Abstract
Clear-cell renal cell carcinoma (ccRCC), the most common subtype of renal cancer, has a poor clinical outcome. A hallmark of ccRCC is genetic loss-of-function of VHL (von Hippel–Lindau) that leads to a highly vascularized tumor microenvironment. Although many ccRCC patients initially respond to antiangiogenic therapies, virtually all develop progressive, drug-refractory disease. Given the role of dysregulated expressions of cytoskeletal and cytoskeleton-regulatory proteins in tumor progression, we performed analyses of The Cancer Genome Atlas (TCGA) transcriptome data for different classes of actin-binding proteins to demonstrate that increased mRNA expression of profilin1 (Pfn1), Arp3, cofilin1, Ena/VASP, and CapZ, is an indicator of poor prognosis in ccRCC. Focusing further on Pfn1, we performed immunohistochemistry-based classification of Pfn1 staining in tissue microarrays, which indicated Pfn1 positivity in both tumor and stromal cells; however, the vast majority of ccRCC tumors tend to be Pfn1-positive selectively in stromal cells only. This finding is further supported by evidence for dramatic transcriptional up-regulation of Pfn1 in tumor-associated vascular endothelial cells in the clinical specimens of ccRCC. In vitro studies support the importance of Pfn1 in proliferation and migration of RCC cells and in soluble Pfn1's involvement in vascular endothelial cell tumor cell cross-talk. Furthermore, proof-of-concept studies demonstrate that treatment with a novel computationally designed Pfn1–actin interaction inhibitor identified herein reduces proliferation and migration of RCC cells in vitro and RCC tumor growth in vivo. Based on these findings, we propose a potentiating role for Pfn1 in promoting tumor cell aggressiveness in the setting of ccRCC.
Keywords: Profilin, renal cancer, actin-binding proteins, Mena, VASP, tumor-associated vascular endothelial cells, small molecule inhibitor, cancer biology, actin, profilin, tumor, endothelial cell, clear-cell renal cell carcinoma
The estimated incidence of and number of deaths from renal cell carcinoma (RCC) in the United States in 2019 are 73,280 and 14,770, respectively (1). The most common subtype, clear-cell RCC (ccRCC), occurs in >75% of RCC patients. Approximately 20–30% of those patients present with metastasis at the time of diagnosis. Another one-third of patients, following initial treatment, develop either local recurrence and/or distant metastases. Disquietingly, the 5-year survival of patients with advanced-stage ccRCC remains only 10% (2–4). A distinguishing hallmark of ccRCC is its highly vascularized tumor microenvironment (TME) arising from the genetic loss of function (LOF) of the tumor-suppressor protein VHL (von Hippel–Lindau; inactivated in >90% of cases of sporadic ccRCC), leading to hypoxia-inducible factor-1 and -2 stabilization and up-regulation of the proangiogenic factor VEGF (4). Although many ccRCC patients initially respond to therapies targeting VEGF or other proangiogenic signaling pathways, very few patients exhibit durable treatment-associated benefits, and virtually all develop progressive, drug-refractory disease (5–7). Deeper molecular understanding of the pathogenesis and progression of ccRCC and its sequelae is expected to lead to the development of novel targeted interventions capable of providing improved clinical benefit.
Dynamic control of the actin cytoskeleton is a key feature of all actin-dependent biological processes that includes cell migration and proliferation in both physiological and pathological contexts. Actin cytoskeletal regulation in cells involves the concerted actions of several major classes of actin-binding proteins (ABPs). Among them, the Pfn (profilin) family of ABPs play a key role in promoting actin polymerization in cells through their nucleotide-exchange activity on G-actin (facilitates ADP-to-ATP exchange on G-actin) and ability to act as a carrier of ATP–G-actin to a number of other actin assembly factors bearing polyproline motifs during F-actin elongation. Pfn's importance in the regulation of actin dynamics and actin-based cellular processes, such as cell migration and proliferation, is well-established in the literature (8–13). Differential proteomic analyses of supernatants from the cultures of RCC cells versus normal kidney epithelial cells supported by RT-PCR and qualitative immunohistochemistry (IHC) analyses of a small number of RCC (14 cases) versus normal kidney tissues provided the first evidence of overexpression of Pfn1 (the major isoform of Pfn family of ABPs) in human ccRCC (14). Proteomic studies further identified Pfn1 to be one of the candidate markers of late stage ccRCC (15, 16). In concordance with these findings, semiquantitative IHC studies performed on a larger cohort of patient samples (384 cases) subsequently established higher Pfn1 expression correlated with shorter overall survival (OS) and progression-free survival (PFS) of ccRCC patients (17). This particular study reported no statistically significant difference in % Pfn1low versus %Pfn1high expression between tumor and matched noncancerous regions of kidney in ccRCC patients. However, because those stratifications were based on percentages of Pfn1-immunoreactive regions on a semiquantitative evaluation scale, an accurate assessment of Pfn1 expression in different cell compartments in tumor versus normal adjacent regions of kidney in ccRCC patients is lacking in the literature. Overall, these studies suggest Pfn1's positive association with advanced disease features and adverse clinical outcomes in the setting of ccRCC. In this study, we first analyzed publicly available transcriptome data sets to identify other ABPs in addition to Pfn1 that have prognostic significance in ccRCC. We further demonstrate dysregulated expression of Pfn1 and a subset of those ABPs queried herein in tumor-associated (TA)–VECs in ccRCC patients. Additional experimental evidence developed in our studies support the role of Pfn1 in promoting proliferation and migration of RCC cells, and VEC-tumor cell cross-talk. Finally, we demonstrate proof-of-concept for a novel Pfn1–actin interaction inhibitor identified herein to reduce RCC cell aggressiveness in vitro and tumor growth in vivo.
Results
Pfn1, CapZ, Cofilin1, Arp3, and ena/VASP are markers of poor prognosis in ccRCC
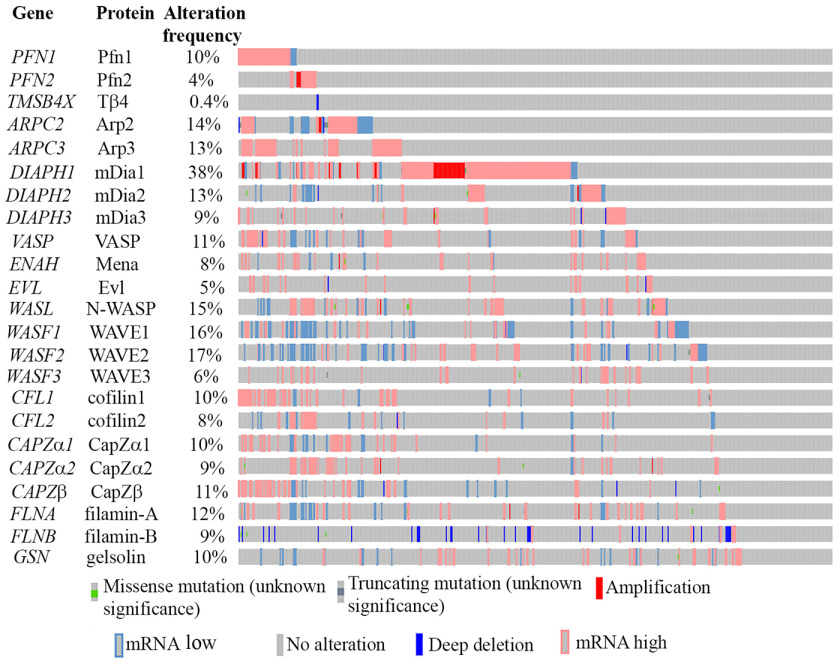
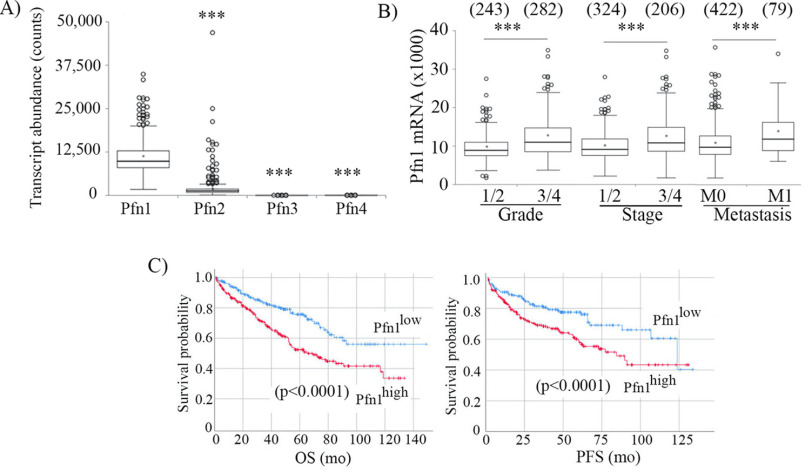
We first queried a The Cancer Genome Atlas (TCGA) transcriptome data set (downloaded from CBioportal) representing all 537 clinical cases of ccRCC to examine alteration characteristics (overexpression, gene amplification, down-regulation, deep-deletion, missense, and/or truncating mutations) of a total of eight major classes of actin cytoskeleton regulatory factors (Fig. 1). We included ABPs that are involved in (i) actin nucleotide exchange and shuttling (Pfn), (ii) G-actin sequestration (thymosin β4; maintains the cellular pool of polymerization-competent ATP-bound G-actin), (iii) actin nucleation (Arp2/3 complex, mammalian diaphanous (mDia)), (iv) activation of actin-nucleating factors (N-WASP (neural Wiskott–Aldrich syndrome protein), WAVE (WASP-associated verprolin homology)), (v) F-actin elongation (mDia and Ena (enabled)/VASP (vasodilator-stimulated phosphoprotein)), (vi) F-actin depolymerization and/or severing (cofilin and gelsolin), (vii) F-actin capping (CapZ), and (viii) F-actin cross-linking (filamin). As for Pfn, we restricted our analyses to Pfn1 and Pfn2 isoforms only because PFN3 transcript was undetectable, and the average transcript abundance of PFN4 (5.98 ± 5.45) was essentially negligible compared with that of either PFN1 (10958.0 ± 4594.6) or PFN2 (1821.1 ± 2996.3, expressed at a 6-fold lower abundance than Pfn1) in ccRCC tumors (Fig. 2A). Genes that are predominantly up-regulated (either through mRNA overexpression or gene amplification) include PFN isoforms (PFN1 and PFN2), ARPC3 (encodes Arp3 subunit of the Arp2/3 complex), DIAPH1 (encodes mDia1; exhibits the highest alteration frequency at 38%), DIAPH3 (encodes mDia3), ENAH (encodes Mena), VASP, EVL, CFL1 (encodes cofilin1), WASL (encodes N-WASP), and various CAPZ subunits. The most consistent down-regulated gene was FLNB (encodes Filamin-B), which involved deep deletion. For the remainder of the queried genes, no significant bias in any particular direction of change was evident. A subset of queried ABPs demonstrated association with disease features and/or clinical outcome in ccRCC (summarized in Table S1). Tumors of advanced Fuhrman grade (3 or 4) and stage (3 or 4) and associated with distant metastasis (M1) exhibited higher values of mRNA expression of PFN1 (but not the minor isoform PFN2), ARPC3, CFL1, CAPZ subunits (CAPZα1 and CAPZβ), and VASP relative to lower grade/stage (1 or 2) and nonmetastatic (M0) tumors, and these differences were statistically significant. Accordingly, higher expressions (more than the median value) of these genes also correlated with lower OS, as well as PFS of ccRCC patients. ENAH and EVL (the other two members of Ena/VASP protein family genes) also exhibited similar trends as seen for VASP with the only exceptions that ENAH expression was not different between M0 and M1 tumors and that high EVL expression only negatively impacted OS in a statistically significant manner (note however that EVL's association with PFS was very close to significance with p value equal to 0.056). Collectively, these data suggest that elevated expressions of PFN1, CFL1, CAPZ, ARPC3, and ENAH/VASP genes are poor prognostic features of ccRCC (disease association and survival plots pertinent to PFN1 are shown in Fig. 2 (B and C); disease association and survival plots for CFL1, CAPZ, ARPC3, and ENAH/VASP genes are shown in Figs. S1 and S2, respectively). We also found that expressions of a subset of ABP-encoding genes represent indicators of better prognosis in ccRCC (Table S1). Specifically, higher expressions of WASL (encodes N-WASP), WASF2 (encodes WAVE2), and GSN (encodes gelsolin) appeared to represent signatures of low-grade/stage and nonmetastatic tumors in association with improved survival (OS and/or PFS) in ccRCC patients. Furthermore, although the mDia1-encoding DIAPH1 gene is frequently overexpressed/amplified in ccRCC, higher expression of DIAPH1 also correlated with improved patient survival. Interestingly, there was no significant trend in DIAPH1 gene expression versus the clinicopathological features of tumors (survival curves for these four genes are shown in Fig. S3).
