Abstract
Academic research plays a key role in identifying new drug targets, including understanding target biology and links between targets and disease states. To lead to new drugs, however, research must progress from purely academic exploration to the initiation of efforts to identify and test a drug candidate in clinical trials, which are typically conducted by the biopharma industry. This transition can be facilitated by a timely focus on target assessment aspects such as target-related safety issues, druggability and assayability, as well as the potential for target modulation to achieve differentiation from established therapies. Here, we present recommendations from the GOT-IT working group, which have been designed to support academic scientists and funders of translational research in identifying and prioritizing target assessment activities and in defining a critical path to reach scientific goals as well as goals related to licensing, partnering with industry or initiating clinical development programmes. Based on sets of guiding questions for different areas of target assessment, the GOT-IT framework is intended to stimulate academic scientists’ awareness of factors that make translational research more robust and efficient, and to facilitate academia–industry collaboration.
Subject terms: Translational research, Target validation
Academic research has a key role in identifying new drug targets, but to lead to new drugs this research must progress to testing drug candidates in clinical trials, which are typically conducted by industry. This Perspective presents a framework to support academic scientists and funders in prioritizing target assessment activities and in defining a critical path to reach scientific goals as well as goals related to licensing, partnering with industry or initiating clinical trials.
Introduction
The discovery and development of first-in-class drugs often begins with the identification of a new drug target, such as an enzyme linked to a disease process. Once a new target has been identified, various strategies are used to provide validation of the target and to support decisions, such as initiating an extensive drug discovery programme, conducting a proof-of-concept trial in humans or partnering with another organization. Importantly, insufficient validation of drug targets at an early stage has been linked to costly clinical failures1 and low drug approval rates2,3. Indeed, it was predicted a decade ago that more effective target validation as well as early proof-of-concept studies could reduce attrition in phase II clinical trials by ~24%, lowering the cost of developing new molecular entities by ~30% (ref.4). As a result, there is common consensus that robust target validation is a crucial part of drug discovery and deserves greater emphasis in order to facilitate the development of new therapies.
Target validation can be understood in numerous ways depending on the context (Box 1), but usually describes the technical evaluation of whether a target has a key role in a disease process and whether pharmacological modulation of the target could be effective in a defined patient population5. As projects on promising targets progress towards the clinical development of new drug candidates, additional aspects become critical. These aspects include the ‘druggability’ of the target, potential target-related safety issues, biomarker research, legal and intellectual property issues, the extent of the unmet medical need in the potential patient populations intended to benefit from the new drug and the commercial potential of the new drug if it successfully reaches the market6. Together, these considerations are defined as target assessment in the context of this Perspective article.
Newly identified drug targets often originate from academic research7,8, with the biopharma industry typically leading drug discovery and development, including the optimization and selection of drug candidates, preclinical safety studies and clinical trials. Consequently, academia and industry need to interact at some point and close the gap between basic research and drug discovery to translate research on new targets into clinically effective products9–15. Several factors critically influence these interactions16,17. From an academic perspective, target assessment is time and resource-consuming18, with limited novelty value compared with target identification, and therefore offers less attractive possibilities to publish scientific articles. In addition, decision-making in academia is often strongly influenced by the administrative framework, such as PhD student contracts or the need for PhD students to submit their thesis after a defined period of time. At the same time, universities are increasingly interested in commercializing the results of academic research, which may require balancing the freedom to publish with protecting and offering access to intellectual property. Furthermore, funders of translational research would like to focus on the most pertinent questions, such as the relevance to human disease therapy and current unmet medical needs. From an industry perspective, selected projects should offer freedom to operate and possibilities to protect intellectual property with the potential for development of effective new therapies. The risks associated with insufficient data quality and the lack of documentation are very well recognized, and if data quality and documentation procedures have inadequate standards, additional resources are required to verify published results19,20.
Recommendations for some aspects of target assessment have been published — for example, AstraZeneca’s ‘5R’ framework, based on the most important technical determinants of project success and pipeline quality identified by the company: the right target, the right patient, the right tissue, the right safety and the right commercial potential21,22. Furthermore, several articles have highlighted indication-specific features, as in the field of cancer research23–26, or discussed the optimal use of translational research tools, such as CRISPR–Cas9 technology27,28 and the application of bioactive small molecules as affinity probes for target validation studies24,29–33.
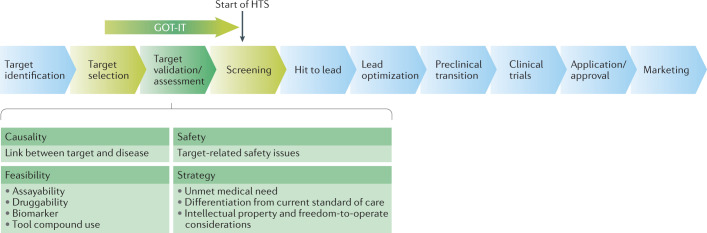
However, a comprehensive and practical set of recommendations that can guide stepwise decision-making when assessing new targets on the path from target identification to regulatory approval of a new drug has yet to be provided. In this context, steps and goals for translational academic scientists and research organizations may include development of biomarkers or tool compounds to understand biological mechanisms, formation of collaborations with an industry partner, founding a biotech start-up company or licensing of early-stage projects to industry16. To establish a structured framework for target assessment, particularly in an academic environment, the GOT-IT (Guidelines On Target Assessment for Innovative Therapeutics) working group was established, funded by the German Federal Ministry of Education and Research (BMBF). The drug discovery experience of the working group participants was complemented by project leaders and industrial mentors from different research areas involved in the BMBF Target Validation for Drug Development programme (Supplementary Table 1). Additional input was obtained from experts within academia, contract research organizations, technology transfer offices, learned societies and industry.
Here, we present the recommendations established by the GOT-IT group, which aim to: enhance awareness of key aspects of target validation and assessment; assist in prioritizing activities for efficient use of resources; help to define project-specific value inflexion points, key activities and milestones relevant for decision-making at an early stage; and support the creation of an attractive data package as the foundation to reach the project goals. Importantly, guidance is also presented on data robustness34 and reproducibility, thereby increasing confidence in the potential of a target and facilitating the transition from purely academic research to rigorous drug discovery. After summarizing the conceptual basis of the recommendations, this article focuses on five key areas of target assessment in which they are applied, concluding with a discussion of key issues in the application of the recommendations.
Box 1 Frequency of target assessment aspects used in academic research.
To determine which characteristics are frequently used in academic publications to validate and assess a newly identified drug target and to determine differences between industrial and academic assessment approaches, a status quo literature analysis was conducted. For this analysis, a set of target assessment elements were pre-specified and their prevalence was analysed in 428 relevant articles from academic research groups, dealing with the process of target validation/assessment (see Supplementary Box 8 for details). Whereas most publications discussed the link between target and disease and identified future patient populations for treatment (85.5%), only a minority of papers included the application of biomarkers (6.1%), target assayability (1.9%), potential safety issues (9.1%), three-dimensional structure discovery (8.6%) or the intellectual property/patent situation (2.1%). However, around half of all articles (53.0%) made use of tool compounds to validate genetic target modulation approaches (see figure, panel a).
Reporting of data quality requirements, such as implementation of the Landis 4 criteria (that is, defining inclusion and exclusion criteria, sample size calculation, blinding and randomization)175 and the validation of research tools (such as cell line authentication or antibody specificity checks), occurred in only a very small percentage of papers (see figure, panel b). Blinding and randomization for in vivo studies were the most frequently reported Landis criteria, at 12.4% and 28.9%, respectively. Only 0.8% of all papers implemented all Landis 4 criteria in their in vivo studies (0% for in vitro studies). These findings are in line with previous reports on in vivo and in vitro research176, indicating that data quality measures to ensure unbiased and robust research outcomes are still not more frequently implemented in studies aiming to validate a potential new drug target.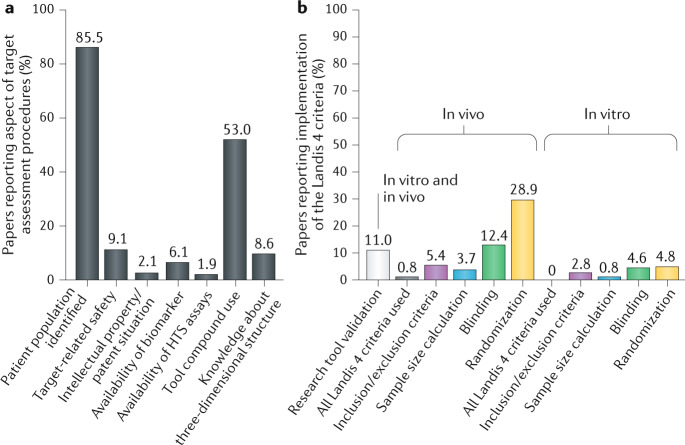
HTS, high-throughput screening.
The GOT-IT critical path concept
Given that there is no single solution for all research areas, institutions and targets, the guidelines for assessing a drug target must be flexible and fit for purpose (for example, applicable within practical limits of time and money). For any selected target or specific research environment, target assessment is not an absolute ‘truth’ and its relative value is strongly dependent on the therapeutic area, the current knowledge and the state of technology. Consequently, best practice must be tailored to individual research and disease fields of interest35. Thus, the GOT-IT recommendations are based on a modular system — the critical path — that allows for sufficient flexibility to address individual project goals and milestones, indication-specific needs and availability of resources.
Definition of assessment blocks
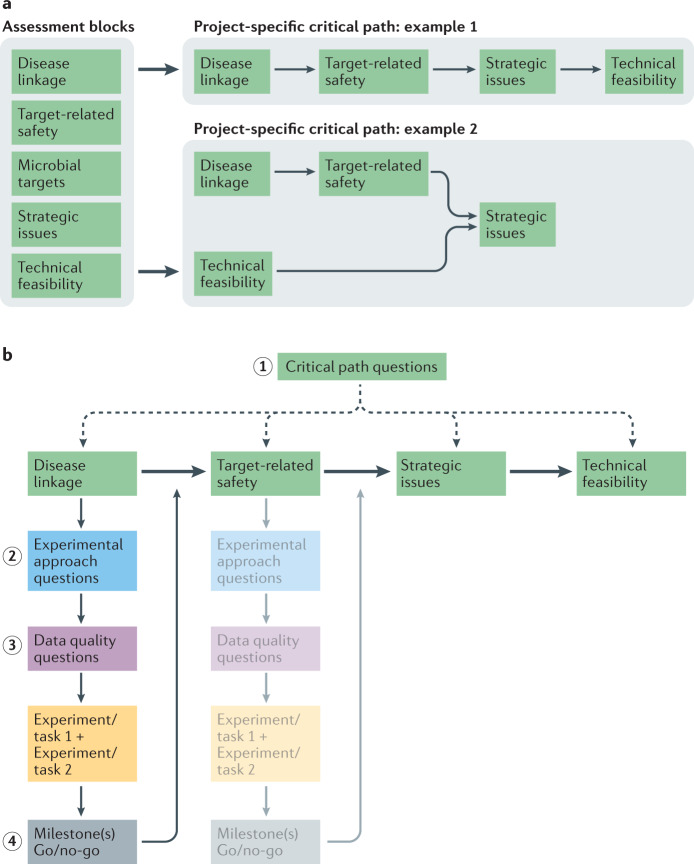
The GOT-IT recommendations categorize and group relevant aspects of practical target validation and assessment into assessment blocks (AB1–AB5) defining the main framework of the project-specific critical path (Fig. 1), similar to the categories characterizing key properties of a good drug target as proposed by Gashaw et al.6:
AB1: target–disease linkage (that is, the causal relationship between target and disease)
AB2: safety aspects (on-target or target-related)
AB3: microbial targets (aspects related to non-human targets)
AB4: strategic issues (for example, clinical needs and commercial potential)
AB5: technical feasibility (including druggability, assayability and biomarker availability)
Fig. 1. Defining the critical path of target assessment.
a | Different target assessment aspects and requirements are grouped into assessment blocks, which can be arranged into a project-specific critical path according to project needs, timelines and available resources. The order of assessment blocks is project-specific and some assessment blocks can run in parallel, might not be relevant or do not apply due to indication-specific characteristics. b | The identification and arrangement of all project-relevant assessment blocks is facilitated by answering a set of critical path questions at the beginning of a project (step 1). As a next step, sets of experimental approach questions help to define specific activities for each selected assessment block (step 2) to identify key experiments to increase confidence in the target of interest. Requirements to ensure data quality/robustness also need to be checked after new assessment block-specific tasks have been identified, especially for decision-enabling processes. For this, a detailed set of data quality questions has been created (step 3). If all milestone requirements and go criteria are fulfilled, a go decision is appropriate and the next assessment block can be addressed (step 4).
Traditionally, academic drug discovery is aimed at better understanding the underlying biology of a disease, and so AB1 and AB2 might be most important16,36. However, depending on the project goals, such as spin-off formation or licensing, AB4 and/or AB5 can also become crucial to increase the value of a project. Consequently, assessment blocks can run in parallel or might not be relevant or applicable to some project goals at all (Fig. 1a). It is also important to note that target assessment approaches may differ depending on whether the target is human or non-human. Thus, the GOT-IT recommendations discuss general aspects as well as aspects that are specific for either human or microbial targets: AB1 and AB2 are most relevant for human targets, whereas AB3 focuses primarily on characteristics related to microbial targets. AB4 and AB5 cover aspects that, in most cases, apply to all indications or research fields.
The relevant assessment blocks can be assembled into a critical path for a specific project (Fig. 2), thereby prioritizing the identified activities that must be successfully completed at different points in order for the project to reach defined decision points. Importantly, building the critical path also facilitates and optimizes resource management, as gaps in resources available to reach individual milestones become evident. It should be noted that the critical path concept used by GOT-IT is not directly related to the one developed by the US Food and Drug Administration (FDA) Critical Path Initiative (CPI), which was launched in 2004 (ref.37) to focus on improving the drug development process by establishing new evaluation tools for more efficient and effective procedures and to streamline clinical trials and manufacturing38,39.
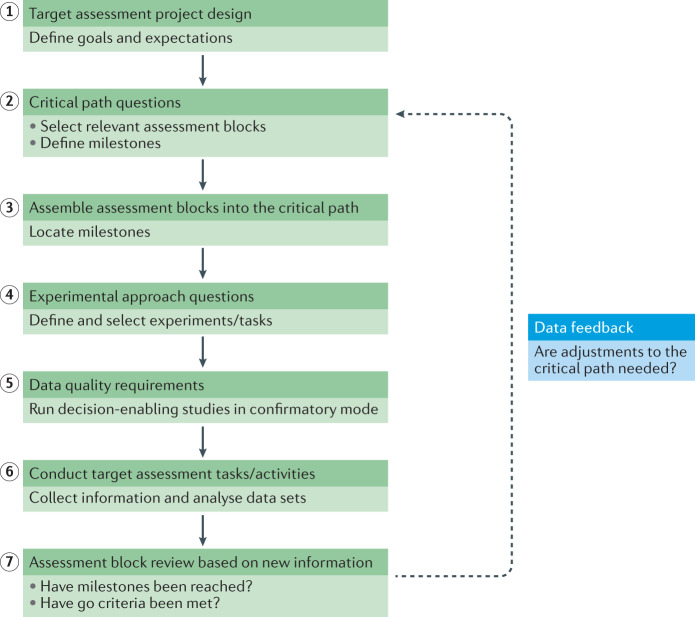
Fig. 2. Workflow of the target assessment process according to the GOT-IT recommendations.
