Abstract
The lung is inhabited by resident alveolar and interstitial macrophages as well as monocytic cells that survey lung tissues. Each cell type plays distinct functional roles under homeostatic and inflammatory conditions, but mechanisms establishing their molecular identities and functional potential remain poorly understood. Here, systematic evaluation of transcriptomes and open chromatin of alveolar macrophages (AMs), interstitial macrophages (IMs) and lung monocytes from two mouse strains enabled inference of common and cell-specific transcriptional regulators. We provide evidence that these factors drive selection of regulatory landscapes that specify distinct phenotypes of AMs and IMs and entrain qualitatively different responses to Toll-like receptor 4 (TLR4) signaling in vivo. These studies reveal a striking divergence in a fundamental innate immune response pathway in AMs and establish a framework for further understanding macrophage diversity in the lung.
Introduction
Innate immune cells, including macrophages, monocytes and dendritic cells, collectively called mononuclear phagocytes (MPs), provide the first line of cellular defense against pathogens. In addition, macrophages play essential roles in diverse developmental and homeostatic processes1. Within the lung, MPs are situated at and between the epithelial and endothelial surfaces, and their bulk gene expression profiles change during development2. Alveolar macrophages (AMs) are derived from embryonic erythromyeloid progenitor cells3, 4 and from fetal liver monocytes5. AMs play key roles in surfactant turnover and in maintaining an anti-inflammatory state that benefits efficient gas exchange6, 7, 8. However, the lung is populated by several subsets of MPs in addition to AMs9, 10. Interstitial macrophages (IMs) proposed to be derived from yolk sac macrophages and later to be replaced by circulating monocytes11 are situated in the lung interstitium around airways, blood vessels and nerves12, 13. In addition, the extensive capillary bed of the lung harbors a marginated pool of monocytes that is retained even after extensive perfusion of the lung14. When the lung is injured or infected, additional monocytes are recruited and can amplify the inflammatory response15, 16, 17.
The tissue environment plays a crucial role in determining the expression and function of transcription factors (TFs) that specify tissue-resident macrophage phenotypes18. Following entry into the developing fetal lung, embryonic macrophages destined to become AMs upregulate Pparg, Klf4, Atf5 and Cebpb19. Consistent with the upregulation of these TFs at the mRNA level, their corresponding DNA recognition motifs are highly enriched in AM enhancers20 and loss of Pparg expression results in defective AM differentiation21, 22. Conversely, rapid changes in enhancer activity states and gene expression are observed following the transfer of resident macrophages to a tissue culture environment23, 24. These findings imply that tissue-derived signals are constantly sensed and integrated at the level of enhancers, which function to maintain tissue-specific programs of macrophage gene expression.
Cell-specific enhancers also function as important sites of action of broadly expressed signal-dependent TFs, exemplified by NF-κB and nuclear hormone receptors25. In principle, the distinct enhancer landscapes of each cell type enable cell-specific transcriptional responses to a common signal, such as lipopolysaccharide (LPS). These context-dependent responses to common signals are likely to be essential to the specific functions of distinct macrophage and monocyte subsets. Here, we use genetic and genomic approaches to investigate mechanisms that regulate gene expression in distinct tissue-resident macrophages in the lung and demonstrate divergent functional responses of these cell subsets to the Gram-negative bacterial endotoxin LPS.
Results
Cell-specific transcriptomes of lung macrophages and monocytes
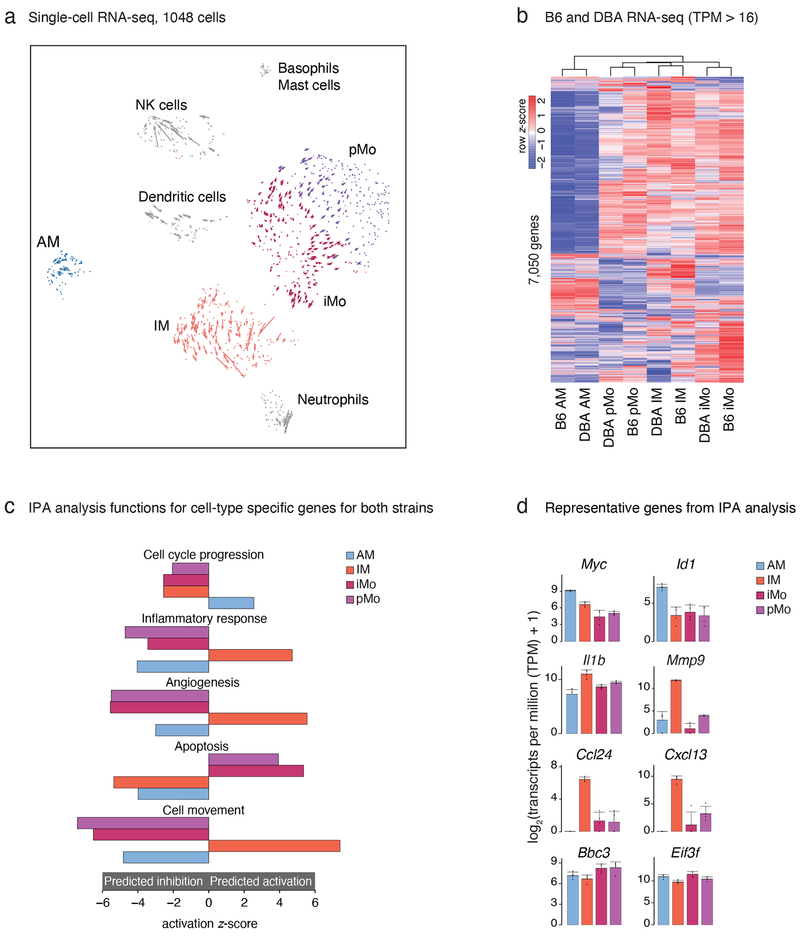
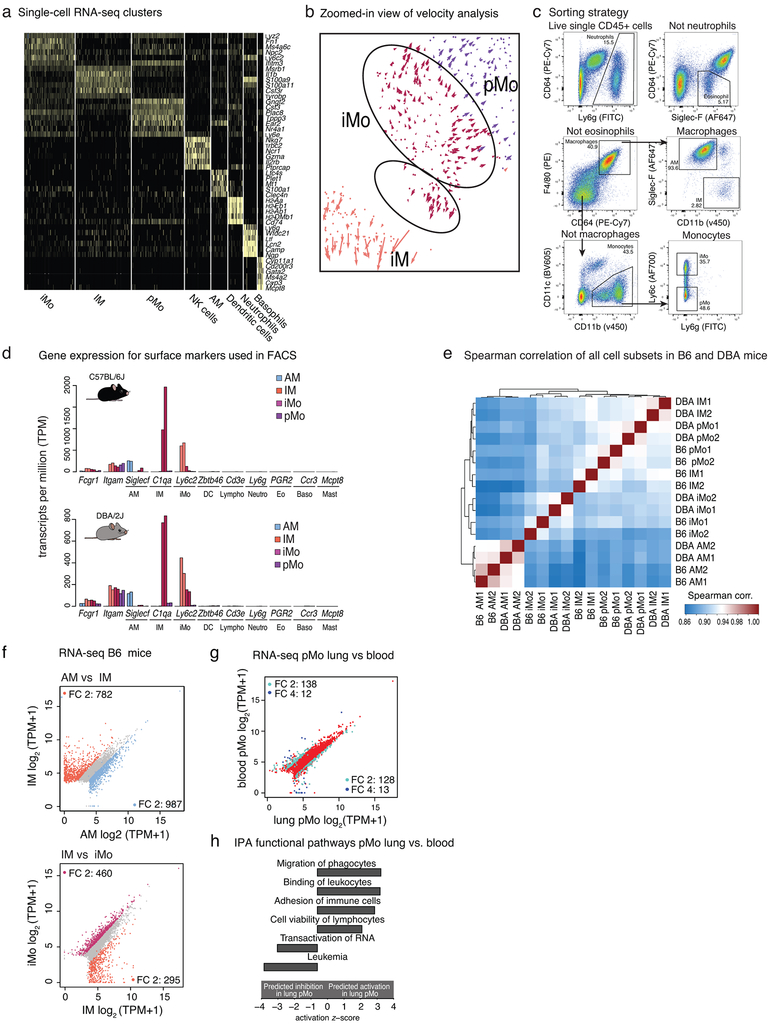
To measure cellular heterogeneity of the lung myeloid compartment with an unbiased approach, we performed single-cell RNA-seq (scRNA-seq). Following extensive perfusion of the lung vasculature to remove circulating blood cells, we isolated resident lung myeloid cells (CD45+ cells depleted of T and B lymphocytes) by fluorescence-activated cell sorting (FACS) from 4 mice and generated single-cell transcriptional profiles for 1,048 cells that passed rigorous quality control. Dimensionality reduction analysis (t-SNE) separated cells into 7 clusters based on their gene expression pattern (Fig.1a and Extended Data Fig.1a). These clusters included AMs with high Ltc4, Plet1, Mt1 expression and IMs with high Il1b and S100a expression. In addition, two populations were related to the major subsets of circulating blood monocytes26, 27; inflammatory monocytes (iMos) with high Ly6c2 expression, and patrolling monocytes (pMos) with Nr4a1 expression.
Figure 1: Transcriptomes of lung mononuclear phagocytes (MPs) at baseline.
a. Single cell RNA-seq, plot shows dimensionality reduction analysis (t-SNE) incorporated with RNA velocity analysis. Cells from the lungs of 3 male and 1 female DBA/2J mice were pooled.
b. Gene expression of lung MP in C57BL/6J (B6) and DBA/2J (DBA) mice. Heat map shows unsupervised hierarchical clustering of genes expressed more than 16 transcripts per million (TPM) in at least one cell type in either B6 or DBA mice. Values are row z-scores.
c. Ingenuity pathway analysis (IPA) of genes differentially expressed between lung MP of B6 and DBA mice. Differentially expressed genes calculated using DESeq2 (FC > 2, FDR < 0.01) were merged and submitted to IPA analysis. A selection of highly significant functions and their activation z-scores are shown in the bar graphs (blue for AM, orange for IM, bordeaux for iMo, purple for pMo). The same color scheme is used throughout the article.
d. Examples of gene expression from the IPA functions depicted in Fig. 1c. Bar graphs represent mean log2 transcripts per million (TPM+1) ± SD. Differential gene expression was calculated using DESeq2.
