Significance
Key members of the Bcl-2 family of proteins that control cell death possess a carboxy-terminal transmembrane segment that contributes to subcellular localization and modulates apoptotic functions. The role of Mcl-1 and specially Bok proteins remains elusive. We reveal that the coexpression of Mcl-1 and Bok TMDs produces an increase in the number of ER mitochondrial-associated membranes (MAMs). Furthermore, our findings provide mechanistic insight into the interaction between Mcl-1 and Bok transmembrane domains that can be envisioned as a target for therapeutic intervention.
Keywords: apoptosis, Bcl-2, Bok, Mcl-1, transmembrane
Abstract
The Bcl-2 protein family comprises both pro- and antiapoptotic members that control the permeabilization of the mitochondrial outer membrane, a crucial step in the modulation of apoptosis. Recent research has demonstrated that the carboxyl-terminal transmembrane domain (TMD) of some Bcl-2 protein family members can modulate apoptosis; however, the transmembrane interactome of the antiapoptotic protein Mcl-1 remains largely unexplored. Here, we demonstrate that the Mcl-1 TMD forms homooligomers in the mitochondrial membrane, competes with full-length Mcl-1 protein with regards to its antiapoptotic function, and induces cell death in a Bok-dependent manner. While the Bok TMD oligomers locate preferentially to the endoplasmic reticulum (ER), heterooligomerization between the TMDs of Mcl-1 and Bok predominantly takes place at the mitochondrial membrane. Strikingly, the coexpression of Mcl-1 and Bok TMDs produces an increase in ER mitochondrial-associated membranes, suggesting an active role of Mcl-1 in the induced mitochondrial targeting of Bok. Finally, the introduction of Mcl-1 TMD somatic mutations detected in cancer patients alters the TMD interaction pattern to provide the Mcl-1 protein with enhanced antiapoptotic activity, thereby highlighting the clinical relevance of Mcl-1 TMD interactions.
The proteins of the Bcl-2 family govern mitochondrial outer membrane permeabilization during apoptotic cell death (1, 2). According to their function and the number of Bcl-2 homology (BH) domains, Bcl-2 members can be classified into three groups: the antiapoptotic proteins (Bcl-2, Bcl-xL, Bcl-w, Mcl-1, A1, and Bcl-B); the proapoptotic BH3-only “sensor” proteins (Bad, Bid, Bim, BMF, Puma, Noxa, etc.); and the proapoptotic “executor” proteins (Bax, Bok, and Bak). Moreover, almost all members of the family have a carboxyl-terminal transmembrane domain (TMD). These proapoptotic and antiapoptotic proteins form a complex network of interactions, both in the cytosol and in the lipid membrane that finally determines cell fate (3). During apoptosis, most interactions occur at intracellular organelle membranes where the Bcl-2 TMDs have been traditionally described as mere membrane anchor domains; however, recent mechanistic studies have established that TMDs can fulfill many other functions. Said studies have provided evidence that Bcl-2 TMDs participate in the retrotranslocation of proapoptotic members from the mitochondria to the cytosol (4), in mitochondrial fusion and fission processes (5–8), and in metabolism modulation (9). Additionally, their homo- and heterointeractions fine tune apoptotic response (9, 10). However, the underlying network of transmembrane interactions and their functional implications regarding the modulation of apoptosis remain unclear.
Antiapoptotic Mcl-1 protein sequesters proapoptotic members (Bax, Bak, and Bok) and interacts with BH3-only proteins (Bid, Bim, Noxa, and Puma) (11–13). Mcl-1 overexpression leads to acquired resistance to antitumoral treatments (14–19). The association of Mcl-1 with tumorigenesis has fostered the development of novel Mcl-1 inhibitors (20) that have recently entered clinical trials (NCT02992483, NCT02979366, and NCT03672695).
The Mcl-1 TMD mediates the subcellular location of the protein (21). In fact, the Mcl-1 protein generally locates to the mitochondria, but TMD deletion leads to a cytosolic distribution. Interestingly, recent reports suggested a putative role for the Mcl-1 TMD as a mitochondrial priming tool (22) and in the modulation of the proapoptotic effector Bok, although the molecular mechanism involved remains unknown (13).
Among in silico approximations, molecular simulations (molecular dynamics [MD]) of TMD dimerization have recently become common due to certain methodological advancements; however, the interactions with the Bcl-2 TMDs have yet to be considered. All-atom (AA) simulations are now employed to probe the stability of preformed dimers and their effects on membrane structure (23). Furthermore, coarse-grained (CG) models can be used to predict dimer structures and dimerization energetics (24, 25).
The oligomerization studies performed in our new work demonstrate that the Mcl-1 TMD forms stable homooligomers at the mitochondrial membrane. The Mcl-1 TMD induces apoptosis and competes with the antiapoptotic function of the full-length (FL) protein. Apoptosis induced by the Mcl-1 TMD requires the presence of the Bok member. In agreement with these findings, the Mcl-1 TMD heterooligomerizes with the Bok TMD and this heterointeraction promotes a shift in the subcellular localization of the Bok TMD from the endoplasmic reticulum (ER) to the mitochondria. Furthermore, our results confirm that the coexpression of Mcl-1 and Bok TMDs produces an increase in the number of ER mitochondrial-associated membranes (MAMs). Together, these results provide evidence for a plausible Mcl-1 TMD-driven Bok TMD movement through the endomembrane compartments, which might account for the functional role of Bok in the mitochondrial membrane. Finally, the introduction of somatic mutations found in the Mcl-1 TMD of cancer patients alters the TMD interaction pattern and produces a protein with higher antiapoptotic activity.
Altogether, our results provide evidence that consolidates the role of Bcl-2 TMDs in the modulation of apoptosis; not only do they contribute to the selection of the pro- or antiapoptotic “partner interactor,” but they also direct the shuffling of Bcl-2 protein members among subcellular compartments. Understanding the functional relevance of Mcl-1 TMD interactions at the molecular level will provide areas of intervention for the development of innovative therapeutic strategies.
Results and Discussion
Mcl-1 TMD Oligomerizes at the Mitochondrial Membrane.