Steps 1–7: the steps of the GOT-IT (Guidelines On Target Assessment for Innovative Therapeutics) workflow to define a project-specific critical path. The dotted arrow indicates an important data feedback mechanism, highlighting the recurrent nature of the data review exercise for each assessment block after a certain set of information has been collected. The design of studies and experiments as well as the definition of goals and expectations, specifying the direction of a translational project, can be facilitated by completing a target assessment project plan at the beginning of a project (see Supplementary Table 2 for a template).
Critical path questions help to prioritize assessment blocks
The process of selecting, positioning and prioritizing assessment blocks within the critical path is supported by a set of critical path questions (CPQs). These CPQs ensure that only relevant assessment blocks are selected. A similar set of research questions was recommended by Moore et al. in planning clinical and translational research40 and is part of the Translational Medicine Guide developed by Merck41.
The goal is to prioritize assessment block-related activities once a target is identified, allocate existing resources (such as money and time) and generate confidence in project progression. Overall, CPQs help to convey an understanding of the complexity of the target assessment process, provide an overview of relevant target assessment aspects and guide scientists’ decisions on the priority a certain assessment block should be given within the critical path. Using the list of CPQs, scientists (and funders) are enabled to detect key gaps and roadblocks in a translational target assessment project and to identify critical areas (‘red flags’) to be addressed at the early stages of a project.
A total of 40 CPQs have been designed to facilitate assembly of assessment blocks into a project-specific critical path (Box 2). It is important to note that when no information is available to answer a specific question or if questions are not relevant for a specific project, the option/answer ‘not applicable’ is justifiable.
Box 2 Critical path questions.
AB1: target–disease linkage (human targets)
Is the target perturbation a cause or consequence of the human disease process?
Is the therapeutic relevance (such as human connection) of models used sufficiently high for decision-making?
Is the target expression pattern known (that is, within the anticipated patient population)?
Is the target manipulation process clinically relevant?
Is the read-out used to detect target-dependent processes disease-relevant?
Is the stimulus used to activate or influence target-dependent processes disease-relevant?
Are the biological consequences of an observed effect size known?
AB2: target-related safety (human targets)
-
8.
Is the target selective and not genetically linked to other diseases (or phenotypes or organ systems)?
-
9.
Is there prior knowledge on safety of the target or reported evidence for the role of the target in a known pathway and/or physiological process that may be harmful if disrupted?
-
10.
Are in vitro or pharmacologically relevant animal models available for safety testing?
-
11.
Do models used for safety testing translate well to humans?
-
12.
Are safety biomarkers available and can adverse effects be monitored and/or predicted by safety biomarkers?
-
13.
Is there sufficient confidence that a necessary safety window has been or can be established?
-
14.
Is the disease life-threatening (at what stage of the disease is the target of relevance)?
-
15.
Is the tissue distribution of the target known (in humans or in animals)?
AB3: microbial targets (non-human targets)
-
16.
Do target properties show desirable features of an antimicrobial drug target (see AB3: microbial targets in text for details)?
-
17.
Can target essentiality be analysed under therapeutically relevant infection conditions?
-
18.
If the functional role of a target is known, does the target modulation/inhibition result in an expected phenotype?
-
19.
Does the phenotype (for example, bacterial viability) upon tool compound use differ compared with the phenotype of genetically modified strains (such as target knockout strains)?
-
20.
Do complementary or alternative pathways or reactions exist (not influenced by the target)?
-
21.
Is the target (or a close homologue) present only in microorganisms, but not in the host?
-
22.
Can target mutants be isolated and do these mutations lead to reduced inhibition of (or binding to) the altered target?
-
23.
Is the target present or essential only in specific species?
AB4: strategic issues (human and non-human targets)
-
24.
Is there an unmet medical need (independent of commercial interest and prevalence of disease)?
-
25.
Differentiation over current standard of care: Is the target either totally novel or addressed using novel technology that promises to be advantageous over previous approaches?
-
26.
Can all activities be conducted without infringing any intellectual property rights?
-
27.
Has or should intellectual property be generated and protected?
-
28.
Is the competitive landscape in the target area known?
-
29.
Have commercial needs been addressed?
-
30.
Has a patient stratification plan been developed?
AB5: technical feasibility (human and non-human targets)
-
31.
Can the target be assayed in a relevant system and are the assay read-outs quantifiable?
-
32.
Are biomarkers available to demonstrate target engagement in patients?
-
33.
Is the target accessible?
-
34.
Is the target modifiable?
-
35.
Are any tool modulators available?
-
36.
Has the likelihood of target-related tolerance development been evaluated?
General questions and data quality questions relevant for all assessment blocks
-
37.
Do decision-enabling data sets (for example, supporting target–disease linkage) originate from studies of high internal validity?
-
38.
Is the quality of tools and reagents available sufficient for decision-making (for example, cell line authentication)?
-
39.
Have documentation requirements been implemented?
-
40.
Are decision-enabling data sets based on converging evidence from independent studies?
Experimental approach questions define tasks for assessment blocks
To identify project-specific activities for each selected assessment block, scientists can use defined sets of experimental approach questions (EAQs), which are based on important experimental target assessment approaches42–56 (Fig. 1b; Supplementary Boxes 1–5). EAQs support evaluation of whether the confidence level achieved is sufficient for decision-making and reaching the next milestone or whether certain aspects of a selected assessment block should be addressed in more detail. In other words, they help determine whether assessment block-related tasks are sufficiently complete for the project to progress to the next assessment block or whether further resources should be invested to deepen understanding of the current activity.
EAQs also help to design high-quality ‘killer experiments’ with the potential to invalidate a target. Thus, EAQs provide the basis for ‘go/no-go’ decisions depending on the results of all experimental data obtained for each individual assessment block57–59.
The lists of EAQs illustrate the types of experiments that can be considered for target assessment. However, investigators are encouraged to adapt the assessment block-specific lists according to their needs and project resources. Similar to the CPQs, not all EAQs need to be taken into account if certain questions are justifiably irrelevant to the project.
Data quality, robustness and documentation requirements
Several steps in drug discovery and development need to be compliant with established GxP-based quality requirements such as good laboratory practice toxicology, but analogous standards for non-regulated areas of drug discovery and target assessment are not available. A specialized set of quality guidelines is needed that specifically focuses on study design, unbiased conduct, statistical analysis and transparent reporting, and that will support academic–industry interactions by aligning quality criteria in preclinical research60. Here, the FAIRsharing initiative, for example, is seeking to make quality proposals across all disciplines more visible to scientists61 and the European Quality in Preclinical Data (EQIPD) initiative has developed a novel preclinical research quality system that can be applied in both academic and industry environments (Table 1).
Table 1.
Examples of tools and resources for supporting target validation and assessment efforts
| Tools/resources | Content/references | URL |
|---|---|---|
| The Cancer Genome Atlas | A database of various human tumour profiles based on DNA, RNA, protein expression levels and epigenetic factors165 | https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga |
| Chemicalprobes.org | A portal that focuses on providing accessible expert advice for how to find and use chemical probes for biomedical research and drug discovery151 | https://www.chemicalprobes.org/ |
| European Lead Factory | A collaborative public–private partnership aiming to deliver innovative drug discovery starting points (including the definition of detailed assay criteria and screening requirements)166,167 | https://www.europeanleadfactory.eu/ |
| EQIPD Quality System | A novel preclinical research quality system that can be applied in both the public and private sectors168 and which has been developed by the European Quality in Preclinical Data (EQIPD) consortium formed by 29 institutions across 8 different countries | https://quality-preclinical-data.eu/ |
| FAIRsharing | A curated, informative and educational resource on data and metadata standards, interrelated to databases and data policies61 | https://fairsharing.org/ |
| Genotype-Tissue Expression project | A project that is building a public resource for tissue-specific gene expression and regulation169 | https://www.gtexportal.org/home/ |
| GOT-IT Expert Platform | A platform that facilitates contacts between academic researchers and industry experts to foster new academia–industry collaborations | http://portal.braincommons.org/public/tools/gotit |
| GWAS Catalog | A curated collection of all published genome-wide association studies enabling investigations to identify causal variants, understand disease mechanisms and establish targets for novel therapies170 | https://www.ebi.ac.uk/gwas/home |
| The Human Protein Atlas | An open-access resource with the aim to map all of the human proteins in cells, tissues and organs171 | https://www.proteinatlas.org/ |
| Mouse Genome Informatics | A collection of mouse mutations, related phenotypes and diseases172 | http://www.informatics.jax.org/ |
| Open Targets Platform | A platform that integrates data on phenotypes, co-localization and prioritization signatures for a given target in relation to a specific disease49 | targetvalidation.org |
| SGC (Chemical Probes) | A collaborative effort to provide a unique collection of probes with their associated data, control compounds and recommendations on their use163,164 | https://www.thesgc.org/chemical-probes |
| SPARK Global Initiative | An international network to promote exchange of expertise and to join forces to evaluate, enrich and advance projects focused on immediate unmet medical need173 | https://sparkglobal.io/ |
Further relevant drug target discovery platforms have been introduced and summarized, for example, by Paananen and Fortino174.
Consequently, questions related to data quality are crucial for various assessment blocks (Fig. 1b; Box 2) and need to be addressed after new assessment block-specific tasks and activities have been identified, especially for decision-enabling processes. To achieve this, an additional detailed set of data quality questions has been created (Box 3).
The data quality questions highlight the importance of increasing the internal validity of key experiments62, including crucial processes such as blinding and randomization, appropriate statistical power analyses and primary end point definitions. They also emphasize the need to establish external validity by multiple independent replicates as well as several orthogonal technologies, which provide greater confidence and converging evidence for the therapeutic relevance of a target.
In addition, a major requirement to ensure robust research outcomes is that researchers routinely question reagent purity, authenticate cell lines, validate antibodies and animal models, and include appropriate controls when planning and conducting an experiment63.
With regard to documentation, guiding principles to make data Findable, Accessible, Interoperable, and Reusable (FAIR) have been designed64. An experimental record should provide sufficient information and level of detail to permit peers to reconstruct and/or repeat a study, based on the information provided, and to compare outcomes65. Good documentation practices include regularly monitored and signature-approved methods for recording, correcting and managing data, documents and records, to ensure the reliability and integrity of information and data. For non-paper-based systems and documents, Title 21 of the Code of Federal Regulations describes the FDA regulations on electronic records. In particular, Part 11 defines the criteria under which electronic records and electronic signatures are considered trustworthy, reliable and essentially equivalent to paper records66. These guidelines can be critical, for instance, when generating intellectual property, and compliance with good documentation practices is required by both the US and the European regulatory authorities (that is, the FDA and the European Medicines Agency)67. In this context, the FDA uses the acronym ALCOA (‘attributable, legible, contemporaneous, original and accurate’) to describe its expectations regarding (electronic) research data66. ALCOA-plus, in addition, places further emphasis on the attributes of being ‘complete, consistent, enduring and available’. Establishing procedures for archiving and safeguarding of documents from the outset of a project and being compliant with the requirements of the regulatory authorities can save time and costs when these documents need to be made available for clinical trial applications or during inspections, for example.
Specific guidance for documentation and the traceability of research data is provided in Supplementary Box 6.
Box 3 Data quality questions.
Internal validity
For in vivo research and relevant in vitro studies: have the Landis 4 criteria (blinding, randomization, sample size calculation, inclusion/exclusion criteria pre-specified) been implemented?
For studies using target knockdown cells, have rescue experiments been performed to control for off-target effects (for example, re-expression of target wild-type and/or mutant version of the target)?
Were critical studies adequately controlled (for example, positive/negative controls included)?
Have ‘reference targets’ (for target invalidation) been used?
Was a statistical analysis plan designed and pre-specified before the beginning of experimental studies?
Has the primary outcome been pre-specified?
Allocation concealment: are procedures in place for protecting the randomization process?
Biological versus technical replicates: have biologically distinct or independent samples been analysed?
External validity and converging evidence
-
9.
Could fundamental experiments be independently replicated in-house?
-
10.
Could fundamental experiments be replicated by independent laboratories?
-
11.
Has more than one model system been used?
-
12.
Has more than one read-out been used?
-
13.
Has more than one method for target manipulation been used?
-
14.
Does tool compound use result in a similar phenotype compared with genetic target modulation?
-
15.
Do cell-free, cell-based and in vivo data show correlation?
-
16.
Have meta-analyses been performed using systems in which the target of interest has been manipulated?
-
17.
Has confirmation of key results by knockout studies (for example, via CRISPR–Cas9) been considered?
Quality of tools and reagents
-
18.
Were cell lines checked for potential contamination and other reagents for expected activity?
-
19.
Have cell line authentication procedures been performed?
-
20.
Have cells and products derived from mammalian cell culture been tested for the presence of Mycoplasma?
-
21.
Have specificity controls for critical antibodies been performed?
-
22.
For target knockout in vitro studies, has a matched (isogenic) pair of control and knockout cell lines been generated?
-
23.
Have plasmids been sequenced and the proteins assayed for confounding contaminants?
-
24.
Small interfering RNA (siRNA)/short hairpin RNA (shRNA) studies: have probes against the target been employed from multiple independent suppliers — with a full provision of positive and negative control probes?
-
25.
siRNA/shRNA studies: have multiple transfection reagents been used (to mitigate off-target effects)?
-
26.
shRNA studies: has a sufficiently high number of stable clones been generated to reproduce the desired effect?
-
27.
siRNA/shRNA studies: does the knockdown efficiency correlate with both biomarker and phenotypic read-outs?
AB1: disease linkage
Major reasons for the failure of new drugs to show efficacy in clinical trials include lack of data demonstrating a causal linkage of the drug target with the disease or a poor understanding of the role of the target in the underlying disease pathophysiology21. Thus, AB1 involves generating evidence that modulation of the target of interest leads to clinically relevant physiological effects (see Box 4 and Supplementary Box 7 for examples).
Box 4 Examples highlighting the importance of various target assessment aspects.
Target–disease linkage
The maternal embryonic leucine zipper kinase (MELK) was implicated as a therapeutic target in human cancer based on correlation data describing elevated levels of MELK RNA expression in human tumours and mouse models177 and an observed association between its increased expression levels and poor clinical prognosis178. MELK-targeted RNAi experiments and a small-molecule MELK inhibitor (OTS167), showing inhibited proliferation in cultured human cancer cells178 and in cell lines/xenografts179, respectively, were used to establish a causal role for MELK in cancer growth.
