Abstract
Extracellular vesicle (EV) is a unified terminology of membrane-enclosed vesicular species ubiquitously secreted by almost every cell type and present in all body fluids. They carry a cargo of lipids, metabolites, nucleic acids and proteins for their clearance from cells as well as for cell-to-cell communications. The exact composition of EVs and their specific functions are not well understood due to the underdevelopment of the separation protocols, especially those from the central nervous system including animal and human brain tissues as well as cerebrospinal fluids, and the low yield of proteins in the separated EVs. To understand their exact molecular composition and their functional roles, development of the reliable protocols for EV separation is necessary. Here we report the methods for EV separation from human and mouse unfixed frozen brain tissues by a sucrose step gradient ultracentrifugation method, and from human cerebrospinal fluids by an affinity capture method. The separated EVs were assessed for morphological, biophysical and proteomic properties of separated EVs by nanoparticle tracking analysis, transmission electron microscopy, and labeled and label-free mass-spectroscopy for protein profiling with step-by-step protocols for each assessment.
Keywords: brain tissue, cerebrospinal fluid, exosome, extracellular vesicles, microvesicle, nanoparticle tracking analysis, transmission electron microscopy, proteome
1. Introduction
Extracellular vesicles (EV) include exosomes (50–150nm), which are secreted into the extracellular space after fusion of multivesicular bodies (MVBs) with plasma membrane; ectosomes/microvesicles (150–1000nm), which are shed from plasma membrane; and apoptotic bodies (1000–5000nm), which are generated from apoptotic cells. EVs are produced by almost all cell types in the central nervous system [1–3]. EVs contain many species of nucleic acids (microRNA, ncRNA, mRNA, DNA among others), lipids, and proteins that can be transferred to recipient cells as a form of cell-to-cell communication [4–10]. Several proteins are reported as markers for exosomes, such as tetraspanin protein (CD9, CD63 and CD81), syntenin-1, TSG101, and major histocompatibility complex (MHC) class I and II [11,12]. In comparison, markers for microvesicles include glycoprotein 1b, actinin-4, heat shock protein (HSP) 90B1, mitofilin, and myosin light chain [11–14]. EVs are found in all body fluids, including blood, urine, saliva and cerebrospinal fluid (CSF) [15,16]. EVs have been extensively investigated by liquid biopsy modality for diagnosis as well as therapeutic vehicles for drug delivery especially in cancer fields, but much less in the neuroscience field [17–21]. We are, however, still in an infancy stage for standardizing the protocols for the separation of EVs from animal and human biospecimens [1,22]. The International Society for Extracellular Vesicles (ISEV) members published a guideline on Minimal Information for Studies of Extracellular Vesicles (MISEV) in 2014 [22], which was updated in 2018 [1]. In the MISEV2018 guideline, they classified the EV separation protocols into four categories based on recovery and specificity, and created a checklist to demonstrate the EV nature and the degree of purity of an EV preparation. Standardization of these protocols for efficient recovery of EVs with a high purity from biological samples, however, has not been achieved yet. In the past 10 years, there has been expanded investigation of physiological and pathological functions of EVs from cell culture or biospecimen. In addition, these studies have focused on EVs in neurodegenerative diseases including Alzheimer’s disease (AD), Huntington’s disease, Prion disease, Parkinson’s disease, and amyotrophic lateral sclerosis. It is now known that brain derived-EVs contain pathogenic proteins, such as tau, amyloid beta (Aβ), superoxide dismutase, transactive response DNA binding protein 43kDa (TDP-43) and α-synuclein [15,23–27]. These EVs interestingly contribute to the cell-to-cell propagation of the disease [23,25,28,29]. EVs containing pathogenic proteins are also detectable in CSF, serum, and plasma from patients with neurodegenerative diseases [30–35]. Investigation of the brain-derived EV cargo, therefore, is important to further understand the progression of neurodegenerative diseases. In addition, the CSF, serum, and plasma-derived EVs are also of particular interest as liquid biopsy samples for diagnosing and monitoring the progression of neurodegenerative diseases. In this report, we describe step-by-step protocols for the assessment of morphological, biophysical and proteomic properties of separated EVs by transmission electron microscopy, nanoparticle tracking analysis and mass-spectroscopy.
2. Preparation for human biospecimen studies
2.1. Preparation of Institutional Review Board (IRB) submission, and the workstation and outfit for minimizing protein contamination
Materials and methods
Equipment:
AirClean 600 Series Ductless chemical workstation
Biological Safety Cabinet
Laboratory supplies:
3M™ 1860Healthcare Particulate Respirator and Surgical Mask (# 18–992 Thermo Fischer Scientific)
Disposable laboratory coat (# 17–100-940 Thermo fisher Scientific)
Disposable Polypropylene Bouffant Cap (# 19–170-900 or 1 Thermo Fisher Scientific)
Disposable Filtering Facepiece Respirator, N95 (# 8511 Thermo Fisher Scientific)
Grey safety goggle (# 19–181-513 Thermo Fisher Scientific)
Regular bleach (# 30966 Clorox)
N-GEN Nitrile Gloves (# 44–101 Genesee Scientific)
Procedure:
2.1.1. Ethics
The research using human subjects will require institutional approval, typically from the Institutional Review Board (IRB). Researchers should confirm or consult the IRB before initiating any research including request of human biospecimens and biological data.
2.1.2. Human biospecimens
Researchers will submit requests to human biospecimen banks including brain tissue banks and blood banks. This normally involves approval of the request by the bank, followed by arrangement of the shipping of the requested materials. The details are beyond the scope of this article. Prior to the contact with biospecimens, they will need pre-screening for infectious agents, especially for the experiments not having fixation or sterilization steps for inactivating the potential infectivity of the biospecimens.
2.1.3. Handling of human biospecimens
Note: For human biospecimens, biosafety level 2 procedures should be implemented for the experiment as follows.
Laboratory practices: Access to the laboratory is restricted when work is being conducted.
Safety equipment: Appropriate personal protective equipment (PPE) is worn, including lab coats, gloves, eye protection and face shields. All procedures that can cause infection from aerosols or splashes are performed within a biological safety cabinet (BSC). An autoclave or an alternative method of decontamination is available.
Facility construction: The laboratory has self-closing doors. A sink and eyewash are readily available.
2.1.4. Preparation for mass-spectroscopy
Wear classic laboratory coat, laboratory gloves, safety goggles, and disposable masks and caps. Always perform the experiment in the AirClean workstation for minimizing the protein contamination.
3. Methods for separation and assessment of human biospecimen-derived EVs
3.1. Protocols for separation of EVs from unfixed frozen human and murine brain tissues
Materials and methods
Reagents:
Unfixed frozen human brain tissue
Note: The brains (1.0g) for this experiment were obtained from the Greater Los Angeles Veteran’s Affairs Hospital and stored at −80°C.
Unfixed frozen murine brain tissue
Note: The whole brain tissues were collected from adult C57BL/6 mouse, snap frozen in liquid nitrogen, and stored at −80°C.
Plastic Petri Dishes (# S33580A Thermo fisher Scientific)
Papain (# LK003178 Worthington-biochemical corporation)
Razor blade (# 12–640 Thermo Fischer Scientific)
Tweezer, Stainless Steel (# 58086-NM Ted Pella Inc)
Earle’s Balanced Salt Solution (EBSS) (# 14155063 Gibco/Thermo Fisher Scientific)
Hibernate E solution (# A1247601 Gibco/Thermo Fisher Scientific)
Halt Protease and Phosphatase Inhibitor Cocktails (# PI78443 Thermo Fisher Scientific)
Fisher Scientific Isotemp Water Bath (Thermo Fisher Scientific)
IKA RW16 Basic Electronic Overhead Stirrer (IKA)
PTFE tissue grinder (standard clearance 0.15–0.25mm, # 89026–384 VWR)
High-speed centrifuge (# 5720R Eppendorf)
High-speed centrifuge (# 5430R Eppendorf)
High-speed centrifuge (# Avanti J-E JA25–50 Beckman Coulter)
Ultracentrifuge (# Optima-XE SW41, Beckman Coulter)
Sterile nylon mesh cell strainers (40-μm pore size, # 22–363-547 Thermo Fisher Scientific) Hydrophilic Polyethersulfone (PES) membrane filter (0.22-μm pore size) (# SLGP033RS EMD Millipore)
BD Luer-Loktm Disposable Syringes without Needles 10-mL (# 309604 Becton Dickinson)
Phosphate buffered saline (PBS, Ca/Mg free, pH 7.4) (# 10–010-049 Gibco/Thermo Fisher Scientific)
Note: The PBS should be filtrated twice with PES membrane filter (0.22-μm pore size).
Sucrose (# S5–3 Thermo Fisher Scientific)
Note: The sucrose should be dissolved and diluted in double-filtered PBS (dfPBS) with PES membrane filter (0.22-μm pore size).
Polyallomer ultracentrifuge tube with 13.2 mL capacity (# 331372 Beckman Coulter)
Olympus 15 mL Centrifuge Tubes (# 28–101 Genesee Scientific)
Olympus 50 mL Conical Centrifuge Tubes (# 28–106 Genesee Scientific)
LoBind Microcentrifuge Tubes 1.5 mL (# 022–43-108–1 Eppendorf)
LoBind Microcentrifuge Tubes 0.5 mL (# 022–43-106–4 Eppendorf)
Polycarbonate Bottle with Screw-On Cap 50 mL (# 357002 Beckman Coulter)
qEV original column (# qEV original/70nm - 5Pack IZON)
qEV Rack (# qEV Rack IZON)
LP Vortex Mixer (# 88880017 Thermo Fisher Scientific)
mySPIN™ 6 Mini Centrifugae (# 75004061 Thermo Fisher Scientific)
ProPette™ Electronic Pipette Controller (#1191K61 Thomas Scientific)
Procedure:
3.1.1. Dissection of unfixed human and mouse frozen brain tissues
Transfer 0.5g of unfixed frozen brain tissue to plastic petri dish on ice
Add 1mL of ice-cold Hibernate E solution to plastic petri dish
-
Chop the brain tissue on ice using a razor blade to generate 0.5mm3 sections
Note: In this protocol, 0.5g of frozen human cortical grey matter or whole frozen mouse brain tissue is used per sample.
-
Transfer the section samples to 15-mL centrifuge tubes and add Hibernate E solution containing 20 units of Papain in Earle’s Balanced Salt Solution (EBSS) up to 3mL
Note: Enzymes such as papain, collagenase, and trypsin are commonly used for dissection. The enzyme should be selected with subsequent analysis.
Incubate in a water bath at 37°C for 15 min by stirring once every 5 min
Place immediately on ice and add 6mL of ice-cold Hibernate E with Halt Protease and Phosphatase Inhibitor Cocktails (1:100)
-
Homogenize gently the dissociated brain tissue samples (20 strokes) with a glass-Teflon homogenizer
Note: This homogenate step may need to utilize a Dounce homogenizer (loose pestle) or no homogenizer to distinguish intracellular vesicle from extracellular vesicle for subsequent analysis.
Filtrate the homogenized tissue samples with sterile nylon mesh filter (40-μm pore size) to new 50-mL tube
3.1.2. Differential centrifugation
-
Centrifuge the tissue samples at 300 × g for 10 min at 4°C using 5720R from Eppendorf
Note: This pellet contains cell debris and nucleus.
Transfer the supernatant to new 15-mL tube
-
Centrifuge at 2,000 × g for 10 min at 4°C using 5720R from Eppendorf
Note: This pellet contains apoptotic bodies and microsomes.
Transfer the supernatant to polycarbonate bottles or 1.5-mL tube
-
Centrifuge at 10,000 × g for 10 min at 4°C using Avanti J-E JA25–50 from Beckman Coulter or 5430R from Eppendorf
Note: This pellet contains Large EVs (microvesicles) and plasma membranes.
