Abstract
CK2 is a ubiquitous Ser/Thr protein kinase involved in the control of various signaling pathways and is known to be constitutively active. In the present study, we identified aryl 2-aminothiazoles as a novel class of CK2 inhibitors, which displayed a non ATP-competitive mode of action and stabilized an inactive conformation of CK2 in solution. Enzyme kinetics studies, STD-NMR, circular dichroism spectroscopy and native mass spectrometry experiments demonstrated that the compounds bind in an allosteric pocket outside the ATP-binding site. Our data, combined with molecular docking studies, strongly suggested that this new binding site was located at the interface between the αC helix and the flexible glycine-rich loop. A first hit optimization led to compound 7, exhibiting an IC50 of 3.4 μM against purified CK2α in combination with a favorable selectivity profile. Thus, we identified a novel class of CK2 inhibitors targeting an allosteric pocket, offering great potential for further optimization into anti-cancer drugs.
Keywords: Kinase inhibitors, protein kinase CK2, enzymatic studies, allosteric modulators, protein NMR
INTRODUCTION
A large fraction of the cellular phosphoproteome relies on the activity of protein kinase CK2,1 a heterotetrameric serine/threonine kinase, formed of α catalytic and β regulatory subunits and involved in the activation of several pro-oncogenic pathways that are critical for cell proliferation, differentiation and survival.2,3 As CK2 is implicated in cancerous transformation, its inhibition proved to be an effective method to induce tumor regression, and the ATP-competitive compound CX-49454 (5-(3-chlorophenylamino)benzo[c][2,6]naphthyridine-8-carboxylic acid, silmitasertib, IC50 = 14 nM) is currently in phase II clinical trials for the treatment of cholangiocarcinoma.5 However, CX-4945 was recently reported to be more potent against several additional kinases such as Clk (Cdc-like kinase) 2 (IC50 = 4 nM),6 as well as Dyrk1A and Dyrk1B (IC50s = 6.8 and 6.4 nM, respectively),7 kinases involved in the regulation of alternative splicing (Clk2 and Dyrk1A), NFAT signaling and neuronal functions (Dyrk1A), and myogenesis (Dyrk1B). This underlines the fact that potent and selective CK2α inhibitors are difficult to obtain through targeting the ATP binding pocket. Therefore, research efforts currently strive to identify inhibitors that exploit other regions of the protein to suppress the catalytic activity of the kinase.8 Only few CK2α inhibitors with a non-ATP competitive mode of action were hitherto reported; among them were azonaphthalene dye–derived compounds; however, the binding site and potential mechanism of inhibition remained elusive.9 Besides, inorganic PolyOxoMetalate complexes (POMs), e.g. [P2Mo18O62]6–, were described as potent allosteric inhibitors of CK2α, exhibiting nanomolar IC50s and great selectivity in a panel of 29 kinases.10 However, POMs lack cellular activity and there is little perspective to develop them further as in vivo active agents.
A small alternative binding pocket, termed αD pocket, was recently identified by Spring and co-workers, located below the ATP binding pocket on the large lobe of the catalytic subunit.11 In their follow-up study, an IC50 of 7 μM against CK2α was reached with the most optimized ligand CAM4712, however, the selectivity over other kinases had dropped during hit optimization.12
In the present study, we describe the identification of a novel active class of non ATP-competitive CK2 inhibitors. Using different methodologies, we provide evidence that the compounds target an alternative binding pocket, distinct from the ATP binding site and from the αD pocket.
RESULTS AND DISCUSSION
Hit identification and analysis of the inhibition mode.
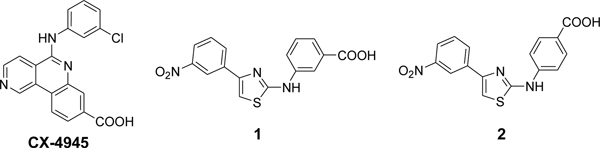
A Virtual Ligand Screening (VLS) campaign13 targeting the α/β interface of CK2 was performed with 2·106 compounds. The first hundred, highest-ranked molecules were evaluated for their inhibitory activity against CK2α using a standard radioactive assay.10,14 Originally designed as a potential protein–protein interaction inhibitor, compound 1 (Chart 1), possessing a 2-aminothiazole scaffold, was identified as the most active hit exhibiting an IC50 value of 27.7 μM for CKα.
Chart 1.
Chemical structures of CX-4945 (silmitasertib) and hit compounds 1,2
To define the biochemical mechanism of action of compound 1, we examined the effects of increasing concentrations of ATP or peptide substrate, and the presence of CK2β on the inhibitory activity of the compound. We found that CK2 inhibition by 1 was independent of the concentrations of both ATP (Figure 1A) and peptide substrate (Figure 1B). More surprisingly, both CK2α and the CK2α2β2 holoenzyme were inhibited to the same extent by 1 (Figure 1C), indicating that the inhibition pattern was also independent of the presence of CK2β. This last result clearly indicated that the binding site of hit compound 1 was not located in the CK2 α/β subunit interface. We then searched for commercially available analogs of 1 and purchased the derivative 2 (Chart 1), having a para acid substitution; it showed an improved IC50 value of 7.0 μM for CK2α.
Figure 1.
Analysis of the inhibition mode of compound 1. A. Effects of increasing ATP concentrations on the inhibition of CK2α activity by compound 1. CK2α (20 ng) was incubated with or without 15 μM of 1 after which its activity was assayed with 200 μM of CK2β–independent peptide substrate in the presence of increasing ATP concentrations. B. Effects of increasing peptide substrate concentrations on the inhibition of CK2 activity by 1. CK2α (20 ng) was incubated with (o open circles) or without (• filled circles) 15 μM of 1 after which its activity was assayed with 100 μM ATP in the presence of increasing concentrations of CK2β–independent peptide substrate. C. Effects of the presence of CK2β on the enzymatic activity. CK2α (20 ng) (• filled circles) or CK2α2β2 (40 ng) (o open circles) were incubated in the presence of increasing concentrations of compound 1 after which CK2 activity was assayed with a CK2β–independent peptide substrate
Lineweaver-Burk inhibition plots indicated that in the presence of a saturating peptide substrate concentration (600 μM), compound 2 could bind to either the CK2–peptide substrate complex or the CK2–ATP–peptide complex showing a mixed-type mechanism of inhibition with respect to ATP (Figure 2A).15 This indicated that compound 2 was not an ATP site–directed inhibitor. Again, the inhibitory potency of compound 2 was similar toward the monomeric CK2α or the CK2α2β2 holoenzyme (Figure 2B), and was barely affected by increasing concentrations of CK2β (Figure 2C). Collectively, the absence of competition by ATP or the CK2β subunit strongly suggested that 2 bound outside the CK2α/CK2β interface and did not compete with ATP, at least not directly; both a type II-like inhibition mode or binding outside the ATP-binding pocket were in agreement with the data obtained this far.
Figure 2.
Analysis of the inhibition mode of compound 2. A. Lineweaver-Burk inhibition plot of human recombinant CK2α by compound 2 at various ATP concentrations. The Ki was determined by plotting the slopes at varying inhibitor concentrations. B. CK2α and CK2α2β2 activity in the presence of increasing concentrations of compound 2. C. Effect of increasing CK2β concentrations on the inhibition of a fixed amounts of CK2α (150 nM) by compound 2 (20 μM).
Hit compound 2 is highly selective.
To further evaluate compound 2, a selectivity profiling against 44 human kinases was performed (Table 1). At 50 μM of 2, which led to a full inhibition of CK2α, only three other kinases from the panel showed a reduction of activity by more than 50 %: EGFR (74 % inhibition), EphA4 (55 %) and pim-1 (54 %). This remarkable selectivity of the non-optimized hit compound argued for an interaction with a non-conserved binding site.
Table 1.
