Significance
HPV+ HNSCC is a growing problem in the United States. We show that HPV16 E6/E7 render HNSCC sensitive to WEE1 inhibition through a CDK1-FOXM1 circuit that drives premature mitosis and requires E6 activities beyond p53 inactivation. Primary HPV+ HNSCCs exhibit high FOXM1 activity, which may underlie their sensitivity to WEE1 inhibition. These data provide a mechanistic rationale for WEE1i incorporation into HPV+ HNSCC treatment.
Keywords: HPV16, WEE1, AZD1775, head and neck cancer, FOXM1
Abstract
Head and neck squamous cell carcinoma (HNSCC) associated with high-risk human papilloma virus (HPV) infection is a growing clinical problem. The WEE1 kinase inhibitor AZD1775 (WEE1i) overrides cell cycle checkpoints and is being studied in HNSCC regimens. We show that the HPV16 E6/E7 oncoproteins sensitize HNSCC cells to single-agent WEE1i treatment through activation of a FOXM1-CDK1 circuit that drives mitotic gene expression and DNA damage. An isogenic cell system indicated that E6 largely accounts for these phenotypes in ways that extend beyond p53 inactivation. A targeted genomic analysis implicated FOXM1 signaling downstream of E6/E7 expression and analyses of primary tumors and The Cancer Genome Atlas (TCGA) data revealed an activated FOXM1-directed promitotic transcriptional signature in HPV+ versus HPV- HNSCCs. Finally, we demonstrate the causality of FOXM1 in driving WEE1i sensitivity. These data suggest that elevated basal FOXM1 activity predisposes HPV+ HNSCC to WEE1i-induced toxicity and provide mechanistic insights into WEE1i and HPV+ HNSCC therapies.
Cyclin-dependent kinases (CDKs) are frequently deregulated in cancers and drive tumorigenesis (1). CDKs and their regulators are also important targets for cancer therapy (2). In addition to orchestrating cell cycle transitions, CDKs regulate cellular checkpoints that sense and respond to genotoxic stresses. CDK1 mediates the G2/M checkpoint that prevents mitotic entry in the face of various insults, while CDK2 controls an S phase checkpoint activated by DNA damage and replication stress (3). While numerous mechanisms regulate cyclin-CDKs, reversible CDK inhibitory phosphorylation is particularly important during checkpoint regulation. WEE1 inhibits CDK1 and CDK2 by phosphorylating their tyrosine 15 (Y15) and threonine 14 (T14) residues, whereas these phosphorylations are removed by the CDC25 phosphatases (3). Checkpoint signaling pathways regulate WEE1/CDC25 activity at many levels, thereby linking CDK inhibitory phosphorylation with cellular responses to genotoxic damage (3, 4). WEE1 also plays important roles in processes in unperturbed cell cycles such as DNA replication and replication stress responses (3).
WEE1 has been intensely studied as a therapeutic target for cancer treatment in preclinical and clinical studies, many of which have used the pharmacologic WEE1 inhibitor, AZD1775 (3, 5). Because WEE1 inhibition compromises both the G2/M and S phase checkpoints, AZD1775 (termed WEE1i, hereafter) has been used as a chemosensitizer, primarily to force cells through cell cycle checkpoints. For example, WEE1i causes cancer cells treated with genotoxic chemotherapy to enter mitosis inappropriately; this is exacerbated by p53 mutations and may lead to consequences such as mitotic catastrophe and apoptosis (5–8). WEE1i also disrupts the S phase checkpoint, and augments DNA damage and causes outcomes such as replication catastrophe when combined with agents that induce replication stress and/or DNA damage (9, 10). WEE1i is also being actively studied in early-stage clinical trials that have primarily focused on advanced solid tumors, both as monotherapy and combined with chemotherapy (3, 5). These trials have clearly demonstrated that WEE1i can be safely administered and have provided early insights into possible efficacy.
Head and neck squamous cell carcinoma (HNSCC) is a major source of cancer mortality worldwide (11). There are two distinct etiologies to HNSCC (11). First, carcinogens such as tobacco and alcohol promote HNSCCs with high mutational burdens, which usually include p53 mutations. Second, high-risk HPVs promote tumorigenesis in the oropharynx, which involves two viral oncoproteins that target tumor suppressor pathways: E6, which inactivates the p53 tumor suppressor protein by instigating its degradation, and E7, which binds to and inactivates the Retinoblastoma protein (pRb) (12). Both types of HNSCC pathogenesis cause widespread cell cycle deregulation, including p53 inactivation (through either mutation or E6 expression). Concordant with the role of cell cycle deregulation in HNSCC pathogenesis, we and others identified WEE1 as a promising target in p53-mutated and HPV+ HNSCC. This sensitivity to WEE1i involves mechanisms in both G1-S and G2-M regulation (8, 13–16). We recently reported a phase I trial of AZD1775 in combination with cisplatin and docetaxel in a neoadjuvant setting against HNSCC (17). Although this study was not designed to ascertain efficacy, all four patients with HPV+ tumors exhibited responses.
Here, we describe a mechanism whereby E6 and E7 sensitize HNSCC cells to WEE1i treatment, in which CDK1 and its substrate FOXM1 mediate the inappropriate mitotic entry of cells with unresolved DNA damage and/or replication stress. Compared with HPV-negative HNSCC, HPV+ tumors are hypersensitive to premature mitosis and DNA damage induced by WEE1i, and the combination of E6 and E7 (hereafter E6/E7) renders HPV-negative HNSCC cells similarly sensitive to WEE1i. While prolonged WEE1i treatment paradoxically caused CDK inhibition in control cells, WEE1i treatment of E6/E7 cells caused sustained CDK1 hyperactivity that drove DNA damage. Gene expression profiling revealed overexpression of FOXM1-target genes in WEE1i-treated E6/E7 HNSCC cells. Moreover, WEE1i caused FOXM1 hyperactivation in HPV+ and E6/E7 cells, which drove premature mitosis and DNA damage. E6 accounted for much of the WEE1i sensitivity observed in HPV+ HNSCC, and this was not always phenocopied by p53 loss. E6-driven WEE1i sensitivity thus involves mechanisms that extend beyond p53 inactivation. Finally, HPV+ tumor samples exhibited FOXM1 pathway activation compared with HPV-negative tumors, and analyses of The Cancer Genome Atlas (TCGA) data revealed that a FOXM1-directed gene expression signature is highly enriched in HPV+ HNSCCs.
In sum, we describe a CDK1-FOXM1–dependent pathway that underlies WEE1i sensitivity in E6/E7 HNSCC cells and suggest that FOXM1-driven mitotic gene expression predisposes HPV+ HNSCCs to premature mitosis, cell cycle disruption, and DNA damage caused by WEE1 inhibition. This FOXM1/CDK1/WEE1 network may thus provide therapeutic approaches to HPV+ HNSCC.
Results
WEE1 Inhibition Causes Premature Mitosis in HPV+ HNSCC Cell Lines.
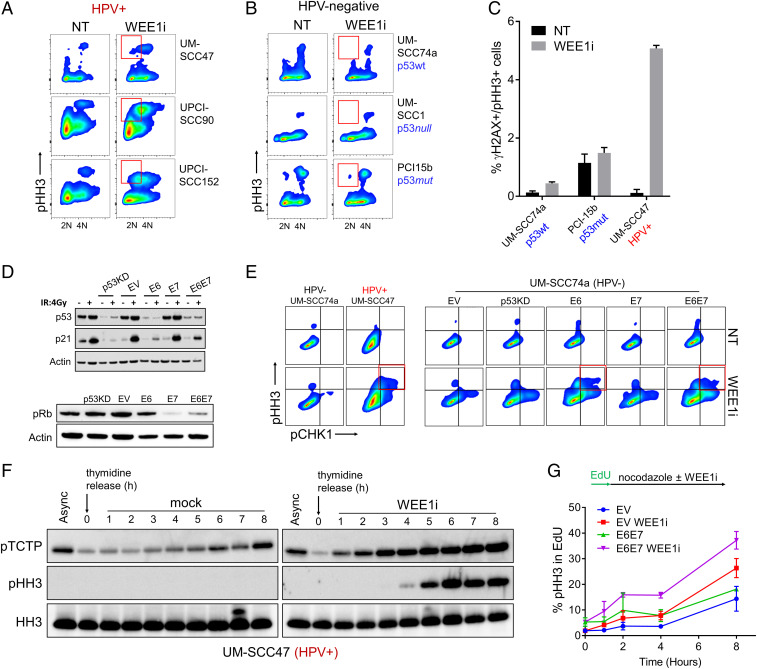
A hallmark of premature mitosis is cells that exhibit both <4N DNA content and histone H3 serine 10 phosphorylation (pHH3), which indicates cells that entered mitosis prior to completing DNA replication (7, 8). Treatment of p53-mutated cancer cells with WEE1i and chemotherapy (but not WEE1i alone) causes premature mitosis (5–7, 18). By using a HNSCC cell line in which we depleted p53, we found that p53 contributes to WEE1i sensitivity through both S phase and mitotic mechanisms and that combined WEE1i/chemotherapy treatment caused increased premature mitosis compared with WEE1i monotherapy (8). To further explore therapeutic opportunities for WEE1i in HNSCC and the role of HPV therein, we compared the effect of WEE1i monotherapy in three HPV+ HSNCC cell lines (UM-SCC47, UPCI-SCC90, and UPCI-SCC152; Fig. 1A) with that in three HPV-negative HNSCC cell lines (UM-SCC74a, UM-SCC1, and PCI-15I; Fig. 1B). WEE1i robustly induced premature mitosis in the HPV+ cell lines but not the HPV-negative cells. Of note, HPV+ HNSCC cell lines have higher WEE1 protein levels relative to HPV-negative HNSCC cells (SI Appendix, Fig. S1A). Combination chemotherapy/WEE1i treatment of p53-mutant cells also leads to cells positive for both pHH3 and the DNA-damage marker γH2AX, indicating cells forced into mitosis with DNA damage (7, 8, 19). WEE1i monotherapy caused increased numbers of γH2AX+/pHH3+ UM-SCC47 cells (HPV+), but not of HPV-negative HNSCC cell lines, regardless of their p53 status (Fig. 1C). While HPV infection itself may activate DNA damage repair factors (20), the basal amount of DNA damage (γH2AX) was not increased in UM-SCC47 cells (HPV+) compared to HPV-negative cells, but WEE1i treatment caused the largest relative increase in γH2AX in HPV+ UM-SCC47 cells (SI Appendix, Fig. S1 B–D). HPV+ HNSCC cells thus appear very sensitive to WEE1i monotherapy-induced premature mitosis, even when compared with p53-mutant (HPV-negative) HNSCC cells.