Filtrate the supernatant through a PES membrane filter (0.22-μm pore size) into a new polyallomer ultracentrifuge tube with 13.2-mL capacity
Add ice-cold Hibernate E with Halt Protease and Phosphatase Inhibitor Cocktails up to 12.5mL
-
Centrifuge at 140,000 × g for 70 min at 4°C using Optima-XE SW41 from Beckman Coulter
Note: Since the high-speed ultracentrifugation can induce the formation of EV aggregates, EV aggregation may be prevented by cushioning 1mL of 1.0M sucrose on the tube bottom.
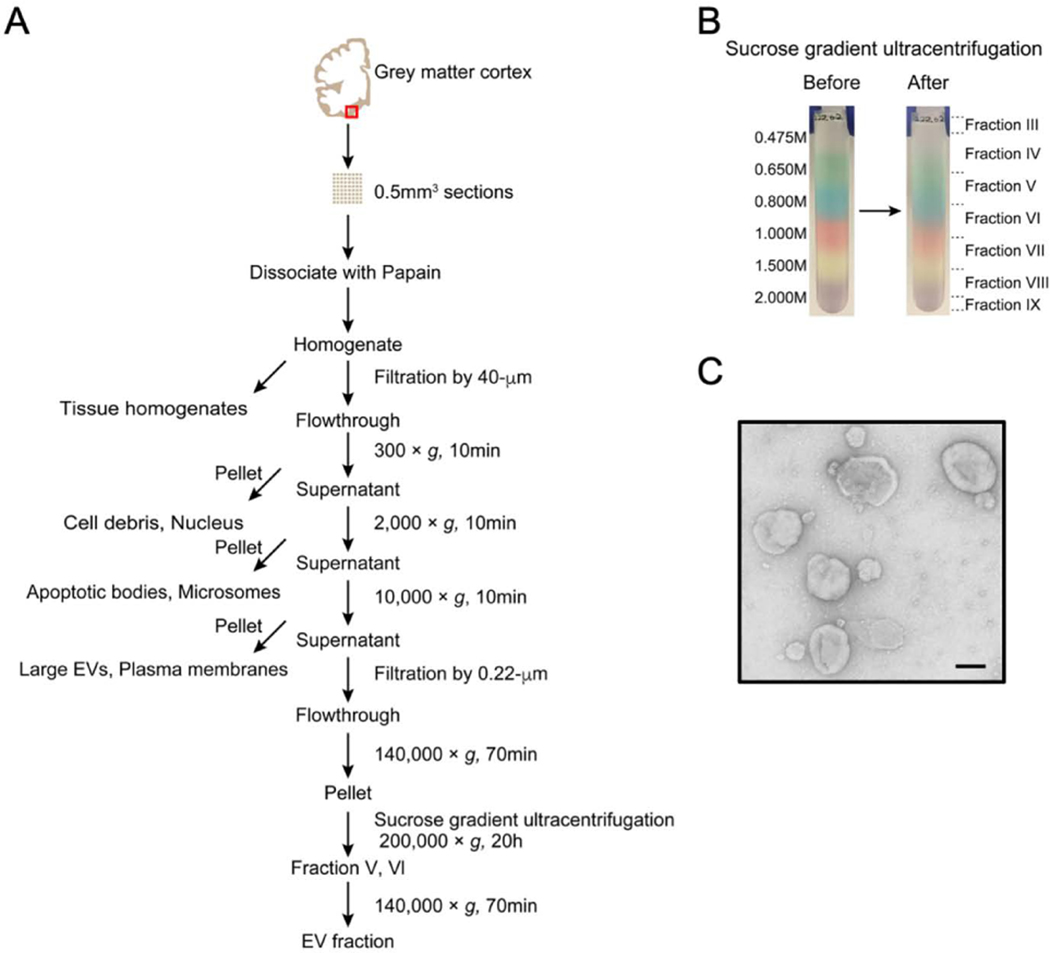
3.1.3. Sucrose gradient ultracentrifugation
After ultracentrifugation (3.1.2.- Step 8), the supernatant is completely discarded, and pellets are resuspended in 2mL of 0.475M of sucrose solution
-
Create the sucrose step gradient with six 2-mL steps starting from 2.0M to 1.5M, 1.0M, 0. 825M, 0.65M, and 0.475M (containing the resuspended pellet) in a polyallomer ultracentrifuge tube
Note: These sucrose solutions were colored with commercially available food coloring to facilitate capture of the EV-rich interphase present between certain steps (Please see Figure 1).
Centrifuge at 200,000 × g (35,000 rpm) for 20 h at 4°C using Optima-XE SW41 from Beckman Coulter
-
Collect in 2mL fractions, except for the first and last fractions, which are 1mL each.
Note: Fraction “III” corresponds to the 1mL removed from the tube, and particles in this area had a buoyant density of approximately 1.08 g/cm3. The area between the first (0.475M) and second (0.650M) step is collected and corresponded to fraction “VI” with a buoyant density of 1.08 – 1.10 g/cm3, while the area between the second (0.650M) and third (0.825M) steps is collected and correspond to fraction “V” with a buoyant density of 1.10 – 1.12 g/cm3, and the interphase between the third (0.825M) and fourth (1.000M) steps correspond to fraction “VI” with a buoyant density of 1.12 – 1.15 g/cm3. Interphase between the fourth (1.000M) and fifth (1.500M) steps, fraction VII; between the fifth (1.500M) and sixth (2.000M) steps, fraction VIII; and the 1mL remaining in the tube, fraction IX (please see Figure 1). The fractions V and VI (1.10 – 1.15 g/cm3) are EV-rich fractions.
Dilute the V and VI fractions separately to a total volume of 12.5mL with dfPBS in a polyallomer ultracentrifuge tube
Centrifuge at 140,000 × g for 70 min at 4°C using Optima-XE SW41 from Beckman Coulter
Discard the supernatant and resuspend the final pellet from each fraction in 30μL of dfPBS
Figure 1. Flowchart of EV separation from unfixed human brain tissue via SG-UC method.
A) Sucrose gradient ultracentrifugation (SG-UC) protocol for separation of EVs from unfixed human frozen brain tissue. See section 3.1 for detailed method. Red square shows gray matter in brain cortex. B) Representative example of sucrose gradient before and after ultracentrifugation. C) Transmission electron microscopy (TEM) image of frozen human brain tissue-derived EVs (pooled fractions V and VI). Scale bar; 100 nm.
3.1.4. Size exclusion chromatography (qEV original column)
-
Equilibrate the qEV original column with at least 30mL of dfPBS buffer
Note: Allow to come to room temperature.
After ultracentrifugation (3.1.2.-step 8), the supernatant is completely discarded, and the pellet is resuspended in 1000μL of dfPBS. Mix by vortexing
Load the 1000μL of the vortexed sample to the qEV original column
-
Add in 3.0mL minus the initial sample volume of dfPBS to top of the column
Note: Don’t load more than 2.0mL at a time. The first 6 fractions (3mL) do not contain vesicles (fraction 1–6).
-
Add 500μL of dfPBS at a time to top of the column
Note: It collect this completely in a tube, and then add 500μL more for the next fraction.
-
Collect 5 vesicle fractions of 500μL of each (fraction 7–11)
Note: It is recommended to measure the particle number and protein concentration of those fractions. EVs elute predominantly into fractions 7, 8 and 9 with removal of ~99.8% of bulk proteins. If higher purity is desired, collect only the earliest peak fraction (fraction 7 and 8).
3.2. Protocols for separation of EVs from human cerebrospinal fluid (CSF)
Materials and Methods
Reagents:
Human cerebrospinal fluid (CSF) specimen
Note: Postmortem CSF specimens (1.0mL, 37 years old female, PMI 9h, pre-screened for infectious diseases, no previous cycle of freezing and thawing) were obtained from University of Maryland Brain and Tissue Bank and stored at −80°C. For identification of 700 EV proteins using 1D-LC-MS/MS (Thermo Scientific Q-Exactive with Waters NanoAcquity UPLC), at least 500μL of human CSF is needed to collect EVs (1.6 × 1010 particles / mL). The sample volume should be empirically optimized for this purpose.
High-speed centrifuge (# 5720R Eppendorf)
Spin-X centrifuge tube filter (cellulose acetate membrane, pore size 0.22-μm) (# CLS8160 Millipore Sigma)
Spin-X centrifuge tube filter (cellulose acetate membrane, pore size 0.45-μm) (# CLS8162 Millipore Sigma)
Halt Protease and Phosphatase Inhibitor Cocktails (# PI78443 Thermo Fisher Scientific)
MagCapture Exosome Isolation kit (# 293–77601 FUJIFILM Wako Pure Chemical Corporation)
Note: This kit can be recovered a maximum of 1–2 × 1010 particles.
LoBind Microcentrifuge Tubes 1.5 mL (# 022–43-108–1 Eppendorf)
DynaMag™-2 Magnet (# 12321D Thermo Fisher Scientific)
LP Vortex Mixer (# 88880017 Thermo Fisher Scientific)
mySPIN™ 6 Mini Centrifugae (# 75004061 Thermo Fisher Scientific)
ProPette™ Electronic Pipette Controller (# 1191K61 Thomas Scientific)
Procedure:
3.2.1. Sample preparation
Thaw human CSF sample on ice
Transfer 600μL of CSF sample to new 1.5mL microcentrifuge tube
Centrifuge the CSF sample at 1,200 × g for 20 min at 4°C using 5720R from Eppendorf
Transfer the supernatant to new 1.5mL tube
Centrifuge at 10,000 × g for 30 min at 4°C using 5720R from Eppendorf
Transfer the supernatant to Spin-X centrifuge tube filter (cellulose acetate membrane, pore size 0.22-μm)
Centrifuge at 3,000 × g for 5 min at 4°C using 5720R from Eppendorf
Transfer the 500μL of flow-through to new low bind tube
Add Halt Protease and Phosphatase Inhibitor Cocktails to CSF sample (1:100)
3.2.2. Affinity capture (MagCapture Exosome isolation kit)
3.2.2.1. Immobilization of EVs Capture
Vortex the streptavidin magnetic beads for 30sec
Transfer 60μL of streptavidin magnetic beads into a Reaction Tube
Add the 500μL of Exosome Capture Immobilizing Buffer to the tube
Place on the magnetic stand, and removed the supernatant after separation of magnetic beads
Resuspend the magnetic bead pellets in 500μL of Exosome Capture Immobilizing Buffer containing 10μL of Biotin labeled Exosome Capture and mix by vortexing
Rotate the tube at 4°C for 10 min
Spin down, place on the magnetic stand, and remove the supernatant after separation of magnetic beads
Wash the magnetic beads with 500μL of Exosome Capture Immobilizing Buffer
Spin down, place on the magnetic stand, and remove the supernatant after separation of magnetic beads
Repeat steps 8–9 twice
3.2.2.2. Affinity reaction
-
11.
Add Exosome Binding Enhancer (1:500) to 500μL of CSF and mix by vortexing
-
12.
Transfer the sample to the immobilized beads tube (3.2.2.1.-step10)
-
13.
Incubate at 4°C for 3h with rotator
-
14.
Spin down, place on the magnetic stand, and remove the supernatant after separation of magnetic beads
-
15.
Wash the bead pellets in 1mL of Washing buffer with Exosome Binding Enhancer (1:500) into the tube and mix by vortexing
-
16.
Spin down, place on the magnetic stand, and remove the supernatant after separation of magnetic beads
-
17.
Repeat steps 15–16 twice
3.2.2.3. Elution of EVs
-
18.
Add 50μL of Exosome Elution Buffer to a tube
-
19.
Allow tube to stand at room temperature for 10 min
-
20.
