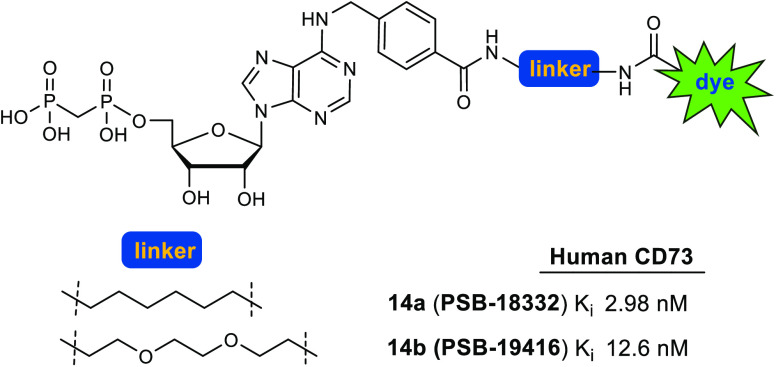
Abstract
Ecto-5′-nucleotidase (CD73) catalyzes the hydrolysis of AMP to anti-inflammatory, immunosuppressive adenosine. It is expressed on vascular endothelial, epithelial, and also numerous cancer cells where it strongly contributes to an immunosuppressive microenvironment. In the present study we designed and synthesized fluorescent-labeled CD73 inhibitors with low nanomolar affinity and high selectivity based on N6-benzyl-α,β-methylene-ADP (PSB-12379) as a lead structure. Fluorescein was attached to the benzyl residue via different linkers resulting in PSB-19416 (14b, Ki 12.6 nM) and PSB-18332 (14a, Ki 2.98 nM) as fluorescent high-affinity probes for CD73. These compounds are anticipated to become useful tools for biological studies, drug screening, and diagnostic applications.
Keywords: Adenosine, CD73, ecto-5′-nucleotidase, fluorescence, immunotherapy, inflammation
Ecto-5′-nucleotidase (ecto-5′-NT, cluster of differentiation 73 (CD73), EC 3.1.3.5) is a cell surface-localized enzyme which dephosphorylates extracellular nucleoside monophosphates, especially AMP, yielding the corresponding nucleoside, mainly adenosine.1−5 It is linked to the cell membrane via a glycosylphosphatidylinositol (GPI) linker but can also be shed and then circulate in the bloodstream and other biological fluids as a soluble enzyme.6,7 CD73 is expressed by subpopulations of T and B lymphocytes,8,9 on endothelial cells4 and stem cells,10 and in high levels by a variety of tumor cells.1,3 The main enzymatic reaction product of CD73, adenosine, leads to the activation of adenosine receptors (subtypes A1, A2A, A2B, A3). A2A and/or A2B adenosine receptors mediate anti-inflammatory, immunosuppressive, angiogenic, pro-metastatic, and proliferative effects and thereby play an important role in cancer progression.4,11−15 In contrast, lowered expression levels of CD73 were reported to be strongly linked to various inflammatory and (auto)immune diseases.8 Atherosclerosis is an inflammatory disease of the endothelial wall, and CD73 deficiency accelerates its progression.16 Upregulation of CD73 activity in brain damage models was considered as an adaptive response leading to an increase in neuroprotective adenosine.17 Recently, CD73 was used as a specific marker molecule of multipotent stromal cells by developing a CD73-enhanced green fluorescent protein (EGFP) reporter mouse that allows the tracking of these cells in vivo.10 This demonstrates the high relevance of CD73 expression levels and the usefulness of tools and probes for monitoring this important enzyme. Moreover, due to the complexity of the entire purinome, which also includes intracellular nucleosides and nucleotides,18 specific marker molecules are urgently required. Fluorescence-labeled small-molecule probes could be particularly useful for elucidating the role of CD73 in various diseases. Such fluorescent ligands for visualizing CD73 and measuring its expression levels under physiological and pathological conditions would be most valuable as diagnostic tools.
Inhibitory CD73 antibodies are currently evaluated in clinical trials for cancer therapy.18,19 Small molecule CD73 inhibitors have also been described, which can be divided into nucleotide and non-nucleotide derivatives (Figure 1).20−29 In 2015, our group published the first CD73 inhibitors with low nanomolar potency, e.g. PSB-12379 (I), a very selective CD73 inhibitor which displays high metabolic stablility.20 This α,β-methylene-ADP- (AOPCP-) derived inhibitor was subsequently optimized resulting in PSB-12489 (II).21 Recently, the first small molecule CD73 inhibitor entered clinical trials, namely the nucleotide analog AB680 (III).23,27 Moreover, compounds combining the diphosphonate partial structure with cytotoxic nucleosides have been reported as CD73 inhibitors, e.g compound IV.28 In addition, non-nucleotide-based inhibitors have been described including sulfoanthraquinone (V), sulfonamide (VI), and sulfonaphthalene (VII) derivatives, all of which showed moderate potency.24−26 Very recently, benzotriazole-based inhibitors (VIII) with potency in the low nanomolar range were reported.29
Figure 1.
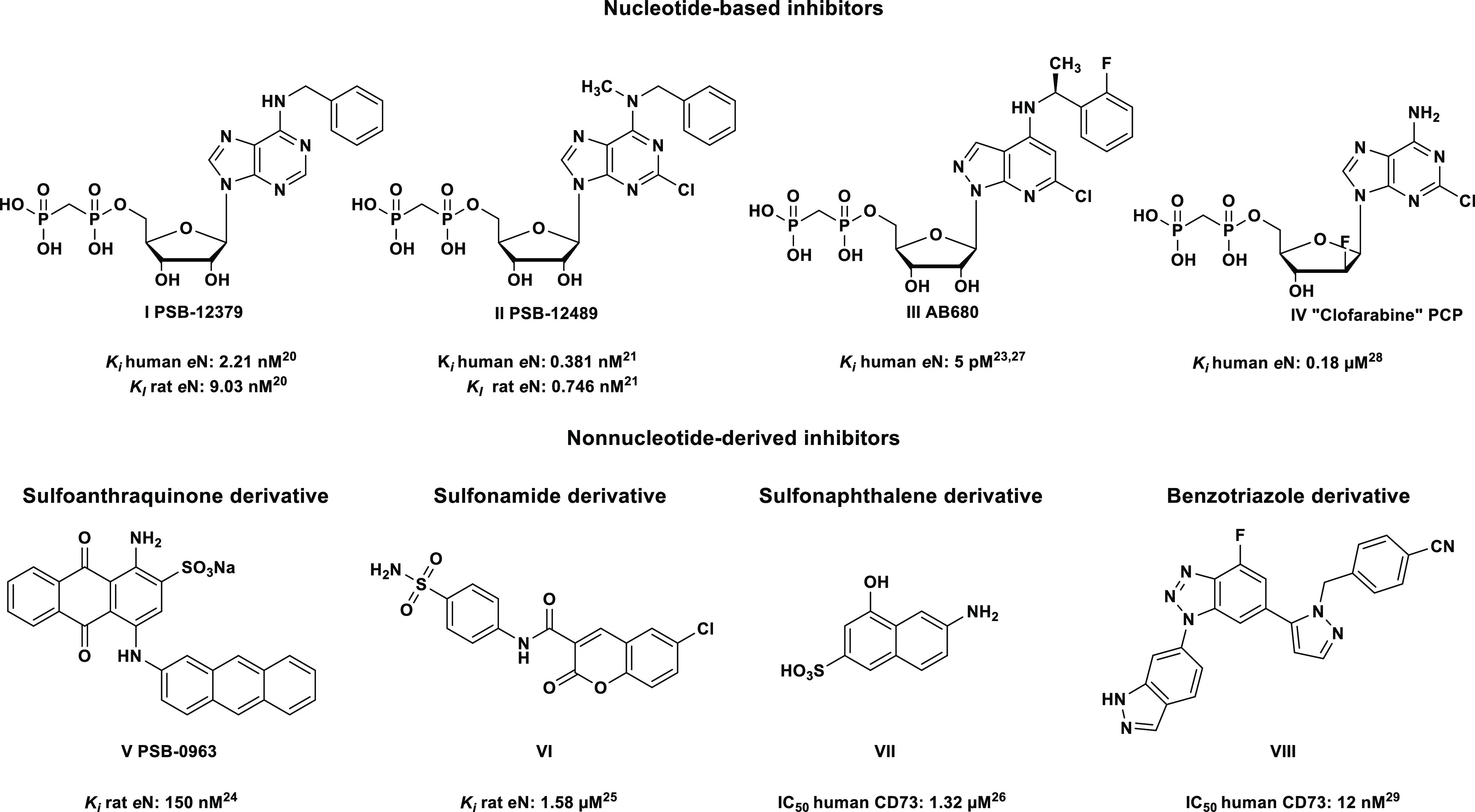
Selected nucleotide- and non-nucleotide-based inhibitors of CD73.