Residual kinase activity in the presence of compound 2 at 50 μM.a
| Protein kinase | Residual activity (%) | Protein kinase | Residual activity (%) | Protein kinase | Residual activity (%) | Protein kinase | Residual activity (%) |
|---|---|---|---|---|---|---|---|
| CK2α | 0 | HIPK2 | 83 | IR, activated | 93 | CaMK1 | 101 |
| EGFR | 26 | MAPK1 | 84 | CDK6/cyclinD3 | 95 | CDK2/cyclinE | 101 |
| EphA4 | 45 | LKB1 | 85 | cSRC | 97 | Met | 102 |
| pim-1 | 46 | PKBα | 85 | FGFR1 | 97 | PAK2 | 102 |
| MUSK | 62 | CDK2/cyclinA | 86 | Fyn | 97 | IKKα | 103 |
| PDGFRα | 65 | PKCα | 86 | HIPK1 | 97 | MEK1 | 103 |
| AMPKα1 | 71 | cKit | 87 | HIPK3 | 98 | Abl | 104 |
| Aurora-B | 74 | PKA | 87 | CDK9/cyclinT1 | 99 | CDK7/cyclinH | 104 |
| Dyrk2 | 78 | CDK1/cyclinB | 89 | Rsk1 | 99 | JAK2 | 104 |
| Aurora-A | 79 | CHK2 | 89 | ROCK-1 | 100 | Lyn | 104 |
| GSK3β | 79 | c-RAF | 92 | Yes | 100 | mTOR | 104 |
Results were obtained with ATP concentration adjusted to the respective Km of each kinase. Duplicate values did not differ by more than 15% in all assays.
Preliminary hit optimization.
Derivatives of compound 2 were synthesized (Scheme 1) to establish preliminary structure–activity–relationships (SAR) (Table 2). The presence of a carboxylic acid function turned out to be important, as it could not be removed (compound 3) or replaced by the methyl ester (4) without significant loss of activity, suggesting that the binding could involve ionic interactions with a basic residue of the protein. The presence of an aryl group at position 4 of the aminothiazole core was found to be crucial, as indicated by the complete lack of activity of the truncated analog 5. While modulation of the electron density of this aromatic ring (compounds 2 and 8) did not substantially change the inhibitory activity compared with that of the unsubstituted congener 6, increasing the lipophilicity by a bromo-substituent significantly enhanced the affinity to reach the sub-micromolar range (Ki = 0.7 μM, compound 7, Table 2). Thus, the preliminary SAR obtained with the 4-phenyl substitutions suggested that binding of this moiety to CK2α was driven by hydrophobic interaction with a lipophilic pocket.
Scheme 1.
Synthesis of aminothiazole derivatives 3-9.a
aReagents and conditions: (a) CS2, Et3N in THF/H2O 1/1, RT, 24h, then I2 in THF, 0°C to RT, 3h. (b) NH4OH 30%, RT, 6h. (c) EtOH, reflux, 3–12h. (d) Cat. H2SO4, MeOH, reflux, 2h (R2 = COOH and R1 = m-nitrophenyl).
Table 2.
Chemical structures and CK2α inhibitory activities of compounds 1–9.
| Compound | CK2α inhibitory activitya |
|---|---|
 |
IC50 = 27.7 μM |
 |
IC50 = 7.0 μM Kib = 1.6 μM |
 |
27 (±10) % inh. at 10 μM |
 |
9 (±7) % inh. at 10 μM |
 |
0 (±6) % inh. at 10 μM |
 |
IC50 = 9.0 μM Kib = 1.1 μM |
 |
IC50 = 3.4 μM Kib = 0.7 μM |
 |
IC50 = 12.0 μM |
 |
IC50 = 14.0 μM |
IC50 values are the mean of ≥2 experiments (SD < 20 %).
Ki values were determined from graphical plotting of the Lineweaver-Burk plot slopes.
Selectivity and non-ATP competitive behavior of the optimized inhibitor 7.
The selectivity of the most potent compound 7, which retained a mixed-type mechanism of inhibition toward ATP (Figure 3), was evaluated against a panel of 32 selected kinases, specifically chosen from closely related kinases of the CMGC family but also including kinases from other branches of the kinome (Table 3). Compound 7 was screened at a concentration of 50 μM, which is approx. 15 times the IC50 value for CK2α. Under these conditions, six other kinases were inhibited by more than 60%: EphA4 (73%), GSK3β (71%), CK1γ1 (69%), ACVR1 (66%), Clk4 (61%) and MLCK (61%). Notably, Clk2 and Dyrk1A, the main off-targets of the ATP-competitive compound CX-4945, were only weakly inhibited (by approximately 30%).
Figure 3.
Lineweaver-Burk inhibition plot of human recombinant CK2α by compound 7 at various concentrations (μM). Ki was determined by plotting the slopes at varying inhibitor concentrations.
Table 3.
Residual kinase activity in the presence of compound 7 at 50 μM.a
| Protein kinase | Residual activity (%) | Protein kinase | Residual activity (%) | Protein kinase | Residual activity (%) |
|---|---|---|---|---|---|
| EphA4 | 27 | Clk2 | 67 | PKCα | 92 |
| GSK3β | 29 | Abl | 69 | MEK1 | 95 |
| CK1γ1 | 31 | Chk2 | 72 | JAK2 | 98 |
| ACVR1 (ALK2) | 34 | TBK1 | 75 | MKNK1 (MNK1) | 101 |
| Clk4 | 39 | CDK1/cyclinB | 78 | PDGFRα | 102 |
| MLCK (MLCK2) | 39 | Dyrk1A | 79 | DAPK3 | 104 |
| STK17A (DRAK1) | 48 | Clk1 | 80 | EGFR | 106 |
| CK2ά | 48 | HIPK3 | 82 | CDK2/cyclinE | 107 |
| GSG2 (Haspin) | 55 | Clk3 | 86 | ROCK1 | 107 |
| pim1 | 57 | RIPK2 | 86 | cKit | 116 |
| HIPK2 | 57 | MuSK | 90 |
Results were obtained with ATP concentration adjusted to the respective Km of each kinase. Duplicate values did not differ by more than 17% in all assays.
The novel inhibitors decrease the thermal stability of CK2α.
To further investigate the non-ATP competitive binding mechanism, the thermal stability of CK2α in the presence of our compounds was compared to its thermal stability in the presence of CX-4945 using circular dichroism (CD) spectroscopy, which allows monitoring the transition to the unfolded state. For these experiments, compounds 6 and 9 were chosen owing to their superior water solubility. An enhancement of the thermal stability was consistently reported for ATP-competitive ligands.16,17 In the presence of compounds 6 or 9, however, we observed a clear reduction of the melting temperature Tm (Table 4, entries 2 and 3), compared with the sole enzyme (Table 4, entry 1). In contrast, the ATP-competitive reference compound CX-4945 stabilized the protein structure, resulting in an increased Tm (Table 4, entry 4). These results added to the evidence that our 2-aminothiazole derivatives did not target the ATP-binding pocket.
Table 4.
Unfolding temperatures of CK2α in the presence of compound 6, 9 or CX-4945, as monitored by thermal denaturation of protein by circular dichroism (TdCD).
| Entry | Sample | Tm (°C) |
|---|---|---|
| 1 | CK2α | 53.2 (± 0.3) |
| 2 | CK2α + 6 (50 μM) | 50.3 (± 0.3) |
| 3 | CK2α + 9 (6.25 μMa) | 52.0 (± 0.2) |
| 4 | CK2α + CX-4945 (6.25 μMa) | 55.4 (± 0.2) |
The midpoint unfolding temperatures (Tm) are averages of three independent experiments (± SD). CK2α concentration was 5 μM.
Concentrations were limited by the compound solubility in methanol.
Experimental evidence that the novel compounds bind outside the ATP binding pocket.