Spin down, place on the magnetic stand, and transfer the supernatant to low binding tube
-
21.
Repeat once (step 18–20)
-
22.
Transfer the total 100μL of EV samples to Spin-X centrifuge tube (cellulose acetate membrane, pore size 0.45-μm)
Note: The eluted EVs might be contaminated with trace amounts of magnetic beads. If the separated EVs are analyzed by NTA, TEM, and proteomics analysis, the eluted EVs should be filtered by using 0.45-μm pore size filter.
-
23.
Centrifuge at 3,000 × g for 5 min at 4°C using 5720R from Eppendorf
-
24.
Transfer the flow-through EV samples to new low binding tube
3.3. Assessment of protein concentration by BCA
Materials and Methods
Reagents:
Pierce BCA protein assay kit (# 23225 Thermo Fisher Scientific)
BSA standards (# 23209 Thermo Fisher Scientific)
Flat Bottom Cell Culture Plates-96 well plate (# 07–200-87 Corning)
TET buffer (50mM Tris-HCl pH 7.5, 2mM EDTA, 1% Triton X-100)
Microplate reader (Biotek synergy MX)
Procedure:
-
Lyse separated EV samples (1:10 for brain-derived EV or 1:1 for plasma-derived EV) with TET buffer before loading into the assay
Note: The separated EV sample to be used: 3μL for brain-derived EV, 15μL for plasma-derived EV. Urea, guanidine or SDS may be used for misfolding proteins.
Keep on ice for 15 min
Dilute BSA standard with the same lysis buffer
Prepare BCA Working Reagent (WR) by mixing 50 parts of BCA Reagent A with 1 part of BCA Reagent B
Add 1:8 ratio of sample to reaction components into a 96-well plate
Cover the plate with lid and incubate at 37°C (for standard BCA) or 60°C (for micro-BCA) for 30 min
Cool the plate to room temperature
-
Read OD562nm by a microplate reader
Note: The working concentration of BCA is 5–2000μg/mL. The samples should be empirically diluted for accurate measurement based on the initial reading of the protein concentration.
3.4. Nanoparticle Tracking Analysis (NTA)
Materials and Methods
Reagents:
DL-Dithiothreitol (# 43815 Sigma)
Phosphate buffered saline (PBS, Ca/Mg free, pH 7.4) (# 10–010-049 Gibco/Thermo Fisher Scientific)
Note: The PBS should be filtrated twice with PES membrane filter (0.22-μm pore size).
Ultrasonic cleaner (Brookstone)
Nanosight 300 machine (Malvern Panalytical Inc)
Nanosight NTA 3.3 software (Malvern Panalytical Inc)
Syringe pump infusion system (Harvard Laboratories/Malvern Panalytical Inc)
BD Tuberculin syringes 1.0-mL (# 309623 Becton Dickinson)
Procedure:
-
Dilute the EV samples in dfPBS to get particles within the target reading range for the Nanosight 300 machine, which is 10–100 particles per frame
Note: For brain-derive EV, the dithiothreitol (final: 0.1mM) is added to EV samples, followed by sonication for 480 sec at level 3 in an ultrasonic cleaner for dissociation of aggregated EVs.
Load the samples using a syringe pump infusion system or manual syringe
-
Set Screen Gain to 1.0 and adjust the Camera Level until the particle in the screen can be clearly seen
Note: The camera Level should be set at the same value during the set of experiment.
Adjust the laser beam position and focus the field so that particles are clearly detectable
Capture five 60-second or 30-second videos for each sample at 21°C constant
-
Set a detection threshold, which has the maximum detection of the particles in the field
Note: The detection threshold should be fixed during the set of experiment.
-
Analyze the data with Nanosight NTA 3.3 software (Malvern Panalytical Inc)
Note: Particle counts are normalized for dilution on the machine, dilution of the final pellet, and starting material of the exosome extraction.
3.5. Transmission electron microscopy (TEM)
Materials and Methods
Reagents:
carbon-coated grid (# CF400-CU EMS www.emsdiasum.com)
Filter paper (# 1 Whatman)
Uranyl formate (# 22451 EMS)
JEOL 1200EX Transmission electron microscope (JEOL Solutions for innovation)
Procedure:
Adsorb 5μL of the EV sample for 1 min to a carbon-coated grid that had been made hydrophilic by a 20-second exposure to a glow discharge (25mA)
Remove the excess liquid with a filter paper
Float briefly the grid on a drop of water to wash away phosphate or salt
Blot on a filer paper
Stain with 0.75% uranyl formate for 15 sec
Remove the excess uranyl formate with a filter paper
Examine the grids in a JEOL 1200EX Transmission electron microscope and images are recorded with an AMT 2k CCD camera
3.6. Mass-spectroscopy for proteomic profiling
3.6.1. Mass spectroscopy for human brain-derived EVs (for Section 4.1.)
Materials and Methods
Reagents:
1M Triethylammonium bicarbonate (TEAB) (# 90114 Thermo fisher Scientific)
Tris (2-carbocyethyl) phosphine (TCEP) (# T2556 Thermo fisher Scientific)
Iodoacetamide (# A39271 Thermo fisher Scientific)
Methanol (# 34860 Millipore Sigma)
Chloroform (# 34854 Millipore Sigma)
HLPC grade Water (# 34877 Millipore Sigma)
Mass spectrometry grade trypsin (# V5280 Promega)
Vacuum centrifugation (# SPD1010 Speedvac system Thermo Fisher Scientific)
High-speed centrifuge (# 5720R Eppendorf)
LoBind Microcentrifuge Tubes 1.5 mL (# 022–43-108–1 Eppendorf)
TMT 10-plex reagents (# 90110 Thermo Fisher Scientific)
Acetonitrile (ACN) (# 34851 Millipore Sigma)
Pierce water LC-MS Grade (# 51140 Thermo Fisher Scientific)
Trifluoroacetic acid (TFA) (# 80457 Millipore Sigma)
Ammonium bicarbonate (# 285099 Millipore Sigma)
Oasis HLB 96-well μElution plate (Waters)
Formic acid (FA) (# F0507 Millipore Sigma)
Q-Exactive Orbitrap mass spectrometer (Thermo Fisher Scientific)
Ultimate 3000 high-performance liquid chromatography (HPLC) (Dionex)
Procedure:
3.6.1.1. Sample Preparation
-
Add 100mM Triethylammonium bicarbonate (TEAB) with 2% SDS to the EV samples (25μg)
Note: The digested peptide samples are injected 2μg per injection to Nano-LC-MS/MS.
Add 200mM Tris (2-carbocyethyl) phosphine (TCEP) to each sample
Incubate at 55°C for 1 h and cool down to room temperature
Alkylate with 375mM iodoacetamide (IAA) for 30 min at room temperature in the dark
Add methanol and vortex, followed by chloroform and mix thoroughly by vortexing
Add ultrapure water and mix by vortexing
Centrifuge at 14,000 × g for 5 min at 4°C
Remove the water/methanol layer and add methanol and mix by vortexing
Centrifuge at 14,000 × g for 5 min at 4°C
Remove the supernatant and dry up the protein by vacuum centrifugation
Re-suspend the pellets in 50mM TEAB
Digest with mass spectrometry grade trypsin in 50mM ammonium bicarbonate (pH 7.5) for 16 h at 37°C (1:40 w/w trypsin-to-protein)
Equilibrate the TMT label Reagents to room temperature
Dissolve anhydrous acetonitrile to each TMT label reagent tube
Add the TMT label Reagent to digested peptides
Incubate the reaction for 1h at room temperature
Add 5% hydroxylamine to each tube and incubate for 15 min to quench the reaction
Combine all samples at equal amounts
Dry up the sample under vacuum centrifugation
Reconstitute by trifluoroacetic acid (TFA) solution
Desalt the samples using Oasis HLB 96-well μElution plate
Elute the labeled peptides using 95% ACN/water
Dry up the eluted peptides in vacuum centrifugation
Re-suspend in 5% ACN/water/0.1% formic acid (FA)
3.6.1.2. Liquid Chromatography (LC)-Tandem Mass Spectrometry (MS/MS) Analysis
Nano-LC-MS/MS analysis is conducted by a Q-Exactive Orbitrap mass spectrometer (Thermo Fisher Scientific) equipped with electrospray ionization (ESI) source and coupled with an Ultimate 3000 high-performance liquid chromatography (HPLC) (Dionex). The TMT labelled peptides were separated on a C18 reverse-phase capillary column (pepMap, 75μm x 150 mm, Thermo Fisher) with a linear gradient of 2–35% acetonitrile in 0.1% formic acid. The top fifteen most abundant ions per scan were selected for MS/MS analysis with a full scan MS ranging 300–1800 m/z) and normalized collision energy (NCE) at 32.
3.6.1.3. Sequence Database Searching and Data Analysis
The Raw data files are processed with Proteome Discoverer (Thermo, version 2.1) and searched using Sequest HT against the UniProt Homo sapiens database for protein identification. The false discovery rate (FDR) at peptide and protein levels < 0.01. TMT tags on lysine residues and peptide N termini and carbamidomethylation of cysteine residues were set as static modifications, while oxidation of methionine residues was set as a variable modification. For protein identification, two missed trypsin cleavage sites were allowed. For TMT-based reporter ion quantitation, we extracted summed signal-to-noise ratio for each TMT channel.
3.6.2. Mass spectroscopy for mouse brain-derived EVs (for Section 4.2.)
Materials and Methods
Reagents:
Dithiothreitol (# 165680010 Fisher Scientific)
Iodoacetamide (# 12270050 Fisher Scientific)
Mass spectrometry grade trypsin (# 90059 Thermo Fisher Scientific)
Formic acid (FA) LC-MS Grade (# A117–50 Fisher Scientific)
Urea (# 327380010 Fisher Scientific)
Ammonium bicarbonate (# 393212500 Fisher Scientific)
Water LC-MS Grade (# 51140 Thermo Fisher Scientific)
Acetonitrile (ACN) LC-MS Grade (# 85188 Thermo Fisher Scientific)
Vacufuge plus (Eppendorf)
Centrifuge (5427R, Eppendorf)
LoBind Microcentrifuge Tubes 1.5 mL (# 022–43-108–1 Eppendorf)
C18 ZipTip (Millipore)
C18 pre-column (3 μm, 75 μm × 2 cm, Thermo Fisher Scientific)
C18 analytical column (2 μm, 75 μm × 50 cm, Thermo Fisher Scientific)
Q-ExactiveHF-X mass spectrometer (Thermo Fisher Scientific)
Easy nanoLC1200 (Thermo Fisher Scientific)
Procedure:
3.6.2.1. Sample Preparation
-
Lyse 8M urea to the EV samples (30μg)
Note: The digested peptide samples are injected 2μg per 1 injection to Nano LC-MS/MS.
Reduce with 2.5mM dithiothreitol for 1 h at 37°C
Alkylate with 5mM iodoacetamide for 30 min at room temperature in the dark
Dilute the samples up to 1M urea
Digest with mass spectrometry grade trypsin in 50mM triethylammonium bicarbonate overnight at 37°C (1:20 w/w trypsin-to-protein)
Desalt the peptides using C-18 ziptip
Dry up the eluted peptides in a vacuum centrifugation
Resuspend in 0.1% formic acid (FA)
3.6.2.2. Liquid Chromatography (LC)-Electrospray Ionization (ESI) Tandem MS (MS/MS) Analysis
Analysis was performed on an Easy nanoLC1200 (Thermo Fisher Scientific) coupled to a Q-Exactive HF-X mass spectrometer (Thermo Fisher Scientific). A 2μg of peptides were loaded onto a C18 pre-column connected to a 50 cm easy spray C18 analytical column (2 μm, 75 μm × 50 cm, Thermo Fisher Scientific) in positive mode by data dependent acquisition (DDA). The mobile phase was 2% ACN with 0.1% FA (A) and 80% ACN with 0.1% FA (B). The gradient profile was 5% B at 0 min, 30% B at 90 min, 100% B from 99 to 116 min, and 2% B at 117 to 120 min. The column oven was 55°C. The full scan range was 300 to 1650 m/z with and AGC of 3e6 followed by 15 MS/MS scans. The normalized collision energy was 28% and dynamic exclusion was set to 40s.
