Abstract
Recent proteomics studies of vertebrate striated muscle have identified lysine acetylation at several sites on actin. Acetylation is a reversible post-translational modification that neutralizes lysine's positive charge. Positively charged residues on actin, particularly Lys326 and Lys328, are predicted to form critical electrostatic interactions with tropomyosin (Tpm) that promote its binding to filamentous (F)-actin and bias Tpm to an azimuthal location where it impedes myosin attachment. The troponin (Tn) complex also influences Tpm's position along F-actin as a function of Ca2+ to regulate exposure of myosin-binding sites and, thus, myosin cross-bridge recruitment and force production. Interestingly, Lys326 and Lys328 are among the documented acetylated residues. Using an acetic anhydride-based labeling approach, we showed that excessive, nonspecific actin acetylation did not disrupt characteristic F-actin–Tpm binding. However, it significantly reduced Tpm-mediated inhibition of myosin attachment, as reflected by increased F-actin–Tpm motility that persisted in the presence of Tn and submaximal Ca2+. Furthermore, decreasing the extent of chemical acetylation, to presumptively target highly reactive Lys326 and Lys328, also resulted in less inhibited F-actin–Tpm, implying that modifying only these residues influences Tpm's location and, potentially, thin filament regulation. To unequivocally determine the residue-specific consequences of acetylation on Tn–Tpm–based regulation of actomyosin activity, we assessed the effects of K326Q and K328Q acetyl (Ac)-mimetic actin on Ca2+-dependent, in vitro motility parameters of reconstituted thin filaments (RTFs). Incorporation of K328Q actin significantly enhanced Ca2+ sensitivity of RTF activation relative to control. Together, our findings suggest that actin acetylation, especially Lys328, modulates muscle contraction via disrupting inhibitory Tpm positioning.
Keywords: actin, lysine acetylation, tropomyosin, thin filament, myosin, in vitro motility, acetylation, muscle physiology
Muscle contraction is driven by cyclical, force-generating interactions between myosin-containing thick and actin-based thin filaments (1, 2). Conversely, relaxation occurs when actomyosin binding is inhibited and/or suppressed by Tn and Tpm on thin filaments, or by the sequestration of myosin heads on thick filaments (1–4). Tn is a trimer composed of inhibitory (TnI), Tpm-binding (TnT), and Ca2+-binding (TnC) subunits (1, 2, 5). Each Tn–Tpm holoregulatory complex associates with seven actin protomers of the thin filament to confer Ca2+ sensitivity of contraction (1, 2). Under low Ca2+, TnI restricts Tpm to the blocked “B-state” position along F-actin where it sterically inhibits myosin binding (1, 2, 5). This effect is attenuated as free intracellular Ca2+ rises and binds to TnC. Ca2+-TnC binding relieves TnI-mediated constraints on Tpm and facilitates its azimuthal movement to the closed “C-state,” partial exposure of myosin-binding sites on actin, and weak myosin binding. The transition of myosin heads from a weak to strong F-actin-bound state induces additional Tpm movement to the open “M-state,” triggering cooperative thin filament activation and force generation. Hence, contraction demands both Tn- and myosin-dependent shifts in Tpm position.
Tpm is an α-helical, coiled-coil dimer consisting of seven tandem “quasi-equivalent,” pseudorepeating motifs that bind adjacent actin monomers (6–8). It self-polymerizes head to tail to form a continuous cable that extends the entire length of F-actin (9–13). Tpm dimers individually bind with weak affinity, orchestrated by favorable electrostatic contacts between residues of each actin protomer and Tpm pseudorepeat, yet the polymerized cable binds with exceedingly high affinity (10, 14–19). This contrasting behavior, described as “gestalt-binding,” bestows on Tpm the requisite ability to easily shift its azimuthal position over the F-actin surface to regulate myosin binding to the thin filament without risk of catastrophic detachment (10, 20). Mutations that disrupt the weak, local binding of Tpm to F-actin, as suggested by distorted, computationally derived F-actin–Tpm electrostatic energy landscapes, may minimally impact F-actin–Tpm global binding affinity yet elicit significant changes in Tpm's ability to properly impede actomyosin binding and cross-bridge cycling (21–23). Therefore, perturbed F-actin–Tpm electrostatic interactions could affect Tpm's preferred binding position and/or its regulatory switching between the B-, C-, and M-states along F-actin and, thus, Tpm-mediated regulation of actomyosin activity.
EM reconstructions and in silico-generated atomic models of Tpm bound to F-actin have identified Tpm's potential default Apo- or “A-state” binding location, where the regulatory strand is positioned such that it sterically hinders myosin binding in the absence of Tn (12, 15, 20, 24). Lys326 and Lys328 of actin are predicted to form highly favorable electrostatic contacts with Tpm and stabilize its native inhibitory configuration (12, 15, 17–19, 25, 26). These associations dominate the computed F-actin–Tpm interaction energy landscape and, together with TnI-actin interactions, are essential for establishing the B-state (5, 12, 15, 22, 27). Modification or mutation of Lys326 or Lys328 on actin or residues in their vicinity has been shown to alter contractile regulation and induce cardiac and skeletal myopathies (21, 22, 28–30).
Recently, 10 distinct lysines of actin were identified as targets of acetylation in independent proteomics assessments of healthy, vertebrate cardiac and skeletal muscle (31, 32). Of those 10 residues, 5, including Lys326 and Lys328, were reported to be acetylated in both studies. Lysine acetylation is a reversible post-translational modification (PTM) where the transfer of an Ac group to the residue's side-chain terminal amine neutralizes its positive charge. Because Lys326 and Lys328 on actin are likely critical for establishing favorable electrostatic interactions with Tpm, “masking” the charges could destabilize F-actin–Tpm binding and/or inhibitory positioning, resulting in increased actomyosin cross-bridge formation and enhanced force production. In fact, mimicking acetylation by replacing the charged lysines at positions 326 and 328 with uncharged glutamines in Drosophila indirect flight muscle (IFM) altered flight ability and stimulated IFM fiber destruction because of excessive, myosin-dependent force generation (30).
In vivo, actin acetylation levels may be distinctly modulated. For example, fluctuations in intracellular concentrations of Ac-CoA stimulate changes in lysine acetylation levels via mass action (33, 34). Alternatively, enzymatic-mediated changes in acetylation require targeting of actin by particular lysine acetylase(s) (KATs) and/or deacetylase(s) (KDACs). Despite evidence implicating specific enzymes that may perform this role in vivo (35–37), to the best of our knowledge no group has provided evidence of successful enzymatic acetylation of purified, sarcomeric actin in vitro. On the other hand, in vitro chemical acetylation of actin is eminently feasible (38–40). For example, Hitchcock-DeGregori et al. discovered, via a competitive labeling method, that Lys326 and Lys328 on F-actin displayed a high propensity for acetylation and were preferentially modified relative to all other lysines (38).
Here, we hypothesize that actin Lys326 and/or Lys328 acetylation decreases Tpm-based inhibition of myosin binding by direct disruption of F-actin–Tpm electrostatic contacts and resultant Tpm mispositioning, which ultimately manifests as enhanced Ca2+ sensitivity in regulated thin filaments. To test our hypothesis, we implemented a two-pronged approach that determined the effects of 1) indiscriminate and probabilistic, but preferential, actin lysine acetylation on global F-actin–Tpm binding and on the inherent inhibitory positioning of Tpm along F-actin in the absence and presence of Tn and 2) residue-specific pseudoacetylation of Lys326 or Lys328 on Ca2+-mediated regulation of RTFs. Our results suggest that although Lys326 and Lys328 acetylation does not affect collective F-actin–Tpm affinity, Lys328 acetylation does lessen Tpm's intrinsic ability to inhibit myosin cross-bridge binding, which contributes to enhanced thin filament Ca2+ sensitivity.
Results
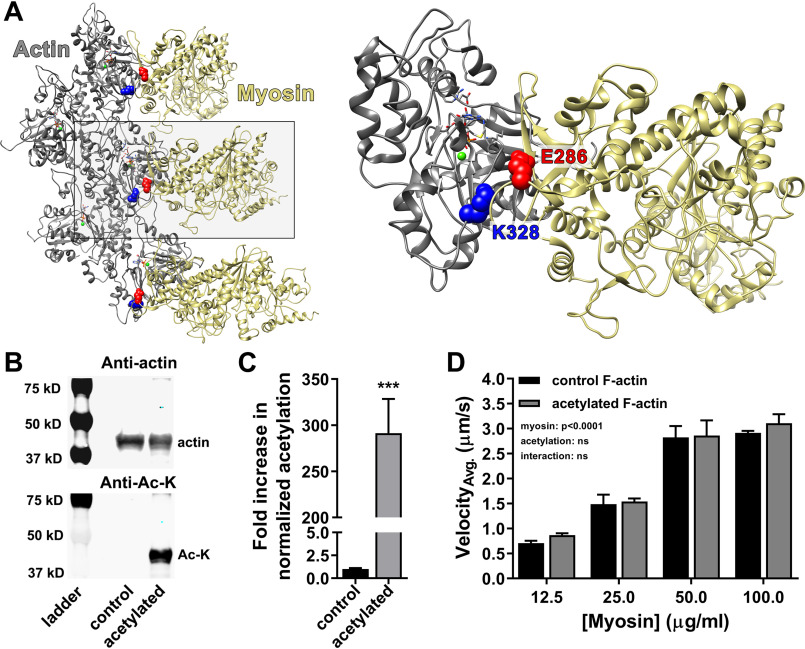
According to the Behrmann et al. (41) F-actin–Tpm–myosin rigor structure, Lys328 of actin forms a salt bridge with Glu286 of myosin (Fig. 1A). Acetylation of Lys328 and masking of its positive charge would predictably disrupt this interaction, which could potentially affect myosin binding and/or cross-bridge cycling. We pan-acetylated F-actin with acetic anhydride to increase actin-normalized lysine acetylation levels ∼290-fold (Fig. 1, B and C, and Fig. S1) and employed in vitro motility assays to assess its gross effect on actomyosin interactions (Fig. 1D). Sliding velocities of control and acetylated F-actin did not significantly differ at any of the myosin concentrations tested, suggesting that, in general, actomyosin cross-bridge cycling is seemingly unaffected by actin acetylation.
Figure 1.
Actin acetylation does not significantly alter myosin-driven F-actin sliding velocity. A, proposed Lys328–Glu286 (blue–red, respectively) actin–myosin (gray–tan, respectively) salt bridge established during strong binding (Protein Data Bank code 4A7f) (41). B, Western blots of actin treated with suprastoichiometric acetic anhydride diluted in acetonitrile (acetylated) revealed increased lysine acetylation relative to actin resuspended in acetonitrile only (control). The blots were probed with anti-actin (top panel) and anti–Ac-lysine (bottom panel; Anti-Ac-K) antibodies. C, anti–Ac-lysine intensities in B were normalized to corresponding anti-actin signals. Actin-normalized lysine acetylation increased ∼290-fold (291 ± 37) relative to control. D, in vitro motility of Alexa 568 Ph-labeled control (black) and acetylated (gray) F-actin at varying myosin concentrations. Average velocities (VelocityAvg.) were not significantly different, suggesting no change in actomyosin cross-bridge cycling caused by actin acetylation (two-way ANOVA; n = 4).