To elucidate the binding region of this series, NMR Saturation Transfer Difference18 (STD) competition experiments were performed between our 2-aminothiazole ligands and the ATP-competitive ligands CX-4945 or AMP-PNP using CK2α protein. Again, compounds 6 and 9 were chosen because of their relatively good solubility in water. Figure 4 shows the STD spectrum of compound 6 bound to CK2α. Upon addition of CX-4945, simultaneous binding of both compounds 6 and CX-4945 was observed (Figure 4A). The absence of competition between these compounds indicated that compound 6 did not bind to the same site as CX-4945. By comparison, the competition between CX-4945 and AMP-PNP was observed in STD experiments that were recorded under similar experimental conditions (Supplementary Figure S1). AMP-PNP occupies a larger space than CX-4945, due its more extended molecular structure. While the binding of AMP-PNP to CK2α was observed with the apo enzyme (Figure 4B), AMP-PNP was not able to bind to CK2α in the presence of 2-aminothiazole 6 (Figure 4B). Identical results were observed for compound 9, as illustrated by Figure 5A. Altogether, our results proved that compounds 6 and 9 did not bind in the region of the ATP binding pocket of CK2α where CX-4945 was shown to interact with in cocrystal structures (PDB entries 3PE119 and 3NGA20). The fact that an apparent competition was observed between the binding of 6 or 9 and AMP-PNP can be interpreted in two ways: either a direct steric clash occurs between the compound and AMP-PNP, or the compound triggers an allosteric change which leads to a partial occlusion of the ATP binding site, e. g. where the terminal phosphate binds, while sparing all areas engaged in CX-4945 binding (these possibilities are investigated further below, cf. section “Binding mode prediction by molecular docking”).
Figure 4.
STD–based evidence for the simultaneous binding of compound 6 and CX-4945 to CK2α, epitope mapping for compound 6. A. 1D and STD NMR experiments of compound 6 (500 μM) recorded alone and in the presence of the kinase inhibitor CX-4945 (100 μM) with 5 μM CK2α; NMR peaks for CX-4945 are labeled by crosses (x). B. 1D and STD NMR experiments of the ATP-binding site inhibitor AMP-PNP (500 μM) recorded alone and in the presence of compound 6 (500 μM) with 5 μM CK2α. C. STD factors measured for compound 6 upon binding to CK2α. D. Representation of the STD factors with a color scale on the chemical structure of 6.
Figure 5.
STD experiments for compound 9 bound to CK2α. 1D and STD NMR experiments of the ATP-binding site inhibitor AMP-PNP (500 μM) recorded alone and in the presence of compound 9 (500 μM) with 5 μM CK2α.
STD experiments can also discriminate parts of the ligand that are buried in the protein structure upon binding from parts of the compound exposed to the solvent, through the so-called STD–based epitope mapping. STD factors were calculated for the protons of compound 6 (Figure 4C). These data indicate that the benzoic acid moiety of 6 was exposed to the solvent, while the proton of the thiazole ring and the para protons of the phenyl ring were buried within the protein (Figure 4D).
Mapping of the binding site by alanine scan mutational analysis.
Our cocrystallization attempts aiming at elucidating the non-ATP competitive binding mode of 2-aminothiazole derivatives with CK2α were not successful. Some crystallization issues were already noticed for non-ATP competitive CK2 inhibitors.9 Therefore, we employed alternative techniques to complement the results from the NMR competition experiments.
In the literature, crystallographic studies21,22,23 and long timescale molecular dynamics simulations24 suggested that the glycine-rich loop is a highly flexible part of CK2α. Indeed, in most CK2α crystal structures, the glycine-rich loop conformation is maintained by two hydrogen bonds linking the Tyr50 of this loop and two lysine residues (Lys74, Lys77) of the basic cluster. Nonetheless, two crystal structures (PDB entries: 3JUH25 and 3FWQ26) were identified, in which the key hydrogen bonds fixing Tyr50 are lacking. Further examination of these structures revealed a hydrophobic pocket between the glycine-rich loop and the αC-helix, delimited by residues Tyr50, Lys71, Val73, Lys74 and Lys77 (Figure 6). We hypothesized that this hydrophobic pocket, in combination with the adjacent lysine residues, could be an allosteric binding site for our aminothiazole inhibitors. Our findings clearly proved that the pocket targeted by our compounds was distinct from the so-called αD pocket, which was located near the αD-helix and delimited by Val162, Asn119, Pro159, Met221, Met137, Ile133, Ile128, Ile124 residues.11
Figure 6.
A. Superposition of four CK2α structures (3PE1, magenta; 3FWQ, blue; 3JUH, cyan; 3Q9Z29; green). The ATP-binding site is occupied by CX-4945 in 3PE1 and by AMP-PNP in 3JUH and 3Q9Z (not shown for clarity). B. A separated view from front or top of each structure is depicted. The pockets were identified using ICM pocket finder and represented with a mesh surface. The putative allosteric pocket is partially available in 3PE1, fully accessible in 3FWQ and 3JUH but completely absent in the conformation crystallized in 3Q9Z.
Interestingly, the basic lysine cluster was addressed by small molecules before, being the target of the acidic part (aspartic or glutamic tri/tetrapeptides) of TBI-derived bisubstrate inhibitors, which bound to both the ATP site and this basic environment.27,28
To investigate more deeply this protein region, nine GST-CK2α single-point alanine mutants were tested in enzymatic assays in the presence of compound 7 (Figure 7 and Supplementary Table S1). Three mutants significantly less sensitive to inhibition by 7 (Lys74Ala, Lys77Ala and His160Ala) were further tested in dose–response experiments. Compared with wild type CK2α, the IC50 values of compound 7 were increased for the Lys74Ala (1.8-fold) and Lys77Ala (3.2-fold) mutants, even considering that the remaining lysine residue 76 (located nearby) might compensate in part for the loss of electrostatic interactions, thus attenuating the drop in binding affinity. Rather unexpectedly, the His160Ala mutant also showed an increased IC50 (2.2 fold) (Figure 7).
Figure 7.
Percentage of inhibition of GST-CK2α wild-type and single alanine mutants in the presence of compound 7 (4 μM). IC50s were determined for GST-CK2α wild-type and mutants with reduced sensitivity toward inhibition (S.D. < 20%). The fold increase in the respective IC50 over that of the wild-type is indicated at the bottom. nd: not determined.
Although His160 is not in close proximity to the putative allosteric pocket, it displayed a direct interaction with the P-loop tip residueTyr50 in both the crystallized inactive states of CK2α, 3FWQ and 4UB7.23 This might indicate that at least in some inactive conformational states, His160 helps to stabilize the position of Tyr50 and hence of the entire glycine-rich loop, thus opening the gate to the allosteric pocket. In contrast, in the active conformation of CK2α, the position of Tyr50 is rather stabilized by H-bonding or cation–π interactions with lysines from the basic cluster, as can be observed e. g. in the crystal structures of PBD entries 3PE1, 3OWJ30 and 3Q9Z. In light of these considerations, the mutation of His160 might diminish the population of inactive conformational states in the equilibrium that can be recognized by our allosteric ligand.
Confirmation of the alternative binding site by competition experiments followed by native mass spectrometry-based detection.
To complete the binding site mapping based on the single alanine mutants scan, we were looking for probe compounds known to bind in the region of the assumed allosteric pocket. To this end, we selected inositol hexakisphosphate (also named IP6), which was reported as a CK2α ligand binding to the named basic cluster, as demonstrated by X-ray studies (Figure 8A).31 Native mass spectrometry was employed as a fast, label-free, and highly accurate biophysical technique to characterize the binding properties of 6 to CK2α. Native mass spectra of CK2α1–335 (deletion mutant construct used in native MS experiments; henceforth referred to as CK2α) were acquired using 10 μM protein in 0.50 M ammonium acetate pH 8.5 and 5% (v/v) DMSO under non-denaturing conditions by nano-electrospray ionization-mass spectrometry (nESI-MS) that preserve non-covalent interactions. CK2α itself produced a well-resolved charge state series, with ions possessing low charges, signifying the presence of native-like folded structures (Figure 8B). CK2α produced three charge states spanning 9+ to 11+ in the m/z range of 3,600–4,500, with the main charge state being 10+. The observed mass (39,879 ± 3 Da) and calculated mass (39,871 Da) from the protein sequence of the CK2α construct are in good agreement, and establish the oligomeric state of CK2α as a monomer, consistent with published structural data (Figure 8B).31 The binding of IP6 to CK2α was evaluated by incubating 10 μM CK2α with 100 μM IP6. Native mass spectrometry showed that CK2α binds IP6 with 1:1 and 1:2 stoichiometry, with 1:1 binding being the predominant stoichiometry, and the 1:2 stoichiometry probably arising from non-specific binding (Figure 8C).32 CK2α binds to 6 with various stoichiometries, possibly reflecting the presence of multiple ligand-binding sites, or non-specific binding (Supplementary Figure S2).28b,32 In the presence of CX-4945, the observation of the CK2α-CX-4945-6 ternary complex peak indicated that 6 binds to CK2α at a site apart from the ATP site (Supplementary Figure S2), thus corroborating the results obtained with NMR STD experiments. To verify our hypothesis that a part of 6 binds to the same basic pocket as IP6, we performed binding competition experiments between CK2α inhibitor 6 and IP6 using native mass spectrometry for detection. In the presence of 6, we observed the dissociation of the CK2α-IP6 complex, while the CK2α−6 complex was formed, as indicated by the disappearance and appearance of the complex peaks, respectively (Figure 8C). In addition, no CK2α-IP6-6 ternary complex peak was found, altogether suggesting that 6 is a competitive ligand of IP6 and binds to the same basic pocket region also including Lys74 and Lys77 (Figure 8A).
