Abstract
Stability of antibody–drug conjugates (ADCs) in mouse serum is one of the critical requirements for the evaluation of ADCs in mouse tumor models. Described herein is a strategy to address the mouse serum instability of uncialamycin linker–payloads through various chemical approaches that involve modification of different parts of the linker and payload. This effort ultimately led to the identification of a m-amide p-aminobenzyl carbamate (MA-PABC) group that resulted in linkers with dramatic improvement of mouse serum stability without affecting the desired proteolytic cleavage.
Keywords: Antibody−drug conjugates, linker, mouse serum, stability, uncialamycin, payload
Antibody–drug conjugates (ADCs) are large and complex molecular entities comprising a tumor-targeting antibody and usually a cytotoxic payload (drug) appended by a chemical linker. With the approval of eight ADCs (Kadcyla, Adcetris, Besponsa, Mylotarg, Polivy, Enhertu, Padcev, and Trodelvy) and more than 60 products under clinical development, they constitute an important modality for anticancer drug development.1 Structurally, each component of an ADC requires a unique set of biochemical properties to make the ADC successful. For example, an ideal antibody should have high binding affinity to the tumor antigen, minimal nonspecific binding, and an efficient internalization process. Similarly, the drug (payload) component should have high potency, a defined mechanism of action, chemical stability, and an amenable handle for attachment of the linker. The linker that connects the antibody to the drug should ideally be stable in circulation and cleavable by intracellular proteases such as cathepsin B inside the cell to release the drug to act on the intended target.2,3 In the absence of linker stability in serum, premature release of the payload can result in systemic toxicity. On the other hand, inefficient cleavage of the linker inside the cell may not produce the intended antitumor activity.4
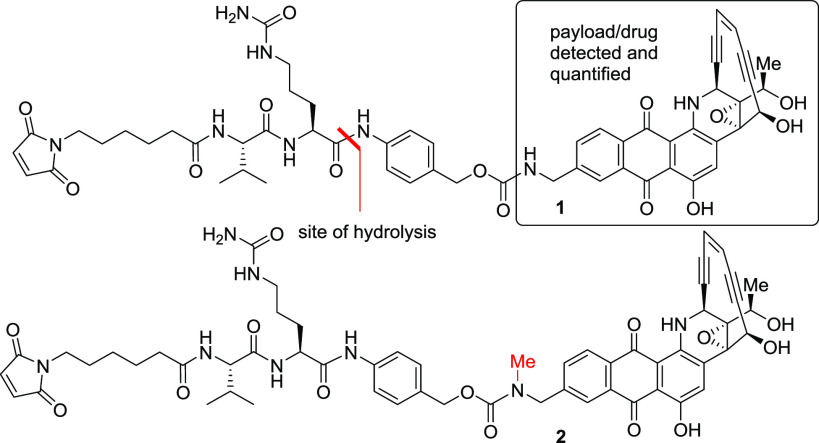
During the preparation of uncialamycin ADCs, we found the instability of the linker–payload in mouse serum to be a major challenge whereby hydrolysis of the dipeptide followed by loss of the p-aminobenzyl spacer group led to release of the highly potent payload (1 in Table 1).5,6 While the linker–payloads were stable in human serum and cleaved by cathepsin B as desired, the undesirable release of the payload in mouse serum could lead to potential systemic toxicity in mouse tumor models. The assessment of linker–payload stability was done by initial conversion of the maleimide group to the N-acetylcysteine (NAC) derivative to avoid any side reactions arising from free maleimide, followed by incubation of the NAC derivative in serum, where the released payload was identified and quantified by LC–MS. The instability of valine-citrulline-based linkers seems to be a general phenomenon and has been widely reported.7,8 Minor chemical modification of the payload through the introduction of a methyl group did not provide any stability advantage, whereas any significant changes to the payload were not attempted because of its favorable potency and stability (2 in Table 1). The mouse serum instability of the linker–payload has been attributed to esterase-mediated amide hydrolysis and subsequent release of the drug.7,8
Table 1. Stability of Initial Linker–Payloadsa.
| reagent | 1 | 2 |
|---|---|---|
| cathepsin B | 100 | 100 |
| human serum | 0 | 0 |
| mouse serum | 80 | 78 |
Values shown are % drug release from the N-acetylcysteine (NAC) derivative in 24 h.
