Abstract
Autism spectrum disorder is a neurodevelopmental disorder characterized by difficulties in social communication and interaction, as well as repetitive and characteristic patterns of behaviour. Although the pathogenesis of autism spectrum disorder is unknown, being overweight or obesity during infancy and low weight at birth are known as risks, suggesting a metabolic aspect. In this study, we investigated adipose tissue development as a pathophysiological factor of autism spectrum disorder by examining the serum levels of adipokines and other metabolic markers in autism spectrum disorder children (n = 123) and typically developing children (n = 92) at 4–12 years of age. Among multiple measures exhibiting age-dependent trajectories, the leptin levels displayed different trajectory patterns between autism spectrum disorder and typically developing children, supporting an adipose tissue-dependent mechanism of autism spectrum disorder. Of particular interest, the levels of fatty acid binding protein 4 (FABP4) were significantly lower in autism spectrum disorder children than in typically developing subjects, at preschool age (4–6 years old: n = 21 for autism spectrum disorder and n = 26 for typically developing). The receiver operating characteristic curve analysis discriminated autism spectrum disorder children from typically developing children with a sensitivity of 94.4% and a specificity of 75.0%. We re-sequenced the exons of the FABP4 gene in a Japanese cohort comprising 659 autism spectrum disorder and 1000 control samples, and identified two rare functional variants in the autism spectrum disorder group. The Trp98Stop, one of the two variants, was transmitted to the proband from his mother with a history of depression. The disruption of the Fabp4 gene in mice evoked autism spectrum disorder-like behavioural phenotypes and increased spine density on apical dendrites of pyramidal neurons, which has been observed in the postmortem brains of autism spectrum disorder subjects. The Fabp4 knockout mice had an altered fatty acid composition in the cortex. Collectively, these results suggest that an ‘adipo-brain axis’ may underlie the pathophysiology of autism spectrum disorder, with FABP4 as a potential molecule for use as a biomarker.
Keywords: early childhood, early diagnostic biomarker, functional variants, adipose tissue, polyunsaturated fatty acid (PUFA)
We discovered lower circulating fatty acid binding protein 4 (FABP4), mainly secreted by the adipose tissue, in children with autism spectrum disorder, and identified functional FABP4 variants in the autism spectrum disorder group. Furthermore, Fabp4 knock-out mice displayed autism spectrum disorder-relevant phenotypes. We propose that the ‘adipo-brain axis’ may underlie the pathophysiology of autism spectrum disorder.
Graphical Abstract
Graphical Abstract.
Introduction
Autism spectrum disorder (ASD) is a neurodevelopmental disorder characterized by persistent difficulties in social communication and social interaction. The prevalence of ASD, now estimated to be 1–2%, is increasing annually (World Health Organization; https://www.who.int/news-room/fact-sheets/detail/autism-spectrum-disorders, last accessed 9 September 2020) (Weintraub, 2011; Lai et al., 2014). Although ASD is thought to be caused by complex interactions between genetic and environmental factors, the detailed mechanism of the pathogenesis of ASD remains unclear (de la Torre-Ubieta et al., 2016; Rylaarsdam and Guemez-Gamboa, 2019).
ASD children show various developmental differences compared with typically developing (TD) children in early childhood. For example, early brain overgrowth at 6‒24 months of age has been frequently reported as a neuroanatomical feature of ASD children (Courchesne et al., 2011). Another study reported on the aberrant development of white matter tracts in the first year of life of ASD children (Wolff et al., 2012). In addition to brain development, ASD children present several characteristics relevant to the lipid metabolism, particularly in early development. Adipose tissue secretes hundreds of bioactive molecules (adipokines) (Luo and Liu, 2016), some of which modulate brain functions (Parimisetty et al., 2016). ASD children show changes in the levels of adipokines, including increased leptin levels (Ashwood et al., 2008; Blardi et al., 2010), decreased adiponectin levels (Fujita-Shimizu et al., 2010) and increased monocyte chemotactic protein-1 (MCP-1) levels (Ashwood et al., 2011) in early childhood.
Fatty acid binding protein 4 (FABP4, also known as adipocyte-FABP or aP2), a member of the FABP family (Zimmerman and Veerkamp, 2002), is thought to form a complex with fatty acids (FAs) (Gillilan et al., 2007) and functions as a chaperone for FAs within cells (Furuhashi and Hotamisligil, 2008). FABP4 was recently identified as a novel adipokine and may play a role as an adipokine in the pathology of metabolic syndromes (Furuhashi and Hotamisligil, 2008; Prentice et al., 2019). In our previous study, FABP4 expression was found to decrease in the scalp hair follicles of patients with schizophrenia (Maekawa et al., 2015). However, to date, the relationship between FABP4 and autism has not been investigated.
In this study, we systematically measured the serum levels of various adipokines, including FABP4, in subjects aged 2–12 years (preschool to elementary school age) and found that serum FABP4 levels were reduced in ASD children at the preschool stage. Based on these results, we further investigated a possible link between the pathogenesis of ASD and the functional disturbance of FABP4 by leveraging human genetics and analysis of gene-deficient mice.
Materials and methods
Human serum samples
All subjects were Japanese living in central or eastern Japan, including the Hokuriku, Chubu, Tokai, Kanto and Tohoku regions. The clinical diagnosis of ASD was made by board-certified child psychiatrists based on the Diagnostic and Statistical Manual of Mental Disorders, fourth edition, text revision (American Psychiatric Association, 2000) using the information of developmental history. The ASD diagnosis was confirmed by a structured developmental interview with the subject’s parents (Autism Diagnostic Interview-Revised) (Lord et al., 1994; Le Couteur et al., 2008; Falkmer et al., 2013). One of the authors (K.J.T.) previously attended an Autism Diagnostic Interview-Revised research training workshop and established research reliability with Weill Cornell Medicine Psychiatry. The ethics committees of the participating institutes approved the study protocols, and written informed consent was obtained from the parents of all participants. The current study was conducted in accordance with the Declaration of Helsinki.
The first sample set included 102 ASD and 87 TD children, which were divided into a preschool-aged group (4–6 years old) (ASD: 18 men, 3 women; mean age, 5.70 ± 0.96 years; TD children: 14 men, 12 women, mean age 6.05 ± 0.64 years) and an elementary school-aged group (7–12 years old) (ASD: 93 men, 9 women, mean age 10.08 ± 1.55 years; TD children: 55 men, 11 women, mean age 10.47 ± 1.70 years) (Supplementary Table 1). The second sample set included 21 ASD (2–4 years old) (14 men, 7 women; mean age, 3.79 ± 0.74 years) and 24 TD children (2–4 years old) (13 men, 11 women; mean age, 3.72 ± 0.82 years) (Supplementary Table 2). Fasting blood samples were collected between 6:00 and 9:00 by venipuncture from all the participants, and the samples were kept at room temperature for 30 min. They were then centrifuged, divided into 200-μl aliquots and stored at −80°C until use.
Measurement of adipokines and metabolic biomarkers in serum samples
Serum FABP4 concentrations were measured using enzyme-linked immunosorbent assay (ELISA) kits (Cayman Chemical, Ann Arbor, MI, USA). Levels of insulin, MCP-1, and leptin were measured using Multiplex Assay kit ‘Human Metabolic Disease Panel’ (Merck Millipore, Darmstadt, Germany). The adiponectin levels were determined using sandwich ELISA kits (R&D Systems Inc., Minneapolis, MN, USA), according to the manufacturer's instructions. The glucose concentrations were measured using a commercial kit (Glu-CII; Wako Pure Chemical Industries, Ltd., Osaka, Japan). The levels of free fatty acid were measured using a Free Fatty Acid Fluorometric Assay Kit (Cayman Chemical). Each serum sample was analysed in duplicate, and the mean value of the two measures was used for analysis.
