Abstract
Introduction
Protein corona (PC) deposition on nanoparticles (NPs) in biological systems contributes to a great extent to NPs’ fates; their targeting potential, the interaction with different biological systems and the subsequent functions. PC – when properly tuned – can serve as a potential avenue for optimization of NPs’ use in cancer therapy.
Methods
Poly-lactic co-glycolic acid (PLGA)-based NPs exhibiting different physicochemical properties were fabricated and characterized. The PC makeup of these NPs were qualitatively and quantitatively analyzed by Western blot and Bradford assay, respectively. The effect of PC on the release of NPs’ cargos and the intracellular uptake into B16F10 melanoma cells has been studied.
Results
The composition of NPs (polymeric PLGA NPs vs lipid-polymer hybrid NPs) and the conjugation of an active targeting ligand (cRGDyk peptide) represented the major determinants of the PC makeup of NPs. The in vitro release of the loaded cargos from the NPs depended on the PC and the presence of serum proteins in the release medium. Higher cumulative release has been recorded in the presence of proteins in the case of peptide conjugated NPs, cNPs, while the unconjugated formulations, uNPs, showed an opposite pattern. NPs intracellular uptake studies revealed important roles of distinct serum and cellular proteins on the extent of NPs’ accumulation in melanoma cells. For example, the abundance of vitronectin (VN) protein from serum has been positively related to the intracellular accumulation of the NPs.
Conclusion
Careful engineering of nanocarriers can modulate the recruitment of some proteins suggesting a potential use for achieving endogenous targeting to overcome the current limitations of targeted delivery of chemotherapeutic agents.
Keywords: nanoparticles, active targeting, passive targeting, endogenous targeting, melanoma, protein corona, intracellular uptake
Introduction
Conventional chemotherapeutic agents possess unfavourable pharmacokinetic (PK) profiles and off-target distribution which contribute to the severe toxicities of these agents.1 The lack of specificity can be largely overcome by optimum engineering of suitable nanocarriers.2 A major determinant of the NPs’ fates upon introduction to the biological environment is the deposition of serum proteins on the NPs’ surfaces. These proteins wrap the NPs with a layer called a protein corona (PC). The composition of this PC evolves over time; for instance, proteins characterized by high affinity towards NPs and less tendency to desorb constitute the hard corona. While proteins with lower affinity and higher tendency to desorb from NPs’ surfaces represent the soft corona and tend to be exchanged with other proteins.3 The hard corona, being the long-lasting layer, contributes to a greater extent to the control of the behaviour and biological fates of NPs.4,5 The composition of PC is largely determined by many NPs’-related factors (e.g. composition, size, shape and surface properties) and environment-related factors (source of proteins, exposure temperature and flow rates).4,6–8 Therefore, each NP formulation has been suggested to possess a PC fingerprint with distinct downstream effects. PC has been shown to alter the passive and active targeting processes. On one hand, some proteins are regarded as opsonins, e.g. immunoglobulins, and complement proteins which mark the NPs to be taken up by the phagocytic cells. Then, NPs eventually end up in the reticuloendothelial system (RES) organs. On the other hand, other proteins act as dysopsonins, e.g. clusterin α and albumin. They prevent deposition of opsonins on NPs and prolong their circulation time giving a better chance for NPs to reach their target organs.5,9–14 In addition, PC can alter the active targeting capacities by masking the targeting moieties and hence preventing recognition by their target receptors.15,16 However, in some studies, PC has been shown to possess harmless or advantageous potential. Therefore, careful design of NPs to recruit specific proteins can represent a good chance for achieving endogenous targeting.15
The physicochemical properties of NPs are important determinants of the their biodistribution, interaction with the target and non-target cells and the subsequent functions.17,18 Therefore, in this study, NPs possessing different physicochemical properties (size, surface charge and composition) have been synthesized. In addition, an Arginine-Glycine-Aspartic acid (RGD)-based peptide, cRGDyk, has been chemically coupled to the NPs. cRGDyk cyclic pentapeptide is a selective inhibitor to αvβ3 integrins that are cell surface proteins overexpressed in melanoma.19–24 The PC of the synthesized NPs has been characterized in terms of the quantity and the abundance of distinct proteins of interest. The role of this PC on the NPs’ behavior in terms of the in vitro release of loaded cargos and the in vitro cellular uptake has then been characterized.
Vitronectin (VN), Complement 3 (C3) protein, Clusterin α (CLU) and Albumin (ALB) are the selected serum proteins of interest in this study. While Filamin 1 (FLN1), Annexin I (ANXA1) and Clathrin HC (CHC) represent the selected cellular proteins. These proteins are selected for the following reasons. VN is selected for its known affinity towards αVβ3 integrin receptors that are overexpressed in melanoma.13,14 C3 protein is an opsonin which has been critically associated with fast clearance and short blood circulation time of NPs after intravenous (IV) administration.5,11–13 In addition, complement peptide receptors, anaphylatoxin receptors, have recently been reported to be overexpressed in melanoma exhibiting a role in immune surveillance.25,26 On the contrary, CLU plays a predominant role as a dysopsonin which, when abundant in PC, can reduce the opsonization process and the RES uptake of the NPs.12,14 Melanoma has also been reported to overexpress CLU isoforms.27,28 In a similar manner, ALB exerts a similar dysopsonin role along with its capacity to initiate receptor-mediate endocytosis and reduce the endolysosomal degradation of NPs after cellular internalization.5,13,29 Moreover, the selected cellular proteins are known to be involved in endocytosis, receptor recycling and NPs’ interaction with cell membrane and cytoplasmic components;13,29 FLN1 is a cytoplasmic protein that links actin filaments to membrane glycoproteins. ANXA1 is present in the basement membrane and extracellular matrix (ECM) and is associated with positive regulation of vesicle fusion and receptor recycling. CHC is a major protein of coated pits and vesicles in the cell membrane.13,29
Materials and Methods
Materials
The mouse monoclonal antibodies (Vitronectin 65/75 antibody (D-8), Complement protein 3 (C3), antibody (B-9), Albumin (ALB) antibody (E-11), Clusterin-α Antibody (A-11), Annexin I Antibody (EH17a), Clathrin HC Antibody (TD.1) and Filamin 1 Antibody (E-3) were purchased from Santa Cruz, Germany. Fluorescein isothiocyanate (FITC) was also purchased from Santa Cruz, Germany. c(RGDyk) peptide 99.59% was purchased from Selleckchem, USA. Female C57 Black 6 (C57BL/6) mice were obtained from the animal house breeds of the German University in Cairo (GUC). The experiment protocol followed ARRIVE guidelines and was approved by the Research Ethics Committee of the Faculty of Pharmacy and Biotechnology, GUC. Each three mice weighing 20–25 g were housed in suitably-sized cages to allow free movements. The cages were exposed to standard conditions of housing with a 12-h light/dark cycle and a temperature of 22°C. All mice received standard laboratory diet and were granted free access to water. The mice were cared for on a daily basis. Mouse serum (MS) was obtained by collecting blood using the retro-orbital bleeding technique under aseptic conditions in specialized serum separator collection tubes. Getting rid of animals’ remains by incineration was done according to the approved animal waste disposal system.
Preparation of uNPs and cNPs
FITC-loaded PLGA NPs were prepared as previously described.30 Among a list of NPs prepared following different formulation and process variables, four formulations were selected for further investigation in this study. Details of the composition of the four selected formulas of the NPs are shown in Table 1.
Table 1.
Composition of the Peptide-Unconjugated NPs (uNPs)
| NPs Formula Type | NPs Formula Code | Amount of PLGA (mg) | Concentration of PLGA (mg/mL) | Additives in the Continuous Phase |
|---|---|---|---|---|
| Polymeric | uNP1 | 50 | 50 | – |
| uNP2 | 25 | |||
| Hybrid | uNP3 | 50 | 0.15% lecithin 4% ethanol |
|
| uNP4 | 25 |
Carbodiimide crosslinking chemistry was employed for the conjugation of cRGDyk peptide to the NPs’ surfaces to yield the corresponding cNPs. The conjugation reaction was performed at PLGA:cRGDyk molar ratio of 1:1.8,30
Particle Size and Surface Charge Assessment
A one mg/mL suspension of NPs was prepared by diluting the final preparation with ultra-pure water (UPW), phosphate-buffered saline (PBS) or MS for the analysis of the particle size and surface charge (Malvern Zetasizer, Nano-ZS, UK). PBS was adjusted to pH = 5.0, 6.8 or 7.4 to simulate the endolysosomal pH, the weakly acidic environment of the tumor tissue or the physiological pH respectively.30–32
Quantification of the NPs’ PC by Bradford Assay
For PC formation, NPs were incubated with PBS adjusted at pH = 5.0, 6.8 or 7.4 in the presence of 10% MS or with 100% MS for 1 h at 37 °C under shaking at 200 strokes per minute to allow the adsorption of serum proteins prior to Bradford protein assay. It is worth mentioning that a 1-h incubation was reported to be enough for the formation of a stable hard corona of NPs; whereas similar amounts of adsorbed proteins were detected over a duration of 5 to 90 minutes.7 For the protein quantification, a modified protocol of Partikel et al33,34 was employed. Eight mg of NPs were hydrolyzed with 500 μL of 0.5 M sodium hydroxide for 15 min at 60 °C and 1200 rpm in a heating dry block (Eppendorf Thermomixer, Germany). For the formation of a protein-dye complex, 1.5 mL of Bradford reagent was added and incubated for a further 10 min. Absorbance was then monitored at 595 nm for the quantification of the adsorbed proteins and compared to a calibration curve of bovine serum albumin (BSA) in the range of 0.01–1 mg/mL.33,34
For the calculation of the total amount of proteins adsorbed per surface area of NPs, density of 1.3 g/cm3,35 and a spherical shape, as estimated in our previous work by the scanning electron microscopy (SEM),30 were assumed.