Figure 8.
MS–based evidence for competitive binding between IP6 and compound 6 in CK2α. A. Crystal structure of CK2α in complex with IP6 (PDB file 3W8L31), with an inset showing interactions with amino acid residues Lys74, Lys77, Asn189 and Arg191. Hydrogen bonding interactions are indicated with blue dashed lines. B. Native mass spectra of monomeric CK2α (10 μM), showing three charge states, in the presence of 5% (v/v) DMSO. C. Competitive effect of 6 (10 μM) on the CK2α-IP6 interaction (10 μM CK2α, 100 μM IP6), with an inset showing a magnification of the 10+ charge state. Addition of 6 caused the dissociation of the CK2α-IP6 complex to yield only CK2α-6 complexes. All charge states are colored and indicated with symbols. The charge states of each species are indicated in the spectra.
Binding mode prediction by molecular docking.
Guided by the above experimental data as a whole, we performed molecular modeling studies to gain insight in a potential binding mode. On the basis of the results from the STD NMR competition experiments, compound 6 was docked simultaneously with either CX-4945 or AMP-PNP into the CK2α structures where the presumed allosteric pocket was most accessible for a small ligand, in PDB entries 3JUH (chain A) and 3FWQ (chain B) (Figure 9). With both PDB coordinates, similar binding poses were predicted for 6, which bound in an allosteric pocket adjacent to the ATP binding site, located between the αC-helix and the glycine–rich loop (cf. Figure 9B). In all cases, basic residues from the αC-helix (Lys74, Lys77 and Arg80) and the activation segment (Arg191) were found to interact with the carboxyl–substituted phenyl ring of 6 via a network of favorable hydrogen bonds and salt bridges or cation–π interactions. Altogether, the depicted binding modes in Figure 9 were fully consistent with the SAR, the pattern of exposed/buried protons as determined by STD-NMR experiments (cf. Figure 4C), the alanine scan mutational analysis (Figure 7), and the competition experiments between IP6 and 6 (Figure 8C). In further agreement with the STD-NMR experiments, the docking simulations indicated that 6 and CX4945 can bind simultaneously to both CK2α structures without mutual interference (Figures 9A, C). In PDB entry 3JUH, CK2α was cocrystallized with AMP-PNP; using this cocrystal structure, 6 was docked in close proximity to the ATP analogue in some poses, but not in all. Figure 9B depicts the binding pose with the closest distance between the two ligands. As indicated by the van der Waals volumes (cyan and orange surfaces of AMP-PNP and 6, respectively, Figure 9B), no significant steric clash with the ATP analogue occurred. The closest distance between the terminal phosphate of AMP-PNP and the benzene ring was 3.57 Å, which is above the sum of the van der Waals radii, hence this binding model rather suggested that simultaneous binding would be possible, conflicting with the rigorous displacement of the AMP-PNP by 6 and 9 that was observed in the STD-NMR experiments (Figures 4B and 5). In addition, it is hardly conceivable that binding of a small ligand to an active CK2α conformation such as in 3JUH would induce a full inhibition of the catalytic activity, given that the ligand does neither compete with ATP binding nor with substrate binding (cf. Figure 1). Contrastingly, in the inactive CK2α structure in 3FWQ, Arg47 and Tyr50 from the glycine–rich loop collapse in the space normally accommodating the α- and γ-phosphate moieties of ATP26 (cf. Figure 9D). Hence, the remaining space is sufficient for the binding of CX-4945 (Figure 9C) but not for AMP-PNP or ATP (Figure 9D). Although is unlikely that one of the binding poses obtained with the 3FWQ or 3JUH coordinates reflects the exact binding mode of our inhibitors, the binding model involving 3FWQ provided a better explanation for the STD-NMR competition experiments of 6 and 9 with AMP-PNP (cf. Figures 4B and 5), as the stabilization of such a CK2α conformation upon binding of an effector would strongly diminish the affinity of ATP for its binding pocket.
Figure 9.
Prediction of the binding mode of 6 in CK2α. To derive a potential binding mode consistent with the STD-NMR experimental results (cf. Figures 4 and 5), molecular docking of 6 was performed in the presence of either CX-4945 or AMP-PNP using MOE. Two different CK2α structures were employed, which showed an accessible alternative binding pocket near the region identified through site directed mutagenesis (Figure 7) and binding competition experiments with IP6 (Figure 8). PDB entry 3JUH (chain A) was used in A and B, and 3FWQ (chain B) in C and D. Relevant residues and structural elements are labeled. In addition, transparent van der Waals surfaces are shown in A and B for AMP-PNP (cyan, in the ATP binding pocket) and for 6 (orange, in the allosteric pocket). In C, the partially occluded ATP binding pocket (with bound CX-4945) and the allosteric pocket (with bound 6) are outlined by a gold surface. In D, the van der Waals surface of AMP-PNP is indicated by a mesh; the molecule was placed in ATP pocket region of 3FWQ following to a superimposition with the CK2α/AMP-PNP cocrystal of 3JUH. The steric clashes with the glycine-rich loop residues Arg47 and Tyr50 are indicated by red circles. In A–D, H-bonds are indicated by blue dashed lines, and CH–π interactions by brown dashed lines, respectively. The grey tubular element denotes the smoothened backbone trace of the glycine–rich loop and adjacent parts of beta sheet β3.
Conclusions
To date, few studies described alternative strategies for the pharmacological inhibition of CK2 through exploiting binding sites outside the ATP pocket8–11,27 and even fewer presented drug-like small molecules.12 Originally designed as protein–protein interaction inhibitors, the aminothiazole derivatives described in this paper exhibited an original allosteric mode of inhibition toward protein kinase CK2. Using a comprehensive set of experiments, including enzyme kinetics studies, STD NMR, circular dichroism studies, alanine scan analysis and competition experiments monitored by native mass spectrometry, we could prove that the novel compounds target an alternative binding pocket outside the ATP site; this novel binding site could be unequivocally mapped to a region between the glycine-rich loop and the αC-helix of CK2α, which was – to the best of our knowledge – not reported previously to be targeted by small molecule inhibitors.
In light of the favorable selectivity profile – even prior to any optimization – and the still small molecule size, we anticipate that the compounds presented herein can be developed further into selective and drug-like leads.
EXPERIMENTAL SECTION
Chemistry.
