Significance
Understanding the tolerance mechanism to inhibiting chemicals can help in the design of more robust microbial strains for use in industrial bioprocesses. Aromatic acids are important bioactive molecules and precursors for many plant natural products. This study focused on analyzing Saccharomyces cerevisiae mutants tolerant to aromatic acids and revealed that regulating the transport of these compounds from the cytosol is the main mechanism to improve tolerance. Using this knowledge, the transport of aromatic acid can be fine tuned to improve the secretion of aromatic compounds and improve the robustness of yeast strains for industrial use.
Keywords: adaptive laboratory evolution, aromatic acid, tolerance, transporter
Abstract
Toxicity from the external presence or internal production of compounds can reduce the growth and viability of microbial cell factories and compromise productivity. Aromatic compounds are generally toxic for microorganisms, which makes their production in microbial hosts challenging. Here we use adaptive laboratory evolution to generate Saccharomyces cerevisiae mutants tolerant to two aromatic acids, coumaric acid and ferulic acid. The evolution experiments were performed at low pH (3.5) to reproduce conditions typical of industrial processes. Mutant strains tolerant to levels of aromatic acids near the solubility limit were then analyzed by whole genome sequencing, which revealed prevalent point mutations in a transcriptional activator (Aro80) that is responsible for regulating the use of aromatic amino acids as the nitrogen source. Among the genes regulated by Aro80, ESBP6 was found to be responsible for increasing tolerance to aromatic acids by exporting them out of the cell. Further examination of the native function of Esbp6 revealed that this transporter can excrete fusel acids (byproducts of aromatic amino acid catabolism) and this role is shared with at least one additional transporter native to S. cerevisiae (Pdr12). Besides conferring tolerance to aromatic acids, ESBP6 overexpression was also shown to significantly improve the secretion in coumaric acid production strains. Overall, we showed that regulating the activity of transporters is a major mechanism to improve tolerance to aromatic acids. These findings can be used to modulate the intracellular concentration of aromatic compounds to optimize the excretion of such products while keeping precursor molecules inside the cell.
Saccharomyces cerevisiae has proven to be a great microbial host for producing compounds of plant origin (1, 2). A frequent hurdle when constructing new microbial cell factories is the toxicity of the final product or some of the intermediary metabolites that can result from introducing heterologous enzyme activities. Compounds with aromatic groups have shown particular toxicity (3–5) and mitigating their inhibition on growth can help accelerate the design and improve the performance of new cell factories.
Aromatic acids, such as coumaric acid, can be used as platform chemicals but are also important precursors for producing plant natural products such as flavonoids (6) and phenols (7). Even at low concentration, coumaric acid and related compounds (i.e., phenylpropanoic acids) are known inhibitors of S. cerevisiae growth (4, 8), making their presence inside the cell a potential issue when engineering the production of natural products that require them as precursors. Given the limited information of how phenylpropanoic acids inhibit growth, using techniques such as adaptive laboratory evolution (ALE) can help generate tolerant strains and further elucidate the genetic basis underlying tolerance phenotypes and how these compounds stress their host (9).
Acid tolerance in S. cerevisiae has been studied in detail for many organic acids and usually requires the cell to up-regulate transporters (e.g., Pdr12) to reduce the concentration of the acid anions inside the cell and proton pumps (Pma1) to restore a neutral pH in the cytosol (reviewed in refs. 10–12). Organic acids are known to exhibit a pH-dependent toxicity because only the protonated form of the acid can cross the membrane, making them more toxic at lower pH (10). Regarding tolerance to aromatic acids in S. cerevisiae, the deletion of PDR12 was shown to induce sensitivity to fusel acids (phenylacetic acid and indoleacetic acid) (13). Furthermore, tolerance to benzoic acid has also been shown to be mediated by Pdr12 (14) or other transporters like Tpo2 (15).