DNA samples
DNA samples were collected from Japanese subjects who were born and living in central Japan separately from the serum samples. ASD was diagnosed based on the Autism Diagnostic Interview-Revised . Control subjects were recruited from volunteers with no present or previous evidence of psychosis during brief interviews by psychiatrists. The DNA sample set comprised 659 subjects with ASD (529 men, 130 women; mean age, 12.13 ± 7.82 years) and 1000 controls (500 men, 500 women; mean age 41.40 ± 14.37 years).
Re-sequencing analysis of FABP4
All coding exons and exon/intron boundaries of the FABP4 gene were screened for polymorphisms by direct sequencing using the BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, CA, USA) and an ABI 3730xl sequencer (Applied Biosystems). Polymorphisms were detected using SEQUENCHER software (Gene Codes Corporation, Ann Arbor, MI, USA) (Balan et al., 2014). The genomic structure of FABP4 (RefSeq: NM_001442.2) was based on the UCSC hg19 draft assembly of the human genome database (http://www.genome.ucsc.edu, last accessed 9 September 2020). The NCBI database (https://www.ncbi.nlm.nih.gov/snp/, last accessed 9 September 2020) was searched for known single nucleotide polymorphisms (SNPs).
Construction of plasmids
Human FABP4 cDNA was obtained by PCR using Human Fetal Brain Marathon-Ready cDNA (Clontech, Mountain View, CA, USA) and the following primer set: forward: 5′-ATTGAATTCATGTGTGATGCTTTTGTA-3′, reverse: 5′-TGAGTCGACTTATGCTCTCTCATAAAC-3′. The amplified cDNA was cloned into the mammalian expression vector pcDNA3 (Invitrogen, Grand Island, NY, USA). The Thr8Ala mutant was created by conventional site-directed mutagenesis. For the bacterial expression construct, a PCR fragment from the pcDNA3-FABP4 construct was inserted into the bacterial expression vector pGEX-6P-3 (GE Healthcare Life Sciences, Tokyo, Japan) using XhoI/BamHI. The structures of the generated plasmids were validated by Sanger sequencing.
Recombinant FABP4 protein
Recombinant FABP4 proteins (WT and Thr8Ala) were obtained as reported previously (Shimamoto et al., 2014). Briefly, the proteins were produced using Escherichia coli BL21 (DE3) (BioDynamics Laboratory Inc., Tokyo, Japan) and purified using glutathione sepharose 4B (GE Healthcare Life Sciences). After the GST moiety was removed by digestion with PreScission protease (GE Healthcare Life Sciences) on the beads, the resulting recombinant proteins were de-lipidated using a Lipidex1000 column (PerkinElmer, Waltham, MA, USA).
Binding assay
The fluorescent FABP ligand 1-anilinonaphthalene-8-sulfonic acid (ANS) was purchased from Cayman Chemical (Ann Arbor, MI, USA). Binding assays using ANS were based on a previously described procedure (Shimamoto et al., 2014). Fluorescence was measured using a multi-label counter (excitation at 355 nm and emission at 460 nm) (Wallac 1420 ARVO MX-2; Perkin Elmer). The dissociation constant (Kd) values were calculated by non-linear regression analysis using GraphPad Prism software (version 8, GraphPad Software, San Diego, CA, USA).
Animals
Fabp4-disrupted mice (gifted from Prof. Hotamışlıgil) were established as described in Hotamisligil et al. (1996) and maintained as a closed colony. The mice were maintained as a closed colony and heterozygotes were intercrossed to produce wild-type (WT) and Fabp4-null mice. The animals were housed in groups of four or five in standard cages in a temperature- and humidity-controlled room with a 12 h light/dark cycle (lights on at 08:00). The animals had free access to standard lab chow (CRF-1) (Charles River formula; purchased from Oriental Yeast, Tokyo, Japan) and tap water. All animal experiments were performed using male animals (7–16 animals/group, depending on the experiment). The experimental procedures were approved by the RIKEN Animal Ethics Committee (permission number: H30-B030139).
Behavioural analyses
The behavioural profiles of the Fabp4 KO mice were assessed using the three-chamber social interaction, Morris water maze, and other behavioural tests at 6 weeks to 6 months, as well as with ultrasonic vocalization tests at postnatal day 5 (P5) to P14. The protocols for behavioural tests were as described in the Supplementary Methods and the literature (Ohnishi et al., 2010; Shimamoto et al., 2014).
Spine analysis
Mice were deeply anaesthetized with isoflurane and perfused transcardially with 4% paraformaldehyde in phosphate-buffered saline (PBS). The brains were trimmed and cut into 200-μm thick coronal sections using a vibratome. Lipophilic dye (DiI, Invitrogen) was coated onto tungsten particles (Bio-Rad, Hercules, CA, USA). DiI-coated particles were delivered to the slices using the Helios Gene Gun system (Bio-Rad). A polycarbonate filter with 8.0-μm pores (Beckton Dickinson, Franklin Lakes, NJ, USA) was inserted between the gun and the preparation to remove clusters of large particles. The density of labelling was regulated via gas pressure (95–105 psi helium) (Mataga et al., 2004). A confocal laser-scanning microscope FV1000 (Olympus, Tokyo, Japan) was used to image the labelled structures. Randomly labelled typical pyramidal neurons were selected from layers II–III and V in the temporal cortex. Images at 0.45-μm steps were acquired and stacked for 3D reconstruction using ImageJ (https://imagej.nih.gov/ij/, last accessed 9 September 2020) and Spiso-3D (mathematical and automated software calculating geometrical parameters of spines) (Mukai et al., 2011). Spines were counted along an apical dendrite for each neuron in eight mice per group. Spine density on apical dendrites was averaged at 50–100-μm from the cell body (Hutsler and Zhang, 2010).
Fatty acid analysis
Tissue samples were homogenized in ice-cold saline, and aliquots were used for lipid analysis. Total lipids were extracted according to the method described by Bligh and Dyer (1959). Total phospholipid fractions were separated by thin-layer chromatography. After transmethylation with HCl-methanol, the FA composition was analysed by gas chromatography (GC-2014; Shimadzu Corporation, Kyoto, Japan) with a DB-225 capillary column (length, 30 m; internal diameter, 0.25 mm; film, 0.25 μm; J&M Scientific, Folsom, CA, USA). The entire system was controlled using gas chromatography software GC-solution version 2.3 (Shimadzu Corporation). Each FA was expressed as a percentage of the total FA content.
Data availability
The data supporting these findings are available upon request.
Statistical analysis
The data were analysed using Prism 8 or SPSS version 16 (SPSS Inc., Chicago, IL, USA). Continuous data are expressed as median and range or mean ± standard error (SE). Outliers were removed based on the 1.5× interquartile range (IQR) rule. Statistical analyses were performed using the Mann–Whitney U test for mean differences and the Spearman rank test for correlation. P < 0.05 was considered significant.
Results
Serum levels of adipokines in childhood
The demographic data of the participants are summarized in Supplementary Table 1. No significant differences in age, intelligence quotient, weight, height, body mass index (Supplementary Table 1) or blood biochemical markers were found between the ASD and TD groups (Supplementary Table 3) in the all-age group and in each specific age group.
