Abstract
Background
Congenital bilateral absence of vas deferens (CBAVD) is an important disease of male infertility, which affects 1%–2% of infertile population. In addition to common mutations of CFTR, copy number variants (CNVs) have also been implicated as one of the pathogenesis of CBAVD. The present study aimed to investigate the genetic contribution of CFTR CNVs in Chinese Han population with CBAVD.
Methods
Two hundred and sixty‐three CBAVD patients were recruited. Genomic DNA was extracted from peripheral blood samples. The Multiplex Ligation‐dependent Probe Amplification assay was performed which targets the region of the CFTR gene.
Results
Among 263 Chinese men affected with CBAVD in this study, 5 (1.90%) patients were detected for copy number variants in the region of CFTR gene (4 of them carried partial deletions and 1 of them carried partial duplication of CFTR gene).
Conclusions
The study showed that the rate of CFTR CNVs in Chinese population with CBAVD were basically consistent with the previous reports. And the study first revealed genetic risk of CNVs of CFTR on a large sample size of CBAVD patients in Chinese Han population, which prompted that it was necessary to detect CNVs of CFTR in Chinese Han people with CBAVD.
Keywords: CFTR, Chinese Han population, Congenital bilateral absence of vas deferens, copy number variants
The study showed that the rate of CFTR CNVs in Chinese CBAVD population were basically consistent with the previous reports. And the study firstly revealed genetic risk of CNVs of CFTR on a large sample size of CBAVD patients in Chinese Han population, which prompted that it was necessary to detect CNVs of CFTR in Chinese Han people with CBAVD.
![]()
1. INTRODUCTION
Congenital bilateral absence of vas deferens (CBAVD) is an important disease of male infertility, accounting for 1%–2% of infertile population (Jequier, Ansell, & Bullimore, 1985; Yu, Chen, Ni, & Li, 2012). CBAVD is characterized by absence of vas deferens and semen abnormality (Practice Committee of the American Society for Reproductive Medicine in collaboration with the Society for Male & Urology, 2018).
Cystic fibrosis (CF) is a well‐known genetic disease, and showed racial differences (it is common in Caucasians but not common in Asians) (Yang, Wang, Zhang, Li, & Wang, 2018). Cystic fibrosis transmembrane conductance regulator gene (CFTR) (OMIM: 602421) defect can cause CF (Zielenski & Tsui, 1995), and Cystic Fibrosis Mutation Database (CFMDB) (https://www.genet.sickkids.on.ca/cftr/app) has been established.
Also, CFTR mutations were closely related to CBAVD (Cai et al., 2019; Chillón et al., 1995), which occurred in more than three quarters of CBAVD patients (Yu et al., 2012). In addition to common mutations (single nucleotide variants or small insertions and deletions), copy number variants (CNVs) (including large deletions and duplications) have also been implicated as one of the pathogenesis of CBAVD (Hantash et al., 2006; Ratbi et al., 2007; Taulan et al., 2007).
However, traditional methods cannot identify all CFTR variants (Férec et al., 2006; Hwang et al., 2018). Currently, Multiplex Ligation‐dependent Probe Amplification (MLPA) is an effective method to detect CNVs (Schrijver, Rappahahn, Pique, Kharrazi, & Wong, 2008; Taulan et al., 2012). Therefore, we used this technique to investigate the genetic contribution of CFTR CNVs in Chinese Han population with CBAVD.
2. MATERIALS AND METHODS
2.1. Ethical compliance
The study was approved by the Research Ethics Committee of Peking Union Medical College Hospital. And all participants signed informed consent forms.