Figure 1.
A graphical summary of alteration of different classes of ABPs assembled from the transcriptome data of 537 ccRCC patients (TCGA data set plotted on cBioportal). Each vertical line represents a clinical case. For these analyses, an absolute value of mRNA expression z score of >1.5 was used as a cutoff for defining overexpression and down-regulation with the mean and variance calculated based on the expression distribution of tumors diploid for any given gene.
Figure 2.
Adverse association of Pfn1 with patient outcome in human ccRCC. A, relative transcript abundance of different variants of Pfn in ccRCC. ***, p < 0.0001. B and C, association of transcript expressions of Pfn1 with disease features (grade, stage, metastasis) (B) and survival (OS and PFS) (C) of ccRCC patients (these data are based on the analyses of TCGA data representing 537 clinical cases). ***, p < 0.0001. Samples with missing stage, grade, and metastasis information were excluded (numbers in parentheses in B denote number of clinical cases in each category). For survival analyses (available for all 537 patients), transcript expression was dichotomized at the median value. Red and blue lines in Kaplan–Meier survival plots indicate higher and lower than median expression, respectively.
Given that PFN1, ARPC3, ENAH/VASP, CFL1, and CAPZ represented poor prognostic indicators in the ccRCC setting, we further analyzed the TCGA transcriptome data set to examine the expression of these select ABP-encoding genes with clinical outcome in chromophobe (n = 63 patients) and papillary cell (n = 292 patients) histological subtypes of RCC. With the exception of ENAH, for which higher expression is correlated with shorter OS (p = 0.009) and PFS (p = 0.009), none of the other ABPs demonstrated any association with patient outcome in the setting of chromophobe RCC. However, we cannot dismiss the possibility that lack of clinical association of at least some of these ABP-encoding genes could be due to the small number of patient samples available for this subtype of RCC in the TCGA database. In the case of papillary cell RCC, higher expression of only ENAH and CAPZα1 genes correlated with statistically significant shorter OS (p values equal to 0.025 and 0.019 for ENAH and CAPZα1, respectively), whereas higher expression of only EVL and ARPC3 genes was associated with diminished PFS (p values equal to 0.04 and 0.05, respectively). Therefore, adverse clinical outcome associated with elevated tumor expression of PFN1, VASP, and CFL1 genes appears to be restricted solely to the clear-cell histological subtype. We focused subsequent studies on Pfn1 because of concordance of our transcriptome-based findings with previous proteomic and semiquantitative IHC-based correlation of Pfn1 expression with advanced disease features and adverse patient outcome from other independent studies (15–17).
Pfn1 is preferentially overexpressed in TA-VECs in ccRCC
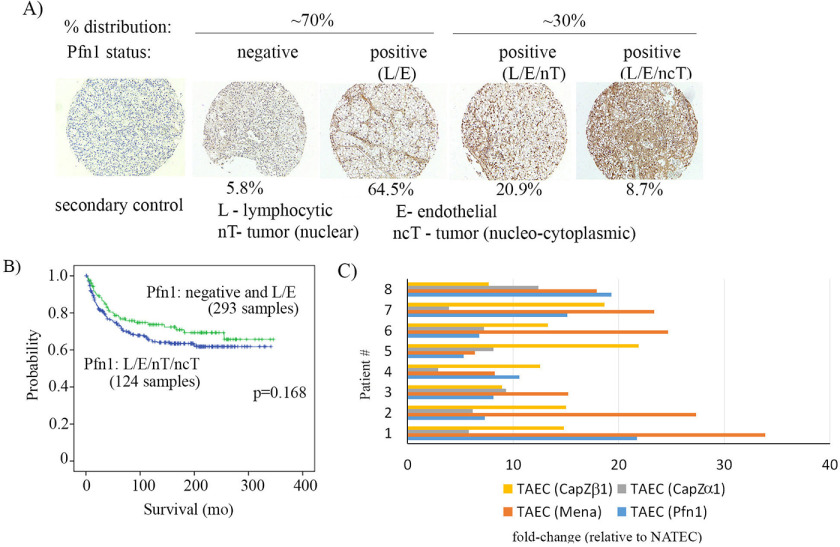
To determine which cells in the ccRCC TME preferentially overexpressed Pfn1, we next performed a qualitative IHC assessment of tissue microarrays (TMAs) containing a large cohort (n = 417) of ccRCC tumors (Fig. 3A), using a Pfn1 antibody that we used for IHC in our previous studies (18, 19). By immunoblot analyses of cell lysates in Pfn-isoform–specific gene knockdown settings, we also previously confirmed that this antibody does not recognize Pfn2 (20). Furthermore, human tumor sections (kidney or ovarian) incubated with secondary antibody alone (i.e. without the presence of Pfn1 antibody) did not result in either chromagen- or fluorescence-based IHC signal, further establishing the specificity of our IHC staining (Fig. S4). Pfn1 IHC of ccRCC TMA revealed three distinct patterns of Pfn1 expression: (i) negligible Pfn1 expression throughout the tumor (5.8% frequency), (ii) strong Pfn1 expression in stromal cells (VEC, lymphocytes) but negligible expression in tumor cells (64.5% frequency), and (iii) strong Pfn1 expression in both stromal and tumor cells (29.7% frequency). In the latter sample cohort, 71% of specimens exhibited a nuclear distribution of Pfn1 expression in tumor cells, with the remaining 29% of specimens characterized by nucleocytoplasmic staining of tumor cells. These findings suggest that the vast majority of ccRCC tumors display a pattern of strong Pfn1 expression selectively in stromal cells within the TME; this is in accord with the general assessment of Minamida et al. (14) in their qualitative evaluation of a limited number of ccRCC samples. Notably, patients with tumors graded for coordinate Pfn1 positivity in all three cellular compartments (VECs, lymphocytes, and tumor cells) tended to have reduced OS when compared with patients with tumors in which only stromal cells were graded as Pfn1-positive (p = 0.168; Fig. 3B). Although difference in the survival characteristics between the two cellular patterns of Pfn1 expressions was not statistically significant, we speculate that may be due to limitations in qualitative characterization of Pfn1 expression in our IHC studies of tumor specimens.
Figure 3.
Expression of Pfn1 in cells within the TME of human ccRCC. A, Pfn1 IHC of ccRCC TMAs showing percentages of frequency of tumors with different expression patterns of Pfn1 (n = 417 tumors) (blue, nuclei). B, Kaplan–Meier survival curves of ccRCC patients with the indicated expression characteristics of Pfn1. C, transcript expression profiling of Pfn1, Mena, and CapZ isoforms in CD34+CD45negCD146+ VEC flow-sorted from the digests of fresh renal tumors and patient-matched normal adjacent tissues (n = 8 patients).
Given the prevalence of a stromal cell–associated signature for Pfn1 overexpression in ccRCC, we next performed gene expression profiling of flow-sorted CD34+CD45negCD146+ VEC isolated from freshly isolated enzymatically digested, ccRCC tumors, which revealed a robust (5.3–21.7-fold) up-regulation in Pfn1 expression in tumor-associated TA-VEC relative to VEC sorted from patient-matched normal adjacent tissue (NAT-VEC; Fig. 3C). Among the other poor prognostic cytoskeletal markers, only Mena, CapZα1, and CapZβ were also determined to be strongly up-regulated in tumor-associated VEC (TA-VEC) versus normal adjacent tissue VEC (NAT-VEC) (Fig. 3C). In contrast, a parallel analysis of flow-sorted CD34negCD45negCD146+ vascular pericytes isolated from tumor versus NAT samples showed no significant differences in the expression of these proteins (data not shown). Because Pfn1 interacts with Mena/VASP proteins and is a major promoter of Mena/VASP-mediated actin assembly and actin-driven cellular processes (21), transcriptional dysregulation of the Pfn1:Mena/VASP cytoskeletal pathway in TA-VEC could represent a bio-signature of poor prognosis in ccRCC.
A previous study correlated elevated serum levels of Pfn1 (sPfn1) with the degree of atherosclerosis in humans (22), suggesting that sPfn1 has potential to serve as a cytoskeletal biomarker in certain disease contexts. We performed exploratory ELISA to compare sPfn1 levels between small cohorts of stage 4/M1 ccRCC patients (n = 29; 8 female, 21 male; median age, 59 years; range, 45–67 years) and age-matched normal human donors (n = 11; 5 male, 6 female; median age, 60 years; range, 48–76 years) as control. The ccRCC patients had completed treatment with high-dose interleukin-2 combined with the autophagy inhibitor hydroxychloroquine. Pfn1 was detectable in both normal donors and ccRCC patients in the ng/ml range. Despite the small sample sizes evaluated, the mean sPfn1 level in ccRCC patients at baseline/pretreatment (29.6 ± 15.3 ng/ml) was ∼2-fold higher (p = 0.0003) versus normal donors (15.83 ± 7.62 ng/ml; Fig. S5). Although the significant overlap of sPfn1 level between normal and patient cohorts precludes us from any definitive conclusion at this point, our preliminary indication of a trend of elevated sPfn1 level in ccRCC patients justifies future studies involving a larger cohort of subjects to examine the potential utility of sPfn1 as a predictive biomarker in ccRCC.
