Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes novel coronavirus disease (COVID-19) outbreak in more than 200 countries around the world. The early diagnosis of infected patients is needed to discontinue this outbreak. The diagnosis of coronavirus infection from radiography images is the fastest method. In this paper, two different ensemble deep transfer learning models have been designed for COVID-19 diagnosis utilizing the chest X-rays. Both models have utilized pre-trained models for better performance. They are able to differentiate COVID-19, viral pneumonia, and bacterial pneumonia. Both models have been developed to improve the generalization capability of the classifier for binary and multi-class problems. The proposed models have been tested on two well-known datasets. Experimental results reveal that the proposed framework outperforms the existing techniques in terms of sensitivity, specificity, and accuracy.
Keywords: COVID-19, SARS-CoV-2, Transfer learning, Chest X-ray, Ensemble models
Introduction
The outbreak of novel coronavirus surprised the whole world. The disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is also known as COVID-19. According to the World Health Organization (WHO), more than fifteen million peoples are infected from this virus across 215 countries (Coronavirus Disease (COVID-19) 2020). 0.63 million deaths with 9.43 million recovered cases are reported globally by 23 July 2020. USA, Brazil, and India are severely affected with 4.1 million, 2.2 million, and 1.3 million active corona cases, respectively.
Due to the communicable nature of this virus and inappropriate treatment, the early detection of an infected person is required to discontinue the coronavirus spread (Oh et al. 2020). The main source for detecting the infected person is symptoms developed in the patients. The infected person may suffer from fever, cough, breathing problem, sore throat, diarrhoea, and headache (COVID-19 symptoms 2020). Loss of smell, tiredness, disappearing of taste, and aches may be found in some patients. However, the presence of COVID-19 symptoms may not be found in some infected persons (Loey et al. 2020). Due to this, the detection of an infected person is a very difficult task. The health facilities of many developed countries have been exhausted due to the rapid increase in corona affected persons. These countries are facing a shortage of corona testing kits and ventilators. Due to this, some countries announced the lockdown to break the chain of coronavirus and safe their population.
Besides the imposition of lockdown, screening of patient is required for isolation and treatment. Initially, real-time reverse transcription-polymerase chain reaction (RT-PCR) method is used for detecting the coronavirus infection in the patients (Corman et al. 2020). RT-PCR is performed on the clinical samples of patients and results are obtained within a few hours to two days. Due to the low sensitivity of RT-PCR and unavailability of kits, radiography imaging techniques can be utilized to detect COVID-19 in the patients (Xie et al. 2020). It has been observed from research articles that chest scans may be suitable for COVID-19 detection (Corman et al. 2020; Xie et al. 2020). Hence, the diagnosis and detection of coronavirus infection in COVID patients can be done through chest radiography techniques. The chest X-ray and chest CT scan are two well-known radiological techniques. Chest X-ray is preferred over the chest CT scan due to easy availability of X-ray machines in hospitals and low ionizing radiation exposure to patients (Yu et al. 2020).
The radiological expert is required to analyze chest X-ray for finding the COVID-19 imaging patterns. However, it is a time-consuming and error-prone task (Cellina et al. 2020). Therefore, the automatic analysis of chest x-ray is required. Recently, deep learning (DL) techniques are widely used in medical imaging for the diagnosis of different diseases (Shen et al. 2017). DL techniques automatically extract the features from the given image without any user intervention (Wang and Xia 2018). Due to this, these techniques are used in the classification of infections in chest X-ray images. Therefore, DL techniques are used for automatic analysis of chest X-ray for detecting the COVID-19 imaging patterns from patient’s images.
The main contribution of this paper is as follows:
-
(i)
An ensemble deep transfer learning models are designed for COVID-19 diagnosis using chest X-ray images.
-
(ii)
The proposed model not only helpful in diagnosing the COVID-19 infected patients but also able to differentiate the COVID-19 from diagnosing pneumonia (i.e., viral and bacterial).
-
(iii)
The proposed framework extracts the features from chest X-ray images using an ensemble deep learning network. Thereafter, the extracted features are applied to classifier for further classification.
-
(iv)
The performance of the proposed framework has been tested on two well-known datasets.
-
(v)
The proposed framework has been compared with the competitive models in terms of various performance metrics such as accuracy, f-measure, area under curve, precision and recall.
The remainder of this paper is structured as follows. The related work is described in Sect. 2. Section 3 presents the proposed approach for COVID-19 detection in infected persons. Experimental results and discussions are given in Sect. 4. The concluding remarks are drawn in Sect. 5.
Related work
Recently, deep learning techniques such as convolutional neural networks (CNN) and transfer learning are widely used for classification of COVID-19 from chest X-rays. Researchers have done a lot of work in the short span of time.
Basu and Mitra (2020) proposed a domain extension transfer learning for detecting the abnormality instigated by COVID-19 in chest X-rays. They used Gradient Class Activation Map for detecting features from X-ray images and validated over NIH chest X-ray dataset. The accuracy obtained from this model is 95.3%. Luz et al. (2020) studied the deep learning architectures for identification of COVID-19. COVIDx dataset is used to demonstrate the efficiency of the proposed model. Flat EfficientNet model achieved 93.9% accuracy with 96.8% sensitivity. Hemdan et al. (2020) developed a novel automatic framework namely COVIDX-Net for identification of coronavirus infection in the patients’ chest X-ray. COVIDX-Net utilized seven different deep learning architectures. COVIDX-Net is tested on only 50 chest X-ray images. Both VGG19 and DenseNet201 provided 90% accuracy on these images. Ozturk et al. (2020) presented an automatic COVID-19 detection model named as DarkCovidNet using chest X-ray. DarkCovidNet is trained on 125 COVID-19 chest images. The classification accuracies obtained from this model are 98.08% and 87.02% for binary and multi-class cases. The main drawback of this model is that the limited number of coronavirus infected X-ray images are used for validation.