2.2. Participants
From 2012 to 2018, 263 CBAVD patients were recruited from Urological Department of Peking Union Medical College Hospital. These patients are all Chinese Han people ages ranging from 21 to 49 years (29 ± 4.85). Patients claimed infertility after a few years of marriage. According to the guidelines published by the American Society for reproductive genetics in 2018 (Practice Committee of the American Society for Reproductive Medicine in collaboration with the Society for Male & Urology, 2018) and previous studies of our research group (Yang et al., 2018), inclusion criteria of CBAVD patients were as follows: impalpable vas deferens, absence of vas deferens by ultrasound, semen abnormality (volume <2.0 ml, pH value <7.2, and fructose <25 μM/ejaculate), normal serum follicle‐stimulating hormone levels, and luteinizing hormone levels, no typical symptoms of CF except for CBAVD.
2.3. DNA extraction
Genomic DNA from 263 CBAVD patients were extracted from peripheral venous blood using QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany).
2.4. MLPA assay
The probes targeted the exon region of CFTR gene (Reference sequence numbers NM_000492.4). The PCR products were analyzed by capillary electrophoresis. The fluorescence data were processed by Applied Biosystems 3730xl DNA Analyzer, and the peak area data were collected. Relative peak area of CFTR probe recognition sequence was compared with the reference gene. Dosage quotient (DQ) was used as the judging basis of CNVs. We considered normal results with values between 0.8 and 1.2. DQ values equal to zero were considered homozygous deletion. DQ values between 0.4 and 0.65 were considered heterozygous deletion, DQ values between 1.3 and 1.65 were considered heterozygous duplication.
2.5. Evaluation of CNVs detected according to American College of Medical Genetics and Genomics (ACMG) guidelines
The CNV interpretation scoring metric put forward by ACMG was used to evaluate the impact on human health. Five aspects were studied: initial assessment of genomic content, overlap with established haploinsufficient regions, number of protein‐coding genes, evaluation of genomic content using public databases and literature, and evaluation of inheritance pattern for patient.
3. RESULTS
3.1. MLPA assay targeted CFTR gene on CBAVD patients
Among 263 Chinese men affected with CBAVD in this study, 5 (1.90%) patients were detected for copy number variants in the region of exons of the CFTR gene, including 1 patient carried heterozygous duplication of exons 1–3 of CFTR (CFTRdup1‐3, Legacy name), 2 patients carried heterozygous deletion of exons 18–20 of CFTR, 1 patient carried heterozygous deletion of exons 14–15 of CFTR, and 1 patient carried heterozygous deletion of exons 22–24 of CFTR (Table 1). In summary, four of them carried partial deletions and 1 of them carried partial duplication of CFTR gene (Figure 1).
TABLE 1.
CNVs of CFTR gene and clinical features in CBAVD patients
| Patient No. | Copy number status of CFTR a | Dosage quotient | Age (years) | Left testicular volume (ml) | Right testicular volume (ml) | Left epididymis | Right epididymis | FSH | LH | PRL | E2 | T |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 217 | Heterozygous deletion of exons 14–15 |
0.64(E14); 0.65(E15) |
31 | 13 | 13 | Normal | Normal | 2.14 | 0.98 | 5.2 | 61.8 | 3 |
| 243 | Heterozygous deletion of exons 18–20 |
0.45(E18); 0.5(E19); 0.47(E20) |
33 | 13 | 13 | Normal | Normal | 5.1 | 3.96 | 16.3 | 40.7 | 3.42 |
| 299 | Heterozygous duplication of exons 1–3 |
1.65(E1); 1.33(E2); 1.41(E3) |
31 | 10 | 10 | Normal | Normal | 3.4 | 5.95 | 10 | 70.7 | 8.39 |
| 324 | Heterozygous deletion of exons 22–24 |
0.45(E22); 0.44(E23); 0.57(E24) |
28 | 10 | 10 | Normal | Normal | 4.4 | 3.65 | 6.52 | 20.8 | 3.93 |
| 357 | Heterozygous deletion of exons 18–20 |
0.45(E18); 0.57(E19); 0.47(E20) |
27 | 15 | 15 | Normal | Normal | 2.83 | 2.19 | 3.59 | 73.4 | 2.94 |
Testicular volume (normal range: 15–25 ml) FSH, Follicle stimulating hormone (normal range: 1.27–12.96 mIU/mL); LH, Luteinizing hormone (normal range: 1.24–8.62 mIU/mL); PRL, Prolactin (normal range: 2.64–13.13 ng/ml); E2, Estradiol (normal range: 20–75 pg/mL);T, Testosterone (normal range: 4–8 ng/mL).
