Abstract
The enzyme β-carotene oxygenase 2 (BCO2) converts carotenoids into more polar metabolites. Studies in mammals, fish, and birds revealed that BCO2 controls carotenoid homeostasis and is involved in the pathway for vitamin A production. However, it is controversial whether BCO2 function is conserved in humans, because of a 4-amino acid long insertion caused by a splice acceptor site polymorphism. We here show that human BCO2 splice variants, BCO2a and BCO2b, are expressed as pre-proteins with mitochondrial targeting sequence (MTS). The MTS of BCO2a directed a green fluorescent reporter protein to the mitochondria when expressed in ARPE-19 cells. Removal of the MTS increased solubility of BCO2a when expressed in Escherichia coli and rendered the recombinant protein enzymatically active. The expression of the enzymatically active recombinant human BCO2a was further improved by codon optimization and its fusion with maltose-binding protein. Introduction of the 4-amino acid insertion into mouse Bco2 did not impede the chimeric enzyme's catalytic proficiency. We further showed that the chimeric BCO2 displayed broad substrate specificity and converted carotenoids into two ionones and a central C14-apocarotendial by oxidative cleavage reactions at C9,C10 and C9',C10'. Thus, our study demonstrates that human BCO2 is a catalytically competent enzyme. Consequently, information on BCO2 becomes broadly applicable in human biology with important implications for the physiology of the eyes and other tissues.
Keywords: Vision, retina, metabolism, dioxygenase, carotenoids, macular pigments, apocarotenoids, β-carotene-oxygenase 2 (BCO2), vision, carotenoid apocarotenoid, BCO2
The beneficial role of carotenoids in human health is currently a subject of intense investigation. Low status of carotenoids is associated with vitamin A deficiency and a number of degenerative diseases including cardiovascular disease, cognitive impairments, and age-related macular degeneration (1–6). A number of potential mechanisms through which carotenoids can benefit human health have been proposed. The most commonly cited is their ability of acting as antioxidants, e.g. as free radical scavengers, in lipophilic environments such as membranes and lipoproteins (7). The blue light filtering properties of carotenoids compose another mechanism of protection of cellular components from the environment. Macular pigments have been chemically identified as the carotenoids lutein, zeaxanthin, and meso-zeaxanthin (3). These carotenoids are enriched in the fovea in primate retinas and confer its yellow color. Hence, the fovea is traditionally known as the macula lutea, or “yellow spot.” The macular pigments can protect the retina against light damage (8, 9) and reduce the adverse impact of light scattering and chromatic aberration, thereby optimizing contrast sensitivity of the retina (10).
Carotenoids must be acquired from the diet within the intestine and are distributed in lipoproteins to target tissues (11, 12). Genetic studies in animals identified candidate genes that affect tissue distribution and accumulation of carotenoids, such as class B scavenger receptors for cellular uptake of carotenoids (13, 14), cellular carotenoid-binding proteins (15, 16), and carotenoid cleavage dioxygenases (CCDs) (17–19). Clinical studies have associated genetic polymorphism in these genes with macular pigment density in the human population (4, 20, 21). Among these genes, the BCO2 gene encodes β-carotene-9',10'-dioxygenase that catalyzes a region-specific oxidative cleavage at the C9',C10' double bond in the central polyene chain of carotenoids (22). Mouse and ferret BCO2 display broad substrate specificity and convert carotenoid substrates, including zeaxanthin and lutein, into apocarotenoid products (23–25). Studies in Bco2 knockout mice showed that the enzyme controls carotenoid homeostasis and is involved in the production of apocarotenoid signaling molecules, including retinoids (25–27). Genetic studies in cow, sheep, and rabbits have identified inactivating mutations in the BCO2 gene that phenotypically display as yellow fat phenotype (28–31).
Five splice variants of human BCO2 have been annotated in the National Center for Biotechnology Information (NCBI) website (Gene code 83875) (Fig. 1). Interestingly, it was shown that all human BCO2 variants possess a GKAA insertion that is caused by the use of an alternate donor splice site during mRNA maturation (Fig. 1A) (32). This insertion is specific for primate BCO2s and is not found in other vertebrates. Previously, it was proposed that inactivity of the BCO2a variant results in macular pigment accumulation in the human retina (32). Biochemical analysis suggested that recombinant human BCO2 protein is enzymatically inactive when expressed in Escherichia coli (33) and that the GKAA insertion rendered recombinant murine BCO2 enzymatically inactive (32). However, studies by others challenged this proposal. A recombinant macaque BCO2 displays enzymatic activity although the corresponding gene possesses the same splice acceptor site polymorphism as the human gene (23). Additionally, expression of recombinant human BCO2 in a bacterial test system revealed enzymatic activity of recombinant human BCO2s (34–36).
Figure 1.
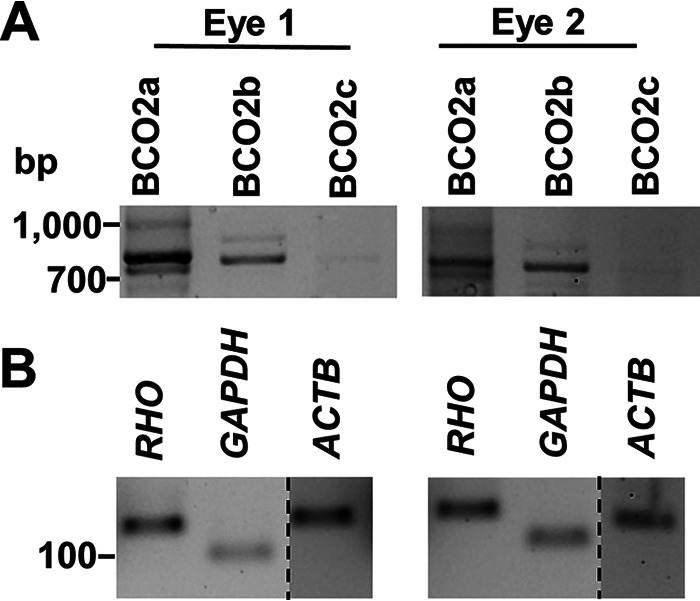
Expression of BCO2 splice variants in human retina. A, RT-PCR analysis of the mRNAs of different human BCO2 splice variants (BCO2a, BCO2b, and BCO2c) in total RNA preparations of two individual human retinas. mRNA encoding BCO2a and BCO2b are expressed in human retina. B, RT-PCR analysis of mRNA expression of Rhodopsin (RHO), GAPDH, and β-Actin (ACTB). Dashed lines indicate where a single gel image was spliced to exclude irrelevant lanes.
The current claim that BCO2, a protein evolutionarily conserved because the split between jawed and jawless vertebrates (34), has become inactive in primates is puzzling. This claim eliminates an important metabolic pathway for carotenoid homeostasis and vitamin A production in human physiology (25, 37). The question whether humans encode an enzymatically active BCO2 protein also is of clinical relevance. Many nutritionists and clinicians advocate for higher carotenoid intake because of their beneficial health effects (3, 5, 38). The establishment of such recommendations is based on the relationships between carotenoid intake, blood, and tissue concentrations. This relationship is not linear and is influenced by many host and environmental factors (11, 39). Additionally, excessive accumulation of carotenoids is a risk factor for detrimental health outcomes in smokers in clinical studies (40, 41). Therefore, we here wished to scrutinize whether humans express an active BCO2 enzyme that can control tissue homeostasis of carotenoids and its metabolites.
Results
Expression of BCO2 splice variant in the human retina
We focused on the three major splice mRNA variants, encoding, respectively, 579- (BCO2a), 545- (BCO2b), and 539- (BCO2c) amino acid long proteins (Fig. S1). Sequence alignment reveals that these proteins differ in their amino-terminal parts but otherwise share sequence identity, including conserved histidine residues for binding of the cofactor ferrous iron (Fig. S1). Additionally, BCO2c displays a 6-amino acid deletion in the coding region when compared with BCO2a and BCO2b (Fig. S1). We performed RT-PCR analysis with total RNA preparations isolated from retinas of human donor eyes with intron-spanning PCR primer sets. These primer sets were designed to specifically amplify each splice variant of the human BCO2 gene. This analysis detected PCR products for the mRNA splice variants encoding BCO2a and BCO2b in both retinas (Fig. 1A). Under the applied conditions, no PCR product for BCO2c was amplified from the RNA preparations of the retinas of the donor eyes. As controls, RT-PCR products for RHO mRNA encoding rhodopsin, GAPDH encoding glyceraldehyde-3-phosphate dehydrogenase, and ACTB encoding β-actin were amplified from the same RNA preparations and resulted in products of the expected sizes (Fig. 1B).
Recombinant human BCO2a is inactive
We found that mRNAs encoding BCO2a and BCO2b were expressed in the human retina. To test whether they encode functional enzymes, we took advantage of a cell-based assay (42). In this assay, BCO2 and other members of the CCD superfamily are expressed in an E. coli strain, which can synthesize carotenoids. This synthesis is achieved by transformation of the bacteria with plasmids harboring genes for carotenoid synthesis from Erwina uredovora (43). The accumulation of zeaxanthin confers a yellow color to the bacteria. Expression of an enzymatically active BCO2 leads to a discoloration of the bacteria because of the cleavage of zeaxanthin into apocarotenoids with blue-shifted spectra and lower absorption coefficient. We employed this assay for the human BCO2a and BCO2b as outlined under “Experimental procedures.” The pellets of the zeaxanthin producing bacteria expressing BCO2a showed a bright yellow color. The pellet of the bacteria expressing BCO2b had a lighter color, indicating an enzymatic turnover of zeaxanthin (Fig. 2A). Zeaxanthin turnover by the BCO2b was confirmed by HPLC analysis (Fig. 2, B and C). In bacteria extracts expressing BCO2b, amounts of β-cryptoxanthin, the direct precursor of zeaxanthin in the carotenoid synthesis pathway, and zeaxanthin were significantly reduced when compared with bacteria expressing BCO2a (Fig. 2, A and B).
Figure 2.