3.6.2.3. Sequence Database Searching and Data Analysis
The raw LC-MS/MS data were searched using MaxQuant (v1.6.7) (Max Planck Institute of Biochemistry) against the Uniprot/Swissprot database for Mus musculus with a 1% false discovery rate (FDR). For protein identification, two missed trypsin cleavage sites were allowed. Carbamidomethylation of cysteine residues was set as a fixed modification, while methionine oxidation and N-terminal acetylation were set as variable modifications.
3.6.3. Human CSF-derived EVs (for Section 4.3.)
Materials and Methods
Reagents:
Acetone (# 179124 Millipore Sigma)
Laemmli sample buffer (# 1610747 Bio-Rad)
10% Mini-PROTEAN® TGX™ Precast Protein Gels (# 456–1033 Bio-Rad)
Distilled water (# 15230001 Thermo Fisher Scientific)
Vacuum centrifugation (# SPD1010 Speedvac system Thermo fisher Scientific)
High-speed centrifuge (# 5720R Eppendorf)
LoBind Microcentrifuge Tubes 1.5 mL (# 022–43-108–1 Eppendorf)
Methanol (# 179337 Millipore Sigma)
Acetic acid (# 320099 Millipore Sigma)
Fixing solution (50% methanol and 7% acetic acid)
Incubation Tray (# L-07037–002 Advansta)
GelCode™ Blue Stain Reagent (# PI24590 Thermo Fisher Scientific)
Razor blade (# 12–640 Thermo Fischer Scientific)
Trifluoroacetic acid (TFA) (# 80457 Millipore Sigma)
Formic acid (FA) (# F0507 Millipore Sigma)
Acetonitrile (ACN) (# 34851 Millipore Sigma)
ProteaseMAX Surfactant, Trypsin Enhancer (# V2071 Promega)
Trypsin Sequencing grade (# 11418475001 Roche/Sigma)
Ammonium bicarbonate (# 285099 Millipore Sigma)
Pierce water LC-MS Grade (# 51140 Thermo Fisher Scientific)
Q-Exactive mass spectrometer (Thermo Fisher Scientific)
NanoAcquity ultra-performance liquid chromatography (UPLC) (Waters Corporation)
Procedure:
3.6.3.1. Sample Preparation
-
Add ice-cold 100% (w/v) acetone to the isolated EV fraction (final concentration: 20% w/v)
Note: The TCA/acetone precipitation is more effective than either TCA or acetone to precipitate proteins.
Incubate for 16 h on −20°C
Centrifuge at 20,000 × g for 15 min at 4°C
Discard the supernatant
Add ice-cold acetone to the pellets
Centrifuge at 20,000 × g for 15 min at 4°C
Discard the supernatant
Repeat (step 5–7)
-
Dry up the pellets at room temperature
Note: The pellets should not be over-dry because it may not dissolve properly.
-
Resuspend the pellet in Laemmli sampling buffer
Note: The pellets directly resuspend in Laemmli sampling buffer for In-gel digestion.
Boil the sample at 95°C for 10 min and cool to room temperature
Apply the samples into a 10% Mini-PROTEAN® TGX™ Precast Protein Gels
Run until 10 mm from the top of the gel
Move the gel to Incubation tray
Fix the gel with fixing solution for 15 min
Rinse the gel with distilled water three time for 5 min
Stain the gel with GelCode™ Blue Stain Reagent for 2 h at room temperature
De-stain the gel in distilled water four times for 15min by soaking
Cut out bands from gel by Razor blade and place the gel slices in 1.5mL centrifuge tube
Add 1mL water and incubate for 30 min
Remove the water with pipette
Incubate in 50mM ammonium bicarbonate/acetonitrile (ACN) (1:1) for 1 h at room temperature
Replace with 100% ACN and incubate until the gel slices turned opaque white
Remove the ACN
Dry up the gel slice in a vacuuming centrifugation using SpeedVac
Reduce 45mM dithiothreitol in 250mM ammonium bicarbonate for 30 min at 50°C and cool down to room temperature
Alkylate 100mM iodoacetamide in 100mM ammonium bicarbonate for 30 min in the dark
Remove all the liquid
Wash the gel twice in water
Replace with 100% acetonitrile
Remove all the liquid and dry up the gel slice in a Vacuum centrifugation
Rehydrate in 50mM ammonium bicarbonate containing 0.01% ProteaseMAX Surfactant and 2ng/μL trypsin
Add bicarbonate buffer to ensure complete submersion of the gel
Incubate for 21 h at 37°C
Transfer the supernatants to 1.5-mL microcentrifuge tube
Dehydrate the gel slices with 80% ACN/1% formic acid (FA)
Transfer the supernatants to same 1.5-mL microcentrifuge tube (step 35)
Dry up the sample in a vacuuming centrifugation
Re-dissolve the pellets with 5% ACN/0.1% trifluoroacetic acid (TFA)
3.6.3.2. Liquid Chromatography (LC)-Electrospray Ionization (ESI) Tandem MS (MS/MS) Analysis
A 2μL injection was loaded by a Waters NanoAcquity Ultra Performance Liquid Chromatography in 5% ACN (0.1% FA) at 4.0μL/min for 4min onto a 100μm I.D. fused-silica pre-column packed with 2 cm of 5 μm (200Å) Magic C18AQ (Bruker-Michrom Inc., MA, USA). Peptides were eluted at 300nL/min from a 75 μm I.D. gravity-pulled analytical column packed with 25 cm of 3μm (100Å) Magic C18AQ particles using a linear gradient from 5–35% of mobile phase B (acetonitrile + 0.1% formic acid) in mobile phase A (water + 0.1% formic acid) over 60mins. Ions were introduced by positive electrospray ionization via liquid junction at 1.4kV into a Thermo Scientific Q Exactive hybrid mass spectrometer. Mass spectra were acquired over m/z 300–1750 at 70,000 resolution (m/z 200) with an AGC target of 1e6, and data-dependent acquisition selected the top 10 most abundant precursor ions for tandem mass spectrometry by HCD fragmentation using an isolation width of 1.6 Da, max fill time of 110ms, and AGC target of 1e5. Peptides were fragmented by a normalized collisional energy of 27, and fragment spectra acquired at a resolution of 17,500 (m/z 200).
3.6.3.3. Sequence Database Searching and Data Analysis
Raw data files were peak processed with Proteome Discoverer (version 1.4, Thermo Scientific, USA) followed by identification using Mascot Server (version 2.5, Matrix Science, UK) against the Homo sapiens (Swiss-Prot) FASTA file (downloaded 10/2018). Search parameters included Trypsin/P specificity, up to 2 missed cleavages, a fixed modification of carbamidomethyl cysteine, and variable modifications of oxidized methionine, pyroglutamic acid for Q, and N-terminal acetylation. Assignments were made using a 10ppm mass tolerance for the precursor and 0.05 Da mass tolerance for the fragments. All non-filtered search results were processed by Scaffold (version 4.4.4, Proteome Software, Inc., USA) utilizing the Trans-Proteomic Pipeline (Institute for Systems Biology, WA, USA) with threshold values set at 86% for peptides (1.0% false-discovery rate [FDR]) and 90% for proteins (2 peptide minimum, 0.2% FDR), and quantitative comparisons made using the iBAQ-quantitation method with all samples normalized by total ion current for the run.
Note: All mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org/cgi/GetDataset) via the PRIDE [36] partner repository with the dataset identifier PXD017273.
4. Results
4.1. Biochemical characterization of EVs separated from unfixed frozen human brain tissue by sucrose gradient ultracentrifugation method
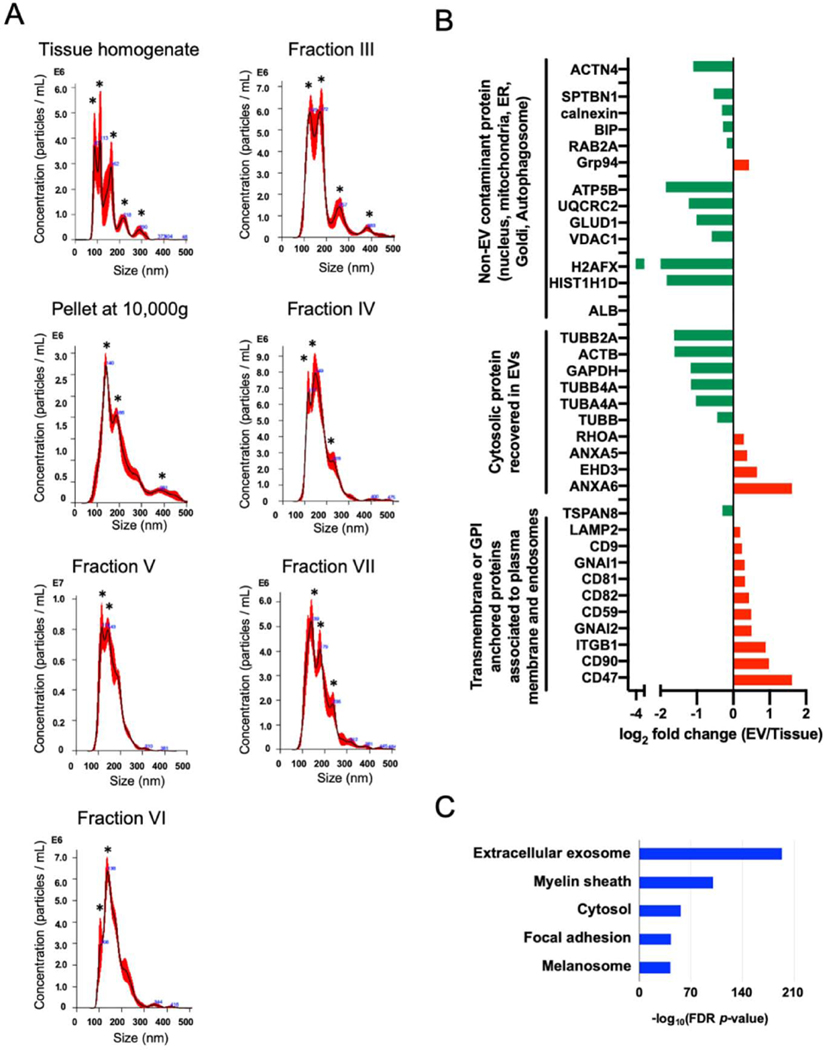
The experimental workflow is summarized in Figure 1A and B. The EVs were separated from unfixed frozen human brain tissue using the discontinuous sucrose gradient ultracentrifugation (SG-UC) method. EVs were analyzed for protein concentration using protein assay (BCA), particle size and number using Nanoparticle tracking analysis (NTA), which determines the size of suspended particles based on their Brownian motion, particle form using Transmission electron microscopy (TEM), and protein profiling using Tandem mass tag (TMT)-based proteomics analysis. The EVs separated from frozen brain tissue was observed cap-sharped morphology by TEM (Figure 1C). Table 1 shows the protein concentration of collected fractions. The fraction VIII and XI were below the detection limit of protein assay. Figure 2A shows the NTA plot of these fractions (tissue homogenate, pellet at 10,000g, and sucrose fraction-III, IV, V, VI, and VII). The tissue homogenate and pellet at 10,000g fractions represented a broad distribution of vesicle with several peaks (asterisk), but the sucrose fraction-V and VI was decreased to a few peaks by SG-UC. The results suggest the EV fraction could separate EV of the same or nearly same size from heterogenous size vesicles of brain tissue by buoyant density. In addition, fractions V and VI (1.10 – 1.15 g/cm3) are the EV fraction in which concentrated EV particles compared to tissue homogenate (Table 1). To check the protein profiling in these fractions, we performed the 10-plex TMT-based quantitative proteomics analysis for the brain tissue homogenates and the separated fractions. We found that these EV fractions were enriched in tetraspanins including CD9 and CD81, annexins, and lysosomal markers such as Lysosome-associated membrane glycoprotein 2 (LAMP2) as compared to tissue homogenates, and diminished in non-EV molecules such as Histone H1 from nucleus, 10kDa heat shock protein (HSPE1) from mitochondria, calnexin from endoplasmic reticulum (ER) and Ras-related protein Rab-2A (RAB2A) from the Golgi apparatus, as listed in MISEV2018 guidelines [1] (Figure 2B and Supplementary Table 1). In addition, these identified proteins were tested by Gene Ontology analysis in The Database for Annotation, Visualization and Integrated Discovery (DAVID) [37,38] for properties pertaining to the ‘cellular component’ (Figure 2C). The EV fraction separated by the SG-UC method was enriched in the Extracellular exosome category. These data indicate that EVs were successfully separated from unfixed frozen brain tissue using discontinuous SG-UC method.