Identification of PC by Sodium Dodecyl Sulfate−Polyacrylamide Gel Electrophoresis (SDS-PAGE) and Western Blot (WB) Analysis
For PC formation, a protocol by Lesniak et al13 was modified as follows;13 a 250 µg/mL of the NPs was prepared in DMEM under serum-free (SF) and MS-rich (SR) conditions. NPs were then incubated for 1 h at 37 °C under shaking at 200 strokes per minute. The NPs suspension was then added to B16F10 cells (seeded at density of 105 and incubated overnight) in a 24-well plate and incubated for a further 3 h to allow the adsorption of cellular proteins on NPs. The cell supernatant was then aspirated from the cell cultures and centrifuged to pellet the NPs. The pelleted NPs were then washed three times with ultrapure water to get rid of the unbound proteins. Finally, the PC layer was desorbed from the NPs by incubation with 50 μL SDS buffer overnight. Centrifugation (45 min, 30,000 x g) was then attempted to separate the desorbed proteins from the NPs’ remains. The proteins in the supernatant were then denatured for 5 min at 95 °C in a heating dry block (Eppendorf Thermomixer, Germany).13
For SDS-PAGE analysis, a similar protocol by Partikel et al was adopted with minor modifications; the samples as well as the protein standard (Color Prestained Protein Standard, Broad Range [11–245 kDa], New England Biolabs, MA, USA or ROTI®Mark TRICOLOR XTRA protein marker [10–310 kDa], Carl Roth, Karlsruhe, Germany), a negative control, and a medium control were applied on an 11% polyacrylamide gel. For the negative controls, UPW was used instead of the SF or SR DMEM. For the medium control, SF or SR DMEM obtained from the cultures were diluted 100x with UPW.33,34
Immunostaining for WBs was performed using the mouse monoclonal primary antibodies; Vitronectin 65/75 antibody (D-8), Complement protein 3 (C3), antibody (B-9), Clusterin-α Antibody (A-11), Annexin I Antibody (EH17a), Clathrin HC Antibody (TD.1) and Filamin 1 Antibody (E-3) (Santa Cruz, Germany), and an alkaline phosphatase conjugated anti-mouse secondary antibody (Promega, Germany) following a standard protocol in our lab.36
Semi-quantification of the relative band intensities of WBs was performed using ImageJ 1.37c software (National Institutes of Health, NIH).
In vitro Release of FITC from uNPs and cNPs
In order to have a valid comparison of the cargo release patterns from different formulations as a function of the release conditions, a protocol by Gomaa et al was adopted with minor modifications. 0.1 mL of the NPs’ suspension (5 mg/mL) was placed in a sealed dialysis bag, immersed in 3 mL of release medium and placed in a shaker incubator (200 strokes/min, 37°C) under aseptic conditions. Seven different types of release media were utilized to evaluate the effect of pH and serum proteins on the NPs’ release behavior. These release media are represented by PBS, pH = 5.0, 6.8 or 7.4 in the presence or absence of 10% MS. Release in 100% MS has also been performed in order to simulate the in vivo conditions of drug release upon systemic administration.37,38 At predefined time points, 0.1-mL samples were withdrawn for the quantification of the fluorescence of the released FITC in a multiwell plate reader (Victor 3V 1420, Perkin Elmer, USA) at excitation/emission wavelength of (485nm/535nm). Amounts of FITC were calculated using a freshly constructed calibration curve of FITC in each of the employed media.38
Evaluation of the in vitro Intracellular Uptake of uNPs and cNPs
For the evaluation of the intracellular uptake of NPs, a modified protocol by Farid et al39 was adopted. Cells were seeded into 96-well plates (104 cells, 100 µL) and incubated overnight under standard culture conditions. A 250 µg/mL of FITC-loaded NPs in SF and SR phenol red-free DMEM was added after aspiration of the old medium and washing of cells with PBS to remove any debris. MS was used instead of the conventional fetal bovine serum (FBS) in order to better correlate the results of the in vitro uptake and the in vivo biodistribution in the tumor-bearing mouse model37 that is attempted in Part II of this study. After a 3-h incubation period, the medium was aspirated, cells were washed twice with PBS and the fluorescence of the internalized FITC-loaded NPs were quantified as previously described.
Sodium azide, NaN3, (15 mM), methyl-β-cyclodextrin, MβCD, (1 mM), sucrose (450 μM) and nystatin (20 μM) were utilized as endocytic inhibitors for the evaluation of the mechanism of endocytic uptake of the NPs under both SF and SR conditions. NaN3 as an ATP depleting agent was utilized to evaluate the energy-dependent uptake pathways. While MβCD, sucrose and nystatin were utilized to evaluate cholesterol-dependent, clathrin-mediated and caveolin-mediated endocytosis respectively. Cells were pretreated with the endocytic inhibitors for 30 min prior to the addition of the NPs.40–42
Evaluation of the Role of Selected Serum and Cellular Proteins on the in vitro Intracellular Uptake of uNPs and cNPs
Monoclonal antibodies against selected serum (VN, C3 protein, CLU and ALB) and cellular proteins (CHC, ANXA1 and FLN1) have been utilized to confirm the relationship between the presence of those proteins in coronas of different NPs and their accumulation in B16F10 melanoma cells. Two approaches have been utilized; first, cells were preincubated with the antibodies (0.1 µg/mL) for 3 h in order to block the corresponding cellular machinery responsible for mediating the uptake of NPs. Second, NPs were premixed with the antibodies (1 ng/mL) prior to their incubation with the cells to allow the antibodies to deposit on NPs’ surfaces and constitute part of their PC.16,43
The following equation was employed for the calculation of the relative intracellular uptake of the NPs in the presence of the antibodies;
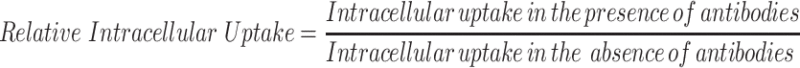 |
(1) |
Statistical Analysis
GraphPad Prism 7.0 (GraphPad Software, Inc, San Diego, CA, USA) was utilized for the statistical analysis of the presented data. All experiments were performed at least three times and data were represented as the mean ± standard error of the mean (SEM). Analysis was performed using One-way ANOVA, Two-way ANOVA Tukey’s Multiple Comparison Test or t-test whenever appropriate at confidence level 95%. Asteriks (*) were utilized to express the levels of statistical significance as follows; (****), (***), (**) and (*) represent (P value < 0.0001), (P value < 0.001), (P value = 0.001 to 0.01) and (P value = 0.01 to 0.05) respectively. The non-significant difference is demonstrated as (ns) with (P value > 0.05).
Results
Physicochemical Characterization of the Formulated NPs
Particle Size and Surface Charge of uNPs and cNPs
All the uNPs maintained similar particle size in all of the employed vehicles with only slight increase in serum (Table 2). Whereas there was an obvious reduction in zeta potential when UPW was replaced by any buffer.
Table 2.
Physicochemical Characteristics of uNPs and cNPs in Different Vehicles
| Formula Name | Sampling Solution | Mean Z-Average ± SD (nm) | PDI ± SD | Mean Zeta Potential ± SD (mV) | Formula Name | Sampling Solution | Mean Z-Average ± SD (nm) | PDI ± SD | Mean Zeta Potential ± SD (mV) |
|---|---|---|---|---|---|---|---|---|---|
| uNP1 | Water | 192±13 | 0.09 ±0.02 | −19.8±1.2 | cNP1 | Water | 244±12 | 0.16 ±0.05 | 10.9±0.9 |
| pH 5.0 | 204±5 | 0.10 ±0.03 | −1.4±1.3 | pH 5.0 | 260±10 | 0.25 ±0.04 | −1.1±0.6 | ||
| pH 6.8 | 207±5 | 0.18 ±0.02 | −0.2±0.9 | pH 6.8 | 245±35 | 0.23 ±0.04 | −0.5±0.8 | ||
| pH 7.4 | 201±5 | 0.10 ±0.01 | −0.5±0.2 | pH 7.4 | 255±9 | 0.17 ±0.19 | −0.8±0.6 | ||
| Serum | 196±22 | 0.12 ±0.10 | −1.2±0.8 | Serum | 136±32*** | 0.28 ±0.02 | −2.4±0.5 | ||
| uNP2 | Water | 144±7 | 0.09 ±0.06 | −17.0±1.3 | cNP2 | Water | 209±3 | 0.10 ±0.01 | 7.0±0.3 |
| pH 5.0 | 147±5 | 0.08 ±0.06 | −0.4±1.5 | pH 5.0 | 208±5 | 0.10 ±0.03 | −0.3±0.2 | ||
| pH 6.8 | 147±3 | 0.07 ±0.05 | −0.7±1.8 | pH 6.8 | 205±10 | 0.12 ±0.01 | −0.5±0.7 | ||
| pH 7.4 | 153±2 | 0.06 ±0.03 | −2.2±0.7 | pH 7.4 | 262±30 | 0.26 ±0.07 | −0.4±0.2 | ||
| Serum | 151±1 | 0.13 ±0.06 | −4.0±3.3 | Serum | 232±2 | 0.14 ±0.03 | −2.0±1.3 | ||
| uNP3 | Water | 254±14 | 0.08 ±0.03 | −19.9±0.4 | cNP3 | Water | 370±22 | 0.32 ±0.04 | −0.9±0.4 |
| pH 5.0 | 270±9 | 0.33 ±0.03 | −1.3±1.5 | pH 5.0 | 393±13 | 0.18 ±0.04 | −0.1+0.8 | ||
| pH 6.8 | 274±7 | 0.21 ±0.04 | −1.3±1.2 | pH 6.8 | 382±20 | 0.27 ±0.25 | −0.3±0.9 | ||
| pH 7.4 | 296±6 | 0.17 ±0.05 | −1.2±0.7 | pH 7.4 | 278±9* | 0.17 ±0.03 | −0.4±0.5 | ||
| Serum | 221±6 | 0.08 ±0.02 | −3.0±0.9 | Serum | 198±57P*** | 0.79 ±0.07 | −2.2±0.6 | ||
| uNP4 | Water | 155±8 | 0.17 ±0.05 | −23±0.9 | cNP4 | Water | 258±34 | 0.34 ±0.06 | −12.3±1.2 |
| pH 5.0 | 162±2 | 0.03±0.01 | −7.3±1.7 | pH 5.0 | 269±9 | 0.25 ±0.01 | −1.5±0.2 | ||
| pH 6.8 | 168±6 | 0.25 ±0.01 | −9.4±0.3 | pH 6.8 | 279±10 | 0.40 ±0.01 | −0.9±0.9 | ||
| pH 7.4 | 175±9 | 0.05 ±0.06 | −7.7±3.4 | pH 7.4 | 291±24 | 0.42 ±0.03 | −1.0±0.8 | ||
| Serum | 175±11 | 0.04 ±0.01 | −5.4±0.9 | Serum | 222±31 | 0.24 ±0.04 | −2.7±0.7 |
Notes: *p value <0.5; ***p value <0.001; (p), polymodal size distribution.