In the present study, we selected nucleotide analogs for fluorescence-labeling, since they show the best combination of high affinity, water-solubility, and metabolic stability. Herein, we describe the synthesis and biological evaluation of the first nucleotide-based, fluorescence-labeled CD73 inhibitors, which exhibit inhibitory potency at low nanomolar concentrations.
Results and Discussion
Design
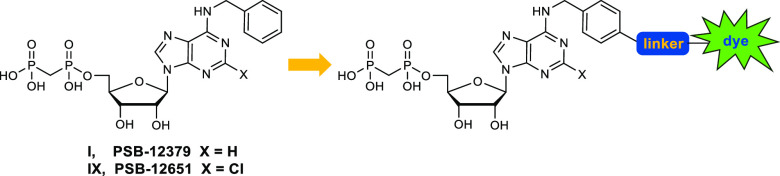
Previously reported X-ray cocrystal structures of human CD73 with nucleotide-based inhibitors along with structure–activity relationship (SAR) studies revealed that bulky, hydrophobic substituents in the N6-position are well tolerated.20−22 Based on these studies, we figured that the best position for attachment of a (relatively large) fluorescent dye would be via a linker to the para-position of the N6-benzyl group in lead structures I and IX (see Figure 2). To optimize the properties of the target compounds, multiple factors have to be taken into account. These include the length of the linker, its lipophilicity, the connection between linker and CD73 inhibitor, and the nature and properties of the fluorophore. For initial labeling studies we selected fluorescein, a strongly fluorescent molecule with an absorption maximum at 492 nm and an emission maximum at 517 nm, which is suitable for biological studies30 and can be applied to living cells because it is not cytotoxic.31,32 Moreover, functionalized fluorescein derivatives are readily available and therefore ideal for initial proof-of-concept studies. Nucleotide analogs I and IX were selected as lead structures to which the fluorescent dye was to be attached (Figure 2).
Figure 2.
Design of a fluorescent probe for CD73 based on PSB-12379 (I)20 and PSB-12651 (IX).21
Chemistry
Our initial targets were fluorescein-labeled compounds 7a and 7b, which we planned to synthesize by convergent approach A (Scheme 1). 2,6-Dichloro-9-(2′,3′,5′-tri-O-acetyl-β-d-ribofuranosyl)purine (1) was subjected to nucleophilic substitution by 4-(aminomethyl)benzoic acid resulting in selective substitution in the 6-position of the purine ring in analogy to conditions described by Bhattarai et al.20
Scheme 1. Original Pathway A to Obtain Compounds 7a and 7b.
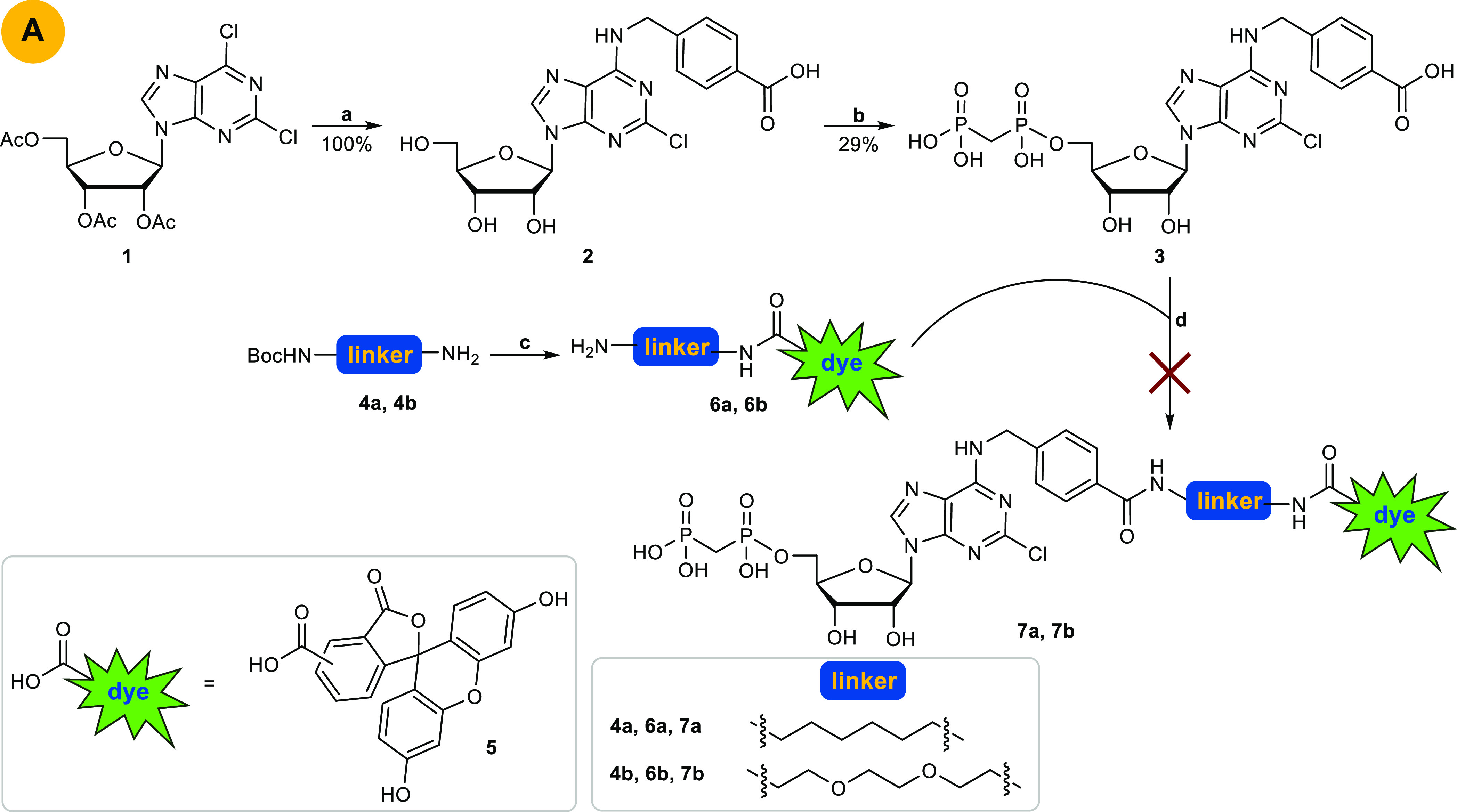
Reagents and conditions: (a) two steps: (i) 4-(aminomethyl)benzoic acid, Et3N, EtOH absolute, reflux, overnight; (ii) sodium methoxide, methanol, rt, 18 h; (b) two steps: (i) methylenebis(phosphonic dichloride), trimethyl phosphate, Ar, 0 °C, 30 min; (ii) triethylammonium bicarbonate (TEAC) buffer pH 7.4–7.6, rt, 1 h; (c) (i) 5, HOBt, DCC, THF, rt, overnight; (ii) 6–8% TFA, DCM, rt, 6 h; (d) HOBt, DCC, THF, rt, overnight.