However, deleting MELK in different cancer cell lines using CRISPR–Cas9 had no beneficial effect180. In these cells, OTS167 was still active, suggesting that the observed cytotoxicity following OTS167 treatment reflected off-target, MELK-independent mechanisms181. Similarly, substantial off-target effects of the short hairpin RNA reagents used were also suggested to have contributed to their anti-proliferative effects182.
Applying the GOT-IT (Guidelines On Target Assessment for Innovative Therapeutics) framework by analysing causality more stringently (critical path question (CPQ) #1: ‘Is the target perturbation a cause or consequence of the human disease process?’) and by focusing on controlling tool compound and small interfering RNA quality (Box 3) could have helped to identify the target’s non-essential nature for cancer cell proliferation early on and could have directed the OTS167 programme to address the poor selectivity for MELK before progressing into expensive clinical proof-of-concept studies183.
Early safety de-risking of a novel target
The relevance of the GOT-IT CPQs in assessing target-related safety concerns can be illustrated by retrospective analysis of the following example, based on CPQ #9: ‘Is there prior knowledge on safety of the target or reported evidence for the role of the target in a known pathway and/or physiological process that may be harmful if disrupted?’
Targeting the receptor tyrosine kinase ErbB2 for cancer treatment leads to target-related cardiotoxicity181. This is due to the role of the ErbB2 pathway in cardiomyocyte differentiation and survival, involving the formation of a neuregulin 1b (NRG1)/ErbB2/ErbB4 complex that is critical for ErbB2 signalling. Whereas a more precise analysis of the involvement of ErbB2 in other tissues may have predicted the toxicity, understanding the role of ErbB2 and the mechanism of this on-target toxicity provided a starting point for the development of alternative drugs targeting different ErbB2 epitopes to allow for the assembly of the NRG1/ErbB2/ErbB4 complex, thereby circumventing the cardiotoxic effect184,185.
Target engagement biomarker use
Target engagement biomarkers enable an initial assessment of beneficial pharmacological activity, support translation from animals to humans and may be used as decision-making tools by providing information on mechanisms of action, dose–responses and efficacy. The need for such biomarkers is addressed by CPQ #32: ‘Are biomarkers available to demonstrate target engagement in patients?’
In the development programme for sitagliptin, the first approved compound for the treatment of type 2 diabetes in a class of drugs that target dipeptidyl peptidase 4 (DPP4), DPP4 enzyme activity was investigated as a target engagement biomarker. It was shown in preclinical studies that 80% inhibition of DPP4 resulted in maximal lowering of blood glucose levels. Similar degrees of DPP4 inhibition in the first human studies were associated with reduced blood glucose levels. These findings helped to design and conduct the subsequent phase II clinical studies and shortened the clinical development time186–188.
Tool compound use
CPQ #35 addresses the availability of tool compounds that modulate the target. The following example illustrates the importance of such compounds for testing a therapeutic hypothesis.
The Traf2 and Nck-interacting kinase (TNIK) was suggested to activate various WNT target genes based on small interfering RNA-mediated downregulation of TNIK levels as well as overexpression of mutant TNIK versions189. However, when a selective ATP-competitive TNIK inhibitor was discovered, the use of this tool compound revealed that small molecule-mediated inhibition of TNIK kinase activity had minimal effects on either WNT-driven transcription or cell viability190. This finding suggested that a scaffolding function of TNIK may be more important for WNT signalling than its kinase activity, and decreased confidence in TNIK kinase inhibition as a therapeutic strategy24.
Converging evidence and causal relationships
Available models and assays for preclinical target validation often inadequately assess causal relationships of direct relevance to humans and fail to provide converging evidence to support or refute a therapeutic hypothesis for the target. Even an apparently robust finding might still be a false positive, and a consistent pattern of bias or systematic error may explain the results68. However, risks due to limitations of observational data (for example, the omission or inconsistent measurement of crucial variables) can be mitigated by using multiple assay systems and models, which increase the chance of assessing the appropriate disease-relevant context. Thus, preclinical data are more readily translated to the clinical situation when they are based on converging evidence from at least two, preferably more, independent experimental procedures, each capturing differing features of the modelled disorder26.
To increase confidence and validity, it is important to differentiate between ‘association’ (for example, a target has altered expression, distribution or activity in diseased samples but does not cause disease development) and ‘causation’ (for example, altered pathophysiology as a result of target modulation)69. Criteria for establishing causal relationships have evolved70, but none of these criteria have absolute value alone, and causal inference often involves a process in which evidence accumulates from multiple sources71. One way to achieve this is by anchoring causation to ‘experiments of nature’, such as naturally occurring human conditions or states in which genetic variations that affect the levels and/or activity of a biological target have a reproducible effect on human physiology72. Such genetic information can complement existing lines of evidence, and can help establish a chain of causality, as a genetic perturbation (such as an inherited mutation in a target protein) is constant from birth and thus precedes the disease state rather than being affected by the disease environment. In the context of drug discovery, experiments of nature may mimic the effect of therapeutic target modulation and provide guidance to estimate dose–response curves72.
Relevance of the model system
Model systems with a clear link to the disease of interest, such as patient-derived primary cells or induced pluripotent stem cells with disease-linked mutations, are usually considered more representative of human physiology than generic cell lines or cellular systems with overexpressed single proteins73. Similarly, the closer an assay read-out is to a clinical end point, the more mechanisms modulated by the target (or modulating the target) with the potential to translate to clinical efficacy can be captured. Project resources should be invested to prioritize development of model systems with greater relevance to human disease.
Most assay systems to study target-relevant aspects require a stimulus for a set period of time to achieve a desired phenotype. The ideal stimulation conditions would be drawn from an accurate understanding of the disorder’s root causes. If such comprehensive knowledge is lacking, highly disease-relevant biological systems that intrinsically contain the appropriate stimulus can be most useful (for example, patient-derived cells incorporating disease-causing genetic alterations)73.
Target manipulation procedures
Functional changes in the target during the pathophysiology of a disease usually provide greater confidence than changes in the target expression levels. Highly selective manipulation processes (such as knockout technologies) are preferable to indirectly or non-selectively induced changes (such as histone deacetylase inhibition). Reversing the phenotype by manipulating the target in a bidirectional manner also provides a high degree of confidence in the functional role of the target. For example, a small interfering RNA/short hairpin RNA rescue experiment provides an elegant way to elucidate a target’s activity and to exclude potential off-target artefacts by re-expressing the target cDNA (as wild-type and/or mutant versions) that is resistant to the silencing reagent25.
Therapeutically relevant magnitude of change
From a clinical perspective, it is important to agree upon the most relevant model system (see above) and define meaningful study end points, which can then be used as reference points for expected effect sizes.
For data analysis, statistical testing based on appropriate thresholds that are set before data collection and interpretation can help to avoid bias and facilitate discrimination between a signal and noise. However, even strict significance thresholds cannot always separate true from false positive findings, and generating converging evidence is generally needed to determine which effect sizes are worth investigating and therapeutically relevant. If an outcome hypothesis is not well-founded and an experiment has low statistical power, there is a greater chance that false positive data will be generated, despite a statistically significant p value (which is often defined as p < 0.05)74. With a large sample size, the p value for an observation can be low even if the effect size is small and biologically irrelevant. Conversely, with small sample sizes or large variability, the p value for an observation could be >0.05, even though the difference is large enough to be biologically relevant75–79. Therefore, it should be critically considered whether the effect sizes observed in an experimental model are of a magnitude that would be clinically meaningful if confirmed in patients.
Selecting the most promising among multiple relevant drug targets
Chronic or complex diseases often develop as a combination of multiple genetic and environmental factors. If several potential drug targets exist, selection and prioritization processes should involve consideration of causality and effect sizes observed upon target modulation. In this context, it has to be acknowledged, however, that some preclinical models cannot be further improved given the current state of technology. This means that two or more targets may ‘score’ equally highly when assessing effect sizes and model relevance, for instance. In this situation, it might actually be beneficial to continue working initially on both targets and to exploit the rare occasion of having found targets with similar robust validation levels.
In addition, focusing purely on the magnitude of change may mean that biologically meaningful targets are overlooked. For example, small effect sizes of individual single-nucleotide polymorphisms in genome-wide association studies do not necessarily dictate low efficacy when the corresponding protein is targeted by a drug: the effect size of single-nucleotide polymorphism(s) in the gene coding for HMG Co-A reductase (HMGCR) is relatively modest, whereas the effect size of statins used as lipid-lowering drugs targeting HMGCR is much larger80,81.
AB2: safety
Toxic effects of drug candidates are a major cause of project terminations, and may result in limitation of dosage and/or limitation to specific patient populations if projects do progress. Safety issues can result from on-target (or target-related) toxic effects caused by modulating the biological function of the target of interest, and/or off-target toxic effects caused by modulation of other targets or pathways by a drug candidate, which are often linked to the properties of the compound tested (such as lack of selectivity or toxic metabolites)82.
In the context of the GOT-IT recommendations, the focus is on the assessment of target-related safety. For antimicrobial drug development, safety aspects related to targets present in the host are discussed in this section and those related to microbial targets are discussed under AB3.
Early identification of potential target-related safety risks and increased understanding of the underlying molecular mechanisms can guide project progression and help develop mitigation strategies21,83. The following activities may be considered to achieve this goal.
Prior knowledge
A first step is to review human and animal genetic databases to identify known loss-of-function or gain-of-function mutations in the target that may be relevant to disease or toxic effects, which may also reflect heterogeneity in the patient population of interest84. In addition, drug approval or clinical trial databases are potential sources of information relevant to toxicity if drugs modulating the target already exist85.
Target expression/tissue distribution
It is important to investigate and understand the gene, RNA and protein expression of the target in humans and in animals, as it can be assumed that the broader the target expression, the higher the risk for adverse effects resulting from administration of a drug that systemically modulates target activity83. Here, differential expression in animal or human disease samples versus healthy controls can facilitate early assessment of putative target-related adverse events. Useful resources to study target expression are listed in Table 1.
Similarly, if a target is highly expressed in organs that are not relevant to the therapeutic goal of target modulation, a critical functional role of the target in these organs also has to be assumed86, raising greater concern over potential adverse effects. Pleiotropic effects of target modulation can also affect multiple organs at different times or lead to more than one phenotype6.
Modulation of the target expression level
Modulation of target expression levels in transgenic animals or in a specific organ and characterization of the model phenotype can help identify potential target-related adverse events. Importantly, a target knockout may cause developmental effects up to embryonic lethality, thus restricting further experimental assessment. It may also lead to a different phenotype to that induced by pharmacological means, as other functions of the protein, such as protein–protein interactions, may be lost87,88. This issue could be addressed with gene knock-in studies in which the wild-type target is replaced by a mutant protein with properties closer to those of pharmacological intervention (for example, kinase-dead mutants). Alternatively, conditional knockout of target genes, both in animal models and in cell culture, has been used recently as a more selective approach to modulation of target expression in drug safety testing89,90.
Tool compound use
Studies with target-specific tool molecules may further help to confirm target-related safety issues observed using genetic approaches. In order to understand the target-related effects, it is an advantage to use highly selective compounds, if available (see also AB5). In addition, inactive enantiomers can be a powerful control to investigate target-related safety, as target-related toxicities should be minimal and other compound structure-related toxic effects should be comparable with those of the active isomer84,86. However, examples of inactive compounds that show structure-related toxicity different from the active enantiomer (such as thalidomide) do exist91.
Pathway analysis and target function
If understanding of the target function is limited, analysis of pathway upstream or downstream components (or specific cellular functions related to the target) can be highly informative. Target-related toxicity can also result from involvement of the target in potentially harmful pathways and/or physiological processes. Consideration of target paralogues, closely related genes or proteins with conserved domains similar to the target of interest may help to identify potential safety issues.
For antiviral therapies based on host targets, it has to be considered that different viruses often hijack the cellular machinery of the host in similar ways. Consequently, modulating these host pathways can inhibit all viruses that depend on that particular function. As a drawback, however, most cellular processes involved in viral replication are also likely to be essential for normal cellular activity, and so can result in side effects when these processes are modulated pharmacologically92.
Human and cross-species translatability
When choosing a model system to study target-related safety aspects, the likely cross-species translation of these toxicities must be considered. Sometimes, genes are only expressed in disease states or are not present/functional in the selected preclinical species93,94. Moreover, the organotypic quality of the model can be critical95. A human primary cell model (such as primary hepatocytes), for example, is more organotypically relevant than data from a human cell line (such as HepG2 cells).
Several other factors can also affect translatability, both in terms of safety and disease relevance. These include epigenetic changes and antigen drift, differences in cell culture conditions, the stage of disease investigated and gender96. For example, cells can differ in their responses to the same test compounds depending on the stimulus, as can the same strain of animal from different breeders. Females and males, both animals and humans, are well known to differ in many respects in their responses to drugs, as has been observed for pain treatments97. Consequently, it is crucial for translatability that the conditions likely to exist in the target human indication are recreated as closely as possible in preclinical studies.
Safety biomarkers
Developing robust safety read-outs/biomarkers is key for assessment of target-related safety aspects and the success of translational drug discovery programmes85. Target engagement markers can be used to identify cases where target occupancy may drive on-target toxicity.
Risk to benefit ratio
Ultimately, toxicities may restrict the potential patient population and define exclusions for certain patient groups (patient stratification). In addition, the risk profile may support modulation of a drug target in certain diseases and patient groups but not in others83. Understandably, the acceptance of target-related undesired effects is considerably higher in life-threatening conditions than in less devastating diseases or disease stages.
AB3: microbial targets
This assessment block addresses general aspects and attributes of non-human drug targets, with special emphasis on bacterial and viral target molecules.
For most antimicrobial research (and other therapeutic areas), drug discovery approaches have typically been divided into two classes: target-based drug discovery and phenotypic drug discovery. The target-based strategy is based on the identification and validation of a molecular target before lead discovery starts. In contrast, phenotypic screening identifies chemical matter that induces desired phenotypic changes in cells or organisms73,98,99, and this approach has been particularly popular in antibacterial drug discovery, given the simplicity and strong translational relevance of antibacterial activity as an assay read-out. Phenotypic drug discovery does not require prior knowledge about the compound’s mode of action. However, retrospective identification and validation of the role of molecular target(s) in the observed phenotypic responses (target deconvolution) can greatly facilitate subsequent target-specific optimization of pharmacological properties through structure–activity relationship studies. In addition, target identification and validation can enable species selectivity as well as target-based side effects to be addressed, thereby potentially reducing later-stage attrition.
In cases where antimicrobial targets have been established, the following aspects may need to be considered.
Microbial target properties
Relevant characteristics of microbial drug targets include: essentiality for microbial growth and survival; pharmacological tractability and accessibility; similarity to related mammalian molecules; presence in important pathogens; potential for the development of resistance; and lack of target-based cross-resistance100–102.