CFTR: Reference sequence numbers NM_000492.4.
FIGURE 1.
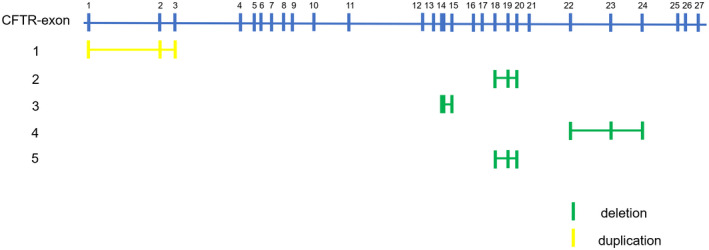
Distribution of copy number variants on CFTR exons. CFTR has 27 exons, 5 patients carried different CNVs. Yellow rectangles symbol duplication. Green rectangles symbol deletion
3.2. Clinical characteristics of five patients with CNVs
The five patients met the inclusion criteria described above. They have impalpable vas deferens but normal testis and epididymis. Their hormone levels are basically normal. See Table 1 for details.
3.3. The impact of CNVs on human health
Three CNVs (CFTRdup1‐3, deletion of exons 22–24 and deletion of exons 18–20) have been reported. Only deletion of exons 14–15 of CFTR was not reported before. Three CNVs (deletion of exons 14–15, deletion of exons 22–24 and deletion of exons 18–20) were evaluated to be pathogenic. CFTRdup1‐3 was evaluated to be uncertain significance. See details in Table 2.
TABLE 2.
CNV Interpretation Scoring Metric of ACMG
| Patient No. | Copy number status | Contain protein‐coding elements (Score) | Overlap with established haploinsufficient genes (Score) a | Number of protein‐coding RefSeq genes (Score) | Analysis of Public Databases and Literature | Inheritance Pattern for Patient (Score) | Total score | Classification c |
|---|---|---|---|---|---|---|---|---|
| 217 | Heterozygous deletion of exons 14–15 | Yes (0) |
Both breakpoints are within CFTR and the exons are in biologically‐relevant transtripts (0.9) |
1 (0) | Skip | Inheritance information is unavailable but the patient has highly specific phenotype of CBAVD(0.3) | 1.2 | Pathogenic |
| 243 | Heterozygous deletion of exons 18–20 | Yes (0) |
Both breakpoints are within CFTR and the exons are in biologically‐relevant transtripts (0.9) |
1 (0) | Skip | Inheritance information is unavailable but the patient has highly specific phenotype of CBAVD(0.3) | 1.2 | Pathogenic |
| 299 | Heterozygous duplication of exons 1–3 | Yes (0) | Both breakpoints are within CFTR and the type is presumed in tandem duplication (0.45) | 1 (0) |
The patient has a highly specific phenotype consistent with CFTR, but the inheritance is unknown (0.1). And the CNV did not overlap with common population variation b |
Inheritance information is unavailable but the patient has highly specific phenotype of CBAVD(0.3) | 0.85 | Uncertain Significance |
| 324 | Heterozygous deletion of exons 22–24 | Yes (0) |
Both breakpoints are within CFTR and the exons are in biologically relevant transtripts (0.9) |
1 (0) | Skip | Inheritance information is unavailable but the patient has highly specific phenotype of CBAVD(0.3) | 1.2 | Pathogenic |
| 357 | Heterozygous deletion of exons 18–20 | Yes (0) |
Both breakpoints are within CFTR and the exons are in biologically relevant transtripts (0.9) |
1 (0) | Skip | Inheritance information is unavailable but the patient has highly specific phenotype of CBAVD(0.3) | 1.2 | Pathogenic |
The haploinsufficiency evaluation of CFTR has been established (https://dosage.clinicalgenome.org/).