Pfn1 is an important regulator of RCC tumor cell proliferation and migration
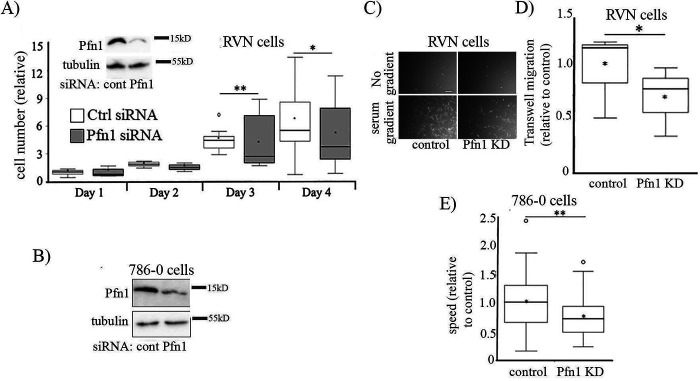
Next, to determine whether Pfn1 has any role in proliferation of ccRCC cells, we first studied the effect of transient silencing Pfn1 expression on proliferation of RVN (a variant of the widely used murine RENCA RCC cell line engineered for VHL gene deletion by CRISPR/Cas9) and 786-0 (a VHL-negative human cell line originally derived from ccRCC clinical specimen) cell lines, respectively, in 2D culture. Note that although the parent RENCA (VHLWT) cell line has been considered poorly reflective of human ccRCC, RVN (VHL−/−) cells overexpress many of the genes associated with aggressive ccRCC in humans and appear to represent an appropriate surrogate of human disease (23, 24). Proliferation of both VHL−/− cell lines was reduced when Pfn1 expression was suppressed (Fig. 4, A and B). Pfn1's interactions with actin and other actin assembly factors (such as Mena/VASP proteins) play important roles in actin polymerization at the leading edge and membrane protrusion (a key step of cell migration) (21, 25). In most physiological contexts, LOF of Pfn1 leads to defects in membrane protrusion and cell migration (8–11). However, in certain cancer cells (breast, hepatic), Pfn1 depletion induces a hypermigratory phenotype (26–30), suggesting that Pfn1's role in cell migration is highly context-specific. To determine whether Pfn1 has a pro- or antimigratory effect in RCC cells, we also studied the effect of silencing Pfn1 expression on chemotactic and random single-cell migration of RVN and 786-0 cells, respectively. Similar to our results with regard to proliferation, Pfn1 depletion attenuated chemotactic and random migration of RVN (Fig. 4, C and D) and 786-0 (Fig. 4E) cells by ∼30 and ∼25%, respectively. Although chemotactic migration of RVN cells was scored 20 h after cell seeding, because our cell proliferation assay did not show any significant difference in cell count between the control and Pfn1 knockdown groups within 24 h after cell seeding (Fig. 4A), reduced Transwell migration in Pfn1 knockdown setting is not due to secondary effect on cell proliferation (this is further underscored by single-cell motility data of 786-0 cells). Collectively, these findings support a Pfn1 dependence for RCC cell migration and proliferation.
Figure 4.
Pfn1 promotes migration and proliferation of RCC cells. Effect of Pfn1 knockdown (KD) on proliferation (A and B) and migration (C–E) of VHL-negative murine RCC (RVN) and human RCC (786-0) cells. Immunoblot insets in A and B demonstrate Pfn1 knockdown (tubulin blot serves as the loading control). Proliferation data are normalized to cell count of control group of cells on day 1. C and D represent Transwell migration data of RVN cells (scale bar in C, 200 μm). E summarizes single-cell random motility data of 786-0 cells. Single-cell migration was quantified using cells transfected with either smart-pool control siRNA (n = 83 cells) or Pfn1 siRNA (n = 75 cells). Triplicate wells were quantified from three individual experiments. *, p < 0.05; **, p < 0.01. Ctrl or cont, control.
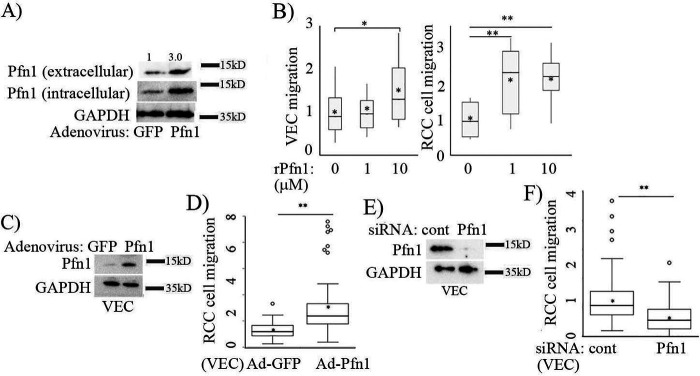
Bidirectional communication between VEC and cancer cells through various cell surface receptors and soluble factors impacts the migratory behavior of cancer cells. Although Pfn1 is mainly an intracellular protein, the presence of Pfn1 in the conditioned media of various types of cultured cells (VECs, glomerular mesangial cells, RCCs, and breast cancer cells) has been documented previously by us and other groups (14, 20, 31). We have also shown that extracellular release of Pfn1 is sensitive to perturbations of actin-sensitive signaling pathways, suggesting that Pfn1 release from cells is an actively regulated process (20). Given our observation of Pfn1 overexpression in TA-VEC, we next analyzed the effect of intracellular elevation of Pfn1 on its extracellular release based on immunoblot analyses of Pfn1 in conditioned media isolated from Pfn1-overexpressing versus control VEC. We observed a dramatic increase in extracellular release of Pfn1 from VEC following transduction with an adenovirus-encoding Pfn1 (Ad-Pfn1; with Ad-GFP transduced cells serving as a control; Fig. 5A). The fold change in the basal extracellular release of Pfn1 (∼3-fold) paralleled the level of gross protein overexpression (∼4.7-fold) based on cell lysate analyses. To determine whether soluble Pfn1 modulates cell migration, we next analyzed the effect of adding recombinant Pfn1 (rPfn1; 1 or 10 μm) to cultures of VEC or RCC cells. Although a concentration range of 1–10 μm is several orders of magnitude higher than our estimated average serum concentration of sPfn1 in ccRCC patients (∼30 ng/ml; this is equivalent to ∼2 nm), we speculate that the basal local concentration of extracellular Pfn1 in a tissue environment is likely much higher than that of sPfn1 in the serum, and in the µM range based on the following reasons. First, we were able to detect extracellular Pfn1 by immunoblot analyses (detection sensitivity is in the ng range) of a few tens of microliters of supernatant in cell culture settings. Second, the local concentration of extracellular Pfn1 can be further augmented by entrapment of proteins by ECM and/or or in the setting of elevated intracellular Pfn1 level as occurs in ccRCC (this is further supported by our data in overexpression setting). We found rPfn1 stimulated the migratory ability of both VEC and RCC cells, although RCC cells exhibited greater responsiveness versus VEC, because 1 μm rPfn1 was sufficient to increase RVN motility by >2-fold, whereas VEC required 10 μm rPfn1 to increase motility by even ∼1.5-fold (Fig. 5B). Furthermore, in a Transwell culture system, RVN cell motility was found to be >2.5-fold greater when allowed to migrate toward conditioned media harvested from the culture of Pfn1-overexpressing VEC versus the same from control VEC (Fig. 5, C and D). Conversely, Transwell migration of RVN cells toward VEC-conditioned media was reduced by ∼50% when Pfn1 expression was silenced in VEC (Fig. 5, E and F). Collectively, these findings suggest that Pfn1 may serve as a paracrine VEC-secreted extracellular regulator of RCC cell migration.
Figure 5.
Effect of modulating Pfn1 expression in VEC on RCC cell migration. A, Pfn1 and GAPDH (loading control) immunoblots showing the effect of adenoviral-mediated Pfn1 overexpression in VEC on extracellular release of Pfn1 as measured in the conditioned media (Ad-GFP, control). B, box-and-whisker plots summarizing the effects of purified rPfn1 protein in culture media on the average speed of VEC and RVN cells in random motility assay (data normalized to control with buffer only; n = 40 cells/group from two independent experiments). C–F, box plots summarizing the effects of conditioned media of Pfn1 overexpressing (D) and knockdown (F) cultures of VEC on chemotactic migration of RVN cells (migration data normalized to control Ad-GFP transduction or siRNA transfection condition). Pfn1 and GAPDH (loading control) immunoblots in C and E confirm Ad-Pfn1– and Pfn1-siRNA–mediated overexpression and knockdown of Pfn1, respectively (data summarized from three experiments). *, p < 0.05; **, p < 0.01.