In this work, we used ALE to study tolerance mechanisms of S. cerevisiae to two aromatic acids: coumaric acid and ferulic acid. Tolerance to either aromatic acid was traced back to mutations in a transcription activator involved in aromatic amino acid (AAA) catabolism (ARO80). Further analysis of possible targets of Aro80 showed that increased expression of the transporter ESPB6 (previously not known to transport aromatic acids) could reproduce the tolerance phenotypes observed. Since Esbp6 is active when AAAs are used as the nitrogen source, we explored its role in these conditions and determined that this transporter can export fusel acids from the cytosol. The role of Esbp6 in fusel acid tolerance partly overlaps with the function of another transporter, Pdr12, but the mechanism of activation between the two differs. While ESBP6 is activated by Aro80 in response to the presence of AAAs, PDR12 is up-regulated by War1 possibly as a response to the presence of fusel acids.
Results
Adaptive Laboratory Evolution and Mutant Screening.
ALE was utilized to generate multiple independent lineages of evolved populations with increased tolerance to the coumaric and ferulic acids. First, the native tolerance of S. cerevisiae to each aromatic acid was determined by measuring growth in medium with different concentrations of each compound. Since organic acids are known to exhibit a pH-dependent toxicity (higher toxicity at low pH), we performed the growth tests and evolution at a pH of 3.5 using citrate-phosphate buffer as described by Kildegaard et al. (16). Both aromatic acids have a maximum solubility close to 1 g/L in the conditions used, and a starting concentration of 0.2 g/L for either acid was enough to apply an adequate level of selection pressure on the growth of S. cerevisiae for the ALE experiments.
To maximize the genetic diversity that could be obtained during the evolutionary paths, five single clones were used as a seed culture to start five independent replicates for each acid. The growth of each replicate was monitored periodically during evolution and the concentration of the acids was slowly increased to encourage incremental increases in fitness (maximum growth rate) by using a strategy similar to a previous publication (17). The experiment was allowed to continue as long as the fitness increased significantly for a certain time period (18). When the fitness appeared to reach a plateau, the ALE experiment was terminated, and an aliquot of each evolved population was plated to isolate individual clones. Overall, all evolution replicates reached the solubility limit for each aromatic acid (1 g/L), which shows the ALE experiments succeeded in producing mutants with high tolerance. It is worth noting that the solubility and relative toxicity of both aromatic acids change with pH. At low pH (3.5), the maximum solubility is low, but their inhibiting effect is quite high. The ALE mutants obtained here can tolerate 1 g/L of either aromatic acid at pH 3.5 but have the potential to tolerate much higher acid concentrations at higher pHs. On average, ALE experiments for coumaric acid tolerance were terminated after 90 transfers and ferulic acid tolerance was evolved for 69 transfers.
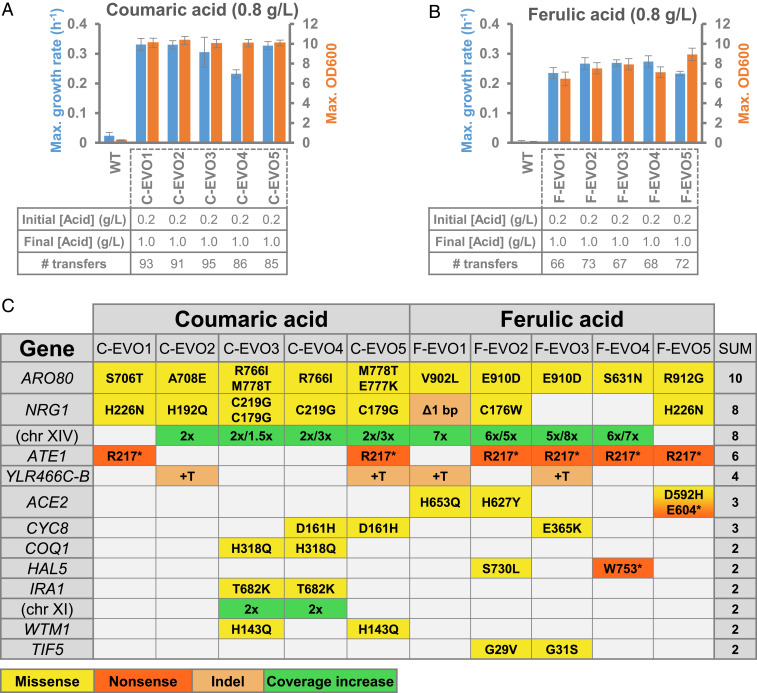
Individual clones were isolated and screened from the independently evolved populations to identify those with increased tolerance and to use for further studying. Within each evolution experiment, we picked 10 individual clones from culture plates and grew them in medium with inhibiting concentrations of each aromatic acid (0.8 g/L). For each set of clones, we chose the 3 with the highest growth rate for further characterization. Fig. 1 A and B shows the average growth rates and maximum optical densities for the 3 best performing clones within each evolution replicate (EVO1-EVO5) in comparison to a reference strain (wild type [WT]). Generally, all selected clones showed a superior performance in terms of increased tolerance and fitness compared to the reference strain. Most of the coumaric acid evolved strains were able to achieve a maximum growth rate above 0.30 h−1, which is considerably higher than the reference strain (WT) but not as high as the reference strain in medium without aromatic acids (0.41 h−1). The only exception was found in the clones isolated from the C-EVO4 population, whose average growth rate was only 0.23 h−1. Consistent with the performance of cells during the preliminary acid inhibition test, the selected ferulic acid evolved clones showed a lower fitness than coumaric acid evolved clones for the same acid concentration.