We measured the serum levels of adipokines which have been reported to be altered in ASD, including leptin, adiponectin and MCP-1 (Ashwood et al., 2008; Blardi et al., 2010; Fujita-Shimizu et al., 2010; Ashwood et al., 2011), except for FABP4, which, to the best of our knowledge, was first examined here. We also measured other metabolic parameters (insulin, glucose and free fatty acid) in the sera of ASD and TD children. The FABP4 and MCP-1 levels were negatively correlated with age in TD children, but not in ASD children (Fig. 1A and B). Leptin levels were positively correlated with age in ASD children but not in TD children (Fig. 1C). Adiponectin and free fatty acid levels were negatively correlated with age (Fig. 1D–G), while insulin and glucose levels were positively correlated with age in both the TD and ASD groups. Leptin level trajectories showed a significant difference between the ASD and TD groups in terms of the Spearman’s correlation coefficients, while the other measures did not (Fig. 1H).
Figure 1.
Serum levels of adipokines and other metabolic parameters in ASD and TD subjects. (A–G) Correlation between age and the levels of adipokines or other metabolic parameters: (A) FABP4, (B) leptin, (C) MCP-1, (D) adiponectin, (E) insulin, (F) glucose and (G) free fatty acids. (H) Comparison of correlation coefficients between ASD and TD groups. (I) FABP4 levels in ASD and TD groups of 4–6 years old. P-values were calculated using two-tailed Mann–Whitney U tests. (J) receiver operating characteristic curve for FABP4 at 4–6 years old. (K) FABP4 levels in ASD and TD groups at 2–4 years old. P-values were calculated using two-tailed Mann–Whitney U tests. (L) receiver operating characteristic curve for FABP4 at 2–4 years old.
Considering the age-dependent changes in the levels of the above molecules, we divided the cohort into two groups: preschool-aged (4–6 years old) and elementary school-aged (7–12 years old) groups, and compared the mean levels of the molecules between the ASD and TD groups (Supplementary Table 1). In the preschool-aged group, the levels of FABP4 in ASD children were significantly lower than those in TD children (Fig. 1H), however, the levels of other molecules did not differ significantly between the ASD and TD groups (Supplementary Fig. 1). In the elementary school-aged group, the levels of MCP-1 and free fatty acid were significantly higher in the ASD group than in the TD group, but the levels of other markers were not significantly different between the ASD and TD groups (Supplementary Fig. 2).
FABP4 as an early biomarker of ASD
Since the early diagnosis and early intervention of ASD are important for prognosis (Lai et al., 2014) and the FABP4 levels in ASD children were low, specifically at the preschool stage (Fig. 1H), we subsequently focused on FABP4 and evaluated its potential as an early diagnostic biomarker. A receiver operating characteristic curve analysis determined the cut-off level at 16.7 ng/ml (Fig. 1I). The sensitivity, specificity, positive predictive values and negative predictive values were 94.4%, 75.0%, 5.6% and 25.0%, respectively. We did not observe any correlations between the levels of FABP4 and the psychological scores on ASD (intelligence quotient and items in Autism Diagnostic Interview-Revised) (Supplementary Table 4). The levels of FABP4 did not differ between the male and female samples (Supplementary Table 5). The serum levels of FABP4 are associated with obesity (Aeberli et al., 2008; Krzystek-Korpacka et al., 2011; Dencker et al., 2017). However, we found no correlations between the levels of FABP4 and physical indices (body mass index, height and weight) (Supplementary Table 6).
To determine whether the serum level of FABP4 could serve as a useful biomarker in the earlier stage, we examined the second set of samples at the toddler stage (2–4 years old), consisting of 21 ASD and 24 TD children (Supplementary Table 2). Again, the FABP4 levels were significantly lower in ASD children than in TD children (Fig. 1J). A receiver operating characteristic curve determined the cut-off level at 12.7 ng/ml. The sensitivity, specificity, positive predictive values and negative predictive values were 81.0%, 71.4%, 19.1% and 28.6%, respectively (Fig. 1K). These results demonstrate that the serum levels of FABP4 could be a versatile diagnostic biomarker for ASD at an early stage (at least 2–6 years old).
Exon resequencing analysis of the FABP4 gene
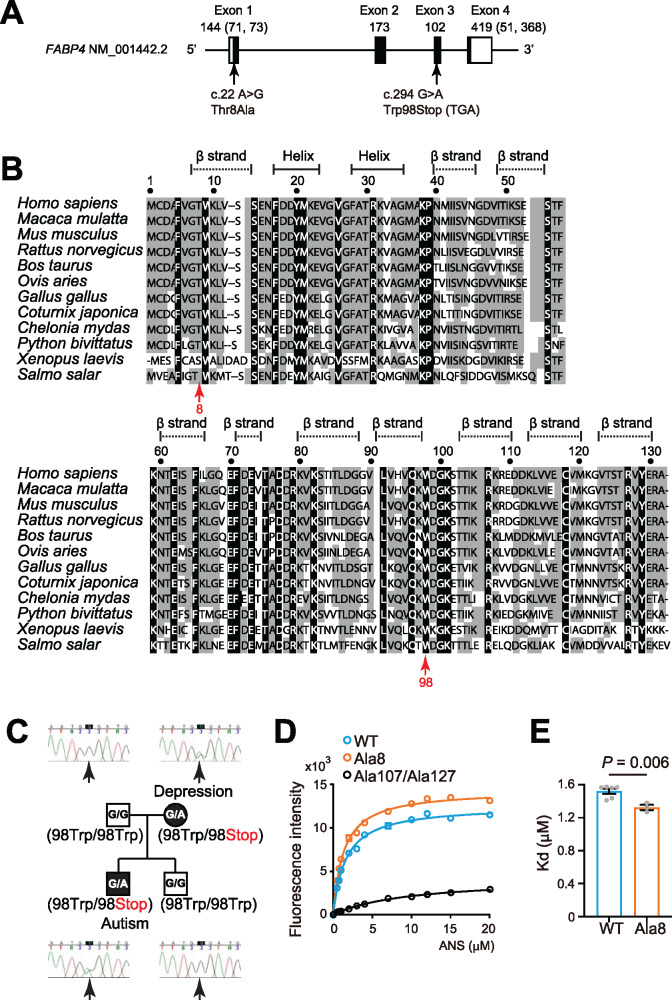
To search for a genetic underpinning of FABP4 abnormality seen in ASD, we conducted an exon resequencing analysis of the FABP4 gene using 659 ASD and 1000 control subjects. We identified one case with the missense mutation Thr8Ala and one case with the nonsense mutation Trp98Stop (Table 1 and Fig. 2A). FABP4 Thr8 is highly conserved among species and is located within a β-strand (Fig. 2B). The in silico web tool ‘PolyPhen2’ (http://genetics.bwh.harvard.edu/pph2/, last accessed 9 September 2020) estimates the possible impact of an amino acid substitution on the structure and function of the protein. This algorithm predicted that the FABP4 Thr8Ala mutation could be ‘possibly damaging’. Pedigree samples were unavailable for this mutation. The FABP4 Trp98Stop mutation would result in the loss of 33 amino acids at the C-terminus, which is highly conserved and includes three β-strands (Fig. 2B) and three amino acids needed for FA binding (Cys118, Arg127 and Tyr129) (BioLiP database: https://zhanglab.ccmb.med.umich.edu/BioLiP/, last accessed 9 September 2020) (Supplementary Table 7). It is also possible that this transcript may be rapidly degraded by the nonsense-mediated mRNA decay system (Maquat, 2005; Behm-Ansmant et al., 2007; Kurosaki and Maquat, 2016). This mutation was transmitted from his mother with a history of depression (Fig. 2C), suggesting that the mutation may co-segregate with psychiatric illnesses. The serum FABP4 level of the proband (12 years old) was 7.3 ng/ml, which is nearly half of the average levels of the TD group (14.4 ng/ml) and the ASD group (14.1 ng/ml) of the similar ages (elementary school ages) (Supplementary Fig. 2). These results suggest that although rare, there are ASD cases where FABP4 gene defects may be involved in the pathogenesis of ASD.