To investigate developmental relationships between lung monocytes and macrophages we performed RNA velocity analysis, which distinguishes between spliced and un-spliced mRNA to predict the ‘future’ state of individual cells28. The directionality vectors associated with each cell suggest that a substantial proportion of iMos are predicted to become more similar to pMos with time (Fig. 1a, Extended Data Fig. 1b). In addition, a smaller sub-cluster of iMos is predicted to differentiate into IMs.
Based on the population structure defined by scRNA-seq, we isolated four subsets (AM, IM, iMo, pMo) using FACS (gating strategy shown in Extended Data Fig. 1c)29. The sorted cells were processed for bulk RNA sequencing (RNA-seq) and for chromatin accessibility by Assay for Transposase-Accessible Chromatin (ATAC-seq)30. Contamination by other cell types was negligible for all MP subsets and Spearman correlation values were high for RNA-seq replicates in any given MP subset (Extended Data Fig. 1d, e). To investigate the impact of genetic variation on lung MP gene signatures during homeostasis we performed these experiments using two strains of mice C57BL/6J (B6) and DBA/2J (DBA) . These strains of mice exhibit 4.4 million single nucleotide polymorphisms (SNPs) and 540,000 insertions and deletions (InDels)31. Phenotypically, they differ in their susceptibility to a spectrum of inflammatory lung diseases32, 33.
Gene expression profiles of lung MP subsets were markedly different at baseline (Fig. 1b). Clustering of RNA-seq data segregated samples by cell type, with strain as a secondary determinant. Comparing lung MP subsets, AMs were most different from all other cell types in both strains. For example, in B6 mice we found 1,769 differentially expressed genes between AMs and IMs (FC > 2, FDR < 0.05) (Extended Data Fig. 1f). Diversity between transcriptional programs for the remaining three cell subsets was less pronounced but still substantial with 1,076 genes differentially expressed between IMs and iMos and 584 genes between iMos and pMos (FC > 2, FDR < 0.05) in B6 mice (Extended Data Fig. 1f). Notably, while lung iMos were very similar to circulating iMos, pMos retained in the lung following extensive perfusion exhibited 266 genes that were differentially expressed (FC > 2, FDR < 0.05) compared to pMos in circulating blood (Extended Data Fig. 1g). Ingenuity pathway analysis (IPA) showed that genes involved in migration and adhesion were more highly expressed in pMos isolated from the lung (Extended Data Fig. 1h).
To infer potential biological meaning of the differentially expressed genes between lung MP subsets, we merged the differentially expressed genes in both C57 or DBA datasets. Using the obtained gene list, we performed IPA (Fig. 1d). AMs were enriched for genes associated with cell cycle progression, supporting previous studies indicating that these cells are self-renewing. For example, Myc, a transcription factor known to play a central role in stimulating cell cycle progression, and Id1 known to play a role in cell growth were expressed highest in AMs (Fig. 1d). Under baseline conditions, IMs were the only lung MP subset enriched in genes positively associated with inflammatory response, exemplified by high expression of Il1b (Fig. 1d). In addition, IMs were enriched in genes related to cell movement (Cxcl13, Ccl24) and angiogenesis (Mmp9) (Fig. 1d). Apoptosis related genes, including Bbc3 and Eif3f, were most enriched in iMos and pMos. Collectively, these cell-specific transcriptomes define the distinct molecular phenotypes of the corresponding lung MPs.
Strain-specific transcriptomes of lung MPs
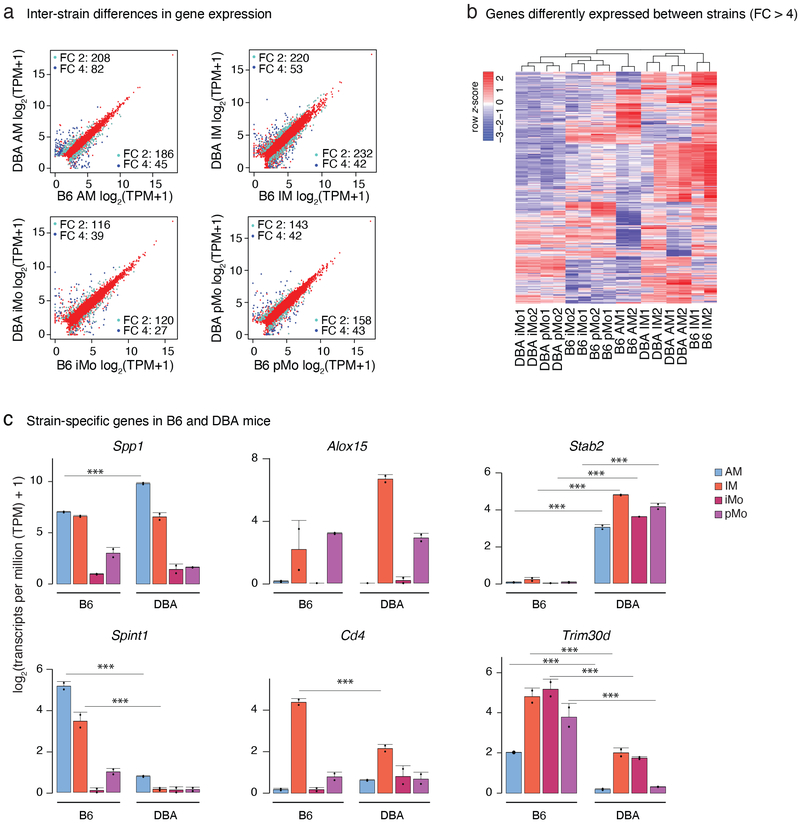
Analysis of gene expression between B6 and DBA mice showed hundreds of mRNA transcripts expressed with at least a two-fold difference between the strains in each lung MP subset (Fig. 2a). Of note, most of the inter-strain differences in gene expression were cell-subset-specific (Fig. 2b, c). For example, Spp1 , the gene encoding osteopontin, a molecule known to recruit cells to inflammatory sites and promote cell survival by regulating apoptosis34 was several fold enriched in AMs from DBA mice (Fig. 2c). Increased expression of Spp1 in response to cigarette smoke is linked to emphysema35. Thus, higher levels in AMs may contribute to the greater sensitivity of DBA mice to cigarette-smoke induced emphysema than B6 mice33. Contrarily, Spint1 a gene encoding a serine protease inhibitor, was much higher expressed in AMs from B6 mice. An example of a gene selectively expressed in DBA IMs is Alox15, encoding for a lipoxygenase implicated in anti-inflammation36. Strain- and cell type-specificity could also be observed for Cd4, which was only expressed in B6 IMs. Ligation of CD4 on monocytes/macrophages modulates gene and cytokine protein expression as well as T cell responses37. Stab2 a transmembrane receptor involved in bacterial scavenging, cell adhesion and angiogenesis38 was only expressed in lung MPs from DBA mice. Conversely, Trim30d, was expressed by lung MPs isolated from B6 mice but absent in DBA mice. These results thereby identify genes that may play important roles in determining the phenotypic diversity of B6 and DBA mice.
Figure 2: Strain-specific transcriptomes of lung mononuclear phagocytes (MPs) at baseline.
a. Comparison of gene expression in B6 and DBA mice for each lung MP subset. Scatter plots are showing genes with transcripts per million (TPM) > 4. Pale blue dots show genes with fold change (FC) 2 or higher, dark blue dots show genes with FC 4 or higher. N=2 for each cell type in both strains.
b. Inter-cellular and inter-strain comparison of highly differentially expressed genes. Heat map shows unsupervised hierarchical clustering of genes with FC 4 or higher between the strains. Values are row z-score.
c. Examples of strain specific genes included in the heat map in Fig. 2b. Bar graphs represent mean log2 transcripts per million (TPM+1) ± SD, N=2. Asterisks indicate statistical significance, * p-adj < 0.05, ** p-adj < 0.01, *** p-adj < 0.001 calculated by DESeq2 and multiple-testing corrected with Benjamini-Hochberg.