Fig. 1.
Single-agent WEE1i causes premature mitosis in HPV+ cell lines via E6 and E7. (A) FACS analysis of pHH3 in three HPV+ HNSCC cell lines ± WEE1i. Red gates in A and B show <4N DNA content. (B) FACS analysis of pHH3 in HPV-negative HNSCC cells (p53 status indicated) ± WEE1i as in A. (C) FACS analysis shows percentages of γH2AX+/pHH3+ cells in HPV+ (UM-SCC47) and HPV-negative (UM-SCC74a and PCI-15b) cell lines ± WEE1i. HPV and/or p53 status is indicated (n = 2). (D) UM-SCC74a cells expressing E6, E7, both E6 and E7 (E6/E7), shRNA against p53, or empty vector control (EV) were constructed. Immunoblots of p53 and p21 in response to ionizing radiation (4Gy) (Upper), or of pRb (Lower). (E) FACS analysis of pHH3 and pCHK1 (serine 345) in HPV-negative vs. HPV+ HNSCC cells ± WEE1i (Left), and in EV, p53KD, E6, E7 and E6/E7 UM-SCC74a cells ± WEE1i (Right). Red gate shows double positive populations. (F) Western blots of UM-SCC47 cells synchronized using 4 mM thymidine for 24 h then released into fresh media ± WEE1i. Async, asynchronous. (G) S-M progression assay. S phase cells were labeled by an EdU (10 μM) pulse for 20 min. Following EdU washout, cells were grown ± WEE1i in presence of nocodazole. pHH3 intensity was measured by FACS at the indicated time points. (n = 4).
HPV Oncogenes Confer Sensitivity of HNSCC Cells to WEE1i-Induced Premature Mitosis.
We next studied how E6 and E7 contribute to the sensitivity of HPV+ HNSCC cells to WEE1 inhibition. To circumvent possible confounding impacts of genetic heterogeneity among different cell lines (Fig. 1 A–C), we constructed an isogenic cell panel by expressing HPV16 E6, E7, or E6/E7 in UM-SCC74a cells (p53-WT, HPV-negative). We compared these cell lines with UM-SCC74a cells in which p53 was stably inactivated by short hairpin RNA (shRNA) (p53KD) (8). p53 shRNA and E6 expression antagonized p53 to a similar extent, as shown by reduced p53 abundance and the lack of p21Cip1 induction by ionizing radiation, compared with E7 or empty vector control cells (EV) (Fig. 1D). WEE1i triggered more premature mitosis in E6 and E6/E7 cells compared with p53KD or EV, as shown by the presence of <4N/pHH3+ cells (SI Appendix, Fig. S2A). We further characterized mitotic entry in WEE1i-treated E6/E7 cells in several ways. First, we examined pHH3+ cells for phosphorylation of the CHK1 protein on serine 345 (pCHK1), which indicates cells that entered mitosis with unresolved replication stress (4). pCHK1+/pHH3+ cells were found in WEE1i-treated HPV+ UM-SCC47 cells but not in HPV-negative UM-SCC74a cells (Fig. 1 E, Left). Similarly, pCHK1+/pHH3+ cells were found in WEE1i-treated E6 and E6/E7 cells, but not in E7, p53KD, or EV cells (Fig. 1 E, Right). The induction of pCHK1 was also confirmed by immunoblotting (SI Appendix, Fig. S1 E and F). E6 and E6/E7 cells also exhibited high levels of γH2AX+/pHH3+ mitotic cells after WEE1i treatment compared with p53KD or EV cells (SI Appendix, Fig. S2A). Polo-like-kinase-1 (PLK1) is normally activated in early G2 after DNA replication is completed (21). We identified cells with high PLK1 activity by phosphorylation of a PLK1 substrate (Translationally Controlled Tumor Protein-pTCTP) (22) and found increased γH2AX in pTCTP-positive E6 and E6/E7 cells treated with WEE1i (SI Appendix, Fig. S2B). E6/E7 thus renders HNSCC sensitive to WEE1i-induced premature mitosis, and this is largely attributable to E6.
Consistent with premature mitosis, WEE1i accelerated the rate at which UM-SCC47 cells (HPV+) synchronized in S phase progressed to M phase, as shown by the accumulation of pTCTP+ and pHH3+ cells after S phase release (Fig. 1F). Brief exposure (1 h) of asynchronous UM-SCC74a to WEE1i also up-regulated mitotic makers in an E6/E7-dependent manner (SI Appendix, Fig. S3A). To examine how E6/E7 expression impacts S-M progression in WEE1i-treated asynchronous cells, we labeled S phase cells with 5-ethynyl-20-deoxyuridine (EdU) and followed their entry into mitosis (pHH3+). WEE1i accelerated S-M progression in both EV and E6/E7 cells but to a greater extent in E6/E7 cells (Fig. 1G). Asynchronous E6 and E6/E7 cells also exhibited an increased mitotic fraction when compared with EV cells as shown by DNA content (SI Appendix, Fig. S3B) and pHH3 staining (SI Appendix, Fig. S3C). Finally, WEE1i increased PLK1 activity (pTCTP) and induced PLK1 expression in E6 and E6/E7 cells (SI Appendix, Fig. S3 D and E).
While E6 and sh-p53 each inactivated p53 to the same extent (Fig. 1D), they led to somewhat different WEE1i-induced responses. For example, pCHK1+/pHH3+ cells (Fig. 1E) and γH2AX+/pHH3+ cells (SI Appendix, Fig. S2A) were more common among WEE1i-treated E6 and E6/E7 cells than among p53KD cells. We also used CRISPR/Cas9 to delete p53 in UM-SCC74a cells (SI Appendix, Fig. S3 F and G) and found that the p53-deleted cells did not significantly induce PLK1, whereas E6/E7 cells did, although they had similar trends (SI Appendix, Fig. S3 H and I). In light of the robust premature mitoses caused by WEE1i in HPV+ compared with p53 mutant HPV-negative cell lines (Fig. 1 A–C), the data suggest that E6-mediated WEE1i sensitivity may involve p53-independent activities in addition to its well-descibed p53-dependent mechanisms (see below and Discussion).
Previous studies found that WEE1i induced apoptosis in HPV+ cell lines through repression of antiapoptotic proteins (16) and, thus, we examined how E6/E7 affect proliferation and apoptosis after WEE1i treatment. We confirmed that HPV+ cell lines exhibited decreased proliferation after WEE1i treatment followed by drug washout compared with UM-SCC74a cells (HPV-negative, p53 WT) (SI Appendix, Fig. S4A). UM-SCC74a cells regained WEE1i sensitivity after E6, E7, and E6/E7 expression, and this sensitivity was greatest in E6/E7 cells (SI Appendix, Fig. S4B). WEE1i also inhibited three-dimensional spheroid growth in ultra-low attachment plates of E6/E7 cells more so than of EV cells, as evidenced by viability and apoptosis (SI Appendix, Fig. S4 C–E). Two additional apoptotic markers (cleaved PARP expression and sub-G1 DNA content) were induced by WEE1i treatment of E6/E7 cells and suppressed by caspase inhibition, further indicating that E6/E7 sensitizes HNSCC cells to WEE1i-mediated apoptosis (SI Appendix, Fig. S4F).
HPV Oncogenes Increase DNA Damage Caused by WEE1i Treatment of HNSCC Cells.
