Significance
Aberrant PD-1 expression is known to inhibit T cell effector activity and plays a pivotal role in tumor immune escape. Since maintaining an appropriate level of PD-1 expression is of great significance, it should be highly regulated at multiple levels. Despite extensive studies to date on this topic, it remains unclear how PD-1 expression is precisely regulated at the posttranslation level both during T cell activation and among the tumor microenvironment. Here, we show that KLHL22 maintains PD-1 protein homeostasis by degrading incompletely glycosylated PD-1, thereby preventing excessive T cell suppression. KLHL22-mediated degradation of PD-1 protein will be ineffective in response to either the tumor microenvironment or treatment with 5-FU, resulting in excessive PD-1 accumulation and reduced T cell activity.
Keywords: PD-1, protein degradation, 5-FU, immune checkpoint block therapy
Abstract
Aberrant programmed cell death protein 1 (PD-1) expression on the surface of T cells is known to inhibit T cell effector activity and to play a pivotal role in tumor immune escape; thus, maintaining an appropriate level of PD-1 expression is of great significance. We identified KLHL22, an adaptor of the Cul3-based E3 ligase, as a major PD-1–associated protein that mediates the degradation of PD-1 before its transport to the cell surface. KLHL22 deficiency leads to overaccumulation of PD-1, which represses the antitumor response of T cells and promotes tumor progression. Importantly, KLHL22 was markedly decreased in tumor-infiltrating T cells from colorectal cancer patients. Meanwhile, treatment with 5-fluorouracil (5-FU) could increase PD-1 expression by inhibiting the transcription of KLHL22. These findings reveal that KLHL22 plays a crucial role in preventing excessive T cell suppression by maintaining PD-1 expression homeostasis and suggest the therapeutic potential of 5-FU in combination with anti–PD-1 in colorectal cancer patients.
Currently, blocking programmed cell death protein 1 (PD-1) and its ligand PD-L1 is one of the most promising approaches to activate antitumor immunity, which has been extremely successful against a diverse range of malignancies (1). PD-1 is expressed on the surface of activated T cells and transduces immunoinhibitory signals, which inhibit T cell proliferation, survival, and effector functions, and therefore remarkably restrains the tumor-specific immune response (2, 3).
Since PD-1 has such a strong influence on T cell function, its expression should be highly regulated at multiple levels (4). Upon T cell activation, a significant increase in PD-1 expression on the surface of T cells was observed (5, 6); this increase is modulated by various transcription factors and signaling pathways, such as nuclear factor of activated T cells (NFAT) (7), activator protein 1 (AP1) (8), T-bet (9), Forkhead box protein O1 (FoxO1) (10), Notch (11), B-lymphocyte maturation protein 1 (Blimp1) (12), and GSK3α/β (13). Moreover, epigenetic regulation, including DNA methylation, may contribute to PD-1 expression by modulating the transcriptional activity of the PDCD1 gene (14, 15).
However, the dynamic regulation of the PD-1 protein at the posttranslational level has not yet been fully investigated. Posttranslational protein regulation is a critical mechanism that tightly controls protein levels and function, which facilitates cell responses to dynamic changes in signals and maintains homeostasis of intra- and extracellular environments (16). Ubiquitination is the predominant posttranslational modification that controls protein level by directing proteins to the proteasome for degradation; this process is involved in the regulation of numerous activities, such as receptor down-regulation, cellular signal transduction, and the immune response (17, 18). It has been reported that FBXO38 targets PD-1 on the cell surface for proteasome pathway degradation after PD-1 internalization, which means that PD-1 is already partially or completely active on T cells (19). However, as PD-1 is the key immune checkpoint factor, the dynamic regulation of protein levels might include multiple aspects, but whether intracellular PD-1 protein could be regulated before it is transported to the cell surface remains unknown.
The other important question in the field is that resistance and low response rate limit the efficiency of immunotherapy despite its status as one of the most promising treatment options for cancer (20, 21). Combining immunotherapy with chemo- and radiotherapy has been proven to elicit responses in certain cancers (22, 23). For example, preclinical trial results show treatment with 5-fluorouracil (5-FU), one of the first-line chemotherapeutic drugs for colorectal cancer (CRC), combined with PD-1 monoclonal antibody, improved overall survival compared with that of chemotherapy alone (24). Nonetheless, the mechanism of how combination therapy elicits a superior effect remains largely unclear. Treatment with 5-FU causes an increase in the expression of tumor-associated antigens, which induces a tumor-specific immune response (25, 26); however, this mechanism is insufficient to explain the differences in the effect of combination therapies among various chemotherapy drugs and tumor types (27).
In this study, we identified KLHL22 as a major PD-1–associated protein that mediates the degradation of PD-1 in T cells before it is transported to the cell surface. Deficiency of KLHL22 leads to aberrant PD-1 expression, which causes excessive T cell suppression and promotes tumor progression. Importantly, treatment with 5-FU could increase PD-1 expression by decreasing the transcription level of KLHL22. Our study reveals that KLHL22 regulates PD-1 expression to maintain appropriate levels of PD-1 and T cell homeostasis.
Results
KLHL22 Is a Major PD-1–Associated Protein.
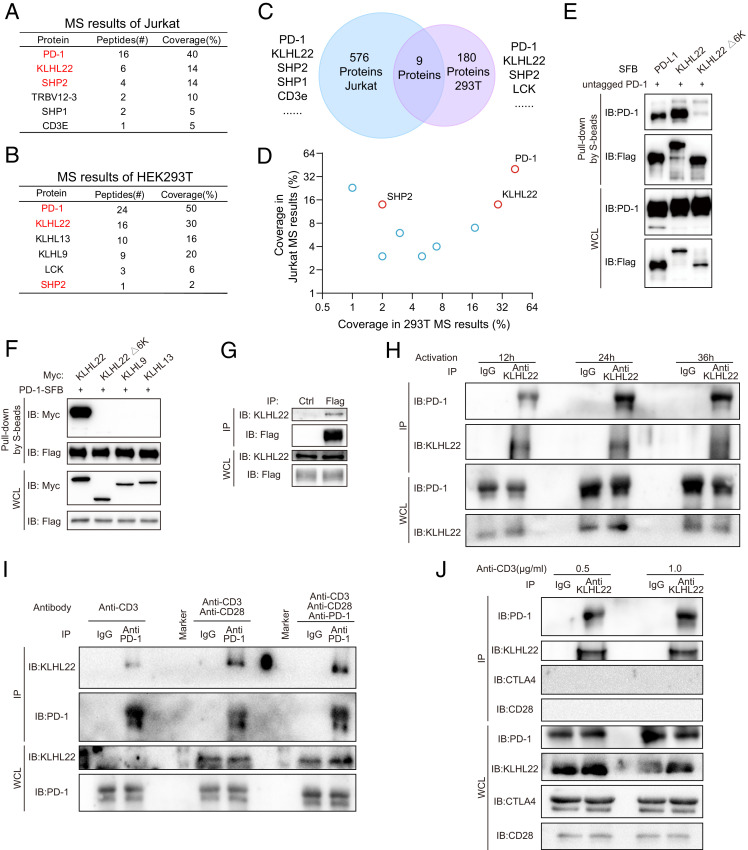
To explore the regulators of PD-1 at the protein level, we performed mass spectrometry (MS) on Jurkat cells stably expressing PD-1–FLAG and tandem affinity purification combined with MS (TAP/MS) on HEK293T cells stably expressing PD-1 with an SFB (S protein-2x FLAG-Streptavidin binding peptide) tag (28) (SI Appendix, Fig. S1A). Among the listed proteins in Jurkat cells (Fig. 1A and Dataset S1), SHP2 and SHP1, which have been proven to directly bind PD-1 protein (29, 30), were identified, suggesting that the results accurately reflect the interactions with PD-1 in cells. Interestingly, KLHL22, a substrate-specific adaptor protein of BTB-CUL3-RBX1 (BCR) E3 protein ligase, was also identified as a major PD-1–associated protein, ranking 19th (top 3.30%) among the 576 proteins. Meanwhile, the TAP/MS results from HEK293T cells identified SHP2 and LCK, both of which have been reported as PD-1–associated proteins (31) (Fig. 1B and Dataset S2). Surprisingly, KLHL22 also appeared on the top of this list, ranking 22nd among 186 proteins, along with KLHL13 and KLHL9. Overlapping the MS results from the two cell lines revealed nine proteins that appeared on both lists, including PD-1 and SHP2 (Fig. 1C and Dataset S3). In addition to PD-1, KLHL22 was the only protein with a coverage rate greater than 10% in both results (Fig. 1D), suggesting that KLHL22 is a major PD-1–associated protein. To further exclude the effects of common contaminants in purification results, we analyzed the MS data in the Crapome database, which stores contaminants generated by the proteomics researchers. We found that KLHL22 appeared very infrequently in these contaminant protein lists (SI Appendix, Fig. S1B), which made it rank even higher in the MS enrichment results after ruling out the possible contaminants.
Fig. 1.