Inference of transcriptional regulators from open chromatin landscapes
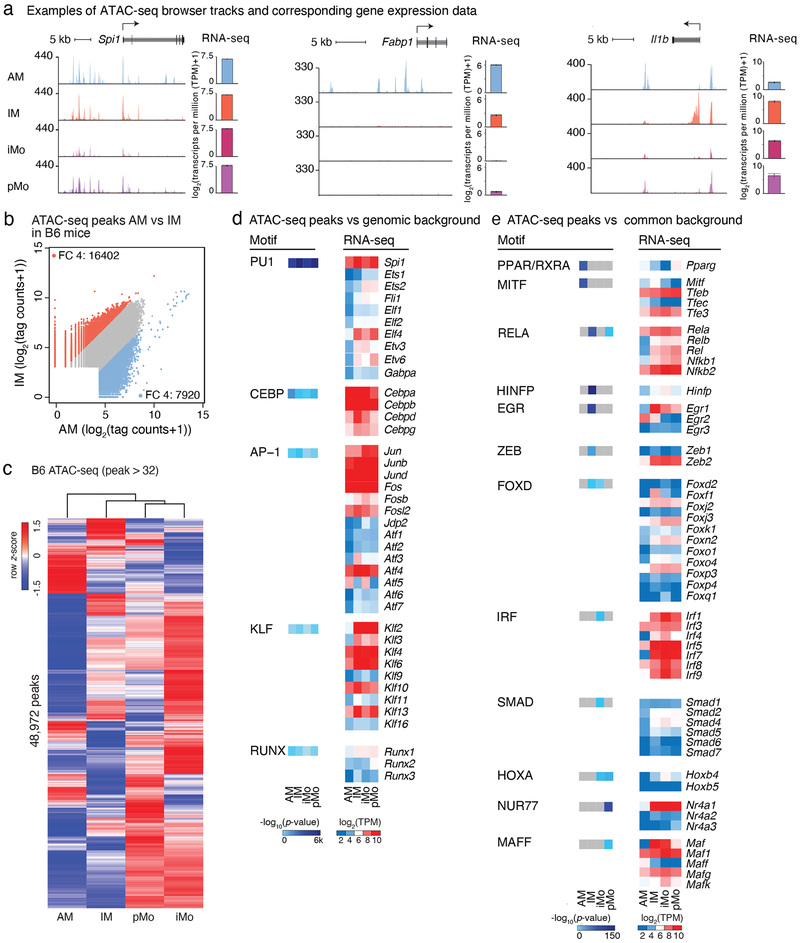
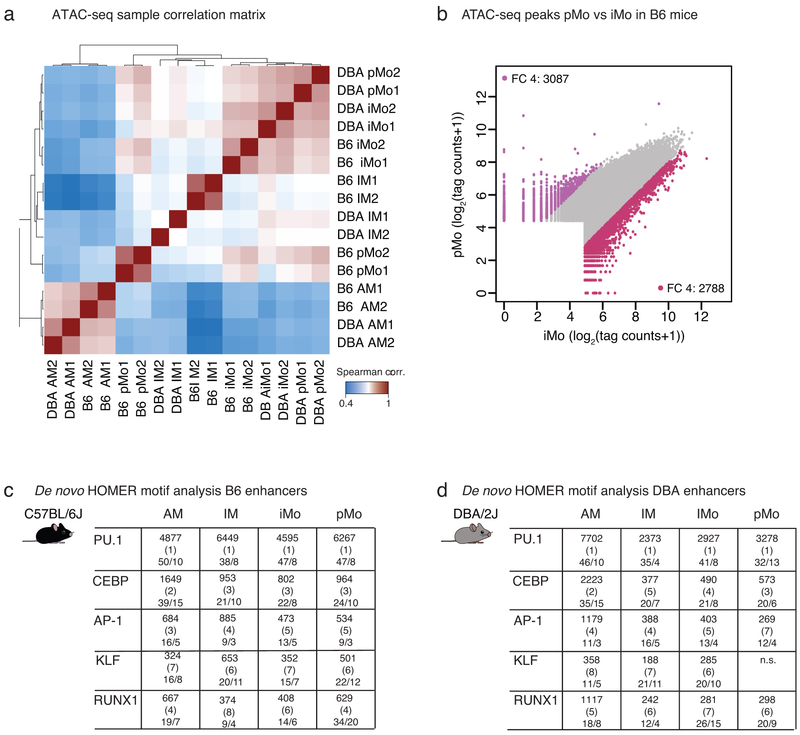
To investigate mechanisms responsible for the cell-subset-specific gene programs, we identified accessible regions of chromatin in lung MPs by performing ATAC-seq (Fig. 3a)30. We found the biological replicates to be highly correlated (Extended Data Fig. 2a). Essential myeloid genes, such as Spi1, encoding for the master TF PU.1, showed a highly overlapping open chromatin pattern in all lung MP subsets (Fig. 3a). In contrast, chromatin regions neighboring the AM-specific Fabp1 gene exhibited accessibility only in AMs. Open chromatin near Il1b, a pro-inflammatory cytokine, showed partial overlap among lung MPs, with a clear IM-specific peak, the cell subset showing the most inflammatory transcriptional pattern at baseline (Fig. 3a). In these examples, gene expression tracked closely with chromatin accessibility.
Figure 3: Epigenetic landscapes of lung mononuclear phagocytes (MPs).
a. Examples of ATAC-seq browser tracks for Spi1, Fabp1 and Il1b for the four lung MP subsets. Included are RNA-seq expression data for the same genes. Bar graphs represent mean log2 transcripts per million (TPM+1) ± SD, N=2.
b. Comparison of open chromatin regions for AM vs IM in B6 mice. Scatter plots are showing log2 tag counts of ATAC-seq peaks, colored dots show ATAC-seq peaks with a fold change (FC) of 4 or higher (blue dots for AM and orange for IM).
c. Heat map of hierarchical clustering of ATAC-seq signal in peaks with more than 32 tag counts in lung MP from B6 mice. Values are row z-scores.
d. HOMER de novo motif analysis of distal regions of open chromatin (> 3 kb from a TSS) using GC matched genomic background (left panel). Gene expression of transcription factor family members corresponding to identified motifs (right panel). N=2.
e. HOMER de novo motif analysis of cell subset specific distal regions of open chromatin (> 3 kb from a TSS) using all common peaks as background (left panel). Gene expression of transcription factor family members corresponding to identified motifs (right panel). N=2.
Genome-wide comparison of ATAC-seq peak tag counts revealed several thousand genomic sites with differentially opened chromatin between lung MP subsets. For example, pairwise comparison of AMs vs IMs revealed 16,402 peaks that displayed at least a 4-fold increase in ATAC-seq peak tag counts in IMs and 7,920 peaks in AMs (Fig. 3b). Comparison of the two lung monocyte subsets identified 3,087 peaks enriched in pMos and 2,788 peaks enriched in iMos (Extended Data Fig. 2b) implying that these cells indeed use distinct gene regulatory networks and a cell-subset-specific TF repertoire.
Clustering of lung MPs based on chromatin accessibility resulted in similar segregation as observed with clustering based on gene expression. AMs were most distant from the other 3 cell subsets and pMos and iMos grouped most closely together (Fig. 3c). To identify candidate TFs that could bind to distant open chromatin regions that would include enhancers, we performed de novo motif enrichment analysis of distal ATAC-seq peaks (> 3 kb from a transcription start site (TSS)). Using GC-matched genomic background in the HOMER39 motif analysis we identified motifs for common myeloid cell lineage factors such as PU.1, C/EBP, AP-1, KLF and RUNX (Fig. 3d). This core set of motifs was highly enriched in all lung MP in both strains (Extended Data Fig. 2c,d), consistent with general roles of TFs recognizing these motifs in establishing macrophage identity.
To increase the power to detect cell-subset-specific TFs, we performed HOMER de novo motif analysis comparing cell-specific ATAC peaks to all ATAC peaks present in any of the four lung MPs (Fig. 3e). This method normalizes out motifs for TFs that are common to the four lung MP subsets and enriches for subset-specific motifs. Using this approach, we identified both known and previously unknown cell-subset-specific TFs. For example, PPAR is one of the top motifs in AMs and NUR77 one of the top motifs in pMos. These findings are in line with previously published data showing that PPARγ is essential for AM development and that NUR77 is a master regulator of patrolling monocytes21, 40. Notably, the RELA motif, recognized by NF-κB, showed highest enrichment in IM enhancers, consistent with the enhanced inflammatory signature of these cells. IRF and SMAD motifs were selectively enriched in iMos, which are recognized by TFs with established roles in regulating pro- and anti-inflammatory programs of gene expression, respectively.
Other enriched motifs in unique lung MP subsets are associated with TFs for which functions in MP subsets have not been described. For example, AMs were enriched in the binding motif for MITF. IM enhancer regions were selectively enriched in HINFP, EGR and ZEB motifs. The MAFF motif was selectively enriched in pMos and is recognized by MAF TFs that form heterodimers with many other TFs, notably the NFE2 family41.
Since TFs of the same family can bind to the same or similar motifs, motif enrichment analysis cannot differentiate between TF family members. To infer likely candidates, we associated enriched motifs with mRNA expression of highly expressed TF family members that could bind these motifs. This approach revealed further similarities and differences among lung MPs. For example, PU.1, encoded by Spi1, was the ETS-domain transcription factor family member with the highest expression in all lung MPs (Fig. 3d). Compared to Spi1, other ETS members showed a more moderate expression level and higher degree of variation between cell subsets. Elf4, a TF previously shown to be important for interferon production and innate immune response against viruses42, showed cell-subset-specificity with highest expression in monocytes (Fig. 3d). Members of the C/EBP, AP-1 and KLF families were highly expressed in general, with some family members showing cell-subset-specificity. Of the five NF-κB proteins that could bind the RELA motif, Rela (also known as p65) and Nfkb2 (encoding for the p52 precursor protein also known as p100) were the most prominent (Fig. 3e). Overall, these findings suggest the identities of TF combinations required for establishing the regulatory landscapes of each lung MP subset.
Transcriptional regulators inferred from effects of genetic variation
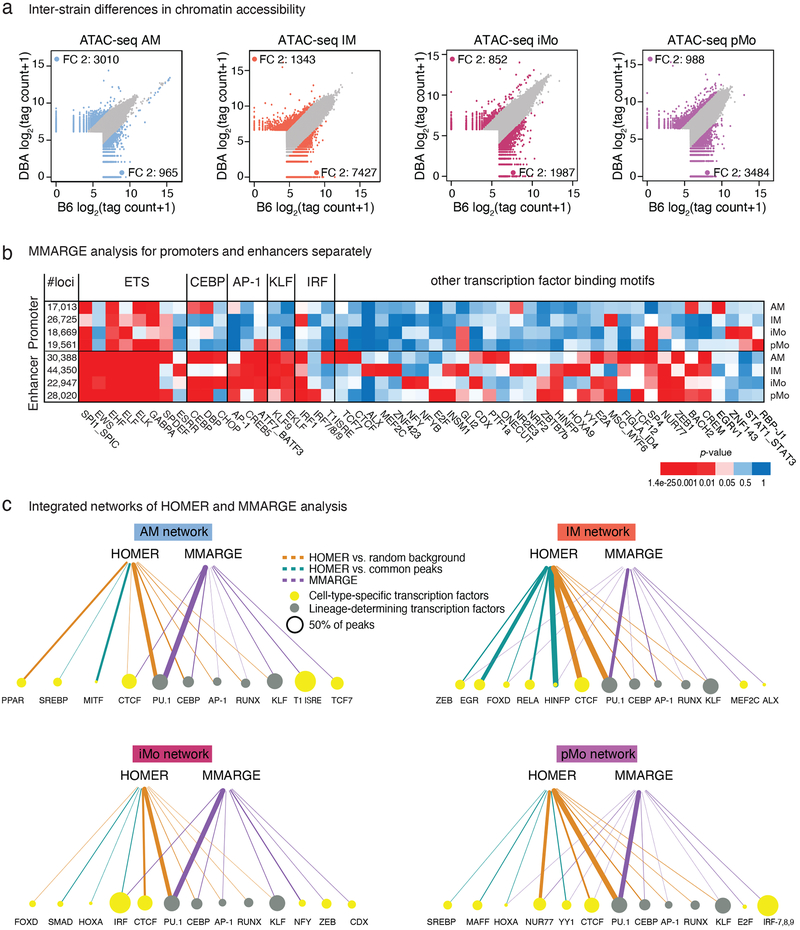
The functional relevance of TF motifs can be assessed by analyzing the impact of motif mutations43, 44. To investigate the functional importance of specific motifs in establishing open chromatin, we analyzed the effects of the natural genetic variation distinguishing B6 and DBA mice31. Comparing open chromatin regions between B6 and DBA mice (Fig. 4a), we found 2,000–8,000 genomic regions that showed at least a 2-fold difference in ATAC-seq signal in lung MP subsets.