Subsequent phosphonylation with methylenebis(phosphonic dichloride) in trimethyl phosphate followed by hydrolysis with aqueous triethylammonium bicarbonate (TEAC) buffer according to a previously reported method33 yielded product 3. The dye was attached to a linker by coupling of 5(6)-carboxyfluorescein (5) with commercially available boc-protected 1,6-hexanediamine (4a) or N-boc-2,2′-(ethylenedioxy)diethylamine (4b) in the presence of dicyclohexylcarbodiimide (DCC) and hydroxybenzotriazole (HOBt) in tetrahydrofuran (THF) followed by boc-deprotection with trifluoroacetic acid (TFA) in dichloromethane (DCM) yielding 6a and 6b.34 The final amide coupling reaction of carboxylic acid 3 with amine 6a or 6b with DCC/HOBt in THF, however, failed to produce 7a and 7b. This was probably due to a competing reaction of the phosphonic acid moiety of nucleotide analog 3 with DCC. Therefore, we decided to alternate the reaction sequence (Scheme 2). In synthetic approach B (see Scheme 2), the fluorescent dye with the attached linker (compound 6a, 6b) was coupled with the carboxybenzyl-substituted nucleoside 2 yielding the fluorescent-labeled adenosine derivatives 8a and 8b. Final phosphonylation of the nucleosides, however, was not successful. A reason for this might be the insufficient solubility of 8a and 8b in trimethyl phosphate. Next, we attached only the linker moiety to N6-carboxybenzyladenosine (2) yielding amino-functionalized nucleoside derivative 9a and 9b. Subsequent phosphonylation provided the nucleotide analogs 10a and 10b (approach C, Scheme 2). Final coupling with 5(6)-carboxyfluorescein 5 to introduce the fluorescent label, however, failed (Scheme 2).
Scheme 2. Pathways B and C to Obtain Compounds 7a and 7b.
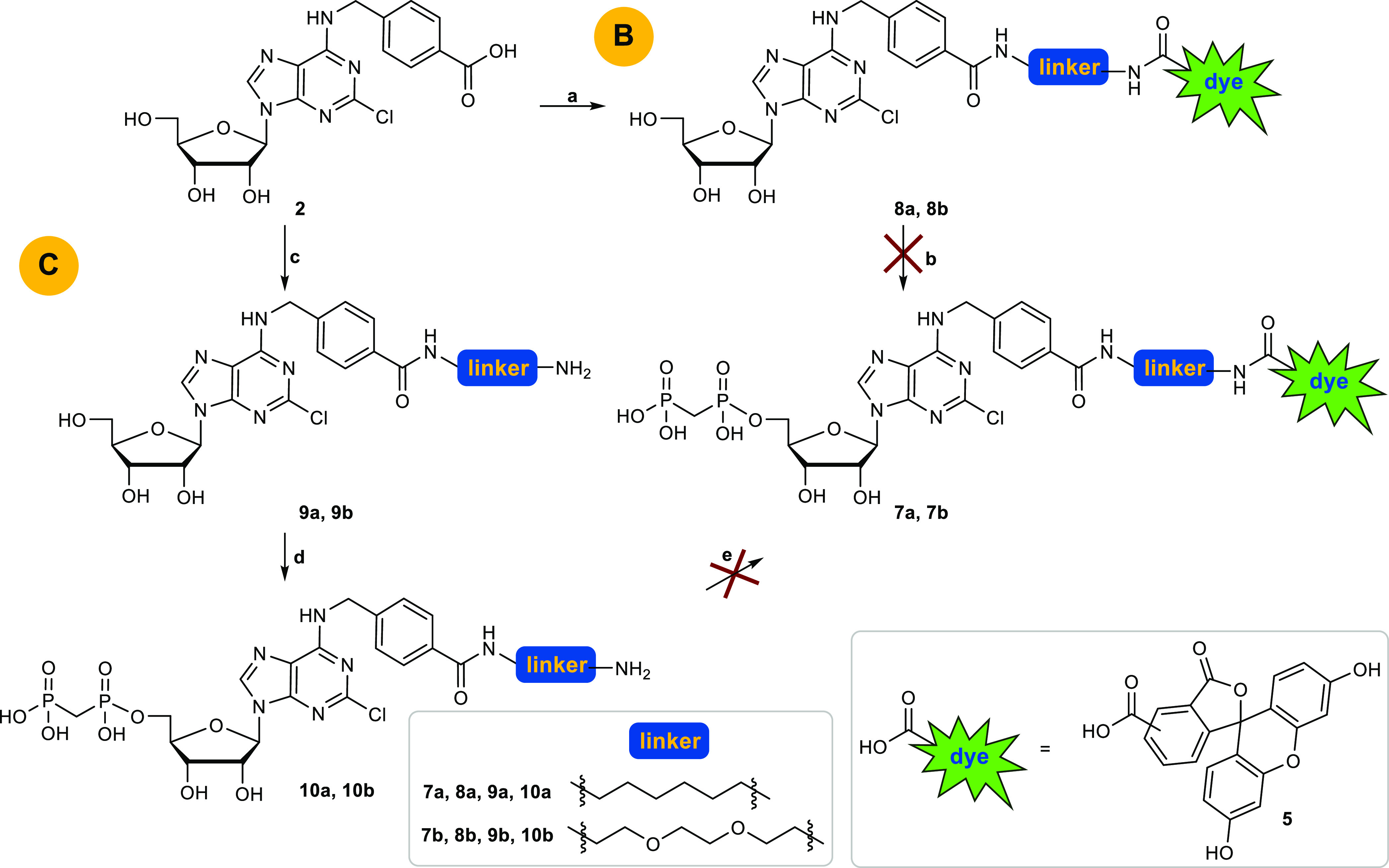
Reagents and conditions: (a) 6a or 6b, HOBt, DCC, THF, rt, overnight; (b) two steps: (i) methylenebis(phosphonic dichloride), trimethyl phosphate, Ar, 0 °C, 30 min; (ii) TEAC buffer, pH 7.4–7.6, rt, 1 h; (c) two steps: (i) 6a or 6b, HOBt, DCC, THF, rt, overnight; (ii) 6–8% TFA, DCM, rt, 6 h; (d) two steps: (i) methylenebis(phosphonic dichloride), trimethyl phosphate, Ar, 0 °C, 30 min; (ii) TEAC buffer, pH 7.4–7.6, rt, 1 h; (e) 5, HOBt, DCC, THF, rt, overnight.
Due to these difficulties, we designed an alternative approach avoiding amide coupling reaction to attach the fluorescent label to the nucleoside or nucleotide derivative. Thus, we decided to precouple the entire 6-substituents of the target purine nucleotide analogs 7a and 7b and introduce it in the very last step by nucleophilic substitution of the 6-chloropurine nucleotide precursor (see Scheme 3). This required phosphonylation of 6-chloro- or 2,6-dichloropurine-β-d-ribonucleoside before introduction of the 6-substituent. However, deprotection of 2,6-dichloro-9-(2′,3′,5′-tri-O-acetyl-β-d-ribofuranosyl)purine (1) with sodium methoxide or ammonia can lead to nucleophilic substitution of the chloro-function at the 6-position of the purine nucleoside. Therefore, we focused on lead structure I (Figure 2) for fluorescent labeling, which lacks the 2-chloro substituent of lead structure IX and corresponding target structures 7a,b.