Several approaches were considered to resolve the mouse serum instability issue, including the use of esterase inhibitors in efficacy studies, use of an esterase knockout mouse model, and the development of site-specific conjugation chemistries.9−11 While potentially useful, each of the approaches posed a unique set of challenges such as potential toxicity associated with esterase inhibitors, unavailability of a knockout mouse tumor model and complexities associated with antibody engineering. At the same time, we believed that a chemical approach to address this issue would be ideal if successful. Although esterases such as Ces1c have been reported to be among the enzymes responsible for such linker hydrolysis,9,12 because of the large number of esterases present in mouse serum and the lack of structural information, a traditional medicinal chemistry approach could not be undertaken. We believed that esterase-mediated hydrolysis might be mitigated through careful and judicious modification of the linker and/or payload. Thus, an empirical approach was taken to systematically modify parts of the linker and the payload with the expectation that such changes would alter the rate of amide hydrolysis (Figure 1).
Figure 1.
Chemical approach to address mouse serum hydrolysis
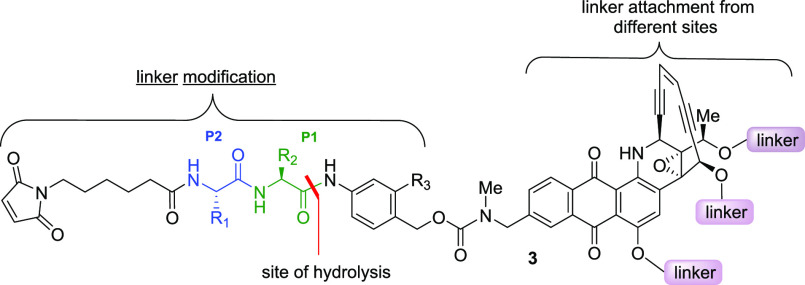
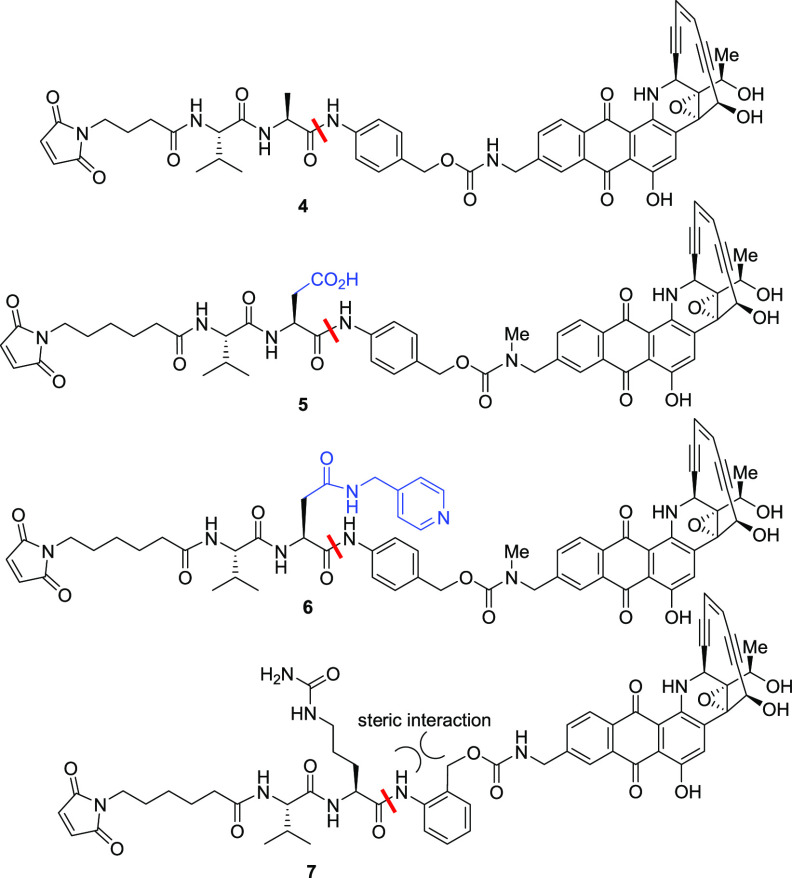
We initiated our chemical approach by modification of the dipeptide linker (Table 2). While most of the dipeptides used in ADCs contain a basic and/or polar amino acid at the P1 position, we were curious to see the effect of its replacement with a neutral or acidic group. Replacing the polar citrulline with neutral/nonpolar alanine in 4 increased the hydrolysis in mouse serum compared with 1 or 2. However, use of negatively charged aspartic acid in 5 significantly reduced both cathepsin B and mouse serum cleavage, suggesting that basic or polar groups are favored in this position. Derivatization of the carboxylic acid with (4-pyridylmethyl)amine, which had been found to assist in reducing hydrolysis in mouse serum in a different chemotype, did not provide any meaningful difference in 6.13 In compound 7, the steric hindrance imposed by the ortho