Figure 4: Effects of natural genetic variation on lung mononuclear phagocyte (MP) open chromatin.
a. Inter-strain comparison of ATAC-seq peak tag counts for the four lung MP subsets. Scatter plots are showing log2 tag counts of ATAC-seq peaks, colored dots show ATAC-seq peaks with fold change (FC) 2 or higher (blue for AM, orange for IM, bordeaux for iMo and purple for pMo).
b. MMARGE analysis of lung MP. Distribution of chromatin accessibility was calculated for all peaks missing a motif in either B6 or DBA mice. Student’s t-test was used to test for significant differences between these two distributions. Heatmap showing p-values for all significant motifs in enhancers (> 3kb away from TSS) and promoters (< 3kb away from TSS). N=2 for each cell type in both strains.
c. Integrated network of HOMER and MMARGE analysis results for each lung MP subset. Node sizes are proportional to the percentage of peaks containing the motif. Lineage-determining transcription factors (TF) are represented with green nodes, other TF with yellow nodes. Edges are proportional to p-values. Edge color represents the analysis method; brown for HOMER with GC matched random background, turquoise for HOMER with common peaks as background, purple for MMARGE.
Effects of SNPs and InDels on open chromatin were assessed using MMARGE (Motif Mutation Analysis for Regulatory Genomic Elements)45. This method evaluates the significance of the relationship of mutations within TF motifs with strain-specific gain or loss of specific chromatin features, such as ATAC-seq signal. For example, if SNPs or InDels within PU.1 recognition motifs in B6 mice are statistically associated with a reduction in local ATAC-seq peaks compared to peaks in DBA mice, the PU.1 motif is scored as contributing to the formation of open chromatin. MMARGE analysis is thus qualitatively different from motif enrichment analysis in that it examines the functional consequence of a mutation within a binding motif on chromatin accessibility.
Application of this method to an annotated library of 287 distinct TF motifs yielded 51 significant motifs across the four lung MP subsets (Fig. 4b). As expected, motifs recognized by key regulators of the macrophage lineage such as PU.1, CEBP, AP-1, and KLF were highly significant in most or all lung MPs. MMARGE analysis also identified motifs in which mutations exerted subset-specific effects. For example, although a generic IRF motif was significant across all subsets, mutations in the motif for T1ISRE selectively affected ATAC peaks specific for AMs, and motifs for IRF-7,8,9 specifically affected pMos. ALX, MEF2C and ZNF423 were motifs specific for IMs. NFY and NFYB mutations were significantly associated with reduced open chromatin in iMos, but not other lung MP subsets (Fig. 4b,c). We also confirmed that mutations in the motif recognized by ZEB led to higher chromatin accessibility, consistent with its reported function as a transcriptional repressor43. When applying MMARGE specifically to open promoter regions in the two mouse strains, a much lower number of motifs were found to be significantly associated with chromatin accessibility. EGR was specific for AMs and MSC for IMs. Mutations in RBP-J and STAT motifs in promoter regions led to significant changes in chromatin accessibility in both monocyte subsets (Fig. 4b).
To integrate the findings of HOMER motif analyses using genomic background and lung MP common background with the functional results of MMARGE, we constructed networks of the most relevant TFs for each cell subset (Fig. 4c). In these networks the node size is proportional to the fraction of open chromatin regions containing the motif and the edge thickness represents the significance (p-value) of this motif within each analysis. Edges are colored according to the motif analysis method; HOMER using random GC background is brown, HOMER using lung MP common background is turquoise and MMARGE is purple.
These networks showed a high overlap of HOMER and MMARGE results for the critical macrophage regulators PU.1, CEBP, AP-1 and KLF. RUNX was the only macrophage lineage-determining TF (LDTF) (gray nodes) that did not show concomitant significance with motif enrichment and functional analysis in all cell subsets. In addition to the five LDTFs, this integrated approach identified additional TFs (yellow nodes) for each cell subset that reached significance in both analyses. For AMs, CTCF reached significance in HOMER and MMARGE analysis, while for IMs ZEB, FOXD, and HINFP were significant. Regarding the two monocyte subsets, IRF was significant for iMos and NUR77, HOXA, YY1 and CTCF for pMos. Taken together, these data provide functional evidence for putative TFs that shape the chromatin landscape in lung MPs and provide evidence for additional TFs that have selective roles in specific subsets of lung MPs that were not identified by conventional motif enrichment analysis.
Divergent responses of lung MPs to acute inflammation
The divergent baseline patterns of gene expression and distinct open chromatin landscapes associated with each lung MP subset suggests that they are programmed to respond to environmental perturbations in a cell-specific manner. To directly test this prediction, we performed RNA-seq analysis of AMs, IMs and iMos following exposure to LPS administered either intraperitoneally (i.p.) or intranasally (i.n.). Inflammation induced by i.p. LPS is a widely accepted model for endotoxin-mediated septic shock/acute respiratory distress syndrome and i.n. LPS for pneumonia46.
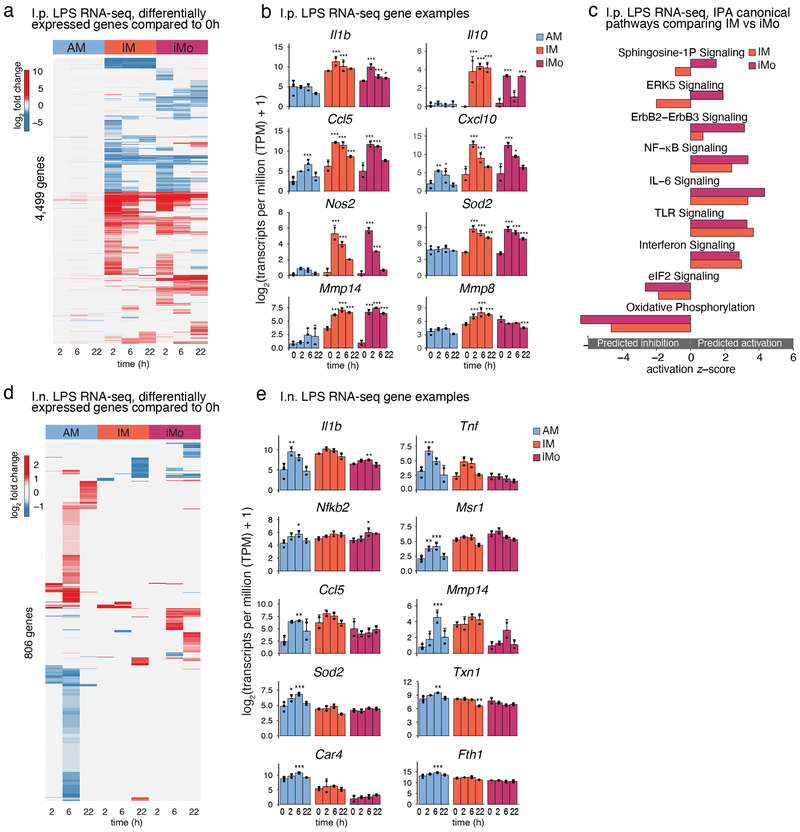
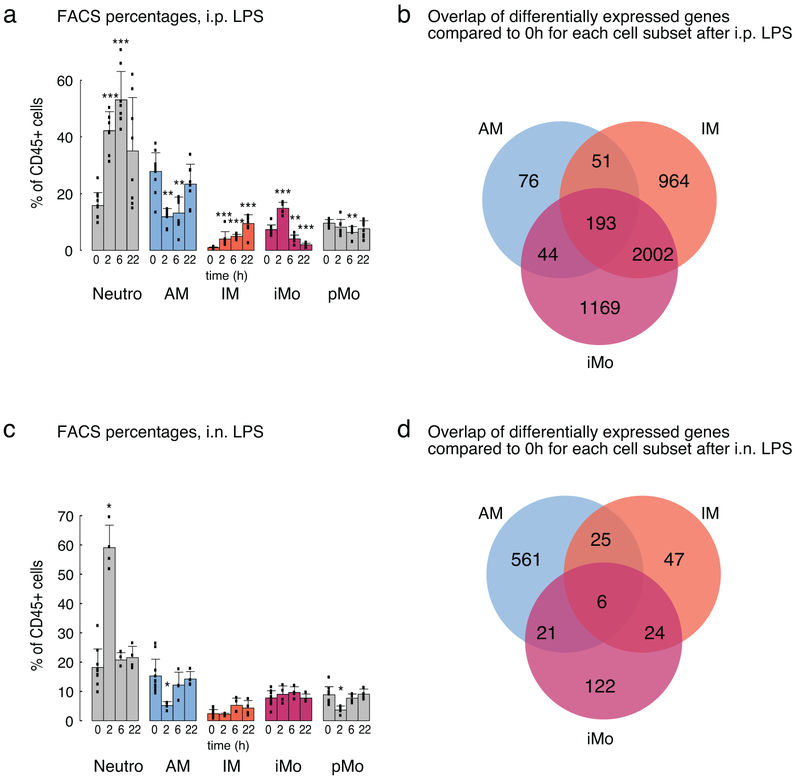
I.p. LPS administration resulted in a prominent inflammatory response in the lung with a significant shift in the composition of myeloid cells 2 h, 6 h and 22 h after LPS administration (Extended Data Fig. 3a). As expected, neutrophils and iMo were rapidly recruited to the lungs. The proportion of IMs gradually increased during the course of inflammation, while AMs showed a temporary relative decrease at 2 h and 6 h after i.p. LPS administration (Extended Data Fig. 3a).