Fig. 1.
Summary of the results from the adaptive laboratory evolution experiments for tolerance to coumaric acid and ferulic acid. Average growth rates (h−1) and maximum optical densities (OD600) achieved by the three best performing mutants isolated from each of the five populations evolved for coumaric acid tolerance (A) and ferulic acid tolerance (B). Error bars represent the SD from the three best mutants (each with three replicates). (C) Table of mutations for genes mutated in more than one population. Information regarding the individual growth rates, maximum optical densities, and lists of all mutations detected can be found in SI Appendix, Tables S1–S4.
In order to understand if the evolved phenotypes relied on degradation of the aromatic acids, the cultures of two evolved clones for each acid were analyzed by high performance liquid chromatography (HPLC). For all four cultivations the concentration of coumaric acid and ferulic acid remained stable at 800 mg/L, indicating that the evolved phenotypes relied on physiological adaptation to tolerate the presence of aromatic acids. Since it has previously been shown that another strain of S. cerevisiae can degrade aromatic acids (19), it was surprising to realize that none of the tested ALE mutants seemed to degrade these compounds.
Whole Genome Sequencing of Evolved Mutants.
To investigate the genetic basis behind the tolerance phenotypes obtained, we performed whole genome sequencing on three mutant clones exhibiting the highest growth rates within each independently evolved population (for a total of 30 clones). Fig. 1C shows an overview of the genes mutated in at least two independently evolved populations (full list provided in SI Appendix, Tables S3 and S4). Among the most common genetic changes, we found mutations in ARO80, NRG1, and an increase (up to eightfold) in read depth for certain sections of chromosome XIV. Mutations in ARO80 seem of crucial importance given their presence in all of the clones sequenced for both aromatic acids. The diversity of mutations found in ARO80 is also a good indicator for the importance of this gene in tolerance to aromatic acids. From a total of 10 independent evolution experiments, we found 9 distinct amino acid substitutions (Fig. 1C) but no cases of nonsense mutations. This suggests that selective pressure for ARO80 mutations is high, but loss of function does not appear to be responsible for the tolerance phenotype. Aro80 is a transcription factor previously shown to activate genes needed for using AAAs as the nitrogen source (20, 21). In the presence of AAAs, Aro80 induces the transcription of genes from the Ehrlich pathway (22), leading to the assimilation of nitrogen from these compounds and excretion of fusel acids and fusel alcohols. Given the similarity of coumaric and ferulic acids to some of the metabolites in the Ehrlich pathway, mutations in ARO80 indicated that genes involved in detoxifying such compounds were probably being activated.
Trying to connect mutations in NRG1 to aromatic acid tolerance is not as straightforward as for ARO80. NRG1 codes for a transcriptional repressor involved in controlling the expression of many glucose-repressed genes (23) and also regulating tolerance to Na+, Li+, and growth in alkaline conditions (24). Although NRG1 was shown to be required for tolerance to other stressing conditions, the reason for it being frequently mutated in aromatic acid tolerant strains is not known.