substituent completely blocked the much-needed cathepsin B cleavage, whereas it was less effective in reducing esterase hydrolysis in mice. A corresponding meta substitution was not attempted because it would not favor the self-immolation of the newly formed m-aminobenzyl carbamate. This study suggested that a neutral or basic amino acid at the P1 position is readily cleaved but acidic and sterically encumbered groups provide more resistance for esterase- or protease-mediated hydrolysis.
Table 2. Effect of P1 Amino Acid Modificationa.
| reagent | 4 | 5 | 6 | 7 |
|---|---|---|---|---|
| cathepsin B | 100 | 5 | 20 (4 h) | 0 |
| human serum | 0 | 0 | 0 | 0 |
| mouse serum | 100 | 72 | 100 | 70 |
Values shown are % drug release from the NAC derivative in 24 h.
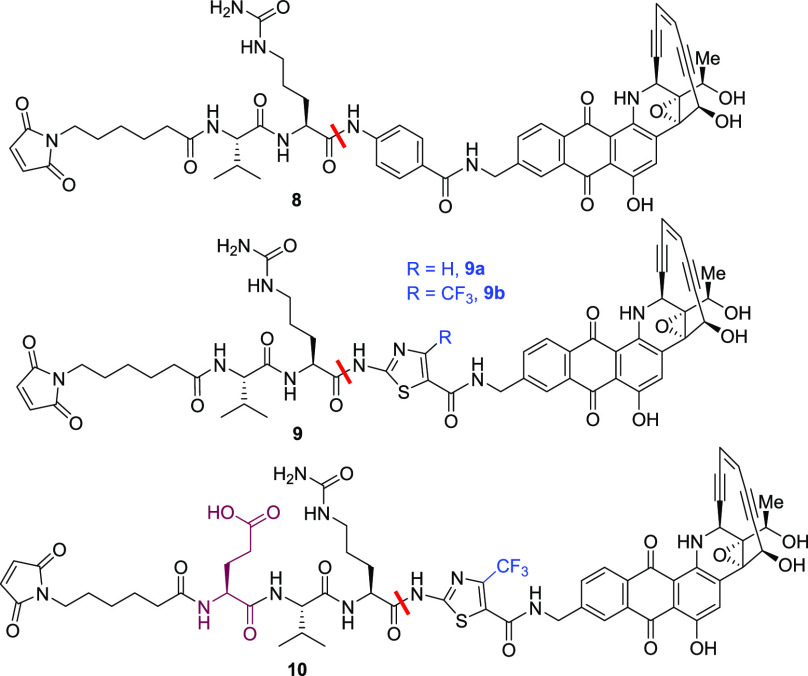
We next focused on an amide class of linker–payloads where self-immolation is not required and cleavage of the amide bond between the P1 amino acid and the p-aminobenzyl carbamate results in direct release of the drug (Table 3). In this series, the prototypical compound 8 was most readily cleaved in mouse serum (100% in 4 h) despite being completely stable in human serum. Keeping the dipeptide unchanged, we replaced the p-aminobenzyl amide with various heterocycles, and among many things attempted, the thiazole amides seemed to be quite promising. Thiazole 9a was much more stable (45% hydrolysis in 24 h) in mouse serum compared with compound 8, but it unexpectedly picked up some human serum hydrolysis (12% in 24 h). Substitution of the thiazole with an electron-withdrawing trifluoromethyl group further reduced the hydrolysis in both human and mouse serum. Finally, the addition of glutamic acid at the P3 position in 9b dramatically improved the stability in mouse serum, but hydrolysis in human serum could not be completely eliminated (10). It is interesting to note that concurrent with our independent work, others also observed that addition of glutamic acid at this position greatly enhances the stability of the linker–payloads in mouse plasma and in the corresponding ADCs in vivo.14 This exercise provided important insights into the structural features of linkers that can reduce esterase hydrolysis.