IMs and iMos exhibited robust and largely overlapping changes in gene expression after i.p. LPS administration (in comparison to baseline expression), whereas AMs showed very modest changes (Fig. 5a,b and Extended Data Fig. 3b). In addition to proinflammatory cytokines (Il1b, Ccl5, Cxcl10), genes for anti-inflammatory cytokines such Il10 and reactive oxygen species scavengers such as Sod2, as well as matrix metallopeptidases (Mmp14, Mmp8), were rapidly induced in IMs and iMos after LPS administration (Fig. 5b).
Figure 5: Unique gene expression signatures for lung mononuclear phagocytes (MPs) during the course of acute lung inflammation induced by intraperitoneal (i.p.) and intranasal (i.n.) LPS administration.
a. Heat map of differentially expressed genes after i.p. LPS administration. Genes with transcripts per million (TPM) > 8 in at least 2 samples were included. Columns represent the mean fold change of gene expression at different time points after LPS administration compared to the baseline of each cell subset. The heat color represents the log2 fold change for 4,499 genes with a significance of FDR < 0.05 in at least one of the time course comparisons. Red represents up-regulated, blue represents down-regulated genes.
b. Examples of genes induced after i.p. LPS administration in AM, IM and iMo. Shown are log2 (TPM+1) ± SD. N=4 for 0h, N=2 for all other time points. Asterisks indicate statistical significance compared to 0h, * p-adj < 0.05, ** p-adj < 0.01, *** p-adj < 0.001 reported by DESeq2 multiple testing corrected with Benjamini-Hochberg method.
c. Ingenuity Pathway Analysis (IPA) showing canonical pathways of genes altered by i.p. LPS administration as compared to each cell’s own baseline. Each bar represents the activation z-score for pathways predicted to be activated (positive z-score) or inhibited (negative z-score).
d. Heat map of differentially expressed genes after i.n. LPS administration. Genes with TPM > 8 in at least 2 samples were included. Columns represent the mean fold change of gene expression at different time points after LPS administration compared to baseline for each cell subset. The heat color represents the log2 fold change for 806 genes with a significance of FDR < 0.05 in at least one of the time course comparisons. Red represents up-regulated, blue represents down-regulated genes.
e. Examples of genes induced after i.n. LPS administration in AM, IM and iMo. Shown are log2 (TPM+1) ± SD. N=4 for 0h, N=2 for all other time points. Asterisks indicates statistical difference compared to 0h, * p-adj < 0.05, ** p-adj < 0.01, *** p-adj < 0.001 calculated by DESeq2 and multiple-testing corrected with Benjamini-Hochberg.
IPA of differentially regulated genes in IMs and iMos compared to their own baseline indicated a pattern that was consistent with rapid activation of TLR4 and downstream signaling pathways (Fig. 5c). Induced pathways included TLR signaling, NF-κB signaling, IL-6 and interferon signaling. Enriched pathways for iMos and IMs were qualitatively similar, but some substantial quantitative differences were noted, including oxidative phosphorylation, eIF2 and ErbB2 signaling. ERK5 signaling and sphingosine-1-phosphate signaling were predicted to be activated in iMos but inhibited in IMs.
I.n. delivery of LPS resulted in a robust recruitment of neutrophils to the lung, similar to the i.p. LPS model (Extended Data Fig. 3c). In contrast to i.p. delivery, AMs were the most responsive lung macrophages, with very few acute changes in gene expression observed in IMs and iMos (Fig. 5d and Extended Data Fig. 3d). In general, the inflammatory response was more dampened in the i.n. LPS response compared to i.p. LPS with fewer number of differentially expressed genes and less pronounced changes compared to baseline values (Fig. 5e). A small set of genes was rapidly induced in AMs that overlapped with rapidly induced genes in IMs including Il1b and Tnf (Fig. 5e). However, the majority of genes responding to LPS in AMs exhibited a maximum response at 6 h, representing a delay in comparison to the maximal response of IMs and iMos, which occurred at 2 h following i.p. administration of LPS (Fig. 5d).
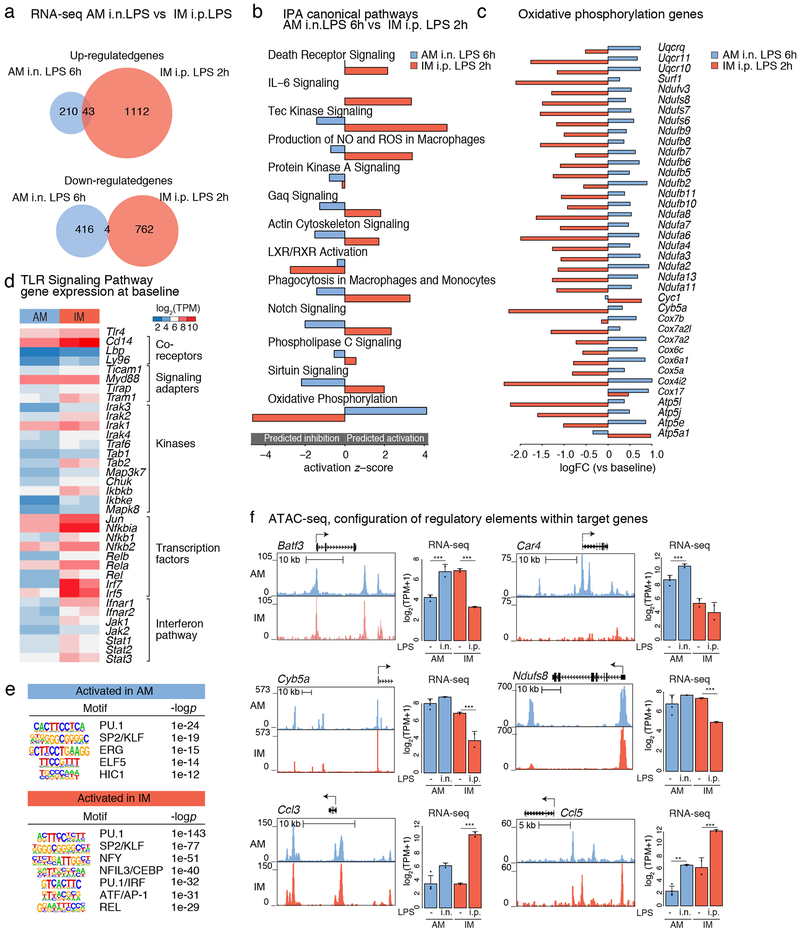
Comparison of genes induced at the time of maximal response in IMs (2 h, i.p. LPS) to genes induced at the time of maximal response in AMs (6 h, i.n. LPS) indicated that of the 253 up-regulated genes in AMs and 1,155 up-regulated genes in IMs, only 43 were in common (Fig. 6a). A similar pattern was observed for down-regulated genes. Remarkably, IPA analysis indicated that the predicted functional consequences of the AM and IM changes in gene expression were in opposite directions for several of the most enriched pathways (Fig. 6b). One of the most striking differences is provided by differential regulation of genes involved in oxidative phosphorylation, which were down-regulated in IMs but up-regulated in AMs (Fig. 6c).
Figure 6: Intranasal (i.n.) LPS administration induces a moderate activation of AMs.
a. Venn diagram of up-regulated (top panel) and down-regulated (bottom panel) genes after 6h i.n. LPS administration in AM versus 2h i.p. LPS administration in IM.
b. Ingenuity Pathway Analysis (IPA) showing canonical pathways of genes altered by 6h i.n. LPS administration in AM (compared to AM baseline) versus 2h i.p. LPS administration in IM (compared to IM baseline). Shown is the z-score for pathways predicted to activated (positive z-score) and inhibited (negative z-score).
c. Log fold change (logFC) (versus baseline) for genes from IPA oxidative phosphorylation pathway in AM after i.n. LPS administration at 6h and in IM after i.p. LPS administration at 2h. N=2 for each cell type.
d. Gene expression shown in log2 transcripts per million (TPM) for genes of the TLR signaling pathway for AM and IM at baseline.
e. HOMER de novo motif analysis results for promoter regions (< 3kb from TSS) of genes activated in AM at 6h after i.n. LPS (top panel) and IM at 2h after i.p. LPS (bottom panel). N=2 for each cell type.
f. UCSC genome browser tracks for ATAC-seq signal (left panel) and bar plots for expression signal (right panel) for genes that exhibit divergent responses to LPS in AMs and IMs. N=2. Shown are log2 (TPM+1) ± SD. Asterisks indicates statistical difference compared to 0h, * p-adj < 0.05, ** p-adj < 0.01, *** p-adj < 0.001 calculated by DESeq2 and multiple-testing corrected with Benjamini-Hochberg.
To explore possible mechanisms, we analyzed the basal expression of genes belonging to the TLR signaling pathway (Fig. 6d). Tlr4, Ly96 (encoding MD2), Cd14, Lbp (both encoding co-receptors for LPS), and the signaling adapter protein encoded by Myd88, are expressed at similar levels in AMs and IMs. However, the majority of genes encoding additional downstream adapters, kinases and transcription factors mediating responses to TLR4 ligation exhibit lower expression in AMs. These genes include the key signaling adapters encoded by Tram1, Ticam1, Tirap and Traf6 and downstream kinases encoded by Mapk8, Ikbkb, Tab1 and Tab2. Genes encoding several major transcriptional targets of these kinases also exhibit lower levels of expression in AMs, including all five family members of the NF-κB family cJun and Irf5 and Irf7. These differences would be predicted to reduce the strength of the response of AMs to LPS, but not result in the observed divergent patterns of this response.
We next analyzed open chromatin in the vicinity (+/− 3 kb) of the transcriptional start sites (TSSs) of genes that exhibited differential responses to i.p. LPS in IMs at 2 h and responses to i.n. LPS in AMs at 6 h (Fig. 6e). In both cases, open chromatin was enriched for motifs recognized by PU.1 and SP2/KLF factors. However, the remaining significant motifs were divergent. Notably, ATAC-seq peaks associated with genes preferentially induced in IMs were enriched for NFY, NFIL3/CEBP, IRF, AP-1 and REL motifs. In contrast, ATAC-seq peaks associated with genes preferentially induced in AMs were enriched for ERG, ELF5 and HIC1 motifs.