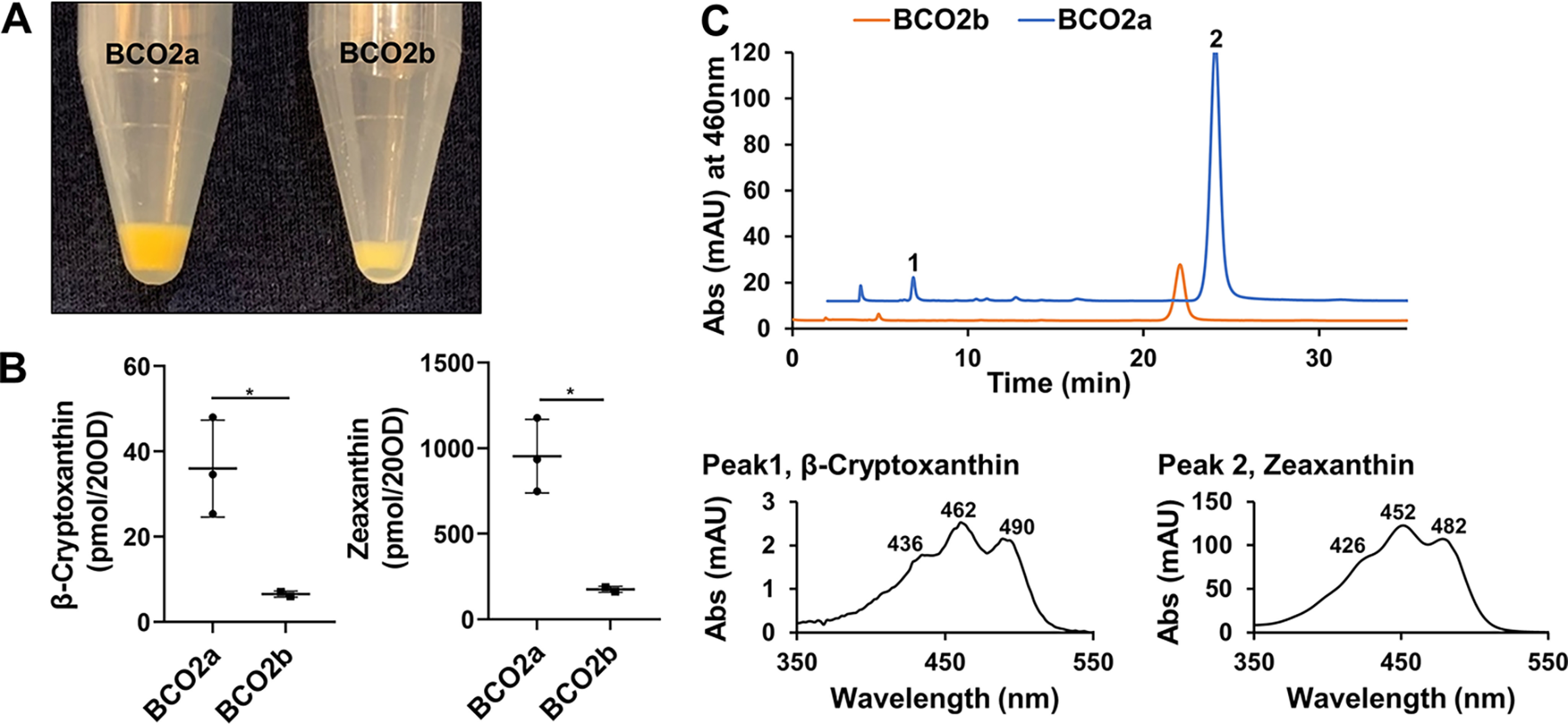
Human BCO2a is enzymatically inactive. Enzymatic activity of human BCO2a and BCO2b were examined in a cell-based assay. Both human isoforms were expressed in E. coli cells engineered to produce zeaxanthin. A, colors of bacteria pellets expressing BCO2a and BCO2b. B, quantification of the amounts of zeaxanthin and β-cryptoxanthin in the bacteria pellets (20 A600). The whisker plot represents mean ± S.E. from three independent experiments. Asterisks represent p values of 0.0404 and 0.0168, respectively. C, representative HPLC traces at 460 nm of lipid extracts from bacteria pellets expressing BCO2b (orange) and BCO2a (blue). Peak 1 corresponds to β-cryptoxanthin and peak 2 corresponds to zeaxanthin. Spectral characteristics of each carotenoid are displayed in the lower panel of C.
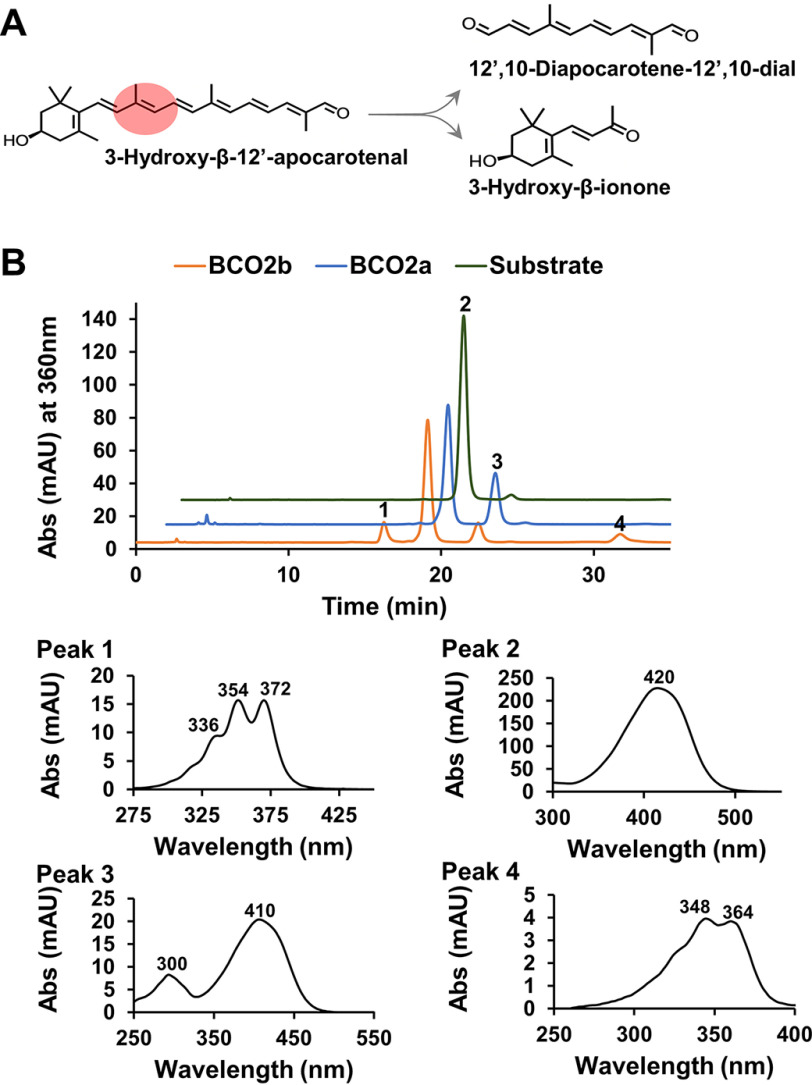
Human BCO2b converts apocarotenoids by oxidative cleavage
We previously established an in vitro enzyme assay to measure mouse BCO2 activity with apocarotenoids as substrates (37). We now applied this assay to human BCO2s using 3-hydroxy-β-12'-apocarotenal as a substrate. Cleavage of the apocarotenoid by BCO2b results in the formation of 3-hydroxy-β-ionone and 12',10-diapocarotene-12',10-dial (Fig. 3A). HPLC analysis revealed that the incubation of protein extracts containing human BCO2b with this substrate yielded a product that eluted earlier (Fig. 3B, peak 1) than 3-hydroxy-β-12'-apocarotenal (Fig. 3B, peak 2) and displayed spectral characteristics of a C12-apocarotene-dialdehyde (Fig. 3B, peak 1). Additionally, we found a product that eluted later at minute 32 with spectral characteristics of the corresponding di-alcohol (Fig. 3B, peak 4). Identical products were obtained when we incubated recombinant mBCO2 with this substrate (Fig. S2). Incubation of human BCO2a with the apocarotenoid did not result in product formation (Fig. 3B). However, we observed an increase of a peak, which resembled a cis-diastereomer of 3-hydroxy-β-12'-apocarotenal. The putative cis-diastereomer was present at lower amounts when we incubated the substrate in buffer alone (Fig. 3B, peak 3). Therefore, we cannot exclude that it is formed by thermal isomerization during the incubation process.
Figure 3.
Human BCO2b cleaves apocarotenoids. A, scheme of the reaction catalyzed by BCO2. 3-Hydroxy-12'-carotenal is cleaved at position C9,C10 into 3-hydroxy-β-ionone and 12',10-diapocarotene-12',10-dial. B, representative HPLC traces at 360 nm of assays with BCO2a (blue), BCO2b (orange), and buffer control (green). 2000 pmol of 3-hydroxy-12'-carotenal were incubated with the respective protein extracts for 12 min. Spectral characteristics of peak 1, 12',10-diapocarotene-12',10-dial; peak 2, 3-hydroxy-12'-carotenal; peak 3, putative cis-stereomer of 3-hydroxy-12'-carotenal, and peak 4, 12',10-diapocarotene-12',10-diol are displayed in the lower panel of B.
Human BCO2s contain a mitochondrial target sequence
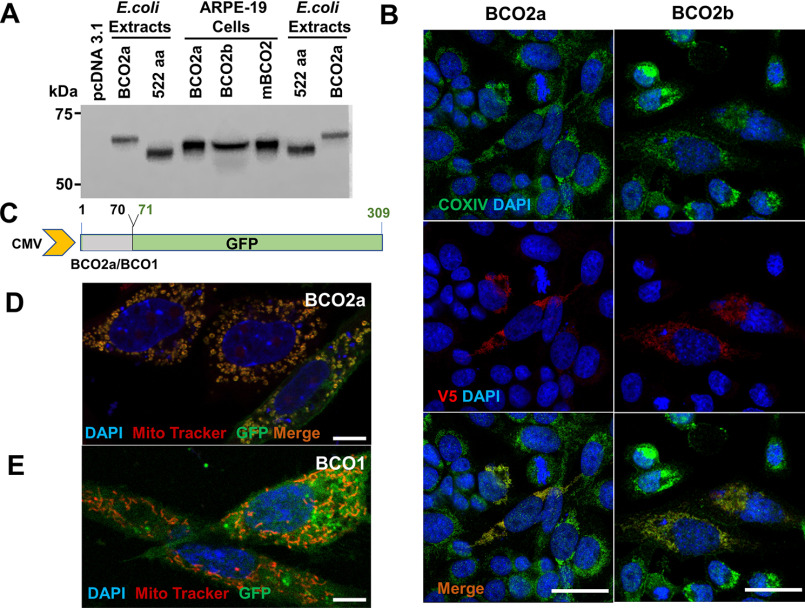
In a previous study, a single 63-kDa BCO2 protein was detected by Western blotting analysis of the human retina protein extracts, likely corresponding to the 579-amino acid long BCO2a (32). In the mouse retina, a 60-kDa BCO2 protein was detected corresponding to the 532-amino acid long murine BCO2 protein (32). However, no controls were included in this study to validate the specificity of the commercial antibody. We and others previously showed that BCO2 is highly expressed in the liver (35, 44). Therefore, we used hepatic protein extracts from WT mice and mice deficient for BCO2 or its closely related BCO1 homolog to validate the commercial antibody. Surprisingly, the antibody detected a band of approximately 50 kDa, independent of the BCO2 genotype of the mouse livers (Fig. S3A). Thus, we next tested it against recombinant BCO2 protein expressed in E. coli. Again, the commercial antibody failed to detect BCO2 but detected an unknown protein of 100 kDa in the bacterial protein extracts (Fig. S3B), demonstrating its ineffectiveness for immunodetection of mammalian BCO2 protein.
We next expressed BCO2a and BCO2b as V5-tagged proteins (70.59 and 66.44 kDa, respectively) in the human retinal pigment epithelium cell line ARPE-19, to study the proteins in a natural cellular environment. Western blotting analysis with an antibody directed against the C-terminal V5 epitopes of the recombinant proteins revealed single bands for the BCO2a and BCO2b (Fig. 4A). Notably, the human BCO2 bands displayed identical electromobility on the SDS-PAGE. Interestingly, the protein migrated with the same size as the V5-tagged mBCO2 protein (65.06 kDa), which we expressed in ARPE-19 cells as a control (Fig. 4A). In contrast, a V5-tagged human BCO2a (69.16 kDa) displayed a significantly different mobility on the same SDS-PAGE when expressed in bacterial cells (Fig. 4A). A recombinant truncated 522-amino acid long human BCO2 migrated with a significantly lower molecular mass when expressed as V5-tagged protein (62.62 kDa), demonstrating that the SDS-PAGE separated proteins in this molecular size range (Fig. 4A).
Figure 4.
Human BCO2 is expressed with a mitochondrial target sequence. A, Western blotting of protein extracts of E. coli and ARPE-19 cells expressing tagged human BCO2 variants. Proteins were detected with anti-V5 antibody. The empty pcDNA3.1 vector was included as control in ARPE-19 cells. An amino-terminal truncated version of human BCO2 (522 aa) was included as a control in E. coli expression analysis. Note that BCO2a displays a slower mobility than that of the 522-amino acid long truncated variant when expressed in E. coli. In ARPE-19 cells, BCO2a displays a similar mobility as BCO2b and mBCO2, indicative of post-translational modification. B, plasmids for the expression of V5-tagged BCO2a and BCO2b were transfected into ARPE-19 cells. Immunostaining was performed with anti-V5 antibody for BCO2a and BCO2b (red), and mitochondria cytochrome oxidase (COXIV) (green). Nuclei were stained with DAPI (blue). Merged images at the bottom show co-localization of BCO2a and COXIV as well as BCO2b and COXIV. Scale bar represents 10 μm. C, scheme of BCO2a-GFP and BCO1-GFP fusion constructs generated using the pcDNA3.1/CT-GFP plasmid. The first 70 amino acids of either BCO2a or BCO1 were fused to GFP downstream of the cytomegalovirus (CMV) promoter. The BCO2a-GFP construct contains the first 70 aa of BCO2a, which includes the N terminus leader sequence, fused to GFP and BCO1-GFP was used as a negative control (D and E). Plasmid constructs for the expression of GFP were transfected into ARPE-19 cells. Cells were stained with MitoTracker to visualize mitochondria. D, fluorescent images shows that signals for mitochondria and BCO2a-GFP are merged. E, fluorescent images shows that signals for mitochondria and BCO1-GFP are located to distinct subcellular regions. Scale bars represent 10 μm in B and 5 μm in D and E.