Table 1.
Assessment of tissue homogenates, pellet at 10,000g, sucrose fraction III, IV, V, VI and VII separated from human brain tissue by SG-UC method
| Tissue homogenates | Pellet at 10,000g | Fraction III | Fraction IV | Fraction V | Fraction VI | Fraction VII | |
|---|---|---|---|---|---|---|---|
| Mode size (nm)a | 113 | 139 | 172 | 148 | 138 | 139 | 119 |
| Particle numbera,b | 1.31E+12 | 3.21E+11 | 5.09E+10 | 1.42E+11 | 1.52E+11 | 6.77E+10 | 1.12E+10 |
| Total protein (μg)b,c | 3672 | 352.1 | 25.6 | 56.1 | 80.3 | 85.1 | 24.3 |
| Particles/proteins (μg) | 3.57E+08 | 9.12E+08 | 1.99E+09 | 2.53E+09 | 1.89E+09 | 7.96E+08 | 4.61E+08 |
Particle number and size of separated EVs were measured by Nanoparticle Tracking Analysis (NTA)
The starting material is 0.5g of human brain cortex grey matter
The total protein levels were measured by BCA assay.
Figure 2. Characterization of EVs separated from unfixed human brain tissues by SG-UC method.
A) NTA plot of mode size and concentration of separated EVs (tissue homogenate, pellets after 10,000g, fraction III, IV, V, VI, VII after SG-UC); The black line shows the fitting curve. Red line represents the error bar. Y axis: EV particle counts [/mL], X axis: EV particle size [nm]. A peak was indicated with black asterisk. B) The bar charts of EV protein or non-EV protein in separated EV. Y axis: EV marker protein or non-EV marker protein, X axis: log2 fold change (EV/brain tissue homogenates). Red bar shows the proteins are enriched in EVs. Green bar shows the proteins are enriched in brain tissue homogenates. C) DAVID (Bioinformatics Resources 6.8) GO terms of Top10 ‘Cellular Components’ with -log10(FDR p-value).
4.2. Comparison of unfixed murine brain-derived EVs separated via sucrose gradient ultracentrifugation and size exclusion chromatography methods
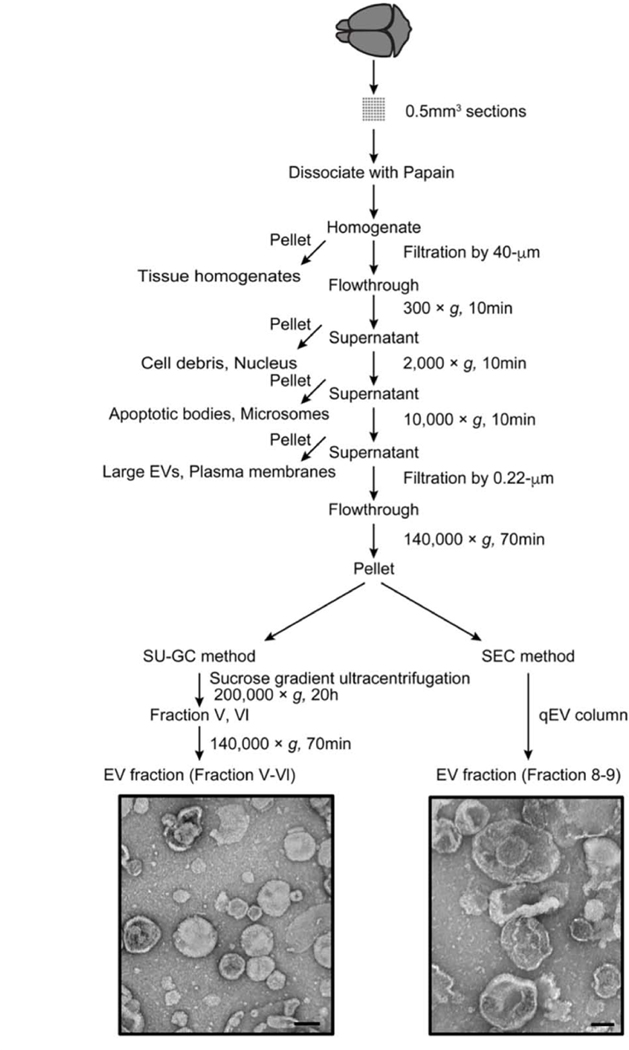
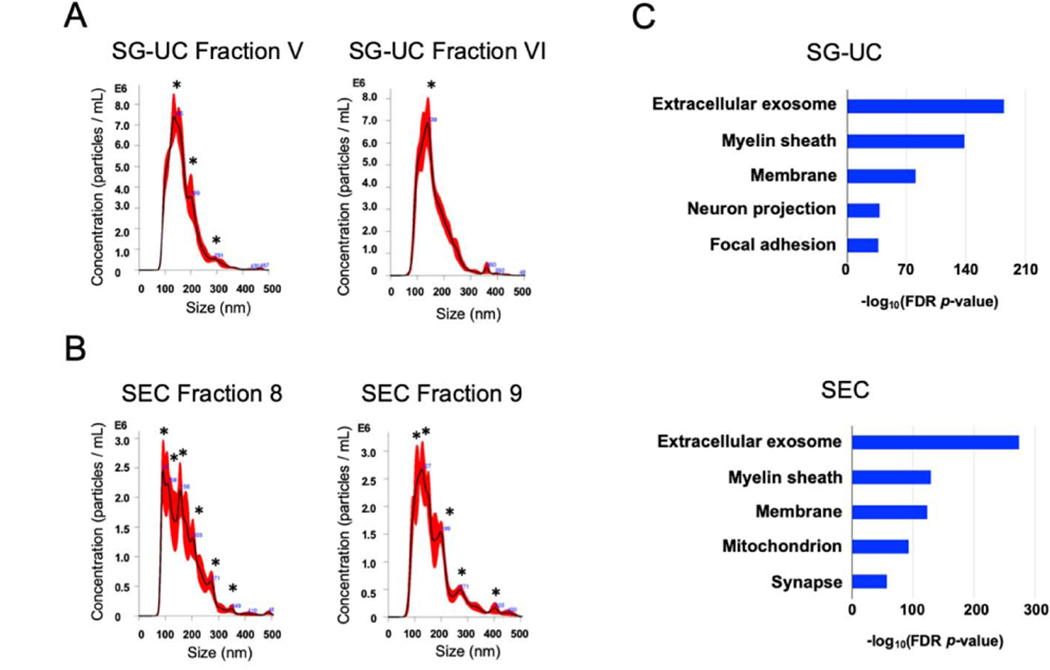
Using the SG-UC protocol, we found that the small vesicles corresponding to exosome size (50–150nm) are also present in other sucrose fractions as determined by NTA (Table 1). These fractions also contain EV markers including CD9 and CD81 as determined by proteomic profiling. To collect these EVs as they migrated into fractions other than V and VI, we decided to test alternative methods instead of the SG-UC. We separated EVs from mouse brain tissue by size exclusion chromatography (SEC method; qEV original column). The SEC method can separate the soluble molecules, high-density lipoprotein (HDL) (5–12 nm), low-density lipoprotein (LDL) (18–25 nm), and intermediate-density lipoprotein (IDL) (25–35 nm) from EV fractions [39]. We compared the EV separation methods based on the particle size, particle counts, protein yield, and proteomic profiling (Figure 3). The particle size and number of separated EVs was determined by NTA, and the protein concentration was determined by BCA (Figure 4A, B and Table 2). The EVs separated via the SEC method had fractions 8 and 9 as the EV fractions. The SEC method had higher EV particle numbers and protein yield compared to the SG-UC method (Table 2). The SEC method, however, showed a broad distribution of vesicles with several peaks (asterisk) as compared to the SG-UC method (Figure 4A and B). In addition, TEM imaging was shown that the EVs separated by the SEC method contain differently sized vesicles, such as small and medium size particles compared to SG-UC method (Figure 3). In addition, a label-free Nano-LC-MS/MS analysis of these EV samples identified 933 and 1823 proteins in the EVs separated by SG-UC and SEC, respectively (Table 2). The SEC method separated EVs were enriched in molecules such as tetraspanins including CD9, CD81 and CD82, and ESCRT proteins including programmed cell death 6-interactting protein (PDCD6IP; Alix) and hepatocyte growth factor-regulated tyrosine kinase substrate (HGS) as compared to the SG-UC method. However, the non-EV molecules, such as MICOS complex subunit MIC60 (IMMT), calnexin, and Endoplasmic reticulum chaperone Bip (BIP) were also detected. This suggests that separated populations may contain overlap of multiple EV sub-species. These identified proteins in the EV samples were tested by DAVID GO analysis for properties pertaining to the ‘cellular component’ (Figure 4C). Both EV fractions separated by SG-UC and SEC methods were enriched the Extracellular exosome category. The SEC method, however, also enriched mitochondrion category as non-EV molecules. The SG-UC method has disadvantage in work time and sample size per run, but it could recover small EVs of the same size with the highest enrichment between two methods (Table 2).
Figure 3. Flowchart of EV separation from unfixed murine brain tissue via SG-UC and SEC methods.
Sucrose gradient ultracentrifugation (SG-UC) or Size exclusion chromatography (SEC) protocols for separating of EVs from unfixed mouse frozen brain tissue. See section 3.1 for detailed methods. TEM image of frozen mouse brain-derived EVs (pooled fractions V and VI for SG-UC and pooled fraction 8 and 9 for SEC). Scale bar; 100 nm.
Figure 4. Characterization of EV separated from unfixed murine brain tissues by SEC and SU-GC methods.
A-B) NTA of EVs separated from unfixed frozen murine brain tissues. The black line shows the best-fitting curve. Red line represents the error bar. Y axis: EV particle counts [/mL], X axis: EV particle size [nm]. A peak was indicated with black asterisk. A) SG-UC fraction V and VI. B) SEC_qEVoriginal column fraction 8 and 9. C) DAVID GO terms of Top10 ‘Cellular Components’ with -log10(FDR p-value).
Table 2.