To rule out the possibility that the lack of an effect on F-actin sliding velocities was because Lys328 remained unmodified, we employed MS to confirm which lysines were acetylated following chemical treatment (Table 1). Specifically, MS detected 22 unique spectral matches to 19 unique tryptic peptide sequences. These corresponded to 12 distinct acetylation sites, most of which are common to multiple actin gene products, including cardiac, skeletal, and cytoplasmic isoforms. Lys328 acetylation was confirmed. Therefore, the absence of an acetylation-induced effect on actomyosin cycling, and specifically F-actin propulsion speeds, could not be attributed to a failure of Lys328 to be chemically modified in vitro.
Table 1.
Chemically acetylated lysine residues on actin
Actin was acetylated with an 80:1 molar ratio of acetic anhydride:actin, digested with trypsin and analyzed by MS as described under “Experimental procedures.” Acetylated peptides were identified by a mass search of in silico trypsinized rabbit skeletal actin (UniProt KB-P68135), allowing for variable modification of lysine residues with a mass shift of 42 kDa (71). Annotated spectra associated with each peptide can be found in the supporting information (Fig. S6)
| Entry | Acetylated peptide sequence | Modifications | Protein accessions | Protein name | Acetylation sitea | Peptide identification probability | Acetyl site localization probabilityb | Best Mascot ion scorec | Mascot identity scored | Observed mass | Actual mass | Charge | Δ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % | % | ppm | |||||||||||
| 1 | (K)DSYVGDEAQSkR(G) | Acetyl (+42) | XP_002722940.1, XP_002718044.1, NP_001095153.1 | Actin, Sk, Ca, and Cy | Lys61 | 100 | 100 | 79.84 | 27.19 | 698.8216 | 1,395.63 | 2 | 1.354 |
| 2 | (R)VAPEEHPTLLTEAPLNPkANR(E) | Acetyl (+42) | XP_002722940.1, XP_002718044.1 | Actin, Sk, and Ca | Lys113 | 100 | 100 | 78.04 | 31.45 | 780.4176 | 2,338.23 | 3 | 1.127 |
| 3 | (R)DLTDYLMkILTER(X) | Acetyl (+42) | XP_002722940.1, XP_002718044.1, NP_001095153.1 | Actin, Sk, Ca, and Cy | Lys191 | 100 | 100 | 51.8 | 30.77 | 551.6243 | 1,651.85 | 3 | 1.044 |
| 4 | (R)DLTDYLmkILTER(X) | Oxidation (+16), acetyl (+42) | XP_002722940.1, XP_002718044.1, NP_001095153.1 | Actin, Sk, Ca and Cy | Lys191 | 100 | 100 | 62.48 | 30.76 | 834.9309 | 1,667.85 | 2 | 1.676 |
| 5 | (K)EkLcYVALDFENEMATAASSSSLEK(S) | Acetyl (+42), carbamidomethyl (+57) | XP_002722940.1, XP_002718044.1 | Actin, Sk, and Ca | Lys215 | 100 | 100 | 109.05 | 31.86 | 945.7747 | 2,834.30 | 3 | 1.139 |
| 6 | (R)cPETLFQPSFIGMESAGIHETTYnSIMk(C) | Carbamidomethyl (+57), deamidated (+1), acetyl (+42) | XP_002722940.1, XP_002718044.1 | Actin, Sk, and Ca | Lys284 | 100 | 100 | 50.58 | 31.39 | 1,077.83 | 3,230.48 | 3 | 4.349 |
| 7 | (R)kDLYANNVMSGGTTmYPGIADR(M) | Acetyl (+42), oxidation (+16) | XP_002722940.1 | Actin and Sk | Lys291 | 100 | 100 | 80.92 | 31.06 | 811.3799 | 2,431.12 | 3 | 1.209 |
| 8 | (R)kDLYANNVmSGGTTmYPGIADR(M) | Acetyl (+42), oxidation (+16), oxidation (+16) | XP_002722940.1 | Actin and Sk | Lys291 | 100 | 100 | 69.9 | 31.14 | 816.7142 | 2,447.12 | 3 | 4.377 |
| 9 | (R)MQkEITALAPSTMK(I) | Acetyl (+42) | XP_002722940.1, XP_002718044.1, NP_001095153.1 | Actin, Sk, Ca, and Cy | Lys315 | 100 | 100 | 56.72 | 31.49 | 795.915 | 1,589.82 | 2 | −0.3323 |
| 10 | (K)EITALAPSTMKIk(I) | Acetyl (+42) | XP_002722940.1, XP_002718044.1, NP_001095153.1 | Actin, Sk, Ca, and Cy | Lys326 | 100 | 100 | 59.07 | 29.24 | 722.9083 | 1,443.80 | 2 | 0.7138 |
| 11 | (K)EITALAPSTMkIkIIAPPER(K) | Acetyl (+42), acetyl (+42) | XP_002722940.1, XP_002718044.1, NP_001095153.1 | Actin, Sk, Ca, and Cy | Lys326, Lys328 | 100 | 100 | 36.84 | 26.92 | 755.0963 | 2,262.27 | 3 | 0.3617 |
| 12 | (K)IkIIAPPER(K) | Acetyl (+42) | XP_002722940.1, XP_002718044.1, NP_001095153.1 | Actin, Sk, Ca, and Cy | Lys328 | 100 | 100 | 51.91 | 25.00 | 539.8353 | 1,077.66 | 2 | 1.067 |
| 13 | (K)IIAPPERkYSVWIGGSILASLSTFQQmWITK(Q) | Acetyl (+42), oxidation (+16) | XP_002722940.1 | Actin and Sk | Lys336 | 100 | 100 | 18.07 | 29.25 | 895.4843 | 3,577.91 | 4 | 1.979 |
| 14 | (K)IIAPPERKYSVWIGGSILASLSTFQQMWITkqEYDEAGPSIVHR(K) | Acetyl (+42), deamidated (+1) | XP_002722940.1 | Actin and Sk | Lys359 | 100 | 100 | 103.97 | 30.22 | 1,009.93 | 5,044.60 | 5 | 4.512 |
| 15 | (R)KYSVWIGGSILASLSTFQqMWITk(Q) | Deamidated (+1), acetyl (+42) | XP_002722940.1 | Actin and Sk | Lys359 | 100 | 100 | 43.34 | 32.46 | 929.8234 | 2,786.45 | 3 | 4.556 |
| 16 | (R)kYSVWIGGSILASLSTFQQmWITKQEYDEAGPSIVHR(K) | Acetyl (+42), oxidation (+16) | XP_002722940.1 | Actin and Sk | Lys336 | 100 | 100 | 56.21 | 33.57 | 857.6372 | 4,283.15 | 5 | 3.015 |
| 17 | (R)KYSVWIGGSILASLSTFQqMWITkQEYDEAGPSIVHR(K) | Deamidated (+1), acetyl (+42) | XP_002722940.1 | Actin and Sk | Lys359 | 100 | 100 | 35.18 | 33.39 | 854.6355 | 4,268.14 | 5 | 3.646 |
| 18 | (K)YSVWIGGSILASLSTFQQMWITkqEYDEAGPSIVHR(K) | Acetyl (+42), deamidated (+1) | XP_002722940.1 | Actin and Sk | Lys359 | 100 | 100 | 56.21 | 34.16 | 1,036.02 | 4,140.05 | 4 | 3.985 |
| 19 | (R)KYSVWIGGSILASLSTFQQmWISkQEYDEAGPSIVHR(K) | Oxidation (+16), acetyl (+42) | XP_002718044.1 | Actin and Ca | Lys359 | 100 | 100 | 35.64 | 33.96 | 1,068.29 | 4,269.13 | 4 | 2.638 |
| 20 | (R)kDLYAnnVLSGGTTMYPGIADR(M) | Acetyl (+42), deamidated (+1), deamidated (+1) | XP_002718044.1 | Actin and Ca | Lys291 | 96 | 100 | 16.94 | 31.74 | 600.7888 | 2,399.13 | 4 | −2.33 |
| 21 | (R)EIVRDIKEkLcYVALDFEqEMATAASSSSLEK(S) | Acetyl (+42), carbamidomethyl (+57), deamidated (+1) | NP_001095153.1 | Actin and Cy | Lys213 | 97 | 100 | 20.47 | 34.17 | 926.7111 | 3,702.82 | 4 | 3.85 |
| 22 | (K)EkLcYVALDFEqEMATAASSSSLEK(S) | Acetyl (+42), carbamidomethyl (+57), deamidated (+1) | NP_001095153.1 | Actin and Cy | Lys213 | 100 | 100 | 63.54 | 32.40 | 950.7781 | 2,849.31 | 3 | 4.832 |
aSite numbering according to the sequence, in vivo, which excludes the first two amino acids that are cleaved post-translationally.
bPosition of Ac moiety assessed using the A-score algorithm of Beausoleil et al. (71).
cIon score = −10log (probability that match is random).
dIdentity score is the score greater than which peptide identity has p < 0.05.
