Abstract
The tricarboxylic acid (TCA) cycle converts the end products of glycolysis and fatty acid β-oxidation into the reducing equivalents NADH and FADH2. Although mitochondrial matrix uptake of Ca2+ enhances ATP production, it remains unclear whether deprivation of mitochondrial TCA substrates alters mitochondrial Ca2+ flux. We investigated the effect of TCA cycle substrates on MCU-mediated mitochondrial matrix uptake of Ca2+, mitochondrial bioenergetics and autophagic flux. Inhibition of glycolysis, mitochondrial pyruvate transport, or mitochondrial fatty acid transport triggered expression of the MCU gatekeeper MICU1 but not the MCU core subunit. Knockdown of mitochondrial pyruvate carrier (MPC) isoforms or expression of the dominant negative mutant MPC1R97W resulted in increased MICU1 protein abundance and inhibition of MCU-mediated mitochondrial matrix uptake of Ca2+. We also found that genetic ablation of MPC1 in hepatocytes and mouse embryonic fibroblasts resulted in reduced resting matrix Ca2+, likely because of increased MICU1 expression but resulted in changes in mitochondrial morphology. TCA cycle substrate–dependent MICU1 expression was mediated by the transcription factor early growth response 1 (EGR1). Blocking mitochondrial pyruvate or fatty acid flux was linked to increased autophagy marker abundance. These studies reveal a mechanism that controls the MCU-mediated Ca2+ flux machinery and that depends on TCA cycle substrate availability. This mechanism generates a metabolic homeostatic circuit that protects cells from bioenergetic crisis and mitochondrial Ca2+ overload during periods of nutrient stress.
One-Sentence Summary:
Nutrient stress triggers transcriptional changes that limit Ca2+ influx into mitochondria through the MCU.
Editor’s Summary:
Rheostat for mitochondrial Ca2+ uptake during stress
Ca2+ entering mitochondria through the mitochondrial Ca2+ uniporter (MCU) normally promotes the generation of ATP through the TCA cycle. Nemani et al. found that nutrient stress initiated a transcriptional program that restricted Ca2+ influx into mitochondria through the MCU to prevent changes in mitochondrial dynamics and bioenergetics. Pharmacological or genetic disruptions in glycolysis or uptake of pyruvate or fatty acids induced the transcription factor EGR1 to increase the expression of the gene encoding MICU1, a gatekeeper for the MCU, resulting in reduced mitochondrial Ca2+ uptake. The authors propose that this response prevents mitochondria from becoming overloaded with Ca2+ and limits bioenergetics crises during nutrient stress.
INTRODUCTION
Metabolic homeostasis is essential not only at the cellular level for processes from cell division, growth, proliferation, differentiation to cell death, but also for tissue and whole organismal development. The three major catabolic pathways of energy production stem from glucose, lipids and proteins. Mutations in components in these pathways present with disrupted energy metabolism in patients leading to organ system damage and sudden death (1). Oxidation of glucose-derived metabolites by mitochondria provide positive feedback mechanisms that subsequently increase uptake of Ca2+ into the matrix, which in turn activate mitochondrial metabolism, further enhancing ATP synthesis (2-4). Although pyruvate is generated through multiple routes in the cytoplasmic milieu, it is transported into mitochondria through a hetero-oligomeric complex of mitochondrial pyruvate carrier MPC1 and MPC2 proteins (5, 6). The critical role of MPCs in pyruvate transport is supported by the finding that a MPC1R97W mutation perturbs pyruvate transport from the cytosol to the mitochondrial matrix, resulting in fatally severe hyper-lactic acidemia and hyper-pyruvicemia (7). Similar symptoms of elevated lactate levels and metabolic acidosis occur in neonates with mutations in the E1a subunit of pyruvate dehydrogenase (PDH) (8-10). Although genetic deletion of MPC results in embryonic lethality in mouse models, several normal and cancer cell lines survive the metabolic stress of loss of MPC through unknown mechanisms (11, 12). Conversely, forced ectopic expression of MPC suppresses tumor growth (13). Additionally, mutations in carnitine palmitoyl transferase (CPT1) that prevent conversion of carnitine to acyl-carnitine for transport into the mitochondria also result in severe metabolic crisis (14-16). Other mutations in mitochondrial carriers that cause deficient or absent transport of substrates into the mitochondrial matrix result in hyperglycemia, brain damage, coma, cardiac arrest and death (17). Metabolic response to nutrients is tightly regulated through nutrient and energy sensing mechanisms. Different organelles respond differently to metabolic stress. The ER has several mechanisms to detect nutrient stress (18-22). On the other hand, mitochondria employ two broad mechanisms of response to nutrient stress: altered mitochondrial Ca2+ dynamics and transcriptional and posttranslational modifications (23-25). Although aberrant energy production is a common denominator, the molecular mechanisms by which mitochondria sense and regulate their activity based on the availability of metabolic substrates remains elusive.
Ca2+ flux across the inner mitochondrial membrane (IMM) regulates cellular bioenergetics, intracellular cytoplasmic Ca2+ signals, and various cell death pathways (26, 27). Ca2+ uptake into the mitochondria is driven by the organelle’s highly negative membrane potential and occurs through a highly selective channel, the mitochondrial calcium uniporter (MCU) (28, 29). Catabolism of carbohydrates, proteins and fatty acids results in the production of substrates that energize mitochondria, facilitating ATP generation. Mitochondrial matrix calcium activates multiple components of the TCA cycle, facilitating the production of reducing equivalents that feed into the electron transport chain (ETC) (27). Thus Ca2+ and metabolites are both necessary to maintain a pool of healthy mitochondria that generate ATP for cellular processes. How Ca2+-dependent mechanisms modulate mitochondria functions during nutrient deficiency, however, are poorly defined.
Here, we showed that mitochondria responded to nutrient stress by transcriptional upregulation of the gene encoding the gatekeeper of the mitochondrial calcium uniporter, MICU1, which was mediated by early growth response 1 (EGR1). The resultant increase in MICU1 expression led to inhibition of MCU-mediated mitochondrial matrix Ca2+ (mCa2+) uptake under nutrient deficient conditions, possibly preventing mCa2+ overload and subsequent cellular death. Furthermore, substrate deficiency impaired mitochondrial bioenergetics and promotes cell survival by up-regulating autophagy.
RESULTS:
Deprivation of mitochondrial substrates by inhibiting metabolic flux induces MCU complex MICU1 protein abundance
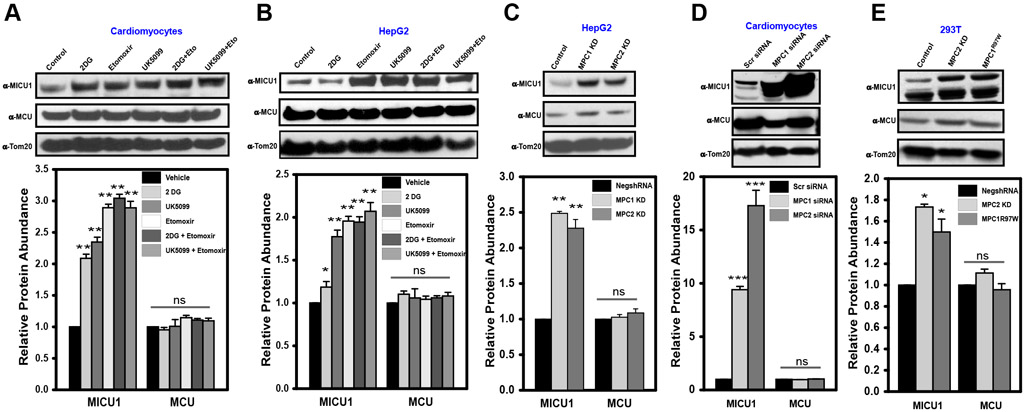
Cytosolic Ca2+ signals determine mitochondrial reducing equivalents and cell survival by altering MCU-mediated mCa2+ uptake (30). To understand whether altering mitochondrial substrates modulated MCU complex abundance, we treated primary cardiomyocytes with inhibitors of key metabolic steps—glycolysis (2-deoxyglucose, 2-DG), mitochondrial fatty acid transport (etomoxir), or mitochondrial pyruvate transport (UK5099). Glycolysis is halted by 2-deoxyglucose (2-DG), etomoxir blocks fatty acid transport by inhibiting carnitine acyltransferase, and UK5099 interrupts mitochondrial pyruvate transport (31-33). Western blot analysis revealed that 2-DG, etomoxir, and UK5099, alone or in combination, significantly increased MICU1 protein abundance without affecting MCU expression (Fig. 1A). Pharmacologic blockade of pyruvate metabolism in the HepG2 hepatocellular carcinoma cell line also increased MICU1 protein abundance without altering MCU protein expression levels after (Fig. 1B). Next, we utilized an shRNA-based strategy to target the pyruvate transporter proteins MPC1 and MPC2 and induce a state of nutrient stress. RNAi-mediated silencing decreased the abundance of MPC1 and MPC2 by at least 80% in primary cardiomyocytes, HEK 293T and HepG2 cells (fig. S1, A to C), leading to increased MICU1 protein expression in all three cell types (Fig. 1, C to E). To further verify the link between mitochondrial pyruvate metabolism and MICU1 expression, we stably reconstituted the human dominant negative MPC mutant (MPC1R97W) in HEK293T cells (fig. S1C). Cells stably expressing MPC1R97W exhibited increased MICU1 protein levels similar to those in MPC siRNA-transfected cells (Fig. 1E). Densitometric analysis revealed a 2- to 15-fold increase in MICU1 protein abundance due to pharmacological or genetic interference in pyruvate metabolism (Fig. 1, A to E). Together, these data reveal that preventing pyruvate and fatty acid entry into mitochondria results in MICU1 up-regulation.
Figure 1: Deprivation of TCA cycle metabolites increases MICU1 protein abundance.
A and B) Primary rat cardiomyocytes (A) and HepG2 cells (B) were exposed to either 2-DG (1mM), etomoxir (10μM), UK5099 (100μM), or combination of 2-DG + etomoxir and UK5099 + etomoxir overnight and immunoblotted for MICU1 and MCU. TOM20 was used as loading control. Bottom panel presents relative protein abundance quantified using Image J. Mean ± SEM from three independent experiments. *P<0.05, **P<0.01, n.s., not significant.
C) MPC1 knockdown (KD) and MPC2 KD HepG2 cells were immunoblotted for MICU1 and MCU. TOM20 was used as loading control. Bottom panel presents relative density of protein abundance quantified using Image J. Mean ± SEM from three independent experiments. **P<0.01, n.s., not significant.