Assessment of EVs separated from unfixed frozen murine brain tissue by SG-UC and SEC methods
| SG-UC Fraction V | SG-UC Fraction Vl | SEC Fraction 8 | SEC Fraction 9 | |
|---|---|---|---|---|
| Mode size (nm)a | 135 | 139 | 90 | 127 |
| Particle numbera,b | 1.06E+10 / 30μL | 1.09E+10 / 30μL | 6.80E+10 / 500μL | 7.80E+10 / 500μL |
| Total protein (μg)b,c | 43.3 | 41.1 | 44.1 | 59.3 |
| Particles/proteins (μg) | 2.44E+08 | 2.65E+08 | 1.54E+09 | 1.31E+09 |
| Protein identificationd | 933 | 1823 | ||
| Extracellular Exosomee | 489 (52.4%) | 827 (45.4%) | ||
Particle number and size of separated EVs were measured by Nanoparticle Tracking Analysis (NTA)
The starting material is 0.5g of human brain cortex grey matter
The total protein levels were measured by BCA assay.
4.3. Characterization of EVs separated from human CSF by affinity capture method
To create the protein profiling of CSF-derived EVs, we separated EVs from CSF by affinity capture (AC method; MagCapture Exosome Isolation kit), which can also separate them from EV fractions by capturing with phosphatidylserine [40], and analyzed the secondary collected EVs by NTA and proteome analysis. The experimental workflow is summarized in Figure 5.
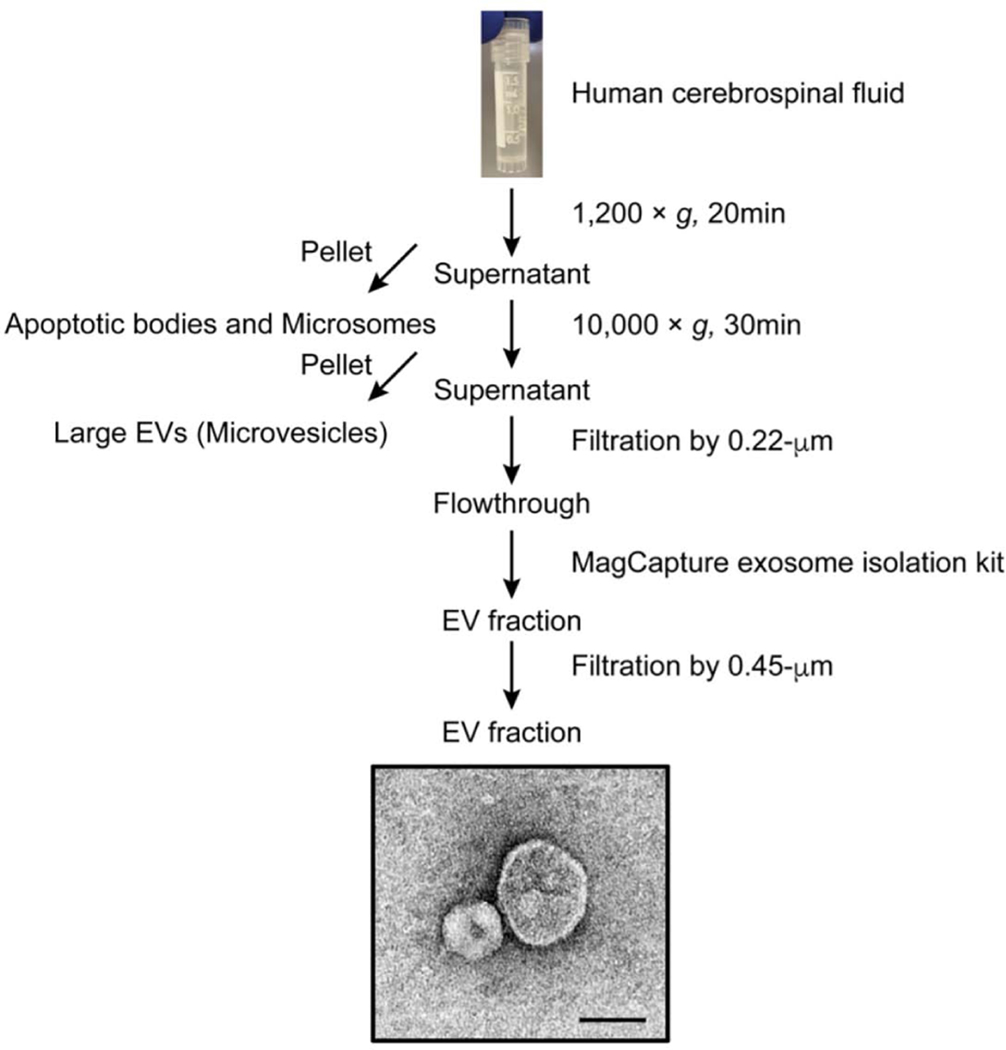
Figure 5. Flowchart of EV separation from human Cerebrospinal fluid via AC method.
Affinity capture (AC) protocol for separating and concentrating of EVs from human CSF. See section 3.1 for detailed methods. TEM image of human CSF-derived EVs. Scale bar; 100 nm.
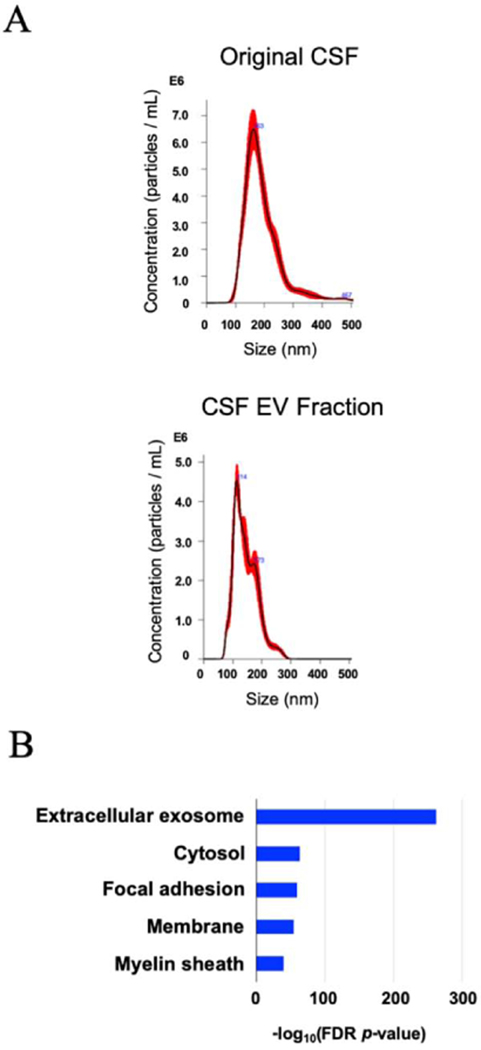
The particle size and number of separated EV were determined by NTA, and the protein concentration was determined by BCA (Figure 6A and Table 3). In addition, the separated EVs from CSF was showed classical EV morphology by TEM (Figure 5). The AD methods could successfully separate and concentrate EV of the same or nearly same size from human CSF (Table 3). Nano-LC-MS/MS-based protein profiling of the CSF-derived EVs identified a total of 700 unique proteins. The EV molecules such as tetraspanin and ESCRT, including PDCD6IP/Alix and syntenin-1 proteins could enrich in the EV fraction separated by AC method.
Figure 6. Characterization of EVs separated from human CSF by AC method.
A) NTA plot of mode size and concentration of separated EVs by AC method; The black line shows the best-fitting curve. Red line represents the error bar. Y axis: EV particle counts [/mL], X axis: EV particle size [nm]. B) DAVID GO terms of Top10 ‘Cellular Components’ with - log10(FDR p-value).
Table 3.
Assessment of EVs separated from human CSF by AC method
| Human CSF | EV Fraction | |
|---|---|---|
| Mode size (nm)a | 162 | 114 |
| Particle numbera,b | 7.9E+9 / 500μL CSF | 2.15E+9 / 100μL |
| Total protein (μg)b,c | 1125.73 | 1.78 |
| Protein identificationd | - | 700 |
| Particles/proteins (μg) | 7.02E+06 | 1.21E+09 |
Particle number and size of separated EVs were measured by Nanoparticle Tracking Analysis (NTA)
The starting material is 500ml of human CSF
The total protein levels were measured by BCA assay
The proteins were identified by LC-MS/MS for EV fraction, but CSF was not analyzed by LC-MS/MS.
In addition, the EV fraction was enriched in Extracellular exosome molecules as determined by DAVID GO (Figure 6B). These results show that this EV separation method may be effective for separating EVs from biospecimens of limited availability for proteomic analysis. We have also separated the EV by UC and SEC method for LC-MS/MS, but we could not collect enough EV for proteomics analysis and the proteins mostly could not be identified.
5. Discussion
Here we described our recently established protocols for the separation of EVs from biospecimens with a specific focus on unfixed brain tissues and CSF. In the past decade, investigation of EV function in neurodegenerative diseases have expanded, and given rise to the need to separate high purity EVs from biospecimens to understand the role of EVs in progression of those diseases, as well as develop the biomarkers for monitoring and diagnostic of progression of those diseases. Previously, the protocols for separation of EV from brain tissues by sucrose or iodixanol density gradient ultracentrifugation have been reported [24–26,41]. Gonzalez et al has described the protocol to enrich EVs from frozen and fresh brain tissues by sucrose density gradient ultracentrifugation for the first time, and shown that the EVs separated from APP transgenic mouse brain tissue are contained neuropathogenic molecule such as full length APP, APP C-terminal fragments and amyloid-β peptide [24]. Hurwitz et al. described the technique for the enrichment of brain-derived EV by iodixanol sedimentation and flotation gradient based isolation techniques [26]. They showed the purity of EVs with less contaminating debris by iodixanol flotation density isolation as compared to sedimentation gradient isolation, along with the utility of this method in downstream proteomic analysis. The comparison of two isolation techniques, however, were not normalized by the total spectral counting in label-free proteomics.
In addition, non-specific EV protein HSC70 was detected in both sedimentation- and flotation-density gradient isolation by western blotting. Thus, we think that there is no obvious difference between sedimentation and flotation-based isolation techniques. Vella et al has reported a protocol to separate small EVs from human frontal cortical tissues using a triple sucrose cushion without homogenization and filtration [41]. The homogenization and filtration may reduce the purity of EVs by causing cell disruption and release of intracellular vesicles such as the mitochondria, endoplasmic reticulum (ER) and Golgi apparatus. In addition, they utilized collagenase type III to dissociate cells from these tissues, and minimize cell disruption. The separated EV fraction without the cell disruption was enriched in proteins related to the exosome biogenesis, the endosomal pathway and endosomal trafficking pathways, and less contamination non-EV molecules. Our method separated EVs from human unfixed brain tissue by papain dissociation, homogenization with standard tissue grinder and sucrose gradient ultracentrifugation. The separated EV fractions are enriched with tetraspanins and ESCRT proteins as compared to tissue homogenates, and less contamination of non-EV molecules such as the nucleus, mitochondria, ER, Golgi-related proteins by TMT-based the proteomics dataset (Figure 2 B and Supplementary Table 1). On other hand, the EV fraction was also enriched myelin sheath-related molecules compared to published proteomics datasets for separation of brain EVs. This result suggests that the EVs separated by this method contained myelin related protein and lipid, presumably from oligodendrocytes [42,43]. In our proteomics data, CD9 and CD81 were also detected in fraction III (1.06 – 1.08 g/cm3), IV (1.08 – 1.10 g/cm3) and VII (1.15 – 1.18 g/cm3). In addition, small EVs were observed in fraction III and IV by NTA, when DTT was added to the EV fraction to dissociate aggregated EVs. Based on the findings, fraction VII may contain the large EVs and aggregated EVs generated by ultracentrifugation [44]. The fraction III and IV, on the other hand, may contain small EVs, LDL, IDL or HDL [39,45]. We separated brain-derived EVs using the SEC method instead of SG-UC after ultracentrifugation because the SEC method can avoid EVs from SG-UC-related aggregation, and could separate EVs from soluble proteins, HDL, LDL and IDL [46], which were frequently contaminated in EVs separated by the DC-UC or SG-UC methods [47–50]. Several reports, however, have shown SEC method failed to separate soluble proteins, apolipoproteins, HDL and LDL from EVs [51–59]. This could be due to the pooling of fractions from 7 to 9 or 10 in those studies. If fractions 7 to 8 or 9 were used, according to manufacturer’s instructions, this could be avoided. The SEC methods can collect more EVs and identify more EV marker molecules than SG-UC method in our study. The SEC method, however, also enriched in non-EV molecules (such as mitochondrion category) and contained more particles at various sizes than conventional EV size of around 100 nm compared to the SG-UC method (Figure 3 and 4). Thus, we suggest that SG-UC method is suitable for EV proteomic analysis. However, the sucrose cushion should be used for ultracentrifugation before SG-UC to avoid aggregation of EVs. In summary, we could collect 166μg of EVs from 0.5g of frozen human cortical grey matter or whole frozen mouse brain tissue using our separation protocol for brain-derived EV. The protein yield is sufficient for NTA, protein assay, TEM, western blot and proteomic profiling by mass-spectroscopy (Table 1 and 2).
