Abstract
Background
Infectious disease predictions models, including virtually all epidemiological models describing the spread of the SARS-CoV-2 pandemic, are rarely evaluated empirically. The aim of the present study was to investigate the predictive accuracy of a prognostic model for forecasting the development of the cumulative number of reported SARS-CoV-2 cases in countries and administrative regions worldwide until the end of May 2020.
Methods
The cumulative number of reported SARS-CoV-2 cases was forecasted in 251 regions with a horizon of two weeks, one month, and two months using a hierarchical logistic model at the end of March 2020. Forecasts were compared to actual observations by using a series of evaluation metrics.
Results
On average, predictive accuracy was very high in nearly all regions at the two weeks forecast, high in most regions at the one month forecast, and notable in the majority of the regions at the two months forecast. Higher accuracy was associated with the availability of more data for estimation and with a more pronounced cumulative case growth from the first case to the date of estimation. In some strongly affected regions, cumulative case counts were considerably underestimated.
Conclusions
With keeping its limitations in mind, the investigated model may be used for the preparation and distribution of resources during the initial phase of epidemics. Future research should primarily address the model’s assumptions and its scope of applicability. In addition, establishing a relationship with known mechanisms and traditional epidemiological models of disease transmission would be desirable.
Keywords: Coronavirus, COVID-19, Communicable diseases, Epidemiologic methods, Forecasting, Statistical models, Public health
Background
Mathematical and simulation models of infectious disease dynamics are essential for understanding and forecasting the development of epidemics [1]. The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has called increased attention to epidemiological modeling both as a method of scientific inquiry and as a tool to inform political decision making [2, 3].
Among epidemiological modeling methods, a distinction between mechanistic and phenomenological approaches is frequently made. While mechanistic approaches model the transmission dynamics based on substantial concepts from biology, virology, infectology, and related disciplines, phenomenological (sometimes termed ‘statistical’) models are looking for a mathematical function that fits observed data well without clear assumptions about the underlying processes [1, 2]. Mechanistic models are usually used to compare possible scenarios and to estimate the relative effects of different interventions rather than to produce precise predictions. On the contrary, phenomenological models are commonly optimized for forecasting. From a broader perspective, mechanistic and phenomenological approaches can be considered as the epidemiological modeling representatives of the long-standing explanation-prediction controversy [4]. It should be noted that although the distinction between these two model classes is instructive and one side usually predominates, most approaches have both mechanistic and phenomenological components, and some are explicitly balanced (so called ‘semi-mechanistic’ or ‘hybrid’ models).
Although the value of any predictive model is ultimately determined by whether it improves critical decision making [5, 6], a rigorous scientific appraisal should also include a comparison of what have been predicted to what have actually happened [1, 7, 8]. Unfortunately, the predictive accuracy of infectious disease predictions models is rarely evaluated during or after outbreaks [7, 8]. Notable exceptions include systematic evaluation of models about the epidemiology of severe acute respiratory syndrome (SARS) [9, 10], influenza [11, 12], ebola [5, 7, 13, 14], dengue [8, 15], foot-and-mouth disease [6], and trachoma [16].
The SARS-CoV-2 pandemic has prompted a large amount of epidemiological modeling efforts, including studies with primarily mechanistic (e.g., references [17–21]) and primarily phenomenological (e.g., references [22, 23]) approaches. According to the knowledge of the author up to October 2020, a truly prognostic evaluation of existing models using new data that had not yet been available at the point of model development is not common practice. In order to start closing this gap, the objective of the present study was to evaluate the predictive accuracy of a phenomenologically oriented model that was calibrated on data up to the end of March 2020 for forecasting the development of the cumulative number of reported SARS-CoV-2 cases in countries and administrative regions worldwide [24].
Methods
Data
As described in detail elsewhere [24], the model was fitted using information on the cumulative number of confirmed SARS-CoV-2 infections in the COVID-19 data repository of the Johns Hopkins University Center for Systems Science and Engineering [25, 26]. Cumulative case count data from 251 countries and administrative regions were used for calibrating the model, with daily time series from the day of the first reported case to 29 March 2020 in each region. For evaluation, data on confirmed cases were extracted from the same database two weeks, one month, and two months after model development (12 April, 29 April, and 29 May 2020). Sufficient information for creating predictions of the most likely number of cases in all investigated countries and administrative regions for any time horizon was made publicly available at the beginning of April 2020 [24].