D) Primary rat cardiomyocytes were transiently silenced for MPC1 and MPC2 using RNAi and immunoblotted for MICU1 and MCU. TOM20 was used as loading control. Bottom panel presents relative protein abundance quantified using Image J. Mean ± SEM from three independent experiments. ***P<0.001, n.s., not significant.
E) HEK293T control, MPC2 KD and stably expressing MPC1R97W cells were immunoblotted for MICU1 and MCU. Tom20 was used as loading control. Bottom panel presents relative protein abundance quantified using Image J. Mean ± SEM from three independent experiments. *P<0.05, n.s., not significant.
Loss of MPC1- and MPC2-dependent MICU1 induction alters basal MCU-mediated mCa2+ uptake
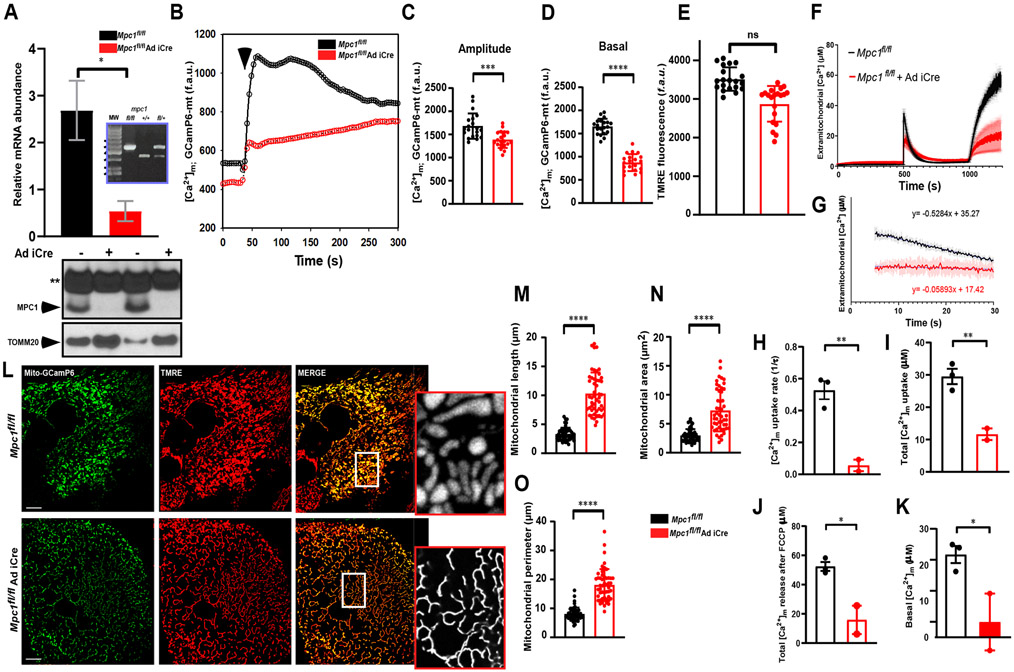
Next, we assessed if the elevated levels of MICU1 protein expression observed after blockade of mitochondrial pyruvate entry played a role in modulating MCU-mediated mCa2+ uptake. MICU1 binds to the MCU and inhibits MCU-mediated Ca2+ uptake in resting cells; loss of MICU1 results in basal matrix mCa2+ accumulation (34-38). To understand how nutrient deficiency alters mitochondrial function, HepG2 cells stably expressing MPC1 and MPC2 shRNA were generated, which exhibited over 80% knockdown efficiency at the mRNA and protein levels (fig. S2, A to D). From the six stable cell lines we observed two scenarios. First, MPC1.1 and MPC1.2 shRNA HepG2 clones expressed reduced levels of MPC1 and MPC2 mRNA, possibly due to a lack of isoform specificity of the shRNA. Second, consistent with previous reports, we observed that loss of one isoform in MPC1.3, MPC2.1, MPC2.2, and MPC2.3 shRNA HepG2 clones resulted in possible destabilization of MPC protein complex and subsequent protein degradation of heteromeric subunit (fig. S2, C and D) (5, 6, 12). For our experiments we utilized MPC1.3 and MPC2.1 stable cell lines. Because MICU1 was elevated under conditions of nutrient deficiency, we asked if these cells exhibited differences in basal matrix Ca2+. Digitonin-permeabilized control and MPC shRNA-expressing cells were exposed to thapsigargin (Tg), a SERCA inhibitor that prevents Ca2+ uptake by the ER, and subsequently with the protonophore CCCP, which depolarizes the mitochondrial membrane potential (ΔΨm) and causes release of matrix Ca2+. CCCP-induced release of matrix Ca2+ was significantly reduced in cells with MPC knockdown (fig. S2, E and F). To validate that the observed decrease in matrix Ca2+ was due to pyruvate deficiency-dependent increases in MICU1 expression, we performed rescue experiments. Reconstitution of MPC1 and MPC2 in MPC shRNA-expressing cells normalized basal matrix Ca2+, as expected (fig. S2, G and H). Because mitochondria shape cytosolic Ca2+ dynamics and loss of the pyruvate carrier resulted in decreased matrix Ca2+, we next tested if cytosolic Ca2+ dynamics were also affected by loss of MPC expression. We observed that ATP-induced cCa2+ elevation was unaffected by knockdown of either MPC1 or MPC2 (fig. S2I and inset), despite significant decreases in mCa2+ uptake (fig. S2, J and K). This loss of ATP-induced mCa2+ uptake was reversed by MPC reconstitution, demonstrating the specificity of the relationship between mitochondrial pyruvate availability and the organelle’s cCa2+ buffering capacity (fig. S2L). To demonstrate that genetic ablation of MPC1 lowered mCa2+ through regulation of MCU activity, MPC1 was deleted using an adenoviral mediated Cre-recombinase (Ad iCre) delivered to MPC1fl/fl hepatocytes (Fig. 2A). Hepatocytes with Cre-mediated deletion of MPC1 exhibited reduced mitochondrial Ca2+ uptake (Fig. 2B and 2C). Additionally, basal mitochondrial Ca2+ was significantly reduced, an observation that suggests that the MCU “gatekeeping” activity was increased, possibly due to upregulation of MICU1 (Fig. 2D). In permeabilized primary hepatocytes, normal ΔΨm was maintained after Cre-mediated loss of MPC1 (Fig. 2E). However, MPC1 deletion significantly reduced mCa2+ uptake after exposure to a bolus of Ca2+ (Fig. 2F). Moreover, MPC1fl/fl + Ad iCre hepatocytes had significantly reduced MCU activity, MCU-mediated Ca2+ uptake rates, and basal matrix [Ca2+] following extramitochondrial Ca2+ addition (Fig. 2, G to K). Mitochondria exhibited a unique morphology upon acute deletion of MPC1 (Fig. 2L). Analyses of mitochondrial length, area and perimeter showed a significant remodeling of mitochondria (Fig. 2M-O). We then asked whether these functional and morphologic changes were conserved across cell types. MPCfl/fl mouse embryonic fibroblasts (MEFs) infected with Ad iCre demonstrated mCa2+ dynamics similar to those found in MPC1 deleted primary hepatocytes (Fig. S3A). Live cell confocal analysis of mCa2+ dynamics revealed that peak and basal [Ca2+]m were significantly lower in MPCfl/fl + Ad iCre MEFs (Fig. S3 B-C). As in primary hepatocytes, MPC deletion did not alter the ΔΨm, in mouse embryonic fibroblasts (fig. S3, D and E). Although mitochondrial morphology was also altered in the MPCfl/fl MEFs after Cre-mediated MPC1 deletion, the remodeling was distinctive compared to primary hepatocytes; the average length, perimeter, and area were significantly reduced (fig. S3 F-H). These studies suggest that the mechanism behind the alteration of MCU-mediated mCa2+ uptake during conditions of nutrient deficiency is conserved across cell types, although the resulting mitochondrial remodeling that occurs under these conditions may be affected by differences in cellular-specific metabolite sensitivities.
Figure 2. MPC1-deficient hepatocytes exhibit reduced MCU-mediated mitochondrial Ca2+ uptake.
A) MPC1 transcript was analyzed by RT-qPCR. Inset depicts genotyping of MPC1 floxed mice. Bottom panel shows the MPC1 protein abundance four days after viral Ad iCre infection. Data from three independent experiments.
B) Mean traces for [Ca2+]m (GCaMP6-mt) fluorescence measured in Mpc1fl/fl or Mpc1fl/fl + Ad iCre hepatocytes. n = 3 independent experiments.
C) Quantification of peak mito-GCaMP6 fluorescence after ionomycin (2.5 μM) stimulation from traces shown in (B).
D) Quantification of basal mito-GCaMP6 fluorescence from traces shown in (B).
E) Quantification of TMRE fluorescence in Mpc1fl/fl and Mpc1fl/fl + Ad iCre hepatocytes.
F) Hepatocytes from Mpc1fl/fl mice were isolated, infected with Ad iCre for five days and Permeabilized with digitonin (40 μg/ml) in intracellular-like media containing thapsigargin (2 μM) and [Ca2+] indicator Fura-2FF (1 μM). After reaching steady-state ΔΨm, a bolus Ca2+ (50 μM) pulse was added at the indicated time-point before the mitochondrial uncoupler CCCP (10 μM) was added. Representative traces of extramitochondrial Ca2+ ([Ca2+]out) clearance by permeabilized hepatocytes (MPC1fl/fl (black) and MPC1fl/fl + Ad iCre (red)). n = 3 independent experiments.
G) Analysis of mitochondrial Ca2+ uptake rate from traces shown in (F).
H) Bar chart represents mitochondrial Ca2+ uptake rate from traces shown in (F).
I) Quantification of total mitochondrial Ca2+ uptake following extramitochondrial Ca2+ (50 μM) pulse addition from traces shown in (F).
J) Total Ca2+m uptake after single Ca2+ pulse was determined in MPCfl/fl (black) and MPC1fl/fl + Ad iCre (red) hepatocytes by recording Ca2+ released by CCCP from traces shown in (F).
K) Quantification of basal matrix Ca2+ from traces shown in (F).
L) Assessment of hepatocyte mitochondrial morphology. 36 hours after mito-GCaMP6 transduction, hepatocytes were stained with TMRE and images were acquired using confocal imaging system. Inset shows higher magnification images. Scale bar, 10 μm. n = 3-6 independent experiments.
M-O) Analyses of mitochondrial length, area, and perimeter from data shown in (L). 50 mitochondria per condition. Mean ± SEM. *P<0.05, **P<0.01, ***P<0.001, n.s., not significant.