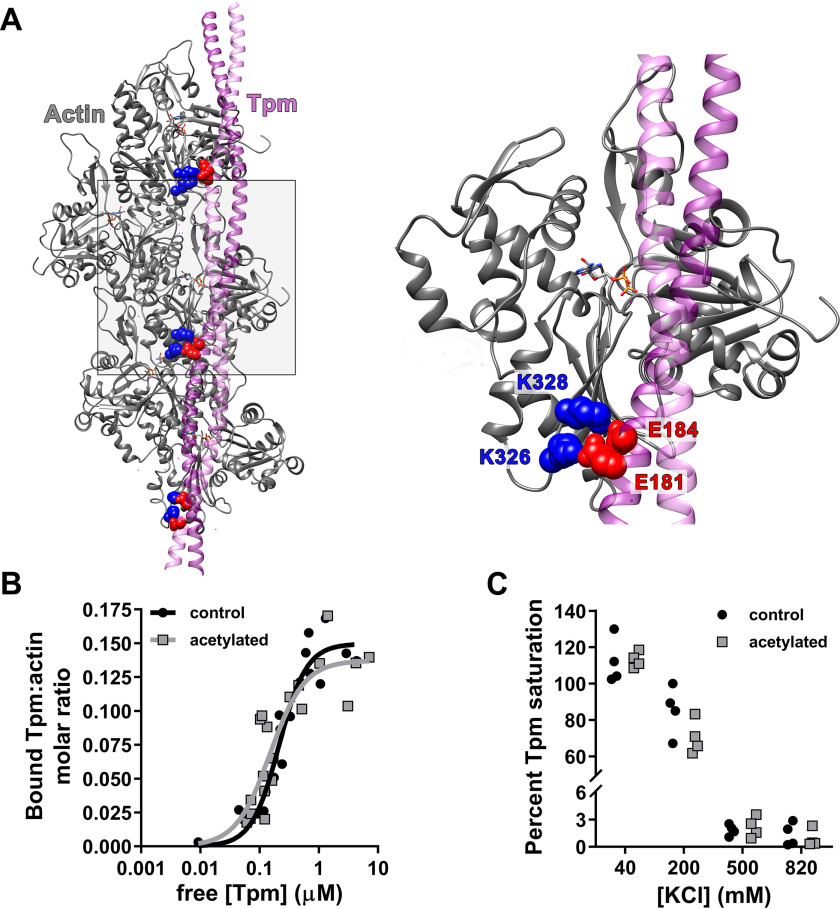
Lys326 and Lys328 of seven consecutive actin protomers are predicted to interact with negatively charged residues of every pseudorepeat along Tpm's entire length (12, 15, 20, 25, 26) (Fig. 2A). Previously, site-directed mutagenesis of Asp25, Glu334, Lys326, and Lys328 of human smooth muscle actin was shown to result in a complete loss of bacterially expressed Tpm binding, which suggests that the lysines may contribute to global F-actin–Tpm binding (19). Therefore, we determined the effect of F-actin acetylation on the binding of tissue-purified cardiac Tpm via co-sedimentation (Fig. S2). We postulated that modifying the preponderance of actin lysines, especially Lys326 and Lys328, would potentially decrease maximum F-actin–Tpm binding (Bmax) and/or affinity (Kd). However, Bmax and Kd of Tpm for control (Bmax = 0.15 ± 0.01; Kd = 0.21 ± 0.03 μm) and acetylated (Bmax = 0.14 ± 0.01; Kd = 0.15 ± 0.04 μm) F-actin did not significantly differ (Fig. 2B). Thus, extensive chemical modification of actin lysines did not appear to alter either the number of available Tpm-binding sites or the affinity of the polymerized Tpm cable for F-actin.
Figure 2.
Actin acetylation does not disrupt global F-actin–Tpm binding. A, purported contacts between Lys326/Lys328 (blue) on actin (gray) and negatively charged Tpm (purple) residues (red) in pseudorepeats 4, 5, and 6 (from bottom to top) (12). B, F-actin–Tpm co-sedimentation data were fit to the Hill equation (y = Bmax × xh/(Kdh + xh)), and no significant differences were found in Bmax or Kd of Tpm for control (black; Bmax = 0.15 ± 0.009; Kd = 0.21 ± 0.03 μm, respectively) versus acetylated (gray; Bmax = 0.14 ± 0.013; Kd = 0.15 ± 0.04 μm, respectively) F-actin. C, saturating amounts of Tpm were mixed with control (black) or acetylated (gray) F-actin at 40, 200, 500, and 820 mm [KCl] and pelleted. At 40 mm, the percentage of Tpm saturation of control (112 ± 6.3%) versus acetylated (113 ± 2.2%) F-actin was equivalent. Increasing [KCl] to 200 mm similarly decreased Tpm binding to control (85.4 ± 6.85%) and acetylated (70.4 ± 4.68%) F-actin, which was nearly fully ablated by 500 mm.
It is well-established that F-actin–Tpm association is highly influenced by buffer ionic strength because of the considerable contribution of electrostatic interactions to binding (42–44). Therefore, actin acetylation may alter the buffer salt sensitivity of Tpm binding because of marked reduction in positively charged lysines, specifically Lys326 and/or Lys328. We performed F-actin–Tpm co-sedimentation assays at varying KCl concentrations to determine whether actin acetylation modifies the effect of buffer salt on F-actin–Tpm binding (Fig. 2C). At 40 and 200 mm KCl, control (112 ± 6.3 and 85.4 ± 6.9%, respectively) and acetylated (113 ± 2.2 and 70.4 ± 4.7%, respectively) F-actin bound roughly equivalent amounts of Tpm, although increasing KCl concentration to 500 mm and above resulted in a nearly complete lack of Tpm binding to both F-actin types. This supports the idea that despite extensive lysine charge neutralization of acetylated F-actin, including Lys326 and Lys328, collective Tpm binding to F-actin remains largely intact.
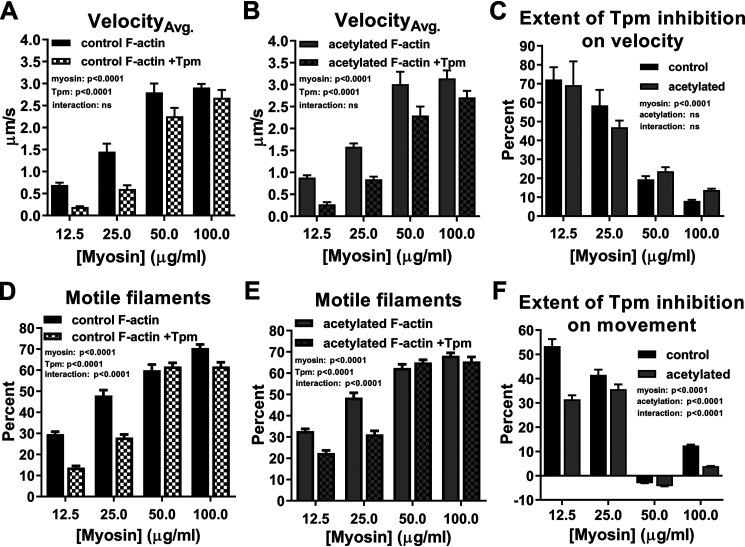
Next, we assessed whether Tpm was located along acetylated F-actin at or near its typical inhibitory A-state position and could, therefore, alter actomyosin cycling and/or the extent of myosin recruitment, which influences muscle contraction (45, 46). We first determined in vitro motility velocities of control and acetylated F-actin and F-actin–Tpm at varying myosin concentrations (Fig. 3, A and B). To facilitate interpretation regarding the degree to which Tpm reduces filament gliding speeds, we normalized control and acetylated F-actin–Tpm velocities at each myosin concentration to their respective F-actin velocities, subtracted normalized values from one, and multiplied by 100 to quantify the percentage of Tpm-based inhibition (Fig. 3C). As observed previously (47–50), Tpm significantly decreased (p < 0.0001) both control (Fig. 3A) and acetylated (Fig. 3B) F-actin velocities; however, actin acetylation had no significant effect on Tpm-mediated reduction of velocity (Fig. 3C). To further assess the effect of Tpm on actomyosin binding, we quantified the percentage of moving control and acetylated actin and actin–Tpm filaments (Fig. 3, D and E). Consistent with an inhibitory effect on velocity (Fig. 3, A–C), Tpm significantly decreased (p < 0.0001) the percentage of motile control (Fig. 3D) and acetylated (Fig. 3E) F-actin. However, in contrast, acetylated F-actin–Tpm movement was significantly elevated (i.e. less inhibited (p < 0.0001)) relative to control (Fig. 3F), whereas the PTM, again (Fig. 1D), had no impact on F-actin movement (Fig. 3, D and E). Therefore, Tpm's intrinsic ability to inhibit myosin recruitment to acetylated F-actin, as opposed to cross-bridge turnover rate, was significantly attenuated compared with control F-actin (Fig. 3, C and F). In addition, the greatest difference (i.e. a 63% relative increase) in acetylated versus control F-actin–Tpm movement occurred at the lowest myosin concentration tested (12.5 µg/ml). Under these conditions, myosin-dependent, propagated movement of Tpm and exposure of proximal binding sites is normally relatively minimal. Thus, Tpm likely remains predominantly in the A-state along control F-actin, and the extent of myosin binding is determined by Tpm's inherent ability to block myosin attachment, which appears compromised following modification of F-actin lysines. Taken together, the data suggest that actin acetylation increases cross-bridge accessibility by impairing Tpm's capacity to adequately impede actomyosin binding when Tpm alone is bound to F-actin. This effect could potentially translate into less inhibited thin filaments in the presence of Tn and Ca2+.
Figure 3.
Actin acetylation reduces Tpm-based inhibition of actomyosin binding. A and B, control (A, black) and acetylated (B, gray) F-actin (solid) and F-actin–Tpm (checkered) velocities significantly increased as a function of myosin concentration, whereas Tpm addition significantly decreased velocities (two-way ANOVA; p < 0.0001; n = 4). C, percentage of decrease in control (black) and acetylated (gray) F-actin velocities observed as a result of Tpm addition revealed that acetylation had no significant effect on Tpm-mediated reduction of velocity (two-way ANOVA; n = 4). D–F, the effects of myosin concentration and Tpm on percentage of moving control (D, black) and acetylated (E, gray) F-actin (solid) and F-actin–Tpm (checkered) mirrored those on velocity, whereas actin acetylation (F, gray) significantly decreased Tpm-based inhibition of filament motion relative to control (black) (two-way ANOVA; p < 0.0001; n = 17–22).
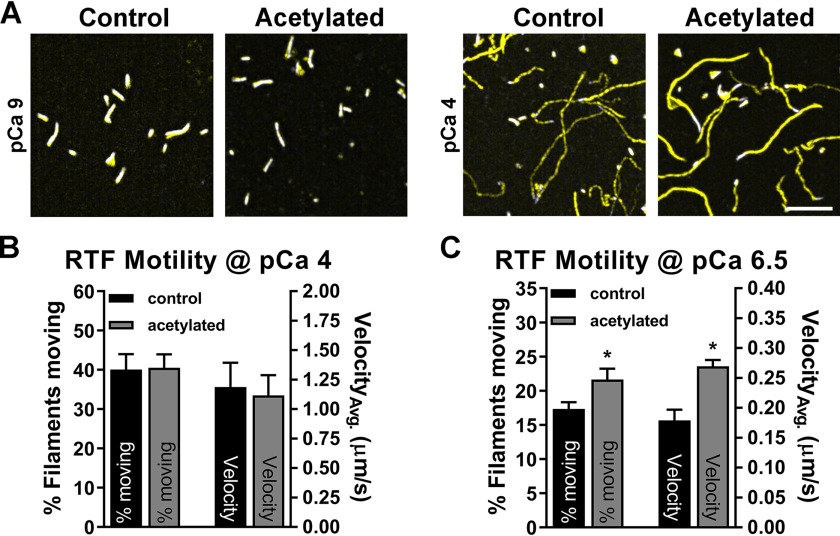
We subsequently assessed the ability of acetylated actin to serve as the backbone of a filament that responds to Ca2+. We reconstituted thin filaments by adding Tn–Tpm to control and acetylated F-actin and determined the velocity and percentage of moving filaments at three distinct Ca2+ levels. Given the more pronounced relative effect of actin acetylation on F-actin–Tpm movement under low myosin concentrations, we elected to measure RTF activation at 25 µg/ml myosin. At low Ca2+ (pCa 9), movement of control and acetylated actin-containing RTFs was limited and discontinuous and, therefore, unquantifiable under our motion criteria (see supporting information) (Fig. 4A). At high Ca2+ (pCa 4), the average velocity and percentage of filaments moving of acetylated actin-containing RTFs (1.11 ± 0.17 μm/s and 40.5 ± 3.4%, respectively) were comparable with control (1.19 ± 0.21 μm/s and 40.1 ± 3.9%, respectively) (Fig. 4, A and B). However, when assessed at a submaximal, activating level of steady-state Ca2+ (pCa 6.5), akin to an amount transiently present in actively contracting human myocytes (51–53), average acetylated versus control actin-containing RTF velocities (0.27 ± 0.01 μm/s versus 0.18 ± 0.02 μm/s, respectively) along with percentage of filaments moving (24.75 ± 1.63% versus 20.11 ± 1.45%, respectively) were significantly elevated (p < 0.05) (Fig. 4C). These results are consistent with enhanced F-actin–Tpm movement (Fig. 3, D–F) and indicate that acetylated actin is capable of forming a Ca2+-responsive thin filament that is less inhibited and hypersensitive to Ca2+.