KLHL22 is a major PD-1–associated protein. (A) PD-1–associated proteins in Jurkat cells were identified by MS. Jurkat cells stably expressing PD-1–FLAG were purified with FLAG-M2 beads and analyzed by MS. Jurkat cells was stimulated by PMA (50 ng/mL) and ionomycin (1 μM) for 12 h. The table lists the selected proteins identified by MS. The full protein list is provided in Dataset S1. (B) PD-1–associated proteins in HEK293T cells were identified by TAP/MS. HEK293T cells stably expressing PD-1–SFB were purified by TAP/MS. The table lists the selected proteins identified by MS. The full protein list is provided in Dataset S2. (C and D) Venn diagram showing the overlap of two MS results. Proteins appearing in the MS results of both the Jurkat PD-1–FLAG cells and HEK293T PD-1–SFB cells. The results from the overlap shown in C are presented in a 2D graph in D. The full protein list is provided in Dataset S3. (E) 6-Kelch repeats in KLHL22 are required for the interaction between KLHL22 and PD-1. HEK293T cells were cotransfected with untagged PD-1 (PD-1 without an artificial tag) and PD-L1-SFB, SFB-KLHL22, or SFB-KLHL22Δ6K. The cell lysates were subjected to pull-down assays with S protein Sepharose and immunoblotted with the indicated antibodies. (F) HEK293T cells were cotransfected with PD-1–SFB and Myc-KLHL22, Myc-KLHL22Δ6K, Myc-KLHL9, or Myc-KLHL13. The cell lysates were subjected to pull-down assays with S-protein Sepharose and immunoblotted with the indicated antibodies. (G) Lysates of Jurkat cells stably expressing PD-1–FLAG were immunoprecipitated with FLAG-M2 beads or protein G beads with IgG and subjected to immunoblotting with the indicated antibodies. Jurkat cells were stimulated by PMA (50 ng/mL) and ionomycin (1 μM) for 12 h. (H) The endogenous interaction of PD-1 and KLHL22 in healthy human PBMCs using KLHL22 antibody pull-down. Human healthy PBMCs lysates were immunoprecipitated with an anti-KLHL22 antibody or IgG and subjected to immunoblotting with the indicated antibodies. PBMCs were stimulated by anti-CD3 (1 μg/mL) and anti-CD28 (2 μg/mL) for 12 h, 24 h, or 36 h. (I) Endogenous PD-1 associates with endogenous KLHL22 in healthy human PBMCs. Human healthy PBMCs lysates were immunoprecipitated with an anti–PD-1 antibody or IgG and subjected to immunoblotting with the indicated antibodies. PBMCs were stimulated by anti-CD3 (1 μg/mL) or anti-CD3 (1 μg/mL)/anti-CD28 (2 μg/mL) for 24 h. The third group was also treated with PD-1 antibody (2 μg/mL) for 24 h. (J) Endogenous KLHL22 associates with endogenous PD-1 in healthy human PBMCs. CD28 and CTLA4 were tested simultaneously and showed negative results. Human healthy PBMCs lysates were immunoprecipitated with an anti-KLHL22 antibody or IgG and subjected to immunoblotting with the indicated antibodies. PBMCs were stimulated by anti-CD3 (0.5 μg/mL or 1 μg/mL) and anti-CD28 (2 μg/mL) for 24 h.
Subsequently, we confirmed the interaction between KLHL22 and PD-1. The intensity of the KLHL22/PD-1 interaction was the same as that of the PD-1/PD-L1 interaction, while the KLHL22Δ6K (6 kelch repeats) mutant, which lacks the substrate recognition motif (32), showed no interaction with PD-1 (Fig. 1E). Meanwhile, KLHL13 and KLHL9, which also were identified in the TAP/MS results of HEK293T cells but not in the MS results of Jurkat cells, showed no direct interaction with PD-1 (SI Appendix, Fig. S1C). It has been reported that KLHL9 and KLHL13 are associated with KLHL22, which might explain why KLHL9 and KLHL13 appeared in the MS results. Reverse coimmunoprecipitation (co-IP) between PD-1 and KLHL22 also showed the same results (Fig. 1F). CTLA4 did not detect an interaction with KLHL22 (SI Appendix, Fig. S1D). Furthermore, IP of stably expressed PD-1–FLAG in Jurkat cells pulled down endogenous KLHL22 (Fig. 1G and SI Appendix, Fig. S1E). To further validate the interaction of KLHL22 and PD-1 in primary cells, we performed IP in peripheral blood mononuclear cells (PBMCs) under different conditions and showed that endogenous PD-1, not CTLA4 nor CD28, interacts with endogenous KLHL22 in PBMCs (Figs. 1 H–J).
Collectively, these data demonstrate that KLHL22 is a major PD-1–interacting protein. In view of the role of KLHL22 in the CUL3/KLHL22/RBX1 E3 complex, these results suggest that KLHL22 is a specific E3 ligase adaptor of PD-1.
Loss of KLHL22 Leads to Up-Regulation of PD-1 at the Protein Level.
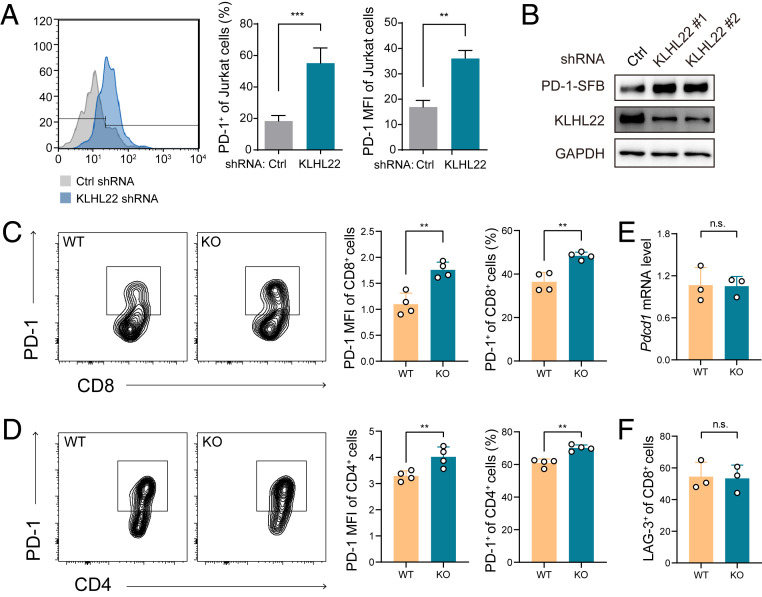
E3 ligase specifically recognizes substrates and targets them for ubiquitination, and a large percentage of ubiquitinated proteins are subject to protein degradation via the proteasome pathway (33). Thus, we first investigated whether KLHL22 targets PD-1 for degradation. Indeed, short-hairpin RNA (shRNA)-mediated knockdown of KLHL22 in activated Jurkat cells led to higher levels of PD-1 on the cell surface (Fig. 2A and SI Appendix, Fig. S2 A and B), and this shRNA did not affect the expression of another E3 ligase of PD-1, FBXO38. The same results were observed when Jurkat cells were treated with the Cullin family inhibitor MLN4924 (34) (SI Appendix, Fig. S2C). We also observed a prominent increase in PD-1 expression in KLHL22-knockdown HEK293T cells, which is consistent with the MS results and suggests that the effect of KLHL22 on PD-1 is independent of the regulatory network of T cells (Fig. 2B). At the same time, KLHL22 knockdown did not affect the mRNA level of PD-1 in activated Jurkat cells, which ruled out the possibility that KLHL22 influences PD-1 expression at the transcriptional level (SI Appendix, Fig. S2D).
Fig. 2.
Loss of KLHL22 leads to up-regulation of PD-1 at the protein level. (A) Depletion of KLHL22 increases PD-1 expression on the surface of Jurkat cells. Jurkat cells infected with lentivirus containing control or KLHL22-specific shRNA were stimulated with anti-CD3 (1 μg/mL) and anti-CD28 (2 μg/mL) for 24 h and subjected to flow cytometry to measure PD-1 expression on the cell surface. n = 3 biological independent samples, **P < 0.01, ***P < 0.001, unpaired Student’s t test. (B) Knockdown of KLHL22 increases PD-1 expression in HEK293T cells stably expressing PD-1–SFB. HEK293T cells stably expressing PD-1–SFB were infected with lentivirus containing control or KLHL22-specific shRNA and subjected to immunoblotting to detect PD-1 expression. (C and D) Cell-surface expression of PD-1 is higher in activated CD8+ (C) and CD4+ (D) T cells from Klhl22 KO mice than in those from WT mice. Naïve T cells from WT and Klhl22 KO mice were stimulated with anti-CD3 (1 μg/mL) and anti-CD28 (2 μg/mL) for 24 h. n = 4 mice per group. **P < 0.01, unpaired Student’s t test. (E) Transcription levels of Pdcd1 in CD3+ T cells from WT and Klhl22 KO mice showed no differences. CD3+ T cells were isolated from lymph nodes, and qRT-PCR analysis was used to measure the mRNA level of Pdcd1. n = 3 mice per group; ns, not significant, unpaired Student’s t test. (F) Cell-surface expression levels of LAG-3 in activated CD8+ T cells from WT and Klhl22 KO mice showed no differences. Naïve T cells from WT and Klhl22 KO mice were stimulated with anti-CD3 and anti-CD28 antibodies for 24 h. n = 3 mice per group; ns, not significant, unpaired Student’s t test.
To determine the relevance of KLHL22 in vivo, we generated Klhl22 knockout (KO) mice. Deletion of Klhl22 did not affect the development or peripheral homeostasis of T cells (SI Appendix, Fig. S2 E–G), and T cells from Klhl22 KO mouse showed approximately the same level of CD44 as that in WT mice after stimulation by anti-CD3 and anti-CD28 antibodies, suggesting that T cell activation was not affected by the deletion of Klhl22 (SI Appendix, Fig. S2H). However, PD-1 levels on the cell surface were increased in both CD8+ T cells and CD4+ T cells from Klhl22 KO mice (Fig. 2 C and D), with no changes in the transcription levels of Pdcd1 (Fig. 2E); these data are consistent with the above results in Jurkat and HEK293T cells. In comparison, the cell surface expression of the other two checkpoint receptors, LAG3 and TIM-3 (30), was not altered (Fig. 2F).
Taken together, these results show that KLHL22 down-regulation increases the protein level of PD-1, suggesting that KLHL22 is the E3 adaptor of PD-1 that mediates its degradation.
KLHL22 Mediates the Degradation of PD-1 before It Is Transported to the Cell Surface.