To further verify the role of MPC in mCa2+ uptake, we measured basal mCa2+ levels in MPC1R97W-expressing cells. A point mutation in MPC1 transforming arginine to tryptophan (R97W), renders defects in mitochondrial pyruvate oxidation. Patients with MPC1R97W mutation exhibit lactic acidosis and hyper-pyruvicemia (7). As expected, MPC1R97W expression lowered mitochondrial matrix Ca2+ content without affecting ΔΨm (Fig. S4, A to D). Collectively, these data suggest that inhibition of mitochondrial pyruvate transport alters MCU-mediated mCa2+ uptake possibly through nutrient stress-induced expression of MICU1.
EGR1 controls the MICU1 promoter and induces MICU1 expression during mitochondrial stress
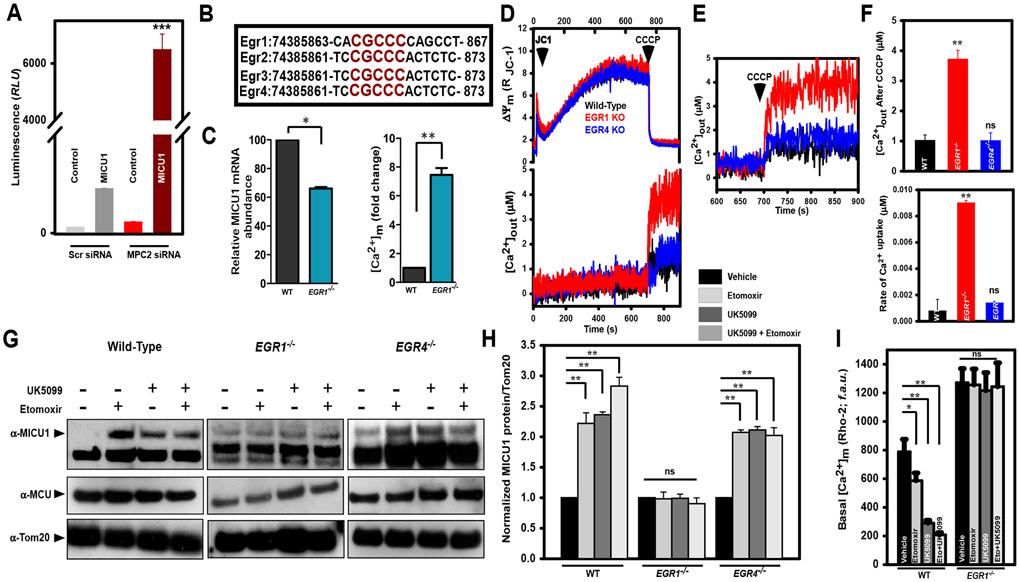
A possible mechanism for MICU1 upregulation during periods of nutritional deficiency is through alteration of MICU1 transcriptional regulation. To test this notion, a proximal MICU1 promoter (−566 to −297 bp) was inserted into a luciferase vector. Silencing of MPC2 expression in HEK 293T cells led to an ~600-fold increase in MICU1 transcription promoter luminescence signal compared to cells expressing a control luciferase vector (Fig. 3A). Bioinformatic analysis of the MICU1 promoter region revealed several putative binding sites for the transcription factor EGR. Early growth response factors (EGRs) are first among the transcription factors to respond to various environmental conditions—such as growth factors, hormones, neurotransmitters, mitogens and metabolites—that functionally control cell proliferation in multiple cell types (39-41). The EGR family consists of four members (EGR1, EGR2, EGR3 and EGR4) with considerable overlap in their consensus DNA binding sequences (Fig. 3B). To test whether EGR1 regulated MICU1 transcription, we utilized primary endothelial cells (ECs) freshly isolated from EGR1−/− mice. EGR1−/− ECs displayed a constitutive loss of MICU1 mRNA expression with concomitant increases in basal mCa2+ levels (Fig. 3C). Because MICU1 is a gatekeeper of MCU at low cCa2+ (below 1.0-2.0 μM), we then tested if EGR1 knockout mitochondria cleared 1 μM Ca2+ through MCU. EGR1−/− ECs but not WT and EGR4−/− ECs rapidly took up Ca2+ after application of a 1 μM Ca2+ bolus without changes in ΔΨm (Fig. 3, D-F). Consistent with our MICU1 luciferase reporter results, EGR1−/− ECs demonstrated an increase in release of mCa2+ upon addition of CCCP (Fig. 3, E and F), suggesting that EGR1 likely induces MICU1 transcription during states of mitochondrial nutrient deficiency. To further confirm that MICU1 expression was regulated by EGR1 rather than EGR4, primary hepatocytes isolated from EGR1−/− and EGR4−/− mice were treated with UK5099 and etomoxir. WT and EGR4−/− hepatocytes, but not EGR1−/− hepatocytes, exhibited a significant upregulation of MICU1 protein that was similar to other cell types (Fig. 3G). Densitometric analysis revealed about 2 to 3-fold increase in the protein levels of MICU1 in the WT and EGR4−/− but not in EGR1−/− hepatocytes (Fig. 3H). RT-qPCR analyses showed cells treated with UK5099 exhibited a significant elevation in MICU1 mRNA transcript (fig. S5). Although MICU1 protein abundance was elevated by blockade of either pyruvate or fatty acid transport into the mitochondria (Fig. 1A to E), it is plausible that Etomoxir-induced MICU1 protein elevation is partly through MICU protein complex stabilization (42). Nevertheless, future studies are warranted to understand the fatty acid dependent MCU activity. Finally, we found that basal mCa2+, was decreased in WT and EGR1−/− hepatocytes (Fig. 3I). These observations reveal that EGR1 is required for nutrient stress-induced MICU1 upregulation for control of mCa2+ uptake.
Figure 3: EGR1 but not EGR4 regulates MICU1 transcription under substrate deficiency.
A) Control and MPC2 siRNA knockdown 293T cells were transfected with MICU1 promoter-luciferase construct and analyzed for luciferase activity. Data represent mean ± SEM (n=3 independent experiments), ***P<0.001.
B) Schematic of the consensus DNA binding region in EGR1, EGR2, EGR3 and EGR4.
C) Left panel presents relative MICU1 mRNA expression in ECs from WT and EGR1−/− animals. Right panel represents quantification of normalized basal matrix mCa2+ in ECs from WT and EGR1−/− animals (n=3 animals for each genotype).
D) Top panel. Representative traces of permeabilized (40 μg/ml digitonin) ECs from WT, EGR1−/− and EGR4−/− mice loaded with 2 μM thapsigargin (Tg) and the ratiometric ΔΨm fluorophore JC-1 and pulsed with 1μM Ca2+ then with 10 μM uncoupler CCCP at 700 s. Bottom panel. Representative traces of permeabilized (40 μg/ml digitonin) ECs from WT, EGR1−/− and EGR4−/− mice loaded with 2 μM thapsigargin and ratiometric Ca2+ indicator Fura2-FF then pulsed with 1μM Ca2+ followed by 10 μM CCCP at 700 s. Data representative of 3 independent experiments.
E) Magnified inset showing matrix mCa2+ released upon stimulation with CCCP.
F) Top panel. Bar graph presents quantification of amount of matrix mCa2+ released upon stimulation with CCCP. Bottom panel. Bar graph presents quantification of rate of mCa2+ uptake with 1μM Ca2+ pulse. Data from 3 independent experiments.
G) Hepatocytes isolated from WT, EGR1−/− and EGR4−/− mice were exposed to either 2-DG (1mM), etomoxir (10μM), UK5099 (100 μM), or combination of 2-DG + etomoxir and UK5099 + etomoxir overnight and immunoblotted for MICU1 and MCU. TOM20 was used as loading control.
H) Bar graph presents relative protein abundance quantified using Image J. Data represent mean ± SEM from 3 independent experiments. **P<0.01, ns= not significant.
I) Bar graph presents basal mCa2+ as measured by Rho-2 fluorescence in hepatocytes isolated from WT and EGR1−/− animals exposed to either 2-DG, etomoxir, UK5099, or combination of 2-DG + etomoxir and UK5099 + etomoxir. Data represent mean ± SEM from 3 independent experiments *P<0.05, **P<0.01, ns= not significant.