The Heterozygous duplication of exons 1–3 did not overlap with common population variation (http://dgv.tcag.ca/).
Pathogenic: 0.99 or more points; Likely Pathogenic: 0.90 to 0.98; Variant of Uncertain Significance: 0.89 to −0.89; Likely Benign: −0.90 to −0.98; Benign: −0.99 or fewer points.
4. DISCUSSION
In the Cystic Fibrosis Mutation Database (www.genet.sickkids.on.ca/) and reports in other database, most of the results we found has been detected. CFTRdup1‐3 was evaluated to be uncertain significance, but T. Casals et al identified CFTRdup1‐3 from two unrelated Spanish CF alleles (Ramos, Gartner, & Casals, 2008). While Hantash FM et al identified the deletion of exons 22–24 of CFTR in a CBAVD patient (Hantash et al., 2006). Chevalier‐Porst and Bozon (2000) identified the deletion of exons 18–20 in a patient who has sweat chloride 82 mmol/l.
And we found a CNV (deletion of exons 14–15 of CFTR) from a CBAVD patients which was not reported before. No identical overlapped variant was found in Database of Genomic Variants (DGV, http://dgv.tcag.ca/dgv/app/home/) and Genome Aggregation Database (GnomAD, http://gnomad.broadinstitute.org/). Thus, this heterozygous deletion of CFTR was considered rare in the general population. CFTR consists of five domains, including two transmembrane‐spanning domains (MSD1 and MSD2), two nucleotide‐binding domains (NBDs) and a regulatory domain (R) (Morales et al., 1996). Exon 14 and exon 15 was located in the regulatory domain and MSD2. 2 MSDs form selective chloride channels, while phosphorylation of R controls channel activity (Sheppard & Welsh, 1999). Deletion of exons 14–15 may be destructive to the function of the CFTR protein. According to the American College of Medical Genetics and Genomics guidelines (Riggs et al., 2020), we thought that deletion of exons 14–15 of CFTR maybe pathogenic (see details in Table 2).
Almost all men with CF have the symptoms of CBAVD (Practice Committee of the American Society for Reproductive Medicine in collaboration with the Society for Male & Urology, 2018), and assessment of patients with CNVs (large deletions and duplications) in our study showed that they have CBAVD typical clinical characteristics. Furthermore, patients with large deletions had more severe outcomes due to reduced function of the protein (Hantash et al., 2006; Taulan et al., 2007). While large duplications also contributed to morbidity (Costantino et al., 2011; Martins et al., 2019; Paracchini et al., 2008; Petrova et al., 2019). So the CNVs found in our study contributed to CBAVD genetically.
CNVs have occurred in both CF and CBAVD. In the world, there are 2% of large deletions or duplications of CFTR which affect from single exons to entire gene (Taulan et al., 2012). In a study of Brazil population, 3 (1.82%) CNVs (CFTRdup2‐3, CFTRdel25‐26, and CFTRdel25‐27‐CTTNBP2) were identified in 165 CF patients (Martins et al., 2019). In a study of Ecuadorian population, five (3.55%) cases were identified with three deletions (deletion of CFTR exon 10, deletion of CFTR exon 12, CFTRdele22,23) in 141 CF patients using microarrays (Ruiz‐Cabezas et al., 2019). large genomic deletions of CFTR (CFTRdele2 and CFTRdele22_24) was identified in 2 (1.10%) of the 182 samples affected with CBAVD analyzed in a study of European population (Taulan et al., 2007).
