Significance
Cyanobacteriochrome (CBCR) photoreceptors leverage light-triggered photoisomerization of bound bilin chromophores to regulate gene expression, directional movement, and cell–cell communication in cyanobacteria. CBCRs responding to far-red light provide an emerging class of small reagents for applications in synthetic biology. We have found CBCRs that use 181,182-dihydrobiliverdin or biliverdin chromophore precursors to sense far-red light. This lineage is distinguished from others by exclusion of phycobilins, the normal CBCR chromophores. Owing to the deep penetrance of far-red light in mammalian tissue and availability of biliverdin therein, this lineage of CBCRs holds great potential for developing synthetic biology tools of biomedical interest.
Keywords: biliprotein, photochromism, bilin-protein interaction, spectral tuning, phylogenic analysis
Abstract
Cyanobacteriochromes (CBCRs) are photoswitchable linear tetrapyrrole (bilin)-based light sensors in the phytochrome superfamily with a broad spectral range from the near UV through the far red (330 to 760 nm). The recent discovery of far-red absorbing CBCRs (frCBCRs) has garnered considerable interest from the optogenetic and imaging communities because of the deep penetrance of far-red light into mammalian tissue and the small size of the CBCR protein scaffold. The present studies were undertaken to determine the structural basis for far-red absorption by JSC1_58120g3, a frCBCR from the thermophilic cyanobacterium Leptolyngbya sp. JSC-1 that is a representative member of a phylogenetically distinct class. Unlike most CBCRs that bind phycocyanobilin (PCB), a phycobilin naturally occurring in cyanobacteria and only a few eukaryotic phototrophs, JSC1_58120g3’s far-red absorption arises from incorporation of the PCB biosynthetic intermediate 181,182-dihydrobiliverdin (181,182-DHBV) rather than the more reduced and more abundant PCB. JSC1_58120g3 can also yield a far-red–absorbing adduct with the more widespread linear tetrapyrrole biliverdin IXα (BV), thus circumventing the need to coproduce or supplement optogenetic cell lines with PCB. Using high-resolution X-ray crystal structures of 181,182-DHBV and BV adducts of JSC1_58120g3 along with structure-guided mutagenesis, we have defined residues critical for its verdin-binding preference and far-red absorption. Far-red sensing and verdin incorporation make this frCBCR lineage an attractive template for developing robust optogenetic and imaging reagents for deep tissue applications.
Nearly all organisms utilize photosensory proteins to detect and respond to the light environment. In many animals, vision is mediated by type II opsin photoreceptors utilizing a retinal chromophore (1). Type I opsins include light-gated proton and ion channels in archaea, bacteria, green algae, and fungi (2, 3). Entrainment of circadian rhythms to light/dark cycles can involve retinal-based opsins (4), flavin-based cryptochromes (5), or light-oxygen-voltage-sensing (LOV) domains (6, 7). LOV domains are also found in phototropins, the photoreceptors controlling plant phototropism (8). Linear tetrapyrrole (bilin)-based phytochrome photoreceptors are widespread in plants, algae, fungi, cyanobacteria, and other bacteria (9, 10). Plant phytochromes use 15,16 double-bond photoisomerization of their bilin chromophores to photoconvert between a dark-adapted 15Z red-absorbing photostate (Pr) and a 15E far-red–absorbing (Pfr) photoproduct, regulating diverse developmental processes including seed germination, chloroplast development, seedling establishment, shade avoidance, and flowering (9, 11, 12).
Members of the phytochrome superfamily are attractive targets for development of optogenetic and imaging tools due to their strong absorption in the near-infrared window that provides optimal light penetrance and minimal interference from cellular components in mammalian tissue (13). Reversible red- and far-red–mediated binding of Arabidopsis thaliana phytochrome B to phytochrome interaction factors has been used to control gene transcription, actin assembly, and GTP-dependent signaling modules using light (14–16). Photoreversible transcriptional control was also achieved in bacteria using the light input domain of cyanobacterial phytochrome 1 (Cph1) from Synechocystis sp. PCC 6803 fused to the receiver output domain of OmpR (17–19). Extensive engineering of bacterial phytochromes (BphPs) has yielded red-absorbing and far-red–emitting fluorescent proteins (20–24). BphPs provide a useful template for such applications because they incorporate more extensively conjugated and a longer wavelength-absorbing biliverdin IXα (BV) chromophore instead of the reduced phycobilins phycocyanobilin (PCB) and phytochromobilin (PΦB) used by cyanobacterial, algal, and plant phytochromes (SI Appendix, Fig. S1A). PCB and PΦB are restricted to oxygenic photosynthetic taxa, whereas BV is present in a wider range of organisms including mammals (9, 25).
Canonical phytochromes, which include BphPs and plant/cyanobacterial phytochromes, are mostly dimeric proteins with a conserved, N-terminal PAS-GAF-PHY tridomain photosensory core module of ∼500 amino acids. Attempts to create smaller, monomeric fluorescent probes based on BphPs have been successful, but the most robust of these have substantially blue-shifted spectra (26). The discovery of cyanobacteriochromes (CBCRs), distantly related bilin-binding GAF-only photoreceptors that sense light ranging from the near UV to the far red (27–36), has proven an exciting opportunity for engineering smaller optogenetic reporters and control elements. Using structure-guided and random mutagenesis approaches, two groups have developed BV-binding CBCRs for fluorescent applications in the far-red spectral region (37, 38). Although these engineered proteins have lower fluorescence quantum yields than engineered BV-binding phycobiliproteins (39–42), fluorescent CBCRs may yet hold promise as probes for superresolution microscopy due to the combination of photo-switchable and long-lasting fluorescence (43). Current far-red–sensing CBCRs (frCBCRs) perhaps hold greater promise as optogenetic actuators and photoacoustic contrast agents that photoconvert under far-red light to yield light-regulated responses. Indeed, CBCR-regulated histidine kinases (cHKs) (18, 19, 44) and CBCR-regulated photoswitchable adenylyl cyclases (cPACs) (45) have been exploited to control gene expression in cells using green/red, UV/green, blue/green, and far-red/red photocycles.
Two phylogenetically distinct clusters of naturally occurring frCBCRs were identified recently. Members of the first cluster of frCBCRs have been reported only from Acaryochloris marina MBIC 11017. These frCBCRs detect far-red light due to their ability to incorporate BV in the absence of PCB (37, 46–49). This frCBCR clade arose within the eXtended Red/Green (XRG) CBCR lineage, and representative members typically exhibit far-red/orange (FR/O) photocycles when bound to BV (15Z λmax ∼ 697 to 702 nm) and red/green photocycles when bound to PCB (47–49). Based on four conserved residues sufficient to confer BV binding (37), we will refer to this frCBCR family as the AmBV4 lineage. Interestingly, A. marina MBIC 11017 possesses an unusually complex pathway for PCB biosynthesis. This cyanobacterium has duplicated the ferredoxin-dependent bilin reductase PcyA, which converts BV into PCB (46, 50, 51). One of these enzymes rapidly turns over BV into PCB; the other is a slower enzyme that accumulates the intermediate 181,182-dihydroBV (181,182-DHBV), which AmBV4 CBCRs are also able to bind (46). The second cluster of frCBCRs instead arose within the greater green/red (GGR) lineage that is evolutionarily distant from the XRG lineage (52). We will refer to these frCBCRs as “Far-Red of the Greater Green/Red” (FRoGGR) CBCRs. FRoGGRs incorporate PCB and exhibit the most red-shifted known absorption in their dark states with PCB-derived chromophores (λmax 725 to 755 nm) but do not bind BV (35).