The identical mobility of BCO2a, BCO2b, and mBCO2 in the ARPE-19 cells suggested that the proteins were post-translationally truncated to yield proteins of similar molecular sizes. To demonstrate that human BCO2a and BCO2b are localized to the mitochondria of ARPE-19 cells, we performed immunohistochemistry. Immunostaining for the V5-tagged protein and confocal imaging revealed that the staining for BCO2a and BCO2b merged with the staining for a mitochondrial marker in this experiment, indicative for its mitochondrial localization (Fig. 4B). To demonstrate more directly that the N-terminal part of BCO2a splice variants serves as MTS, we cloned the sequence encoding the first 70 amino acids of BCO2a in front of the gene encoding a GFP (Fig. 4C). As control, we inserted the sequence encoding the first 70 amino acids of the N-terminal part of human BCO1 into the same GFP reporter vector (Fig. 4C). In contrast to BCO2, the human BCO1 protein resides in the cytoplasm and is not transported to mitochondria (45). In both plasmids, GFP is expressed as fusion protein with an N-terminal part of either BCO1 or BCO2. We transfected the plasmids into ARPE-19 cells and microscopically analyzed the subcellular localization of GFP. The GFP expressed with the N-terminal BCO2a sequence displayed a mitochondrial localization as indicated by co-localization with MitoTracker (Fig. 4D). In contrast, the GFP expressed with the N-terminal BCO1 sequence displayed a cellular distribution as expected for the cytoplasmic BCO1 protein (Fig. 4E).
Removal of mitochondria targeting sequence results in enzymatic activity of BCO2a
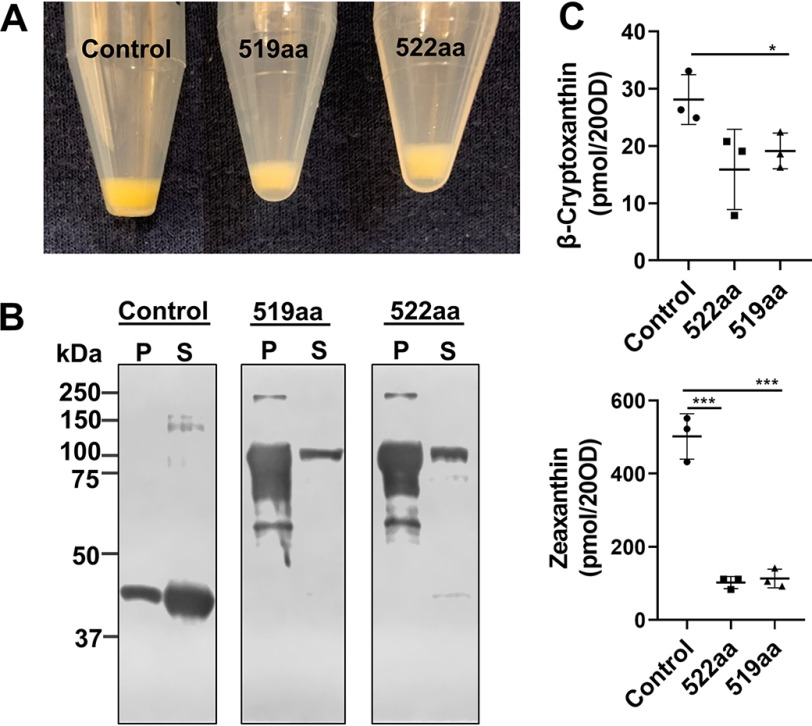
We observed that recombinant BCO2a was largely insoluble when expressed as pre-protein in E. coli cells (Fig. S4). Thus, we applied three modifications to the expression strategy: (i) we removed the mitochondrial targeting sequence from the BCO2a pre-protein (Fig. S5); (ii) we optimized the codon usage to bacterial expression systems; and (iii) we cloned the synthetic gene into the pMAL vector and expressed it as a maltose-binding protein (MBP) fusion. The latter strategy significantly improved the stability of recombinant mBCO2 and yielded higher amounts of soluble protein (Fig. S6). Using the established cell-based assay, the generated BCO2 proteins were expressed in zeaxanthin-producing E. coli cells. When compared with the pMAL vector control, the recombinant 522- and 519-amino acid-long human BCO2 proteins showed enzymatic activity as indicated by a color shift from yellow to pale yellow (Fig. 5A). When compared with the control cells expressing MBP, cells expressing the two BCO2 fusion proteins displayed significantly lower levels of zeaxanthin and its metabolic precursor β-cryptoxanthin (Fig. 5C). To demonstrate that the proteins were expressed in soluble form, an antibody directed against MBP was used for their detection. The Western blotting showed a sizeable amount of the MBP fusion BCO2 proteins in the soluble fraction (Fig. 5B), even though the majority of the protein still existed in the insoluble fraction upon cell lysis (Fig. 5B).
Figure 5.
Removal of the mitochondrial leader sequence renders BCO2a enzymatically active. Enzymatic activity of the synthetic 519- and 522-aa long human BCO2 proteins expressed as maltose-binding protein fusions. The vector expressing the maltose-binding protein was used as control. Both human isoforms were expressed in E. coli cells engineered to produce zeaxanthin. A, colors of bacteria pellets expressing maltose-binding protein (control), 519 aa, and 522 aa. B, Western blotting of protein extracts expressing maltose-binding protein (control), 519 aa, and 522 aa. 10 µg of insoluble (P) and soluble (S) protein fraction was separated per lane. C, quantification of the amounts of zeaxanthin and β-cryptoxanthin in bacteria (20 A600) expressing maltose-binding protein (control), 519 aa, and 522 aa. Values give mean ± S.E. *, indicates significance in unpaired t test for β-cryptoxanthin levels between 519 aa and control p < 0.0441. ***, represent significance for zeaxanthin levels between 522 aa and control p < 0.0004 and 519 aa and control p < 0.0006.
GKAA insertion does not abolish BCO2's enzymatic activity
Human BCO2 also contains a 4-amino acid (GKAA) insertion (Fig. S1, position 169-172 in BCO2a), which is absent in rodents (Fig. S1). It was reported that the insertion abolishes enzymatic activity when introduced in mBCO2, but a detailed analysis of the biochemical properties of the chimeric mouse protein was not provided in this study (32). To determine the effects of the GKAA insertion on BCO2 enzymatic activity, we generated mBCO2 with a GKAA insertion and expressed it as a pMAL fusion protein. As a control, the mBCO2 was PCR amplified and cloned into the same vector. We first applied the cell-based assay to compare enzymatic activity of mBCO2 and mBCO2-GKAA. In cells expressing each plasmid the color shift occurred as a result of zeaxanthin cleavage (Fig. S7A). Western blotting confirmed that both mBCO2 and the chimeric mBCO2-GKAA existed in significant amounts in the soluble fraction (Fig. S7B). After carotenoid extraction, zeaxanthin turnover was confirmed using HPLC analysis (Fig. S7C). In both lipid extracts of mouse BCO2 and mBCO2-GKAA, carotenoids were at the limit of detection level. In contrast, bacteria expressing the MBP control displayed large amounts of β-cryptoxanthin (Fig. S7C, peak 1) and zeaxanthin (Fig. S7C, peak 2).
We next compared enzymatic properties of mBCO2 and the chimeric mBCO2. For this purpose, we purified the recombinant proteins by affinity chromatography (Fig. S8). To characterize whether the GKAA insertion altered enzymatic properties of BCO2, we determined turnover rates with increasing concentrations of 3-hydroxy-β-12'-apocarotenal and 50 μg of purified enzyme (Fig. 6A). We followed the reaction spectroscopically by measuring the decrease in absorption at 420 nm as outlined under “Experimental procedures.” 3-Hydroxy-β-12'-apocarotenal cleavage at position C9,C10 into 12',10-diapocarotene-12',10-dial was also confirmed by HPLC analysis (Fig. S2). The chimeric BCO2 and mBCO2 displayed Michaelis-Menten kinetics under the applied condition. Although mBCO2 displayed a higher Michalis constant for the substrate (36 versus 16 μm), it demonstrated higher maximal velocity for the substrate when compared with the chimeric BCO2 enzyme (782 versus 306 pmol min−1) (Fig. 6A). From these parameters we calculated the turnover rate (kcat) to be 0.8 and 0.4 s−1 for mBCO2 and mBCO2-GKAA, respectively. Thus, our analysis revealed that the GKAA insertion did not abolish the catalytic activity of the mouse protein as previously reported by others (32, 33).
Figure 6.
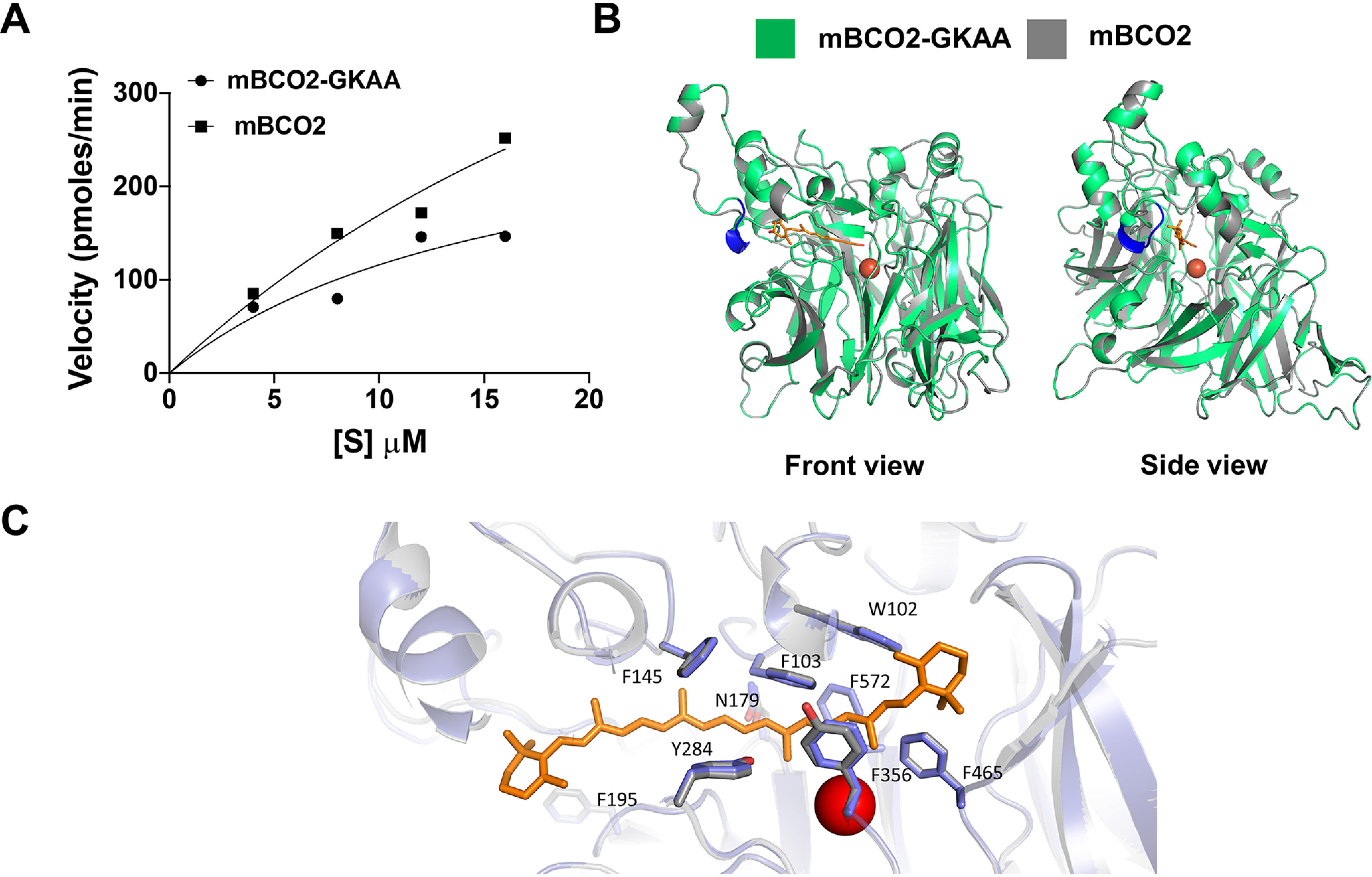
Effect of GKAA insertion on catalytic activity and structure of BCO2. A, purified mBCO2-GKAA and mBCO2 (50 µg/assay each) were incubated in the presence of increasing amounts of 3-hydroxy-β-12'-carotenal as described under “Experimental procedures.” Velocity versus concentration graph demonstrates that both mBCO2 and mBCO2-GKAA displayed Michaelis-Menten kinetics under applied conditions with Vmax value of 306 pmol min−1 and Km value of 16 μm for mBCO2-GKAA and Vmax value of 783 pmol min−1 and a Km value of 36 μm for mBCO2. B, homology modeling of mBCO2-GKAA (green) and mBCO2 (gray) were done using the structure of CCD of candidatus N. devanaterra as a template. Front view shows that the GKAA insertion is present on the surface of BCO2 and away from active site. Side view shows that the GKAA does not hinder docked apocarotenoid molecule in the active site. GKAA insertion, blue; apocarotenoid molecule, orange; Fe2+, red circle. C, magnified view of the substrate tunnels of human (purple) and mBCO2 (gray). The numbering of the amino acids refers to the human enzyme. The ferrous iron in the active center is indicated in red. A docked carotenoid molecule is indicated in orange.
