2) and assayed at absorbance of 532 nm. All these experiments were conducted in accordance with manufacturers’ instruction. Triplicates of each sample were detected. Statistical significance was determined using SPSS with two-tailed unpaired Student’s t-tests.
Tissue Embedding and H&E Staining
Fresh islet tissue was placed in 10% formalin prepared in advance. It was dehydrated with alcohol, made transparent with xylene, and embedded in paraffin. Paraffin blocks were cut into 5–8 μm slices, then scalded with hot water. Slices were dried and stained with H&E. Next, samples were dehydrated with alcohol and sealed. Stained slides were imaged on an Aperio ScanScope CS2 at 20× magnification (Leica Biosystems).
Total RNA Extraction and Preparation of RNA-Seq Libraries
Total RNA was isolated with Trizol reagent (Invitrogen Life Technologies). Concentration, quality, and integrity were detected using spectrophotometry (NanoDrop; Thermo Fisher Scientific). Three micrograms of RNA (3 µg) was used as input material for RNA-sample preparations. Sequencing libraries were generated using a TruSeq RNA sample-preparation kit (Illumina, San Diego, CA, USA). Briefly, mRNA was purified using poly-T oligo–attached magnetic beads. Then, fragmentation was carried out with divalent cations under elevated temperature. First-strand cDNA was synthesized using random oligonucleotides and SuperScript II, followed by second-strand cDNA using DNA polymerase I and RNase H. After adenylation of the 3′ ends of the DNA fragments, Illumina PE adapter oligonucleotides were ligated to prepare for hybridization. To select cDNA fragments of the preferred 200 bp length, the library fragments were purified using the AMPure XP system (Beckman Coulter, Beverly, CA, USA). DNA with ligated adaptor molecules on both ends were enriched with Illumina PCR primer cocktail in a 15-cycle PCR process.
Real-Time qPCR Analysis
Total hepatic RNA from frozen liver was extracted using an RNeasy Mini kit (74104; Qiagen, Hilden, Germany) and treated with DNase I (M0303L; NEB, Ipswich, MA, USA). cDNA was reverse-transcribed with oligo-dT primers with a transcriptor first-strand cDNA-synthesis kit (04896866001; Roche, Basel, Switzerland). A real-time qPCR assay was performed using a FastStart Essential DNA Green Master kit (6402712001; Roche) in a LightCycler 480 II system (Roche). Sequences of primers and cycling conditions for tubulin were forward 5ʹ-TACCCTCGCATCCACTTCCCT-3ʹ and reverse 5ʹ-CGCTTGGTCTTGATGGTGGCA-3ʹ. Each sampling point was run in triplicate.22 Threshold cycle (CT) values of positive amplicons were analyzed using the ΔΔCT method in order to acquire a relative fold increase calibrated to the housekeeping gene and normalized to the control.Statistically significance was determined using SPSS with two-tailed unpaired Student’s t-tests.
Prediction of Ast-Related Targets
Ast-related targets were predicted based on similarity to known drug–target interactions via the BATMAN database (http://bionet.ncpsb.org/batman-tcm/index.php/home), Comparative Toxicogenomics Database (CTD; http://ctdbase.org), and STITCH (http://stitch.embl.de) database. In BATMAN, the prediction was considered a binary-classification issue. Predicted candidate targets (including known targets) with scores not <20 for each ingredient were presented and used for further bioinformatic analyses. In the CTD, hierarchical interaction–type vocabulary is used to describe the common physical, regulatory, and biochemical interactions between chemicals and genes. Confidence scores supported by STITCH served as a reference to mark high-confidence interactions between compounds and protein modules.23
Prediction of DM-Related Target Analysis with GeneCards
DM-related targets were predicted on GeneCards, which is a searchable and integrative database providing user-friendly and comprehensive information on all annotated and predicted human genes (https://www.genecards.org).
GO, KEGG Analysis, and Network Construction
GO analysis was performed on STRING, a database of known and predicted protein–protein interactions (https://string-db.org/cgi/info.pl). KEGG analysis for key Ast targets was collected on DAVID 6.7 (https://david.ncifcrf.gov). For deeper understanding of the relationships among Ast, target proteins, and DM, networks were constructed and analyzed with Cytoscape 3.7.2.
Validation of Binding Capacity Between Ast and Key Targets by Molecular Docking
We used SwissDock (http://www.swissdock.ch) to validate binding capacity between Ast and the targets.24 This is a web service to predict molecular interactions that may occur between a target protein and a small molecule.24 Crystal-structure information of the key targets was obtained from PDB (http://www.rcsb.org). Docking results between Ast and key targets were analyzed on SwissDock to validate binding properties. The closer the binding of the target protein and Ast is, the more energy they release and the lower the energy of ΔG. Therefore, we chose the lowest ΔG value for demonstrating binding patterns between Ast and target-protein chains.