Pfn1–actin interaction inhibitor reduces RCC cell aggressiveness
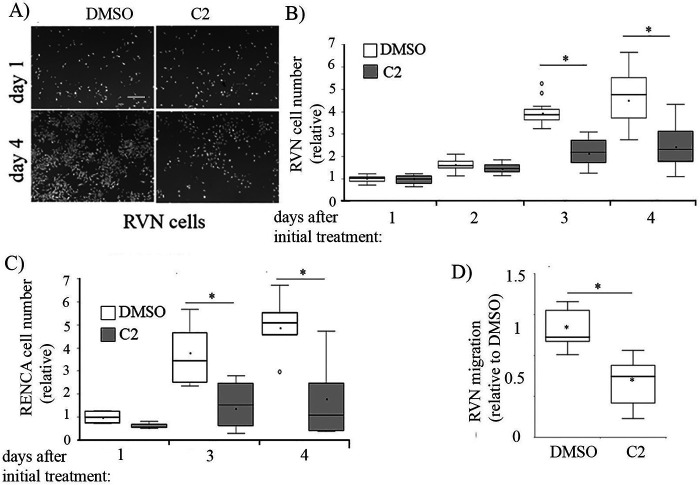
Using a computationally guided biochemical screen, we recently identified two structurally similar first-generation small molecule antagonists of the Pfn1–actin interaction (C1: 8-(3-hydroxyphenyl)-10-(4-methylphenyl)-2,4,5,6,7,11,12-heptaazatricyclo[7.4.0.0³,7]trideca-1(13),3,5,9,11-pentaen-13-ol], and C2: 8-(3-hydroxyphenyl)-10-phenyl-2,4,5,6,7,11,12-heptaazatricyclo[7.4.0.0³,7]trideca-1(13),3,5,9,11-pentaen-13-ol]) (32). Essentially, in a pyrene-based biochemical actin polymerization assay, these compounds reversed Pfn1's inhibitory effect on actin polymerization but did not affect actin polymerization on their own (i.e. in the absence of Pfn1). Note that unlike its role in promoting actin polymerization in cells, Pfn1 reduces actin polymerization in this biochemical assay because of its inhibition of actin nucleation and pointed-end growth of actin filaments. We further demonstrated that these compounds are noncytotoxic (at concentrations up to 100 μm) and elicit various phenotypes in normal cells including reduced levels of actin polymerization, slower migration, and reduced proliferation when applied in the concentration range of 50–100 μm, consistent with the loss of function of Pfn1. Whether tumor cell migration and proliferation are also susceptible to small molecule inhibition of the Pfn1–actin interaction had not previously been investigated. Therefore, we first tested the effect of compound C2 on the proliferation and migration of RCC cells. We observed that the proliferation of both VHL-positive RENCA and VHL-deficient RVN cells in 2D cultures was significantly reduced in the presence of 50 μm C2 treatment versus control DMSO-treated cells (Fig. 6, A and C). The antiproliferative effects of C2 appeared to be more prominent on RENCA than RVN cells, possibly because of the intrinsically higher proliferative capacity of RVN cells versus RENCA cells. Serum-induced chemotactic migration of RVN cells was also dramatically reduced (∼60%) following treatment with C2 (Fig. 6D). Because in proliferation assay C2 treatment did not affect cell count within 24 h of cell seeding, a 60% reduction in Transwell migration in response to C2 treatment is not due to any secondary effect on cell proliferation. Interestingly, C2-induced inhibition of RVN cell migration was more robust than that achieved in a Pfn1 knockdown setting. A potential reason underlying this observation is discussed later.
Figure 6.
Effect of small molecule inhibitor of Pfn1–actin interaction C2 on RCC cell proliferation and migration. A–C, box plots summarizing the effect of treatment with 50 µm C2 versus DMSO (control) on proliferation of VHL-negative RVN cells (B; A shows representative images of DAPI-stained cultures on indicated days with scale bar indicating 200 μm), and VHL-positive RENCA cells (C). C2 was added either on the day of seeding (D0, or RVN cells) or 1 day after seeding (D1, for RENCA cells) and was replenished either daily (for RVN cells) or every alternate day (for RENCA cells) during the culture period. Proliferation data are normalized to cell count of the control (DMSO-treated) group of cells. D, box plot summarizing the effect of C2 versus DMSO treatment on serum-induced chemotactic migration of RVN cells (results of three independent experiments). *, p < 0.05.
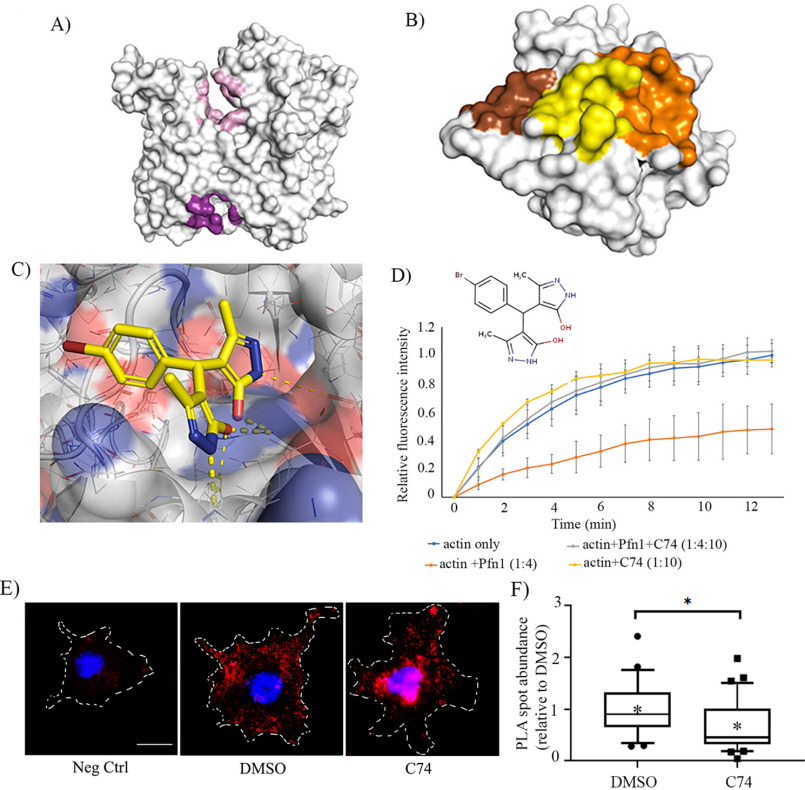
Encouraged by these results, we set up a follow-on screen based on the structure of C2 with the goal of identifying novel inhibitors of Pfn1–actin interaction with improved potency. This screen was purposely designed to not be constrained by the putative C2-binding site. Instead, whole protein docking of C2 on both actin and Pfn1 was performed to generate pharmacophore models and screen for new compounds (Fig. 7, A–C). A total of 67 small molecules were biochemically screened that identified C74 (4-[(4-bromophenyl)(5-hydroxy-3-methyl-1H-pyrazol-4-yl)methyl]-3-methyl-1H-pyrazol-5-ol) as another compound that was structurally distinct from C2 and was able to reverse Pfn1's effect on actin polymerization in vitro but did not significantly alter actin polymerization on its own (Fig. 7D). Live-dead staining of RVN cells treated with different doses (0–50 μm) of C74 revealed ∼100% cell viability, suggesting that C74 is not cytotoxic at least up to 50 μm concentration (Fig. S6A); this was further confirmed by morphological assessment of both VEC and RVN cells (Fig. S6B). Similar to our previously reported finding for C2 (32), C74-treated cells often exhibited stellate morphology, a phenotype indicative of reduced spreading ability; this is consistent with Pfn1's general role in membrane protrusion.
Figure 7.
Identification of C74 as a novel inhibitor of Pfn1–actin interaction. A–C, computationally identified putative binding sites. A, the two binding sites identified by docking our initial hit compound (C2) to actin (actin ATP-binding site shown in pink, and actin–Pfn1 site1 shown in purple). B, the three binding sites identified through docking the initial hit at the Pfn1–actin binding interface. Pfn1–actin site 1 (yellow), site 2 (brown), and site 3 (orange) are shown. C, C74 shown in its highest scoring pose in actin–Pfn1 site 1. This was identified by optimizing the pose of the molecule using a convolutional neural network. Favorable polar contacts are shown via yellow dotted lines. The convolutional neural network exhibits a stronger preference for these interactions compared with the empirical Vina scoring function. D, pyrene–actin polymerization assay curves for the indicated experimental conditions recorded for 15 min after addition of the polymerization buffer. Each time point represents the means ± standard deviation values of the fluorescence intensity of polymerized pyrene–actin relative to the maximum fluorescence intensity for the actin alone condition (data are summarized from three experiments). The numbers in parentheses indicate relative concentrations of actin, recombinant GST–Pfn1, and C74. The actual concentrations of actin and Pfn1 were 10 and 40 μm, respectively. C74 was added at 100 μm (Pfn1:C74 = 1:2.5) concentration. E, representative fluorescence images of Pfn1–actin PLA spots (red) in VEC following overnight treatment of either DMSO (control) or 25 μm C74 (nuclei are counter-stained with DAPI). An image of the negative control (Neg Ctrl) PLA stain (where one of the primary antibodies is omitted) is shown alongside for comparison. Dashed lines indicate boundaries of the cells (scale bar, 10 μm). F, a box-and-whisker plot displaying the number of PLA spots in C74-treated cells relative to the mean value scored for the DMSO control (n = 25 cells/group pooled from two independent experiments). *, p < 0.05.
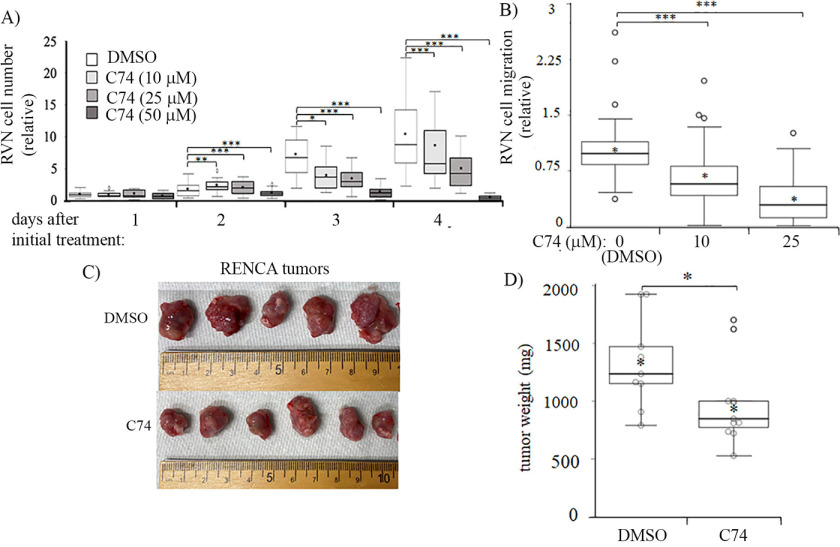
Next, as a proof-of-concept test for C74's ability to inhibit Pfn1–actin interaction in cells, we performed proximity ligation assay (PLA) in DMSO versus C74-treated VEC as we had previously done for C2 (32). Because PLA signal is extremely sensitive to the distance between the interacting molecules concerned, we reasoned that PLA assay is more efficient in capturing the direct Pfn1–actin complex (either exclusive or part of a small complex involving other binding partners) or at best small indirect complexes than in capturing those involving indirect interactions as a part of large protein complexes. Consistent with the pyrene–actin assay results, C74 treatment substantially reduced (by ∼50%) Pfn1–actin interaction in VEC (Fig. 7, E and F). Subsequent functional assays revealed that treatment with C74 reduced serum-induced chemotactic migration and proliferation of RVN cells in a dose-dependent manner; importantly, C74 was able to elicit these effects at a substantially lower range of concentrations (10–25 μm) than needed for C2 suggesting an improvement in efficacy (Fig. 8, A and B). Because the off-target effect is often a general concern for small molecules, we also evaluated the effect of C74 (or DMSO as control) on proliferation of RVN cells that were pretransfected with either control- or Pfn1-siRNA. Note that we previously showed that silencing Pfn1 does not alter the expression of Pfn2 and vice versa (20). We found that C74 treatment reduced the proliferation of control but not Pfn1 knockdown cells (Fig. S7), suggesting that the anti-proliferative effect of C74 is Pfn1-dependent.