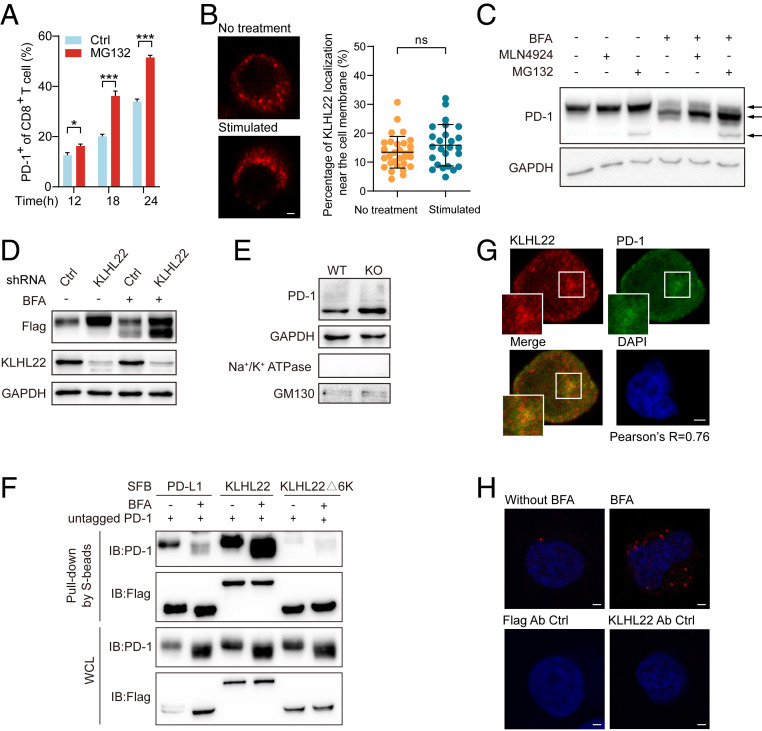
As a transmembrane protein, PD-1 might be degraded at different stages of the protein synthesis and transport processes (35, 36). We found that PD-1 could be degraded by the ubiquitin-proteasome pathway at multiple stages of its existence (Fig. 3A and SI Appendix, Fig. S3A). Thus, we next explored at which stage PD-1 was ubiquitinated and degraded by KLHL22. Since KLHL22 is not located on the cell membrane regardless of the T cell activation state (Fig. 3B), we inferred that PD-1 might be recognized by KLHL22 before its translocation to the cell surface.
Fig. 3.
KLHL22 mediates the degradation of PD-1 before it is transported to the cell surface. (A) Cell-surface levels of PD-1 of CD8+ T cell in activated healthy human PBMCs with or without MG132 treatment were detected by flow cytometry. PBMCs were stimulated with anti-CD3 (1 μg/mL), anti-CD28 (2 μg/mL) and treated with MG132 (1 μM) for indicated hours. n = 3 repeats, *P < 0.05, ***P < 0.001, unpaired Student’s t test. (B) KLHL22 does not localize to the cell membrane in healthy human PBMCs with or without stimulation. PBMCs were incubated in the presence or absence of anti-CD3 (1 μg/mL), anti-CD28 (2 μg/mL), and subjected to immunostaining with KLHL22 antibodies. (Scale bar, 2 μm.) On the right, no significant difference in cell membrane localization of KLHL22 before and after activation. Immunofluorescence staining for KLHL22 was performed in PBMCs. MATLAB was used to identify KLHL22 near membrane location in multiple pictures and quantify the percentage of KLHL22 localization near the cell membrane. ns, not significant, unpaired Student’s t test. (C) Simultaneous treatment with MLN4924 and BFA increased the levels of incompletely glycosylated PD-1. HEK293T cells stably expressing untagged PD-1 were treated with BFA (1 µM), MLN4924 (1 µM), and MG132 (1 µM) as indicated for 12 h. The three arrows on the right side of the figure indicate fully glycosylated PD-1 (top), incompletely glycosylated PD-1 (middle), and newly synthesized PD-1 (bottom). (D) KLHL22 down-regulation leads to more accumulation of incompletely glycosylated PD-1 than fully glycosylated PD-1. The expression level of PD-1 in HEK293T cells subjected to BFA treatment and/or KLHL22 shRNA lentivirus infection was detected. HEK293T cells stably expressing PD-1–SFB were infected with lentivirus containing control or KLHL22-specific shRNA and incubated in the presence or absence of BFA (1 µM, 12 h). Cells were then subjected to immunoblotting with the indicated antibodies. (E) The amount of cytoplasmic PD-1 is higher in CD3+ T cells from Klhl22 KO mice cells than in those from WT mice. The Golgi apparatus was extracted from CD3+ T cells from WT and Klhl22 KO mice and subjected to immunoblotting to detect the protein level of PD-1 in the Golgi apparatus. Na+/K+ ATPase served as a cell membrane marker, whereas GM130 served as a Golgi apparatus marker. (F) KLHL22 has a higher affinity for incompletely glycosylated PD-1 than for fully glycosylated PD-1. HEK293T cells stably expressing untagged PD-1 were transfected with the indicated plasmids in the presence or absence of BFA (1 µM, 12 h), and cell lysates were subjected to pull-down assays with S-protein Sepharose and immunoblotted with the indicated antibodies. (G) PD-1 colocalizes with KLHL22 in the cytoplasm in Jurkat cells. Colocalization of PD-1 and endogenous KLHL22 in Jurkat cells stably expressing PD-1–FLAG was confirmed by immunostaining with anti-KLHL22 and anti-FLAG antibodies. Jurkat cells were stimulated with PMA (50 ng/mL 12 h)/ionomycin (1 µM 12 h) and treated with BFA (1 µM, 6 h). Pearson’s r = 0.76. (Scale bar, 2 μm.) (H) PLA was used to detect the colocalization of PD-1 and endogenous KLHL22 in Jurkat PD-1–FLAG stable cell lines in the presence or absence of BFA (1 µM, 6 h). Using of only one antibody (anti-KLHL22 or anti-FLAG) served as the negative control groups. (Scale bar, 2 μm.)
To determine at which stage PD-1 could be degraded by KLHL22, we used brefeldin A (BFA), which inhibits protein transport from the endoplasmic reticulum (ER) to the Golgi apparatus (37). As predicted, we found that a low molecular mass (∼40 kDa) band of PD-1 emerged after BFA treatment, which represents a form of PD-1 with incomplete glycosylation (Fig. 3C). Simultaneous treatment with BFA and MLN4924 led to a higher level of incompletely glycosylated PD-1 (Fig. 3C). We confirmed this phenomenon with [35S] pulse-chase experiment (SI Appendix, Fig. S3 B and C). Importantly, in the absence of KLHL22 expression, the amount of accumulated incompletely glycosylated PD-1 was greater than that of fully glycosylated PD-1 (Fig. 3D). These results indicated that PD-1 could be degraded by KLHL22 before it becomes fully mature and reaches the cell surface. To further verify this phenomenon, we extracted Golgi apparatuses from CD3+ T cells in the lymph nodes of Klhl22 KO and WT mice. With mature PD-1 on the cell membrane cleared, the amount of incompletely glycosylated PD-1 was higher in Klhl22 KO cells than in WT cells (Fig. 3E), which indicates that KLHL22 mediates PD-1 degradation before it is completely glycosylated and translocated to the cell surface.
Next, we verified whether KLHL22 interacts with incompletely glycosylated PD-1. As expected, we found that KLHL22 rather than KLHL9 and KLHL13 has a stronger interaction with incompletely glycosylated PD-1, while PD-L1 shows a great decrease of interaction with incompletely glycosylated PD-1, consistent with the interaction between PD-1 and PD-L1 depends on glycosylation (38) (Fig. 3F and SI Appendix, Fig. S3D).
We then confirmed the colocalization of PD-1 and KLHL22 by immunofluorescence. After BFA treatment, as PD-1 expressed on the cell membrane was removed, strong colocalization of PD-1 and KLHL22 was observed in the cytoplasm (Fig. 3G). More importantly, the colocalization of KLHL22 and PD-1 was significantly increased in Jurkat cells treated with BFA as indicated by the Duolink proximity ligation assay, further suggesting that KLHL22 better recognizes incompletely glycosylated PD-1 than mature PD-1 in vivo. (Fig. 3H).
These data demonstrate that KLHL22 recognizes incompletely glycosylated PD-1 and mediates its degradation before it reaches the cell surface.
PD-1 Could Be Ubiquitinated by KLHL22/CUL3/RBX1 In Vivo.
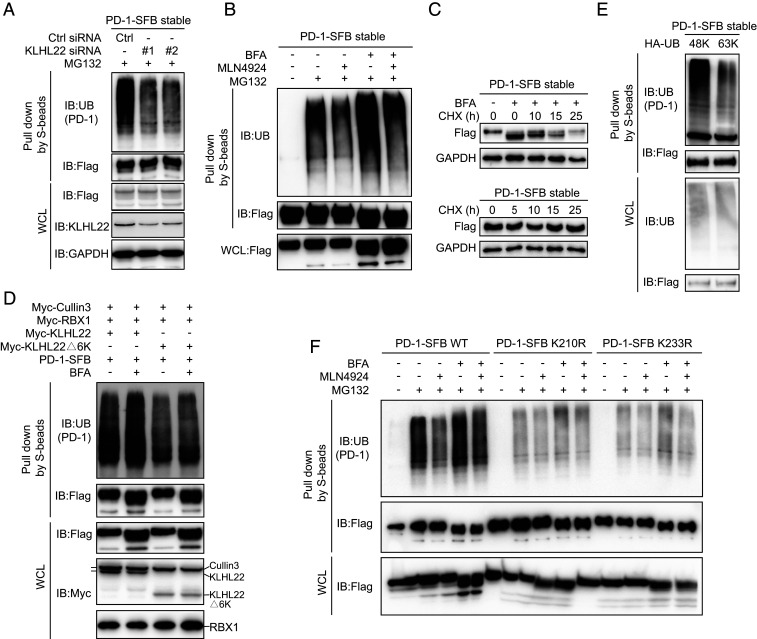
KLHL22 is a substrate-specific adaptor protein of the BCR E3 ligase complex that recognizes substrates and mediates their ubiquitination (32, 39). Thus, we first tested the ubiquitination of PD-1. As predicted, PD-1 ubiquitination was detected after MG132 treatment, and KLHL22 knockdown significantly decreased the level of ubiquitinated PD-1 (Fig. 4A). Interestingly, the level of ubiquitination of incompletely glycosylated PD-1 was higher than that of mature PD-1, and MLN4924 attenuated the ubiquitination of incompletely glycosylated PD-1 (Fig. 4B). Consistent with these results, compared with mature PD-1, incompletely glycosylated PD-1 had a faster turnover rate (Fig. 4C).