Model
A hierarchical logistic model was fit to observed data [24]. The logistic part of the model was based on the ecological concept of self-limiting population growth [27] and used a formulation with five parameters [28], controlling the expected final case count at the end of the outbreak (parameter a), the maximum speed of reaching the expected final case count (parameter b), the approximate time point of the transition of the outbreak from an accelerating to a decelerating dynamic (parameter c), the case count at the beginning of the outbreak (parameter d), and the degree of asymmetry between the accelerating and decelerating phases of the outbreak (parameter g). The predicted number of cumulative case counts in region i at day t from the first reported case was estimated as
with log-normally distributed errors.
The hierarchical part of the model was inspired by random-effect meta-analysis assuming that the parameters of the logistic equation are similar, but not necessarily identical, across the investigated regions [29, 30]. This was implemented by restricting the parameters of the logistic equation to follow a normal distribution in the population of regions. With respect to interpretation, this means that the model was based on the hypothesis, that the pandemic runs a similar course in all countries and regions, even though they are expected to differ to a certain degree regarding the number of cases in their first report, the expected final case count, the time point and speed of the accelerating and decelerating phases of the outbreak, as well as the time point, extent, and effects of control measures.
Estimation
The statistical procedures and program code are described in detail elsewhere [24]. Computations were performed in a Bayesian framework using Markov chain Monte Carlo sampling methods in WinBUGS version 1.4.3 [31]. Parameter estimates were given uninformative priors, and results were obtained from three independent Markov chains with a total of 60,000 iterations and a thinning rate of 60, after dropping 40,000 burn-in simulations.
Evaluation metrics
For evaluating each individual estimate i at time point t, four measures were calculated.
The difference between logarithmic predicted and observed counts (“error in logs”, EIL) was defined as
with ln being the natural logarithm, and npred and nobs being the predicted and the observed cumulative case counts, respectively.
The absolute error in logs (AIEL) was calculated as
The percentage error (PE) was calculated as
and the absolute percentage error (APE) as
Summary estimates of predictive accuracy across all k regions at a given time point t are listed in the following.
The root mean squared error in logs (RMSE) was defined as
and the mean absolute percentage error (MAPE) was calculated as
The coefficient of determination R2t was additionally determined from a linear model regressing the logarithmic observed values on the logarithmic predictions with the intercept fixed at zero. Furthermore, the intraclass correlation coefficient ICC(3,1)t was calculated for quantifying the level of absolute agreement between predicted and observed values from a two-way mixed-effects model [32]. Bootstrapping was used with 1000 samples to create 95% confidence intervals for summary estimates of predictive accuracy.
Factors associated with accuracy
In order to identify factors associated with the accuracy of the predictions, the AEIL was regressed on the number of available data points, the difference in the logarithm of the first and the last case count at the moment of estimation (as a proxy for progress of the epidemic), and their interaction term. Estimates are reported with 95% parametric confidence intervals.
Furthermore, strongly affected regions (defined by a minimum of 10,000 cases at the forecasted time point) with the most extreme under- and overestimation were identified to gain additional qualitative insights on model performance.
Results
Data
In 251 regions, the number of available data points at estimation ranged from 2 to 68 with a median of 25 and a mean of 31.48 days. The cumulative number of reported cases at the point of the first non-zero count ranged from 1 to 444 with a median of 1 and a mean of 4.09 across regions. The cumulative number of reported cases at model estimation (29 March 2020) ranged from 1 to 140,886 with a median of 139 and a mean of 2869.