Among 263 Chinese men affected with CBAVD in our study, 5 (1.90%) patients were detected for CNVs in the region of exons of the CFTR gene. Refer to previous studies, four (1.80%) large rearrangements (two partial deletions (exons 17a–18 and 22–23), a complete deletion and a partial duplication (exons 11–13)) of CFTR were found in 222 CBAVD patients in a French study using semi‐quantitative fluorescent multiplex PCR (QFM‐PCR) assay (Ratbi et al., 2007). A large deletion of CFTR (deletion of exons 22–24) in one (2.08%) of the 48 CBAVD patients was identified in an American study (Hantash et al., 2006). Detection rate of CNVs basically consistent with previous studies suggested that the use of the MLPA technique in our study was effective for the detection of CNVs. Given to the research mentioned above, the results showed less obvious racial differences in CNVs of CFTR in different CBAVD populations.
However, Yuan et al. in 2019 showed that two CFTR gene mutations in 58.33% (42/72) of Congenital absence of vas deferens (CAVD) patients, only one mutation in 18.06% (13/72) and no mutation in 23.61% (17/72). And they did not find deletions or duplications using MLPA analysis on CFTR gene in 30 CAVD patients (with one/no mutation) (Yuan et al., 2019). In 2020, Wang et al. found 15 CFTR mutations in 15 CBAVD patients among 38 samples (Wang et al., 2020), but they did not detect CNVs. CNV screening was performed on some samples of CAVD in these two studies, but no CNV was found. Our study was conducted in a large sample of cases, but CNV was found only in 1.9% of the samples. As there are less CNV in CBAVD, CNV may not be found in small samples. And to our best knowledge, this study first reported CNVs of CFTR on a large sample size of CBAVD patients in Chinese population.
Our results also showed that the detection of CNVs of CFTR would be significant for genetic counseling in Chinese Han population with CBAVD. Some mutations may exist in patients without CFTR abnormalities using traditional detection methods. Traditional testing also means comprehensive screening, such as F508del and 100‐mutation panels. However 10%–40% of the mutations cannot be identified by common methods (Practice Committee of the American Society for Reproductive Medicine in collaboration with the Society for Male & Urology, 2018). Before the treatment of assisted reproduction, genetic factors of the patients with normal spermatogenesis should be analyzed, so as to provide better reproductive guidance (Meng et al., 2001). But given to relatively low positive rate (1.90% in our study, no CFTR CNVs found in other Chinese studies (Wang et al., 2020; Yuan et al., 2019) and the conditions for detecting CNVs in previous studies (Hantash et al., 2006; Ratbi et al., 2007; Taulan et al., 2007), we recommended that CNV screening should be performed in CBAVD patients with one/no mutation.
It is worth noting that adhesion G‐protein coupled receptor G2 (ADGRG2) gene (OMIM: 300572) was also related to CBAVD with X‐linked inheritance pattern (Patat et al., 2016). Yuan et al. in 2019 identified 2 novel ADGRG2 mutations in 10 CBAVD patients without CFTR mutations (Yuan et al., 2019) and Wang et al. identified one ADGRG2 mutations in 38 CBAVD patients (Wang et al., 2020) in Chinese populations. Also, other genes may also be associated with CBAVD, for example, SLC9A3 (OMIM: 182307) deletion occurred in some Taiwanese CBAVD patients without common CFTR mutations (Wang et al., 2017).
In our study, we found that it was feasible to use the MLPA assay to test the CNVs (large deletions and duplications) of CFTR in our patients. Our results showed that CNVs of CFTR contributed to CBAVD genetically. And 1.90% CBAVD patients carried CNVs of exons of CFTR in Chinese Han populations, which was basically consistent with the previous reports. While this study first revealed genetic risk of CNVs of CFTR on a large sample size of CBAVD patients in Chinese Han population, and prompted that it was necessary to detect CNVs of CFTR in Chinese Han people with CBAVD.