Table 1.
Polymorphisms identified in the FABP4 gene
| Exon | Nucleotide change | Amino acid change | dbSNP ID | Autism (n = 516) | Control (n = 1000) |
|---|---|---|---|---|---|
| c.1-6C>T | rs1585812890* | 0 | 1 | ||
| 1 | c.22A>G | Thr8Ala | rs1585812863* | 1 | 0 |
| 3 | c.294G>A | Trp98Stop | rs150131014* | 1 | 0 |
| 4 | c.399 + 11G>A | rs138590127* | 0 | 3 |
Asterisks indicate newly identified SNPs. They were registered in the dbSNP (https://www.ncbi.nlm.nih.gov/snp/, last accessed 9 September 2020).
Figure 2.
Exon resequencing analysis of the FABP4 gene. (A) Gene structure of human FABP4. Exon is shown as a square, with the coding regions in black and the untranslated regions in white. Polymorphisms identified in ASD are indicated by arrows. (B) Amino acid sequence alignment of FABP4 between multiple species. Black boxes indicate amino acids conserved from Atlantic salmon (Salmo salar) to humans. The grey boxes indicate partially conserved amino acids. The variants identified in ASD are shown using arrows, with the amino acid positions. (C) Family structure of the proband with the FABP4 Trp98Stop mutation. The black symbols represent individuals with the FABP4 Trp98Stop mutation. The squares and circles indicate men and women, respectively. (D) ANS was incubated with recombinant FABP4 proteins and the intensity of fluorescence was recorded (excitation filter: 355 nm, emission filter: 460 nm). The data are representative of independent experiments. (E) Kd value obtained from ANS assays.
Binding property of the FABP4 mutant to the fluorescent ligand ANS
To investigate the biochemical effects of the FABP4 Thr8Ala variant, we measured the binding potential of the recombinant FABP4 mutant protein to 1-anilinonaphthalene-8-sulfonic acid (ANS). ANS is a fluorescent dye that binds with a high affinity to the ligand-binding pocket of the FABP proteins. In addition to the WT and the Thr8Ala mutant, we prepared a double mutant, FABP4 Arg107Ala/Arg127Ala, as a binding pocket-destructive mutant (Shimamoto et al., 2014). We were able to purify these proteins to an almost homogeneous level (Supplementary Fig. 3). We performed titration analyses of their affinities to the ANS (Fig. 2D). The dissociation constant (Kd) of the double mutant (FABP4 Ala107/Ala127) to ANS was significantly higher than that of WT. In contrast, the Kd value of the Ala8 mutant to ANS was significantly lower than that of WT (Fig. 2E), demonstrating that the missense mutation increases the binding affinity to ANS, and probably to natural ligands such as fatty acids. However, the in vivo nature of the FABP4 Trp98Stop and Thr8Ala variants remains elusive.
Expression of FABP4 in postmortem brains
It has been reported that FABP4 is expressed in the human brain during both childhood (Human Brain Transcriptome, http://hbatlas.org/, last accessed 9 September 2020) and adulthood (Supplementary Fig. 4) (Maekawa et al., 2015). We examined the expression levels of FABP4 in postmortem brain samples from autistic subjects in Broadmann area 9 (control: n = 10, ASD: n = 10), Broadmann area 21 (control: n = 14, ASD: n = 14), Broadmann area 40 (control: n = 13, ASD: n = 14) and dorsal raphe nucleus (control: n = 8, ASD: n = 8) (Supplementary Table 8). However, the expression of FABP4 was unchanged between the ASD and control groups in any brain region (Supplementary Fig. 4B). These results may correspond to the results in a Japanese cohort in which there were no significant differences in the FABP4 serum levels between the ASD and TD groups at the elementary school age. The ages of the postmortem brain samples ranged from elementary school ages and above. Extended brain samples, in terms of both age span and sample size, will need to be examined in a future study.
ASD-like behavioural phenotypes of Fabp4 KO mice
To investigate the effects of a functional disturbance of FABP4, we evaluated the behaviours of Fabp4 KO mice. The Fabp4 KO mice were healthy with no visible abnormalities in terms of growth or morphology. In the three-chamber test (Fig. 3A), a reduced exploratory behaviour toward a stranger mouse was observed in Fabp4 KO mice (Fig. 3B). In the Morris water maze test (Fig. 3C), Fabp4 KO mice exhibited reduced spatial learning and memory, represented by the lack of a clear increase in the time spent in the target quadrant (platform) or the number of times the target platform was crossed after training (Fig. 3D and E). This result may be related to the impaired visuospatial ability reported in individuals with ASD (McGrath et al., 2012; Habib et al., 2019). We did not observe significant differences in a reversal learning test between the Fabp4 KO and WT mice (data not shown), which evaluates a perseveration trait usually under the condition of non-impaired spatial learning ability. To evaluate the ability of verbal communication in Fabp4 KO mice, we examined the ultrasonic vocalization of the pups when they were separated from the dam at P5, P7, P10 and P14. While we found no significant differences in the number of calls between the WT littermates and Fabp4 KO mice at any stage (Fig. 3F), Fabp4 KO mice showed significant increases in the duration of total calls (Fig. 3G and H) and in several subtype calls (Supplementary Fig. 5A and B) at P5. We found that the ratio of calls with a peak frequency maximum above 90 kHz was elevated in Fabp4 KO mice compared to their WT littermates at P5 (Fig. 3I and Supplementary Fig. 5C). These findings are consistent with the report that crying in infants with ASD has relatively higher frequencies (Esposito et al., 2017). No significant differences were observed in the other behavioural batteries (Supplementary Table 9). Collectively, Fabp4 KO mice showed some behavioural phenotypes of ASD.
Figure 3.
Behavioural analyses in Fabp4 KO mice. (A, B) Three-chamber social behaviour test. (A) Schematic diagram of the equipment used in the three-chamber social behaviour test. (B) Left panel shows the total time spent in the stranger mouse area and the novel object area. Right panel shows the number of intrusions into the stranger mouse area and the novel object area. P-values were calculated using two-tailed Mann–Whitney U tests. (C, D) Morris water maze test. (C) The Morris water maze examination schedule. After 5 days of training, a probe test was performed. AL = adjacent left; AR = adjacent right (upper panel); OQ = opposite quadrant; TQ = target quadrant. In the lower panel, a representative example of the behavioural patterns exhibited by mice of each genotype during the 10 min of the study is shown. (D) The upper panel shows the total length of the stay (s) in each quadrant area during the probe test. The lower panel shows the number of times the quadrant area was crossed over during the probe test. P-values were calculated using one-way ANOVA, followed by post hoc Tukey’s test. (E–H) Ultrasonic vocalization analysis of Fabp4 KO pups. (E) Total number of calls at postnatal day 5 (P5), P7, P10 and P14. (F) Detailed spectral features of a call. (G) Average of duration of calls at P5. P-values were calculated using two-tailed Mann–Whitney U tests. (H) Ratio of calls above 90 Hz at P5. P-values were calculated using two-tailed Mann–Whitney U tests.