However, it is important to note that essentiality of targets can be conditional and context-dependent (especially for targets involved in metabolism) and can vary across species, cell types and growth conditions103,104. Thus, essentiality needs to be considered under therapeutically relevant infection conditions. When pursuing microbial targets, a further challenge is that the desired spectrum of activity for certain drugs can be limited if the target is not conserved across related bacteria or viruses, which has recently been shown for antivirals developed to treat hepatitis C virus infection105.
In some cases, inhibiting the synthesis of virulence or pathogenesis factors may present a target for antimicrobial strategies. This approach does not primarily focus on killing the infectious agent but on supporting the immune system to neutralize pathogens by disarming their lethal weapons, and reducing evolutionary pressure for the emergence of resistance106,107.
Targeted gene modification
To validate a microbial gene product as a drug target, the transient or permanent abrogation of its function should result in a loss of microbial infectivity, growth and/or proliferation, and viability under disease-relevant conditions (pathogenicity). Target mutations can be used to investigate the resistance-associated mechanism by transferring resistant mutations back to the parental strain. Conditional expression systems enable simultaneous assessment of multiple conditional mutants and regulation of target gene expression in response to a stimulus. Infection models based on conditional expression systems may help to clarify whether a target is required for initiating or maintaining an infection.
Assuming that a compound that inhibits the target is available, overexpression of a bacterial or parasite target should raise its minimum inhibitory concentration (MIC), whereas downregulation of the target expression level should reduce its MIC. However, results can be obscured if the inhibitor shows polypharmacology by modulating more than one target108,109.
Furthermore, target mutants preventing interaction with a compound should reduce compound inhibition of the altered target and increase the MIC. With targets encoded by single genes, these mutations can be identified by isolating the mutants and subsequent mutation mapping. As a specificity control, the target gene can be replaced with a known insensitive version.
Pathway analysis and virus–host interactions
Pathways involving targets for antimicrobials are generally essential for microbial viability and often related to macromolecular synthesis. Disruption of one of these pathways should significantly impair microbial survival under disease-relevant conditions, if no alternative pathway or target with redundant function exists. If a macromolecular synthesis pathway is affected specifically, incorporation of precursors of pathway end products should be inhibited preferentially110. In addition, it has to be ensured that the target is essential not only for microbial survival in vitro but also in the nutrient-rich human body in vivo, where host reaction products may be used to bypass a targeted biosynthesis pathway111.
To study specific virus–host interactions at the molecular level, cellular model systems that reflect the phenotype of the healthy organism are required. Although primary cells are most representative of the tissue they were isolated from, their use is limited by their short lifespan, challenges in expression of exogenous genes and an unnatural strong interferon-response to viral infection as cells approach senescence. Thus, it is critical to immortalize cells in a way that preserves the integrity of innate signalling pathways, including intact interferon-dependent as well as interferon-independent antiviral responses112.
Safety
One key question is whether or not a close human target homologue exists. If it does, then it is critical to identify a therapeutic window that would allow progression of development113. In this context, most emerging viral infections are acute, therefore limiting the issue of adverse effects, as shorter treatment periods may be sufficient102.
A further aspect to consider, especially for antibacterial drugs, is that many targets are common (similar) in most bacteria strains, and beneficial bacterial populations such as those in the gut may also be affected by pharmacological intervention.
Target location and expression
In general, the target should be expressed in infection-relevant cells and tissues. The permeability and efflux characteristics of various bacterial strains, especially Gram-negative bacteria, are critical determinants of antibacterial activity. So, for Gram-negative bacteria, extracellular or periplasmic targets are preferable, as compound uptake across both the outer and inner membranes as well as efflux by bacterial pumps are issues100,109.
Even though a target is expressed in different species, it may be essential in only one organism but dispensable in others due to bypass reactions or redundant enzyme equipment111. However, if the target enzyme is known to be present or essential in only a specific species, then lack of activity of a compound against other species can provide supportive evidence for the specificity of action100.
Resistance
There are several ways by which microbes can develop drug resistance. Target-related resistance mechanisms include target mutation/modification to alter drug binding and emergence of alternative (bypass) mechanisms to circumvent target functions. Thus, a single enzyme as a target may increase the likelihood of rapid resistance selection. In this case, the potential for emergence and the frequency of single-step spontaneous resistance owing to mutations in the target should be monitored (for example, through serial passaging experiments)100,109,113,114. For antibacterials, off-target resistance mechanisms such as enhanced drug efflux processes will also need to be taken into consideration (see AB5, assayability).
AB4: strategic issues
Progression of research on new targets towards the clinic often involves establishing start-up companies or partnerships with industry to get access to the resources and expertise required for drug development115. Thus, AB4 deals with strategic aspects of target assessment that are important for investors and industry partners, including intellectual property associated with validated targets, the extent of unmet medical needs for diseases in which the target is relevant and the competitor landscape, which all influence the commercial potential for a drug that modulates the new target.
Intellectual property
A validated target provides an opportunity to file a new patent on a target-based treatment approach, thereby creating intellectual property116. Importantly, however, a patent cannot be granted if an invention was previously known or used by others, or published anywhere in the world, including poster presentations and grant abstracts/applications (if published).
Obtaining a composition of matter patent — a very strong form of patent protection — is only possible if small-molecule compounds or biologics that modulate the target can be identified117. Alternatively, a patent application for the drug target itself can be considered, which would usually contain reach-through claims (that is, the application of an agonist/antagonist of target X to treat disease Y)118 or screening/assay claims (that is, a method of screening for molecules that agonize or antagonize target X)117. Reach-through claims have limited value, however, as US and European courts have ruled against patents that claim the use of any possible hypothetical compound/antibodies against the target of interest, without direct supporting data118. In contrast, screening assay claims are more likely to be granted, given that a novel screening assay developed by the patent applicant can be included in the patent to exemplify the claims. The protection through screening assay claims, nevertheless, is also rather weak as any deviation from the protected protocol is not covered and filed patents are (generally) published after 18 months116,119.
Evaluating the existing intellectual property landscape is also important. A freedom-to-operate analysis investigates the ability to develop and market a new (target-based) treatment approach, including the development of an assay to identify target modulators, without infringing the valid and enforceable intellectual property rights of third parties120. Several options can be considered if freedom to operate cannot be established due to existing patents that pose a potential obstacle to commercialization. These options include seeking alternative ways to develop the product outside the scope of a blocking patent; obtaining a licence under the blocking patent; opposing/invalidating blocking patents; and adjusting timing based on the expiration date of a blocking patent or considering the territorial restrictions of a blocking patent121.
Unmet medical needs
Although ‘unmet medical need’ is a widely used term in the healthcare sector, no standardized definition and method is available to assess and compare the medical needs addressed by different drug development projects. One way to investigate the unmet medical need is to clearly define the area of clinical interest at the beginning of a project. A new target-related treatment approach then fulfils an unmet medical need if it addresses a condition within this indication that is not adequately covered by existing treatments. The scale of unmet medical need can be assessed by determining the mortality, symptom and disease burden, side effects, treatment inconvenience, patient perception and time spent in disease122, amongst others.
A rationale is needed for the expectation that modulation of the target drives differentiation from current standard(s) of care. Ideally, a target is either totally novel or being addressed using novel technology that promises to be advantageous over previous approaches; for example, allosteric modulators that could be more selective than catalytic site inhibitors and thereby have a better safety profile.
Competitive landscape
The way in which a new treatment approach differs from existing medicines is often overlooked in academic drug discovery projects. However, for a project on a new target to be attractive to investors or industry partners, the competitive edge needs to be objectively evaluated. This exercise should provide information on how a potential new drug modulating the target can be differentiated from existing or future competitors and what type of data should be obtained to best demonstrate value. Differentiation may be in terms of specificity, reduced or narrower toxicity, or broader or greater efficacy, and should also consider patients’ and physicians’ satisfaction with existing therapeutics, as well as potential treatments that are in the pipeline from other companies.
To determine the viability of possible differentiation strategies to compete in the market, a SWOT analysis can be included in the competitive landscape assessment, which identifies and evaluates the strengths, weaknesses, opportunities and threats associated with a certain target-based product.
Evaluation of commercial potential and needs
Evaluation of the commercial potential of an early-phase drug R&D project by investors or potential industry partners is typically based on standard financial methodology, such as calculation of the net present value or risk-adjusted net present value123,124, which may be unfamiliar to academic researchers. More simply, however, commercial needs are often linked to a reasonable market size based on the unmet medical need and the range and location of patients eligible for therapy. In this respect, it might be beneficial to broaden the potential therapeutic uses by looking for additional indications in which the target might also play a role. Further factors to consider include the competitive position of the treatment approach, the reimbursement environment, the costs to reach critical decision points, the time to launch and the overall probability of technical and regulatory success for the programme21.
For universities, an important question is how to progress with the development of a new target-based approach once the target is experimentally validated, in order to best position it, for example, for licensing to industry. The institution should ensure that technology transfer and legal offices understand the balance between confidentiality and the freedom to publish on one side and offering simplified access to intellectual property on the other. This balancing act can involve confidential disclosure agreements, material transfer agreements and sponsored research agreements16. The evaluation of these factors will be facilitated by a strategic dialogue between all stakeholders involved.
AB5: feasibility
The potential for an innovative and promising molecular target to provide the basis for a successful drug discovery programme is affected by aspects related to the technical feasibility of the project, such as the ‘druggability’ and ‘assayability’ of a target, and biomarker availability, which may need to be taken into account early on when designing the critical path.
Target druggability
To qualify as druggable6,16, a target must be accessible to the therapeutic molecule and a measurable biological reaction must be provoked as a consequence of the drug interaction with the target. Current approaches to evaluate protein modifiability often consist of methods using sequence-related properties as well as three-dimensional structures of the target of interest drawn from crystallography and computer-based conformational assays. These structural models for the target and/or close homologues can later facilitate structure-based design methods and target engagement studies125–128. Furthermore, the location of a target in a specific organ influences the target accessibility and needs to be taken into account when designing screening assays or modulating compounds.
If target activation is required for disease modulation, it has to be clarified how this can be best achieved, for example, by inhibition of auto-inhibitory domains, by activation of a cell-surface receptor through ligand binding sites or by exploiting allosteric regulatory mechanisms. In the case that splice variants of the target exist, these may differ from the full-length protein with regard to its activity, expressed protein domains, cellular location and tissue distribution as well as affinity for drug compounds129, potentially affecting target treatment options.
In general, when a new target is being selected for a potential drug discovery programme, early insights into which therapeutic modality is most promising and whether the protein has a small-molecule binding site or, alternatively, an accessible antibody epitope is important130. To increase the number of druggable targets, innovative approaches have been developed, such as proteolysis targeting chimeric (PROTAC) molecules, which bind to the target of interest and, via another moiety, recruit intracellular E3 ligases with the aim to proteolytically degrade the target protein, thereby addressing previously undruggable target classes or protein–protein interactions131–134. Hence, the druggable space can be further broadened by applying novel technologies, which promises additional drug targets in the future.
Assayability
To support a screening programme for a suitable lead candidate, assays for target binding and/or function need to be available. In general, the majority of assays in use are based on either whole-cell systems or on purified target proteins (biochemical assays), directly measuring the target engagement of the drug candidates under development.
For human targets, subsequent translation of data from immortal cell lines to primary human cells is often crucial to ensure that the effects observed are indeed disease-relevant. Furthermore, three-dimensional cell cultures and high-content cellular imaging can be employed for screening assays in order to shrink the gap between in vitro cell models and real tissues135,136.
For microbial targets, limitations of biochemical assays (such as lack of information on the impact of bacterial drug efflux pumps) can be circumvented by monitoring phenotypic changes such as growth arrest in intact pathogens in response to drug candidates. In the simplest case, microbial growth directly results in an easily detectable signal (such as changes in absorption, fluorescence or bioluminescence). More complex techniques using tools such as microfluidics, fluorescence polarization activity assays or mass spectrometry have been developed that enable rapid and large-scale analysis and allow the screening of compounds for their ability to specifically interact with a given drug target, or use MIC shift assays to detect hypersensitive or resistant mutants and thereby control for compound specificity137–141.
For viruses, the assay read-out could be the virus titre, the expression level of viral proteins in infected cells or the cell viability. For viral load tests, it is generally advisable to aim for at least a 2–3 log10 drop in viral titre to reflect an acceptable overall viral load/copy number reduction. Follow-up assays are usually required to confirm the mode of action and establish that identified compounds are not acting by compromising host cell functions. Other key factors, which can influence the assay robustness and relevance, include the method applied to prepare the virus stock (such as using mosquito cells for flavivirus production to recapitulate the natural cycle of infectivity between mosquitoes and humans) and the choice of host cells142.
Whatever the assay format or the species origin of the target, several factors need to be considered when developing screening assays143,144, for example, the inclusion of known target ligands to confirm that the assay pharmacology is predictive for the disease state; reproducibility of the assay across assay plates and screening days for the duration of the entire drug discovery programme; size selection of microtitre plates, assay reagents and assay volumes to minimize the costs of the assay; assay quality, determined by the Z′ factor, which is based on the signal window and the variance of high and low signals145; compatibility of the assay with the solvent concentration used to store the intended compound library; and the availability of secondary and confirmatory screens to confirm effects of hit compounds after having established reliable dose–response curves in the primary assay.
Development and use of tool compounds
Tool compounds can be small molecules, peptides or antibodies that may not possess the properties of a lead compound, but have sufficient potency, biochemical and cellular selectivity, and cell permeability for their use to increase confidence in the relationship between engagement of the target and the observed phenotype24,31–33. A high-quality tool compound offers an ideal opportunity to test the therapeutic hypothesis with an agent that has an efficacy profile close to that of the desired clinical candidate, yet without needing to complete the often challenging, final-stage optimization steps of drug compounds, including the pharmacokinetic profile, drug–drug interactions and safety parameters31,146.
In general, when comparing the effects of tool compounds with genetic target knockout approaches, it has to be considered that a knockout removes all target functions, including enzymatic activity and scaffolding, whereas 100% modulation of all target functions with a tool compound is unlikely. Another difference is related to the timing of target modulation. Tool compounds can be added or removed, and can therefore readily be applied to illuminate the effects of target modulation at a specific point in a disease process. This is in contrast to genetic knockouts, although approaches for conditional silencing of gene expression are available147.
If tool compounds are used to support validation of a certain target for a specific disease, in vivo experiments should confirm that the concentration in relevant cells and tissues is adequate and linked to its efficacy. Ideally, this exposure–efficacy relationship can be investigated by measurement of target engagement inside the cell using, for instance, positron emission tomography ligands, activity-based proteomic profiling or other target engagement biomarkers21,24,31.