Figure 4.
Reduced Tpm-based inhibition persists in acetylated actin-containing RTFs. A, representative images illustrating immeasurable (pCa 9) and equivalent (pCa 4) movement of control and acetylated actin-containing RTFs by overlaying the first frame of a 17–20-s movie (white) with a summative image of total motion (yellow). Scale bar, 7 μm. B, percentage of filaments moving (left; n = 9) and average velocities (right; n = 2) of control (black; 40.1 ± 3.9% and 1.19 ± 0.21 μm/s, respectively) and acetylated (gray; 40.5 ± 3.4% and 1.11 ± 0.17 μm/s, respectively) RTFs at pCa 4 were indistinguishable (two-tailed t test). C, percentage of filaments moving (left; n = 7) and average velocities (right; n = 2) of acetylated (24.75 ± 1.63% and 0.27 ± 0.01 μm/s, respectively) RTFs were significantly greater than control (20.11 ± 1.45% and 0.18 ± 0.02 μm/s, respectively) at pCa 6.5 (two-tailed t test; *, p < 0.05).
Because Lys50, Lys61, Lys315, Lys326, and Lys328 are commonly observed actin acetylation sites in vertebrates (31, 32), they may be the most physiologically relevant. As previously noted, Lys326 and Lys328 are critical for establishing Tpm's inhibitory state in vitro and in vivo (12, 15, 20, 22, 30); accordingly, it stands to reason that acetylation of these residues specifically would have the greatest contribution to Tpm mispositioning and increased actomyosin accessibility. Therefore, we attempted to more precisely resolve the effects of modifying Lys326 and Lys328 by altering the acetylation reaction to favorably target these residues (Fig. S3). Based on previous measures of relative reactivities of actin lysines (38), we proceeded under the premise that performing the acetylation reaction with substoichiometric amounts of acetic anhydride would preferentially target Lys326 and Lys328, and thus, any observed changes in Tpm-dependent inhibition of myosin binding could predominantly be attributed to modifying these residues. F-actin–Tpm in vitro motility was performed following treatment of actin with either sub- or suprastoichiometric amounts of acetic anhydride (Fig. 5A). Substoichiometric acetic anhydride treatment resulted in significantly greater (p < 0.001) F-actin–Tpm movement relative to control. Furthermore, movement of F-actin–Tpm containing substoichiometric-treated actin was statistically equivalent to F-actin–Tpm movement containing suprastoichiometric-treated actin (Fig. 5A). Therefore, actin acetylation with substoichiometric levels of acetic anhydride that favors Lys326 and Lys328 acetylation recapitulates the impact of actin treated with suprastoichiometric levels of acetic anhydride, which is less discriminate. This might indicate that Lys326 and/or Lys328 acetylation preferentially contributes to the observed increases in the percentage of F-actin–Tpm movement and RTF movement and velocity, which is in agreement with corroborating structural and in silico modeling data of the F-actin–Tpm interface (12, 15, 17–19, 25, 26). Hence, actin Lys326 and/or Lys328 modification specifically may modulate regulation of filaments replete with Tn–Tpm complexes.
Figure 5.
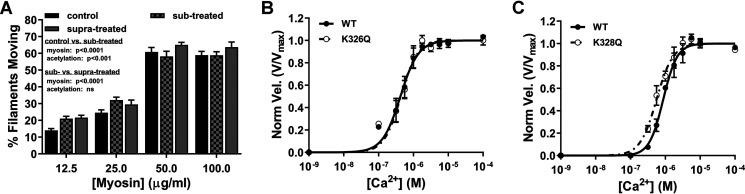
Lys328 pseudoacetylation enhances RTF Ca2+ sensitivity. A, treatment of actin with a substoichiometric amount of acetic anhydride (checkered gray) significantly increased percentage of actin–Tpm filaments moving relative to control (black) (two-way ANOVA; p < 0.001; n = 9–12), whereas substoichiometric-treated F-actin–Tpm movement was not significantly different from suprastoichiometric-treated (gray) (two-way ANOVA, n = 10–12). B and C, velocity-normalized plots of Ca2+-dependent activation of K326Q- and K328Q-containing RTFs relative to respective WT controls. K326Q and K328Q maximum velocities (Vmax = 4.1 ± 0.1 and 3.3 ± 0.06 μm/s, respectively) did not significantly differ from respective internal WT controls (Vmax = 4.2 ± 0.09 and 3.3 ± 0.06 μm/s, respectively). B, Ca2+-dependent activation of K326Q-containing RTFs was equivalent to WT control actin-containing RTFs as revealed by no significant differences in Ca2+ sensitivity ([Ca2+]50 = 0.43 ± 0.029 versus 0.42 ± 0.027 μm, respectively) or cooperativity (h = 1.8 ± 0.25 versus 1.7 ± 0.21, respectively). C, although there was no significant change in cooperativity of K328Q-containing RTFs (h = 1.8 ± 0.21) relative to WT control (h = 2.2 ± 0.22), K328Q-containing RTF Ca2+ sensitivity ([Ca2+]50 = 0.56 ± 0.032 μm) was significantly increased relative to WT control ([Ca2+]50 = 0.87 ± 0.044 μm) (p < 0.0001; n = 4).
Drosophila serves as a vehicle for transgenic actin production that can be purified in ample amounts needed for in vitro studies (22, 49, 54). The Act88F IFM actin isoform shares 92% identity with human vertebrate skeletal actin, and previous in vitro studies have confirmed that the two behave indistinguishably in vitro (22, 55). Similarly, the Act57B cardiac actin isoform is 93% identical to human vertebrate cardiac actin. To unambiguously assess the site-specific impact of Lys326 or Lys328 modification on Ca2+-dependent thin filament regulation, we created transgenic Drosophila fly lines that express WT, K326Q, or K328Q Ac-mimetic (charge-neutralizing, yet sterically similar) Act57B actin in the IFM (30). Transgenic actin comprised roughly 15% of total IFM actin (Fig. S4), which was isolated and reconstituted into thin filaments using vertebrate cardiac Tn and Tpm. Ca2+ responsiveness of Drosophila actin-containing RTFs was determined via regulated in vitro motility at 100 µg/ml myosin as described previously (22). Maximum velocities of RTFs containing transgenic pseudoacetylated actin did not deviate more than 3% from respective transgenic WT actin-containing RTFs (Fig. 5, B and C). Furthermore, K326Q actin-containing RTFs exhibited no change in Ca2+ sensitivity ([Ca2+]50 = 0.43 ± 0.029 μm) or cooperativity (h = 1.8 ± 0.25) relative to WT ([Ca2+]50 = 0.42 ± 0.027 μm and h = 1.7 ± 0.21) (Fig. 5B). However, K328Q actin-containing RTFs were significantly hypersensitive (p < 0.0001) to Ca2+ ([Ca2+]50 = 0.56 ± 0.032 μm) relative to WT ([Ca2+]50 = 0.87 ± 0.044 μm), with no change in cooperativity (h = 1.8 ± 0.21 and 2.2 ± 0.22, respectively) (Fig. 5C). Increased Ca2+ sensitivity in K328Q actin-containing RTFs is noteworthy for three reasons: 1) it is consistent with our previous data indicating increased myosin binding to acetylated F-actin–Tpm and RTFs at submaximal Ca2+ (Figs. 3 and 4); 2) it suggests that Lys328 acetylation alone is sufficient to cause Tpm mispositioning, and therefore less Ca2+ is required for RTF activation; and 3) it reveals the potential for Lys328 acetylation specifically to modulate and enhance thin filament Ca2+ sensitivity in vivo.
Discussion
This is the first study to assess the in vitro effects of actin acetylation on Tpm- and Tn–Tpm-mediated inhibition of actomyosin binding and cycling activity. Interestingly, in vitro chemical acetylation of F-actin appeared to have little effect on Tpm binding as determined by co-sedimentation assays. Because a previous study reported that the simultaneous substitution of actin Asp25, Glu334, Lys326, and Lys328, with alanines abolished F-actin–Tpm binding (19), we anticipated, at a minimum, a decrease in global Tpm binding affinity. However, no such reduction was observed under our experimental conditions. This discrepancy might be due to one or more of the following factors. First, negative effects on Tpm binding caused by alanine substitution–induced perturbations in the F-actin–Tpm topological landscape might exceed those elicited by either glutamine substitutions or Ac group transfer and charge neutralization. Second, relatively few positively charged Lys326 and/or Lys328 residues may be sufficient for collective Tpm binding. Third, disparities resulting from expression of recombinant rat striated α-Tpm and human smooth muscle actin compared with tissue-purified bovine cardiac Tpm and rabbit skeletal actin, as employed here, could differentially affect Tpm binding to the modified actins (16, 19, 20, 56, 57). Fourth, positively charged arginines on actin potentially minimize the loss in binding caused by lysine charge neutralization. Last, negatively charged Asp25 and/or Glu334 are also important contributors to global F-actin–Tpm binding affinity. Discriminating among these and additional possible mechanisms will require further investigation.
According to the results obtained using our in vitro motility protocol, acetylation had no effect on rabbit skeletal F-actin sliding speeds across all concentrations of rabbit skeletal myosin tested. This corroborates the notion that the modification does not overtly impact actomyosin interactions and cross-bridge turnover rate in the absence of Tpm (Fig. 1D). In the presence of the regulatory strand, however, the number of motile actin–Tpm filaments significantly increased relative to control (Fig. 3, D–F), and the increase was significant under conditions that favored pan-acetylation of actin lysines or, potentially, under that which favored modification of only Lys326 and Lys328 (38) (Fig. 5A). These findings added credence to the thought that Lys326 and/or Lys328 modification might mitigate the ability of Tpm to inherently block myosin binding to actin.
