Abstract
Intervertebral disc degeneration (IDD) is the most common cause of low-back pain. Accumulating evidence indicates that the expression profiling of noncoding RNAs (ncRNAs), including microRNAs (miRNAs), circular RNAs (circRNAs), and long noncoding RNAs (lncRNAs), are different between intervertebral disc tissues obtained from healthy individuals and patients with IDD. However, the roles of ncRNAs in IDD are still unclear until now. In this review, we summarize the studies concerning ncRNA interactions and regulatory functions in IDD. Apoptosis, aberrant proliferation, extracellular matrix degradation, and inflammatory abnormality are tetrad fundamental pathologic phenotypes in IDD. We demonstrated that ncRNAs are playing vital roles in apoptosis, proliferation, ECM degeneration, and inflammation process of IDD. The ncRNAs participate in underlying mechanisms of IDD in different ways. MiRNAs downregulate target genes’ expression by directly binding to the 3′-untranslated region of mRNAs. CircRNAs and lncRNAs act as sponges or competing endogenous RNAs by competitively binding to miRNAs and regulating the expression of mRNAs. The lncRNAs, circRNAs, miRNAs, and mRNAs widely crosstalk and form complex regulatory networks in the degenerative processes. The current review presents novel insights into the pathogenesis of IDD and potentially sheds light on the therapeutics in the future.
Keywords: Apoptosis, Cell proliferation, Extracellular matrix degeneration, Intervertebral disc degeneration, Inflammation, Noncoding-RNA, Nucleus pulposus cell
Background
Intervertebral disc degeneration (IDD) is the most common cause of low-back pain, which affects over 70% of people at some points of their whole lifetime [1–3]. However, due to the poor understandings of the pathogenesis of the disorder, few treatment regimens have been put forward, and none of the current clinical interventions for IDD has been confirmed as efficient and radical treatment modalities [3–5]. Therefore, an in-depth investigation of the regulatory machinery of IDD is urgently needed in the present.
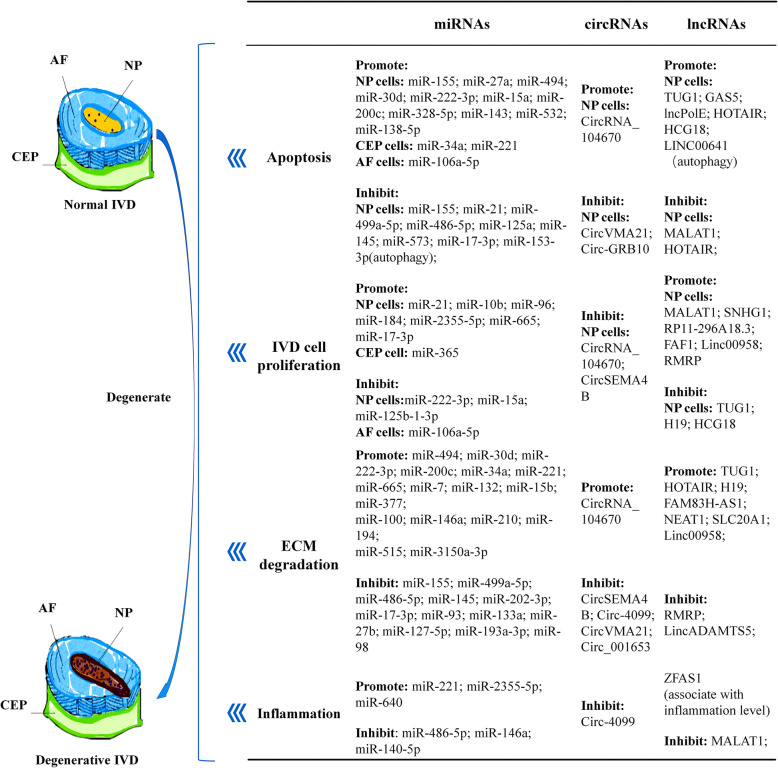
Intervertebral disc (IVD) can be divided into three morphologically distinct regions, i.e., the sandwiched central nucleus pulposus (NP), peripheral annulus fibrosus (AF), and cranial or caudal cartilaginous endplate (CEP) (Fig. 1). During the process of IDD, the apoptosis of IVD cells is abnormally increased with the cells aberrantly clustering, dysregulation of extracellular matrix (ECM) proteins (abnormally synthesized and/or degraded), and excessive expression of inflammatory factors which accelerate the formation of inflammatory microenvironment/niche and eventually violate the adjacent IVD cells [6–10]. These pathophysiological processes result in a vicious circle of progressive aggravation of degeneration.
Fig. 1.
Noncoding RNAs involved in multiple pathological processes of IDD development (apoptosis, ECM degradation, cell proliferation, and inflammation)
Emerging evidence reveals that genetic and environmental factors are both influencing factors of IDD, whereas genetic factors seem to be the outweighed one. Notably, a multitude of genetic factors, implicating in the underlying regulatory mechanisms, are dysregulated in IDD, especially the noncoding RNAs (ncRNAs) [6, 11, 12]. NcRNAs consist of a large family of RNAs without coding function and outcome as cellular effectors, i.e., proteins. So far, the identified ncRNAs in homo sapiens include miRNAs, circRNAs, lncRNAs, and emerging small RNAs. The expression profiling of ncRNAs of IDD samples is significantly different from those from healthy ones, reflected by differentially expressed levels and types of ncRNAs unraveled by microarray and/or sequencing analyses. It is suggested that ncRNAs are playing vital roles in apoptosis, proliferation, ECM degeneration, and inflammation process of IDD [12–17]. Owing to that, we established the coding-noncoding SuperSeries Datasets as GSE67567 in human IDD, including lncRNAs, mRNAs, and circRNAs, and miRNAs datasets as GSE19943, GSE63492, GSE56081, and GSE67566, as well as studies from other investigators (Table 1). Given the scarcity of studies summarizing the research progress of ncRNAs in IDD, we designed and conducted a review across the published papers [13, 17, 18, 20–25]. In the current work, the state-of-art research advance and therapeutic potentials concerning the regulatory roles of miRNAs/circRNAs/lncRNAs in degenerated discs of human or animal models were summarized and discussed (Fig. 1).
Table 1.
mRNA and ncRNA expression profiles of IDD deposited in gene expression omnibus
| Data accession number | Types of RNA profiling | Platform | BioProject | Samples | Control set | Publication year | Contributors |
|---|---|---|---|---|---|---|---|
| GSE19943 | miRNAs |
Exiqon human miRCURY LNA™ microRNA Array V11.0 |
PRJNA120173 | 3 control (scoliosis) vs. 3 degenerative nucleus pulposus (NP) cell samples, extracted from NP tissue without cultures | 2011 | Wang et al. [18] | |
| GSE45856 | miRNAs |
miRCURY LNA microRNA Array, 6th generation - hsa, mmu & rno |
PRJNA196506 | 3 control (traumatic normal) vs. 3 degenerative IVD tissues using TRIspin method | 2013 | Zhao et al. [19] | |
| GSE56081 |
mRNAs LncRNAs |
Arraystar Human LncRNA microarray V2.0 (Agilent_033010 Probe Name version) |
PRJNA242356 | 5 control (cadaveric normal) vs. 5 degenerative NP tissues using TRIspin method | 2014 | Wan et al. [13] | |
| GSE63492 | miRNAs |
ExiqonmiRCURY LNA microRNA Array miRBase v18.0 |
PRJNA268036 | 5 control (cadaveric normal) vs. 5 degenerative NP tissues using TRIspin method | 2016 | Lan et al. [20] | |
| GSE67566 | circRNAs |
Agilent-069978 Arraystar Human CircRNA microarray V1 |
PRJNA280274 | 5 control (cadaveric normal) vs. 5 degenerative NP tissues using TRIspin method | 2016 | Lan et al. [20] | |
| GSE67567 | Noncoding RNA SuperSeries |
GPL15314 Arraystar Human LncRNA microarray V2.0 (Agilent_033010 Probe Name version) |
PRJNA280271 | In combination | 5 control (cadaveric normal) vs. 5 degenerative NP tissues | 2016 | Lan et al. [20] |
| GSE153761 | lncRNAs, mRNAs and circRNAs |
Agilent-078298 human ceRNA array V1.0 4X180K |
PRJNA643990 | GSM4653870 | 3 control (traumatic normal) vs. 3 degenerative cartilage endplate of cervical disc | 2020 | Yuan et al. |
| GSM4653871 | |||||||
| GSM4653872 | |||||||
| GSM4653873 | |||||||
| GSM4653874 | |||||||
| GSM4653875 |
The regulatory mechanism of miRNAs in IDD
The expression profile and molecular mechanisms of miRNAs in IDD
Accumulating evidence indicates that the miRNA expression profile in IDD cases is significantly different from those in the controls. In 2011, we presented the first line of evidence on miRNA expression profiling in IDD, using scoliosis NP tissues as control. Twenty-nine differentially expressed miRNAs were identified, with 6 upregulated and 23 downregulated [18]. Thereafter, the emerging molecules as miRNAs catch the attention of global researchers, manifesting as an increasing number of published studies. Subsequently, Zhao et al. compared the expression profile of miRNAs between IDD and spinal cord injury patients in 2014. Twenty-six miRNAs were downregulated in the IDD group, while 25 upregulated [19]. Further investigation revealed these dysregulated miRNAs controlled several signaling pathways, which are pivotal in the pathogenesis of IDD, such as Wnt [19, 26, 27], phosphoinositide 3-kinase/Akt (PI3K/Akt) [19, 28], and mitogen-activated protein kinase (MAPK) [19, 29], etc. Consistently, Hu et al. demonstrated that among the 253 miRNAs detected both in IDD and scoliosis samples, three were downregulated and six were upregulated in degenerative samples. The downstream targets were predicted to be genes or proteins associated with degeneration, such as drosophila mothers against decapentaplegic protein family member 4 (SMAD4), which play important roles in cell-cycle-related pathways [30].
Complementary base sequence endows miRNAs the ability to bind the 3′untranslated region (3′UTR) of particular mRNA. The binding of miRNAs and mRNAs results in a decreased expression of the target proteins [31, 32], while most of them are hub proteins, which play a crucial role in essential pathways associated with degeneration. Thus, miRNAs indirectly control the pathological processes in disc degeneration. The IDD-related miRNAs are presented in Table 2.
Table 2.