Previously, protocols for separation of EVs from human CSF using ultracentrifugation, high-resolution flow cytometric sorting or size exclusion chromatography in downstream proteomic analysis have been reported [30,60–66]. Chiasserini et al have reported the EV were separated from 6mL of human CSF by ultracentrifugation and a sucrose cushion, and they represent the largest and most comprehensive dataset by proteomics analysis [61]. The separated EVs were highly enriched with the brain cell-type specific molecules and EV molecules such as the Alix, syntenin, and Rab proteins. This method, however, needs a very large volumes of CSF per sample and lengthy procedure lasting 16–24 h. This is unsuitable for scaling up larger population of the human study. Pieragostino et al on the other hand, reported EVs could be directly separated from 100μ of CSF using high-resolution flow cytometry sorting, and characterized the protein profiling of EVs by mass spectrometry [63]. This method, if reproducible, could be suitable for the larger clinical studies and can avoids any preparation steps, which may reduce the yield. One caveat is that the size of CSF-derived EVs is in the range 333–1326 nm, which we didn’t observe in our TEM study of CSF-derived EVs. The actual size of FACS-isolated EVs should be independently validated. The AC method can separate small EVs compared with FACS and does not require large volume of CSF samples. In our proteomics data, the AC method could enrich EV molecules such as tetraspanins and ESCRT proteins, but serum proteins such as an albumin and apolipoprotein were also identified. This is minor contamination, as Patel et al reported that EVs separated from culture media by the AC method were of good quality to remove the serum protein contamination for proteomic analysis [52]. In summary, using the AC method, high purity EVs can be separated from as low as 500 μL of CSF within 5 h for proteomics profiling (Table 3). In addition, multiple clinical samples (20–30 samples) can be processed at one time.
In conclusion, it is important to develop protocol standard operation procedure for EV separation from brain tissue and CSF samples for future development of EV-based diagnostics, and to investigate the biological function for progression of neurodegenerative disorders using a large volume of human biospecimens. The protocols we provided for separation of EVs from unfixed brain tissue and CSF samples and mass spectroscopy as a downstream application may respond to the urgent unmet needs.
Supplementary Material
Highlights.
This review presents the step-by-step protocols for EV separation methods from brain tissue
This review describes EV separation methods from human cerebrospinal fluids
This review compares the quality control measures of separation protocols by biophysical and proteomic characterizations
Acknowledgments:
The author thanks M. Ericsson (Electron Microscopy Facility, Harvard Medical School) for electron microscopic imaging services, K. Takamatsu-Yukawa (Ikezu lab members) for experimental supports, J. D. Leszyk and S. Shaffer (Mass Spectrometry Facility, Massachusetts Medical School) for mass-spectroscopy services.
Funding: This work is in part funded by Alzheimer’s Association AARF-9550302678 (SM) and DVT-14–320835 (TI), BrightFocus Foundation (A2016551S), Cure Alzheimer’s Fund (TI), NIH RF1AG054199 (TI), NIH R56AG057469 (TI), NIH R01AG054672 (TI) and NIH R21NS104609 (TI).
Footnotes
Conflicts of Interest: The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- [1].Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. , Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines, J Extracell Vesicles. 7 (2018) 1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].You Y, Ikezu T, Emerging roles of extracellular vesicles in neurodegenerative disorders, Neurobiol. Dis. 130 (2019) 104512. doi: 10.1016/j.nbd.2019.104512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Delpech J-C, Herron S, Botros MB, Ikezu T, Neuroimmune Crosstalk through Extracellular Vesicles in Health and Disease, Trends in Neurosciences. 42 (2019) 361–372. doi: 10.1016/j.tins.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Taylor AR, Robinson MB, Gifondorwa DJ, Tytell M, Milligan CE, Regulation of heat shock protein 70 release in astrocytes: role of signaling kinases, Dev Neurobiol. 67 (2007) 1815–1829. doi: 10.1002/dneu.20559. [DOI] [PubMed] [Google Scholar]
- [5].Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, et al. , Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells, Nat. Cell Biol. 10 (2008) 619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- [6].Castellana D, Zobairi F, Martinez MC, Panaro MA, Mitolo V, Freyssinet J-M, et al. , Membrane microvesicles as actors in the establishment of a favorable prostatic tumoral niche: a role for activated fibroblasts and CX3CL1-CX3CR1 axis, Cancer Res. 69 (2009) 785–793. doi: 10.1158/0008-5472.CAN-08-1946. [DOI] [PubMed] [Google Scholar]
- [7].Frühbeis C, Fröhlich D, Kuo WΜ, Amphornrat J, Thilemann S, Saab AS, et al. , Neurotransmitter-triggered transfer of exosomes mediates oligodendrocyte-neuron communication, PLoS Biol. 11 (2013) e1001604. doi: 10.1371/journal.pbio.1001604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Record M, Carayon K, Poirot M, Silvente-Poirot S, Exosomes as new vesicular lipid transporters involved in cell-cell communication and various pathophysiologies, Biochim. Biophys. Acta. 1841 (2014) 108–120. doi: 10.1016/j.bbalip.2013.10.004. [DOI] [PubMed] [Google Scholar]
- [9].Hoshino A, Costa-Silva B, Shen T-L, Rodrigues G, Hashimoto A, Tesic Mark M, et al. , Tumour exosome integrins determine organotropic metastasis, Nature. 527 (2015) 329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tominaga N, Kosaka N, Ono M, Katsuda T, Yoshioka Y, Tamura K, et al. , Brain metastatic cancer cells release microRNA-181c-containing extracellular vesicles capable of destructing blood-brain barrier, Nat Commun. 6 (2015) 6716–12. doi: 10.1038/ncomms7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kowal J, Arras G, Colombo M, Jouve M, Morath JΜ, Primdal-Bengtson B, et al. , Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes, Proc. Natl. Acad. Sci. U.S.a. 113 (2016) E968–77. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhang H, Freitas D, Kim HS, Fabijanic K, Li Z, Chen H, et al. , Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation, Nat. Cell Biol. 20 (2018) 332–343. doi: 10.1038/s41556-018-0040-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bruschi M, Granata S, Santucci L, Candiano G, Fabris A, Antonucci N, et al. , Proteomic Analysis of Urinary Microvesicles and Exosomes in Medullary Sponge Kidney Disease and Autosomal Dominant Polycystic Kidney Disease, Clin J Am Soc Nephrol. 14 (2019) 834–843. doi: 10.2215/CJN.12191018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Haraszti RA, Didiot M-C, Sapp E, Leszyk J, Shaffer SA, Rockwell HE, et al. , High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources, J Extracell Vesicles. 5 (2016) 32570. doi: 10.3402/jev.v5.32570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].DeLeo AM, Ikezu T, Extracellular Vesicle Biology in Alzheimer’s Disease and Related Tauopathy, J Neuroimmune Pharmacol. 13 (2018) 292–308. doi: 10.1007/s11481-017-9768-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Simpson RJ, Lim JW, Moritz RL, Mathivanan S, Exosomes: proteomic insights and diagnostic potential, 6 (2009) 267–283. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- [17].Murphy DE, de Jong OG, Brouwer M, Wood MJ, Lavieu G, Schiffelers RM, et al. , Extracellular vesicle-based therapeutics: natural versus engineered targeting and trafficking, Exp. Mol. Med. 51 (2019) 32–12. doi: 10.1038/s12276-019-0223-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Karttunen J, Heiskanen M, Lipponen A, Poulsen D, Pitkänen A, Extracellular Vesicles as Diagnostics and Therapeutics for Structural Epilepsies, International Journal of Molecular Sciences 2017, Vol. 18, Page 612. 20 (2019) 1259. doi: 10.3390/ijms20061259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kannan A, Philley JV, Hertweck KL, Ndetan H, Singh KΜ, Sivakumar S, et al. , Cancer Testis Antigen Promotes Triple Negative Breast Cancer Metastasis and is Traceable in the Circulating Extracellular Vesicles, Sci Rep 9 (2019) 11632–12. doi: 10.1038/s41598-019-48064-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Branscome H, Paul S, Khatkar Μ, Kim Y, Barclay RA, Pinto DO, et al. , Stem Cell Extracellular Vesicles and their Potential to Contribute to the Repair of Damaged CNS Cells, J Neuroimmune Pharmacol. 36 (2019) 301–18. doi: 10.1007/s11481-019-09865-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].McKiernan J, Donovan MJ, Margolis E, Partin A, Carter B, Brown G, et al. , A Prospective Adaptive Utility Trial to Validate Performance of a Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer in Patients with Prostate-specific Antigen 2–10ng/ml at Initial Biopsy, Eur. Urol. 74 (2018) 731–738. doi: 10.1016/j.eururo.2018.08.019. [DOI] [PubMed] [Google Scholar]
- [22].Lötvall J, Hill AF, Hochberg F, Buzás EI, Di Vizio D, Gardiner C, et al. , Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles, J Extracell Vesicles. 3 (2014) 26913. doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Asai H, Ikezu S, Tsunoda S, Medalla M, Luebke J, Haydar T, et al. , Depletion of microglia and inhibition of exosome synthesis halt tau propagation, Nat. Neurosci. 18 (2015) 1584–1593. doi: 10.1038/nn.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Perez-Gonzalez R, Gauthier SA, Kumar A, Levy E, The exosome secretory pathway transports amyloid precursor protein carboxyl-terminal fragments from the cell into the brain extracellular space, J. Biol. Chem. 287 (2012) 43108–43115. doi: 10.1074/jbc.M112.404467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Silverman JM, Christy D, Shyu CC, Moon K-M, Fernando S, Gidden Z, et al. , CNS-derived extracellular vesicles from superoxide dismutase 1 (SOD1)G93A ALS mice originate from astrocytes and neurons and carry misfolded SOD1, J. Biol. Chem. 294 (2019) 3744–3759. doi: 10.1074/jbc.RA118.004825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hurwitz SN, Sun L, Cole KY, Ford CR, Olcese JM, Meckes DG, An optimized method for enrichment of whole brain-derived extracellular vesicles reveals insight into neurodegenerative processes in a mouse model of Alzheimer’s disease, J. Neurosci. Methods. 307 (2018) 210–220. doi: 10.1016/j.jneumeth.2018.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sproviero D, La Salvia S, Giannini M, Crippa V, Gagliardi S, Bernuzzi S, et al. , Pathological Proteins Are Transported by Extracellular Vesicles of Sporadic Amyotrophic Lateral Sclerosis Patients, Front Neurosci. 12 (2018) 12093. doi: 10.3389/fnins.2018.00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sardar Sinha M, Ansell-Schultz A, Civitelli L, Hildesjö C, Larsson M, Lannfelt L, et al. , Alzheimer’s disease pathology propagation by exosomes containing toxic amyloid-beta oligomers, Acta Neuropathologica. 136 (2018) 41–56. doi: 10.1007/s00401-018-1868-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ngolab J, Trinh I, Rockenstein E, Mante M, Florio J, Trejo M, et al. , Brain-derived exosomes from dementia with Lewy bodies propagate α-synuclein pathology, Acta Neuropathol Commun. 5 (2017) 46–10. doi: 10.1186/s40478-017-0445-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Muraoka S, Jedrychowski MΜ, Tatebe H, DeLeo AM, Ikezu S, Tokuda T, et al. , Proteomic Profiling of Extracellular Vesicles Isolated From Cerebrospinal Fluid of Former National Football League Players at Risk for Chronic Traumatic Encephalopathy, Front Neurosci. 13 (2019) 1059. doi: 10.3389/fnins.2019.01059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Stuendl A, Kunadt M, Kruse N, Bartels C, Moebius W, Danzer KM, et al. , Induction of α-synuclein aggregate formation by CSF exosomes from patients with Parkinson’s disease and dementia with Lewy bodies, Brain. 139 (2016) 481–494. doi: 10.1093/brain/awv346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Feneberg E, Steinacker Μ, Lehnert S, Schneider A, Walther Μ, Thal DR, et al. , Limited role of free TDP-43 as a diagnostic tool in neurodegenerative diseases, Amyotroph Lateral Scler Frontotemporal Degener. 15 (2014) 351–356. doi: 10.3109/21678421.2014.905606. [DOI] [PubMed] [Google Scholar]