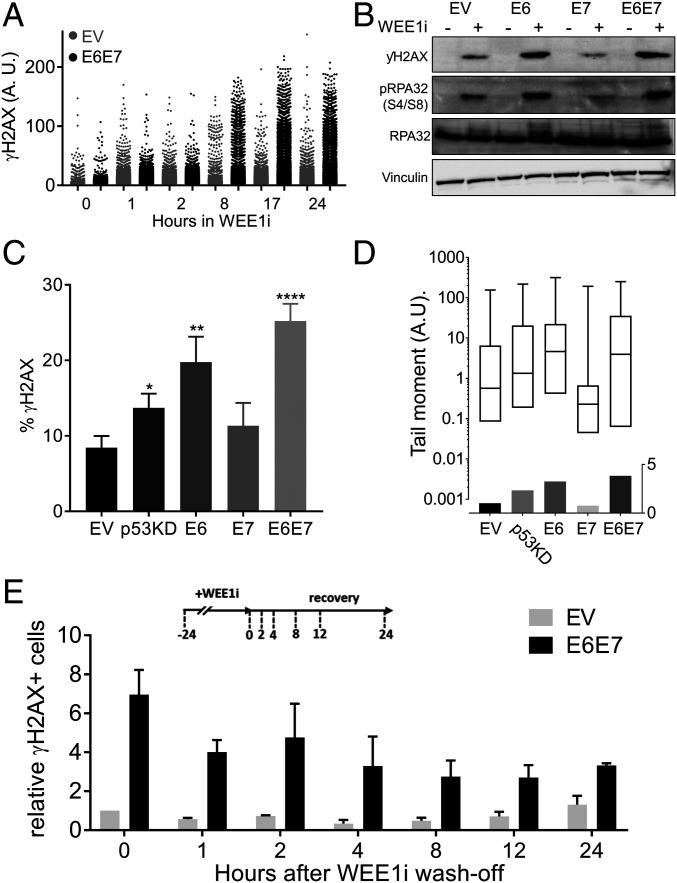
We next examined the DNA damage kinetics after WEE1i and found that E6/E7 cells accumulated γH2AX more rapidly, and to a higher extent, than did EV cells (Fig. 2A). WEE1i-induced DNA damage was primarily mediated by E6 and was highest in E6/E7 cells (Fig. 2 B and C). In addition to γH2AX, we examined two other markers of DNA breaks: RPA32 hyperphosphorylation (serine 4/serine 8) and DNA fragmentation in alkaline comet assays. RPA32 hyperphosphorylation marks double-strand breaks and was elevated in WEE1i-treated E6 and E6/E7 cells (Fig. 2B). Alkaline comet assays also revealed increased DNA breaks in WEE1i-treated E6, p53KD, and E6/E7 cells, and these were significantly elevated in E6/E7 cells compared with p53KD cells (Fig. 2D). γH2AX-positive cells also persisted longer in E6/E7 cells after WEE1i removal relative to EV cells (Fig. 2E).
Fig. 2.
HPV16 E6/E7 sensitize UM-SCC74a cells to WEE1i-induced DNA damage. (A) QIBC analysis of γH2AX in EV and E6/E7 UM-SCC74a cells treated with WEE1i as indicated. (B) Western blots of γH2AX and pRPA32 (serine 4/serine 8) in EV, E6, E7, and E6/E7 cells ± WEE1i. (C) Percentages of γH2AX in WEE1i-treated EV, p53KD, E6, E7, and E6/E7 cells, measured by FACS (n > 5) *P < 0.05, **P < 0.01, and ****P < 0.00001. (D). Alkaline comet assays in WEE1i-treated EV, p53KD, E6, E7, and E6/E7 cells shown as tail moment (Upper) or fold increase above control cells (Lower). (E) QIBC analysis of pan-nuclear γH2AX in EV vs. E6/E7 UM-SCC74a cells after release from WEE1i as indicated, (n = 2).
E6/E7 Regulate the Dynamics of CDK Activity following WEE1 Inhibition.
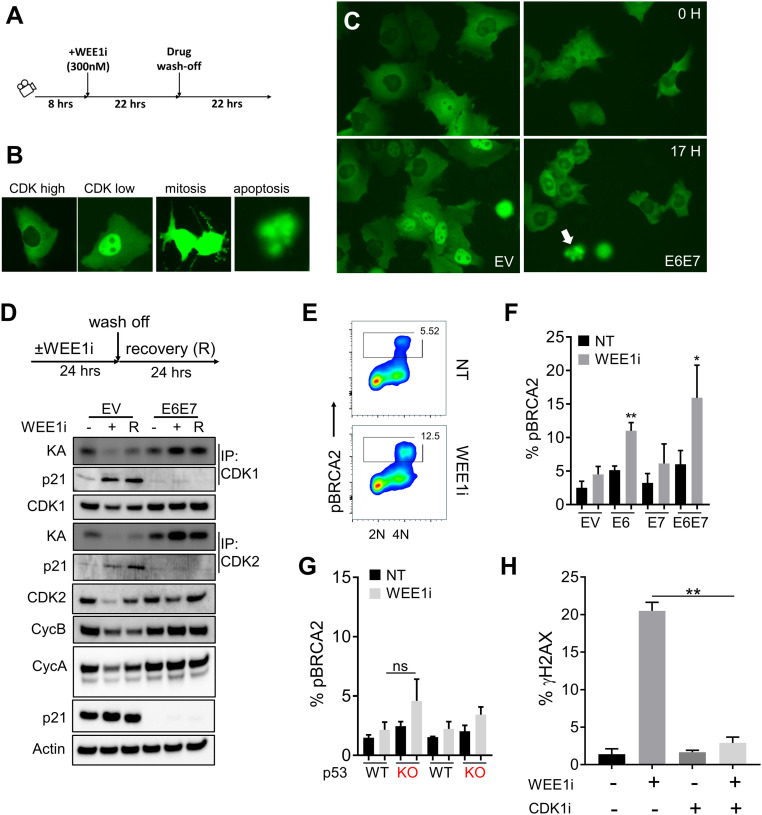
We used a CDK1/2 biosensor (cytoplasmic localization = high CDK activity; nuclear localization = low CDK activity) to examine how E6/E7 affects CDK activity in vivo after WEE1i treatment and recovery (Fig. 3 A and B) (22, 23). As expected, WEE1i caused an initial increase in CDK activity in EV cells that lasted up to 8 h (Movie S1). Surprisingly, this transient surge of CDK activity was followed by prolonged CDK inhibition throughout the remainder of WEE1i treatment and recovery (Movie S1 and Fig. 3C). Thus, rather than simply functioning as a CDK activator, WEE1i paradoxically suppressed CDK activity after prolonged treatment. In contrast, E6/E7 cells did not exhibit this biphasic response but instead exhibited sustained CDK hyperactivity throughout the WEE1i treatment and recovery periods (Movie S2 and Fig. 3C). These observations were confirmed with CDK-associated kinase assays. CDK1/2 activity decreased during and after prolonged WEE1i treatment (24 h) in EV cells and was associated with increased binding of the p21 CDK inhibitor to both CDKs (Fig. 3D). In contrast, E6/E7 cells exhibited CDK1/2 hyperactivity throughout the WEE1i treatment and washout period (Fig. 3D). As expected, E6/E7 expression eliminated p21 induction, resulting in sustained CDK hyperactivity during and after WEE1i treatment (Fig. 3D). Prolonged WEE1i treatment of EV cells thus triggers a p53/p21-dependent response that suppresses CDK activity, and this response is abrogated by E6-mediated p53 degradation.
Fig. 3.
Aberrant CDK1 activation by WEE1i in E6 and E6/E7 cells is genotoxic. (A) Experimental design. DHB-mVenus CDK1/2 reporter in EV or E6/E7 cells treated as indicated was monitored by live cell imaging. (B) Examples of CDK high, CDK low, mitotic, and apoptotic cells are shown. (C) Examples of EV and E6/E7 cells expressing CDK1/2 reporter after exposure to WEE1i as indicated. Pairs of images of each cell type represent the same field taken 17 h apart. Note high nuclear signal (low CDK1/2) in EV cells at 17 h of WEE1i vs. low nuclear signal (high CDK1/2) in E6/E7 cells. White arrow indicates apoptotic cell morphology. (D) CDK1/2 kinase activity (KA) was measured by in vitro kinase assays of immunoprecipitated (IP) cyclin/CDK complexes as indicated. EV and E6/E7 cells were treated ± WEE1i and released as indicated. Protein extracts were isolated and analyzed for protein abundance and kinase activity. (E) FACS analysis of CDK1-mediated pBRCA2 (serine 3291) in WEE1i-treated E6/E7 (predominantly G2/M) cells. (F) Percentages of pBRCA2+ cells analyzed by FACS in EV, E6, E7, and E6/E7 cells ± WEE1i (n = 2). (G) Percentages of pBRCA2+ cells in p53 +/+ and p53 −/− UM-SCC74a cells ± WEE1i; ns, not signficant. (H) Percentages of γH2AX analyzed by FACS in UM-SCC74a E6/E7 cells treated ± WEE1i and/or CDK1i as indicated for 3 h (n = 3). *P < 0.05, **P < 0.01.