Togaçar et al. (2020) used a fuzzy colour technique for converting the original chest X-ray dataset into a structured dataset. Thereafter, the structured dataset is converted into a stacked dataset using image stacking technique. MobileNetV2 and SqueezeNet are used for identification of COVID-19 patterns in the images. The classification accuracy obtained from MobileNetV2 and SqueezeNet is 98.25% and 97.81%, respectively. Tuncer et al. (2020) proposed a novel method for detection of COVID-19 pattern in chest X-ray images. Residual Exemplar Local Binary Pattern (ResExLBP) is used for feature extraction. After that, the important features are selected through iterative ReliefF. These selected features are applied to five well-known classifiers. This approach is tested on 321 chest X-ray images and achieved a 99% success rate for SVM classifier. Das et al. (2020a, b) presented an automatic detection of coronavirus infection from chest X-ray images using deep learning technique. The classification accuracy obtained from the proposed model is 97.40%. Wang and Wong (2020) developed a COVID-Net framework for detection of COVID-19. COVID-Net attained 92.4% accuracy in classifying normal, pneumonia, and COVID-19 classes. COVID-Net provides better performance than the VGG-19 and ResNet-50 architecture.
Mahmud et al. (2020) proposed a CovXNet for detecting COVID-19 patterns from the patient’s chest X-rays. The predications obtained from CovXNet are further optimized using the stacking algorithm. Their model is tested over 915 images. The classification accuracies obtained from CovXNet are 97.4% and 89.6% for binary and three-class cases, respectively. Apostolopoulos and Mpesiana (2020) presented an automatic detection of COVID-19 from X-ray images using five pre-trained deep learning architectures. The pre-trained architectures are VGG-19, MobileNet, Inception, Xception, and Inception_ResNet_V2. The accuracies obtained from VGG-19 are 98.75% and 93.48% for binary and multiclass cases, respectively.
Rahimzadeh and Attar (2020) presented the hybridization of Xception and ResNet50_V2 for identification of COVID-19 patterns in the chest X-ray images. Their model is tested on 6054 images and attained classification accuracy of 91.4%. Shelke et al. (2020) proposed a classification model to analyze and diagnosis of COVID-19 using chest X-ray images. The accuracy obtained from DenseNet-161 is 98.9%. However, they have tested on a very small dataset that consists of only twenty-two X-ray images. Narin et al. (2020) studied the three pre-trained deep learning architectures namely ResNet50, Inception_V3, and Inception-ResNet_V2 for detection of coronavirus infection in the chest X-ray images. The COVID-19 detection accuracy achieved from ResNet50 is 98%.
Abbas et al. (2020) proposed a DeTraC model for detection of COVID-19 from the chest X-ray images. They used a pre-trained CNN model for deep feature extraction. Thereafter, class decomposition is used to extract the local structure from the data distribution. Gradient descent method is used for classification. The classification results are further refined through a class composition layer of DeTraC. DeTraC achieved an accuracy of 95.12% over 105 chest X-ray images. Chouhan et al. (2020) proposed an ensemble approach for pre-trained deep learning architectures. They used AlexNet, DenseNet121, Inception_V3, ResNet18 and GoogleNet architectures for COVID-19 classification. Their ensemble model achieved the classification accuracy of 96.4%. Das et al. (2020a, 2020b) presented a truncated InceptionNet model for screening of COVID-19 from the chest X-ray images. The model is tested on three publicly available chest X-ray datasets.
Proposed ensemble deep transfer leanring model
In this section, the detail description of the proposed ensemble deep learning model is presented.
Motivation
The availability of large-scale well-annotated datasets especially in image classification supports better feature extraction and results owning to a distinct boundary between various classes and enough data to train on. Hence, the models trained on such diverse data are generic enough to learn and represent class boundaries even in the case of medical data where there is a contingency for high generalization error due to the unbalanced and limited availability of dataset. Transfer Learning, is thus, utilizing the feature extraction capabilities of pre-trained models on huge datasets like ImageNet (Deng et al. 2009) that can capture the class boundaries well. Transfer learning not only eliminates the prerequisite for a huge dataset but also provides quick results in this pandemic situation. This motivates us to utilize these techniques in our proposed deep learning framework for classification of COVID-19.
Ensemble learning
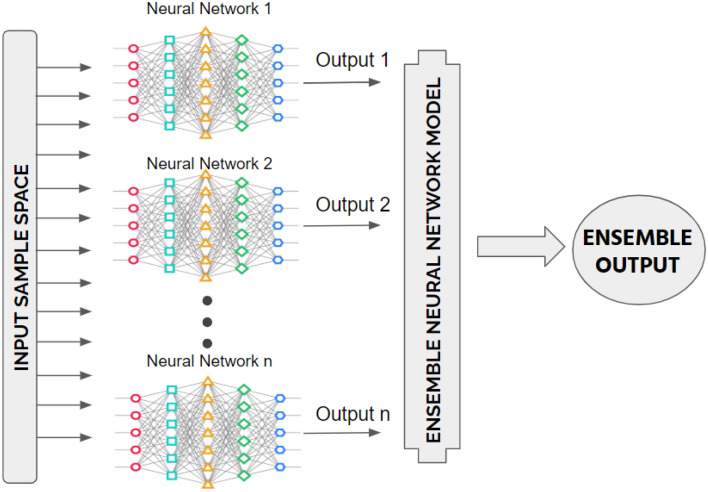
To reduce the errors in detection, an ensemble deep transfer learning model is proposed by combining the outputs of different independently trained neural networks (in our case, transfer learning models) as shown in Fig. 1. An ensemble of transfer learning networks may be a robust approach by reducing errors. It produces optimal results from the combined networks with the least possible errors. After data-preprocessing, the convolutional neural network architecture is built using pre-trained models. The core components of the employed models are described in the preceding subsections.
Fig. 1.
Block diagram of an ensemble learning model by considering number of artificial neural networks
Layers of deep CNN
Pooling layers
Pooling layers are used for multi-scale analysis and reducing the input data (image) size for feature reduction. Max Pooling and Average Pooling layers are most widely utilized in CNN. The average pooling layer ( is used in the proposed architecture, which can be mathematically defined as (Wang et al. 2018):
| 1 |
where and denote pooling region and k cardinal number, respectively. denotes the activation set in defined as:
| 2 |
Here, .
Fully connected layer and Softmax
The fully connected layers (FC) have full connections to the neurons. The inputs to FC are multiplied with FC weight matrix to produce the multiplication result. In the proposed architecture, a fully connected dense layer is used with Softmax activation function for multi-class classification.
The working of the dense layer with Softmax activation (Wang et al. 2018) can be described through the concept of conditional probability. Softmax function () is the probability of a sample belonging to class and defined as:
| 3 |
In Eq. 3, is the class probability and the total number of classes is given by and the class probability is . Equation 3 can also be redefined as:
| 4 |
Here,
| 5 |
Dropout
Dropouts are added to regularize the convolutional neural networks by randomly setting the outgoing neurons at the hidden layers 0 at each training iteration. The dropped neurons have no contribution to the forward pass or back-propagation while training phase. This leads to different architecture being used for each forward pass step despite having shared weights. This assists in avoiding over-fitting in the model. In this paper, the dropouts are set to 0.2 and 0.3 for the three-class classification problem (Krizhevsky et al. 2012).