Direct analysis of open chromatin patterns of genes exhibiting differential responses in AMs exposed to i.n. LPS and IMs exposed to i.p. LPS revealed a continuum ranging from nearly identical patterns to strongly cell-type-specific patterns (Fig. 6f). For example, while Batf and Cyb5a are induced in AMs but repressed in IMs, the open chromatin environment is nearly indistinguishable in the two cell subsets. A similar pattern of open chromatin is also observed for Ccl3, which is conversely induced in IMs compared to AMs. In contrast, open chromatin environments in the vicinity of Car4 and Ndufs8, which are selectively induced in AMs, and Ccl5, which is induced in IMs, exhibit substantial differences in their local ATAC-seq profiles, consistent with the results of motif enrichment analysis. Therefore, AMs and IMs exhibit qualitatively different responses to LPS in vivo associated with differences in the TLR4 signaling pathway at baseline and the open chromatin landscapes of TLR4 target genes.
Discussion
Although lung MPs reside in the same organ, they exist in distinct microenvironments and perform different functions. Consistent with this, transcriptomic analysis of major subsets of lung MPs identified by scRNA-seq indicates substantial differences in gene expression under homeostatic conditions. For each subset, natural genetic variation distinguishing B6 and DBA mice resulted in hundreds of significant changes in gene expression, the majority of which were cell-subset-specific. These differences are most likely explained primarily by effects of genetic variation on cell-specific regulatory elements43 and contribute to the phenotypic diversity of B6 and DBA mice. The delineation of the specific patterns of open chromatin in each cell-subset and the influence of genetic variation enabled inference of major classes of TFs underlying their development and function, some of which have not previously been suggested to play roles in lung macrophages. It will therefore be of interest to perform loss of function experiments and identify signaling pathways that are necessary for the expression and activities of these factors within each macrophage subset.
The most striking findings to emerge from the present studies were the qualitatively different responses of AMs and IMs to i.p. and i.n. LPS. At one level, these divergent responses can be easily understood based on anatomic compartmentalization of the signal. Remarkably, the direct responses of IMs to i.p. LPS and AMs to i.n. LPS are mostly divergent, in many cases leading to predictions of opposite biological outcomes. Analysis of the open chromatin landscapes of genes that exhibit divergent responses to LPS in AMs and IMs suggest at least two underlying mechanisms. A subset of genes exhibit distinct patterns of open chromatin, consistent with differences in enriched motifs for differentially regulated genes as a whole. In these cases, the accessible landscapes for signal-dependent TFs, such as NF-κB, are different. Cell-specific responses to TLR4 ligation of genes with distinct patterns of open chromatin are likely to result at least in part from the corresponding genomic regions functioning as cell-specific regulatory elements.
A second subset of genes exhibiting AM- or IM-specific responses to LPS were observed to have indistinguishable patterns of local open chromatin. While it is possible that cell-specific regulation is determined by more distal regulatory elements for some of these genes, our findings are more consistent with the possibility that the mechanisms by which TLR4 ligation regulates gene expression at the signaling level are substantially different in AMs vs IMs. The reduced expression of genes encoding components of the TLR4 signaling pathway in AMs as compared to IMs is consistent with an attenuated response to LPS in comparison to IMs, but does not explain genes that are activated in an AM-specific manner. Instead, our findings suggest that TLR4 couples to alternative, as yet undefined, signaling molecules in AMs to activate a divergent set of transcriptional activators and/or repressors. This mechanism would provide an alternative explanation for cell-specific differences in responses of genes exhibiting similar patterns of open chromatin, through the recruitment of different classes of signal-dependent factors that are not involved in the selection of open chromatin itself. Moving forward, it will be of substantial interest to better understand mechanisms underlying distinct responses of AMs and IMs to TLR4 ligation and more generally to decipher their cell-type specific responses in disease models .
Methods
Animal model
Animals.
We obtained C57BL/6J and DBA/2J mice from the Jackson Laboratory. We established breeding colonies for both strains in our vivarium and housed them in adjacent cages. Male mice, 8–10 weeks old were used for experiments. All protocols were approved by the University of California, San Diego Animal Care and Use Committee. Mice were housed in cages, maximum 4 mice/cage, in a 12 h/12 h light/dark cycle with free access to food and water.
For baseline experiments we used the lungs of two mice/per sample, we performed >5 independent experiments and representative data of two biological replicates are used for anlaysis. The different lung MP subsets used for final analysis derive from multiple mice for both strains.
Acute lung inflammation, acute respiratory distress model.
C57BL/6J mice were injected intraperitoneally with LPS (Escherichia coli O55:B5, Sigma-Aldrich) suspended in 50 μl of PBS at 1 mg/kg concentration. Control mice were injected with 50 μl PBS intraperitoneally. Mice were euthanized with carbon dioxide 2 h, 6 h and 22 h after LPS administration and lungs were harvested for lung MP isolation. 2 mice per time point were pooled and 2 biological replicates were performed at each timepoint equaling 4 mice/time point. 3 independent experiments were performed.
Acute lung inflammation, pneumonia model.
C57BL/6J mice were lightly anesthetized with isoflurane and 10 μg of LPS (Escherichia coli O55:B5, Sigma-Aldrich) suspended in 50 μl PBS was administered intranasally. Control mice were intranasally instilled with 50 μl PBS. Mice were euthanized with carbon dioxide 2 h, 6 h and 22 h after LPS administration and lungs were harvested for lung MP isolation. 2 mice per time point were pooled and 2 biological replicates were performed at each timepoint equaling 4 mice/time point. 3 independent experiment were performed.
Lung MP isolation
Mice were euthanized with carbon dioxide. Lungs were perfused with PBS trough the right ventricle until no blood was visible in the tissue. Lungs were removed, minced and placed in a digestion buffer containing collagenase type 4 (1.6 mg/ml; Roche), DNase type 1 (50 units/ml; Roche) and Flavoperidol (1 μM, Sigma-Aldrich) in RPMI 1640 medium. Lungs were incubated at 37 °C for 15 min while on a rotator. Red blood cells were lysed using RBC lysis buffer (eBioscience) at 4 °C for a 5 min incubation. The obtained cell suspension was filtered using a 70-micron filter, washed and cells were resuspended in PBS. Cells were then incubated with anti-Fc blocking mAb (anti CD16/32, clone 93, BioLegend) and viability dye Zombie Aqua (BioLegend) for 10 min. After this, cells where stained with an antibody cocktail containing APC Cy7-conjugated CD45, PE-conjugated F4/80, BV605-conjugated CD11c, v450-conjugated CD11b, PE Cy7-conjugated CD64, AF700-conjufgated Ly6c, FITC-conjugated Ly6g, AF647-conjugated SiglecF mAb for 30 min. All labeled cells were passed through a FACS Aria II flow cytometer (BD Biosciences) and after proper gating 4 subsets of lung MPs were sorted29. Data were analyzed with FlowJo 8.3.3 software (TreeStar Inc.). The antibodies used for FACS are listed in Supplementary Table 1.
Single-cell RNA-sequencing
Lungs of 2-week old DBA/2J mice (3 males and 1 female) were harvested and single cell suspension of lung cells were prepared as described above. Using FACS we isolated lung myeloid cells by gating on CD45+ cells and excluding T cells, B cells and γ/δT cells using antibodies against TCRβ chain, CD19 and TCR γ/δ chain. The antibodies used for FACS are listed in Supplementary Table 2.
Sorted lung myeloid cells were suspended in PBS containing 0.04% molecular biology grade non-acetylated BSA at a concentration of 1,000 cells/μL. The purity of the sorted cells as well as the sorted cell number were verified. Using the 10X GemCode technology single cells were captured with barcoded beads in a droplet emulsion for the RT reaction. After obtaining 10X barcoded cDNA, libraries were prepared following the standard protocol of 10X Single Cell 3′ v1. The libraries were sequenced on one lane of a Hi-Seq 4000 (Illumina).
RNA-sequencing library preparation
Baseline samples of sorted lung MPs from C57BL/6J and DBA/2J mice were used. Sequencing libraries were prepared from polyA enriched mRNA as previously described47. PolyA enriched mRNA was fragmented, in 2x Superscript III first-strand buffer with 10 mM DTT (Invitrogen), by incubation at 94 °C for 9 min, then immediately chilled on ice before the next step. The 10 μL of fragmented mRNA, 0.5 μL of Random primer (Invitrogen), 0.5 μL of Oligo dT primer (Invitrogen), 0.5 μL of SUPERase-In (Ambion), 1 μL of dNTPs (10 mM) and 1 μL of DTT (10 mM) were heated at 50 °C for 3 min. At the end of incubation, 5.8 μL of water, 1 μL of DTT (100 mM), 0.1 μL Actinomycin D (2 μg/μL), 0.2 μL of 1% Tween-20 (Sigma) and 0.2 μL of Superscript III (Invitrogen) were added and incubated in a PCR machine using the following conditions: 25 °C for 10 min, 50 °C for 50 min, and a 4 °C hold. The product was then purified with RNAClean XP beads according to manufacture’s instruction and eluted with 10 μL nuclease-free water. The RNA/cDNA double-stranded hybrid was then added to 1.5 μL of Blue Buffer (Enzymatics), 1.1 μL of dUTP mix (10 mM dATP, dCTP, dGTP and 20 mM dUTP), 0.2 μL of RNAse H (5 U/μL), 1.05 μL of water, 1 μL of DNA polymerase I (Enzymatics) and 0.15 μL of 1% Tween-20. The mixture was incubated at 16 °C for 1 h. The resulting dUTP-marked dsDNA was purified using 28 μL of Sera-Mag Speedbeads (Thermo Fisher Scientific), diluted with 20% PEG8000, 2.5M NaCl to final of 13% PEG, eluted with 40 μL EB buffer (10 mM Tris-Cl, pH 8.5) and frozen −80°C. The purified dsDNA (40 μL) underwent end repair by blunting, A-tailing and adapter ligation as previously described39 using barcoded adapters (NextFlex, Bioo Scientific). Libraries were PCR-amplified for 14–16 cycles, size selected using 10% PAGE/TBE gels, eluted and quantified with Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific), and sequenced on a Hi-Seq 4000 (Illumina) for 51 cycles or a NextSeq 500 (Illumina) for 75 cycles.