Increases in coverage of chromosome XIV were used to pinpoint a gene related to the tolerance phenotype by looking at the intersection of coverage increases. Computing the intersection between the DNA regions affected in coumaric and ferulic acid tolerance mutants revealed a 35-kb segment that is common to all mutants (spanning open reading frames [ORFs] from YNL123W to YNL141W). We decided to focus our interest in this mutated region by comparing the documented targets of Aro80 (retrieved from Yeastract) (25) with regions of chromosome XIV that are affected by a coverage increase. From this approach, it was concluded that there is one gene that met the criteria (SI Appendix, Table S5). The gene in question is ESBP6 (YNL025C) and codes for a protein that has similarity to monocarboxylate permeases. This gene was shown to be induced by Aro80 in the presence of AAAs (21) but its role has never been uncovered. Its cellular localization is not clear according to YeastRGB (26), but one high throughput dataset suggested that Esbp6 is located in the cell periphery (27). Furthermore, an overexpression mutant of ESBP6 was shown to confer tolerance to 6% lactic acid (28).
ATE1, ACE2, and CYC8 are also mutated in multiple evolution lineages. Mutations in ACE2 were shown to prevent yeast cells from separating from each other after cell division, which leads to the formation of agglomerates (29, 30). Even though the inner cells inside the agglomerates may benefit from some protection against harsh conditions, we have previously observed that mutations in ACE2 contribute very little to improving the growth rate in the presence of organic acids (31). Regarding the mutations in CYC8, we previously showed that mutations in this gene can confer tolerance to pimelic acid (31). However, the exact mechanism leading to the tolerance was never uncovered. Finally, mutations in ATE1 were found to be the same (R217*) in all mutants where they were present. This is an indication that this mutation probably arose in the seed culture used to start the ALE experiments and is not relevant for the tolerance phenotypes.
Reverse Engineering of Mutations to Determine Causality.
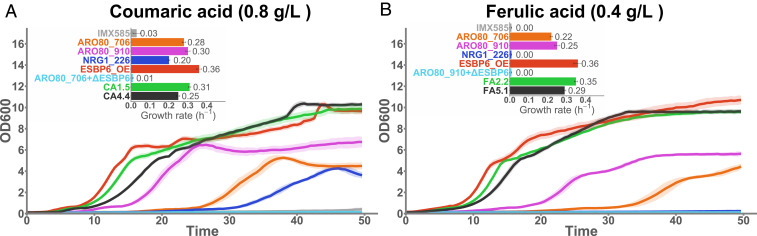
In order to find the genetic causes for the tolerance phenotypes, we implemented a selection of the most frequent mutations in a strain with a clean background. To test the contribution of mutations in ARO80, we selected two different point mutations found in this gene: one mutation taken from a lineage evolved in coumaric acid (ARO80S706T found in C-EVO1) and another present in two lineages evolved in ferulic acid (ARO80E910D found in F-EVO2 and F-EVO3). Fig. 2 shows the growth curves for ARO80S706T (ARO80_706) and ARO80E910D (ARO80_910) mutants in the presence of inhibiting concentrations of coumaric acid or ferulic acid. Both point mutations tested for ARO80 confer significant tolerance to coumaric and ferulic acid in comparison to a reference strain (IMX585). However, it is clear from Fig. 2 that the mutant ARO80E910D reaches the stationary phase faster than ARO80S706T. Although the maximum growth rates do not differ much between the two ARO80 mutants tested, the length of the lag phase is shorter for ARO80E910D. When we compare the ARO80 mutants to the ALE mutants where each mutation was taken from (CA1.5 and FA2.2), we see that mutations in ARO80 are not enough to replicate the full tolerance phenotype, especially in terms of the lag phase length and capacity to grow after the diauxic shift.
Fig. 2.