Impaired mitochondrial pyruvate flux promotes lactate accumulation and perturbs bioenergetics
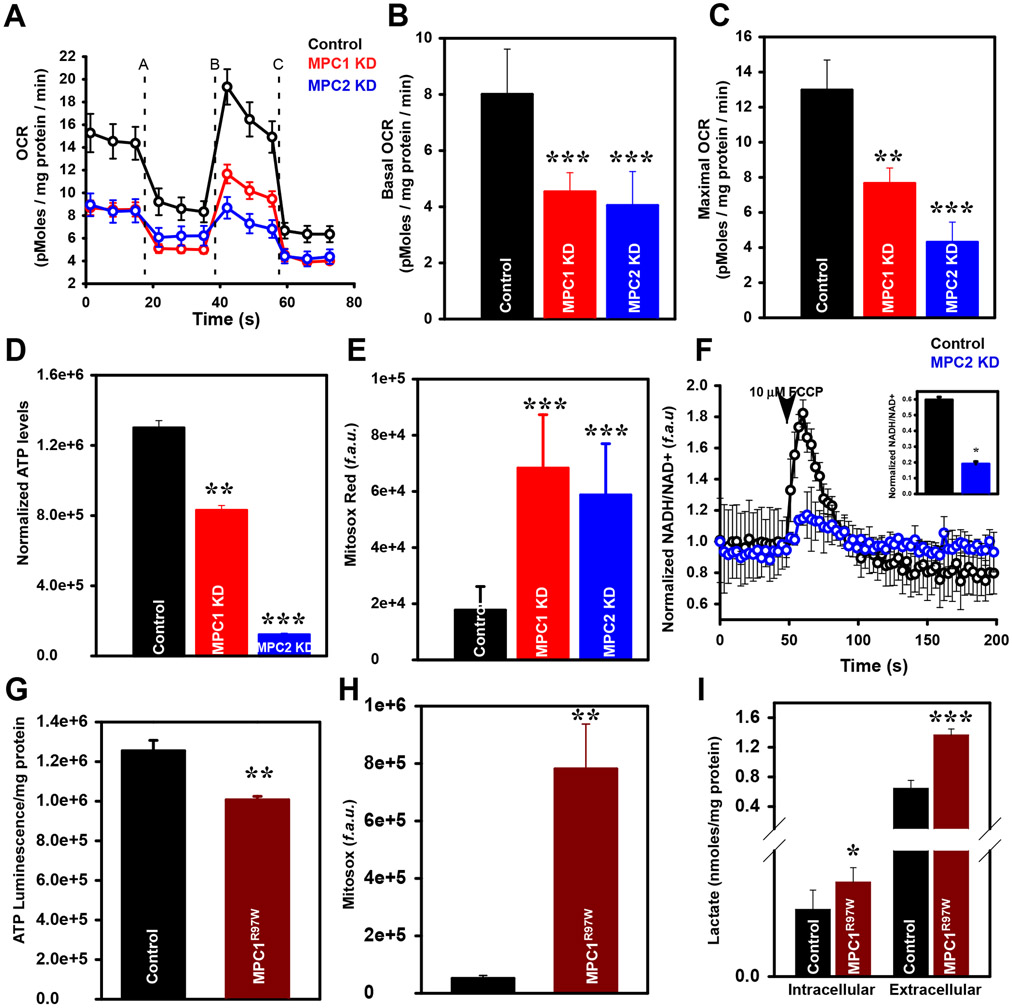
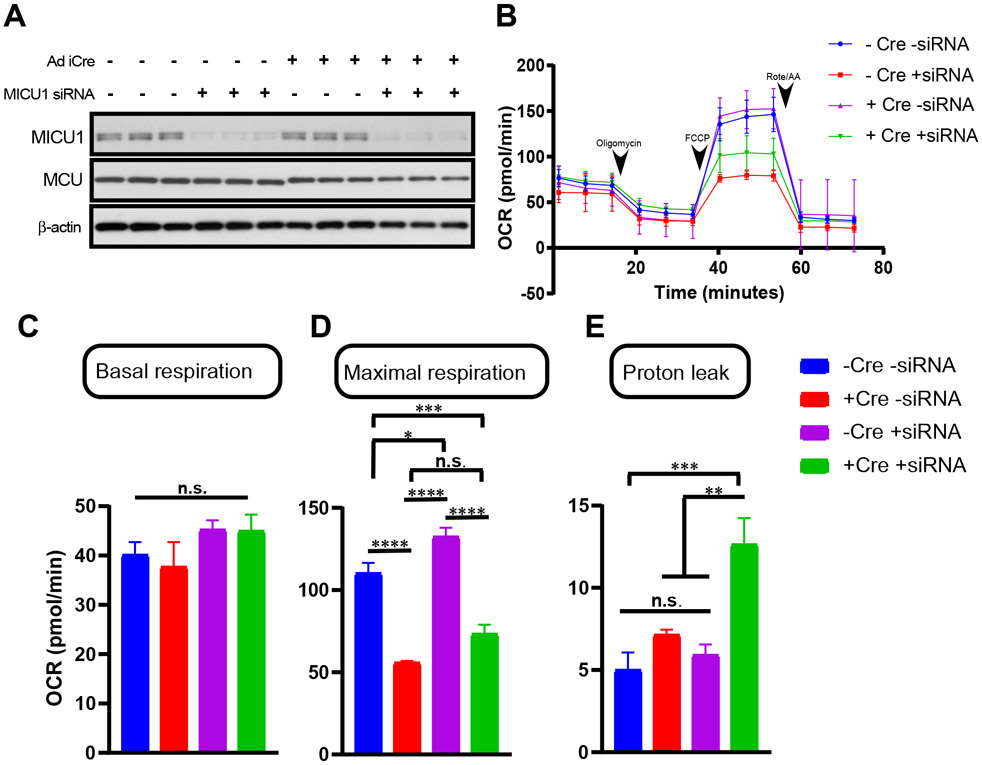
To examine the link between pyruvate flux and MICU1-mediated MCU complex function, we measured mitochondrial bioenergetic parameters. Basal and maximal oxygen consumption were significantly decreased after knockdown of either MPC1 or MPC2 (Fig. 4, A to C). Additionally, MPC knockdown decreased total cellular ATP levels and increased levels of mROS (Fig. 4, D and E). Because pyruvate metabolic flux generates NADH, we next assessed mitochondrial NADH synthesizing capabilities. To measure the dynamic change of NADH/NAD+ ratio, control and MPC2 KD HepG2 cells were transiently transfected with a NADH sensor, peredox-mCherry. Stimulation with the protonophore FCCP rapidly dissipates ΔΨm, leading to maximization of electron transport chain complex activity. As expected, FCCP rapidly induced an increase in NADH in control cells, but MPC2 knockdown greatly attenuated FCCP-induced NADH production (Fig. 4F and inset), consistent with lower pyruvate and reduced basal mCa2+. MPC1R97W mutant cells showed reduced cellular ATP levels and increased mitochondrial ROS (Fig. 4, G and H). Additionally, intracellular and extracellular lactate levels were elevated in MPC1R97W-expressing cells compared to control cells (Fig. 4I), suggesting conversion of the accumulated cytosolic pyruvate to lactate. Together, these data suggest that the blockade of pyruvate entry into mitochondria results in bioenergetic crisis, possibly due to increased MICU1 expression. To demonstrate that MICU1 plays a role in MPC1/2 knockdown-mediated bioenergetic crisis, hepatocytes were harvested from control MPCfl/fl mice and those injected with Ad iCre virus. siRNA targeting MICU1 was then added to the hepatocytes to create four conditions: −Cre/−siRNA, −Cre/+siRNA, +Cre/−siRNA, +Cre/+siRNA (Fig. 5A), and OCR was measured (Fig. 5, B-E). As expected, MPC1 deletion resulted in marked reduction of maximal OCR (Figure 5, B and D). Intriguingly, MICU1 silencing alone showed significant increase in OCR (Fig. 5, B and D). Silencing of MICU1 when MPC1 was deleted caused a partial recovery of maximal OCR (Fig. 5B and D). Together, these results support the notion that increased MICU1 expression under conditions of mitochondrial pyruvate deprivation controls mitochondrial bioenergetics.
Figure 4: Loss of mitochondrial pyruvate flux perturbs cellular bioenergetics.
A) Measurement of oxygen consumption rate (OCR) in WT, MPC1KD and MPC2 KD HepG2 cells. After basal OCR was measured, oligomycin (1 μM) (A), FCCP (1 μM) (B), and rotenone (100 μM) + Antimycin A (100 μM) (C) were added as indicated.
B and C) Bar graph presents mean basal and maximal OCR. Mean ± SEM, (n=10 biological replicates for each group); ***P<0.001, **P<0.01
D) Bar graph presents cellular ATP levels in WT, MPC1 KD and MPC2 KD HepG2 cells. Mean ± SEM (n=8 biological replicates for each group); ***P<0.001, **P<0.01
E) Quantification of MitoSOX Red fluorescence in WT, MPC1 KD and MPC2 KD HepG2 cells. Mean ± SEM (n=8 biological replicates for each group); ***P<0.001, **P<0.01
F) Mean traces of cellular NADH/NAD+ fluorescence in WT, MPC1 KD and MPC2 KD HepG2 cells. Inset: Peak NADH/NAD+ fluorescence. Mean ± SEM (n=4 biological replicates for each group); *P<0.05
G) Bar graph presents cellular ATP levels in HepG2 cells stably expressing FLAG-tagged empty vector and MPC1R97W plasmids. Mean ± SEM (n=8 biological replicates for each group); **P<0.01
H) Quantification of MitoSOX Red fluorescence in HepG2 cells stably expressing FLAG-tagged empty vector and MPC1R97W plasmids. Mean ± SEM (n=3 biological replicates for each group); **P<0.01
I) Quantification of intracellular and extracellular lactate levels in HepG2 cells stably expressing FLAG-tagged empty vector and MPC1R97W plasmids. Mean ± SEM (n=3 biological replicates for each group); ***P<0.001, *P<0.05
Figure 5. MICU1 alters mitochondrial oxygen consumption rate in MPC1 deleted hepatocytes.
A) Assessment of MICU1 protein abundance in MPC1fl/fl hepatocytes following Ad iCre transduction and MICU1 specific siRNA. Protein abundance assessed in the following four groups: Mpc1fl/fl -siRNA, Mpc1fl/fl + Ad iCre -siRNA, Mpc1fl/fl + MICU1siRNA, Mpc1fl/fl + Ad iCre + MICU1siRNA (n=3 independent experiments).
B) Measurement of oxygen consumption rate (OCR) in Mpc1fl/fl -siRNA, Mpc1fl/fl + Ad iCre -siRNA, Mpc1fl/fl + MICU1siRNA, Mpc1fl/fl + Ad iCre + MICU1siRNA hepatocytes. After basal OCR was measured, oligomycin (1 μM) (A), FCCP (2 μM) (B), and rotenone (0.5 μM) + Antimycin A (0.5 μM) were added as indicated (n=3 independent experiments).
C-E) Bar graphs present mean basal OCR (C), maximal OCR (D), and proton leak (E). Mean ± SEM (n=7 biological replicates for each group); *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001, n.s. not significant.