Experimentally verified miRNAs associated with IDD
| MiRNA | Expression | Target(s) | Functions | Publication year | References |
|---|---|---|---|---|---|
| MiR-155 | ↓ |
FADD, caspase-3 ERK1/2, TCF7L2, MMP-16 |
↓NP cell apoptosis ↓ECM degradation |
2011 2016–2018 |
[18] |
| MiR-21 |
↓* ↑ |
PTEN PTEN, PDCD4 PTEN |
↓NP cell apoptosis ↑NP cell proliferation ↑ECM degradation |
2018 2014,2016 2018 |
[36] [38] |
| MiR-27a | ↑ | PI3K | ↑NP cell apoptosis | 2013 | [39] |
| MiR-499a-5p | ↓ | SOX4 |
↓NP cell apoptosis ↓ECM degradation |
2019 2019 |
[40] [40] |
| MiR-494 | ↑ |
SOX9, JunD SOX9 |
↑NP cell apoptosis ↑ECM degradation |
2015,2017 2017 |
[41] |
| MiR-30d | ↑ | SOX9 |
↑NP cell apoptosis ↑ECM degradation |
2018 2018 |
[43] [43] |
| MiR-222-3p | ↑ | CDKN1B |
↑NP cell apoptosis ↓NP cell proliferation ↑ECM degradation |
2019 2019 2019 |
[44] [44] [44] |
| MiR-15a | ↑ | MAP3K9 |
↑NP cell apoptosis ↓NP cell proliferation |
2017 2017 |
[45] [45] |
| MiR-486-5p | ↓ | FOXO1 |
↓NP cell apoptosis ↓ECM degradation ↓inflammation |
2019 2019 2019 |
[46] [46] [46] |
| MiR-200c | ↑ | XIAP |
↑NP cell apoptosis ↑ECM degradation |
2018 2018 |
[23] [23] |
| MiR-328-5p | ↑ | ERBB2 | ↑NP cell apoptosis | 2018 | [47] |
| MiR-34a | ↑ |
GDF5 Bcl-2 |
↑ECM degradation ↑CEP cell apoptosis |
2016 2015 |
[48] [7] |
| MiR-143 | ↑ | Bcl-2 | ↑NP cell apoptosis | 2017 | [49] |
| MiR-532 | ↑ | Bcl-9 | ↑NP cell apoptosis | 2018 | [50] |
| MiR-125a | ↓ | TP53INP1 | ↓NP cell apoptosis | 2016 | [51] |
| MiR-221 | ↑ |
ERα FOXO3, TRPS1 BMP-Smad pathway |
↑CEP cell apoptosis ↑ECM degradation ↑inflammation ↓chondrogenesis ↓AF cell osteogenic differentiation |
2018 2018 2018 2018 2016 |
[52] [52] [52] [53] [54] |
| MiR-138-5p | ↑ | SIRT1 | ↑NP cell apoptosis | 2016 | [55] |
| MiR-145 | ↓ | ADAM17 |
↓NP cell apoptosis ↓ECM degradation |
2019 2019 |
[56] [56] |
| MiR-573 | ↓ | Bax | ↓NP cell apoptosis | 2019 | [57] |
| MiR-153-3p | ↓ | ATG5 | ↓NP cell autophagy | 2019 | [58] |
| MiR-106a-5p | ↑ | ATG7 |
↑AF cell apoptosis ↓AF cell proliferation |
2019 2019 |
[8] [8] |
| MiR-10b | ↑ | HOXD10 | ↑NP cell proliferation | 2013 | [59] |
| MiR-96 | ↑ | ARID2 | ↑NP cell proliferation | 2017 | [60] |
| MiR-184 | ↑ | GAS1 | ↑NP cell proliferation | 2017 | [61] |
| MiR-2355-5p | ↑ | ERFFI1 |
↑NP cell proliferation ↑inflammation |
2019 2019 |
[62] [62] |
| MiR-365 | ↓ | HDAC4 | ↑CEP cell proliferation | 2019 | [63] |
| MiR-125b-1-3p | ↑ | TSHZ3 | ↓NP cell proliferation | 2018 | [64] |
| MiR-665 | ↑ | GDF5 |
↑NP cell proliferation ↑ECM degradation |
2018 2018 |
[65] [65] |
| MiR-7 | ↑ | GDF5 | ↑ECM degradation | 2016 | [66] |
| MiR-132 | ↑ | GDF5 | ↑ECM degradation | 2017 | [67] |
| MiR-15b | ↑ | SMAD3 | ↑ECM degradation | 2017 | [68] |
| MiR-20a | ↑ | ANKH | ↑CEP chondrocyte calcification | 2016 | [69] |
| MiR-377 | ↑ | ADAMTS5 | ↑ECM degradation | 2013 | [70] |
| MiR-202-3p | ↓ | MMP1 | ↓ECM degradation | 2019 | [71] |
| MiR-17-3p | ↓ | MMP2 |
↓ECM degradation ↓NP cell apoptosis ↑NP cell proliferation |
2018 2018 2018 |
[72] [72] [72] |
| MiR-93 | ↓ | MMP3 | ↓ECM degradation | 2015 | [73] |
| MiR-133a | ↓ | MMP9 | ↓ECM degradation | 2016 | [74] |
| MiR-27b | ↓ | MMP13 | ↓ECM degradation | 2016 | [75] |
| MiR-127-5p | ↓ | MMP13 | ↓ECM degradation | 2017 | [76] |
| MiR-193a-3p | ↓ | MMP14 | ↓ECM degradation | 2016 | [77] |
| MiR-98 | ↓ | IL-6 | ↓ECM degradation | 2016 | [78] |
| MiR-100 | ↑ | FGFR3 | ↑ECM degradation | 2015 | [79] |
| MiR-146a | Not clear** | TRAF6 |
↑ECM degradation ↓inflammation |
2015 2015,2017 |
[22] |
| MiR-210 | ↑ | ATG7 | ↑ECM degradation | 2017 | [81] |
| MiR-194 | ↑ | CHSY1/2/3 | ↑ECM degradation | 2017 | [82] |
| MiR-515 | ↑ | CHSY1/2/3 | ↑ECM degradation | 2017 | [82] |
| MiR-3150a-3p | ↑ | ACAN | ↑ECM degradation | 2018 | [83] |
| MiR-640 | ↑ | LRP1,β-catenin, EP300 | ↑inflammation | 2019 | [84] |
| MiR-140-5p | ↓ | TLR4 | ↓inflammation | 2018 | [85] |
The expression, targets, and functions of miRNAs related to IDD were displayed in Table 2. “↓” represents downregulation, while “↑” represents upregulation
*Decrease in apoptotic NP cells.**It is reported that miR-146a is significantly downregulated in the PBMCs of IDD patients, but its expression in NP cells is unclear [80]
In summary, 49 miRNAs were reported with a relationship to IDD, among the total number of 38,589 miRNAs of Homo sapiens, according to miRBase Release 22.1 (http://www.mirbase.org/). Whereas studies have been focused on intra-cellular miRNAs, cell-free miRNAs emerge as potential novel biomarkers for a variety of human diseases. Recently, exRNA Atlas has been proposed across human biofluids, which is also essential in the regulation of IDD [86].
The roles of miRNAs in IVD cell apoptosis
Accumulating evidence shows that several miRNAs function as inducers or inhibitors in the apoptosis of IVD cells via specific target genes or pathways [87]. For instance, downregulated miR-155 was suggested triggering the Fas-mediated apoptosis by disinhibiting FADD and CASP-3 in NP cells [18]. Similarly, the expression of miR-21 [36], miR-499a-5p [40], miR-486-5p [46], miR-125a [51], miR-145 [56], and miR-573 [57] are decreased in IDD, which act as apoptosis inhibitors via binding to the 3′UTRs of mRNAs of PTEN, SOX4, FOXO1, TP53INP1, ADAM17, and Bax, respectively. In contrast to these findings, miRNAs such as miR-27a [39], miR-494 [41, 42], miR-30d [43], miR-222-3p [44], miR-15a [45], miR-143 [49], miR-532 [50], miR-138-5p [55] in NP cells, miR-106a-5p [8] in AF cells, and miR-34a [7] and miR-221 [52] in CEP cells, display potential pro-apoptotic effects in IDD, via inhibiting the expression of downstream hub proteins in several pathways. Apart from the aforementioned mechanisms, miRNAs, i.e., miR-153-3p, participating in the autophagy, also contributes to the disc degeneration eventually [58].
In summary, there are eight miRNAs acting as inhibitors of apoptosis in IDD, whereas eleven miRNAs act as promoters of apoptosis.
The roles of miRNAs in IVD cell proliferation
Cell number in healthy human IVDs is limited and sparsely distributed. However, the cells were reported to proliferate into clusters in IDD [88]. In this complex pathophysiological process, multiple miRNAs acting as vital indirect regulators in IVD cell proliferation can be employed as biomarkers. For example, in NP cells, the aberrant overexpression of miR-21 increases the proliferation level of degenerated NP cells by downregulating PDCD4 and PTEN. Thus, the disinhibition effect increased the phosphorylation level of c-Jun and AKT proteins, which could induce cell proliferation. Liu et al. found that miR-21 knockdown reversed cell proliferation, while Ly294002, an AKT inhibitor, reversing the effect induced by miR-21. These results indicate that miR-21 is a potential biomarker and therapeutic target of IDD [21, 37].
Besides, overexpression of miR-10b [59], miR-96 [60], miR-184 [61], miR-2355-5p [62], and miR-665 [65] could also promote the proliferation of degenerated NP cells via targeting PTEN/PDCD4, HOXD10, ARID2, GAS1, ERFFI1, and GDF5, while upregulation of miR-222-3p [44], miR-15a [45], and miR-125b-1-3p [64] had an opposite effect by inhibiting the expression of CDKN1B, MAP3K9, and TSHZ3. These downstream genes regulate NP cell proliferation by controlling crucial pathways, such as RhoC-Akt pathway [59], PTEN/AKT pathway [21, 37], ARID2/AKT signaling [60], and activating/deactivating molecular molecules like AKT [21, 37, 61]. Among them, miR-222-3p promotes the proliferation of IVD cells and accelerates the apoptosis and ECM degradation via the same pathway [44]. In accordance with this, miR-15a [45], miR-106a-5p [8] and miR-17-3p [72] have a similar effect, which limits their application as therapeutic targets.
In addition to NP cells, miR-106a-5p [8] in AF cells and miR-365 [63] in CEP cells are also associated with cell proliferation, by inhibiting the proliferation level via ATG7 and increasing proliferation via HDAC4, respectively.
Collectively, there were 12 miRNAs involved in IVD cell proliferation, with eight miRNAs promoting proliferation and four miRNAs inhibiting proliferation.