Low input RNA-sequencing library preparation
Lung MP subsets sorted after i.p. LPS or i.n. LPS administration were used. Sequencing libraries were prepared according to the Smart-seq2 method48 with some modifications. 500–5000 FACS isolated cells in Trizol (Thermo Fisher) were used as starting material. RNA was extracted with the Direct-zol MicroPrep kit (Zymo Research) with on-column DNAseI treatment. 10 μL purified RNA was mixed with 5.5 μL of SMARTScribe 5X First-Strand Buffer (Clontech), 1 μL polyT-RT primer (2.5 μM, 5′-AAGCAGTGGTATCAACGCAGAGTAC(T30)VN, 0.5 μL SUPERase-IN (Ambion), 4 μL dNTP mix (10 mM, Invitrogen), 0.5 μL DTT (20 mM, Clontech) and 2 μL Betaine solution (5 M, Sigma), incubated 50 °C 3 min. 3.9 μL of first strand mix, containing 0.2 μL 1% Tween-20, 0.32 μL MgCl2 (500 mM), 0.88 μL Betaine solution (5 M, Sigma), 0.5 μL (5 M, Sigma) SUPERase-IN (Ambion) and 2 μL SMARTScribe Reverse Transcriptase (100 U/μL Clontech) was added and incubated one cycle 25 °C 3 min, 42 °C 60 min. 1.62 μL template switch (TS) reaction mix containing 0.8 μL biotin-TS oligo (10 μM, Biotin-5′-AAGCAGTGGTATCAACGCAGAGTACATrGrG+G-3′), 0.5 μL SMARTScribe Reverse Transcriptase (100 U/μL Clontech) and 0.32 μL SMARTScribe 5X First-Strand Buffer (Clontech) was added, then incubated at 50 °C 2 min, 42 °C 80 min, 70 °C 10 min. 14.8 μL second strand synthesis, pre-amplification mix containing 1 μL pre-amp oligo (10 μM, 5′AAGCAGTGGTATCAACGCAGAGT-3′), 8.8 μL KAPA HiFi Fidelity Buffer (5X, KAPA Biosystems), 3.5 μL dNTP mix (10 mM, Invitrogen) and 1.5 μL KAPA HiFi HotStart DNA Polymerase (1U/μL, KAPA Biosystems), was added, then amplified by PCR: 95 °C 3 min, 8 cycles 98 °C 20 sec, 67 °C 15 sec and 72 °C 6 min, final extension 72 °C 5 min. The synthesized dsDNA was purified using Sera-Mag Speedbeads (Thermo Fisher Scientific) with final 8.4% PEG8000, 1.1M NaCl, then eluted with 13 μL UltraPure water (Invitrogen). The product was quantified by Qubit dsDNA High Sensitivity Assay Kit (Invitrogen) and libraries were prepared using the Nextera DNA Sample Preparation kit (Illumina). Tagmentation mix containing 11 μL 2X Tagment DNA Buffer and 1 μL Tagment DNA Enzyme was added to 10 μL purified DNA, then incubated at 55 °C for 15 min. 6 μL Nextera Resuspension Buffer (Illumina) was added and incubated at 21 °C for 5 min. Tagmented DNA was purified using Sera-Mag Speedbeads (Thermo Fisher Scientific) with final 7.8% PEG8000, 0.98M NaCl, then eluted with 25 μL UltraPure water (Invitrogen). Final enrichment amplification was performed with Nextera primers, adding 1 μL Ad1 (100 μM, universal no barcode), 1 μL unique Ad2.n (100 μM, N7xx) barcoding primers and 27 μL NEBNext High-Fidelity 2X PCR Master Mix (New England BioLabs), then amplified by PCR: 72 °C 5 min, 98 °C 30 sec, 8–13 cycles 98 °C 10 sec, 63 °C 30 sec, and 72 °C 1 min. Libraries were purified using 1.5 volumes of SpeedBeads in 2.5 M NaCl, 20% PEG8000, further size selected using 10% PAGE/TBE gels, eluted and quantified with Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific), pooled and sequenced on a NextSeq 500 (Illumina) for 76 cycles or a Hi-Seq 4000 (Illumina) for 51 cycles.
ATAC-seq library preparation
Sorted cells in FACS Buffer were pelleted by centrifugation at 400g for 6 min at 4 °C in a pre-cooled fixed-angle microfuge. Supernatant was carefully removed and cell pellet was resuspend in 47.5 μl lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL CA-630), 2.5 μl of TDE1 (Cat# FC-121–1030, Illumina) by pipetting. Transposition reactions were incubated at 37 °C for 30 min. Transposed DNA was purified using a Zymo ChIP DNA Clean & Concentrator kit (Cat# D5205) and purified DNA was eluted in 10 μl elution buffer (10 mM Tris-HCl, pH 8). Transposed fragments were amplified and purified as described previously30 with modified primers.
Data analysis
Single-cell RNA-sequencing.
We used the R package Seurat 49 (version 2.3.4) to perform quality control and clustering analysis. Unique molecular identifiers (UMI) that barcode each individual mRNA molecule within a cell during reverse transcription were used to remove PCR duplicates. Cells expressing <300 or >2000 genes were filtered out to exclude non-cells or cell aggregates. We also filtered out cells that contained a percentage of mitochondrial UMI reads 5%. After quality control, 1,048 cells were included in subsequent analysis. Seurat standard pipeline was used for normalization and scaling, and first 11 PCs from PCA were used for t-Distributed Stochastic Neighbor Embedding (t-SNE) dimensionality reduction. The shared nearest neighbor (SNN) modularity optimization based clustering algorithm was used to group cells into 8 clusters with resolution=0.8. Velocyto28 a python package was used to predict the migration of cells.
RNA-sequencing analysis.
FASTQ files were processed to assess quality by determining general sequencing bias, clonality and adapter sequence contamination. RNA sequencing reads were aligned to the individual genomes for the mouse strain (based on mm10 reference genome) of origin using STAR50. Gene expression levels were calculated using HOMER39 by counting all strand specific reads within exons. Only the most abundant transcripts, including multiple alternative variants, were selected for each gene, and the genes whith a length smaller than 250bp were removed. Transcripts per million (TPM) were used to evaluate the correlation among replicates. Differential gene expression was calculated using DESeq251 to assess both biological and technical variability between experiments. Unsupervised hierarchical clustering was used to cluster the gene expression in the heatmaps. To identify functional enrichment of genes we used Ingenuity Pathway Analysis (IPA) software (Qiagen).
ATAC-sequencing analysis.
FASTQ files were mapped to individual genomes for the mouse strain (based on mm10 reference genome) with Bowtie252 and shifted back to reference coordinates for DBA/2J strain. Peaks were called with HOMER (findPeaks) using parameters “-style factor -minDist 200 -size 200”. After merging these peaks, correlations among replicates from the same cell subset (and the same LPS treatment time point, if applicable) were evaluated by correlation using tag counts. The two most highly correlated samples were used for identifying the most robust peaks using the irreproducible discovery rate (IDR) method. For this step, peaks were called with HOMER’s findPeaks, using parameters “-L 0 -C 0 -fdr 0.9 -minDist 200 -size 200”. For baseline analysis, peaks were merged with HOMER’s mergePeaks and annotated with HOMER’s annotatePeaks.pl using the merged IDR tag directories. For the LPS experiments, peaks were merged with HOMER’s mergePeaks using parameters “-d 100”. The raw tags of all samples which had reasonable correlation were quantified with HOMER (annotatePeaks.pl) using parameter “-noadj”. For each cell subset, these raw tags which contained at least 16 tags in at least 2 samples were used to identify differentially bounded peaks by DESeq2. De novo motif analysis was performed on peaks 3kb away from closest TSS using a modified version of HOMER’s findMotifsGenome.pl from MMARGE. For LPS analysis, peaks 3kb away from the closest TSS were used and motif analysis was performed with HOMER’s findMotifsGenome.pl with default parameters.
Motif Mutation Analysis for Regulatory Genomic Elements (MMARGE)45.
To model the impact of a motif on chromatin accessibility, MMARGE’s pairwise comparison was used. In brief, peaks from both strains per cell type were merged with HOMER’s mergedPeaks and annotated with normalized tag counts. These files were scanned for motifs and the distribution of chromatin accessibility is calculated for all peaks missing a motif in either C57BL/6J or DBA/2J mice. To calculate significance, a Student’s t-test is performed between these two distributions.
Statistical analyses
Differences in gene expression between different cell subsets and between the two mouse strains were calculated by DESeq2 and multiple-testing was corrected with the Benjamini-Hochberg procvedure. Data are presented as mean ± SD. Differences in percentage of different subsets of myeloid cells in the lung after i.p. and i.n. LPS administration were tested using the non-parametric Wilcoxon signed-rank test with Bonferroni correction. Data are presented as mean ± SD.
Data availability statement
The sequencing data reported in this article are deposited in GEO with the following accession number GSE136916. Additional data supporting the presented findings are available in the manuscript and upon request from the corresponding author.
Extended Data
Extended Data Fig. 1. Single-cell RNA-seq clusters, FACS sorting strategy and quality control of FACS and RNA-seq results.
a. Single-cell RNA-seq gene expression in clusters used to determine cluster identities. Heat map is representing expression values for the most significant genes in each cluster. Cells from the lungs of 3 male and 1 female DBA/2J mice were pooled.
b. Zoomed-in view of velocity analysis for single-cell RNA-seq from Fig. 1a.
c. Flow cytometry analysis and sorting strategy to obtain subsets of lung mononuclear phagocytes (MPs).
d. Validation of sorting strategy with gene expression in sorted lung MPs in C57BL/6J (B6) mice (top panel) and DBA/2J (DBA) mice (bottom panel). Bars represent transcripts per million (TPM) for one mouse. Two replicates are shown.
e. Spearman correlation heat map of all RNA-seq replicates, N=2 for each cell type in both mouse strains.
f. Comparison of gene expression for AM vs. IM and iMo vs. pMo in B6 mice. Scatter plots are showing genes with TPM > 16. Blue dots for AM, orange dots for IM and bordeaux dots for iMo show genes with fold change (FC) 2 or higher.
g. Comparison of gene expression in pMo isolated from lung vs pMo isolated from circulating blood. Scatter plots show genes with TPM > 16. Pale blue dots show genes with FC 2 or higher, dark blue dots show genes with FC 4 or higher.
h. Ingenuity pathway analysis (IPA) functional pathways for genes differentially regulated in pMo isolated from lung vs pMo isolated from circulating blood.