Upon Peptide Conjugation at PLGA: cRGDyk (P:R) molar ratio of 1:1, an obvious increase in the particles size was observed when all cNPs were analyzed in UPW as compared to their un-conjugated counterparts. Regarding the surface charge, all cNPs showed a less negative zeta potential most prominently in the polymeric cNP1 and cNP2 which showed a reversed charge from negative to positive indicative of successful conjugation when compared to uNP1 and cNP2, respectively. Looking more closely to the effect of serum proteins on particle size, a reduction in particle size could be observed most prominently in cNP1 and cNP3.
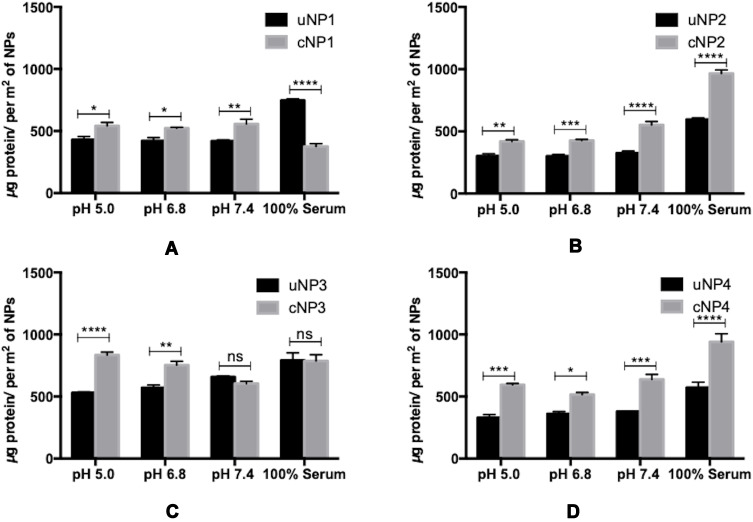
Quantification of the Adsorbed Proteins on uNPs and cNPs
The quantities of the adsorbed proteins on NPs’ surfaces expressed as µg protein per m2 of NPs are shown in Figure 1. The factors affecting the amount of adsorbed proteins could be summarized into NPs-related (composition and peptide conjugation) and medium-related factors (pH and serum concentration) as follows. NPs’ composition and the peptide conjugation status contributed to a great extent to the amount of the adsorbed proteins on NPs’ surfaces. It could generally be observed that hybrid formulations, NP3 and NP4, exhibited higher quantities of proteins in their coronas than their polymeric counterparts, NP1 and NP2 respectively. In addition, peptide conjugation increased the amount of the adsorbed proteins per surface area with the exception of cNP1, which showed opposite results. This observation coincides with the decrease in its particle size from 196 nm to 136 nm upon peptide conjugation as shown in Table 2. This decrease in size is accompanied by an increase in surface area.
Figure 1.
Quantities of the adsorbed proteins on uNPs and cNPs in µg/m2 of uNP1 and cNP1 (A), uNP2 and cNP2 (B), uNP3 and cNP3 (C) and uNP4 and cNP4 (D). (****), (***), (**) and (*) represent (P value < 0.0001), (P value < 0.001), (P value = 0.001 to 0.01) and (P value = 0.01 to 0.05) respectively. The non-significant difference is demonstrated as (ns) with (P value > 0.05).
On the other hand, the medium-related parameters contributed to a lesser extent. The amount of the adsorbed proteins on different uNPs or cNPs was independent on the pH of the medium in all of the formulations except for the hybrid NP3 in both of its forms. The amount of adsorbed proteins on the hybrid NP3 increased with pH in its unconjugated form, uNP3, and showed an opposite pattern in its peptide-conjugated form, cNP3. Lastly, as the percentage of serum increased from 10% to 100%, the amount of the adsorbed proteins increased in most of the formulations except cNP1, which exhibited a significantly smaller particle size in 100% serum in comparison to lower serum levels (Table 2).
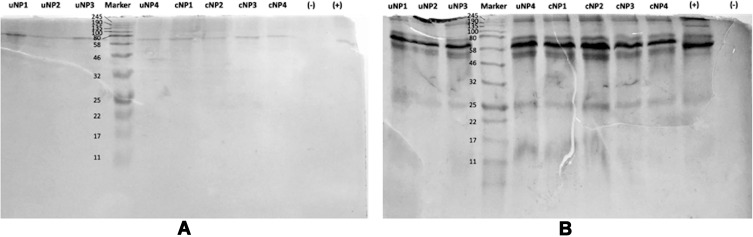
Qualitative Analysis of the PC on uNPs and cNPs
As indicated by the molecular weight (MWt) in SDS-PAGE (Figure 2), the protein band observed at MWt between 58–80 kDa, most probably denotes albumin. Albumin has a MWt of 66 kDa and is characterized by its highest abundance in serum. The intensities of the protein bands observed for NPs incubated with B16.F10 melanoma cells in SF conditions (Figure 2A) are much lower than NPs incubated in SR conditions (Figure 2B). It could also be observed that proteins with large molecular weight > 46 kDa are more abundant in all samples than those with smaller MWt.
Figure 2.
SDS-PAGE of the PC of uNPs and cNPs incubated with B16F10 melanoma cells in SF (A) and SR (B) conditions. Color Prestained Protein Standard, Broad Range (11–245 kDa) is used as a standard marker.
Abbreviations: -, negative control; +, medium control.
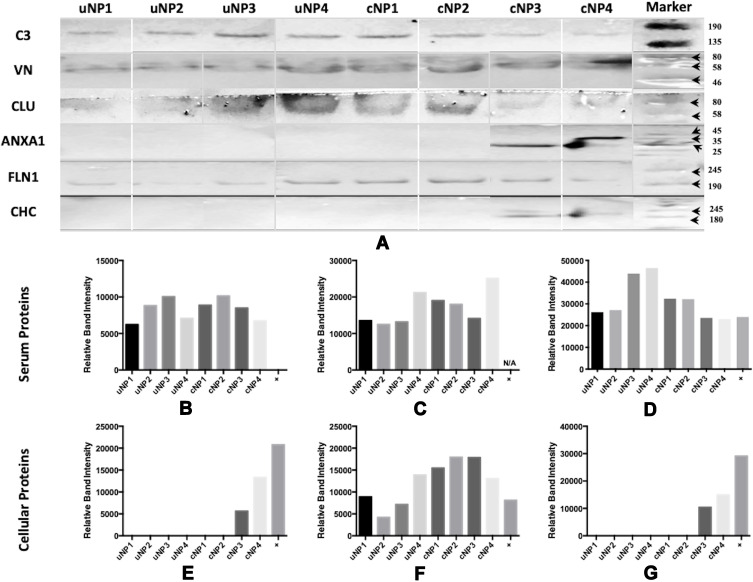
All samples of uNPs and cNPs incubated with melanoma cells in SF conditions, showed no signals in WB analysis. Otherwise, WB analysis of serum proteins (C3 protein, VN and CLU) and cellular proteins (FLN1, ANXA1 and CHC) obtained from uNPs and cNPs incubated with melanoma cells in SR conditions are shown in Figure 3A. In addition, the abundance of the selected proteins (Figure 3B–G) obtained by semi-quantification of the relative band intensities was observed to be mastered by the NPs’ composition, particle size, surface charge and the peptide conjugation status as follows.
Figure 3.
Western blot of the adsorbed PC on uNPs and cNPs after incubation with B16F10 melanoma cells in SR conditions (A) and Relative band intensities (obtained from single WBs) of C3 protein (B), VN (C), CLU (D), ANXA1 (E), FLN1 (F) and CHC (G). Bound antibodies were stained by the NBT/BcIP technique. Color Prestained Protein Standard, Broad Range (11–245 kDa), is used as a standard protein marker for VN, C3, CLU and FLN1 and ROTI®Mark TRICOLOR XTRA (10–310 kDa) is used as a standard protein marker for ANXA1 and CHC.
Relative Abundance of Serum Proteins
It is observed that C3 protein, absent from the medium control (data not shown), is enriched by all the NPs to different extents showing least abundance in uNP1 and NP4 in both of its unconjugated and conjugated forms (Figure 3B). Whereas, VN level was generally higher for the cNPs than uNPs and recorded the highest intensity in cNP4 (Figure 3C). CLU showed higher abundance in the hybrid NPs of the uNPs and the polymeric ones of the cNPs (Figure 3D).
Relative Abundance of Cellular Proteins
The level of the adsorbed FLN1 in NPs’ PC was generally higher in the case of cNPs except for NP4. Lowest levels of FLN1 were recorded for uNP2 and uNP3 (Figure 3F). ANXA1 and CHC were detected in the PC of cNP3 and cNP4 only (Figure 3E and G respectively). Two bands of ANXA1 were observed in the medium control (Supplemental materials, Section 1.1, Figure S1), corresponding to two isoforms of the protein. These isoforms were differently integrated into the PC of cNP3 and cNP4 (Figure 3A).