5. CONCLUSIONS
The study showed that 1.90% (5/263) CBAVD patients carried CNVs of exons of CFTR in Chinese Han populations by MLPA, suggesting that the rate of CFTR CNVs in Chinese CBAVD population were basically consistent with the previous reports. And this study first revealed genetic risk of CNVs of CFTR on a large sample size of CBAVD patients in Chinese Han population, which prompted that it was necessary to detect CNVs of CFTR in Chinese Han people with CBAVD.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
All authors contributed to the study conception and design. Material preparation were performed by Chengquan Ma and Tengyan Li, data collection and analysis were performed by Binbin Wang, Hongjun Li, and Jing Wang. The first draft of the manuscript was written by Chengquan Ma and Ruyi Wang, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
ACKNOWLEDGMENT
Thank all the authors for their contributions in this research.
Chengquan Ma and Ruyi Wang contributed equally to this work.
Funding information
This project was supported by the National Natural Science Foundation of China (Grant No. 81671448 and Grant No. 81871152).
Contributor Information
Hongjun Li, Email: lihongjun@pumch.cn.
Binbin Wang, Email: wbbahu@163.com.
REFERENCES
- Cai, H. , Qing, X. , Niringiyumukiza, J. D. , Zhan, X. , Mo, D. , Zhou, Y. , & Shang, X. (2019). CFTR variants and renal abnormalities in males with congenital unilateral absence of the vas deferens (CUAVD): A systematic review and meta‐analysis of observational studies. Genetics in Medicine, 21(4), 826–836. 10.1038/s41436-018-0262-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier‐Porst, F. , & Bozon, D. (2000). Retrieved from www.genet.sickkids.on.ca/MutationDetailPage.external?sp=1106
- Chillón, M. , Casals, T. , Mercier, B. , Bassas, L. , Lissens, W. , Silber, S. , … Estivill, X. (1995). Mutations in the cystic fibrosis gene in patients with congenital absence of the vas deferens. The New England Journal of Medicine, 332(22), 1475–1480. 10.1056/nejm199506013322204 [DOI] [PubMed] [Google Scholar]
- Costantino, L. , Rusconi, D. , Claut, L. , Colombo, C. , Novara, F. , Paracchini, V. , … Seia, M. (2011). A wide methodological approach to identify a large duplication in CFTR gene in a CF patient uncharacterised by sequencing analysis. Journal of Cystic Fibrosis, 10(6), 412–417. 10.1016/j.jcf.2011.06.007 [DOI] [PubMed] [Google Scholar]
- Férec, C. , Casals, T. , Chuzhanova, N. , Macek, M. , Bienvenu, T. , Holubova, A. , … Chen, J.‐M. (2006). Gross genomic rearrangements involving deletions in the CFTR gene: Characterization of six new events from a large cohort of hitherto unidentified cystic fibrosis chromosomes and meta‐analysis of the underlying mechanisms. European Journal of Human Genetics : EJHG, 14(5), 567–576. 10.1038/sj.ejhg.5201590 [DOI] [PubMed] [Google Scholar]
- Hantash, F. M. , Milunsky, A. , Wang, Z. , Anderson, B. , Sun, W. , Anguiano, A. , & Strom, C. M. (2006). A large deletion in the CFTR gene in CBAVD. Genetics in Medicine, 8(2), 93–95. 10.1097/01.gim.0000200945.54234.d7 [DOI] [PubMed] [Google Scholar]
- Hwang, K. , Smith, J. F. , Coward, R. M. , Penzias, A. , Bendikson, K. , Butts, S. , … Vernon, M. (2018). Evaluation of the azoospermic male: A committee opinion. Fertility and Sterility, 109(5), 777–782. 10.1016/j.fertnstert.2018.01.043 [DOI] [PubMed] [Google Scholar]