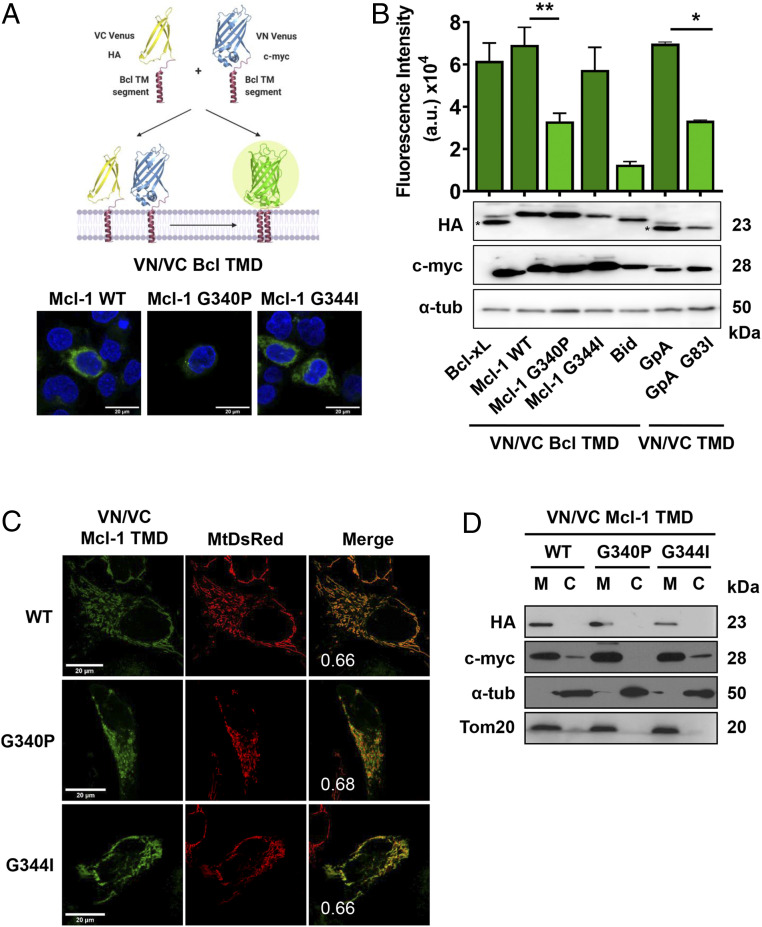
We first analyzed the self-association of Mcl-1 TMDs in nonapoptotic HCT116 human colorectal carcinoma cells by biomolecular fluorescence complementation (BiFC) assays (Fig. 1A). In these assays, the interaction of the Mcl-1 TMD fused to the N- and C-terminal fragments of the Venus fluorescent protein (VN and VC, respectively) reconstitutes the full-length protein. The appearance of green fluorescence at the same level previously described for Bcl-xL (10) suggests the existence of Mcl-1 TMD homointeractions (Fig. 1B). In these experiments, we employed the Glycophorin A (GpA) TMD as a well-known transmembrane dimer (26, 27), the dimer disruptive mutant GpA G83I (28), and the alpha 6 helix of Bid (Bid TMD) (29) as negative controls for oligomer formation. We observed equivalent protein expression levels for each construct (Fig. 1 B, Bottom). Proper membrane insertion of the control constructs was corroborated in maltose complementation assays using the ToxR system (30) (SI Appendix, Fig. S1).
Fig. 1.
Mcl-1 TMD homooligomerizes in the mitochondrial membrane. (A) Venus reconstitution depends on interactions established between Bcl TMDs. Self-association of the wild-type Mcl-1 TMD and single amino acid substitution variants measured by BiFC in HCT116 cells. Representative images of fluorescent signal in the confocal microscope are shown. Blue signal corresponds to Hoescht staining and green signal to Venus reconstitution. The Bcl-xL TMD and the GpA TMD homooligomerization served as positive controls and the GpA G83I and Bid TMD as negative controls. Asterisk (*) indicates predicted size. (B) Graphical representation of the Venus fluorescent quantification by spectrophotometer. Error bars represent the mean ± SEM, n = 6. Significant differences compared to the Mcl-1 WT TMD analyzed using Dunnett’s multiple comparison test (95% CI). *P < 0.05, **P < 0.01. Chimeric protein expression of VN (c-myc) and VC (HA) constructs compared in the Bottom; α-tubulin used as a loading control. Asterisks indicate the mobility expected for Bcl-xL and Bid constructs. (C) Confocal images of HeLa cells expressing MtDsRed marker and transfected with VN and VC Mcl-1 TMD constructs. Homooligomers formation was observed in the green channel. Colocalizations are shown in yellow with the PCC. (D) Subcellular fractionation of HCT116 cells transfected with VN (c-myc) and VC (HA) TMD constructs was controlled using Tom20 (mitochondrial fraction, M) and α-tubulin (cytosol, C).
Next, we studied the subcellular distribution of Mcl-1 TMD-driven homointeractions by confocal microscopy colocalization studies of the Venus protein with the Discosoma sp. red fluorescent protein (DsRed) fused to the mitochondrial signaling sequence of preornithine transcarbamylase (MtDsRed) (31) (Fig. 1C). Confocal microscopy images confirmed the mitochondrial localization of the Mcl-1 TMD with a Pearson’s correlation coefficient (PCC) of 0.66. Subcellular fractionation assays (Fig. 1D) and carbonate extraction experiments (SI Appendix, Fig. S2A) corroborated the presence and the insertion of the Mcl-1 TMD in the same fraction as the outer mitochondrial membrane protein Tom20.
BiFC assays require the identification of disruptive mutations or deletions to corroborate protein–protein interaction specificity (32). We searched for “hot spots” in the TMD sequence that could form the packing surface. In transmembrane protein segments, in contrast to soluble proteins, the dihedral angles of glycines fall into the alpha-helical region of the Ramachandran plot (33) and are frequently found in transmembrane helix–helix packing interfaces where they form part of the interaction domain (33–36). The analysis of the Mcl-1 TMD sequence revealed that Gly-340 and Gly-344 form part of a GxxxG motif that is frequently used by membrane proteins to establish oligomerization (G340VGAG344LAYLIR) (37–40) thereby, they could behave as hot spots in the mutational analysis. We designed two mutant versions of the Mcl-1 TMD protein, where glycine-340 was substituted by proline (Mcl-1 G340P TMD) and glycine-344 substituted by isoleucine (Mcl-1 G344I TMD). We found almost identical protein expression and mitochondrial association ability of both mutants when compared to the wild-type protein, as confirmed by subcellular fractionation and colocalization studies with MtDsRed (Fig. 1 C and D). Thus, the changes observed in the reconstitution of the fluorescent protein can be exclusively attributed to changes in the oligomerization ability of the mutants. Fluorescence quantification of the Mcl-1 G340P TMD mutant revealed a significant decrease in the signal that confirmed the loss of oligomerization and demonstrated the specificity of the Mcl-1 TMD packing interaction (Fig. 1B). The insertion capability of the Mcl-1 TMD mutants was also confirmed by carbonate extraction experiments (SI Appendix, Fig. S2A). The Mcl-1 G344I TMD did not affect oligomerization, indicating that this G344 is not a key player for maintaining the homooligomeric structure (see also molecular simulations below and SI Appendix, Fig. S3. Detection of Mcl-1 full-length homooligomers in BiFC assays that were disrupted in the Mcl-1 FL G340P mutant confirmed the functional relevance of this interaction (SI Appendix, Fig. S2B).
Altogether, these results confirm that, as for the Bcl-2 and Bcl-xL TMDs (10), the Mcl-1 TMD forms homooligomers in the mitochondrial membranes of nonapoptotic cells.
Mcl-1 TMD Induces Cell Death by Competition with Full-Length Mcl-1 Protein.
