Abstract
The route of transmission of Novel SARS-CoV-2 virus is ambiguous. In this regard we planned a study to find out SARS-CoV-RNA shedding in various clinical samples of 9 COVID-19 positive patients. SARS-CoV-RNA was detected in nasal swab (NS), throat swab (TS) and faecal sample but was not detected in serum and urine samples. We also report that SARS-CoV-2-RNA persisted in faeces for >20 days. Persistence of faecal RNA might impose challenge in infection control and the disease may spread to household contacts if discharged. Perineal cleaning and hygiene may be advised at the time of vaginal delivery.
Keywords: SARS CoV-2, Clinical samples, Clinical features, Faecal RNA persistence, Perineal cleaning
Introduction
The SARS-CoV-2 virus had originated from Wuhan, China, in December 2019 and was declared as Public Health Emergency of International concern by World Health Organization (WHO) [1]. There is currently no drug treatment and vaccine approved for SARS-CoV-2 virus [2]. Although, there is some literatures on presence of SARS-CoV-RNA in pulmonary and extra-pulmonary clinical samples such as blood, urine and faeces, but still the route of transmission of this new SARS-CoV-2 virus is still not clear and more studies are needed to know the mode of spread [[3], [4], [5], [6], [7], [8]]. In this view, we carried out study to determine the SARS-CoV-2 viral shedding in various clinical specimens.
Material and method
Total nine COVID-19 patients admitted in King Georges Medical University (KGMU), Lucknow, Uttar Pradesh (UP), India were enrolled in the study. Written informed consent was taken by participants. The study is cleared institutional ethical committee. Nasal swab (NS), Throat swab (TS), serum, urine and stool specimens were collected from all patients every alternate day up to 4 weeks after admission. The specimens were collected following universal standard precautions and transported to microbiology laboratory at 2–8 °C. All clinical samples were tested for SARS-CoV-2-RNA by RT-PCR. Total viral nucleic acid was extracted from clinical sample using PureLink DNA/RNA mini kit (Invitrogen, USA) as per manufacturer's instructions and processed for real time PCR. Human Rnase P gene was tested in all samples to check the quality of samples and to validate the process of nucleic acid extraction. Real time RT-PCR assays were performed on Quantstudio 7 (Life Technologies, USA) using Superscript III One-Step Real- Time PCR kit (Cat no. 11732088, ThermoFisher Scientific, USA). The real time PCR was performed as per protocol described by Corman et al., 2020. First the sample was tested by 2 stage protocol screening and confirmation. Human Rnase P gene was tested in all samples to check the quality of samples and to validate the process of nucleic acid extraction. SARS-CoV-2-E-gene and Rnase P was used as for screening while SARS-CoV-2- RdRp and ORF1b was used in as the confirmatory assay. Patients' convalescent sample was only tested using SARS-CoV-2-E-gene with sample quality check (Rnase P). Convalescent sample was tested every alternate day up to 4 weeks.
Results
Age of COVID-19 cases ranged from 2.6 yrs to 53 yrs with majority of adult males. Only one child of 2.6yrs was enrolled in the study, rest were adult cases i.e. four males and three females. We did not observe dissimilarity of virus shedding pattern in both the gender category. Although, faecal RNA persisted longer in the child for more than 20 days. SARS-CoV-RNA was detected in nasal swab (NS), throat swab (TS) and fecal sample but was absent in serum and urine samples of COVID-19 cases. The virus persisted for minimum of 9 day to maximum of 24 days in our patients. According to our study, SARS-CoV-2 RNA clearance from nasal and throat convalescent samples was earlier than faecal sample. Throat has the fastest clearance followed by Nasal swab and stool at the end. The fecal RNA was detected for another 3–5 days even though NS and TS sample's RNA were negative as shown in Fig. 1 . Faecal RNA initially had low viral load (high Ct value) but later increased (high viral load; low Ct value) gradually. The observation of NS and TS sample RNA was reverse of stool sample as shown in Fig. 2 .
Fig. 1.
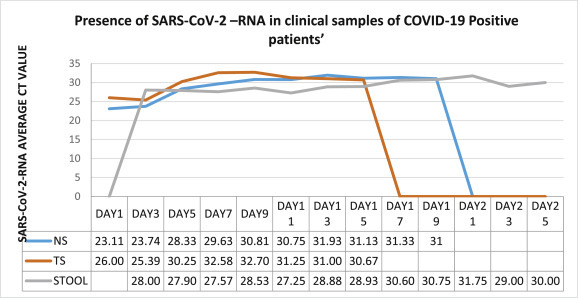
SARS-CoV-2 (represented in form of threshold cycle (CT)) viral load in different clinical samples of COVID-19 positive patients.
Fig. 2.
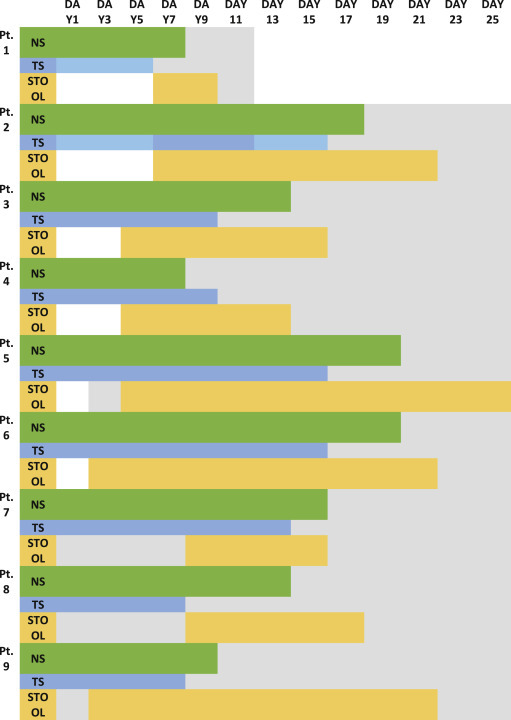
Persistence of SARS-CoV-2 viremia followed over 4 weeks in different clinical samples.