Hyperacetylation of actin did not adversely affect thin filament stability or trigger disassembly because increasing actin acetylation ∼290-fold did not hamper in vitro reconstitution of or render RTFs unresponsive to Ca2+. Acetylated actin-containing RTF motility was completely absent at pCa 9 (Fig. 4A) and comparable with control RTFs at pCa 4 (Fig. 4, A and B). Therefore, at steady state, and in the presence of low or maximal Ca2+ (58–60), acetylation did not seemingly impact Tn's ability to bind F-actin–Tpm and establish the thin filament regulatory B- or M-states. However, at submaximal, activating pCa 6.5, both the percentage of motile and average velocity of acetylated actin-containing RTFs were significantly higher than control (Fig. 4C), consistent with reduced inhibition of myosin binding. Thus, despite a loss in positive surface charge, acetylated actin was capable of reconstituting into, and forming the molecular backbone of, a Ca2+-responsive thin filament that evidently is hypersensitive to Ca2+ as indicated by increased myosin recruitment and propulsion speed at pCa 6.5.
Based on F-actin–Tpm results supporting the idea that Lys326 and/or Lys328 actin acetylation was largely responsible for attenuating Tpm-based inhibition (Fig. 5A), we reconstituted thin filaments containing Ac-mimetic K326Q or K328Q actin and tested the residue-specific effects of the modification on Ca2+ regulation of motility by measuring maximum sliding speeds, Ca2+ sensitivity, and cooperativity of activation (Fig. 5, B and C). We found that RTFs containing ∼15% K328Q pseudoacetylated actin (30) were hypersensitive to Ca2+ (Fig. 5C). Enhanced Ca2+ sensitivity is consistent with increased acetylated F-actin–Tpm movement (Fig.'s 3, 5A) and enhanced activation of acetylated actin-containing RTFs at intermediate Ca2+ (Fig. 4C). Mechanistically, each result can be explained by either an actin acetylation- or K328Q-induced loss in Tpm-based inhibition of myosin binding.
Although Lys326 acetylation is not disqualified as a modifier of thin filament Ca2+ sensitivity, our results suggest that relative to Lys328, Lys326 acetylation is, at the very least, significantly less impactful when present in RTFs at the tested amount. This is supported by our previous in vivo results that evaluated the specific effects of K326Q and K328Q Ac-mimetic actin on Drosophila flight ability and IFM morphology (30). Flies that ectopically expressed K328Q actin in IFMs, at roughly the same proportion present in RTFs here, were rendered flightless because of muscle destruction caused by severe, myosin-dependent hypercontraction. On the other hand, whereas the flight ability of K326Q-containing flies was significantly reduced, gross morphological structure of the IFMs was preserved. Therefore, when present at comparable levels in muscle, K328Q elicited a far more severe phenotype than K326Q actin, which suggests that the relative influence of Lys328 pseudoacetylation on the regulation of striated muscle contraction exceeds that of Lys326. Our in vitro study corroborates the in vivo data and reveals a potential molecular mechanism to explain the exacerbated K328Q muscular phenotype. Relative to WT and K326Q, Lys328 pseudoacetylation of actin increases myosin binding to thin filaments because of elevated Ca2+ sensitivity and a greater loss in Tpm-based inhibition. These effects likely result in dysregulation of force production in vivo, which elicits destruction of IFMs; muscles that are particularly sensitive to perturbation.
Previous work has demonstrated a correlation between myofilament and myofibrillar protein acetylation levels and muscle function (35, 37); however, individual contributions from the various modified proteins to overall behavior remain, for the most part, unknown. One study reported that enzyme-mediated lysine acetylation of striated muscle myosin in vitro increases actomyosin affinity and F-actin sliding velocity, suggesting an acetylation-dependent enhancement in cross-bridge cycling (61). In contrast, our data suggest that actin acetylation does not appreciably affect actomyosin activity except when Tpm is present. Recently, we found that introduction of cardiac TnI Ac-mimetic K132Q decreased Ca2+ sensitivity of both RTFs and isolated myofibrils (62). In comparison, K328Q actin increased RTF Ca2+ sensitivity. Future work should focus on how acetylation of other known targets in muscle, e.g. TnT, TnC, titin, etc., individually and in combination, affect actomyosin activity and muscle performance (32).
Given both the Ac reactivity and functional importance of actin Lys328 (30, 38), it will be important to determine whether its acetylation status is sensitive to changes in cellular Ac-CoA levels, either physiological (e.g. feeding/fasting) or pathological (e.g. heart failure (63, 64)). Alternatively, increased/decreased KAT or KDAC enzymatic activity could alter levels of the PTM via a residue-specific mechanism. Although a number of small-molecule compounds have been shown to affect Ac-CoA tissue levels by inhibiting Ac-CoA carboxylase (65–68), discovery of specific KAT and/or KDAC activators/inhibitors that preferentially target sarcomeric actin, and in particular Lys326 and/or Lys328, has been absent.
Ultimately, it will be informative to assess the site-specific stoichiometry of acetylation on actin, both in vitro and in vivo. Preliminary assessments by targeted parallel reaction monitoring MS are qualitatively consistent with Lys326 and Lys328 as highly reactive acetylation sites on actin,1 as reported previously (30, 38). However, rigorous absolute quantitation at all previously reported acetylation sites on actin is technically challenging because of the presence of easily oxidized cysteine and methionine residues flanking many of the acetylation sites (Table 1). Nevertheless, site-specific actin Ac-peptide assays are in development.
In conclusion, notwithstanding the apparent marginal impact of actin acetylation on global F-actin–Tpm binding affinity and stoichiometry, it reduced Tpm's ability to block actomyosin interactions and sensitized RTFs bathed in a submaximal, activating amount of Ca2+. Site-specific mimicry of acetylation at the reactive residue, Lys328 on actin, is sufficient to recapitulate the Ca2+ sensitization of RTF activation. We submit that the physiological acetylation status of Lys328 and its susceptibility to change, in the context of disease, warrant further scrutiny because the PTM may serve as a potent modulator of striated muscle contraction.
Experimental procedures
Molecular modeling
Actin–myosin–Tpm and actin–Tpm structures were generated using Chimera version 1.9 (69) derived from Protein Data Bank codes 4A7f (41) and 9PDB file provided in Orzechowski et al. (12).
Drosophila stocks and husbandry
All flies were raised at 25 °C on a standard cornmeal–yeast–sucrose–agar medium. The Act88F-Gal4 line was a gift from Dr. Richard M. Cripps (University of New Mexico, Albuquerque, NM, USA), and flies containing one of the UAS-Act57B actin transgenes (UAS-Act57BGFP.WT, UAS-Act57BWT, UAS-Act57BK326Q, or UAS-Act57BK328Q) were generated as described previously (30). Virgin Act88F-Gal4 female flies were crossed with males containing one of the UAS-Act57B transgenes to create progeny that overexpressed transgenic Act57BWT, Act57BK326Q, or Act57BK328Q actin specifically in the IFM.
Protein sources, preparation, and quantitation
Lyophilized rabbit skeletal actin was obtained from Cytoskeleton (catalog no. AKL95). Psoas muscle myosin was purified (70) from white New Zealand rabbits provided by Dr. David Kass (Johns Hopkins University). Tissue-purified porcine cardiac Tpm was provided by Dr. Jeff Moore (University of Massachusetts–Lowell), and bovine cardiac isoforms of Tpm and Tn were provided by Dr. Larry Tobacman (University of Illinois at Chicago). Pools containing ectopically expressed Act57BWT, Act57BK326Q, or Act57BK328Q actin were purified from Drosophila IFMs according to previously published protocols (22, 49, 54).
In vitro actin acetylation and quantitation of lysine acetylation levels
In vitro actin acetylation was performed as described in Hitchcock-DeGregori et al. (38). 10 mg/ml globular (G-) actin, rehydrated as per manufacturer's instructions in 5 mm Tris-HCl, pH 8.0, 0.2 mm CaCl2, 0.2 mm ATP, 5% (w/v) sucrose, and 1% (w/v) dextran, was distributed into 10 µl aliquots, flash-frozen in liquid nitrogen, and stored at −80 °C. Thawed G-actin was diluted 10-fold in low-salt buffer (25 mm KCl, 4 mm MgCl2, 1 mm EGTA, 25 mm imidazole, pH 7.2, 10 mm DTT, 1 mm ATP) and polymerized for ∼1 h at room temperature. Stock acetic anhydride (Sigma–Aldrich lot no. SHBJ2093) was diluted 1:70 (4-fold ratio of total moles of acetic anhydride:total actin lysines) or 1:5600 (0.05 total mol of acetic anhydride:total actin lysines) in 100% acetonitrile and added to 96 µl of 1 mg/ml F-actin via addition of four 0.3 µl aliquots every 3 min with continuous agitation. Actins were stored at 4 °C overnight and dialyzed twice in the cold room against 1 liter of buffer (100 or 200 mm KCl, 5 mm MgCl2, 5 mm Tris-HCl, pH 7.5) for at least 4 h to remove unreacted acetic anhydride.
Dialyzed acetylated actin was prepared in 4× lithium dodecyl sulfate sample buffer (Invitrogen lot no. 1920134), and ∼3 µg was loaded onto a 4–15% gradient SDS-PAGE gel (Bio-Rad catalog no. 456-1086). Proteins were transferred to a nitrocellulose membrane, blocked for 1 h in 1:1 mixture of TBS with 0.1% Tween 20 (Sigma lot no. SLBD6080V) and blocking buffer (LI-COR catalog no. 927-40000), and probed with anti–Ac-lysine (Cytoskeleton catalog no. AAC-03) antibodies, diluted 1:750 in TBS, for 1 h at room temperature or overnight in the cold room. Following four or five washes in TBS with 0.1% Tween 20 (TBST), Ac-lysine–bound primary antibodies were conjugated to 1:10,000 green secondary antibodies (LI-COR Biosciences) via 1 h incubation at room temperature. After numerous TBST washes, the membranes were exposed to a 1:7500 diluted anti-actin primary antibody (Proteintech catalog no. 20536-1-AP) at room temperature for 1 h, washed, and conjugated to 1:10,000 diluted red secondary antibodies (LI-COR Biosciences). Unconjugated antibodies were washed, and membranes were imaged on a LI-COR Odyssey fluorescence imager. Anti–Ac-lysine intensities were normalized to actin, and the average fold increase in actin-normalized lysine acetylation was determined for 9–11 individual measures.
Sample peptide preparation, chromatography, MS, and data analysis
48 µl of 1 mg/ml F-actin (25 mm KCl, 4 mm MgCl2, 1 mm EGTA, 25 mm imidazole, pH 7.2, 10 mm DTT, 1 mm ATP) were aliquoted into two separate tubes. Each tube was mixed with either 100% acetonitrile or acetonitrile containing 1:140 (v/v) diluted stock acetic anhydride (Sigma–Aldrich lot no. SHBJ2093) via addition of four 0.3 µl aliquots every 3 min under constant mixing. Total moles of acetic anhydride added to the latter mixture was four times the number of actin lysines. F-actin samples were left at 4 °C overnight, and the following day ∼5 µg total protein were dried down, redissolved in 50 µl of 20 mm ammonium bicarbonate (pH 8.0), then reduced with 5 µl of 50 mm DTT at 55 °C for 1 h, and alkylated with 5 µl 50 mm iodoacetamide in the dark for 15 min. Enzymatic cleavage was performed with MS-grade trypsin/lys-C (Promega) at 37 °C overnight. The peptides were desalted with stage tips constructed from Empore C18 material (3M Corporation).