Rectified linear units (ReLU)
The activation function of ReLU can be defined as:
| 6 |
The training time of ReLU is significantly lesser than sigmoidal functions. It is also helpful to overcome gradient-based training and poor performance in neural networks due to the widespread saturation. In comparison to the sigmoid function, ReLU reduces the training time by fastening the convergence of stochastic gradient descent (Wang et al. 2018).
Sigmoid function
The sigmoid function (also referred to as logistic function) is simply used to predict the probability of a given output. The sigmoid function gives desirable results for the probability of [0,1]. The sigmoid produces a flexible S-shaped curve or the “Sigmoid” curve. It can be mathematically defined as:
| 7 |
Here, is the input.
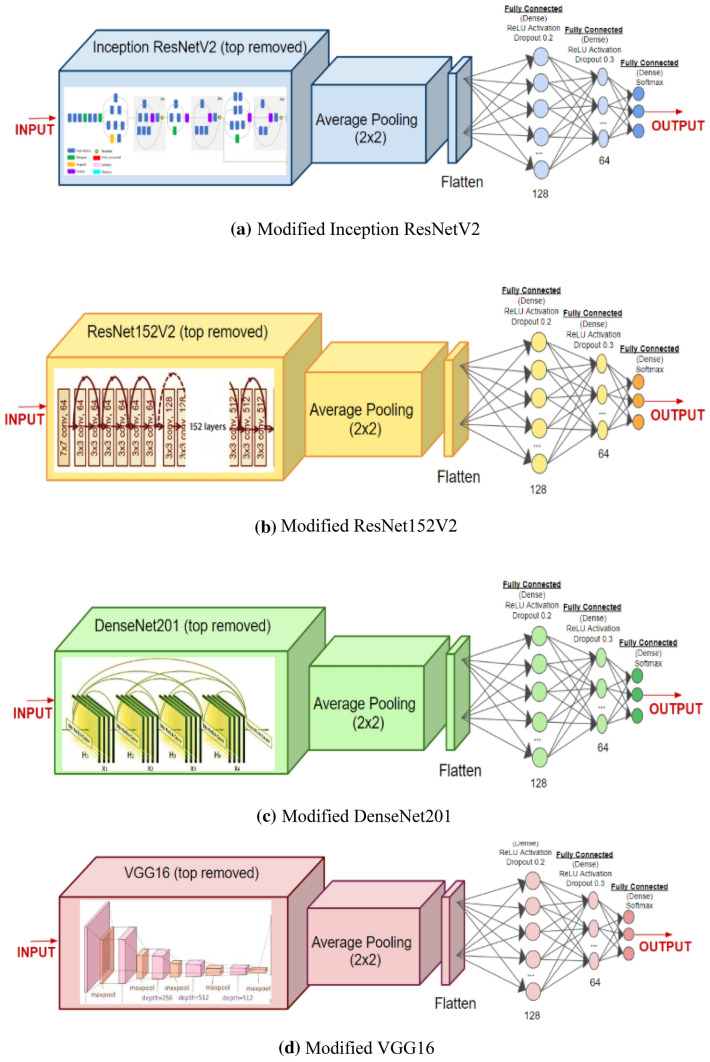
Architectures of deep transfer learning models
Figure 2 shows the architectures of DenseNet201 (Huang et al. 2017), ResNet152V2 (He et al. 2016), InceptionResNetV2 (Szegedy et al. 2017), and VGG16 (Simonyan and Zisserman 2014). The neurons for the first dense layer are 64 in place of 128 for binary class classification problem. The fine-tuned pre-trained model with several layers is used for feature extraction. In Dense Layer, Softmax activation function is introduced for three-class classification problem. However, Sigmoid activation is used for binary classification The models are trained for 300 epochs with batch size set to 16. Adam optimizer is used for fine-tuning of the models. To prevent overfitting, regularization is achieved by using an early stopping criterion.
Fig. 2.
Architectures of modified deep transfer learning models a Inception ResNetV2, b ResNet152V2, c DenseNet201, d VGG16
Ensemble deep transfer learning model for three-class classification problem
The above-mentioned transfer learning classifiers motivated us to propose the ensemble stacked classifier. The ensemble of deep CNNs may prove to be a powerful technique for better results as it works on the concept of combining the decisions obtained from various models (Siwek et al. 2009; Islam and Zhang 2018). Owning to the stochastic nature of deep CNNs, each neural network architecture learns some specific patterns from other neural networks. The ensemble technique provides better boosting accuracy and feature extraction capability (Alshazly et al. 2019).
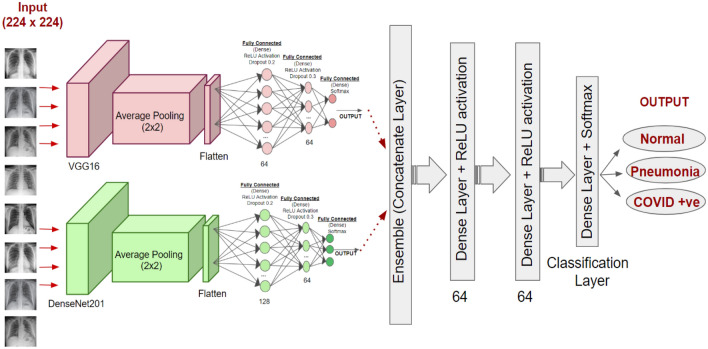
Figure 3 shows the architecture of the proposed ensemble model for three-class classification problem. The ensemble is done through a concatenation of two deep learning models namely, VGG16 and DenseNet201. InceptionRestNetV2 and ResNet152V2 are not considered for the ensemble process due to their relatively low performance for the three-class problem as reported in the literature. During the hyper-parameter tuning, fully connected layers (64 neurons each with 0.2 and 0.1 dropouts) having Softmax activation function have been added for three-class classification.
Fig. 3.
Proposed ensemble deep transfer learning model by using the modified VGG16 and DenseNet201 models for multi-classification
The base neural networks have been frozen to preserve ImageNet weights during the training phase. The architecture added after ensemble is trained on Dataset 2. The ensemble neural network has been trained for 10 epochs with a batch size of 64. The learning rate is set to 0.00001.