- [33].Winston CN, Goetzl EJ, Akers JC, Carter BS, Rockenstein EM, Galasko D, et al. , Prediction of conversion from mild cognitive impairment to dementia with neuronally derived blood exosome protein profile, Alzheimers Dement (Amst). 3 (2016) 63–72. doi: 10.1016/j.dadm.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Shi M, Liu C, Cook TJ, Bullock KM, Zhao Y, Ginghina C, et al. , Plasma exosomal α-synuclein is likely CNS-derived and increased in Parkinson’s disease, Acta Neuropathologica. 128 (2014) 639–650. doi: 10.1007/s00401-014-1314-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yang Y, Keene CD, Peskind ER, Galasko DR, Hu S-C, Cudaback E, et al. , Cerebrospinal Fluid Particles in Alzheimer Disease and Parkinson Disease, J. Neuropathol. Exp. Neurol. 74 (2015) 672–687. doi: 10.1097/NEN.0000000000000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, et al. , The PRIDE database and related tools and resources in 2019: improving support for quantification data, Nucleic Acids Res. 47 (2019) D442-D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Huang DW, Sherman BT, Lempicki RA, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources, Nat Protoc. 4 (2009) 44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- [38].Huang DW, Sherman BT, Lempicki RA, Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists, Nucleic Acids Res. 37 (2009) 1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Karimi N, Cvjetkovic A, Jang SC, Crescitelli R, Hosseinpour Feizi MA, Nieuwland R, et al. , Detailed analysis of the plasma extracellular vesicle proteome after separation from lipoproteins, Cell. Mol. Life Sci. 75 (2018) 2873–2886. doi: 10.1007/s00018-018-2773-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Nakai W, Yoshida T, Diez D, Miyatake Y, Nishibu T, Imawaka N, et al. , A novel affinity-based method for the isolation of highly purified extracellular vesicles, Sci Rep. 6 (2016) 33935. doi: 10.1038/srep33935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Vella LJ, Scicluna BJ, Cheng L, Bawden EG, Masters CL, Ang C-S, et al. , A rigorous method to enrich for exosomes from brain tissue, J Extracell Vesicles. 6 (2017) 1348885. doi: 10.1080/20013078.2017.1348885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Krämer-Albers E-M, Bretz N, Tenzer S, Winterstein C, Möbius W, Berger H, et al. , Oligodendrocytes secrete exosomes containing major myelin and stress-protective proteins: Trophic support for axons? Proteomics Clin Appl. 1 (2007) 1446–1461. doi: 10.1002/prca.200700522. [DOI] [PubMed] [Google Scholar]
- [43].Bakhti M, Winter C, Simons M, Inhibition of myelin membrane sheath formation by oligodendrocyte-derived exosome-like vesicles, J. Biol. Chem. 286 (2011) 787–796. doi: 10.1074/jbc.M110.190009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Nordin JZ, Lee Y, Vader Μ, Mäger I, Johansson HJ, Heusermann W, et al. , Ultrafiltration with size-exclusion liquid chromatography for high yield isolation of extracellular vesicles preserving intact biophysical and functional properties, Nanomedicine. 11 (2015) 879–883. doi: 10.1016/j.nano.2015.01.003. [DOI] [PubMed] [Google Scholar]
- [45].Morozov VA, Lagaye S, Hepatitis C virus: Morphogenesis, infection and therapy, World J Hepatol. 10 (2018) 186–212. doi: 10.4254/wjh.v10.i2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Williams ΜT, Superko HR, Haskell WL, Alderman EL, Blanche ΜJ, Holl LG, et al. , Smallest LDL particles are most strongly related to coronary disease progression ii men, Arterioscler. Thromb. Vasc. Biol. 23 (2003) 314–321. doi: 10.1161/01.atv.0000053385.64132.2d. [DOI] [PubMed] [Google Scholar]
- [47].Lobb RJ, Becker M, Wen SW, Wong CSF, Wiegmans AΜ, Leimgruber A, et al. Optimized exosome isolation protocol for cell culture supernatant and human plasma, Extracell Vesicles. 4 (2015) 27031. doi: 10.3402/jev.v4.27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Muller L, Hong C-S, Stolz DB, Watkins SC, Whiteside TL, Isolation of biologically-active exosomes from human plasma, J. Immunol. Methods. 411 (2014) 565. doi: 10.1016/j.jim.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Böing AN, van der Pol E, Grootemaat AE, Coumans FAW, Sturk A, Nieuwland R, Single-step isolation of extracellular vesicles by size-exclusion chromatography, J Extracell Vesicles. 3 (2014) 23430. doi: 10.3402/jev.v3.23430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Buschmann D, Kirchner B, Hermann S, Märte M, Wurmser C, Brandes F, et al. , Evaluation of serum extracellular vesicle isolation methods for profiling miRNAs by next-generation sequencing, J Extracell Vesicles. 7 (2018) 1481321. doi: 10.1080/20013078.2018.1481321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].An M, Wu J, Zhu J, Lubman DM, Comparison of an Optimized Ultracentrifugation Method versus Size-Exclusion Chromatography for Isolation of Exosomes from Human Serum, J. Proteome Res. 17 (2018) 3599–3605. doi: 10.1021/acs.jproteome.8b00479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Patel GK, Khan MA, Zubair H, Srivastava SK, Khushman M, Singh S, et al. , Comparative analysis of exosome isolation methods using culture supernatant for optimum yield, purity and downstream applications, Sci Rep. 9 (2019) 5335. doi: 10.1038/s41598-019-41800-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Stranska R, Gysbrechts L, Wouters J, Vermeersch Μ, Bloch K, Dierickx D, et al. , Comparison of membrane affinity-based method with size-exclusion chromatography for isolation of exosome-like vesicles from human plasma, J Transl Med. 16 (2018) 1–9. doi: 10.1186/s12967-017-1374-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Oeyen E, Van Mol K, Baggerman G, Willems H, Boonen K, Rolfo C, et al. , Ultrafiltration and size exclusion chromatography combined with asymmetrical-flow field-flow fractionation for the isolation and characterisation of extracellular vesicles from urine, J Extracell Vesicles. 7 (2018) 1490143. doi: 10.1080/20013078.2018.1490143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Baranyai T, Herczeg K, Onódi Z, Voszka I, Modos K, Marton N, et al. , Isolation of Exosomes from Blood Plasma: Qualitative and Quantitative Comparison of Ultracentrifugation and Size Exclusion Chromatography Methods, PLoS ONE. 10 (2015) e0145686. doi: 10.1371/journal.pone.0145686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Simonsen JB, What Are We Looking At? Extracellular Vesicles, Lipoproteins, or Both? Circ. Res. 121 (2017) 920–922. doi: 10.1161/CIRCRESAHA.117.311767. [DOI] [PubMed] [Google Scholar]
- [57].Hong C-S, Funk S, Muller L, Boyiadzis M, Whiteside TL, Isolation of biologically active and morphologically intact exosomes from plasma of patients with cancer, J Extracell Vesicles. 5 (2016) 29289. doi: 10.3402/jev.v5.29289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Welton JL, Webber JΜ, Botos L-A, Jones M, Clayton A, Ready-made chromatography columns for extracellular vesicle isolation from plasma, J Extracell Vesicles. 4 (2015) 27269. doi: 10.3402/jev.v4.27269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Vogel R, Coumans FAW, Maltesen RG, Böing AN, Bonnington KE, Broekman ML, et al. , A standardized method to determine the concentration of extracellular vesicles using tunable resistive pulse sensing, J Extracell Vesicles. 5 (2016) 31242. doi: 10.3402/jev.v5.31242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Street JM, Barran ΜE, Mackay CL, Weidt S, Balmforth C, Walsh TS, et al. , Identification and proteomic profiling of exosomes in human cerebrospinal fluid, J Transl Med. 10 (2012) 5. doi: 10.1186/1479-5876-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Chiasserini D, van Weering JRT, Piersma SR, Pham TV, Malekzadeh A, Teunissen CE, et al. , Proteomic analysis of cerebrospinal fluid extracellular vesicles: a comprehensive dataset, J Proteomics. 106 (2014) 191–204. doi: 10.1016/j.jprot.2014.04.028. [DOI] [PubMed] [Google Scholar]
- [62].Lee J, McKinney KQ, Pavlopoulos AJ, Han MH, Kim S-H, Kim HJ, et al. , Exosomal proteome analysis of cerebrospinal fluid detects biosignatures of neuromyelitis optica and multiple sclerosis, Clin. Chim. Acta. 462 (2016) 118–126. doi: 10.1016/j.cca.2016.09.001. [DOI] [PubMed] [Google Scholar]
- [63].Pieragostino D, Lanuti Μ, Cicalini I, Cufaro MC, Ciccocioppo F, Ronci M, et al. , Proteomics characterization of extracellular vesicles sorted by flow cytometry reveals a disease-specific molecular cross-talk from cerebrospinal fluid and tears in multiple sclerosis, J Proteomics. 204 (2019) 103403. doi: 10.1016/j.jprot.2019.103403. [DOI] [PubMed] [Google Scholar]
- [64].Welton JL, Loveless S, Stone T, von Ruhland C, Robertson NΜ, Clayton A, Cerebrospinal fluid extracellular vesicle enrichment for protein biomarker discovery in neurological disease; multiple sclerosis, J Extracell Vesicles. 6 (2017) 1369805. doi: 10.1080/20013078.2017.1369805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Hayashi N, Doi H, Kurata Y, Kagawa H, Atobe Y, Funakoshi K, et al. , Proteomic analysis of exosome-enriched fractions derived from cerebrospinal fluid of amyotrophic lateral sclerosis patients, Neuroscience Research. (2019). doi: 10.1016/j.neures.2019.10.010. [DOI] [PubMed] [Google Scholar]
- [66].Manek R, Moghieb A, Yang Z, Kumar D, Kobessiy F, Sarkis GA, et al. , Protein Biomarkers and Neuroproteomics Characterization of Microvesicles/Exosomes from Human Cerebrospinal Fluid Following Traumatic Brain Injury, Mol. Neurobiol. 55 (2018) 6112–6128. doi: 10.1007/s12035-017-0821-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.