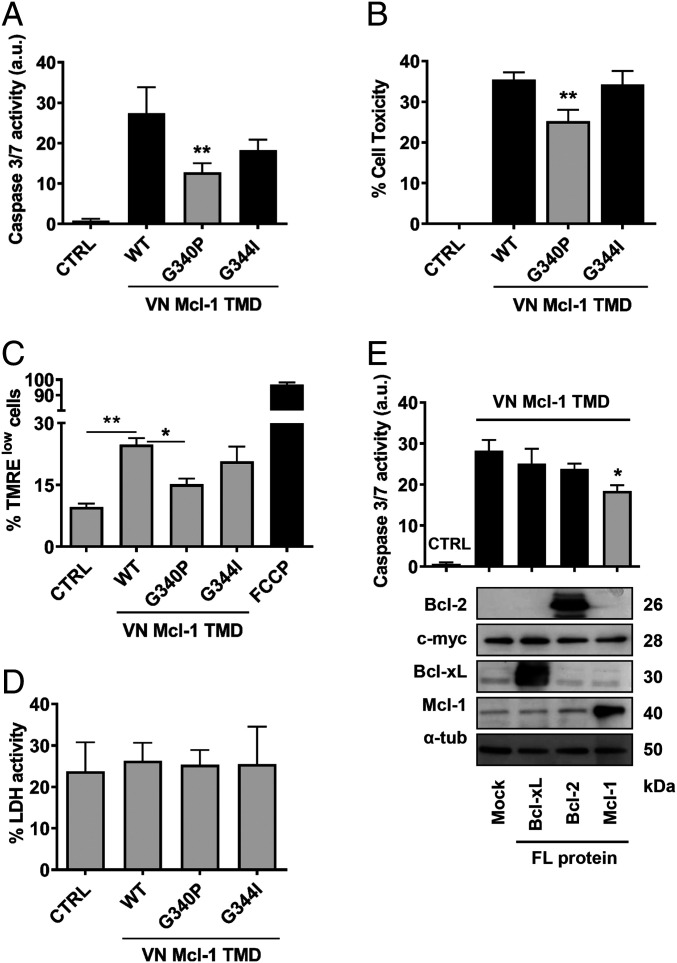
The challenge of working with membrane proteins has hindered their study and limited the number of resolved three-dimensional structures; however, accumulating evidence has established that oligomerization, mediated by interactions between transmembrane segments, can control the function of the whole protein (41, 42). Thus, after demonstrating that the Mcl-1 TMD forms oligomers, we asked whether these interactions played any role in the modulation of apoptosis. To this end, we measured caspase 3/7 activation and cell viability in HCT116 cells overexpressing the Mcl-1 TMD. Interestingly, those constructs that homooligomerized, such as the Mcl-1-WT TMD and the mutant Mcl-1 G344I TMD, induced caspase 3/7 activation and cell death. Furthermore, we found a significant reduction in the apoptosis-inducing ability of the nondimerizing mutant Mcl-1 G340P TMD (Fig. 2 A and B). We obtained comparable results in flow cytometry studies with the tetramethylrhodamine ethyl ester (TMRE) dye that accumulates in active polarized mitochondria. In these studies, the presence of the Mcl-1 TMD prompted the loss of mitochondrial polarization in 25% of cells (Fig. 2C). We observed similar percentages in the case of cells transfected with the Mcl-1 G344I TMD mutant, although this percentage decreased to around 15% in the presence of the Mcl-1 G340P TMD. We failed to observe necrotic behavior in any case, as demonstrated by the absence of lactate dehydrogenase (LDH) activity in cell culture supernatants (Fig. 2D). These results underscore the relevance of lateral Mcl-1 TMD interactions to the modulation of apoptosis.
Fig. 2.
Mcl-1 TMD induces cell death. (A) Caspase 3/7 activity and cell toxicity (B) induced by the VN Mcl-1 WT TMD and single amino acid mutants were analyzed in cytosolic extracts of HCT116 cells 16 h after transfection. CTRL means nontransfected cells. Error bars represent the mean ± SEM, n = 3. P value, according to Dunnett’s test, displayed. **P < 0.01. Mitochondrial polarization status (C) and LDH activity (D) in the above-described conditions. The mitochondrial uncoupler FCCP was used as a positive control for mitochondrial depolarization. (E) HCT116 cells were transfected with Mcl-1, Bcl-xL, or Bcl-2 FL proteins. After 24 h, the VN Mcl-1 TMD was expressed for 16 h, and caspase 3/7 activity measured in cytosolic extracts. CTRL refers to nontransfected cells and mock condition refers to cells transfected with the corresponding empty vector for FL expression proteins. Error bars represent the mean ± SEM, n = 5. *P < 0.05. Chimeric protein expression of the VN Mcl-1 TMD (c-myc) and Mcl-1, Bcl-xL or Bcl-2 FL proteins are shown in the Bottom using α-tubulin as a loading control.
Next, we attempted to elucidate whether the ability of the Mcl-1 TMD to induce apoptosis derived from its competition with the endogenous full-length Mcl-1 protein (Mcl-1 FL), by impeding its antiapoptotic functions in the cell and inducing apoptosis. To analyze this possibility, we studied the effect of overexpressing Mcl-1 FL protein on Mcl-1 TMD-induced apoptosis. We found a significant reduction in the ability of the Mcl-1 TMD to activate caspase 3/7 when coexpressed with the Mcl-1 FL protein (Fig. 2E); in contrast, no reduction was observed following the coexpression of other antiapoptotic proteins (e.g., Bcl-2 FL or Bcl-xL FL). We confirmed the overexpression of full-length proteins by Western blotting (Fig. 2 E, Bottom).
Overall, these results suggest the existence of lateral transmembrane interaction specificity between different members of the Bcl-2 protein family.
Mcl-1 TMD Induces Apoptosis in a Bax/Bak-Independent and Bok-Dependent Manner.
The antiapoptotic function of Mcl-1 has been attributed to the establishment of heterointeractions with different proapoptotic members of the Bcl-2 protein family. Among these partners are the apoptotic effectors Bax, Bak, and Bok, and several BH3 only proteins, which include Bim and Bid (43–45). To elucidate the relevance of the Mcl-1 TMD in the modulation of the proapoptotic Bcl-2 effector function, we first analyzed the ability of the Mcl-1 TMD to form heterooligomers with the Bax, Bak, and Bok TMDs by BiFC. Interestingly, the Mcl-1 TMD only formed heterooligomers with the Bok TMD (Fig. 3A). These results agree with recent reports in which the lack of the Mcl-1 TMD does not change the specificity of Bak interactions (45), suggesting that Mcl-1 utilizes a different molecular mechanism to modulate the activity of proapoptotic Bak and Bok proteins.
Fig. 3.
The Mcl-1 TMD apoptosis induction is Bok dependent. (A) Oligomerization analysis of the Mcl-1 TMD and TMDs from the proapoptotic proteins Bax, Bak, and Bok measured by BiFC in HCT116 cells. On the Left, representative confocal images of the BiFC assay are shown. The graph represents the quantification of the complement fluorescent measurements. Error bars represent the mean ± SEM, n = 4. P value, according to Dunnett’s test, displayed. *P < 0.05. The bottom section of the graph shows the protein expression level of the constructs. Caspase 3/7 activity (B) and mitochondrial membrane potential changes (C) induced by the VN Mcl-1 WT TMD and single amino acid variants was analyzed in Bax−/−Bak−/− HCT116 cells after 16 h transfection. CTRL refers to nontransfected cells. Error bars represent the mean ± SEM, n = 4. *P < 0.05. c-myc expression was analyzed for all constructs (Bottom). α-Tubulin used as a loading control. Caspase 3/7 activity was analyzed in WT (D) and Bax−/−Bak−/− (E) HCT116 cells transfected with VN Mcl-1 TMD or VN ΔTMD after 24 h of Bok silencing (siRNA Bok, siBok; siRNA Random, siRand). Error bars represent the mean ± SEM, n = 4. P value, according to Sidak’s test, displayed. **P < 0.01. n.s., not significant. Protein expression was monitored by Western blotting using α-tubulin as a loading control.
The lack of interaction between the Mcl-1 TMD and those from Bak and Bax raised the possibility that cell death induced by the Mcl-1 TMD remained independent of the presence of those proapoptotic full-length proteins. To address this question, we transfected the Mcl-1 TMD and both G340P and G344I mutants in Bax−/− Bak−/− HCT116 cells. In all cases, transfection produced comparable levels of caspase 3/7 activity and similar changes in mitochondrial membrane potential to those observed in WT HCT116 cells (compare Fig. 3B to Fig. 2A and Fig. 3C to Fig. 2C), thereby confirming our hypothesis. Next, we assessed whether Bok down-regulation affected Mcl-1 TMD-induced cell death. Gene silencing of endogenous Bok protein strongly reduced caspase 3/7 activity in both, WT and Bax−/− Bak−/− HCT116 cells transfected with the Mcl-1 TMD, while we did not observe significant changes in the presence of a random siRNA (Fig. 3 D and E). Knockdown experiments with stably expressing shRNA for Bok HCT116 cells reproduced cell death protection (SI Appendix, Fig. S4 A and B). These results confirm that Mcl-1 TMD-induced cell death depends on the presence of the proapoptotic effector Bok.
