Abstract
The initial greening of angiosperms involves light activation of photoreceptors that trigger photomorphogenesis, followed by the development of chloroplasts. In these semi‐autonomous organelles, construction of the photosynthetic apparatus depends on the coordination of nuclear and plastid gene expression. Here, we show that the expression of PAP8, an essential subunit of the plastid‐encoded RNA polymerase (PEP) in Arabidopsis thaliana, is under the control of a regulatory element recognized by the photomorphogenic factor HY5. PAP8 protein is localized and active in both plastids and the nucleus, and particularly required for the formation of late photobodies. In the pap8 albino mutant, phytochrome‐mediated signalling is altered, degradation of the chloroplast development repressors PIF1/PIF3 is disrupted, HY5 is not stabilized, and the expression of the photomorphogenesis regulator GLK1 is impaired. PAP8 translocates into plastids via its targeting pre‐sequence, interacts with the PEP and eventually reaches the nucleus, where it can interact with another PEP subunit pTAC12/HMR/PAP5. Since PAP8 is required for the phytochrome B‐mediated signalling cascade and the reshaping of the PEP activity, it may coordinate nuclear gene expression with PEP‐driven chloroplastic gene expression during chloroplast biogenesis.
Keywords: ArabidopsisPEP/PAPs, biogenesis, chloroplast, photobodies, photomorphogenesis
Subject Categories: Development & Differentiation, Plant Biology,
The plastidial RNA polymerase subunit PAP8 promotes chloroplast biogenesis in response to light via phytochrome B regulation of both skotomorphogenic and photomorphogenic factors in Arabidopsis thaliana.
Introduction
Chloroplasts are the organelles conducting photosynthesis in plants and green algae (Jarvis & Lopez‐Juez, 2013). In angiosperms, the perception of light is essential to trigger photomorphogenesis, during which the photosynthetic organelles differentiate from chlorophyll‐free proplastids. In contrast, seedlings sheltered from light, perform skotomorphogenesis, a dark‐specific developmental programme triggering hypocotyl elongation giving the shoot apex a chance to reach light. Meanwhile, the apical hook is preserved, maintaining non‐developing cotyledons downward and close, serving as a protective shield for the quiescent shoot apical meristem. At the cellular level, the cotyledons generate chlorophyll‐free etioplasts incapable of performing photosynthesis (Liebers et al, 2017). The lack of chloroplast development in the dark can be regarded as a way to optimize the use of limited resources stored in the seed to efficiently reach the surface. Then, illumination of the cotyledons causes the conversion of the phytochrome photoreceptors into an active state launching the photomorphogenic programme (Solymosi & Schoefs, 2010) and reviewed in Ref. Hernandez‐Verdeja et al (2020). This programme involves also major morphological changes in the seedling including the repression of hypocotyl elongation and the opening of the cotyledons that are then rapidly engaged in chloroplast biogenesis (Pogson et al, 2015).
As remnant of their endosymbiotic origin, plastids possess their own genetic system, which contributes to the construction of the photosynthetic apparatus after illumination (Jarvis & Lopez‐Juez, 2013). A plastid‐encoded RNA polymerase (PEP) is required for proper transcription of photosynthesis genes encoded by the plastid genome. This PEP complex is composed of a prokaryotic core of four distinct bacterial‐like subunits (α2, β, β′, β″) surrounded by (at least) 12 additional nuclear‐encoded subunits of eukaryotic origin (Pfannschmidt et al, 2015) known as PEP‐associated proteins (PAPs). The association of PAPs to the prokaryotic core is strictly induced by light through the action of phytochromes (Pfannschmidt & Link, 1994; Yang et al, 2019; Yoo et al, 2019). Importantly, the PAP association to the core of the PEP appears to be one important bottleneck of chloroplast formation since genetic inactivation of any PAP results in albinism (Pfalz & Pfannschmidt, 2013). The genes for PAPs appear to represent a regulatory unit that exhibits very similar co‐expression profiles albeit the involved genes encode proteins that belong to very different functional classes that could not be predictably united before. They all exhibit a basal expression in the dark followed by a rapid and transient peak after light exposure strongly suggesting a connection of their expression to the light regulation network (Liebers et al, 2018).
In the dark, photomorphogenesis is actively inhibited by the negative regulatory module CONSTITUTIVE PHOTOMORPHOGENIC/DE‐ETIOLATED/FUSCA (COP/DET/FUS) (Sullivan et al, 2003). In particular, the E3 ubiquitin ligase COP1 was shown to destabilize two basic domain/leucine zipper (bZIP) transcription factors known to initiate photomorphogenesis (ELONGATED HYPOCOTYL 5, HY5 and its homologous protein HYH) (Osterlund et al, 2000; Holm et al, 2002). Upon illumination, the cytosolic pool of inactive phytochrome B (PHYBPr) is converted into an active state (PHYBPfr), triggering its translocation into the nucleus (Yamaguchi et al, 1999; Chen et al, 2003) where it physically interacts with PHYTOCHROME‐INTERACTING FACTORS (PIFs) (Huq et al, 2004) leading to the emergence of a mutual negative feedback loop (Leivar & Monte, 2014). PIFs belong to a subset of the basic helix–loop–helix (bHLH) superfamily of transcription factors. Four of them in particular (PIF1, P1F3, P1F4 and P1F5) collectively act with some redundancy, as transcriptional repressors of photomorphogenesis in the dark (Leivar et al, 2008). They are destabilized by light upon their interaction with the photoactivated phytochrome molecules (Al‐Sady et al, 2006) leading to a de‐repression of the photomorphogenic programme (Jiao et al, 2007). The antagonistic role of phytochromes and the different PIFs is responsible for complex adaptive developmental responses of the seedling to changes in their light environment including de‐etiolation and shade avoidance. PIF1 and PIF3, in particular, were identified as having a predominant role in chloroplast development (Stephenson et al, 2009). In the pursuit of greening, the phytochrome‐mediated light signalling represses the COP1‐mediated destabilization of HY5, thereby leading to its accumulation. Stabilized HY5 can then initiate expression of photomorphogenic factors (Lee et al, 2007). Meanwhile, light exposure triggers the transcriptional activation of GOLDEN2‐LIKE 1 and 2 (GLK1 and GLK2) transcription factors that are responsible for the proper expression of nuclear photosynthesis genes (Waters & Langdale, 2009; Waters et al, 2009). The action of phytochrome within the nucleus was visualized using a GFP tag (PHYB‐GFP or PBG) revealing that phytochrome B translocates into the nucleus and then aggregates into specific speckles within the nuclear matrix (Yamaguchi et al, 1999). Early speckles are numerous and small, while later speckles become larger and less abundant without changing the phytochrome content that remains rather stable. Late speckle formation, also designated “late photobodies”, requires the presence of HEMERA (HMR), a dually localized protein present in the nucleus and in plastids (Chen et al, 2010; Nevarez et al, 2017), that is able to physically interact with phytochromes (Galvao et al, 2012). In plastids, HMR is known as pTAC12/PAP5 representing a member of the PAP family that is essential for chloroplast biogenesis since genetic inactivation of the protein blocks plastid differentiation leading to albinism (Pfalz et al, 2006, 2015). For PAPs, different functions can be predicted from their amino acid sequences, but their precise roles either as single subunits or in complex are not yet understood. PAP8 is one of the most enigmatic members among the PAPs, as its deduced amino acid sequence does not harbour any known functional domain although recent findings suggest a role as a transcriptional enhancer of the PEP complex (Ding et al, 2019). Separate genetic studies based on the mapping of quantitative trait loci in natural accessions of Arabidopsis revealed that the overlapping loci ESPRESSO (Swarup et al, 1999) and LIGHT1 (Borevitz et al, 2002), both corresponding to PAP8, are responsible for major variation in natural circadian rhythms or hypocotyl elongation across a wide range of environments (Loudet et al, 2008). ESPRESSO transcripts cycle in a diurnal pattern and LIGHT1 responds to various wavelengths and has a significant epistatic interaction with LIGHT2/PHYB. A separate transcript analysis revealed that PAP8 responds to temperature (Danilova et al, 2018a) and phyto‐hormones (Danilova et al, 2018b). In the following, PAP8 will be used as the gene name for At1g21600 also corresponding to ESPRESSO, LIGHT1 or pTAC6.
Here, we show that PAP8 is a dually localized nucleo‐plastidic protein with a nuclear pool important for the proper timing of chloroplast biogenesis. In particular PAP8 interacts with HMR, it is essential for phytochrome‐mediated signal transduction, PIF1 and PIF3 degradation, HY5 stabilization and GLK transcript accumulation indicating that it represents a novel key regulator of the light‐signalling network.
Results
PAP8 plays an essential role in chloroplast biogenesis
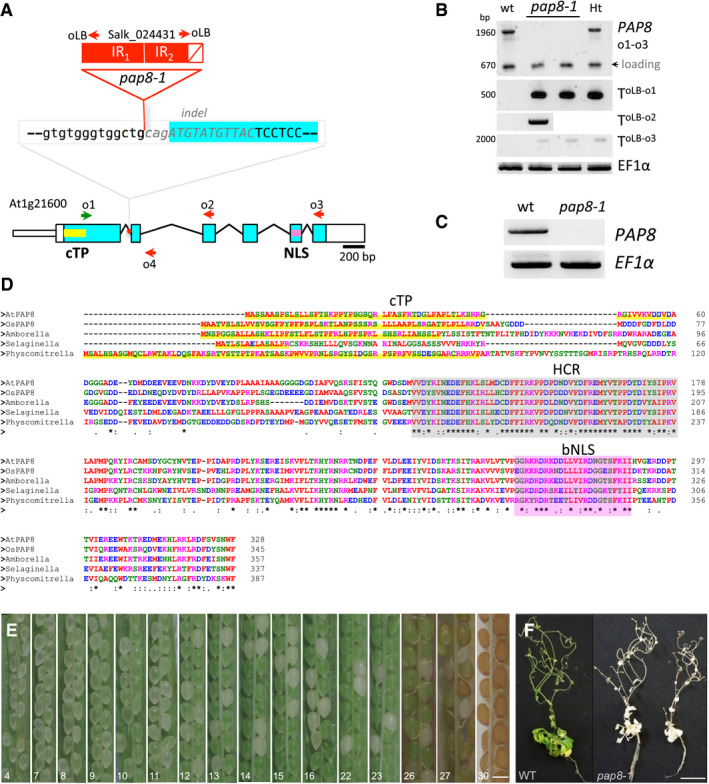
PAP8 was identified by targeted proteomics as pTAC6, a component of the transcriptionally active chromosome of plastids, a biochemically defined DNA‐protein structure capable of performing faithful transcription of plastid genes in vitro (Pfalz et al, 2006). The T‐DNA insertion line “SALK_024431” of Arabidopsis, referred to as the pap8‐1 mutant in this study, displayed an albino phenotype with a strong depletion of PEP‐dependent photosynthesis transcripts, a reduced pigments accumulation and developmentally arrested plastids (Pfalz et al, 2006; Appendix Fig S1). An orthologous protein of pTAC6 was then isolated from a highly purified Sinapis alba PEP complex and subsequently renamed PAP8 as being a bona fide component of this PEP complex (Steiner et al, 2011). The pap8‐1 allele corresponds to the insertion of an inverted repeat of the T‐DNA into the first intron of the gene (Fig 1A). Amplicon sequencing, after PCR‐based genotyping (Fig 1B), showed that 11 bp of the second exon are missing so that the open reading frame (ORF) is destroyed notwithstanding possible events of T‐DNA splicing. Besides, a PAP8 transcript spanning the insertion point could not be detected with RT–PCR in the homozygous pap8‐1 mutant (Fig 1C), indicating that pap8‐1 is a genuine null allele. The conceptually translated protein sequence is found in the terrestrial green lineage starting from mosses to Eudicots (Fig 1D), though absent in ferns, gymnosperms and a few basal angiosperms. A predicted N‐terminal chloroplast transit peptide (cTP) rapidly diverged while a highly conserved region (HCR) of unknown function seems to be under a strong selection pressure, as it is almost unchanged since the last common ancestor of all terrestrial plants. Hence, the sporophytic lethality of the pap8 mutant triggers the assumption that the protein had brought an important function to the green lineage in its way to conquer dry lands, and then became essential to Eudicots and Monocots.
Figure 1. Genetic analysis of the mutant pap8‐1 .
-
AStructure of the PAP8 locus, blue boxes: exons, lines: introns. Red box: inserted T‐DNA as inverted repeats (IR1/IR2) in the first intron. White box with a diagonal red line: deletion at the left border of IR2 and part of second exon (italicized grey sequence). Green and red arrows represent forward and reverse primers, respectively, as o1: oPAP8_rtp_F; o2: oPAP8_E3_R; o3: oPAP8_rtp_R; o4:op8i2_R; and oLB: oLBb1.3.
-
BPCR performed on genomic DNA with indicated primers as shown: o1: oPAP8_rtpF, o2: oPAP8_E3R, o3: oPAP8_rtpR, oLB: oLBb1.3, EF1α: ELONGATION FACTOR 1α, WT: wild type, pap8‐1: homozygous albino plant, Ht: heterozygous green plant; T: T‐DNA, arrowhead: 670‐bp contaminant amplification product used as loading control.
-
CRT–PCR on wild type and pap8‐1 homozygous plants grown in the dark for 3 days followed with 72‐h growth under white light to allow greening of the wild type; EF1α used as control.
-
DSequence alignment of predicted full‐length orthologous PAP8 protein found in representatives of major phylogenetic clades Arabidopsis thaliana, At1g21600; Oryza sativa Indica, EEC67529.1; Amborella trichopoda, XP_006827378.1; Selaginella moellendorffii, XP_002976643.2; Physcomitrella patens, XP_024396032.1. cTP, chloroplast transit peptide as predicted with ChloroP1.1 (www.cbs.dtu.dk) underlined in yellow; HCR shaded in grey, highly conserved region. (*), (:) or (.), conserved, strongly similar or weakly similar amino acid properties (standards from www.uniprot.org). Amino acids colours as in Clustal Omega (red (AVFPMILW): small + hydrophobic [includes aromomatic − Y]); blue (DE): acidic; magenta (RHK): basic; green (STYHCNGQ): hydroxly + sulfhydryl + amine + G.) bNLS, bipartite NLS as predicted with NLS mapper (http://nls-mapper.iab.keio.ac.jp).