In this report, we characterize a third group of frCBCRs. Typified by the far-red/red (FR/R) CBCR JSC1_58120g3 from Leptolyngbya sp. strain JSC-1 that was recently used to engineer a far-red–responsive cPAC (45), members of this frCBCR lineage exclude phycobilins and incorporate BV or 181,182-DHBV to yield sensors with far-red absorption maxima (λmax 712 to 728 nm). This monophyletic class of CBCRs also arose within the XRG lineage but is phylogenetically distinct from the AmBV4 class of frCBCRs. High-resolution crystal structures of JSC1_58120g3 identify a conserved proline residue near the bilin A-ring that we show to be essential for exclusion of phycobilins with reduced A-rings. Due to their intrinsic ability to bind BV and their red-shifted FR/R photocycles relative to AmBV4 proteins, we propose that these frCBCRs will be excellent candidates for far-red optogenetic and/or far-red–imaging applications.
Results
Phylogenetic and Spectroscopic Analyses Define a Previously Uncharacterized frCBCR Lineage.
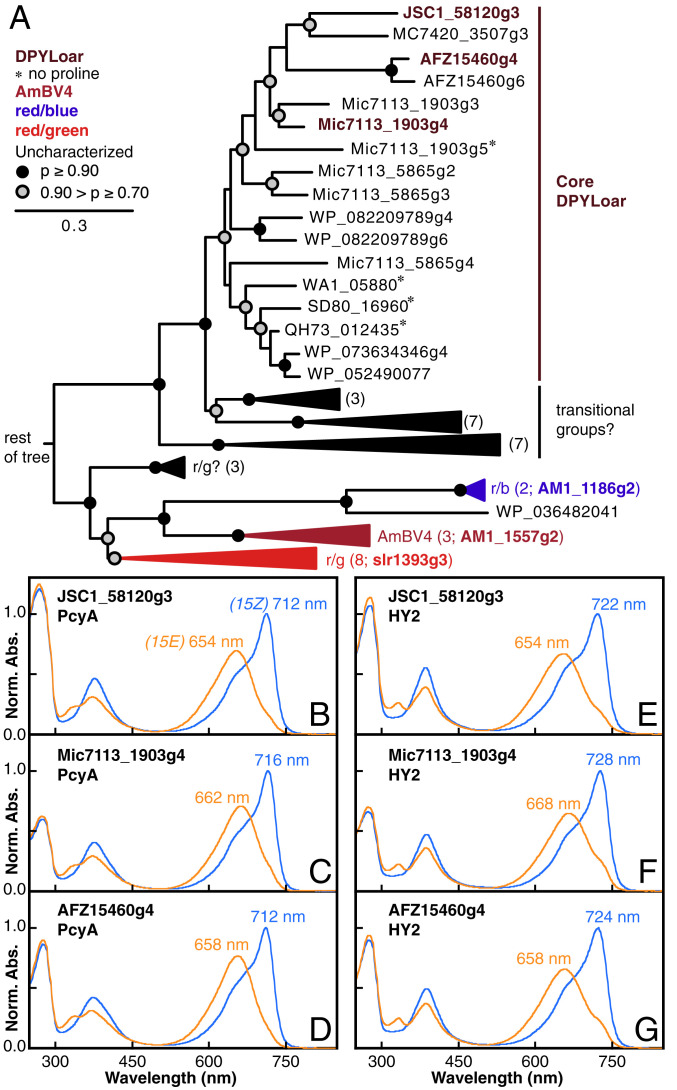
In previous work, we noted that recombinant JSC1_58120g3 exhibited an unusually red-shifted far-red/red photocycle [712/654 nm (45)]. This CBCR is one of three arranged in tandem in a CBCR-regulated methyl-accepting chemotaxis protein (cMCP), an architecture consistent with known phototaxis sensors (53, 54). Neither of the adjacent domains, JSC1_58120g2 and JSC1_58120g4, exhibited similar behavior. Instead, these CBCRs exhibited UV/blue and red/green photocycles with phycoviolobilin (PVB) and PCB chromophores, respectively (SI Appendix, Fig. S1C). BLAST searches (55) using JSC1_58120g3 as a query identified a number of CBCR sequences with characteristic amino acid substitutions relative to canonical red/green CBCRs, with other residues conserved among the two groups (SI Appendix, Fig. S2A). Phylogenetic analysis established that JSC1_58120g3 and related proteins comprise a clade that arose within the XRG lineage. These proteins are distinct from the AmBV4 family (Fig. 1A and SI Appendix, Fig. S2B). As members of the XRG lineage, JSC1_58120g3 and other members of this lineage are also not closely related to the previously identified PCB-binding FRoGGR cluster that evolved within the greater green/red (GGR) lineage of CBCRs (52).
Fig. 1.
A newly identified lineage of far-red photosensors. (A) A partially collapsed phylogenetic tree is shown for the DPYLoar lineage described in this study. Parentheses next to collapsed branches indicate the number of CBCRs found on that branch, as well as a characterized CBCR from the branch, if available. Full tree available in SI Appendix, Fig. S2B. (B–D) Dark-adapted (blue, 15Z) and photoproduct (orange, 15E) absorption spectra are shown for JSC1_58120g3 (B), Mic7113_1903g4 (C), and AFZ15460g4 (D) expressed with PCB biosynthetic enzymes. (E–G) Spectra are shown as in B–D for proteins expressed with PΦB biosynthetic enzymes.
We next expressed two additional proteins within this lineage in Escherichia coli cells engineered for PCB or PΦB synthesis. Both Mic7113_1903g4 and AFZ15460g4 yielded photoactive CBCRs with FR/R photocycles similar to those of JSC1_58120g3 (Fig. 1 B–G and SI Appendix, Table S1). Dark-state spectra for each CBCR obtained from PΦB-producing cell lines were red-shifted by ∼10 nm relative to those obtained from PCB expression lines (Fig. 1: compare B–D with E–G). However, this bilin-dependent shift was considerably less for the photoproduct states. Taken together, these data indicate that this clade encompasses a previously uncharacterized lineage of FR/R sensors.
Members of This Lineage of frCBCRs Possess Atypical Chromophores.
To determine the molecular basis of spectral tuning in this family, we first measured the absorption spectra of the three FR/R CBCRs following denaturation in 6 M guanidinium-HCl (SI Appendix, Fig. S3A). The denatured far-red–absorbing dark state of JSC1_58120g3 was photochemically inactive, while the denatured red-absorbing photoproduct was significantly blue shifted and was still photoactive (SI Appendix, Fig. S3B). Based on the known behavior of bilins under denatured conditions (56, 57), these results establish 15Z and 15E configurations for the chromophore in the far-red–absorbing and red-absorbing states, respectively. Typically, chromophore-binding CBCRs expressed in PCB- or PΦB-producing cells retain PCB or PΦB adducts or carry out isomerization at the C5 methine bridge (31, 33). Absorption maxima of the three CBCRs in this lineage under denaturing conditions were red shifted relative to those of red/green CBCRs incorporating PCB or PΦB used as standards (NpR6012g4 and NpR5113g2: SI Appendix, Fig. S3 B–F). Unexpectedly, denatured spectra for all three frCBCRs from PΦB-producing cells resembled spectra for BV-bound AM1_C0023g2 (47), while those from PCB-producing cells were intermediate between PCB- and BV-adduct standards, similar to a recently reported population with bound 181,182–DHBV (46). Denaturation disrupts bilin-protein interactions responsible for spectral tuning of the bilin chromophore in the native protein context (30, 31, 58), so these results indicate that the bilin chromophores of this new frCBCR lineage differ from those of the PCB and PΦB adducts of similarly expressed CBCR standards (32, 59).
Members of This Lineage of frCBCRs Bind Verdins Rather than A-Ring–Reduced Phycobilins.