Individual estimates of predictive accuracy
The probability density function of the percentage error (PE) at the day of estimation as well at the forecasts after two weeks, one month, and two months, respectively, is displayed in Fig. 1. At the day of estimation, the median relative error indicated an average underestimation of the cumulative case count by about one third across regions. The relative error distribution was rather narrow, with only a tenth of predictions showing an underestimation exceeding − 62.8% and none of the predictions having more than 36.9% error. Across forecasts, the median percentage error was always less than 20%, although an overestimation by more than two hundred percent was observed in 7.2, 19.1, and 19.5% of the cases at the two weeks, one months, and two months forecasts, respectively. The proportion of regions with an underestimation exceeding minus two thirds (− 66.6%) was 12.4, 19.5, and 28.7% at the two weeks, one months, and two months forecasts, respectively.
Fig. 1.
Probability density function of the percentage error at different forecast horizons. The solid line shows the median, the dashed lines show the first and third quartiles, and the dotted lines show the first and ninth deciles. The x-axis is trimmed at 2.5
The calibration plots suggest an increasing number of regions for which case counts are substantially under- or overestimated with increasing length of the forecast period (Fig. 2). Nevertheless, a strong positive association between predicted and observed case counts is apparent even after two months.
Fig. 2.
Calibration plots at different forecast horizons. Points refer to regions. The solid black line indicates no prediction error, the blue area indicates a prediction error by a factor of two or less, and the green area indicates a prediction error by a factor of ten or less. Both axes are log-transformed
Summary estimates of predictive accuracy
All parameters show an increasing amount of error with increasing length of the forecast period (Table 1). The MAPE shows that, on average, estimates are off by more than one hundred, two hundred, and four hundred percent at the two weeks, one month, and two months forecasts, respectively. The coefficient of determination indicates a very strong relative association between predicted and observed case counts, and the intraclass correlation coefficient suggests that the level of absolute agreement is excellent after two weeks and still high after one month, but sinks to a moderate level after two months.
Table 1.
Summary estimates of predictive accuracy
| RMSE (95% CI) |
MAPE (95% CI) |
R2 (95% CI) |
ICC (95% CI) |
|
|---|---|---|---|---|
| Day of estimation | 0.640 (0.577 to 0.707) | 0.323 (0.295 to 0.356) | 0.989 (0.986 to 0.992) | 0.984 (0.979 to 988) |
| Two weeks forecast | 0.900 (0.803 to 1.05) | 1.085 (0.673 to 2.598) | 0.980 (0.971 to 0.984) | 0.935 (0.905 to 0.950) |
| One month forecast | 1.393 (1.271 to 1.546) | 2.133 (1.600 to 2.953) | 0.958 (0.948 to 0.966) | 0.828 (0.777 to 0.866) |
| Two months forecast | 1.958 (1.791 to 2.157) | 4.250 (2.907 to 6.735) | 0.931 (0.914 to 0.943) | 0.679 (0.606 to 0.748) |
RMSE root mean squared error in logarithmic case counts, MAPE mean absolute percentage error in case counts, R2 coefficient of determination, ICC intraclass correlation, CI confidence interval
Factors associated with accuracy
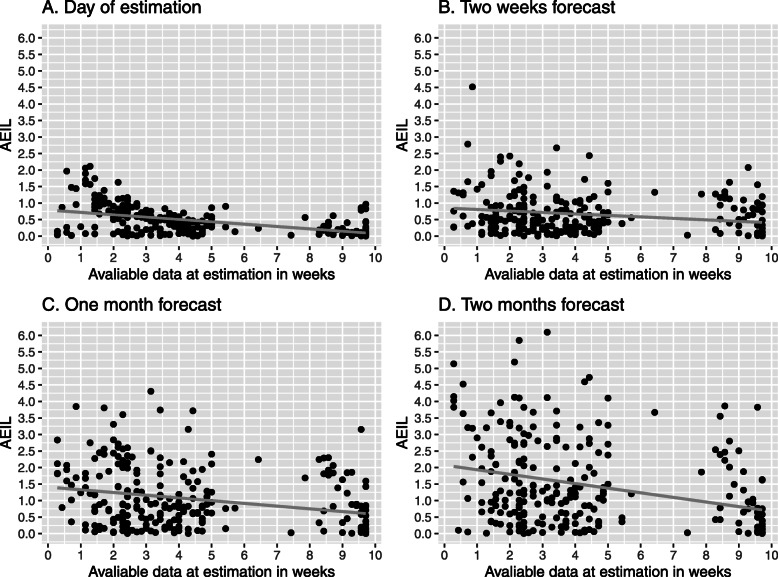
Visual analysis suggests that a larger number of available data points at estimation (Fig. 3) and a more extensive growth of the logarithmic case counts from the first reported case until estimation (Fig. 4) are associated with a lower prediction error. This is confirmed by regression analyses indicating statistically significant associations that are becoming stronger with increasing forecast horizon (Table 2). These two factors have also a multiplicative effect, as indicated by the statistically significant interaction term.