The roles of miRNAs in ECM degradation and inflammation
Generally, IVD cells play an essential role in secreting ECM components like collagens and proteoglycans to maintain IVD’s structural stability and resist mechanical loads [89, 90]. However, in IDD, the unbalance between synthesis and degradation of ECM makes the IVD unrenewable and degenerative, especially in NP tissues [91]. MiRNAs modulate the degradation of ECM by regulating the expression of essential enzymes such as matrix metalloproteinases (MMPs) or cytokines such as interleukins.
It is reported that inhibition of miR-665 [65], miR-7 [66], miR-132 [67], and miR-34a [48] effectively attenuate ECM degradation in degenerative NP tissues by directly upregulating the expression of growth differentiation factor-5 (GDF5), which can inhibit the expression of ECM catabolic factors, such as MMP and ADAMTS4, and upregulating the production of anabolic proteins, such as type II collagen and aggrecan.
A series of miRNA, i.e., miR-202-3p [71], miR-17-3p [72], miR-93 [73], miR-133a [74], miR-27b [75], miR-127-5p [76], miR-193a-3p [77], and miR-155 [33] are significantly downregulated in degenerative NP tissues, with their expression levels reversely correlated with the grade of IDD, which induce type II collagen synthesis via directly suppressing the expression levels of MMP1, MMP2, MMP3, MMP9, MMP13, MMP14, and MMP16, respectively, whereas overexpression of miRNAs mentioned above can stop and reverse the degradative process, indicating that they are potential biomarkers and therapeutic targets of IDD.
In addition, two different protective mechanisms of miR-155 have been clarified in ECM degradation. Ye et al. have shown that the knockdown of miR-155 results in decreased expressions of collagen II and glycosaminoglycan by increasing the expression of ERK1/2 [34]. Sun et al. have reported that an essential transcription factor, TCF7L2, which acts as an activator in the process of chondrocyte matrix degradation through p65/NF-κB signaling, was repressed by miR-155 [35].
Wang et al. discovered that miR-21 is upregulated in IDD tissues and positively correlated with the degradation grade, which indicates miR-21 cannot only inhibit NP cell apoptosis and promote proliferation as mentioned above, but also promote ECM degradation through repressing the PTEN/AKT/mTOR signaling pathway [38]. SRY-related high-mobility group box (SOX)-4 and SOX9 are respectively targeting molecules of miR-499a-5p [40], miR-494 [41], and miR30d [43], by repressing the apoptosis of NP cells and ECM degradation.
As well, a number of miRNAs can affect the process of ECM degradation, including miR-222-3p [44], miR-486-5p [46], miR-221 [52, 53], miR-145 [56], and miR-98 [78] in NP tissues, miR-221 [54] in AF tissues, and miR-20a [69] in CEP tissues. The expression, targets, and functions of these miRNAs are listed in Table 2.
Apart from apoptosis, proliferation, and ECM degradation, inflammation responses and inflammatory cytokines are also regarded as crucial factors in the pathogenesis of IDD [92]. miRNAs associated with the production of inflammation cytokines, such as miR-486-5p [46], miR-221 [52], miR-2355-5p [62], miR-146a [22, 80], miR-640 [84], and miR-140-5p [85] are also listed in Table 2 and can also be used as therapeutic targets of IDD. In general, there are six reported miRNAs pertaining to inflammation during IDD in various subparts of the IVDs via a multitude of targeting genes, affecting a variety of inflammatory cytokines. Three deregulated miRNAs (miR-140-5p targeting TLR4 [85], miR-486-5p targeting FOXO1 [46] and miR-146a targeting TRAF6 [22, 80]; all studying in NP cells) are associated with decreased levels of inflammation, whereas three miRNAs (miR-221 targeting ERα in CEP cells [52], miR-640 targeting LRP1, β-catenin and EP300 in NP and AF cells [84], and miR-2355-5p targeting ERFFI1 in NP cells [62]) are linked with increased levels of inflammation during IDD.
The regulatory mechanism of circRNAs in IDD
The profile and mechanism of circRNAs in IDD
CircRNAs are a group of single-stranded RNAs with loop structures, which act as competing endogenous RNAs (ceRNAs) and restore the functions of specific genes by sponging miRNAs [17, 93]. A specific miRNA could be sponged by various distinct circRNAs, forming a circRNA-miRNA-mRNA interaction network [20, 47, 72]. Thus, circRNAs seems like a critical regulator in gene expression.
We presented the first line of evidence of circRNAs expression profiling in human IDD In 2016. We found 636 differentially expressed circRNAs in human lumbar IVDs, with 354 upregulated and 282 downregulated [20]. Zou and colleagues indicated that many genes regulated by circRNAs are playing crucial roles in the pathogenesis of IDD, via over 15 signaling pathways, such as Wnt and integrin signaling pathways. Pairs of host genes and circRNA can be divided into four categories according to their profile: circRNA and its host genes downregulated, circRNA and its host genes upregulated, circRNA downregulated and its host genes upregulated, and circRNA upregulated and its host genes downregulated [93].
Several experiments were conducted to investigate the differences between the profile of circRNAs in degenerative IVDs and that in normal IVDs. Wang et al. have provided another line of evidence that 72 circRNAs were upregulated by more than two-fold in degenerative NP tissues [94]. Following this, another team identified there were 7294 circRNAs aberrantly expressed (3724 upregulated, 3570 downregulated, fold change > 2) in degenerative NP cells [17]. Recently, Li et al. reviewed the results from related publications from 2016 to 2019 and confirmed that the profile in IDD patients is different from that in the control group, with the number of upregulated circRNAs ranging from 51 to 3724, and the number of downregulated circRNAs ranging from 21 to 3570 [15].
As the dynamic development of miRBase reflecting novel findings in miRNAs, multiple circRNA databases have been proposed with changing numbers and updated findings as well. So far, there are hundreds of human circRNAs reported (148 in chondrocytes and 104 in osteocytes) [95].
The roles of circRNAs in IDD
Specific circRNA can indirectly regulate apoptosis, proliferation, and ECM degradation by modulating the level of functional miRNA, contributing to the disc degeneration. Specifically, CircVMA21 [23], Circ-GRB10 [47], and CircRNA_104670 [72] are involved in apoptosis regulation. CircRNA_104670 [72] and CircSEMA4B [96] are related to NP cell proliferation. CircVMA21 [23], Circ-4099 [94], CircSEMA4B [96], and CircRNA_104670 [72] are associated with ECM degradation.
As shown in Table 3, miR-200c accelerates the apoptosis of NP cell and ECM degradation via inhibiting XIAP, whereas CircVMA21 alleviates the negative effect of sponging miR-200c. However, in degenerative IVD tissues, the expression of CircVMA21 is repressed, resulting in aberrant higher level of miR-200c and IDD [23]. Circ-GRB10 is downregulated in degenerative NP tissues. Transient overexpression of GRB10 could attenuate the apoptosis of NP cells by sequestering miR-328-5p and leading to the activation of genes associated with proliferation via the ErbB pathway [47].
Table 3.
Experimentally verified circRNAs associated with IDD
| CircRNA | Expression | Target miRNA | Functions | Publication year | References |
|---|---|---|---|---|---|
| CircVMA21 | ↓ | miR-200c |
↓NP cell apoptosis ↓ECM degradation |
2018 2018 |
[23] [23] |
| Circ-GRB10 | ↓ | miR-328-5p | ↓NP cell apoptosis | 2018 | [47] |
| CircSEMA4B | ↓ | miR-431 |
↓NP cell proliferation ↓ECM degradation |
2018 2018 |
[96] [96] |
| CircRNA_104670 | ↑ | miR-17-3p |
↑NP cell apoptosis ↓NP cell proliferation ↑ECM degradation |
2018 2018 2018 |
[72] [72] [72] |
| Circ-4099 | ↑ | miR-616-5p |
↓ECM degradation ↓Inflammation |
2018 2018 |
[94] [94] |
The expression, targets, and functions of circRNAs related to IDD were displayed in Table 3. “↓” represents downregulation, while “↑” represents upregulation
Besides, overexpression of circSEMA4B could inhibit NP cells from proliferation and ECM degradation induced by IL-1β via indirectly rescuing SFRP1 or GSK-3β in Wnt signaling from miR-431 [96]. Song et al. found that upregulated CircRNA_104670 accelerates apoptosis and inhibits cell proliferation and collagen II synthesis in NP cells via circRNA_104670/miR-17-3p/MMP2 network [72]. In contrast, upregulated Circ-4099 acts as a protective factor by disinhibiting the expression of collagen II and aggrecan and downregulating the synthesis of the pro-inflammatory factors such as IL-1β, TNF-α, and PGE2 by sponging miR-616-5p. The expression data, targeted miRNAs, and functions of circRNA in IDD are listed in Table 3.
The regulatory mechanism of lncRNAs in IDD
The profile and mechanism of lncRNAs in IDD
LncRNAs are a group of ncRNAs with more than 200 nucleotides. LncRNAs take the role of ceRNAs (as circRNAs) or small interfering RNA (siRNAs) and participate in the lncRNA/circRNA/miRNA/mRNA network as transcriptional regulators [97]. They regulate gene expression or control the signaling pathways by competitively sponging and inactivating specific miRNAs [12, 13]. Some lncRNAs even regulate the activity or stability of proteins by directly interacting with them [98, 99]. Investigations indicate that lncRNAs exert their regulatory function in various ways (i.e., reducing the methylation level of the promoter region may accelerate the expression of specific lncRNAs in IVD cells [100]). Therefore, the aberrant expression of lncRNAs will cause the degeneration of IVD cells and result in the development of IDD.
Ample evidence indicates that the profile of lncRNAs in degenerative IVDs is totally different from those in normal IVDs. In 2014, we reported the first expression profiling of lncRNAs in human IDD by using the same human lumbar IVD samples as circRNAs. One hundred sixteen lncRNAs (with 67 upregulated and 49 downregulated) and 260 mRNAs were differentially expressed in degenerative samples with an absolute fold change greater than ten [13]. Among the deregulated lncRNAs in IDD, HOTAIR (NR_003716) is the top downregulated lncRNAs (fold change, 148.53; P < 0.001) [13]. Later, Zhao et al. reported that 1530 of 1854 differential expressed lncRNAs might have 6386 potential target genes, whereas Han et al. reported 632 lncRNAs are differentially expressed in IDD tissues among 40,716 detected lncRNAs [101, 102]. Li and colleagues reviewed the articles related to expression profiles of lncRNAs and summarized the number of differentially expressed lncRNAs. The number of upregulated lncRNAs is ranging from 67 to 2234, while the downregulated ones ranging from 49 to 938 [97]. These results indicate that lncRNAs could modulate the destiny of NP cells in IDD and be transformed into screening biomarkers or therapeutic targets.