We were fortunate to obtain well-diffracting crystals of JSC1_58120g3 in the far-red–absorbing state isolated from cells engineered to produce either PCB (PDB ID 6XHH) or PΦB (PDB ID 6XHG). For the PCB-expressed protein, crystals adopted P1 symmetry with two monomers per asymmetric unit. The crystal structure was determined at 1.5-Å resolution by molecular replacement using a polyalanine model of the red-absorbing dark state of AnPixJg2 (PDB ID 3W2Z), which refined to R-factor/R-free of 19.4/23.1% (SI Appendix, Table S2). For the PΦB-expressed protein, crystals also adopted P1 symmetry with two monomers per asymmetric unit. This structure was determined at 2.3-Å resolution by molecular replacement using the PCB-expressed protein as search model and was refined to R-factor/R-free of 23.1/26.4% (SI Appendix, Table S2).
As expected, both JSC1_58120g3 structures adopted GAF folds similar to those of other XRG CBCRs in the 15Z state, such as AnPixJg2 (PDB ID 3W2Z) (60, 61), AnPixJg2_BV4 (5ZOH) (37), NpR6012g4 (6BHN) (62), and slr1393g3 (5DFX) (63). In all these structures, the bilin chromophore is sandwiched between the central β-sheet (strands β1–β6) and helices α3 and α4, with the “backside” helices α1, α2, and α5 bundled on the opposite side of the central β-sheet (SI Appendix, Fig. S4A). JSC1_58120g3 contains a 16-residue insertion between strand β4 and helix α4 that takes on a random coil structure (brick red arrow, SI Appendix, Fig. S4A) and packs against strand β4 and helix α5, but this loop feature is not conserved in the new frCBCR lineage. JSC1_58120g3 also lacks a small loop immediately preceding strand β3 (teal arrow, SI Appendix, Fig. S4A) that is present in other known XRG CBCR structures.
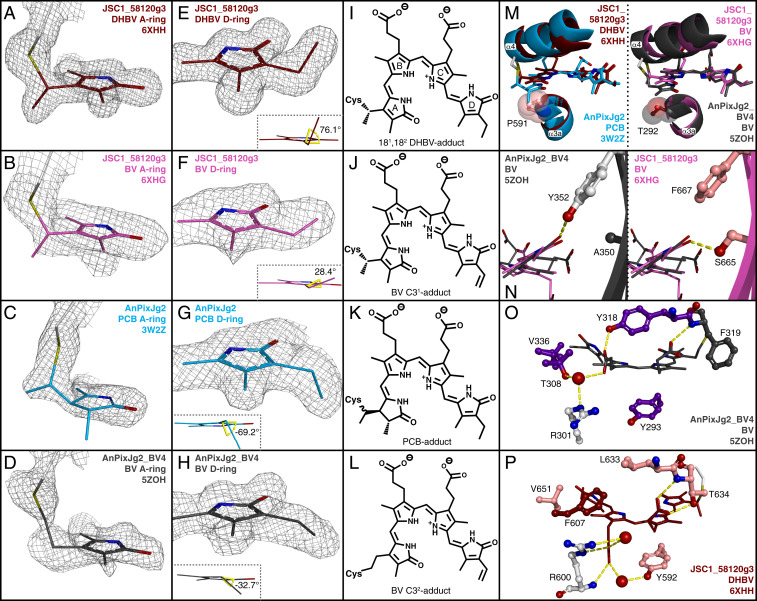
It also is notable that the chromophore moieties in both JSC1_58120g3 structures exhibit altered electron density relative to that of AnPixJg2 in a manner consistent with trigonal planar geometry at C2 and C3 of the A pyrrole ring and the presence of a 2,3–double bond (Fig. 2 A and B). This contrasts with a tetrahedral geometry in the PCB adduct of AnPixJg2 (Fig. 2 C and K). Electron density also shows that the chromophore is attached to Cys636 through a thioether linkage to the C31 carbon in both structures (Fig. 2 A, B, I, and J). This is in contrast with the C32 thioether attachment to BV seen in the AnPixJg2_BV4 structure (Fig. 2 D and L) (37) and in most structures of BphPs and CBCRs with BV chromophores (64–69). Known exceptions include an engineered fluorescent variant of NpR3784 that exhibits C31 attachment of BV with a C31-C32 double bond and a similar variant of Rhodopseudomonas palustris BphP1 that possesses thioether linkages to both C31 and C32 of BV (24, 38).
Fig. 2.
JSC1_58120g3 crystal structures. (A–D) Chromophore A-ring models and electron densities for JSC1_58120g3 coexpressed with PcyA (A) or with HY2 (B), PCB adduct of AnPixJg2 (C), and BV adduct of AnPixJg2_BV4 (D). (E–H) Chromophore D-ring models and electron densities are shown as in A–D. (Insets in E–H) C19-C18-C181-C182 dihedral angles. (I–L) Chemical representations of chromophore adducts as determined from A–H. (M) Alignments depicting helix α4 and chromophore position shift. Protein mainchain is depicted in ribbon view with Pro591 and Thr292 sidechains shown as space-filling spheres. JSC1_58120g3-DHBV is brick red, AnPixJg2 teal, JSC1_58120g3-BV magenta, and AnPixJg2_BV4 charcoal. DPYLoar-conserved residues are colored by atom with carbon in salmon. XRG-conserved residues are colored by atom with carbon in light gray. (N) Orientation of BV chromophore D-ring with XRG-conserved β6 residues (Left) or DPYLoar-conserved β6 residues (Right). (O) Chromophore arrangement and protein contacts in AnPixJg2_BV4. Colored as in M, and AmBV4-conserved residues are colored by atom with carbon in violet. Water molecules depicted as red spheres. (P) Chromophore arrangement and equivalent protein contacts in JSC1_58120g3-DHBV.
The two JSC1_58120g3 structures exhibit differences in the geometry of their C18 side chains. For the PCB-expressed protein, the C19-C18-C181-C182 dihedral angle is 76.1° (Fig. 2E), indicating the presence of an ethyl group at the C18 position like that of the AnPixJg2 chromophore (Fig. 2G). The C19-C18-C181-C182 dihedral angle in the crystal obtained from the PΦB expression system measures just 28.4° (Fig. 2F), consistent with the presence of a vinyl substituent at the C18 position like that of the AnPixJg2_BV4 chromophore (Fig. 2H). Taken together with the A-ring geometry, these data support the conclusion that 181,182–DHBV and BV were the precursors of the respective covalent bilin adducts in JSC1_58120g3 rather than PCB and PΦB (Fig. 2 I–L). This result is surprising, because the more oxidized verdins 181,182–DHBV and BV are expected to be present only in small amounts relative to the phycobilin products PCB and PΦB because 181,182–DHBV is a transient intermediate in the conversion of BV to PCB by PcyA (50, 51) and because BV remains tightly bound to heme oxygenase until it is converted to PΦB by HY2 (SI Appendix, Fig. S1A). Thus, JSC1_58120g3 discriminates against PCB and PΦB, preferring their verdin biosynthetic precursors. The similarity between denatured spectra of JSC1_58120g3, Mic7113_1903g4, and AFZ15460g4 (SI Appendix, Fig. S3 E and F) demonstrates that this behavior is a conserved feature of this frCBCR lineage.
Conserved Sequence Elements Directly Interact with the Chromophore.