Fig. 3.
Association of the amount of available data at estimation and predictive accuracy (AEIL) at different forecast horizons. AEIL = absolute difference between logarithmic predicted and observed case counts. Points refer to regions. The grey line corresponds to a linear smoothing curve
Fig. 4.
Association of growth in logarithmic case counts until estimation and predictive accuracy (AEIL) at different forecast horizons. AEIL = absolute difference between logarithmic predicted and observed case counts. Points refer to regions. The grey line corresponds to a linear smoothing curve
Table 2.
Linear regression coefficients for factors associated with prediction accuracy (AEIL)
| Number of data points in weeks (95% CI) |
Growth in logarithmic case counts until estimation (95% CI) |
Interaction term (95% CI) |
|
|---|---|---|---|
| Day of estimation | −0.077*** (− 0.114 to − 0.040) | −0.016 (− 0.055 to 0.023) | 0.002 (− 0.005 to 0.009) |
| Two weeks forecast | −0.073* (− 1.304 to − 0.015) | −0.100** (− 1.614 to − 0.039) | 0.011* (0.000 to 0.022) |
| One month forecast | − 0.131** (− 0.216 to − 0.046) | −0.145** (− 0.235 to − 0.054) | 0.017* (0.001 to 0.034) |
| Two months forecast | −0.242*** (− 0.361 to − 0.124) | −0.242*** (− 0.368 to − 0.117) | 0.032** (0.010 to 0.055) |
AEIL = absolute difference between logarithmic predicted and observed case counts; CI = confidence interval; *p < .050; **p < .010; ***p < .001
Strongly affected regions (a minimum of 10,000 cases) with extreme under- or overestimation of the cumulative case counts are presented in Table 3. Among the listed regions, the extent of underestimation was considerable (an EIL below − 1.6, roughly corresponding to an underestimation by a factor of five) at the one and two months forecasts, with most regions being located in Asia. Among strongly affected regions, overestimation was rather moderate (an EIL below 0.7, roughly corresponding to an overestimation by a factor of two) in most cases. Substantial overestimation (an EIL between 0.7 and 1.6) was present in Austria and Switzerland at the one and two months forecasts and in the United States at the one month forecast. No strongly affected region with a considerable overestimation (EIL above 1.6) was identified.
Table 3.
Most extreme under- or overestimation for regions with a minimum number of 10,000 cases
| Underestimation | Overestimation | |||
|---|---|---|---|---|
| Region | EIL | Region | EIL | |
| Day of estimation | Belgium | −0.565 | Hubei, China | 0.022 |
| United States of America | −0.444 | Germany | 0.020 | |
| Netherlands | −0.422 | NA | NA | |
| Switzerland | −0.322 | NA | NA | |
| Italy | −0.301 | NA | NA | |
| Two weeks forecast | Belgium | −1.274 | Austria | 0.657 |
| Sweden | −1.171 | Quebec, Canada | 0.498 | |
| Russia | −0.939 | Switzerland | 0.399 | |
| France | −0.651 | United States of America | 0.336 | |
| Iran | −0.556 | Germany | 0.096 | |
| One month forecast | Belarus | −3.719 | Austria | 1.281 |
| Qatar | − 3.159 | Switzerland | 0.889 | |
| Singapore | −3.155 | United States of America | 0.714 | |
| India | −2.301 | Quebec, Canada | 0.638 | |
| Russia | −2.290 | Portugal | 0.402 | |
| Two months forecast | Bangladesh | −6.097 | Austria | 1.398 |
| Belarus | −4.730 | Switzerland | 1.012 | |
| Qatar | −4.597 | United States of America | 0.399 | |
| Kuwait | −4.104 | Israel | 0.358 | |
| India | −3.864 | Portugal | 0.302 | |
EIL difference between logarithmic predicted and observed case counts, NA not applicable
Discussion
In the present study, a hierarchical logistic model was used to predict cumulative counts of confirmed SARS-CoV-2 cases in 251 countries and administrative regions with two weeks, one month, and two months forecasting horizons in the early phase of the pandemic. Several metrics were used to evaluate predictions visually and statistically. In summary, case counts could be predicted in the majority of the regions with a surprising accuracy. In spite of the facts that at the time of estimation (29 March 2020) only about one month’s data were available on average in each region, and that most regions were at the very beginning of the epidemic, a massive difference between forecast and observation was rather the exception than the rule. Summary metrics of predictive accuracy suggested very strong prognostic validity the model for a horizon of two weeks, substantial accuracy after one month, and still notable, although markedly lower, accuracy after two months. This is in good agreement with studies finding that the horizon for reasonable epidemiological predictions covers a few weeks at most [7, 15].