Ensemble deep transfer learning model for two-class classification
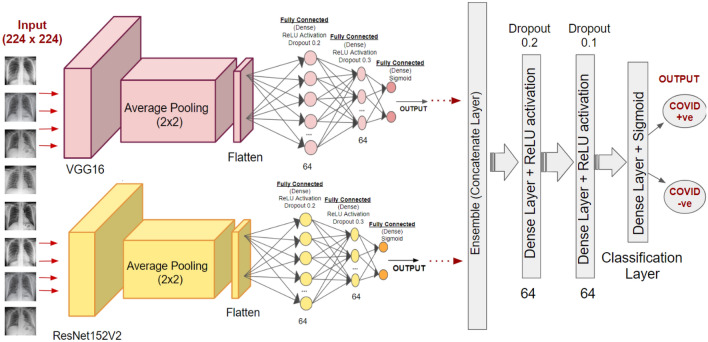
The four modified architectures of pre-trained models as depicted in Fig. 2 (using Sigmoid in the last Dense Layer instead of Softmax) are trained on the first dataset. Similar to the three-class ensemble model, all the four transfer learning models, i.e., modified DenseNet201, Inception ResNetV2, ResNet152V2, and VGG16 are initially trained and evaluated individually. The best two of four models are further ensembled in the proposed architecture. A similar architecture is followed post ensemble with two dense layers (64 neurons each and dropouts) for further classification. However, a dense layer with Sigmoid activation is introduced for final binary classification to obtain probabilities in the range of [0,1] for two classes namely, COVID positive and COVID negative. Figure 4 shows the architecture of the proposed model for binary classification.
Fig. 4.
Proposed ensemble framework (architecture) for binary classification
The proposed model is trained for 20 epochs while keeping the base models are frozen. The batch size and learning rate are set to 64 and 0.00001, respectively. The entire pre-processing, augmentation, and model training is performed through TensorFlow framework.
Performance analysis
In order to validate the performance of the proposed model, they have been compared with recently developed transfer learning models such as VGG16, ResNet152V2, InceptionResnetV2, andDenseNet201. Experimental results and discussion are mentioned in the preceding subsections.
Dataset
Two different datasets are used for validation of the proposed models. The first dataset has been obtained from Kaggle datasets resource (Asraf 2020). It consists of X-Ray-scans that have COVID positive, COVID negative and Pneumonia subjects. The COVID positive and COVID negative from the mentioned dataset is utilized for binary classification.
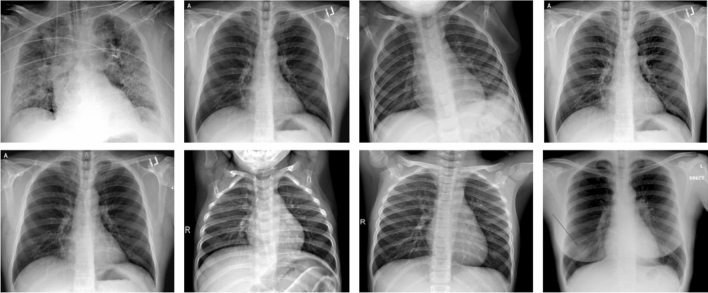
The second dataset is created by researchers from the University of Dhaka and Qatar University along with medical practitioners and collaborators (Chowdhury et al. 2020). It consists of 1203 Chest X-ray scans on COVID, normal, and pneumonia subjects (viral as well as bacterial pneumonia). This dataset is used for multi-class classification. Due to the lack of data corresponding to the COVID class, image data from (Darshan 2020) is also combined for making it 401 images corresponding to each class. Samples images taken from two datasets are shown in Fig. 5.
Fig. 5.
Sample X-ray images for binary and multi-classification
Data preparation and preprocessing
For better performance of the proposed system, X-ray images are resized to 224 × 224 × 3 (RGB). It is essential to augment data for better generalization capability. As neural networks have millions of parameters, the data must be in proportional orders for a good learning capacity. To make up for the lack of data available, the training and validation data is augmented by horizontal flipping, vertical flipping, rotation by 45° and slant angle (0.2) for sheer transformation.
Augmentation is also carried out on the validation dataset as our model may be validated through the variety of inputs. The models not only learn from synthetically modified data but are also validated on augmented data. To ensure a uniform data distribution, image normalization has been carried out by dividing the images by the number of channels, i.e., 255 to achieve the normalized data in the range of [0, 1]. This will ensure better convergence during the training of the neural network.
The last step is data splitting for training, validation, and testing. For both classification problems, the split is initially done through 15% of data for testing and remaining data (85%) is again split. Among this data, 68% for model training and 17% for model validation. The model learns and assigns weights from the training data due to this more weightage has been assigned from the total dataset. However, a slight variation in results may be noticed if the training proportion of data is decreased or increased.
Performance metrics
The performance of the proposed models has been tested for classification. The confusion-matrix based quantitative metrics are Accuracy , Sensitivity , Specificity Precision , Recall , and F1-Score Accuracy can be defined as the overall performance of the model by quantifying the correctly predicted labels. can be mathematically explained as (Basavegowda and Dagnew 2020, Osterland and Weber 2019, and Ghosh et al. 2020):
| 8 |
and denote how accurately a classifier predicts the positive and negative labels quantifying the true positive rate and the true negative rate respectively. and can be explained as in Eq. 9 and Eq. 10, respectively (Gupta et al. 2019):
| 9 |
| 10 |
The exactness of results can be evaluated using Precision . Precision gives the agreement of data labels with the positive labels given by the classifier (Please refer to Eq. 11). Similarly, Recall , also known as Sensitivity (as defined in Eq. 9) evaluates the completeness of the classifier. Higher the R-value lower would be false negatives as can be defined as (Wang et al. 2019 and Wiens 2019):
| 11 |
| 12 |
F1-Score quantifies the data distributions by giving a combined score of and as in Eq. 13
| 13 |
In Eqs. 8–13, TP defines the true positive subject, i.e., a subject who is COVID-19 +ve and the model has correctly classified the same as COVID-19 +ve. Similarly, TN is the truly predicted negative case. Whereas, FP AND FN are the incorrect predictions by the model corresponding to COVID-19 +ve and COVID-19 -ve subjects. The proposed model has also been evaluated using a multiclass confusion matrix for three classes that are COVID-19 (+ve), Normal and Pneumonia (see Fig. 6).
Fig. 6.