CDK1 activation by WEE1i was also demonstrated by BRCA2 phosphorylation on serine 3291 (pBRCA2) (24), which was increased in WEE1i-treated E6/E7 and E6 cells (Fig. 3 E and F). Compared with E6 cells, pBRCA2 levels were not significantly increased in WEE1i-treated p53−/− cells (Fig. 3G). Finally, pharmacologic CDK1 inhibition abrogated WEE1i-induced γH2AX accumulation in E6/E7 cells (Fig. 3H). These data support a model in which sustained CDK1 hyperactivity in E6/E7 cells after WEE1i treatment drives DNA damage. The lack of CDK1 hyperactivity in p53−/− cells again highlights the different responses to WEE1i of E6-expressing cells compared to cells with exclusive p53 inactivation and reinforces the causal role of CDK1 hyperactivity in DNA damage.
FOXM1 Activation by WEE1i Drives Premature Mitosis in E6/E7 Cells.
To identify the mechanism(s) driving premature mitosis in WEE1i-treated E6/E7 cells, we profiled the expression using an 84-gene array in mock and WEE1i-treated E6/E7 and control (hTERT) oral keratinocytes (SI Appendix, Fig. S5A). A subset of 25 genes was highly expressed in WEE1i-treated E6/E7 cells (SI Appendix, Fig. S5A and Table S1), and transcription factor (TF) motif analysis using Enrichr (25) revealed that binding sites for FOXM1 and E2F were the most enriched motifs among these genes (P values 5.19e-09 and 2.99e-07 respectively, SI Appendix, Fig. S5B and Table S2). RT-qPCR confirmed E6/E7-specific induction of CCNB1, a canonical FOXM1 target gene, upon WEE1i treatment in UM-SCC74a cells (SI Appendix, Fig. S5C), and similar findings were seen with PLK1, another FOXM1 target gene (SI Appendix, Fig. S2E). In contrast, CCNB1 and PLK1 were not induced by WEE1i in p53-deleted UM-SCC74a cells (SI Appendix, Figs. S5D and S3I). Unlike FOXM1, two E2F target genes (CCNA2 and CCNE1) were insensitive to WEE1i treatment in E6/E7 UM-SCC74a cells (SI Appendix, Fig. S5C). These analyses suggested that WEE1i treatment of E6/E7 cells increases FOXM1 activity. Of note, CCNE1 expression in WEE1i-treated E6/E7 HNSCC cells did not correspond to the changes seen in E6/E7 keratinocytes (SI Appendix, Fig. S5A).
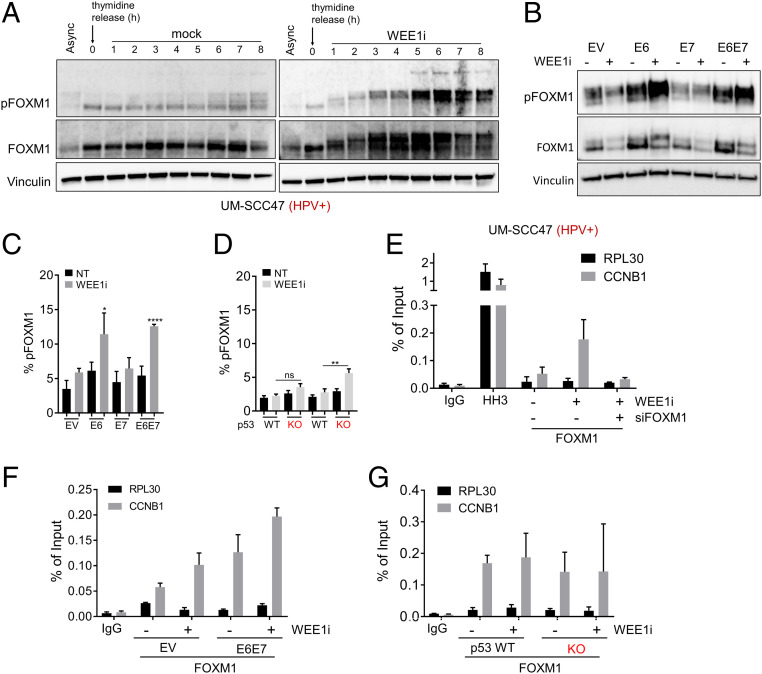
FOXM1 is a master transcriptional regulator of mitotic genes (21, 26, 27), and CDK1 directly phosphorylates FOXM1, which increases its activity by recruiting FOXM1 to promoters of its target genes (28). CDK1 is thus both a FOXM1 target gene and activator, leading to a mutually reinforcing CDK1-FOXM1 circuit. We hypothesized that CDK1-driven FOXM1 hyperactivity contributes to WEE1i-induced premature mitosis in HPV+ and E6/E7 cells. We first assessed FOXM1 activity following WEE1i treatment in synchronized HPV+ UM-SCC47 cells (Fig. 4A) and found dramatic FOXM1 hyperphosphorylation, as indicated by an anti-phospho-FOXM1 antibody that detects the activating CDK1-mediated phosphorylation (threonine 600, pFOXM1) and by the mobility shift of the FOXM1 protein (29). WEE1i treatment did not increase pFOXM1 in HPV-negative cells, while E6 expression recapitulated the WEE1i-dependent pFOXM1 induction in HPV+ HNSCC, as shown by both fluorescence-activated cell sorting (FACS) and immunoblotting assays of asynchronous cells (Fig. 4 B and C). In contrast, WEE1i-mediated induction of pFOXM1 was rather modest in p53−/− cells (Fig. 4D). We also determined that WEE1i treatment causes increased FOXM1 occupancy at the CCNB1 promoter in HPV+ UM-SCC47 cells (Fig. 4E). Accordingly, more FOXM1 occupancy at the CCNB1 promoter was also found in WEE1i-treated E6/E7 cells than in EV cells (Fig. 4F) but not in in p53−/− cells compared to their respective controls (Fig. 4G). FOXM1 and CDK1 were thus closely correlated in the isogenic cell panel; in both cases, their activities were higher in E6 cells than in E7 or p53 KO cells and highest in E6/E7 cells.
Fig. 4.
Hyperactivation of FOXM1 mitotic switch by WEE1i in HPV+ cells via E6/E7. (A) Western blots of UM-SCC47 cell lysates. Cells were synchronized in thymidine for 24 h then released into fresh media ± WEE1i as indicated. (B) Western blots of asynchronous EV, E6, E7, and E6/E7 UM-SCC74a cell lysates ± WEE1i. (C) FACS analysis of FOXM1-phosphothreonine 600 (pFOXM1) in EV, E6, E7, and E6/E7 ± WEE1i. (D) FACS analysis of pFOXM1 in p53−/− vs. p53+/+ clones ± WEE1i. (E) ChIP-qPCR of FOXM1 at CCNB1 promoter in WEE1i-treated UM-SCC47 cells. Enrichment of FOXM1 is absent in FOXM1-depleted cells. No enrichment at RPL30 promoter (negative control). (F) ChIP-qPCR of FOXM1 at CCNB1 promoter in EV vs. E6/E7 UM-SCC74a E6/E7 cells ± WEE1i. Data are shown as fold increase relative to untreated EV cells. (G) ChIP-qPCR of FOXM1 at CCNB1 promoter in p53+/+ and p53 −/− UM-SCC74a cells. Data are shown as fold increase relative to untreated p53+/+ cells. *P < 0.05, **P < 0.01, and ****P < 0.00001.
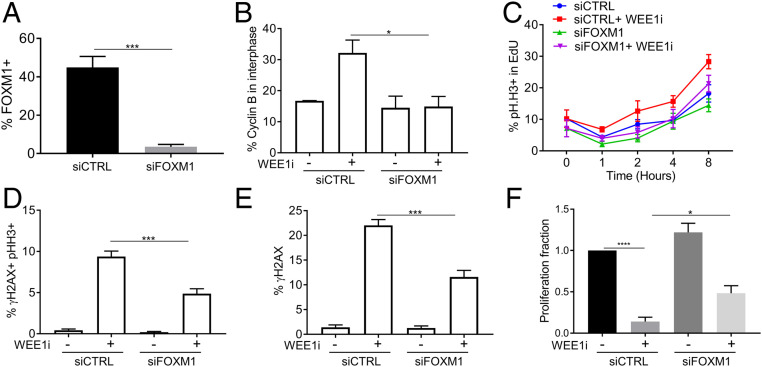
Previous studies found that ATR prevents untimely cyclin B expression in S phase and prevents premature mitosis caused by premature activation of CDK1-FOXM1 circuitry (29). We thus determined if FOXM1 drives induction of cyclin B upon WEE1 inhibition in E6/E7 cells and found that small interfering RNA (siRNA)-mediated FOXM1 silencing (Fig. 5A) prevented WEE1i-induced premature cyclin B accumulation in interphase cells (Fig. 5B and SI Appendix, Fig. S6A). FOXM1 depletion did not affect WEE1 abundance (SI Appendix, Fig. S6B) or asynchronous cell cycle kinetics (SI Appendix, Fig. S6C). FOXM1 silencing reduced the S-M phase acceleration in E6/E7 cells caused by WEE1i treatment (Fig. 5C) and also reduced WEE1i-induced premature mitoses (γH2AX/pHH3+ cells, Fig. 5D) and total DNA damage (γH2AX+ cells, Fig. 5E). siFOXM1 rescued the proliferative defect caused by WEE1i to almost the same extent as its impact on DNA damage (Fig. 5F). Because this was a long-term proliferation assay, whereas FOXM1 knockdown was transiently mediated by siRNA, this experiment likely underestimates the FOXM1 dependancy of proliferative arrest. These data support the idea that FOXM1-driven cyclin B expression in E6E7 cells plays a crucial role in their WEE1i sensitivty.