MPC-mediated pyruvate flux controls autophagy
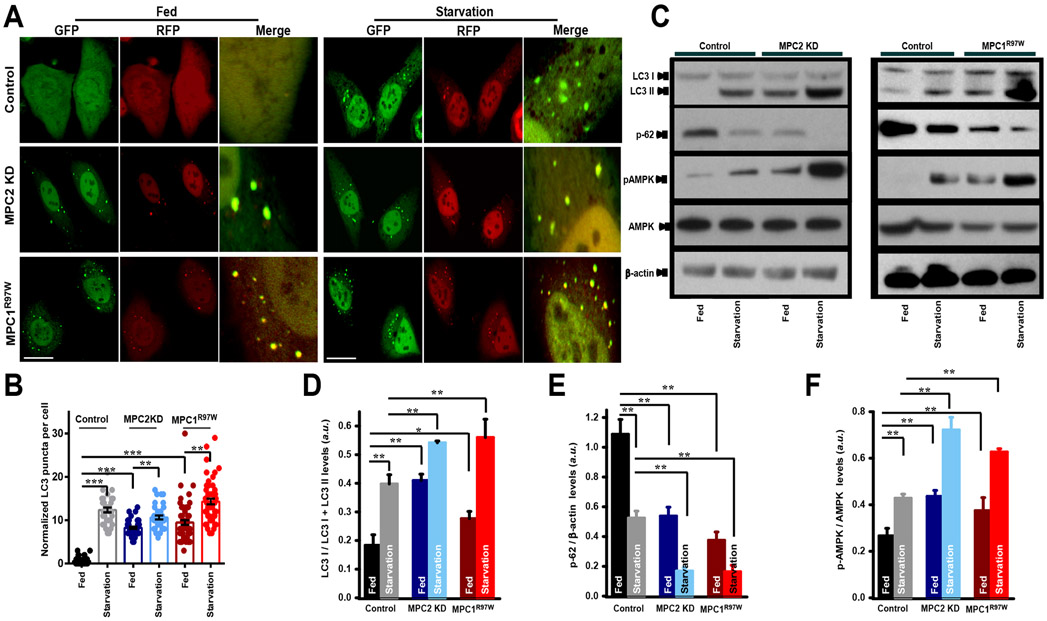
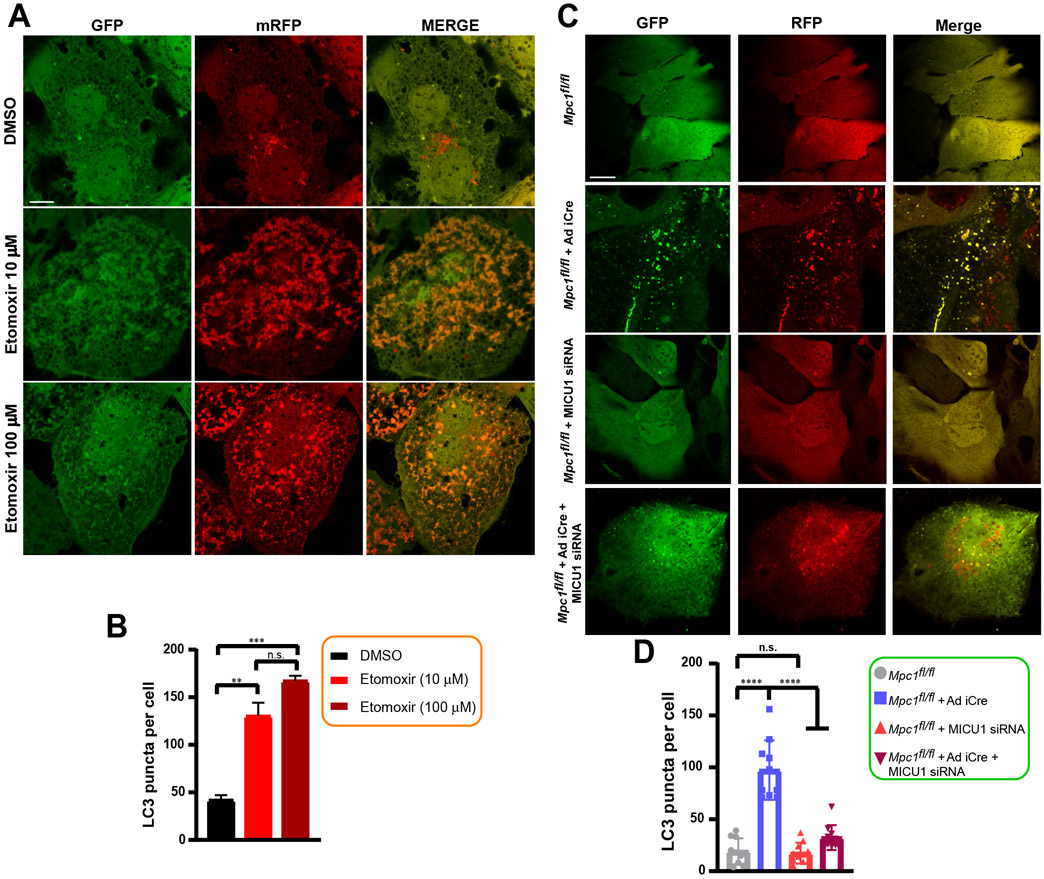
Having observed that loss of mitochondrial pyruvate transport machinery detrimentally affected cellular energy metabolism, we next examined if alternative modes of survival were being utilized by the cell. Autophagy is a conserved catabolic process of degrading selective dysfunctional cellular components and organelles under conditions of nutrient deprivation and energy crisis (43, 44). When energy demand exceeds supply, dysfunctional mitochondria are targeted to the autophagosome and degraded by lysosomes. Thus, eliminating damaged mitochondria is a survival mechanism during nutrient stress. Autophagy is a dynamic process that includes formation of autophagosomes, fusion of autophagosomes and lysosomes, and finally, degradation in lysosomes (45). To investigate whether autophagy was increased during perturbed mitochondrial pyruvate transport, autophagic flux analysis was performed in cells lacking functional MPCs and transduced with the tandem LC3-GFP-RFP reporter that allows for detection of each step of the autophagic flux process. Green puncta indicates autophagosome formation and red puncta indicates autophagolysosome formation. In the acidic environment of lysosomes, GFP signal is quenched. Under fed conditions, whereas nominal levels of LC3 puncta were observed in control cells, an increase in LC3 signal (red and green puncta) was observed in cells expressing either MPC2 shRNA or the MPC1R97W mutant (Fig. 6A), suggesting that even under growth-optimal conditions, loss of MPC led to increased autophagic flux corresponding with reduced cellular energy production. Starvation led to increased levels of puncta in all the cell types (Fig. 6A). Quantification of the number of LC3 autophagosomes revealed a significant increase in the number of LC3 puncta in cells expressing either MPC2 shRNA or MPC1R97W under basal conditions (Fig. 6, A and B). Starvation resulted in similar numbers of autophagosomes in all cell types (Fig. 6, A and B). Complementary to the imaging data, lysates from cells expressing MPC2 shRNA or MPC1R97W showed enhanced LC3I processing to LC3II and increased degradation of p62 under normal conditions compared to control cells (Fig. 6, C to F). We next asked whether pharmacologic blockade of fatty acid flux by etomoxir enhanced autophagic flux. Similar to MPC2 knockdown and MPC1R97W mutant overexpression conditions, the number of LC3 puncta were increased following etomoxir treatment in WT hepatocytes (Fig. 7, A and B). Finally, we examined whether genetic deletion of MPC1 and/or KD of MICU1 altered the autophagic flux process. Hepatocytes were infected with Ad iCre virus, transfected with MICU1 siRNA (generating −Cre/−siRNA, −Cre/+siRNA, +Cre/−siRNA, and +Cre/+siRNA cells), and infected with adenovirus expressing tandem fluorescent-tagged LC3 (mRFP-GFP-LC3) (Fig. 7C). mRFP-LC3 puncta were imaged and quantified to analyze the degree of autophagic flux within cells. +Cre cells had more puncta than −Cre cells, indicating a link between MPC deletion and induction of autophagy (Fig. 7D). The highest number of puncta observed was within the +Cre/−siRNA condition, in which MPC was deleted and MICU1 was unaltered. In contrast, MPC deletion and MICU1 silencing resulted in a significant reduction of LC3 puncta, suggesting that MPC deletion promotes autophagic signaling partly through MICU1 induction and MCU activity. Collectively, these results establish a link between alterations in MCU-mediated mCa2+ uptake and increases autophagic flux during periods of metabolite scarcity and nutrient stress.
Figure 6: Loss of mitochondrial pyruvate flux upregulates autophagic flux.
A) Representative confocal images of mRFP-GFP tandem fluorescent-tagged LC3 (tfLC3) in WT, MPC2 KD and MPC1R97W HeLa cells before and after 24 hours starvation (n=4 cells for each group). Scale bar, 20 μm.
B) Quantification of autophagy performed as normalized LC3 puncta. Mean ± SEM (n=4 cells for each group); ***P < 0.001
C) Western blot analysis of cell lysates from WT, MPC2 KD and MPC1R97W HeLa cells before and after starved for 16h. The cell lysates were probed for LC3, p-AMPK, AMPK and p62 antibodies, β-actin served as the loading control. (n=3 independent experiments)
D) Bar graph presents quantification of LC3-I/LC3-I+LC3-II. Mean ± SEM (n=3 independent experiments); **P<0.01, *P<0.05
E) Bar graph presents quantification of p62. Mean ± SEM (n=3 independent experiments); **P<0.01
F) Bar graph presents quantification of p-AMPK/AMPK. Mean ± SEM (n=3 independent experiments); **P<0.01
Figure 7: Pharmacologic blockade of fatty acid flux and genetic ablation of MPC1 elicit autophagic flux.
A) Representative confocal images of mRFP-GFP tandem fluorescent-tagged LC3 (tfLC3) in WT hepatocytes treated with etomoxir (10 or 100 μM) for 24 hours. Scale bar, 10 μm.
B) Quantification of autophagy performed as normalized LC3 puncta. Mean ± SEM (n=3 independent experiments) **P<0.01, ***P<0.001, n.s. not significant.
C) Representative confocal images of mRFP-GFP tandem fluorescent-tagged LC3 (tfLC3) in Mpc1fl/fl -siRNA, Mpc1fl/fl + Ad iCre -siRNA, Mpc1fl/fl + MICU1 siRNA, Mpc1fl/fl + Ad iCre + MICU1 siRNA hepatocytes. Scale bar, 10 μm.
D) Quantification of autophagy performed as normalized LC3 puncta. Mean ± SEM (n= 3 independent experiments). LC3 puncta was quantified from 10 cells per group. ****P<0.0001, n.s. not significant.
DISCUSSION
The ability to sense and respond to stress conditions is a key feature at cellular level that drives evolution and facilitates growth (46). This study revealed an unexpected role of a mCa2+ sensing protein in mediating responses to mitochondrial substrate availability. MICU1 protein abundance was induced by both blocking pyruvate and fatty acid transport into the mitochondria. Our analysis of MCU-mediated mCa2+ uptake under conditions of metabolic stress indicated that EGR1-mediated MICU1 upregulation prevented basal mCa2+ accumulation and reduced mitochondrial bioenergetics. Conversely, deletion of MPC caused an increase in cellular lactate and in oxidative stress. Functionally, we found that blockade of MPC-dependent pyruvate flux enhanced autophagy. Our findings suggest that substrate-dependent control of mCa2+ uptake could determine metabolic switching and cell survival under conditions of nutritional scarcity.