Table 3. Stability of Amide Linker–Payloadsa.
| reagent | 8 | 9a | 9b | 10 |
|---|---|---|---|---|
| cathepsin B | 100 | 100 | 100 | 100 |
| human serum | 0 | 12 | 5 | 4 |
| mouse serum | 100 (4 h) | 45 | 18 | 6 |
Values shown are % drug release from the NAC derivative in 24 h.
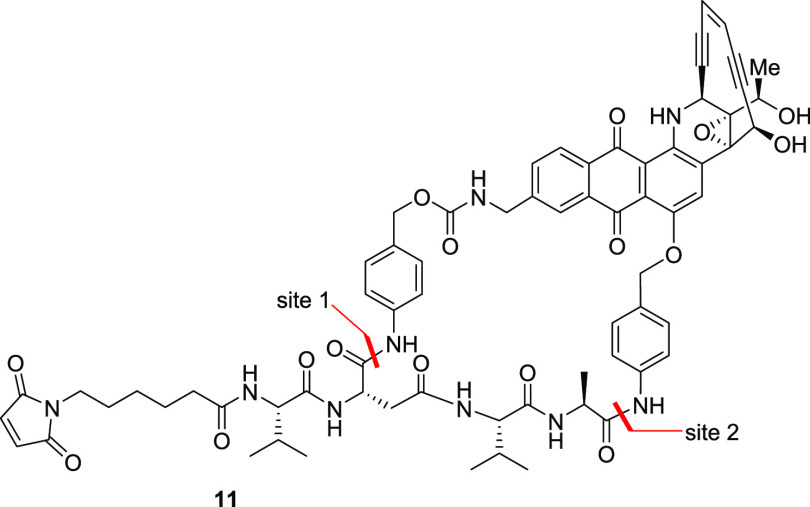
Since appending the linker to different parts of the drug, such as on the amines described here or the phenol,6 led to an incremental increase in mouse serum stability, we thought that a combination of these two approaches might provide an additive effect. Thus a macrocyclic system was designed in which a tetrapeptide linker would be used to connect both the benzylamine and the phenol (Table 4). Having such a macrocyclic system would potentially block the accessibility of esterase by virtue of steric hindrance, but we also suspected that it might reduce the necessary intracellular proteolytic hydrolysis. While we found that macrocyclic compound 11 showed a reduction in mouse serum hydrolysis, it unfortunately blocked the much needed cathepsin B cleavage. Interestingly, despite being more sterically encumbered, the amide bond next to the carbamate was cleaved first, followed by hydrolysis of the ether amide, ultimately releasing the free drug as observed by LC–MS.
Table 4. Stability of the Macrocyclic Analoguea.
| reagent | 11 |
|---|---|
| cathepsin B | 6 |
| human serum | 0 |
| mouse serum | 73 |
Values shown are % drug release from the NAC derivative in 24 h.