-
EHalf‐open siliques of a heterozygous plant showing the embryo greening; scale bar equals 250 μm. The given number is the position rank of the silique from top to bottom of the inflorescence presenting the segregation of homozygous and heterozygous seeds based on their ability to transiently develop chloroplasts.
-
FMutant Rescue: WT and two representative pap8‐1 plants were grown in vitro using sucrose and low white light intensity (of 10 μmol m−2 s−1); scale bar equals 20 mm.
Source data are available online for this figure.
In all orthologous proteins, a nuclear localization signal (NLS) could be predicted, pointing to a possible localization of the protein inside the nucleus. Functional complementation (Fig EV1) was carried out following the strategy presented in Appendix Fig S2. The full‐length coding sequence of PAP8 driven by 1.1 kb of its own promoter (pPAP8::PAP8; Appendix Table S1) could fully restore the greening of the mutant with a chlorophyll content undistinguishable from that of the wild type (Fig EV1D and E). Heterozygotes were phenotypically undistinguishable from wild type except within the developing silique where one quarter of the embryos were unable to green (Fig 1E) following, without gametic distortion, Mendel's ratio for the segregation of recessive alleles (Appendix Fig S1B). Mutant homozygotes, however, were albino and sporophytic lethal, with a strong reduction in cotyledon size (Appendix Fig S1C). pap8‐1 dies quickly after light exposure unless grown in vitro on a carbon source in dimmed light (Fig 1F). Albeit their heterotrophic growth, plants pursued a rather normal development until reproduction. PAP8 is, therefore, a specific factor essential for chloroplast biogenesis without noticeably affecting other plastid functions non‐related to photosynthesis or the apparent photomorphogenic programme that is associated with de‐etiolated plants (such as ceased hypocotyl elongation, apical hook unfolding and cotyledon opening).
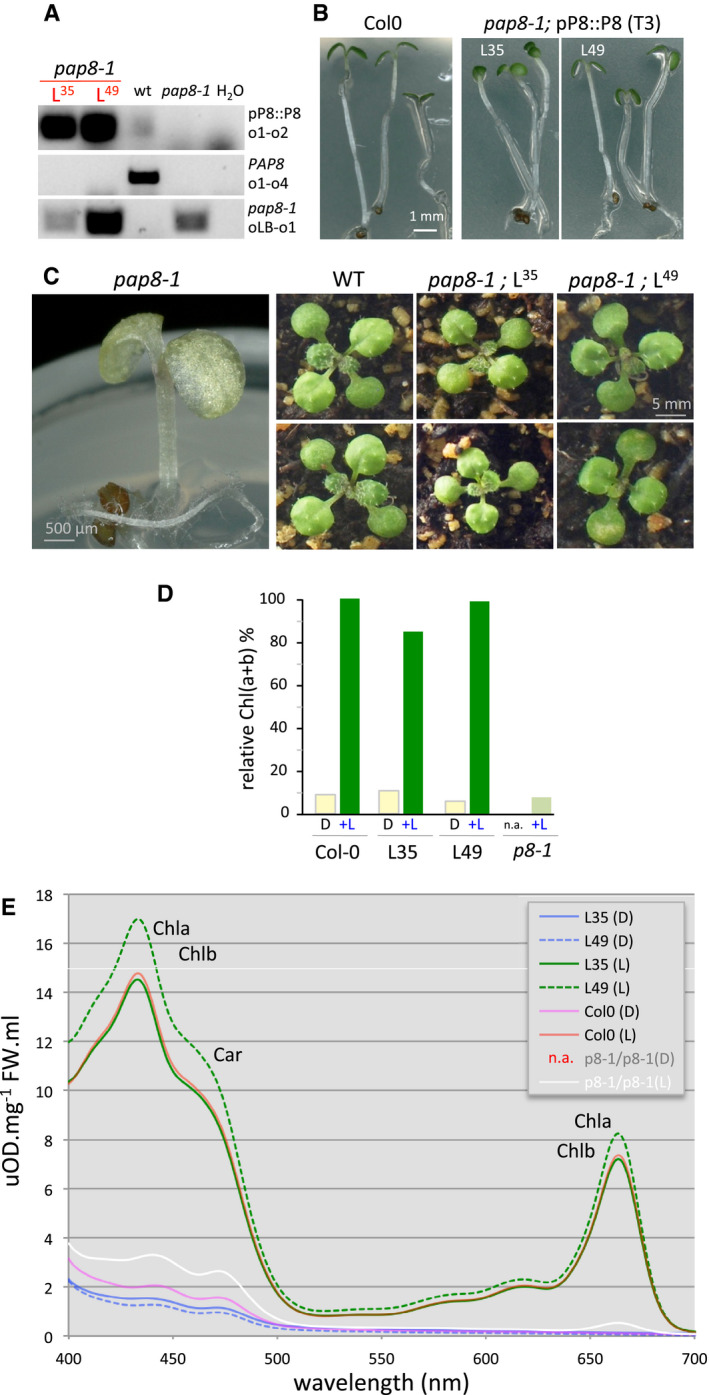
Figure EV1. (Related to Fig 1) Functional complementation of pap8‐1 .
-
APCR on genomic DNA; L35, L49: Two independent “pBB389” transgenic lines; primers are the same as in Fig 1B and o4: op8i2_R.
-
BGreening assay on wild type and rescued pap8‐1 homozygous plants from third generation transgenic lines (T3) grown in vitro 3 days in the dark followed with a 30‐h light treatment. L35 and L49 are two independent rescued lines.
-
CPhenotypes of pap8‐1 homozygous plant grown in vitro, and two representative plants of wild type or pap8‐1/pP8::PAP8 (line L35 or line L49) grown on soil.
-
DContent of total chlorophylls (Chl(a+b)) normalized to fresh weight and relative to wild type in the given genotypes grown in the dark (D) or grown in the dark followed with 30 h of white light treatment (+L); n.a. not applicable.
-
ESpectrophotometric analysis of pigments: absorption spectra of acetone‐soluble extracts from seedling grown in vitro 3 days in the dark (D) or 3 days in the dark plus 30 h of white light (L) Col‐0, wild type; p8‐1/p8-1, homozygous mutant pap8‐1; L35 and L49, two lines of pap8‐1/pPAP8::PAP8; n.a., not applicable. Absorbance was normalized to fresh weight (FW); Chla, chlorophyll a; Chlb, chlorophyll b; Car, carotenoids.
The PAP8 promoter involves typical light‐responsive cis‐elements
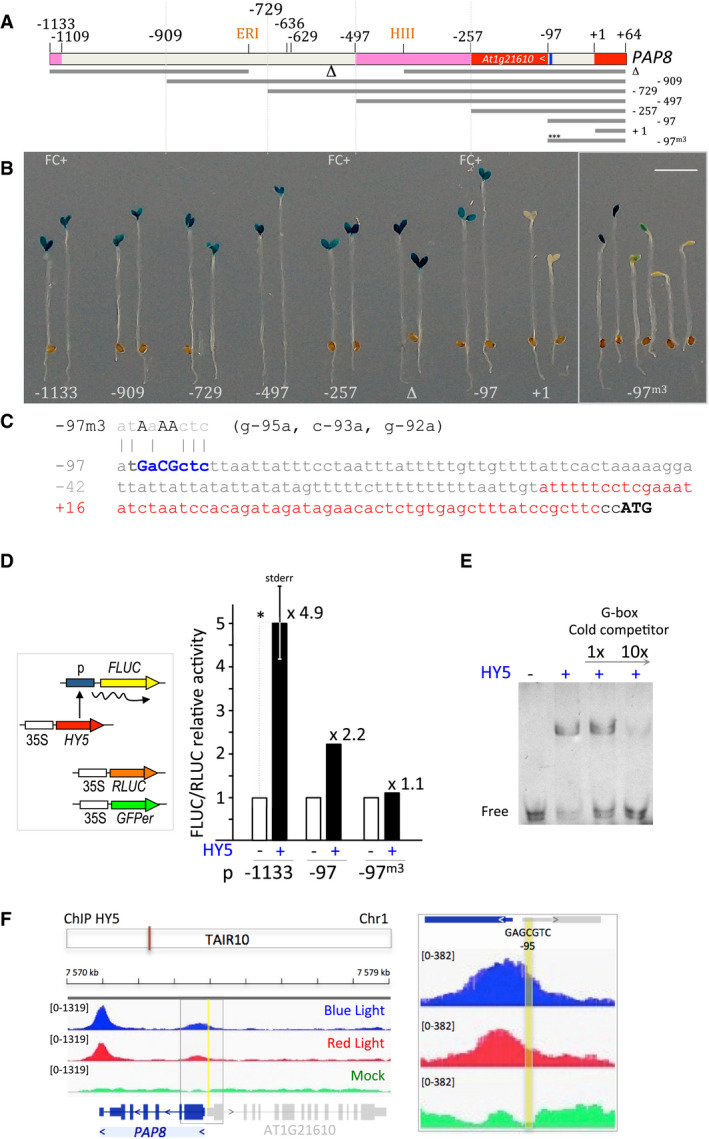
PAP genes are co‐regulated, at least for a significant part of their transcriptional response (Pfannschmidt et al, 2015; Liebers et al, 2018); as a canonical example, the promoter activity of PAP8 is transitorily specific to tissues with photosynthetic potential such as the cotyledons and leaf primordia. It is first restricted to the epidermis during skotomorphogenesis, induced in the palisade after light exposure and then slowly diminished (Liebers et al, 2018). Searching for cis‐regulatory elements by a deletion series of the PAP8 promoter, a short sequence starting at −97 from the transcriptional initiation start (tis) was found to be sufficient to retain cotyledon specificity while a construct starting at position +1 completely lost its reporter activity (Fig 2A and B). The two short versions of the promoter (−257 and −97) driving PAP8 expression were able to complement pap8‐1 (Appendix Table S1). Within the 97‐bp region (Fig 2C), a nearly palindromic element (GAcGCTC) was predicted to be a putative non‐symmetrical element recognized by proteins with basic leucine zipper domains (bZIP). Site‐directed mutagenesis of this element resulted in a disturbed GUS expression (Fig 2B). Using PlantPAN3 (Chow et al, 2019), three bona fide elements for bZIP transcription factors (TF) were predicted in both strands of the DNA (Appendix Fig S3 and Table S2). Interestingly, the two bZIP TFs, HY5 and HYH are known to be involved in the early steps of photomorphogenesis (Holm et al, 2002; Li et al, 2017). Hence, a few bZIP TFs, TGA2 as the best prediction according to the two elements found on the plus strand, HY5 and HYH as educated guesses, and bZIP60 as an out‐group related to stress response (Iwata et al, 2008) were tested in a dual‐luciferase reporter assay (Appendix Fig S4). HY5 proved to be the most efficient, enhancing transcriptional activity of the long (−1,133 bp) PAP8 promoter region by more than fivefold over the control (Fig 2D). For the shorter though functional −97‐bp promoter, HY5 promoted transcriptional activity with a twofold increase while a 3‐bp replacement in the core of the element yielded significantly reduced activation. Moreover, recombinant HY5 was able to specifically bind the cis‐regulatory element in vitro (Fig 2E) in strength comparable to that of the canonical G‐box element used as competitor (Yoon et al, 2006). In addition, the release of chromatin‐immuno‐precipitation (ChIP) sequencing data using “GFP” antibody on a hy5/HY5::HY5‐YFP genetic background (Hajdu et al, 2018) allowed the detection of HY5 on the 5′‐region containing the identified regulatory element and the 3′‐region of PAP8 after blue light or red light exposure (Fig 2F). While the expression of PAPs is essential for greening, hy5 mutants display slight greening defects indicating that functional redundancies and compensations occur in the regulation of its target genes (Gangappa & Botto, 2016). For example, the paralogous transcription factor HYH (Holm et al, 2002) is also active on the PAP8 promoter (Appendix Fig S4). In conclusion, ChIP and EMSA indicate that HY5 can bind the PAP8 promoter and that it can activate the promoter in a heterologous system, but given that no expression changes were seen in a hy5‐1 mutant, possibly due to functional redundancy, the ChIP‐seq/EMSA/transactivation data remain to be challenged in more sophisticated genetic backgrounds. Moreover, the epidermal specificity of the PAP‐promoter activity during skotomorphogenesis may result from a separate pathway linked to cell identity in relation to development. In this context though, it is of interest to note that PHYB promoter activity in the dark shows a pattern similar to that of the PAP8 promoter (Somers & Quail, 1995). It is, thus, unlikely that the −97‐element is solely responsible for the transcriptional regulation of PAP8. Future investigations will focus on the network that may regulate PAP8 and the PAPs in general. It would be of great interest to test (i) the role of the 3′‐UTR element of PAP8, where HY5 is also sitting, and (ii) whether the PIFs play a role in the dark‐dependent expression of the PAPs in the epidermal cell layer and/or as repressors in the palisade.
Figure 2. HY5 is a potential regulator of PAP8 expression.
-
APAP8 promoter deletion strategy. ERI, EcoRI site; HIII, HindIII site; indicated positions are given relative to the transcription start noted as +1; red boxes represent untranslated regions; pink boxes, ORF of an upstream gene; nearly palindromic element is given in blue. −97m3: mutated promoter as described in C with three mutations (m3) indicated with “***”.
-
BTwo or six (−97m3) representative primary transformants expressing GUS under the given PAP8 promoter version; FC+, the corresponding promoters were tested positive in functional complementation of the mutant pap8‐1. Scale bar equals 3 mm.
-
CProximal PAP8 promoter region; m3, 3‐bp substitutions within the −97‐bp promoter; 5′‐UTR in red; ATG, start codon of PAP8.
-
DDual‐luciferase reporter assay; Renilla luciferase (Rluc) used as internal control and GFPer used as control for the transfected area; the promoters driving Firefly luciferase (Fluc) were transfected in onion epidermis cells without or with constitutively expressed HY5. The Fluc/Rluc activity was set to 1 for the minus‐HY5 control; mean ± standard error corresponding to 3 replicates; photon counts are given in source data. *ε‐test = 3.43 > 1.96 corresponding to P‐value < 0.001.