Increased spine density in Fabp4 KO mice
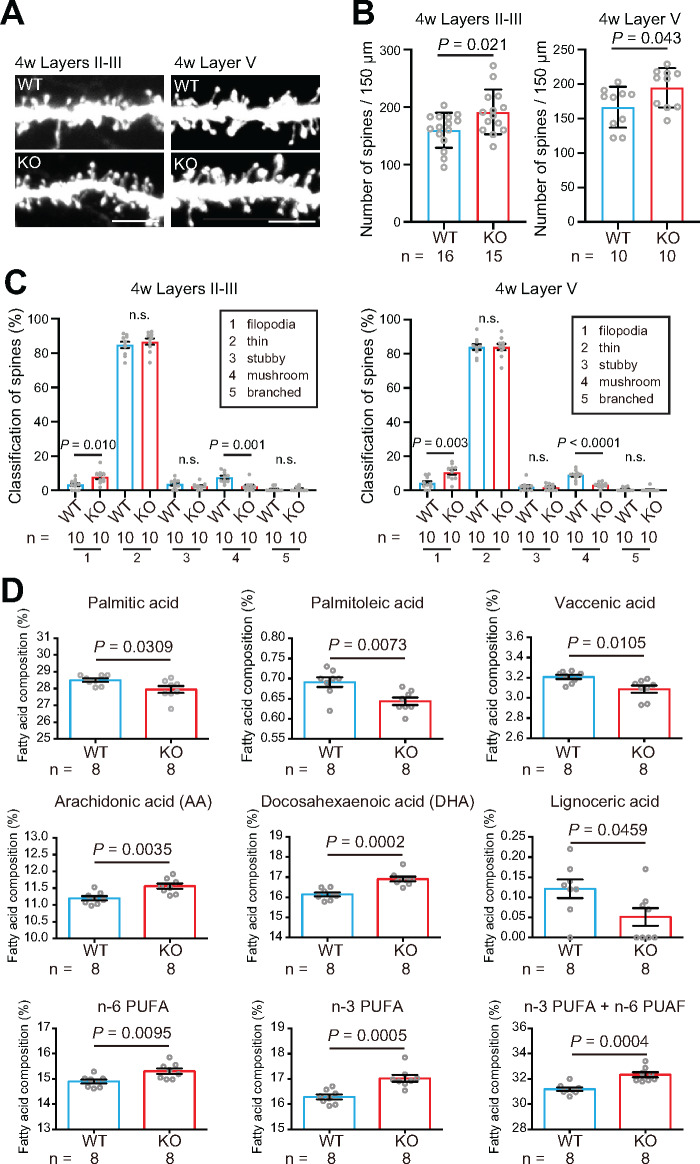
The spine density on apical dendrites of pyramidal neurons has been found to be increased in layers II–III and layer V of the temporal lobe in individuals with ASD (Hutsler and Zhang, 2010). To investigate the effects of Fabp4 loss-of-function, we analysed spine density in the brains of 4-week-old Fabp4 KO mice. The spine density on apical dendrites of pyramidal neurons was significantly increased in layers II–III and layer V in the temporal cortex of Fabp4 KO mice compared to that of their WT littermates (Fig. 4A and B). We obtained similar results in 8-week-old Fabp4 KO mice (Supplementary Fig. 6). These results indicate that Fabp4 KO mice also emulate ASD-like phenotypes with respect to spine histology. Next, we morphologically classified the dendritic spines into five categories: filopodia, thin, stubby, mushroom and branched. Four-week-old Fabp4 KO mice showed significantly fewer mushroom spines (mature form) with a concomitant increase in the number of filopodia spines (immature) (Fig. 4C), suggesting that a systemic loss of Fabp4 affects the maturation of spines in the brain. However, spine analysis of brain regions other than the temporal cortex is required.
Figure 4.
Spine density and FA composition of the cortex of Fabp4 KO mice. (A, B) Spine density on apical dendrites in layers II–III and layer V of the temporal cortex at 4 weeks. P-values were calculated using unpaired t-tests. Scale bar: 5 μm (C) Morphological classification of spines on apical dendrites (filopodia, thin, stubby, mushroom and branched). P-values were calculated using unpaired t-tests. (D) FA composition in the cortex at 4 weeks. P-values were calculated using two-tailed Mann–Whitney U tests. PUFA, polyunsaturated fatty acid.
Abnormalities in the fatty acid composition of the brain and peripherals in Fabp4 KO mice
To investigate the effects of Fabp4 loss-of-function on the FA composition in the cortex, we measured the FAs in the phospholipids in the cortex of 4-week-old Fabp4 KO mice. The percentage ratios of arachidonic acid (20:4n − 6) and docosahexaenoic acid (22:6n − 3), and the percentage of n − 6 polyunsaturated fatty acid, n − 3 polyunsaturated fatty acid and total polyunsaturated fatty acid (n − 3 polyunsaturated fatty acid + n − 6 polyunsaturated fatty acid) in total FAs were found to increase in the cortex of Fabp4 KO mice. On the other hand, the percentages of palmitic acid (16:0), palmitoleic acid (16:1 n − 7), vaccenic acid (18:1 n − 7) and lignoceric acid (24:0) in total FAs were decreased in the cortex of Fabp4 KO mice (Fig. 4D). No significant differences were observed in the percentages of other FAs in total FAs (Supplementary Fig. 7).
We also examined the FA composition in the erythrocyte membrane fractions (RBC) and plasma. The percentages of linoleic acid (18:2n − 6, LA) and alpha-linolenic acid (18:3n − 3) in the total FAs were increased and the percentages of docosatetraenoic acid (22:4n − 6) and nervonic acid (24:1n − 9) in the total FAs were decreased in Fabp4 KO mice (Supplementary Table 10). The percentage of docosapentaenoic acid (22:5n − 3) in the total FAs was increased in the plasma of the gene-deficient mice (Supplementary Table 10).
Collectively, the loss-of-function of the Fabp4 gene can elicit changes in the FA composition in the phospholipids both, in the brain and the peripherals, suggesting that Fabp4/FABP4 may play a role in lipid dynamics. However, the altered FA species varied between tissues.
Discussion
In the present study, we focused on the ‘adipo-brain axis’ as a potential pathophysiology of ASD by examining the serum levels of adipokines and other metabolic markers. Genome-wide association studies have repeatedly reported on the overlap between genetic risk variants in ASD and obesity (Bachmann-Gagescu et al., 2010; Walters et al., 2010; Shinawi et al., 2011; Lee et al., 2012; Sharma et al., 2013; Cortes and Wevrick, 2018; Grove et al., 2019). In addition, a low birth weight has been reported to increase the risk of ASD (Schendel and Bhasin, 2008; Lampi et al., 2012).
We found here that the age-dependent trajectories of adipokine levels were different between ASD and TD children. In particular, the FABP4 levels were found to be significantly lower in ASD children than in TD children in early childhood (2–6 years old). In addition, we have obtained data for the potential of FABP4 as a pathogenetic underpinning factor and an early diagnostic biomarker for ASD.
Regarding the mechanistic role of FABP4 in ASD, we performed exon resequencing analysis of this gene. As a result, two novel and functional variants of the FABP4 gene were identified, although they were rare. In a previous large-scale exome sequencing study, a de novo missense variant in FABP4 was also found in an individual with ASD (Satterstrom et al., 2020). Moreover, in the same study, 102 risk genes of ASD were identified by exome sequencing, most of which were involved in synaptic functions and gene expression regulation (Satterstrom et al., 2020). The protein–protein interaction network between FABP4 and these 102 candidates revealed that FABP4 interacts with CREBBP, MED13L, PTEN and NCOA1 (Supplementary Fig. 8), which in turn interact with other proteins. This suggests a potential role of FABP4-related network in the manifestation of ASD pathogenesis/phenotype, at least in a subset of ASD.