Results
Prediction of Ast Targets
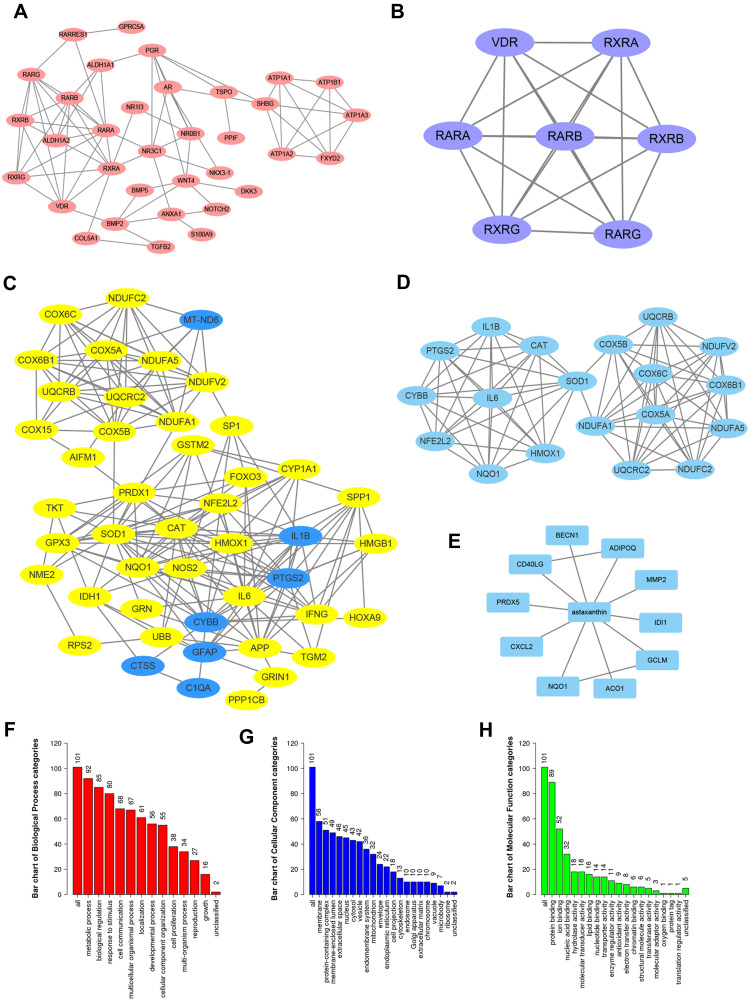
Potential Ast targets were predicted with three different approaches. Firstly, we collected 43 potential protein targets of Ast using BATMAN, from which 34 genes showed protein–protein interaction (PPI) networks on Cytoscape (Figure 2A). The key proteins from the 34 targets were analyzed using the cluster function. These were VDR, RXRA, RARA, RARB, RXRB, RXRG, and RARG (Figure 2B). On the CTD, we found 68 potential targets of Ast, of which 47 targets were on PPI networks and 19 clustered (Figure 2C, and D). Eleven targets were collected using the STITCH database (Figure 2E). In total, 21 repeated genes were removed, while the remaining 101 targets were subjected to GO analysis. Significantly enriched GO terms with adjusted p<0.05 are shown in the figures. We found that the metabolic process was the most enriched, with 92 targets on biological process analysis (Figure 2F). In cellular component categories, 58 targets were involved in membranes (Figure 2G), which is consistent with the special structure features and function of Ast. In the molecular function categories, targets were mostly enriched in protein binding (Figure 2H).
Figure 2.
Ast-target networks. (A) PPI network of potential Ast-target analysis predicted with BATMAN. (B) Key proteins from BATMAN-predicted targets analyzed by the cluster function of Cytoscape software. (C) PPI network of potential Ast-target analysis predicted with CTD database. (D) Key proteins from CTD predicted targets analyzed by the cluster function of Cytoscape software. (E) PPI network of potential Ast-target analysis predicted with STRING. (F–H) GO enrichment analysis of combined Ast targeted genes from BATMAN and CTD using STRING: number of genes enriched in biological process (F), cellular component (G), and molecular function (H).
Potential Targets of DM Analysis Through GeneCards
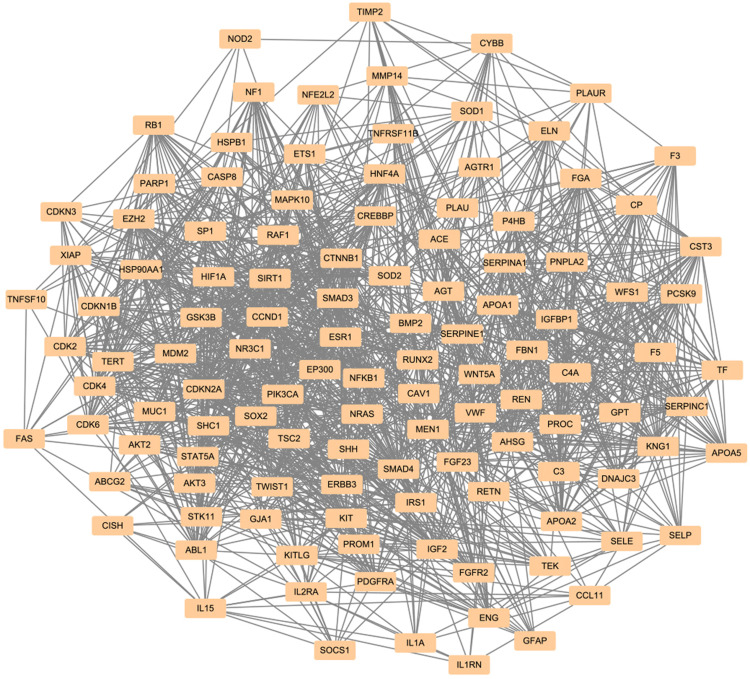
In order to find whether the Ast targets were involved in DM-related genes, GeneCards was used to predict potential targets of the disease. The term “diabetes mellitus” was used, and 13,784 targets were collected. In sum, 118 clustered genes out of 13,784 targets were found using Cytoscape 3.7.2 (Figure 3).
Figure 3.
Disease-target network of DM. The dark-yellow nodes represent DM-related targets.
Ast Treatment Reduced TC and TG Levels and Induced Insulin Secretion in DM Rats
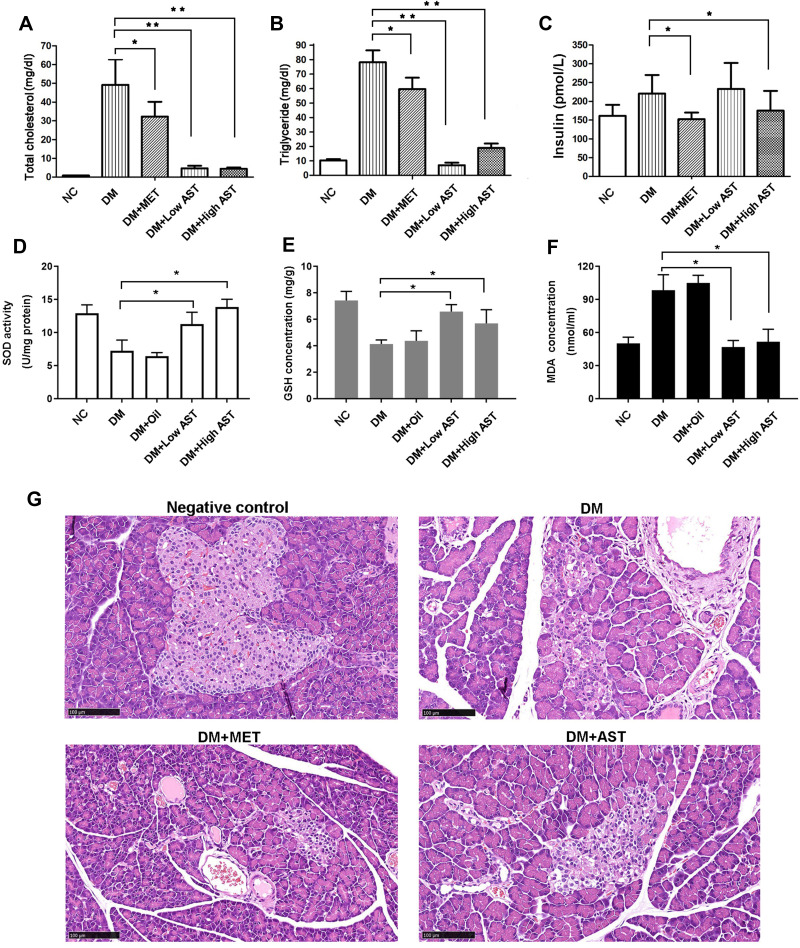
To confirm the effects of Ast in regulating TC, TG, and insulin levels, we built the DM rat model with STZ. The effects of Ast were examined in the negative-control, DM-model, DM with Met, and DM groups with low-dose (200 mg/kg) and high-dose (300 mg/kg) Ast. There was no significant difference in body weight among Ast-treated DM rats, the DM group, and the Met-treated group (Supplemental Figure 1). TC and TG levels in both low-dose (200 mg/kg) and high-dose (300 mg/kg) Ast-treated groups decreased significantly compared with DM rats (Figure 4A and B). High-dose Ast showed more effectiveness on insulin secretion than low-dose Ast (Figure 4C). These results confirmed that Ast treatment reduced TC and TG levels and induced insulin secretion in DM rats.