Figure 8.
Demonstration of C74's ability to reduce RCC cell proliferation in vitro and tumor growth in vivo. A and B, box plots summarizing the results of indicated doses of C74 on proliferation (A) and serum-induced chemotactic migration of RVN cells (B). **, p < 0.01; ***, p < 0.001. For proliferation experiments, C74 was added on the day of cell seeding (D0) and replenished daily during the culture period. C and D, effect of in vivo administration of C74 versus DMSO on tumor growth (C, representative tumors; D, quantification) from subcutaneously implanted RENCA cells in BALB/c mice (data summarized from nine and ten DMSO- and C74-treated mice pooled from two independent experiments).
Lastly, to determine whether C74 has any effect on tumor growth in vivo, we performed an exploratory study in which we established subcutaneous tumors by transplantation of RENCA cells in syngeneic Balb/c mice and injected either C74 (dosage, 16 mg/kg) or equivalent volume of DMSO (vehicle) dissolved in saline directly at the tumor cell inoculation site daily over a course of 19 days starting from day 1. In pilot studies, we a priori confirmed that daily administration of C74 (at 16 mg/kg) for 14 days in BALB/c mice did not lead to weight loss (Fig. S8) or significant changes in serum levels of kidney/liver enzymes and waste products (Table S2), suggesting that C74 does not elicit any widespread toxicity in vivo (Fig. S8 and Table S2). Based on the tumor weight measurements, the average end-point tumor burden of C74-treated animals (978.8±366.0 mg; n = 10 animals) was found to be significantly lower than that of DMSO-treated animals (1327.5±397.5 mg; n = 9 animals) (Fig. 8, C and D). Collectively, these results demonstrate proof of concepts of the ability of small molecule inhibitor of Pfn1–actin interaction to diminish RCC cell aggressiveness in vitro and in vivo.
Discussion
Although previous studies correlated elevated Pfn1 expression to features of advanced-stage disease and adverse patient outcomes in the setting of ccRCC (15–17), whether and how Pfn1 dysregulation might promote ccRCC disease progression remains unknown. In this study, we report several novel findings. Specifically, we for the first time provide evidence for: (i) dramatic Pfn1 up-regulation in TA-VECs in human ccRCC, (ii) the ability of Pfn1 to modulate RCC tumor cells (either histologically validated human ccRCC cells or murine VHL-knockout RCC cells as a surrogate of the human ccRCC disease) migration through both intrinsic and extrinsic (i.e. paracrine) formats, and (iii) diminished aggressiveness of RCC cells in response to novel small molecule inhibitor of the Pfn1–actin interaction. Collectively, these novel findings suggest that Pfn1 may not only represent a cogent tissue biomarker of clinical prognosis in ccRCC patients but also indicate a potential causality between Pfn1 dysregulation and tumor progression. In addition to Pfn1, we also report several other important actin-regulatory proteins including Arp3, cofilin1, Mena, VASP, Evl, and CapZ as poor prognostic markers of ccRCC. Our findings related to Arp3 are consistent with defective directional migration and invasive growth of RCC cells in response to LOF of Brk1 (a component of WAVE/SCAR –Arp2/3-mediated actin nucleation pathway that impairs Arp2/3 localization to cellular protrusions when down-regulated) (33). Previous studies have also provided evidence for cofilin1 overexpression in RCC (34), cofilin1 serving as a late stage marker of ccRCC (16), increased apoptotic cell death in RCC cells after silencing of slingshot phosphatase (a positive regulator of cofilin) (35), and AGPTL3-induced inhibition of RCC cell migration via a VASP-linked pathway (36). When taken together, these findings, along with our TCGA data, suggest dysregulation of both actin assembly and disassembly (or limiting) pathways likely contributes to ccRCC progression.
Given our findings for the predominance of dysregulated Pfn1 expression in tumor-associated stromal cells rather than in tumor cells, it is conceivable that Pfn1 promotes tumor progression in ccRCC primarily through its paracrine action as a (VEC) secreted/elaborated product within the TME. At least two possible scenarios can be envisioned. First, we demonstrate in this study that intracellular Pfn1 elevation is accompanied by increased extracellular release of Pfn1 in VEC and stimulatory action of extracellular Pfn1 on cell (VEC and RCC) migration, resonating with previously reported findings in the context of proliferation of mesangial (31) and vascular smooth muscle cells (22). These data suggest that Pfn1 also has the ability to serves as an extracellular agent, irrespective of the cell type of origin. Although this aspect of Pfn1 function is highly intriguing and has not been explored previously, it raises a potential scenario for extracellular Pfn1 serving as a paracrine or autocrine signaling mediator in biological contexts, perhaps released when cells are stressed or damaged, and partly contributing to elevated sPfn1 in the serum of ccRCC patients. The elucidation of mechanism(s) underlying extracellular release of Pfn1 and whether extracellular Pfn1 acts through a cell surface receptor or via internalization into target cells to modulate biologic end points require further investigation.
Second, angiogenesis is a cardinal feature of ccRCC progression. VEGF-induced site-specific phosphorylation of Pfn1 in VEC has been previously linked to positive regulation of Pfn1–actin interaction and angiocrine factor production (through hypoxia-inducible factor stabilization), leading to stimulation of angiogenesis in the context of tissue-repair and glioblastoma progression (11, 37). Consistent with those findings, we previously demonstrated that Pfn1 dependence for VEC migration, proliferation, and angiogenesis (9, 10, 38). Therefore, it is conceivable that Pfn1 up-regulation in TA-VEC may promote tumor progression in ccRCC partly through stimulating tumor angiogenesis. Pfn1 cooperates with Mena/VASP proteins (critical regulators of actin cytoskeletal structure) to regulate actin polymerization at the leading edge of migrating cells to promote cell migration (21, 25, 39). VASP participates in filopodial protrusion and adhesion, driving tip cell migration during angiogenesis (40). Inactivating the signaling pathway that stimulates VASP function suppresses VEGF-dependent angiogenesis in vivo (41). Mena's role in angiogenesis has not yet been directly examined; endothelial cell (over)expression of Mena in colorectal and salivary gland tumors suggests its potential relevance in tumor angiogenesis/progression (42, 43). Interestingly, a previous study showed that Pfn1 and VASP are coordinately up-regulated in VEC during angiogenesis in vitro (although this study did not examine Mena expression) (44); whether this occurs in vivo remains unknown. Because our study provides the first evidence for coordinated up-regulation of Pfn1 and Mena (also a prognostic indicator of ccRCC) in TA-VEC in situ, we speculate that the synergistic action of Pfn1 and Mena may promote ccRCC angiogenesis and disease progression, an issue that we hope to address in future studies.
Third, tumor-associated blood vessels are also structurally abnormal, characterized by vascular leakiness and impaired perfusion. These features not only facilitate intravasation and dissemination of tumor cells leading to metastasis, but they support hypoxia that coordinately conditions cancer cells for aggressive behavior, while enforcing a state of immunosuppression in the TME (marked by enhanced recruitment and expansion of immunosuppressive T-cells (myeloid-derived suppressor cells and regulatory T-cells) and reduced cytotoxic T-cell infiltration) (45, 46). We previously showed that Pfn1 depletion in VEC confers increased resistance to VEGF-induced junctional disruption in vitro (10). Increased Pfn1 expression has also been linked to compromised barrier function of retinal VEC and microvascular leakage in vivo (47, 48). Therefore, it is conceivable that Pfn1 up-regulation in TA-VEC may exacerbate the hyperpermeability of tumor-associated blood vessels, resulting in an overall immunosuppressive TME. Previous studies have shown that global haploinsufficiency of Pfn1 reduces macrophage infiltration in the vascular intima in certain disease settings (atherosclerosis, diabetes) (49, 50). A recent study has also demonstrated Pfn1's ability to inhibit migration and cytotoxic action of T-cells in vitro (51). Hence, it will be interesting to explore in future studies whether Pfn1 dysregulation in immune cells and VEC could directly and/or indirectly impact the infiltration of immune cell subpopulations and their effector functions/survival within the TME, at least partially contributing to disease progression.
Finally, in this study, we report identification of a novel small molecule inhibitor of the Pfn1–actin interaction (C74) and the abilities of this inhibitor as well as a previous generation inhibitor (C2) to substantially reduce migration and proliferation of RCC cells in cell culture settings. In fact, both types of small molecule inhibitors reduced cell migration more robustly than the levels achieved in the Pfn1 knockdown setting. Pfn1 promotes cell migration through its actin interaction. However, Pfn1 also binds to membrane phosphoinositides. We previously showed that Pfn1–phosphoinositide interaction inhibits cell migration through suppressing phosphatidylinositol 3,4-bisphosphate–dependent membrane recruitment of certain promigratory protein complexes, resulting in a hypermigratory phenotype of breast cancer cells in Pfn1-deficient condition (26). The competing actions of Pfn1 on cell migration through its actin versus phosphoinositide interaction can be a potential reason underlying stronger inhibition of cell migration by small molecule–mediated intervention of Pfn1–actin interaction than by Pfn1 depletion. Although human Pfn2 has 62% sequence identity with Pfn1, the interface with actin is almost entirely conserved between these two variants of Pfn with only a few analogous variations in the residues (Ser84 (Pfn1) versus Thr84 (Pfn2) and Glu82 (Pfn1) versus Asp82 (Pfn2)). Our model predicts that the residues near C74 on Pfn1 are Arg74, Asp75, Ser76, and Glu82. Therefore, without further experimental validation, we cannot rule out the possibility of C74's ability to co-target Pfn2–actin interaction. Because Pfn3 and Pfn4 have <40% sequence identity with Pfn1, and the crystal structure of either of these two forms of Pfn in a complex with actin is not available to date, we are unable to comment on the likelihood of C74's targeting to these variants of Pfn.
However, our expression analyses demonstrated that the average transcript abundances of Pfn2 is 6-fold lower than that of Pfn1 and that the other two minor isoforms are either not expressed at all or expressed at a virtually negligible level compared with that of Pfn1 in ccRCC tumors. Therefore, unless C74 co-targets Pfn2–actin interaction and at a much higher affinity than that involving Pfn1, it is likely that Pfn1 is the primary target of C74 in RCC cells, a scenario that is indirectly supported by our experimental data showing Pfn1 dependence for the antiproliferative action of C74 on RVN cells in vitro. Our exploratory studies also demonstrate an initial proof of concept of the ability of this inhibitor to reduce tumor growth in vivo. Although our serum chemistry panel data argue against toxic effects of C74, we cannot absolutely rule out the possibility of off-target effects of our compound in vivo. Our in vitro proof-of-concept data showing C74's abilities to reduce Pfn1 interaction in cells and cell proliferation in a Pfn1-dependent manner are suggestive of biological mechanism of action of C74 at least partly involving Pfn1. However, we acknowledge that additional in vivo studies involving the actin-binding mutant of Pfn1 (these studies are beyond the scope of the present work) are needed for definitive proof of target specificity of our compound.