Fig. 4.
KLHL22 mediates polyubiquitination-directed degradation of incompletely glycosylated PD-1. (A) PD-1 ubiquitination is inhibited by KLHL22 depletion. Control or KLHL22-specific siRNA was transfected into HEK293T cells stably expressing PD-1–SFB in the presence of MG132 (1 µM 24 h). Cell lysates were subjected to pull-down assays by S-protein Sepharose and immunoblotted with the indicated antibodies. (B) PD-1 ubiquitination is inhibited upon MLN4924 treatment. HEK293T cells stably expressing PD-1–SFB were treated with BFA (1 µM), MLN4924 (1 µM), and MG132 (1 µM) as indicated for 12 h, and cell lysates were subjected to pull-down assays with S-protein Sepharose and immunoblotted with anti-FLAG and antiubiquitin antibodies. (C) Incompletely glycosylated PD-1 is unstable in vivo. PD-1–SFB stable cells were incubated in medium containing 10 μg/mL cycloheximide (CHX) in the presence or absence of BFA (1 µM) for the indicated time. Western blotting was carried out using the indicated antibodies. (D) The KLHL22/CUL3/RBX1 complex ubiquitinates PD-1 in vivo. CUL3, RBX1, and either KLHL22 or KLHL22Δ6K were overexpressed in HEK293T cells stably expressing PD-1–SFB, and cell lysates were subjected to pull-down assays with S-protein Sepharose and immunoblotted with the indicated antibodies. (E) Only 48K ubiquitin can be conjugated to PD-1. For ubiquitination mutants transfected into HEK293T cells stably expressing PD-1–SFB, all lysine’s were mutated to arginine except Lys48 (48K) or Lys63 (63K). Cells were treated with MG132 (1 µM, 24 h). Ubiquitination of PD-1 was detected by immunoblotting with antiubiquitin antibody. Cell lysates were subjected to pull-down assays with S-protein Sepharose and immunoblotted with antiubiquitin and anti-FLAG antibodies. (F) The ubiquitination of PD-1 on K210R and K233R is significantly reduced. PD-1–SFB (WT), PD-1–SFB (K210R), or PD-1–SFB (K233R) was transfected into HEK293T cells treated with BFA (1 µM), MLN4924 (1 µM) and MG132 (1 µM) as indicated for 12 h. The resulting cell lysates were subjected to pull-down assays with S-protein Sepharose and immunoblotted with the indicated antibodies.
Furthermore, we found that the KLHL22/CUL3/RBX1 complex distinctively increased the PD-1 ubiquitination level (Fig. 4D), and BFA treatment plus transfection with WT KLHL22 but not KLHL22Δ6K caused a significant increase in PD-1 ubiquitination compared with the levels in the untreated groups, which confirmed that KLHL22 mediates the ubiquitination of incompletely glycosylated PD-1. We also assessed the levels of different ubiquitin variants (48K, 63K), except that the corresponding point remained a lysine while all of the other sites were mutated to arginines. Only the 48K ubiquitin variant could interact with PD-1 (Fig. 4E). The 48K ubiquitination chain mediates protein degradation by the proteasome pathway (40), which is consistent with the experimental results. Furthermore, to identify ubiquitination sites of PD-1, we used UbPred (41) to predict possible lysine residues of PD-1. Both K210 and K233 showed a great possibility to be ubiquitinated. Then we performed a ubiquitination assay with BFA and MLN4924 treatment and confirmed that K210 and K233 were both ubiquitination sites of incompletely glycosylated PD-1 (Fig. 4F).
Therefore, we demonstrate that the KLHL22/CUL3/RBX1 complex specifically mediates PD-1 ubiquitination and promotes its degradation.
KLHL22 Enhances Tumor Immunity by Degrading PD-1.
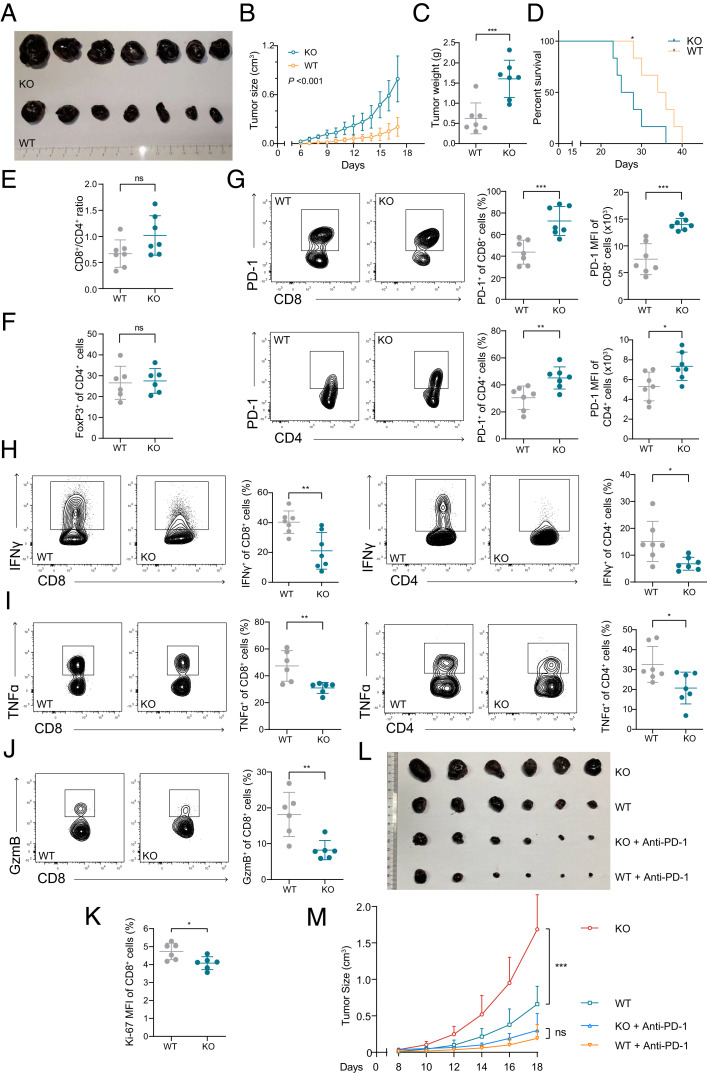
Due to the vital role of PD-1 in tumor immunosuppression (42, 43), mouse models of subcutaneous melanoma and CRC were used to explore the role of KLHL22 in tumor immunity. Klhl22 KO mice inoculated with tumors had faster tumor progression (Fig. 5 A–C and SI Appendix, Fig. S4 A and B) and shorter overall survival (Fig. 5D) than did WT mice.
Fig. 5.
KLHL22 regulates T cell antitumor immunity. (A–C) Klhl22 KO mice showed faster tumor progression. WT and Klhl22 KO mice were subcutaneously injected with B16F10 melanoma cells. Tumor growth was monitored over a period of 17 d. The tumors were isolated from mice killed on day 17 and measured. n = 7 mice per group, ***P < 0.001, (B) two-way ANOVA, (C) unpaired Student’s t test. (D) Klhl22 KO mice inoculated with tumors have a shorter survival time. WT mice and Klhl22 KO mice were subcutaneously injected with B16F10 melanoma cells, and the survival was assessed in WT and Klhl22 KO mice. n = 7 mice per group, *P < 0.05, log-rank (Mantel–Cox) test. (E and F) The CD8+/CD4+ ratio (E) and regulatory T cell ratio (F) of tumor-infiltrating T cells were not significantly different in tumor tissues from WT mice and those from Klhl22 KO mice. Tumor-infiltrating T cells from WT and Klhl22 KO mice were isolated on day 17 and subjected to flow cytometry to measure the CD4/CD8 ratio and T cell ratio. n = 7 mice per group; ns, not significant, unpaired Student’s t test. (G) PD-1 expression in tumor-infiltrating T cells from Klhl22 KO mice is significantly higher than that in cells from WT mice. Tumor-infiltrating T cells from WT and Klhl22 KO mice were isolated on day 17 and subjected to flow cytometry. n = 7 mice per group, *P < 0.05, **P < 0.01, ***P < 0.001, unpaired Student’s t test. (H–J) Cytokine production by tumor-infiltrating CD8+ and CD4+ T cells from Klhl22 KO mice is inhibited. Tumor-infiltrating T cells from WT and Klhl22 KO mice were isolated on day 17 and subjected to flow cytometry. n = 7 or 6 mice per group, *P < 0.05, **P < 0.01, unpaired Student’s t test. (K) The proliferation of tumor-infiltrating CD8+ T cells was decreased in Klhl22 KO mice. Tumor-infiltrating T cells from WT and Klhl22 KO mice were isolated on day 17 and subjected to flow cytometry. n = 6 mice per group, *P < 0.05, **P < 0.01, unpaired Student’s t test. (L and M) PD-1 antibody treatment dramatically diminished the difference in tumor growth between WT and KO mice where there was no statistically significant difference. WT and Klhl22 mice were subcutaneously injected with B16F10 melanoma cells. Tumor growth was monitored over a period of 18 d. Mice were intraperitoneally injected with PBS or anti–PD-1 (RMP1-14, 200 μg per mouse, dissolved in PBS) every 3 d (three times in total) 8 d after B16F10 inoculation and tumor sizes were recorded every 2 d afterward (n = 6). ns, not significant, ***P < 0.001, two-way ANOVA.
Klhl22 deficiency did not affect the ratio of CD8+ and CD4+ T cells or the percentage of Treg cells in the tumor microenvironment (Fig. 5 E and F). However, the levels of PD-1 on the cell surface of tumor-infiltrating CD8+ T cells and CD4+ T cells were remarkably higher in Klhl22 KO mice than in WT mice (Fig. 5G and SI Appendix, Fig. S4C). The numbers of tumor-infiltrating CD8+ and CD4+ T cells producing the effector cytokines IFN-γ, TNF, and granzyme B were also significantly decreased in Klhl22 KO mice (Figs. 5 H–J), and the proliferation of CD8+ T cells was also impaired (Fig. 5K). These results indicate a greater extent of CD8+ and CD4+ T cell-excessive suppression in the tumor microenvironments in Klhl22 KO mice.