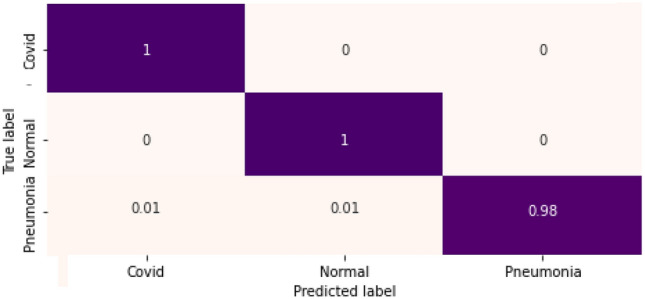
Confusion matrix for three-class classification
Experimentation 1: three class-classification
Table 1 shows performance evaluation of the proposed approach with VGG16, ResNet152V2, InceptionResnetV2, andDenseNet201. It is observed from Table 1 that the above-mentioned models achieve comparable results. DenseNet201 attained the best testing accuracy of 96.68%, which is better than VGG16, ResNet152V2, and InceptionResnetV2. The proposed ensemble technique outperforms the pre-trained models with a testing accuracy of 99.21%. The proposed architecture provides better generalization capability than the other models. The ensemble technique takes a minimum of 6 mi for the training phase and a few seconds for the testing phase. Each class namely COVID-19 (+ve), normal, and pneumonia achieve high precision values. It means that the correct positive predictions out of all the subjects can be classified as positive.
Table 1.
Test evaluation of three class classifiers
| Model/Metric | Label | Precision | Recall | F1-Score | Accuracy(overall) (%) |
|---|---|---|---|---|---|
| VGG16 | Covid19 | 0.95 | 0.98 | 0.97 | |
| Normal | 0.94 | 1.00 | 0.97 | ||
| Pneumonia | 0.98 | 0.88 | 0.93 | ||
| [Macro average] | 0.96 | 0.96 | 0.96 | 95.70 | |
| ResNet152V2 | Covid19 | 0.92 | 1.00 | 0.96 | |
| Normal | 0.95 | 0.98 | 0.96 | ||
| Pneumonia | 0.99 | 0.92 | 0.95 | ||
| [Macro average] | 0.95 | 0.96 | 0.96 | 95.58 | |
| InceptionResnetV2 | Covid19 | 0.95 | 1.00 | 0.98 | |
| Normal | 0.95 | 0.95 | 0.95 | ||
| Pneumonia | 0.95 | 0.90 | 0.92 | ||
| [Macro average] | 0.95 | 0.95 | 0.95 | 95.02 | |
| DenseNet201 | Covid19 | 0.95 | 1.00 | 0.98 | |
| Normal | 0.95 | 1.00 | 0.98 | ||
| Pneumonia | 1.00 | 0.90 | 0.95 | ||
| [Macro average] | 0.97 | 0.97 | 0.97 | 96.68 | |
| Proposed Ensemble | Covid19 | 0.98 | 1.00 | 0.99 | |
| (VGG16 + DenseNet) | Normal | 0.99 | 1.00 | 0.99 | |
| Pneumonia | 1.00 | 0.98 | 0.99 | ||
| [Macro average] | 0.99 | 0.99 | 0.99 | 99.21 |
Final precision, recall, F1-score and accuracy (macro average values) of the proposed ensemble model for three-class classification are highlighted in bold
The precision for pneumonia subjects is one indicating that there are zero false-positive predictions. The macro average (unweighted mean per label) precision, recall, and F1-Score for the proposed framework are 0.99, which justifies the validity of the model.
Figure 6 shows the computed confusion matrix for three-class classification. Figure 7 shows the training and validation loss for the proposed model. The model converges well, reaching peak accuracy while keeping the training as well as validation losses minimal. It is found that the training and validation losses are not far from approximately similar values. Our model is not over-fitted for this dataset.
Fig. 7.
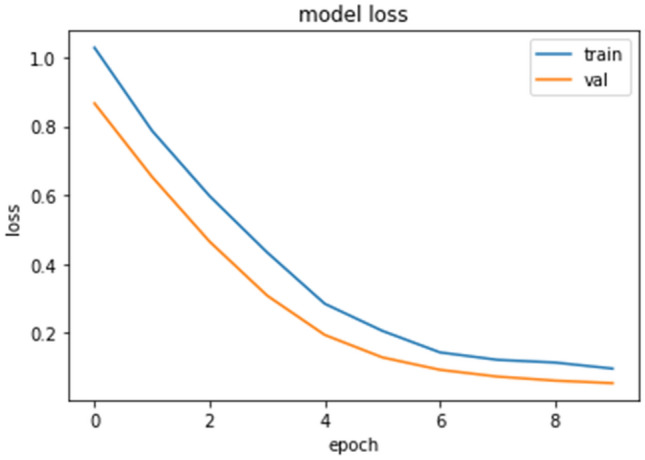
Loss plot for the proposed ensemble (three class)
Figure 8 depicts the Area Under the receiver curve (AUC) for the proposed ensemble model. It is observed that a perfect AUC = 1 is achieved for COVID-19 (+ve) subject and normal subjects, indicating the model has a perfect sense of separability of the said, two classes. For pneumonia, the score is 0.99 is achieved, which is appreciable, however, for a critical situation like COVID-19, efforts to increase it can be made.
Fig. 8.
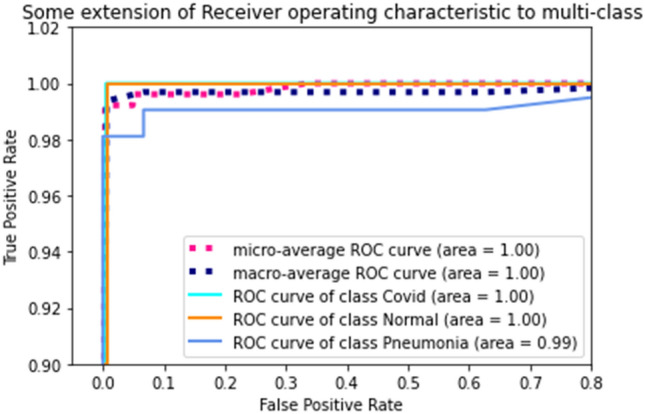
Area under ROC (AUC) for the proposed ensemble, depicting the specific AUC for each class along with the micro-average and macro-average overall AUC
Experimentation 2: two class-classification
For the binary classification, the four learning models are individually trained and achieved accuracies of 95.12% (VGG16), 94.61% (ResNet152V2), 93.07% (InceptionResNetV2) and 94.10% (DenseNet201). The training time is calculated to be approximately 10 min for the proposed architecture. Both VGG16 and ResNet152V2 achieved the highest accuracy. However, there is still scope of improvement (precision and sensitivity scores) by considering the sensitive problem. For this, the ensemble model is introduced, which is utilized in the multi-class classification problem. The proposed ensemble architecture consists of VGG16 and ResNet152V2 that surpassed the basing of models by achieved a 1.2% (roughly) higher accuracy. The proposed architecture attained 0.959 precision and accuracy of 96.15% indicating the correctness of predicted COVID and normal subjects.