To corroborate the genetic contribution of FABP4 to the pathogenesis of ASD, we analysed phenotypes of Fabp4 gene-disrupted mice. Fabp4 KO mice exhibited deficits in social behaviour (Fig. 3A and B), in accordance with previous studies in other ASD mouse models (e.g. Shank3 haploinsufficient mice, Syngap1 haploinsufficient mice and Chd8 haploinsufficient mice) (Bozdagi et al., 2010; Berryer et al., 2016; Katayama et al., 2016). Fabp4 KO mice displayed an increased density of immature spines. It is worth noting that this spine phenotype is similar to the phenotype of Fmr1 KO mice modelling ASD (Galvez and Greenough, 2005). Collectively, these results indicate that the Fabp4 KO mice displayed ASD-like features and may therefore be useful as a new mouse model for ASD.
The cortex of Fabp4 KO mice showed abnormalities in terms of FA composition, including significant increases in docosahexaenoic acid and arachidonic acid. Arachidonic acid and docosahexaenoic acid are known to function as endogenous ligands of the nuclear receptor peroxisome proliferator-activated receptor (Ppar) (Harmon et al., 2011), the activation of which has been reported to increase spine density by regulating downstream genes (Brodbeck et al., 2008; Patel et al., 2018). The over-activation of Ppar elicited by an increase in the arachidonic acid and docosahexaenoic acid contents in the cortex of Fabp4 KO animals may result in abnormal spine formation. On the other hand, palmitic acid levels were decreased in the cortex of Fabp4 KO mice. palmitic acid is involved in the S-palmitoylation of multiple synaptic proteins (Zareba-Koziol et al., 2018). Thus, the reduction of palmitic acid in the cortex of Fabp4 KO mice may also play a role in abnormal spine formation.
The dysregulated transport of FAs from the blood to the brain may play a crucial role in the abnormal composition of FA in Fabp4 KO mice, and potentially in ASD pathophysiology. It has been reported that FABP5, another member of the FABP superfamily, is expressed in brain endothelial cells and is involved in the uptake and subsequent blood–brain barrier (BBB) transport of docosahexaenoic acid (Pan et al., 2015). FABP4 is expressed in the same cell population (Elmasri et al., 2009). Therefore, we speculate that FABP4 may also contribute to the transport of FA across the BBB and play a role in ASD pathogenesis.
Our previous studies showed that genetic variations of the brain-expressed FABP genes (FABP3, FABP5 and FABP7) are involved in the pathogenesis of schizophrenia and ASD (Watanabe et al., 2007; Iwayama et al., 2010; Maekawa et al., 2010; Shimamoto et al., 2014). Some variants in these genes affected ligand-binding properties (Shimamoto et al., 2014). The current study also found that a missense variant of FABP4 affects ligand-binding properties. Thus, we speculate that peripheral FABP4 may contribute to the pathogenesis of ASD in an indirect manner by altering the fatty acid composition in the brain.
With respect to the lowered serum levels of FABP4 in ASD, environmental factors are likely to play a substantial role, given the rarity of functional variants and no significant genome-wide association study signal in the FABP4 gene locus (Grove et al., 2019). It has been shown that low birth weight, prematurity and ‘small for gestational age’ increase the risk for ASD (Schendel and Bhasin, 2008; Lampi et al., 2012). In a mouse model, an abnormal premature leptin surge was observed in offspring that had experienced malnutrition in utero (Yura et al., 2005). Considering the links of abnormal in utero growth to ASD and to aberrant adipose tissue development, it would be an important future issue to retrospectively examine whether ASD children with low FABP4 levels suffered from malnutrition in foetal stage.
There are several limitations to the present study. First, the sample size used to evaluate the serum concentration of FABP4 protein was relatively small in the current study. Therefore, testing the FABP4 levels in a larger sample size should help us to clarify the clinical or physiological nature of ASD with lowered FABP4. Second, our results were obtained only from Japanese sample sets. Since there are differences in adipose tissue development between different races (Heymsfield et al., 2016), the examination of other ethnic samples, as well as with larger sizes, will be needed to establish serum FABP4 levels as a reliable and universal biomarker. Third, the mechanism for the existence of a time window of lowered FABP4 levels in ASD remains unknown. Fourth, regarding the role of FABP4, the details of the ‘adipo-brain axis’ have yet to be elucidated. To address the third and fourth issues, the generation and analysis of conditional knockout mice of Fabp4 in a spatio (tissue-specific)-temporal (developmental stage-specific) manner will be helpful. It will also be interesting to study whether the alterations in FA composition observed in Fabp4 KO mice are also found in the brains of ASD patients. Lastly, aberrant dendrite morphology was reported in the postmortem brain from subjects with ASD (Raymond et al., 1995; Mukaetova-Ladinska et al., 2004) and in the model animals of ASD including Fmr1 KO mice (Thomas et al., 2008), CGG knock-in mice with expanded CGG repeats in the 5'-UTR of the Fmr1 gene (Berman et al., 2012) and Mecp2-defiecient mice (Belichenko et al., 2009; Nguyen et al., 2012). Therefore, detailed morphological analysis of dendrites and spines in the different cortical regions of Fabp4 KO mice remains an important issue for the future study.
In summary, in this study, the development of adipose tissue was found to be dysregulated in ASD children, as exemplified by the differential trajectories of serum adipokine levels between ASD and TD subjects. Importantly, this indicated that FABP4 may be a novel player in ASD pathogenesis and a useful biomarker for ASD at early stages, shaping the role of ‘adipo-brain axis’ in ASD (Fig. 5). Although the replication of the present finding will be necessary, it would also be useful to perform a prospective cohort study on newborns to examine whether the levels of FABP4 at birth can be used to predict the future manifestation of ASD.
Figure 5.
Illustration of ‘adipo-brain axis’ leveraged by FABP4 for a pathogenesis of ASD. AA = arachidonic acid; BBB = blood–brain barrier; DHA = docosahexaenoic acid; PA = palmitic acid.
Supplementary material
Supplementary material is available at Brain Communications online.
Supplementary Material
Acknowledgements
We thank the participants for clinical analyses in this study. We thank Dr. Keisuke Wakusawa at Department of Rehabilitation, Miyagi Children’s Hospital, Drs. Daisuke Kurita and Tomoyasu Wakuda at Department of Psychiatry, Hamamatsu University School of Medicine and Dr. Manabu Makinodan for their help to collect serum samples. We thank Professor Gökhan S. Hotamışlıgil for the gift of Fabp4 KO mice. We also thank Dr. Takashi Namba for helpful comments on the manuscript. We are grateful to the Support Unit for Bio-Material Analysis and Animal Resources Development, Research Resources Division, RIKEN Center for Brain Science, for sequencing service and animal care.
Funding
This work was supported by the Strategic Research Program for Brain Sciences from AMED (Japan Agency for Medical Research and Development) under Grant Numbers JP19dm0107129 (M.M.) and JP19dm0107083 (T.Y.), by JSPS KAKENHI under Grant Number JP18K07578 (M.M.) and by the Grant-in-Aid for Scientific Research on Innovative Areas from the MEXT under Grant Number JP18H05435 (T.Y.). In addition, this study was supported by grants from the SENSHIN Medical Research Foundation (M.M.).
Competing interests
The authors report no competing interests.