Figure 4.
Ast functions in regulating lipid metabolism and antioxidation activity in DM rats. Tail-vein blood (~50 μL) from overnight-fasted or ad libitum–fed rats was collected. Changes in total cholesterol (A), triglycerides (B), and insulin (C) in rats treated with or without Ast or Met were detected using tail-vein blood. SOD activity (D), GSH concentration (E), and MDA concentration (F) were examined using liver tissue from rats treated with or without Ast or olive oil. In all tests, values are means of ten rats. Statistically significant differences were determined using SPSS with two-tailed unpaired Student’s t-tests (*p<0.05; **p<0.01). (G) Representative images of H&E staining of fresh pancreas. Scale bar 100 μm.
Ast Treatment Reduced MDA Levels and Induced SOD and GSH Activity in DM Rats
SOD is an antioxidant enzyme that through catalysis produces hydrogen peroxide and oxygen.25 GSH is a small-molecule scavenger that can remove O2–, H2O2, and LOOH. MDA is produced from oxygen free radicals attacking polyunsaturated fatty acids in the membrane, causing lipid peroxidation. Quantitative analysis of SOD, GSH, and MDA was done. The results are shown in Figure 4D–F. We found that SOD activity in DM rats was only ~50%–60% of the high- or low-dose Ast rats (Figure 4D). The level of GSH was also reduced in DM rats, while Ast treatment significantly increased it (Figure 4E). MDA levels was highly raised in DM rats, though reduced in Ast rats (Figure 4F). All significant differences were versus DM-model data. Taken together, all these results indicate that oxidation levels in DM rats decreased significantly and antioxidant status improved with Ast supplementation.
Ast Treatment Helped Alleviate Islet-Cell Injury in DM Rats
The effects of Ast in regulating islet-cell status in DM rats were detected withH&E staining. Results showed that STZ caused extensive damage in islet cells of DM rats, including characteristic lipid vacuolization (Figure 4G). After Met treatment, cells in islets showed less vacuolation than in DM rats. Meanwhile, following low-dose Ast treatment, islet cells became less vacuolar than both DM and Met-treated rats (Figure 4G). These data suggested that Ast treatment helped alleviate islet-cell vacuolation in DM rats.
RNA-Seq Analysis of Ast- and Met-Treated DM Rats
For further detection of genes related to DM rats receiving Ast treatment, RNA-seq analysis was conducted. With an Illumina HiSeq sequencer, after filtering low-quality reads and removing reads aligned to either rRNA or tRNA, 503,515,926 clean reads from liver tissue of negative-control rats (128,184,131 reads), DM-model rats (128,407,964 reads), and DM rats given Ast (116,628,228 reads) or Met (130,295,600 reads) were selected for further analysis (Supplemental Table S1). The raw data of Illumina reads are shown in the NCBI Sequence Read Archive browser (http://ncbi.nlm.nih.gov/sra, accession PRJNA644021). There was ~95.96% of reads for each sample mapped to the Genome Rattus_norvegicus.Rnor_6.0.dna.toplevel, fa. About 90.57% were aligned with unique genes without ambiguity. Gross comparisons against untreated rats (negative controls) were shown in Figure 5A. All transcripts that were differentially expressed in DM rats compared to negative-control rats are shown in Supplemental Table S2.
Figure 5.
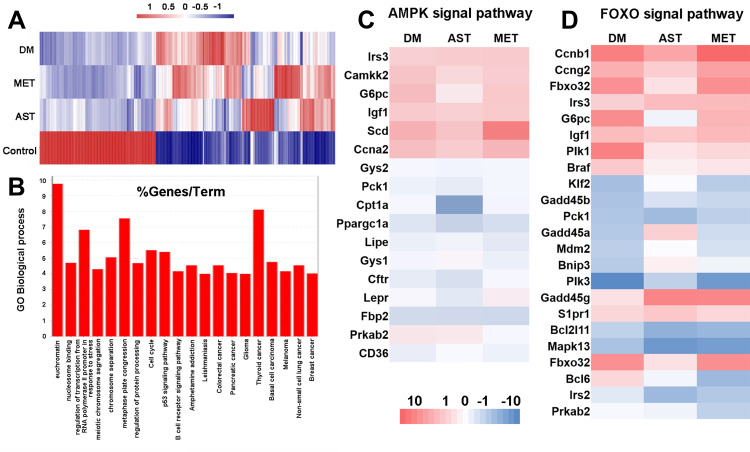
RNA-seq analyses of differentially expressed transcripts in negative-control, Ast-treated, Met-treated, and positive-control DM rats. (A) Heat maps for cluster display of all differentially expressed transcripts. Red, upregulated genes; blue, downregulated genes. All comparisons were made relative to gene-expression levels of the negative-control rats. (B) GO biological process analysis of all the log2FC ≥2 genes in the Ast vs DM comparison group using the STRING database. (C, D) Heat maps for cluster display of AMPK-signaling pathway (C) and FOXO-signaling pathway.