As a word of warning, there are many unsuitable compounds that have been widely used based on their purported activities against particular targets but do not meet critical potency and specificity criteria (such as observed efficacy due to polypharmacology), and conclusions drawn from their use must be made with reservation24. The selection of chemical probes is rarely based on objective assessment of all potential compounds and is often affected by historical and commercial bias32,148. Thus, principles defining a quality chemical probe have been developed33,149,150 (see also Supplementary Box 5). For example, to control for any off-target activity of the tool compound, a structurally orthogonal chemical probe to enable cross-validation studies as well as an inactive close analogue as a negative control (for example, an inactive enantiomer) are highly recommended31. Initiatives have been created to make available a large number of innovative high-quality probes generated by pharmaceutical companies and to provide community-driven support32,151 (see also Table 1).
Target-related tolerance to drug treatment
Unless drug treatment is acute, the development of target-related tolerance (that is, a loss of efficacy with repeated drug exposure) may be an important factor to consider as it can influence translational success or failure. There are various mechanisms that may potentially trigger such loss of effect, such as a decrease in the target availability for drug binding (for example, by affecting target receptor internalization through a change in its post-translational modification pattern152), a decrease in target sensitivity153 or a decrease in the ability of a target receptor to produce a downstream response (for example, due to a switch in G proteins binding to GPCRs)153. In addition, the turnover rate of a protein target can influence treatment options, and covalent inhibitors may be advantageous if the target has a long half-life in cells, leading to a potentially lower dosing schedule129.
In autoimmunity, the therapeutic induction of tolerance to autoantigens (for example, by surface receptor-mediated generation of regulatory T cells) may play a critical role for drug discovery154. Target supersensitivity can also occur, as with central dopamine receptors in some central nervous system disorders155, necessitating adjustments to drug dosing.
It is therefore advisable to include and potentially prioritize studies using repeated exposure of suitable tool compounds in the early phases of drug discovery when assessing a target, in particular when the desired mode of therapeutic action relies on stimulation or activation of the target (although there are also examples of inhibitors and antagonists that induce tolerance development)156–158.
Biomarker identification and development
Biomarkers enable a direct correlation to be drawn between target engagement and measurements of drug efficacy, and may provide surrogates for clinically meaningful end points that are useful in clinical trials, particularly early in development. In this respect, the early identification of mechanistic biomarkers able to confirm the mode of action can be critical to assess a target. A biomarker that detects a potent modulation/engagement of the target or target-dependent pathway, but without any therapeutic benefit, questions the validity of the target/pathway as a new treatment approach6.
A key step is to identify biomarkers that robustly reflect the same physiological outcomes induced by target modulation in preclinical models as in humans, thereby helping to translate target-related activities from animal to human159. Importantly, many biomarkers simply indicate disease-related observations without a key causal role31. Thus, the availability of cross-species, easily accessible, accurately measurable, validated biomarkers is crucial to the success of a translational programme, and failure to identify strategies for biomarker development may lead to early roadblocks within drug discovery programmes. Similar points are also relevant for microbial targets160.
Applying the GOT-IT workflow
Establishing a project plan
Entering drug discovery projects with a strategic plan that clearly outlines goals and expectations, milestones and go/no-go decision points helps to reduce operational risks and facilitates resource management16,36,161,162. Even at this early stage, several key factors for project success can be established (at least tentatively), such as identifying collaboration partners, target indication(s) and medical needs, as well as relevant biological test systems, tool compounds and biomarkers. Using this information, the criteria and requirements for transition from one assessment block to the next stage of the critical path will be defined. Thus, such a target assessment project plan summarizes at the very beginning how molecular targets are being assessed for entry into a drug discovery pipeline and what data sets need to be generated, taking into account the objectives of each individual translational project (Fig. 2, step 1 of the workflow). To support the project planning and design phase, a target assessment project plan template is provided in Supplementary Table 2.
Based on the target assessment project plan template, the next steps of the GOT-IT workflow ensure, using the CPQs provided, that all relevant assessment blocks are selected and assembled into the critical path (Fig. 2, steps 2–3). The EAQs support the identification of critical tasks and activities for each assessment block (Fig. 2, step 4), and the data quality questions help ensure that decision-enabling studies have a sufficiently high level of internal/external validity (Fig. 2, step 5). Examples of the use of CPQs and EAQs are shown in Boxes 4 and 5 (see also Supplementary Box 7).
It is important to stress, however, that not all steps can be pre-planned before starting a translational project and that target assessment is not a process written in stone. Essentially, the evaluation of assessment blocks is not only guided by predefined milestones and go/no-go decision points but also by a repeated data review and feedback process once a new set of information is available or a new data set has been generated (Fig. 2, steps 6–7). For this decision-making and reviewing process, helpful questions include: ‘Do key study results obtained meet go/no-go-decision criteria pre-specified in the target assessment project plan? Does the quality of evidence level attained enable go/no-go decisions? Based on the key study results obtained, are any critical path adjustments required? Is the selection and order of assessment blocks still relevant and meaningful?’.
Box 5 Examples of target assessment related to antimicrobial drugs.
Host-targeted antiviral drugs
A highly pathogenic novel coronavirus, severe acute respiratory syndrome-associated coronavirus 2 (SARS-CoV-2), emerged in 2019 as the cause of coronavirus disease 2019 (COVID-19)191. Confirmation of the viral use of human proteins, such as the transmembrane protease serine subtype 2 (TMPRSS2) and the angiotensin-converting enzyme 2 (ACE2) for viral entry, helped to quickly identify relevant and potentially promising molecular drug targets in the host192,193.
However, focusing on GOT-IT (Guidelines On Target Assessment for Innovative Therapeutics) critical path question (CPQ) #23: ‘Is the target present or essential only in specific species?’ revealed that ACE2 may be expressed in mice only in the tongue and skin, but not in the lung (as in humans)194, suggesting that the expression pattern of ACE2 in mammals is species-specific. In addition, several critical differences in the sequence of human ACE2 and mouse or rat ACE2 have been detected195. These findings may explain why mouse or rat ACE2 proteins have a low affinity for the S protein of SARS-CoV-2 (ref.196) and hamper the use of rodent models for SARS-CoV-2 studies.
Given the importance of animal models that mimic the human disease for developing novel therapeutics (related to CPQ #17: ‘Can target essentiality be analysed under therapeutically relevant infection conditions?’), the GOT-IT framework would prioritize investment in developing murine models that better reflect the interaction of the human target and drug candidates; for example, by using CRISPR–Cas9 knock-in technology to generate mouse models expressing human ACE2 (ref.197). The high priority of generating clinically relevant model systems (see CPQ #2: ‘Is the therapeutic relevance (such as human connection) of models used sufficiently high for decision-making?’) is also supported by findings that ACE2 is embedded in a complex and tightly regulated physiological system198, influenced by multiple factors such as gender199, co-medication200 and underlying pathophysiological conditions such as hypertension201.
Furthermore, target-related safety issues for the use of drugs such as ACE2 inhibitors in COVID-19 patients with hypertension need to be evaluated (see Supplementary Box 3 and CPQ #21).
Resistance to antibacterial drugs
Bacterial type I signal peptidases (SPases) can be inhibited by arylomycins and related lipoglycopeptide antibiotics. However, for drug development, only a prohibitively narrow spectrum of activity was originally found for these molecules as they did not show any activity against Gram-positive as well as Gram-negative bacteria tested202–204.
Applying the GOT-IT approach would have identified EAQ #3: ‘Is the target accessible to the inhibiting agent?’ and CPQ #22: ‘Can target mutants be isolated and do these mutations lead to reduced inhibition of (or binding to) the altered target?’ as the critical questions for SPase-focused projects.
Given that in Gram-negative bacteria the catalytic domain of SPase is located in the periplasmic space, target accessibility and insufficient penetration of the outer membrane could have been one reason for the observed lack of activity202,203. However, membrane penetration cannot explain the resistance of Gram-positive bacteria and addressing EAQ #3 indeed showed that arylomycins do have some intrinsic ability to penetrate the outer membrane.
Subsequent studies focusing on CPQ #22 revealed that Staphylococcus epidermidis, which is sensitive to the arylomycins, develops resistance via mutations in SPase and that analogous mutations are naturally present in many inherently resistant bacteria, such as Escherichia coli or Pseudomonas aeruginosa205. This analysis led to the identification of various bacterial species lacking these specific SPase mutations and, therefore, showing sensitivity to arylomycins.
So, understanding the molecular mechanism of drug resistance increased the activity spectrum of the arylomycins to unexpected bacterial species and could translate into new therapies to address the growing threat of multidrug-resistant infections205.
Bypass mechanisms and species specificity for antibacterial drugs
The relevance of CPQ #20: ‘Do complementary or alternative pathways or reactions exist (not influenced by the target)?’ and CPQ #23: ‘Is the target present or essential only in specific species?’ is illustrated by studies of the molecular target FabI (enoyl-acyl carrier protein (ACP) reductase) expressed by Staphylococcus aureus, Streptococcus pneumoniae and Haemophilus influenzae. This target was the basis for the fatty acid biosynthesis inhibitor programme, the only surviving lead series originating from a large screening campaign by GSK206. However, it was later found that this target was not suitable for the desired broad-spectrum approach as several other bacterial species do not rely on the enoyl-ACP reductase function of FabI due to the presence of other, distinct isoforms such as FabK, FabL and FabV, which can catalyse the same reaction and which vary in sensitivity to fatty acid biosynthesis inhibitors111,207–209.
Limitations and responsibilities of project leaders
The main motivation for the development of target assessment guidelines is to help scientists identify relevant activities to generate as much critical information as possible, in order to provide confidence in project progression and the commitment of additional resources. However, it must be recognized that there is no single best or correct approach, as too many project-specific elements exist that cannot be covered by general target assessment guidelines.
Although the GOT-IT recommendations support the identification of value inflexion points for each project, scientists are ultimately responsible for deciding when a ‘quality of evidence’ level is reached to enable go/no-go decisions and to move on to the next activity or assessment block. In other words, investigators must decide on the point at which further gains in knowledge no longer justify any additional investment of resources.
In this context, scientists need to be willing to design and to run ‘killer experiments’ that increase confidence in the target of interest, but also to determine a ‘de-risking’ process, potentially invalidating a target. Here, a common feature of academic drug discovery is the lack of definitive criteria for either pursuing or terminating a project. These need to be implemented to review and balance the decisions made, while carefully scrutinizing results and ideas59. The go/no-go approach is certainly unusual in an academic environment and may create challenges in the way projects are conducted. Academic scientists usually aim at finding a solution without weighing results against resources spent, whereas translational research requires a balanced cost–effect evaluation and a clear path forward to operationalize the basic knowledge obtained in understanding target–disease relationships. To complement existing research expertise and to increase capabilities, it is, therefore, highly advisable for academic project leaders to establish collaborations with experts early within (for example, with medicinal chemists) or outside (for example, with contract research organizations and biotech companies) the academic environment. To facilitate these interactions, the GOT-IT Expert Platform has been designed (Table 1).
Conclusions
The GOT-IT recommendations provide a structured approach to assemble an optimal path between the starting point (for example, identification of a potential new target) and the achievement of project-specific goals (for example, spin-off formation or initiation of a drug discovery project), and should help to increase the value of a translational programme, irrespective of the indication or target selected. The modular design of the GOT-IT recommendations allows scientists, funders and other stakeholders to focus on those target-related aspects that are most relevant for their respective goals and objectives. This also provides an opportunity to highlight the strengths and achievements of academic drug discovery projects and to generate compelling arguments to facilitate partnering with industry and/or support efforts to gain further investment.
Importantly, the recommendations not only focus on establishing a causal link between the target and the disease of interest but also raise awareness about technical and commercial aspects related to the target. These factors are important for typical project progression into clinical development, but are either often overlooked or not considered until later in the process, which may not be optimal (Fig. 3). Indeed, the full assessment of these aspects can change the direction of a translational project, help to invest resources in the most economic manner and increase the project value as well as the likelihood of successfully reaching project goals and milestones.
Fig. 3. Position of the GOT-IT recommendations in the drug discovery value chain.
The GOT-IT (Guidelines On Target Assessment for Innovative Therapeutics) recommendations cover and raise awareness for experimental, technical or translational/commercial activities within the drug discovery value chain that assess the potential therapeutic benefit of a molecular drug target (green areas). Guidance is also provided to support selection and prioritization of the most promising drug targets, up to the point where the first hit compounds are identified using available screening assays (light green areas). The GOT-IT recommendations do not address chemical and compound-related aspects, including hit/lead compound optimization, absorption, distribution, metabolism and excretion (ADME) studies, intellectual property and freedom-to-operate aspects of the compound, its physicochemical properties, drug–drug interactions, compound safety and so on, which are also critical for the success for drug development but are beyond the scope of this article (light blue areas). HTS, high-throughput screening.
The recommendations also direct activities and support decision-making by taking into account not only which experiments have been chosen and prioritized but also how they have been designed and executed, as robust and transparent data sets form the foundation for a successful project outcome. Here, the GOT-IT recommendations aim to ensure that the quality of evidence surrounding the target of interest is scientifically, technically and commercially compelling. This includes data quality requirements, which can protect against potential sources of bias and violations of good research practice during study design, conduct, analysis, reporting and storage. In this context, the EQIPD Quality System (Table 1) provides solutions and support for scientists and translational research projects on how to enhance the robustness, rigour and validity of research data. Several more resources to improve research tools and processes, based on collaborative efforts, are freely available for academic scientists to use when assessing a new target (Table 1). For example, the Structural Genomics Consortium (SGC), a public–private partnership focusing on pre-competitive protein-based research, facilitates new drug discovery programmes by generating knowledge and reagents that can be applied to validate therapeutic targets. SGC chemical probes are open-access reagents for the biomedical research community with no restrictions on their application151,163,164.
Such research community activities, together with the GOT-IT recommendations, promise to help to generate high-quality data sets, increase trust between partners and provide a strong foundation for subsequent drug discovery programmes. This could lead to substantial new therapeutic opportunities based on the many molecular drug targets that are still to be identified, validated and assessed.
Supplementary information
Acknowledgements
This work was supported by the German Federal Ministry of Education and Research (BMBF) programme Targetvalidierung für die pharmazeutische Wirkstoffentwicklung (Funding reference numbers: 16GW0192K and 16GW0193). The GOT-IT Team thanks all members of the BMBF-Target Validation Initiative for fruitful discussions and their help in producing the guidelines — especially A. Bera, H. Brötz-Oesterhelt, K.P. Koller, R. Kirsch, R. Metternich, H. Meyer, K. Ohlsen, P. Sennhenn and W. Wohlleben for providing critical input on the manuscript and the critical path concept. A full list of all members is provided in Supplementary Table 1. The authors thank A. Grünweller for providing important comments on the manuscript and K. Rakesh (née Sharma) von Manteuffel for her contribution in analysing the target validation/assessment literature. The GOT-IT team thank L. Lancashire and Cohen Veterans Bioscience as a partner for developing the GOT-IT Expert Platform as well as L. Leonhardt, A. Dwertmann and the VDI/VDE Innovation + Technik GmbH for their support and guidance during the project funding period. M.C.J.H. and M.J.P. (now retired) are members of the LOEWE Research Centre for Translational Medicine and Pharmacology, funded by the State of Hessen. A.B., M.J.P. and U.D. are members of the Preclinical Data Forum, a network of the European College of Neuropsychopharmacology and Cohen Veterans Bioscience, and the GOT-IT project outline originated from the network discussions.