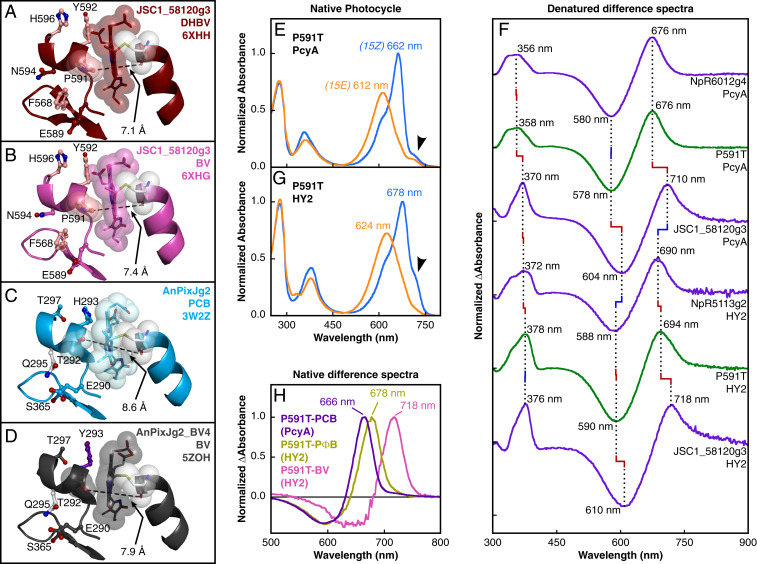
Conserved residues known to be important for chromophore positioning in XRG CBCRs, including W588, D590, and H637 (61), all maintain typical interactions with the chromophore in both JSC1_58120g3 structures (SI Appendix, Fig. S5 A and B). The conserved β6 tyrosine of XRG CBCRs is replaced in JSC1_58120g3 by Phe667. The tyrosine phenolic moiety normally interacts with the D-ring carbonyl (37, 61–63), but instead Ser665 of JSC1_58120g3 supplies a similar interaction from the other face of the D-ring (Fig. 2N). Ser665 and Phe667 are conserved in the JSC1_58120g3 cluster (SI Appendix, Fig. S2A). Helix α4 contains the canonical chromophore-binding residue Cys636. Both this helix and the covalently attached chromophore are substantially shifted in both JSC1_58120g3 structures relative to AnPixJg2, AnPixJg2_BV4 (Fig. 2M), and slr1393g3 (SI Appendix, Fig. S4 A and B). This shift causes the JSC1_58120g3 chromophores to sit at an angle in the binding pocket relative to the chromophores in AnPixJg2 and AnPixJg2_BV4, and to a lesser extent relative to that in slr1393g3. Relative to AnPixJg2, the chromophore in both JSC1_58120g3 structures is shifted such that the D-ring is closer to the central β-sheet, similar to the chromophores in AnPixJg2_BV4 and slr1393g3 (SI Appendix, Fig. S4B). Valine (JSC1_58120g3, AnPixJg2_BV4) or threonine (slr1393g3) residues on β5 appear to accommodate this shift toward the β-sheet (Fig. 2 O and P and SI Appendix, Fig. S5H), compared to a bulkier isoleucine in AnPixJg2 and many other XRG CBCRs. The overall shift in chromophore and helix α4 appears to correlate with another conserved residue in this cluster, Pro591. Structural superpositions indicate that Pro591 would clash with the chromophore A-rings were it present in AnPixJg2 and AnPixJg2_BV4 (Fig. 2M). Similarly, aligning the C32-linked BV from AnPixJg2_BV4 into JSC1_58120g3 would result in a clash between Pro591 and the A-ring as well as clashes between β5 and β6 residues and the D-ring (SI Appendix, Fig. S4 C–F).
Closer inspection of Pro591 reveals that it contributes to a reduced binding pocket width surrounding the A-ring. In JSC1_58120g3 structures, there is a 7.1- or 7.4-Å gap, compared with 8.6 Å in AnPixJg2 with PCB adduct or 7.9 Å in AnPixJg2_BV4 (Fig. 3 A–D). We reasoned that such a restricted binding pocket could provide a structural basis for preventing phycobilin chromophore incorporation due to the bulkier A-rings of PCB and PΦB caused by tetrahedral geometry at C2 and C3 (Fig. 2K). To test this, we used site-directed mutagenesis to replace Pro591 with Thr, the equivalent residue in AnPixJg2. The P591T variant of JSC1_58120g3 readily incorporated chromophore when expressed with PcyA and exhibited a substantially blue-shifted R/O photocycle compared to wild type (WT) (Fig. 3E). Denatured difference spectra for this variant closely resembled PCB and were distinct from WT JSC1_58120g3 expressed with PcyA (Fig. 3F). Likewise, P591T JSC1_58120g3 expressed with HY2 exhibited a blue-shifted R/O photocycle relative to wild-type and denatured spectra resembled PΦB (Fig. 3 F and G). Both P591T preparations retained FR shoulders in their native spectra (Fig. 3 E and G), probably indicating residual verdin chromophore(s). In P591T expressed with HY2, the FR shoulder is substantial enough to extract separate PΦB and BV populations from the native difference spectra (Fig. 3H). Together, these results indicate that Pro591 is necessary for phycobilin exclusion in this lineage, but not for verdin affinity. Based on the Asp-Pro-Tyr-Leu consensus sequence of the motif containing this proline and the preference of these CBCRs for verdin chromophores with “oxidized A-rings (oar),” we designate this lineage “DPYLoar.”
Fig. 3.
Phycobilin exclusion by Pro591. (A–D) View of the A-ring–binding pocket and nearby residues with opening width labeled for JSC1_58120g3-DHBV (A), JSC1_58120g3-BV (B), AnPixJg2 (C), and AnPixJg2_BV4 (D). Mainchain is depicted in ribbon view with Pro591/Thr292, chromophore, and primary cysteine represented as space-filling spheres. Coloring as in Fig. 2. (E) Dark-adapted 15Z (blue) and photoproduct 15E (orange) absorbance spectra are shown for P591T JSC1_58120g3 expressed with PcyA. Arrowhead denotes FR shoulder. (F) Stacked 15Z-15E difference spectra for denatured proteins compare P591T and WT JSC1_58120g3 to standards for PCB adduct (NpR6012g4) and PΦB adduct (NpR5113g2). (G) Spectra are shown for P591T JSC1_58120g3 expressed with HY2 as in E. (H) Native 15Z-15E difference spectra for P591T-PCB (violet) and estimated P591T-PΦB (bronze) and P591T-BV (pink) populations.
Distinct Mechanisms for BV Binding in AmBV4 and DPYLoar Families.
Four residues conserved within the AmBV4 lineage are sufficient to confer BV affinity to multiple XRG CBCRs (37). The full set of four residues (Tyr293, Thr308, Tyr318, Val336 in AnPixJg2_BV4, Fig. 2O) was determined to be essential for efficient incorporation of BV into a model red/green CBCR, yet DPYLoar CBCRs retain only two of these residues (Tyr592 and Val651 in JSC1_58120g3: Fig. 2P). The other residues are equivalent to Phe607 and Leu633. Introduction of Val336 or Val651 relative to canonical red/green CBCRs replaces larger Ile/Leu residues, apparently accommodating the shift of the chromophore toward the β-sheet in both lineages. However, the remaining three positions appear to perform divergent roles in DPYLoar and AmBV4 CBCRs. Tyr293 in AnPixJg2_BV4 forms only hydrophobic interactions with the chromophore B-ring, while Tyr592 in JSC1_58120g3 forms a water-mediated polar contact with the C-ring propionate.