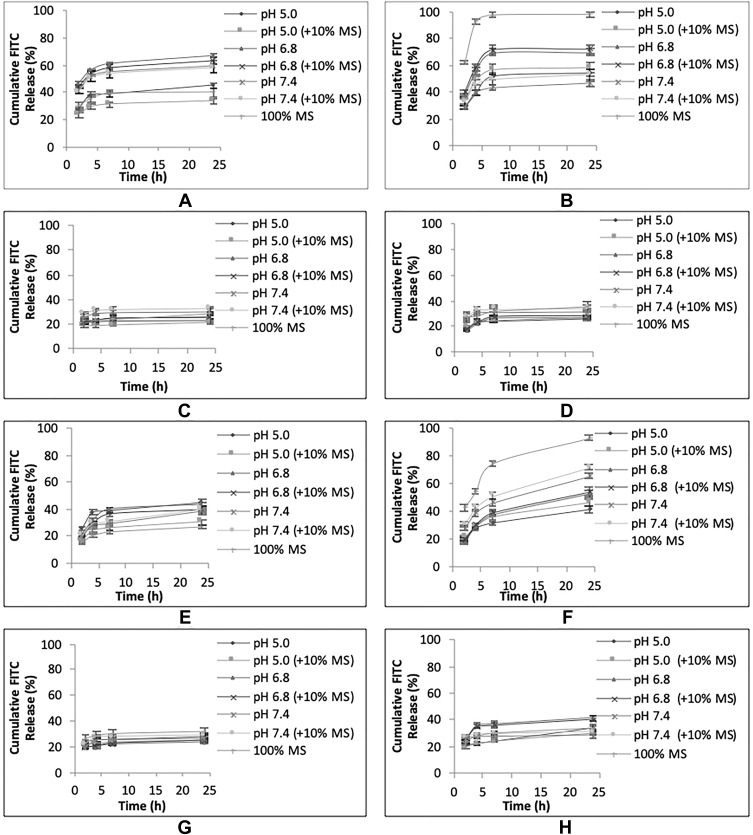
In vitro Release of the Loaded Cargo, FITC Dye, from uNPs and cNPs
The effect of pH, peptide conjugation and serum proteins on the release of the encapsulated FITC is elaborated in Figures 4 and in the supplemental materials, Section 1.2, Figure S2. In addition, a discussion of the relationship between the cumulative released % of FITC and the amount of adsorbed proteins obtained by Bradford assay (Figure 1) is established.
Figure 4.
In vitro release of FITC from uNPs and cNPs in different release media. The cumulative FITC release profiles of uNP1 and cNP1 (A, B), uNP2 and cNP2 (C, D), uNP3 and cNP3 (E, F), uNP4 and cNP4 (G, H).
Effect of pH, Peptide Conjugation and Serum Proteins on the Release of the Encapsulated FITC
uNP1 and cNP1 (Figure 4A and B)
In uNP1, the highest cumulative released % of FITC was observed at pH 6.8. In addition, MS caused a slight reduction in the cumulative released % of FITC coinciding with a slight enlargement of its particle size reported previously in Table 2.
In cNP1, the highest cumulative released % of FITC was observed at pH 6.8 as well second to 100% MS. Whereas MS, which caused a marked reduction in particle size, markedly increased the release showing almost 100% release of the loaded cargo at 6 h.
Upon comparison of uNP1 and cNP1 to evaluate the role of peptide conjugation on the in vitro release (Figure S2A), it could be observed that peptide conjugation markedly increased the release in MS, and to a lesser extent at pH 5.0 and pH 6.8. An opposite trend was witnessed at higher pH 7.4.
uNP2 and cNP2 (Figure 4C and D)
Release is independent on pH, the peptide conjugation and the presence of MS.
uNP3 and cNP3 (Figure 4E and F)
In uNP3, no marked difference in the release was observed as a function of pH. On the other hand, the presence of MS slightly decreased the release similar to uNP1.
In cNP3, the slowest release was observed at pH 5.0, then increased as pH increased from 6.8 to 7.4. In the meanwhile, MS caused a marked increase in the released % showing 70% in the first 6 hours (N.B. cNP3 has shown a multimodal size distribution in serum, Table 2).
Upon comparison of uNP3 and cNP3 (Figure S2C), it could be observed that cNP3 shows a higher release pattern than uNP3 at all the employed pH ranges.
uNP4 and cNP4 (Figure 4G and H)
Release is independent on the peptide conjugation and the presence of serum proteins. Regarding the effect of pH, no difference was observed for uNP4 at different pH values; however, release from cNP4 at pH 6.8 was significantly faster than pH 5.0 and 7.4 similar to cNP1 and cNP3.
Collectively, the large formulations (cNP1 and cNP3) showed faster release in 100% MS than the smaller ones (cNP2 and cNP4) regardless of their composition. In addition, peptide conjugation caused a general increase in release most evidently in cNP3; while the pH-dependent release of FITC varied from one formulation to another according to the physicochemical properties of NPs; the particle size, the composition and the peptide conjugation status.
The Relationship Between the Release of FITC and the Amount of Adsorbed Proteins
In the polymeric formulation, NP1, there was an inverse proportionality between the quantity of the adsorbed proteins (Figure 1A) and the extent of FITC release after 24 h (Figure S2A). In the meanwhile, the release of FITC from the polymeric formulation, NP2, (Figure S2B) was independent on the amount of the adsorbed proteins (Figure 1B).
On the other hand, there was a positive relationship between the amount of the adsorbed proteins (Figure 1C) and the cumulative release of FITC from the hybrid formulation, NP3, at low pH 5.0 and 6.8 (and Figure S2C). However, this trend was not extrapolatable to pH 7.4 and 100% MS where both forms of NP3 exhibited similar amount of the adsorbed proteins but different release patterns (N.B cNP3 exhibited small particle size at pH 7.4 and in 100% serum).
A positive relationship between the amount of the adsorbed proteins (Figure 1D) and the cumulative release of FITC in the hybrid NP4 was recorded only at pH 6.8 (Figure S2D).
To sum up, the effect of the amount of the adsorbed proteins on cargo release depended on the composition of the NPs showing an inverse relationship in the case of the polymeric NPs and direct in the case of the hybrid ones.
Intracellular Uptake of uNPs and cNPs in B16F10 Melanoma Cells in SF and SR Conditions
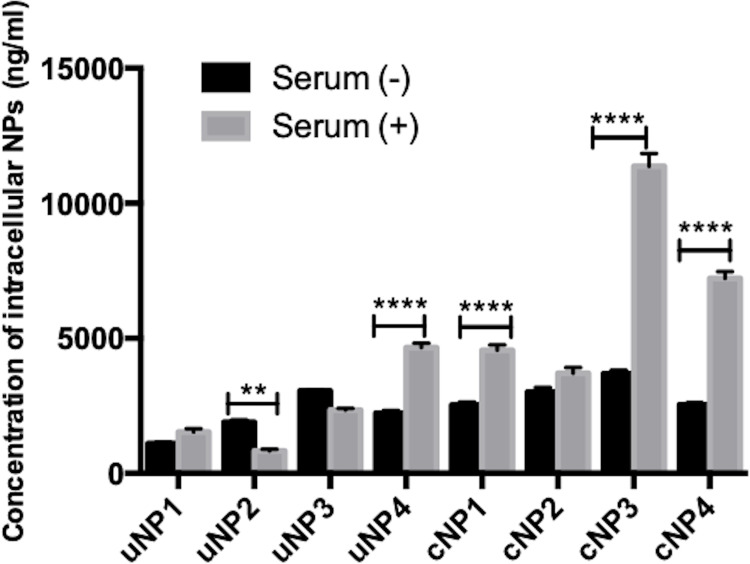
It could be observed from the in vitro intracellular uptake studies of NPs in B16F10 melanoma cells (Figure 5), that peptide conjugation increased the accumulation of all NPs’ formulations more prominently in the SR conditions. Highest levels of NPs accumulation were recorded in the case of the hybrid cNPs, cNP3 and cNP4, in SR conditions.
Figure 5.
Intracellular uptake of uNPs and cNPs in B16F10 melanoma cells after 3-h incubation under both SF and SR conditions. (****) and (**) represent (P value < 0.0001) and (P value = 0.001 to 0.01) respectively.
In addition, upon comparison of the uptake of each formulation in SF and SR conditions, it could be observed that the presence of serum proteins in the culture medium increased the uptake of uNP4 and all cNPs. However, the presence of serum proteins reduced or caused no significant change in the uptake of the rest of uNPs. The mechanism of NPs uptake has been studied and the results are elaborated in the supplemental materials, Section 1.3, Figure S3.