The transcriptome of Ast-treated DM rats was then compared to the DM-model group. In the Ast-treated group, expression was induced for 601 genes and repressed for 228 genes (Figure 5A, Supplemental Table S3). With KEGG analysis, we found expression of nine genes involved in the endocytosis pathway was induced (Table 1). Seven genes involved in the cell-cycle pathway were induced (Table 1). Also, we noticed that the SRC proto-oncogene involved in cell adhesion was induced with a 1.53-fold change. We then analyzed repressed genes in Ast-treated rats compared to the DM group. Six genes involved in the TNF-signaling pathway were downregulated (Table 2). Eight genes involved in the MAPK-signaling pathway were downregulated, and fold changes are shown in Table 2. We also conducted GO biological process analysis for all the log2FC ≥2 genes (139 in total) involved in the Ast vs DM comparison (Figure 5B). The top three enriched processes were for euchromatin, thyroid cancer, and metaphase-plate congression. Taken together, these results provide showed that Ast had a comprehensive effect in regulating extracellular to intracellular signal-transduction effects in DM rats.
Table 1.
Upregulated genes involved in endocytosis and cell-cycle pathways in Ast-treated DM Rats compared to DM rats
| Pathway | Gene | Description | Log2FC |
|---|---|---|---|
| Ast vs DM | |||
| Endocytosis | CBL | Cbl proto-oncogene | 1.91 |
| RT1-M2 | RT1 class Ib, locus M2 | 1.81 | |
| ARRB1 | Arrestin β1 | 1.71 | |
| GRK3 | G protein–coupled receptor kinase 3 | 1.59 | |
| DAB2 | DAB2 clathrin adaptor protein | 1.44 | |
| ASAP2 | ArfGAP with SH3 domain, ankyrin repeat, and PH domain 2 | 1.43 | |
| MVB12B | Multivesicular body subunit 12B | 1.34 | |
| CSF1R | Colony stimulating factor 1 receptor | 1.21 | |
| ARAP1 | ArfGAP with RhoGAP domain, ankyrin repeat, and PH domain 1 | 1.18 | |
| Cell cycle | ESPL1 | Extra spindle pole bodies like 1 separase | 3.91 |
| TTK | Ttk protein kinase | 3.30 | |
| CDK6 | Cyclin-dependent kinase 6 | 2.25 | |
| MCM5 | Minichromosome maintenance complex component 5 | 2.09 | |
| MCM6 | Minichromosome maintenance complex component 6 | 2.07 | |
| MCM3 | Minichromosome maintenance complex component 3 | 1.97 | |
| PRKDC | Protein kinase, DNA-activated, catalytic polypeptide | 1.15 |
Table 2.
Downregulated genes involved in MAPK- and TNF-signaling pathways in Ast-treated DM rats compared to DM rats
| Pathway | Gene | Description | Log2FC |
|---|---|---|---|
| Ast vs DM | |||
| MAPK signaling | HSPA1B | Heat-shock protein family A (Hsp70) 1B | −3.54 |
| NTRK1 | Neurotrophic receptor tyrosine kinase 1 | −3.14 | |
| FOS | FBJ osteosarcoma oncogene | −2.16 | |
| GADD45G | Growth arrest and DNA damage–inducible γ | −2.09 | |
| DUSP8 | Dual-specificity phosphatase 8 | −1.72 | |
| DUSP5 | Dual-specificity phosphatase 5 | −1.39 | |
| MAPK8IP1 | Mitogen-activated protein kinase 8 interacting protein 1 | −1.11 | |
| ANAPC11 | Anaphase-promoting complex subunit 11 | −1.02 | |
| TNF signaling | JUN | Jun proto-oncogene AP1 transcription factor subunit | −2.67 |
| SOCS3 | Suppressor of cytokine signaling 3 | −2.03 | |
| MAP2K6 | Mitogen-activated protein kinase kinase 6 | −1.92 | |
| JUNB | JunB proto-oncogene AP1 transcription factor subunit | −1.66 | |
| BCL3 | B-cell CLL/lymphoma 3 | −1.21 | |
| TNFRSF1B | TNF-receptor superfamily 1B | −1.08 |
The transcriptomes of Ast-treated DM rats were compared to Met-treated DM groups. Results showed that expression of 187 genes in Ast group was upregulated, while that of 153 genes was downregulated (Figure 5A, Supplemental Table S4). Cluster analysis showed that five genes involved in PPAR signaling were induced. Three genes from the cAMP-signaling pathway were also upregulated. In the AMPK-signaling pathway, four genes were induced. In contrast, genes that belonged to the steroid-biosynthesis and MAPK-signaling pathways were downregulated. For the steroid-biosynthesis pathway, eight genes were repressed. Furthermore, there were eight genes downregulated in the MAPK-signaling pathway (Table 3). Taken together, these results suggested that Ast had a more significant effect in promoting transmembrane conductance and calcium homeostasis, while Met produced greater inhibition of steroid biosynthesis and inflammation-related genes.
Table 3.
Cluster analysis of differentially expressed genes in Ast-treated DM rats compared to Met-treated groups
| Pathway | Name | Description | Log2FC |
|---|---|---|---|
| Ast vs Met | |||
| PPAR signaling | FABP2 | Fatty acid–binding protein 2 | 1.39 |
| GK | Glycerol kinase | 1.38 | |
| PLTP | Phospholipid-transfer protein | 1.10 | |
| APOA1 | Apolipoprotein A1 | 1.13 | |
| cAMP signaling | CACNA1C | Calcium voltage-gated channel subunit α1C | 2.11 |
| PLD1 | Phospholipase D1 | 1.44 | |
| ATP2B2 | ATPase plasma membrane Ca2+ transporting 2 | 1.43 | |
| AMPK signaling | G6PC | Glucose-6-phosphatase catalytic subunit | 1.83 |
| PFKFB1 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 | 1.31 | |
| LEPR | Leptin receptor | 1.24 | |
| CFTR | Cystic fibrosis transmembrane-conductance regulator | 1.12 | |
| Steroid biosynthesis | SQLE | Squalene epoxidase | −2.20 |
| MSMO1 | Methylsterol monooxygenase 1 | −2.07 | |
| CYP51 | Cytochrome P450, family 51 | −2.05 | |
| HSD17B7 | 17β-hydroxysteroid dehydrogenase 7 | −1.94 | |
| LSS | Lanosterol synthase (2,3-oxidosqualene-lanosterolcyclase) | −1.76 | |
| TM7SF2 | Transmembrane 7 superfamily member 2 | −1.30 | |
| EBP | Emopamil-binding protein (sterol isomerase) | −1.28 | |
| SC5D | Sterol-C5-desaturase | −1.10 | |
| MAPK signaling | NTRK1 | Neurotrophic receptor tyrosine kinase 1 | −2.75 |
| FOS | FBJ osteosarcoma oncogene | −2.39 | |
| HSPA1B | Heat-shock protein family A (Hsp70) 1B | −2.38 | |
| JUN | Jun proto-oncogene, AP1 transcription-factor subunit | −1.75 | |
| GADD45A | Growth arrest and DNA damage–inducible 45α | −1.73 | |
| MAP2K6 | Mitogen-activated protein kinase kinase 6 | −1.36 | |
| SRF | Serum response factor | −1.25 | |
| DUSP8 | Dual-specificity phosphatase 8 | −1.16 |
We also investigated differentially expressed genes involved in the AMPK- and FOXO-signaling pathways with heat-map analysis (Figure 5C and D). We found that expression of SCD, involved in the AMPK-signaling pathway, which contributes to the biosynthesis of membrane phospholipids, cholesterol esters, and TGs, was downregulated to 1.8, while this was 3.2 in DM rats.26 Expression of IRS3, involved in both the AMPK- and FOXO-signaling pathways and upstream of AKT, was much higher in Ast-treated DM rats than DM rats. We also observed that CD36, which functions in mitochondrial β-oxidation and free fatty–acid transport, was expressed at a much higher level in Ast-treated DM rats than DM control rats. Taken together, these results indicated that Ast had a comprehensive effect on regulating AMPK and FOXO signaling.