The experimental results reveal that the proposed ensemble technique attains high specificity rates. It means that there would be no false-positive predictions, which directly implies a lesser obligation on the healthcare system. This will aid in the correct usage of testing kits and health-care facilities by reaching those who genuinely require it.
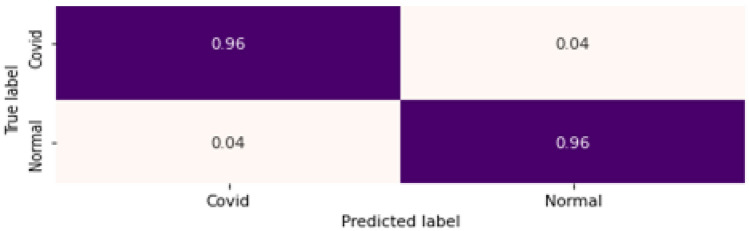
To evaluate the proposed binary classifier, the confusion matrix and the loss plot is given in Figs. 9 and 10, respectively. 4% false predictions are observed (positive as well as negative) with a 96% true predication for COVID-19(+ve) as well as normal subjects. The model fits well, as can be seen from the loss plot, avoiding any overfitting, as clear from the minimum (~ 0) difference between the training and validation error.
Fig. 9.
Confusion Matrix for binary classification
Fig. 10.
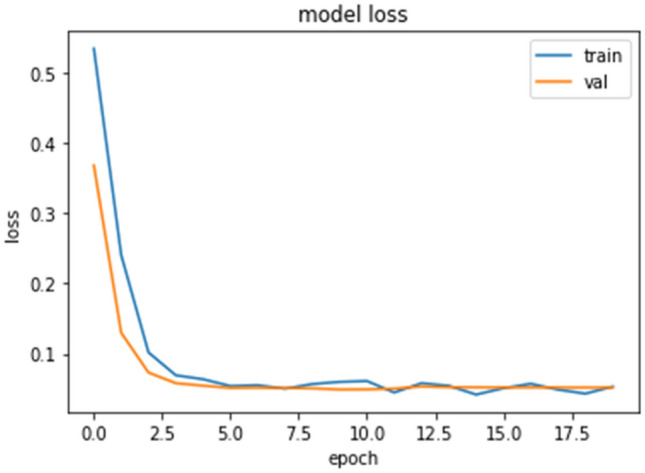
Loss plot for binary classification (ensemble)
The Area Under the receiver curve (AUC) is also computed as shown in Fig. 11 for all binary classifiers implemented. It can be observed that the proposed ensemble performs better than the individual base models giving good separability between the two classes. This also supports the fact that the ensemble process results in better-generalized models resulting in efficient frameworks. Although, a 0.1% difference is seen between the AUC of both base models (VGG16 and ResNet152V2) and the proposed ensemble, in medical research, 0.1% increase in the model quality is critical.
Fig. 11.
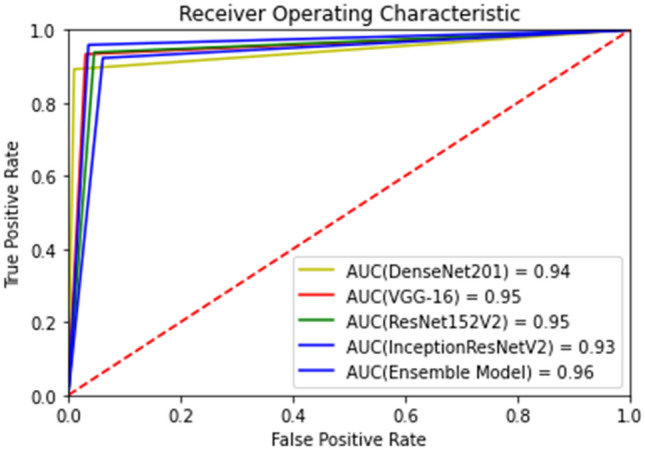
Area under ROC (AUC) for the 5 models implemented for binary classification
Comparative analysis
It can be established from the results that deep learning framework can significantly assist in the detection of COVID-19 subjects using X-ray images by giving a low cost and rapid solution for the diagnosis of COVID-19. Table 3 shows the performance evaluation of the proposed binary classification technique with state-of-the-art techniques. Waheed et al. (2020)proposed a framework an appreciable accuracy of 95% however, the false negative is the rate was 10%, which is menacing for the specific problem in hand. The proposed architectures by Hemdan et al. (2020) and Sethy and Behera (2020) achieved good results with high sensitivity and specificity values, the data utilized was extremely limited, which may indicate reduced generalization. To overcome these possible inhibitions, we utilized a larger data set with augmentation and well as tried to reduce the false predictions. Considering, the statistical metrics in Table 2, the proposed ensemble surpassed the state-of-the-art concerning better generalization as well as by minimizing the false predictions achieving an overall classification accuracy of 96.15%.
Table 3.
Performance evaluation of different deep learning techniques on the binary dataset
| State-of-the-art | Accuracy (%) |
|---|---|
| ResNet50 + SVM (Sethy and Behera 2020) | 95.38 |
| CovidGAN (Waheed et al. 2020) | 95 |
| COVIDX-Net (Hemdan et al. 2020) | 90 |
| CAAD (Zhang et al. 2020) | 72.77 |
| CNN trained from scratch (Maghdid et al. 2020) | 94 |
| Proposed ensemble model | 96.15 |
Accuracy of the proposed ensemble model is highlighted to emphasize on the highest accuracy value with respect to the other techniques that have been utilized for binary classification
Table 2.