Fig. 5.
FOXM1 mediates premature mitosis upon WEE1 inhibition in E6/E7 cells. UM-SCC74a-E6/E7 cells were transfected with nontargeting siRNA or siRNA against FOXM1 for 40 h. (A) FACS analysis of total FOXM1 ± FOXM1 siRNA. (B) FACS analysis of cyclin B in interphase E6/E7 cells. Examples of cyclin B interphase gating in SI Appendix, Fig. S6A. (C) S-M progression assays performed as in Fig. 1 using E6/E7 cells ± WEE1i ± FOXM1 depletion. FACS analysis of γH2AX and pHH3 (D) and γH2AX (E) ± WEE1i ± FOXM1 depletion. (F) Quantification of change in proliferation in WEE1i-treated cells relative to mock-treated E6/E7 cells ± WEE1i ± FOXM1 depletion. *P < 0.05, ***P < 0.001, and ****P < 0.00001.
FOXM1 Activity Is Up-Regulated in HPV+ HNSCC.
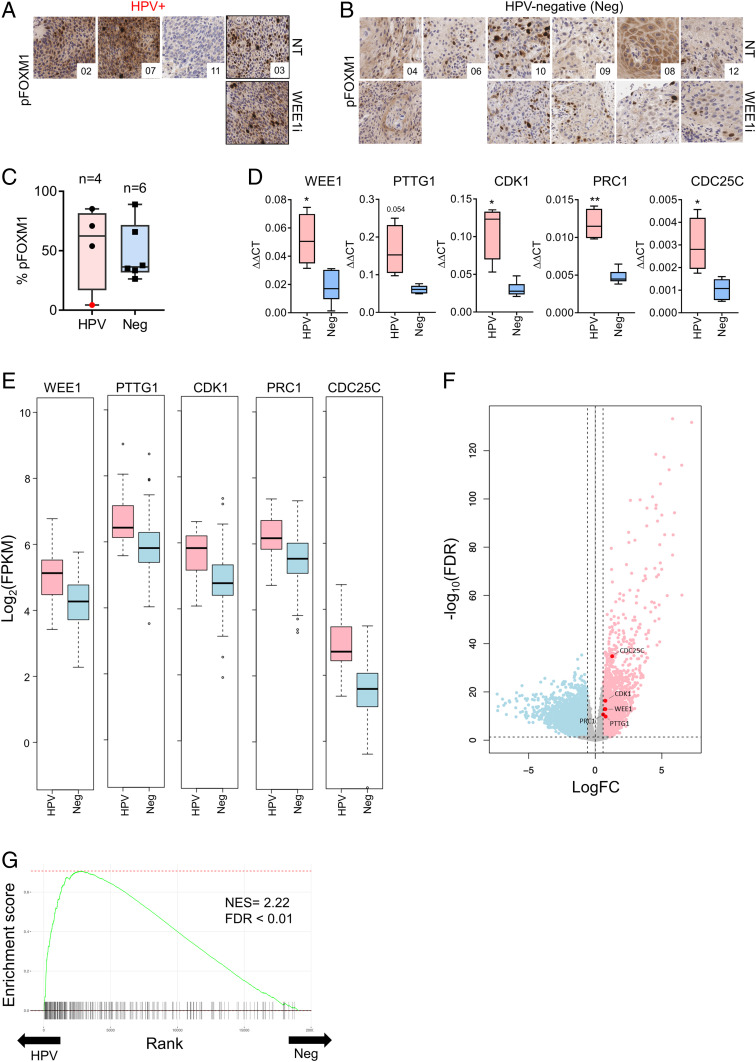
To assess the FOXM1 pathway in primary HNSCCs, we examined pFOXM1 in serial biopsies obtained from patients with HNSCC who were treated with WEE1i and chemotherapy in a phase I study, four of whom had HPV+ tumors (10). Posttreatment biopsies were obtained 8 h after neoadjuvant WEE1i, although only a single HPV+ post-WEE1i biopsy was available for this study. Three of four HPV+ tumors had high nuclear pFOXM1 staining pre-WEE1i treatment (Fig. 6 A and C), and this was sustained post-WEE1i treatment in the single evaluable HPV+ patient (Fig. 6A). In contrast, pFOXM1 levels were lower in HPV-negative tumor biopsies before and after treatment (Fig. 6 B and C). Of note, the HPV+ tumor sample that lacked pFOXM1 staining also carries somatic mutations in the PIK3CA and AKT genes (10), which are required upstream regulators of FOXM1 expression (30). We found that FOXM1-target genes were overexpressed in E6/E7 keratinocytes (SI Appendix, Table S2) in HPV+ vs. HPV-negative pretreatment tumor biopsies (CDC25C, PRC1, CDK1, PTTG1, and WEE1) (Fig. 6D). While limited by the small number of patient samples, these data support the idea that FOXM1 activity is high in HPV+ HNSCC and agree with the finding that E6 expression up-regulates FOXM1 in oropharyngeal cancer (24, 25).
Fig. 6.
Aberrant FOXM1 activation in HPV+ vs. HPV-negative HNSCCs. (A) pFOXM1 immunohistochemical (IHC) staining in pre-WEE1i and post-WEE1i treatment tumor biopsies from WEE1i phase I clinical trial of HPV+ patients (40×). (B) Staining as in A for HPV-negative (Neg) patients at 40×. (C) Quantification of % positive pFOXM1 nuclear areas in pretreatment biopsies from A and B. Patient 11 (with PIK3CA and AKT mutations) shown in red. (D) Gene expression analysis of representative FOXM1 target genes using a custom RT2 Profiler qPCR array of tumor biopsies from the phase I trial (17). (E) Analysis of FOXM1 target genes expression in HPV+ vs. HPV-negative HNSC tumors from TCGA. HNSC TCGA RNA-seq data are represented as log2 of fragments per kilobase of transcript per million mapped reads from RNA-seq analysis. (F) A volcano plot showing up-regulated (red; n = 2,037) and down-regulated (blue; n = 2,218) genes in HPV+ HNSCC tumors relative to HPV-negative tumors (FDR < 0.05 and 1.5× fold-change) in the HNSC TCGA cohort. (G) Fast gene set enrichment analysis (fgsea) showing the enrichment of FOXM1-target genes among HPV+ HNSCC transcriptomes of the HNSC TCGA cohort. The FOXM1 gene signature (FISCHER_FOXM1) had the second highest normalized enrichment score of all of the gene sets tested (NES = 2.22, FDR < 0.01) in the HNSC TCGA cohort. *P < 0.05, **P < 0.01. NES, normalized enrichment score; FDR, false discovery rate.
To extend these pilot data, we examined TCGA head and neck cancer cohort RNA-seq data from the Genomic Data Commons (31) and again found that the FOXM1-target genes studied above were overexpressed in HPV+ HNSCC vs. HPV-negative HNSCC (Fig. 6 F and G). Remarkably, unbiased gene set enrichment analysis (GSEA) using the fgsea algorithm revealed that a signature composed of FOXM1-target genes (FISCHER_FOXM1; 268 genes; ref. 32) was very highly enriched in HPV+ tumors compared with HPV-negative tumors (second most enriched gene set of 1,329 curated gene signatures tested, adjusted P value = 0.0059) (Fig. 6H and SI Appendix, Table S3). Moreover, the vast majority of the top 35 curated gene sets enriched in HPV+ tumors involved cell cycle processes, DNA replication/repair, and checkpoints. These data provide further evidence of increased FOXM1 pathway activation in HPV+ HNSCC and highlight the profound cell cycle deregulation that occurs in HPV+ HNSCC.