3-((4-(3-nitrophenyl)thiazol-2-yl)amino)benzoic acid (1) and 4-((4-(3-nitrophenyl)thiazol-2-yl)amino)benzoic acid (2) were purchased from Sigma-Aldrich. All commercially available chemicals (phenylthiourea, 2-bromo-3’-nitro-acetophenone, 1,2-dichloro-1-ethoxyethane, 2-bromoacetophenone, 2-bromo-1-(3-bromophenyl)ethanone), 2-bromo-1-(3-methoxyphenyl) ethanone, 2-bromo-1-(pyridin-2-yl)ethanone hydrobromide and solvents were purchased from Sigma-Aldrich, Acros Organics, Fischer scientific or Alfa Aesar and were used without further purification. Reactions were monitored by Thin Layer Chromatography (TLC Silica gel 60 F254) purchased from Merck and observed under UV light (254 nm). Purification by semi-preparative HPLC was carried out on an Agilent 1200 series HPLC system (Agilent Technologies) using an Agilent C18 column (30 × 100 mm/10 μm) as stationary phase and a gradient of water and acetonitrile as eluent. 1H and 13C NMR spectra were obtained in dimethylsulphoxide-d6 on a Bruker DRX-500 NMR spectrometer, operating at 500 MHz for the 1H and at 125 MHz for the 13C at 300K, using residual signal of deuterated NMR solvent as internal reference.33 Chemical shifts are reported in parts per million (ppm), multiplicity of the signals are indicated by lower-case letters (singlet s, doublet d, triplet t, quadruplet q, multiplet m, broad singlet br s, or combination of letters). DEPT135 was used to determine carbon multiplicity. Analytical HPLC was performed using a SpectraSYSTEM™ (ThermoFisher) with a Macherey-Nagel C18 column (3 × 125 mm/5 μm). HPLC purities were determined by UV absorption at 254 nm. ElectroSpray Ionisation (ESI) low resolution mass spectra were measured on a Finnigan Surveyor MSQ Plus mass spectrometer (ThermoFisher). Melting points (mp) of the solids were determined by SMP3 Melting Point Apparatus from Bibby Sterling. High-resolution mass spectra (HRMS) were recorded on a Bruker MicroTOF Q mass spectrometer.
4-Thioureidobenzoic acid.
This compound was prepared according to a described procedure.34 Carbon disulfide (1.32 mL, 22 mmol) was slowly added to a mixture of 4-aminobenzoic acid (1.0 g, 7.3 mmol,), THF (5 mL), H2O (5 mL) and Et3N (1.82 mL, 18 mmol). The resulting mixture was stirred at RT for 24h. Then iodine (1.94 g, 7.7 mmol) in THF (5 mL) was added dropwise at 0°C and mixture was stirred for 3h at RT. A 1M HCl aqueous solution (7.5 mL) and Na2SO3 (184 mg, 1.46 mmol) were added and the stirring was continued for 15 min. The aqueous layer was extracted with EtOAc (3 × 50 mL). Combined organic layers were washed twice with brine, dried on Na2SO4, filtered and concentrated to give the isothiocyanate intermediate as a slightly yellow solid, which was converted to the thiourea derivatives by stirring it at RT in NH4OH (30 %) for 6h. After removal of the insoluble parts by filtration on a sintered-glass funnel, filtrate was concentrated to afford the title product as an off-white solid (1.43 g, 7.3 mmol, quantitative yield). LC-UV purity = 80%. The compound was not ionized by ESI. 1H-NMR (500 MHz, DMSO-d6): δ (ppm) 7.56 – 7.58 (m, 2H), 7.80 – 7.82 (m, 2H). 13C-NMR (125.7 MHz, DMSO-d6): δ (ppm) 116.9, 120.9, 129.3, 130.6, 169.2, 181.0. mp = 189–191°C (in accordance with literature: 194°C).35
General procedure for the synthesis of 2-aminothiazole analogs 3,5–9.
An ethanolic solution of monobromo/dichloro reagent (2’-bromo-acetophenone derivative or 1,2-dichloro-1-ethoxyethane) and aryl thiourea derivative at equimolar ratio (unless otherwise indicated) adjusted with respect to LC-UV purity was heated at 80°C for 3–12h (reaction monitoring by TLC). The mixture was then cooled down to RT and water (twice more than EtOH v/v) was added. The mixture was stirred for 30 min, the suspension was then filtrated. The obtained solid was washed twice with water to afford the product as a solid. When necessary, a further purification was performed by (i) recrystallization or washing with boiling MeCN or (ii) flash chromatography with cyclohexane/acetone or (iii) semi-preparative HPLC to obtain the product with LC-UV purity higher than 95%.
[4-(3-Nitro-phenyl)-thiazol-2-yl]-phenyl-amine (3).
The reaction was carried out according to the general procedure. Scale: 2-bromo-3’-nitro-acetophenone (161 mg, 0.66 mmol), phenylthiourea (100 mg, 0.66 mmol), absolute EtOH (5 mL). After removal of the solvents, water (10 mL) and MeOH (2 mL) were added. The pH of the solution was adjusted to 9 with solid Na2CO3 and then stirred for 1h. After filtration and washing of the residual solid with water (3 × 5 mL), the title product was obtained as a yellow solid (196 mg, 0.66 mmol, quantitative yield); mp = 136–138 °C. LC-UV purity = 99%. LCMS (ESI+) m/z = 298.06 for [M+H]+. 1H-NMR (500 MHz, DMSO-d6): δ (ppm) 6.99 (t, J = 6.9 Hz, 1H), 7.36 (t, J = 6.6 Hz, 2H), 7.65 (s, 1H), 7.71 (m, 3H), 8.14 (d, J = 7.3 Hz, 1H), 8.35 (d, J = 6.9 Hz, 1H), 8.69 (s, 1H), 10.39 (s, 1H). 13C-NMR (125.7 MHz, DMSO-d6): δ (ppm) 105.6 (CH), 116.9 (2 × CH), 119.8 (CH), 121.4 (CH), 121.9 (CH), 129.0 (2 × CH), 130.1 (CH), 131.7 (CH), 135.9 (C), 140.9 (C), 147.6 (C), 148.2 (C), 163.5 (C). HRMS (ESI+): m/z calcd for C15H12N3O2S [M+H]+: 298.0645; found: 298.0633.
4-[4-(3-Nitro-phenyl)-thiazol-2-ylamino]-benzoic acid methyl ester (4).
Three drops of 95% H2SO4 were added to a mixture of 4-[4-(3-nitro-phenyl)-thiazol-2-ylamino]-benzoic acid 2 (46 mg, 0.14 mmol) in MeOH (5 mL) and the mixture was then heated at reflux for 2h. Na2CO3 1 M was added until pH = 10–11 and MeOH was removed under vacuum. The obtained solid was filtrated and washed with water (2 × 5 mL) to afford the title product as a yellow solid (49 mg, 0.14 mmol, quantitative yield); mp = 242–244 °C. LC-UV purity = 97%. LCMS (ESI+) m/z = 356.01 for [M+H]+. 1H-NMR (500 MHz, DMSO-d6): δ (ppm) 3.82 (s, 3H), 7.79 (m, 4H), 7.96 (s, 2H), 8.17 (s, 1H), 8.40 (s, 1H), 8.71 (s, 1H), 10.83 (s, 1H). 13C-NMR (125.7 MHz, DMSO-d6): δ (ppm) 51.7 (CH3), 107.0 (CH), 116.1 (2 × CH), 119.9 (CH), 121.8 (C), 122.1 (CH), 130.3 (CH), 130.6 (2 × CH), 131.9 (CH), 135.7 (C), 144.9 (C), 147.8 (C), 148.3 (C), 162.6 (C), 165.8 (C). HRMS (ESI+): m/z calcd for C17H14N3O4S [M+H]+: 356.0700; found: 356.0695.
4-(Thiazol-2-ylamino)-benzoic acid (5).
The reaction was carried out according to the general procedure. Scale: 1,2-dichloro-1-ethoxyethane (50 μL, 0.41 mmol), 4-thioureidobenzoic acid (80 mg, 0.41 mmol), absolute EtOH (5 mL). Purification by semi-preparative HPLC (H2O/MeCN/ 0.1% TFA: from 79/20/1 to 0/99/1 in 40 min) to afford ethyl 4-(thiazol-2-ylamino)benzoate as a white solid (23 mg, 0.09 mmol, 23% yield). LC-UV purity = 96%. LCMS (ESI+) m/z = 249.01 for [M+H]+.The ethyl 4-(thiazol-2-ylamino)benzoate (21 mg, 0.085 mmol) obtained was directly hydrolyzed in a mixture of THF (2 mL) and aqueous 0.5 M NaOH (2.3 mL, 1.15 mmol) at reflux for 4h. Aqueous 1 M HCl was added until pH = 1 and then solvents were removed under vacuum. Crude product was purified by semi-preparative HPLC (H2O/MeCN + 0.1% TFA: from 90/10 to 10/90 in 40 min) to afford the title product as a white solid (16 mg, 0.07 mmol, 86% yield); mp = 270–272 °C. LC-UV purity > 99%. LCMS (ESI+) m/z = 220.78 for [M+H]+. 1H-NMR (500 MHz, DMSO-d6): δ (ppm) 7.02 (d, J = 3.8 Hz, 1H), 7.33 (d, J = 3.8 Hz, 1H), 7.72 (d, J = 8.5 Hz, 2H), 7.88 (d, J = 8.8 Hz, 2H), 10.59 (s, 1H). 13C-NMR (125.7 MHz, DMSO-d6): δ (ppm) 109.7 (CH), 115.7 (2 × CH), 122.5 (C), 130.7 (2 × CH), 138.8 (CH), 144.9 (C), 162.9 (C), 166.9 (C). HRMS (ESI+): m/z calcd for C10H9N2O2S [M+H]+: 221.0379; found: 221.0369.