-
EElectromobility shift assay of a probe corresponding to the near palindromic PAP8 element (GAcGCTC) with recombinant HY5 protein; a probe containing a canonical G‐box element (CACGTG) recognized by HY5 was used as cold competitor.
-
FIntegrative genomics viewer (IGV) images of the chromatin immunoprecipitation (ChIP) sequencing data30 at the PAP8 locus; TAIR10, annotation according to the Arabidopsis thaliana information resource orange box indicates the PAP8 locus. ChIP on hy5‐ks50; 35S:HY5‐YFP exposed to blue light or red light using GFP antibody and compared with mock corresponding to ChIP control experiment done without antibody. Each treatment is presented as track overlay of triplicates: the read count is given within the “group autoscale” range in brackets. Close up on the 5′‐UTR region centred on the −95‐promoter element in yellow.
Source data are available online for this figure.
PAP8 functions in plastids and in the nucleus
PAP8 displays a predicted chloroplast transit peptide (cTP) of 59 amino acids and a predicted bipartite nuclear localization signal (NLS) comprising 26 amino acids (Pfannschmidt et al, 2015), and therefore may belong to a group of dually localized proteins present in nuclei and plastids (Krause et al, 2012). Both predicted targeting sequences are simultaneously functional since a translational fusion of PAP8‐GFP (Fig 3A) displayed a signal in nucleus and plastids of transiently transfected onion cells (Fig 3B). A polyclonal serum was raised against the recombinant PAP8 protein corresponding to its mature form without its predicted cTP. The specificity of the serum was validated in planta using the mutant pap8‐1 and the recombinant protein (rP8, Appendix Fig S5A). PAP8 is largely enriched in the sub‐cellular fraction corresponding to sedimented organelles (mostly nuclei and plastids) obtained from 5‐day‐old Arabidopsis seedlings (Appendix Fig S5B). PAP8 was then detected both in the nucleus and in the plastid fractions obtained from seedlings either grown in the dark or under white light (Fig 3C) confirming the dual localization of PAP8. The distribution of PAP8 between the nucleus and the corresponding plastid‐type fraction is changed after light exposure. In etiolated seedlings, PAP8 was found mainly in the nucleus with traces in etioplasts (EP), while in photomorphogenic seedlings PAP8 was strongly enriched in chloroplasts (CP). Notably, both fractions (nuclei and plastids) displayed a signal of the same apparent molecular weight and similar to that of the designed ∆cTP recombinant protein suggesting that the nuclear fraction contains the processed version of the protein originating from plastids where the cleavage of the pre‐sequence occurs during import.
Figure 3. PAP8 is dually localized in plastids and nucleus.
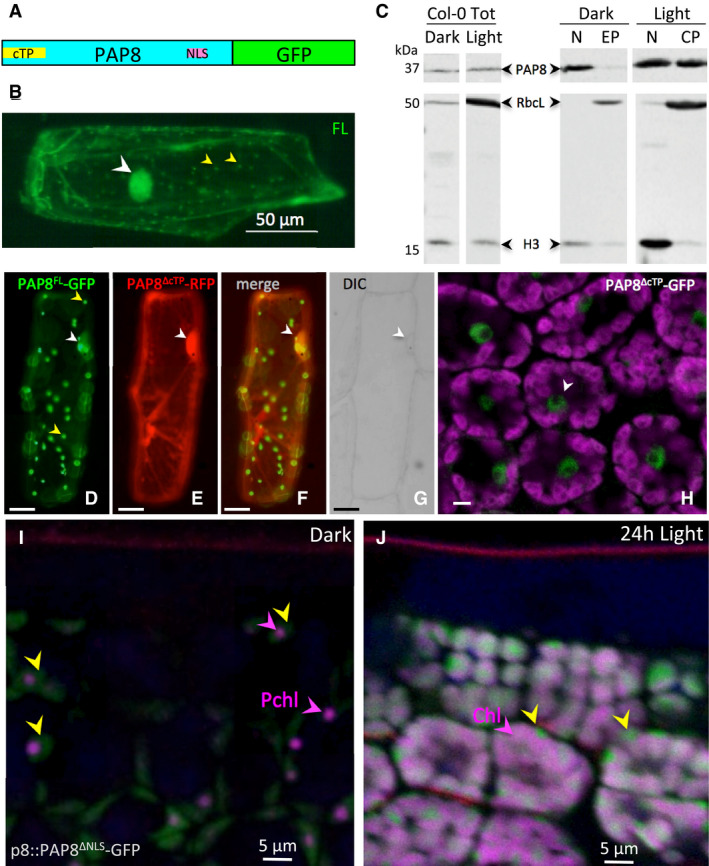
-
ASchematic illustration of domain structure of Arabidopsis PAP8 fused to GFP. cTP, chloroplast transit peptide; NLS, nuclear localization signal.
-
BTransiently expressed PAP8FL‐GFP (full‐length coding sequence of PAP8 fused to GFP) in onion epidermal cells displays a dual localization in the nucleus (white arrowhead) and in plastids (yellow arrowheads).
-
CImmuno‐blots for the detection of PAP8 in total protein extracts (Col‐0 Tot) the nuclear fraction (N) and the plastidic fractions (EP, etioplast; CP, chloroplast) of etiolated (dark) or photomorphogenic (light) Arabidopsis seedlings by immuno‐Western blotting using a PAP8 antiserum (PAP8). As control, a mixture of antisera raised against histone 3 (H3) and the large subunit of ribulose 1, 5‐bisphosphate carboxylase/oxygenase (RbcL) was used to evaluate reciprocal contaminations. The lanes are extracted from the same blot.
-
D–GCo‐expression analysis of PAP8FL‐GFP (D) with PAP8ΔcTP‐RFP (E) in onion epidermal cells; (F) green and red channels merged; yellow arrowheads show plastids; white arrowheads show nuclei as observed with DIC, differential interference contrast in (G); FL, PAP8 full‐length ORF; ΔcTP, deletion of the cTP; scale bars equal 20 μm.
-
HConfocal imaging on Arabidopsis cotyledons expressing PAP8ΔcTP‐GFP, details of a few palisade cells (see Appendix Fig S7B for a view in a cross section between the two cotyledons); magenta, auto‐fluorescence of the chloroplast; the white arrowhead shows a nucleus; scale bar equals 5 μm.
-
I, JConfocal imaging on Arabidopsis cotyledons stably expressing pP8::PAP8ΔNLS‐GFP; ΔNLS, deletion of the NLS (I) during skotomorphogenesis and (J) after 24‐h light; yellow arrowheads show the GFP signal; the picture is a merge of different channels: GFP in green, Pchl,: protochlorophyllide or Chl: chlorophyll in magenta marked with arrowheads, and propidium iodide, showing the waxy cuticle in red, the empty space correspond to the layer of highly vacuolated epidermal cells.
Source data are available online for this figure.
To investigate this, PAP8 localization was artificially uncoupled using a mutation strategy. Variants of PAP8‐GFP lacking the cTP (∆cTP), the NLS (∆NLS), both signals (∆∆) or containing a mutated NLS with five neutral substitutions of the positively charged amino acids within the NLS (NLSm5) were cloned. In transiently transfected onion cells (Fig 3D–G) and in Arabidopsis thaliana lines with stable expression, PAP8∆cTP‐GFP displayed nuclear accumulation (Figs 3E and H, and EV2A and B), whereas the ∆NLS and the NLSm5 variants were strictly restricted to plastids (Figs 3I and J, and 4A and B, and EV2A and C) indicating that the cTP supports chloroplast import and that nuclear localization depends on its NLS. Thus, the PAP8 sub‐cellular localization can be controlled in transgenic plants using the different targeting signals, and the corresponding transgene can therefore be assessed for functionality in pap8‐1. Hence, PAP8 variants fused or not to GFP, as indicated, were expressed under the constitutive promoter CaMV35S or its own promoter pP8 (pPAP8‐1133), in wild type or in pap8‐1. In contrast to pP8::PAP8, all genetic constructions with the GFP tag were unable to yield functional complementation (Fig EV3). Since GFP may very likely impose a steric hindrance to the function of PAP8, only protein accumulation and sub‐cellular localization were tested using the fluorescent marker. In addition, the greening of plants expressing part or full‐length sequence of PAP8 under 35S promoter was strongly altered with no regard to its functionality or its proper localization (Fig EV3B), suggesting that overexpression or miss‐expression of the transgene with part of the PAP8 sequence might sequester a component of unknown nature (protein, RNA or else) that affects the biogenesis or stability of the chloroplast within the cell.
Figure EV2. (Related to Fig 3) Sub‐cellular localization of PAP8 variants.
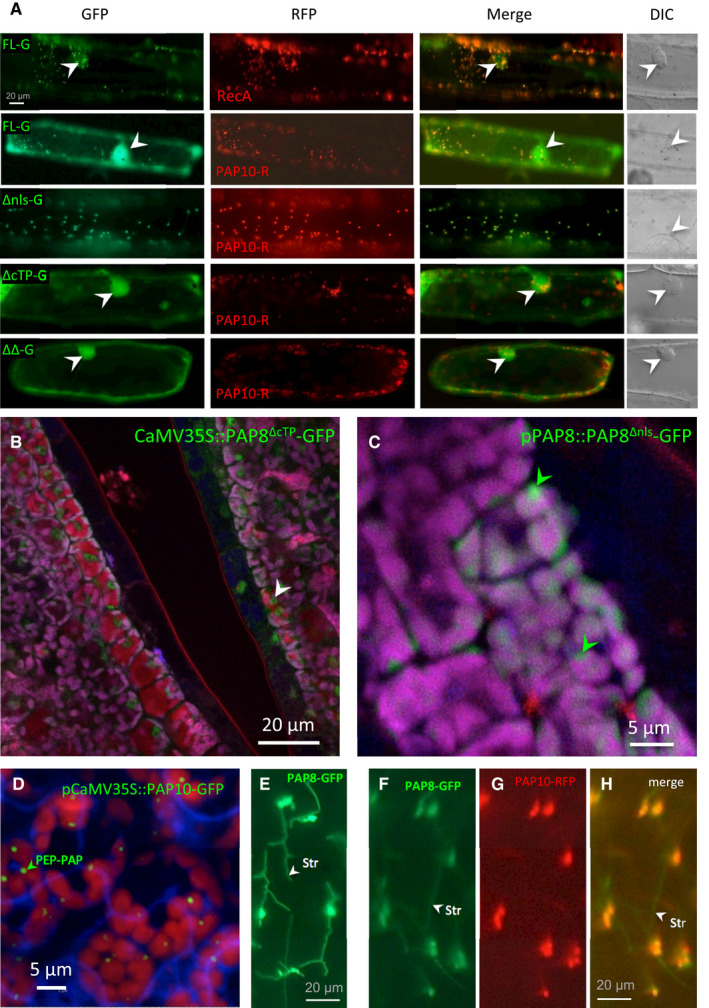
-
ATransient assay in onion epidermal cells, pictures are given at the same scale. FL, full‐length ORF; −G, translational fusion with GFP; ΔNLS, deletion of the NLS; ΔcTP, deletion of the cTP, ΔΔ, deletion of both the NLS and the cTP. Onion cell co‐transfected with the corresponding variant fused to GFP and a plastid control fused to RFP (PAP10, PAP10‐RFP or RecA, RecA‐RFP) Merge, merged channels; DIC, differential interference contrast microscopy pictures to reveal the position of the nucleus within the cell when fluorescent nuclei were observed (marked with white arrowheads).
-
B, CConfocal imaging of stably expressed CaMV35S::PAP8∆cTP‐GFP (B) or pPAP8::PAP8∆nls‐GFP (C) in cotyledons of Arabidopsis thaliana; white arrowheads indicate nuclei; green arrowheads indicate sub‐plastidial localization. Observations similar to (C) were recorded for pPAP8::PAP8FL‐GFP.
-
DConfocal imaging of stably expressed CaMV35S::PAP10‐GFP in cotyledons of Arabidopsis thaliana showing the PEP‐PAP complex; the green arrowhead indicates the putative location of PEP‐PAP complexes within one chloroplast.
-
E–HPAP8‐GFP in stromules of onion epidermal cells expressed alone (E) or in co‐localization with PAP10‐GFP (F–H); stromules (str, white arrowheads) are only marked by PAP8‐GFP.
Source data are available online for this figure.
Figure 4. PAP8 is functional in both the chloroplast and the nucleus.
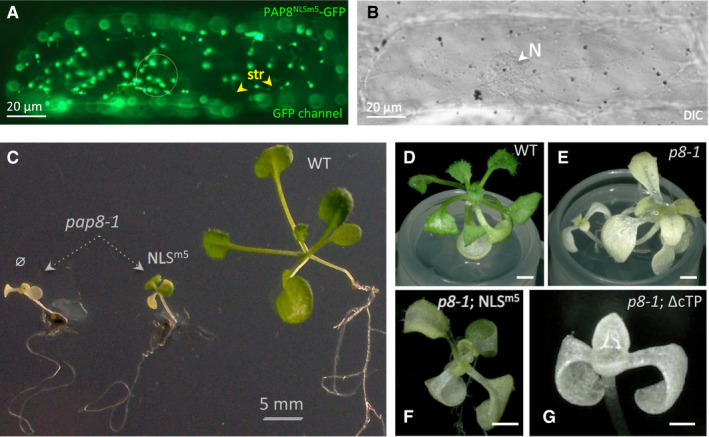
-
A, B(A) Transient expression of PAP8NLSm5‐GFP mutated in the NLS as described in Fig EV4A. The circle marks the position of the nucleus (N) as observed with DIC in (B); yellow arrowheads show stromules (str). Scale bars equal 20 μm.
-
C–GPictures of representative genotypes obtained in the functional complementation test of pap8‐1 using pP8::PAP8‐NLSm5 (C, F) or pP8::PAP8ΔcTP (G) both constructions without GFP tags; ø indicates pap8‐1 without any PAP8 transgene. (D–G) Pictures using Keyence technology of plants with genotypes as labelled; transgenes expressed using the 1.1‐kb PAP8 promoter. Scale bars equal 1 mm.