Test evaluation of two-class classifiers
| Model/Metric | Precision | Recall | F1-Score | Sensitivity | Specificity | Accuracy (%) |
|---|---|---|---|---|---|---|
| VGG16 | 0.945 | 0.969 | 0.952 | 0.969 | 0.933 | 95.12 |
| ResNet152V2 | 0.939 | 0.953 | 0.946 | 0.953 | 0.938 | 94.61 |
| InceptionresNetV2 | 0.924 | 0.938 | 0.931 | 0.938 | 0.923 | 93.07 |
| DenseNet201 | 0.901 | 0.989 | 0.943 | 0.989 | 0.892 | 94.10 |
| Proposed ensemble | 0.959 | 0.964 | 0.961 | 0.964 | 0.958 | 96.15 |
Final precision, recall, F1-score, sensitivity, specificity and accuracy of the proposed ensemble model for binary classification are highlighted in bold
Table 4 shows the performance comparison of the proposed technique for multi-classification with five state-of-the-art techniques. Although the false predictions in MobileNetV2 (Apostolopoulos and Mpesiana 2020) and Dark COVID-Net (Ozturk et al. 2020) were slightly lower than the proposed model, however, the model lacked with an approximately 5% and 12% lower accuracy. Islam et al. (2020) proposed a novel approach using CNN as a feature extractor and LSTM for detection and their approach closely follows behind our proposed ensemble framework with an accuracy of 97%. Hence, the proposed ensemble for the multi-classification task outperformed various other approaches with a final classification accuracy of 99.21%.
Table 4.
Performance evaluation of different deep learning techniques on the multi-class dataset
| State-of-the-art | Accuracy (%) |
|---|---|
| Dark COVID-Net (Ozturk et al. 2020) | 87.02 |
| MobileNetV2 (Apostolopoulos and Mpesiana 2020) | 94.72 |
| CORO-Net (Khan et al. 2020) | 95 |
| CNN + LSTM (Islam et al. 2020) | 97 |
| Covid-Net (Wang and Wong 2020) | 93.3 |
| Proposed ensemble model | 99.21 |
Accuracy of the proposed ensemble model is highlighted to emphasize on the highest accuracy value with respect to the other techniques that have been utilized for three-class classification
Conclusions and future scope
In this paper, the ensemble models using deep convolutional neural networks are proposed for the detection of COVID-19 from chest X-ray images. The proposed model not only detect COVID-19 infection from infected persons but is also able to detect COVID-19 from normal and pneumonia (Viral and Bacterial) persons. The proposed models are tested over two well-known datasets. They achieved the accuracies of 99.21% and 96.15% for three class and binary classification, respectively. The proposed model may be leveraged by health officials and researchers to accelerate the prediction of COVID-19 in this global pandemic situation.
There are some limitations of the proposed framework, which can be improved in the future. A limited patient dataset is available that eventually impacts the training and learning capacity of the proposed models. In future, more patient data can be leveraged specifically COVID-19 subjects that can improve the feature extraction capabilities of the proposed model. In the current pandemic situation, a deeper analysis is required for differentiating the COVID-19, viral pneumonia and bacterial pneumonia. The work can also be extended by adding risk and survival prediction of prospective/confirmed COVID-19 patients that can be helpful in the healthcare planning and management strategies.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abbas A, Abdelsamea MM, Gaber MM (2020) Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. arXiv preprint arXiv:2003.13815 [DOI] [PMC free article] [PubMed]
- Alshazly H, Linse C, Barth E, Martinetz T. Ensembles of deep learning models and transfer learning for ear recognition. Sensors. 2019;19(19):4139. doi: 10.3390/s19194139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolopoulos ID, Mpesiana TA. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020;3:1. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asraf A (2020) COVID19_Pneumonia_Normal_Chest_Xray(PA)_Dataset, June 29, 2020. https://www.kaggle.com/amanullahasraf/covid19-pneumonia-normal-chest-xraypa-dataset
- Basavegowda HS, Dagnew G. Deep learning approach for microarray cancer data classification. CAAI Trans Intell Technol. 2020;5(1):22–33. doi: 10.1049/trit.2019.0028. [DOI] [Google Scholar]
- Basu S, Mitra S (2020) Deep Learning for screening COVID-19 using chest X-Ray Images. arXiv preprint arXiv:2004.10507
- Cellina M, Orsi M, Toluian T, Valenti Pittino C, Oliva G. False negative chest X-Rays in patients affected by COVID-19 pneumonia and corresponding chest CT findings. Radiography. 2020;26(3):e189–e194. doi: 10.1016/j.radi.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouhan V, Singh SK, Khamparia A, Gupta D, Tiwari P, Moreira C, Damaševičius R, De Albuquerque VHC. A novel transfer learning based approach for pneumonia detection in chest X-ray images. Appl Sci. 2020;10(2):559. doi: 10.3390/app10020559. [DOI] [Google Scholar]
- Chowdhury MEH, Rahman T, Khandakar A, Mazhar R, Kadir MA, Mahbub ZB, Islam KR et al (2020) Can AI help in screening viral and COVID-19 pneumonia? arXiv preprint arXiv:2003.13145
- Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronavirus Disease (COVID-19) (2020) Events as They Happen (WHO). https://www.who.int/emergencies/diseases/novel-coronavirus-2019/events-as-they-happen. Accessed 15 July 2020
- COVID-19 symptoms (2020) https://www.who.int/health-topics/coronavirus#tab=tab_3. Accessed 15 July 2020
- Darshan (2020) COVID-19 detection X-ray dataset. March 22, 2020. https://www.kaggle.com/darshan1504/covid19-detection-xray-dataset
- Das D, Santosh KC, Pal U. Truncated inception net: COVID-19 outbreak screening using chest X-rays. Phys Eng Sci Med. 2020;43:915–925. doi: 10.1007/s13246-020-00888-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das NN, Kumar N, Kaur M, Kumar V, Singh D. Automated deep transfer learning-based approach for detection of COVID-19 infection in chest X-rays. IRBM. 2020 doi: 10.1016/j.irbm.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Dong W, Socher R, Li LJ, Li K, Fei-Fei L (2009) Imagenet: A large-scale hierarchical image database. In: 2009 IEEE conference on computer vision and pattern recognition. Ieee, pp 248–255
- Ghosh S, Shivakumara P, Roy P, Pal U, Lu T. Graphology based handwritten character analysis for human behaviour identification. CAAI Trans Intell Technol. 2020;5(1):55–65. doi: 10.1049/trit.2019.0051. [DOI] [Google Scholar]