Glossary
- ASD =
autism spectrum disorder
- FA =
fatty acid
- FABP4 =
fatty acid binding protein 4
- MCP-1 =
monocyte chemotactic protein-1
- TD =
typically developing
References
- Aeberli I, Beljean N, Lehmann R, l'Allemand D, Spinas GA, Zimmermann MB. The increase of fatty acid-binding protein aP2 in overweight and obese children: interactions with dietary fat and impact on measures of subclinical inflammation. Int J Obes 2008; 32: 1513–20. [DOI] [PubMed] [Google Scholar]
- Ashwood P, Krakowiak P, Hertz-Picciotto I, Hansen R, Pessah IN, Van de Water J. Associations of impaired behaviors with elevated plasma chemokines in autism spectrum disorders. J Neuroimmunol 2011; 232: 196–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwood P, Kwong C, Hansen R, Hertz-Picciotto I, Croen L, Krakowiak P, et al. Brief report: plasma leptin levels are elevated in autism: association with early onset phenotype? J Autism Dev Disord 2008; 38: 169–75. [DOI] [PubMed] [Google Scholar]
- Bachmann-Gagescu R, Mefford HC, Cowan C, Glew GM, Hing AV, Wallace S, et al. Recurrent 200-kb deletions of 16p11.2 that include the SH2B1 gene are associated with developmental delay and obesity. Genet Med 2010; 12: 641–7. [DOI] [PubMed] [Google Scholar]
- Balan S, Iwayama Y, Maekawa M, Toyota T, Ohnishi T, Toyoshima M, et al. Exon resequencing of H3K9 methyltransferase complex genes, EHMT1, EHTM2 and WIZ, in Japanese autism subjects. Mol Autism 2014; 5: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Kashima I, Rehwinkel J, Sauliere J, Wittkopp N, Izaurralde E. mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett 2007; 581: 2845–53. [DOI] [PubMed] [Google Scholar]
- Belichenko PV, Wright EE, Belichenko NP, Masliah E, Li HH, Mobley WC, et al. Widespread changes in dendritic and axonal morphology in Mecp2-mutant mouse models of Rett syndrome: evidence for disruption of neuronal networks. J Comp Neurol 2009; 514: 240–58. [DOI] [PubMed] [Google Scholar]
- Berman RF, Murray KD, Arque G, Hunsaker MR, Wenzel HJ. Abnormal dendrite and spine morphology in primary visual cortex in the CGG knock-in mouse model of the fragile X premutation. Epilepsia 2012; 53: 150–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berryer MH, Chattopadhyaya B, Xing P, Riebe I, Bosoi C, Sanon N, et al. Decrease of SYNGAP1 in GABAergic cells impairs inhibitory synapse connectivity, synaptic inhibition and cognitive function. Nat Commun 2016; 7: 13340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blardi P, de Lalla A, Ceccatelli L, Vanessa G, Auteri A, Hayek J. Variations of plasma leptin and adiponectin levels in autistic patients. Neurosci Lett 2010; 479: 54–7. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol 1959; 37: 911–7. [DOI] [PubMed] [Google Scholar]
- Bozdagi O, Sakurai T, Papapetrou D, Wang X, Dickstein DL, Takahashi N, et al. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Mol Autism 2010; 1: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodbeck J, Balestra ME, Saunders AM, Roses AD, Mahley RW, Huang Y. Rosiglitazone increases dendritic spine density and rescues spine loss caused by apolipoprotein E4 in primary cortical neurons. Proc Natl Acad Sci USA 2008; 105: 1343–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes HD, Wevrick R. Genetic analysis of very obese children with autism spectrum disorder. Mol Genet Genomics 2018; 293: 725–36. [DOI] [PubMed] [Google Scholar]
- Courchesne E, Campbell K, Solso S. Brain growth across the life span in autism: age-specific changes in anatomical pathology. Brain Res 2011; 1380: 138–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre-Ubieta L, Won HJ, Stein JL, Geschwind DH. Advancing the understanding of autism disease mechanisms through genetics. Nat Med 2016; 22: 345–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dencker M, Danielson A, Karlsson MK, Wollmer P, Andersen LB, Thorsson O. Total body fat, abdominal fat, body fat distribution and surrogate markers for health related to adipocyte fatty acid-binding protein (FABP4) in children. J Pediatr Endocrinol Metab 2017; 30: 375–82. [DOI] [PubMed] [Google Scholar]
- Elmasri H, Karaaslan C, Teper Y, Ghelfi E, Weng M, Ince TA, et al. Fatty acid binding protein 4 is a target of VEGF and a regulator of cell proliferation in endothelial cells. FASEB J 2009; 23: 3865–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G, Hiroi N, Scattoni ML. Cry, baby, cry: expression of distress as a biomarker and modulator in autism spectrum disorder. Int J Neuropsychopharmacol 2017; 20: 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkmer T, Anderson K, Falkmer M, Horlin C. Diagnostic procedures in autism spectrum disorders: a systematic literature review. Eur Child Adolesc Psychiatry 2013; 22: 329–40. [DOI] [PubMed] [Google Scholar]
- Fujita-Shimizu A, Suzuki K, Nakamura K, Miyachi T, Matsuzaki H, Kajizuka M, et al. Decreased serum levels of adiponectin in subjects with autism. Prog Neuropsychopharmacol Biol Psychiatry 2010; 34: 455–8. [DOI] [PubMed] [Google Scholar]
- Furuhashi M, Hotamisligil GS. Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat Rev Drug Discov 2008; 7: 489–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez R, Greenough WT. Sequence of abnormal dendritic spine development in primary somatosensory cortex of a mouse model of the fragile X mental retardation syndrome. Am J Med Genet A 2005; 135A: 155–60. [DOI] [PubMed] [Google Scholar]
- Gillilan RE, Ayers SD, Noy N. Structural basis for activation of fatty acid-binding protein 4. J Mol Biol 2007; 372: 1246–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove J, Ripke S, Als TD, Mattheisen M, Walters RK, Won H, et al. ; Autism Spectrum Disorder Working Group of the Psychiatric Genomics Consortium. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet 2019; 51: 431–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib A, Harris L, Pollick F, Melville C. A meta-analysis of working memory in individuals with autism spectrum disorders. PLoS One 2019; 14: e0216198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon GS, Lam MT, Glass CK. PPARs and lipid ligands in inflammation and metabolism. Chem Rev 2011; 111: 6321–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymsfield SB, Peterson CM, Thomas DM, Heo M, Schuna JM. Why are there race/ethnic differences in adult body mass index-adiposity relationships? A quantitative critical review. Obes Rev 2016; 17: 262–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil GS, Johnson RS, Distel RJ, Ellis R, Papaioannou VE, Spiegelman BM. Uncoupling of obesity from insulin resistance through a targeted mutation in aP2, the adipocyte fatty acid binding protein. Science 1996; 274: 1377–9. [DOI] [PubMed] [Google Scholar]
- Hutsler JJ, Zhang H. Increased dendritic spine densities on cortical projection neurons in autism spectrum disorders. Brain Res 2010; 1309: 83–94. [DOI] [PubMed] [Google Scholar]
- Iwayama Y, Hattori E, Maekawa M, Yamada K, Toyota T, Ohnishi T, et al. Association analyses between brain-expressed fatty-acid binding protein (FABP) genes and schizophrenia and bipolar disorder. Am J Med Genet B 2010; 153B: 484–93. [DOI] [PubMed] [Google Scholar]
- Katayama Y, Nishiyama M, Shoji H, Ohkawa Y, Kawamura A, Sato T, et al. CHD8 haploinsufficiency results in autistic-like phenotypes in mice. Nature 2016; 537: 675–9. [DOI] [PubMed] [Google Scholar]
- Krzystek-Korpacka M, Patryn E, Bednarz-Misa I, Mierzchala M, Hotowy K, Czapinska E, et al. Circulating adipocyte fatty acid-binding protein, juvenile obesity, and metabolic syndrome. J Pediatr Endocrinol Metab 2011; 24: 921–8. [DOI] [PubMed] [Google Scholar]