Figure EV3. (Related to Fig 3) PAP8 and its variants fused to GFP .
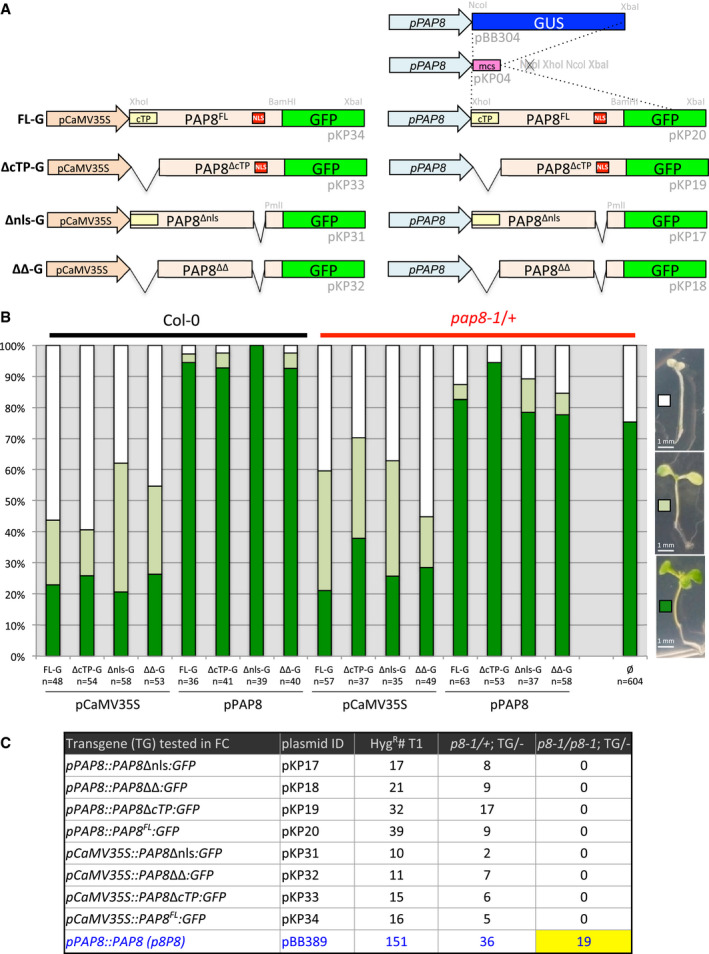
-
ASchematic illustration of the GFP‐fused PAP8 variants used for sub‐cellular localization and functional complementation tests FL‐G, PAP8 full‐length ORF translationally fused to GFP; Δnls‐G, deletion of the NLS (nuclear localization signal), ORF fused to GFP; ΔcTP‐G, deletion of the cTP (chloroplastic transit peptide), ORF fused to GFP; ΔΔ‐G combined deletion of NLS and cTP in the ORF fused to GFP. The variants are expressed under the control of the CaMV35S promoter or under the native PAP8‐1.1-kb promoter. Plasmid identification and relevant restriction sites for cloning are given in light grey (see Appendix Table S1); mcs, multiple cloning site.
-
BTransgenic lines obtained with the constructions described above. Three phenotypic classes have been recorded corresponding to albino (white squares), pale green (light green squares) and green plants (green squares). Col‐0, wild type; pap8‐1/+, heterozygous mixture; n, total number of recorded plants; ø, no transgene.
-
CFunctional complementation output. Hygromycin‐resistant plants were transferred on soil and grown under long‐day conditions (16‐h light/8-h dark; ˜ 70 μmol m−2 s−1) at 21°C and 60% humidity. Genomic DNA was isolated from true leaves and used for genotyping. The presence of the pap8‐1 allele was confirmed using the primer ortpF/oLBb1.3. PAP8 wild‐type allele tested with ortpF/op8i2_R. The insertion of the transgene of interest was tested with oPAP8_rtp_F/oE3_R. The number of the tested plants (HygR#T1), the number of double heterozygous plants (p8‐1/+; TG/−) and the number of sesqui‐mutant plants (p8‐1/p8-1; TG/−) are depicted for each construction; p8P8 presented as positive control. None of the 39 T1 plants with pP8::PAP8‐GFP were photosynthetic and homozygous pap8‐1. Therefore, two doubly heterozygous (pap8‐1/+; TG/−) expressing GFP were tested for their segregation pattern (Line 1: 34% albino, n = 169, and Line 2: 24% albino, n = 199) and compared to that of pap8‐1/+ (28% albinos, n = 99). In the absence of statistical difference between the samples (ε = 0.102≪1.96 for α = 0.05; ε‐test, Fisher Yates), PAP8‐GFP was declared not functional as opposed to PAP8.
Source data are available online for this figure.
In wild type, the GFP signal of pP8::PAP8ΔNLS‐GFP increased in chloroplast sub‐domains during the transition from dark to light (Fig 3I and J). Therefore, protein accumulation follows the promoter PAP8 induction in the palisade cells (Liebers et al, 2018) and is consistent with the immune detection of the native PAP8 in sub‐cellular fractions. In contrast to the fluorescently tagged PAP10 that does not contain a predicted NLS and show a sharp and distinct localization (Fig EV2D), a wider signal of PAP8‐GFP indicates that foci slowly appear after light exposure, while part of the pool remains in the stroma. The foci, specifically marked with PAP10, may correspond to the assembly of the prokaryotic PEP core complexes with the eukaryotic PAPs. The stroma localization of PAP8‐GFP was confirmed by the PAP8‐GFP signal transiently observed in stromules of onion cells, while the PAP10‐RFP signal is absent from these stromules (Fig EV2E–H). Therefore, PAP8 may be set free from the PEP‐PAP complex allowing for re‐localization in the nucleus. Whether this release is allowed through saturation of the complex or a change in its affinity remains unknown to date.
Since GFP‐tagged PAP8 could not rescue the mutant, untagged PAP8 variants were tested in functional hemi‐complementation (Figs 4 and EV4). In contrast to the ΔcTP variant unable to cross the plastidial envelope and unable to rescue the albinism (Fig 4C and G), the NLSm5 variant (Fig EV4A) could restore the greening of the mutant albeit with strong delays in growth (Figs 4C–G and EV4) suggesting that the chloroplast‐localized PAP8NLSm5 carries its chloroplast function for the greening but that in the absence of the nucleus‐localized pool the timing of chloroplast biogenesis is altered, with substantial consequences on the timing of light‐controlled development. Therefore, PAP8, through its nuclear pool, may carry a function related to the light‐signalling response.
Figure EV4. (Related to Fig 4) Hemi‐complementation test in pap8‐1 .
-
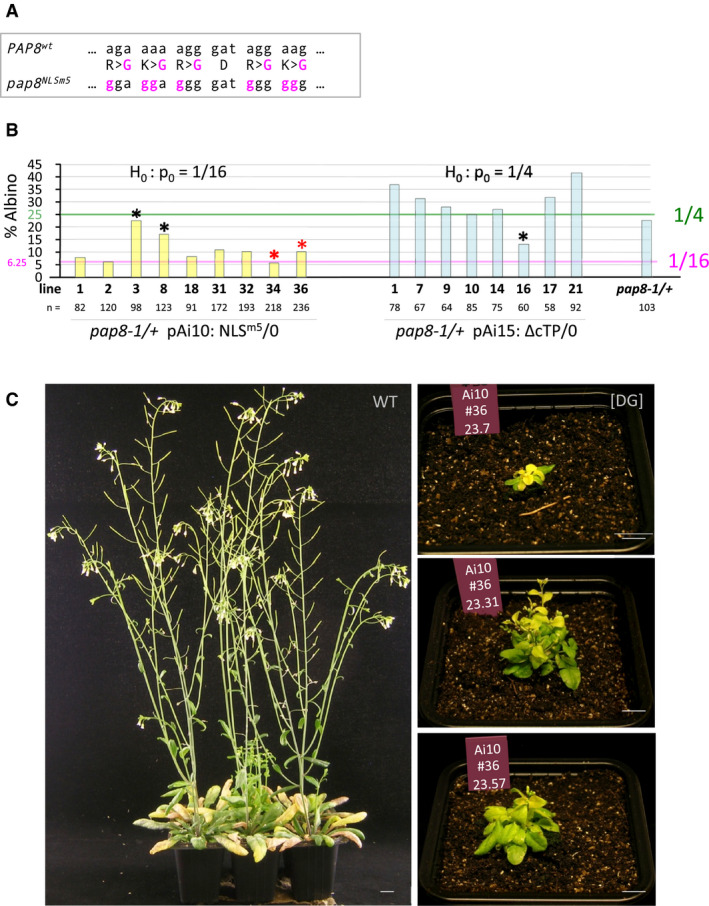
AGenetic alterations of the pap8 NLSm5 allele. Altered codons and corresponding changes in amino acids (central line) are in magenta; the sign “>” indicates aa replacement at the same position.
-
BBar graph representing the albino segregation ratios in doubly heterozygous transgenic lines (pap8-1/+; TG/−) obtained with pPAP8::PAP8NLSm5 (NLSm5: pAi10) and pPAP8::PAP8∆cTP (∆cTP: pAi15). pap8‐1/+ used as control; n, total number of recorded plants. Segregation pattern were tested using ε‐test with null hypothesis set to p0 = 1/16 (greening complementation) for NLSm5 and p0 = 1/4 (no complementation) for ∆cTP (see data source); black *, outliers correspond to the samples that did not pass the statistical test; red * indicate chosen genotypes for follow‐up studies.
-
CPhenotype of pap8‐1 transformed with pPAP8::PAP8NLSm5. WT, 5‐week-old Col‐0 control; [DG], delayed greening phenotype observed for the partial rescue of pap8‐1 mutant expressing PAP8NLSm5 under its endogenous promoter; pictures depict three 15‐week-old plants in which the alteration of the greening corresponds to the emergence of white leaves that slowly acquire the photosynthetic apparatus; plants #7, #31 and #57 are siblings of the same genotype (Ai10#34). Scale bars equal 10 mm.
Source data are available online for this figure.
PAP8 mediates phytochrome signalling
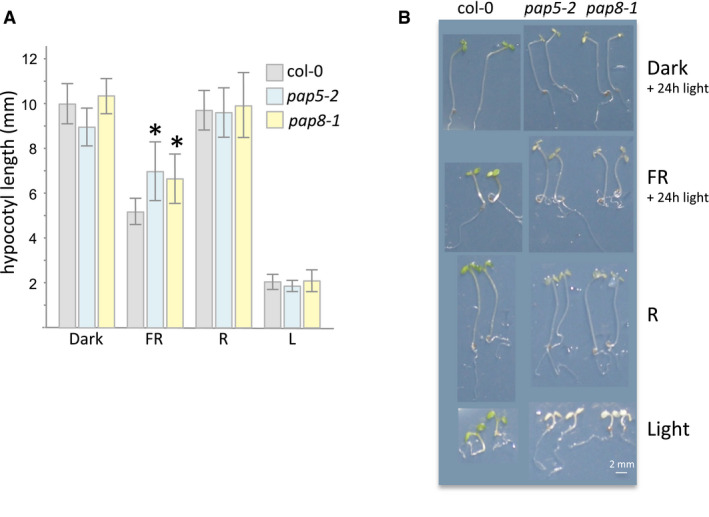
To test this assumption, different light qualities were applied to the plants. Although, in our in vitro growing conditions, pap8‐1 responded normally to red and white lights with proper de‐etiolation (cotyledon and apical hook opening), far‐red light treatment yielded a significantly reduced repression of hypocotyl length, a phenotype similar to that of hmr‐2/pap5‐2 (Chen et al, 2010; Fig EV5).
Figure EV5. Hypocotyl growth experiment.
-
AHypocotyl length measurements, given as mean ± SD, of genotypes grown under different light sources. Individual measurements and sample sizes are given in source data. Dark, true dark treatment; FR, far‐red light (low fluence approx. 10 μmol m−2 s−1; peak at 730 nm ± 10 nm); R, red light (8 μmol m−2 s−1 peak at 660 nm ± 10 nm); L, white light 30 μmol m−2 s−1. Whereas pap5‐2 and pap8‐1 are statistically undistinguishable using δ‐test for the comparison of the mean (Fisher Yates δ = 0.791 ≪ Uα = 0.05 = 1.96, not significant at α set to 0.05), both pap5‐2 and pap8‐1 show significant hypocotyl differences compared with wild type (δ = 5.374; δ = 5.061 respectively so that both *P‐values < 10−6).
-
BImages of representative seedlings.
Source data are available online for this figure.