Although most predictions were fairly accurate, some were still considerably off. They were most likely to be found in regions with a lower amount of available data at the date of estimation and/or with a more limited growth between the date of the first case and the date of estimation. In general, underestimation seems to be somewhat more pronounced than overestimation, particularly in strongly affected regions (i.e., with cumulative case counts above 10,000 at the point of validation). The strongly affected regions for which the model provided too low predictions included several countries in which mitigation strategies might have been less effective than in other regions, as suggested by the only slowly or not at all decelerating cumulative case growth curves at the beginning of June 2020 (e.g., India, Bangladesh, Qatar). On the other hand, the strongly affected regions with a substantial overestimation of cumulative case counts are characterized by an extremely successful mitigation of the initial phase of the epidemic (mainly Austria and Switzerland). Hence, predictive errors are likely to be closely related to one of the central assumptions of the model, i.e., that timing, extent, and effectiveness of control measures are comparable across regions. Obviously, the forecasts based on the presented model are likely to reach their limits in regions that deviate too strongly from the average case. As shifting individual estimates towards the group mean is also a statistical property of hierarchical models [33], extreme cases are likely to fall outside the scope of validity of the presented approach. As the variation in the course of epidemic trajectories among regions is likely to increase with time, the similarity assumption is expected to become more and more problematic with an ongoing epidemic. In consequence, generalizing the presented findings beyond the initial phase of epidemics is not warranted.
A notable feature of the model that it provides predictions without any reference to measures taken to control the epidemic. This “ignorance” towards interventions, paired with fairly accurate predictions, may be misinterpreted as evidence of dispensability of the mitigation and containment measures implemented in most countries. However, it is far more likely that the key model assumption suggesting similarity of the course of the epidemic and of the control measures taken across regions in the early phase of the epidemic holds to a substantial extent. In cases when it does not, model performance is very poor, as discussed above. Bringing these issue together, the hierarchical structure of the model appears to have both benefits and risks: sufficiently accurate predictions for a large number of regions even at a very early stage of the epidemic come with the price of considerably erroneous predictions for atypical regions. Consequently, if used with the aim of generating locally applicable predictions for a particular region, forecasts may be improved by using data from comparable regions with a higher probability than from rather dissimilar regions [34].
The presented evaluation study has several limitations. First, the case counts were not standardized in any form. Expressing them as cumulative incidence rates (e.g., per 100,000 persons) is likely to have increased homogeneity across regions and enhanced interpretability. As it has been shown in a specific analysis of the development of the SARS-CoV-2 epidemic in German federal states, standardization has rendered using log-transformation of case counts for homogenization superfluous [35]. Second, in the present study uncertainty of the predictions remained unconsidered, although measures of uncertainty, such as reliability and sharpness, can be just as important for forecasting as bias [7]. Third, predictions only at selected time points were analyzed, and it cannot be excluded that choosing other time points would have led to different results. Nevertheless, the general pattern of findings is unlikely to have changed substantially.