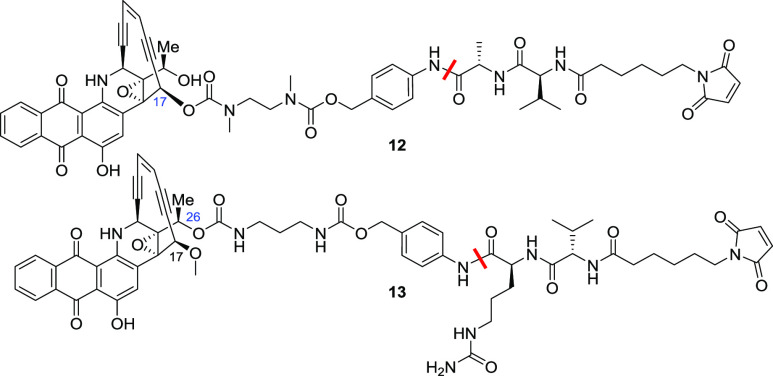
Next, we explored the attachment of linker using two alcohols that were first chemically differentiated (Table 5). The alcohols needed an amine handle, which was generated through the attachment of diamine carbamates capable of self-immolation via internal cyclization to finally release the alcohols once the PABA–C linker was cleaved.15 Both linker–payloads 12 and 13 were cleaved by cathepsin B and were stable in human serum, but they were readily hydrolyzed in the mouse serum, indicating that such an approach was not helpful in reducing mouse serum hydrolysis.
Table 5. Stability of Alcohol-Linked Compoundsa.
| reagent | 12 | 13 |
|---|---|---|
| cathepsin B | 100 | 100 |
| human serum | 0 | 0 |
| mouse serum | 80 | 78 |
Values shown are % drug release from the NAC derivative in 24 h.
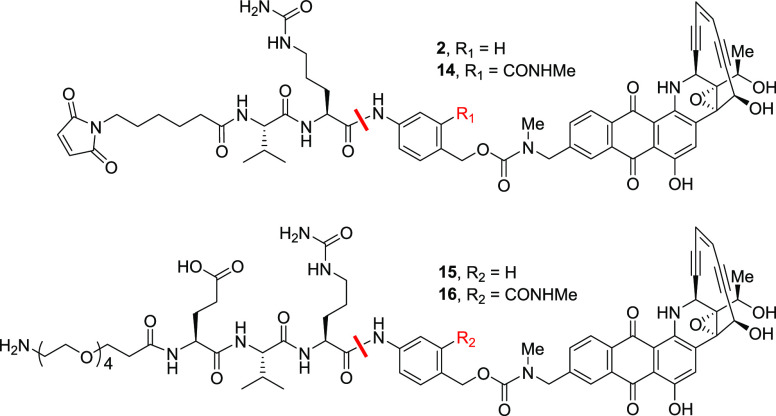
During the course of these studies, we observed that both esterase- and protease-mediated hydrolysis of the dipeptide linker was quite sensitive to steric effects around the amide bond under consideration (examples 7 and 11). As opposed to the substituents at the ortho position of the p-aminobenzyl carbamate, we thought that those at the meta position would be better tolerated. We envisioned that it would be ideal if the substituents were tunable for steric and electronic effects and assisted in increasing the hydrophilicity of the linker–payload with a highly lipophilic payload such as uncialamycin. Such criteria could be fulfilled by incorporating an amide at the meta position of the p-aminobenzyl carbamate. With this hypothesis, we assembled 14, our first linker–payload incorporating an m-amide (MA-PABC) linker (Table 6). As a control, the unsubstituted linker 2 was used, which was completely hydrolyzed in mouse serum to release the payload in 24 h. Incubation of the new MA-PABC linker 14 with cathepsin B showed that it was efficiently cleaved by cathepsin B as desired, indicating that substitution at the meta position of the PABC is better tolerated, as predicted. The MA-PABC linker–payload was also stable in human serum over 24 h.
Table 6. Stability of m-Amide Linker–Payloadsa.
| reagent | 2 | 14 | 15 | 16 |
|---|---|---|---|---|
| cathepsin B | 100 | 100 | 100 | 100 |
| human serum | 0 | 0 | 0 | 0 |
| mouse serum | 100 | 50 | 31 | 7 |
Values shown are % drug release in 24 h. Compounds 2 and 14 were used as NAC derivatives.
Encouragingly, the m-amide 14 was only 50% hydrolyzed in mouse serum in 24 h, unlike compound 2. We were curious to see whether the increase in stability due to various structural changes in linkers was additive. When the glutamic acid was added in linker–payload 15, mouse serum hydrolysis was substantially reduced to 31%. When both glutamic and the MA-PABC were combined in analogue 16, hydrolysis in mouse serum decreased dramatically to 7% over 24 h. An important additional advantage of having glutamic acid was the enhancement of the aqueous solubility of the uncialamycin linker–payloads, which were quite challenging for conjugation with the antibody because of the hydrophobicity imposed by the payload. Thus, by combining the best attributes of the linkers from different structure–activity relationship studies, we were able to design linkers with ideal stability profiles.