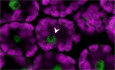
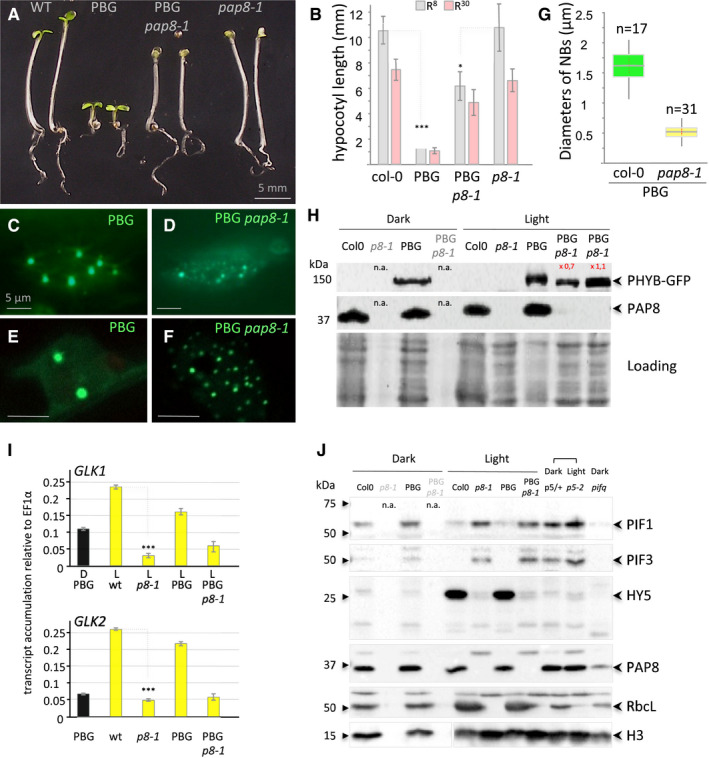
Stable overexpression of a phytochrome PHYB‐GFP (PBG) is known to mediate hypersensitivity of Arabidopsis seedlings to red light (8–30 μmol m−2 s−1, Fig 5A and B) leading to a significant inhibition of hypocotyl elongation when compared to WT (Yamaguchi et al, 1999). After introducing PBG into the pap8‐1 mutant background, however, this PBG effect was largely lost, indicating that PAP8 plays a role in the PHYB‐mediated light response. This lack of physiological response correlates with the retention of small PBG speckles in pap8‐1 corresponding to the absence of late photobodies in comparison with wild type (Fig 5C–G). The change in the photobodies patterning is not due to a change in PBG accumulation as tested by immune detection of the GFP tag in the different genetic backgrounds (Fig 5H). Late photobodies are known to be associated with the targeted degradation of PIFs (Leivar & Monte, 2014), the key regulators in the phytochrome signalling network. This pointed to an active role of PAP8 in the light‐induced gene expression programme as illustrated with the strong defects in the accumulation of GLK1, and GLK2 transcripts in pap8‐1 (Fig 5I) thus interrupting the light‐induced expression of photosynthesis‐associated nuclear genes (PhANGs) (Waters & Langdale, 2009; Oh & Montgomery, 2014). The defect in GLKs transcript accumulation was also observed in pap7‐1 (Grubler et al, 2017) accounting for the albino syndrome of the pap mutants where the expression of PhANGs is strongly altered.
Figure 5. PAP8 is essential for the PHYB‐mediated light induction of photomorphogenesis.
-
APhenotypes of given genotypes subjected to 5 days of illumination at 8 μmol m−2 s−1 660‐nm red light. PBG, pCaMV35S::PHYB‐GFP transformed in pap8‐1/+ (among 25 lines selected for GFP expression see data source; 2 doubly heterozygous pap8‐1/+; PBG/− lines #6 and #7 segregated the photobodies alteration with the albinism).
-
BHypocotyl length, given as the mean ± SD, of plants grown as in (A) (R8, grey bars) or at 30 μmol m−2 s−1 660‐nm red light (R30, pink bars) showing partial insensitivity of pap8‐1 to the PBG overexpression. Measurements are given in source data; in the order of the graph n equals (50, 133, 58, 43, 36, 112, 25, 60) δ‐test (comparison of the mean) R8: δPBG/PBGp8‐1 = 20.38 corresponding to P‐value < 10−31 δwt/p8-1 = 0.5 < 1.96 not significant at α set to 0.05).
-
C–GNuclear accumulation of PBG observed under GFP excitation in the given genotypes. (C, D) Epi‐fluorescence microscopy. (E, F) Confocal microscopy showing the size of the nuclear bodies. Scale bars equal 5 μm. (G) Box plot (Min, 1st quartile, median as the central band, 3rd quartile, max) on the diameter of the nuclear bodies (NBs); n equals the number of records.
-
HImmuno‐blots using a GFP antibody or a PAP8 antibody showing, respectively, the levels of PHYB‐GFP and PAP8 in the given genotypes grown in the dark for 3 days or in light; n.a., not applicable as the pap8‐1 mutant can only be visually distinguished from wild type after light exposure; 2 lines (L#06 and L#07) for PBG/pap8‐1 were tested. Coomassie blue staining presented as loading; signals were quantified using ImageJ.
-
IRT–qPCR analysis on wild type, pap8‐1, PBG and PBG pap8‐1. Seedlings were grown in the dark (D) or under white light (L, 30 μmol m−2 s−1); levels of transcripts are given relative to EF1α; error bars correspond to standard errors on technical triplicates, and the dark sample is the wild‐type PBG line. δ‐test (comparison of the mean) ***P < 10−72.
-
JImmuno‐blots showing the levels of PIF1, PIF3 and HY5 in given genotypes grown in the Dark or light condition as noted: p5/+, mix of an heterozygous pap5‐2 siblings progeny undistinguishable from wild type; p5‐2, pap5‐2 and pifq, quadruple pif1‐1 pif3‐3 pif4‐2 pif5‐3 mutant; histone H3 (H3), RbcL and PAP8 were used as controls; n.a., not applicable.
Source data are available online for this figure.
The light‐induced destabilization of PIF1 and PIF3 is altered in pap8‐1 and PBG/pap8‐1, conversely the light‐induced stabilization of HY5 does not occur in pap8‐1 (Fig 5J). Interestingly, these molecular phenotypes in pap8‐1 are very similar to those observed in pap5‐2 used as control. Therefore, PAP8 supports the degradation of PIF1 and PIF3 and stabilizes HY5 with no effect caused by the presence of PBG. In the light‐signalling cascade, PIFs are known to act upstream of HY5 and in a reciprocal negative feedback loop with PHYB (Leivar & Monte, 2014). This indicates that the alteration of the signalling in pap8‐1 (the albino block depicted in Fig 8) acts upstream of PIF1 and PIF3 by specifically blocking the HY5 to GLK pathway without altering the de‐etiolating pathway: The apical hook and the cotyledons can open. The nature of the block remains unknown; it could be due to direct functional alteration of a PAP nuclear sub‐complex in which PAP8 and PAP5 may act co‐ordinately and dependently, or due to an upstream retrograde signal coming from the challenged pap8‐deficient chloroplast (Martin et al, 2016). Concerning the growth of the hypocotyl, the situation remains complex whether PBG is considered or not.
Therefore, PAP8 is important for the proper expression of GLKs. Should this occur through the nuclear function of PAP8, directly or through a PAP8‐containing complex, this would simply explain the delayed greening and growth observed in the partially rescued phenotype of the PAP8NLSm5 variant, in which nuclear PAP8 is absent. Should the expression of GLK1 be controlled by the state of the plastids through a distinct molecular pathway, this would then be an indirect consequence of the pap8‐1 phenotype and more generally of the pap albino syndrome. Future research will probably help solving this conundrum.
PAP8 physically interacts with HMR/PAP5
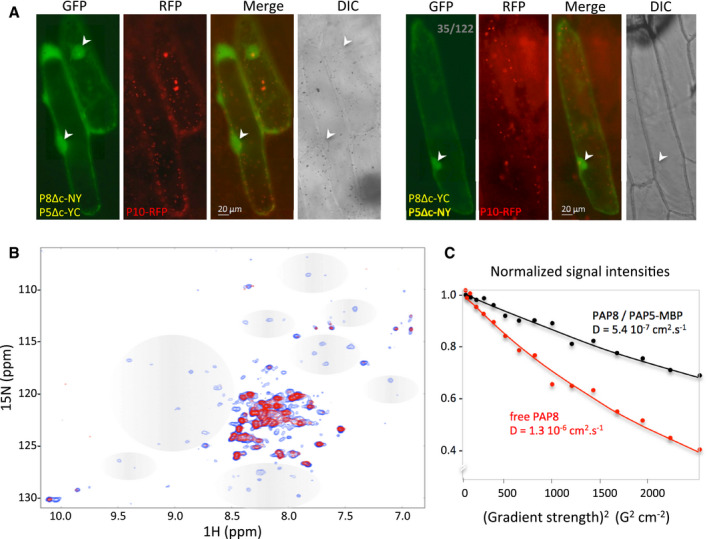
The cellular distribution of PBG and other defects in pap8‐1 are highly similar to those of hmr‐2/pap5‐2. HMR/PAP5 is a nucleo‐plastidic protein identified to be (i) important for the initiation of photomorphogenesis (Chen et al, 2010; Qiu et al, 2015) and (ii) a component of the chloroplast PEP complex (Steiner et al, 2011; Nevarez et al, 2017). Although yeast two‐hybrid studies did not report any interaction between the two proteins (Arsova et al, 2010; Gao et al, 2011; Yu et al, 2013), bimolecular fluorescence complementation technology (BiFC, Fig 6A, Appendix Fig S6 for control experiments) revealed that PAP8∆cTP and HMR/PAP5∆cTP could together restore split YFP fluorescence indicating that they get in close proximity within the nucleoplasm.
Figure 6. PAP8 interacts with pTAC12/HMR/PAP5.
-
ABimolecular fluorescence complementation tests using in combination PAP8ΔcTP‐NY (P8Δc‐NY) with PAP5ΔcTP‐YC (P5Δc‐YC) or PAP8ΔcTP‐YC (P8Δc‐YC) with PAP5ΔcTP‐NY (P5Δc‐NY); PAP10‐RFP (P10‐RFP) was used as internal positive control for transfection; the ratio in grey depicts the number of green‐fluorescent cells over red‐fluorescent cells. Arrowheads indicate nuclei. See Appendix Fig S6 for control experiments; transgenes expressed under CaMV35S promoter.
-
BOverlay of 1H‐15N correlation 2D NMR spectra of free 15N‐labelled PAP8 alone (blue) or in complex with PAP5 (red). Grey areas depict changes in signals in the PAP8 spectrum.
-
C15N‐Filtered diffusion ordered spectroscopy‐NMR measurements to PAP8. Exponential decay curves of PAP8 in the absence or in the presence of MBP‐PAP5 are shown in red and black, respectively. The units on the y‐axis are normalized values of the integrals of the signal measured in the amide proton region.
Source data are available online for this figure.
The 1H‐15N‐correlation NMR spectrum of PAP8 showed two populations of peaks according to their intensities and frequency distributions (blue signal, Fig 6B). About 40 peaks of high intensities present in a narrow frequency range in the proton dimension (8.0–8.5 ppm) correspond to very dynamic and flexible regions of the protein, with a short apparent rotational correlation time (τ c = 2.5 ns, measured at 300 K in a [15N,1H]‐TRACT experiment). The other population of lower intensity peaks with a large frequency distribution corresponds to well‐folded domains (τ c = 17 ns). The interaction with PAP5 was tested in a second 1H‐15N‐correlation NMR spectrum with unlabelled PAP5‐MBP (red signal). The flexible regions of PAP8 were not affected, neither in chemical shift nor in dynamics (τ c ≃ 2.5 ns), and only weakly in intensity. Hence, these flexible regions are not involved in the interaction. By contrast, the low‐intensity peaks from the structured region did not appear when PAP5‐MBP was added even after a 14‐fold longer experiment (13 h versus 53 min for the free 15N‐PAP8 spectrum) indicating that PAP5‐MPB interacts with the structured region of PAP8.
The robust NMR signals of the PAP8 flexible residues permitted translational diffusion measurements in 15N‐filtered DOSY spectra with a selective detection of the amide protons bound to the PAP8‐15N atoms avoiding perturbations by the unlabelled protein. The diffusion rate of free PAP8 was significantly larger than that of the mixture with PAP5‐MBP, indicating again that both proteins interact with each other (Fig 6C). The control experiment using MBP yielded super‐imposable spectra and identical PAP8 translational diffusion, indicating that MBP is not involved in the interaction between PAP8 and PAP5‐MPB. The Kd corresponding to the interaction between PAP8 and PAP5 was estimated in the range of 50–100 μM according to a sub‐stoichiometric titration experiment (Appendix Fig S7; Williamson, 2013).
HMR/PAP5 physically interacts with PAP8 through a well‐structured region. Hence, these physical properties reinforce the assumption that PAP8 and PAP5 might form a nuclear complex. It is very likely that additional components could stabilize the unstructured region of PAP8 and enhance its affinity to PAP5 allowing BiFC detection in vivo. Should such a nuclear complex exist, the proteins could work cooperatively in an interdependent fashion like it is already hypothesized for the PEP complex in the chloroplast. Consequently, individual mutant phenotypes, such as those displaying the PAPs syndrome, would resemble each other, as it is observed for pap8‐1 and pap5‐2, e.g., in the formation of the photobodies. It is very likely that both mutant alleles display an epistasis where the lack of one gene function masks the lack of the second gene function. Whether this is true in the chloroplast and in the nucleus remains to be addressed.
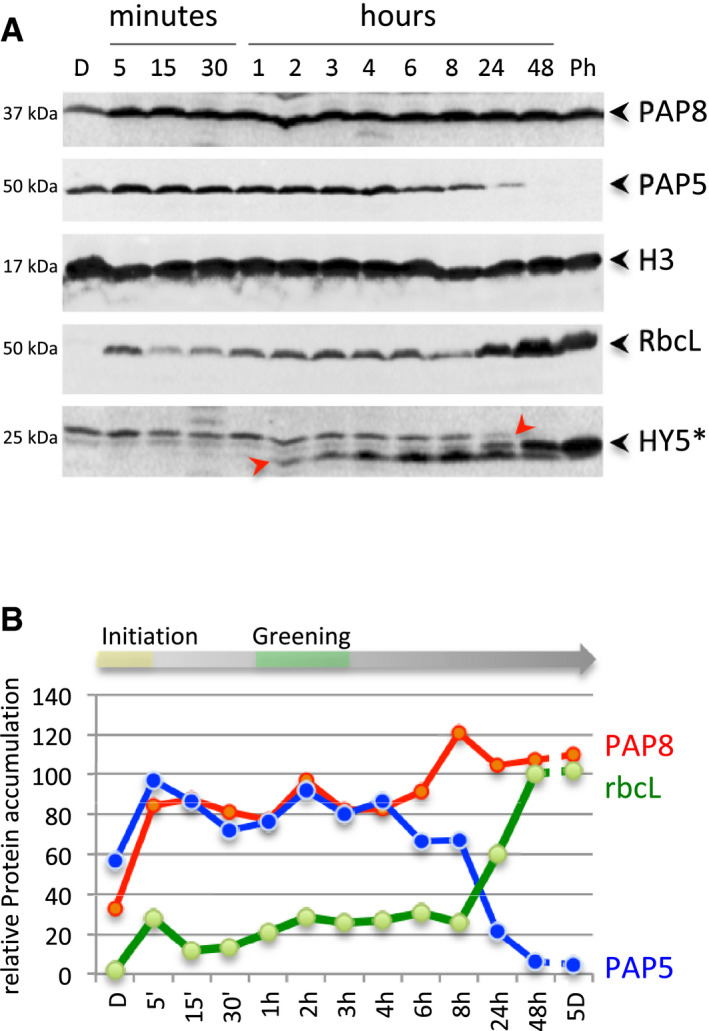
The light‐induced PAP‐dependent setting of the chloroplastic function prompted a question of timing. Therefore, kinetics of protein accumulation were established using Western blots (Fig 7). The amount of immune‐detected PAP8 and PAP5 in dark‐grown seedlings rises to nearly their maximum amount within 5 min after light exposure (Fig 7A and B), while the stabilization of HY5 takes a few hours to be detected, closely followed with the rise of the ribulose‐1,5‐bisphosphate carboxylase/oxygenase. The accumulation of the PAPs in early chloroplasts is therefore much faster than other tested components responding to light, eventually placing the reshaping of the PEP‐PAP complex within the organelle as one of the earliest molecular events in chloroplast biogenesis.