Real-Time qPCR Analysis to Confirm RNA-Seq Analysis
To verify mRNA-expression levels obtained on RNA-seq analysis, real-time qPCR was performed using RNA extracted from the livers of DM-model rats and Ast-treated DM rats. Sequences of the primers used for qPCR are shown in Supplemental Table S5. Twelve related genes were analyzed for their expression: UNC5C, SRGAP1, SEMA5A, SRC, PLXNA3, SEMA4D, SLIT3, SEMA3C, SMO, SSH1, SEMA6A, and SSH2. As expected, all these genes were enhanced by Ast treatment, which was consistent with the RNA-seq results (Supplemental Figure 2).
Intersecting Targets Identified and Molecular Docking
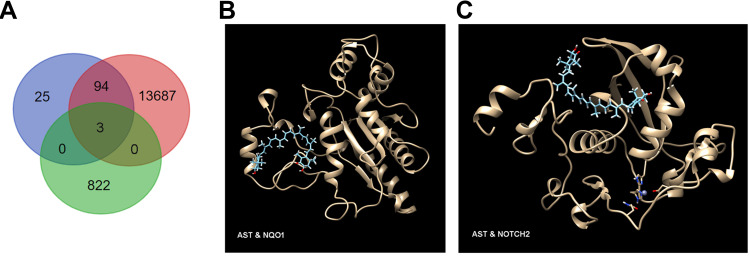
Three intersecting targets were identified from data obtained from Ast targets, DM targets, and RNA-seq analysis using Bioinformatics & Evolutionary Genomics (http://bioinformatics.psb.ugent.be/webtools/Venn) (Figure 6A): Col5A1, which functions in binding to insulin and has recently been identified as a new lipid species–associated loci associated with cardiovascular diseasesrisk,27 Nqo1 a phase II enzyme that serves in the maintenance of cellular defense against oxidative stress and redox homeostasis,28 and Notch2, which plays a crucial role in marginal-zone B-cell development.
Figure 6.
Identification of three intersecting targets. Targets were identified with Venn analysis (A). Molecular docking of NQO1 (B) and NOTCH2 (C) with Ast. The golden structure represents the target protein, with Ast structure in cyan.
To further certify binding capacity between Ast and the targets, molecular docking with SwissDock was performed. As Col5A1 lacks the crystal-structure information for docking, we performed docking analysis between Ast and Nqo1 and Ast and Notch2. (Figure 6B and C). The docking pattern with the lowest ΔG value was chosen, as it showed the most closely binding capability.24 For Nqo1 binding, the lowest ΔG was −9.07 kcal/mol, while the lowest ΔG was −8.52 kcal/mol for Notch2 binding, indicating the specific binding possibility between Ast and key targets for DM treatment.
Pathway Analysis and Network Construction
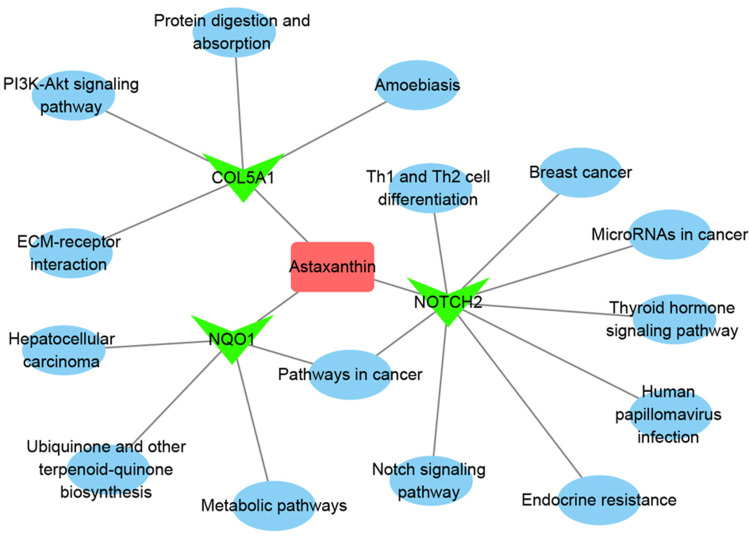
For understanding the regulated networks, related pathways were then collected with DAVID. Results showed that Nqo1 was involved in ubiquinone and other terpenoid–quinone biosynthesis, metabolic pathways, pathways in cancer, and hepatocellular carcinoma pathways. Col5A1 takes part in protein digestion and absorption, ECM-receptor interaction, the PI3K–Akt signaling pathway, and amoebiasis pathways. Notch2 functions in endocrine resistance, the Notch-signaling pathway, TH1- and TH2-cell differentiation, the thyroid hormone–signaling pathway, human papillomavirus infection, cancer, and microRNAs in cancer and breast cancer pathways. Based on pathway analysis, the entire Ast targets–pathway network was constructed with Cytoscape 3.7.2. As shown in Figure 7, the interaction network had 20 nodes and 19 edges. The red, green, and blue colors show Ast, target proteins, and pathways, respectively. These results suggested that Ast targets a variety of pathways to form a network of systemic pharmacological effects to reduce the oxidative stress caused by DM.
Figure 7.
The Ast target–pathway network. The pink rectangle represents Ast, magenta triangle targets, and blue circles related pathways.