The C-ring propionate in both JSC1_58120g3 structures maintains an orientation orthogonal to the C-ring plane, as in BV in AnPixJg2_BV4, but the propionate is positioned on the α-face rather than the β-face of the chromophore. The C-ring propionate positioning in JSC1_58120g3 is tethered by direct and water-mediated polar contacts to both sidechain and backbone of Arg600 and also by a water-mediated polar contact to the sidechain of Tyr592 (Fig. 2P); the resulting conformation removes a potential clash with Phe607 or interaction with Leu633, negating the apparent roles of Thr308 and Tyr318 in AnPixJg2_BV4 (Fig. 2O). Phe607 is shifted toward the chromophore relative to the position of Phe308 in AnPixJg2 and occupies some of the space vacated by the C-ring propionate (SI Appendix, Fig. S5 C–F). This movement may be influenced by a water molecule that inserts between β5 and the loop preceding β4. The backbone of Ala653, conserved in DPYLoars and found on β5, coordinates this water and replaces a proline conserved in XRG CBCRs (SI Appendix, Fig. S5 C–F). This alters the position of β4, pushing Phe607 closer to the chromophore. Two nearby Tyr residues on α3 and β5 that are broadly conserved in XRG CBCRs are replaced in DPYLoar CBCRs with Phe601 and Cys649, perhaps to alleviate clashes with repositioned Phe607. Notably, the arrangement of the chromophore and some surrounding residues in JSC1_58120g3 structures appears to be most similar to the red-absorbing 15Z state structure of slr1393g3 (SI Appendix, Figs. S4B and S5 G and H). Given that slr1393g3 is intermediate between AnPixJg2 and DPYLoars in the current phylogeny (SI Appendix, Fig. S2B), this suggests that some of the observed structural rearrangements relative to AnPixJg2 were acquired before the transition to phycobilin exclusion.
An additional set of conserved DPYLoar residues is distal to the chromophore, but they apparently form a lineage-specific interaction network. Arg654, Asp566, Tyr567, and Asp570 (on strands β1–2) and Glu589, Asn594, and Gln602 (preceding and within helix α3) form a network of polar contacts that link the two regions together in a manner not seen in other CBCR structures (SI Appendix, Fig. S6 A and B). These interactions pack around Phe568, another conserved DPYLoar residue that replaces smaller Ser/Thr residues commonly found in XRG CBCRs. This locks Phe568 in a strained backbone configuration and orients the sidechain as a prop against the phycobilin-excluding Pro591 residue. The conserved nature of many of these residues and their positioning in proximity to Pro591 implicate a potential supporting role in phycobilin exclusion or in chromophore incorporation generally.
Discussion
We report the discovery of DPYLoar CBCRs, the third known group of frCBCRs and the first such group exhibiting preferential incorporation of 181,182-DHBV. Unlike previously described FRoGGR CBCRs that use phycobilins but not verdins (35) and AmBV4 CBCRs that use either type of bilin (37, 47, 48), DPYLoar CBCRs generate far-red/red photocycles (Fig. 1 B–G) by excluding phycobilin chromophores in favor of more conjugated verdins. Until recently, 181,182-DHBV was thought to be a transient intermediate formed during PCB synthesis by cyanobacterial PcyA (50, 51) and had only been identified as an authentic chromophore precursor in some cryptophyte phycobiliproteins (70, 71). The first report of 181,182–DHBV adducts in CBCRs used AmBV4 CBCRs coexpressed with an atypical PcyA enzyme from A. marina MBIC11017 that accumulates 181,182–DHBV at higher levels (46). We here demonstrate 181,182–DHBV binding in DPYLoar CBCRs after coexpression in the standard system used to incorporate PCB into dozens of phytochromes and CBCRs (32, 52, 59, 72, 73) through a combination of denaturation analysis (SI Appendix, Fig. S3 B–F) and a 1.5-Å crystal structure of JSC1_58120g3 in which the chromophore is well resolved (Fig. 2 A and E). DPYLoar CBCRs expressed with PΦB-producing HY2 instead attach the BV chromophore, as confirmed by denaturation (SI Appendix, Fig. S3 B–F) and by a 2.3-Å structure of JSC1_58120g3 (Fig. 2 B and F). AmBV4 CBCRs are able to bind BV to produce FR/O photocycles only in the absence of PCB (47–49), making DPYLoar CBCRs the only known lineage that bind verdin chromophores even when expressed with PcyA or HY2. In JSC1_58120g3, this behavior requires a conserved proline residue that constrains the A-ring–binding pocket (Fig. 3 A–D), with a P591T variant of JSC1_58120g3 restoring PCB binding (Fig. 3 E–H). It is possible that slight changes in structural context within the DPYLoar lineage would allow for other residues to participate in PCB exclusion as well.
DPYLoar CBCRs are unusually far-red– and red-shifted compared to non-FRoGGR CBCRs (SI Appendix, Fig. S6C). Natural and engineered AmBV4 CBCRs with bound BV range from 697 to 713 nm and from 605 to 638 nm in the 15Z and 15E photostates, respectively, while DPYLoar CBCRs range from 722 to 728 nm and from 654 to 662 nm with the same chromophore (SI Appendix, Table S1). P591T retains a more red-shifted 15Z state than most XRG CBCRs, although the native chromophore is not as red shifted relative to denatured bilin as are those of wild-type DPYLoar CBCRs (SI Appendix, Fig. S6C). This indicates that Pro591 also may contribute to 15Z tuning, perhaps via the substantial chromophore positional shift that it appears to cause (Fig. 2M and SI Appendix, Fig. S4B).
The protein-chromophore interactions responsible for spectral tuning of the 15E photostate are not well established in the absence of a photoproduct structure, but comparison to other XRG CBCRs offers some insight. Mutational studies have identified residues involved in photoproduct tuning (59, 74), and structures of green-absorbing 15E PCB chromophores of both NpR6012g4 and slr1393g3 (SI Appendix, Fig. S6E) reveal strongly out-of-plane A- and D-rings relative to the B- and C-rings (62, 63). Native “twisted” and denatured CBCRs fall on separate trendlines when a 15Z-to-15E blue shift is plotted against a 15Z peak wavelength for different chromophores (SI Appendix, Fig. S6D). Variants with reduced ability to blue shift the 15E chromophore cluster toward the denatured trendline (35, 59, 74), possibly indicating a less twisted chromophore in such variants. DPYLoar CBCRs consistently cluster along the denatured/relaxed trendline regardless of the chromophore adduct. By contrast, natural and engineered AmBV4 CBCRs appear to possess more twisted PCB adducts than their corresponding BV adducts. Indeed, known verdin-bound CBCRs predominantly fall along the relaxed trendline; that is, verdin adducts may be intrinsically less twisted in their 15E photostate. The improved A-ring conjugation of verdins may disfavor the twisted A-ring geometry observed in green-absorbing 15E states of the corresponding PCB adducts (SI Appendix, Fig. S6F), resulting in a more “relaxed” geometry. However, both PCB and PΦB adducts of P591T JSC1_58120g3 remain on the relaxed trendline, indicating that some additional structural mechanism in DPYLoar CBCRs may reduce or prevent twisting. It is possible that the conserved residues Phe571 and Phe644, equivalent to conserved residues involved in 15E blue shift in typical XRG CBCRs (62, 74), are unable to perform their usual roles due to the altered chromophore positioning in DPYLoars. Alternatively, the network of polar contacts and Phe568 linking β1-β2 with α3 (SI Appendix, Fig. S6A) could serve to rigidify a region that undergoes substantial conformational change upon photoconversion in typical XRGs (62, 63) (SI Appendix, Fig. S6 G and H). Preventing this rearrangement may interfere with the nearby A-ring twist, resulting in a reduced blue shift. Further mutagenesis experiments and a photoproduct structure will help to elucidate this issue.
Although both AmBV4 and DPYLoar classes of frCBCRs arose within the XRG lineage, they are phylogenetically distinct and thus represent an independent evolution of efficient BV incorporation (Fig. 1A and SI Appendix, Fig. S2B). DPYLoar CBCRs have only two of the four amino acids that confer BV affinity on the AmBV4 lineage (Fig. 2 O and P). It is not immediately clear if additional conserved DPYLoar residues play a role in BV affinity, and additional mutagenesis experiments will be required. Furthermore, DPYLoar CBCRs have the unique ability to exclude phycobilin chromophores via a conserved proline that restricts binding-pocket width at the A-ring (Fig. 3 A–G). This conserved proline may also provide the basis for the unusual C31 attachment to verdins by altering chromophore position such that C32 attachment is sterically unfavorable (SI Appendix, Fig. S4 C–F). However, this is difficult to confirm without a P591T-verdin structure or NMR data because we currently lack spectral signatures for attachment at C31 vs. C32. Regardless, DPYLoar CBCRs are clearly a phylogenetically and functionally distinct third class of frCBCRs.