Figure 7. Temporal resolution of protein content in seedlings during the dark‐to‐light transition.
-
AImmuno‐blots showing the levels of PAP8, PAP5, histone H3 (H3), Rubisco (RbcL) and HY5; D, dark; Ph, photomorphogenic growth conditions from germination on. *, HY5 is detected as a modified form (+6 kDa); the arrows indicate the two different post‐translational modified forms of HY5 for which accumulation occurs upon light exposure.
-
BRelative protein contents normalized to histone H3 during the transition from skotomorphogenesis to photomorphogenesis, we propose a phase of photo‐initiation corresponding to the 5 min of light allowing PAP8 and PAP5 to rise to nearly 100% of their maximum. The first macroscopic signs of greening are indicated around 3 h, while the photosynthetic apparatus bursts at 8 h to be fully accumulated at 48 h.
Source data are available online for this figure.
Concluding Remarks
This study revealed that PAP8 represents a novel regulatory component that links photomorphogenesis and chloroplast biogenesis through its dual localization. PAP8, therefore, is a novel member of the nucleo‐plastidic protein family involved in chloroplast biogenesis (Yang et al, 2019; Yoo et al, 2019). It is proposed that the nuclear fraction of PAP8 is essential to properly transduce the light signal from photoactivated PHYB to the expression of GLK1, one of the master regulators of nuclear photosynthesis genes. Taken together, the results presented in this study prompted a model of PAP8 action within the transition from skotomorphogenesis to photomorphogenesis (Fig 8). A dark operating unknown transcription factor (TF?) allows the production of PAP8 in the epidermal cells where it mostly accumulates in the nucleus (Fig 3C) in a form identical in size to the processed plastid form, suggesting that it passes through the plastid for removal of the transit sequence. Alternatively, a processed PAP8 may reach the nucleus without entering the chloroplast via an unknown mechanism. This would, however, require the removal of the transit sequence outside of the plastid by a yet unidentified protease activity. This possibility is, however, unlikely, as it would compromise the plastid targeting of other nuclear‐encoded plastid proteins. Moreover, mutations of the NLS restrict PAP8 to plastids, indicating that alternative splicing or alternative initiation of translation that could potentially remove the cTP is also unlikely to occur, as these mechanisms would ultimately produce a cytosolic protein. Once in the nucleus, PAP8 may interact with HMR/PAP5 in a PAP nuclear sub‐complex PAP‐NSC. Upon light exposure, rapid photo‐converted PHYB requires the PAP‐NSC to transduce the signal to PIF1 and PIF3 for their COP1‐mediated degradation. At this early stage, a non‐cell‐autonomous signal such as the one that operates for de‐etiolating hypocotyls may then allow the signal to invade the palisade tissue where PIFs are destabilized and HY5 is stabilized escaping COP1‐mediated degradation. In turn, HY5 may activate the PAP8 promoter (and potentially other PAP promoters) in cells with a fate associated with photosynthesis, allowing for the assembly of the PEP‐PAP complex, itself necessary for the expression of the photosynthesis‐associated plastid genes (PhAPGs). HY5 was also found on the chromatin associated with both GLK1 and GLK2 (Hajdu et al, 2018) and could therefore activate them directly or indirectly through other light‐responsive factors. In turn GLKs, under GUN1‐mediated retrograde control (Tokumaru et al, 2017), activate the photosynthesis‐associated nuclear genes (PhANGs). Both PhANGs and PhAPGs participate safely in the build‐up of the photosynthetic apparatus (PS). In the absence of PAP8, PIFs are less degraded, HY5 is not stabilized and the GLK pathway does not operate. Concomitantly, PAP8 (marked as absent) cannot assemble in the PEP‐PAP complex and PEP‐dependent genes are not correctly expressed. In consequence, chloroplasts do not differentiate leading to the albino syndrome. Whether PAP8 is a positively acting factor in retrograde signalling remains to be investigated; however, it is tempting to speculate that it mediates the GUN1‐controlled retrograde signal(s). A recent study reported the physical interaction of plastid PAP8 and GUN1 (Tadini et al, 2016) putting both proteins into a common biological context. Future studies will focus on the understanding of the role of PAP8 in retrograde signalling and the connection to the light‐signalling network promoting chloroplast biogenesis.
Figure 8. Model for the connection of HY5 to the GLK1 genetic pathways through the action of PAP8 during the dark‐to‐light transition.
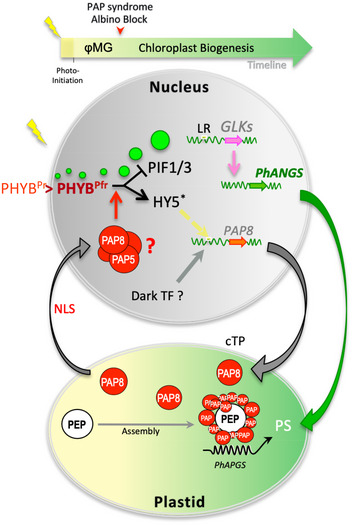
The yellow spark represents the initial exposure of the seedling to light. Photo‐initiation is mediated by PAP8 and PAP5 rapid accumulation in early photomorphogenesis (φMG): the Pr state of phytochrome B (PHYBPr) is converted in the Pfr state (PHYBPfr), which enters the nucleus, accumulates in early photobodies (small green discs) that regroup in late photobodies (large green discs) while promoting the destabilization of PIFs (PIF1 and PIF3 in particular). The red arrow in the timeline represents the albino block observed in pap8‐1 that may represent a key feature of the PAP syndrome. The dark induces ubiquitination and degradation of HY5Ub impeding recognition of a cis‐regulatory element in the promoter of PAP8 (yellow box). Light induces transcriptional activation of PAP8 in palisade cells likely through the HY5 pathway (dashed yellow arrow on PAP8 promoter); this part of the model is supported by transient assays, in vitro experiments, and in vivo genome‐wide ChIP sequencing data. The cTP pre‐sequence of PAP8 allows plastid import and then PAP8 assembly within the PEP‐PAP complex. Using an unknown trafficking route such as travelling across the plastid envelope (?), part of the processed PAP8 pool is found in the nucleus where its action could be necessary for the PHYB‐mediated transcriptional activation of GLK1 directly through HY530 (dashed yellow arrow) or other light‐responsive factors (LR), which in turn can activate the photosynthetic‐associated nuclear genes (PhANGs) concomitantly to the (PEP‐PAP)‐driven expression of the photosynthetic‐associated plastid genes (PhAPGs) essential for the building of the photosynthetic apparatus (PS) in the functional chloroplast.
Materials and Methods
Accessions
TAIR (http://www.arabidopsis.org/)—PAP8/pTAC6: At1g21600; PAP5/HMR/pTAC12: At2g34640; PAP10/TrxZ: At3g06730; GLK1: At2g20570; HY5: At5g11260; PHYB: At2g18790; EF1α: At5g60390.
HYH: At3g17609; TGA2: At5g06950; bZIP60: At1g42990; LAF1: At4g25560; PIL1: At2g46970; ELIP2: At4g14690 PSY: At5g17230.
Statistical analysis
Percentages were compared using ε‐test, whereas mean values were compared using δ‐test; statistical values were confronted to the table of normal distribution (Fisher Yates: Statistical tables for biological, agricultural and medical research (Oliver and Boyd, Edinburgh)) with α set to 0.05 or to retrieve P‐values.
Biological materials
Arabidopsis thaliana seeds, pap8‐1: SALK_024431 (N524431), and Col‐0: SALK_6000, were obtained from The European Arabidopsis Stock Centre NASC. Escherichia coli DH5α strain (lacZ‐ΔM15 Δ(lacZYA‐argF) U169 recA1 endA1 hsdR17(rK‐mK+) supE44 thi‐1 gyrA96 relA1) was used for cloning. Agrobacterium tumefaciens strain C58C1 pMP90 was used for transgenesis. Rosetta™2 (DE3) (Novagen) cells were used for protein production with pBB543 (HY5‐H6), pAG21d (PAP8ΔcTP‐H6), pAG08 (H6‐MBP‐PAP5‐H6) or pETM40 (MBP); see Appendix Table S1 for details.
Plant transformation
Electrocompetent Agrobacterium were transformed with binary plasmids containing our transgene (see Appendix Table S1) (antibiotics: gentamycin rifampicin and spectinomycin for the plasmid carrying the transgene). Strains were then used for floral dip infiltration of the significant genotypes (medium: 2.2 g MS salts, 1 ml Gamborg's 1,000× B5 vitamins, 0.5% sucrose, 44 nM benzyl amino purine, 300 μl/l Silwet L‐77). Sporophytic lethal pap8‐1 was used as the progeny of a heterozygous plant; transgenic plants were then selected to carry the mutant allele pap8‐1 (yielding albino plants in the progeny) and to carry the selection marker using the corresponding antibiotic.
Growth conditions
Plants were grown on 1/2 MS media, sucrose and 0.8% agar. Seeds were imbibed and stratified for 2 days at 4°C, before growth at 21°C for 3 days in darkness. Afterwards, plants were transferred to continuous white light (30 μmol m−2 s−1). For kinetics and organelle fractionations, wild type was grown on MS medium without sucrose at 18°C. For pharmacological rescues of pap8‐1, imbibed seeds were spread in sterile plastic boxes, containing ½ MS media with 3% sucrose. After stratification seen above, seeds were transferred to continuous white light (10 μmol m−2 s−1) at 21°C for 7 days, before a shift to short day conditions (8‐h light/16‐h darkness) in the same light until robust rosette plants were developed. Afterwards, plants were shifted to long‐day conditions (16‐h light/8‐h darkness) in order to induce flowering. Hypocotyl length was measured using ImageJ on pictures of agar plates after light treatments as indicated. For the pap genotypes grown in the dark or far‐red light, an additional 24‐h growth under white light was necessary to pick the homozygotes present at a ratio of ¼. The plate was then compared to the picture to map and mark each mutant otherwise unrecognizable. True dark treatment was done after imbibition (2–3 h under white light) by a 5‐min far‐red treatment (30 μmol m−2 s−1) and then wrapped in aluminium foil and placed in the dark at 21°C. Pigments were analysed by spectroscopy in 80% acetone. The chlorophyll content was normalized to the fresh weight corresponding to 70–110 mg seedlings and calculated using published formula (Porra et al, 1989).
Gene expression and protein sub‐cellular localization
Transient expression in onion cells (bulb sliced to ~ 16 cm2) was conducted using the Biolistic PDS 1000/He Particle Delivery System (Bio‐Rad) (1,100 psi, 10 cm travelling distance) with DNA onto 1 μm gold particles (Seashell Technology™) following instructions. After 16–40 h in the dark at 24°C, the epidermis was peeled and observed by fluorescence microscopy with a Nikon AxioScope equipped with FITC filters and an AxioCam MRc camera. Pictures acquired with Nikon's Zen software. Confocal microscopy was performed on a Leica TCS SP2 or a Zeiss LSM800. Protein localization of stably transformed plants was examined on cotyledons or hypocotyls.
Luciferase assay
Onion epidermal cells were transfected by micro‐projectile bombardment with 0.5 μg Kar6 (p35S::GFPer) 0.2 μg pRLC (Renilla luciferase), 1.5 μg of the luciferase reporter construct (pProm‐Luc) and 1.5 μg of the trans‐activating construct (p35S::TF), kept dark 20 h, 21°C and ground in liquid nitrogen. GFP was used to restrict the transfected area for protein extraction in 1 ml of PBLuc buffer (200 mM NaPO4, pH 7, 4 mM EDTA, 2 mM DTT, 5% glycerol, 10 mg l−1 BSA and 1 mM PMSF) and assayed using the Dual‐Luciferase® Reporter Assay System (Promega). Experiments were normalized to negative control set at 1.
Electromobility shift assay
The DNA probe for the PAP8 promoter (oP8Box_F, GgataccaaaaatGAcGCTCttaattatttcc; oP8Box_R, ggaaataattaaGAGCgTCatttttggtatc) or the cold probe containing a canonical G‐box (GbH5_F, GttctagtgtatcagaCACGTGtcgacaaactggtgg; GbH5_R, ccaccagtttgtcgaCACGTGtctgatacactagaa) was generated by annealing single‐stranded oligonucleotides (with a protruding G on one 5′‐end) in annealing buffer (10 mM Tris pH 7.5, 150 mM NaCl and 1 mM EDTA). 4 pmol of dsDNA was labelled by end filling with 8 pmol Cy3‐dCTP and 1 unit of Klenow fragment for 1 h at 37°C, followed by enzyme inactivation at 65°C for 10 min. For each reaction, 10 nM fluorescent dsDNA was incubated with the protein in 20 μl binding buffer (10 mM HEPES pH 7.5, 1 mM spermidine, 1% glycerol, 14 mM EDTA pH 8, 0.3 mg ml−1 BSA, 0.25% CHAPS, 28 ng μl−1 fish sperm DNA (Roche) and 3 mM TCEP). After 15‐min incubation on ice, binding reactions were loaded onto native 6% polyacrylamide gels 0.5× TBE and electrophoresed at 90 V for 90 min at 4°C. Gels were scanned on a ChemidocXRS ™ Imaging System (Bio‐Rad).