To predict whether the GKAA insertion affected substrate specificity of BCO2, we performed molecular modeling using a crystal structure of an archaeal CCD from candidatus Nitrosotalea devanaterra (NdCCD) (46). This protein shows a similar arrangement of amino acid side residues as vertebrate CCDs in the substrate tunnel and its structure was resolved with a bound carotenoid substrate (46). As shown in Fig. 6B, GKAA insertion is present on the surface of BCO2 and away from active site. Side view of the model shows that the GKAA insertion did not hinder docking of 3-hydroxy-β-12'-apocarotenal into the substrate tunnel of mBCO2 and the chimeric mBCO2 (Fig. 6A). Furthermore, modeling revealed that the arrangement of amino acids in the substrate tunnel is well conserved between mBCO2 and human BCO2 (Fig. 6C). To demonstrate that the structural predictions translate into catalytic activity, we tested several carotenoid substrates with mouse BCO2 and the its chimeric counterpart. In a first set of assays we incubated the proteins with detergent micelles that contained 3(R),3'(R)-β,β-carotene-diol, 3(R),3'(S)-β,β-carotene-diol, and 3(S),3'(S)-β,β-carotene-diol. After the reaction, carotenoids and apocarotenoids were extracted and separated by HPLC analysis. All three enantiomers were converted by the recombinant mBCO2 and its chimeric variant (Fig. 7 and Fig. S9). The major cleavage products were 10',10-diapocarotene-10,10'-dial and 3-hydroxy-β-ionone. Minor cleavage product was 3-hydroxy-β-apo-10'-carotenal. The identity of the different cleavage products of zeaxanthin enantiomers was confirmed by the spectral characteristics as well as by the specific masses of these compounds by LC–MS analysis (Fig. 7 and Fig. S9).
Figure 7.
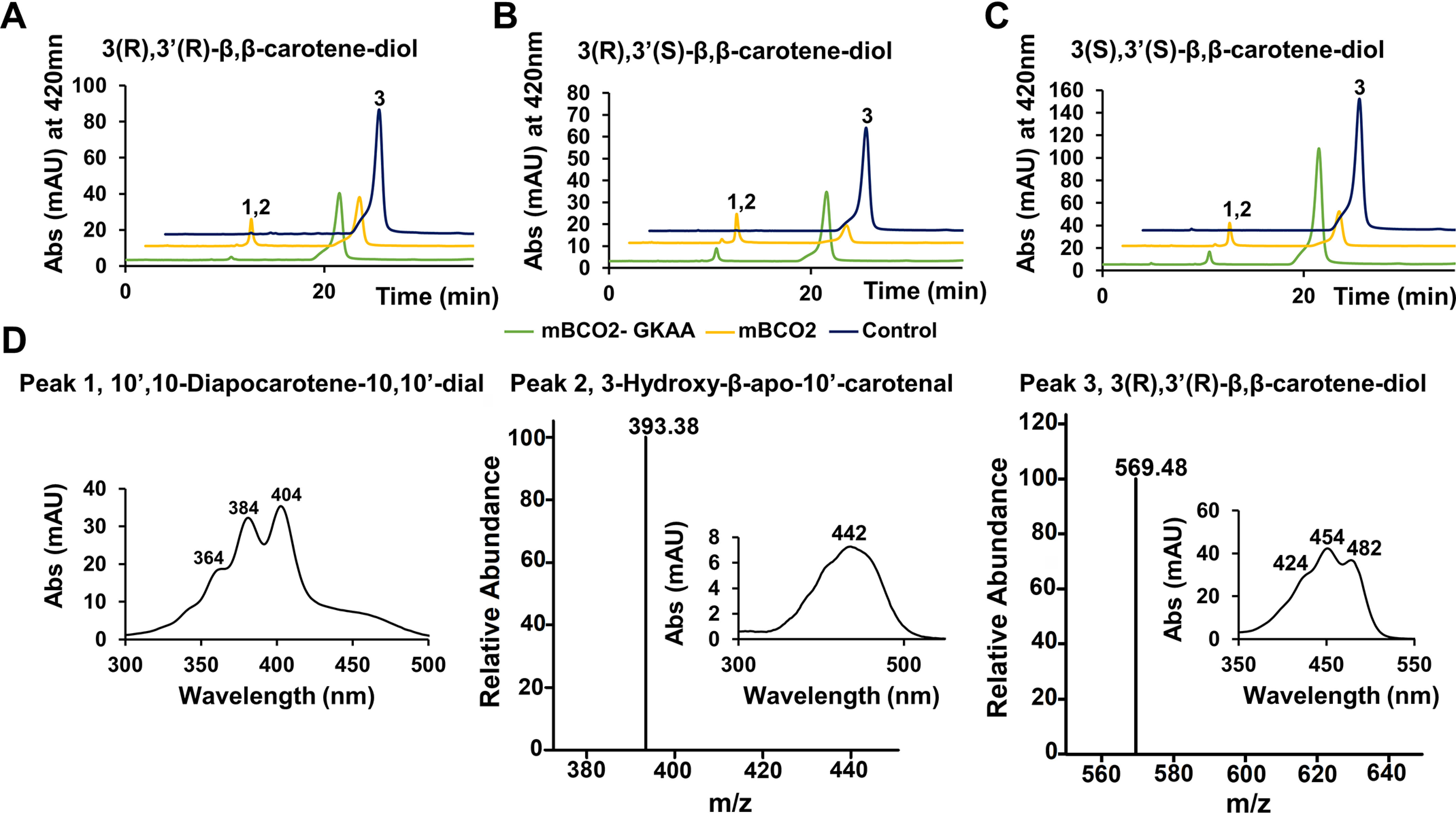
Conversion of zeaxanthin enantiomers by BCO2. Purified (50 µg) mBCO2 (yellow traces) and mBCO2-GKAA (green traces) were incubated with substrate. Buffer incubations (navy traces) served as control. The reactions were carried out for 10 min. Lipids were extracted and subjected to HPLC analysis. The figure presents HPLC traces at 420 nm for the assays with (A) 3(R),3'(R)-β,β-carotene-diol, (B) 3(R),3'(S)-β,β-carotene-diol, and (C) 3(S),3'(S)-β,β-carotene-diol. D, representative spectra of peak 1, 10',10-diapocarotene-10',10-dial (left), peak 2, 3-hydroxy-β-apo-10'-carotenal with corresponding representative MS-MS spectra (middle), and peak 3, 3(R),3'(R)-β,β-carotene-diol with the corresponding MS spectra (right). Note that the spectral characteristics of the zeaxanthin enantiomers are identical.
We used mouse liver to isolate oxidized zeaxanthin metabolites, including ε,ε-carotene-3,3′-dione and β,ε-3-hydroxy-carotene-3'-one. We and others (25, 47) previously showed that these oxidation products accumulate in mouse tissues upon zeaxanthin supplementation. We then employed the isolated zeaxanthin metabolites in tests for enzymatic activity with recombinant mouse BCO2 variants. As shown in Fig. 8, mBCO2 and mBCO2-GKAA readily converted these oxidized metabolites into apocarotenoids. Thus, the GKAA insertion had no effect on the binding and turnover of a large variety of zeaxanthin derivatives that naturally exits in the retina and other mammalian tissues (47–49).
Figure 8.