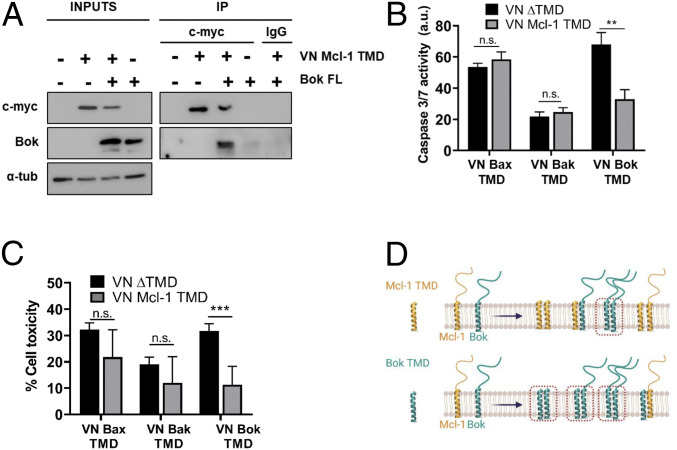
To confirm the existence of protein-protein interactions between the Mcl-1 TMD and Bok, we performed coimmunoprecipitation experiments in cells overexpressing both proteins. For this purpose, we transfected HCT116 cells with the myc-tagged VN Mcl-1 TMD and full-length Bok (Bok FL) constructs and immunoprecipitated with an anti-myc antibody. The Mcl-1 TMD displayed strong coimmunoprecipitation with the Bok FL protein, confirming the existence of interactions between these proteins (Fig. 4A). Coimmunoprecipitation between both full-length proteins was also observed (SI Appendix, Fig. S4C). Interestingly, cotransfection of Flag-Bok FL with Mcl-1 G340P FL or Mcl-1 ΔTMD produced a decrease in Bok and Mcl-1 protein levels (SI Appendix, Fig. S4D). As the protein stability of both Bok and Mcl-1 is low (46), these results suggest a role for transmembrane interactions in maintaining protein stability. Next, we addressed the functional relevance of the heterointeraction between Mcl-1 and Bok TMDs regarding the modulation of apoptosis. Interestingly, overexpression of the isolated Bok TMD induced a robust caspase 3/7 activation that the presence of the Mcl-1 TMD counteracted (Fig. 4 B and C). The concurrent presence of both TMDs is probably causing the formation of TMD heterodimers that are not competent for the induction of cell death (Fig. 4D).
Fig. 4.
The Mcl-1 TMD immunoprecipitates Bok FL protein. (A) Bok FL and the VN Mcl-1 TMD were coexpressed in HCT116 cells. Immunoprecipitates were probed by Western blotting with anti-Bok and anti-c-myc antibodies. IgG nonrelated was used as negative antibody control. (B) The proapoptotic effect of the Bok TMD is partially inhibited by the presence of the Mcl-1 TMD, as measured by caspase 3/7 activity and cell toxicity assays (C) in HCT116 cells cotransfected with VN Bcl TMD constructs. Error bars represent the mean ± SEM, n = 3. **P < 0.01; ***P < 0.001. n.s., not significant. (D) Scheme of putative heterointeractions formed upon transfection of Mcl-1 TMD (Upper) or Bok TMD (Lower) in cells. Red squares highlight complexes that could be involved in cell death induction.
On the contrary, we did not observe any changes in cell death induced by Bax or Bak TMDs following Mcl-1 TMD coexpression. In fact, cell death induction by Bok TMD is independent of Bax and Bak and depends on endogenous Bok (SI Appendix, Fig. S4 E and F).
Altogether, these results reinforce both the specificity and the functional relevance of the interaction between Bok protein and the Mcl-1 TMD.
Molecular Simulation Studies of Mcl-1 TMD Membrane Interactions.
To obtain detailed molecular insights into the putative interactions between Mcl-1 and Bok TMDs within the membrane, we performed multiscale MD simulations of both homo- and heterodimers. First, we constructed alpha-helical peptides in the atomistic resolution (SI Appendix, Table S1) and equilibrated them in a lipid bilayer (47). Next, we transformed the peptide structures into CG resolution and generated 1,000 initial structures of the two peptides (24). We then embedded the peptides in a lipid bilayer (48) and allowed them to dimerize. We used a development version of the upcoming release of the MARTINI model (49), which accounts for the excessive aggregation of membrane proteins (50). Then, we clustered the dimer structures and fine grained the centroids of the two most populated clusters (for each homo- and heterodimer). These fine-grained dimers were embedded in lipid bilayers containing 150 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC) molecules using the CHARMM-GUI server (51), and finally simulated for 1 µs each. We then analyzed the simulation outputs for stability and for structural details such as the interacting amino acids and the crossing angles between the TM domains in the dimer.
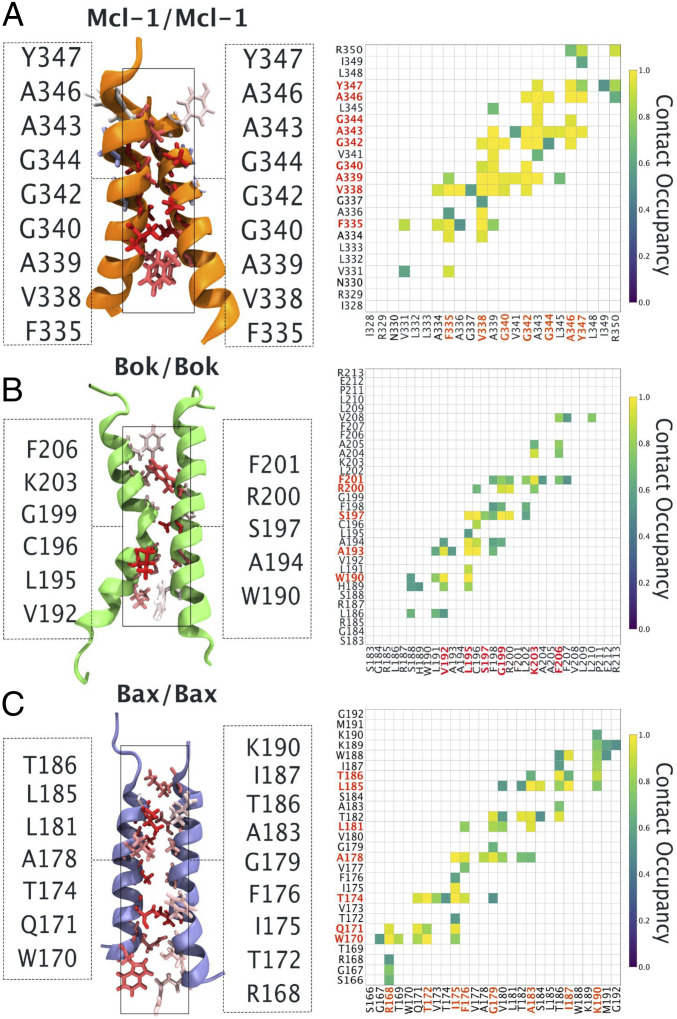
Results from these studies demonstrated that all TMDs homodimers (Mcl-1–Mcl-1, Bok–Bok, and Bax–Bax) formed stable dimers within 1 µs of the simulation time, as evidenced by the time evolution of distances (Fig. 5 and SI Appendix, Fig. S5) (52). There existed two modes of binding for each homodimer. The first binding mode (more populated, called cluster 1) is characterized by 1) larger overall tilt of the dimer with respect to the lipid bilayer, and 2) smaller crossing angle between monomers forming a dimer. The second binding mode (less populated, called cluster 2) is characterized by 1) smaller overall tilt of the dimer, and 2) a larger crossing angle between monomers forming a dimer. The differences in the average helical content between these two modes are negligible, with the exception of Bax–Bax homodimer, where the difference in the average helical content between two binding modes reaches around 5% (SI Appendix, Tables S1–S3). The changes in the transmembrane helical tilt between the different binding modes could reflect the existence of different functional states of the protein. This has been identified in other TMDs, such as those found in the light-sensitive receptor rhodopsin, where changes in helical conformation and interhelical motions determine the operating mechanism of the photoreceptor (53).