ER and the mitochondria are major metabolite sensing organelles (18, 20, 47-49). Proper protein folding in the ER is an energy consuming process. Conditions of nutrient stress limit energy production, thus halting the protein folding process. The ER senses an accumulation of unfolded proteins and triggers an unfolded protein response (UPR) that induces the transcription of a large set of nuclear-encoded genes to either degrade the unfolded proteins or to increase the capacity for protein folding. Thus, the ER senses nutrient deprivation and upregulates the UPR accordingly (49, 50). The mitochondrion, on the other hand, responds to nutrient stress by undergoing numerous posttranslational modifications of proteins and altering its dynamics (47). Metabolic changes in mitochondria cause broad changes in nuclear gene expression through retrograde mitochondria-to-nuclear signaling referred to as the mitochondrial stress response. Our data describes a mechanism by which MICU1 protein levels were elevated upon mitochondrial nutrient deprivation in various cell types, so that MICU1 may serve as a sensor of mitochondrial substrate levels (Fig. 1A to E, ). MICU1 is characterized as the gatekeeper of the mitochondrial calcium uniporter complex, limiting Ca2+ entry into the mitochondria under resting conditions (34). Consequently, increased MICU1 levels coincide with decreased basal resting matrix Ca2+, thereby protecting the cells from Ca2+ overload under conditions of metabolite scarcity (Fig. 2, B and F). To dissect the mechanisms of the mitochondrial metabolite-Ca2+ homeostasis axis, we utilized genetic and pharmacologic tools to silence and inhibit the mitochondrial pyruvate carrier proteins. The mitochondrial pyruvate transport machinery is comprised of MPC1 and MPC2 which form a hetero-oligomeric complex. Loss of one isoform results in loss of the other subunit, halting pyruvate flux into mitochondria. Genetic deletion of either MPC1 or MPC2 in mice results in embryonic lethality at E11 - E13.5, likely due to decreased mitochondrial oxidative phosphorylation activity (11, 51). Our results showed that cells in which MPC2 was silenced had a more severe phenotype compared to cells in which MPC1 was silenced, possibly due to the homo-oligomeric nature of MPC2. This original observation has been confirmed by a study using purified recombinant human MPC1 and MPC2 reconstituted in lipid vesicles. Whereas MPC2 homo-oligomers efficiently transport pyruvate at a rate similar to that of MPC1-MPC2 hetero-oligomers, MPC1 homo-oligomers do not transport pyruvate into the proteoliposomes (52). Disease-associated mutations in the mitochondrial pyruvate carrier cause severe neonatal metabolic acidosis with growth retardation leading to sudden death (7). Our findings reveal that loss of pyruvate entry into the mitochondrial matrix either due to silencing of MPC or due to mutations in MPC (MPC1R97W) led to elevated MICU1 protein levels. Luciferase reporter assays revealed MICU1 was transcriptionally elevated by EGR1 but not EGR4 (Fig. 3A and G).
Mitochondria rapidly adapt to changes in energy supply and demand and alter their capacity for and efficiency of ATP generation. Substrates such as pyruvate, succinate, and malate feed into the TCA cycle producing reducing equivalents that in turn supply the electron transport chain (ETC) for ATP generation. Loss of pyruvate flux into mitochondria led to a reduction in basal and maximal OCR and total cellular ATP levels and an increase in mROS levels. Production of reducing equivalents (NADH) was severely perturbed in cells with MPC knockdown compared to the control, revealing severe cellular bioenergetic crisis (Fig. 4 F). Additionally, we observed an increase in autophagy in the nutrient deprived cells, as well as in cells treated with pharmacologic inhibitors of TCA precursor oxidation and genetic deletion of MPC1 (Fig 6, A to F and 7A to D). In conclusion, we describe a mechanism by which mitochondria respond to metabolite deficiency through induction of MICU1, which controls mCa2+ uptake and bioenergetics. Thus, increased MICU1 levels protect the cells from Ca2+-overload and cell death in periods of nutritional deficiency.
MATERIALS AND METHODS
Cell line culture
HeLa, HEK 293T and HepG2 cells were cultured using low-glucose DMEM (GIBCO, Life Technologies) containing 10% (v/v) fetal bovine serum and 1% (v/v) penicillin/ streptomycin, with or without 2μg/ml puromycin. HepG2 and HEK293T cells (5x105 /well) grown in 6 well plates were transduced with lentiviruses expressing shRNA for NegShRNA, MPC1 (Sigma # SHCLNV-NM_016098) and MPC2 (Sigma #SHCLNV-NM_015415). HEK293T cells were transfected with FLAG-tagged MPC1R97W. Two days post transduction, the cells were selected with puromycin (2 μg/ml) or blasticidin (2 μg/ml) for 6–10 d and expanded.
Mouse models and primary cell culture
MPC1fl/fl mice originally were obtained from Eric Taylor (University of Iowa), where they were generated on a C57Bl/6N background, backcrossed through C57Bl/6J. MPC1fl/fl mice were maintained through homozygous breeding under standard housing conditions. EGR1−/− and EGR4−/− animals were previously generated (53-56). All animal experiments were approved by UT Health San Antonio’s and Temple University’s IACUC and followed AAALAC guidelines.
Primary mouse hepatocytes were isolated using a two-step collagenase perfusion technique with slight modifications (57, 58). In brief, mouse liver was sequentially perfused with perfusion medium-I (DPBS containing 10mM HEPES, 0.05% w/v KCl, 5mM glucose, 200 μM EDTA, pH 7.4) and perfusion medium-II (DPBS containing 30mM HEPES, 0.05% w/v KCL, 5mM Glucose, 1mM CaCl2, pH 7.4) containing collagenase D (400μg/ml). Liver lobes were dissected and dissociated, and crude hepatocyte preparation was passed through a gauze mesh filter (100μM diameter). The preparation was centrifuged at 50×g for 2 min to pellet parenchymal hepatocytes. The hepatocytes were washed five times with perfusion medium-II and plated in culture dishes in Williams E medium (supplemented with 1% (v/v) antibiotic-antimycotic solution (Gibco), 1% (v/v) 200mM L-glutamine, 1% (v/v) non-essential amino acids, and 10% (v/v) heat-inactivated fetal bovine serum). After four to six hours, the culture medium was replaced. MPCfl/fl hepatocytes were subsequently treated with Ad iCre (Vector BioLabs, Cat#1045N) using an MOI of 10-20. Mouse pulmonary microvascular endothelial cells (MPMVECs) were isolated from the lungs of wild-type, EGR1−/− and EGR4−/− mice as described elsewhere (59). Briefly, freshly harvested mouse lungs were treated with collagenase, and endothelial cells were isolated with magnetic beads coated with monoclonal antibody to CD144 (BD Biosciences Pharmingen). MPMVECs were propagated in DMEM complete media supplemented with 15% (v/v) FBS, nonessential amino acids, and penicillin/streptomycin. Cells that were not bound to the beads during MPMVEC isolation were plated separately and grown as murine lung fibroblasts (MLFs). MLFs were transduced with Ad-Cre-GFP virus (50 MOI). The loss of EGR1 and EGR4 were confirmed by western blotting.
Ventricular cardiomyocytes from one to two-day-old rat hearts (NRVMs) were prepared as previously described (60, 61). NRVMs were cultured in Ham’s F-10 supplemented with 5% fetal bovine serum (FBS) and penicillin/streptomycin (100 U/ml) at 37°C in a 95% air/5% CO2 humidified atmosphere for 4 days. NRVMs were transfected with indicated siRNAs and experiments were performed after 72 hr.
MPCfl/fl mouse embryonic fibroblasts (MEFs) were isolated by harvesting embryos from pregnant MPCfl/fl mice 13-14 days post-coitum (62, 63). Pregnant mice were anesthetized with isoflurane and killed by cervical dislocation. Embryos were immediately dissected and removed from the uterine cavity. Harvested embryos were rinsed in 70% (v/v) ethanol and placed in separate covered petri dishes containing 10 mL of 0.25% trypsin-EDTA. Heads of embryos were removed and used for genotypic confirmation. Embryonic tissues were then minced by chopping with a razor blade, and the resulting solution was incubated at 37°C for 15 minutes. The cell suspension was then transferred to a conical tube and fresh MEF growth media (DMEM supplemented with 10% (v/v) FBS, 1% (v/v) 200 mM L-glutamine, 1% antibiotic-antimycotic solution) was added to deactivate the trypsin. Cells were pelleted at 1000 rpm for 5 min and resuspended in fresh MEF growth medium and plated in T-75 flasks overnight. The following day, media was changed, and cells were allowed to expand before trypsinization, replating, and use in experiments.
Plasmids and antibodies
Peredox-mCherry (64) was purchased. Antibodies specific for MPC1 (Cell Signaling Technology, Cat # 14462S; 1:1000), MPC2 (Sigma, Cat# HPA056091; 1:500) LC3 (Sigma, Cat# L7543; 1:10000), p62 (Cell Signaling Technology, Cat# 5114; 1:1000), P-AMPK (Cell Signaling Technology; 1:1,000), AMPK (Cell Signaling Technology; 1:1000), β-actin (Santa Cruz Biotechnology Inc, Cat# sc-47778 HRP; 1:10000), FLAG (1:2000; Monoclonal anti-FLAG M2-peroxidase, Sigma), Tom 20 (Santa Cruz Biotechnology Inc, Cat# sc-11415). Antibodies specific for MICU1 were generated in-house (anti-rabbit MICU1) (30, 58, 65, 66).
RT-qPCR analysis
The knockdown expression was assessed by RT-qPCR. Briefly, total RNA was isolated from HepG2 Neg shRNA, MPC1 KD and MPC2 KD cells, and primary hepatocytes using the RNeasy Mini Kit (Qiagen, Valencia, CA) in accordance with the manufacturer’s instructions. Total RNA (1 μg) was reverse transcribed with iScript cDNA synthesis kit (BioRad). Real-time qPCR was performed with the gene specific Taqman qPCR primers as per the manufacturer’s instructions. The relative mRNA abundance was normalized to WT . (7300 Real Time PCR system RQ study software ; Applied Biosystems, Carlsbad, CA).
Matrix Ca2+ and ΔΨm measurement in permeabilized cells
Mitochondrial Ca2+ uptake and ΔΨm were determined by simultaneous monitoring of cytosolic Ca2+ with Fura-2FF (0.5 μM; Life Technologies) and ΔΨm with the lipophilic cationic dye 5,5′,6,6′-tetrachloro-3,3′-tetraethylbenzimidazolcarbocyanine (JC-1; 800 nM; Life Technologies). After trypsinization and neutralization, cells were pelleted at 200 × g for 5 min, resuspended in 20 mL of phosphate-buffered saline, and pelleted at 150 × g for 2 min. 8 × 106 cells were resuspended in 1.5 mL ICM buffer containing 40 μg/ml digitonin to permeabilize the cells, protease inhibitors (EDTA-free complete tablets; Roche Applied Science, Indianapolis, IN), 2 μM thapsigargin to block the SERCA pump, and 2 mM succinate to energize mitochondria (67). Fura-2FF (1 μM) was used as a cytosolic Ca2+ indicator. After 20 s of data recording, JC-1 was added. At 1200 s or 400 s CCCP, 10 μM, was added. Fluorescence was measured using a dual-wavelength spectrofluorometer (PTI) with 490-nm excitation and 535-nm emission for monomeric JC-1 and 570/595 nm for the J-aggregate. ΔΨm was calculated as the ratio of J-aggregate and the monomer (68).