Cloning
Minipreps were performed using Qiagen kits and DNA in‐gel purification using GeneClean III Kit (MP Biomedicals). All cloning PCRs were done using Phusion™ High‐Fidelity DNA Polymerase (Thermo Scientific). PAP8 full‐length open reading frame was amplified from cDNA prepared with germinating seedlings using primers as described in Appendix Table S1 and cloned in TA cloning vectors (pGem‐T, Promega). Translational fusions of PAP8‐GFP were obtained using XhoI BamHI fragments inserted into pEZS‐NL (Carnegie institution, Stanford): PAP8 and ∆cTP or XhoI‐to‐PmlI fragments for ∆NLS and ∆cTP/∆NLS after PCR fragment cloning using oPAP8ΔNLS_PmlI. ΔNLS fragments were generated using the endogenous PmlI site at the 3′‐end of the NLS and the PCR‐based insertion of another PmlI site at the 5′‐end; then, NLS was clipped off using PmlI and backbone ligation. NLSm5 was generated using PCR‐based site‐directed mutagenesis (see Appendix Table S1 for primers). For plant transgenesis, ORFs were cloned into pBB304e (pPAP8::GUS10) or derivatives of pART27 (Blanvillain et al, 2011).
Mutant characterization and RT–PCR
gDNA preparation: leaf tissues were ground in 1.5‐ml reaction tubes and then homogenized in 400 μl of EB buffer (200 mM Tris–HCl pH 7.5, 250 mM NaCl, 25 mM EDTA, 0.5% SDS). After 5 min at 10,000 g, 400 μl of supernatant was added to 400 μl isopropanol. After 10 min at 10,000 g, the pellet was washed with 750 μl of EtOH 80% and then dried. DNA was then suspended in 50 μl of water. The PCR was done with indicated primers. For RT–PCR, the RNeasy Plant Minikit (Qiagen) was used; RNA samples were treated with RNAse‐free DNase. The RTs were performed using 2 μg of RNA, SuperScript IV VILO kit (invitrogen), dT18 primer, 1st‐strand buffer and RNase inhibitor. The RT programme was set at 65°C for 5 min, 5°C for 1 min and then after addition of RT mix, 42°C for 50 min and 70°C for 10 min. The PCR was done with indicated primers on 0.5 μl of cDNA. The absence of genomic DNA was checked by PCR on EF1α. oPAP8_rtp_F, tggtggtgatggagatatcg; oPAP8_rtp_R, tttgagacactgaagtctcg; op8i2_R, aaggaagtctcagaacaacgc; oLBb1.3, attttgccgatttcggaac; oE3_R, tagtcactcattgcacatcg; EF1α: F, caggctgattgtgctgttcttatcat; R, cttgtagacatcctgaagtggaaga. GLK1: Frtpcr, cacatgaacgcttcttcaacg; Rrtpcr, tgtagctctggtgtccaatcc. qPCR on GLK1 and GLK2 was performed using Power SYBR Green Master Mix (Thermo Fisher Scientific). Primer sequences were designed with Quantprime. Their efficiencies were between 90 and 110%, and they did not amplify genomic DNA. oEF1α_qF, tgagcacgctcttcttgctttca; oEF1α_qR, tgtaacaagatggatgccaccacc; oGLK1_qF, ttctaccgccatgcctaatccg; oGLK1_qR, actggcggtgctctaaatctcg; oGLK2_qF, agcatcggtgttcccacaagac; oGLK2_qR, tcgagggatgaatgtcgatggg.
Protein production
HY5. Rosetta2 cells were grown overnight in 50 ml LB with 100 μg ml−1 of carbenicillin and 34 μg ml−1 of chloramphenicol at 37°C. 1 l of LB + antibiotics was then inoculated and cultivated at 37°C to 0.1 OD600. At 0.6 OD600, the temperature was decreased to 16°C and 0.5 mM of isopropyl β‐D‐1‐thiogalactopyranoside was added. After an overnight induction, cells were harvested by centrifugation at 5,500 g, for 25 min, at 4°C. The cell pellet was resuspended in 30 ml of lysis buffer: 50 mM Tris–HCl, pH 8.0, 0.5 M NaCl, 20 mM imidazol pH 8.0 with a Complete™ Protease inhibitor Cocktail Tablet (Roche). The lysate was centrifuged at 15,000 g, for 40 min, at 4°C. The purification was performed at 20°C. After filtration, the supernatant was applied onto a NiNTA column in 50 mM Tris–HCl, pH 8, 0.5 M NaCl, 20 mM imidazol pH 8. HY5 was eluted in 50 mM Tris–HCl, pH 8, 0.5 M NaCl and 300 mM imidazol. After dialysis in 50 mM HEPES pH 7, 0.5 M NaCl, HY5 was concentrated using an Amicon Ultra 15‐ml centrifugal filter and a 10‐kDa‐membrane cut‐off before loading on a Superdex 75 10/30 and eluted with 25 mM HEPES pH 7, 50 mM NaCl. PAP8. Production as above except that 10 mM β‐mercaptoethanol was added to lysis and elution buffers; 1 mM DTT added to dialysis buffer. PAP8 was loaded on a Superdex 200 10/30 and eluted with 10 mM Tris–HCl pH 8, 50 mM NaCl, 5 mM DTT. Rabbit polyclonal antibodies against PAP8 were produced by ProteoGenix. In Western blots, PAP8 is detected at ~ 38 kDa, which is 7 kDa larger than the theoretical MW of processed PAP8 (31.1 kDa) without its transit peptide (6.2 kDa). With the MW correction on negative charges (D/E) using the linear correlation of Guan et al (2015) (equation y = 276.5x − 31.33, where x is the ratio of acidic AA (D+E) and y the average of delta MW in Da per AA), the high occurrence of D+E in PAP8 (58/269 = 0.216) then causes a calculated retardation of 7.6 kDa, which explains the apparent MW of PAP8 observed in Western blot. 15 N‐PAP8 was produced in minimum medium M9 containing 1 g l−1 15NH4Cl (M9‐15N). 5 ml of LB + antibiotics was inoculated with cells containing pAG21d. After 10 h of growth, 1 ml was added to 100 ml of M9‐15N + antibiotics. At 2 OD600 (16 h), the culture was centrifuged at 4,000 g and the pellet was used to inoculate 1 l of M9 + antibiotics. Culture, induction and purification were done as described for PAP8. MBP‐PAP5 . Culture and overexpression were done as for PAP8; 100 μg ml−1 of kanamycin was used instead of carbenicillin. 20 mM β‐mercaptoethanol was added to lysis buffer. The lysate was centrifuged at 15,000 g, for 40 min, at 4°C. The purification was performed at 20°C. The supernatant was applied onto an amylose column in 20 mM Tris–HCl pH 7.5, 200 mM NaCl, 1 mM EDTA and 10 mM β‐mercaptoethanol. MBP‐PAP5 was eluted in 20 mM Tris–HCl pH 7.5, 200 mM NaCl, 1 mM EDTA, 10 mM β‐mercaptoethanol and 10 mM maltose. MBP‐PAP5 was then concentrated with an Amicon Ultra 15‐ml centrifugal filter and a 30‐kDa‐membrane cut‐off before loading on a HiLoad 16/60 Superdex 200 and eluted with 10 mM Tris–HCl pH 8.0, 50 mM NaCl and 5 mM DTT. MBP. Culture and overexpression in LB followed the same procedure than for PAP5. The cell pellet was suspended in 30 ml lysis buffer. The lysate was centrifuged at 15,000 g, for 40 min, at 4°C. The purification was performed at 20°C. The supernatant was applied onto an amylose column in 20 mM Tris–HCl pH 7.5, 200 mM NaCl and 1 mM EDTA. MBP was eluted in 20 mM Tris–HCl pH 7.5, 200 mM NaCl, 1 mM EDTA and 10 mM maltose. MBP was then concentrated with an Amicon Ultra 15‐ml centrifugal filter and a 10‐kDa‐membrane cut‐off before loading on a HiLoad 16/60 Superdex 75 and eluted with 10 mM Tris–HCl pH 8.0, 150 mM NaCl and 10% glycerol. The pools containing pure HY5, PAP8, PAP5 or MBP were concentrated using Amicon centrifugal filter.
Protein extraction and Western immuno‐detection
Five‐day‐old Arabidopsis (50 mg, approx. 100 seedlings) are collected and homogenized using a Precellys™ tissue homogenizer (3 × 20 s, 9,300 g, break 30 s) in 100 μl of denaturing extraction buffer (DEB: Tris–HCl 100 mM pH 6.8, Urea 8 M, EDTA/EGTA 10 mM, DTT 10 mM, protease inhibitor (Roche) 1 tablet per 10 ml and 100‐μl glass beads (diameter 4–6 mm). The samples are centrifuged (10 min, 4°C, 9,300 g). The total soluble protein samples (TSP) are titrated by Bradford assay before mixing in Laemmli buffer (Tris–HCl 100 mM pH 6.8, glycerol 10%, SDS 2%, DTT 50 mM, bromophenol blue 0.25%) and heated 10 min at 80°C. TSP were separated by SDS–PAGE and transferred on nylon membrane (Bio‐Rad). The membrane was blocked in TBS, Tween 0.1% and non‐fat dry milk 5% w/v. The membrane was probed in TBS‐Tween 0.1%, with different primary antibodies against PAP8 (this study), PAP5 (PhytoAB, Ref. PHY0389), Histone H3 (Agrisera, Ref. AS10710), RbcL (Agrisera, Ref. AS03037), HY5 (PhytoAB, Ref. PHY0264), PHYB (PhytoAB, Ref. PHY0750), PIF1 (PhytoAB, Ref. PHY0830) and PIF3 (PhytoAB, Ref. PHY0063); dilutions H3, RbcL: 1/10,000; others: 1/5,000. Membranes were washed (five times, 5 min in a TBS‐Tween 0.1%); secondary antibody, goat anti‐rabbit conjugated with a horse radish peroxidase was used at a dilution of 1/5,000. Signal was detected using a chemiluminescent substrate (Bio‐Rad, ECL Kit).
Organelles fractionation
Five‐day‐old Arabidopsis seedlings, exposed to light or dark, were homogenized in liquid N2. The powder was dissolved in a cold native extraction buffer (NEB: Tris–HCl 100 mM pH 7.4, glycerol 25%, KCl 20 mM, EDTA 2 mM, MgCl2 2.5 mM, sucrose 250 mM, DTT 5 mM, protease inhibitor Roche™, 1 tablet per 50 ml) at a ratio of 1:3 (w/v). The extract was filtered through three layers of miracloth and one layer of nylon (100 μm) centrifuged (10 min at 1,500 g, 4°C). The supernatant was deposited on percoll 80% and centrifuged (swinging rotor, 5 min at 2,300 g, 4°C) to remove the pellet of starch. The supernatant was loaded on 35% percoll and centrifuged (swinging rotor, 5 min at 2,300 g, 4°C) to separate swimming plastids from the pellet of nuclei. The nuclei were washed two times with a plastid‐lysis buffer (NEB + 2% triton) followed with centrifugation (5 min at 1,500 g, 4°C). Fractions, corresponding to plastids or nuclei, were suspended in DEB shaken on a vortex (10 min at 4°C) before centrifugation (10 min at 9,300 g, 4°C). The TSP were subjected to Western blot analysis as above.
NMR spectroscopy
The 1H‐15N spectrum of the 15N‐labelled PAP8 alone was recorded at 300 K using a BEST‐TROSY experiment on a protein sample concentrated at 100 μM in 10 mM Tris buffer containing 50 mM NaCl and 5 mM DTT in a 95:5% H2O:D2O solvent at pH 8.0 for 49 min. The same experiment was performed on 15N‐labelled PAP8 at a concentration of 88 μM in the presence of an unlabelled PAP5‐MPB construct in stoichiometric conditions increasing the number of scans to reach an experimental time of 13 h. The control experiment involving 15N‐PAP8 and unlabelled MPB also in stoichiometric conditions was performed to check the interaction assumption between the two proteins. The following NMR experiments 1H‐15N BEST‐TROSY (Favier et al, 2011), TRACT (to estimate the global correlation time) (Lee et al, 2006) and DOSY experiments (for measuring the translational diffusion) (Morris & Johnson, 1992) were recorded on a Bruker AVANCE™ III spectrometer operating at 1H frequency of 700 MHz and equipped with a triple resonance pulsed field gradient cryo‐probe.
Author contributions
TP, DC and RB designed research. ML, F‐XG, AI, KP, LC, MC, RR, FC, DC and RB performed research. DC, TP and RB analysed data. EBE contributed mass spectrometry data. AF and PG contributed NMR data. TP and RB wrote the paper with contributions from ML, FXG, DC, AF, PG and EBE. All authors approved the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Source Data for Expanded View and Appendix
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Acknowledgements
We acknowledge the platforms of the Grenoble Instruct‐ERIC Centre (ISBG; UMS 3518 CNRS‐CEA‐UGA‐EMBL) within the Grenoble Partnership for Structural Biology (PSB). Platform access was supported by FRISBI (ANR‐10‐INBS‐05‐02) and GRAL, a project of the University Grenoble Alpes Graduate School (Ecoles Universitaires de Recherche) CBH‐EUR‐GS (ANR‐17‐EURE‐0003). IBS acknowledges integration into the Interdisciplinary Research Institute of Grenoble (IRIG, CEA). The work was supported by the Agence National de la Recherche (grant PepRegulChloro3D), the Deutsche Forschungsgemeinschaft to T.P. (PF323‐5) and the AGIR programme of Université Grenoble‐Alpes (UGA) to R.B. The project received further support by institutional grants to the Laboratoire de Physiologie Cellulaire et Végétale by Labex Grenoble Alliance of Integrated Structural Biology (GRAL) and ANR‐17‐EURE‐0003. We thank F Barneche for the ChIP‐seq analysis at the PAP8 locus; E Monte and G Toledo‐Ortiz for pifq seeds; A Guerrero‐Criado, R Toutain and S Coveley for their help at the bench. We thank E Thevenon in the Parcy Lab for advice on fluorescent labelling EMSA. We thank S Lerbs‐Mache for critical reading. We express our gratitude in memory of D Grunwald for his help on confocal imaging.