Fig. 5.
Homodimer atomistic molecular dynamics simulations. Left shows the average structures of (A) Mcl-1, (B) Bok, and (C) Bax homodimers based on cluster 2 structures. Dimerization interfaces are shown illustrating the contribution to the interaction for each residue with a contribution >0.5. It is defined as the sum of the occupancy of contacts for each residue normalized to the maximum value of each system and is represented with the colored licorice draw method using the blue-white-red color scale, where red indicates large values. Right shows the corresponding average amino acid–amino acid occupancy of contact. Contact occupancies of less than 0.5 were removed for clarity. Contact occupancies equal to 1.0 correspond to the situation where given amino acids were constantly in contact. The average was calculated over time and over three repetitions of each system. Two amino acids were considered to be in contact if any of their atoms were closer than 6 Å.
The first binding mode showed a less broad network of stable interpeptide contacts in the atomistic simulations, indicating a more dynamic interface. Instead, the second binding mode displayed more interpeptide contacts that were stably present in the atomistic simulations (Fig. 5A depicts the residues involved in homodimer contacts). In the second binding mode, the introduction of the G340P mutation (SI Appendix, Fig. S3A) in the Mcl-1 TMD homodimer led to the loss of stable interactions. In contrast, the inclusion of the G344I mutation (SI Appendix, Fig. S3B) maintained a similar dimerization interface as the Mcl-1 WT TMD homodimer. These results agree with experimental studies, thus suggesting that the simulation is able to provide an accurate molecular view (snapshots in Fig. 5) on the dimer structures probed by the experiments. Based on molecular simulations the residue G342, is located at the dimer interface. As expected, the mutant Mcl-1 G342P TMD reduced dimer association confirming its relevance for homodimer formation (SI Appendix, Fig. S2C).
In the case of the Mcl-1/Bok TMD heterodimers, the majority of simulations led to the formation of a stable heterodimer, with one exception where the dimer dissociated (SI Appendix, Fig. S5). However, in the case of the Mcl-1/Bax TMD heterodimer, the majority of the heterodimers dissociated within 600 ns of the simulation (SI Appendix, Fig. S5; see detailed explanation in SI Appendix, Fig. S6). Based on the high contact occupancies and low interaction energies, we identified: V192, L195, L202, and F206 as residues strongly involved in the stabilization of the Mcl-1/Bok interface (SI Appendix, Fig. S6A).
The stability of the Mcl-1 TMD dimers changed from most stable to least stable in the following order (SI Appendix, Fig. S5): Mcl-1 TMD homodimer > Mcl-1/Bok TMDs heterodimer >> Mcl-1/Bax TMDs heterodimer, again, in close agreement with the oligomerization results obtained in the BiFC experimental assays for these interactions (Fig. 3A).
Mcl-1 TMD Tethers the Bok TMD to the Mitochondria.
The role of Bok in the modulation of apoptosis remains controversial (54). Bok mainly localizes to the ER (55); however, several reports claim a role for Bok in mitochodrial outer membrane permeabilization (MOMP) (46, 56), which would suggest the translocation of Bok from the ER to the mitochondria to exert its proapoptotic function. While many studies have revealed that Mcl-1 interacts with Bok, contradictory results exist regarding the ability of Mcl-1 to counteract Bok-induced apoptosis (13, 46, 57–59). To describe the role of Bok and Mcl-1 TMDs in the modulation of apoptosis, we analyzed the interactions among their TMDs in our BiFC assay. Interestingly, Bok and Mcl-1 TMDs form heterooligomers that are disrupted in the Mcl-1 G340P TMD mutant but not affected in G344I TMD (Fig. 6A). Molecular simulations of the heterodimer point in the same direction locating the G340 residue near the heterodimer interaction interface while the G344I is orientated toward the lipid milieu (Fig. 6B).
Fig. 6.
The Mcl-1 TMD translocates the Bok TMD to the mitochondria. (A) Heterooligomerization analysis of the Mcl-1 WT TMD and single amino acid variants with the Bok TMD measured by BiFC in HCT116 cells. (B) Left shows the average structures of Mcl-1–Bok TMD heterodimers based on cluster 2 structures. Right shows the corresponding average amino acid–amino acid occupancy of contact. (C) Confocal images of HeLa cells stained with red ER tracker (Left) or expressing MtDsRed marker (Right) and transfected with VN and VC Bok TMD constructs. Oligomers were observed in the green channel. Merge panels show colocalizations with the PCC. (D) Confocal images of HeLa cells expressing MtDsRed marker and transfected with the VN Mcl-1 TMD plus the VC Bok TMD constructs. As above, oligomers were observed in the green channel. (E) Graph shows average PCC in the mitochondria in C and D experimental conditions. Bars represent mean ± SEM (n = 86 cells per condition); P value, according to Student t test, displayed ***P < 0.001. (F) Representative confocal images and quantification of in situ PLA targeting VDAC1 and IP3R interactions in HeLa cells transfected with VN + VC ΔTMD constructs or VN Mcl-1 TMD and VC Bok TMD as indicated. The experiment was performed three times; error bars represent the mean ± SEM; ***P < 0.001.
We then analyzed the oligomeric state of the Bok TMD by cotransfecting the VC and VN Bok TMD in HeLa cells. Confocal microscopy studies provided evidence that Bok TMD homooligomers formed and colocalized preferentially within the ER with a PCC of 0.94 (Fig. 6C). This observation contrasts with the subcellular localization of the Mcl-1 TMD, which mainly localizes to the mitochondria (Fig. 1C); hence, the differential localization of Mcl-1 and Bok TMD homooligomers challenges the results obtained from the coimmunoprecipitation experiments and functional assays.
Interestingly, the introduction of both constructs in the BiFC system (VN Mcl-1 TMD and VC Bok TMD) proved the heterointeraction between Bok and Mcl-1, in agreement with our MD data, and revealed that this interaction takes place in the mitochondria (Fig. 6 D and E). This result suggests a putative role for the Mcl-1 TMD in the facilitation of the Bok TMD movement from the ER to the mitochondria to exert its function. We hypothesize that a second signaling event would be necessary to activate Bok and induce cell death once in the mitochondria.
Thus, we reasoned that the presence of both TMDs must favor an increase in contact sites between both organelles. To evaluate this possibility, we analyzed the ability of Mcl-1/Bok TMDs heterodimers to change the number of ER-MAMs within the cell. To visualize and quantify endogenous ER-MAMs in cells, we employed a recently developed proximity ligation assay (PLA) (60). For this purpose, we incubated HeLa cells with antibodies targeting the VDAC1 and IP3R1 proteins from the outer mitochondrial and ER membranes, respectively. When compared to control conditions, the expression of Mcl-1/Bok TMDs significantly increased the number of ER-MAMs (from 152 ± 65 to 232 ± 64) (Fig. 6F). This result confirms that Mcl-1 and Bok TMDs participate in endomembrane fusion events taking place between the mitochondria and ER and could explain the transference of the Bok TMD from the ER to the mitochondria to exert its proapoptotic function.