Evaluation of the Role of Selected Serum and Cellular Proteins on the in vitro Intracellular Uptake of uNPs and cNPs
First Approach: B16F10 Melanoma Cells Pretreated with the Antibodies
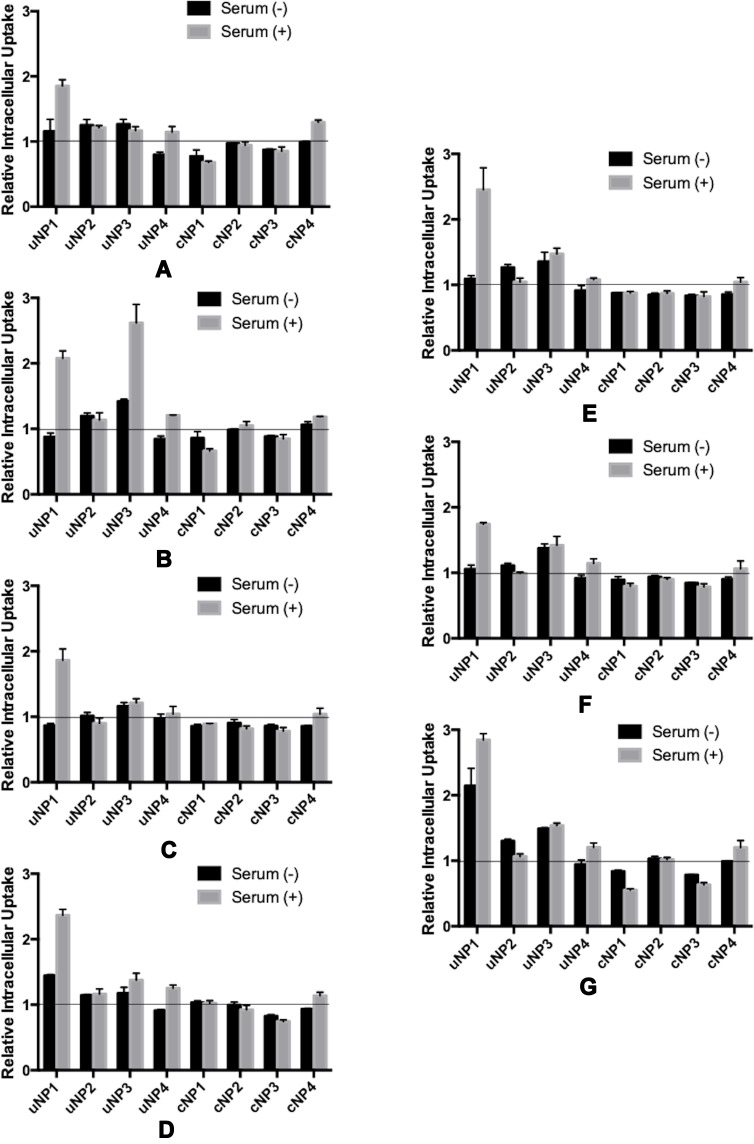
A positive relationship between the abundance of the selected proteins in PC of NPs and the intracellular uptake could be observed as a decrease in NPs’ intracellular accumulation upon pretreating the cells with the corresponding antibody prior to NPs’ incubation and vice versa.
The Effect of the Selected Serum Proteins
The effect of serum proteins (VN, C3 protein, CLU and ALB) on NPs’ uptake is more prominent in the presence of MS (Figures 6A–D and the supplemental materials, section 1.4, Figure S4). It could generally be observed that serum proteins analyzed in this section exhibited a positive relationship with the intracellular uptake in the case of cNPs and a negative relationship in the case of uNPs. In addition, the behavior of cNP4 resembled the uNPs more than cNPs.
Figure 6.
Relative Intracellular Uptake upon pretreating B16F10 melanoma cells with antibodies against selected serum (A–D) and cellular (E–G) proteins prior to incubation with the uNPs and cNPs; VN (A), C3 protein (B), CLU (C), ALB (D), CHC (E), ANXA1 (F), and FLN1 (G).
The Effect of the Selected Cellular Proteins
The effect of cellular proteins (CHC, ANXA1 and FLN1) on NPs’ intracellular uptake could be summarized as follows (Figures 6E–G and the supplemental materials, section 1.4, Figure S4). Similar to serum proteins, the cellular proteins in question were negatively related with the uptake of uNPs and cNP4; but positively related with the uptake of the rest of cNPs in both SF and SR conditions. In addition, the most prominent effect was recorded for uNP1 and uNP3 in the presence of MS and the least effect was observed with uNP2.
In summary, all the tested serum and cellular proteins negatively related with the intracellular uptake of uNPs and cNP4 and positively related with the intracellular uptake of the rest of cNPs.
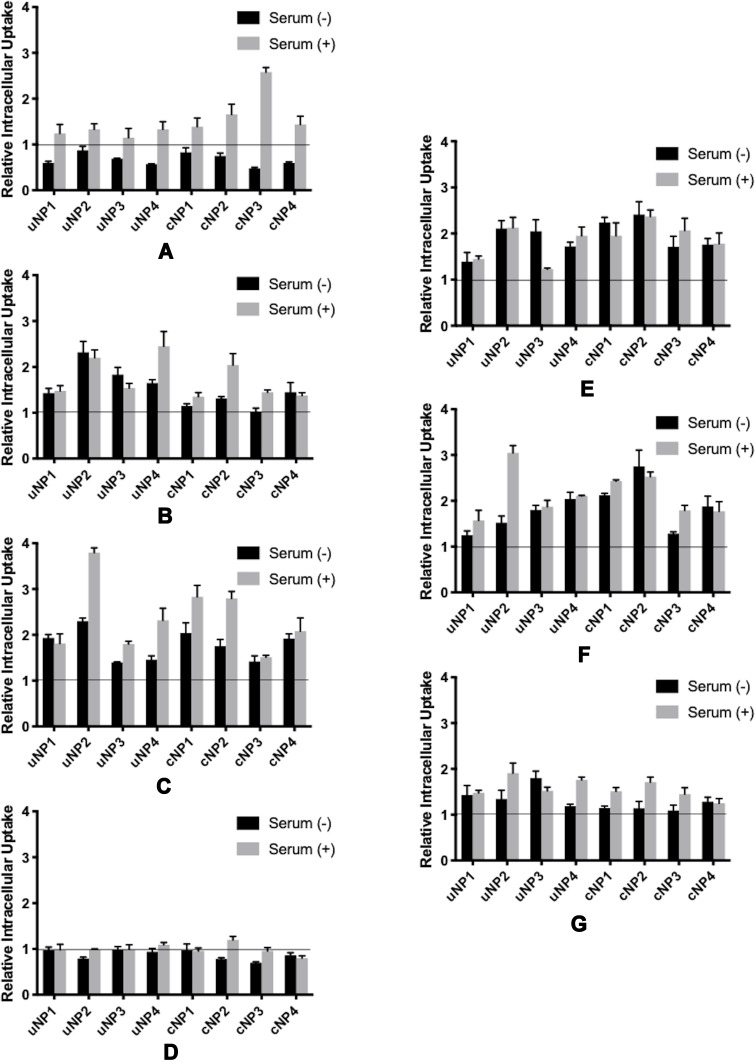
Second Approach: NPs Premixed with the Antibodies
Premixing the NPs with the antibodies could enable their deposition on the NPs’ surfaces to constitute part of the PC.
The Effect of the Selected Serum Proteins
In the case of VN monoclonal antibody, it was observed that NPs’ accumulation in the cells decreased in the SF conditions (Figures 7A and S5). While an opposite pattern was observed in the SR conditions. C3 protein antibody caused more accumulation of NPs in both SF and SR conditions with an augmented increase in the accumulation in uNP4 and cNP2 under SR conditions (Figures 7B and S5). Similarly, CLU antibody increased the NPs’ uptake more prominently in SR conditions (Figures 7C and S5). On the other hand, ALB antibody exhibited minimal effect on the extent of the uptake of NPs (Figures 7D and S5).
Figure 7.
Relative Intracellular Uptake upon premixing the uNPs and cNPs with antibodies against selected serum (A–D) and cellular (E–G) proteins prior to their incubation with B16F10 melanoma cells; VN (A), C3 protein (B), CLU (C), ALB (D), CHC (E), ANXA1 (F), and FLN1 (G).
The Effect of the Selected Cellular Proteins
Antibodies against cellular proteins (CHC, ANXA1 and FLN1) exhibited an enhancement in the uptake of the NPs (Figures 7E–G and S5).
Discussion
In this study, we have been concerned with understanding the factors affecting the formation of PC on NPs exhibiting different characterization (particle size, particle composition, surface charge and targeting ligand conjugation status). The role of the quantity and identity of the adsorbed proteins in determining the behavior and fate of the NPs has also been studied. The relationship between the abundance of distinct proteins in the PC of NPs, the in vitro drug release and the in vitro intracellular uptake of NPs in B16.F10 melanoma cells has also been investigated.