The high BV affinity and red-shifted photocycles of DPYLoars relative to AmBV4 CBCRs make them of great interest for optogenetic and imaging applications in mammalian tissue. In this regard, a functional FR/R light-actuated adenylyl cyclase has already been developed by replacing the blue/green CBCR GAF domain of the cPAC Mic7113_2205 with JSC1_58120g3, normally part of a cMCP (45). In the same study, the green/red CBCR GAF domain of the RcaE cHK also successfully replaced the blue/green Mic7113_2205 CBCR (45), indicating that CBCRs can be made to regulate diverse output domains with the proper fusion. Notably, individual CBCR domains tend to be monomeric in solution (60, 75–77); even a CBCR-regulated diguanylyl cyclase exhibits only transient dimerization even though its catalytic activity requires dimerization (78). The lack of stable dimer formation in CBCRs may limit optogenetic applications involving photo-inducible dimerization, but a forthcoming study reports engineering of photo-specific binding proteins for the red- and green-absorbing photostates of PCB-bound AM1_C0023g2, an AmBV4 CBCR (79). Further engineering and exploration of the DPYLoar family may yield higher BV affinity, improved signal actuators, further red-shifted sensitivity, far-red fluorescence, or perhaps even photoinducible dimerization for applications in deep-tissue optogenetics and imaging via fluorescent or photoacoustic probes.
All DPYLoar CBCRs identified thus far exist in tandem arrays with one or several other CBCRs N-terminal to a signal output domain (SI Appendix, Fig. S1B). Most members of the DPYLoar lineage are found in cMCP putative phototaxis sensors, although some are found in cHKs. Such cMCP-signaling molecules are often associated with phototaxis (53, 54, 80, 81), eliciting movement either toward or away from perceived light. Although it has not been experimentally determined, it is possible that the directionality of phototaxis induced by DPYLoar cMCPs may depend on the ability of the host cyanobacterium to harvest far-red light for photosynthesis. For example, JSC1_58120 may induce positive far-red phototaxis in Leptolyngbya JSC-1, which can harvest far-red light under the FaRLiP response (82–85). There may also be interplay between phototaxis and light-harvesting strategy, such as positive far-red phototaxis when FaRLiP is activated rather than negative phototaxis under other light conditions.
Our work also underscores the importance of in-depth characterization of even well-established CBCR subfamilies. Despite broad interest in biologically encoded far-red sensors, the DPYLoar CBCRs had hidden in plain sight as an apparently unremarkable branch of the XRG lineage that now can be recognized as having the notable ability to exclude phycobilins. Given that PcyA is ubiquitous in cyanobacteria and may be essential (86), the physiological chromophore precursor for DPYLoar proteins is almost certainly 181,182–DHBV, itself generally viewed as a transient intermediate with little physiological relevance. The ability of CBCRs to bind 181,182–DHBV even in the presence of a fully functional PcyA is cause to re-evaluate chromophore assignments in a subset of red/green CBCRs that exhibit far-red shoulders (32, 59). Previous reports have proposed that such populations are likely representative of noncovalently attached PCB based on denatured spectra that were intermediate between PCB and BV adducts. However, we have here shown that such spectra may also be consistent with 181,182–DHBV adducts. Should such CBCRs in fact exhibit substantial affinity for 181,182–DHBV, it is possible that the presence of such a far-red shoulder when expressed with PcyA could provide a rapid means to identify cryptic affinity for BV.
Materials and Methods
Detailed materials and methods are provided in SI Appendix, Supplementary Methods. This includes information on phylogenetic analysis, expression and purification of DPYLoar CBCRs, spectroscopic techniques, crystallization, and structure determination.
Supplementary Material
Acknowledgments
We thank Rei Narikawa and Keiji Fushimi for helpful discussions. Genomic DNA for Leptolyngbya sp. JSC1 was a gift from Professor Don Bryant. This work is based on research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences (NIGMS) from the NIH (P30 GM124165). The Pilatus 6 M detector on the 24-ID-C beam line is funded by a NIH-Office of Research Infrastructure Programs High-End Instrumentation grant (S10 RR029205). This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Use of the Stanford Synchrotron Radiation Lightsource, Stanford Linear Accelerator Center National Accelerator Laboratory, is supported by the DOE, Office of Science, Office of Basic Energy Sciences under Contract DE-AC02-76SF00515. The Stanford Synchrotron Radiation Light Source Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the NIH NIGMS (including P41GM103393). This work was supported by Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science, DOE Grant DOE DE-FG02-09ER16117 (to J.C.L.) and by NIH Grants GM068552 (to J.C.L.) and T32-GM07377 (to M.V.M.). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2016047117/-/DCSupplemental.
Data Availability.
Data for this work have been deposited in the RCSB PDB under PDB ID codes 6XHH (181,182-DHBV adduct) (87) and 6XHG (BV adduct) (88). All study data are included in the article and supporting information.
References
- 1.Jacobs G. H., Photopigments and the dimensionality of animal color vision. Neurosci. Biobehav. Rev. 86, 108–130 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Grote M., Engelhard M., Hegemann P., Of ion pumps, sensors and channels:–Perspectives on microbial rhodopsins between science and history. Biochim. Biophys. Acta Bioenerg. 1837, 533–545 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Brown L. S., Fungal rhodopsins and opsin-related proteins: Eukaryotic homologues of bacteriorhodopsin with unknown functions. Photochem. Photobiol. Sci. 3, 555–565 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Lazzerini Ospri L., Prusky G., Hattar S., Mood, the circadian system, and melanopsin retinal ganglion cells. Annu. Rev. Neurosci. 40, 539–556 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaves I., et al. , The cryptochromes: Blue light photoreceptors in plants and animals. Annu. Rev. Plant Biol. 62, 335–364 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Losi A., Gärtner W., Solving blue light riddles: New lessons from flavin-binding LOV photoreceptors. Photochem. Photobiol. 93, 141–158 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Glantz S. T., et al. , Functional and topological diversity of LOV domain photoreceptors. Proc. Natl. Acad. Sci. U.S.A. 113, E1442–E1451 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christie J. M., Phototropin blue-light receptors. Annu. Rev. Plant Biol. 58, 21–45 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Rockwell N. C., Lagarias J. C., A brief history of phytochromes. ChemPhysChem 11, 1172–1180 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anders K., Essen L. O., The family of phytochrome-like photoreceptors: Diverse, complex and multi-colored, but very useful. Curr. Opin. Struct. Biol. 35, 7–16 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Pham V. N., Kathare P. K., Huq E., Phytochromes and phytochrome interacting factors. Plant Physiol. 176, 1025–1038 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bae G., Choi G., Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 59, 281–311 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Weissleder R., A clearer vision for in vivo imaging. Nat. Biotechnol. 19, 316–317 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Levskaya A., Weiner O. D., Lim W. A., Voigt C. A., Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461, 997–1001 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimizu-Sato S., Huq E., Tepperman J. M., Quail P. H., A light-switchable gene promoter system. Nat. Biotechnol. 