Mcl-1 TMDs: From Bench to Bedside.
In the hope of translating our results from basic research to the clinical setting, we identified amino acid changes within the Mcl-1 TMD in cancer patients by searching the catalog of somatic mutations in cancer (COSMIC) (61). Mutation impact scores on COSMIC are calculated by Functional Analysis Through Hidden Markov Models-Multiple Kernel Learning (FATHMM-MKL) (62) where only scores ≥0.7 are classified as pathogenic. We found that the frequency of mutations in this protein region is very low. However, we encounter two mutations, an A339T mutant from a malignant melanoma patient with a pathogenic score of 0.93 and a L348V mutant from a lung adenocarcinoma patient with a pathogenic score of 0.79 (SI Appendix, Table S4) that could give us the clues to understand the Mcl-1 TMD cellular functions. Because Mcl-1 overexpression in hematologic malignancies and solid tumors is associated with poor prognosis and resistance to treatment, these highly conserved protein regions represent good intervention points for future drug development.
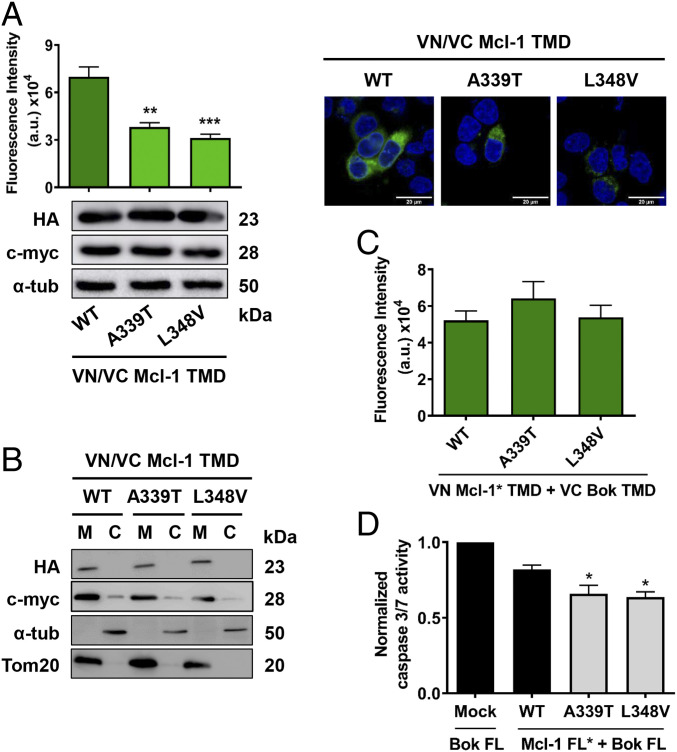
We first introduced these mutations into Mcl-1 TMD BiFC constructs and then analyzed their ability to influence the homooligomerization state of the protein. Our analysis revealed that patient-derived mutations significantly reduced the ability of the Mcl-1 TMD to form homooligomers (Fig. 7A), although the protein was correctly located in the mitochondrial membrane (Fig. 7B) and their membrane insertion capability was conserved (SI Appendix, Fig. S7).
Fig. 7.
Somatic mutations of the Mcl-1 TMD affect protein functionality. (A) Oligomerization analysis of Mcl-1 mutant TMDs measured by BiFC in HCT116 cells. Error bars represent the mean ± SEM, n = 4 (**P < 0.01; ***P < 0.001). Confocal images of BiFC and Western blotting analysis of protein expression of the constructs are also included. (B) Subcellular fractionation of HCT116 cells transfected with VN (c-myc) and VC (HA) mutant TMD constructs (mitochondrial fraction, M, and cytosol, C). (C) Heterooligomerization analysis of the Mcl-1 mutants TMD with Bok TMD measured by BiFC in HCT116 cells. (D) Caspase 3/7 activity induced by Bok FL plus mock plasmid, Mcl-1 WT FL, or mutant FL was analyzed in cytosolic extracts from HCT116 cells 16 h after transfection. Error bars represent the mean ± SEM, n = 4; *P < 0.05.
Analysis of the heterointeractions with the Bok TMD reflected a maintained level or, indeed, a slight increase in Mcl-1/Bok TMD heterointeractions (Fig. 7C). Experimental results for mutations of residues involved in the Mcl-1 TMD homodimer interface are in good agreement with molecular simulations (SI Appendix, Fig. S8 for detailed explanation).
As the isolated Mcl-1 TMDs behave as antagonists of the FL protein, we expected the opposite behavior in patients. In fact, the introduction of the mutations into the Mcl-1 protein generated proteins (Mcl-1 A339T FL and Mcl-1 L348V FL) that display a greater protective effect upon apoptosis induction by Bok than the wild-type protein (Fig. 7D and SI Appendix, Fig. S9). Thus, an enhanced antiapoptotic behavior could provide a survival advantage to tumor cells containing mutations in the Mcl-1 TMD and contribute to cancer progression.
Altogether, our results highlight the relevance of Mcl-1/Bok TMD interactions for apoptosis modulation. We have found that mutations in these protein regions could provide survival advantages in tumors. It is important to note that the identification of transmembrane interaction interfaces implicated in cell death modulation provides a point of intervention to foster drug development.
Discussion
The proapoptotic function of Bok and its modulation by the antiapoptotic protein Mcl-1 have been the subject of broad discussion in the literature. Physical interactions between Mcl-1 and Bok were first described by double hybrid experiments from a rat ovarian cDNA library (57). Since then, reports have identified these heterointeractions through coimmunoprecipitation experiments (46), although other attempts have failed to detect them (55). Reports have demonstrated that Mcl-1 does not influence Bok-mediated apoptosis (46), while others provide evidence for inhibition (13). Finally, research has established the ability of Bok to induce apoptosis in the absence of Bax and Bak; however, similar studies have also demonstrated the requirement for these proapoptotic effectors (55, 56). To fully elucidate the role of these proteins, and since their interactions take place in the intracellular membranes, this study focused on understanding the role of transmembrane regions on the equilibria that determine cell fate. The formation of homooligomers and the cell death induced by the C-terminal region of Mcl-1 (177 to 350) was recently described by Hellmuth and Stemmann (63) as part of the control mechanisms for the correct spindle assembly checkpoint. Here, we report that the apoptosis-inducing capability can be attributed to the Mcl-1 TMD region and is Bok dependent (SI Appendix, Figs. S3 and S4). Bok is mainly localized at the ER and Golgi membranes, but a fraction of the protein localizes at the mitochondrial outer membrane (MOM) and at the contact sites between the ER and mitochondria (46, 55). Here, we show that Mcl-1 TMD interacts and tethers Bok TMD to the mitochondria (Figs. 3 and 6 and SI Appendix, Fig. S4), thereby suggesting that Mcl-1 could maintain a fraction of Bok stable in an off state at the mitochondria as the appropriate cell death stimulus reaches the cell. In fact, our results suggest that the lack of a functional Mcl-1 TMD decreases Bok protein stability (SI Appendix, Fig. S4).