Discussion
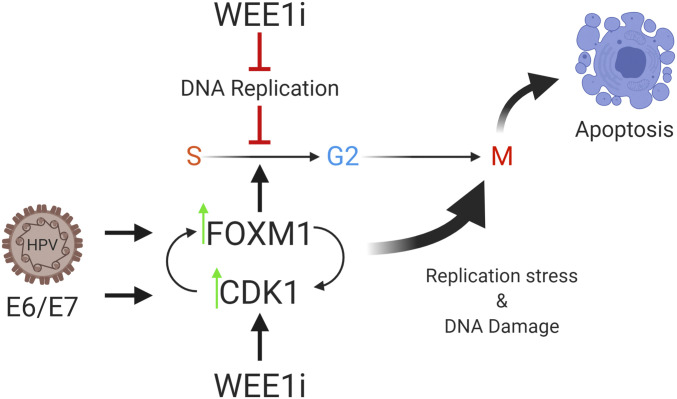
We describe a mechanism that confers WEE1i sensitivity to HPV+ HNSCC that involves: 1) premature mitosis, 2) DNA damage, 3) persistent CDK hyperactivity, and 4) elevated FOXM1 activity. Both CDK1 and FOXM1 play causal roles in driving DNA damage and premature mitosis in WEE1i-treated HPV+ HNSCC. Because CDK1 is both a FOXM1 target gene and activator, the data support a model in which activation of a mutually reinforcing FOXM1-CDK1 switch drives premature mitosis and DNA damage (Fig. 7). Previous studies have described premature mitosis resulting from WEE1i and chemotherapy combinations (but not WEE1i alone), and it was proposed that the role of genotoxic chemotherapy is to allow the accumulation of promitotic factors by delaying S phase (5–7). In contrast, we find that WEE1i is sufficient for robust induction of premature mitosis in HPV+ HNSCC cells and suggest that CDK1/FOXM1-driven promitotic gene expression allows WEE1i to promote premature mitosis without additional cell cycle perturbations. Analyses of tumor biopsies and public TCGA datasets revealed that the FOXM1 pathway is markedly activated in HPV+ HNSCC, which may predispose HNSCC to these effects caused by WEE1i monotherapy.
Fig. 7.
HPV-mediated FOXM1 aberrant activation sensitizes HPV+ cells to WEE1i-mediated premature mitosis. HPV E6/E7 expression up-regulates CDK1/FOXM1 activity, thereby disrupting normal cell cycle control, leading to WEE1 activation, which acts as a brake against mitotic abnormalities. Created with BioRender.
The relative sensitivity of HPV+ vs. HPV-negative HNSCC to WEE1i is not commonly seen with other antineoplastic agents. There is no difference in the cisplatin sensitivity of HPV+ vs. HPV-negative cells (33), and an unbiased screen employing >1,000 compounds to identify those with selective toxicity to HPV+ HNSCC found WEE1i as one of the top scoring hits, whereas radiomimetic drugs such as platinum and doxorubicin were not (34). Moreover, while ATR inhibition sensitized both HPV-negative and HPV+ HNSCC to cisplatin treatment, the antitumor effect of ATR monotherapy was limited in either group (35). Our findings (SI Appendix, Fig. S4 A–F) add to a growing body of literature on the specificity of the WEE1i antitumor potential for HPV+ HNSCC (13, 16, 34).
While naturally occurring HPV+ HNSCCs express both E6 and E7, we used an isogenic cell panel to evaluate the roles of E6 and E7 separately in WEE1i sensitivity. Both oncoproteins contribute to WEE1i sensitivity in HPV+ HNSCC, but E6 primarliy accounted for several WEE1i sensitivity phenotypes, including premature mitosis, DNA damage, and CDK1/FOXM1 hyperactivation. However, combined E6/E7 expression enabled the greatest consequences of WEE1i treatment with respect to premature mitosis with DNA damage (SI Appendix, Fig. S2), apoptosis (SI Appendix, Fig. S4), mitotic fraction (SI Appendix, Fig. S3 B and C), DNA damage (Fig. 2 C and D), and FOXM1 activity (SI Appendix, Figs. S3E and S5C and Fig. 4C). HPV+ tumors are defective in homologous recombination (HR) and Fanconi anemia (FA) repair pathways (36, 37), loss of which confer sensitivity to WEE1 inhibition (19). On its own, E7 promoted WEE1i-induced cell death through an unknown mechanism, although this was also less than was seen with E6/E7. Overall, the data indicate that E6 and E7 coordinately contribute to WEE1i sensitivity in HPV+ HNSCC.
E6 likely confers WEE1i sensitivity through p53-dependent and p53-independent functions. For example, WEE1i treatment of p53-WT HNSCC cells for longer than ∼8 h caused prolonged CDK inhibition that correlated with increased p21 binding to CDK1 and CDK2 (Fig. 3 C and D and Movie S1). E6 expression abrogated p21 expression and allowed persistent WEE1i-induced CDK1/2 activation, which presumably reflects E6-mediated p53 degradation (Fig. 3 C and D and Movie S2). The finding that prolonged WEE1i treatment in cells with intact p53 function caused CDK inhibition rather than CDK activation has important implications for clinical strategies employing WEEi and is reminiscent of findings in primary mouse embryonic fibroblasts treated with WEE1i (38). However, while p53-p21 loss is likely required for sustained CDK1 hyperactivity following WEE1i treatment, it is likely not sufficient, since CDK1 activity is not significantly increased in p53−/− cells, compared with E6 cells (Fig. 3 F and G; see below). Increased CDK1 activity in E6 cells compared with p53−/− cells may partly reflect increased CCNB1 expression (SI Appendix, Fig. S5C), which agrees with the reported E6-induced CDK1 activation independently of p53 function (39).
p53 inactivation is a recurring theme for HNSCC irrespective of the mode of p53 deficiency (i.e., mutational or HPV-inactivated). Indeed, previous studies from ourselves and others have shown that p53 loss sensitizes cancer cells to WEE1i through mechanisms that impact the S and G2/M checkpoints and cell cycle progression (8, 13–15). Here, we find that relative to E6/E7, p53 loss leads to limited premature mitosis (SI Appendix, Fig. S2), modestly increased mitotic fraction (SI Appendix, Fig. S3C), and DNA breaks (Fig. 2 C and D) in cells treated with WEE1i. However, p53 loss had less impact than E6 expression in HNSCC cells after WEE1i treatment in several respects, including: 1) reduced premature mitosis and mitotic fraction (Fig. 1E and SI Appendix, Figs. S2A and S3 B and C), 2) less CDK1 activation (Fig. 3 G and H), and 3) reduced FOXM1 activation (Fig. 4 D–F and SI Appendix, Figs. S3 H and I and S5D). These data suggest that in addition to its role in p53 degradation, E6 may have important p53-independent activities that contribute to WEE1i sensitivity in HPV+ HNSCC cells, although these mechanisms require further study. Because most HSNCCs are p53-deficient, both p53-dependent and p53-independent E6 functions may have clinically relevant roles in future HNSCC regimens.
FOXM1 activation by CDK1-directed and PLK1-directed phosphorylation initiates a promitotic transcriptional program (27, 28, 40). Depletion of FOXM1 prevents CDK1-driven premature mitosis and DNA damage caused by ATR inhibition in normal cells (29), demonstrating the critical role of tightly controlled FOXM1 activity. We similarly find that FOXM1 contributes to CDK1-driven premature mitosis in WEE1i-treated HPV+ HNSCC. Because WEE1 is itself a FOXM1 target and overexpressed in HPV+ HNSCCs relative to HPV-negative HNSCCs (41, 42), it may serve as a brake to guard against mitotic abnormalities. Our findings that FOXM1 activity and mitotic gene expression are elevated in HPV+ HNSCC further reinforce the importance of this pathway and cell cycle deregulation in cancers associated with HPV. HPV may up-regulate FOXM1 activity in many ways. For example, FOXM1 is a known target of p53-mediated repression (43), which is disrupted by E6. Both E6 and E7 can also promote FOXM1 activity via p53-independent mechanisms, such as by disrupting the repressive DREAM (dimerization partner, RB-like, E2F and multivulval class B) pathway at many levels (44), which antagonizes FOXM1 activity. Moreover, E6 expression directly up-regulates FOXM1 activity via a GRHL2/NKX2-1 axis (45, 46). Finally, both mitotic kinases that activate FOXM1 are themselves up-regulated by HPV: E6 increases CDK1 activity, whereas E7 promotes mitotic entry via aberrant PLK1 activation (39, 47, 48). The increased FOXM1 transcriptional program that we find in HPV+ HNSCC likely reflects these many ways in which HPV+ impacts FOXM1 activity.
Our results naturally prompt the question of where WEE1 inhibition might hold most clinical relevance in the evolving HNSCC therapeutic landscape. Clinical investigation in this disease is typically dichotomized into treatment approaches for 1) locally advanced disease wherein the intent of treatment is cure and a combination of surgery and/or radiation without systemic therapy is utilized, and 2) recurrent/metastatic disease, which is approached with palliative intent using systemic agents. The understanding that HPV+ HNSCC is a distinct pathologic and prognostic entity within HNSCC has led to efforts to study the HPV+ subset separately (or at least as a stratification factor) in clinical trials for both curative and palliative settings. Our findings support a potential role for WEE1 inhibitors in both treatment scenarios, in which agents of this unique mechanism of action are absent from the existing therapeutic armamentaria.