Tryptic peptides were subjected to nano-reversed phase HPLC coupled to MS/MS. Chromatography was conducted on a 75 μm × 250 mm column that was packed in-house into a PicoFrit self-pack (New Objective) with ReproSil-Pur C18-AQ (3 μm particles, 120 Å pores; Dr. Maisch, GmbH) and eluted with a 120 min linear two-solvent gradient (solvent A: 0.1% (v/v) formic acid (Suprapur, EMD Millipore) with in-house deionized, reverse-osmosed water; solvent B: 0.1% formic acid in 80% (v/v) acetonitrile (Fisher Scientific) into an Orbitrap Fusion Lumos mass spectrometer equipped with the Easy-IC (internal calibration; Thermo Scientific) option. Data-dependent MS/MS acquisition was employed using a 3 s cycle time between MS scans. The resolution was 120,000 for MS and 30,000 for MS/MS. Signal targets were 2 × 106 ions in 60 s for MS and 5 × 104 ions in 54 s for MS/MS.
Acquired masses were searched against the RefSeq83 Oryctolagus cuniculus database using Mascot version 2.6.2 (Matrix Science), as implemented in Proteome Discoverer 1.4 (Thermo Scientific) with lysine acetylation (+42 Da), deamidation of asparagine and glutamine (+1 Da), and oxidation of methionine (+16 Da) included as variable search parameters. Carbamidomethyl cysteine (+57 Da) was set as a static search parameter. The precursor ion mass tolerance was set to 5 ppm, whereas 0.02 Da was used for MS/MS. Three missed cleavages were permitted. Data from Proteome Discoverer (1% false discovery rate) were imported into Scaffold Q+ v4.1.0 (Proteome Software) for conversion to mzIdentML and subsequent data inspection in Scaffold PTM v3.3.0 (Proteome Software). Lysine acetylation site confidence was assessed using Scaffold PTM's implementation of the A score (71).
F-actin–Tpm co-sedimentation
Dialyzed control and acetylated F-actin were diluted ∼5-fold in buffer that yielded a final composition of 200 mm KCl, 2 mm MgCl2, 10 mm Tris-HCl, pH 7.5, and 10 mm DTT. Stock Tpm concentration (65 μm) was determined via Bradford colorimetric assay (Bio-Rad), and 25 µl of F-actin (∼3.5 µg) was mixed with minute volumes of porcine cardiac Tpm in duplicate at the following Tpm:actin molar ratios: 1:16, 1:15, 1:12, 1:10, 7:1, 1:5, 1:4, 1:3, 1:2, 1:1, 1.5:1, and 2:1. Mixtures were placed at room temperature for 30 min, then on ice for a minimum of 2 h, and spun down in a TLA-100 ultracentrifuge rotor at 100,000 rpm (436,000 × g) for 20 min at 4 °C. 12 µl of supernatant were added to 3 µl of 4× lithium dodecyl sulfate sample buffer (Invitrogen lot no. 1920134), whereas 15 µl of 1× sample buffer was added to the pellets. The samples were heated to ∼90 °C for 3–5 min, centrifuged, loaded on a 4–15% gel (Bio-Rad catalog no. 456-1086), and run at 100 V for 1.5 h. The gels were stained overnight in G-250 Coomassie (Bio-Rad catalog no. 161-0786) and destained in double distilled water. In addition to assessment of F-actin–Tpm binding at “medium” ionic strength (i.e. 200 mm KCl), co-sedimentation experiments were equivalently performed under low (40 mm KCl, 4.2 mm MgCl2, 0.8 mm EGTA, 20 mm imidazole, 1 mm Tris-HCl, pH 7.5, and 10 mm DTT) and high (500 mm KCl, 2 mm MgCl2, 5 mm Tris-HCl, pH 7.5, 10 mm DTT) ionic strengths at 1:3 and 1:4 Tpm:actin molar ratios. KCl concentration was further increased to 820 mm (2 mm MgCl2, 5 mm Tris-HCl, pH 7.5, 10 mm DTT), and F-actin–Tpm binding was determined at 1:2, 1:1, 1.5:1, and 2:1 Tpm:actin ratios.
Determination of F-actin–Tpm affinity
Coomassie-stained gels were scanned using a LI-COR Odyssey fluorescence imager, and band intensities were quantified using ImageStudio software. Known amounts of Tpm (0.5–6 µg) were used to plot a standard intensity curve to determine µg of Tpm in pellet and supernatant fractions and converted to moles (67 kDa). Negligible amounts of Tpm were found in pellet fractions in the absence of F-actin (Fig. S5). To determine moles of actin, relative amounts present in pellet and supernatant fractions were calculated by dividing intensities of each band by their sum. The amount of actin in pellet and supernatant fractions was then calculated by multiplying by total actin and converted to moles (42 kDa). Bound Tpm:actin molar ratios were plotted against free Tpm concentrations, and the data were fit to the Hill equation (y = Bmax × xh/(Kdh + xh)) to derive Bmax and Kd of Tpm for control and acetylated actin. Significant differences in fit parameters were assessed via extra sum of squares F-tests, with p < 0.05 deemed significant. The percentage of saturation of F-actin–Tpm binding was defined as the molar ratio of Tpm to actin in the pellet times 700.
F-actin, F-actin–Tpm, and RTF in vitro motility
A portion of dialyzed F-actin was diluted in low-salt buffer to ∼1 μm and fluorescently labeled overnight via equimolar addition of Alexa 568 phalloidin (Ph) (Thermo Fisher catalog no. A12380). In vitro motility of control and acetylated Alexa 568 Ph-labeled F-actin was performed at myosin concentrations between 12.5 and 100 µg/ml at 30 °C, pH 7.2, and an ionic strength of 37 mm. Briefly, myosin was introduced into a flow cell and allowed 2 min to bind a nitrocellulose-coated coverslip. The surface was blocked with 2 mg/ml BSA, myosin “dead heads” were nonreversibly bound to unlabeled actin filaments, and enzymatically active myosin was bound to <10 nm Alexa 568 Ph-labeled F-actin. Fluorescent actin was excited and imaged using an X-CITE 120 LED lamp and 531/40 filter on an Olympus IX73 microscope. Emitted light was captured at 593/40 and detected on a Hamamatsu Flash 4LT EMCCD camera, operated with HCI imaging software. The videos were converted to multipage TIFs and imported into ImageJ for processing. Duplicate F-actin motility assessments from two separate suprastoichiometric acetylation reactions and one substoichiometric acetylation reaction were obtained in parallel with control F-actins. Videos from four to eight areas of each flow cell were recorded at 1–10 fps for 20 frames total, and the velocities of moving filaments were measured via automated tracking by ImageJ plugin wrmtrack (72). Average velocities and errors from two technical replicate experiments per biological replicate were calculated for filaments classified as movers (see supporting information). Total filaments analyzed per myosin concentration ranged between 250 and 2000.
Alexa 568 Ph-labeled F-actin was also incubated with 300 nm bovine cardiac Tpm on ice for at least 30 min prior to F-actin–Tpm motility. Motility buffer also included 150 nm Tpm to maintain Tpm saturation of F-actin. Average velocities and percent filaments moving of control and acetylated F-actin–Tpm samples were normalized to respective F-actin velocities and percentage of filaments moving at each myosin concentration. F-actin–normalized F-actin–Tpm motility parameters were subtracted from one and multiplied by 100 to determine the percentage of Tpm-based inhibition at each myosin concentration. The effect of acetylation on Tpm-based inhibition was assessed via two-way ANOVA with p < 0.05 considered significant.
RTF motility at 25 µg/ml myosin was identically performed as F-actin–Tpm, except that 300 and 150 nm bovine cardiac Tn was added to F-actin–Tpm and Tpm-Ca2+-containing motility buffers (50 mm KCl), respectively. Significant differences in average velocities and the percentage of moving filaments at pCa 4 and 6.5 between control and acetylated actin-containing RTFs were determined via two-tailed t tests with p < 0.05 considered significant. Ca2+-regulated motility of RTFs containing Drosophila-derived, transgenic Act57B actin at 100 µg/ml myosin concentration was performed as described previously (22). Briefly, average RTF velocities calculated from two biological and two technical replicate experiments (n = 4) at each distinct [Ca2+] were pooled and weighted based on the number of filaments analyzed for each of four replicate experiments. Pooled, raw velocity data were plotted as a function of [Ca2+] and fit to the Hill equation to derive the Vmax value for each RTF type. Average velocities at each [Ca2+] were then normalized to each corresponding, Hill equation–derived Vmax, and pooled normalized velocity versus [Ca2+] data were refit to the Hill equation. Significant differences in [Ca2+]50 and cooperativity were determined via extra sum of squares F-tests between the fits.
Data availability
All data, associated protocols, methods, and sources of materials are available in the main text or in the supporting information. Additionally, the MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (73) partner repository with the data set identifier PXD020732 and PXD019276.
Supplementary Material
Acknowledgments
We thank Dr.'s J. Moore (University of Massachusetts–Lowell) and L. S. Tobacman (University of Illinois at Chicago) for providing tissue-purified Tpm and Meera C. Viswanathan (Johns Hopkins University) and Dr. Anna Blice-Baum (Johns Hopkins University) for expert technical assistance. MS on acetylated actin was conducted by the Johns Hopkins University Mass Spectrometry Facility, for which we thank Robert O'Meally and Dr. Robert N. Cole.
This article contains supporting information.
Author contributions—W. S., D. B. F., and A. C. conceptualization; W. S., A. M., and D. B. F. data curation; W. S., D. B. F., and A. C. formal analysis; W. S., D. B. F., and A. C. validation; W. S., A. M., D. B. F., and A. C. investigation; W. S., D. B. F., and A. C. visualization; W. S., D. B. F., and A. C. methodology; W. S., A. M., D. B. F., and A. C. writing-original draft; W. S., D. B. F., and A. C. writing-review and editing; D. B. F. and A. C. resources; D. B. F. software; D. B. F. and A. C. supervision; D. B. F. and A. C. funding acquisition; D. B. F. and A. C. project administration.
Funding and additional information—This work was supported by National Institutes of Health Grants R01HL134821 (to D. B. F.) and R01HL124091 (to A. C.), American Heart Association Grants 18TPA34170575 (to D. B. F.) and 17POST33630159 (to W. S.), and a Johns Hopkins University Catalyst Award (to D. B. F.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Anthony Cammarato and D. Brian Foster, unpublished observations.