We speculate at least two experimental issues could have blunted the in vivo efficacy of the compound in our studies. First, direct injection of the compound at the tumor site without any carrier could have promoted rapid clearance of the compound from the injection site. Second, targeted delivery of the compound geared toward specific cell type(s) might be a preferred approach. These limitations need to be overcome in future studies. We also need to expand our in vivo studies in an orthotopic setting to determine whether Pfn1 inhibition retards the progression of pre-established tumors and improves animal survival. This not only will conceptually justify Pfn1 as an interventional target in ccRCC but also will motivate future efforts to experimentally characterize the inhibitor's binding mode to facilitate medicinal chemistry optimization for improved potency and drug-like properties for potential translation into the clinic.
Materials and methods
Cell culture, transfection, and viral transduction
Murine VHL-deleted variant of RENCA (referred to as RVN cells) (19) cells were cultured in DMEM with 10% (v/v) FBS and 1% (v/v) antibiotics. Human VHL-negative 786-0 cells (ATCC) were cultured in RPMI 1640 medium supplemented with 10% (v/v) FBS and antibiotics. HmVEC-1, a widely used immortalized human dermal microvascular EC line (ATCC; CRL-3243, referred to as VEC), were cultured in MCDB-131 (Life Technologies) growth medium supplemented with 10% (v/v) FBS, 1% (v/v) antibiotics, 10 ng/ml epidermal growth factor, 1 µg/ml hydrocortisone, and 10 mm l-glutamine. For adenovirus infection, the cells were plated, allowed to attach, and then infected with adenovirus encoding either GFP (Ad-GFP) or Pfn1 (Ad-Pfn1) at a multiplicity of infection of 500 for 24 h, before further incubation for an additional 48 h in fresh culture medium prior to use. For knockdown studies, the cells were transfected with 50 nm of either smart-pool control or Pfn1 siRNA for 72 h as previously reported (20). For cytotoxicity assessment, we stained cells with a live/dead assay kit (Life Technologies, Inc.) following the manufacturer's protocol, as we previously described (20).
Cell migration and proliferation assays
For single-cell migration experiments, RVN, VEC, and 786-0 cells were plated in 24-well plates coated with type I collagen (Millipore) overnight. Where indicated, purified Pfn1 protein (Abcam, ab87760) was added to the culture medium (at either 1 or 10 μm concentration) for several hours prior to time-lapse imaging. Time-lapse images of randomly migrating cells were collected using a 10× objective for 120 min at 1-min time intervals using cellSens (Olympus Life Science) software. The centroid of the cell nucleus was tracked using ImageJ, and the average speed of migration was computed on a per-cell basis as before (21). For Transwell migration experiments, 25,000 RVN cells were plated in triplicate in the upper chamber of 8.0-μm Transwell plates in the serum-free medium and allowed to migrate either toward a 10% FBS gradient or VEC-conditioned media established in the lower chamber for 20 h (control wells contained serum-free medium in the lower chamber as well). Nonmigrating cells in the upper chamber were removed by swabbing; transmigrated cells were fixed in 3.7% formaldehyde and then stained with DAPI. The images were acquired at 10 random fields at 10× magnification for enumeration of nuclei using ImageJ. For cell proliferation assay, 1000 RCC cells were plated per well in 96-well plates using quadruplicate determinations, with proliferation assessed by time-course analyses of DAPI-stained nuclei based on images acquired over three to five random fields/well.
Conditioned media
Conditioned media were collected from the culture dishes following overnight incubation of cells in serum-free medium. The collected media were filtered (0.45-μm size) and concentrated using a 10-kDa cutoff filter. The concentrate was reconstituted with 2× Laemmli sample buffer and boiled before being analyzed by gel electrophoresis.
Proximity–ligation assay
The PLA was performed using the Duolink kit with anti-mouse plus and anti-rabbit minus probes (Sigma, DUO9210), using two primary antibodies of different species targeting either Pfn1 (Abcam, 1:200, ab124904, rabbit) or actin (BD Biosciences, 1:100, 612656, mouse), and the PLA images were analyzed following protocols we previously described (32). As a negative control, one of the primary antibodies was omitted from the reaction.
Protein extraction and immunoblotting
Cell lysates were prepared by a modified RIPA buffer (25 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% (v/v) Nonidet P-40, 5% (v/v) glycerol), 1 mm EDTA, 50 mm NaF, 1 mm sodium pervanadate, and protease inhibitors supplemented with 6× sample buffer diluted to 1× with the final SDS concentration in the lysis buffer equivalent to 2%. Working dilutions for the various antibodies were monoclonal Pfn1 (Abcam, ab124904; 1:3000), monoclonal Tubulin (Sigma, T9026, 1:3000), and monoclonal GAPDH (DSHB, DSHB-hGAPDH-2G7; 1:100).
Immunohistochemistry
For Pfn1 IHC of ccRCC TMA, tissue sections were deparaffinized, and rehydrated tissue sections were incubated overnight with the primary anti-Pfn1 antibody (clone EPR6304, catalog no. ab124904, Abcam, dilution 1:100). For immunodetection of the primary antibody, a secondary biotin-labeled anti-rabbit antibody (catalog no. ab97049, Abcam) and streptavidin-peroxidase conjugates (catalog no. 11089153001, Roche) were used. The staining was detected with the Pierce DAB substrate kit (catalog no. 34002, ThermoFisher). The slides were counterstained with hematoxylin before dehydration and mounting. In negative control experiments, primary antibody was omitted from the staining of either ccRCC or ovarian (kindly provided by Dr. Jamie Lesnock, Magee Hospital, Pittsburgh, PA, USA) tumor samples.
ELISA
Serum Pfn1 was detected with a commercial ELISA kit (LSbio, catalog no. LS-F24009), with recombinant GST-Pfn1 used for constructing the calibration curve (sensitivity, 46.8 pg/ml). Bacterial expression and purification of GST-Pfn1 have been previously described (32). Serum was diluted 10–20-fold to ensure the absorbance reading for Pfn1 within the linear range of the calibration curve. Blood samples of ccRCC patients and normal subjects were collected following the guidelines of approved institutional review board protocols at the University of Pittsburgh abiding by the Helsinki Principles.
RCC TMA
The ccRCC TMA was obtained from the tissue bank of the National Center for Tumor Diseases at the University of Heidelberg (52) and used in accordance to the regulations of the tissue bank, as well as under approval of the Ethics Committee of the University of Heidelberg School of Medicine abiding by the declaration of Helsinki Principle. RCC specimens from 417 patients were included in the final analysis. The TMA specimen were collected between 1990 and 2005 with a median follow-up time of 112.7 months (range, 0.6–345.9 months). Clinicopathological features were as follows: grade, G1/G2 (n = 355, 85.1%), G3/4 (n = 57, 13.7%), and G unknown (n = 5, 1.2%); stage, pT1/T2 (n = 280, 67.2%), pT3/T4 (n = 135, 32.4%), and pTx (n = 2, 0.5%); lymph node involvement, c/pN0 (n = 393, 94.3%) and N+ (n = 24, 5.8%); and distant metastasis, cM0 (n = 357, 85.6%) and M+ (n = 60, 14.4%).
Transcriptome analysis of flow-sorted TA-VEC and NAT-VEC
Surgically excised ccRCC tumor and tumor-adjacent normal kidney tissues were minced, digested with collagenase, passed through 70-mm mesh filter and Ficoll density gradient before being stained with fluorescent-labeled antibodies against human CD34, CD45, and CD146 (all from BD Biosciences) and DAPI (Sigma–Aldrich). Stained cells were then sorted using a biocontained FACSAria cell sorters (in the Department of Immunology's Unified Flow Cytometry Facility, University of Pittsburgh) based on phenotype: i.e. CD34+CD45negCD146+ VEC and CD34negCD45negCD146+ pericytes. RNA was isolated from the sorted cells and analyzed using Affymetrix U133 Plus 2.0 Array chips as previously described (53).
Pfn1–Actin interaction inhibitor identification
Whole protein docking was performed using smina (54), with the exhaustiveness parameter set to 50 and the autobox_ligand parameter set equal to the target protein. Actin monomers, stripped of nonprotein atoms, were extracted from Protein Data Bank entries 2BTF and 4JHD. The profilin monomer was extracted from entry 2BTF, and then, to get a broader sampling of conformations to address the lack of pockets at the protein–protein interface, an AMBER molecular dynamics simulation was run for 100 ns using the ff15ipq force field and TIP3P water. The three most diverse conformations, as determined by backbone RMSD with respect to the starting conformation and each other, were selected as additional structures for whole protein docking. Docking C2 to these five structures (two actin and three Pfn1) identified six high-scoring binding sites, one of which was the original putative C2-binding site. Using the docked C2 conformation as reference, we manually selected pharmacophore features for each binding site and screened the MolPort library of commercially available compounds using Pharmit (55), an online resource for interactive structure-based virtual screening. Pharmit returns hit compounds aligned to the query pharmacophore. These conformations were then energy-minimized using the AutoDock Vina (56) scoring function with respect to the receptor structure and rescored using our convolutional neural network (57) scoring function. We then clustered all hit molecules for each binding site with the cluster_mols plugin for PyMOL and selected the top scoring molecule for each scoring method in each cluster (if available). This resulted in 67 selected hits that were further tested in pyrene-based actin polymerization biochemical assay as described in our previous study (32).
In vivo tumorigenesis
In 6–7-week-old BALB/cJ mice, 1 × 106 RENCA cells were implanted subcutaneously in a 1:1 PBS:Matrigel (R&D Systems, Minneapolis, MN, USA) mixture, supplemented with 81 µg of C74 or DMSO control. Intratumoral injections were then given daily with either C74 (16 mg/kg) or control dissolved in saline through day 19 post-implantation. On day 20, the mice were sacrificed, and the harvested tumors were weighed. For toxicological studies, C74 (at 16 mg/kg) was administered through an intraperitoneal route in normal 6–7-week-old BALB/cJ mice daily for 2 weeks. At the end, blood was collected by cardiac puncture before the mice were sacrificed. Serum samples prepared from blood were submitted to IDEXX Bioanalytics (Sacramento, CA, USA) for comprehensive chemical analyses. All animal experiments were performed in compliance with the guidelines of the Institutional Animal Care and Use committee of the University of Pittsburgh.