- Jequier, A. M. , Ansell, I. D. , & Bullimore, N. J. (1985). Congenital absence of the vasa deferentia presenting with infertility. Journal of Andrology, 6(1), 15–19. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/3918979 [PubMed] [Google Scholar]
- Martins, R. D. S. , Campos Junior, M. , dos Santos Moreira, A. , Marques Zembrzuski, V. , da Fonseca, A. C. P. , Abreu, G. D. M. , … de Cabello, G. M. K. (2019). Identification of a novel large deletion and other copy number variations in the CFTR gene in patients with Cystic Fibrosis from a multiethnic population. Molecular Genetics & Genomic Medicine, 7(7), e00645 10.1002/mgg3.645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, M. V. , Black, L. D. , Cha, I. , Ljung, B. M. , Pera, R. A. , & Turek, P. J. (2001). Impaired spermatogenesis in men with congenital absence of the vas deferens. Human Reproduction, 16(3), 529–533. 10.1093/humrep/16.3.529 [DOI] [PubMed] [Google Scholar]
- Morales, M. M. , Carroll, T. P. , Morita, T. , Schwiebert, E. M. , Devuyst, O. , Wilson, P. D. , … Cutting, G. R. (1996). Both the wild type and a functional isoform of CFTR are expressed in kidney. American Journal of Physiology‐Renal Physiology, 270(6), F1038–F1048. 10.1152/ajprenal.1996.270.6.F1038 [DOI] [PubMed] [Google Scholar]
- Paracchini, V. , Seia, M. , Coviello, D. , Porcaro, L. , Costantino, L. , Capasso, P. , … Colombo, C. (2008). Molecular and clinical features associated with CFTR gene rearrangements in Italian population: Identification of a new duplication and recurrent deletions. Clinical Genetics, 73(4), 346–352. 10.1111/j.1399-0004.2007.00957.x [DOI] [PubMed] [Google Scholar]
- Patat, O. , Pagin, A. , Siegfried, A. , Mitchell, V. , Chassaing, N. , Faguer, S. , … Bieth, E. (2016). Truncating mutations in the adhesion G protein‐coupled receptor G2 gene ADGRG2 cause an X‐linked congenital bilateral absence of vas deferens. American Journal of Human Genetics, 99(2), 437–442. 10.1016/j.ajhg.2016.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova, N. V. , Marakhonov, A. V. , Vasilyeva, T. A. , Kashirskaya, N. Y. , Ginter, E. K. , Kutsev, S. I. , & Zinchenko, R. A. (2019). Comprehensive genotyping reveals novel CFTR variants in cystic fibrosis patients from the Russian Federation. Clinical Genetics, 95(3), 444–447. 10.1111/cge.13477 [DOI] [PubMed] [Google Scholar]
- Practice Committee of the American Society for Reproductive Medicine in collaboration with the Society for Male, R., & Urology . (2018). Evaluation of the azoospermic male: A committee opinion. Fertility and Sterility, 109(5), 777–782. 10.1016/j.fertnstert.2018.01.043 [DOI] [PubMed] [Google Scholar]
- Ramos, L. , Gartner, J. G. S. , & Casals, T. (2008). Retrieved from http://www.genet.sickkids.on.ca/MutationDetailPage.external?sp=1664
- Ratbi, I. , Legendre, M. , Niel, F. , Martin, J. , Soufir, J.‐C. , Izard, V. , … Girodon, E. (2007). Detection of cystic fibrosis transmembrane conductance regulator (CFTR) gene rearrangements enriches the mutation spectrum in congenital bilateral absence of the vas deferens and impacts on genetic counselling. Human Reproduction (Oxford, England), 22(5), 1285–1291. 10.1093/humrep/dem024 [DOI] [PubMed] [Google Scholar]