Scheme 3. Successful Pathway D to Obtain 14a and 14b.
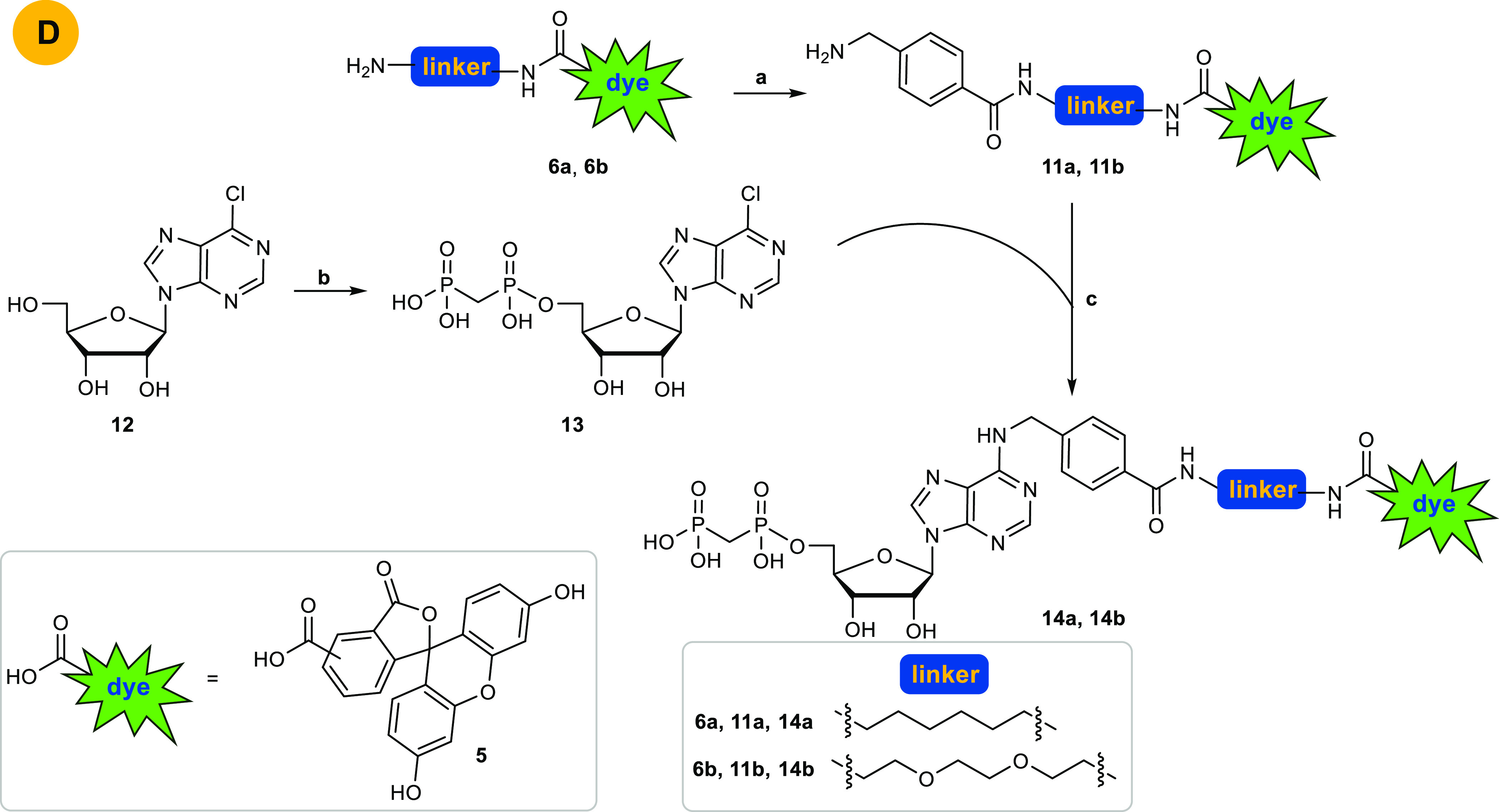
Reagents and conditions: (a) (i) 4-(aminomethyl)benzoic acid, HOBt, DCC, THF, rt, overnight; (ii) 6–8% TFA, DCM, rt, 6 h; (b) two steps: (i) methylenebis(phosphonic dichloride), trimethyl phosphate, Ar, 0 °C, 30 min; (ii) TEAC buffer, pH 7.4–7.6, rt, 1 h; (c) Et3N, EtOH absolute, reflux, overnight.
In the first step, the linker-dye conjugates 6a and 6b were coupled to 4-(boc-aminobenzyl)benzoic acid which was preactivated using DCC and HOBt in THF. The boc-group was subsequently cleaved using TFA.34 Purification by RP-HPLC yielded the desired amines 11a and 11b. In parallel, commercially available 6-chloro-β-d-ribofuranosylpurine (12) was phosphonylated using methylenebis(phosphonic dichloride) in trimethyl phosphate followed by hydrolysis with aqueous TEAC buffer according to a previously reported method.33 Purification by RP-HPLC to remove inorganic phosphates gave the desired nucleotide analog 13. In the next step, nucleophilic substitution of 13 by the amines 11a or 11b in absolute ethanol in the presence of triethylamine was performed under reflux conditions in analogy to published procedures.20 Final purification by RP-HPLC gave the desired products 14a and 14b in yields of 50% and 49%, respectively (last reaction step). The structures of the synthesized fluorescent nucleotide analogs were confirmed by 1H-, 13C-, and 31P NMR spectroscopy, in addition to LC/ESI-(UV)MS analysis performed in both positive and negative mode, which confirmed a purity of greater than 95% for both compounds.