Discussion
We reported SARS-CoV-2 RNA early clearance from nasal and throat convalescent samples on comparison to faecal sample; similar observation was found by Zhang W et al. from China [6]. Another study from China reported SARS-CoV-2 RNA in NS/TS, but they did not found the virus in stool and urine, however they observed persistence of SARS-CoV-2-RNA in nasal swab for longer duration and of higher viremia in comparison to throat swab Cai J et al., 2020 reported similar findings by demonstrating the fecal viral RNA in up to 30% of patients from day 5 after onset and up to 4–5 weeks in moderate cases [4,5]. The finding was conspicuous in one of our pediatric patient (case 8) where NS and TS sample SARS-CoV-2 RNA was only present for 9 days post illness and the faecal RNA persisted for >20 days. A similar finding was documented by Cai J et al., 2020 [4].
It is a matter of concern that COVID-19 patients were discharged from hospital on the basis of symptoms resolution and negative NS/TS swabs samples as their faecal samples are still positive for SARS-CoV-2 RNA. Although, the viability of SARS-CoV-2 virus in faecal samples yet to be explored on viral culture; therefore the significance of oral-faecal route remains undetermined. To add further, sewage water stability of this SARS-CoV-2 enveloped virus is still a query that needs to be studied.
In view of current data of faecal shedding of SARS-CoV-2, discharged patients could be advised to follow all sanitation and personal hygiene practices along with regular cleaning and decontamination of toilet and fomites in order to protect household contacts Strict hand hygiene practices should be followed. This applies to all convalescing patients, but particularly to convalescent children. In the vaginal deliveries, perineal hygiene and care should be advised [9].
Author contribution
Shantanu Prakash: Concepts, Design, Definition of intellectual content, Literature searchm, Clinical studies, Experimental studies, Data acquisition, Manuscript preparation, Manuscript editing, Manuscript review. Suruchi Shukla: Concepts, Design, Definition of intellectual content, Literature search, Clinical studies, Experimental studies, Manuscript preparation, Manuscript editing, Manuscript review. Hricha Mishra: Experimental studies, Data analysis, Statistical analysis. Om Prakash: Experimental studies, Manuscript preparation. Danish N Khan: Experimental studies, Data analysis Statistical analysis. Ajay Pandey: Experimental studies. D. Himanshu Reddy: Definition of intellectual content, Experimental studies, Manuscript editing, Manuscript review. Amita Jain: , Design, Definition of intellectual content, Literature search, Clinical studies, Experimental studies, Data acquisition, Manuscript preparation, Manuscript editing, Manuscript review, Guarantor.
Declaration of competing interest
None.
Acknowledgement & Funding
The financial support received from the Indian Council of Medical Research (ICMR), New Delhi Grant 83rd ECM IIA/P9 is acknowledged.
References
- 1.WHO coronavirus disease (COVID-19) dashboard. https://covid19.who.int/
- 2.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020 Jan;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C., Zimmer T., Thiel V., Janke C., Guggemos W., Seilmaier M., Drosten C., Vollmar P., Zwirglmaier K., Zange S., Wölfel R., Hoelscher M. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020 Mar 5;382(10):970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai J., Xu J., Lin D., Yang Z., Xu L. A Case Series of children with 2019 novel coronavirus infection: clinical and epidemiological features. Clin Infect Dis. 2020 Sep 12;71(6):1547–1551. doi: 10.1093/cid/ciaa198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., Guo Q., Song T., He J., Yen H.L., Peiris M., Wu J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020 Mar 19;382(12):1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang W., Du R.H., Li B., Zheng X.S., Yang X.L., Hu B., Wang Y.Y., Xiao G.F., Yan B., Shi Z.L., Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microb Infect. 2020 Feb 17;9(1):386–389. doi: 10.1080/22221751.2020.1729071. PMID: 32065057; PMCID: PMC7048229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lescure F.X., Bouadma L., Nguyen D., Parisey M., Wicky P.H., Behillil S., Gaymard A., Bouscambert-Duchamp M., Donati F., Le Hingrat Q., Enouf V., Houhou-Fidouh N., Valette M., Mailles A., Lucet J.C., Mentre F., Duval X., Descamps D., Malvy D., Timsit J.F., Lina B., van-der-Werf S., Yazdanpanah Y. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect Dis. 2020 Jun;20(6):697–706. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020 Apr;20(4):411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parasa S., Desai M., Thoguluva Chandrasekar V. Prevalence of gastrointestinal symptoms and fecal viral shedding in patients with coronavirus disease 2019: a systematic review and meta-analysis. JAMA Netw Open. 2020;3(6) doi: 10.1001/jamanetworkopen.2020.11335. [DOI] [PMC free article] [PubMed] [Google Scholar]