The forecasting model itself has some weaknesses as well [24, 35]. Most importantly, it models the reported rather than the true number of cases and therefore can be subject to different forms of testing and reporting bias. Considerable improvement regarding this point can realistically be expected first when regional findings form well-conducted epidemiological studies become available. Second, using cumulative rather than new case counts for modeling can lead to serious errors [36]. Another major limitation of the model is that it works only as long as the conditions of the epidemic remain largely unchanged in each region, i.e., within a single epidemic wave with fairly constant testing and reporting practices and without serious disruptions. This issue could perhaps be addressed by using dynamic (time-dependent) rather than fixed (time-invariant) model parameters [37]. Finally, the primarily phenomenological nature of the model warns to be careful with interpretation [38–40] and calls for integration with mechanistic components, in order to create a hybrid approach that is capable of producing widely generalizable conclusions [41].
Conclusions
As stated by one of the most prominent epidemiologist of the SARS-CoV-2 pandemic, Neil Ferguson, models are “not crystal balls” [3]. However, without rigorous scientific evaluation, they run the risk of becoming one, characterized not by correct predictions but by obscurity. Some state that epidemiological forecasting is “more challenging than weather forecasting” [42], and complexity of modeling and reliance on assumptions make it difficult to assess the trustworthiness of models based solely on their inherent structure. Just like we trust weather forecasts that prove to be accurate by experience, empirical comparison of modeling predictions with actual observations should become an essential step of epidemiological model evaluation.
Acknowledgements
Not applicable.
Abbreviations
- AEIL
Absolute error in logs
- APE
Absolute percentage error
- EIL
Error in logs
- ICC
Intraclass correlation coefficient
- MAPE
Mean absolute percentage error
- PE
Percentage error
- RMSE
Root mean squared error in logs
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
Author’s contributions
LK designed and performed the study, analyzed the data, interpreted the results, and wrote the manuscript. The author(s) read and approved the final manuscript.
Funding
The study was not externally funded. Open Access funding enabled and organized by Projekt DEAL.
Availability of data and materials
The datasets generated and/or analysed during the current study are available in the 2019 Novel Coronavirus COVID-19 (2019-nCoV) Data Repository of the Johns Hopkins University Center for Systems Science and Engineering, https://github.com/CSSEGISandData/COVID-19.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Heesterbeek H, Anderson RM, Andreasen V, Bansal S, De Angelis D, Dye C, et al. Modeling infectious disease dynamics in the complex landscape of global health. Science. 2015;347:aaa4339. doi: 10.1126/science.aaa4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holmdahl I, Buckee C. Wrong but useful - what Covid-19 epidemiologic models can and cannot tell us. N Engl J Med. 2020;383:303–305. doi: 10.1056/NEJMp2016822. [DOI] [PubMed] [Google Scholar]
- 3.Adam D. Special report: the simulations driving the world’s response to COVID-19. Nature. 2020;580:316–318. doi: 10.1038/d41586-020-01003-6. [DOI] [PubMed] [Google Scholar]
- 4.Shmueli G. To explain or to predict? Stat Sci. 2010;25:289–310. doi: 10.1214/10-STS330. [DOI] [Google Scholar]
- 5.Li S-L, Bjørnstad ON, Ferrari MJ, Mummah R, Runge MC, Fonnesbeck CJ, et al. Essential information: uncertainty and optimal control of Ebola outbreaks. PNAS. 2017;114:5659–5664. doi: 10.1073/pnas.1617482114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Probert WJM, Jewell CP, Werkman M, Fonnesbeck CJ, Goto Y, Runge MC, et al. Real-time decision-making during emergency disease outbreaks. PLoS Comput Biol. 2018;14:e1006202. doi: 10.1371/journal.pcbi.1006202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Funk S, Camacho A, Kucharski AJ, Lowe R, Eggo RM, Edmunds WJ. Assessing the performance of real-time epidemic forecasts: a case study of Ebola in the Western area region of Sierra Leone, 2014-15. PLoS Comput Biol. 2019;15:e1006785. doi: 10.1371/journal.pcbi.1006785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johansson MA, Reich NG, Hota A, Brownstein JS, Santillana M. Evaluating the performance of infectious disease forecasts: a comparison of climate-driven and seasonal dengue forecasts for Mexico. Sci Rep. 2016;6:33707. [DOI] [PMC free article] [PubMed]