In Table 2, particle size and surface charge analysis for uNPs and cNPs showed that peptide conjugation causes a slight particle size enlargement and an obvious reduction in the negativity of the surface charge. In the case of NP1 and NP2, peptide conjugation caused a reversal of the surface charge as evidenced by the positive values for zeta potential. Possible explanations for this change in the zeta potential values include the involvement of the negatively-charged carboxyl groups on PLGA in the carbodiimide crosslinking reaction, in addition to the positivity imparted by the arginine groups abundant in the utilized cRGDyk peptide. This variation could qualitatively prove the success of peptide conjugation.44–47 Similarly, the decrease in zeta potential of all the NPs’ formulations in PBS could be attributed to the electrolyte effect. In previous studies, NaCl and MgCl2 were reported to cause neutralization of the negatively-charged carboxyl groups of PLGA.48,49
The formation of PC layer on the surfaces of uNPs and cNPs produces opposite effects on particle size. PC formation increases the particle size of uNPs which could be explained by virtue of the formation of a coating layer around the NPs as previously reported.4,50 On the other hand, a reduction in the particle size was observed in most cNPs, most significantly in the case of cNP1 and cNP3 which showed a reduction from 244 to 136 nm and from 370 to 198 nm respectively in serum. This could be attributed to the loss of the NPs’ integrity and the interruption of the cohesive forces in the polymeric core of the NPs by the adsorbed proteins as previously discussed by Abstiens et al.51 Regarding the surface charge of the NPs in the presence of serum, all NPs maintained slightly negative zeta potential independent of the original zeta potential of the NPs in UPW. These findings are consistent with previous reports.8,13,50,52
The quantity of the proteins in the PC of the hybrid NPs was observed to be higher than the polymeric ones. This could be explained on the basis of the difference in surface properties of the NPs,53,54 the fluidity of the NPs conferred by the lecithin coat,50 the difference in the degree of hydrophobicity5 or to the difference in surface roughness and particle curvature.55,56 In addition, this could be explained by virtue of the abundance of different functional groups in the lecithin coat that could offer more interaction with serum proteins.52 Similarly, the amount of the adsorbed proteins on NPs’ surfaces has been shown to be higher for the cRGDyk-conjugated over the unconjugated ones. This is consistent with previous reports which showed that as the amount of the targeting ligand molecules increased, the amount and the types of the adsorbed serum proteins increased, independent of the particle size.6,8,57 The dependence of the quantity of the adsorbed proteins on pH has been shown to be governed by the NPs’ properties. The pattern of increase or decrease of the amount of proteins with increasing the pH was different according to the tagging status. And in consistency with previous results, higher serum concentration coincides with higher amount of adsorbed proteins on NPs.4
NPs incubated with cells have been reported to adsorb PC of different nature than NPs incubated with serum away from the cells. This occurs when NPs interact with different cellular proteins in the cytosol, the cell membrane or the endocytic and exocytic vesicles.13,29 All samples of uNPs and cNPs incubated with melanoma cells under SF conditions showed no signals in the WB analysis. This makes sense for the selected serum proteins because they are absent or present in trace amounts in the SF experiments. However, for the cellular proteins, the absence of signals could in part be due to a low detection limit or due to adsorption of a different profile of cellular proteins not tested in this study. In the meanwhile, the abundance of the selected serum and cellular proteins calculated from the relative band intensities for uNPs and cNPs incubated with melanoma cells under SR conditions has shown that deposition of proteins from serum on NPs’ surfaces did not prevent interaction of NPs with the cellular proteins. In addition, the particle composition and the peptide conjugation status contributed to a great extent to the abundance of the selected proteins in consistency with findings reported earlier.53,55 C3 protein was observed to be enriched on all the NPs despite being absent from the medium control. This is a common finding for PLGA-based NPs; in a previous study, NPs could enrich some serum proteins that were under the detection limit in the positive control (FBS).5,50 The abundance of proteins with molecular weight > 46 kDa is also consistent with previous reports.50
In this study, the effect of the pH of the dissolution medium on the release of FITC from the NPs was investigated. PBS pH 7.4, 6.8 and 5.0 was used to resemble pH of blood, the tumor microenvironment and the endolysosomal compartment respectively.22 In literature, cargo release from PLGA NPs was shown to increase with the decrease in pH; whereas PLGA polymer degrades faster at lower pH.22,58 However, in our study, this was not the case in all the formulations, indicating that cargo release is multifactorial and is more complicated than to be controlled only by the pH of the release medium. The amount of the adsorbed proteins on NPs’ surfaces,59 the concentration of serum proteins in the release medium,4,50 the composition of the NPs60 and the peptide conjugation status61 contribute to a greater extent to the cargo release patterns.
One interesting parameter is the formation of PC on NPs’ surfaces whose effect on cargo release has been proven to be double-faceted. On one hand, proteins can add an extra shield to the cargo release and hence slow down the release rate and attenuate the burst release.50,59 On the other hand, proteins can disrupt NPs’ integrity and cause their disassembly, promoting the leaching of the loaded cargo.4,51 In addition, proteins in the PC can have high affinity to the loaded cargo molecules facilitating their escape from the NPs’ core.51 The later phenomenon explains the obtained results whereby cNPs which exhibited higher quantities of the adsorbed proteins in their PC, have shown faster release rates than their peptide unconjugated counterparts. In addition, the presence of a PC layer could disrupt the lipid coat in the hybrid NPs, NP3 and NP4, hindering its ability to sustain the release of the loaded cargo as previously reported.4,30,51,62
In the in vitro uptake results, the higher extent of NPs’ accumulation upon peptide conjugation, especially for the hybrid formulations (cNP3 and cNP4) could be attributed to both the role of cRDGyk in anchoring the NPs to the integrin receptors bringing about higher accumulation tendencies24 and the advantage of the lipid coat in increasing the cellular levels of the NPs.62 This observation could also be related to the zeta potential of cNPs; it was previously discussed in this work (Table 2) that peptide conjugation reduced the overall surface charge of the NPs. This could possibly reduce the repulsive forces between the negative surface charge of the NPs and the negative charge of the cell membrane.63–65 In addition, the mechanism of the intracellular uptake has proven to be a very important determinant of the accumulation of the NPs and is dependent on the NPs’ properties.66,67 Peptide conjugation was observed to change the mechanism of endocytic uptake of NPs; whereas cNPs were found to exhibit more dependence on the caveolin-dependent rather than clathrin-dependent uptake pathway (Figure S3). These findings come in harmony with previously reported investigations.44
Regarding the role of PC on NPs’ uptake, it was reported that the adsorption of proteins on NPs’ surfaces could attenuate the extent of intracellular accumulation of the NPs, be unharmful or rather beneficial.68 Formation of a PC on NPs’ surfaces could decrease NPs’ uptake by decreasing their anchorage to the cell membrane by inducing a decrease in the surface free energy.13,15 In addition, excess serum proteins in the culture medium could saturate the membrane receptors involved in NPs’ uptake.4 Alternatively, a high extent of accumulation of cNPs in B16F10 melanoma cells in the presence of serum proteins was also observed and was consistent with previous reports.7,15 Cyclic RGD peptide was reported to maintain its integrins binding affinity after protein adsorption resulting in increased cellular accumulation of NPs, even at low conjugation density. This observation was more prominent in large-sized NPs which possessed multiple binding sites.8 In addition, some proteins in PC of NPs could facilitate receptor-mediated endocytosis. For example, VN which showed highest relative abundance in the PC of cNP4 could have contributed to its increased accumulation in the cells.6,15,52,56 Similarly, CHC, which was exclusively abundant in cNP3 and cNP4, could have contributed to the higher intracellular accumulation of these formulations as well.13
From the view of the relative intracellular uptake results, a positive relationship between the selected cellular and serum proteins and the intracellular uptake of NPs was observed in both experimental settings with some exceptions. This could be explained on the basis of the functions of the selected proteins as follows. VN, as a ligand for the integrin receptor overly expressed on melanoma cells, can provide new binding sites for the NPs to the cells bringing about higher internalization of NPs.6,52,56 C3 protein can also bind to the anaphylatoxin or the complement peptide receptors recently reported to be overexpressed on a number of cancer cells including B16F10 melanoma cells.25,26 Similarly, melanoma is characterized by overexpression of CLU isoforms which correlate with malignancy and progression.27,28 In addition, ALB is reported to possess a role in increasing NPs’ accumulation by facilitating receptor-mediated endocytosis5 and reducing their degradation in the endolysosomal compartment.6 Moreover, the selected cellular proteins are known to be involved in endocytosis, receptor recycling and NPs’ interaction with the cell membrane and the cytoplasmic components.13,29
On the contrary, the negative relationship observed in the case of VN when the antibody was premixed with the NPs in the absence of MS could raise the concern that the adsorbed proteins may need assistance from other proteins, which are absent in SF conditions, to produce their intended effect on the cells as reported in earlier studies.15
Regarding the negative relationship observed in the case of uNPs when the cells were pretreated with the antibodies, further investigation on the orientation of the selected proteins on NPs’ surfaces and the subsequent role on the interaction with the cells is needed. Some proteins, upon adsorption on some NPs’ surfaces, change their structure and subsequently their function. They could eventually be recognized by different receptors.12 In addition, the interaction between different proteins in the same PC need to be thoroughly investigated. The shielding effect of some proteins present in the PC exerted over other ones has also been suggested. This effect could make some proteins in PC unrecognizable by their corresponding receptors.4,54
Finally, the resemblance between the behavior of cNP4 and the uNPs with respect to both the serum and cellular proteins, upon pretreating the cells with the antibodies, could probably be driven by the loss of the peptide-functionalized lipid coat upon adsorption of PC that has been reported to cause disassembly of some NPs.46,51
Conclusion
NPs tend to form PCs of a different nature according to the exposure conditions between the proteins and the NPs; and coronas of a different nature guide the NPs to different fates. Curious investigation of the role of distinct proteins in directing the NPs towards achieving better therapeutic outcomes has evolved the concept of endogenous targeting. Careful engineering of nanocarriers can modulate the recruitment of some proteins suggesting a potential use for overcoming the current limitations of targeted drug delivery. In this part of the study, NPs’ interaction with the cells has only been tested in vitro under specific conditions. In this setting, VN serum protein has shown to be a potential serum protein, that when sufficiently abundant in PC of NPs, could enhance NPs’ accumulation inside the targeted melanoma cells. However, this experimental setting which studies NPs’ interaction with melanoma cells at defined time point (3 h incubation) does not take into account that both NPs internalization and elimination are dynamic processes, i.e. the internalized NPs could exit the cell via different mechanisms and can be re-internalized back. In addition, NPs’ behavior in culture conditions does not necessarily match their behavior in vivo. The absence of an adequate correlation between the in vitro cellular uptake of the NPs and the in vivo tumor accumulation could be explained based on the major hurdles that face the NPs upon in vivo administration, e.g. the uptake by the RES organs or the failure of deep penetration into the target tissues in the presence of densely packed structures of the tumor tissue. Therefore, in Part II of this study, thorough investigation of the kinetics of NPs’ uptake and elimination and the active and passive targeting capabilities of the NPs in melanoma-bearing mouse model is carried out to propose solid bases for future PC modulation to optimize the endogenous targeting of the NPs.
Disclosure
The authors report no conflicts of interest for this work.