Phenotypic characterization of the reverse engineered mutants in the presence of the aromatic acids. (A) Growth curves and maximum growth rates of a reference strain (IMX585) in comparison to five reverse engineered strains (ARO80S706T, ARO80E910D, NRG1H226N, ESBP6_OE, and ARO80S706T+ΔESBP6) and two evolved mutants tolerant to coumaric acid (CA1.5 and CA4.4). Cells were grown in 96-well plates with a volume of 250 μL in medium with 0.8 g/L of coumaric acid. (B) Growth curves and maximum growth rates of a reference strain (IMX585) in comparison to five reverse engineered strains (ARO80S706T, ARO80E910D, NRG1H226N, ESBP6_OE, and ARO80E910D+ΔESBP6) and two evolved mutants tolerant to ferulic acid (FA2.2 and FA5.1). Cells were grown in 96-well plates with a volume of 250 μL in medium with 0.4 g/L of ferulic acid. All cultivations were performed for three biological replicates (each with three technical replicates) and the average OD600s are shown as solid lines with the interval encompassing the SD represented as a shaded area. The maximum growth rates represent the average of individually calculated growth rates for all replicates and the errors bars represent the SD.
Frequent mutations in NRG1 in both coumaric and ferulic acid tolerant mutants provided justification to test if point mutations in this gene were contributing to the tolerance phenotypes. Since the mutation NRG1H226N appeared in mutants for both aromatic acids, we constructed a mutant displaying this mutation (NRG1_226) and tested its growth in the presence of coumaric acid or ferulic acid (Fig. 2). In coumaric acid, the strain NRG1_226 performs better than the control strain, but it shows a long lag phase and lower maximum growth rate (0.20 h−1) than the evolved clones and ARO80 mutants. In ferulic acid, no growth was detected, even at 0.4 g/L. These findings suggested that the effect of NRG1 is minor on its own and could be dependent on the presence of additional mutations to confer a significant fitness improvement.
The increase in read coverage for chromosome XIV would be difficult to reverse engineer given the length of DNA involved. Nevertheless, the presence of a target of Aro80 in the affected areas suggested that a single gene could be the reason behind this genetic alteration. To test this hypothesis, we introduced an extra copy of ESBP6 under the control of a strong promoter (PTEF1) in a clean background strain, resulting in strain ESBP6_OE. As shown in Fig. 2, ESBP6_OE grows robustly in medium with inhibiting amounts of coumaric or ferulic acid. In comparison to all ALE mutants tested, the strain ESBP6_OE matches or outperforms them in terms of maximum growth rate and length of the lag phase. To evaluate if ESBP6 was responsible for the tolerance exhibited by ARO80 mutants, we deleted this gene in each of the ARO80 single mutant background (strains ARO80_706+ΔESBP6 and ARO80_910+ΔESBP6). The growth profiles of both strains showed that knocking out ESBP6 completely abolishes the tolerance phenotype observed for ARO80S706T and ARO80E910D, revealing that Esbp6 has an essential role in tolerance to aromatic acids.
Overall, testing the impact of each mutation in a clean background strain revealed that the tolerance phenotypes evolved here were likely caused by the up-regulation of ESBP6. This was achieved via evolution by point mutations in the transcription factor ARO80 that controls ESBP6 and by an increase in the number of copies of the ESBP6 locus in the genome.
ESBP6’s Role in S. cerevisiae.
Testing individual mutations from tolerant mutants (Fig. 2) revealed that ESBP6 overexpression resulted in the highest tolerance to aromatic acids. Although the native function of ESBP6 was unclear, the expression of this gene was shown to be induced in the presence of AAAs by the transcription factor Aro80 (21). When S. cerevisiae uses AAAs as the nitrogen source, Aro80 induces the expression of genes from the Ehrlich pathway. Among the compounds involved in the Ehrlich pathway, fusel acids (phenylacetic acid, 4-hydroxyphenylacetic acid, and indoleacetic acid) are the most similar to coumaric and ferulic acids. The exact way fusel acids are excreted to the medium is not known, but the presence of the ABC transporter Pdr12 was shown to be required for tolerance to phenylacetic acid (PAA) and indoleacetic acid (IAA) (13).