Statistics
Statistical tests were performed with either one-way analysis of variance followed by Tukey's post hoc test or nonparametric Mann–Whitney test with Hodges–Lehmann estimate for p value estimation (for TCGA data), when appropriate. A Kruskal–Wallis test was used for ranked analysis of small samples. For survival analyses, a log rank test was performed. Differences exhibiting p < 0.05 were considered as statistically significant. The data are displayed either as box plots or box-and-whisker plots. For both plots, asterisks represent the means. The middle line and the upper and lower hinges of the box represent the median and 75th and 25th percentiles of data, respectively. For box plots, the data display spans from 10th to 90th percentile of the data values with outliers indicated, whereas the whiskers in box-and-whisker plots represent the maximum and minimum values.
Data availability
All data are available in the article. Curated transcriptome data for various clinical subtypes of RCC were downloaded from the cBioportal website (RRID:SCR_014555).
Supplementary Material
This article contains supporting information.
Author contributions—A. A., D. G., J. S., Y. W., R. M., A. K., S. D., and W. J. S. data curation; A. A., D. G., P. F., J. S., Y. W., R. M., A. K., S. D., DK, and W. J. S. formal analysis; A. A., D. G., S. D., and W. J. S. validation; A. A., D. G., P. F., J. S., and PR investigation; A. A., D. G., P. F., W. J. S., and PR writing-original draft; D. G., P. F., and D. K. methodology; P. F., J. M., A. D., L. W., D. K., and W. J. S. resources; M. T. L., D. K., W. J. S., and P. R. conceptualization; M. T. L., D. K., and P. R. supervision; M. T. L. and P. R. funding acquisition; M. T. L. and P. R. project administration; M. T. L., D. K., W. J. S., and P. R. writing-review and editing.
Funding and additional information—This work was supported by the University of Pittsburgh Hillman Cancer Center Developmental Funding Program Grants 1R01 CA181450-01 and R01CA206012 (to M. T. L.); the Cardiovascular Training T32 Grant Program and Achievement Rewards for College Scientists funding (to A. A.); a National Cancer Center fellowship (to D. G.); Department of Defense Grant W81XWH-19-1-0768 (to P. R.); NCI, National Institutes of Health Grant CA108607 (to P. R.); and NIGMS, National Institutes of Health Grant GM108340-04 (to D. K.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- RCC
- renal cell carcinoma
- ccRCC
- clear-cell RCC
- TCGA
- The Cancer Genome Atlas
- VEC
- vascular endothelial cell
- TME
- tumor microenvironment
- LOF
- loss of function
- VEGF
- vascular endothelial growth factor
- ABP
- actin-binding protein
- IHC
- immunohistochemistry
- OS
- overall survival
- PFS
- progression-free survival
- TA
- tumor-associated
- TMA
- tissue microarray
- NAT
- normal adjacent tissue
- PLA
- proximity ligation assay
- FBS
- fetal bovine serum
- Ad
- adenovirus
- DAPI
- 4′,6-diamino-2-phenylindole
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase.
References
- 1. Siegel R. L., Miller K. D., and Jemal A. (2019) Cancer statistics, 2019. CA Cancer J. Clin. 69, 7–34 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 2. Jonasch E., Gao J., and Rathmell W. K. (2014) Renal cell carcinoma. BMJ 349, g4797 10.1136/bmj.g4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Juengel E., Krueger G., Rutz J., Nelson K., Werner I., Relja B., Seliger B., Fisslthaler B., Fleming I., Tsaur I., Haferkamp A., and Blaheta R. A. (2016) Renal cell carcinoma alters endothelial receptor expression responsible for leukocyte adhesion. Oncotarget 7, 20410–20424 10.18632/oncotarget.7804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pichler R., and Heidegger I. (2017) Novel concepts of antiangiogenic therapies in metastatic renal cell cancer. Memo 10, 206–212 10.1007/s12254-017-0344-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goel S., Duda D. G., Xu L., Munn L. L., Boucher Y., Fukumura D., and Jain R. K. (2011) Normalization of the vasculature for treatment of cancer and other diseases. Physiol. Rev. 91, 1071–1121 10.1152/physrev.00038.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bergers G., and Hanahan D. (2008) Modes of resistance to anti-angiogenic therapy. Nat. Rev. 8, 592–603 10.1038/nrc2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Welti J., Loges S., Dimmeler S., and Carmeliet P. (2013) Recent molecular discoveries in angiogenesis and antiangiogenic therapies in cancer. J. Clin. Invest. 123, 3190–3200 10.1172/JCI70212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ding Z., Bae Y. H., and Roy P. (2012) Molecular insights on context-specific role of profilin-1 in cell migration. Cell Adh. Migr. 6, 442–449 10.4161/cam.21832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ding Z., Gau D., Deasy B., Wells A., and Roy P. (2009) Both actin and polyproline interactions of profilin-1 are required for migration, invasion and capillary morphogenesis of vascular endothelial cells. Exp. Cell Res. 315, 2963–2973 10.1016/j.yexcr.2009.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ding Z., Lambrechts A., Parepally M., and Roy P. (2006) Silencing profilin-1 inhibits endothelial cell proliferation, migration and cord morphogenesis. J. Cell Sci. 119, 4127–4137 10.1242/jcs.03178 [DOI] [PubMed] [Google Scholar]
- 11. Fan Y., Arif A., Gong Y., Jia J., Eswarappa S. M., Willard B., Horowitz A., Graham L. M., Penn M. S., and Fox P. L. (2012) Stimulus-dependent phosphorylation of profilin-1 in angiogenesis. Nat. Cell Biol. 14, 1046–1056 10.1038/ncb2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kullmann J. A., Neumeyer A., Gurniak C. B., Friauf E., Witke W., and Rust M. B. (2011) Profilin1 is required for glial cell adhesion and radial migration of cerebellar granule neurons. EMBO Rep. 13, 75–82 10.1038/embor.2011.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Böttcher R. T., Wiesner S., Braun A., Wimmer R., Berna A., Elad N., Medalia O., Pfeifer A., Aszódi A., Costell M., and Fässler R. (2009) Profilin 1 is required for abscission during late cytokinesis of chondrocytes. EMBO J. 28, 1157–1169 10.1038/emboj.2009.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Minamida S., Iwamura M., Kodera Y., Kawashima Y., Ikeda M., Okusa H., Fujita T., Maeda T., and Baba S. (2011) Profilin 1 overexpression in renal cell carcinoma. Int. J. Urol. 18, 63–71 10.1111/j.1442-2042.2010.02670.x [DOI] [PubMed] [Google Scholar]
- 15. Masui O., White N. M., DeSouza L. V., Krakovska O., Matta A., Metias S., Khalil B., Romaschin A. D., Honey R. J., Stewart R., Pace K., Bjarnason G. A., Siu K. W., and Yousef G. M. (2013) Quantitative proteomic analysis in metastatic renal cell carcinoma reveals a unique set of proteins with potential prognostic significance. Mol. Cell. Proteomics 12, 132–144 10.1074/mcp.M112.020701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Neely B. A., Wilkins C. E., Marlow L. A., Malyarenko D., Kim Y., Ignatchenko A., Sasinowska H., Sasinowski M., Nyalwidhe J. O., Kislinger T., Copland J. A., and Drake R. R. (2016) Proteotranscriptomic analysis reveals stage specific changes in the molecular landscape of clear-cell renal cell carcinoma. PLoS One 11, e0154074 10.1371/journal.pone.0154074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Karamchandani J. R., Gabril M. Y., Ibrahim R., Scorilas A., Filter E., Finelli A., Lee J. Y., Ordon M., Pasic M., Romaschin A. D., and Yousef G. M. (2015) Profilin-1 expression is associated with high grade and stage and decreased disease-free survival in renal cell carcinoma. Hum. Pathol. 46, 673–680 [DOI] [PubMed] [Google Scholar]
- 18. Gau D. M., Lesnock J. L., Hood B. L., Bhargava R., Sun M., Darcy K., Luthra S., Chandran U., Conrads T. P., Edwards R. P., Kelley J. L., Krivak T. C., and Roy P. (2015) BRCA1 deficiency in ovarian cancer is associated with alteration in expression of several key regulators of cell motility: a proteomics study. Cell Cycle 14, 1884–1892 10.1080/15384101.2015.1036203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chakraborty S., Jiang C., Gau D., Oddo M., Ding Z., Vollmer L., Joy M., Schiemann W., Stolz D. B., Vogt A., Ghosh S., and Roy P. (2018) Profilin-1 deficiency leads to SMAD3 upregulation and impaired 3D outgrowth of breast cancer cells. Br. J. Cancer 119, 1106–1117 10.1038/s41416-018-0284-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Joy M., Gau D., Castellucci N., Prywes R., and Roy P. (2017) The myocardin-related transcription factor MKL co-regulates the cellular levels of two profilin isoforms. J. Biol. Chem. 292, 11777–11791 10.1074/jbc.M117.781104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gau D., Veon W., Shroff S. G., and Roy P. (2019) The VASP-profilin1 (Pfn1) interaction is critical for efficient cell migration and is regulated by cell-substrate adhesion in a PKA-dependent manner. J. Biol. Chem. 294, 6972–6985 10.1074/jbc.RA118.005255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Caglayan E., Romeo G. R., Kappert K., Odenthal M., Südkamp M., Body S. C., Shernan S. K., Hackbusch D., Vantler M., Kazlauskas A., and Rosenkranz S. (2010) Profilin-1 is expressed in human atherosclerotic plaques and induces atherogenic effects on vascular smooth muscle cells. PLoS One 5, e13608 10.1371/journal.pone.0013608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hu J., Schokrpur S., Archang M., Hermann K., Sharrow A. C., Khanna P., Novak J., Signoretti S., Bhatt R. S., Knudsen B. S., Xu H., and Wu L. (2018) A non-integrating lentiviral approach overcomes Cas9-induced immune rejection to establish an immunocompetent metastatic renal cancer model. Mol. Ther. Methods Clin. Dev. 9, 203–210 10.1016/j.omtm.2018.