4-(4-Phenyl-thiazol-2-ylamino)-benzoic acid (6).
The reaction was carried out according to the general procedure. Scale: 2-bromo-acetophenone (102 mg, 0.51 mmol), 4-thioureidobenzoic acid (100 mg, 0.51 mmol), absolute EtOH (5 mL). Purification was performed by washing with boiling MeCN (5 mL) and hot filtration to afford the title product as a grey solid (70 mg, 0.24 mmol, 58% yield); mp = 256–258 °C. LC-UV purity = 95%. LCMS (ESI+) m/z = 615.83 for [2M+Na]+. 1H-NMR (500 MHz, DMSO-d6): δ (ppm) 7.33 (t, J = 6.9 Hz, 1H), 7.44 (t, J = 6.9 Hz, 3H), 7.84 (d, J = 8.5 Hz, 2H), 7.95 (d, J = 7.9 Hz, 4H), 10.70 (br s, 1H). 13C-NMR (125.7 MHz, DMSO-d6): δ (ppm) 104.1 (CH), 115.9 (2 × CH), 122.8 (C), 125.7 (2 × CH), 127.7 (CH), 128.6 (2 × CH), 130.8 (2 × CH), 134.3 (C), 144.9 (C), 150.2 (C), 162.2 (C), 167.0 (C). HRMS (ESI+): m/z calcd for C16H13N2O2S [M+H]+: 297.0692; found: 297.0686.
4-[4-(3-Bromo-phenyl)-thiazol-2-ylamino]-benzoic acid (7).
The reaction was carried out according to the general procedure. Scale: 2-bromo-1-(3-bromophenyl)ethanone (100 mg, 0.36 mmol), 4-thioureidobenzoic acid (106 mg, 0.54 mmol), absolute EtOH (5 mL). Purification was performed by recrystallization in MeCN (5 mL) to afford the title product as a yellow solid (75 mg, 0.20 mmol, 56% yield); mp = 281–283°C. LC-UV purity = 95%. LCMS (ESI+) m/z = 374.62/376.65 for [M+H]+. 1H-NMR (500 MHz, DMSO-d6): δ (ppm) 7.42 (t, J = 7.9 Hz, 1H), 7.52 (d, J = 7.1 Hz, 1H), 7.62 (s, 1H), 7.79 (d, J = 8.8 Hz, 2H), 7.93 – 7.98 (m, 3H), 8.11 (t, J = 1.6 Hz, 1H), 10.73 (s, 1H), 12.60 (br s, 1H). 13C-NMR (125.7 MHz, DMSO-d6): δ (ppm) 105.7 (CH), 115.9 (2 × CH), 122.1 (C), 122.9 (C), 124.7 (CH), 128.0 (CH), 130.3 (CH), 130.8 (2 × CH), 130.9 (CH), 136.5 (C), 144.7 (C), 148.4 (C), 162.4 (C), 166.9 (C). HRMS (ESI+): m/z calcd for C16H12BrN2O2S [M+H]+: 374.9797; found: 374.9789.
4-[4-(3-Methoxy-phenyl)-thiazol-2-ylamino]-benzoic acid (8).
The reaction was carried out according to the general procedure. Scale: 2-bromo-3’-methoxy-acetophenone (152 mg, 0.66 mmol,), 4-thioureidobenzoic acid (100 mg, 0.51 mmol), absolute EtOH (10 mL). Purification was performed by washing with boiling MeCN (5 mL) and hot filtration to give a solid which was then purified by flash chromatography (cyclohexane/acetone 5/5) to afford the title product as a yellow solid (29 mg, 0.09 mmol, 17% yield); mp = 246–248°C. LC-UV purity = 96%. LCMS (ESI+) m/z = 327.10 for [M+H]+. 1H-NMR (500 MHz, DMSO-d6): δ (ppm) 3.83 (s, 3H), 6.91 (dd, J = 8.20, 2.21 Hz, 1H), 7.36 (t, J = 8.20 Hz, 1H), 7.48 (s, 2H), 7.53 (d, J = 7.88 Hz, 1H), 7.81 (d, J = 8.83 Hz, 2H), 7.94 (d, J = 8.83 Hz, 2H), 10.69 (s, 1H), 12.56 (br s, 1H). 13C-NMR (125.7 MHz, DMSO-d6): δ (ppm) 55.0 (CH3), 104.5 (CH), 111.1 (CH), 113.2 (CH), 115.8 (CH), 118.1 (CH), 122.8 (C), 129.7 (CH), 130.8 (CH), 135.6 (C), 144.8 (C), 150.0 (C), 159.5 (C), 162.1 (C), 167.0 (C). HRMS (ESI+): m/z calcd for C17H15N2O3S [M+H]+: 327.0798; found: 327.0786.
4-(4-Pyridin-2-yl-thiazol-2-ylamino)-benzoic acid (9).
The reaction was carried out according to the general procedure. Scale: 2-bromo-1-(pyridin-2-yl)ethanone hydrobromide (171 mg, 0.61 mmol), 4-thioureidobenzoic acid (100 mg, 0.51 mmol), absolute EtOH (10 mL). After water (5 mL) addition to the mixture, NH4OH (30%) was added until complete dissolution. The pH of the solution was then adjusted to 2–3 by addition of aqueous HCl (1M) and the mixture was stirred for 30 min. The obtained solid was then filtrated and washed with water (3 × 5 mL) to afford the title product as a yellow solid (85 mg, 0.28 mmol, 56% yield); mp = 274–276°C. LC-UV purity = 97%. LCMS (ESI+) m/z = 298.07 for [M+H]+. 1H-NMR (500 MHz, DMSO-d6): δ (ppm) 7.83 (t, J = 5.5 Hz, 1H), 7.95 (m, 4H), 8.35, (br s, 1H), 8.50 (m, 2H), 8.79 (d, J = 5.4 Hz, 1H), 11.24 (br s, 1H). 13C-NMR (125.7 MHz, DMSO-d6): δ (ppm) 114.8 (CH), 116.6 (2 × CH), 123.4 (CH), 124.7 (CH), 130.7 (2 × CH), 143.1 (CH), 144.2 (C), 144.6 (CH), 145.9 (C), 163.5 (C), 167.0 (C). HRMS (ESI+): m/z calcd for C15H12N3O2S [M+H]+: 298.0645; found: 298.0631.
Biology.
Radiometric kinase assays.
The CK2 radiometric kinase assay was performed as previously described.10 Production and purification of GST-rhCK2α mutants were carried out as described in the paper of Moucadel et al.,9 and inhibitors were tested following the CK2 radiometric kinase assay procedure. ATP concentrations were 100 μM if not stated otherwise.
Temperature-dependent circular dichroism (TdCD).