Competing interests
M.J.P. is a member of the Scientific Advisory Boards of EpiEndo Pharmaceuticals, Cyclone Therapeutics and aidCURE GmbH. A.B. is an employee and/or shareholder of PAASP GmbH, PAASP US LLC, EXCIVA GmbH, Synventa LLC and RITEC Pharma. The other authors declare no competing financial interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41573-020-0087-3.
References
- 1.Bunnage ME. Getting pharmaceutical R&D back on target. Nat. Chem. Biol. 2011;7:335–339. doi: 10.1038/nchembio.581. [DOI] [PubMed] [Google Scholar]
- 2.Hay M, Thomas DW, Craighead JL, Economides C, Rosenthal J. Clinical development success rates for investigational drugs. Nat. Biotechnol. 2014;32:40–51. doi: 10.1038/nbt.2786. [DOI] [PubMed] [Google Scholar]
- 3.Dowden H, Munro J. Trends in clinical success rates and therapeutic focus. Nat. Rev. Drug Discov. 2019;18:495–496. doi: 10.1038/d41573-019-00074-z. [DOI] [PubMed] [Google Scholar]
- 4.Paul SM, et al. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nat. Rev. Drug Discov. 2010;9:203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- 5.Blake, R. A. in High Content Screening: A Powerful Approach to Systems Cell Biology and Drug Discovery (eds Taylor, D. L., Haskins, J. R. & Giuliano, K. A.) 367–377 (Humana, 2006).
- 6.Gashaw I, Ellinghaus P, Sommer A, Asadullah K. What makes a good drug target? Drug Discov. Today. 2011;16:1037–1043. doi: 10.1016/j.drudis.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Frearson J, Wyatt P. Drug discovery in academia — the third way? Expert Opin. Drug Discov. 2010;5:909–919. doi: 10.1517/17460441.2010.506508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Everett JR. Academic drug discovery: current status and prospects. Expert Opin. Drug Discov. 2015;10:937–944. doi: 10.1517/17460441.2015.1059816. [DOI] [PubMed] [Google Scholar]
- 9.Loregian A, Palù G. How academic labs can approach the drug discovery process as a way to synergize with big pharma. Trends Microbiol. 2013;21:261–264. doi: 10.1016/j.tim.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 10.Tralau-Stewart CJ, Wyatt CA, Kleyn DE, Ayad A. Drug discovery: new models for industry–academic partnerships. Drug Discov. Today. 2009;14:95–101. doi: 10.1016/j.drudis.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Edwards A. Reproducibility: team up with industry. Nature. 2016;531:299–301. doi: 10.1038/531299a. [DOI] [PubMed] [Google Scholar]
- 12.Rosenblatt M. How academia and the pharmaceutical industry can work together: the president’s lecture, annual meeting of the American Thoracic Society, San Francisco, California. Ann. Am. Thorac. Soc. 2013;10:31–38. doi: 10.1513/AnnalsATS.201209-075PS. [DOI] [PubMed] [Google Scholar]
- 13.Frye SV. Drug discovery in academic institutions. Hematol. Am. Soc. Hematol. Educ. Program. 2013;2013:300–305. doi: 10.1182/asheducation-2013.1.300. [DOI] [PubMed] [Google Scholar]
- 14.Verkman AS. Drug discovery in academia. Am. J. Physiol. Cell Physiol. 2004;286:C465–C474. doi: 10.1152/ajpcell.00397.2003. [DOI] [PubMed] [Google Scholar]
- 15.Yu HWH. Bridging the translational gap: collaborative drug development and dispelling the stigma of commercialization. Drug Discov. Today. 2016;21:299–305. doi: 10.1016/j.drudis.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 16.Dahlin JL, Inglese J, Walters MA. Mitigating risk in academic preclinical drug discovery. Nat. Rev. Drug Discov. 2015;14:279–294. doi: 10.1038/nrd4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergauer T, Ruppert T, Essioux L, Spleiss O. Drug target identification and validation: global pharmaceutical industry experts on challenges, best strategies, innovative precompetitive collaboration concepts, and future areas of industry precompetitive research and development. Ther. Innov. Regul. Sci. 2016;50:769–776. doi: 10.1177/2168479016651298. [DOI] [PubMed] [Google Scholar]
- 18.Strovel, J. et al. in Assay Guidance Manual (eds Sittampalam, G. S. et al.) (Eli Lilly & Company and National Center for Advancing Translational Sciences, 2004). [PubMed]
- 19.Begley CG, Ellis LM. Drug development: raise standards for preclinical cancer research. Nature. 2012;483:531–533. doi: 10.1038/483531a. [DOI] [PubMed] [Google Scholar]
- 20.Prinz F, Schlange T, Asadullah K. Believe it or not: how much can we rely on published data on potential drug targets? Nat. Rev. Drug Discov. 2011;10:712. doi: 10.1038/nrd3439-c1. [DOI] [PubMed] [Google Scholar]
- 21.Cook D, et al. Lessons learned from the fate of AstraZeneca’s drug pipeline: a five-dimensional framework. Nat. Rev. Drug Discov. 2014;13:419–431. doi: 10.1038/nrd4309. [DOI] [PubMed] [Google Scholar]
- 22.Morgan P, et al. Impact of a five-dimensional framework on R&D productivity at AstraZeneca. Nat. Rev. Drug Discov. 2018;17:167–181. doi: 10.1038/nrd.2017.244. [DOI] [PubMed] [Google Scholar]
- 23.Stock JK, Jones NP, Hammonds T, Roffey J, Dillon C. Addressing the right targets in oncology: challenges and alternative approaches. J. Biomol. Screen. 2015;20:305–317. doi: 10.1177/1087057114564349. [DOI] [PubMed] [Google Scholar]
- 24.Blagg J, Workman P. Chemical biology approaches to target validation in cancer. Curr. Opin. Pharmacol. 2014;17:87–100. doi: 10.1016/j.coph.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 25.Cortés-Cros M, Schmelzle T, Stucke VM, Hofmann F. The path to oncology drug target validation: an industry perspective. Methods Mol. Biol. 2013;986:3–13. doi: 10.1007/978-1-62703-311-4_1. [DOI] [PubMed] [Google Scholar]
- 26.Kaelin WG. Common pitfalls in preclinical cancer target validation. Nat. Rev. Cancer. 2017;17:425–440. doi: 10.1038/nrc.2017.32. [DOI] [PubMed] [Google Scholar]
- 27.Lu Q, et al. Applications of CRISPR genome editing technology in drug target identification and validation. Expert Opin. Drug Discov. 2017;12:541–552. doi: 10.1080/17460441.2017.1317244. [DOI] [PubMed] [Google Scholar]
- 28.Moore JD. The impact of CRISPR–Cas9 on target identification and validation. Drug Discov. Today. 2015;20:450–457. doi: 10.1016/j.drudis.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 29.Sim DS, Kauser K. In vivo target validation using biological molecules in drug development. Handb. Exp. Pharmacol. 2016;232:59–70. doi: 10.1007/164_2015_17. [DOI] [PubMed] [Google Scholar]
- 30.Leslie BJ, Hergenrother PJ. Identification of the cellular targets of bioactive small organic molecules using affinity reagents. Chem. Soc. Rev. 2008;37:1347–1360. doi: 10.1039/b702942j. [DOI] [PubMed] [Google Scholar]
- 31.Bunnage ME, Chekler ELP, Jones LH. Target validation using chemical probes. Nat. Chem. Biol. 2013;9:195–199. doi: 10.1038/nchembio.1197. [DOI] [PubMed] [Google Scholar]
- 32.Arrowsmith CH, et al. The promise and peril of chemical probes. Nat. Chem. Biol. 2015;11:536–541. doi: 10.1038/nchembio.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frye SV. The art of the chemical probe. Nat. Chem. Biol. 2010;6:159–161. doi: 10.1038/nchembio.296. [DOI] [PubMed] [Google Scholar]
- 34.Erdogan BR, Michel MC. Building robustness into translational research. Handb. Exp. Pharmacol. 2019;257:1–13. doi: 10.1007/164_2019_283. [DOI] [PubMed] [Google Scholar]
- 35.Chadwick AT, Segall MD. Overcoming psychological barriers to good discovery decisions. Drug Discov. Today. 2010;15:561–569. doi: 10.1016/j.drudis.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 36.Ellinger B, Gribbon P. Risk mitigation in academic drug discovery. Expert Opin. Drug Discov. 2016;11:333–336. doi: 10.1517/17460441.2016.1152257. [DOI] [PubMed] [Google Scholar]
- 37.US Food and Drug Administration. FDA’s Critical Path Initiativehttps://wayback.archive-it.org/7993/20180125035414/https:/www.fda.gov/ScienceResearch/SpecialTopics/CriticalPathInitiative/ucm076689.htm (2016).
- 38.Woodcock J. Paving the critical path of drug development: the CDER perspective. Nat. Rev. Drug Discov. 2014;13:783–784. doi: 10.1038/nrd4435. [DOI] [PubMed] [Google Scholar]
- 39.Woodcock J, Woosley R. The FDA critical path initiative and its influence on new drug development. Annu. Rev. Med. 2008;59:1–12. doi: 10.1146/annurev.med.59.090506.155819. [DOI] [PubMed] [Google Scholar]
- 40.Moore CG, Carter RE, Nietert PJ, Stewart PW. Recommendations for planning pilot studies in clinical and translational research. Clin. Transl. Sci. 2011;4:332–337. doi: 10.1111/j.1752-8062.2011.00347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dolgos H, et al. Translational Medicine Guide transforms drug development processes: the recent Merck experience. Drug Discov. Today. 2016;21:517–526. doi: 10.1016/j.drudis.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 42.Thomsen SK, Gloyn AL. Human genetics as a model for target validation: finding new therapies for diabetes. Diabetologia. 2017;60:960–970. doi: 10.1007/s00125-017-4270-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vasaikar S, Bhatia P, Bhatia PG, Chu Yaiw K. Complementary approaches to existing target based drug discovery for identifying novel drug targets. Biomedicines. 2016;4:27. doi: 10.3390/biomedicines4040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sahota T, Danhof M, Della Pasqua O. Pharmacology-based toxicity assessment: towards quantitative risk prediction in humans. Mutagenesis. 2016;31:359–374. doi: 10.1093/mutage/gev081. [DOI] [PubMed] [Google Scholar]
- 45.Andrade EL, et al. Non-clinical studies required for new drug development — part I: early in silico and in vitro studies, new target discovery and validation, proof of principles and robustness of animal studies. Braz. J. Med. Biol. Res. 2016;49:e5644. doi: 10.1590/1414-431X20165644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carney S. Target validation. Drug Discov. Today. 2012;17:S1–S2. doi: 10.1016/j.drudis.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 47.Chen XP, Du GH. Target validation: a door to drug discovery. Drug Discov. Ther. 2007;1:23–29. [PubMed] [Google Scholar]
- 48.Frigault MM, Barrett JC. Is target validation all we need? Curr. Opin. Pharmacol. 2014;17:81–86. doi: 10.1016/j.coph.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 49.Koscielny G, et al. Open Targets: a platform for therapeutic target identification and validation. Nucleic Acids Res. 2017;45:D985–D994. doi: 10.1093/nar/gkw1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mullard A. Reliability of ‘new drug target’ claims called into question. Nat. Rev. Drug Discov. 2011;10:643–644. doi: 10.1038/nrd3545. [DOI] [PubMed] [Google Scholar]
- 51.Ohlmeyer M, Zhou M-M. Integration of small-molecule discovery in academic biomedical research. Mt. Sinai J. Med. 2010;77:350–357. doi: 10.1002/msj.20197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Plata-Salamán CR, Ilyin SE. Considerations for target validation and industrial approaches. Methods Mol. Biol. 2007;361:299–309. doi: 10.1385/1-59745-208-4:299. [DOI] [PubMed] [Google Scholar]
- 53.Sioud M. Main approaches to target discovery and validation. Methods Mol. Biol. 2007;360:1–12. doi: 10.1385/1-59745-165-7:1. [DOI] [PubMed] [Google Scholar]
- 54.Smith C. Drug target validation: hitting the target. Nature. 2003;422:341. doi: 10.1038/422341a. [DOI] [PubMed] [Google Scholar]
- 55.Sweis RF. Target (in)validation: a critical, sometimes unheralded, role of modern medicinal chemistry. ACS Med. Chem. Lett. 2015;6:618–621. doi: 10.1021/acsmedchemlett.5b00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vandamme D, Minke BA, Fitzmaurice W, Kholodenko BN, Kolch W. Systems biology-embedded target validation: improving efficacy in drug discovery. Wiley Interdiscip. Rev. Syst. Biol. Med. 2014;6:1–11. doi: 10.1002/wsbm.1253. [DOI] [PubMed] [Google Scholar]
- 57.Jones LH. An industry perspective on drug target validation. Expert Opin. Drug Discov. 2016;11:623–625. doi: 10.1080/17460441.2016.1182484. [DOI] [PubMed] [Google Scholar]
- 58.Jekunen A. Decision-making in product portfolios of pharmaceutical research and development — managing streams of innovation in highly regulated markets. Drug Des. Devel. Ther. 2014;8:2009–2016. doi: 10.2147/DDDT.S68579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scannell JW, Bosley J. When quality beats quantity: decision theory, drug discovery, and the reproducibility crisis. PLoS ONE. 2016;11:e0147215. doi: 10.1371/journal.pone.0147215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jasny BR, et al. Fostering reproducibility in industry–academia research. Science. 2017;357:759–761. doi: 10.1126/science.aan4906. [DOI] [PubMed] [Google Scholar]
- 61.Sansone S-A, et al. FAIRsharing as a community approach to standards, repositories and policies. Nat. Biotechnol. 2019;37:358–367. doi: 10.1038/s41587-019-0080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kimmelman J, Mogil JS, Dirnagl U. Distinguishing between exploratory and confirmatory preclinical research will improve translation. PLoS Biol. 2014;12:e1001863. doi: 10.1371/journal.pbio.1001863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williams M. Reagent validation to facilitate experimental reproducibility. Curr. Protoc. Pharmacol. 2018;81:e40. doi: 10.1002/cpph.40. [DOI] [PubMed] [Google Scholar]
- 64.Wilkinson MD, et al. The FAIR guiding principles for scientific data management and stewardship. Sci. Data. 2016;3:1–9. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perrier L, et al. Research data management in academic institutions: a scoping review. PLoS ONE. 2017;12:e0178261. doi: 10.1371/journal.pone.0178261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rattan AK. Data integrity: history, issues, and remediation of issues. PDA J. Pharm. Sci. Technol. 2018;72:105–116. doi: 10.5731/pdajpst.2017.007765. [DOI] [PubMed] [Google Scholar]
- 67.Kumar Krishan. Good Documentation Practices (GDPs) in pharmaceutical industry. J. Anal. Pharm. Res. 2017;4:00100. [Google Scholar]
- 68.Ciesielski TH, et al. Diverse convergent evidence in the genetic analysis of complex disease: coordinating omic, informatic, and experimental evidence to better identify and validate risk factors. BioData Min. 2014;7:10. doi: 10.1186/1756-0381-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Decher N, Netter MF, Streit AK. Putative impact of RNA editing on drug discovery. Chem. Biol. Drug Des. 2013;81:13–21. doi: 10.1111/cbdd.12045. [DOI] [PubMed] [Google Scholar]
- 70.Hill AB. The environment and disease: association or causation? 1965. J. R. Soc. Med. 2015;108:32–37. doi: 10.1177/0141076814562718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Phillips CV, Goodman KJ. The missed lessons of Sir Austin Bradford Hill. Epidemiol. Perspect. Innov. 2004;1:3. doi: 10.1186/1742-5573-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Plenge RM, Scolnick EM, Altshuler D. Validating therapeutic targets through human genetics. Nat. Rev. Drug. Discov. 2013;12:581–594. doi: 10.1038/nrd4051. [DOI] [PubMed] [Google Scholar]
- 73.Vincent F, et al. Developing predictive assays: the phenotypic screening ‘rule of 3’. Sci. Transl. Med. 2015;7:293ps15. doi: 10.1126/scitranslmed.aab1201. [DOI] [PubMed] [Google Scholar]
- 74.Motulsky HJ. Common misconceptions about data analysis and statistics. Naunyn. Schmiedebergs Arch. Pharmacol. 2014;387:1017–1023. doi: 10.1007/s00210-014-1037-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Button KS, et al. Power failure: why small sample size undermines the reliability of neuroscience. Nat. Rev. Neurosci. 2013;14:365–376. doi: 10.1038/nrn3475. [DOI] [PubMed] [Google Scholar]
- 76.Sullivan GM, Feinn R. Using effect size — or why the P value is not enough. J. Grad. Med. Educ. 2012;4:279–282. doi: 10.4300/JGME-D-12-00156.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wasserstein RL, Schirm AL, Lazar NA. Moving to a world beyond “p < 0.05”. Am. Stat. 2019;73:1–19. [Google Scholar]
- 78.Dirnagl U. The p value wars (again) Eur. J. Nucl. Med. Mol. Imaging. 2019;46:2421–2423. doi: 10.1007/s00259-019-04467-5. [DOI] [PubMed] [Google Scholar]
- 79.Ioannidis JPA. The importance of predefined rules and prespecified statistical analyses: do not abandon significance. JAMA. 2019;321:2067–2068. doi: 10.1001/jama.2019.4582. [DOI] [PubMed] [Google Scholar]
- 80.Floris M, Olla S, Schlessinger D, Cucca F. Genetic-driven druggable target identification and validation. Trends Genet. 2018;34:558–570. doi: 10.1016/j.tig.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Teslovich TM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guengerich FP. Mechanisms of drug toxicity and relevance to pharmaceutical development. Drug. Metab. Pharmacokinet. 2011;26:3–14. doi: 10.2133/dmpk.dmpk-10-rv-062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Roberts RA. Understanding drug targets: no such thing as bad news. Drug Discov. Today. 2018;23:1925–1928. doi: 10.1016/j.drudis.2018.05.028. [DOI] [PubMed] [Google Scholar]
- 84.Rudmann DG. On-target and off-target-based toxicologic effects. Toxicol. Pathol. 2013;41:310–314. doi: 10.1177/0192623312464311. [DOI] [PubMed] [Google Scholar]
- 85.Hornberg JJ, et al. Exploratory toxicology as an integrated part of drug discovery. Part I: why and how. Drug Discov. Today. 2014;19:1131–1136. doi: 10.1016/j.drudis.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 86.Xu, J. J. & Urban, L. Predictive Toxicology in Drug Safety (Cambridge Univ. Press, 2010).