In patients with recurrent/metastatic HNSCC, regardless of HPV status, immune checkpoint inhibitors (ICIs) are approved as first-line treatment as a single agent (for a biomarker-enriched PDL1+ population) or in combination with chemotherapy (49). ICIs are also indicated as single agents after progression on a platinum chemotherapy given in the curative or recurrent/metastatic setting (50, 51). Yet even in the PDL1+ population, the reported overall response rate to first-line single agent immune checkpoint inhibitor is ∼19%, with a median overall survival of 12.3 mo (49). The therapeutic standard for patients who progress or do not benefit from ICIs is not established, and WEE1 inhibition represents a promising option for this group in need of improved oncologic outcomes. In these patients where maintenance of quality of life is of critical importance, an active, enterically administered, single agent would be attractive both from a standpoint of reduced toxicity and ease of administration. Additionally, preliminary studies suggest that WEE1 inhibitors may augment the efficacy of ICIs that could translate to improved response rates and overall survival (52, 53). A combination strategy of WEE1 and immune checkpoint inhibition is currently being explored in an early phase clinical trial (NCT02617277).
In the curative intent scenario, recently completed and ongoing clinical trials have focused on deescalation of the standard curative intent concurrent chemoradiation approach in HPV+ HNSCC. Two landmark clinical trials demonstrated the superiority of cisplatin-based concurrent chemoradiation compared to cetuximab-based radiation therapy, affirming this as a standard for locally advanced disease (54, 55). Despite this, the staggering acute and late toxicities of cisplatin-based chemoradiation are well recognized (56). There remains a critical need to identify better-tolerated systemic agents that may spare toxicity, potentially allowing reduced doses of radiation without compromising outcomes. It is in this scenario that WEE1 inhibition may play a key role in upfront curative therapy, perhaps as a radiosensitizer that could replace more toxic chemotherapy agents given concurrently with radiation. Ongoing clinical trials in locally advanced disease (NCT03028766, NCT02585973) are expected to shed light on the safety and efficacy of this agent used in combination with definitive and postoperative radiation.
In summary, our results provide mechanistic insights into the efficacy of WEE1 inhibition in HPV+ HNSCC and support the continued investigation of WEE1 inhibitors in this patient subset, perhaps with even broader applicability to the larger group of HPV-related malignancies.
Methods
Cell Lines, Vectors, RNAi, and Chemicals.
The HNSCC cell lines, UM-SCC47, UM-SCC74a, UM-SCC1, PCI-15B, UPCI-SCC90, and UPCI-SCC152 were cultured in Dulbecco’s Modified Eagle Media containing 10% fetal bovine serum, 1% l-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. Retroviruses for LXSN, E6, E7, and E6/E7 were produced as per standard protocol (37) and were transduced into UM-SCC74a cells. Following G418 selection (1 mg/mL), expression of HPV oncoproteins was functionally validated and early passage cells were cryopreserved. The OKF4-hTERT and OKF4-E6/E7 cell lines were cultured in Keratinocyte-SFM medium supplemented with pen-strep, glutamine, and nonessential amino acids (Thermo Fisher Scientific). Cells were used within 3 mo after thawing and were routinely tested for mycoplasma contamination prior to cryopreservation or upon thawing.
Stable knockdown of p53 in UM-SCC74a cells was described previously (8). CRISPR lentiviral vector plasmid pLenti-Cas9 is a derivative of pLenti-CRISPRv1, containing Cas9 and puromycin resistance gene. TP53 single targeting guide sgTP53_3 is a gift from William Hahn, Dana–Farber Cancer Institute, Boston, MA (Addgene plasmid no. 78164) (57). siRNAs against p21 (CDKN1A) were from Qiagen (FlexiTube GeneSolution GS1026), and FOXM1-targeting siRNA pool was from Dharmacon (FOXM1 SMARTpools ON-TARGETplus Dharmacon L-009762-00-0005). Nontargeting control siRNA was from Qiagen (AllStars Negative Control siRNA, 1027280). siRNA transfections were performed using Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher Scientific) according to manufacturer’s protocol.
WEE1i AZD1775 was provided by AstraZeneca through a collaborative agreement and was used at 300 nmol/L unless otherwise noted. RO-3306 (CDK1i) was used at 5 μmol/L, EdU at 10 μM, 5-bromo-2-deoxyuridine at 10 μM, and nocodazole at 40 ng/L, all of which were purchased from Sigma-Aldrich. Caspase inhibitor Q-VD-OPH (Selleckchem) was used at 25 nmol/L.
S-M Progression Assay.
To determine the fraction of cells transitioning from S phase into mitosis, we adapted the S-M QIBC protocol (29) to FACS. Briefly, cells were pulse labeled with 10 μM EdU for 20 min, washed, and grown in the presence of WEE1i and 40 ng/L nocodazole for up to 8 h. Cells were fixed at the indicated time points for FACS analysis according to the protocol described above, but with the modification of including a Click-iT step for EdU incorporation according to the manufacturer’s protocol (Thermo Fisher Scientific).
Live Cell Reporter for CDK2 Activity.
UM-SCC74a EV and E6/E7 cells were transduced with the CDK reporter mVenus-DNA helicase B (DHB) (amino acids 994–1087) (gift from Sabrina Spencer, University of Colorado, Boulder, CO) (58) and sorted on a FACSAria II flow cytometer (BD), and correct expression patterns were validated by live-cell microscopy. For time-lapse microscopy of CDK-reporter EV and E6/E7 cells, images were acquired every 12 min using a Nikon Ti Live microscope with 20× (N.A. 0.4) air objective at 37 °C and 5% CO2. Images were analyzed using ImageJ. Cells were seeded 24 h before imaging. WEE1i (300 nmol/L) was added after 22 h of image collection, media was carefully replaced with fresh media, and imaging continued for an additional 22 h.
Chromatin Immunoprecipitation Assays.
Chromatin immunoprecipitation (ChIP) was conducted using the SimpleChIP Enzymatic Chromatin IP Kit, Agarose Beads (Cell Signaling Technologies) according to the manufacturer’s protocol with 4 × 106 cells and 2 μL of FOXM1 antibody (Bethyl). Histone H3 and Rabbit IgG were used as positive and negative controls, respectively. To detect FOXM1 occupancy at the Cyclin B (CCNB) promoter, ChIP DNA was amplified by qPCR with forward and reverse primers specific to the CCNB promoter and analyzed using the 2-ΔΔCT method. Where indicated, FOXM1 was depleted by siRNA for a total of 40 h.
RT2 Profiler PCR Array.
Custom RT2 profiler PCR array (Qiagen) was performed as described previously (8) using RNA extracted from OKF4-hTERT and OKF4-E6/E7 cells treated for 17 h ± 300 nmol/L WEE1i. Data are shown as ΔΔCT.
Immunoblotting, immunofluorescence, RT-qPCR, proliferation, and flow cytometry assays were performed as per standard protocols (8) and are detailed in SI Appendix, Supplemental Experimental Procedures. All antibodies used are listed in detail in SI Appendix, Table S4. Oligonucleotides used in qPCR and ChIP are listed in SI Appendix, Table S5.
Statistical Analyses.
All statistical tests were conducted with GraphPad Prism version 7.0. The results are presented as the mean ± SD or the mean ± SEM of at least three experiments unless stated otherwise and were evaluated using an unpaired Student’s t test (two-tailed; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.00001).
Supplementary Material
Acknowledgments
We thank Julio Vazquez and David McDonald from Fred Hutch Scientific Imaging for their help with QIBC analyses. We also acknowledge support from the Fred Hutch Shared Resources Histopathology, Genomics & Bioinformatics, and Flow Cytometry Cores. This work was supported by NIH/National Cancer Institute Grants R01 CA215647 (to B.E.C. and J.M.S.) and R01 CA193808 (to B.E.C.), and Seattle Translational Tumor Research programmatic investment grant (to E.M., B.E.C., and D.A.G.). D.A.G. was also supported by R35 CA209979. B.E.C. holds the Rosput Reynolds Endowed Chair. This research was also supported by the Shared Resources of the Fred Hutch/University of Washington Cancer Consortium Grant P30 CA015704.
Footnotes
Competing interest statement: B.E.C. is the founder and equity holder of Coho Therapeutics, a startup company that is focused on protein degradation and is completely unrelated to the work presented in this paper.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2013921117/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information.