Cytosolic and mitochondrial Ca2+ dynamics
HepG2 negative shRNA (NegshRNA), MPC1 knockdown and MPC2 knockdown cells were grown on 25-mm glass coverslips for 48 h and loaded with 2 μM Rhod-2 AM (50 min) and 5 μM Fluo-4 AM (30 min) (Life Technologies) in extracellular medium (69, 70). After 1 min of baseline recording, agonist (ATP, 10 μM) was added, and confocal images were recorded every 3 s (510 Meta; Carl Zeiss, Thornwood, NY) at 488- and 561-nm excitation using a 63× oil objective to simultaneously monitor cytoplasmic and mitochondrial Ca2+ dynamics. Images were analyzed and quantified using ImageJ (National Institutes of Health, Bethesda, MD) and custom-made software (Spectralyzer, Elmsford, NY).
Confocal analysis of mitochondrial Ca2+ and ΔΨm
Freshly harvested primary murine hepatocytes were cultured on collagen coated glass coverslips and infected with an adenovirus construct of mito-GCaMP6 for 24 hours. The next day, cells were incubated with TMRE (50 nM) for 30 min at 37°C. Images were acquired using a Leica SP8 confocal microscope using a 100× oil objective at excitation wavelengths of 488 nm and 556 nm. After 30 sec baseline recording, ionomycin (2.5 μM) was added and images collected every 3 seconds. Images were quantified for mito-GCaMP6 and TMRE (50 nM) fluorescence using LASX software (Leica) and plotted as arbitrary units in GraphPad Prism 6 software. Mitochondrial length, perimeter, and surface area were quantified using ImageJ and plotted in GraphPad Prism.
Immunoblotting
HepG2 cells, HEK 293T cells, cardiomyocytes, and hepatocytes (from WT, MPCfl/fl, EGR1−/− and EGR4−/− mice) were treated with or without the inhibitors 2DG (1mM), etomoxir (10μM), UK5099 (100μM) or all three overnight and lysed using RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.25% deoxycholic acid, 1 mM EDTA, 1% NP-40, protease inhibitor cocktail (Complete; Roche and 1 mM PMSF)). Equal amounts of protein were separated on 4-12% Bis-Tris polyacrylamide gel, transferred to a PVDF membrane, and probed with corresponding antibodies as specified.
Mitochondrial ROS measurement
Mitochondrial superoxide was measured by using the mitochondrial oxygen free radical indicator MitoSOX Red (molecular probes; Invitrogen) as described previously (70, 71). Briefly, cells grown on 0.2% gelatin coated glass coverslips were loaded with 5 μM MitoSOX Red for 30 min, and coverslips were mounted in an open perfusion microincubator (PDMI-2; Harvard Apparatus) at 37°C and imaged. Confocal (510 Meta; Carl Zeiss, Inc.) images were obtained at 561 nm excitation by using a 63× oil objective. Images were analyzed, and the mean MitoSOX Red fluorescence was quantified by using Image J software (NIH).
Oxygen consumption rate
Oxygen consumption rate (OCR) was measured in MPCfl/fl primary hepatocytes, HepG2 control, MPC1 knockdown and MPC2 knockdown at 37°C in an XF96 extracellular flux analyzer (Seahorse Bioscience). Cells (ECs; 2×104, cardiomyocytes; 4× 103, hepatocytes; 6 x 104) were sequentially exposed to oligomycin, FCCP, and rotenone plus antimycin A (72).
Total cellular ATP
Total ATP abundance was assessed using CellTiter-Glo luminescent assay, and luminescence was measured using the Infinite M1000 PRO plate reader (, Tecan).
Luciferase assay
HEK293 cells transfected with scrambled or MPC2 siRNA grown on 6 well plates were transfected with either MICU1 promoter reporter plasmid or control vector using Mirus LT-1 transfection reagent (Mirus) as per the manufacturer’s instructions. 48 hr post-transfection, cells were lysed (Promega) and the luciferase activity was detected using Bright-Glo Luciferase Assay System (Promega) as per the manufacturer’s instruction. Values were normalized with untreated controls and the relative luciferase activity (RLU) was plotted using GraphPad PRISM software version 5.0 (GraphPad).
NADH measurement
HepG2 NegshRNA and MPC2 KD cells (1×105 cells) were transiently transfected with peredox m-Cherry. Cells were imaged 48 hr post transfection at 37°C. Confocal images were acquired at 405 nm and 561 nm excitation every 3s using Carl Zeiss 710 Meta NLO. Images were analyzed using ZEN 2010 software and plotted with SigmaPlot. (73).
Autophagy
HeLa NegshRNA, MPC2 knockdown, MPC1R97W and primary hepatocyges were plated on 0.2% gelatin coated cover slips. Cells were infected with adenovirus expressing mRFP-GFP-LC3. After 36 hours of infection confocal images were obtained at 561 and 488 nm excitation using a 40× oil objective (510 Meta; Carl Zeiss, Inc.) and analyzed and quantified using ZEN 2010 software. Autophagic flux was determined by evaluating the punctate pattern of GFPLC3/ mRFP as described earlier (74). For primary hepatocyte autophagic flux analysis, images were acquired using a Leica SP8 confocal microscope using a 100× oil objective at excitations of 488 nm and 556 nm. Images were analyzed using LASX software (Leica) and plotted in GraphPad Prism 6 software.
Statistical analysis
Data were expressed as the mean ± SEM. Statistical significance was evaluated via Student’s unpaired t test, one-way and two-way ANOVA. P <0.05 was considered statistically significant. All experiments were conducted at least three times unless otherwise specified. Data were plotted either with Sigma Plot 11.0 software or GraphPad Prism version 6 software.
Supplementary Material
Figure S1: MPC1 and MPC2 abundance following RNAi-mediated knockdown.
Figure S2: Silencing MPC alters mitochondrial Ca2+ uptake in a MICU1-dependent manner.
Figure S3. MPC1-deficient MEFs exhibit reduced MCU-mediated mitochondrial Ca2+ uptake and altered mitochondrial morphology.
Figure S4: Pyruvate carrier mutations result in decreased matrix Ca2+ in a MICU1-dependent manner.
Figure S5: Pharmacologic blockade of fatty acid and pyruvate flux by etomoxir and UK5099 enhances MICU1 mRNA abundance.
Acknowledgements:
We thank Gary Yellen (Harvard Medical School) for sharing Peredox plasmid construct. We also thank Eric Taylor for sharing MPC1fl/fl mice.
Funding: This research was funded by the National Institutes of Health (R01GM109882, R01HL086699, R01HL142673, and 1S10RR027327 to M.M. This work was partly supported by DOD/DHP-CDMRP PR181598P-1 to MM and KS. N.N., is supported by the AHA fellowships (17PRE33660720). S.P is supported by NIH K99/R00 fellowship (1K99HL138268-01). Z.D. is supported by China Scholarship Council (No.201403170252). B.T.E. is supported by both the National Center for Advancing Translational Sciences and the National Institute of General Medical Sciences of the NIH under award numbers TL1 TR002647 and K12 GM11726.
Footnotes
Competing Interests: The authors declare that they have no competing interests.
Data and Materials Availability: All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
REFERENCES AND NOTES
- 1.Vafai SB, Mootha VK, Medicine. A common pathway for a rare disease? Science 342, 1453–1454 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Clapham DE, Calcium signaling. Cell 80, 259–268 (1995). [DOI] [PubMed] [Google Scholar]
- 3.Pagliarini DJ, Rutter J, Hallmarks of a new era in mitochondrial biochemistry. Genes Dev 27, 2615–2627 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams GS, Boyman L, Chikando AC, Khairallah RJ, Lederer WJ, Mitochondrial calcium uptake. Proc Natl Acad Sci U S A 110, 10479–10486 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herzig S et al. , Identification and Functional Expression of the Mitochondrial Pyruvate Carrier. Science, (2012). [DOI] [PubMed] [Google Scholar]
- 6.Bricker DK et al. , A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science 337, 96–100 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brivet M et al. , Impaired mitochondrial pyruvate importation in a patient and a fetus at risk. Molecular genetics and metabolism 78, 186–192 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Awata H, Endo F, Tanoue A, Kitano A, Matsuda I, Characterization of a point mutation in the pyruvate dehydrogenase E1 alpha gene from two boys with primary lactic acidaemia. Journal of inherited metabolic disease 17, 189–195 (1994). [DOI] [PubMed] [Google Scholar]
- 9.De Meirleir L, Lissens W, Vamos E, Liebaers I, Pyruvate dehydrogenase (PDH) deficiency caused by a 21-base pair insertion mutation in the E1 alpha subunit. Human genetics 88, 649–652 (1992). [DOI] [PubMed] [Google Scholar]
- 10.Brown GK, Pyruvate dehydrogenase E1 alpha deficiency. Journal of inherited metabolic disease 15, 625–633 (1992). [DOI] [PubMed] [Google Scholar]
- 11.Vigueira PA et al. , Mitochondrial pyruvate carrier 2 hypomorphism in mice leads to defects in glucose-stimulated insulin secretion. Cell reports 7, 2042–2053 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vanderperre B, Bender T, Kunji ER, Martinou JC, Mitochondrial pyruvate import and its effects on homeostasis. Curr Opin Cell Biol 33, 35–41 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Schell JC et al. , A role for the mitochondrial pyruvate carrier as a repressor of the Warburg effect and colon cancer cell growth. Molecular cell 56, 400–413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee BH et al. , Atypical manifestation of carnitine palmitoyltransferase 1A deficiency: hepatosplenomegaly and nephromegaly. Journal of pediatric gastroenterology and nutrition 60, e19–22 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Bennett MJ, Santani AB, in GeneReviews((R)), Adam MP et al. , Eds. (Seattle (WA: ), 1993). [Google Scholar]