The roles of lncRNAs in IDD
The roles of lncRNAs in IDD can be divided into four main categories according to their functions (apoptosis, cell proliferation, ECM degradation, inflammation) as well. A specific lncRNA can have two or more functions simultaneously.
Chen et al. found that overexpression of TUG1 in degenerative NP samples accelerates cell apoptosis, via upregulating the levels of Bax&caspase-3 (the latter are pro-apoptotic factors) in Wnt1/β-catenin pathway and downregulating the levels of Bcl-2, an anti-apoptotic factor. In addition, the increased level of TUG1 also deteriorates the degradation of ECM by breaking the expression balance in the ECM-degrading and anti-ECM-degrading genes [24]. Both GAS5 and lncPolE are overexpressed in degenerative IVD samples, displaying similar roles in apoptosis. While GAS5 increases the apoptosis by binding to miR-155, lncPolE negatively regulates PolE [100, 103].
Emerging evidence suggests that autophagy is an essential process in IDD and has a close relationship with apoptosis. Zhang and colleagues reported that overexpression of HOTAIR accelerates NP cell apoptosis via stimulating cell autophagy [104]. On the contrary, Shao et al. indicated that downregulated HOTAIR expression inhibits cell apoptosis via the Notch signaling pathway by sponging miR-34a-5p. In other words, the overexpression of HOTAIR reduces NP cell apoptosis [105]. On account of the incompatible viewpoints, further investigations are needed to clarify the real effects of HOTAIR in apoptosis. In addition to HOTAIR, LINC00641 accelerates cell autophagy by sponging miR-153-3p, which can inactivate autophagy-related gene 5 (ATG5) [58].
Aberrant cell proliferation is another core pathogenesis in IDD. SNHG1 promotes NP cell proliferation via sponging miR-326, and downregulated miR-326 disinhibits NP cell proliferation by inactivating PCNA and cyclin D1 expression. Similarly, RP11-296A18.3/miR-138/HIF1A, RMRP/miR-206/PCNA, H19/miR-22/LEF1/Wnt/β-catenin signaling, and HCG18/miR-146a-5p/TRAF6/NF-κB axis can also increase or decrease the level of proliferation, respectively [10, 25, 106–108]. Targeting extracellular signal-regulated kinase (Erk) and miR-146a-5p/TRAF6/NF-κB axis, respectively, lncRNA FAF1 and HCG18 modulate the ratio of synthesis-phase cells among all the cells in NP tissue [109].
H19, Linc00958, and SLC20A1 have been reported to upregulate ECM degradation via sponging miRNAs [107, 109, 110]. It is noteworthy that H19 plays a role as a competitor to LEF1 for binding miR-22, regulating Wnt/β-catenin pathway [107]. Linc00958 and NEAT1 exert their function by increasing the expression of MMPs via upregulating SMAD and inhibiting the synthesis of aggrecan and collagen-II in the ERK/MAPK pathway, respectively [109, 111]. Wei et al. demonstrated that decreased FAM83H-AS1 in IDD results in ECM degeneration, by targeting Notch1 and Hes1 [112]. While Linc-ADAMTS5, interacting with splicing factor proline/glutamine-rich (SFPQ), which induces the down expression of ADAMTS5, alleviates the ECM deterioration process [113].
Inflammatory cytokines and inflammatory cytokine-related lncRNAs are also involved in IDD. Several members of the interleukin family, such as IL-1 and IL-6, were widely noted as pro-inflammatory factors, giving rise to the degeneration of ECM and apoptosis of IVD cells. In vitro studies showed that overexpression of MALAT1 attenuates IL-1 and IL-6 induced inflammation by sponging miR-503, displaying a protective effect on IVD cell [114]. Besides, ZFAS1 is linked with inflammatory cytokine levels in IDD. Since the positive correlation between the intensity of inflammatory and severity of degeneration, ZFAS1 is regarded as a sensitive predictor of IDD [115].
LncRNAs related to the modulation and prediction of IDD are listed in Table 4.
Table 4.
Experimentally verified lncRNAs associated with IDD
| LncRNA | Expression | Target(s) | Functions | Publication year | References |
|---|---|---|---|---|---|
| TUG1 | ↑ | Wnt1/β-catenin, Bax& caspase-3 |
↑NP cell apoptosis ↓NP cell proliferation ↑ECM degradation |
2017 2017 2017 |
[24] [24] [24] |
| GAS5 | ↑ | miR-155 | ↑NP cell apoptosis | 2019 | [103] |
| LncPolE | ↑ | PolE | ↑NP cell apoptosis | 2019 | [100] |
| HOTAIR |
↑ ↓ |
AMPK/mTOR/ULK1 miR-34a-5p |
↑NP cell apoptosis ↑ECM degradation ↓NP cell apoptosis |
2020 2020 2019 |
[104] [104] [105] |
| MALAT1 | ↓ | miR-503 |
↓NP cell apoptosis ↑NP cell proliferation ↓Inflammation |
2017 2017 2017 |
[114] [114] [114] |
| LINC00641 | ↑ | miR-153-3p | ↑NP cell autophagy | 2019 | [58] |
| SNHG1 | ↑ | miR-326 | ↑NP cell proliferation | 2018 | [106] |
| RP11-296A18.3 | ↑ | miR-138 |
↑NP cell proliferation ↑ECM synthesis |
2017 2017 |
[10] [10] |
| H19 | ↑ | miR-22 |
↓NP cell proliferation ↑ECM degradation |
2018 2018 |
[107] [107] |
| FAF1 | ↑ | Erk | ↑NP cell proliferation | 2018 | [116] |
| FAM83H-AS1 | ↑ | Notch1 |
↑NP cell growth ↑ECM degradation |
2019 2019 |
[112] [112] |
| HCG18 | ↑ | miR-146a-5p |
↑NP cell apoptosis ↓NP cell proliferation |
2017 2017 |
[108] [108] |
| Linc00958 | ↑ | miR-203 |
↑NP cell proliferation ↑ECM degradation |
2019 2019 |
[109] [109] |
| RMRP | ↑ | miR-206 |
↑NP cell proliferation ↓ECM degradation |
2018 2018 |
[25] [25] |
| NEAT1 | ↑ | ERK1/2, MAPK | ↑ECM degradation | 2018 | [111] |
| SLC20A1 | ↑ | miR-31-5p | ↑ECM degradation | 2019 | [110] |
| Linc-ADAMTS5 | ↑ | SFPQ | ↓ECM degradation | 2017 | [113] |
| ZFAS1 | ↑ | Associate with inflammation level | 2019 | [115] |
The expression, targets, and functions of lncRNAs related to IDD were displayed in Table 4. “↓” represents downregulation, while “↑” represents upregulation
Conclusion
In recent years, a large number of investigations have depicted a bright future for ncRNAs, which play roles as delicate regulators in the pathogenesis of IDD. The lncRNA/circRNA/miRNA/mRNA networks and the widespread crosstalks between the RNAs provide us another way to recognize and understand the pathogenesis of IDD [19]. A number of aberrantly expressed RNAs have been regarded as early diagnostic biomarkers or useful therapeutic targets. Moreover, novel materials and technologies, such as injectable hydrogel or nanoparticle which is loadable for small RNAs [117], genetic technologies and stem cell-based therapies [118, 119], are developing rapidly, making it possible to interfere the RNA expression inside IVD cells.
The rapid development of high throughput biotechnological tools greatly facilitates the studies for ncRNAs in IDD. The most common biotechnological approaches are microarray analysis for specific ncRNAs and/or sequencing technologies. Following successful RNA isolation and quality control, ncRNA expression in IDD can be detected via developed microarray chips with known covered ncRNA numbers and types according to corresponding ncRNA database versions. Alternatively, ncRNA expression in IDD can be studied using next-generation sequencing platforms following reverse transcription to cDNA. Thereafter, sequencing data can be mapped to human genomic version (the updated version as GRCh38) and annotated into various subtypes of ncRNAs, with the pros of uncovering novel ncRNAs and cons as introducing errors/mutations during reverse transcription. A combined exploration of both biotechnologies might overcome the cons and improve the studies of ncRNAs in IDD. Novel sequencing technologies are needed for direct sequencing of RNAs and omitting the reverse transcription step. In addition, there are triple common tools/techniques for ncRNA studies following screening. First, RT-PCR tool aims for the detection of expression levels of ncRNAs. Second, bioinformatics and online software tools apply for ncRNA function, target, and interaction predictions. Third, in vitro modulation (upregulation and repression) designates for target and function validations.
However, we are still facing with lots of challenges. Lack of knowledge about the overall view of the ncRNA networks makes it challenging to identify the key nodes to interfere with. The roles of tRNAs and emerging small RNAs, i.e., small nucleolar RNAs (snoRNAs) and PIWI-interacting RNAs (piRNAs), which may be equally important in IDD, remain unclear and deserve thorough studies. Stem cells, such as mesenchymal stem cells (MSCs), have already been used in IVD degeneration therapies for assisting tissue regeneration and exosome secretion, which contains miRNAs to improve microenvironment. However, the inflammatory milieu of IVDs is tough for MSCs to survive in degenerative IVD tissues [120, 121]. Thus, improvement in tissue engineering techniques is urgently needed in seed cell implanting [122]. Future studies should keep focusing on the molecular mechanisms of crosstalk among ncRNAs, especially novel snoRNAs, piRNAs, and tRNAs, and seek feasible ways in seed cell implantation, nanoparticles containing RNA molecules or engineered tissues to interfere the hub nodes in the regulatory network. With the issues solved, research advances in the regulatory machinery of ncRNAs will provide the medical community with a brighter future for IDD therapies.