References
- 1.Pérez-Herrero E, Fernández-Medarde A. Advanced targeted therapies in cancer: drug nanocarriers, the future of chemotherapy. Eur J Pharm Biopharm. 2015;93(March):52–79. doi: 10.1016/j.ejpb.2015.03.018 [DOI] [PubMed] [Google Scholar]
- 2.Wakaska RR. Passive and active targeting in tumor microenvironment. Int J Drug Dev Res. 2017;9(June):37–41. [Google Scholar]
- 3.Yu Q, Zhao L, Guo C, Yan B, Su G. Regulating protein corona formation and dynamic protein exchange by controlling nanoparticle hydrophobicity. Front Bioeng Biotechnol. 2020;8:210. doi: 10.3389/fbioe.2020.00210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Francia V, Yang K, Deville S, Reker-Smit C, Nelissen I, Salvati A. Corona composition can affect the mechanisms cells use to internalize nanoparticles. ACS Nano. 2019;13(10):11107–11121. doi: 10.1021/acsnano.9b03824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gossmann R, Fahrländer E, Hummel M, Mulac D, Brockmeyer J, Langer K. Comparative examination of adsorption of serum proteins on HSA- and PLGA-based nanoparticles using SDS–PAGE and LC–MS. Eur J Pharm Biopharm. 2015;93:80–87. doi: 10.1016/J.EJPB.2015.03.021 [DOI] [PubMed] [Google Scholar]
- 6.Xiao W, Gao H. The impact of protein corona on the behavior and targeting capability of nanoparticle-based delivery system. Int J Pharm. 2018;552(1–2):328–339. doi: 10.1016/j.ijpharm.2018.10.011 [DOI] [PubMed] [Google Scholar]
- 7.Palchetti S, Pozzi D, Capriotti AL, et al. Influence of dynamic flow environment on nanoparticle-protein corona: from protein patterns to uptake in cancer cells. Colloids Surf B Biointerfaces. 2017;153:263–271. doi: 10.1016/j.colsurfb.2017.02.037 [DOI] [PubMed] [Google Scholar]
- 8.Su G, Jiang H, Xu B, Yu Y, Chen X. Effects of protein corona on active and passive targeting of cyclic RGD peptide-functionalized PEGylation nanoparticles. Mol Pharm. 2018;15(11):5019–5030. doi: 10.1021/acs.molpharmaceut.8b00612 [DOI] [PubMed] [Google Scholar]
- 9.Weber C, Simon J, Mailänder V, Morsbach S, Landfester K. Preservation of the soft protein corona in distinct flow allows identification of weakly bound proteins. Acta Biomater. 2018;76:217–224. doi: 10.1016/j.actbio.2018.05.057 [DOI] [PubMed] [Google Scholar]
- 10.Liu N, Tang M, Ding J. The interaction between nanoparticles-protein corona complex and cells and its toxic effect on cells. Chemosphere. 2020. doi: 10.1016/j.chemosphere.2019.125624 [DOI] [PubMed] [Google Scholar]
- 11.Hu Z, Zhang H, Zhang Y, Wu R, Zou H. Nanoparticle size matters in the formation of plasma protein coronas on Fe3O4 nanoparticles. Colloids Surf B Biointerfaces. 2014;121:354–361. doi: 10.1016/j.colsurfb.2014.06.016 [DOI] [PubMed] [Google Scholar]
- 12.Simon J, Müller LK, Kokkinopoulou M, et al. Exploiting the biomolecular corona: pre-coating of nanoparticles enables controlled cellular interactions. Nanoscale. 2018;10(22):10731–10739. doi: 10.1039/C8NR03331E [DOI] [PubMed] [Google Scholar]
- 13.Lesniak A, Fenaroli F, Monopoli MP, Åberg C, Dawson KA, Salvati A. Effects of the presence or absence of a protein corona on silica nanoparticle uptake and impact on cells. ACS Nano. 2012;6(7):5845–5857. doi: 10.1021/nn300223w [DOI] [PubMed] [Google Scholar]
- 14.Cagliani R, Gatto F, Bardi G. Protein adsorption: a feasible method for nanoparticle functionalization? Materials. 2019;12(12):1991. doi: 10.3390/ma12121991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen D, Ganesh S, Wang W, Amiji M. The role of surface chemistry in serum protein corona-mediated cellular delivery and gene silencing with lipid nanoparticles. Nanoscale. 2019;11(18):8760–8775. doi: 10.1039/c8nr09855g [DOI] [PubMed] [Google Scholar]
- 16.Tonigold M, Simon J, Estupiñán D, et al. Pre-adsorption of antibodies enables targeting of nanocarriers despite a biomolecular corona. Nat Nanotechnol. 2018;13:9. doi: 10.1038/s41565-018-0171-6 [DOI] [PubMed] [Google Scholar]
- 17.Strobel C, Oehring H, Herrmann R, Förster M, Reller A, Hilger I. Fate of cerium dioxide nanoparticles in endothelial cells: exocytosis. J Nanopart Res. 2015;17:206. doi: 10.1007/s11051-015-3007-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rafiei P, Haddadi A. Docetaxel-loaded PLGA and PLGA-PEG nanoparticles for intravenous application: pharmacokinetics and biodistribution profile. Int J Nanomedicine. 2017;12:935–947. doi: 10.2147/IJN.S121881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yameen B, Choi WI, Vilos C, Swami A, Shi J, Farokhzad OC. Insight into nanoparticle cellular uptake and intracellular targeting. J Control Release. 2014;190:485–499. doi: 10.1016/j.jconrel.2014.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye Y, Chen X. Integrin targeting for tumor optical imaging. Theranostics. 2011;1:102–126. doi: 10.7150/thno/v01p0102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sims LB, Curtis LT, Frieboes HB, Steinbach-Rankins JM. Enhanced uptake and transport of PLGA-modified nanoparticles in cervical cancer. J Nanobiotechnology. 2016;14:33. doi: 10.1186/s12951-016-0185-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu L, Hu Q, Cheng L, et al. CRGDyK modified PH responsive nanoparticles for specific intracellular delivery of doxorubicin. Acta Biomater. 2016;30:285–298. doi: 10.1016/j.actbio.2015.11.037 [DOI] [PubMed] [Google Scholar]
- 23.Danhier F, Breton AL, Préat V. RGD-based strategies to target alpha(v) beta(3) integrin in cancer therapy and diagnosis. Mol Pharm. 2012;9(11):2961–2973. doi: 10.1021/mp3002733 [DOI] [PubMed] [Google Scholar]
- 24.Alipour M, Baneshi M, Hosseinkhani S, et al. Recent progress in biomedical applications of RGD-based ligand: from precise cancer theranostics to biomaterial engineering: a systematic review. J Biomed Mater Res A. 2020;108(4):839–850. doi: 10.1002/jbm.a.36862 [DOI] [PubMed] [Google Scholar]
- 25.Roumenina LT, Daugan MV, Petitprez F, Sautès-Fridman C, Fridman WH. Context-dependent roles of complement in cancer. Nat Rev Cancer. 2019;698–715. doi: 10.1038/s41568-019-0210-0 [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Zhang H, He Y-W. The complement receptors C3aR and C5aR are a new class of immune checkpoint receptor in cancer immunotherapy. Front Immunol. 2019;101574. doi: 10.3389/fimmu.2019.01574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shannan B, Seifert M, Leskov K, et al. Clusterin (CLU) and melanoma growth: CLU is expressed in malignant melanoma and 1,25-dihydroxyvitamin D3 modulates expression of CLU in melanoma cell lines in vitro. Anticancer Res. 2006;26(4 A):2707–2716. [PubMed] [Google Scholar]
- 28.Ming X, Bao C, Hong T, et al. Clusterin, a novel DEC1 target, modulates DNA damage-mediated cell death. Mol Cancer Res. 2018;16(11):1641–1651. doi: 10.1158/1541-7786.MCR-18-0070 [DOI] [PubMed] [Google Scholar]
- 29.Slowing II, Vivero-Escoto JL, Zhao Y, et al. Exocytosis of mesoporous silica nanoparticles from mammalian cells: from asymmetric cell-to-cell transfer to protein harvesting. Small. 2011;7(11):1526–1532. doi: 10.1002/smll.201002077 [DOI] [PubMed] [Google Scholar]
- 30.Sebak AA, Gomaa II, ElMeshad AN, AbdelKader MH. Targeted photodynamic-induced singlet oxygen production by peptide-conjugated biodegradable nanoparticles for treatment of skin melanoma. Photodiagnosis Photodyn Ther. 2018;23:181–189. doi: 10.1016/j.pdpdt.2018.05.017 [DOI] [PubMed] [Google Scholar]
- 31.Gao N, Xing C, Wang H, et al. PH-responsive dual drug-loaded nanocarriers based on poly (2-Ethyl-2-Oxazoline) modified black phosphorus nanosheets for cancer chemo/photothermal therapy. Front Pharmacol. 2019;10(MAR). doi: 10.3389/fphar.2019.00270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bardania H, Shojaosadati SA, Kobarfard F, et al. Encapsulation of eptifibatide in RGD-modified nanoliposomes improves platelet aggregation inhibitory activity. J Thromb Thrombolysis. 2017;43(2):184–193. doi: 10.1007/s11239-016-1440-6 [DOI] [PubMed] [Google Scholar]
- 33.Partikel K, Korte R, Stein NC, et al. Effect of nanoparticle size and PEGylation on the protein corona of PLGA nanoparticles. Eur J Pharm Biopharm. 2019;141:70–80. doi: 10.1016/j.ejpb.2019.05.006 [DOI] [PubMed] [Google Scholar]
- 34.Partikel K, Korte R, Mulac D, Humpf H-U, Langer K. Serum type and concentration both affect the protein-corona composition of PLGA nanoparticles. Beilstein J Nanotechnol. 2019;10:1002–1015. doi: 10.3762/bjnano.10.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panariti A, Miserocchi G, Rivolta I. The effect of nanoparticle uptake on cellular behavior: disrupting or enabling functions? Nanotechnol Sci Appl. 2012;5(1):87–100. doi: 10.2147/NSA.S25515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Breitinger U, Farag NS, Ali NKM, Breitinger HGA. Patch-clamp study of hepatitis C P7 channels reveals genotype-specific sensitivity to inhibitors. Biophys J. 2016;110(11):2419–2429. doi: 10.1016/j.bpj.2016.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gomaa I, Sebak A, Afifi N, Abdel-Kader M. Liposomal delivery of ferrous chlorophyllin: a novel third generation photosensitizer for in vitro PDT of melanoma. Photodiagnosis Photodyn Ther. 2017;18:162–170. doi: 10.1016/j.pdpdt.2017.01.186 [DOI] [PubMed] [Google Scholar]