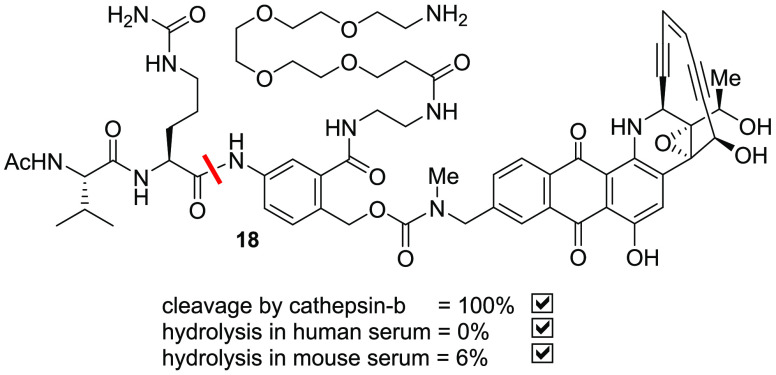
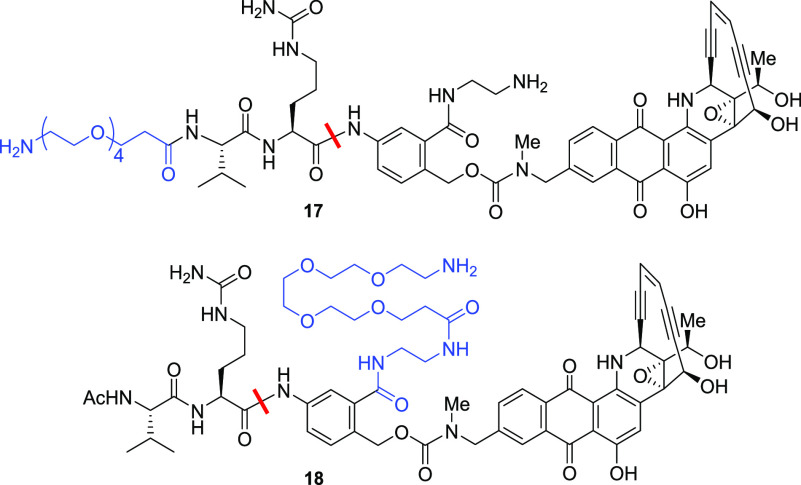
Perhaps the most impressive set of results were obtained from compound 17, in which the amide N-methyl group was elaborated to an N-(2-aminoethyl) group (Table 7). This compound was practically stable in mouse serum over 24 h (3% hydrolysis) without compromising any stability in human serum or cleavability by cathepsin B. The free aminoethyl group in 17 could be further derivatized with groups such as poly(ethylene glycol)s to enhance the overall aqueous solubility for bioconjugation, and those linkers exhibited a stability profile comparable to that of 17.16 Interestingly, transposition of the primary amine conjugation handle from the dipeptide N-terminus to the MA-PABC provided a completely new series of linker–payloads 18 with desirable stability in human/mouse serum and cleavage by cathepsin B. When linker–payloads 18 were conjugated to antibodies using bacterial transglutaminase chemistry and the resulting ADCs were taken for mouse serum stability studies, any residual hydrolysis of the linker–payloads was eliminated, and the ADCs were completely stable without affecting their stability profile in human serum or cathepsin B. More importantly, these ADCs displayed antigen-dependent antitumor activity that will be communicated in detail in a separate paper.
Table 7. Stability of m-Amide Linker–Payloadsa.
| reagent | 17 | 18 |
|---|---|---|
| cathepsin B | 100 | 100 |
| human serum | 0 | 0 |
| mouse serum | 3 | 6 |
Values shown are % drug release from the linker–payload in 24 h.