- PTM
- post-translational modification
- Ac
- Acetyl
- Tpm
- tropomyosin
- Tn
- troponin
- F-
- filamentous
- RTF
- reconstituted thin filament
- TnI
- inhibitory subunit of troponin
- TnT
- tropomyosin-binding subunit of troponin
- TnC
- calcium-binding subunit of troponin
- B-state
- blocking positional state of tropomyosin
- C-state
- closed positional state of tropomyosin
- M-state
- myosin-induced positional state of tropomyosin
- A-state
- “Apo” positional state of tropomyosin
- IFM
- indirect flight muscle
- KAT
- lysine acetylase
- KDAC
- lysine deacetylase
- G-
- globular
- TBS
- Tris-buffered saline
- TBST
- Tris-buffered saline with 0.1% Tween 20
- Ph
- phalloidin
- ANOVA
- analysis of variance.
References
- 1. Gordon A. M., Homsher E., and Regnier M. (2000) Regulation of contraction in striated muscle. Physiol. Rev. 80, 853–924 10.1152/physrev.2000.80.2.853 [DOI] [PubMed] [Google Scholar]
- 2. Lehman W. (2016) Thin filament structure and the steric blocking model. Compr. Physiol. 6, 1043–1069 10.1002/cphy.c150030 [DOI] [PubMed] [Google Scholar]
- 3. McNamara J. W., Li A., Dos Remedios C. G., and Cooke R. (2015) The role of super-relaxed myosin in skeletal and cardiac muscle. Biophys. Rev. 7, 5–14 10.1007/s12551-014-0151-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Trivedi D. V., Adhikari A. S., Sarkar S. S., Ruppel K. M., and Spudich J. A. (2018) Hypertrophic cardiomyopathy and the myosin mesa: viewing an old disease in a new light. Biophys. Rev. 10, 27–48 10.1007/s12551-017-0274-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yamada Y., Namba K., and Fujii T. (2020) Cardiac muscle thin filament structures reveal calcium regulatory mechanism. Nat. Commun. 11, 153 10.1038/s41467-019-14008-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hitchcock-DeGregori S. E., and An Y. (1996) Integral repeats and a continuous coiled coil are required for binding of striated muscle tropomyosin to the regulated actin filament. J. Biol. Chem. 271, 3600–3603 10.1074/jbc.271.7.3600 [DOI] [PubMed] [Google Scholar]
- 7. Hitchcock-DeGregori S. E., Song Y., and Greenfield N. J. (2002) Functions of tropomyosin's periodic repeats. Biochemistry 41, 15036–15044 10.1021/bi026519k [DOI] [PubMed] [Google Scholar]
- 8. Singh A., and Hitchcock-DeGregori S. E. (2007) Tropomyosin's periods are quasi-equivalent for actin binding but have specific regulatory functions. Biochemistry 46, 14917–14927 10.1021/bi701570b [DOI] [PubMed] [Google Scholar]
- 9. Lorenz M., Poole K. J., Popp D., Rosenbaum G., and Holmes K. C. (1995) An atomic model of the unregulated thin filament obtained by X-ray fiber diffraction on oriented actin–tropomyosin gels. J. Mol. Biol. 246, 108–119 10.1006/jmbi.1994.0070 [DOI] [PubMed] [Google Scholar]
- 10. Holmes K. C., and Lehman W. (2008) Gestalt-binding of tropomyosin to actin filaments. J. Muscle Res. Cell Motil. 29, 213–219 10.1007/s10974-008-9157-6 [DOI] [PubMed] [Google Scholar]
- 11. Li X. E., Orzechowski M., Lehman W., and Fischer S. (2014) Structure and flexibility of the tropomyosin overlap junction. Biochem. Biophys. Res. Commun. 446, 304–308 10.1016/j.bbrc.2014.02.097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Orzechowski M., Li X. E., Fischer S., and Lehman W. (2014) An atomic model of the tropomyosin cable on F-actin. Biophys. J. 107, 694–699 10.1016/j.bpj.2014.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schmidt W. M., Lehman W., and Moore J. R. (2015) Direct observation of tropomyosin binding to actin filaments. Cytoskeleton (Hoboken) 72, 292–303 10.1002/cm.21225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tobacman L. S. (2008) Cooperative binding of tropomyosin to actin. Adv. Exp. Med. Biol. 644, 85–94 10.1007/978-0-387-85766-4_7 [DOI] [PubMed] [Google Scholar]
- 15. Li X. E., Tobacman L. S., Mun J. Y., Craig R., Fischer S., and Lehman W. (2011) Tropomyosin position on F-actin revealed by Em reconstruction and computational chemistry. Biophys. J. 100, 1005–1013 10.1016/j.bpj.2010.12.3697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heald R. W., and Hitchcock-DeGregori S. E. (1988) The structure of the amino terminus of tropomyosin is critical for binding to actin in the absence and presence of troponin. J. Biol. Chem. 263, 5254–5259 [PubMed] [Google Scholar]
- 17. Barua B., Pamula M. C., and Hitchcock-DeGregori S. E. (2011) Evolutionarily conserved surface residues constitute actin binding sites of tropomyosin. Proc. Natl. Acad. Sci. U.S.A. 108, 10150–10155 10.1073/pnas.1101221108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barua B. (2013) Periodicities designed in the tropomyosin sequence and structure define its functions. Bioarchitecture 3, 51–56 10.4161/bioa.25616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Barua B., Fagnant P. M., Winkelmann D. A., Trybus K. M., and Hitchcock-DeGregori S. E. (2013) A periodic pattern of evolutionarily conserved basic and acidic residues constitutes the binding interface of actin-tropomyosin. J. Biol. Chem. 288, 9602–9609 10.1074/jbc.M113.451161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lehman W., Orzechowski M., Li X. E., Fischer S., and Raunser S. (2013) Gestalt-binding of tropomyosin on actin during thin filament activation. J. Muscle Res. Cell Motil. 34, 155–163 10.1007/s10974-013-9342-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rynkiewicz M. J., Prum T., Hollenberg S., Kiani F. A., Fagnant P. M., Marston S. B., Trybus K. M., Fischer S., Moore J. R., and Lehman W. (2017) Tropomyosin must interact weakly with actin to effectively regulate thin filament function. Biophys. J. 113, 2444–2451 10.1016/j.bpj.2017.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Viswanathan M. C., Schmidt W., Rynkiewicz M. J., Agarwal K., Gao J., Katz J., Lehman W., and Cammarato A. (2017) Distortion of the actin A-triad results in contractile disinhibition and cardiomyopathy. Cell Rep. 20, 2612–2625 10.1016/j.celrep.2017.08.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Orzechowski M., Moore J. R., Fischer S., and Lehman W. (2014) Tropomyosin movement on F-actin during muscle activation explained by energy landscapes. Arch. Biochem. Biophys. 545, 63–68 10.1016/j.abb.2014.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. von der Ecken J., Müller M., Lehman W., Manstein D. J., Penczek P. A., and Raunser S. (2015) Structure of the F-actin–tropomyosin complex. Nature 519, 114–117 10.1038/nature14033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brown J. H., Zhou Z., Reshetnikova L., Robinson H., Yammani R. D., Tobacman L. S., and Cohen C. (2005) Structure of the mid-region of tropomyosin: bending and binding sites for actin. Proc. Natl. Acad. Sci. U.S.A. 102, 18878–18883 10.1073/pnas.0509269102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Greenfield N. J., Huang Y. J., Swapna G. V., Bhattacharya A., Rapp B., Singh A., Montelione G. T., and Hitchcock-DeGregori S. E. (2006) Solution NMR structure of the junction between tropomyosin molecules: implications for actin binding and regulation. J. Mol. Biol. 364, 80–96 10.1016/j.jmb.2006.08.033 [DOI] [PubMed] [Google Scholar]
- 27. Schmidt W., and Cammarato A. (2020) The actin “A-triad's” role in contractile regulation in health and disease. J. Physiol. 598, 2897–2908 10.1113/JP276741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Papadaki M., Holewinski R. J., Previs S. B., Martin T. G., Stachowski M. J., Li A., Blair C. A., Moravec C. S., Van Eyk J. E., Campbell K. S., Warshaw D. M., and Kirk J. A. (2018) Diabetes with heart failure increases methylglyoxal modifications in the sarcomere, which inhibit function. JCI Insight 3, e121264 10.1172/jci.insight.121264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meng T., Bu W., Ren X., Chen X., Yu J., Eckenhoff R. G., and Gao W. D. (2016) Molecular mechanism of anesthetic-induced depression of myocardial contraction. FASEB J. 30, 2915–2925 10.1096/fj.201600290RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Viswanathan M. C., Blice-Baum A. C., Schmidt W., Foster D. B., and Cammarato A. (2015) Pseudo-acetylation of K326 and K328 of actin disrupts Drosophila melanogaster indirect flight muscle structure and performance. Front. Physiol. 6, 116 10.3389/fphys.2015.00116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lundby A., Lage K., Weinert B. T., Bekker-Jensen D. B., Secher A., Skovgaard T., Kelstrup C. D., Dmytriyev A., Choudhary C., Lundby C., and Olsen J. V. (2012) Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep. 2, 419–431 10.1016/j.celrep.2012.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Foster D. B., Liu T., Rucker J., O'Meally R. N., Devine L. R., Cole R. N., and O'Rourke B. (2013) The cardiac acetyl-lysine proteome. PLoS One 8, e67513 10.1371/journal.pone.0067513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pougovkina O., Te Brinke H., Ofman R., van Cruchten A. G., Kulik W., Wanders R. J., Houten S. M., and de Boer V. C. (2014) Mitochondrial protein acetylation is driven by acetyl-CoA from fatty acid oxidation. Hum. Mol. Genet. 23, 3513–3522 10.1093/hmg/ddu059 [DOI] [PubMed] [Google Scholar]
- 34. Wagner G. R., and Payne R. M. (2013) Widespread and enzyme-independent Nε-acetylation and Nε-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J. Biol. Chem. 288, 29036–29045 10.1074/jbc.M113.486753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gupta M. P., Samant S. A., Smith S. H., and Shroff S. G. (2008) HDAC4 and PCAF bind to cardiac sarcomeres and play a role in regulating myofilament contractile activity. J. Biol. Chem. 283, 10135–10146 10.1074/jbc.M710277200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Waltregny D., Glénisson W., Tran S. L., North B. J., Verdin E., Colige A., and Castronovo V. (2005) Histone deacetylase HDAC8 associates with smooth muscle α-actin and is essential for smooth muscle cell contractility. FASEB J. 19, 966–968 10.1096/fj.04-2303fje [DOI] [PubMed] [Google Scholar]
- 37. Jeong M. Y., Lin Y. H., Wennersten S. A., Demos-Davies K. M., Cavasin M. A., Mahaffey J. H., Monzani V., Saripalli C., Mascagni P., Reece T. B., Ambardekar A. V., Granzier H. L., Dinarello C. A., and McKinsey T. A. (2018) Histone deacetylase activity governs diastolic dysfunction through a nongenomic mechanism. Sci. Transl. Med. 10, eaao0144 10.1126/scitranslmed.aao0144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hitchcock-De Gregori S. E., Mandala S., and Sachs G. A. (1982) Changes in actin lysine reactivities during polymerization detected using a competitive labeling method. J. Biol. Chem. 257, 12573–12580 [PubMed] [Google Scholar]