To further clarify that the effect of Klhl22 KO is through regulation of PD-1, we performed anti–PD-1 checkpoint blockade assay to treat subcutaneous implanted tumors in WT and Klhl22 KO mice. As indicated, PD-1 antibody resulted in significant tumor suppression, and importantly, anti–PD-1 therapy diminished the difference in tumor growth between WT and Klhl22 KO mice (Fig. 5 L and M), suggesting that the difference in the tumor progression rate between WT and KO mice is mainly determined by the difference in PD-1 expression.
These results demonstrate that KLHL22 deficiency could lead to excessive suppression of tumor-infiltrating T cells by up-regulating PD-1 expression.
T Cell Activation Signal and Tumor Microenvironment Regulate KLHL22 Expression.
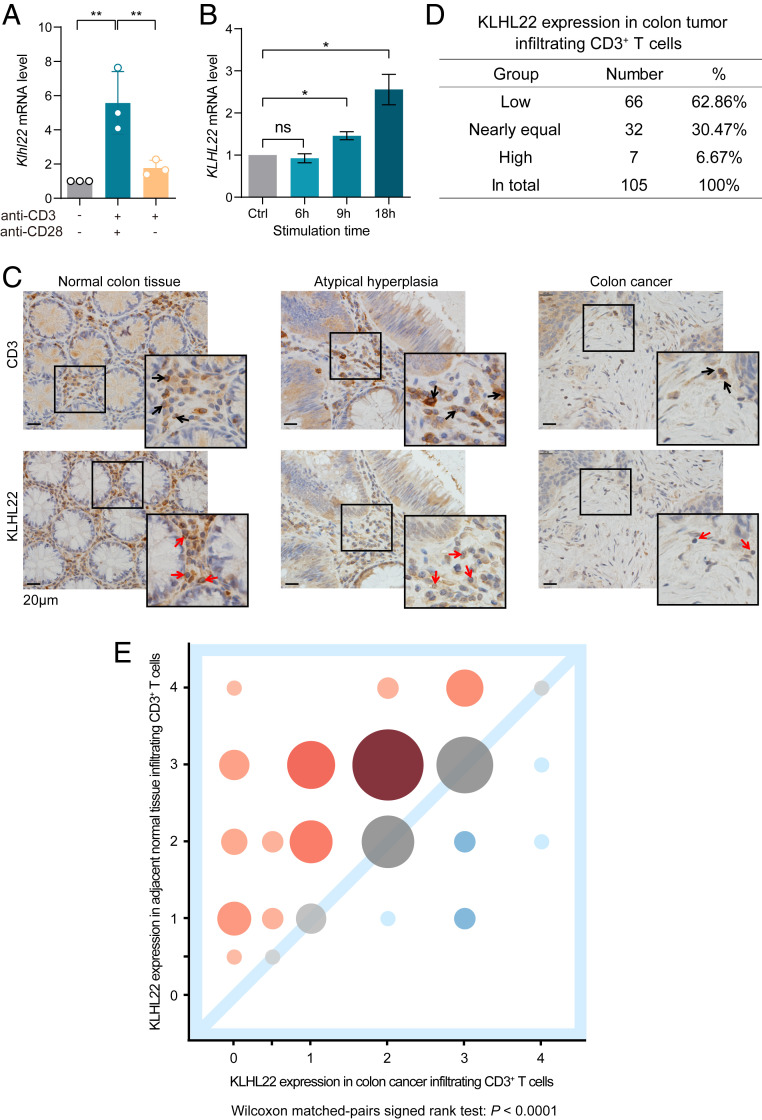
Based on the above results, KLHL22 targets PD-1 and regulates its expression before it reaches the cell surface. Since PD-1 expression is strikingly up-regulated during T cell receptor (TCR) activation (5, 6), maintaining PD-1 homeostasis by regulating PD-1 expression within an appropriate range is critical. Therefore, we wondered whether KLHL22 might also be regulated by TCR activation signals to maintain T cell homeostasis. To address this, we detected the mRNA level of KLHL22 in quiescent and activated T cells and observed a significant increase in the KLHL22 mRNA level after T cell activation (Fig. 6A). The same results were obtained in Jurkat cells via activation with anti-CD3/anti-CD28 (Fig. 6B). Considering that the PD-1 level of T cells increased in Klhl22 KO mice compared with that in WT mice (Fig. 2 C and D), we believe that up-regulation of KLHL22 expression by T cell activation signals can maintain the level of PD-1 within an appropriate range in KLHL22-proficient cells.
Fig. 6.
T cell activation and the tumor microenvironment regulate KLHL22 expression. (A) The transcription levels of Klhl22 in mouse CD3+ T cells increase dramatically upon T cell activation. CD3+ T cells were isolated from the lymph nodes of C57BL6 WT mice and stimulated with anti-CD3 (2 µg/mL) and/or anti-CD28 (4 µg/mL); the cells were then subjected to total RNA extraction and qRT-PCR analysis. n = 3 independent biological samples per group, **P < 0.01, unpaired Student’s t test. (B) The transcription levels of KLHL22 increase over time in activated Jurkat cells. Jurkat cells were stimulated with PMA (50 µg/mL) and ionomycin (1 µM) for the indicated time and then subjected to total RNA extraction and qRT-PCR analysis. n = 3 independent biological samples per group; ns, not significant, *P < 0.05, unpaired Student’s t test. (C) The level of KLHL22 protein was markedly decreased in tumor-infiltrating T cells of CRC patients. Immunohistochemical staining of KLHL22 and CD3e was performed using a colorectal tissue microarray of CRC patients. CD3e staining was used to mark CD3+ T cells, and KLHL22 staining was used to measure KLHL22 expression in CD3+ T cells. Selected samples from the tissue microarrays show KLHL22 expression in CD3+ T cells. Normal colon tissue (Left), atypical hyperplasia (Center), and colon cancer (Right). The number of atypical hyperplasia samples was too low to be included in the subsequent analyses. The 2× magnified image is displayed in the lower right corner. (Scale bar, 20 μm.) (D) Semiquantitative analysis of KLHL22 levels in CRC patients’ CD3+ T cells. Correlation analyses of KLHL22 expression between tumor-infiltrating CD3+ T cells and CD3+ T cells that infiltrated normal adjacent tissues. (E) Bubble chart showing the individual quantities of samples at different levels. Samples with low KLHL22 expression in tumor-infiltrating T cells are in the upper left quadrant of the graph, represented by red bubbles. Samples with high KLHL22 expression in tumor-infiltrating T cells are in the lower right quadrant of the graph, represented by blue bubbles. Larger bubbles indicate a larger number of samples; smaller bubbles are paler in color. Wilcoxon matched-pairs signed rank test: P < 0.0001.
Interestingly, we found that stimulating T cells with anti-CD3 alone led to a mild increase in KLHL22 expression compared with that in response to costimulation with anti-CD3 and anti-CD28 (Fig. 6A). Coincidently, tumors often lack sufficient costimulation and cytokine stimulation (44), and tumor-infiltrating T cells have much higher PD-1 expression in these conditions (45). Therefore, we speculated that KLHL22 might be down-regulated in tumor-infiltrating T cells, which contributes to the uncontrolled PD-1 expression on cells in the tumor microenvironment.
More strikingly, the level of KLHL22 protein was markedly decreased in tumor-infiltrating T cells of CRC patients compared to that in adjacent normal tissue-infiltrating T cells (Fig. 6 C–E and SI Appendix, Fig. S5). Notably, we observed lower KLHL22 expression in tumor-infiltrating T cells than in adjacent normal tissue-infiltrating T cells in 66 of 105 (62.86%) patients, no significant difference in 32 (30.47%) patients, and increased KLHL22 expression in tumor-infiltrating T cells in only 7 (6.67%) patients (Fig. 6D). These results confirm that KLHL22 is down-regulated in tumor-infiltrating T cells.
Taken together, these results suggest that increased KLHL22 expression after T cell stimulation is a mechanism for maintaining the appropriate levels of PD-1 expression, and disruption of this mechanism in the tumor microenvironment is one of the reasons for the extremely high levels of PD-1 expression and T cell suppression.
5-FU Increases PD-1 Expression by Inhibiting KLHL22 Transcription.