Acknowledgements
Not applicable
Abbreviations
- 3′UTR
3′-Untranslated region
- ADAMTS
A disintegrin and metalloprotease with thrombospondin motifs
- AF
Annulus fibrosus
- CDKN1B
Cyclin-dependent kinase inhibitor 1B
- CEP
Cartilaginous endplate
- CeRNAs
Competing endogenous RNAs
- CircRNA
Circular RNA
- Col I
Type I collagen
- Col II
Type II collagen
- ECM
Extracellular matrix
- ERα
Estradiol receptor
- FADD
Fas-associated death domain-containing protein
- FGFR3
Fibroblast growth factor receptor-3
- HOXD10
Homeobox D10
- IDD
Intervertebral disc degeneration
- IL-1β
Interleukin-1β
- IVD
Intervertebral disc
- lncRNA
Long noncoding RNA
- MAPK
Mitogen-activated protein kinase
- miRNAs
MicroRNAs
- MMPs
Matrix metalloproteinases
- NcRNAs
Noncoding RNAs
- NP
Nucleus pulposus
- PDCD4
Programmed cell death 4
- PI3K
Phosphoinositide 3-kinase
- PTEN
Phosphate and tension homology deleted on chromosome ten
- SAA1
Serum amyloid A1
- SMAD4
Mothers against decapentaplegic protein family member 4
- SOX
SRY-related high-mobility group box
- TGF-β
Transforming growth factor-β
- TNF-α
Tumor necrosis factor-α
- TRAIL
Tumor necrosis factor-related apoptosis-inducing ligand
Authors’ contributions
HQW conceived the study. HYG, MKG, and FS investigated and retrieved the published papers. HYG and MKG analyzed data. HYG wrote the original draft. ZYW review and editing the manuscript. All authors have read and agreed to the published version of the manuscript.
Authors’ information
Not applicable
Funding
This research was funded by the Chinese National Natural Science Foundation Grants, grant number No.81572182. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Availability of data and materials
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hao-Yu Guo, Ming-Ke Guo and Zhong-Yuan Wan contributed equally to this work.
Contributor Information
Hao-Yu Guo, Email: 835042285@qq.com.
Ming-Ke Guo, Email: guomk@163.com.
Zhong-Yuan Wan, Email: wanzhongyuan@yeah.net.
Fang Song, Email: wzyaisf@126.com.
Hai-Qiang Wang, Email: drwanghq@163.com, Email: hqwang@sntcm.edu.cn.
References
- 1.Frymoyer JW, Cats-Baril WL. An overview of the incidences and costs of low back pain. Orthop Clin North Am. 1991;22(2):263–271. [PubMed] [Google Scholar]
- 2.Nakamura M, Nishiwaki Y, Ushida T, Toyama Y. Prevalence and characteristics of chronic musculoskeletal pain in Japan. J Orthop Sci. 2011;16(4):424–432. doi: 10.1007/s00776-011-0102-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakamura M, Toyama Y, Nishiwaki Y, Ushida T. Prevalence and characteristics of chronic musculoskeletal pain in Japan: a second survey of people with or without chronic pain. J Orthop Sci. 2014;19(2):339–350. doi: 10.1007/s00776-013-0525-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Konovalov NA, Nazarenko AG, Asyutin DS, Zelenkov PV, Onoprienko RA, Korolishin VA, Cherkiev IU, Martynova MA, Zakirov BA, Timonin SY, et al. Modern treatments for degenerative disc diseases of the lumbosacral spine. A literature review. Zh Vopr Neirokhir Im N N Burdenko. 2016;80(4):102–108. doi: 10.17116/neiro2016804102-108. [DOI] [PubMed] [Google Scholar]
- 5.Lee CK, Langrana NA. A review of spinal fusion for degenerative disc disease: need for alternative treatment approach of disc arthroplasty? Spine J. 2004;4(6 Suppl):173S–176S. doi: 10.1016/j.spinee.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Adams MA, Roughley PJ. What is intervertebral disc degeneration, and what causes it? Spine (Phila Pa 1976) 2006;31(18):2151–2161. doi: 10.1097/01.brs.0000231761.73859.2c. [DOI] [PubMed] [Google Scholar]
- 7.Chen H, Wang J, Hu B, Wu X, Chen Y, Li R, Yuan W. MiR-34a promotes Fas-mediated cartilage endplate chondrocyte apoptosis by targeting Bcl-2. Mol Cell Biochem. 2015;406(1–2):21–30. doi: 10.1007/s11010-015-2420-4. [DOI] [PubMed] [Google Scholar]
- 8.Hai B, Ma Y, Pan X, Yong L, Liang C, He G, Yang C, Zhu B, Liu X. Melatonin benefits to the growth of human annulus fibrosus cells through inhibiting miR-106a-5p/ATG7 signaling pathway. Clin Interv Aging. 2019;14:621–630. doi: 10.2147/CIA.S193765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vergroesen PP, Kingma I, Emanuel KS, Hoogendoorn RJ, Welting TJ, van Royen BJ, van Dieen JH, Smit TH. Mechanics and biology in intervertebral disc degeneration: a vicious circle. Osteoarthr Cartil. 2015;23(7):1057–1070. doi: 10.1016/j.joca.2015.03.028. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Lv G, Li J, Wang B, Zhang Q, Lu C. LncRNA-RP11-296A18.3/miR-138/HIF1A pathway regulates the proliferation ECM synthesis of human nucleus pulposus cells (HNPCs) J Cell Biochem. 2017;118(12):4862–4871. doi: 10.1002/jcb.26166. [DOI] [PubMed] [Google Scholar]
- 11.Mayer JE, Iatridis JC, Chan D, Qureshi SA, Gottesman O, Hecht AC. Genetic polymorphisms associated with intervertebral disc degeneration. Spine J. 2013;13(3):299–317. doi: 10.1016/j.spinee.2013.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu J, Zhang X, Gao W, Hu H, Wang X, Hao D. lncRNA/circRNAmiRNAmRNA ceRNA network in lumbar intervertebral disc degeneration. Mol Med Rep. 2019;20(4):3160–3174. doi: 10.3892/mmr.2019.10569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wan ZY, Song F, Sun Z, Chen YF, Zhang WL, Samartzis D, Ma CJ, Che L, Liu X, Ali MA, et al. Aberrantly expressed long noncoding RNAs in human intervertebral disc degeneration: a microarray related study. Arthritis Res Ther. 2014;16(5):465. doi: 10.1186/s13075-014-0465-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Ni H, Zhao Y, Chen K, Li M, Li C, Zhu X, Fu Q. Potential role of lncRNAs in contributing to pathogenesis of intervertebral disc degeneration based on microarray data. Med Sci Monit. 2015;21:3449–3458. doi: 10.12659/MSM.894638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Z, Chen X, Xu D, Li S, Chan MTV, Wu WKK. Circular RNAs in nucleus pulposus cell function and intervertebral disc degeneration. Cell Prolif. 2019;52(6):e12704. doi: 10.1111/cpr.12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang C, Wang WJ, Yan YG, Xiang YX, Zhang J, Tang ZH, Jiang ZS. MicroRNAs: new players in intervertebral disc degeneration. Clin Chim Acta. 2015;450:333–341. doi: 10.1016/j.cca.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 17.Wang S, Sun J, Yang H, Zou W, Zheng B, Chen Y, Guo Y, Shi J. Profiling and bioinformatics analysis of differentially expressed circular RNAs in human intervertebral disc degeneration. Acta Biochim Biophys Sin Shanghai. 2019;51(6):571–579. doi: 10.1093/abbs/gmz036. [DOI] [PubMed] [Google Scholar]
- 18.Wang HQ, Yu XD, Liu ZH, Cheng X, Samartzis D, Jia LT, Wu SX, Huang J, Chen J, Luo ZJ. Deregulated miR-155 promotes Fas-mediated apoptosis in human intervertebral disc degeneration by targeting FADD and caspase-3. J Pathol. 2011;225(2):232–242. doi: 10.1002/path.2931. [DOI] [PubMed] [Google Scholar]
- 19.Zhao B, Yu Q, Li H, Guo X, He X. Characterization of microRNA expression profiles in patients with intervertebral disc degeneration. Int J Mol Med. 2014;33(1):43–50. doi: 10.3892/ijmm.2013.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lan PH, Liu ZH, Pei YJ, Wu ZG, Yu Y, Yang YF, Liu X, Che L, Ma CJ, Xie YK, et al. Landscape of RNAs in human lumbar disc degeneration. Oncotarget. 2016;7(39):63166–63176. doi: 10.18632/oncotarget.11334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen B, Huang SG, Ju L, Li M, Nie FF, Zhang Y, Zhang YH, Chen X, Gao F. Effect of microRNA-21 on the proliferation of human degenerated nucleus pulposus by targeting programmed cell death 4. Braz J Med Biol Res. 2016;49(6):e5020. [DOI] [PMC free article] [PubMed]
- 22.Gu SX, Li X, Hamilton JL, Chee A, Kc R, Chen D, An HS, Kim JS, Oh CD, Ma YZ, et al. MicroRNA-146a reduces IL-1 dependent inflammatory responses in the intervertebral disc. Gene. 2015;555(2):80–87. doi: 10.1016/j.gene.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng X, Zhang L, Zhang K, Zhang G, Hu Y, Sun X, Zhao C, Li H, Li YM, Zhao J. Circular RNA VMA21 protects against intervertebral disc degeneration through targeting miR-200c and X linked inhibitor-of-apoptosis protein. Ann Rheum Dis. 2018;77(5):770–779. doi: 10.1136/annrheumdis-2017-212056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J, Jia YS, Liu GZ, Sun Q, Zhang F, Ma S, Wang YJ. Role of LncRNA TUG1 in intervertebral disc degeneration and nucleus pulposus cells via regulating Wnt/beta-catenin signaling pathway. Biochem Biophys Res Commun. 2017;491(3):668–674. doi: 10.1016/j.bbrc.2017.07.146. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Peng L, Gong X, Zhang X, Sun R, Du J. LncRNA-RMRP promotes nucleus pulposus cell proliferation through regulating miR-206 expression. J Cell Mol Med. 2018;22(11):5468–5476. doi: 10.1111/jcmm.13817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hiyama A, Yokoyama K, Nukaga T, Sakai D, Mochida J. A complex interaction between Wnt signaling and TNF-alpha in nucleus pulposus cells. Arthritis Res Ther. 2013;15(6):R189. doi: 10.1186/ar4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiyama A, Yokoyama K, Nukaga T, Sakai D, Mochida J. Response to tumor necrosis factor-alpha mediated inflammation involving activation of prostaglandin E2 and Wnt signaling in nucleus pulposus cells. J Orthop Res. 2015;33(12):1756–1768. doi: 10.1002/jor.22959. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, Hu Z, Hao J, Shen J. Low-intensity pulsed ultrasound stimulates the extracellular matrix synthesis of human degenerative nucleus pulposus cells via activating PI3K/Akt pathway. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2013;29(1):34–38. [PubMed] [Google Scholar]