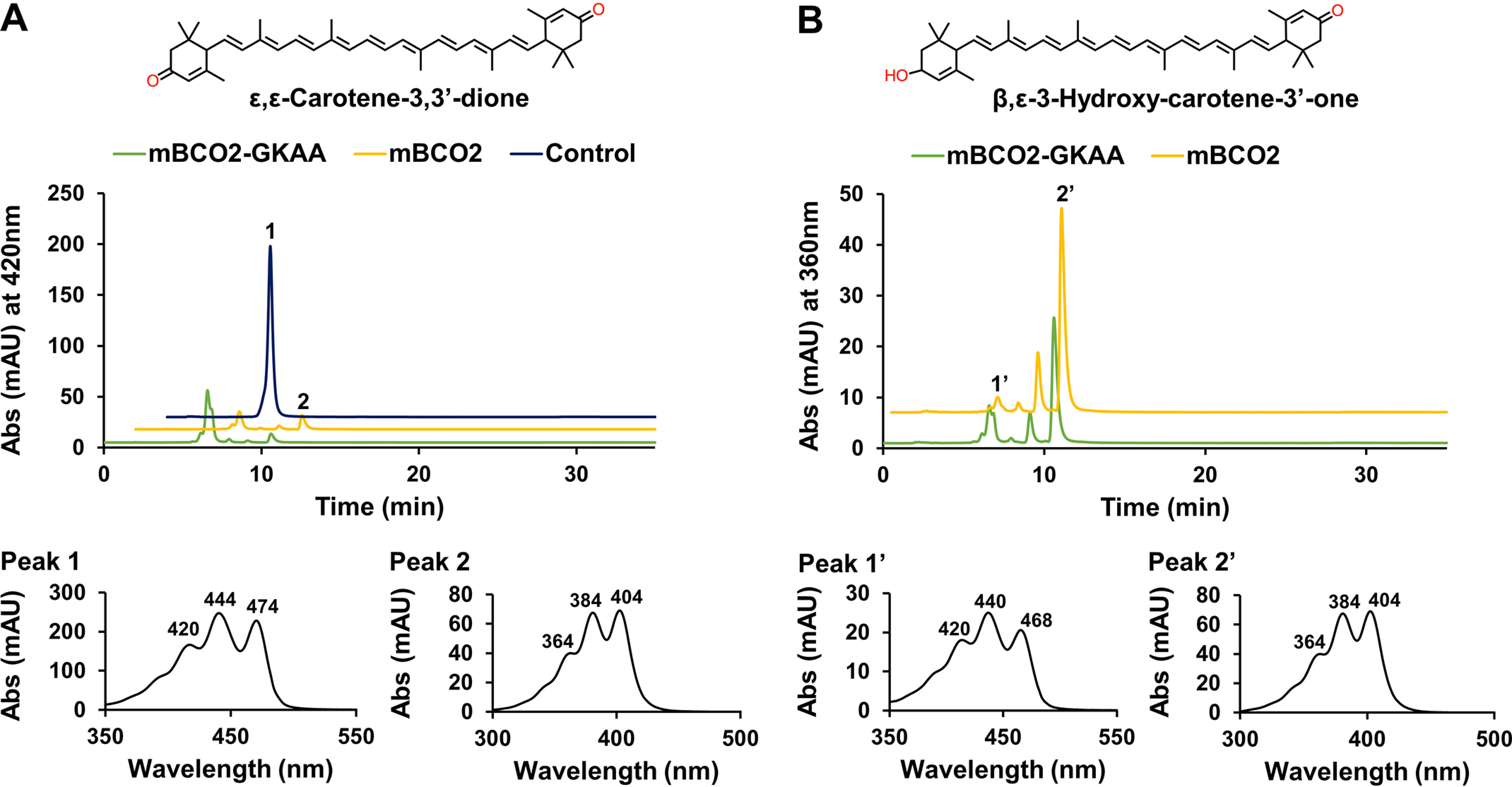
Conversion of oxidized zeaxanthin metabolites by BCO2. Purified (50 µg) mBCO2 (yellow traces) and mBCO2-GKAA (green traces) were incubated with oxidized zeaxanthin metabolites isolated from mouse liver. Buffer incubations (navy trace) served as controls. The reactions were carried out for 10 min. Lipids were extracted and subjected to HPLC analysis. The figure presents HPLC traces, respectively, at 420 and 360 nm, for the reactions with (A) assays with ε,ε-carotene-3,3'-dione (upper panel) and spectral characteristics of peak 1, ε,ε-carotene-3,3'-dione and peak 2, 10',10-diapocarotene-10',10-dial (lower panel), and (B) assays with 3-hydroxy-β,ε-carotene-3'-one (upper panel) and spectral characteristics of peak 1, 3-hydroxy-β,ε-carotene-3'-one and peak 2, 10',10-diapocarotene-10',10-dial (lower panel).
Discussion
Human genomes encode three members of the CCD superfamily of nonheme iron oxygenases (50). These structurally conserved enzymes catalyze reactions at the aqueous lipid interface and convert their substrates into more polar products. The challenging biochemistry of CCDs had led to many conflicting results about these enzymes' functions. For instance, RPE65, the first member of the vertebrate CCD family member to be molecularly identified (51), was initially characterized as retinoid-binding protein rather than an enzyme catalyst (52, 53). Other studies even questioned the existence of BCO1 enzyme for vitamin A production (54). Moreover, two reports concluded that the human BCO2 gene encodes an enzymatically inactive protein (32, 33), indicating that BCO2 functions in carotenoid catabolism and vitamin A production are not conserved in primates. We here showed that human BCO2 variants are expressed as pre-proteins with MTS. Removal of this sequence from BCO2a rendered the recombinant enzyme soluble and active. We further show that a primate-specific GKAA insertion does not impede substrate specificity and catalytic activity of a chimeric mBCO2 enzyme. Thus, we demonstrate that BCO2 biochemistry and physiological functions are evolutionarily well conserved in the primate lineage.
Improving the expression of recombinant CCDs
A critical requirement for the biochemical characterization of CCDs is a robust and reliable expression system for the recombinant proteins. We here established protocols to express human BCO2 in soluble and enzymatically active forms. With the pTrc expression system, we initially confirmed that BCO2a (579 aa) is inactive when expressed in zeaxanthin producing bacteria. However, human BCO2b (545 aa) was active in this test system. We provided evidence that inactivity of BCO2a is caused by the MTS. We showed that this N-terminal sequence targets a GFP reporter into mitochondria of ARPE-19 cells. In contrast, a GFP fused to the N-terminal part of BCO1 was targeted to the cytoplasm. We also provided evidence that the MTS of BCO2 is removed during mitochondrial import because BCO2a, BCO2b, and mBCO2 displayed the same molecular size when expressed in ARPE-19. In contrast, BCO2a migrated with a significantly higher molecular weight when expressed in bacterial cells where this processing does not take place.
Removal of the MTS increased solubility of the recombinant BCO2 and resulted in a catalytically active enzyme. This activity was further enhanced by the fusion of the BCO2 ORF with MBP that further improved the solubility of the protein. Similarly, biochemical characterization of ferret, chicken, and macaque BCO2 were previously achieved with N terminally truncated proteins (23, 33, 55). Only human BCO2b (545 aa) and mBCO2 (532 aa) (25) displayed enzymatic activity when expressed with their MTS. However, both proteins exhibit a significantly shorter MTS than BCO2a, macaque, chicken, and ferret BCO2.
Primate BCO2s also exhibit a 4-amino acid long residue GKAA insertion at position 169-172 (numbering refers to BCO2a), which is not present in other mammalian BCO2s. It was proposed that this insertion inactivates the human enzyme by sterically interfering with substrate binding (32). Moreover, it was reported that introduction of these 4 amino acids into mBCO2 inactivates the protein (32). To investigate the putative structural changes brought on by the GKAA insertion, we modeled BCO2s using the recently solved crystal of NdCCD (46). Upon homology modeling, no marked structural difference between primate and rodent BCO2s were observed. The GKAA insertion did localize away from the substrate tunnel and did not pose an obstruction to the substrate tunnel. Additionally, we showed the atomic structure of the substrate tunnel is well conserved between the human and mouse enzymes. Test for enzymatic activity revealed that chimeric mBCO2 is catalytically competent and displayed broad substrate specificity for zeaxanthin enantiomers and oxidation products. Kinetic tests with an apocarotenoid substrate showed that the chimeric BCO2 enzyme exhibited a lower turnover rate when compared with mBCO2 (kcat 0.4 × s−1 versus 0.9 × s−1). In general, the very low turnover rates of BCO2 make it very challenging to characterize this type of enzyme. In enzyme assays, relatively large amounts of proteins must be incubated to produce apocarotenoid product concentrations that become detectable in colorimetric assays. The lower extinction coefficient of the apocarotenoid products and its chemical instability also complicates its detection. These problems likely contributed to the difficulties in characterizing human BCO2 in previous studies. Improved expression methods, as presented here, eventually overcame these challenges. Ultra-HPLC and more sensitive detection systems for the apocarotenoid products may further improve biochemical assays in futures studies.
Implications for carotenoid biology
Through our demonstration that human BCO2 is a catalytically competent enzyme, all of the prior physiological information on BCO2 becomes applicable in human biology. The BCO2-dependent catabolism of carotenoids provides an explanation for the observation that serum and tissue levels of xanthophyll reaches steady-state levels during supplementation (56). Additionally, it provides a mechanism for the catabolism of oxidized carotenoid metabolites. We and others have shown that these compounds are produced from carotenoids and that their accumulation is associated with adverse health effects in adult vertebrates (25, 57, 58) and their offspring (59, 60). We also showed that BCO2 protected human cell lines from oxidative stress induced by carotenoids (59). Oxidized carotenoids also exist in the human retina and their occurrence is associated with blue light, superoxide anions, and free radicals (48, 61, 62). As we showed here, BCO2 readily converts oxidized zeaxanthin metabolites into two ionones and a C14-apocarotendial by oxidative cleavage at positions C9,C10 and C9',C10' of the parent carotenoids. Conversion of lipids to more polar products is a textbook example for the catabolism of lipophilic xenobiotics. One can assume that the formed apocarotenoid products are available for further enzymatic modifications that eventually lead to their excretion from the body. Thus, our studies indicate that BCO2's role as scavenger of carotenoids and guardian of lipid homeostasis is well conserved in humans.
A previous study reported that BCO2a is the major transcript of the BCO2 gene in the retina of the human eyes (32). We here detected both BCO2a and BCO2b transcripts in RNA preparation of retinas of human donor eyes, indicating that individual differences exist in the expression of BCO2 splice variants. A recent study showed that BCO2 mRNA is highly expressed in the peripheral retina and at a low level in the central retina (63), consistent with the distribution of macula pigments. It remains to be investigated whether the different BCO2 splice variants display a differential expression in central and peripheral parts of the retina. However, our finding that BCO2a and BCO2b mRNA encode mature proteins of similar molecular sizes, indicate that there is no functional difference between these splice variants.
Differential expression of the BCO2 gene may contribute to carotenoid biology in mammals. For instance, rodents with low carotenoid serum and tissue levels express BCO2 in the gastrointestinal tract (22, 64). In contrast, BCO2 expression is very low in the gastrointestinal tract of humans (44). This difference indicates that rodents catabolize absorbed carotenoids in the gut, whereas humans absorb them intact for the distribution to peripheral tissues. In chicken, a polymorphism in the Bco2 promoter region defines skin color (17) and variability in Bco2 expression contributes to sexual dichromatism across finches (65). In lizards, skin ornamentation is also dependent on BCO2 expression (66). The identification of the molecular components that regulate BCO2 mRNA expression will provide answers to the molecular basis of this diversity in vertebrate BCO2 biology.
In conclusion, we established methods to express recombinant human BCO2 in an enzymatically active form. Removal of a MTS from the recombinant protein was instrumental in this endeavor. We also showed that a GKAA insertion did not render mouse BCO2 enzyme inactive or impeded its broad substrate specificity. Thus, we provide evidence that major vertebrate pathways for vitamin A production and carotenoid homeostasis are well conserved during primate evolution.
Experimental procedures
Materials
All chemicals, unless otherwise stated, were purchased from Fisher and Sigma Aldrich. Dulbecco's modified Eagle's medium, fetal bovine serum, and TRIzol RNA isolation reagent were obtained from Invitrogen. The anti-mouse and anti-rabbit horseradish peroxidase-conjugated secondary antibodies were purchased from Promega (Madison, WI). RIPA buffer was purchased from Cell Signaling Technology (Danvers, MA). Carotenoids and apocarotenoids were a gift from DSM (Sisseln, Switzerland). Plasmid and RNA isolation kits were purchased from Qiagen (Hilden, Germany).
Human retina samples
Human retina samples were gifted by Dr. Min Hyung Kang (Ophthalmology, Case Western Reserve University). Donor eyes were acquired from Lions Gift of Sight (formerly referred to as Minnesota Lions Eye Bank), St. Paul, MN. They were obtained with the written consent of the donor or donor's family for medical research in compliance with the Declaration of Helsinki. Lions Gift of Sight is licensed by the Eye Bank Association of America (accreditation number 0015204) and accredited by the FDA (FDA Established Identifier 3000718538). The retina was dissected and used for total RNA preparations using TRIzol RNA isolation reagent (Invitrogen).
RT-PCR analysis of human BCO2 expression
The following forward primers were used for RT-PCR analysis of human BCO2 expression: 5′-AGGAGTCATTCTGCCACTGC-3′ for BCO2a (579 aa), 5′-GACATGGCAACTGGATCTTGAAGG-3′ for BCO2b (545 aa), and 5′-CAACTGGATCTTGAAGGATGAG-3′ for BCO2c (539 aa). The reverse primer used for all human BCO2 isoforms was 5′-TCTCTGGAGGAACCCGAATAACC-3′. RHO, GAPDH, and ACTB were used as positive standards. RHO forward and reverse primers were: 5′-CCATCATGGGCGTTGCCTTC-3′ and 5′-TCGTTGTTGACCTCCGGCTT-3′, respectively. GAPDH forward and reverse primers were: 5′-ACAACTTTGGTATCGTGGAAGG-3′ and 5′-GCCATCACGCCACAGTTTC-3′, respectively. ACTB forward and reverse primers were: 5′-CTGGAACGGTGAAGGTGACA-3′ and 5′-AAGGGACTTCCTGTAACAATGCA-3′, respectively.