Extended Data Fig. 2. ATAC-seq quality control and HOMER de novo motif analysis.
a. Spearman correlation heat map of all ATAC-seq replicates, N=2 for each cell type in both mouse strains.
b. Comparison of open chromatin regions for iMo vs pMo in B6 mice. Scatter plot shows log2 tag counts of ATAC-seq peaks, colored dots show ATAC-seq peaks with fold change (FC) 4 or higher, bordeaux for iMo and purple for pMo.
c. HOMER de novo motif enrichment analysis for transcription factor (TF) binding sites in distal regions of open chromatin (> 3 kb from a TSS) likely representing enhancers using GC matched background for B6 mice. Boxes display −log10 p-values for enrichment of the motif, rank order in parenthesis and percentage of motif occurrence in peaks vs background, N=2 for each cell type in both mouse strains.
d. HOMER de novo motif enrichment analysis for TF binding sites in distal regions of open chromatin (> 3 kb from a TSS) likely representing enhancers using GC matched background for DBA mice. Boxes display −log10 p-values for enrichment of the motif, rank order in parenthesis and percentage of motif occurrence in peaks vs background, N=2 for each cell type in both mouse strains.
Extended Data Fig. 3. Flow cytometry and ingenuity pathway analysis (IPA) of lung mononuclear phagocytes (MPs) after LPS administration.
a. Flow cytometry analysis of lung MP subsets and neutrophils after i.p. LPS administration. Bars represent the average percentage of given cell subset out of CD45+ leukocytes ± SD. Three independent experiments were pooled, 0h N=11, for all other time points N=8. Non-parametric Wilcoxon signed-rank test with Bonferroni correction was used, * p < 0.05, ** p < 0.01, *** p < 0.001.
b. Venn diagram of 4,499 differentially expressed genes in AM, IM and iMo after i.p. LPS administration.
c. Flow cytometry analysis of lung MP subsets and neutrophils after i.n. LPS administration. Bars represent the average percentage of given cell subset out of CD45+ leukocytes ± SD. Two independent experiments were pooled, 0h N= 11, for all other time points N=4. Non-parametric Wilcoxon signed-rank test with Bonferroni correction was used, * p < 0.05, ** p < 0.01, *** p < 0.001.
d. Venn diagram of 806 differentially expressed genes in AM, IM and iMo after i.n. LPS administration.
Supplementary Material
Acknowledgments
These studies were supported by NIH grants DK091183 and DK063491. E.S. was supported by NIH grant K08HL140198. C.E.R. was supported by NIH grants R00HL123485 and R01HL147187. L.S.P. was supported by NIH grants HL086324, HL126703, HL143256 and the Gerber Foundation (1823-3830). We thank T. Rombaldo for assistance with FACS, J. Collier for technical assistance and L. van Ael for help with preparation of the manuscript. Sequencing of RNA-seq and ATAC-seq libraries was conducted at the IGM Genomics Center, University of California, San Diego, La Jolla, CA.
Footnotes
Declaration of interests
The authors declare no competing interests.
References
- 1.Pollard JW Trophic macrophages in development and disease. Nat Rev Immunol 9, 259–270 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ardini-Poleske ME et al. LungMAP: The Molecular Atlas of Lung Development Program. American journal of physiology. Lung cellular and molecular physiology 313, L733–L740 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gomez Perdiguero E et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 518, 547–551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoeffel G et al. C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity 42, 665–678 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guilliams M et al. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. The Journal of experimental medicine 210, 1977–1992 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck-Schimmer B et al. Alveolar macrophages regulate neutrophil recruitment in endotoxin-induced lung injury. Respiratory research 6, 61 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hussell T & Bell TJ Alveolar macrophages: plasticity in a tissue-specific context. Nat Rev Immunol 14, 81–93 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Schneider C et al. Alveolar macrophages are essential for protection from respiratory failure and associated morbidity following influenza virus infection. PLoS Pathog 10, e1004053 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen M et al. Lung Single-Cell Signaling Interaction Map Reveals Basophil Role in Macrophage Imprinting. Cell 175, 1031–1044 e1018 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Reyfman PA et al. Single-Cell Transcriptomic Analysis of Human Lung Provides Insights into the Pathobiology of Pulmonary Fibrosis. American journal of respiratory and critical care medicine 199, 1517–1536 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan SY & Krasnow MA Developmental origin of lung macrophage diversity. Development 143, 1318–1327 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakarov S et al. Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science 363 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Gibbings SL et al. Three Unique Interstitial Macrophages in the Murine Lung at Steady State. American journal of respiratory cell and molecular biology 57, 66–76 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tatham KC et al. Intravascular donor monocytes play a central role in lung transplant ischaemia-reperfusion injury. Thorax 73, 350–360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maus UA et al. Monocytes are potent facilitators of alveolar neutrophil emigration during lung inflammation: role of the CCL2-CCR2 axis. Journal of immunology 170, 3273–3278 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Misharin AV et al. Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. The Journal of experimental medicine 214, 2387–2404 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jardine L et al. Lipopolysaccharide inhalation recruits monocytes and dendritic cell subsets to the alveolar airspace. Nature communications 10, 1999 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.T’Jonck W, Guilliams M & Bonnardel J Niche signals and transcription factors involved in tissue-resident macrophage development. Cell Immunol 330, 43–53 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mass E et al. Specification of tissue-resident macrophages during organogenesis. Science 4238, epub (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lavin Y et al. Tissue-Resident Macrophage Enhancer Landscapes Are Shaped by the Local Microenvironment. Cell 159, 1312–1326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneider C et al. Induction of the nuclear receptor PPAR-gamma by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nature immunology 15, 1026–1037 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Baker AD et al. PPARgamma regulates the expression of cholesterol metabolism genes in alveolar macrophages. Biochem Biophys Res Commun 393, 682–687 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Gosselin D et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 159, 1327–1340 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gosselin D et al. An environment-dependent transcriptional network specifies human microglia identity. Science 356 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang DX & Glass CK Towards an understanding of cell-specific functions of signal-dependent transcription factors. J Mol Endocrinol 51, T37–50 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geissmann F et al. Development of monocytes, macrophages, and dendritic cells. Science 327, 656–661 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mildner A et al. Genomic Characterization of Murine Monocytes Reveals C/EBPbeta Transcription Factor Dependence of Ly6C(−) Cells. Immunity 46, 849–862 e847 (2017). [DOI] [PubMed] [Google Scholar]
- 28.La Manno G et al. RNA velocity of single cells. Nature 560, 494–498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Misharin AV, Morales-Nebreda L, Mutlu GM, Budinger GR & Perlman H Flow cytometric analysis of macrophages and dendritic cell subsets in the mouse lung. American journal of respiratory cell and molecular biology 49, 503–510 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buenrostro JD, Wu B, Chang HY & Greenleaf WJ ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol 109, 21 29 21–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keane TM et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477, 289–294 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitehead GS, Burch LH, Berman KG, Piantadosi CA & Schwartz DA Genetic basis of murine responses to hyperoxia-induced lung injury. Immunogenetics 58, 793–804 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartalesi B et al. Different lung responses to cigarette smoke in two strains of mice sensitive to oxidants. The European respiratory journal 25, 15–22 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Rittling SR Osteopontin in macrophage function. Expert Rev Mol Med 13, e15 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Shan M et al. Cigarette smoke induction of osteopontin (SPP1) mediates T(H)17 inflammation in human and experimental emphysema. Sci Transl Med 4, 117ra119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russell CD & Schwarze J The role of pro-resolution lipid mediators in infectious disease. Immunology 141, 166–173 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhen A et al. CD4 ligation on human blood monocytes triggers macrophage differentiation and enhances HIV infection. J Virol 88, 9934–9946 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adachi H & Tsujimoto M FEEL-1, a novel scavenger receptor with in vitro bacteria-binding and angiogenesis-modulating activities. J Biol Chem 277, 34264–34270 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas GD et al. Deleting an Nr4a1 Super-Enhancer Subdomain Ablates Ly6C(low) Monocytes while Preserving Macrophage Gene Function. Immunity 45, 975–987 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kannan MB, Solovieva V & Blank V The small MAF transcription factors MAFF, MAFG and MAFK: current knowledge and perspectives. Biochim Biophys Acta 1823, 1841–1846 (2012). [DOI] [PubMed] [Google Scholar]
- 42.You F et al. ELF4 is critical for induction of type I interferon and the host antiviral response. Nature immunology 14, 1237–1246 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Link VM et al. Analysis of Genetically Diverse Macrophages Reveals Local and Domain-wide Mechanisms that Control Transcription Factor Binding and Function. Cell 173, 1796–1809 e1717 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heinz S et al. Effect of natural genetic variation on enhancer selection and function. Nature 503, 487–492 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Link VM, Romanoski CE, Metzler D & Glass CK MMARGE: Motif Mutation Analysis for Regulatory Genomic Elements. Nucleic Acids Res (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matute-Bello G et al. An official American Thoracic Society workshop report: features and measurements of experimental acute lung injury in animals. American journal of respiratory cell and molecular biology 44, 725–738 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oishi Y et al. SREBP1 Contributes to Resolution of Pro-inflammatory TLR4 Signaling by Reprogramming Fatty Acid Metabolism. Cell Metab 25, 412–427 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
References (Methods only)
- 48.Picelli S et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods 10, 1096–1098 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data reported in this article are deposited in GEO with the following accession number GSE136916. Additional data supporting the presented findings are available in the manuscript and upon request from the corresponding author.