- 87.Knight ZA, Shokat KM. Chemical genetics: where genetics and pharmacology meet. Cell. 2007;128:425–430. doi: 10.1016/j.cell.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 88.Mellor HR, Bell AR, Valentin J-P, Roberts RRA. Cardiotoxicity associated with targeting kinase pathways in cancer. Toxicol. Sci. 2011;120:14–32. doi: 10.1093/toxsci/kfq378. [DOI] [PubMed] [Google Scholar]
- 89.Leonhard WN, et al. Salsalate, but not metformin or canagliflozin, slows kidney cyst growth in an adult-onset mouse model of polycystic kidney disease. EBioMedicine. 2019;47:436–445. doi: 10.1016/j.ebiom.2019.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lea JD, Clarke JI, McGuire N, Antoine DJ. Redox-dependent HMGB1 isoforms as pivotal co-ordinators of drug-induced liver injury: mechanistic biomarkers and therapeutic targets. Antioxid. Redox Signal. 2016;24:652–665. doi: 10.1089/ars.2015.6406. [DOI] [PubMed] [Google Scholar]
- 91.Fischer ES, et al. Structure of the DDB1–CRBN E3 ubiquitin ligase in complex with thalidomide. Nature. 2014;512:49–53. doi: 10.1038/nature13527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee SM-Y, Yen H-L. Targeting the host or the virus: current and novel concepts for antiviral approaches against influenza virus infection. Antiviral Res. 2012;96:391–404. doi: 10.1016/j.antiviral.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vamathevan JJ, et al. Minipig and beagle animal model genomes aid species selection in pharmaceutical discovery and development. Toxicol. Appl. Pharmacol. 2013;270:149–157. doi: 10.1016/j.taap.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 94.Bhoumik P, Del Rio-Espinola A, Hahne F, Moggs J, Grenet O. Translational safety genetics. Toxicol. Pathol. 2017;45:119–126. doi: 10.1177/0192623316675064. [DOI] [PubMed] [Google Scholar]
- 95.Hartung T, et al. Toward good in vitro reporting standards. ALTEX. 2019;36:3–17. doi: 10.14573/altex.1812191. [DOI] [PubMed] [Google Scholar]
- 96.Parnham MJ, Geisslinger G. Pharmacological plasticity—how do you hit a moving target? Pharmacol. Res. Perspect. 2019;7:e00532. doi: 10.1002/prp2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dance A. Why the sexes don’t feel pain the same way. Nature. 2019;567:448–450. doi: 10.1038/d41586-019-00895-3. [DOI] [PubMed] [Google Scholar]
- 98.Sams-Dodd F. Target-based drug discovery: is something wrong? Drug Discov. Today. 2005;10:139–147. doi: 10.1016/S1359-6446(04)03316-1. [DOI] [PubMed] [Google Scholar]
- 99.Lee JA, Berg EL. Neoclassic drug discovery: the case for lead generation using phenotypic and functional approaches. J. Biomol. Screen. 2013;18:1143–1155. doi: 10.1177/1087057113506118. [DOI] [PubMed] [Google Scholar]
- 100.Silver LL. Appropriate targets for antibacterial drugs. Cold Spring Harb. Perspect. Med. 2016;6:a030239. doi: 10.1101/cshperspect.a030239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Silver LL. Multi-targeting by monotherapeutic antibacterials. Nat. Rev. Drug. Discov. 2007;6:41–55. doi: 10.1038/nrd2202. [DOI] [PubMed] [Google Scholar]
- 102.Everts M, Cihlar T, Bostwick JR, Whitley RJ. Accelerating drug development: antiviral therapies for emerging viruses as a model. Annu. Rev. Pharmacol. Toxicol. 2017;57:155–169. doi: 10.1146/annurev-pharmtox-010716-104533. [DOI] [PubMed] [Google Scholar]
- 103.Larrimore KE, Rancati G. The conditional nature of gene essentiality. Curr. Opin. Genet. Dev. 2019;58–59:55–61. doi: 10.1016/j.gde.2019.07.015. [DOI] [PubMed] [Google Scholar]
- 104.Murima P, McKinney JD, Pethe K. Targeting bacterial central metabolism for drug development. Chem. Biol. 2014;21:1423–1432. doi: 10.1016/j.chembiol.2014.08.020. [DOI] [PubMed] [Google Scholar]
- 105.Asselah T, Boyer N, Saadoun D, Martinot-Peignoux M, Marcellin P. Direct-acting antivirals for the treatment of hepatitis C virus infection: optimizing current IFN-free treatment and future perspectives. Liver Int. 2016;36(Suppl. 1):47–57. doi: 10.1111/liv.13027. [DOI] [PubMed] [Google Scholar]
- 106.Mühlen, S. & Dersch, P. in How to Overcome the Antibiotic Crisis: Facts, Challenges, Technologies and Future Perspectives (eds Stadler, M. & Dersch, P.) 147–183 (Springer International, 2016).
- 107.Belete TM. Novel targets to develop new antibacterial agents and novel alternatives to antibacterial agents. Hum. Microbiome J. 2019;11:100052. [Google Scholar]
- 108.Silver LL. Challenges of antibacterial discovery. Clin. Microbiol. Rev. 2011;24:71–109. doi: 10.1128/CMR.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jackson N, Czaplewski L, Piddock LJV. Discovery and development of new antibacterial drugs: learning from experience? J. Antimicrob. Chemother. 2018;73:1452–1459. doi: 10.1093/jac/dky019. [DOI] [PubMed] [Google Scholar]
- 110.Singh NK, Selvam SM, Chakravarthy P. T-iDT: tool for identification of drug target in bacteria and validation by Mycobacterium tuberculosis. Silico Biol. 2006;6:485–493. [PubMed] [Google Scholar]
- 111.Brötz-Oesterhelt H, Sass P. Postgenomic strategies in antibacterial drug discovery. Future Microbiol. 2010;5:1553–1579. doi: 10.2217/fmb.10.119. [DOI] [PubMed] [Google Scholar]
- 112.Hare D, Collins S, Cuddington B, Mossman K. The importance of physiologically relevant cell lines for studying virus–host interactions. Viruses. 2016;8:297. doi: 10.3390/v8110297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Singh SB, Young K, Silver LL. What is an ‘ideal’ antibiotic? Discovery challenges and path forward. Biochem. Pharmacol. 2017;133:63–73. doi: 10.1016/j.bcp.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 114.Ioerger TR, et al. Identification of new drug targets and resistance mechanisms in Mycobacterium tuberculosis. PLoS ONE. 2013;8:e75245. doi: 10.1371/journal.pone.0075245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gehr S, Garner CC. Rescuing the lost in translation. Cell. 2016;165:765–770. doi: 10.1016/j.cell.2016.04.043. [DOI] [PubMed] [Google Scholar]
- 116.Reschen, R. Making the most of academic drug target discoveries. Oxford University Innovationhttps://innovation.ox.ac.uk/wp-content/uploads/2014/10/Making-the-most-of-academic-drug-target-discoveries.pdf (2014).
- 117.DDW. Patenting the Tools of Drug Discoveryhttps://www.ddw-online.com/business/p148569-patenting-the-tools-of-drug-discovery.html (2007).
- 118.Bohrer RA. Reach-through claims for drug target patents: Rx for pharmaceutical policy. Nat. Biotechnol. 2008;26:55–56. doi: 10.1038/nbt0108-55. [DOI] [PubMed] [Google Scholar]
- 119.USPTO. USPTO Will Begin Publishing Patent Applicationshttps://www.uspto.gov/about-us/news-updates/uspto-will-begin-publishing-patent-applications (2000).
- 120.Zhang YP. Around the block. Bioentrepreneur. 2010;28:1239–1241. [Google Scholar]
- 121.Nonaka, H. in FTO (Freedom to Operate) in the Pharmaceutical Industry 16–33 (Nomos Verlagsgesellschaft, 2018).
- 122.Vennemann M, et al. Future unmet medical need as a guiding principle for pharmaceutical R&D. Drug Discov. Today. 2019;24:1924–1929. doi: 10.1016/j.drudis.2019.06.004. [DOI] [PubMed] [Google Scholar]
- 123.Svennebring AM, Wikberg JE. Net present value approaches for drug discovery. Springerplus. 2013;2:140. doi: 10.1186/2193-1801-2-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Stewart JJ, Allison PN, Johnson RS. Putting a price on biotechnology. Nat. Biotechnol. 2001;19:813–817. doi: 10.1038/nbt0901-813. [DOI] [PubMed] [Google Scholar]
- 125.Surade S, Blundell TL. Structural biology and drug discovery of difficult targets: the limits of ligandability. Chem. Biol. 2012;19:42–50. doi: 10.1016/j.chembiol.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 126.Egner U, Hillig RC. A structural biology view of target drugability. Expert Opin. Drug Discov. 2008;3:391–401. doi: 10.1517/17460441.3.4.391. [DOI] [PubMed] [Google Scholar]
- 127.Perola E, Herman L, Weiss J. Development of a rule-based method for the assessment of protein druggability. J. Chem. Inf. Model. 2012;52:1027–1038. doi: 10.1021/ci200613b. [DOI] [PubMed] [Google Scholar]
- 128.Dunn, I. S. Searching for Molecular Solutions: Empirical Discovery and its Future (Wiley, 2010).