- 29.Miao D, Zhang L. Leptin modulates the expression of catabolic genes in rat nucleus pulposus cells through the mitogen-activated protein kinase and Janus kinase 2/signal transducer and activator of transcription 3 pathways. Mol Med Rep. 2015;12(2):1761–1768. doi: 10.3892/mmr.2015.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu P, Feng B, Wang G, Ning B, Jia T. Microarray based analysis of gene regulation by microRNA in intervertebral disc degeneration. Mol Med Rep. 2015;12(4):4925–4930. doi: 10.3892/mmr.2015.4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guancial EA, Bellmunt J, Yeh S, Rosenberg JE, Berman DM. The evolving understanding of microRNA in bladder cancer. Urol Oncol. 2014;32(1):41 e31–41 e40. doi: 10.1016/j.urolonc.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshino H, Seki N, Itesako T, Chiyomaru T, Nakagawa M, Enokida H. Aberrant expression of microRNAs in bladder cancer. Nat Rev Urol. 2013;10(7):396–404. doi: 10.1038/nrurol.2013.113. [DOI] [PubMed] [Google Scholar]
- 33.Zhang WL, Chen YF, Meng HZ, Du JJ, Luan GN, Wang HQ, Yang MW, Luo ZJ. Role of miR-155 in the regulation of MMP-16 expression in intervertebral disc degeneration. J Orthop Res. 2017;35(6):1323–1334. doi: 10.1002/jor.23313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye D, Dai L, Yao Y, Qin S, Xie H, Wang W, Liang W. miR-155 inhibits nucleus pulposus cells’ degeneration through targeting ERK 1/2. Dis Markers. 2016;2016:6984270. doi: 10.1155/2016/6984270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun J, Hong J, Sun S, Wang X, Peng Y, Zhou J, Huang Y, Li S, Chen W, Li C, et al. Transcription factor 7-like 2 controls matrix degradation through nuclear factor kappaB signaling and is repressed by microRNA-155 in nucleus pulposus cells. Biomed Pharmacother. 2018;108:646–655. doi: 10.1016/j.biopha.2018.09.076. [DOI] [PubMed] [Google Scholar]
- 36.Cheng X, Zhang G, Zhang L, Hu Y, Zhang K, Sun X, Zhao C, Li H, Li YM, Zhao J. Mesenchymal stem cells deliver exogenous miR-21 via exosomes to inhibit nucleus pulposus cell apoptosis and reduce intervertebral disc degeneration. J Cell Mol Med. 2018;22(1):261–276. doi: 10.1111/jcmm.13316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu H, Huang X, Liu X, Xiao S, Zhang Y, Xiang T, Shen X, Wang G, Sheng B. miR-21 promotes human nucleus pulposus cell proliferation through PTEN/AKT signaling. Int J Mol Sci. 2014;15(3):4007–4018. doi: 10.3390/ijms15034007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang WJ, Yang W, Ouyang ZH, Xue JB, Li XL, Zhang J, He WS, Chen WK, Yan YG, Wang C. MiR-21 promotes ECM degradation through inhibiting autophagy via the PTEN/akt/mTOR signaling pathway in human degenerated NP cells. Biomed Pharmacother. 2018;99:725–734. doi: 10.1016/j.biopha.2018.01.154. [DOI] [PubMed] [Google Scholar]
- 39.Liu G, Cao P, Chen H, Yuan W, Wang J, Tang X. MiR-27a regulates apoptosis in nucleus pulposus cells by targeting PI3K. PLoS One. 2013;8(9):e75251. doi: 10.1371/journal.pone.0075251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun JC, Zheng B, Sun RX, Meng YK, Wang SM, Yang HS, Chen Y, Shi JG, Guo YF. MiR-499a-5p suppresses apoptosis of human nucleus pulposus cells and degradation of their extracellular matrix by targeting SOX4. Biomed Pharmacother. 2019;113:108652. doi: 10.1016/j.biopha.2019.108652. [DOI] [PubMed] [Google Scholar]
- 41.Kang L, Yang C, Song Y, Zhao K, Liu W, Hua W, Wang K, Tu J, Li S, Yin H, et al. MicroRNA-494 promotes apoptosis and extracellular matrix degradation in degenerative human nucleus pulposus cells. Oncotarget. 2017;8(17):27868–27881. doi: 10.18632/oncotarget.15838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang T, Li P, Ma X, Tian P, Han C, Zang J, Kong J, Yan H. MicroRNA-494 inhibition protects nucleus pulposus cells from TNF-alpha-induced apoptosis by targeting JunD. Biochimie. 2015;115:1–7. doi: 10.1016/j.biochi.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 43.Lv J, Li S, Wan T, Yang Y, Cheng Y, Xue R. Inhibition of microRNA-30d attenuates the apoptosis and extracellular matrix degradation of degenerative human nucleus pulposus cells by up-regulating SOX9. Chem Biol Interact. 2018;296:89–97. doi: 10.1016/j.cbi.2018.09.010. [DOI] [PubMed] [Google Scholar]
- 44.Liu J, Yu J, Jiang W, He M, Zhao J. Targeting of CDKN1B by miR-222-3p may contribute to the development of intervertebral disc degeneration. FEBS Open Bio. 2019;9(4):728–735. doi: 10.1002/2211-5463.12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cai P, Yang T, Jiang X, Zheng M, Xu G, Xia J. Role of miR-15a in intervertebral disc degeneration through targeting MAP3K9. Biomed Pharmacother. 2017;87:568–574. doi: 10.1016/j.biopha.2016.12.128. [DOI] [PubMed] [Google Scholar]
- 46.Chai X, Si H, Song J, Chong Y, Wang J, Zhao G. miR-486-5p inhibits inflammatory response, matrix degradation and apoptosis of nucleus pulposus cells through directly targeting FOXO1 in intervertebral disc degeneration. Cell Physiol Biochem. 2019;52(1):109–118. doi: 10.33594/000000008. [DOI] [PubMed] [Google Scholar]
- 47.Guo W, Zhang B, Mu K, Feng SQ, Dong ZY, Ning GZ, Li HR, Liu S, Zhao L, Li Y, et al. Circular RNA GRB10 as a competitive endogenous RNA regulating nucleus pulposus cells death in degenerative intervertebral disk. Cell Death Dis. 2018;9(3):319. doi: 10.1038/s41419-017-0232-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu W, Zhang Y, Feng X, Li S, Gao Y, Wang K, Song Y, Yang S, Tu J, Shao Z, et al. Inhibition of microRNA-34a prevents IL-1beta-induced extracellular matrix degradation in nucleus pulposus by increasing GDF5 expression. Exp Biol Med (Maywood) 2016;241(17):1924–1932. doi: 10.1177/1535370216657444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao K, Zhang Y, Kang L, Song Y, Wang K, Li S, Wu X, Hua W, Shao Z, Yang S, et al. Epigenetic silencing of miRNA-143 regulates apoptosis by targeting BCL2 in human intervertebral disc degeneration. Gene. 2017;628:259–266. doi: 10.1016/j.gene.2017.07.043. [DOI] [PubMed] [Google Scholar]
- 50.Sun Z, Jian Y, Fu H, Li B. MiR-532 downregulation of the Wnt/beta-catenin signaling via targeting Bcl-9 and induced human intervertebral disc nucleus pulposus cells apoptosis. J Pharmacol Sci. 2018;138(4):263–270. doi: 10.1016/j.jphs.2018.10.007. [DOI] [PubMed] [Google Scholar]
- 51.Ma JF, Zang LN, Xi YM, Yang WJ, Zou D. MiR-125a Rs12976445 polymorphism is associated with the apoptosis status of nucleus pulposus cells and the risk of intervertebral disc degeneration. Cell Physiol Biochem. 2016;38(1):295–305. doi: 10.1159/000438630. [DOI] [PubMed] [Google Scholar]
- 52.Sheng B, Yuan Y, Liu X, Zhang Y, Liu H, Shen X, Liu B, Chang L. Protective effect of estrogen against intervertebral disc degeneration is attenuated by miR-221 through targeting estrogen receptor alpha. Acta Biochim Biophys Sin Shanghai. 2018;50(4):345–354. doi: 10.1093/abbs/gmy017. [DOI] [PubMed] [Google Scholar]
- 53.Penolazzi L, Lambertini E, Bergamin LS, Roncada T, De Bonis P, Cavallo M, Piva R. MicroRNA-221 silencing attenuates the degenerated phenotype of intervertebral disc cells. Aging (Albany NY) 2018;10(8):2001–2015. doi: 10.18632/aging.101525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yeh CH, Jin L, Shen F, Balian G, Li XJ. miR-221 attenuates the osteogenic differentiation of human annulus fibrosus cells. Spine J. 2016;16(7):896–904. doi: 10.1016/j.spinee.2016.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang B, Wang D, Yan T, Yuan H. MiR-138-5p promotes TNF-alpha-induced apoptosis in human intervertebral disc degeneration by targeting SIRT1 through PTEN/PI3K/Akt signaling. Exp Cell Res. 2016;345(2):199–205. doi: 10.1016/j.yexcr.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 56.Zhou J, Sun J, Markova DZ, Li S, Kepler CK, Hong J, Huang Y, Chen W, Xu K, Wei F, et al. MicroRNA-145 overexpression attenuates apoptosis and increases matrix synthesis in nucleus pulposus cells. Life Sci. 2019;221:274–283. doi: 10.1016/j.lfs.2019.02.041. [DOI] [PubMed] [Google Scholar]
- 57.Wang R, Wen B, \: miR-573 regulates cell proliferation and apoptosis by targeting Bax in nucleus pulposus cells. Cell Mol Biol Lett 2019, 24:2. [DOI] [PMC free article] [PubMed]
- 58.Wang XB, Wang H, Long HQ, Li DY, Zheng X. LINC00641 regulates autophagy and intervertebral disc degeneration by acting as a competitive endogenous RNA of miR-153-3p under nutrition deprivation stress. J Cell Physiol. 2019;234(5):7115–7127. doi: 10.1002/jcp.27466. [DOI] [PubMed] [Google Scholar]