All single Far-UV-CD spectra were recorded using a Jasco-1500 spectropolarimeter (Gross-Umstadt, Germany). All spectra were recorded using a 1 mm path-length quartz cell, 1 nm bandwidth, 100 nm/min scan speed and 3 accumulations, within a wavelength range of 195 to 250 nm. GST-CK2α protein was diluted at 5 μM with 10 mM KH2PO4 and 50 mM Na2SO4, pH 7.4. Ligand binding was tested at 50 μM for compound 6, 6.25 μM for compound 9 and 6.25 μM for CX-4945 with a final methanol concentration of 0.8 % (v/v). Spectra recorded in the absence of protein were subtracted from the experimental spectra. Temperature ramping experiments were performed from 20 °C to 86 °C, and prior to each recording point, the sample was allowed to equilibrate for 30 seconds at ± 0.1 °C from the desired temperature. Midpoint unfolding temperatures (Tm) were determined using Thermal Denaturation multi-Analysis software.
Protein-ligand interactions by NMR.
NMR experiments were recorded on a 600 MHz spectrometer at 293 K in PBS pH 7.3, 10 mM NaCl, MgCl2 2 mM, 10% D2O. 1D experiments were performed using an excitation sculpting pulse sequence. Saturation Transfer Difference was performed with on-resonance protein saturation at 0.6 ppm using 2 seconds saturation time. 7-Azaindole was used as a reference compound to test the STD-competition with CX-4945. STD experiments were performed using 5 μM CK2α protein in the presence of 4 mM MgCl2. STD factors were measured as previously reported,18 and the values were normalized by assigning the value 100 % to the highest STD factor. Concentrations of the molecules AMP-PNP, 7-azaindole and compound 6 were 500 μM, and the concentration of CX-4945 was 100 μM.
Native mass spectrometry.
All spectra were acquired using a Synapt HD mass spectrometer (Waters) modified for studying high masses and externally calibrated using cesium iodide. Micro Bio–Spin 6 chromatography columns (Bio–Rad) were used to exchange into 0.50 M ammonium acetate solution pH 8.5. CK2α (10 μM) was incubated with ligands (10 μM CX-4945, 100 μM IP6, 10–40 μM 6) at a final concentration of 5% (v/v) DMSO. The protein or protein–ligand solution (2.5 μL) was electrosprayed from a borosilicate emitter (Thermo Scientific) for sampling. Typical conditions employed were capillary voltage of 1.6–2.0 kV, cone voltage of 30–90 V, collision voltage of 10 V, with backing pressure 3 mbar and source temperature of 20 °C. MassLynx 4.1 was used for data acquisition and processing.
Molecular modeling and docking studies.
Structures of the catalytic subunit of the human CK2 enzyme were retrieved from the Pocketome database.36 Including the multiple chains within each structure, there was a total of 72 structures. Candidate ligand docking locations were delineated as shown in Figure 6, one comprising the canonical ATP binding site, and another the proposed allosteric binding site. Prior to docking, the kinase structures were completed by rebuilding and optimization of the missing side-chains and the hydrogen atoms in the presence of cocrystallized ligands (if any). The putative allosteric pocket was most accessible for small ligands in 1JUH (chain A) and 3FWQ (chain B). Molecular docking of compound 6 to these pockets was performed using MOE, basically as previously described.37 For the double docking procedures, ligand 1 (CX-4945 or AMP-PNP) was docked in the ATP binding site (except for AMP-PNP in 3JUH, where the cocrystal complex could be used), then compound 6 was defined as ligand and docked in the putative allosteric pocket. In most cases, the pocket side chains were defined as flexible (5 Å cut off).
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by grants from the “Institut National de la Santé et de la Recherche Médicale”, the “Ligue Nationale et Régionale contre le Cancer”, the “Commissariat à l’Energie Atomique” and the University of Grenoble Alpes and the Espoir Foundation. We gratefully acknowledge the financial support given by “Région Rhône-Alpes” (grant “ARC 1 Santé” 12-008707-01). Dr. Benoît Bestgen would like to thank the “Cancéropôle Lyon Rhône-Alpes Auvergne (CLARA)” for a fellowship “Mobilité Jeunes Chercheurs” (INCA Grant N°2011-097). This work is partially supported by NIH grants R01 GM071872 (RA) and R01 AI118985 (IKu).
Prof. M. Le Borgne would like to acknowledge CLARA, “Université France Allemagne” (UFA) and “Région Rhône-Alpes” for the emergence of ChemBioInteract network. We thank MENRT and EDISS for the PhD fellowship of Benoît Bestgen in collaboration between University Lyon 1 and Saarland University as well as ISPB for an ATER year contract for B. Bestgen.
ABBREVIATIONS USED
- CK2
Casein Kinase 2
- STD
Saturation-Transfer Difference
- Clk
CDC-like kinase
- NFAT
Nuclear Factor of Activated T-cells
- POM
PolyOxoMetalate
- VLS
Virtual Ligand Screening
- Tm
melting temperature
- TdCD
Temperature–dependent circular dichroism
- AMP-PNP
Adenylyl-imidodiphosphate
- IP6
inositol hexakisphosphate
- TBI
4,5,6,7-tetrabromo-1H-benzimidazole
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information. The Supporting Information is available free of charge on the ACS Publications Website at DOI: (to be inserted by publisher).
Figures S1 and S2
1H and 13C NMR spectra of compounds 3-9.
HPLC/UV/MS chromatograms for compounds 6 and 7.
REFERENCES
- (1).Meggio F; Pinna LA One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003, 17, 349–368. [DOI] [PubMed] [Google Scholar]
- (2).Litchfield DW Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem J. 2003, 369, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Protein kinase CK2; Pinna LA, Ed.; Wiley-Blackwell: Ames, Iowa, USA, 2013; 460 p. [Google Scholar]
- (4).Siddiqui-Jain A; Drygin D. ; Streiner N. ; Chua P. ; Pierre F; O’Brien SE; Bliesath J; Omori M; Huser N; Ho C; Proffitt C; Schwaebe MK; Ryckman DM; Rice WG; Anderes K. CX-4945, an orally bioavailable selective inhibitor of protein kinase CK2, inhibits prosurvival and angiogenic signaling and exhibits antitumor efficacy. Cancer Res. 2010, 70, 10288–10298. [DOI] [PubMed] [Google Scholar]
- (5).CX-4945 is now evaluated as a synergistic treatment of cholangocarcinoma with gemcitabine and cisplatin (ClinicalTrials.gov identifier: NCT02128282).