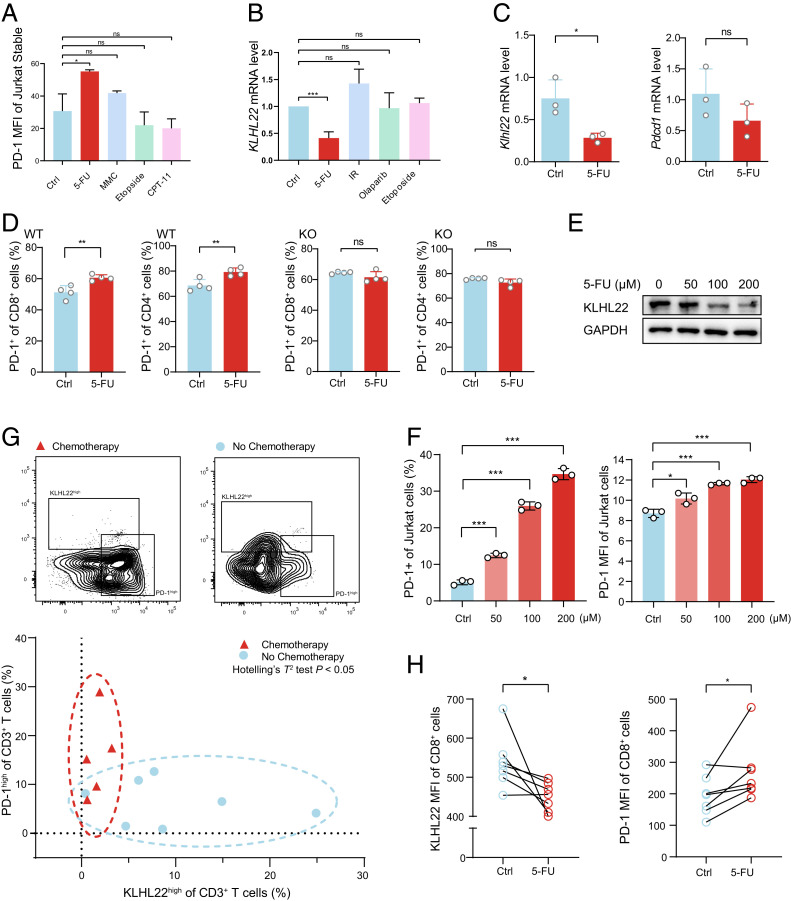
There is a large body of clinical and experimental evidence describing better outcomes with the combination of PD-1 monoclonal antibody and chemotherapy versus chemotherapy alone, but the mechanism is not thoroughly clear (23). Previous work from our laboratory revealed that compared to other chemotherapy drugs, 5-FU can significantly increase PD-1 expression (Fig. 7A). Therefore, we wondered whether chemotherapy drugs such as 5-FU might regulate KLHL22 levels and subsequently affect PD-1 expression. Accordingly, we tested several chemotherapy drugs as well as radiotherapy in Jurkat cells and found that only 5-FU specifically down-regulated KLHL22 expression at the transcriptional level, with the other chemotherapy and radiotherapy regimens showing little effect (Fig. 7B). Notably, 5-FU can enhance PD-1 expression without increasing its mRNA levels (Fig. 7C), and PD-1 expression in T cells was increased after 5-FU treatment in WT mice but not in Klhl22 KO mice (Fig. 7D), further illustrating that down-regulating KLHL22 plays a key role in 5-FU–mediated increases in PD-1 expression. Moreover, 5-FU repressed KLHL22 protein and up-regulated PD-1 expression in a dose-dependent manner (Fig. 7 E and F). All of these data suggest that down-regulating KLHL22 plays a critical role in 5-FU-induced up-regulation of PD-1 expression.
Fig. 7.
5-FU increases the expression of PD-1 by decreasing KLHL22 mRNA levels. (A) Treatment with 5-FU rather than other chemotherapeutic drugs increases PD-1 protein levels. Jurkat cells stably expressing PD-1–FLAG were stimulated with PMA (50 ng/mL 24 h) and ionomycin (1 µM 24 h) in the presence of 5-FU (100 µM 24 h), Etoposide (10 µM, 6 h), MMC (100 nM, 24 h), or CPT-11(10 µM, 24 h), as indicated. Cell-surface levels of PD-1 were measured by flow cytometry. ns, not significant, *P < 0.05, unpaired Student’s t test. (B) 5-FU treatment represses KLHL22 transcription. Jurkat cells were stimulated with PMA (50 ng/mL 24 h) and ionomycin (1 µM 24 h) in the presence of 5-FU (100 µM, 24 h), Etoposide (10 µM, 6 h), Olaparib (10 µM, 24 h), or irradiation (1 Gr) as indicated, and the KLHL22 transcription level was measured by qRT-PCR. ns, not significant, ***P < 0.001, unpaired Student’s t test. (C) 5-FU treatment represses only Klhl22 mRNA levels. CD3+ T cells were freshly isolated from the lymph nodes of WT mice and stimulated with anti-CD3 (1 μg/mL, 24 h) and anti-CD28 (2 μg/mL, 24 h) in the presence or absence of 5-FU (100 µM, 24 h). Klhl22 and Pdcd1 transcription levels were measured by qRT-PCR. ns, not significant, *P < 0.05, unpaired Student’s t test. (D) Deletion of KLHL22 abolishes the effect of 5-FU on PD-1 expression. CD3+ T cells were freshly isolated from the lymph nodes of WT or Klhl22 KO mice and stimulated with anti-CD3 (1 μg/mL, 24 h) and anti-CD28 (2 μg/mL, 24 h) in the presence or absence of 5-FU (100 µM, 24 h). Flow cytometry was used to measure PD-1 levels on the surface of CD8+ or CD4+ T cells. ns, not significant, **P < 0.01, unpaired Student’s t test. (E) 5-FU represses KLHL22 expression in a dose-dependent manner. Jurkat cells were stimulated with PMA (50 ng/mL 24 h) and ionomycin (1 µM 24 h) and treated with a concentration gradient of 5-FU. KLHL22 protein levels were measured by Western blotting. (F) 5-FU increases PD-1 expression in a dose-dependent manner. Jurkat cells were stimulated with PMA (50 ng/mL 24 h) and ionomycin (1 µM 24 h) and treated with a concentration gradient of 5-FU. Cell-surface expression of PD-1 was measured by flow cytometry, *P < 0.05, ***P < 0.001, unpaired Student’s t test. (G) PD-1 is expressed at high levels and KLHL22 is expressed at low levels in patients who received chemotherapy containing 5-FU. Patients were divided into two groups based on whether they had received 5-FU (chemotherapy and no chemotherapy). Tumor-infiltrating lymphocytes were isolated from fresh tumor samples and subjected to flow cytometry analysis to measure cell-surface expression of PD-1 and intracellular expression of KLHL22 in CD3+ T cells. Each point in the figure represents the KLHL22 and PD-1 expression levels of a single patient. Data points corresponding to the chemotherapy group are concentrated in the left region of the figure (red circle), indicating the grouping of patients with low KLHL22 expression and high PD-1 expression, while the no chemotherapy group is concentrated in the lower region of the figure (blue circle), indicating the grouping of patients with high KLHL22 expression and relatively low PD-1 expression. n = 5 patients received chemotherapy and n = 7 patients has not received chemotherapy. Hotelling’s T2 test: P < 0.05. (H) After 5-FU treatment, CD3+ T cells showed reduced KLHL22 expression and increased PD-1 expression compared to that of untreated cells from the same individual patient. PBMCs were isolated from the CRC patients’ blood and divided into two equal volumes. Both were treated with PMA (50 ng/mL 12 h) and ionomycin (1 µM 12 h) in the presence or absence of 5-FU (5-FU and Ctrl, respectively). Flow cytometry analysis was used to measure cell-surface expression on PD-1 and intracellular expression of KLHL22. n = 7 patients, *P < 0.05, paired Student’s t test.
To answer how 5-FU affects KLHL22, we performed a luciferase assay and identified a small region (R3: from −637 bp to transcription start site) of KLHL22 promoter that was sufficient to drive KLHL22 transcription and its transcriptional activity was significantly decreased when treated with 5-FU (SI Appendix, Fig. S6). These results indicate that 5-FU can regulate KLHL22 expression by inhibiting its transcription.
Importantly, in the tumor-infiltrating T cells from CRC patients who were treated with chemotherapy, including 5-FU, PD-1 is highly expressed; in contrast, KLHL22 is expressed at low levels compared to those in tumor-infiltrating T cells from patients who did not receive chemotherapy (Fig. 7G). This suggests that 5-FU treatment increases PD-1 expression in tumor-infiltrating T cells by reducing KLHL22 expression.
To directly demonstrate the effect of 5-FU on the KLHL22–PD-1 axis in human T cells, PBMCs from CRC patients were extracted and treated with 5-FU. Treated CD8+ T cells showed reduced KLHL22 expression and increased PD-1 expression compared to untreated cells from the same individual patient (Fig. 7H).
Taken together, these results show that treatment with 5-FU may lead to a decrease in KLHL22 and an increase in PD-1 expression, which suggests that increased PD-1 expression is responsible for the limited efficacy of 5-FU.
Discussion
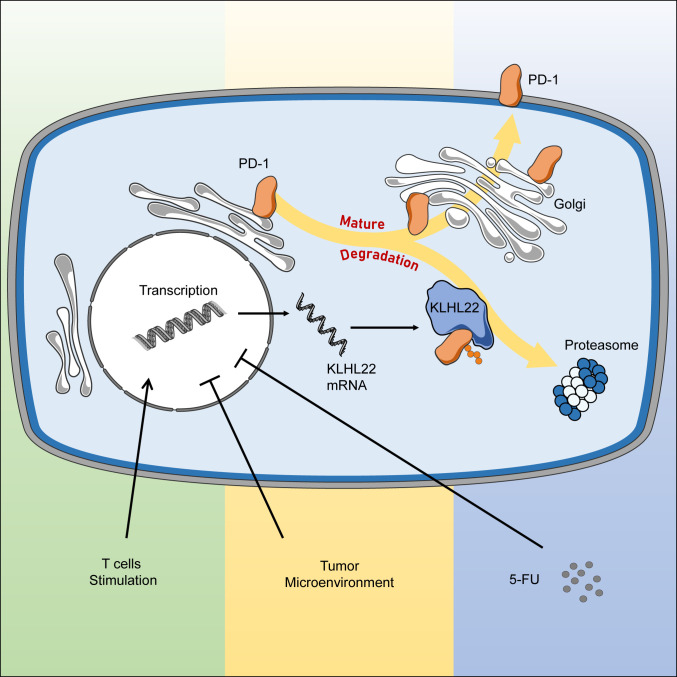
In this study, we performed multiple affinity-coupled MS analyses to identify the regulator of PD-1 and verified that KLHL22 is a major PD-1–interacting protein that binds with high affinity. KLHL22 recognizes incompletely glycosylated PD-1 and mediates its degradation by the proteasome pathway before PD-1 is transported to the cell surface. Upon T cell activation, KLHL22 is up-regulated, which consequently controls the amount of PD-1 on the cell surface and maintains its expression in an appropriate range, preventing excessive suppression of T cells. Under conditions of KLHL22 deficiency or deregulation, such as factors from the tumor microenvironment and treatment with 5-FU, excessive PD-1 accumulation could inhibit T cell activity (Fig. 8).
Fig. 8.