- 16.Choi JS et al. , Novel Mutations in the CPT1A Gene Identified in the Patient Presenting Jaundice as the First Manifestation of Carnitine Palmitoyltransferase 1A Deficiency. Pediatric gastroenterology, hepatology & nutrition 19, 76–81 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmieri F, Diseases caused by defects of mitochondrial carriers: a review. Biochimica et biophysica acta 1777, 564–578 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Elouil H et al. , Acute nutrient regulation of the unfolded protein response and integrated stress response in cultured rat pancreatic islets. Diabetologia 50, 1442–1452 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Gonzales JC et al. , Chemical induction of the unfolded protein response in the liver increases glucose production and is activated during insulin-induced hypoglycaemia in rats. Diabetologia 51, 1920–1929 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mandl J, Meszaros T, Banhegyi G, Hunyady L, Csala M, Endoplasmic reticulum: nutrient sensor in physiology and pathology. Trends in endocrinology and metabolism: TEM 20, 194–201 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Klec C et al. , Presenilin-1 Established ER-Ca(2+) Leak: a Follow Up on Its Importance for the Initial Insulin Secretion in Pancreatic Islets and beta-Cells upon Elevated Glucose. Cell Physiol Biochem 53, 573–586 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Klec C et al. , Glycogen Synthase Kinase 3 Beta Controls Presenilin-1-Mediated Endoplasmic Reticulum Ca(2)(+) Leak Directed to Mitochondria in Pancreatic Islets and beta-Cells. Cell Physiol Biochem 52, 57–75 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gomes LC, Di Benedetto G, Scorrano L, During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nature cell biology 13, 589–598 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toyama EQ et al. , Metabolism. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science (New York, N.Y.) 351, 275–281 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Egan DF et al. , Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331, 456–461 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berridge MJ, Bootman MD, Roderick HL, Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol 4, 517–529 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Glancy B, Balaban RS, Role of mitochondrial Ca2+ in the regulation of cellular energetics. Biochemistry 51, 2959–2973 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baughman JM et al. , Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature 476, 341–345 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Stefani D, Raffaello A, Teardo E, Szabo I, Rizzuto R, A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature 476, 336–340 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shanmughapriya S et al. , Ca2+ signals regulate mitochondrial metabolism by stimulating CREB-mediated expression of the mitochondrial Ca2+ uniporter gene MCU. Science signaling 8, ra23 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halestrap AP, The mitochondrial pyruvate carrier. Kinetics and specificity for substrates and inhibitors. The Biochemical journal 148, 85–96 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Combs DJ, Reuland DS, Martin DB, Zelenock GB, D'Alecy LG, Glycolytic inhibition by 2-deoxyglucose reduces hyperglycemia-associated mortality and morbidity in the ischemic rat. Stroke 17, 989–994 (1986). [DOI] [PubMed] [Google Scholar]
- 33.Lopaschuk GD, Wall SR, Olley PM, Davies NJ, Etomoxir, a carnitine palmitoyltransferase I inhibitor, protects hearts from fatty acid-induced ischemic injury independent of changes in long chain acylcarnitine. Circ Res 63, 1036–1043 (1988). [DOI] [PubMed] [Google Scholar]
- 34.Mallilankaraman K et al. , MICU1 Is an Essential Gatekeeper for MCU-Mediated Mitochondrial Ca2+ Uptake that Regulates Cell Survival. Cell 151, 630–644 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffman NE et al. , MICU1 motifs define mitochondrial calcium uniporter binding and activity. Cell reports 5, 1576–1588 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gottschalk B et al. , MICU1 controls cristae junction and spatially anchors mitochondrial Ca(2+) uniporter complex. Nature communications 10, 3732 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Csordas G et al. , MICU1 controls both the threshold and cooperative activation of the mitochondrial Ca(2)(+) uniporter. Cell Metab 17, 976–987 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamer KJ, Mootha VK, MICU1 and MICU2 play nonredundant roles in the regulation of the mitochondrial calcium uniporter. EMBO reports 15, 299–307 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gashler A, Sukhatme VP, Early growth response protein 1 (Egr-1): prototype of a zinc-finger family of transcription factors. Prog Nucleic Acid Res Mol Biol 50, 191–224 (1995). [DOI] [PubMed] [Google Scholar]
- 40.Kaufmann K, Bach K, Thiel G, The extracellular signal-regulated protein kinases Erk1/Erk2 stimulate expression and biological activity of the transcriptional regulator Egr-1. Biological chemistry 382, 1077–1081 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Thiel G, Cibelli G, Regulation of life and death by the zinc finger transcription factor Egr-1. Journal of cellular physiology 193, 287–292 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Shanmughapriya S et al. , FOXD1-dependent MICU1 expression regulates mitochondrial activity and cell differentiation. Nature communications 9, 3449 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mizushima N, Levine B, Cuervo AM, Klionsky DJ, Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lum JJ, DeBerardinis RJ, Thompson CB, Autophagy in metazoans: cell survival in the land of plenty. Nature Reviews Molecular Cell Biology 6, 439–448 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Mizushima N, Yoshimori T, Levine B, Methods in Mammalian Autophagy Research. Cell 140, 313–326 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Badyaev AV, Stress-induced variation in evolution: from behavioural plasticity to genetic assimilation. Proceedings. Biological sciences 272, 877–886 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao AW, Canto C, Houtkooper RH, Mitochondrial response to nutrient availability and its role in metabolic disease. EMBO Mol Med 6, 580–589 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Efeyan A, Comb WC, Sabatini DM, Nutrient-sensing mechanisms and pathways. Nature 517, 302–310 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaufman RJ et al. , The unfolded protein response in nutrient sensing and differentiation. Nature reviews. Molecular cell biology 3, 411–421 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Kaufman RJ, Orchestrating the unfolded protein response in health and disease. The Journal of clinical investigation 110, 1389–1398 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vanderperre B et al. , Embryonic Lethality of Mitochondrial Pyruvate Carrier 1 Deficient Mouse Can Be Rescued by a Ketogenic Diet. PLoS genetics 12, e1006056 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagampalli RSK et al. , Human mitochondrial pyruvate carrier 2 as an autonomous membrane transporter. Scientific reports 8, 3510 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tourtellotte WG, Nagarajan R, Bartke A, Milbrandt J, Functional compensation by Egr4 in Egr1-dependent luteinizing hormone regulation and Leydig cell steroidogenesis. Molecular and cellular biology 20, 5261–5268 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee SL, Tourtellotte LC, Wesselschmidt RL, Milbrandt J, Growth and differentiation proceeds normally in cells deficient in the immediate early gene NGFI-A. J Biol Chem 270, 9971–9977 (1995). [DOI] [PubMed] [Google Scholar]
- 55.Tourtellotte WG, Nagarajan R, Auyeung A, Mueller C, Milbrandt J, Infertility associated with incomplete spermatogenic arrest and oligozoospermia in Egr4-deficient mice. Development 126, 5061 (1999). [DOI] [PubMed] [Google Scholar]
- 56.Lee SL et al. , Luteinizing hormone deficiency and female infertility in mice lacking the transcription factor NGFI-A (Egr-1). Science 273, 1219–1221 (1996). [DOI] [PubMed] [Google Scholar]
- 57.Li WC, Ralphs KL, Tosh D, Isolation and culture of adult mouse hepatocytes. Methods Mol Biol 633, 185–196 (2010). [DOI] [PubMed] [Google Scholar]
- 58.Tomar D et al. , Blockade of MCU-Mediated Ca(2+) Uptake Perturbs Lipid Metabolism via PP4-Dependent AMPK Dephosphorylation. Cell reports 26, 3709–3725 e3707 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Milovanova T et al. , Lung endothelial cell proliferation with decreased shear stress is mediated by reactive oxygen species. American journal of physiology. Cell physiology 290, C66–76 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Brinks H et al. , Level of G protein-coupled receptor kinase-2 determines myocardial ischemia/reperfusion injury via pro- and anti-apoptotic mechanisms. Circ Res 107, 1140–1149 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vagnozzi RJ et al. , Inhibition of the cardiomyocyte-specific kinase TNNI3K limits oxidative stress, injury, and adverse remodeling in the ischemic heart. Science translational medicine 5, 207ra141 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Durkin ME, Qian X, Popescu NC, Lowy DR, Isolation of Mouse Embryo Fibroblasts. Bio Protoc 3, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lindsten T et al. , The combined functions of proapoptotic Bcl-2 family members bak and bax are essential for normal development of multiple tissues. Mol Cell 6, 1389–1399 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hung YP, Albeck JG, Tantama M, Yellen G, Imaging cytosolic NADH-NAD(+) redox state with a genetically encoded fluorescent biosensor. Cell Metab 14, 545–554 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tomar D et al. , MCUR1 Is a Scaffold Factor for the MCU Complex Function and Promotes Mitochondrial Bioenergetics. Cell reports 15, 1673–1685 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dong Z et al. , Mitochondrial Ca(2+) Uniporter Is a Mitochondrial Luminal Redox Sensor that Augments MCU Channel Activity. Mol Cell 65, 1014–1028 e1017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hawkins BJ et al. , S-glutathionylation activates STIM1 and alters mitochondrial homeostasis. The Journal of cell biology 190, 391–405 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Irrinki KM et al. , Requirement of FADD, NEMO, and BAX/BAK for Aberrant Mitochondrial Function in Tumor Necrosis Factor Alpha-Induced Necrosis. Molecular and cellular biology 31, 3745–3758 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mallilankaraman K et al. , MCUR1 is an essential component of mitochondrial Ca2+ uptake that regulates cellular metabolism. Nature cell biology 14, 1336–1343 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Madesh M et al. , Selective role for superoxide in InsP3 receptor-mediated mitochondrial dysfunction and endothelial apoptosis. The Journal of cell biology 170, 1079–1090 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mukhopadhyay P et al. , Simultaneous detection of apoptosis and mitochondrial superoxide production in live cells by flow cytometry and confocal microscopy. Nat Protoc 2, 2295–2301 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Doonan PJ et al. , LETM1-dependent mitochondrial Ca2+ flux modulates cellular bioenergetics and proliferation. The FASEB Journal 28, 4936–4949 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shanmughapriya S et al. , SPG7 is an Essential and Conserved Component of the Mitochondrial Permeability transition Pore. Biophysical journal 110, 309a–310a (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hariharan N, Zhai P, Sadoshima J, Oxidative Stress Stimulates Autophagic Flux During Ischemia/Reperfusion. Antioxidants & Redox Signaling 14, 2179–2190 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: MPC1 and MPC2 abundance following RNAi-mediated knockdown.
Figure S2: Silencing MPC alters mitochondrial Ca2+ uptake in a MICU1-dependent manner.
Figure S3. MPC1-deficient MEFs exhibit reduced MCU-mediated mitochondrial Ca2+ uptake and altered mitochondrial morphology.
Figure S4: Pyruvate carrier mutations result in decreased matrix Ca2+ in a MICU1-dependent manner.
Figure S5: Pharmacologic blockade of fatty acid and pyruvate flux by etomoxir and UK5099 enhances MICU1 mRNA abundance.