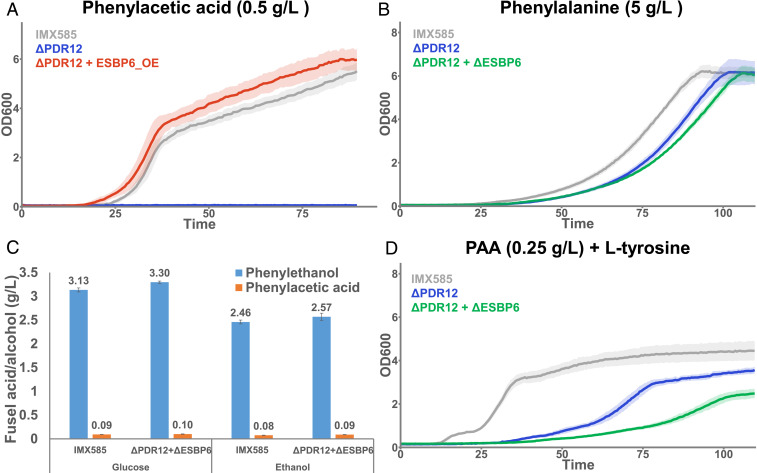
In order to understand if Esbp6 has a role in fusel acid tolerance, we compared the growth of a PDR12 deletion mutant with and without the overexpression of ESBP6 (ΔPDR12+ESBP6_OE and ΔPDR12, respectively) in medium with PAA or IAA. Fig. 3A shows that in the presence of 0.5 g/L of PAA, the PDR12 deletion mutant (ΔPDR12) exhibits a growth disability when compared to the reference strain (IMX585), confirming the essentiality of Pdr12 in these conditions. When ESBP6 is overexpressed on a ΔPDR12 background (ΔPDR12 + ESBP6_OE), growth is restored to wild-type levels (Fig. 3A). The observation that Esbp6 can compensate for Pdr12 absence shows that they have overlapping roles in tolerance to aromatic acids.
Fig. 3.
Influence of PDR12/ESBP6 on fusel acid tolerance and AAA metabolism. (A) Growth curves in medium with 0.5 g/L of phenylacetic acid for a reference strain (IMX585) in comparison to a PDR12 deletion mutant (ΔPDR12) and a double mutant lacking PDR12 and displaying the overexpression of ESBP6 (ΔPDR12+ESBP6_OE). Cells were grown in 96-well plates with a volume of 250 μL in medium with 0.5 g/L of phenylacetic acid. (B) Growth curves in medium with 5 g/L of l-phenylalanine as nitrogen source for a reference strain (IMX585) in comparison to a PDR12 deletion mutant (ΔPDR12) and a double deletion mutant lacking PDR12 and ESBP6 (ΔPDR12+ΔESBP6). Cells were grown in 96-well plates with a volume of 250 μL in medium with 5 g/L of l-phenylalanine as nitrogen source and 20 g/L of ethanol as carbon source. (C) Extracellular concentration of phenylacetic acid and phenylethanol in cultures of a reference strain (IMX585) and a double deletion mutant (ΔPDR12+ΔESBP6) grown in medium with 20 g/L of glucose or 10 g/L of ethanol as carbon source. Cells were grown in shake flasks with a volume of 25 mL in medium with the mentioned nitrogen and carbon sources. Error bars represent the SD from three biological replicates. (D) Growth curves in medium with 0.25 g/L of phenylacetic acid and 0.5 g/L of l-tyrosine as the nitrogen source for a reference strain (IMX585) in comparison to a PDR12 deletion mutant (ΔPDR12) and a double deletion mutant lacking PDR12 and ESBP6 (ΔPDR12+ΔESBP6). Cells were grown in 96-well plates with a volume of 250 μL in medium with 0.5 g/L of l-tyrosine as the nitrogen source and 0.25 g/L of phenylacetic acid. All growth curves were performed for three biological replicates (each with two technical replicates) and the average OD600s are shown as solid lines with the interval encompassing the SD represented as a shaded area.