The amplification and overexpression of Mcl-1 have been reported in several cancer types and correlates with poor prognosis and response to treatments (19, 64–67). Most Mcl-1 inhibitors in preclinical and clinical development target the BH3 binding domain of antiapoptotic Bcl-2 proteins to activate Bax/Bak-dependent cell death (68). However, this inhibition strategy could be inefficient for interactions taking place between Mcl-1 and Bok that appear to be mainly mediated by the transmembrane regions with no relevant contributions of the cytosolic BH3 binding domain (13). Our results suggest that drugs targeting heterointeractions of the Mcl-1/Bok TMDs could represent an effective strategy to release Bok and induce cell death in tumor cells.
Materials and Methods
BiFC TMD Assays.
The Mcl-1, Bcl-2, Bid, Bax, Bak, and Bok TMDs were cloned at the C-terminal end of the Venus VN-terminal (1-155, I152L) and VC-terminal (155-238, A206K). Mutations of BiFC TMDs were obtained using the Stratagene Quikchange II kit. HCT116 cells were cotransfected with 0.5 μg of VN and VC DNA constructs using TurboFect (Thermo Scientific). Fluorescence emission was measured after 16 h transfection in phosphate-buffered saline (PBS) using 96-well black plates and a Wallac 1420 Workstation (ʎexc 510 nm and ʎem 535 nm).
PLA Assay.
Transfected HeLa cells were fixed with 4% paraformaldehyde (PFA) for 10 min and permeabilized with 0.1% Triton X-100 for 10 min at room temperature. Following a blocking step, cells were incubated with VDAC1 (ab14734, Abcam) and IP3R (ab5804, Abcam) antibodies overnight at 4 °C. Hybridization of antibodies using the plus and minus PLA probes and subsequent ligation and amplification reactions were performed according to the manufacturer’s protocol (Sigma-Aldrich). Images were obtained using a Leica SP8 confocal microscope. Sixty transfected cells per condition were quantified using FiJi software.
Supplementary Material
Acknowledgments
We thank Profs. R. Youle and B. Vogelstein for kindly providing the HCT116 cells; Prof. J. V. Esplugues for kindly providing the HeLa MtDsRed cells; and Prof. T. Kaufmann for kindly providing the shControl and shBok HCT116 cells. This work was supported by Spanish Ministry of Economy and Competitiveness Grant SAF2017-84689-R; Generalitat Valenciana Grant PROMETEO/2019/065. E.L. was supported by Generalitat Valenciana Grant ACIF/2016/019. F.L., M.J., and W.K. wish to thank the CSC–IT Center for Science (Espoo, Finland). M.J. acknowledges the Emil Aaltonen Foundation for support.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008885117/-/DCSupplemental.
Data Availability.
Simulations data have been deposited in Zenodo. 1) Monomeric simulations: https://zenodo.org/record/4012224; and 2) dimerization simulations: https://doi.org/10.5281/zenodo.4022900.
References
- 1.Kluck R. M., Bossy-Wetzel E., Green D. R., Newmeyer D. D., The release of cytochrome c from mitochondria: A primary site for Bcl-2 regulation of apoptosis. Science 275, 1132–1136 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Birkinshaw R. W., Czabotar P. E., The BCL-2 family of proteins and mitochondrial outer membrane permeabilisation. Semin. Cell Dev. Biol. 72, 152–162 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Kale J., Osterlund E. J., Andrews D. W., BCL-2 family proteins: Changing partners in the dance towards death. Cell Death Differ. 25, 65–80 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edlich F., et al. , Bcl-x(L) retrotranslocates Bax from the mitochondria into the cytosol. Cell 145, 104–116 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ausman J., et al. , Ceramide-induced BOK promotes mitochondrial fission in preeclampsia. Cell Death Dis. 9, 298 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morciano G., et al. , Intersection of mitochondrial fission and fusion machinery with apoptotic pathways: Role of Mcl-1. Biol. Cell 108, 279–293 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Morciano G., et al. , Mcl-1 involvement in mitochondrial dynamics is associated with apoptotic cell death. Mol. Biol. Cell 27, 20–34 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schulman J. J., et al. , Bok regulates mitochondrial fusion and morphology. Cell Death Differ. 26, 2682–2694 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams A., et al. , The non-apoptotic action of Bcl-xL: Regulating Ca(2+) signaling and bioenergetics at the ER-mitochondrion interface. J. Bioenerg. Biomembr. 48, 211–225 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andreu-Fernández V., et al. , Bax transmembrane domain interacts with prosurvival Bcl-2 proteins in biological membranes. Proc. Natl. Acad. Sci. U.S.A. 114, 310–315 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang T., Glazko A. J., Effect of an adenosine deaminase inhibitor on the uptake and metabolism of arabinosyl adenine (Vidarabine) by intact human erythrocytes. Res. Commun. Chem. Pathol. Pharmacol. 14, 127–140 (1976). [PubMed] [Google Scholar]
- 12.Chen L., et al. , Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol. Cell 17, 393–403 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Stehle D., et al. , Contribution of BH3-domain and transmembrane-domain to the activity and interaction of the pore-forming Bcl-2 proteins Bok, Bak, and Bax. Sci. Rep. 8, 12434 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernández-Marrero Y., Spinner S., Kaufmann T., Jost P. J., Survival control of malignant lymphocytes by anti-apoptotic MCL-1. Leukemia 30, 2152–2159 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Lin J., Fu D., Dai Y., Lin J., Xu T., Mcl-1 inhibitor suppresses tumor growth of esophageal squamous cell carcinoma in a mouse model. Oncotarget 8, 114457–114462 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sieghart W., et al. , Mcl-1 overexpression in hepatocellular carcinoma: A potential target for antisense therapy. J. Hepatol. 44, 151–157 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Campbell K. J., et al. , MCL-1 is a prognostic indicator and drug target in breast cancer. Cell Death Dis. 9, 19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng C., Yang F., Wang J., FBXO4 inhibits lung cancer cell survival by targeting Mcl-1 for degradation. Cancer Gene Ther. 24, 342–347 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Lee E. F., et al. , BCL-XL and MCL-1 are the key BCL-2 family proteins in melanoma cell survival. Cell Death Dis. 10, 342 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fletcher S., MCL-1 inhibitors–Where are we now (2019)? Expert Opin. Ther. Pat. 29, 909–919 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Akgul C., Moulding D. A., White M. R., Edwards S. W., In vivo localisation and stability of human Mcl-1 using green fluorescent protein (GFP) fusion proteins. FEBS Lett. 478, 72–76 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Andreu-Fernández V., et al. , Peptides derived from the transmembrane domain of Bcl-2 proteins as potential mitochondrial priming tools. ACS Chem. Biol. 9, 1799–1811 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Grau B., et al. , The role of hydrophobic matching on transmembrane helix packing in cells. Cell Stress 1, 90–106 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wassenaar T. A., et al. , High-throughput simulations of dimer and trimer assembly of membrane proteins. The DAFT approach. J. Chem. Theory Comput. 11, 2278–2291 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Lelimousin M., Limongelli V., Sansom M. S., Conformational changes in the epidermal growth factor receptor: Role of the transmembrane domain investigated by coarse-grained metaDynamics free energy calculations. J. Am. Chem. Soc. 138, 10611–10622 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemmon M. A., et al. , Glycophorin A dimerization is driven by specific interactions between transmembrane alpha-helices. J. Biol. Chem. 267, 7683–7689 (1992). [PubMed] [Google Scholar]
- 27.Mineev K. S., et al. , Dimeric structure of the transmembrane domain of glycophorin a in lipidic and detergent environments. Acta Naturae 3, 90–98 (2011). [PMC free article] [PubMed] [Google Scholar]
- 28.Lemmon M. A., Flanagan J. M., Treutlein H. R., Zhang J., Engelman D. M., Sequence specificity in the dimerization of transmembrane alpha-helices. Biochemistry 31, 12719–12725 (1992). [DOI] [PubMed] [Google Scholar]