20, 1041–1044 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Leung D. W., Otomo C., Chory J., Rosen M. K., Genetically encoded photoswitching of actin assembly through the Cdc42-WASP-Arp2/3 complex pathway. Proc. Natl. Acad. Sci. U.S.A. 105, 12797–12802 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levskaya A., et al. , Synthetic biology: Engineering Escherichia coli to see light. Nature 438, 441–442 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Tabor J. J., Levskaya A., Voigt C. A., Multichromatic control of gene expression in Escherichia coli. J. Mol. Biol. 405, 315–324 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidl S. R., Sheth R. U., Wu A., Tabor J. J., Refactoring and optimization of light-switchable Escherichia coli two-component systems. ACS Synth. Biol. 3, 820–831 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Shu X., et al. , Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science 324, 804–807 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Filonov G. S., et al. , Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat. Biotechnol. 29, 757–761 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu D., et al. , An improved monomeric infrared fluorescent protein for neuronal and tumour brain imaging. Nat. Commun. 5, 3626 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu D., et al. , A naturally monomeric infrared fluorescent protein for protein labeling in vivo. Nat. Methods 12, 763–765 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baloban M., et al. , Designing brighter near-infrared fluorescent proteins: Insights from structural and biochemical studies. Chem. Sci. (Camb.) 8, 4546–4557 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Auldridge M. E., Forest K. T., Bacterial phytochromes: More than meets the light. Crit. Rev. Biochem. Mol. Biol. 46, 67–88 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Shcherbakova D. M., et al. , Bright monomeric near-infrared fluorescent proteins as tags and biosensors for multiscale imaging. Nat. Commun. 7, 12405 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikeuchi M., Ishizuka T., Cyanobacteriochromes: A new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria. Photochem. Photobiol. Sci. 7, 1159–1167 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Ulijasz A. T., et al. , Cyanochromes are blue/green light photoreversible photoreceptors defined by a stable double cysteine linkage to a phycoviolobilin-type chromophore. J. Biol. Chem. 284, 29757–29772 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song J.-Y., et al. , Near-UV cyanobacteriochrome signaling system elicits negative phototaxis in the cyanobacterium Synechocystis sp. PCC 6803. Proc. Natl. Acad. Sci. U.S.A. 108, 10780–10785 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rockwell N. C., Martin S. S., Feoktistova K., Lagarias J. C., Diverse two-cysteine photocycles in phytochromes and cyanobacteriochromes. Proc. Natl. Acad. Sci. U.S.A. 108, 11854–11859 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rockwell N. C., Martin S. S., Gulevich A. G., Lagarias J. C., Phycoviolobilin formation and spectral tuning in the DXCF cyanobacteriochrome subfamily. Biochemistry 51, 1449–1463 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Rockwell N. C., Martin S. S., Lagarias J. C., Red/green cyanobacteriochromes: Sensors of color and power. Biochemistry 51, 9667–9677 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Rockwell N. C., Martin S. S., Lagarias J. C., Mechanistic insight into the photosensory versatility of DXCF cyanobacteriochromes. Biochemistry 51, 3576–3585 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Hirose Y., et al. , Green/red cyanobacteriochromes regulate complementary chromatic acclimation via a protochromic photocycle. Proc. Natl. Acad. Sci. U.S.A. 110, 4974–4979 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rockwell N. C., Martin S. S., Lagarias J. C., Identification of cyanobacteriochromes detecting far-red light. Biochemistry 55, 3907–3919 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Fushimi K., Narikawa R., Cyanobacteriochromes: Photoreceptors covering the entire UV-to-visible spectrum. Curr. Opin. Struct. Biol. 57, 39–46 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Fushimi K., et al. , Rational conversion of chromophore selectivity of cyanobacteriochromes to accept mammalian intrinsic biliverdin. Proc. Natl. Acad. Sci. U.S.A. 116, 8301–8309 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliinyk O. S., Shemetov A. A., Pletnev S., Shcherbakova D. M., Verkhusha V. V., Smallest near-infrared fluorescent protein evolved from cyanobacteriochrome as versatile tag for spectral multiplexing. Nat. Commun. 10, 279 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodriguez E. A., et al. , A far-red fluorescent protein evolved from a cyanobacterial phycobiliprotein. Nat. Methods 13, 763–769 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding W.-L., et al. , Small monomeric and highly stable near-infrared fluorescent markers derived from the thermophilic phycobiliprotein, ApcF2. Biochim. Biophys. Acta Mol. Cell Res. 1864, 1877–1886 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Ding W.-L., et al. , Far-red acclimating cyanobacterium as versatile source for bright fluorescent biomarkers. Biochim. Biophys. Acta Mol. Cell Res. 1865, 1649–1656 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Hou Y.-N., et al. , Bright near-infrared fluorescence bio-labeling with a biliprotein triad. Biochim. Biophys. Acta Mol. Cell Res. 1866, 277–284 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Zhang J., et al. , Fused-gene approach to photoswitchable and fluorescent biliproteins. Angew. Chem. Int. Ed. 49, 5456–5458 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Ramakrishnan P., Tabor J. J., Repurposing Synechocystis PCC6803 UirS-UirR as a UV-violet/green photoreversible transcriptional regulatory tool in E. coli. ACS Synth. Biol. 5, 733–740 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Blain-Hartung M., et al. , Cyanobacteriochrome-based photoswitchable adenylyl cyclases (cPACs) for broad spectrum light regulation of cAMP levels in cells. J. Biol. Chem. 293, 8473–8483 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyake K., et al. , Functional diversification of two bilin reductases for light perception and harvesting in unique cyanobacterium Acaryochloris marina MBIC 11017. FEBS J., (2020). [DOI] [PubMed] [Google Scholar]
- 47.Fushimi K., et al. , Photoconversion and fluorescence properties of a red/green-type cyanobacteriochrome AM1_C0023g2 that binds not only phycocyanobilin but also biliverdin. Front. Microbiol. 7, 588 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Narikawa R., et al. , A biliverdin-binding cyanobacteriochrome from the chlorophyll d-bearing cyanobacterium Acaryochloris marina. Sci. Rep. 5, 7950 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Narikawa R., Fushimi K., Ikeuchi Ni-Ni-Win, M.,. Red-shifted red/green-type cyanobacteriochrome AM1_1870g3 from the chlorophyll d-bearing cyanobacterium Acaryochloris marina. Biochem. Biophys. Res. Commun. 461, 390–395 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Tu S.-L., Rockwell N. C., Lagarias J. C., Fisher A. J., Insight into the radical mechanism of phycocyanobilin-ferredoxin oxidoreductase (PcyA) revealed by X-ray crystallography and biochemical measurements. Biochemistry 46, 1484–1494 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Frankenberg N., Lagarias J. C., Phycocyanobilin:ferredoxin oxidoreductase of Anabaena sp. PCC 7120: Biochemical and spectroscopic. J. Biol. Chem. 278, 9219–9226 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Rockwell N. C., Martin S. S., Lagarias J. C., Identification of DXCF cyanobacteriochrome lineages with predictable photocycles. Photochem. Photobiol. Sci. 14, 929–941 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Campbell E. L., et al. , Genetic analysis reveals the identity of the photoreceptor for phototaxis in hormogonium filaments of Nostoc punctiforme. J. Bacteriol. 197, 782–791 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Y., et al. , Phototaxis in a wild isolate of the cyanobacterium Synechococcus elongatus. Proc. Natl. Acad. Sci. U.S.A. 115, E12378–E12387 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Altschul S. F., et al. , Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao K.-H., Haessner R., Cmiel E., Scheer H., Type I reversible photochemistry of phycoerythrocyanin involves Z/E-isomerization of α-84 phycoviolobilin chromophore. Biochim. Biophys. Acta Bioenerg. 1228, 235–243 (1995). [Google Scholar]