Discussion
DM causes both psychological and physical distress to patients and their families.5 Hyperglycemia, insulin resistance, and excess fatty acids in DM patients have been reported to increase oxidative stress and glycation end products, leading to vascular inflammation, vasoconstriction, thrombosis, and atherogenesis symptoms.29 Ast is a safe and natural product with no side effects when consumed with food.30–32 Ast works well in reducing oxidative stress in DM rats.8 It is thus considered to be a good candidate for clinical studies. In this work, we identified three Ast protein targets — DM, Col5A1, Nqo1, and Notch2 — by RNA-seq and network-pharmacy analysis. The three target proteins were involved in metabolic pathways, protein digestion and absorption, endocrine resistance, the Notch-signaling pathway and other physiological processes. A systematic overview was built for understanding the possible regulatory mechanisms of Ast in reducing oxidative stress induced by DM.
Based on previous reports, overexpression of SCDin the AMPK-signaling pathway in the normal liver of mouse and rat strains is genetically susceptible to hepatocarcinogenesis, as it causes abnormal biosynthesis of membrane phospholipids, cholesterol esters, and TGs.33 With RNA-seq analysis, we found SCDexpression in Ast-treated DM rats was much lower than in DM control rats (Figure 5C). This result could explain why Ast functions in reducing TC and TG levels in the DM rats efficiently, in that Ast inhibits SCD expression to balance TC and TG levels in DM patients. Furthermore, expression of the IRS3gene, which is upstream of AKT in the insulin-signaling pathway, was much higher in Ast-treated DM rats than DM rats (Figure 5C and D). This is consistent with a previous study showing that Ast treatment significantly enhanced insulin-stimulated phosphorylation of the Akt protein in high fat diet–fed mice.12 Accordingly, Ast functions in promoting both transcription of IRS3and phosphorylation of the downstream protein Akt, which together contributed to the improved insulin sensitivity in the Ast-treated groups. We also observed that the CD36 gene, which functions in mitochondrial β-oxidation and free fatty–acid transport in the AMPK-signaling pathway, was expressed at much higher levels in the Ast-treated DM rats than the DM control rats (Figure 5C). It may help in mitochondrial biogenesis and muscle remodeling, which is consistent with Nishida et al.12
Col5A1 is associated with insulin resistance and is overexpressed in pathways related to integrin cell-surface interactions and insulin signaling in obese women.34 Expression of Col5A1 mRNA in Ast-treated rats was induced 1.21-fold compared with DM rats (Supplemental Table S3). By targeting Ast, the high mRNA level of Col5A1 may play a crucial role in promoting insulin-release balance and relieving insulin resistance. As such, Col5A1 may have a positive effect on regulating insulin resistance and insulin signaling. Furthermore, with the help of Col5A1, Ast might easily colocalize with integrin and react with free radicals at the inner cell surface and also cross the cell membrane to protect it from oxidative stress. Whether there was direct interaction between Ast and Col5A1 needs to be checked in future.
Nqo1 belongs to the family of phase II enzymes, which play essential roles in the maintenance of cellular defense against oxidative stress and redox homeostasis.28 It is regulated byNrf2.35,36 Expression of Nqo1 is induced by oxidative stress.37 Nqo1 is involved in cellular antioxidant defense and plays a pivotal role in the detoxification of quinine. It protects cells against oxidative stress.38 In type 2 DM rats, repeated low-dose radiation reduces testicular cell apoptosis and oxidative stress by inducing Nrf2 and Nqo1.39 According to the RNA-seq analysis, the expression level of Nqo1 was 1.06-fold (log2FC) in Ast-treated DM rats of the DM-model group (Supplemental Table S3). Combined with the prediction of Ast binding to Nqo1 (Figure 6), all this gives us a clue that Ast may directly bind and induce the expression of Nqo1, which leads to low-dose radiation and reduces testicular cell apoptosis and oxidative stress.
The marginal zone of NOD mice is enlarged and has been considered to contribute to autoimmune DM.40–42 Notch2 plays an essential role in marginal-zone B-cell development. On the surface of B cells, Notch2 interacts with its ligands at low affinity in the marginal sinus.37,43–48 Ast treatment induced the expression of Notch2 mRNA to 1.21 times that of the DM-model group. This induction pattern of Ast binding Notch2 might function in maintaining the normal size of marginal-zone B cells and inhibit autoimmune DM to some extent.
Organisms are composed of complex molecular and protein-regulation networks. By interacting with a variety of proteins, compound molecules can play multiple roles in related signal-transduction pathways, thus regulating the process of diseases and complications. Ast is predicted to target a variety of targets and pathways to form a network of systemic pharmacological effects to reduce oxidative stress in DM patients. Although we have provided some clues that Ast might interact with particular targets, more research needs to be carried out to confirm the wide and reliable clinical application of Ast in DM research.
Conclusion
Three Ast protein targets — Col5A1, Nqo1, and Notch2 — were identified. Molecular docking showed a possible interaction of Ast with Nqo1 or Notch2. Ast targets–pathway networks showed that the three target proteins were related to metabolic pathways, protein digestion and absorption, endocrine resistance, the Notch-signaling pathway and other physiological processes. Therefore, Ast is predicted to target a variety of pathways to form a network of systemic pharmacological effects to reduce oxidative stress in DM patients.
Acknowledgments
The authors wish to express their sincere gratitude to the laboratory partners who patiently accompanied the entire experimental process.
Funding Statement
This work was supported by the Modern Aquaecology and Health Aquaculture Innovation Team of Tianjin (grant TD13-5089) and the Scientific Program of Tianjin City (16ZXZYNC00120 and 14ZCZDSY00053).
Abbreviations
Ast, astaxanthin; DM, diabetes mellitus; CTD, Comparative Toxicogenomics Database ROS, reactive oxygen species; TC, total cholesterol; TG, triglyceride; STZ, streptozotocin; GSH, glutathione; H&E, hematoxylin–eosin; NOD, no-obesity diabetes.
Ethics Approval
This research was conducted in accordance with international principles for laboratory animal use and care and the guidelines of the Animal Care Committee at the Faculty of Medicine, Tianjin University (China), and was approved by the Animal Care Committee of the Faculty of Medicine, Tianjin University.
Disclosure
The authors declare that they have no conflicts of interest.