- (6).Kim H; Choi K; Kang H; Lee S-Y; Chi S-W; Lee M-S; Song J; Im D; Choi Y; Cho S. Identification of a novel function of CX-4945 as a splicing regulator. PloS One 2014, 9, e94978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Kim H; Lee K-S; Kim A-K; Choi M; Choi K; Kang M; Chi S-W; Lee M-S; Lee J-S; Lee S-Y; Song W-J; Yu K; Cho S. A chemical with proven clinical safety rescues Down-syndrom-related phenotypes in through DYRK1A inhibition. Dis. Models Mech. 2016, 9, 839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Prudent R; Cochet C. New protein kinase CK2 inhibitors: jumping out of the catalytic box. Chem. Biol. 2009, 16, 112–120. [DOI] [PubMed] [Google Scholar]
- (9).Moucadel V; Prudent R; Sautel CF; Teillet F; Barette C; Lafanechere L; Receveur-Brechot V; Cochet C. Antitumoral activity of allosteric inhibitors of protein kinase CK2 Oncotarget 2011, 2, 997–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Prudent R; Moucadel V; Laudet B; Barette C; Lafanechere L; Hasenknopf B; Li J; Bareyt S; Lacote E; Thorimbert S; Malacria M; Gouzerh P; Cochet C. Identification of polyoxometalates as nanomolar noncompetitive inhibitors of protein kinase CK2. Chem. Biol. 2008, 15, 683–692. [DOI] [PubMed] [Google Scholar]
- (11).De Fusco C; Brear P; Iegre J; Hadje Georgiou K; Sore HF; Hyvönen M; Spring DR A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066. Bioorg. Med. Chem. 2017, 25, 3471–3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Iegre J; Brear P; De Fusco C; Yoshida M; Mitchell SL; Rossmann M; Carro L; Sore HF; Hyvönen M; Spring DR Second-generation CK2α inhibitors targeting the αD pocket. Chem. Sci. 2018, 9, 3041–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Kufareva I; Abagyan R. Strategies to overcome the induced fit effects in molecular docking. In From Computational Biophysics to Systems Biology (CBSB08), Neumann Institute for Computing Series vol. 40; Hansmann UHE; Meinke JH; Mohanty S; Nadler W; Zimmermann O. Eds.; John von Neumann Institute for Computing: Jülich, 2008; pp 1–6. [Google Scholar]
- (14).Lopez-Ramos M; Prudent R; Moucadel V; Sautel CF; Barette C; Lafanechere L; Mouawad L; Grierson D; Schmidt F; Florent J-C; Filippakopoulos P; Bullock AN; Knapp S; Reiser J-B; Cochet C. New potent dual inhibitors of CK2 and Pim kinases: discovery and structural insights. FASEB J. 2010, 24, 3171–3185. [DOI] [PubMed] [Google Scholar]
- (15).a) For a literature precedent of 4-(thiazol-5-yl)benzoic acid derivatives as ATP-competitive CK2 inhibitors, see:Hou Z; Nakanishi I; Kinoshita T; Takei Y; Yasue M; Misu R; Suzuki Y; Nakamura S; Kure T; Ohno H; Murata K; Kitaura K; Hirasawa A; Tsujimoto G; Oishi S; Fujii N. Structure-based design of novel potent protein kinase CK2 (CK2) inhibitors with phenyl-azole scaffolds. J. Med. Chem. 2012, 55, 2899–2903. [DOI] [PubMed] [Google Scholar]; b) Ohno H; Minamiguchi D; Nakamura S; Shu K; Okazaki S; Honda M; Misu R; Moriwaki H; Nakanishi S; Oishi S; Kinoshita T; Nakanishi I; Fujii N. Structure–activity relationship study of 4-(thiazol-5-yl)benzoic acid derivatives as potent protein kinase CK2 inhibitors. Bioorg. Med. Chem. 2016, 24, 1136–1141. [DOI] [PubMed] [Google Scholar]
- (16).Kroe RR; Regan J; Proto A; Peet GW; Roy T; Landro LD; Fuschetto NG; Pargellis CA; Ingraham RH Thermal denaturation: a method to rank slow binding, high-affinity P38alpha MAP kinase inhibitors. J. Med. Chem. 2003, 46, 4669–4675. [DOI] [PubMed] [Google Scholar]
- (17).Fedorov O; Marsden B; Pogacic V; Rellos P; Müller S; Bullock AN; Schwaller J; Sundström M; Knapp S. A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 20523–20528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Mayer M; Meyer B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J. Am. Chem. Soc. 2001, 123, 6108–6117. [DOI] [PubMed] [Google Scholar]
- (19).Battistutta R; Cozza G; Pierre F; Papinutto E; Lolli G; Sarno S; O’Brien SE; Siddiqui-Jain A; Haddach M; Anderes K; Ryckman DM; Meggio F; Pinna LA Unprecedented selectivity and structural determinants of a new class of protein kinase CK2 inhibitors in clinical trials for the treatment of cancer. Biochemistry 2011, 50, 8478–8488. [DOI] [PubMed] [Google Scholar]
- (20).Ferguson AD; Sheth PR; Basso AD; Paliwal S; Gray K; Fischmann TO; Le HV Structural basis of CX-4945 binding to human protein kinase CK2. FEBS Lett. 2011, 585, 104–110. [DOI] [PubMed] [Google Scholar]
- (21).Niefind K; Raaf J; Issinger O-G Protein kinase CK2: from structures to insights. Cell. Mol. Life Sci. 2009, 66, 1800–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Gouron A; Milet A; Jamet H. Conformational flexibility of human casein kinase catalytic subunit explored by metadynamics. Biophys. J. 2014, 106, 1134–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Guerra B; Bischoff N; Bdzhola VG; Yarmoluk SM; Issinger O-G; Golub AG; Golub AG; Niefind K. A note of caution on the role of halogen bonds for protein kinase/inhibitor recognition suggested by high- and low-salt CK2α complex structures. ACS Chem. Biol. 2015, 10, 1654–1660. [DOI] [PubMed] [Google Scholar]
- (24).Srivastava A; Hirota T; Irle S; Tama F. Conformational dynamics of human protein kinase CK2α and its effect on function and inhibition. Proteins 2018, 86, 344–353. [DOI] [PubMed] [Google Scholar]
- (25).Yde CW; Ermakova I; Issinger O-G; Niefind K. Inclining the purine base binding plane in protein kinase CK2 by exchanging the flanking side-chains generates a preference for ATP as a cosubstrate. J. Mol. Biol. 2005, 347, 399–414. [DOI] [PubMed] [Google Scholar]
- (26).Raaf J; Issinger O-G; Niefind K. First inactive conformation of CK2α, the catalytic subunit of protein kinase CK2. J. Mol. Biol. 2009, 386, 1212–1221. [DOI] [PubMed] [Google Scholar]
- (27).Cozza G; Zanin S; Sarno S; Costa E; Girardi C; Ribaudo G; Salvi M; Zagotto G; Ruzzene M; Pinna LA Design, validation and efficacy of bisubstrate inhibitors specifically affecting ecto-CK2 kinase activity Biochem. J. 2015, 471, 415–430. [DOI] [PubMed] [Google Scholar]
- (28).a) For other strategies of bidentate CK2α inhibitors, targeting simultaneously the ATP-binding site and a vicinal pocket, see:Enkvist E; Viht K; Bischoff N; Vahter J; Saaver S; Raidaru G; Issinger O-G; Niefind K; Uri A. A subnanomolar fluorescent probe for protein kinase CK2 interaction studies. Org. Biomol. Chem. 2012, 10, 8645–8653. [DOI] [PubMed] [Google Scholar]; b) Brear P; De Fusco C; Hadje Georgiou K; Francis-Newton NJ; Stubbs CJ; Sore HF; Venkitaraman AR; Abell C; Spring DR; Hyvönen M. Specific inhibition of CK2α from an anchor outside the active site. Chem. Sci. 2016, 7, 6839–6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Papinutto E; Ranchio A, Lolli G; Pinna LA; Battistuta R. Structural and functional analysis of the flexible regions of the catalytic α-subunit of protein kinase CK2. J. Struct. Biol. 2012, 177, 382–391. [DOI] [PubMed] [Google Scholar]
- (30).Prudent R; Moucadel V; Nguyen C-H; Barette C; Schmidt F; Florent J-C; Lafanechère L; Sautel CF; Duchemin-Pelletier E; Spreux E; Filhol O; Reiser J-B; Cochet C. Antitumor activity of pyridocarbazole and benzopyridoindole derivatives that inhibit protein kinase CK2. Cancer Res. 2010, 70, 9865–9874. [DOI] [PubMed] [Google Scholar]
- (31).Lee W-K; Son SH; Jin B-S; Na J-H; Kim S-Y; Kim K-H; Kim EE; Yu YG; Lee HH Structural and functional insights into the regulation mechanism of CK2 by IP6 and the intrinsically disordered protein Nopp140. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 19360–19365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Wang W, Kitova EN; Klassen JS Nonspecific protein–carbohydrate complexes produced by nanoelectrospray ionization: factors influencing their formation and stability. Anal. Chem. 2005, 77, 3060–3071. [DOI] [PubMed] [Google Scholar]
- (33).Gottlieb HE, Kotlyar V, Nudelman A. NMR chemical shifts of common laboratory solvents as trace impurities. J. Org. Chem. 1997, 62, 7512–7515. [DOI] [PubMed] [Google Scholar]
- (34).Nakano S, Saito D. Method for producing isothiocyanate compound having carboxyl group. US20100312000A1, 2010. [Google Scholar]
- (35).Guha-Sircar SS, Patnaik KK Preparation of antibacterials from organomercurials. J. Indian Chem. Soc. 1950, 27, 535–538. [Google Scholar]
- (36).Kufareva I, Ilatovskiy AV, Abagyan R. Pocketome: an encyclopedia of small-molecule binding sites in 4D. Nucleic Acids Res. 2012, 40, Database issue, D535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).ElHady AK, Abdel-Halim M, Abadi AH, Engel M. Development of selective Clk1 and −4 inhibitors for cellular depletion of cancer-relevant Proteins. J. Med. Chem. 2017, 60, 5377–5391. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.