- Riggs, E. R. , Andersen, E. F. , Cherry, A. M. , Kantarci, S. , Kearney, H. , Patel, A. , … Martin, C. L. (2020). Technical standards for the interpretation and reporting of constitutional copy‐number variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genetics in Medicine, 22(2), 245–257. 10.1038/s41436-019-0686-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz‐Cabezas, J. C. , Barros, F. , Sobrino, B. , García, G. , Burgos, R. , Farhat, C. , … Paz‐y‐Miño, C. (2019). Mutational analysis of CFTR in the Ecuadorian population using next‐generation sequencing. Gene, 696, 28–32. 10.1016/j.gene.2019.02.015 [DOI] [PubMed] [Google Scholar]
- Schrijver, I. , Rappahahn, K. , Pique, L. , Kharrazi, M. , & Wong, L.‐J. (2008). Multiplex ligation‐dependent probe amplification identification of whole exon and single nucleotide deletions in the CFTR gene of Hispanic individuals with cystic fibrosis. The Journal of Molecular Diagnostics, 10(4), 368–375. 10.2353/jmoldx.2008.080004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard, D. N. , & Welsh, M. J. (1999). Structure and function of the CFTR chloride channel. Physiological Reviews, 79, S23–S45. 10.1152/physrev.1999.79.1.S23 [DOI] [PubMed] [Google Scholar]
- Taulan, M. , Girardet, A. , Guittard, C. , Altieri, J.‐P. , Templin, C. , Beroud, C. , … Claustres, M. (2007). Large genomic rearrangements in the CFTR gene contribute to CBAVD. BMC Medical Genetics, 8, 22 10.1186/1471-2350-8-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taulan, M. , Viart, V. , Theze, C. , Guittard, C. , Altieri, J.‐P. , Templin, C. , … des Georges, M. (2012). Identification of a novel duplication CFTRdup2 and functional impact of large rearrangements identified in the CFTR gene. Gene, 500(2), 194–198. 10.1016/j.gene.2012.03.043 [DOI] [PubMed] [Google Scholar]
- Wang, H. , An, M. , Liu, Y. , Hu, K. , Jin, Y. , Xu, S. , … Lu, M. (2020). Genetic diagnosis and sperm retrieval outcomes for Chinese patients with congenital bilateral absence of vas deferens. Andrology, 8(5), 1064–1069. 10.1111/andr.12769 [DOI] [PubMed] [Google Scholar]
- Wang, Y. Y. , Lin, Y. H. , Wu, Y. N. , Chen, Y. L. , Lin, Y. C. , Cheng, C. Y. , & Chiang, H. S. (2017). Loss of SLC9A3 decreases CFTR protein and causes obstructed azoospermia in mice. PLoS Genetics, 13(4), e1006715 10.1371/journal.pgen.1006715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, B. , Wang, X. , Zhang, W. , Li, H. , & Wang, B. (2018). Compound heterozygous mutations in CFTR causing CBAVD in Chinese pedigrees. Molecular Genetics & Genomic Medicine, 6(6), 1097–1103. 10.1002/mgg3.486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J. , Chen, Z. , Ni, Y. , & Li, Z. (2012). CFTR mutations in men with congenital bilateral absence of the vas deferens (CBAVD): A systemic review and meta‐analysis. Human Reproduction, 27(1), 25–35. 10.1093/humrep/der377 [DOI] [PubMed] [Google Scholar]
- Yuan, P. , Liang, Z. K. , Liang, H. , Zheng, L. Y. , Li, D. , Li, J. , … Wang, W. J. (2019). Expanding the phenotypic and genetic spectrum of Chinese patients with congenital absence of vas deferens bearing CFTR and ADGRG2 alleles. Andrology, 7(3), 329–340. 10.1111/andr.12592 [DOI] [PubMed] [Google Scholar]
- Zielenski, J. , & Tsui, L.‐C. (1995). Cystic fibrosis: Genotypic and phenotypic variations. Annual Review of Genetics, 29, 777–807. 10.1146/annurev.ge.29.120195.004021 [DOI] [PubMed] [Google Scholar]