- 39. Szilagyi L., and Lu R. C. (1982) Changes of lysine reactivities of actin in complex with myosin subfragment-1, tropomyosin and troponin. Biochim. Biophys. Acta 709, 204–211 10.1016/0167-4838(82)90462-9 [DOI] [PubMed] [Google Scholar]
- 40. Hitchcock-De Gregori S. E. (1982) Study of the structure of troponin-I by measuring the relative reactivities of lysines with acetic anhydride. J. Biol. Chem. 257, 7372–7380 [PubMed] [Google Scholar]
- 41. Behrmann E., Müller M., Penczek P. A., Mannherz H. G., Manstein D. J., and Raunser S. (2012) Structure of the rigor actin–tropomyosin–myosin complex. Cell 150, 327–338 10.1016/j.cell.2012.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Landis C. A., Bobkova A., Homsher E., and Tobacman L. S. (1997) The active state of the thin filament is destabilized by an internal deletion in tropomyosin. J. Biol. Chem. 272, 14051–14056 10.1074/jbc.272.22.14051 [DOI] [PubMed] [Google Scholar]
- 43. Korman V. L., and Tobacman L. S. (1999) Mutations in actin subdomain 3 that impair thin filament regulation by troponin and tropomyosin. J. Biol. Chem. 274, 22191–22196 10.1074/jbc.274.32.22191 [DOI] [PubMed] [Google Scholar]
- 44. Butters C. A., Willadsen K. A., and Tobacman L. S. (1993) Cooperative interactions between adjacent troponin–tropomyosin complexes may be transmitted through the actin filament. J. Biol. Chem. 268, 15565–15570 [PubMed] [Google Scholar]
- 45. Rome L. C., Cook C., Syme D. A., Connaughton M. A., Ashley-Ross M., Klimov A., Tikunov B., and Goldman Y. E. (1999) Trading force for speed: why superfast crossbridge kinetics leads to superlow forces. Proc. Natl. Acad. Sci. U.S.A. 96, 5826–5831 10.1073/pnas.96.10.5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Månsson A., Rassier D., and Tsiavaliaris G. (2015) Poorly understood aspects of striated muscle contraction. Biomed. Res. Int. 2015, 245154 10.1155/2015/245154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gerson J. H., Bobkova E., Homsher E., and Reisler E. (1999) Role of residues 311/312 in actin–tropomyosin interaction in vitro motility study using yeast actin mutant E311A/R312A. J. Biol. Chem. 274, 17545–17550 10.1074/jbc.274.25.17545 [DOI] [PubMed] [Google Scholar]
- 48. VanBuren P., Palmiter K. A., and Warshaw D. M. (1999) Tropomyosin directly modulates actomyosin mechanical performance at the level of a single actin filament. Proc. Natl. Acad. Sci. U.S.A. 96, 12488–12493 10.1073/pnas.96.22.12488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Viswanathan M. C., Schmidt W., Franz P., Rynkiewicz M. J., Newhard C. S., Madan A., Lehman W., Swank D. M., Preller M., and Cammarato A. (2020) A role for actin flexibility in thin filament-mediated contractile regulation and myopathy. Nat. Commun. 11, 2417 10.1038/s41467-020-15922-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Barua B., Winkelmann D. A., White H. D., and Hitchcock-DeGregori S. E. (2012) Regulation of actin–myosin interaction by conserved periodic sites of tropomyosin. Proc. Natl. Acad. Sci. U.S.A. 109, 18425–18430 10.1073/pnas.1212754109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Beuckelmann D. J., Näbauer M., and Erdmann E. (1992) Intracellular calcium handling in isolated ventricular myocytes from patients with terminal heart failure. Circulation 85, 1046–1055 10.1161/01.cir.85.3.1046 [DOI] [PubMed] [Google Scholar]
- 52. Piacentino V. 3rd, Weber C. R., Chen X., Weisser-Thomas J., Margulies K. B., Bers D. M., and Houser S. R. (2003) Cellular basis of abnormal calcium transients of failing human ventricular myocytes. Circ. Res. 92, 651–658 10.1161/01.RES.0000062469.83985.9B [DOI] [PubMed] [Google Scholar]
- 53. Smith G. L., and Eisner D. A. (2019) Calcium buffering in the heart in health and disease. Circulation 139, 2358–2371 10.1161/CIRCULATIONAHA.118.039329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Razzaq A., Schmitz S., Veigel C., Molloy J. E., Geeves M. A., and Sparrow J. C. (1999) Actin residue Glu93 is identified as an amino acid affecting myosin binding. J. Biol. Chem. 274, 28321–28328 10.1074/jbc.274.40.28321 [DOI] [PubMed] [Google Scholar]
- 55. Bing W., Razzaq A., Sparrow J., and Marston S. (1998) Tropomyosin and troponin regulation of wild type and E93K mutant actin filaments from Drosophila flight muscle: charge reversal on actin changes actin-tropomyosin from on to off state. J. Biol. Chem. 273, 15016–15021 10.1074/jbc.273.24.15016 [DOI] [PubMed] [Google Scholar]
- 56. Li X. E., Lehman W., Fischer S., and Holmes K. C. (2010) Curvature variation along the tropomyosin molecule. J. Struct. Biol. 170, 307–312 10.1016/j.jsb.2009.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li X. E., Holmes K. C., Lehman W., Jung H., and Fischer S. (2010) The shape and flexibility of tropomyosin coiled coils: implications for actin filament assembly and regulation. J. Mol. Biol. 395, 327–339 10.1016/j.jmb.2009.10.060 [DOI] [PubMed] [Google Scholar]
- 58. Berchtold M. W., Brinkmeier H., and Müntener M. (2000) Calcium ion in skeletal muscle: its crucial role for muscle function, plasticity, and disease. Physiol. Rev. 80, 1215–1265 10.1152/physrev.2000.80.3.1215 [DOI] [PubMed] [Google Scholar]
- 59. Kuo I. Y., and Ehrlich B. E. (2015) Signaling in muscle contraction. Cold Spring Harb. Perspect. Biol. 7, a006023 10.1101/cshperspect.a006023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chung J. H., Biesiadecki B. J., Ziolo M. T., Davis J. P., and Janssen P. M. (2016) Myofilament calcium sensitivity: role in regulation of in vivo cardiac contraction and relaxation. Front. Physiol. 7, 562 10.3389/fphys.2016.00562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Samant S. A., Pillai V. B., Sundaresan N. R., Shroff S. G., and Gupta M. P. (2015) Histone deacetylase 3 (HDAC3)–dependent reversible lysine acetylation of cardiac myosin heavy chain isoforms modulates their enzymatic and motor activity. J. Biol. Chem. 290, 15559–15569 10.1074/jbc.M115.653048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lin Y. H., Schmidt W., Fritz K. S., Jeong M. Y., Cammarato A., Foster D. B., Biesiadecki B. J., McKinsey T. A., and Woulfe K. C. (2020) Site-specific acetyl-mimetic modification of cardiac troponin I modulates myofilament relaxation and calcium sensitivity. J. Mol. Cell. Cardiol. 139, 135–147 10.1016/j.yjmcc.2020.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Foster D. B., Liu T., Kammers K., O'Meally R., Yang N., Papanicolaou K. N., Talbot C. C. Jr., Cole R. N., and O'Rourke B. (2016) Integrated omic analysis of a guinea pig model of heart failure and sudden cardiac death. J. Proteome Res. 15, 3009–3028 10.1021/acs.jproteome.6b00149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bedi K. C. Jr., Snyder N. W., Brandimarto J., Aziz M., Mesaros C., Worth A. J., Wang L. L., Javaheri A., Blair I. A., Margulies K. B., and Rame J. E. (2016) Evidence for intramyocardial disruption of lipid metabolism and increased myocardial ketone utilization in advanced human heart failure. Circulation 133, 706–716 10.1161/CIRCULATIONAHA.115.017545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tong L., and Harwood H. J. Jr. (2006) Acetyl-coenzyme A carboxylases: versatile targets for drug discovery. J. Cell. Biochem. 99, 1476–1488 10.1002/jcb.21077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Svensson R. U., Parker S. J., Eichner L. J., Kolar M. J., Wallace M., Brun S. N., Lombardo P. S., Van Nostrand J. L., Hutchins A., Vera L., Gerken L., Greenwood J., Bhat S., Harriman G., Westlin W. F., et al. (2016) Inhibition of acetyl-CoA carboxylase suppresses fatty acid synthesis and tumor growth of non-small-cell lung cancer in preclinical models. Nat. Med. 22, 1108–1119 10.1038/nm.4181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kim C. W., Addy C., Kusunoki J., Anderson N. N., Deja S., Fu X., Burgess S. C., Li C., Ruddy M., Chakravarthy M., Previs S., Milstein S., Fitzgerald K., Kelley D. E., and Horton J. D. (2017) Acetyl CoA carboxylase inhibition reduces hepatic steatosis but elevates plasma triglycerides in mice and humans: a bedside to bench investigation. Cell Metab. 26, 576 10.1016/j.cmet.2017.08.011 [DOI] [PubMed] [Google Scholar]
- 68. Lawitz E. J., Coste A., Poordad F., Alkhouri N., Loo N., McColgan B. J., Tarrant J. M., Nguyen T., Han L., Chung C., Ray A. S., McHutchison J. G., Subramanian G. M., Myers R. P., Middleton M. S., et al. (2018) Acetyl-CoA carboxylase inhibitor GS-0976 for 12 weeks reduces hepatic de novo lipogenesis and steatosis in patients with nonalcoholic steatohepatitis. Clin. Gastroenterol. Hepatol. 16, 1983–1991.e3 10.1016/j.cgh.2018.04.042 [DOI] [PubMed] [Google Scholar]
- 69. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 70. Kron S. J., and Spudich J. A. (1986) Fluorescent actin filaments move on myosin fixed to a glass surface. Proc. Natl. Acad. Sci. U.S.A. 83, 6272–6276 10.1073/pnas.83.17.6272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Beausoleil S. A., Villén J., Gerber S. A., Rush J., and Gygi S. P. (2006) A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24, 1285–1292 10.1038/nbt1240 [DOI] [PubMed] [Google Scholar]
- 72. Nussbaum-Krammer C. I., Neto M. F., Brielmann R. M., Pedersen J. S., and Morimoto R. I. (2015) Investigating the spreading and toxicity of prion-like proteins using the metazoan model organism C. elegans. J. Vis. Exp. 95, 52321 10.3791/52321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., Inuganti A., Griss J., Mayer G., Eisenacher M., Perez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S., et al. (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47, D442–D450 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Groth A. C., Fish M., Nusse R., and Calos M. P. (2004) Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166, 1775–1782 10.1534/genetics.166.4.1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Brand A. H., and Perrimon N. (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data, associated protocols, methods, and sources of materials are available in the main text or in the supporting information. Additionally, the MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (73) partner repository with the data set identifier PXD020732 and PXD019276.