BCO2 plasmid constructs for E. coli expression
Untagged full-length human BCO2a (579 aa) and human BCO2b (545 aa) isoforms (Fig. 1A) were previously generated and cloned into the pTrcHis2 expression vectors (Invitrogen) (35). Synthetic codon usage optimized human BCO2 variants, 522 and 519 aa, were custom made by GenScript (Piscataway, NJ). These synthetic BCO2 variants were subcloned into the pMAL-c5x vector (New England Biolabs, Ipswich, MA). The forward primers used included: 5'-ACCACCGTTGAGGAAGCGC-3′ for the 522-aa isoform and 5′-GAGGAAGCGCCGCGTGGTATC-3' for the 519-aa isoform. The reverse primer used for both isoforms was 5'-AAAACCTGCAGGAATCGGGATAAAGGTACCATGAAAG-3' (Invitrogen). 522- and 519-aa variants were cloned using the XmnI and SbfI restriction sites. Mouse BCO2 was cloned into pMAL-c5x vector (New England Biolabs) using the forward primer, 5′-AAAGAAGGAATTGTTGGGACCGAAGCAGAGCCTGCCATGC-3′ and the reverse primer, 5′-AAAACCTGCAGGGATAGGCACAAAGGTGCCATGGAACC-3′ into the XmnI and SbfI restriction sites. A chimeric mouse BCO2 with the 4-amino acid insertion, GKAA, was custom made by GenScript (Piscataway, NJ). Mouse BCO2 with the GKAA insert was cloned into the pMAL-c5x vector using BamHI and HindIII as the restriction sites. The sequences of all plasmid constructs were verified by Sanger sequencing.
Western blotting
Proteins (10-20 μg) were separated on 6–12% SDS-PAGE gels using the Bio-Rad Mini gel system and transferred onto polyvinylidene fluoride membranes (Roche, Basel, Switzerland). Polyvinylidene fluoride membranes were blocked with 5% skim milk prepared in Tris-buffered saline (pH 7.4) containing 0.1% Tween for 1 h and then probed with either commercial anti-BCO2 (Proteintech Group, catalog number 14323-1-AP) or anti-V5 antibody (Invitrogen, catalog number R960-25), or anti–maltose-binding protein antibody (New England Biolabs, catalog number E8032S) (1:1000 dilution) overnight at 4 °C, followed by incubation with the appropriate horseradish peroxidase-conjugated secondary antibody before being visualized with the enhanced ECL chemiluminescence detection system (Pierce or Pharmacia).
Cell-based BCO2 enzymatic activity
XL1-Blue strain of E. coli, engineered to produce the zeaxanthin (43), was transformed with the different BCO2 expression plasmids. Zeaxanthin producing E. coli cells were grown in lysogeny broth (LB) medium at 37 °C. Once an optical density of 1 absorbance unit at 600 nm (1 A600) was achieved, bacterial cell cultures were transferred to 27 °C for 1 h to achieve production of zeaxanthin. To induce expression of BCO2 isoforms, isopropyl β-d-1-thiogalactopyranoside was added to a final concentration of 0.3 mm. Ferrous sulfate at the concentration of 25 mg/liter was added at the time of induction. Expression was continued at 16 °C for 24 h. Cells were harvested by centrifugation at 4000 × g for 15 min at 4 °C. Carotenoid extraction from the bacterial pellet was performed under dim red light (<600 nm). A bacteria pellet (20 A600) was dissolved in 250 μl of PBS buffer. Then 400 μl of acetone, 400 μl of diethyl ether, and 100 μl of petroleum ether was used to extract carotenoids. Phase separation was achieved by centrifugation at 4000 × g for 30 s. The organic phase was collected and the extraction was repeated. The combined organic phases were collected and dried in a SpeedVac (Eppendorf, Hauppauge, NY). Lipids were dissolved in 150 μl of hexane:ethyl acetate (70:30, v/v) and subjected to HPLC analysis.
Production of recombinant BCO2 in bacteria
BL21 E. coli cells were transformed with the respective BCO2 plasmid constructs. E. coli cells were grown in LB medium at 37 °C. Once an optical density of 1 absorbance unit (AU) at 600 nm was achieved, expression of BCO2 isoforms were induced with a final concentration of 0.3 mm isopropyl β-d-1-thiogalactopyranoside and transferred to 16 °C for 24 h. Ferrous sulfate at the concentration of 25 mg/liter was added at the time of induction. After expression, cells were collected by centrifugation at 4000 × g for 15 min at 4 °C and snap frozen at −80 °C until further use. Cell pellets of a 500-ml culture were thawed on ice. The pellet was dissolved in enzyme assay buffer (20 mm Tricine, 150 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, 0.5 mm TCEP, pH 7.4). Then, 20 μl of lysozyme solution (10 mg/ml) was added. After mixing, the solution was incubated for 30 min on ice. Following incubation, the solution was sonicated four times at 35% intensity for 1 min with a 1-min break period between each sonication period using the Q125 Sonicator (QSonica, Newtown, CT). The lysed bacteria were subjected to centrifugation (20,000 × g) for 1 h at 4 °C to separate soluble and insoluble fractions (mBCO2-GKAA). MBP affinity chromatography was used to purify murine BCO2-MBP fusion proteins (mBCO2 and mBCO2-GKAA). Briefly, amylose resin was packed into a 2.5 × 10-cm column. 5 column volumes of column buffer (20 mm Tricine, 150 mm NaCl, 0.5 mm TCEP, pH 7.4) was used to wash the column. Crude soluble fraction was then loaded onto the column at a flow rate of 2 ml min−1. The column was then washed with 12 column volumes of cold column buffer. Column buffer made with the addition of 10 mm maltose was used to elute the fusion protein. Protein-containing fractions were then pooled together and analyzed by SDS-PAGE gel for purity.
Cell-free BCO2 enzyme assay
For enzyme assays with 3-hydroxy-β-12'-carotenal, 100 μl of soluble cell lysate was combined with 100 μl of enzyme assay buffer containing 0.2% (v/v) Triton X-100. Alternatively, 50 μg of purified BCO2 protein was incubated in enzyme assay buffer containing 0.2% (v/v) Triton X-100. In standard assays substrate 3-hydroxy-β-12'-carotenal dissolved in ethanol was added to reach a final concentration of 10 μm. In kinetic analysis with purified mBCO2, different concentrations of substrate (4–16 μm final concentration) were used. Assays with carotenoids were performed as previously described (23). Assays were incubated at 37 °C on a thermomixer (Eppendorf, Hamburg, Germany) under constant shaking (600 rpm). In standard tests, the reaction was stopped after 12 min by adding 400 μl of acetone, 400 μl of diethyl ether, 100 μl of petroleum ether, and lipids were extracted as described above and subjected to HPLC analyses. For kinetic analysis with mouse BCO2 variants, the reaction was upscaled to 500 μl and the decrease of substrate was followed in a spectrophotometer in real time (Shimadzu, Kyoto, Japan) at 420 nm. After the reaction, product formation was verified by lipid extraction and HPLC analysis.
HPLC analysis and MS analysis
Extracted lipids were separated by HPLC performed on a 1200 Agilent HPLC series equipped with a diode array detector and normal-phase Agilent Zorbax silica column (4.6 mm ID × 150 mm with 5 μm packing; Agilent, Santa Clara, CA). Chromatographic separation was achieved with isocratic flow of 30% ethyl acetate in hexane at a flow rate of 1.4 ml/min. The system was scaled with known amounts of authentic zeaxanthin, β-crytoxanthin, and 3-hydroxy-12'-β-apocarotenal standards, which were acquired from DSM (Sisseln, Switzerland). The products of BCO2 enzymatic activity were identified based on their elution time, characteristic UV-visible spectra, and molecular masses. For MS analysis, separated lipids extracted from enzymatic reactions were separated as described above and directed into LTQ linear ion trap mass spectrometer equipped with an atmospheric pressure chemical ionization probe (Thermo Fisher Scientific). The MS signal acquisition was performed in positive ionization mode and the MS parameters were optimized for all-trans-retinol.
BCO2 transfection and expression in ARPE19 cells
We used pcDNA3.1 expression plasmid for the 579- (BCO2a) and 545- (BCO2b) amino acid long human BCO2 proteins as well as the 532-amino acid mouse BCO2 (mBCO2). The construction of these plasmids has been previously described (35). To confirm the function of human BCO2's MTS, we used the pcDNA3.1/CT-GFP (Invitrogen) expression plasmid to generate a BCO2a-GFP fusion construct that encoded the first 70 amino acids of BCO2a fused to GFP. The forward and reverse primers used for this were: 5′-AGGATGGTTTTTCGAGTCTTTCTCC-3′ and 5′-TAGCAGAGATGCCCCGTGGAGCC-3′, respectively. A BCO1-GFP construct was used as a control and generated in the same manner as BCO2a-GFP. The forward and reverse primers used were: 5′-AGGATGGATATAATATTTGGCAGGAATAG-3′ and 5′-TCCTGTAATAGACTTCACCGTCTC-3′, respectively. ARPE-19 cells were maintained in Dulbecco's modified Eagle's medium F-12 with 10% fetal bovine serum and 10 units/ml of penicillin-streptomycin antibiotics, at 37 °C with 5% CO2. For transfection, cells were grown to 50–70% confluence and then transfected with purified plasmid using TransIT-X2® Transfection Reagent (Mirus Bio). At 24 h post-transfection, cells were either used for in situ immunocytochemistry or protein isolation. To isolate total protein, cells were scrapped then centrifuged at 300 × g for 5 min at 4 °C. Cell pellets were dissolved in Nonidet P-40 lysis buffer and incubated on ice for 10 min. The solution was then centrifuged again at 10,000 × g for 10 min at 4 °C. The supernatant was stored at −80 °C for further analysis.
Confocal imaging
Transfected ARPE-19 cells grown on polylysine-treated 8-chamber slides glass coverslips were washed and fixed with freshly prepared solution of 4% paraformaldehyde in PBS (137 mm NaCl, 2.7 mm KCl, 4.3 mm Na2HPO4, and 1.4 mm KH2PO4, pH 7.3) for 30 min at room temperature. Cells were washed and permeabilized with 0.1% Triton X-100 (Roche Applied Science) in PBS and blocked with 4% normal goat serum (Sigma) in PBS-T (blocking buffer). Cells were then incubated overnight at 4 °C in blocking buffer containing mouse anti-V5 serum (to detect BCO2; Invitrogen) and rabbit anti-COX IV serum (Cell Signaling, Boston, MA) at 1:200 dilution. Cells were then washed and incubated with anti-mouse and anti-rabbit secondary antibody conjugated to Alexa 594 and Alexa 488 (Life Technologies, Grand Island, NY), respectively, diluted 1:500 in blocking buffer at room temperature for 1 h. DAPI was used to stain nuclei. For staining of mitochondria, transfected ARPE-19 cells were incubated with 200 nm MitoTracker® Orange CMTMRos (Molecular Probes, Invitrogen) for 30 min in PBS at 37 °C, 5% CO2 in CO2 incubator. Cells were then washed with PBS and fixed in freshly prepared solution of 4% paraformaldehyde in PBS for 30 min at room temperature. Cells were then washed with PBS and mounted with DAPI fluormount (SouthernBiotech). All confocal images were acquired with a Leica TCS SP8 HyVolution2 confocal microscope by using a multiline argon laser (excitation 488 and 594 nm) or a 405-diode laser (excitation 405) with a ×63 C-Apochromat, NA 1.2-O objective.
Homology modeling of mouse BCO2 (mBCO2) and humanized mouse BCO2 (mBCO2-GKAA)
Swiss modeling was used to build a homology model for the different BCO2 enzymes (67). Then, carotenoid and apocarotenoids were modeled into the substrate-binding cavity using NdCCD structure (PDB accession number 6VCH) (46) and Coot software. PyMol software was used to align structures and create figures.