Enzyme Inhibition Assays
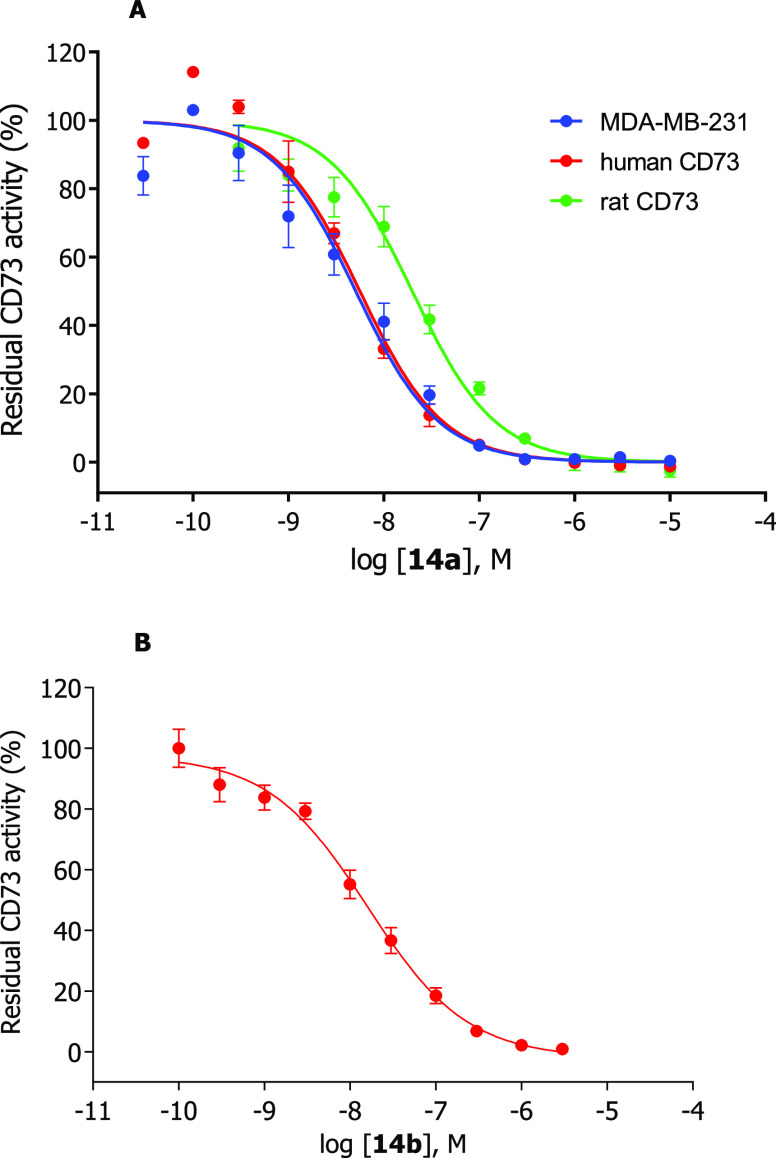
Interaction of the fluorescent probes 14a and 14b with CD73 was determined in radiometric enzyme assays using [3H]AMP as a substrate.35 After incubation, the remaining substrate [3H]AMP can be precipitated with lanthanum chloride and removed by filtration. The enzymatic product [3H]adenosine is collected in scintillation vials and scintillation cocktail is added, followed by quantification using liquid scintillation counting. Different CD73 preparations were employed, including rat and human recombinant soluble CD73 expressed in Spodoptera frugiperda 9 (Sf9) insect cells and membrane preparations of triple-negative breast cancer cells (MDA-MB-231) which natively express CD73.36,37 Full concentration-inhibition curves were determined (Figure 3), and Ki values were calculated from the obtained IC50 values using the Cheng–Prusoff equation.38
Figure 3.
Concentration-inhibition curves of 14a (A) and 14b (B) at human recombinant soluble CD73 (red circles), human membrane-bound CD73 natively expressed in the triple negative breast cancer cell line MDA-MB-231 (blue circles), and rat recombinant soluble CD73 (green circles). Data points are means ± standard error (SEM) from 3 separate experiments. Determined Ki values for 14a: MDA-MB-231 cells, 4.59 nM; human soluble CD73, 2.98 nM; rat soluble CD73, 26.0 nM. Determined Ki value for 14b: human soluble CD73, 12.6 nM.
Fluorescent probe 14b showed a Ki value of 12.6 nM at soluble human CD73, which is in the same range as that of lead compound I (2.21 nM).20 Fluorescence-labeled CD73 inhibitor 14a (Ki 2.98 nM) was 4-fold more potent than 14b. This shows that the nature of the linker has an influence on the affinity: the lipophilic alkyl linker in 14a is better tolerated than the more polar ethylene glycol linker in 14b. Another reason for the difference in potency might be due to the linker length of 14a consisting of 6 carbon atoms, whereas the longer linker in 14b consists of 8 atoms. This leads to the conclusion that a short lipophilic linker is well tolerated by CD73. The more potent compound 14a was additionally evaluated at soluble CD73 from rat and at membrane preparations of triple-negative breast cancer cells (MDA-MB-231), which natively express CD73. The compound displayed a similar Ki value (Ki 4.59 nM) at membrane preparations of MDA-MB-231 cells as at soluble CD73. This is in agreement with previous observations that AOPCP derivatives and analogs, which act as competitive CD73 inhibitors, showed virtually the same inhibitory potency at the soluble as at the membrane-anchored enzyme. At rat CD73, the Ki-value was, however, 8-fold higher (Ki 26.0 nM), indicating species differences between rat and human CD73. The most potent fluorescent CD73 inhibitor 14a was subsequently tested for selectivity versus human ecto-nucleoside triphosphate diphosphohydrolases (ecto-NTPDases), which cleave extracellular nucleoside tri- and diphosphates such as ATP and ADP yielding AMP. The fluorescent CD73 inhibitor 14a was found to be inactive at all four subtypes, NTPDase1 (CD39), NTPDase2 (CD39L1), NTPDase3 (CD39L3), and NTPDase8 at a high concentration of 50 μM (for details see SI). This is in line with previous results showing that N6-substituted AOPCP derivatives typically display high selectivity for CD73.20−22 Moreover, compound 14a neither activated nor inhibited the G protein-coupled ADP-activated human P2Y12 receptor, which is highly expressed on blood platelets, at a high concentration of 10 μM (for details see SI).
Fluorescence Microscopy
Human breast adenocarcinoma cells (MDA-MB-231) were costained with anti-CD73 antibody, together with fluorescence-labeled compounds 14a (Figure 4A) and 14b (Figure 4B). This confocal microscopy analysis confirmed our previous data on high cell-surface expression of CD73 on the cancer cells studied,37,39 and further demonstrated the ability of the novel fluorescent inhibitors to bind to CD73. Our previous studies have also shown the abundant expression of CD73 in the sensory neuroretina and other ocular structures.7,40 Therefore, in another set of experiments we evaluated the CD73-binding ability of 14a and 14b by using mouse eye as a native source of the enzyme. To evaluate the specificity of this binding, eyes were also dissected from CD73–/– mice created by targeted gene disruption, as described previously.41,42 First, the distribution of CD73 activity in the mouse retina was evaluated in situ by a lead (Pb) precipitation method. Employing AMP as a substrate revealed the presence of high AMP-specific brown staining in the rod-and-cones-containing photoreceptor layer in CD73+/+, but not in the CD73–/– eye preparation (Figure 5A). Free-floating eye sections were then costained with antibodies against CD73 and vimentin, in combination with fluorescent CD73 inhibitors. High-resolution immunofluorescence imaging of the medial area of the wild-type retina revealed high CD73 immunoreactivity throughout the entire processes of the photoreceptor cells with the highest expression levels in the outer segment discs and synaptic bodies. A similar staining pattern was observed during incubation of the samples with the fluorescent CD73 inhibitors 14a (Figure 5B) and 14b (Figure 5C). Strikingly, no CD73- and 14a/14b-specific fluorescence signals were detected in CD73–/– eyes (Figure 5B and C, lower panels), thus confirming high specificity of the tested compounds capable of binding selectively to CD73. Notably, the eyes were also costained with vimentin serving as a marker of the intermediate filaments in glial Müller cells, which span across the entire thickness of the neural retina in both CD73+/+ and CD73–/– eyes (see Figure 5).
Figure 4.
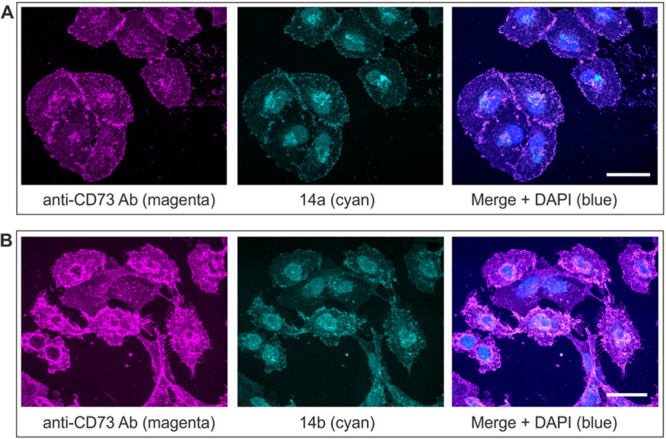
Fluorescence imaging of CD73+ MDA-MB-231 cells treated with the fluorescent CD73 inhibitors. MDA-MB-231 cells grown on cover slips were stained with rabbit anti-human ecto-5′-nucleotidase/CD73 antibody (h5NT-1L)43 and subsequently incubated with the appropriate second-stage antibody (Alexa Fluor 633 goat anti-rabbit IgG), together with fluorescence-labeled CD73 inhibitors 14a (A) and 14b (B), as indicated. The right-hand panels show the merged images with nuclei counterstained with blue fluorescent 4′,6-diamidine-2′-phenylindole dihydrochloride (DAPI); scale bars: 40 μm.