References
- 1.Ambati RR, Phang SM, Ravi S, Aswathanarayana RG. Astaxanthin: sources, extraction, stability, biological activities and its commercial applications–a review. Mar Drugs. 2014;12:128–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Higuera-Ciapara I, Felix-Valenzuela L, Goycoolea FM. Astaxanthin: a review of its chemistry and applications. Crit Rev Food Sci Nutr. 2006;46:185–196. doi: 10.1080/10408690590957188 [DOI] [PubMed] [Google Scholar]
- 3.Guerin M, Huntley ME, Olaizola M. Haematococcus astaxanthin: applications for human health and nutrition. Trends Biotechnol. 2003;21:210–216. doi: 10.1016/S0167-7799(03)00078-7 [DOI] [PubMed] [Google Scholar]
- 4.Yuan JP, Peng J, Yin K, Wang JH. Potential health‐promoting effects of astaxanthin: a high‐value carotenoid mostly from microalgae. Mol Nutr Food Res. 2011;55:150–165. [DOI] [PubMed] [Google Scholar]
- 5.Chatterjee S, Khunti K, Davies MJ. Type 2 diabetes. Lancet. 2017;389:2239–2251. [DOI] [PubMed] [Google Scholar]
- 6.Kitade H, Chen G, Ni Y, Ota T. Nonalcoholic fatty liver disease and insulin resistance: new insights and potential new treatments. Nutrients. 2017;9:387. doi: 10.3390/nu9040387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan T, Zhao Y, Zhang X, Lin X. Astaxanthin inhibits acetaldehyde-induced cytotoxicity in SH-SY5Y cells by modulating Akt/CREB and p38MAPK/ERK signaling pathways. Mar Drugs. 2016;14:56. doi: 10.3390/md14030056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sila A, Kamoun Z, Ghlissi Z, et al. Ability of natural astaxanthin from shrimp by-products to attenuate liver oxidative stress in diabetic rats. Pharmacol Rep. 2015;67:310–316. doi: 10.1016/j.pharep.2014.09.012 [DOI] [PubMed] [Google Scholar]
- 9.Dryden M. Reactive oxygen species: a novel antimicrobial. Int J Antimicrob Agents. 2018;51:299–303. doi: 10.1016/j.ijantimicag.2017.08.029 [DOI] [PubMed] [Google Scholar]
- 10.Makni M, Fetoui H, Gargouri NK, Garoui EM, Zeghal N. Antidiabetic effect of flax and pumpkin seed mixture powder: effect on hyperlipidemia and antioxidant status in alloxan diabetic rats. J Diabetes Complications. 2011;25:339–345. doi: 10.1016/j.jdiacomp.2010.09.001 [DOI] [PubMed] [Google Scholar]
- 11.Uchiyama K, Naito Y, Hasegawa G, Nakamura N, Takahashi J, Yoshikawa T. Astaxanthin protects β-cells against glucose toxicity in diabetic db/ db mice. Redox Rep. 2002;7:290–293. doi: 10.1179/135100002125000811 [DOI] [PubMed] [Google Scholar]
- 12.Nishida Y, Nawaz A, Kado T, et al. Astaxanthin stimulates mitochondrial biogenesis in insulin resistant muscle via activation of AMPK pathway. J Cachexia Sarcopenia Muscle. 2020;11:241–258. doi: 10.1002/jcsm.12530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakano M, Onodera A, Saito E, et al. Effect of astaxanthin in combination with α-tocopherol or ascorbic acid against oxidative damage in diabetic ODS rats. J Nutr Sci Vitaminol (Tokyo). 2008;54:329–334. doi: 10.3177/jnsv.54.329 [DOI] [PubMed] [Google Scholar]
- 14.Nishigaki I, Rajendran P, Venugopal R, Ekambaram G, Sakthisekaran D, Nishigaki Y. Cytoprotective role of astaxanthin against glycated protein/iron chelate‐induced toxicity in human umbilical vein endothelial cells. Phytother Res. 2010;24:54–59. doi: 10.1002/ptr.2867 [DOI] [PubMed] [Google Scholar]
- 15.Naito Y, Uchiyama K, Aoi W, et al. Prevention of diabetic nephropathy by treatment with astaxanthin in diabetic db/ db mice. Biofactors. 2004;20:49–59. doi: 10.1002/biof.5520200105 [DOI] [PubMed] [Google Scholar]
- 16.Kim YJ, Kim YA, Yokozawa T. Protection against oxidative stress, inflammation, and apoptosis of high-glucose-exposed proximal tubular epithelial cells by astaxanthin. J Agric Food Chem. 2009;57:8793–8797. [DOI] [PubMed] [Google Scholar]
- 17.Manabe E, Handa O, Naito Y, et al. Astaxanthin protects mesangial cells from hyperglycemia‐induced oxidative signaling. J Cell Biochem. 2008;103:1925–1937. doi: 10.1002/jcb.21583 [DOI] [PubMed] [Google Scholar]
- 18.Zheng J, Wu M, Wang H, et al. Network pharmacology to unveil the biological basis of health-strengthening herbal medicine in cancer treatment. Cancers. 2018;10(11):461. doi: 10.3390/cancers10110461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun Y, Oberley LW, Li Y. A simple method for clinical assay of superoxide dismutase. Clin Chem. 1988;34:497–500. doi: 10.1093/clinchem/34.3.497 [DOI] [PubMed] [Google Scholar]
- 20.Buege J, Aust S. Microsomal lipid peroxidation methods enzymol 52: 302–310, Find this article online 1978. [DOI] [PubMed]
- 21.Ellman GL. Tissue sulfhydryl groups. Arch Biochem Biophys. 1959;82(1):70–77. doi: 10.1016/0003-9861(59)90090-6 [DOI] [PubMed] [Google Scholar]
- 22.Roy A, Volgin D, Baby S, Kubin L, Lahiri S. Activation of HIF-1 alpha mRNA by hypoxia in isolated rat carotid body. FASEB J. 2002;16(5):A817–A817. [DOI] [PubMed] [Google Scholar]
- 23.Kuhn M, Szklarczyk D, Franceschini A, Von Mering C, Jensen LJ, Bork P. STITCH 3: zooming in on protein–chemical interactions. Nucleic Acids Res. 2012;40:D876–D880. doi: 10.1093/nar/gkr1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grosdidier A, Zoete V, Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011;39:W270–W277. doi: 10.1093/nar/gkr366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson WT, Johnson LAK, Lukaski HC. Serum superoxide dismutase 3 (extracellular superoxide dismutase) activity is a sensitive indicator of Cu status in rats. J Nutr Biochem. 2005;16:682–692. doi: 10.1016/j.jnutbio.2005.03.009 [DOI] [PubMed] [Google Scholar]