- 59.Yu X, Li Z, Shen J, Wu WK, Liang J, Weng X, Qiu G. MicroRNA-10b promotes nucleus pulposus cell proliferation through RhoC-Akt pathway by targeting HOXD10 in intervetebral disc degeneration. PLoS One. 2013;8(12):e83080. doi: 10.1371/journal.pone.0083080. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 60.Tao B, Yi J, Huang C, Xu W, Qin C, Chen L, Chen J, Gao Y, Wang R. microRNA96 regulates the proliferation of nucleus pulposus cells by targeting ARID2/AKT signaling. Mol Med Rep. 2017;16(5):7553–7560. doi: 10.3892/mmr.2017.7560. [DOI] [PubMed] [Google Scholar]
- 61.Li W, Wang P, Zhang Z, Wang W, Liu Y, Qi Q. MiR-184 regulates proliferation in nucleus pulposus cells by targeting GAS1. World Neurosurg. 2017;97:710–715. doi: 10.1016/j.wneu.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 62.Guo Y, Tian L, Liu X, He Y, Chang S, Shen Y. ERRFI1 inhibits proliferation and inflammation of nucleus pulposus and is negatively regulated by miR-2355-5p in intervertebral disc degeneration. Spine (Phila Pa 1976) 2019;44(15):E873–E881. doi: 10.1097/BRS.0000000000003011. [DOI] [PubMed] [Google Scholar]
- 63.Zheng Q, Li XX, Xiao L, Shao S, Jiang H, Zhang XL, Sun LY, Xu HG. MicroRNA-365 functions as a mechanosensitive microRNA to inhibit end plate chondrocyte degeneration by targeting histone deacetylase 4. Bone. 2019;128:115052. doi: 10.1016/j.bone.2019.115052. [DOI] [PubMed] [Google Scholar]
- 64.Meng X, Zhu Y, Tao L, Zhao S, Qiu S. MicroRNA-125b-1-3p mediates intervertebral disc degeneration in rats by targeting teashirt zinc finger homeobox 3. Exp Ther Med. 2018;15(3):2627–2633. doi: 10.3892/etm.2018.5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tan H, Zhao L, Song R, Liu Y, Wang L. microRNA-665 promotes the proliferation and matrix degradation of nucleus pulposus through targeting GDF5 in intervertebral disc degeneration. J Cell Biochem. 2018;119(9):7218–7225. doi: 10.1002/jcb.26888. [DOI] [PubMed] [Google Scholar]
- 66.Liu W, Zhang Y, Xia P, Li S, Feng X, Gao Y, Wang K, Song Y, Duan Z, Yang S, et al. MicroRNA-7 regulates IL-1beta-induced extracellular matrix degeneration by targeting GDF5 in human nucleus pulposus cells. Biomed Pharmacother. 2016;83:1414–1421. doi: 10.1016/j.biopha.2016.08.062. [DOI] [PubMed] [Google Scholar]
- 67.Liu W, Xia P, Feng J, Kang L, Huang M, Wang K, Song Y, Li S, Wu X, Yang S, et al. MicroRNA-132 upregulation promotes matrix degradation in intervertebral disc degeneration. Exp Cell Res. 2017;359(1):39–49. doi: 10.1016/j.yexcr.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 68.Kang L, Yang C, Yin H, Zhao K, Liu W, Hua W, Wang K, Song Y, Tu J, Li S, et al. MicroRNA-15b silencing inhibits IL-1beta-induced extracellular matrix degradation by targeting SMAD3 in human nucleus pulposus cells. Biotechnol Lett. 2017;39(4):623–632. doi: 10.1007/s10529-016-2280-3. [DOI] [PubMed] [Google Scholar]
- 69.Liu MH, Sun C, Yao Y, Fan X, Liu H, Cui YH, Bian XW, Huang B, Zhou Y. Matrix stiffness promotes cartilage endplate chondrocyte calcification in disc degeneration via miR-20a targeting ANKH expression. Sci Rep. 2016;6:25401. doi: 10.1038/srep25401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsirimonaki E, Fedonidis C, Pneumaticos SG, Tragas AA, Michalopoulos I, Mangoura D. PKCepsilon signalling activates ERK1/2, and regulates aggrecan, ADAMTS5, and miR377 gene expression in human nucleus pulposus cells. PLoS One. 2013;8(11):e82045. doi: 10.1371/journal.pone.0082045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shi C, Wu L, Lin W, Cai Y, Zhang Y, Hu B, Gao R, Im HJ, Yuan W, Ye X, et al. MiR-202-3p regulates interleukin-1beta-induced expression of matrix metalloproteinase 1 in human nucleus pulposus. Gene. 2019;687:156–165. doi: 10.1016/j.gene.2018.11.056. [DOI] [PubMed] [Google Scholar]
- 72.Song J, Wang HL, Song KH, Ding ZW, Ma XS, Lu FZ, Xia XL, Wang YW, Fei Z, Jiang JY. CircularRNA_104670 plays a critical role in intervertebral disc degeneration by functioning as a ceRNA. Exp Mol Med. 2018;50(8):94. doi: 10.1038/s12276-018-0125-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jing W, Jiang W. MicroRNA-93 regulates collagen loss by targeting MMP3 in human nucleus pulposus cells. Cell Prolif. 2015;48(3):284–292. doi: 10.1111/cpr.12176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu YQ, Zhang ZH, Zheng YF, Feng SQ. Dysregulated miR-133a mediates loss of type II collagen by directly targeting matrix metalloproteinase 9 (MMP9) in human intervertebral disc degeneration. Spine (Phila Pa 1976) 2016;41(12):E717–E724. doi: 10.1097/BRS.0000000000001375. [DOI] [PubMed] [Google Scholar]
- 75.Li HR, Cui Q, Dong ZY, Zhang JH, Li HQ, Zhao L. Downregulation of miR-27b is involved in loss of type II collagen by directly targeting matrix metalloproteinase 13 (MMP13) in human intervertebral disc degeneration. Spine (Phila Pa 1976) 2016;41(3):E116–E123. doi: 10.1097/BRS.0000000000001139. [DOI] [PubMed] [Google Scholar]
- 76.Hua WB, Wu XH, Zhang YK, Song Y, Tu J, Kang L, Zhao KC, Li S, Wang K, Liu W, et al. Dysregulated miR-127-5p contributes to type II collagen degradation by targeting matrix metalloproteinase-13 in human intervertebral disc degeneration. Biochimie. 2017;139:74–80. doi: 10.1016/j.biochi.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 77.Ji ML, Zhang XJ, Shi PL, Lu J, Wang SZ, Chang Q, Chen H, Wang C. Downregulation of microRNA-193a-3p is involved in invertebral disc degeneration by targeting MMP14. J Mol Med (Berl) 2016;94(4):457–468. doi: 10.1007/s00109-015-1371-2. [DOI] [PubMed] [Google Scholar]
- 78.Ji ML, Lu J, Shi PL, Zhang XJ, Wang SZ, Chang Q, Chen H, Wang C. Dysregulated miR-98 contributes to extracellular matrix degradation by targeting IL-6/STAT3 signaling pathway in human intervertebral disc degeneration. J Bone Miner Res. 2016;31(4):900–909. doi: 10.1002/jbmr.2753. [DOI] [PubMed] [Google Scholar]
- 79.Yan N, Yu S, Zhang H, Hou T. Lumbar disc degeneration is facilitated by MiR-100-mediated FGFR3 suppression. Cell Physiol Biochem. 2015;36(6):2229–2236. doi: 10.1159/000430187. [DOI] [PubMed] [Google Scholar]
- 80.Lv F, Huang Y, Lv W, Yang L, Li F, Fan J, Sun J. MicroRNA-146a ameliorates inflammation via TRAF6/NF-kappaB pathway in intervertebral disc cells. Med Sci Monit. 2017;23:659–664. doi: 10.12659/MSM.898660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang C, Zhang ZZ, Yang W, Ouyang ZH, Xue JB, Li XL, Zhang J, Chen WK, Yan YG, Wang WJ. MiR-210 facilitates ECM degradation by suppressing autophagy via silencing of ATG7 in human degenerated NP cells. Biomed Pharmacother. 2017;93:470–479. doi: 10.1016/j.biopha.2017.06.048. [DOI] [PubMed] [Google Scholar]
- 82.Hu B, Xu C, Tian Y, Shi C, Zhang Y, Deng L, Zhou H, Cao P, Chen H, Yuan W. Inflammatory microRNA-194 and -515 attenuate the biosynthesis of chondroitin sulfate during human intervertebral disc degeneration. Oncotarget. 2017;8(30):49303–49317. doi: 10.18632/oncotarget.17571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang B, Guo W, Sun C, Duan HQ, Yu BB, Mu K, Guan YY, Li Y, Liu S, Liu Y, et al. Dysregulated MiR-3150a-3p promotes lumbar intervertebral disc degeneration by targeting aggrecan. Cell Physiol Biochem. 2018;45(6):2506–2515. doi: 10.1159/000488269. [DOI] [PubMed] [Google Scholar]
- 84.Dong W, Liu J, Lv Y, Wang F, Liu T, Sun S, Liao B, Shu Z, Qian J. miR-640 aggravates intervertebral disc degeneration via NF-kappaB and WNT signalling pathway. Cell Prolif. 2019;52(5):e12664. doi: 10.1111/cpr.12664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang Q, Weng Y, Jiang Y, Zhao S, Zhou D, Xu N. Overexpression of miR-140-5p inhibits lipopolysaccharide-induced human intervertebral disc inflammation and degeneration by downregulating Toll-like receptor 4. Oncol Rep. 2018;40(2):793–802. doi: 10.3892/or.2018.6488. [DOI] [PubMed] [Google Scholar]
- 86.Murillo OD, Thistlethwaite W, Rozowsky J, Subramanian SL, Lucero R, Shah N, Jackson AR, Srinivasan S, Chung A, Laurent CD, et al. exRNA atlas analysis reveals distinct extracellular RNA cargo types and their carriers present across human biofluids. Cell. 2019;177(2):463–477. doi: 10.1016/j.cell.2019.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ma X, Lin Y, Yang K, Yue B, Xiang H, Chen B. Effect of lentivirus-mediated survivin transfection on the morphology and apoptosis of nucleus pulposus cells derived from degenerative human disc in vitro. Int J Mol Med. 2015;36(1):186–194. doi: 10.3892/ijmm.2015.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mern DS, Beierfuss A, Thome C, Hegewald AA. Enhancing human nucleus pulposus cells for biological treatment approaches of degenerative intervertebral disc diseases: a systematic review. J Tissue Eng Regen Med. 2014;8(12):925–936. doi: 10.1002/term.1583. [DOI] [PubMed] [Google Scholar]
- 89.Chen WK, Yu XH, Yang W, Wang C, He WS, Yan YG, Zhang J, Wang WJ. lncRNAs: novel players in intervertebral disc degeneration and osteoarthritis. Cell Prolif. 2017;50(1):e12313. [DOI] [PMC free article] [PubMed]