Data availability
All data are included within the manuscript and supporting information.
Supplementary Material
Acknowledgments
The Leica TCS SP8 confocal used for these studies was made available in the SOM Light Microscopy Imaging Core supported by National Institutes of Health Grant ORIP S10-OD024996. Technical assistance was provided by Richard Lee. We thank Dr. Min Hyung Kang (Ophthalmology, Case Western Reserve University) for sharing retinal samples for RNA isolation. We thank Dr. Adrian Wyss (DSM) for the gift of 3-hydroxy-β-12-apocarotenal and zeaxanthin enantiomers.
This article contains supporting information.
Author contributions—L. D. T., S. B., V. M. P., R. S., N. K., M. G., P. D. K., and J. v. L. data curation; L. D. T., S. B., V. M. P., R. S., N. K., M. G., P. D. K., and J. v. L. formal analysis; L. D. T. investigation; L. D. T., P. D. K., and J. v. L. writing-original draft; V. M. P., R. S., N. K., M. G., P. D. K., and J. v. L. methodology; V. M. P., R. S., and N. K. project administration; N. K., M. G., P. D. K., and J. v. L. funding acquisition; M. G., P. D. K., and J. v. L. conceptualization.
Funding and additional information—This work was supported by National Eye Institute Grants EY020551, EY028121, and EY007157 (to J. V. L.), EY023948 (to M. G.), and P30 Core Grant EY011373 and Department of Veterans Affairs Grant I01BX004939 (to P. D. K.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- CCD
- carotenoid cleavage dioxygenase
- BCO1
- β-carotene oxygenase 1
- BCO2
- β-carotene oxygenase 2
- RPE65
- retinal pigment epithelium-specific 65-kDa protein
- RHO
- rhodopsin
- ACTB
- β-actin
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- MBP
- maltose-binding protein
- MTS
- mitochondrial targeting sequence
- NdCCD
- Nitrosotalea devanaterra carotenoid cleavage dioxygenase
- TCEP
- tris(2-carboxyethyl)phosphine hydrochloride
- aa
- amino acid(s)
- DAPI
- 4′,6-diamidino-2-phenylindole
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- AU
- absorbance unit.
References
- 1. von Lintig J., Moon J., and Babino D. (2020) Molecular components affecting ocular carotenoid and retinoid homeostasis. Prog. Retin. Eye Res. 100864 10.1016/j.preteyeres.2020.100864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Miller A. P., Coronel J., and Amengual J. (2020) The role of β-carotene and vitamin A in atherogenesis: evidences from preclinical and clinical studies. Biochim. Biophys. Acta 1865, 158635 10.1016/j.bbalip.2020.158635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bernstein P. S., Li B., Vachali P. P., Gorusupudi A., Shyam R., Henriksen B. S., and Nolan J. M. (2016) Lutein, zeaxanthin, and meso-zeaxanthin: the basic and clinical science underlying carotenoid-based nutritional interventions against ocular disease. Prog. Retin. Eye Res. 50, 34–66 10.1016/j.preteyeres.2015.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mares J. (2016) Lutein and zeaxanthin isomers in eye health and disease. Annu. Rev. Nutr. 36, 571–602 10.1146/annurev-nutr-071715-051110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Grune T., Lietz G., Palou A., Ross A. C., Stahl W., Tang G., Thurnham D., Yin S. A., and Biesalski H. K. (2010) β-Carotene is an important vitamin A source for humans. J. Nutr. 140, 2268S–2285S 10.3945/jn.109.119024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Johnson E. J. (2014) Role of lutein and zeaxanthin in visual and cognitive function throughout the lifespan. Nutr. Rev. 72, 605–612 10.1111/nure.12133 [DOI] [PubMed] [Google Scholar]
- 7. Krinsky N. I., and Johnson E. J. (2005) Carotenoid actions and their relation to health and disease. Mol. Aspects Med. 26, 459–516 10.1016/j.mam.2005.10.001 [DOI] [PubMed] [Google Scholar]
- 8. Widjaja-Adhi M. A. K., Ramkumar S., and von Lintig J. (2018) Protective role of carotenoids in the visual cycle. FASEB J. 32, fj201800467R 10.1096/fj.201800467R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barker F. M. 2nd, Snodderly D. M., Johnson E. J., Schalch W., Koepcke W., Gerss J., and Neuringer M. (2011) Nutritional manipulation of primate retinas, V: effects of lutein, zeaxanthin, and n-3 fatty acids on retinal sensitivity to blue-light-induced damage. Invest. Ophthalmol. Vis. Sci. 52, 3934–3942 10.1167/iovs.10-5898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hammond B. R. Jr., Fletcher L. M., and Elliott J. G. (2013) Glare disability, photostress recovery, and chromatic contrast: relation to macular pigment and serum lutein and zeaxanthin. Invest. Ophthalmol. Vis. Sci. 54, 476–481 10.1167/iovs.12-10411 [DOI] [PubMed] [Google Scholar]
- 11. von Lintig J., Moon J., Lee J., and Ramkumar S. (2020) Carotenoid metabolism at the intestinal barrier. Biochim. Biophys. Acta 1865, 158580 10.1016/j.bbalip.2019.158580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harrison E. H. (2019) Mechanisms of transport and delivery of vitamin A and carotenoids to the retinal pigment epithelium. Mol. Nutr. Food Res. 63, e1801046 10.1002/mnfr.201801046 [DOI] [PubMed] [Google Scholar]
- 13. Kiefer C., Sumser E., Wernet M. F., and Von Lintig J. (2002) A class B scavenger receptor mediates the cellular uptake of carotenoids in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 99, 10581–10586 10.1073/pnas.162182899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Toomey M. B., Lopes R. J., Araujo P. M., Johnson J. D., Gazda M. A., Afonso S., Mota P. G., Koch R. E., Hill G. E., Corbo J. C., and Carneiro M. (2017) High-density lipoprotein receptor SCARB1 is required for carotenoid coloration in birds. Proc. Natl. Acad. Sci. U.S.A. 114, 5219–5224 10.1073/pnas.1700751114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sakudoh T., Sezutsu H., Nakashima T., Kobayashi I., Fujimoto H., Uchino K., Banno Y., Iwano H., Maekawa H., Tamura T., Kataoka H., and Tsuchida K. (2007) Carotenoid silk coloration is controlled by a carotenoid-binding protein, a product of the yellow blood gene. Proc. Natl. Acad. Sci. U.S.A. 104, 8941–8946 10.1073/pnas.0702860104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li B., Vachali P., Frederick J. M., and Bernstein P. S. (2011) Identification of StARD3 as a lutein-binding protein in the macula of the primate retina. Biochemistry 50, 2541–2549 10.1021/bi101906y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eriksson J., Larson G., Gunnarson U., Bed'hom B., Tixier-Boichard M., Strömstedt L., Wright D., Jungerius A., Vereijken A., Randi E., Jensen P., and Anderson L. (2008) Identification of the yellow skin gene reveals a hybrid origin of the domestic chicken. PLoS Genet. 4, e1000010 10.1371/journal.pgen.1000010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hessel S., Eichinger A., Isken A., Amengual J., Hunzelmann S., Hoeller U., Elste V., Hunziker W., Goralczyk R., Oberhauser V., von Lintig J., and Wyss A. (2007) CMO1 deficiency abolishes vitamin A production from β-carotene and alters lipid metabolism in mice. J. Biol. Chem. 282, 33553–33561 10.1074/jbc.M706763200 [DOI] [PubMed] [Google Scholar]
- 19. Toews D. P., Taylor S. A., Vallender R., Brelsford A., Butcher B. G., Messer P. W., and Lovette I. J. (2016) Plumage genes and little else distinguish the genomes of hybridizing warblers. Curr. Biol. 26, 2313–2318 10.1016/j.cub.2016.06.034 [DOI] [PubMed] [Google Scholar]
- 20. Borel P., de Edelenyi F. S., Vincent-Baudry S., Malezet-Desmoulin C., Margotat A., Lyan B., Gorrand J. M., Meunier N., Drouault-Holowacz S., and Bieuvelet S. (2011) Genetic variants in BCMO1 and CD36 are associated with plasma lutein concentrations and macular pigment optical density in humans. Ann. Med. 43, 47–59 10.3109/07853890.2010.531757 [DOI] [PubMed] [Google Scholar]
- 21. Meyers K. J., Johnson E. J., Bernstein P. S., Iyengar S. K., Engelman C. D., Karki C. K., Liu Z., Igo R. P. Jr., Truitt B., Klein M. L., Snodderly D. M., Blodi B. A., Gehrs K. M., Sarto G. E., Wallace R. B., et al. (2013) Genetic determinants of macular pigments in women of the carotenoids in age-related eye disease study. Invest. Ophthalmol. Vis. Sci. 54, 2333–2345 10.1167/iovs.12-10867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kiefer C., Hessel S., Lampert J. M., Vogt K., Lederer M. O., Breithaupt D. E., and von Lintig J. (2001) Identification and characterization of a mammalian enzyme catalyzing the asymmetric oxidative cleavage of provitamin A. J. Biol. Chem. 276, 14110–14116 10.1074/jbc.M011510200 [DOI] [PubMed] [Google Scholar]
- 23. Babino D., Palczewski G., Widjaja-Adhi M. A., Kiser P. D., Golczak M., and von Lintig J. (2015) Characterization of the role of β-carotene 9,10-dioxygenase in macular pigment metabolism. J. Biol. Chem. 290, 24844–24857 10.1074/jbc.M115.668822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mein J. R., Dolnikowski G. G., Ernst H., Russell R. M., and Wang X. D. (2011) Enzymatic formation of apo-carotenoids from the xanthophyll carotenoids lutein, zeaxanthin and β-cryptoxanthin by ferret carotene-9',10'-monooxygenase. Arch. Biochem. Biophys. 506, 109–121 10.1016/j.abb.2010.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Amengual J., Lobo G. P., Golczak M., Li H. N., Klimova T., Hoppel C. L., Wyss A., Palczewski K., and von Lintig J. (2011) A mitochondrial enzyme degrades carotenoids and protects against oxidative stress. FASEB J. 25, 948–959 10.1096/fj.10-173906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Amengual J., Widjaja-Adhi M. A., Rodriguez-Santiago S., Hessel S., Golczak M., Palczewski K., and von Lintig J. (2013) Two carotenoid oxygenases contribute to mammalian provitamin A metabolism. J. Biol. Chem. 288, 34081–34096 10.1074/jbc.M113.501049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Costabile B. K., Kim Y. K., Iqbal J., Zuccaro M. V., Wassef L., Narayanasamy S., Curley R. W. Jr., Harrison E. H., Hussain M. M., and Quadro L. (2016) β-Apo-10'-carotenoids modulate placental microsomal triglyceride transfer protein expression and function to optimize transport of intact β-carotene to the embryo. J. Biol. Chem. 291, 18525–18535 10.1074/jbc.M116.738336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Berry S. D., Davis S. R., Beattie E. M., Thomas N. L., Burrett A. K., Ward H. E., Stanfield A. M., Biswas M., Andersmit-Udy A. E., Oxley P. E., Barnett J. L., Pearson J. F., van der Does Y., Macgibbon A. H., Spelman R. J., et al. (2009) Mutation in bovine β-carotene oxygenase 2 affects milk color. Genetics 182, 923–926 10.1534/genetics.109.101741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tian R., Pitchford W. S., Morris C. A., Cullen N. G., and Bottema C. D. (2010) Genetic variation in the β,β-carotene-9',10'-dioxygenase gene and association with fat colour in bovine adipose tissue and milk. Anim. Genet. 41, 253–259 10.1111/j.1365-2052.2009.01990.x [DOI] [PubMed] [Google Scholar]