Figure 5.
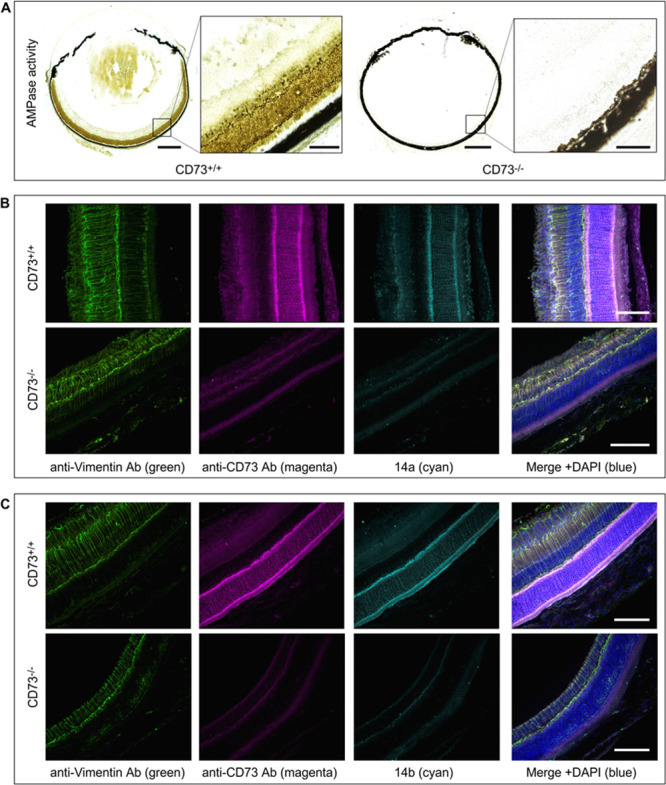
Evidence for selective binding of fluorescent CD73 inhibitors to CD73 in mouse retina. Eyeballs from wild-type (CD73+/+) and knockout CD73–/– mice were enucleated and subsequently subjected to enzyme histochemistry (A), and immunofluorescence staining (B, C) as described in the Supporting Information. (A) AMPase activity was assayed by incubating the eye cryosections from CD73+/+ (left-hand images) and CD73–/– (right-hand) mice with 1 mM AMP, followed by microscopic detection of the nucleotide-derived inorganic phosphate (Pi) as a brown precipitate. (B, C) Free-floating vibratome-cut sections of CD73+/+ (upper panels) and CD73–/– (lower panels) eyes were costained with rabbit anti-rat CD73 (rNu9L-I5)43 and chicken anti-vimentin antibodies and subsequently incubated with the appropriate isotype-matched fluorochrome-conjugated second-stage antibodies, together with fluorescence-labeled CD73 inhibitors 14a and 14b, as indicated. The right-hand panels show the merged images with nuclei counterstained with DAPI; scale bars: 500 μm (A), and 80 μm (A, inset, B, C).
Metabolic Stability
In order to investigate potential enzymatic degradation of 14a, its metabolic stability was investigated in human liver microsomes. These experiments revealed a high metabolic stability with an internal clearance (Clint) of 12 μL/min/mg protein corresponding to a half-life of 116 min (for details see the SI).
Cytotoxicity
Compounds 14a and 14b were additionally tested for potential cytotoxicity in CD73-expressing cell lines, namely the lung cancer cell line A54944 and the triple-negative breast cancer cell line MDA-MB-23121 at concentrations of 1000 nM corresponding to >300-fold of the Ki value of 14a and >80-fold of the Ki value of 14b, respectively. Neither cell proliferation nor viability were affected by the compounds. Even concentrations of up to 10,000 nM, studied in MDA-MB-231 cells, did not affect cell proliferation (for details see the SI). This shows that the fluorescent-labeled CD73 inhibitors should be safe for use in living systems.
Conclusions
Novel fluorescent-labeled AOPCP-derived compounds capable of selectively binding to cell surface CD73 and potently inhibiting its catalytic activity were designed and prepared in high purity. A synthetic route that allowed their fast preparation in good yields was successfully established. The fluorescent compounds showed low nanomolar Ki values, with the most potent ligand being 14a with a Ki value of 2.98 nM. Both 14a and 14b were successfully applied for fluorescence labeling of CD73 in triple negative human breast cancer cells, as well as photoreceptor CD73 in the mouse eye. These compounds represent the first fluorescent small-molecular probes for labeling CD73 of humans and rodents and may become useful diagnostic tools for this pathologically important protein which represents a promising novel drug target.
Acknowledgments
We thank Marion Schneider for LCMS and NMR analyses and Sabine Terhart-Krabbe for NMR spectra.
Glossary
Abbreviations
- ADP
adenosine diphosphate
- AMP
adenosine monophosphate
- AOPCP
adenosine-5′-O-[(phosphonomethyl)phosphonic acid]
- ATP
adenosine triphosphate
- Boc
tert-butyloxycarbonyl
- CD73
ecto-5′-nucleotidase
- Clint
internal clearance
- DAD
diode array detector
- DCC
N,N′-dicyclohexylcarbodiimide
- DCM
dichloromethane
- DCU
dicyclohexylurea
- DMAP
4-dimethylaminopyridine
- DMF
dimethylformamide
- DMSO
dimethylsulfoxide
- GPI
glycosylphosphatidylinositol
- HOBt
1-hydroxybenzotriazole
- HPLC
high performance liquid chromatography
- LC/ESI-MS
HPLC analysis coupled to electrospray ionization mass spectrometry
- NMR
nuclear magnetic resonance
- PBS
phosphate-buffered saline
- PSB
Pharmaceutical Sciences Bonn
- SAR
structure–activity relationship
- SEM
standard error of the mean
- t1/2
half-life
- TEAC
triethylammonium hydrogencarbonate
- TFA
trifluoroacetic acid
- THF
tetrahydrofuran
- TLC
thin layer chromatography
- UV
ultraviolet
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.0c00391.
Synthetic procedures; 1H, 13C, and spectral data of compounds 2, 3, 6a, 6b, 8a, 8b, 9a, 9b, 11a, and 11b; 1H, 13C, and 31P spectral data of 10a, 10b, 13, 14a, and 14b; yields of nucleosides and nucleotides; optimized reaction conditions; description of pharmacological assays; selectivity studies versus NTPDases1, -2, -3, -8 and P2Y12; metabolic stability experiments; cytotoxicity experiments; absorption and emission spectra of 14a and 14b in different solvents. (PDF)
Author Contributions
‡ The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript. C.C.S. and G.R. contributed equally.
We acknowledge financial support by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation), Project-ID: 335447717, SFB 1328. R.M.I. is grateful for a scholarship by the government of Sudan.
The authors declare the following competing financial interest(s): C.E.M. has given scientific advice to Arcus Biosciences Inc., a publicly traded biotechnology company.