In summary, we investigated several chemical approaches to reduce mouse-serum-mediated hydrolysis of uncialamycin linker–payloads; these included a modified dipeptide, a self-immolating group, generation of novel macrocyclic linkers, and attachment of the linker to different parts of the payload. These efforts led to the discovery of trifluoromethylthiazole amides that greatly reduced mouse-serum-mediated hydrolysis of dipeptide linker–payloads; however, payload release in human serum could not be completely eliminated. Further efforts resulted in the development of linker–payloads containing valine-citrulline17 with a novel self-immolating m-amide p-aminobenzyl carbamate (MA-PABC) group that were resistant to hydrolysis in serum yet were efficiently cleaved by intracellular proteases as desired.
Acknowledgments
The authors thank Catherine Bolger for proofreading and Pavel Strop for providing constructive feedback on the manuscript.
Glossary
Abbreviations
- ADC
antibody–drug conjugate
- MA-PABC
m-amide p-aminobenzyl carbamate
- NAC
N-acetylcysteine
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.0c00325.
Synthesis and stability studies of the linker–payloads (PDF)
Author Contributions
The manuscript was written through contributions of all authors. All of the authors approved the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Leung D.; Wurst J. M.; Liu T.; Martinez R. M.; Datta-Mannan A.; Feng Y. Antibody Conjugates-Recent Advances and Future Innovations. Antibodies 2020, 9, 2. 10.3390/antib9010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senter P. D.; Sievers E. L. The Discovery and development of brentuximab vedotin for use in relapsed hodgkin lymphoma and systemic anaplastic large cell lymphoma. Nat. Biotechnol. 2012, 30, 631–637. 10.1038/nbt.2289. [DOI] [PubMed] [Google Scholar]
- Lambert J. M.; Chari R. V. J. Ado-Trastuzumab Emtansine (TDM1): An Antibody–Drug Conjugate (ADC) for HER2-Positive Breast Cancer. J. Med. Chem. 2014, 57, 6949–6964. 10.1021/jm500766w. [DOI] [PubMed] [Google Scholar]
- For a recent review of ADC linkers, see:; Jain N.; Smith S. W.; Ghone S.; Tomczuk B. Current ADC linker chemistry. Pharm. Res. 2015, 32, 3526–3540. 10.1007/s11095-015-1657-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Nicolaou K. C.; Lu M.; Mandal D.; Gangwar S.; Chowdari N. S.; Poudel Y. B.. Derivatives of Uncialamycin, Methods of Synthesis and Their Use as Antitumor Agents. US 9,777,013, 2017.; b Nicolaou K. C.; Lu M.; Mandal D.; Gangwar S.; Chowdari N. S.; Poudel Y. B.. Derivatives of Uncialamycin, Methods of Synthesis and Their Use as Antitumor Agents. US 10,233,192, 2019.
- Poudel Y. B.; Rao C.; Kotapati S.; Deshpande M.; Thevanayagam L.; Pan C.; Cardarelli P.; Chowdari N.; Kaspady M.; Samikannu R.; Kuppusamy P.; Saravanakumar P.; Arunachalam P. N.; Deshpande S.; Rangan V.; Rampulla R.; mathur A.; Vite G. D.; Gangwar S. Design, synthesis and biological evaluation of phenol-linked uncialamycin antibody–drug conjugates. Bioorg. Med. Chem. Lett. 2020, 30, 126782. 10.1016/j.bmcl.2019.126782. [DOI] [PubMed] [Google Scholar]
- Elgersma R. C.; Coumans R. G.; Huijbregts T.; Menge W. M.; Joosten J. A.; Spijker H. J.; de Groot F. M.; van der Lee M. M.; Ubink R.; van den Dobbelsteen D. J.; Egging D. F.; Dokter W. H.; Verheijden G. F.; Lemmens J. M.; Timmers C. M.; Beusker P. H. Design, Synthesis, and Evaluation of Linker-Duocarmycin Payloads: Towad Selection of HER2-Trageting Antiboduody-Drug Conjugate SYD985. Mol. Pharmaceutics 2015, 12, 1813–1835. 10.1021/mp500781a. [DOI] [PubMed] [Google Scholar]