- 30. Vage D. I., and Boman I. A. (2010) A nonsense mutation in the β-carotene oxygenase 2 (BCO2) gene is tightly associated with accumulation of carotenoids in adipose tissue in sheep (Ovis aries). BMC Genet. 11, 10 10.1186/1471-2156-11-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Strychalski J., Brym P., Czarnik U., and Gugołek A. (2015) A novel AAT-deletion mutation in the coding sequence of the BCO2 gene in yellow-fat rabbits. J. Appl. Genet. 56, 535–537 10.1007/s13353-015-0290-9 [DOI] [PubMed] [Google Scholar]
- 32. Li B., Vachali P. P., Gorusupudi A., Shen Z., Sharifzadeh H., Besch B. M., Nelson K., Horvath M. M., Frederick J. M., Baehr W., and Bernstein P. S. (2014) Inactivity of human β,β-carotene-9',10'-dioxygenase (BCO2) underlies retinal accumulation of the human macular carotenoid pigment. Proc. Natl. Acad. Sci. U.S.A. 111, 10173–10178 10.1073/pnas.1402526111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dela Seña C., Sun J., Narayanasamy S., Riedl K. M., Yuan Y., Curley R. W. Jr., Schwartz S. J., and Harrison E. H. (2016) Substrate specificity of purified recombinant chicken β-carotene 9',10'-oxygenase (BCO2). J. Biol. Chem. 291, 14609–14619 10.1074/jbc.M116.723684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Poliakov E., Soucy J., Gentleman S., Rogozin I. B., and Redmond T. M. (2017) Phylogenetic analysis of the metazoan carotenoid oxygenase superfamily: a new ancestral gene assemblage of BCO-like (BCOL) proteins. Sci. Rep. 7, 13192 10.1038/s41598-017-13521-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Palczewski G., Amengual J., Hoppel C. L., and von Lintig J. (2014) Evidence for compartmentalization of mammalian carotenoid metabolism. FASEB J. 28, 4457–4469 10.1096/fj.14-252411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim Y. S., Yeom S. J., and Oh D. K. (2011) Production of beta-apo-10'-carotenal from β-carotene by human β-carotene-9',10'-oxygenase expressed in E. coli. Biotechnol. Lett. 33, 1195–1200 10.1007/s10529-011-0556-1 [DOI] [PubMed] [Google Scholar]
- 37. Kelly M. E., Ramkumar S., Sun W., Colon Ortiz C., Kiser P. D., Golczak M., and von Lintig J. (2018) The biochemical basis of vitamin A production from the asymmetric carotenoid β-cryptoxanthin. ACS Chem. Biol. 13, 2121–2129 10.1021/acschembio.8b00290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Eggersdorfer M., and Wyss A. (2018) Carotenoids in human nutrition and health. Arch. Biochem. Biophys. 652, 18–26 10.1016/j.abb.2018.06.001 [DOI] [PubMed] [Google Scholar]
- 39. Bohn T., Desmarchelier C., Dragsted L. O., Nielsen C. S., Stahl W., Ruhl R., Keijer J., and Borel P. (2017) Host-related factors explaining interindividual variability of carotenoid bioavailability and tissue concentrations in humans. Mol. Nutr. Food Res. 61, 1600685 10.1002/mnfr.201600685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Omenn G. S., Goodman G., Thornquist M., Grizzle J., Rosenstock L., Barnhart S., Balmes J., Cherniack M. G., Cullen M. R., and Glass A. and (1994) The β-carotene and retinol efficacy trial (CARET) for chemoprevention of lung cancer in high risk populations: smokers and asbestos-exposed workers. Cancer Res. 54, 2038s–2043s [PubMed] [Google Scholar]
- 41. Omenn G. S., Goodman G. E., Thornquist M. D., Balmes J., Cullen M. R., Glass A., Keogh J. P., Meyskens F. L., Valanis B., Williams J. H., Barnhart S., and Hammar S. (1996) Effects of a combination of β carotene and vitamin A on lung cancer and cardiovascular disease. N. Engl. J. Med. 334, 1150–1155 10.1056/NEJM199605023341802 [DOI] [PubMed] [Google Scholar]
- 42. von Lintig J., and Vogt K. (2000) Filling the gap in vitamin A research: molecular identification of an enzyme cleaving β-carotene to retinal. J. Biol. Chem. 275, 11915–11920 10.1074/jbc.275.16.11915 [DOI] [PubMed] [Google Scholar]
- 43. Misawa N., Nakagawa M., Kobayashi K., Yamano S., Izawa Y., Nakamura K., and Harashima K. (1990) Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J. Bacteriol. 172, 6704–6712 10.1128/jb.172.12.6704-6712.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lindqvist A., He Y. G., and Andersson S. (2005) Cell type-specific expression of β-carotene 9',10'-monooxygenase in human tissues. J. Histochem. Cytochem. 53, 1403–1412 10.1369/jhc.5A6705.2005 [DOI] [PubMed] [Google Scholar]
- 45. Kowatz T., Babino D., Kiser P., Palczewski K., and von Lintig J. (2013) Characterization of human β,β-carotene-15,15'-monooxygenase (BCMO1) as a soluble monomeric enzyme. Arch. Biochem. Biophys. 539, 214–222 10.1016/j.abb.2013.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Daruwalla A., Zhang J., Lee H. J., Khadka N., Farquhar E. R., Shi W., von Lintig J., and Kiser P. D. (2020) Structural basis for carotenoid cleavage by an archaeal carotenoid dioxygenase. Proc. Natl. Acad. Sci. U.S.A. 117, 19914–19925 10.1073/pnas.2004116117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nagao A., Maoka T., Ono H., Kotake-Nara E., Kobayashi M., and Tomita M. (2015) A 3-hydroxy β-end group in xanthophylls is preferentially oxidized to a 3-oxo ε-end group in mammals. J. Lipid Res. 56, 449–462 10.1194/jlr.P055459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Khachik F., Bernstein P. S., and Garland D. L. (1997) Identification of lutein and zeaxanthin oxidation products in human and monkey retinas. Invest. Ophthalmol. Vis. Sci. 38, 1802–1811 [PubMed] [Google Scholar]
- 49. Khachik F., de Moura F. F., Zhao D. Y., Aebischer C. P., and Bernstein P. S. (2002) Transformations of selected carotenoids in plasma, liver, and ocular tissues of humans and in nonprimate animal models. Invest. Ophthalmol. Vis. Sci. 43, 3383–3392 [PubMed] [Google Scholar]
- 50. Daruwalla A., and Kiser P. D. (2020) Structural and mechanistic aspects of carotenoid cleavage dioxygenases (CCDs). Biochim. Biophys. Acta 1865, 158590 10.1016/j.bbalip.2019.158590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hamel C. P., Tsilou E., Pfeffer B. A., Hooks J. J., Detrick B., and Redmond T. M. (1993) Molecular cloning and expression of RPE65, a novel retinal pigment epithelium-specific microsomal protein that is post-transcriptionally regulated in vitro. J. Biol. Chem. 268, 15751–15757 [PubMed] [Google Scholar]
- 52. Gollapalli D. R., Maiti P., and Rando R. R. (2003) RPE65 operates in the vertebrate visual cycle by stereospecifically binding all-trans-retinyl esters. Biochemistry 42, 11824–11830 10.1021/bi035227w [DOI] [PubMed] [Google Scholar]
- 53. Mata N. L., Moghrabi W. N., Lee J. S., Bui T. V., Radu R. A., Horwitz J., and Travis G. H. (2004) Rpe65 is a retinyl ester binding protein that presents insoluble substrate to the isomerase in retinal pigment epithelial cells. J. Biol. Chem. 279, 635–643 10.1074/jbc.M310042200 [DOI] [PubMed] [Google Scholar]
- 54. Hansen S., and Maret W. (1988) Retinal is not formed in vitro by enzymatic central cleavage of β-carotene. Biochemistry 27, 200–206 10.1021/bi00401a030 [DOI] [PubMed] [Google Scholar]
- 55. Hu K. Q., Liu C., Ernst H., Krinsky N. I., Russell R. M., and Wang X. D. (2006) The biochemical characterization of ferret carotene-9',10'-monooxygenase catalyzing cleavage of carotenoids in vitro and in vivo. J. Biol. Chem. 281, 19327–19338 10.1074/jbc.M512095200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hartmann D., Thürmann P. A., Spitzer V., Schalch W., Manner B., and Cohn W. (2004) Plasma kinetics of zeaxanthin and 3'-dehydro-lutein after multiple oral doses of synthetic zeaxanthin. Am. J. Clin. Nutr. 79, 410–417 10.1093/ajcn/79.3.410 [DOI] [PubMed] [Google Scholar]
- 57. Lim J. Y., Liu C., Hu K. Q., Smith D. E., and Wang X. D. (2018) Ablation of carotenoid cleavage enzymes (BCO1 and BCO2) induced hepatic steatosis by altering the farnesoid X receptor/miR-34a/sirtuin 1 pathway. Arch. Biochem. Biophys. 654, 1–9 10.1016/j.abb.2018.07.007 [DOI] [PubMed] [Google Scholar]
- 58. Wu L., Guo X., Lyu Y., Clarke S. L., Lucas E. A., Smith B. J., Hildebrand D., Wang W., Medeiros D. M., Shen X., and Lin D. (2017) Targeted metabolomics reveals abnormal hepatic energy metabolism by depletion of β-carotene oxygenase 2 in mice. Sci. Rep. 7, 14624 10.1038/s41598-017-15222-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lobo G. P., Isken A., Hoff S., Babino D., and von Lintig J. (2012) BCDO2 acts as a carotenoid scavenger and gatekeeper for the mitochondrial apoptotic pathway. Development 139, 2966–2977 10.1242/dev.079632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Spiegler E., Kim Y. K., Hoyos B., Narayanasamy S., Jiang H., Savio N., Curley R. W. Jr., Harrison E. H., Hammerling U., and Quadro L. (2018) β-Apo-10'-carotenoids support normal embryonic development during vitamin A deficiency. Sci. Rep. 8, 8834 10.1038/s41598-018-27071-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bernstein P. S., Khachik F., Carvalho L. S., Muir G. J., Zhao D. Y., and Katz N. B. (2001) Identification and quantitation of carotenoids and their metabolites in the tissues of the human eye. Exp. Eye Res. 72, 215–223 10.1006/exer.2000.0954 [DOI] [PubMed] [Google Scholar]
- 62. Khachik F., Spangler C. J., Smith J. C. Jr., Canfield L. M., Steck A., and Pfander H. (1997) Identification, quantification, and relative concentrations of carotenoids and their metabolites in human milk and serum. Anal. Chem. 69, 1873–1881 10.1021/ac961085i [DOI] [PubMed] [Google Scholar]
- 63. Voigt A. P., Whitmore S. S., Flamme-Wiese M. J., Riker M., Wiley L. A., Tucker B. A., Stone E. M., Mullins R. F., and Scheetz T. E. (2019) Molecular characterization of foveal versus peripheral human retina by single-cell RNA sequencing. Exp. Eye Res. 184, 234–242 10.1016/j.exer.2019.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Raghuvanshi S., Reed V., Blaner W. S., and Harrison E. H. (2015) Cellular localization of β-carotene 15,15' oxygenase-1 (BCO1) and β-carotene 9',10' oxygenase-2 (BCO2) in rat liver and intestine. Arch. Biochem. Biophys. 572, 19–27 10.1016/j.abb.2014.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chen N. (2020) A gene for color differences between sexes. Science 368, 1185–1186 10.1126/science.abc2242 [DOI] [PubMed] [Google Scholar]
- 66. Andrade P., Pinho C., Pérez I. L. d. G., Afonso S., Brejcha J., Rubin C. J., Wallerman O., Pereira P., Sabatino S. J., Bellati A., Pellitteri-Rosa D., Bosakova Z., Bunikis I., Carretero M. A., Feiner N., et al. (2019) Regulatory changes in pterin and carotenoid genes underlie balanced color polymorphisms in the wall lizard. Proc. Natl. Acad. Sci. U.S.A. 116, 5633–5642 10.1073/pnas.1820320116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F. T., de Beer T. A. P., Rempfer C., Bordoli L., Lepore R., and Schwede T. (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 10.1093/nar/gky427 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included within the manuscript and supporting information.