- 90.Zhang F, Zhao X, Shen H, Zhang C. Molecular mechanisms of cell death in intervertebral disc degeneration (review) Int J Mol Med. 2016;37(6):1439–1448. doi: 10.3892/ijmm.2016.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Le Maitre CL, Pockert A, Buttle DJ, Freemont AJ, Hoyland JA. Matrix synthesis and degradation in human intervertebral disc degeneration. Biochem Soc Trans. 2007;35(Pt 4):652–655. doi: 10.1042/BST0350652. [DOI] [PubMed] [Google Scholar]
- 92.Wuertz K, Vo N, Kletsas D, Boos N. Inflammatory and catabolic signalling in intervertebral discs: the roles of NF-kappaB and MAP kinases. Eur Cell Mater. 2012;23:103–119. doi: 10.22203/ecm.v023a08. [DOI] [PubMed] [Google Scholar]
- 93.Zou F, Ding Z, Jiang J, Lu F, Xia X, Ma X. Confirmation and preliminary analysis of circRNAs potentially involved in human intervertebral disc degeneration. Mol Med Rep. 2017;16(6):9173–9180. doi: 10.3892/mmr.2017.7718. [DOI] [PubMed] [Google Scholar]
- 94.Wang H, He P, Pan H, Long J, Wang J, Li Z, Liu H, Jiang W, Zheng Z. Circular RNA circ-4099 is induced by TNF-alpha and regulates ECM synthesis by blocking miR-616-5p inhibition of Sox9 in intervertebral disc degeneration. Exp Mol Med. 2018;50(4):27. doi: 10.1038/s12276-018-0056-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Maass PG, Glazar P, Memczak S, Dittmar G, Hollfinger I, Schreyer L, Sauer AV, Toka O, Aiuti A, Luft FC, et al. A map of human circular RNAs in clinically relevant tissues. J Mol Med (Berl) 2017;95(11):1179–1189. doi: 10.1007/s00109-017-1582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang X, Wang B, Zou M, Li J, Lu G, Zhang Q, Liu F, Lu C. CircSEMA4B targets miR-431 modulating IL-1beta-induced degradative changes in nucleus pulposus cells in intervertebral disc degeneration via Wnt pathway. Biochim Biophys Acta Mol basis Dis. 2018;1864(11):3754–3768. doi: 10.1016/j.bbadis.2018.08.033. [DOI] [PubMed] [Google Scholar]
- 97.Li Z, Li X, Chen C, Li S, Shen J, Tse G, Chan MTV, Wu WKK. Long non-coding RNAs in nucleus pulposus cell function and intervertebral disc degeneration. Cell Prolif. 2018;51(5):e12483. doi: 10.1111/cpr.12483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20(11):1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yao Y, Li J, Wang L. Large intervening non-coding RNA HOTAIR is an indicator of poor prognosis and a therapeutic target in human cancers. Int J Mol Sci. 2014;15(10):18985–18999. doi: 10.3390/ijms151018985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li X, Lou Z, Liu J, Li H, Lei Y, Zhao X, Zhang F. Upregulation of the long noncoding RNA lncPolE contributes to intervertebral disc degeneration by negatively regulating DNA polymerase epsilon. Am J Transl Res. 2019;11(5):2843–2854. [PMC free article] [PubMed] [Google Scholar]
- 101.Han Z, Wang J, Gao L, Wang Q, Wu J. Aberrantly expressed messenger RNAs and long noncoding RNAs in degenerative nucleus pulposus cells co-cultured with adipose-derived mesenchymal stem cells. Arthritis Res Ther. 2018;20(1):182. doi: 10.1186/s13075-018-1677-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhao B, Lu M, Wang D, Li H, He X. Genome-wide identification of long noncoding RNAs in human intervertebral disc degeneration by RNA sequencing. Biomed Res Int. 2016;2016:3684875. doi: 10.1155/2016/3684875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang Y, Song Q, Huang X, Chen Z, Zhang F, Wang K, Huang G, Shen H. Long noncoding RNA GAS5 promotes apoptosis in primary nucleus pulposus cells derived from the human intervertebral disc via Bcl2 downregulation and caspase3 upregulation. Mol Med Rep. 2019;19(3):2164–2172. doi: 10.3892/mmr.2019.9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhan S, Wang K, Xiang Q, Song Y, Li S, Liang H, Luo R, Wang B, Liao Z, Zhang Y, et al. lncRNA HOTAIR upregulates autophagy to promote apoptosis and senescence of nucleus pulposus cells. J Cell Physiol. 2020;235(3):2195–2208. doi: 10.1002/jcp.29129. [DOI] [PubMed] [Google Scholar]
- 105.Shao T, Hu Y, Tang W, Shen H, Yu Z, Gu J. The long noncoding RNA HOTAIR serves as a microRNA-34a-5p sponge to reduce nucleus pulposus cell apoptosis via a NOTCH1-mediated mechanism. Gene. 2019;715:144029. doi: 10.1016/j.gene.2019.144029. [DOI] [PubMed] [Google Scholar]
- 106.Tan H, Zhao L, Song R, Liu Y, Wang L. The long noncoding RNA SNHG1 promotes nucleus pulposus cell proliferation through regulating miR-326 and CCND1. Am J Physiol Cell Physiol. 2018;315(1):C21–C27. doi: 10.1152/ajpcell.00220.2017. [DOI] [PubMed] [Google Scholar]
- 107.Wang X, Zou M, Li J, Wang B, Zhang Q, Liu F, Lu G. LncRNA H19 targets miR-22 to modulate H2 O2 -induced deregulation in nucleus pulposus cell senescence, proliferation, and ECM synthesis through Wnt signaling. J Cell Biochem. 2018;119(6):4990–5002. doi: 10.1002/jcb.26738. [DOI] [PubMed] [Google Scholar]
- 108.Xi Y, Jiang T, Wang W, Yu J, Wang Y, Wu X, He Y. Long non-coding HCG18 promotes intervertebral disc degeneration by sponging miR-146a-5p and regulating TRAF6 expression. Sci Rep. 2017;7(1):13234. doi: 10.1038/s41598-017-13364-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhao K, Zhang Y, Yuan H, Zhao M, Zhao D. Long noncoding RNA LINC00958 accelerates the proliferation and matrix degradation of the nucleus pulposus by regulating miR-203/SMAD3. Aging (Albany NY) 2019;11(23):10814–10825. doi: 10.18632/aging.102436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yang Y, Zhong Z, Zhao Y, Ren K, Li N. LincRNA-SLC20A1 (SLC20A1) promotes extracellular matrix degradation in nucleus pulposus cells in human intervertebral disc degeneration by targeting the miR-31-5p/MMP3 axis. Int J Clin Exp Pathol. 2019;12(9):3632–3643. [PMC free article] [PubMed] [Google Scholar]
- 111.Ruan Z, Ma H, Li J, Liu H, Jia H, Li F. The long non-coding RNA NEAT1 contributes to extracellular matrix degradation in degenerative human nucleus pulposus cells. Exp Biol Med (Maywood) 2018;243(7):595–600. doi: 10.1177/1535370218760774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wei R, Chen Y, Zhao Z, Gu Q, Wu J. LncRNA FAM83H-AS1 induces nucleus pulposus cell growth via targeting the Notch signaling pathway. J Cell Physiol. 2019;234(12):22163–22171. doi: 10.1002/jcp.28780. [DOI] [PubMed] [Google Scholar]
- 113.Wang K, Song Y, Liu W, Wu X, Zhang Y, Li S, Kang L, Tu J, Zhao K, Hua W, et al. The noncoding RNA linc-ADAMTS5 cooperates with RREB1 to protect from intervertebral disc degeneration through inhibiting ADAMTS5 expression. Clin Sci (Lond) 2017;131(10):965–979. doi: 10.1042/CS20160918. [DOI] [PubMed] [Google Scholar]
- 114.Zhang H, Li J, Duan D, She W, Wang L, Zhang F. The role of lncRNA MALAT1 in intervertebral degenerative disc disease. Int J Clin Exp Pathol. 2017;10(10):10611–10617. [PMC free article] [PubMed] [Google Scholar]
- 115.Deng RY, Hong T, Li CY, Shi CL, Liu C, Jiang FY, Li J, Fan XM, Feng SB, Wang YF. Long non-coding RNA zinc finger antisense 1 expression associates with increased disease risk, elevated disease severity and higher inflammatory cytokines levels in patients with lumbar disc degeneration. Medicine (Baltimore) 2019;98(52):e18465. doi: 10.1097/MD.0000000000018465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mi D, Cai C, Zhou B, Liu X, Ma P, Shen S, Lu W, Huang W. Long noncoding RNA FAF1 promotes intervertebral disc degeneration by targeting the Erk signaling pathway. Mol Med Rep. 2018;17(2):3158–3163. doi: 10.3892/mmr.2017.8237. [DOI] [PubMed] [Google Scholar]
- 117.Feng G, Zha Z, Huang Y, Li J, Wang Y, Ke W, Chen H, Liu L, Song Y, Ge Z. Sustained and bioresponsive two-stage delivery of therapeutic miRNA via Polyplex micelle-loaded injectable hydrogels for inhibition of intervertebral disc fibrosis. Adv Healthc Mater. 2018;7(21):e1800623. doi: 10.1002/adhm.201800623. [DOI] [PubMed] [Google Scholar]
- 118.Ma CJ, Liu X, Che L, Liu ZH, Samartzis D, Wang HQ. Stem cell therapies for intervertebral disc degeneration: immune privilege reinforcement by Fas/FasL regulating machinery. Curr Stem Cell Res Ther. 2015;10(4):285–295. doi: 10.2174/1574888X10666150416114027. [DOI] [PubMed] [Google Scholar]
- 119.Wang HQ. Bring stem cell therapies to cure intervertebral disc degeneration to the forefront. Curr Stem Cell Res Ther. 2015;10(4):284. doi: 10.2174/1574888X1004150513162605. [DOI] [PubMed] [Google Scholar]
- 120.Clarke LE, Richardson SM, Hoyland JA. Harnessing the potential of mesenchymal stem cells for IVD regeneration. Curr Stem Cell Res Ther. 2015;10(4):296–306. doi: 10.2174/1574888X10666141202112638. [DOI] [PubMed] [Google Scholar]
- 121.Krock E, Rosenzweig DH, Haglund L. The inflammatory milieu of the degenerate disc: is mesenchymal stem cell-based therapy for intervertebral disc repair a feasible approach? Curr Stem Cell Res Ther. 2015;10(4):317–328. doi: 10.2174/1574888X10666150211161956. [DOI] [PubMed] [Google Scholar]
- 122.Gantenbein B, Illien-Junger S, Chan SC, Walser J, Haglund L, Ferguson SJ, Iatridis JC, Grad S. Organ culture bioreactors--platforms to study human intervertebral disc degeneration and regenerative therapy. Curr Stem Cell Res Ther. 2015;10(4):339–352. doi: 10.2174/1574888X10666150312102948. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article [and its supplementary information files].