- Gupta B, Tiwari M, Lamba SS. Visibility improvement and mass segmentation of mammogram images using quantile separated histogram equalisation with local contrast enhancement. CAAI Trans Intell Technol. 2019;4(2):73–79. doi: 10.1049/trit.2018.1006. [DOI] [Google Scholar]
- He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
- Hemdan E, Shouman M, Karar M (2020) COVIDX-Net: a framework of deep learning classifiers to diagnose COVID-19 in X-ray images. https://arxiv.org/abs/2003.11055
- Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708
- Islam J, Zhang Y. Brain MRI analysis for Alzheimer’s disease diagnosis using an ensemble system of deep convolutional neural networks. Brain Informat. 2018;5(2):2. doi: 10.1186/s40708-018-0080-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MZ, Islam MM, Asraf A. A combined deep CNN-LSTM network for the detection of novel coronavirus (COVID-19) using X-ray images. Inform Med Unlocked. 2020;20:100412. doi: 10.1016/j.imu.2020.100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AI, Shah JL, Bhat MM. Coronet: a deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput Methods Progr Biomed. 2020;196:105581. doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. In: Advances in neural information processing systems, pp 1097–1105
- Loey M, Smarandache F, Khalifa NEM. Within the lack of chest COVID-19 X-ray dataset: a novel detection model based on GAN and deep transfer learning. Symmetry. 2020;12(4):651. doi: 10.3390/sym12040651. [DOI] [Google Scholar]
- Luz EJS, Silva PL, Silva R, Silva L, Moreira G, Menotti D (2020) Towards an effective and efficient deep learning model for COVID-19 patterns detection in X-ray Images. CoRR. arXiv:2004.05717
- Maghdid HS, Asaad AT, Ghafoor KZ, Sadiq AS, Khan MK (2020) Diagnosing COVID-19 pneumonia from X-ray and CT images using deep learning and transfer learning algorithms. arXiv preprint arXiv:2004.00038
- Mahmud T, Rahman MA, Fattah SA. CovXNet: a multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization. Comput Biol Med. 2020;122:103869. doi: 10.1016/j.compbiomed.2020.103869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narin A, Kaya C, Pamuk Z (2020) Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. arXiv preprint arXiv:2003.10849 [DOI] [PMC free article] [PubMed]
- Oh Y, Park S, Ye JC (2020) Deep learning covid-19 features on cxr using limited training data sets. IEEE Trans Med Imaging 39(8) [DOI] [PubMed]
- Osterland S, Weber J. Analytical analysis of single-stage pressure relief valves. Int J Hydromech. 2019;2(1):32–53. doi: 10.1504/IJHM.2019.098951. [DOI] [Google Scholar]
- Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Acharya UR. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. 2020;121:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimzadeh M, Attar A. A modified deep convolutional neural network for detecting COVID-19 and pneumonia from chest X-ray images based on the concatenation of Xception and ResNet50V2. Informat Med Unlocked. 2020;19:100360. doi: 10.1016/j.imu.2020.100360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethy PK, Behera SK. Detection of coronavirus disease (covid-19) based on deep features. Preprints. 2020;2020030300:2020. [Google Scholar]
- Shelke A, Inamdar M, Shah V, Tiwari A, Hussain A, Chafekar T, Mehendale N. Chest X-ray classification using Deep learning for automated COVID-19 screening. Radiol Imaging. 2020 doi: 10.1101/2020.06.21.20136598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D, Guorong W, Suk H-I. Deep learning in medical image analysis. Annu Rev Biomed Eng. 2017;19:221–248. doi: 10.1146/annurev-bioeng-071516-044442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556
- Siwek K, Osowski S, Szupiluk R. Ensemble neural network approach for accurate load forecasting in a power system. Int J Appl Math Comput Sci. 2009;19(2):303–315. doi: 10.2478/v10006-009-0026-2. [DOI] [Google Scholar]
- Szegedy C, Ioffe S, Vanhoucke V, Alemi AA (2017) Inception-v4, inception-resnet and the impact of residual connections on learning. In: Thirty-first AAAI conference on artificial intelligence
- Toğaçar M, Ergen B, Cömert Z. COVID-19 detection using deep learning models to exploit Social Mimic Optimization and structured chest X-ray images using fuzzy color and stacking approaches. Comput Biol Med. 2020;121:103805. doi: 10.1016/j.compbiomed.2020.103805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuncer T, Dogan S, Ozyurt F. An automated residual exemplar local binary pattern and iterative ReliefF based corona detection method using lung X-ray image. Chemometr Intell Lab Syst. 2020;203:104054. doi: 10.1016/j.chemolab.2020.104054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waheed A, Goyal M, Gupta D, Khanna A, Al-Turjman F, Pinheiro PR. Covidgan: data augmentation using auxiliary classifier gan for improved covid-19 detection. IEEE Access. 2020;8:91916–91923. doi: 10.1109/ACCESS.2020.2994762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wong A (2020) COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. arXiv preprint arXiv:2003.09871 [DOI] [PMC free article] [PubMed]
- Wang H, Xia Y (2018) Chestnet: A deep neural network for classification of thoracic diseases on chest radiography. arXiv preprint arXiv:1807.03058 [DOI] [PubMed]
- Wang S-H, Phillips P, Sui Y, Liu B, Yang M, Cheng H. Classification of Alzheimer’s disease based on eight-layer convolutional neural network with leaky rectified linear unit and max pooling. J Med Syst. 2018;42(5):85. doi: 10.1007/s10916-018-0932-7. [DOI] [PubMed] [Google Scholar]
- Wang R, Yu H, Wang G, Zhang G, Wang W. Study on the dynamic and static characteristics of gas static thrust bearing with micro-hole restrictors. Int J Hydromech. 2019;2(3):189–202. doi: 10.1504/IJHM.2019.102893. [DOI] [Google Scholar]
- Wiens T. Engine speed reduction for hydraulic machinery using predictive algorithms. Int J Hydromech. 2019;2(1):16–31. doi: 10.1504/IJHM.2019.098949. [DOI] [Google Scholar]
- Xie X, Zhong Z, Zhao W, Zheng C, Wang F, Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020;1:200343–200343. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Kang C, Guttery DS, Kadry S, Chen Y, Zhang YD (2020) ResNet-SCDA-50 for breast abnormality classification. In: IEEE/ACM transactions on computational biology and bioinformatics [DOI] [PubMed]
- Zhang J, Xie Y, Li Y, Shen C, Xia Y (2020) Covid-19 screening on chest x-ray images using deep learning based anomaly detection. arXiv preprint arXiv:2003.12338