- 129.Bunnage ME, Gilbert AM, Jones LH, Hett EC. Know your target, know your molecule. Nat. Chem. Biol. 2015;11:368–372. doi: 10.1038/nchembio.1813. [DOI] [PubMed] [Google Scholar]
- 130.Marsden CJ, et al. The use of antibodies in small-molecule drug discovery. J. Biomol. Screen. 2014;19:829–838. doi: 10.1177/1087057114527770. [DOI] [PubMed] [Google Scholar]
- 131.Arkin MR, Tang Y, Wells JA. Small-molecule inhibitors of protein–protein interactions: progressing toward the reality. Chem. Biol. 2014;21:1102–1114. doi: 10.1016/j.chembiol.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ni D, Liu N, Sheng C. Allosteric modulators of protein–protein interactions (PPIs) Adv. Exp. Med. Biol. 2019;1163:313–334. doi: 10.1007/978-981-13-8719-7_13. [DOI] [PubMed] [Google Scholar]
- 133.Yesbolatova A, Tominari Y, Kanemaki MT. Ligand-induced genetic degradation as a tool for target validation. Drug Discov. Today Technol. 2019;31:91–98. doi: 10.1016/j.ddtec.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 134.Pettersson M, Crews CM. Proteolysis targeting chimeras (PROTACs) — past, present and future. Drug Discov. Today Technol. 2019;31:15–27. doi: 10.1016/j.ddtec.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pampaloni F, Ansari N, Stelzer EHK. High-resolution deep imaging of live cellular spheroids with light-sheet-based fluorescence microscopy. Cell Tissue Res. 2013;352:161–177. doi: 10.1007/s00441-013-1589-7. [DOI] [PubMed] [Google Scholar]
- 136.Cautain B, et al. High-content screening of natural products reveals novel nuclear export inhibitors. J. Biomol. Screen. 2014;19:57–65. doi: 10.1177/1087057113501389. [DOI] [PubMed] [Google Scholar]
- 137.De La Fuente R, Sonawane ND, Arumainayagam D, Verkman AS. Small molecules with antimicrobial activity against E. coli and P. aeruginosa identified by high-throughput screening. Br. J. Pharmacol. 2006;149:551–559. doi: 10.1038/sj.bjp.0706873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kaminski TS, Scheler O, Garstecki P. Droplet microfluidics for microbiology: techniques, applications and challenges. Lab Chip. 2016;16:2168–2187. doi: 10.1039/c6lc00367b. [DOI] [PubMed] [Google Scholar]
- 139.Schug KA, et al. Direct affinity screening chromatography-mass spectrometry assay for identification of antibacterial agents from natural product sources. Anal. Chim. Acta. 2012;713:103–110. doi: 10.1016/j.aca.2011.11.038. [DOI] [PubMed] [Google Scholar]
- 140.Liu X, Chen Y, Fierke CA. A real-time fluorescence polarization activity assay to screen for inhibitors of bacterial ribonuclease P. Nucleic Acids Res. 2014;42:e159. doi: 10.1093/nar/gku850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.DeVito JA, et al. An array of target-specific screening strains for antibacterial discovery. Nat. Biotechnol. 2002;20:478–483. doi: 10.1038/nbt0502-478. [DOI] [PubMed] [Google Scholar]
- 142.Warren TK, et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hughes JP, Rees S, Kalindjian SB, Philpott KL. Principles of early drug discovery. Br. J. Pharmacol. 2011;162:1239–1249. doi: 10.1111/j.1476-5381.2010.01127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.European Lead Factory. Requirements for Drug Target Proposalshttps://www.europeanleadfactory.eu/how-submit/drug-target-assays/requirements (2016).
- 145.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 146.Garbaccio RM, Parmee ER. The impact of chemical probes in drug discovery: a pharmaceutical industry perspective. Cell Chem. Biol. 2016;23:10–17. doi: 10.1016/j.chembiol.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 147.O’Connor CJ, Laraia L, Spring DR. Chemical genetics. Chem. Soc. Rev. 2011;40:4332–4345. doi: 10.1039/c1cs15053g. [DOI] [PubMed] [Google Scholar]
- 148.Antolin AA, et al. Objective, quantitative, data-driven assessment of chemical probes. Cell Chem. Biol. 2018;25:194–205.e5. doi: 10.1016/j.chembiol.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Workman P, Collins I. Probing the probes: fitness factors for small molecule tools. Chem. Biol. 2010;17:561–577. doi: 10.1016/j.chembiol.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cohen P. Guidelines for the effective use of chemical inhibitors of protein function to understand their roles in cell regulation. Biochem. J. 2010;425:53–54. doi: 10.1042/BJ20091428. [DOI] [PubMed] [Google Scholar]
- 151.Müller S, et al. Donated chemical probes for open science. eLife. 2018;7:e34311. doi: 10.7554/eLife.34311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Meng X, et al. FBXO38 mediates PD-1 ubiquitination and regulates anti-tumour immunity of T cells. Nature. 2018;564:130–135. doi: 10.1038/s41586-018-0756-0. [DOI] [PubMed] [Google Scholar]
- 153.Walsh, C. T. & Schwartz-Bloom, R. D. Pharmacology: Drug Actions and Reactions (CRC, 2004).
- 154.Rayner F, Isaacs JD. Therapeutic tolerance in autoimmune disease. Semin. Arthritis Rheum. 2018;48:558–562. doi: 10.1016/j.semarthrit.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 155.Seeman P. All roads to schizophrenia lead to dopamine supersensitivity and elevated dopamine D2high receptors. CNS Neurosci. Ther. 2011;17:118–132. doi: 10.1111/j.1755-5949.2010.00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bespalov A, Müller R, Relo A-L, Hudzik T. Drug tolerance: a known unknown in translational neuroscience. Trends Pharmacol. Sci. 2016;37:364–378. doi: 10.1016/j.tips.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 157.Costa LG, Schwab BW, Murphy SD. Tolerance to anticholinesterase compounds in mammals. Toxicology. 1982;25:79–97. doi: 10.1016/0300-483x(82)90021-x. [DOI] [PubMed] [Google Scholar]
- 158.Löscher W, Schmidt D. Experimental and clinical evidence for loss of effect (tolerance) during prolonged treatment with antiepileptic drugs. Epilepsia. 2006;47:1253–1284. doi: 10.1111/j.1528-1167.2006.00607.x. [DOI] [PubMed] [Google Scholar]
- 159.Wendler A, Wehling M. The translatability of animal models for clinical development: biomarkers and disease models. Curr. Opin. Pharmacol. 2010;10:601–606. doi: 10.1016/j.coph.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 160.Nora D, Salluh J, Martin-Loeches I, Póvoa P. Biomarker-guided antibiotic therapy — strengths and limitations. Ann. Transl. Med. 2017;5:208. doi: 10.21037/atm.2017.04.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pritchard JF, et al. Making better drugs: decision gates in non-clinical drug development. Nat. Rev. Drug Discov. 2003;2:542–553. doi: 10.1038/nrd1131. [DOI] [PubMed] [Google Scholar]
- 162.Herschel M. Portfolio decisions in early development. Pharm. Med. 2012;26:77–84. [Google Scholar]
- 163.Lee WH. Open access target validation is a more efficient way to accelerate drug discovery. PLoS Biol. 2015;13:e1002164. doi: 10.1371/journal.pbio.1002164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gileadi O, et al. The scientific impact of the structural genomics consortium: a protein family and ligand-centered approach to medically-relevant human proteins. J. Struct. Funct. Genomics. 2007;8:107–119. doi: 10.1007/s10969-007-9027-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cancer Genome Atlas Research Network et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Karawajczyk A, Orrling KM, de Vlieger JSB, Rijnders T, Tzalis D. The European lead factory: a blueprint for public–private partnerships in early drug discovery. Front. Med. 2016;3:75. doi: 10.3389/fmed.2016.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kingwell K. European lead factory hits its stride. Nat. Rev. Drug Discov. 2016;15:221–222. doi: 10.1038/nrd.2016.64. [DOI] [PubMed] [Google Scholar]
- 168.Bespalov A, et al. Introduction to the EQIPD quality system. OSF Prepr. 2020 doi: 10.31219/osf.io/ng32b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lonsdale J, et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Buniello A, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Uhlen M, et al. Towards a knowledge-based human protein atlas. Nat. Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 172.Bello SM, Smith CL, Eppig JT. Allele, phenotype and disease data at mouse genome informatics: improving access and analysis. Mamm. Genome. 2015;26:285–294. doi: 10.1007/s00335-015-9582-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Parrish MC, Tan YJ, Grimes KV, Mochly-Rosen D. Surviving in the valley of death: opportunities and challenges in translating academic drug discoveries. Annu. Rev. Pharmacol. Toxicol. 2019;59:405–421. doi: 10.1146/annurev-pharmtox-010818-021625. [DOI] [PubMed] [Google Scholar]
- 174.Paananen J, Fortino V. An omics perspective on drug target discovery platforms. Brief. Bioinform. 2019 doi: 10.1093/bib/bbz122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Landis SC, et al. A call for transparent reporting to optimize the predictive value of preclinical research. Nature. 2012;490:187–191. doi: 10.1038/nature11556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.The NPQIP Collaborative group Did a change in Nature journals’ editorial policy for life sciences research improve reporting? BMJ Open Science. 2019;3:e000035. doi: 10.1136/bmjos-2017-000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gray D, et al. Maternal embryonic leucine zipper kinase/murine protein serine-threonine kinase 38 is a promising therapeutic target for multiple cancers. Cancer Res. 2005;65:9751–9761. doi: 10.1158/0008-5472.CAN-04-4531. [DOI] [PubMed] [Google Scholar]
- 178.Wang Y, et al. MELK is an oncogenic kinase essential for mitotic progression in basal-like breast cancer cells. eLife. 2014;3:e01763. doi: 10.7554/eLife.01763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Chung S, Nakamura Y. MELK inhibitor, novel molecular targeted therapeutics for human cancer stem cells. Cell Cycle. 2013;12:1655–1656. doi: 10.4161/cc.24988. [DOI] [PubMed] [Google Scholar]
- 180.Lin A, Giuliano CJ, Sayles NM, Sheltzer JM. CRISPR/Cas9 mutagenesis invalidates a putative cancer dependency targeted in on-going clinical trials. eLife. 2017;6:e24179. doi: 10.7554/eLife.24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lin A, et al. Off-target toxicity is a common mechanism of action of cancer drugs undergoing clinical trials. Sci. Transl. Med. 2019;11:eaaw8412. doi: 10.1126/scitranslmed.aaw8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wang Y, et al. Correction: MELK is an oncogenic kinase essential for mitotic progression in basal-like breast cancer cells. eLife. 2018;7:e36414. doi: 10.7554/eLife.36414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.McDonald IM, Graves LM. Enigmatic MELK: the controversy surrounding its complex role in cancer. J. Biol. Chem. 2020;295:8195–8203. doi: 10.1074/jbc.REV120.013433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Fedele C, Riccio G, Malara AE, D’Alessio G, De Lorenzo C. Mechanisms of cardiotoxicity associated with ErbB2 inhibitors. Breast Cancer Res. Treat. 2012;134:595–602. doi: 10.1007/s10549-012-2103-8. [DOI] [PubMed] [Google Scholar]
- 185.Blomme EAG, Will Y. Toxicology strategies for drug discovery: present and future. Chem. Res. Toxicol. 2016;29:473–504. doi: 10.1021/acs.chemrestox.5b00407. [DOI] [PubMed] [Google Scholar]
- 186.Krishna R, Herman G, Wagner JA. Accelerating drug development using biomarkers: a case study with sitagliptin, a novel DPP4 inhibitor for type 2 diabetes. AAPS J. 2008;10:401–409. doi: 10.1208/s12248-008-9041-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Durham TB, Blanco M-J. Target engagement in lead generation. Bioorg. Med. Chem. Lett. 2015;25:998–1008. doi: 10.1016/j.bmcl.2014.12.076. [DOI] [PubMed] [Google Scholar]
- 188.Wagner JA. Strategic approach to fit-for-purpose biomarkers in drug development. Annu. Rev. Pharmacol. Toxicol. 2008;48:631–651. doi: 10.1146/annurev.pharmtox.48.113006.094611. [DOI] [PubMed] [Google Scholar]
- 189.Mahmoudi T, et al. The kinase TNIK is an essential activator of Wnt target genes. EMBO J. 2009;28:3329–3340. doi: 10.1038/emboj.2009.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ho K-K, et al. Discovery of 4-phenyl-2-phenylaminopyridine based TNIK inhibitors. Bioorg. Med. Chem. Lett. 2013;23:569–573. doi: 10.1016/j.bmcl.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 191.Zhu N, et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Shang J, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hoffmann M, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sun K, Gu L, Ma L, Duan L. Atlas of ACE2 gene expression in mammals reveals novel insights in transmisson of SARS-Cov-2. Preprint at. bioRxiv. 2020 doi: 10.1101/2020.03.30.015644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Komatsu T, et al. Molecular cloning, mRNA expression and chromosomal localization of mouse angiotensin-converting enzyme-related carboxypeptidase (mACE2) DNA Seq. 2002;13:217–220. doi: 10.1080/1042517021000021608. [DOI] [PubMed] [Google Scholar]
- 196.Zhai X, et al. Comparison of SARS-CoV-2 spike protein binding to human, pet, farm animals, and putative intermediate hosts ACE2 and ACE2 receptors. Preprint at. bioRxiv. 2020 doi: 10.1101/2020.05.08.084061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Sun S-H, et al. A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe. 2020;28:124–133.e4. doi: 10.1016/j.chom.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Bourgonje AR, et al. Angiotensin-converting enzyme-2 (ACE2), SARS-CoV-2 and pathophysiology of coronavirus disease 2019 (COVID-19) J. Pathol. 2020;251:228–248. doi: 10.1002/path.5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Gemmati D, et al. COVID-19 and individual genetic susceptibility/receptivity: role of ACE1/ACE2 genes, immunity, inflammation and coagulation. Might the double X-chromosome in females be protective against SARS-CoV-2 compared to the single X-chromosome in males? Int. J. Mol. Sci. 2020;21:3474. doi: 10.3390/ijms21103474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Li Y, Zhou W, Yang L, You R. Physiological and pathological regulation of ACE2, the SARS-CoV-2 receptor. Pharmacol. Res. 2020;157:104833. doi: 10.1016/j.phrs.2020.104833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Perrotta F, Matera MG, Cazzola M, Bianco A. Severe respiratory SARS-CoV2 infection: does ACE2 receptor matter? Respir. Med. 2020;168:105996. doi: 10.1016/j.rmed.2020.105996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Kulanthaivel P, et al. Novel lipoglycopeptides as inhibitors of bacterial signal peptidase I. J. Biol. Chem. 2004;279:36250–36258. doi: 10.1074/jbc.M405884200. [DOI] [PubMed] [Google Scholar]
- 203.Roberts TC, Smith PA, Cirz RT, Romesberg FE. Structural and initial biological analysis of synthetic arylomycin A2. J. Am. Chem. Soc. 2007;129:15830–15838. doi: 10.1021/ja073340u. [DOI] [PubMed] [Google Scholar]
- 204.Schimana J, et al. Arylomycins A and B, new biaryl-bridged lipopeptide antibiotics produced by Streptomyces sp. Tü 6075. I. Taxonomy, fermentation, isolation and biological activities. J. Antibiot. 2002;55:565–570. doi: 10.7164/antibiotics.55.565. [DOI] [PubMed] [Google Scholar]
- 205.Smith PA, et al. Optimized arylomycins are a new class of Gram-negative antibiotics. Nature. 2018;561:189–194. doi: 10.1038/s41586-018-0483-6. [DOI] [PubMed] [Google Scholar]
- 206.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 207.Marrakchi H, et al. Characterization of Streptococcus pneumoniae enoyl-(acyl-carrier protein) reductase (FabK) Biochem. J. 2003;370:1055–1062. doi: 10.1042/BJ20021699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Zhu L, Lin J, Ma J, Cronan JE, Wang H. Triclosan resistance of Pseudomonas aeruginosa PAO1 is due to FabV, a triclosan-resistant enoyl-acyl carrier protein reductase. Antimicrob. Agents Chemother. 2010;54:689–698. doi: 10.1128/AAC.01152-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Moir DT. Identification of inhibitors of bacterial enoyl-acyl carrier protein reductase. Curr. Drug Targets Infect. Disord. 2005;5:297–305. doi: 10.2174/1568005054880154. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.