- 26.Shanklin J, Whittle E, Fox BG. Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry. 1994;33:12787–12794. doi: 10.1021/bi00209a009 [DOI] [PubMed] [Google Scholar]
- 27.Tabassum R, Rämö JT, Ripatti P, et al. Genetic architecture of human plasma lipidome and its link to cardiovascular disease. Nat Commun. 2019;10:1–14. doi: 10.1038/s41467-019-11954-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hasanvand D, Amiri I, Soleimani Asl S, Saidijam M, Shabab N, Artimani T. Effects of CeO2 nanoparticles on the HO-1, NQO1, and GCLC expression in the testes of diabetic rats. Can J Physiol Pharmacol. 2018;96:963–969. doi: 10.1139/cjpp-2017-0784 [DOI] [PubMed] [Google Scholar]
- 29.Henning RJ. Type-2 diabetes mellitus and cardiovascular disease. Future Cardiol. 2018;14:491–509. doi: 10.2217/fca-2018-0045 [DOI] [PubMed] [Google Scholar]
- 30.Rao AR, Baskaran V, Sarada R, Ravishankar GA. In vivo bioavailability and antioxidant activity of carotenoids from microalgal biomass—A repeated dose study. Food Res Int. 2013;54:711–717. [Google Scholar]
- 31.Ranga Rao A, Raghunath Reddy RL, Baskaran V, Sarada R, Ravishankar GA. Characterization of microalgal carotenoids by mass spectrometry and their bioavailability and antioxidant properties elucidated in rat model. J Agric Food Chem. 2010;58:8553–8559. doi: 10.1021/jf101187k [DOI] [PubMed] [Google Scholar]
- 32.Stewart JS, Lignell Å, Pettersson A, Elfving E, Soni MG. Safety assessment of astaxanthin-rich microalgae biomass: acute and subchronic toxicity studies in rats. Food Chem Toxicol. 2008;46:3030–3036. doi: 10.1016/j.fct.2008.05.038 [DOI] [PubMed] [Google Scholar]
- 33.Falvella FS, Pascale RM, Gariboldi M, et al. Stearoyl-CoA desaturase 1 (Scd1) gene overexpression is associated with genetic predisposition to hepatocarcinogenesis in mice and rats. Carcinogenesis. 2002;23:1933–1936. doi: 10.1093/carcin/23.11.1933 [DOI] [PubMed] [Google Scholar]
- 34.Arner P, Sahlqvist A-S, Sinha I, et al. The epigenetic signature of systemic insulin resistance in obese women. Diabetologia. 2016;59:2393–2405. doi: 10.1007/s00125-016-4074-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakamura BN, Lawson G, Chan JY, et al. Knockout of the transcription factor NRF2 disrupts spermatogenesis in an age-dependent manner. Free Radic Biol Med. 2010;49:1368–1379. doi: 10.1016/j.freeradbiomed.2010.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wajda A, Łapczuk J, Grabowska M, et al. Nuclear factor E2-related factor-2 (Nrf2) expression and regulation in male reproductive tract. Pharmacol Rep. 2016;68:101–108. doi: 10.1016/j.pharep.2015.07.005 [DOI] [PubMed] [Google Scholar]
- 37.Philbrook NA, Winn LM. Sub-chronic sulforaphane exposure in CD-1 pregnant mice enhances maternal NADPH quinone oxidoreductase 1 (NQO1) activity and mRNA expression of NQO1, glutathione S-transferase, and glutamate-cysteine ligase: potential implications for fetal protection against toxicant exposure. Reprod Toxicol. 2014;43:30–37. [DOI] [PubMed] [Google Scholar]
- 38.Dinkova-Kostova AT, Talalay P. NAD(P)H: quinone acceptor oxidoreductase 1 (NQO1), a multifunctional antioxidant enzyme and exceptionally versatile cytoprotector. Arch Biochem Biophys. 2010;501:116–123. doi: 10.1016/j.abb.2010.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Y, Kong C, Chen X, et al. Repetitive exposure to low-dose X-irradiation attenuates testicular apoptosis in type 2 diabetic rats, likely via Akt-mediated Nrf2 activation. Mol Cell Endocrinol. 2016;422:203–210. doi: 10.1016/j.mce.2015.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marino E, Batten M, Groom J, et al. Marginal-zone B-cells of nonobese diabetic mice expand with diabetes onset, invade the pancreatic lymph nodes, and present autoantigen to diabetogenic T-cells. Diabetes. 2008;57:395–404. doi: 10.2337/db07-0589 [DOI] [PubMed] [Google Scholar]
- 41.Stolp J, Mariño E, Batten M, et al. Intrinsic molecular factors cause aberrant expansion of the splenic marginal zone B cell population in nonobese diabetic mice. J Immunol. 2013;191:97–109. doi: 10.4049/jimmunol.1203252 [DOI] [PubMed] [Google Scholar]
- 42.Bashratyan R, Sheng H, Regn D, Rahman MJ, Dai YD. Insulinoma‐released exosomes activate autoreactive marginal zone‐like B cells that expand endogenously in prediabetic NOD mice. Eur J Immunol. 2013;43:2588–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saito T, Chiba S, Ichikawa M, et al. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity. 2003;18:675–685. doi: 10.1016/S1074-7613(03)00111-0 [DOI] [PubMed] [Google Scholar]
- 44.Tanigaki K, Han H, Yamamoto N, et al. Notch–RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nat Immunol. 2002;3:443–450. [DOI] [PubMed] [Google Scholar]
- 45.Hozumi K, Negishi N, Suzuki D, et al. Delta-like 1 is necessary for the generation of marginal zone B cells but not T cells in vivo. Nat Immunol. 2004;5:638–644. doi: 10.1038/ni1075 [DOI] [PubMed] [Google Scholar]
- 46.Hampel F, Ehrenberg S, Hojer C, et al. CD19-independent instruction of murine marginal zone B-cell development by constitutive Notch2 signaling. Blood. 2011;118:6321–6331. [DOI] [PubMed] [Google Scholar]
- 47.Witt CM, Won W-J, Hurez V, Klug CA. Notch2 haploinsufficiency results in diminished B1 B cells and a severe reduction in marginal zone B cells. J Immunol. 2003;171:2783–2788. doi: 10.4049/jimmunol.171.6.2783 [DOI] [PubMed] [Google Scholar]
- 48.Tan JB, Xu K, Cretegny K, et al. Lunatic and manic fringe cooperatively enhance marginal zone B cell precursor competition for delta-like 1 in splenic endothelial niches. Immunity. 2009;30:254–263. doi: 10.1016/j.immuni.2008.12.016 [DOI] [PubMed] [Google Scholar]