- 38.Schöttler S, Klein K, Landfester K, Mailänder V. Protein source and choice of anticoagulant decisively affect nanoparticle protein corona and cellular uptake. Nanoscale. 2016;8(10):5526–5536. doi: 10.1039/c5nr08196c [DOI] [PubMed] [Google Scholar]
- 39.Farid MM, Hathout RM, Fawzy M, Abou-Aisha K. Silencing of the scavenger receptor (class b - type 1) gene using SiRNA-loaded chitosan nanoparticles in a HepG2 cell model. Colloids Surf B Biointerfaces. 2014;123:930–937. doi: 10.1016/j.colsurfb.2014.10.045 [DOI] [PubMed] [Google Scholar]
- 40.Díaz-Moscoso A, Vercauteren D, Rejman J, et al. Insights in cellular uptake mechanisms of PDNA-polycationic amphiphilic Cyclodextrin Nanoparticles (CDplexes). J Control Release. 2010;143(3):318–325. doi: 10.1016/j.jconrel.2010.01.016 [DOI] [PubMed] [Google Scholar]
- 41.Ding L, Yao C, Yin X, et al. Size, shape, and protein corona determine cellular uptake and removal mechanisms of gold nanoparticles. Small. 2018;14(42):1801451. doi: 10.1002/smll.201801451 [DOI] [PubMed] [Google Scholar]
- 42.Li Y, Monteiro-Riviere NA. Mechanisms of cell uptake, inflammatory potential and protein corona effects with gold nanoparticles. Nanomedicine. 2016;11(24):3185–3203. doi: 10.2217/nnm-2016-0303 [DOI] [PubMed] [Google Scholar]
- 43.Prozeller D, Pereira J, Simon J, Mailänder V, Morsbach S, Landfester K. Prevention of dominant IgG adsorption on nanocarriers in IgG-enriched blood plasma by clusterin precoating. Adv Sci. 2019;6(10):1802199. doi: 10.1002/advs.201802199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao H, Yang Z, Zhang S, et al. Ligand modified nanoparticles increases cell uptake, alters endocytosis and elevates glioma distribution and internalization. Sci Rep. 2013;3(1):2534. doi: 10.1038/srep02534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graf N, Bielenberg DR, Kolishetti N, et al. Αvβ 3 integrin-targeted PLGA-PEG nanoparticles for enhanced anti-tumor efficacy of a Pt(IV) prodrug. ACS Nano. 2012;6(5):4530–4539. doi: 10.1021/nn301148e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jack HCM, Kaushal S, Tran Cao HS, et al. Half-Antibody functionalized lipid-polymer hybrid nanoparticles for targeted drug delivery to Carcinoembryonic Antigen (CEA) presenting pancreatic cancer cells. Mol Pharm. 2010;7(3):914–920. doi: 10.1021/mp900316a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Imanparast F, Paknejad M, Faramarzi MA, Kobarfard F, Amani A, Doosti M. Potential of MZD7349-conjugated PLGA nanoparticles for selective targeting of vascular cell-adhesion molecule-1 in inflamed endothelium. Microvasc Res. 2016;106:110–116. doi: 10.1016/j.mvr.2016.04.003 [DOI] [PubMed] [Google Scholar]
- 48.Gossmann R, Langer K, Mulac D, Antopolsky M. New perspective in the formulation and characterization of Didodecyldimethylammonium Bromide (DMAB) stabilized Poly(Lactic-Co-Glycolic Acid) (PLGA) nanoparticles. PLoS One. 2015;10(7):e0127532. doi: 10.1371/journal.pone.0127532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van De Ven H, Vandervoort J, Weyenberg W, Apers S, Ludwig A. Mixture designs in the optimisation of PLGA nanoparticles: influence of organic phase composition on β-aescin encapsulation. J Microencapsul. 2012;29(2):115–125. doi: 10.3109/02652048.2011.630108 [DOI] [PubMed] [Google Scholar]
- 50.Al-Ahmady ZS, Hadjidemetriou M, Gubbins J, Kostarelos K. Formation of protein corona in vivo affects drug release from temperature-sensitive liposomes. J Control Release. 2018;276:157–167. doi: 10.1016/j.jconrel.2018.02.038 [DOI] [PubMed] [Google Scholar]
- 51.Abstiens K, Maslanka Figueroa S, Gregoritza M, Goepferich AM. Interaction of functionalized nanoparticles with serum proteins and its impact on colloidal stability and cargo leaching. Soft Matter. 2019;15(4):709–720. doi: 10.1039/C8SM02189A [DOI] [PubMed] [Google Scholar]
- 52.Ritz S, Schöttler S, Kotman N, et al. Protein corona of nanoparticles: distinct proteins regulate the cellular uptake. Biomacromolecules. 2015;16(4):1311–1321. doi: 10.1021/acs.biomac.5b00108 [DOI] [PubMed] [Google Scholar]
- 53.Pozzi D, Caracciolo G, Capriotti AL, et al. Surface chemistry and serum type both determine the nanoparticle-protein corona. J Proteomics. 2015;119. doi: 10.1016/j.jprot.2015.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mirshafiee V, Kim R, Park S, Mahmoudi M, Kraft ML. Impact of protein pre-coating on the protein corona composition and nanoparticle cellular uptake. Biomaterials. 2016;75:295–304. doi: 10.1016/j.biomaterials.2015.10.019 [DOI] [PubMed] [Google Scholar]
- 55.Gunawan C, Lim M, Marquis CP, Amal R. Nanoparticle–Protein Corona Complexes govern the biological fates and functions of nanoparticles. J Mater Chem B. 2014;2(15):2060. doi: 10.1039/c3tb21526a [DOI] [PubMed] [Google Scholar]
- 56.Chen D, Ganesh S, Wang W, Amiji M. Plasma protein adsorption and biological identity of systemically administered nanoparticles. Nanomedicine. 2017;12(17):2113–2135. doi: 10.2217/nnm-2017-0178 [DOI] [PubMed] [Google Scholar]
- 57.Xiao W, Xiong J, Zhang S, Xiong Y, Zhang H, Gao H. Influence of ligands property and particle size of gold nanoparticles on the protein adsorption and corresponding targeting ability. Int J Pharm. 2018;538(1–2):105–111. doi: 10.1016/j.ijpharm.2018.01.011 [DOI] [PubMed] [Google Scholar]
- 58.Chai F, Sun L, He X, et al. Doxorubicin-loaded poly (lactic-co-glycolic acid) nanoparticles coated with chitosan/alginate by layer by layer technology for antitumor applications. Int J Nanomedicine. 2017;12:1791–1802. doi: 10.2147/IJN.S130404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Behzadi S, Serpooshan V, Sakhtianchi R, et al. Protein corona change the drug release profile of nanocarriers: the “overlooked” factor at the nanobio interface. Colloids Surf B Biointerfaces. 2014;123:143–149. doi: 10.1016/j.colsurfb.2014.09.009 [DOI] [PubMed] [Google Scholar]
- 60.Godara S, Lather V, Kirthanashri SV, Awasthi R, Pandita D. Lipid-PLGA hybrid nanoparticles of paclitaxel: preparation, characterization, in vitro and in vivo evaluation. Mater Sci Eng C. 2020;109:110576. doi: 10.1016/j.msec.2019.110576 [DOI] [PubMed] [Google Scholar]
- 61.Wang G, Wang Z, Li C, et al. RGD peptide-modified, paclitaxel prodrug-based, dual-drugs loaded, and redox-sensitive lipid-polymer nanoparticles for the enhanced lung cancer therapy. Biomed Pharmacother. 2018;106:275–284. doi: 10.1016/j.biopha.2018.06.137 [DOI] [PubMed] [Google Scholar]
- 62.Sebak AA. Limitations of PEGylated nanocarriers: unfavourable physicochemical properties, biodistribution patterns and cellular and subcellular fates. Int J Appl Pharm. 2018;10(5):6–12. doi: 10.22159/ijap.2018v10i5.27568 [DOI] [Google Scholar]
- 63.Hui Voon S, Tiew SX, Kue CS, et al. Chitosan-coated poly(lactic-co-glycolic acid)- diiodinated boron-dipyrromethene nanoparticles improve tumor selectivity and stealth properties in photodynamic cancer therapy. J Biomed Nanotechnol. 2016;12:1431–1452. doi: 10.1166/jbn.2016.2263 [DOI] [PubMed] [Google Scholar]
- 64.Oh N, Park J-H-H. Endocytosis and exocytosis of nanoparticles in mammalian cells. Int J Nanomedicine. 2014;9(SUPPL.1):51–63. doi: 10.2147/IJN.S26592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Albanese A, Tang PS, Chan WCW. The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annu Rev Biomed Eng. 2012;14(1):1–16. doi: 10.1146/annurev-bioeng-071811-150124 [DOI] [PubMed] [Google Scholar]
- 66.Wu M, Guo H, Liu L, Liu Y, Xie L. Size-dependent cellular uptake and localization profiles of silver nanoparticles. Int J Nanomedicine. 2019;14:4247–4259. doi: 10.2147/IJN.S201107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ekkapongpisit M, Giovia A, Follo C, Caputo G, Isidoro C. Biocompatibility, endocytosis, and intracellular trafficking of mesoporous silica and polystyrene nanoparticles in ovarian cancer cells: effects of size and surface charge groups. Int J Nanomedicine. 2012;7:4147–4158. doi: 10.2147/IJN.S33803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cui Y, Song X, Li S, et al. The impact of receptor recycling on the exocytosis of αvβ3 integrin targeted gold nanoparticles. Oncotarget. 2017;8(24):38618–38630. doi: 10.18632/oncotarget.16955 [DOI] [PMC free article] [PubMed] [Google Scholar]