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schokrpur S., Hu J., Moughon D. L., Liu P., Lin L. C., Hermann K., Mangul S., Guan W., Pellegrini M., Xu H., and Wu L. (2016) CRISPR-mediated VHL knockout generates an improved model for metastatic renal cell carcinoma. Sci. Rep. 6, 29032 10.1038/srep29032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rotty J. D., Wu C., Haynes E. M., Suarez C., Winkelman J. D., Johnson H. E., Haugh J. M., Kovar D. R., and Bear J. E. (2015) Profilin-1 serves as a gatekeeper for actin assembly by Arp2/3-dependent and -independent pathways. Dev. Cell 32, 54–67 10.1016/j.devcel.2014.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bae Y. H., Ding Z., Das T., Wells A., Gertler F., and Roy P. (2010) Profilin1 regulates PI(3,4)P2 and lamellipodin accumulation at the leading edge thus influencing motility of MDA-MB-231 cells. Proc. Natl. Acad. Sci. U.S.A. 107, 21547–21552 10.1073/pnas.1002309107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bae Y. H., Ding Z., Zou L., Wells A., Gertler F., and Roy P. (2009) Loss of profilin-1 expression enhances breast cancer cell motility by Ena/VASP proteins. J. Cell. Physiol. 219, 354–364 10.1002/jcp.21677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ding Z., Joy M., Bhargava R., Gunsaulus M., Lakshman N., Miron-Mendoza M., Petroll M., Condeelis J., Wells A., and Roy P. (2014) Profilin-1 downregulation has contrasting effects on early vs. late steps of breast cancer metastasis. Oncogene 33, 2065–2074 10.1038/onc.2013.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shen K., Xi Z., Xie J., Wang H., Xie C., Lee C. S., Fahey P., Dong Q., and Xu H. (2016) Guttiferone K suppresses cell motility and metastasis of hepatocellular carcinoma by restoring aberrantly reduced profilin 1. Oncotarget 7, 56650–56663 10.18632/oncotarget.10992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu N., Zhang W., Yang Y., Liang Y. L., Wang L. Y., Jin J. W., Cai X. M., and Zha X. L. (2006) Profilin 1 obtained by proteomic analysis in all-trans retinoic acid-treated hepatocarcinoma cell lines is involved in inhibition of cell proliferation and migration. Proteomics 6, 6095–6106 10.1002/pmic.200500321 [DOI] [PubMed] [Google Scholar]
- 31. Tamura M., Yanagihara N., Tanaka H., Osajima A., Hirano T., Higashi K., Yamada K. M., Nakashima Y., and Hirano H. (2000) Activation of DNA synthesis and AP-1 by profilin, an actin-binding protein, via binding to a cell surface receptor in cultured rat mesangial cells. J. Am. Soc. Nephrol. 11, 1620–1630 [DOI] [PubMed] [Google Scholar]
- 32. Gau D., Lewis T., McDermott L., Wipf P., Koes D., and Roy P. (2018) Structure-based virtual screening identifies a small-molecule inhibitor of the profilin 1–actin interaction. J. Biol. Chem. 293, 2606–2616 10.1074/jbc.M117.809137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Escobar B., de Cárcer G., Fernández-Miranda G., Cascón A., Bravo-Cordero J. J., Montoya M. C., Robledo M., Cañamero M., and Malumbres M. (2010) Brick1 is an essential regulator of actin cytoskeleton required for embryonic development and cell transformation. Cancer Res. 70, 9349–9359 10.1158/0008-5472.CAN-09-4491 [DOI] [PubMed] [Google Scholar]
- 34. Unwin R. D., Craven R. A., Harnden P., Hanrahan S., Totty N., Knowles M., Eardley I., Selby P. J., and Banks R. E. (2003) Proteomic changes in renal cancer and co-ordinate demonstration of both the glycolytic and mitochondrial aspects of the Warburg effect. Proteomics 3, 1620–1632 10.1002/pmic.200300464 [DOI] [PubMed] [Google Scholar]
- 35. Lu X., Boora U., Seabra L., Rabai E. M., Fenton J., Reiman A., Nagy Z., and Maher E. R. (2014) Knockdown of Slingshot 2 (SSH2) serine phosphatase induces Caspase3 activation in human carcinoma cell lines with the loss of the Birt–Hogg–Dubé tumour suppressor gene (FLCN). Oncogene 33, 956–965 10.1038/onc.2013.27 [DOI] [PubMed] [Google Scholar]
- 36. Zhao T., Liang X., Chen J., Bao Y., Wang A., Gan X., Lu X., and Wang L. (2019) ANGPTL3 inhibits renal cell carcinoma metastasis by inhibiting VASP phosphorylation. Biochem. Biophys. Res. Commun. 516, 880–887 10.1016/j.bbrc.2019.06.120 [DOI] [PubMed] [Google Scholar]
- 37. Fan Y., Potdar A. A., Gong Y., Eswarappa S. M., Donnola S., Lathia J. D., Hambardzumyan D., Rich J. N., and Fox P. L. (2014) Profilin-1 phosphorylation directs angiocrine expression and glioblastoma progression through HIF-1α accumulation. Nat. Cell Biol. 16, 445–456 10.1038/ncb2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gau D., Vignaud L., Allen A., Guo Z., Sahel J., Boone D., Koes D., Guillonneau X., and Roy P. (2020) Disruption of profilin1 function suppresses developmental and pathological retinal neovascularization. J. Biol. Chem. 295, 9618–9629 10.1074/jbc.RA120.012613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gertler F. B., Niebuhr K., Reinhard M., Wehland J., and Soriano P. (1996) Mena, a relative of VASP and Drosophila Enabled, is implicated in the control of microfilament dynamics. Cell 87, 227–239 10.1016/S0092-8674(00)81341-0 [DOI] [PubMed] [Google Scholar]
- 40. Fischer R. S., Lam P. Y., Huttenlocher A., and Waterman C. M. (2019) Filopodia and focal adhesions: an integrated system driving branching morphogenesis in neuronal pathfinding and angiogenesis. Dev. Biol. 451, 86–95 10.1016/j.ydbio.2018.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Koika V., Zhou Z., Vasileiadis I., Roussos C., Finetti F., Monti M., Morbidelli L., and Papapetropoulos A. (2010) PKG-I inhibition attenuates vascular endothelial growth factor-stimulated angiogenesis. Vasc. Pharmacol. 53, 215–222 10.1016/j.vph.2010.08.004 [DOI] [PubMed] [Google Scholar]
- 42. Gurzu S., Jung I., Prantner I., Ember I., Pávai Z., and Mezei T. (2008) The expression of cytoskeleton regulatory protein Mena in colorectal lesions. Rom. J. Morphol. Embryol. 49, 345–349 [PubMed] [Google Scholar]
- 43. Gurzu S., Krause M., Ember I., Azamfirei L., Gobel G., Feher K., and Jung I. (2012) Mena, a new available marker in tumors of salivary glands?. Eur. J. Histochem. 56, e8 10.4081/ejh.2012.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Salazar R., Bell S. E., and Davis G. E. (1999) Coordinate induction of the actin cytoskeletal regulatory proteins gelsolin, vasodilator-stimulated phosphoprotein, and profilin during capillary morphogenesis in vitro. Exp. Cell Res. 249, 22–32 10.1006/excr.1999.4460 [DOI] [PubMed] [Google Scholar]
- 45. Martin J. D., Seano G., and Jain R. K. (2019) Normalizing function of tumor vessels: progress, opportunities, and challenges. Annu. Rev. Physiol. 81, 505–534 10.1146/annurev-physiol-020518-114700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shigeta K., Datta M., Hato T., Kitahara S., Chen I. X., Matsui A., Kikuchi H., Mamessier E., Aoki S., Ramjiawan R. R., Ochiai H., Bardeesy N., Huang P., Cobbold M., Zhu A. X., et al. (2020) Dual programmed death receptor-1 and vascular endothelial growth factor receptor-2 blockade promotes vascular normalization and enhances antitumor immune responses in hepatocellular carcinoma. Hepatology 71, 1247–1261 10.1002/hep.30889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ding H., Chen B., Lu Q., and Wang J. (2018) Profilin-1 mediates microvascular endothelial dysfunction in diabetic retinopathy through HIF-1α–dependent pathway. Int. J. Clin. Exp. Pathol. 11, 1247–1255 [PMC free article] [PubMed] [Google Scholar]
- 48. Lu Q., Lu P., Chen W., Lu L., and Zheng Z. (2018) ANGPTL-4 induces diabetic retinal inflammation by activating Profilin-1. Exp. Eye Res. 166, 140–150 10.1016/j.exer.2017.10.009 [DOI] [PubMed] [Google Scholar]
- 49. Romeo G., Frangioni J. V., and Kazlauskas A. (2004) Profilin acts downstream of LDL to mediate diabetic endothelial cell dysfunction. FASEB J. 18, 725–727 10.1096/fj.03-0841fje [DOI] [PubMed] [Google Scholar]
- 50. Romeo G. R., Moulton K. S., and Kazlauskas A. (2007) Attenuated expression of profilin-1 confers protection from atherosclerosis in the LDL receptor null mouse. Circ. Res. 101, 357–367 10.1161/CIRCRESAHA.107.151399 [DOI] [PubMed] [Google Scholar]
- 51. Schoppmeyer R., Zhao R., Cheng H., Hamed M., Liu C., Zhou X., Schwarz E. C., Zhou Y., Knörck A., Schwär G., Ji S., Liu L., Long J., Helms V., Hoth M., et al. (2017) Human profilin 1 is a negative regulator of CTL mediated cell-killing and migration. Eur. J. Immunol. 47, 1562–1572 10.1002/eji.201747124 [DOI] [PubMed] [Google Scholar]
- 52. Zhou X., Li J., Marx C., Tolstov Y., Rauch G., Herpel E., Macher-Goeppinger S., Roth W., Grüllich C., Pahernik S., Hohenfellner M., and Duensing S. (2015) Uncoupling of PUMA expression and apoptosis contributes to functional heterogeneity in renal cell carcinoma: prognostic and translational implications. Transl. Oncol. 8, 480–486 10.1016/j.tranon.2015.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Weinstein A. M., Chen L., Brzana E. A., Patil P. R., Taylor J. L., Fabian K. L., Wallace C. T., Jones S. D., Watkins S. C., Lu B., Stroncek D. F., Denning T. L., Fu Y. X., Cohen P. A., and Storkus W. J. (2017) Tbet and IL-36γ cooperate in therapeutic DC-mediated promotion of ectopic lymphoid organogenesis in the tumor microenvironment. Oncoimmunology 6, e1322238 10.1080/2162402X.2017.1322238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Koes D. R., Baumgartner M. P., and Camacho C. J. (2013) Lessons learned in empirical scoring with smina from the CSAR 2011 benchmarking exercise. J. Chem. Inf. Model. 53, 1893–1904 10.1021/ci300604z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sunseri J., and Koes D. R. (2016) Pharmit: interactive exploration of chemical space. Nucleic Acids Res. 44, W442–W448 10.1093/nar/gkw287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Trott O., and Olson A. J. (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ragoza M., Hochuli J., Idrobo E., Sunseri J., and Koes D. R. (2017) Protein–ligand scoring with convolutional neural networks. J. Chem. Inf. Model. 57, 942–957 10.1021/acs.jcim.6b00740 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the article. Curated transcriptome data for various clinical subtypes of RCC were downloaded from the cBioportal website (RRID:SCR_014555).