- 9.Hsieh Y-H, Cheng Y-S. Real-time forecast of multiphase outbreak. Emerg Infect Dis. 2006;12:122–127. doi: 10.3201/eid1201.050396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou G, Yan G. Severe acute respiratory syndrome epidemic in Asia. Emerg Infect Dis. 2003;9:1608–1610. doi: 10.3201/eid0912.030382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biggerstaff M, Alper D, Dredze M, Fox S, Fung IC-H, Hickmann KS, et al. Results from the centers for disease control and prevention’s predict the 2013–2014 influenza season challenge. BMC Infect Dis. 2016;16:357. doi: 10.1186/s12879-016-1669-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsieh Y-H, Fisman DN, Wu J. On epidemic modeling in real time: an application to the 2009 novel a (H1N1) influenza outbreak in Canada. BMC Res Notes. 2010;3:283. doi: 10.1186/1756-0500-3-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chowell G, Viboud C, Simonsen L, Merler S, Vespignani A. Perspectives on model forecasts of the 2014–2015 Ebola epidemic in West Africa: lessons and the way forward. BMC Med. 2017;15:42. doi: 10.1186/s12916-017-0811-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pell B, Kuang Y, Viboud C, Chowell G. Using phenomenological models for forecasting the 2015 Ebola challenge. Epidemics. 2018;22:62–70. doi: 10.1016/j.epidem.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 15.Reich NG, Lauer SA, Sakrejda K, Iamsirithaworn S, Hinjoy S, Suangtho P, et al. Challenges in real-time prediction of infectious disease: a case study of dengue in Thailand. PLoS Negl Trop Dis. 2016;10:e0004761. doi: 10.1371/journal.pntd.0004761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu F, Porco TC, Amza A, Kadri B, Nassirou B, West SK, et al. Short-term forecasting of the prevalence of trachoma: expert opinion, statistical regression, versus transmission models. PLoS Negl Trop Dis. 2015;9:e0004000. doi: 10.1371/journal.pntd.0004000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferguson N, Laydon D, Nedjati Gilani G, Imai N, Ainslie K, Baguelin M, et al. Impact of non-pharmaceutical interventions (NPIs) to reduce COVID19 mortality and healthcare demand. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hellewell J, Abbott S, Gimma A, Bosse NI, Jarvis CI, Russell TW, et al. Feasibility of controlling COVID-19 outbreaks by isolation of cases and contacts. Lancet Glob Health. 2020;8:e488–e496. doi: 10.1016/S2214-109X(20)30074-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kucharski AJ, Russell TW, Diamond C, Liu Y, Edmunds J, Funk S, et al. Early dynamics of transmission and control of COVID-19: a mathematical modelling study. Lancet Infect Dis. 2020;20:553–558. doi: 10.1016/S1473-3099(20)30144-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395:689–697. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koo JR, Cook AR, Park M, Sun Y, Sun H, Lim JT, et al. Interventions to mitigate early spread of SARS-CoV-2 in Singapore: a modelling study. Lancet Infect Dis. 2020;20:678–688. doi: 10.1016/S1473-3099(20)30162-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roosa K, Lee Y, Luo R, Kirpich A, Rothenberg R, Hyman JM, et al. Real-time forecasts of the COVID-19 epidemic in China from February 5th to February 24th, 2020. Infect Dis Model. 2020;5:256–263. doi: 10.1016/j.idm.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.IHME COVID-19 health service utilization forecasting team, Murray CJ. Forecasting COVID-19 impact on hospital bed-days, ICU-days, ventilator-days and deaths by US state in the next 4 months. medRxiv. 2020; 2020.03.27.20043752.
- 24.Kriston L. Projection of cumulative coronavirus disease 2019 (COVID-19) case growth with a hierarchical logistic model. Bull World Health Organ COVID-19 Open Preprints. Published 7 April 2020. 10.2471/BLT.20.257386.