- Kurosaki T, Maquat LE. Nonsense-mediated mRNA decay in humans at a glance. J Cell Sci 2016; 129: 461–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MC, Lombardo MV, Baron-Cohen S. Autism. Lancet 2014; 383: 896–910. [DOI] [PubMed] [Google Scholar]
- Lampi KM, Lehtonen L, Tran PL, Suominen A, Lehti V, Banerjee PN, et al. Risk of autism spectrum disorders in low birth weight and small for gestational age infants. J Pediatr 2012; 161: 830–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Couteur A, Haden G, Hammal D, McConachie H. Diagnosing autism spectrum disorders in pre-school children using two standardised assessment instruments: the ADI-R and the ADOS. J Autism Dev Disord 2008; 38: 362–72. [DOI] [PubMed] [Google Scholar]
- Lee AW, Hengstler H, Schwald K, Berriel-Diaz M, Loreth D, Kirsch M, et al. Functional inactivation of the genome-wide association study obesity gene neuronal growth regulator 1 in mice causes a body mass phenotype. PLoS One 2012; 7: e41537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C, Rutter M, Le Couteur A. Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord 1994; 24: 659–85. [DOI] [PubMed] [Google Scholar]
- Luo L, Liu M. Adipose tissue in control of metabolism. J Endocrinol 2016; 231: R77–R99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa M, Iwayama Y, Arai R, Nakamura K, Ohnishi T, Toyota T, et al. Polymorphism screening of brain-expressed FABP7, 5 and 3 genes and association studies in autism and schizophrenia in Japanese subjects. J Hum Genet 2010; 55: 127–30. [DOI] [PubMed] [Google Scholar]
- Maekawa M, Yamada K, Toyoshima M, Ohnishi T, Iwayama Y, Shimamoto C, et al. Utility of scalp hair follicles as a novel source of biomarker genes for psychiatric illnesses. Biol Psychiatry 2015; 78: 116–25. [DOI] [PubMed] [Google Scholar]
- Maquat LE. Nonsense-mediated mRNA decay in mammals. J Cell Sci 2005; 118: 1773–6. [DOI] [PubMed] [Google Scholar]
- Mataga N, Mizuguchi Y, Hensch TK. Experience-dependent pruning of dendritic spines in visual cortex by tissue plasminogen activator. Neuron 2004; 44: 1031–41. [DOI] [PubMed] [Google Scholar]
- McGrath J, Johnson K, Ecker C, O'Hanlon E, Gill M, Gallagher L, et al. Atypical visuospatial processing in autism: insights from functional connectivity analysis. Autism Res 2012; 5: 314–30. [DOI] [PubMed] [Google Scholar]
- Mukaetova-Ladinska EB, Arnold H, Jaros E, Perry R, Perry E. Depletion of MAP2 expression and laminar cytoarchitectonic changes in dorsolateral prefrontal cortex in adult autistic individuals. Neuropathol Appl Neurobiol 2004; 30: 615–23. [DOI] [PubMed] [Google Scholar]
- Mukai H, Hatanaka Y, Mitsuhashi K, Hojo Y, Komatsuzaki Y, Sato R, et al. Automated analysis of spines from confocal laser microscopy images: application to the discrimination of androgen and estrogen effects on spinogenesis. Cereb Cortex 2011; 21: 2704–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen MV, Du F, Felice CA, Shan X, Nigam A, Mandel G, et al. MeCP2 is critical for maintaining mature neuronal networks and global brain anatomy during late stages of postnatal brain development and in the mature adult brain. J Neurosci 2012; 32: 10021–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T, Watanabe A, Ohba H, Iwayama Y, Maekawa M, Yoshikawa T. Behavioral analyses of transgenic mice harboring bipolar disorder candidate genes, IMPA1 and IMPA2. Neurosci Res 2010; 67: 86–94. [DOI] [PubMed] [Google Scholar]
- Pan Y, Scanlon MJ, Owada Y, Yamamoto Y, Porter CJ, Nicolazzo JA. Fatty acid-binding protein 5 facilitates the blood-brain barrier transport of docosahexaenoic acid. Mol Pharmaceutics 2015; 12: 4375–85. [DOI] [PubMed] [Google Scholar]
- Parimisetty A, Dorsemans A-C, Awada R, Ravanan P, Diotel N, Lefebvre d’Hellencourt C. Secret talk between adipose tissue and central nervous system via secreted factors-an emerging frontier in the neurodegenerative research. J Neuroinflammation 2016; 13:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D, Roy A, Kundu M, Jana M, Luan C-H, Gonzalez FJ, et al. Aspirin binds to PPARa to stimulate hippocampal plasticity and protect memory. Proc Natl Acad Sci USA 2018; 115: E7408–E17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice KJ, Saksi J, Hotamisligil GS. Adipokine FABP4 integrates energy stores and counterregulatory metabolic responses. J Lipid Res 2019; 60: 734–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond GV, Bauman ML, Kemper TL. Hippocampus in autism: a Golgi analysis. Acta Neuropathol 1995; 91: 117–9. [DOI] [PubMed] [Google Scholar]
- Rylaarsdam L, Guemez-Gamboa A. Genetic causes and modifiers of autism spectrum disorder. Front Cell Neurosci 2019; 13: 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An JY, et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell 2020; 180: 568–84 e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schendel D, Bhasin TK. Birth weight and gestational age characteristics of children with autism, including a comparison with other developmental disabilities. Pediatrics 2008; 121: 1155–64. [DOI] [PubMed] [Google Scholar]
- Sharma JR, Arieff Z, Gameeldien H, Davids M, Kaur M, van der Merwe L. Association analysis of two single-nucleotide polymorphisms of the RELN gene with autism in the South African population. Genet Test Mol Biomarkers 2013; 17: 93–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto C, Ohnishi T, Maekawa M, Watanabe A, Ohba H, Arai R, et al. Functional characterization of FABP3, 5 and 7 gene variants identified in schizophrenia and autism spectrum disorder and mouse behavioral studies. Hum Mol Genet 2014; 23: 6495–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinawi M, Sahoo T, Maranda B, Skinner SA, Skinner C, Chinault C, et al. 11p14.1 microdeletions associated with ADHD, autism, developmental delay, and obesity. Am J Med Genet A 2011; 155: 1272–80. [DOI] [PubMed] [Google Scholar]
- Thomas CC, Combe CL, Dyar KA, Inglis FM. Modest alterations in patterns of motor neuron dendrite morphology in the Fmr1 knockout mouse model for fragile X. Int J Dev Neurosci 2008; 26: 805–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RG, Jacquemont S, Valsesia A, de Smith AJ, Martinet D, Andersson J, et al. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature 2010; 463: 671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A, Toyota T, Owada Y, Hayashi T, Iwayama Y, Matsumata M, et al. Fabp7 maps to a quantitative trait locus for a schizophrenia endophenotype. PLoS Biol 2007; 5: e297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub K. Autism counts. Nature 2011; 479: 22–4. [DOI] [PubMed] [Google Scholar]
- Wolff JJ, Gu H, Gerig G, Elison JT, Styner M, Gouttard S, et al. the IBIS Network. Differences in white matter fiber tract development present from 6 to 24 months in infants with autism. Am J Psychiatry 2012; 169: 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yura S, Itoh H, Sagawa N, Yamamoto H, Masuzaki H, Nakao K, et al. Role of premature leptin surge in obesity resulting from intrauterine undernutrition. Cell Metab 2005; 1: 371–8. [DOI] [PubMed] [Google Scholar]
- Zareba-Koziol M, Figiel I, Bartkowiak-Kaczmarek A, Wlodarczyk J. Insights into protein S-palmitoylation in synaptic plasticity and neurological disorders: potential and limitations of methods for detection and analysis. Front Mol Neurosci 2018; 11:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman AW, Veerkamp JH. New insights into the structure and function of fatty acid-binding proteins. Cell Mol Life Sci 2002; 59: 1096–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting these findings are available upon request.