Working model. The biogenesis of PD-1 contains multiple steps of transportation and modification along the ER–Golgi–plasma membrane trafficking axis. KLHL22 is a major interacting protein of PD-1 and recognizes incompletely glycosylated PD-1, subsequently ubiquitinating and degrading PD-1 before it is transported to the cell surface. T cell activation upon TCR stimulation simultaneously promotes PD-1 and KLHL22 expression. KLHL22 degrades incompletely glycosylated PD-1 and maintains PD-1 homeostasis, preventing excessive suppression of T cells. KLHL22 deficiency, as well as deregulation of KLHL22 in response to the tumor microenvironment or 5-FU treatment, leads to excessive accumulation of PD-1 on the T cell surface and the repression of the antitumor immunity activity of T cells.
PD-1 is a key checkpoint inhibitory receptor that alters the function of T cells (2). PD-1 is absent in resting T cells but is expressed on activated T cells through the TCR and other cytokine receptors (4). Up-regulation of PD-1 is a natural consequence of T cell activation and is necessary for termination of the immune response (46). However, excessive PD-1 expression excessively inhibits T cell activity. Thus, it is important to maintain PD-1 expression within an appropriate range. Transcription of PD-1 is tightly regulated in several ways, and both the activation and repression of PD-1 transcription are under the control of T cell activation (4). Compared with the transcriptional regulation of PDCD1, the mechanism by which PD-1 protein is stabilized and/or degraded largely remains unclear. It has been reported that FBXO38 specifically degrades PD-1 on the cell surface via internalization and subsequent Lys48-linked polyubiquitination (19). FBXO38-mediated PD-1 ubiquitination occurs 2 d after T cell activation, which means that PD-1 has already partially or completely played its role in the immune checkpoint by that point. However, it is unknown how to prevent excessive accumulation of PD-1 before it activates. Here, we report that when T cells are activated, KLHL22 expression is also increased and that KLHL22 could mediate PD-1 degradation before it reaches the cell surface, thus preventing PD-1 overexpression-induced immune suppression. Besides KLHL22 and FBXO38 are responsible for the regulation of PD-1 at different stages, they also do have the potential to play unique roles in different cells, which may require further study.
KLHL22 is a substrate-specific adaptor protein of BCR E3 protein ligase. The Cullin-RING E3 ligases (CRLs), which contain a Cullin scaffold protein and a RING-finger protein, comprise the largest family of E3 ubiquitin ligases (47). Within the Cul3-based ligase complex, a BTB-domain–containing protein and, in most instances, a KLHL protein, are required for recognizing specific substrates (48). Recently, the Cullin3/KLHL22 complex has been proven to participate in diverse cellular mechanisms, including mitotic division and tumorigenesis, suggesting the significance of the Cullin3/KLHL22-mediated ubiquitin-proteasome pathway in regulating vital protein levels and cell functions (32, 39). However, the function of the Cullin3/KLHL22 complex in T cells has not yet been investigated. In this study, we demonstrate that PD-1 specifically interacts with KLHL22 and that Cullin3/KLHL22 regulates PD-1 protein levels during its maturation by targeting incompletely glycosylated PD-1, which mediates its ubiquitination and subsequent degradation by the proteasome. KLHL22 KO mice show excessive PD-1 accumulation and a faster tumor progression. Anti–PD-1 therapy in KLHL22 KO and WT mice suggests that the difference in the tumor progression rate between WT and KO mice is mainly determined by the difference in PD-1 expression. But whether excessive accumulation of PD-1 is the only factor that makes a faster tumor progression of KLHL22 KO mice needs future work, as we cannot exclude that the anti–PD-1 therapy works independently of KLHL22.
Plasma membrane proteins play vital roles in cellular biological functions. The biogenesis of membrane proteins involves multiple steps of transportation and modification along the ER–Golgi–plasma membrane trafficking axis (49). As one of the most dominant posttranslational modifications of plasma membrane proteins, glycosylation primarily begins in the ER and is completed in the Golgi apparatus, resulting in a gradual increase in the molecular weight of the target protein (50). During this maturation process, membrane proteins can be targeted to degradation at different steps (35, 36). Our discovery reveals an intrinsic mechanism targeting PD-1 for degradation before it becomes fully mature and functional, which could rapidly regulate the expression level of PD-1 on the cell surface and help maintain T cell homeostasis.
Tumor-infiltrating T cells have much higher PD-1 expression, which is one of the reasons for the decline of tumor-infiltrating T cells (45). KLHL22 expression is down-regulated in tumor-infiltrating T cells, suggesting that KLHL22-regulated PD-1 expression is uncontrolled in tumor-infiltrating T cells. FBXO38 is also down-regulated in tumor-infiltrating T cells (19). These results indicated that disordered protein regulation is an important contributor to the high expression of PD-1 in tumor-infiltrating T cells. Both tumors and chronic inflammation can induce high PD-1 expression on T cells (51); thus, we explored the role of KLHL22 in the tumor microenvironment. Future work will focus on the regulation of KLHL22 and its roles in chronic inflammation.
Immunotherapy is one of the most promising treatment options for cancer (1). However, many patients still fail to respond, and some cancers are refractory to these therapies (21). To overcome these difficulties, combining traditional therapies with immunotherapy has been proposed and is highly anticipated (23). Nonetheless, the mechanism of how the combination therapy elicits a superior effect compared to traditional therapy alone remains largely unclear, which is an obstacle to developing new treatment strategies or improving existing ones. 5-FU is the first-line treatment for CRC and other types of cancer, including breast cancer, anal cancer, esophageal cancer, stomach cancer, pancreatic cancer, and so on (52). There are several ongoing clinical trials involving PD-1 monoclonal antibodies combined with 5-FU, and although some have preliminarily achieved better results, there is still much room for improvement (24). 5-FU treatment causes an increase in the levels of tumor-associated antigens, which induces a tumor-specific immune response (26), but this mechanism is insufficient to explain the difference in the effects of the combination therapy between the various chemotherapeutic drugs and tumor types. We provide evidence for a new model in which 5-FU down-regulates KLHL22 at the transcriptional level, which leads to PD-1 accumulation at the T cell surface; this might limit the therapeutic effect on tumors.
It is interesting to note that in CRC patients assessed in our study, all five cases that treated with 5-FU showed relatively lower KLHL22 protein levels and relatively higher PD-1 protein levels. Furthermore, individual differences in KLHL22 expression in patients might lead to differences in the efficacy of combination therapy. Based on this, KLHL22 expression in T cells may be a potential indicator of the efficacy of combination therapy. Although we confirmed that 5-FU affects KLHL22 at the transcriptional level, we still do not clearly know the mechanism, which will be explored in future studies.
In conclusion, our data reveal an important role of KLHL22 in maintaining the appropriate level of PD-1 expression and regulating tumor immunity. KLHL22 prevents excessive PD-1 accumulation on the cell surface by degrading PD-1 during its maturation. KLHL22-mediated degradation of PD-1 protein levels will be ineffective in response to either the tumor microenvironment or treatment with 5-FU, resulting in excessive PD-1 accumulation and reduced T cell activity. Overall, our study reveals KLHL22 plays a crucial role in maintaining PD-1 expression homeostasis and preventing excessive T cell suppression.
Materials and Methods
TAP/MS.
HEK293T cells stably expressing PD-1–SFB were lysed with NETN buffer on ice for 20 min. Cell lysates were centrifuged at 20,000 × g for 15 min, and the supernatants were incubated with Streptavidin Sepharose beads (GE Healthcare Lifesciences) for 2 h at 4 °C. The resin was washed three times with NETN buffer and eluted twice with elution buffer (2 mg/mL biotin, 20 mM Tris⋅HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, and 0.5% Nonidet P-40) for 6 h at 4 °C. The eluates were combined and then incubated with S protein agarose (Merck Millipore) for 4 h at 4 °C. The S protein agarose were washed three times with NETN buffer, and proteins bound to the S protein agarose were eluted with 40 μL of 1× SDS loading buffer, separated by SDS/PAGE, and then visualized by Coomassie blue staining. The indicated panels were cut and sent to the Analytical Instrumentation Center of Peking University for MS analysis.
Mice.
The Klhl22 KO mouse was created using a CRISPR/Cas9-mediated genome-editing system from Cyagen Biosciences; exon 4 was selected as the target site. After coinjection of the guide RNA and Cas9 mRNA into fertilized eggs of C57BL/6 mice, mutant lines with a 2,211 base deletion and 2 base insertion were generated. The founders were genotyped by PCR followed by DNA sequencing analysis. All Klhl22 KO mice used were obtained from heterozygous breeding. For animal experiments with Klhl22 KO mice, littermate controls with normal Klhl22 expression were used. Animals were randomly allocated to experimental groups, and all KLHL22 mice used in this study were male mice of matched age. The mice were maintained in specific pathogen-free facilities with a standard 12-h light/dark schedule and provided standard chow and water ad libitum at the Department of Laboratory Animal Science of Peking University. Animal experiments in this study were performed in accordance with the Guidelines of Peking University Animal Care and Use Committee.
Mouse Tumor Models.
Age- and sex-matched Klhl22 KO and WT mice were selected. Next, 1 × 106 B16F10 or MC38 tumor cells were subcutaneously injected into the flanks of the mice. Tumor growth was measured daily or every other day using a caliper, and the tumor volume was calculated using the following formula: Tumor volume = length × width2/2. The animals were killed when detectable tumor burden approximated 10% of total body weight.
For the anti–PD-1 therapy, tumor-bearing mice were inoculated intraperitoneally with anti–PD-1 (RMP1-14, 200 μg per mouse, dissolved in PBS) or PBS on days 8, 11, and 14.
To analyze the phenotype of tumor-infiltrating T cells, mice were killed on day 16 or 17 after tumor cell injection. Tumor tissues were cut into pieces, resuspended in 3 mL of tumor digestion buffer, and rotated at 37 °C for 1 h. Leukocytes were isolated by density-gradient centrifugation using Ficoll. Then, tumor-infiltrating leukocytes were stained using fluorescently labeled antibodies for different markers, such as CD4, CD8, PD-1, and CD44. For intracellular molecular measurement, cells were first stained with cell-surface protein antibody and then fixed, permeabilized, and stained with fluorescently labeled antibodies. To measure the effector function of T cells, cells were stimulated with phorbol myristate acetate (PMA; 50 ng/mL) and ionomycin (1 µM) for 6 h at 37 °C and then analyzed by intracellular staining for IFN-γ, TNF-α, and granzyme B.