- 29.García-Sáez A. J., et al. , Peptides derived from apoptotic Bax and Bid reproduce the poration activity of the parent full-length proteins. Biophys. J. 88, 3976–3990 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berger B. W., et al. , Consensus motif for integrin transmembrane helix association. Proc. Natl. Acad. Sci. U.S.A. 107, 703–708 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gotoh T., Terada K., Oyadomari S., Mori M., hsp70-DnaJ chaperone pair prevents nitric oxide- and CHOP-induced apoptosis by inhibiting translocation of Bax to mitochondria. Cell Death Differ. 11, 390–402 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Kerppola T. K., Bimolecular fluorescence complementation: Visualization of molecular interactions in living cells. Methods Cell Biol. 85, 431–470 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Javadpour M. M., Eilers M., Groesbeek M., Smith S. O., Helix packing in polytopic membrane proteins: Role of glycine in transmembrane helix association. Biophys. J. 77, 1609–1618 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duarte J. M., Biyani N., Baskaran K., Capitani G., An analysis of oligomerization interfaces in transmembrane proteins. BMC Struct. Biol. 13, 21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teese M. G., Langosch D., Role of GxxxG motifs in transmembrane domain interactions. Biochemistry 54, 5125–5135 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Orzáez M., Lukovic D., Abad C., Pérez-Payá E., Mingarro I., Influence of hydrophobic matching on association of model transmembrane fragments containing a minimised glycophorin A dimerisation motif. FEBS Lett. 579, 1633–1638 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Arbely E., Granot Z., Kass I., Orly J., Arkin I. T., A trimerizing GxxxG motif is uniquely inserted in the severe acute respiratory syndrome (SARS) coronavirus spike protein transmembrane domain. Biochemistry 45, 11349–11356 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Bi G., et al. , SOBIR1 requires the GxxxG dimerization motif in its transmembrane domain to form constitutive complexes with receptor-like proteins. Mol. Plant Pathol. 17, 96–107 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faingold O., Cohen T., Shai Y., A GxxxG-like motif within HIV-1 fusion peptide is critical to its immunosuppressant activity, structure, and interaction with the transmembrane domain of the T-cell receptor. J. Biol. Chem. 287, 33503–33511 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yano Y., et al. , GXXXG-mediated parallel and antiparallel dimerization of transmembrane helices and its inhibition by cholesterol: Single-pair FRET and 2D IR studies. Angew. Chem. Int. Ed. Engl. 56, 1756–1759 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan L., et al. , Higher-order clustering of the transmembrane anchor of DR5 drives signaling. Cell 176, 1477–1489.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwon B., Lee M., Waring A. J., Hong M., Oligomeric structure and three-dimensional fold of the HIV gp41 membrane-proximal external region and transmembrane domain in phospholipid bilayers. J. Am. Chem. Soc. 140, 8246–8259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willis S. N., et al. , Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 19, 1294–1305 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Germain M., Duronio V., The N terminus of the anti-apoptotic BCL-2 homologue MCL-1 regulates its localization and function. J. Biol. Chem. 282, 32233–32242 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Hockings C., et al. , Mcl-1 and Bcl-xL sequestration of Bak confers differential resistance to BH3-only proteins. Cell Death Differ. 25, 721–734 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Llambi F., et al. , BOK is a non-canonical BCL-2 family effector of apoptosis regulated by ER-associated degradation. Cell 165, 421–433 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Javanainen M., Equilibration simulations of TM domains of Bcl-2 proteins Mcl-1, Bok, and Bax. Zenodo. 10.5281/zenodo.4012224. Deposited 2 September 2020. [DOI]
- 48.Javanainen M., Universal method for embedding proteins into complex lipid bilayers for molecular dynamics simulations. J. Chem. Theory Comput. 10, 2577–2582 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Marrink S. J., Tieleman D. P., Perspective on the martini model. Chem. Soc. Rev. 42, 6801–6822 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Javanainen M., Martinez-Seara H., Vattulainen I., Excessive aggregation of membrane proteins in the Martini model. PLoS One 12, e0187936 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee J., et al. , CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 12, 405–413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lolicato F., Kulig W., Dimerization simulations of TM domains of Bcl-2 proteins: Mcl-1, Bok, and Bax. Zenodo. 10.5281/zenodo.4022900. Deposited 10 September 2020. [DOI]
- 53.Ren Z., Ren P. X., Balusu R., Yang X., Transmembrane helices tilt, bend, slide, torque, and unwind between functional states of rhodopsin. Sci. Rep. 6, 34129 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haschka M. D., Villunger A., There is something about BOK we just don’t get yet. FEBS J. 284, 708–710 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Echeverry N., et al. , Intracellular localization of the BCL-2 family member BOK and functional implications. Cell Death Differ. 20, 785–799 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Einsele-Scholz S., et al. , Bok is a genuine multi-BH-domain protein that triggers apoptosis in the absence of Bax and Bak. J. Cell Sci. 129, 2213–2223 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Hsu S. Y., Kaipia A., McGee E., Lomeli M., Hsueh A. J., Bok is a pro-apoptotic Bcl-2 protein with restricted expression in reproductive tissues and heterodimerizes with selective anti-apoptotic Bcl-2 family members. Proc. Natl. Acad. Sci. U.S.A. 94, 12401–12406 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.D’Orsi B., et al. , Bok is not pro-apoptotic but suppresses poly ADP-ribose polymerase-dependent cell death pathways and protects against excitotoxic and seizure-induced neuronal injury. J. Neurosci. 36, 4564–4578 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Onyeagucha B., et al. , Novel post-transcriptional and post-translational regulation of pro-apoptotic protein BOK and anti-apoptotic protein Mcl-1 determine the fate of breast cancer cells to survive or die. Oncotarget 8, 85984–85996 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alam M. S., Proximity ligation assay (PLA). Curr. Protoc. Immunol. 123, e58 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tate J. G., et al. , COSMIC: The catalogue of somatic mutations in cancer. Nucleic Acids Res. 47, D941–D947 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shihab H. A., et al. , An integrative approach to predicting the functional effects of non-coding and coding sequence variation. Bioinformatics 31, 1536–1543 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hellmuth S., Stemmann O., Separase-triggered apoptosis enforces minimal length of mitosis. Nature 580, 542–547 (2020). [DOI] [PubMed] [Google Scholar]
- 64.Beroukhim R., et al. , The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shah K., Huang H., Bradbury N. A., Li C., White C., Mcl-1 promotes lung cancer cell migration by directly interacting with VDAC to increase mitochondrial Ca2+ uptake and reactive oxygen species generation. Cell Death Dis. 5, e1482 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Song K.-A., et al. , Increased synthesis of MCL-1 protein underlies initial survival of EGFR-mutant lung cancer to EGFR inhibitors and provides a novel drug target. Clin. Cancer Res. 24, 5658–5672 (2018). [DOI] [PubMed] [Google Scholar]
- 67.Wen Q., et al. , Elevated expression of mcl-1 inhibits apoptosis and predicts poor prognosis in patients with surgically resected non-small cell lung cancer. Diagn. Pathol. 14, 108 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hird A. W., Tron A. E., Recent advances in the development of Mcl-1 inhibitors for cancer therapy. Pharmacol. Ther. 198, 59–67 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Simulations data have been deposited in Zenodo. 1) Monomeric simulations: https://zenodo.org/record/4012224; and 2) dimerization simulations: https://doi.org/10.5281/zenodo.4022900.