Supplementary Material
References
- Colgan S. P.; Eltzschig H. K.; Eckle T.; Thompson L. F. Physiological roles for ecto-5′-nucleotidase (CD73). Purinergic Signalling 2006, 2, 351–360. 10.1007/s11302-005-5302-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yegutkin G. G. Enzymes involved in metabolism of extracellular nucleotides and nucleosides: functional implications and measurement of activities. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 473–497. 10.3109/10409238.2014.953627. [DOI] [PubMed] [Google Scholar]
- Cekic C.; Linden J. Purinergic regulation of the immune system. Nat. Rev. Immunol. 2016, 16, 177–192. 10.1038/nri.2016.4. [DOI] [PubMed] [Google Scholar]
- Antonioli L.; Blandizzi C.; Pacher P.; Haskó G. Immunity, inflammation and cancer: a leading role for adenosine. Nat. Rev. Cancer 2013, 13, 842–857. 10.1038/nrc3613. [DOI] [PubMed] [Google Scholar]
- Baqi Y. Ecto-nucleotidase inhibitors: recent developments in drug discovery. Mini-Rev. Med. Chem. 2015, 15, 21–33. 10.2174/1389557515666150219115141. [DOI] [PubMed] [Google Scholar]
- Knapp K.; Zebisch M.; Pippel J.; El-Tayeb A.; Müller C. E.; Sträter N. Crystal structure of the human ecto-5′-nucleotidase (CD73): insights into the regulation of purinergic signaling. Structure 2012, 20, 2161–2173. 10.1016/j.str.2012.10.001. [DOI] [PubMed] [Google Scholar]
- Zeiner J.; Loukovaara S.; Losenkova K.; Zuccarini M.; Korhonen A. M.; Lehti K.; Kauppinen A.; Kaarniranta K.; Müller C. E.; Jalkanen S.; Yegutkin G. G. Soluble and membrane-bound adenylate kinase and nucleotidases augment ATP-mediated inflammation in diabetic retinopathy eyes with vitreous hemorrhage. J. Mol. Med. 2019, 97, 341–354. 10.1007/s00109-018-01734-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonioli L.; Pacher P.; Vizi E. S.; Haskó G. CD39 and CD73 in immunity and inflammation. Trends Mol. Med. 2013, 19, 355–367. 10.1016/j.molmed.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard B.; Longhi M. S.; Robson S. C.; Stagg J. The ectonucleotidases CD39 and CD73: novel checkpoint inhibitor targets. Immunol. Rev. 2017, 276, 121–144. 10.1111/imr.12528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbach M.; Kimura K.; Luis T. C.; Fuegemann C. J.; Woll P. S.; Hesse M.; Facchini R.; Rieck S.; Jobin K.; Reinhardt J.; Ohneda O.; Wenzel D.; Geisen C.; Kurts C.; Kastenmüller W.; Hölzel M.; Jacobsen S. E. W.; Fleischmann B. K. In vivo labeling by CD73 marks multipotent stromal cells and highlights endothelial heterogeneity in the bone marrow niche. Cell Stem Cell 2018, 22, 262–276. 10.1016/j.stem.2018.01.008. [DOI] [PubMed] [Google Scholar]
- Allard B.; Allard D.; Buisseret L.; Stagg J. The adenosine pathway in immuno-oncology. Nat. Rev. Clin. Oncol. 2020, 1–19. 10.1038/s41571-020-0382-2. [DOI] [PubMed] [Google Scholar]
- Wang L.; Fan J.; Thompson L. F.; Zhang Y.; Shin T.; Curiel T. J.; Zhang B. CD73 has distinct roles in nonhematopoietic and hematopoietic cells to promote tumor growth in mice. J. Clin. Invest. 2011, 121, 2371–2382. 10.1172/JCI45559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.; Liao X.; Yu J.; Zhou P. Role of CD73 in disease: promising prognostic indicator and therapeutic target. cor. Curr. Med. Chem. 2018, 25, 2260–2271. 10.2174/0929867325666180117101114. [DOI] [PubMed] [Google Scholar]
- Corbelini P. F.; Figueiró F.; das Neves G. M.; Andrade S.; Kawano D. F.; Oliveira Battastini A. M.; Eifler-Lima V. L. Insights into ecto-5′-nucleotidase as a new target for cancer therapy: a medicinal chemistry study. Curr. Med. Chem. 2015, 22, 1776–1792. 10.2174/0929867322666150408112615. [DOI] [PubMed] [Google Scholar]
- Iqbal J. Ectonucleotidases: potential target in drug discovery and development. Mini-Rev. Med. Chem. 2019, 19, 866–869. 10.2174/138955751911190517102116. [DOI] [PubMed] [Google Scholar]
- Buchheiser A.; Ebner A.; Burghoff S.; Ding Z.; Romio M.; Viethen C.; Lindecke A.; Köhrer K.; Fischer J. W.; Schrader J. Inactivation of CD73 promotes atherogenesis in apolipoprotein E-deficient mice. Cardiovasc. Res. 2011, 92, 338–347. 10.1093/cvr/cvr218. [DOI] [PubMed] [Google Scholar]
- Kovács Z.; Dobolyi A.; Kékesi K. A.; Juhász G. 5′-nucleotidases, nucleosides and their distribution in the brain: pathological and therapeutic implications. Curr. Med. Chem. 2013, 20, 4217–4240. 10.2174/0929867311320340003. [DOI] [PubMed] [Google Scholar]
- Boison D.; Yegutkin G. G. Adenosine metabolism: emerging concepts for cancer therapy. Cancer Cell 2019, 36, 582–596. 10.1016/j.ccell.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intervention Dynamic Trial Listing Page. Available at https://www.cancer.gov/about-cancer/treatment/clinical-trials/intervention/anti-cd73-monoclonal-antibody-medi9447, Published Online: April 14, 2020, last accessed on 2020-04-14.