Human Cancer and Blood Samples.
CRC tissues and blood samples from patients were obtained at Peking University People’s Hospital, Beijing, China. Tumor tissues and adjacent normal tissues were freshly isolated and digested. Healthy human PBMCs were donated by healthy laboratory members. Tumor-infiltrating leukocytes and PBMCs were enriched by density-gradient centrifugation and stained with anti-CD3, anti-CD8, anti-KLHL22 and anti–PD-1 antibodies. The study protocol was approved by the Clinical Research Ethics Committee of Peking University People’s Hospital and complied with all relevant ethical regulations. The entire experiment was carried out under ethical principles such as the Declaration of Helsinki and International Ethical Guidelines for Biomedical Research Involving Human Subjects.
Statistics Analysis.
Quantitative data are presented as the means ± SD. Prism 8 (GraphPad) and SPSS was used for the statistical analysis. P < 0.05 was considered statistically significant. For normalization, individual data points were normalized to the mean of the WT (or control) sample per experiment, and data from multiple experiments were then pooled. Information on the statistical tests used, precise P values, sample sizes, and n values are provided in each figure caption.
Supplementary Material
Acknowledgments
We thank Wenling Han (Peking University) for the gift of Jurkat cells stably expressing PD-1; Li Su and Centre of Medical and Health Analysis (Peking University) for technical support of flow cytometry; Chen Chen (University College London) for help with drawing; Liu Yang (Peking University) for help with statistical analysis; and all members in J.W.’s laboratory and Wei Wang’s laboratory for insightful discussion and technical assistance. This study was supported by grants from the National Key R&D Program of China (2017YFA0503900 and 2016YFC1302100), the National Natural Science Foundation of China (81672981, 81872282, 81972240, and 31872735), Beijing Municipal Natural Science Foundation (7182082), Clinical Medicine Plus X-Young Scholars Project of Peking University (PKU2018LCXQ01), and Special Projects for Strengthening Basic Research of Peking University (BMU2019JC006).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. N.L. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2004570117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Topalian S. L., et al. , Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 366, 2443–2454 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keir M. E., Butte M. J., Freeman G. J., Sharpe A. H., PD-1 and its ligands in tolerance and immunity. Annu. Rev. Immunol. 26, 677–704 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishimura H., et al. , Developmentally regulated expression of the PD-1 protein on the surface of double-negative (CD4-CD8-) thymocytes. Int. Immunol. 8, 773–780 (1996). [DOI] [PubMed] [Google Scholar]
- 4.Bally A. P. R., Austin J. W., Boss J. M., Genetic and epigenetic regulation of PD-1 expression. J. Immunol. 196, 2431–2437 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agata Y., et al. , Expression of the PD-1 antigen on the surface of stimulated mouse T and B lymphocytes. Int. Immunol. 8, 765–772 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Vibhakar R., Juan G., Traganos F., Darzynkiewicz Z., Finger L. R., Activation-induced expression of human programmed death-1 gene in T-lymphocytes. Exp. Cell Res. 232, 25–28 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Oestreich K. J., Yoon H., Ahmed R., Boss J. M., NFATc1 regulates PD-1 expression upon T cell activation. J. Immunol. 181, 4832–4839 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao G., Deng A., Liu H., Ge G., Liu X., Activator protein 1 suppresses antitumor T-cell function via the induction of programmed death 1. Proc. Natl. Acad. Sci. U.S.A. 109, 15419–15424 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kao C., et al. , Transcription factor T-bet represses expression of the inhibitory receptor PD-1 and sustains virus-specific CD8+ T cell responses during chronic infection. Nat. Immunol. 12, 663–671 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Staron M. M., et al. , The transcription factor FoxO1 sustains expression of the inhibitory receptor PD-1 and survival of antiviral CD8(+) T cells during chronic infection. Immunity 41, 802–814 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathieu M., Cotta-Grand N., Daudelin J.-F., Thébault P., Labrecque N., Notch signaling regulates PD-1 expression during CD8(+) T-cell activation. Immunol. Cell Biol. 91, 82–88 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Lu P., et al. , Blimp-1 represses CD8 T cell expression of PD-1 using a feed-forward transcriptional circuit during acute viral infection. J. Exp. Med. 211, 515–527 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor A., et al. , Glycogen synthase kinase 3 inactivation drives T-bet-mediated downregulation of co-receptor PD-1 to enhance CD8(+) Cytolytic T cell responses. Immunity 44, 274–286 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Youngblood B., et al. , Chronic virus infection enforces demethylation of the locus that encodes PD-1 in antigen-specific CD8(+) T cells. Immunity 35, 400–412 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McPherson R. C., et al. , Epigenetic modification of the PD-1 (Pdcd1) promoter in effector CD4(+) T cells tolerized by peptide immunotherapy. eLife 3, e03416 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu J.-M., Li C.-W., Lai Y.-J., Hung M.-C., Posttranslational modifications of PD-L1 and their applications in cancer therapy. Cancer Res. 78, 6349–6353 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hershko A., Ciechanover A., The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Zhang J., et al. , Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature 553, 91–95 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng X., et al. , FBXO38 mediates PD-1 ubiquitination and regulates anti-tumour immunity of T cells. Nature 564, 130–135 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Yarchoan M., Hopkins A., Jaffee E. M., Tumor mutational burden and response rate to PD-1 inhibition. N. Engl. J. Med. 377, 2500–2501 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma P., Hu-Lieskovan S., Wargo J. A., Ribas A., Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168, 707–723 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng L., et al. , Irradiation and anti-PD-L1 treatment synergistically promote antitumor immunity in mice. J. Clin. Invest. 124, 687–695 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahoney K. M., Rennert P. D., Freeman G. J., Combination cancer immunotherapy and new immunomodulatory targets. Nat. Rev. Drug Discov. 14, 561–584 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Shahda S., et al. , A phase II study of pembrolizumab in combination with mFOLFOX6 for patients with advanced colorectal cancer. J. Clin. Oncol. 35, 3541 (2017). [Google Scholar]
- 25.Hao Y.-B., Yi S.-Y., Ruan J., Zhao L., Nan K.-J., New insights into metronomic chemotherapy-induced immunoregulation. Cancer Lett. 354, 220–226 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Chen G., Emens L. A., Chemoimmunotherapy: Reengineering tumor immunity. Cancer Immunol. Immunother. 62, 203–216 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tintelnot J., Stein A., Immunotherapy in colorectal cancer: Available clinical evidence, challenges and novel approaches. World J. Gastroenterol. 25, 3920–3928 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bai Y., et al. , C1QBP promotes homologous recombination by stabilizing MRE11 and controlling the assembly and activation of MRE11/RAD50/NBS1 complex. Mol. Cell 75, 1299–1314.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Chemnitz J. M., Parry R. V., Nichols K. E., June C. H., Riley J. L., SHP-1 and SHP-2 associate with immunoreceptor tyrosine-based switch motif of programmed death 1 upon primary human T cell stimulation, but only receptor ligation prevents T cell activation. J. Immunol. 173, 945–954 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Yokosuka T., et al. , Programmed cell death 1 forms negative costimulatory microclusters that directly inhibit T cell receptor signaling by recruiting phosphatase SHP2. J. Exp. Med. 209, 1201–1217 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hui E., et al. , T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition. Science 355, 1428–1433 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beck J., et al. , Ubiquitylation-dependent localization of PLK1 in mitosis. Nat. Cell Biol. 15, 430–439 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng N., Shabek N., Ubiquitin ligases: Structure, function, and regulation. Annu. Rev. Biochem. 86, 129–157 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Soucy T. A., et al. , An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Avci D., Lemberg M. K., Clipping or extracting: Two ways to membrane protein degradation. Trends Cell Biol. 25, 611–622 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Schmidt O., et al. , Endosome and Golgi-associated degradation (EGAD) of membrane proteins regulates sphingolipid metabolism. EMBO J. 38, e101433 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klausner R. D., Donaldson J. G., Lippincott-Schwartz J., Brefeldin A., Brefeldin A: Insights into the control of membrane traffic and organelle structure. J. Cell Biol. 116, 1071–1080 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee H.-H., et al. , Removal of N-linked glycosylation enhances PD-L1 detection and predicts anti-PD-1/PD-L1 therapeutic efficacy. Cancer Cell 36, 168–178.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen J., et al. , KLHL22 activates amino-acid-dependent mTORC1 signalling to promote tumorigenesis and ageing. Nature 557, 585–589 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Pickart C. M., Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Radivojac P., et al. , Identification, analysis, and prediction of protein ubiquitination sites. Proteins 78, 365–380 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zou W., Wolchok J. D., Chen L., PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci. Transl. Med. 8, 328rv324 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wartewig T., Ruland J., PD-1 tumor suppressor signaling in T cell lymphomas. Trends Immunol. 40, 403–414 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Sharma P., Allison J. P., The future of immune checkpoint therapy. Science 348, 56–61 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Thompson R. H., et al. , PD-1 is expressed by tumor-infiltrating immune cells and is associated with poor outcome for patients with renal cell carcinoma. Clin. Cancer Res. 13, 1757–1761 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Salmaninejad A., et al. , PD-1/PD-L1 pathway: Basic biology and role in cancer immunotherapy. J. Cell. Physiol. 234, 16824–16837 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Petroski M. D., Deshaies R. J., Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Pintard L., Willems A., Peter M., Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. EMBO J. 23, 1681–1687 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guna A., Hegde R. S., Transmembrane domain recognition during membrane protein biogenesis and quality control. Curr. Biol. 28, R498–R511 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Xu C., Ng D. T., Glycosylation-directed quality control of protein folding. Nat. Rev. Mol. Cell Biol. 16, 742–752 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Wherry E. J., Kurachi M., Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 15, 486–499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McQuade R. M., Stojanovska V., Bornstein J. C., Nurgali K., Colorectal cancer chemotherapy: The evolution of treatment and new approaches. Curr. Med. Chem. 24, 1537–1557 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.