Similar tests performed in the presence of IAA showed that at a concentration of 0.1 g/L, the deletion of PDR12 does not affect growth (SI Appendix, Fig. S1A). This observation contradicts a previous report (13) about the role of PDR12 in tolerance to IAA, but it should be noted that a different media formulation and method for growth evaluation were used. Even though the inactivation of PDR12 showed little effect on tolerance to IAA, overexpressing ESBP6 was still enough to improve growth in comparison to the reference strain (SI Appendix, Fig. S1A), suggesting a certain role for this gene in tolerance to this fusel acid. Besides PAA and IAA, we also tested the effect of 4-hydroxyphenylacetic acid on the growth of S. cerevisiae, but no toxicity was detected up to 2 g/L, which is in accordance with the lower toxicity of this fusel acid to S. cerevisiae (13).
Since PDR12 and ESBP6 were shown to be important for fusel acid tolerance, we then tested if single and double disruption of these genes affects growth of S. cerevisiae when growing on AAAs. Fig. 3B shows that when growing on l-phenylalanine (precursor of PAA), even the simultaneous inactivation of ESBP6 and PDR12 (strain ΔPDR12+ΔESBP6) seems to have little effect. This could be a consequence of two different aspects: the amount of fusel acids produced is below the toxicity level needed to observe a phenotype; and there are additional transporters capable of excreting fusel acids to the medium. To test the first alternative, the amount of PAA and phenylethanol (PE) excreted by cells growing on l-phenylalanine as the nitrogen source was quantified. As shown in Fig. 3C, independently of the carbon source or strain used, the major Ehrlich pathway byproduct excreted is PE. Although a small quantity of PAA is still produced, the amount is much lower than what is needed to induce toxicity. In fermentative conditions the production of PE was expected because the high NADH/NAD+ promoted the production of reduced byproducts. By using ethanol as a carbon source we expected that the growth in full oxidative conditions would promote fusel acid production. However, the results in Fig. 3C showed that the effect of the carbon source on the amount of fusel acid produced was quite low and that factors other than the cell’s oxidative state may be necessary for forcing the production of fusel acids. It is worth noting that all our tests were performed at low pH (3.5), which favors the reentry of any excreted carboxylic acids into the cell.
Given the low levels of PAA produced by cultures grown with AAAs as the nitrogen source, the role of ESBP6 in these conditions could not be directly confirmed. In order to force a response, we added PAA to cultures grown with l-tyrosine or l-tryptophan as nitrogen sources. Adding 0.5 g/L of PAA was enough to completely abolish growth of a PDR12 deletion strain when l-tyrosine or l-tryptophan were provided as the nitrogen source (SI Appendix, Fig. S1B). However, by reducing the concentration of PAA to 0.25 g/L, we could see that a PDR12 deletion mutant (ΔPDR12) could grow in the presence of l-tyrosine (Fig. 3D) but slower than the reference strain (IMX585). Furthermore, when ESBP6 was deleted in addition to PDR12 (ΔPDR12+ΔESBP6) we observed that the resulting mutant was still viable but its growth was reduced in comparison to the PDR12 deletion mutant (Fig. 3D). These results indicate that when l-tyrosine is used as the nitrogen source Pdr12 is the main source of tolerance to PAA, but Esbp6 also contributes to it. It is worth noting that the basal growth displayed by the double deletion mutant suggests that other genes may also be contributing to tolerance to PAA.
Besides the growth tests on PAA, we also tried to confirm the roles of PDR12 and ESBP6 on IAA tolerance in the presence of AAA. As shown in SI Appendix, Fig. S1C, growth on 0.1 g/L of IAA with l-tryptophan as the nitrogen source confirmed that the role of Pdr12 in tolerance to this fusel acid seems minor (SI Appendix, Fig. S1C). However, when ESBP6 is also inactivated there is a clear reduction in growth. This observation confirms that for the fusel acid IAA, Esbp6 plays a more important role than Pdr12.
ESBP6 Influence on Coumaric Acid Production Strains.