- Bhattarai S.; Freundlieb M.; Pippel J.; Meyer A.; Abdelrahman A.; Fiene A.; Lee S.-Y.; Zimmermann H.; Yegutkin G. G.; Sträter N.; El-Tayeb A.; Müller C. E. α,β-Methylene-ADP (AOPCP) derivatives and analogues: development of potent and selective ecto-5′-nucleotidase (CD73) inhibitors. J. Med. Chem. 2015, 58, 6248–6263. 10.1021/acs.jmedchem.5b00802. [DOI] [PubMed] [Google Scholar]
- Bhattarai S.; Pippel J.; Meyer A.; Freundlieb M.; Schmies C.; Abdelrahman A.; Fiene A.; Lee S.-Y.; Zimmermann H.; El-Tayeb A.; Yegutkin G. G.; Sträter N.; Müller C. E. X-ray co-crystal structure guides the way to subnanomolar competitive ecto-5′-nucleotidase (CD73) inhibitors for cancer immunotherapy. Adv. Therap. 2019, 2, 1900075. 10.1002/adtp.201900075. [DOI] [Google Scholar]
- Bhattarai S.; Pippel J.; Scaletti E.; Idris R.; Freundlieb M.; Rolshoven G.; Renn C.; Lee S.-Y.; Abdelrahman A.; Zimmermann H.; El-Tayeb A.; Müller C. E.; Sträter N. 2-substituted α,β-methylene-ADP derivatives: potent competitive ecto-5′-nucleotidase (CD73) inhibitors with variable binding modes. J. Med. Chem. 2020, 63, 2941–2957. 10.1021/acs.jmedchem.9b01611. [DOI] [PubMed] [Google Scholar]
- Bowman C. E.; da Silva R. G.; Pham A.; Young S. W. An exceptionally potent inhibitor of human CD73. Biochemistry 2019, 58, 3331–3334. 10.1021/acs.biochem.9b00448. [DOI] [PubMed] [Google Scholar]
- Baqi Y.; Lee S.-Y.; Iqbal J.; Ripphausen P.; Lehr A.; Scheiff A. B.; Zimmermann H.; Bajorath J.; Müller C. E. Development of potent and selective inhibitors of ecto-5′-nucleotidase based on an anthraquinone scaffold. J. Med. Chem. 2010, 53, 2076–2086. 10.1021/jm901851t. [DOI] [PubMed] [Google Scholar]
- Ripphausen P.; Freundlieb M.; Brunschweiger A.; Zimmermann H.; Müller C. E.; Bajorath J. Virtual screening identifies novel sulfonamide inhibitors of ecto-5′-nucleotidase. J. Med. Chem. 2012, 55, 6576–6581. 10.1021/jm300658n. [DOI] [PubMed] [Google Scholar]
- Iqbal J.; Saeed A.; Raza R.; Matin A.; Hameed A.; Furtmann N.; Lecka J.; Sévigny J.; Bajorath J. Identification of sulfonic acids as efficient ecto-5′-nucleotidase inhibitors. Eur. J. Med. Chem. 2013, 70, 685–691. 10.1016/j.ejmech.2013.10.053. [DOI] [PubMed] [Google Scholar]
- Lawson K. V.; Kalisiak J.; Lindsey E. A.; Newcomb E.; Leleti M. R.; Debien L.; Rosen B. R.; Miles D. H.; Sharif E. U.; Jeffrey J.; Tan J. B. L.; Chen A.; Zhao S.; Xu G.; Fu L.; Jin L.; Park T. W.; Berry W.; Moschütz S.; Scaletti E. R.; Sträter N.; Walker N. P.; Young S. W.; Walters M. J.; Schindler U.; Powers J. P.. Discovery of AB680 - a potent and selective inhibitor of CD73. J. Med. Chem. 2020, 10.1021/acs.jmedchem.0c00525. [DOI] [PubMed] [Google Scholar]
- Dumontet C.; Peyrottes S.; Rabeson C.; Cros-Perrial E.; Géant P. Y.; Chaloin L.; Jordheim L. P. CD73 inhibition by purine cytotoxic nucleoside analogue-based diphosphonates. Eur. J. Med. Chem. 2018, 157, 1051–1055. 10.1016/j.ejmech.2018.08.035. [DOI] [PubMed] [Google Scholar]
- Beatty J. W.; Lindsey E. A.; Thomas-Tran R.; Debien L.; Mandal D.; Jeffrey J. L.; Tran A. T.; Fournier J.; Jacob S. D.; Yan X.; Drew S. L.; Ginn E.; Chen A.; Pham A. T.; Zhao S.; Jin L.; Young S. W.; Walker N. P.; Leleti M. R.; Moschütz S.; Sträter N.; Powers J. P.; Lawson K. V. Discovery of Potent and Selective Non-Nucleotide Small Molecule Inhibitors of CD73. J. Med. Chem. 2020, 63, 3935–3955. 10.1021/acs.jmedchem.9b01713. [DOI] [PubMed] [Google Scholar]
- Guilbault G. G., Ed. Practical Fluorescence; Dekker: New York, 1990. [Google Scholar]
- O’Goshi K.-I.; Serup J. Safety of sodium fluorescein for in vivo study of skin. Skin Res. Technol. 2006, 12, 155–161. 10.1111/j.0909-752X.2006.00147.x. [DOI] [PubMed] [Google Scholar]
- Alford R.; Simpson H. M.; Duberman J.; Hill G. C.; Ogawa M.; Regino C.; Kobayashi H.; Choyke P. L. Toxicity of organic fluorophores used in molecular imaging: literature review. Mol. Imaging 2009, 8, 341–354. 10.2310/7290.2009.00031. [DOI] [PubMed] [Google Scholar]
- Kalek M.; Jemielity J.; Stepinski J.; Stolarski R.; Darzynkiewicz E. A direct method for the synthesis of nucleoside 5′-methylenebis(phosphonate)s from nucleosides. Tetrahedron Lett. 2005, 46, 2417–2421. 10.1016/j.tetlet.2005.02.069. [DOI] [Google Scholar]
- Horatscheck A.; Wagner S.; Ortwein J.; Kim B. G.; Lisurek M.; Beligny S.; Schütz A.; Rademann J. Benzoylphosphonate-based photoactive phosphopeptide mimetics for modulation of protein tyrosine phosphatases and highly specific labeling of SH2 domains. Angew. Chem., Int. Ed. 2012, 51, 9441–9447. 10.1002/anie.201201475. [DOI] [PubMed] [Google Scholar]
- Freundlieb M.; Zimmermann H.; Müller C. E. A new, sensitive ecto-5′-nucleotidase assay for compound screening. Anal. Biochem. 2014, 446, 53–58. 10.1016/j.ab.2013.10.012. [DOI] [PubMed] [Google Scholar]
- Junker A.; Renn C.; Dobelmann C.; Namasivayam V.; Jain S.; Losenkova K.; Irjala H.; Duca S.; Balasubramanian R.; Chakraborty S.; Börgel F.; Zimmermann H.; Yegutkin G. G.; Müller C. E.; Jacobson K. A. Structure-activity relationship of purine and pyrimidine nucleotides as ecto-5′-nucleotidase (CD73) inhibitors. J. Med. Chem. 2019, 62, 3677–3695. 10.1021/acs.jmedchem.9b00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losenkova K.; Zuccarini M.; Karikoski M.; Laurila J.; Boison D.; Jalkanen S.; Yegutkin G. G. Compartmentalization of adenosine metabolism in cancer cells and its modulation during acute hypoxia. J. Cell Sci. 2020, 133, jcs241463 10.1242/jcs.241463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung-Chi C.; Prusoff W. H. Relationship between the inhibition constant (KI) and the concentration of inhibitor which causes 50% inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973, 22, 3099–3108. 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- Virtanen S. S.; Kukkonen-Macchi A.; Vainio M.; Elima K.; Harkonen P. L.; Jalkanen S.; Yegutkin G. G. Adenosine inhibits tumor cell invasion via receptor-independent mechanisms. Mol. Cancer Res. 2014, 12, 1863–1874. 10.1158/1541-7786.MCR-14-0302-T. [DOI] [PubMed] [Google Scholar]
- Loukovaara S.; Sandholm J.; Aalto K.; Liukkonen J.; Jalkanen S.; Yegutkin G. G. Deregulation of ocular nucleotide homeostasis in patients with diabetic retinopathy. J. Mol. Med. 2017, 95, 193–204. 10.1007/s00109-016-1472-6. [DOI] [PubMed] [Google Scholar]
- Thompson L. F.; Eltzschig H. K.; Ibla J. C.; Van De Wiele C. J.; Resta R.; Morote-Garcia J. C.; Colgan S. P. Crucial role for ecto-5′-nucleotidase (CD73) in vascular leakage during hypoxia. J. Exp. Med. 2004, 200, 1395–1405. 10.1084/jem.20040915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yegutkin G. G.; Auvinen K.; Rantakari P.; Hollmen M.; Karikoski M.; Grenman R.; Elima K.; Jalkanen S.; Salmi M. Ecto-5′-nucleotidase/CD73 enhances endothelial barrier function and sprouting in blood but not lymphatic vasculature. Eur. J. Immunol. 2015, 45, 562–573. 10.1002/eji.201444856. [DOI] [PubMed] [Google Scholar]
- http://ectonucleotidases-ab.com/documents/human5-nucleotidase.pdf, last accessed on 2020-08-28.
- Zhu J.; Zeng Y.; Li W.; Qin H.; Zhe L.; Shen D.; Gu D.; Huang J.; Liu Z. CD73/NT5E is a target of miR-30a-5p and plays an important role in the pathogenesis of non-small cell lung cancer. Mol. Cancer 2017, 16, 34. 10.1186/s12943-017-0591-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.