The EMBO Journal (2020) 39: e104941
Contributor Information
Thomas Pfannschmidt, Email: t.pfannschmidt@botanik.uni-hannover.de.
Robert Blanvillain, Email: robert.blanvillain@univ-grenoble-alpes.fr.
Data availability
This study includes no data deposited in external repositories.
References
- Al‐Sady B, Ni W, Kircher S, Schafer E, Quail PH (2006) Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome‐mediated degradation. Mol Cell 23: 439–446 [DOI] [PubMed] [Google Scholar]
- Arsova B, Hoja U, Wimmelbacher M, Greiner E, Ustun S, Melzer M, Petersen K, Lein W, Bornke F (2010) Plastidial thioredoxin z interacts with two fructokinase‐like proteins in a thiol‐dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. Plant Cell 22: 1498–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanvillain R, Wei S, Wei P, Kim JH, Ow DW (2011) Stress tolerance to stress escape in plants: role of the OXS2 zinc‐finger transcription factor family. EMBO J 30: 3812–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz JO, Maloof JN, Lutes J, Dabi T, Redfern JL, Trainer GT, Werner JD, Asami T, Berry CC, Weigel D et al (2002) Quantitative trait loci controlling light and hormone response in two accessions of Arabidopsis thaliana . Genetics 160: 683–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Schwab R, Chory J (2003) Characterization of the requirements for localization of phytochrome B to nuclear bodies. Proc Natl Acad Sci USA 100: 14493–14498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Galvao RM, Li M, Burger B, Bugea J, Bolado J, Chory J (2010) Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell 141: 1230–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C‐N, Lee T‐Y, Hung Y‐C, Li G‐Z, Tseng K‐C, Liu Y‐H, Kuo P‐L, Zheng H‐Q, Wen‐Chi Chang W‐C (2019) PlantPAN3.0: a new and updated resource for reconstructing transcriptional regulatory networks from ChIP‐seq experiments in plants. Nucleic Acids Res 47: D1155–D1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilova MN, Kudryakova NV, Andreeva AA, Doroshenko AS, Pojidaeva ES, Kusnetsov VV (2018a) Differential impact of heat stress on the expression of chloroplast‐encoded genes. Plant Physiol Biochem 129: 90–100 [DOI] [PubMed] [Google Scholar]
- Danilova MN, Andreeva AA, Doroshenko AS, Kudryakova NV, Kuznetsov V, Kusnetsov VV (2018b) Phytohormones regulate the expression of nuclear genes encoding the components of the plastid transcription apparatus. Dokl Biochem Biophys 478: 25–29 [DOI] [PubMed] [Google Scholar]
- Ding S, Zhang Y, Hu Z, Huang X, Zhang B, Lu Q, Wen X, Wang Y, Lu C (2019) mTERF5 acts as a transcriptional pausing factor to positively regulate transcription of chloroplast psbEFLJ. Mol Plant 12: 1259–1277 [DOI] [PubMed] [Google Scholar]
- Favier A, Brutscher B, Favier AB (2011) Recovering lost magnetization: polarization enhancement in biomolecular NMR. J Biomol NMR 49: 9–15 [DOI] [PubMed] [Google Scholar]
- Galvao RM, Li M, Kothadia SM, Haskel JD, Decker PV, Van Buskirk EK, Chen M (2012) Photoactivated phytochromes interact with HEMERA and promote its accumulation to establish photomorphogenesis in Arabidopsis . Genes Dev 26: 1851–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa SN, Botto JF (2016) The multifaceted roles of HY5 in plant growth and development. Mol Plant 9: 1353–1365 [DOI] [PubMed] [Google Scholar]
- Gao ZP, Yu QB, Zhao TT, Ma Q, Chen GX, Yang ZN (2011) A functional component of the transcriptionally active chromosome complex, Arabidopsis pTAC14, interacts with pTAC12/HEMERA and regulates plastid gene expression. Plant Physiol 157: 1733–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubler B, Merendino L, Twardziok SO, Mininno M, Allorent G, Chevalier F, Liebers M, Blanvillain R, Mayer K, Lerbs‐Mache S et al (2017) Light and plastid signals regulate different sets of genes in the albino mutant pap7‐1. Plant Physiol 175: 1203–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Zhu Q, Huang D, Zhao S, Jan Lo L, Peng J (2015) An equation to estimate the difference between theoretically predicted and SDS PAGE‐displayed molecular weights for an acidic peptide. Sci Rep 5: 13370 10.1038/srep13370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdu A, Dobos O, Domijan M, Balint B, Nagy I, Nagy F, Kozma‐Bognar L (2018) ELONGATED HYPOCOTYL 5 mediates blue light signalling to the Arabidopsis circadian clock. Plant J 96: 1242–1254 [DOI] [PubMed] [Google Scholar]
- Hernandez‐Verdeja T, Vuorijoki L, Strand A (2020) Emerging from the darkness: interplay between light and plastid signaling during chloroplast biogenesis. Physiol Plant 169: 397–406 [DOI] [PubMed] [Google Scholar]
- Holm M, Ma LG, Qu LJ, Deng XW (2002) Two interacting bZIP proteins are direct targets of COP1‐mediated control of light‐dependent gene expression in Arabidopsis . Genes Dev 16: 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Al‐Sady B, Hudson M, Kim C, Apel K, Quail PH (2004) Phytochrome‐interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941 [DOI] [PubMed] [Google Scholar]
- Iwata Y, Fedoroff NV, Koizumi N (2008) Arabidopsis bZIP60 is a proteolysis‐activated transcription factor involved in the endoplasmic reticulum stress response. Plant Cell 20: 3107–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P, Lopez‐Juez E (2013) Biogenesis and homeostasis of chloroplasts and other plastids. Nat Rev Mol Cell Biol 14: 787–802 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW (2007) Light‐regulated transcriptional networks in higher plants. Nat Rev Genet 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Krause K, Oetke S, Krupinska K (2012) Dual targeting and retrograde translocation: regulators of plant nuclear gene expression can be sequestered by plastids. Int J Mol Sci 13: 11085–11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Hilty C, Wider G, Wüthrich K (2006) Effective rotational correlation times of proteins from NMR relaxation interference. J Magn Reson 178: 72–76 [DOI] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH (2008) Multiple phytochrome‐interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E (2014) PIFs: systems integrators in plant development. Plant Cell 26: 56–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Zheng L, Zhang J, Lv Y, Liu J, Wang X, Palfalvi G, Wang G, Zhang Y (2017) Characterization and functional analysis of four HYH splicing variants in Arabidopsis hypocotyl elongation. Gene 619: 44–49 [DOI] [PubMed] [Google Scholar]
- Liebers M, Grubler B, Chevalier F, Lerbs‐Mache S, Merendino L, Blanvillain R, Pfannschmidt T (2017) Regulatory shifts in plastid transcription play a key role in morphological conversions of plastids during plant development. Front Plant Sci 8: 23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebers M, Chevalier F, Blanvillain R, Pfannschmidt T (2018) PAP genes are tissue‐ and cell‐specific markers of chloroplast development. Planta 248: 629–646 [DOI] [PubMed] [Google Scholar]
- Loudet O, Michael TP, Burger BT, Le Metté C, Mockler TC, Weigel D, Chory J (2008) A zinc knuckle protein that negatively controls morning‐specific growth in Arabidopsis thaliana . Proc Natl Acad Sci USA 105: 17193–17198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Leivar P, Ludevid D, Tepperman JM, Quail PH, Monte E (2016) Phytochrome and retrograde signalling pathways converge to antagonistically regulate a light‐induced transcriptional network. Nat Commun 7: 11431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KF, Johnson CS (1992) Diffusion‐ordered two‐dimensional nuclear magnetic resonance spectroscopy. J Am Chem Soc 114: 3139–3141 [Google Scholar]
- Nevarez PA, Qiu Y, Inoue H, Yoo CY, Benfey PN, Schnell DJ, Chen M (2017) Mechanism of dual targeting of the phytochrome signaling component HEMERA/pTAC12 to plastids and the nucleus. Plant Physiol 173: 1953–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Montgomery BL (2014) Phytochrome‐dependent coordinate control of distinct aspects of nuclear and plastid gene expression during anterograde signaling and photomorphogenesis. Front Plant Sci 5: 171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light‐regulated development of Arabidopsis . Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmuller R (2006) pTAC2, ‐6, and ‐12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18: 176–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Pfannschmidt T (2013) Essential nucleoid proteins in early chloroplast development. Trends Plant Sci 18: 186–194 [DOI] [PubMed] [Google Scholar]
- Pfalz J, Holtzegel U, Barkan A, Weisheit W, Mittag M, Pfannschmidt T (2015) ZmpTAC12 binds single‐stranded nucleic acids and is essential for accumulation of the plastid‐encoded polymerase complex in maize. New Phytol 206: 1024–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T, Link G (1994) Separation of two classes of plastid DNA‐dependent RNA polymerases that are differentially expressed in mustard (Sinapis alba L.) seedlings. Plant Mol Biol 25: 69–81 [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T, Blanvillain R, Merendino L, Courtois F, Chevalier F, Liebers M, Grubler B, Hommel E, Lerbs‐Mache S (2015) Plastid RNA polymerases: orchestration of enzymes with different evolutionary origins controls chloroplast biogenesis during the plant life cycle. J Exp Bot 66: 6957–6973 [DOI] [PubMed] [Google Scholar]
- Pogson BJ, Ganguly D, Albrecht‐Borth V (2015) Insights into chloroplast biogenesis and development. Biochim Biophys Acta 1847: 1017–1024 [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophyll‐a and chlorophyll‐b extracted with four different solvents – verification of the concentration of chlorophyll standards by atomic‐absorption spectroscopy. Biochim Biophys Acta 275: 384–394 [Google Scholar]
- Qiu Y, Li M, Pasoreck EK, Long L, Shi Y, Galvao RM, Chou CL, Wang H, Sun AY, Zhang YC et al (2015) HEMERA couples the proteolysis and transcriptional activity of PHYTOCHROME INTERACTING FACTORs in Arabidopsis photomorphogenesis. Plant Cell 27: 1409–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solymosi K, Schoefs B (2010) Etioplast and etio‐chloroplast formation under natural conditions: the dark side of chlorophyll biosynthesis in angiosperms. Photosynth Res 105: 143–166 [DOI] [PubMed] [Google Scholar]
- Somers DE, Quail PH (1995) Temporal and spatial expression patterns of PHYA and PHYB genes in Arabidopsis . Plant J 7: 413–427 [DOI] [PubMed] [Google Scholar]
- Steiner S, Schroter Y, Pfalz J, Pfannschmidt T (2011) Identification of essential subunits in the plastid‐encoded RNA polymerase complex reveals building blocks for proper plastid development. Plant Physiol 157: 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson PG, Fankhauser C, Terry MJ (2009) PIF3 is a repressor of chloroplast development. Proc Natl Acad Sci USA 106: 7654–7659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JA, Shirasu K, Deng XW (2003) The diverse roles of ubiquitin and the 26S proteasome in the life of plants. Nat Rev Genet 4: 948–958 [DOI] [PubMed] [Google Scholar]
- Swarup K, Alonso‐Blanco C, Lynn JR, Michaels SD, Amasino RM, Koornneef M, Millar AJ (1999) Natural allelic variation identifies new genes in the Arabidopsis circadian system. Plant J 20: 67–77 [DOI] [PubMed] [Google Scholar]
- Tadini L, Pesaresi P, Kleine T, Rossi F, Guljamow A, Sommer F, Muhlhaus T, Schroda M, Masiero S, Pribil M et al (2016) GUN1 controls accumulation of the plastid ribosomal protein S1 at the protein level and interacts with proteins involved in plastid protein homeostasis. Plant Physiol 170: 1817–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokumaru M, Adachi F, Toda M, Ito‐Inaba Y, Yazu F, Hirosawa Y, Sakakibara Y, Suiko M, Kakizaki T, Inaba T (2017) Ubiquitin‐proteasome dependent regulation of the GOLDEN2‐LIKE 1 transcription factor in response to plastids signals. Plant Physiol 173: 524–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Langdale JA (2009) The making of a chloroplast. EMBO J 28: 2861–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA (2009) GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis . Plant Cell 21: 1109–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson MP (2013) Using chemical shift perturbation to characterise ligand binding. Prog Nucl Magn Reson Spectrosc 73: 1–16 [DOI] [PubMed] [Google Scholar]
- Yamaguchi R, Nakamura M, Mochizuki N, Kay SA, Nagatani A (1999) Light‐dependent translocation of a phytochrome B‐GFP fusion protein to the nucleus in transgenic Arabidopsis . J Cell Biol 145: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang EJ, Yoo CY, Liu J, Wang H, Cao J, Li FW, Pryer KM, Sun TP, Weigel D, Zhou P et al (2019) NCP activates chloroplast transcription by controlling phytochrome‐dependent dual nuclear and plastidial switches. Nat Commun 10: 2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CY, Pasoreck EK, Wang H, Cao J, Blaha GM, Weigel D, Chen M (2019) Phytochrome activates the plastid‐encoded RNA polymerase for chloroplast biogenesis via nucleus‐to‐plastid signaling. Nat Commun 10: 2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon MK, Shin J, Choi G, Choi BS (2006) Intrinsically unstructured N‐terminal domain of bZIP transcription factor HY5. Proteins 65: 856–866 [DOI] [PubMed] [Google Scholar]
- Yu QB, Lu Y, Ma Q, Zhao TT, Huang C, Zhao HF, Zhang XL, Lv RH, Yang ZN (2013) TAC7, an essential component of the plastid transcriptionally active chromosome complex, interacts with FLN1, TAC10, TAC12 and TAC14 to regulate chloroplast gene expression in Arabidopsis thaliana . Physiol Plant 148: 408–421 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Source Data for Expanded View and Appendix
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Data Availability Statement
This study includes no data deposited in external repositories.