References
- 1.Malumbres M., Barbacid M., Cell cycle, CDKs and cancer: A changing paradigm. Nat. Rev. Cancer 9, 153–166 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Otto T., Sicinski P., Cell cycle proteins as promising targets in cancer therapy. Nat. Rev. Cancer 17, 93–115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elbæk C. R., Petrosius V., Sørensen C. S., WEE1 kinase limits CDK activities to safeguard DNA replication and mitotic entry. Mutat. Res. 819-820, 111694 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Sørensen C. S., Syljuåsen R. G., Safeguarding genome integrity: The checkpoint kinases ATR, CHK1 and WEE1 restrain CDK activity during normal DNA replication. Nucleic Acids Res. 40, 477–486 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geenen J. J. J., Schellens J. H. M., Molecular pathways: Targeting the protein kinase Wee1 in cancer. Clin. Cancer Res. 23, 4540–4544 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Hirai H., et al. , Small-molecule inhibition of Wee1 kinase by MK-1775 selectively sensitizes p53-deficient tumor cells to DNA-damaging agents. Mol. Cancer Ther. 8, 2992–3000 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Aarts M., et al. , Forced mitotic entry of S-phase cells as a therapeutic strategy induced by inhibition of WEE1. Cancer Discov. 2, 524–539 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Diab A., et al. , Multiple defects sensitize p53-deficient head and neck cancer cells to the WEE1 kinase inhibition. Mol. Cancer Res. 17, 1115–1128 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beck H., et al. , Cyclin-dependent kinase suppression by WEE1 kinase protects the genome through control of replication initiation and nucleotide consumption. Mol. Cell. Biol. 32, 4226–4236 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toledo L. I., et al. , ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 155, 1088–1103 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Chow L. Q. M., Head and neck cancer. N. Engl. J. Med. 382, 60–72 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Galloway D. A., Laimins L. A., Human papillomaviruses: Shared and distinct pathways for pathogenesis. Curr. Opin. Virol. 14, 87–92 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moser R., et al. , Functional kinomics identifies candidate therapeutic targets in head and neck cancer. Clin. Cancer Res. 20, 4274–4288 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kao M., Green C., Sidorova J., Méndez E., Strategies for targeted therapy in head and neck squamous cell carcinoma using WEE1 inhibitor AZD1775. JAMA Otolaryngol. Head Neck Surg. 143, 631–633 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osman A. A., et al. , Wee-1 kinase inhibition overcomes cisplatin resistance associated with high-risk TP53 mutations in head and neck cancer through mitotic arrest followed by senescence. Mol. Cancer Ther. 14, 608–619 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka N., et al. , Wee-1 kinase inhibition sensitizes high-risk HPV+ HNSCC to apoptosis accompanied by downregulation of MCl-1 and XIAP antiapoptotic proteins. Clin. Cancer Res. 21, 4831–4844 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Méndez E., et al. , A phase I clinical trial of AZD1775 in combination with neoadjuvant weekly docetaxel and cisplatin before definitive therapy in head and neck squamous cell carcinoma. Clin. Cancer Res. 24, 2740–2748 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirai H., et al. , MK-1775, a small molecule Wee1 inhibitor, enhances anti-tumor efficacy of various DNA-damaging agents, including 5-fluorouracil. Cancer Biol. Ther. 9, 514–522 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Aarts M., et al. , Functional genetic screen identifies increased sensitivity to WEE1 inhibition in cells with defects in Fanconi anemia and HR pathways. Mol. Cancer Ther. 14, 865–876 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kono T., et al. , Activation of DNA damage repair factors in HPV positive oropharyngeal cancers. Virology 547, 27–34 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindqvist A., Rodríguez-Bravo V., Medema R. H., The decision to enter mitosis: Feedback and redundancy in the mitotic entry network. J. Cell Biol. 185, 193–202 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemmens B., et al. , DNA replication determines timing of mitosis by restricting CDK1 and PLK1 activation. Mol. Cell 71, 117–128.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hahn A. T., Jones J. T., Meyer T., Quantitative analysis of cell cycle phase durations and PC12 differentiation using fluorescent biosensors. Cell Cycle 8, 1044–1052 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krajewska M., et al. , Forced activation of Cdk1 via wee1 inhibition impairs homologous recombination. Oncogene 32, 3001–3008 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Chen E. Y., et al. , Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sadasivam S., Duan S., DeCaprio J. A., The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev. 26, 474–489 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laoukili J., et al. , FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat. Cell Biol. 7, 126–136 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Chen Y.-J., et al. , A conserved phosphorylation site within the forkhead domain of FoxM1B is required for its activation by cyclin-CDK1. J. Biol. Chem. 284, 30695–30707 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saldivar J. C., et al. , An intrinsic S/G2 checkpoint enforced by ATR. Science 361, 806–810 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penke L. R., et al. , FOXM1 is a critical driver of lung fibroblast activation and fibrogenesis. J. Clin. Invest. 128, 2389–2405 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grossman R. L., et al. , Toward a shared vision for cancer genomic data. N. Engl. J. Med. 375, 1109–1112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischer M., Grossmann P., Padi M., DeCaprio J. A., Integration of TP53, DREAM, MMB-FOXM1 and RB-E2F target gene analyses identifies cell cycle gene regulatory networks. Nucleic Acids Res. 44, 6070–6086 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Busch C. J., et al. , Similar cisplatin sensitivity of HPV-positive and -negative HNSCC cell lines. Oncotarget 7, 35832–35842 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghasemi F., et al. , High-throughput testing in head and neck squamous cell carcinoma identifies agents with preferential activity in human papillomavirus-positive or negative cell lines. Oncotarget 9, 26064–26071 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leonard B. C., et al. , ATR inhibition sensitizes HPV- and HPV+ head and neck squamous cell carcinoma to cisplatin. Oral Oncol. 95, 35–42 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wallace N. A., et al. , High-risk alphapapillomavirus oncogenes impair the homologous recombination pathway. J. Virol. 91, e01084-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khanal S., Galloway D. A., High-risk human papillomavirus oncogenes disrupt the Fanconi anemia DNA repair pathway by impairing localization and de-ubiquitination of FancD2. PLoS Pathog. 15, e1007442 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szmyd R., et al. , Premature activation of Cdk1 leads to mitotic events in S phase and embryonic lethality. Oncogene 38, 998–1018 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang W., et al. , Role of Cdk1 in the p53-independent abrogation of the postmitotic checkpoint by human papillomavirus E6. J. Virol. 89, 2553–2562 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu Z., et al. , Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nat. Cell Biol. 10, 1076–1082 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Slebos R. J. C., et al. , Gene expression differences associated with human papillomavirus status in head and neck squamous cell carcinoma. Clin. Cancer Res. 12, 701–709 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Lohavanichbutr P., et al. , Genomewide gene expression profiles of HPV-positive and HPV-negative oropharyngeal cancer: Potential implications for treatment choices. Arch. Otolaryngol. Head Neck Surg. 135, 180–188 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barsotti A. M., Prives C., Pro-proliferative FoxM1 is a target of p53-mediated repression. Oncogene 28, 4295–4305 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Engeland K., Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM. Cell Death Differ. 25, 114–132 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen P.-M., et al. , Up-regulation of FOXM1 by E6 oncoprotein through the MZF1/NKX2-1 axis is required for human papillomavirus-associated tumorigenesis. Neoplasia 16, 961–971 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen W., et al. , Human papillomavirus 16 E6 induces FoxM1B in oral keratinocytes through GRHL2. J. Dent. Res. 97, 795–802 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson D. A., et al. , The human papillomavirus-16 E6 oncoprotein decreases the vigilance of mitotic checkpoints. Oncogene 15, 3025–3035 (1997). [DOI] [PubMed] [Google Scholar]
- 48.Spardy N., et al. , Human papillomavirus 16 E7 oncoprotein attenuates DNA damage checkpoint control by increasing the proteolytic turnover of claspin. Cancer Res. 69, 7022–7029 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burtness B. et al.; KEYNOTE-048 Investigators , Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): A randomised, open-label, phase 3 study. Lancet 394, 1915–1928 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Cohen E. E. W. et al.; KEYNOTE-040 investigators , Pembrolizumab versus methotrexate, docetaxel, or cetuximab for recurrent or metastatic head-and-neck squamous cell carcinoma (KEYNOTE-040): A randomised, open-label, phase 3 study. Lancet 393, 156–167 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Ferris R. L., et al. , Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 375, 1856–1867 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun L., et al. , WEE1 kinase inhibition reverses G2/M cell cycle checkpoint activation to sensitize cancer cells to immunotherapy. OncoImmunology 7, e1488359 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Friedman J., et al. , Inhibition of WEE1 kinase and cell cycle checkpoint activation sensitizes head and neck cancers to natural killer cell therapies. J. Immunother. Cancer 6, 59 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gillison M. L., et al. , Radiotherapy plus cetuximab or cisplatin in human papillomavirus-positive oropharyngeal cancer (NRG oncology RTOG 1016): A randomised, multicentre, non-inferiority trial. Lancet 393, 40–50 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mehanna H. et al.; De-ESCALaTE HPV Trial Group , Radiotherapy plus cisplatin or cetuximab in low-risk human papillomavirus-positive oropharyngeal cancer (De-ESCALaTE HPV): An open-label randomised controlled phase 3 trial. Lancet 393, 51–60 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Machtay M., et al. , Factors associated with severe late toxicity after concurrent chemoradiation for locally advanced head and neck cancer: An RTOG analysis. J. Clin. Oncol. 26, 3582–3589 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hong A. L., et al. , Integrated genetic and pharmacologic interrogation of rare cancers. Nat. Commun. 7, 11987 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spencer S. L., et al. , The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell 155, 369–383 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.