- Dal Corso A.; Cazzamalli S.; Gébleux R.; Mattarella M.; Neri D. Protease-Cleavable Linkers Modulate the Anticancer Activity of Noninternalizing Antibody–Drug Conjugates. Bioconjugate Chem. 2017, 28, 1826–1833. 10.1021/acs.bioconjchem.7b00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubink R.; Dirksen H. C.; Rouwette M.; Bos E. S.; Janssen I.; Egging D. F.; et al. 181 Unraveling the Interaction between Carboxylesterase 1c and the Antibody–Drug Conjugate SYD985: Improved Translational PK/PD by Using Ces1c Knockout Mice. Mol. Cancer Ther. 2018, 17, 2389–2398. 10.1158/1535-7163.MCT-18-0329. [DOI] [PubMed] [Google Scholar]
- Strop P.; Delaria K.; Foletti D.; Witt J. M.; Hasa-Moreno A.; Poulsen K.; Casas M. G.; Dorywalska M.; Farias S.; Pios A.; Lui V.; Dushin R.; Zhou D.; Navaratnam T.; Tran T. T.; Sutton J.; Lindquist K. C.; Han B.; Liu S. H.; Shelton D. L.; Pons J.; Rajpal A. Site-specific conjugation improves therapeutic index of antibody drug conjugates with high drug loading. Nat. Biotechnol. 2015, 33, 694–696. 10.1038/nbt.3274. [DOI] [PubMed] [Google Scholar]
- Ohri R.; Bhakta S.; Fourie-O’Donohue A.; Dela Cruz-Chuh J.; Tsai S. P.; Cook R.; Wei B.; Ng C.; Wong A. W.; Bos A. B.; Farahi F.; Bhakta J.; Pillow T. H.; Raab H.; Vandlen R.; Polakis P.; Liu Y.; Erickson H.; Junutula J. R.; Kozak K. R. High-Throughput Cysteine Scanning to Identify Stable Antibody Conjugation Sites for Maleimide- and Disulfide-Based Linkers. Bioconjugate Chem. 2018, 29, 473–485. 10.1021/acs.bioconjchem.7b00791. [DOI] [PubMed] [Google Scholar]
- Dorywalska M.; Dushin R.; Moine L.; Farias S. E.; Zhou D.; Navaratnam T.; Lui V.; Hasa-Moreno A.; Casas M. G.; Tran T.-T.; et al. Molecular Basis of Valine-Citrulline-PABC Linker Instability in Site-Specific ADCs and Its Mitigation by Linker Design. Mol. Cancer Ther. 2016, 15, 958–970. 10.1158/1535-7163.MCT-15-1004. [DOI] [PubMed] [Google Scholar]
- Unpublished results.
- Anami Y.; Yamazaki C. M.; Xiong W.; Gui X.; Zhang N.; An Z.; Tsuchikama K. Glutamic acid-valine-citrulline linkers ensure stability and efficacy of antibody–drug conjugates in mice. Nat. Commun. 2018, 9, 2512. 10.1038/s41467-018-04982-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabat D.; Lode H. N.; Pertl U.; Reisfeld R. A.; Rader C.; Lerner R. A.; Barbas C. F. III In vivo activity in a catalytic antibody-prodrug system: Antibody catalyzed etoposide prodrug activation for selective chemotherapy. Proc. Natl. Acad. Sci. U. S. A. 2001, 98, 7528–7533. 10.1073/pnas.131187998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon R. P.; Bovee T. D.; Doronina S. O.; Burke P. J.; Hunter J. H.; Neff-LaFord H. D.; et al. Reducing hydrophobicity of homogeneous antibody–drug conjugates improves pharmacokinetics and therapeutic index. Nat. Biotechnol. 2015, 33, 733–735. 10.1038/nbt.3212. [DOI] [PubMed] [Google Scholar]
- Dubowchik G. M.; Firestone R. A.; Padilla L.; Willner D.; Hofstead S. J.; Mosure K.; Knipe J. O.; Lasch S. J.; Trail P. A. Cathepsin B-labile dipeptide linkers for lysosomal release of doxorubicin from internalizing immunoconjugates: model studies of enzymatic drug release and antigen-specific in vitro anticancer activity. Bioconjugate Chem. 2002, 13, 855–869. 10.1021/bc025536j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.