- 57.Zhao K.-H., Scheer H., Type I and type II reversible photochemistry of phycoerythrocyanin α-subunit from Mastigocladus laminosus both involve Z, E isomerization of phycoviolobilin chromophore and are controlled by sulfhydryls in apoprotein. Biochim. Biophys. Acta Bioenerg. 1228, 244–253 (1995). [Google Scholar]
- 58.Ishizuka T., et al. , The cyanobacteriochrome, TePixJ, isomerizes its own chromophore by converting phycocyanobilin to phycoviolobilin. Biochemistry 50, 953–961 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Rockwell N. C., Martin S. S., Gan F., Bryant D. A., Lagarias J. C., NpR3784 is the prototype for a distinctive group of red/green cyanobacteriochromes using alternative Phe residues for photoproduct tuning. Photochem. Photobiol. Sci. 14, 258–269 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Narikawa R., Muraki N., Shiba T., Ikeuchi M., Kurisu G., Crystallization and preliminary X-ray studies of the chromophore-binding domain of cyanobacteriochrome AnPixJ from Anabaena sp. PCC 7120. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65, 159–162 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Narikawa R., et al. , Structures of cyanobacteriochromes from phototaxis regulators AnPixJ and TePixJ reveal general and specific photoconversion mechanism. Proc. Natl. Acad. Sci. U.S.A. 110, 918–923 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lim S., et al. , Correlating structural and photochemical heterogeneity in cyanobacteriochrome NpR6012g4. Proc. Natl. Acad. Sci. U.S.A. 115, 4387–4392 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu X., et al. , Structural elements regulating the photochromicity in a cyanobacteriochrome. Proc. Natl. Acad. Sci. U.S.A. 117, 2432–2440 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang X., Kuk J., Moffat K., Crystal structure of Pseudomonas aeruginosa bacteriophytochrome: Photoconversion and signal transduction. Proc. Natl. Acad. Sci. U.S.A. 105, 14715–14720 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang X., Kuk J., Moffat K., Conformational differences between the Pfr and Pr states in Pseudomonas aeruginosa bacteriophytochrome. Proc. Natl. Acad. Sci. U.S.A. 106, 15639–15644 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang X., Ren Z., Kuk J., Moffat K., Temperature-scan cryocrystallography reveals reaction intermediates in bacteriophytochrome. Nature 479, 428–432 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wagner J. R., Brunzelle J. S., Forest K. T., Vierstra R. D., A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature 438, 325–331 (2005). [DOI] [PubMed] [Google Scholar]
- 68.Burgie E. S., Zhang J., Vierstra R. D., Crystal structure of Deinococcus phytochrome in the photoactivated state reveals a cascade of structural rearrangements during photoconversion. Structure 24, 448–457 (2016). [DOI] [PubMed] [Google Scholar]
- 69.Otero L. H., et al. , Structure of the full-length bacteriophytochrome from the plant pathogen Xanthomonas campestris provides clues to its long-range signaling mechanism. J. Mol. Biol. 428, 3702–3720 (2016). [DOI] [PubMed] [Google Scholar]
- 70.Wedemayer G. J., Kidd D. G., Wemmer D. E., Glazer A. N., Phycobilins of cryptophycean algae. Occurrence of dihydrobiliverdin and mesobiliverdin in cryptomonad biliproteins. J. Biol. Chem. 267, 7315–7331 (1992). [PubMed] [Google Scholar]
- 71.Harrop S. J., et al. , Single-residue insertion switches the quaternary structure and exciton states of cryptophyte light-harvesting proteins. Proc. Natl. Acad. Sci. U.S.A. 111, E2666–E2675 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gambetta G. A., Lagarias J. C., Genetic engineering of phytochrome biosynthesis in bacteria. Proc. Natl. Acad. Sci. U.S.A. 98, 10566–10571 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rockwell N. C., et al. , Eukaryotic algal phytochromes span the visible spectrum. Proc. Natl. Acad. Sci. U.S.A. 111, 3871–3876 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rockwell N. C., Martin S. S., Gulevich A. G., Lagarias J. C., Conserved phenylalanine residues are required for blue-shifting of cyanobacteriochrome photoproducts. Biochemistry 53, 3118–3130 (2014). [DOI] [PubMed] [Google Scholar]
- 75.Rockwell N. C., Martin S. S., Lim S., Lagarias J. C., Ames J. B., Characterization of red/green cyanobacteriochrome NpR6012g4 by solution nuclear magnetic resonance spectroscopy: A hydrophobic pocket for the C15-E,anti chromophore in the photoproduct. Biochemistry 54, 3772–3783 (2015). [DOI] [PubMed] [Google Scholar]
- 76.Lim S., et al. , Photoconversion changes bilin chromophore conjugation and protein secondary structure in the violet/orange cyanobacteriochrome NpF2164g3′ [corrected]. Photochem. Photobiol. Sci. 13, 951–962 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cornilescu C. C., et al. , Dynamic structural changes underpin photoconversion of a blue/green cyanobacteriochrome between its dark and photoactivated states. J. Biol. Chem. 289, 3055–3065 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Blain-Hartung M., Rockwell N. C., Lagarias J. C., Light-regulated synthesis of cyclic-di-GMP by a bidomain construct of the cyanobacteriochrome Tlr0924 (SesA) without stable dimerization. Biochemistry 56, 6145–6154 (2017). [DOI] [PubMed] [Google Scholar]
- 79.Jang J., McDonald S., Uppalapati M., Wooley G. A., Green, orange, red, and far-red optogenetic tools derived from cyanobacteriochromes. bioRxiv, 10.1101/769422 (2019). [DOI] [Google Scholar]
- 80.Yoshihara S., Suzuki F., Fujita H., Geng X. X., Ikeuchi M., Novel putative photoreceptor and regulatory genes required for the positive phototactic movement of the unicellular motile cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 41, 1299–1304 (2000). [DOI] [PubMed] [Google Scholar]
- 81.Bhaya D., Takahashi A., Grossman A. R., Light regulation of type IV pilus-dependent motility by chemosensor-like elements in Synechocystis PCC6803. Proc. Natl. Acad. Sci. U.S.A. 98, 7540–7545 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gan F., et al. , Extensive remodeling of a cyanobacterial photosynthetic apparatus in far-red light. Science 345, 1312–1317 (2014). [DOI] [PubMed] [Google Scholar]
- 83.Gan F., Shen G., Bryant D. A., Occurrence of far-red light photoacclimation (FaRLiP) in diverse cyanobacteria. Life (Basel) 5, 4–24 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gan F., Bryant D. A., Adaptive and acclimative responses of cyanobacteria to far-red light. Environ. Microbiol. 17, 3450–3465 (2015). [DOI] [PubMed] [Google Scholar]
- 85.Zhao C., et al. , RfpA, RfpB, and RfpC are the master control elements of far-red light photoacclimation (FaRLiP). Front. Microbiol. 6, 1303 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Alvey R. M., Biswas A., Schluchter W. M., Bryant D. A., Effects of modified phycobilin biosynthesis in the cyanobacterium Synechococcus sp. strain PCC 7002. J. Bacteriol. 193, 1663–1671 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Moreno M. V., Rockwell N. C., Fisher A. J., Lagarias J. C., Far-red absorbing dark state of JSC1_58120g3 with bound 181, 182 dihydrobiliverdin IXα (DHBV), the native chromophore precursor. RCSB PDB. 10.2210/pdb6XHH/pdb. Deposited 9 July 2020. [DOI]
- 88.Moreno M. V., Rockwell N. C., Fisher A. J., Lagarias J. C., Far-red absorbing dark state of JSC1_58120g3 with bound biliverdin IXα (BV). RCSB PDB. 10.2210/pdb6XHG/pdb. Deposited 9 July 2020. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data for this work have been deposited in the RCSB PDB under PDB ID codes 6XHH (181,182-DHBV adduct) (87) and 6XHG (BV adduct) (88). All study data are included in the article and supporting information.