- 25.Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johns Hopkins University Center for Systems Science and Engineering. 2019 Novel Coronavirus COVID-19 (2019-nCoV) Data Repository. 2020. https://github.com/CSSEGISandData/COVID-19. Accessed 1 Jun 2020.
- 27.Kingsland S. The refractory model: the logistic curve and the history of population ecology. Q Rev Biol. 1982;57:29–52. doi: 10.1086/412574. [DOI] [Google Scholar]
- 28.Gottschalk PG, Dunn JR. The five-parameter logistic: a characterization and comparison with the four-parameter logistic. Anal Biochem. 2005;343:54–65. doi: 10.1016/j.ab.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 29.Riley RD, Higgins JPT, Deeks JJ. Interpretation of random effects meta-analyses. BMJ. 2011;342:d549. doi: 10.1136/bmj.d549. [DOI] [PubMed] [Google Scholar]
- 30.Kriston L. Dealing with clinical heterogeneity in meta-analysis. Assumptions, methods, interpretation. Int J Meth Psych Res. 2013;22:1–15. doi: 10.1002/mpr.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lunn DJ, Thomas A, Best N, Spiegelhalter D. WinBUGS - a Bayesian modelling framework: concepts, structure, and extensibility. Stat Comput. 2000;10:325–337. doi: 10.1023/A:1008929526011. [DOI] [Google Scholar]
- 32.Shrout PE, Fleiss JL. Intraclass correlations: uses in assessing rater reliability. Psychol Bull. 1979;86:420–428. doi: 10.1037/0033-2909.86.2.420. [DOI] [PubMed] [Google Scholar]
- 33.Diez R. A glossary for multilevel analysis. J Epidemiol Community Health. 2002;56:588–594. doi: 10.1136/jech.56.8.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kriston L, Meister R. Incorporating uncertainty regarding applicability of evidence from meta-analyses into clinical decision making. J Clin Epidemiol. 2014;67:325–334. doi: 10.1016/j.jclinepi.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 35.Kriston L. Aktuelle Entwicklung der kumulativen Inzidenz bestätigter SARS-CoV-2-Infektionen und infektionsbedingter Todesfälle in Deutschland. [Modeling the cumulative incidence of SARS-CoV-2 cases and deaths in Germany]. [German]. OSF Preprints. Published 5 May 2020. 10.31219/osf.io/q2yw5.
- 36.King AA. Domenech de Cellès M, Magpantay FMG, Rohani P. Avoidable errors in the modelling of outbreaks of emerging pathogens, with special reference to Ebola. Proc Biol Sci. 2015;282:20150347. [DOI] [PMC free article] [PubMed]
- 37.Scarpino SV, Petri G. On the predictability of infectious disease outbreaks. Nat Commun. 2019;10:898. doi: 10.1038/s41467-019-08616-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.May RM. Uses and abuses of mathematics in biology. Science. 2004;303:790–793. doi: 10.1126/science.1094442. [DOI] [PubMed] [Google Scholar]
- 39.Razum O, Becher H, Kapaun A, Junghanss T. SARS, lay epidemiology, and fear. Lancet. 2003;361:1739–1740. doi: 10.1016/S0140-6736(03)13335-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jewell NP, Lewnard JA, Jewell BL. Caution warranted: using the Institute for Health Metrics and Evaluation Model for predicting the course of the COVID-19 pandemic. Ann Intern Med. 2020;173:226–227. doi: 10.7326/M20-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kriston L. Machine learning’s feet of clay. J Eval Clin Pract. 2020;26:373–375. doi: 10.1111/jep.13191. [DOI] [PubMed] [Google Scholar]
- 42.Moran KR, Fairchild G, Generous N, Hickmann K, Osthus D, Priedhorsky R, et al. Epidemic forecasting is messier than weather forecasting: the role of human behavior and internet data streams in epidemic forecast. J Infect Dis. 2016;214(Suppl 4):S404–S408. doi: 10.1093/infdis/jiw375. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the 2019 Novel Coronavirus COVID-19 (2019-nCoV) Data Repository of the Johns Hopkins University Center for Systems Science and Engineering, https://github.com/CSSEGISandData/COVID-19.