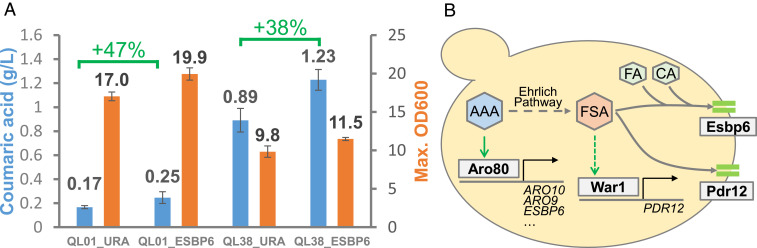
Since Esbp6 confers tolerance to several aromatic acids, we tested if overexpressing its coding gene would also improve the secretion of this class of compounds. Using strains engineered to overproduce coumaric acid, we quantified the amount of secreted product with and without the overexpression of ESBP6. The production tests were carried in the same conditions as the evolution experiments (low pH) to maximize the potential effect of improving product secretion. Fig. 4A shows the impact of overexpressing ESBP6 on two coumaric acid production strains, QL01 and QL38, constructed and characterized elsewhere (32). We selected two strains with varying levels of coumaric acid production to observe if this would affect our results. As shown in Fig. 4A, the coumaric acid production levels increased almost by the same amount in the lower and higher producing strains (47% increase vs. 38% increase). The consistent improvement indicates that increased levels of Esbp6 can promote the export of coumaric acid from the cell and can be used as a strategy to potentially reduce the level of aromatic acids in the cytosol. Although low pH conditions were optimal to observe the effect of ESBP6 on coumaric acid production, the titers of coumaric acid were considerably lower than previously reported for experiments run at pH 6.0 (32). The medium pH affects the relative abundance of protonated organic acids and consequently their ability to enter the cell by diffusion. At low pH, besides secreting the coumaric acid produced biosynthetically, the cell must also secrete the increased flow of coumaric acid reentering the cell by diffusion. Besides boosting the export of coumaric acid, the overexpression of ESBP6 also improved cell growth by 17% in both strains. These results show that besides increasing the secretion of coumaric acid, ESBP6 can reduce the stress burden on the cell and improve the biomass yield.
Fig. 4.
Influence of ESBP6 overexpression on coumaric acid production. (A) Coumaric acid titer and maximum OD600 in strains with (QL01_ESBP6 and QL38_ESBP6) and without (QL01_URA and QL38_URA) the overexpression of ESBP6. Error bars represent the SD from three biological replicates. (B) Schematic illustration of the interaction between aromatic acids, the transcription factors Aro80 and War1, and the transporters Esbp6 and Pdr12. In the presence of AAAs, Aro80 activates the transcription of genes necessary for degrading these compounds, which leads to the production of fusel acids and fusel alcohols. Fusel acid presence putatively induces the transcription of PDR12 (through War1) and both Pdr12 and Esbp6 are responsible for excreting these compounds to the medium. Abbreviations: AAA, aromatic amino acids (l-phenylalanine, l-tyrosine, and l-tryptophan); FSA, fusel acids (phenylacetic acid, 4-hydroxyphenylacetic acid, and indoleacetic acid); CA, coumaric acid; and FA, ferulic acid.
Discussion
ALE and subsequent analysis of mutations has been an indispensable tool to study metabolic pathways and general physiology of eukaryotic cells. Here, using ALE we have generated mutant strains tolerant to two aromatic acids, coumaric and ferulic acids, at levels near their solubility limit. By reverse engineering selected mutations, we showed that increasing the expression of the transporter gene ESBP6 was a main mechanism to tolerate aromatic acids that arose during the ALE experiments. In most of the tolerant mutants, the likely up-regulation of Esbp6 was associated with nine different mutations in ARO80 and duplication of segments from chromosome XIV that included the ESBP6 locus.
Since Esbp6 is activated in the presence of AAAs, we investigated its potential role in medium containing AAAs as nitrogen source and in the presence of fusel acids (aromatic acids derived from AAA). This analysis showed that Esbp6 had a role in tolerance to fusel acids, but its importance is difficult to pinpoint because of redundant transporters present in S. cerevisiae and hurdles in designing experiments where the cell is forced to produce fusel acids instead of fusel alcohols. The overlap in function between Esbp6 and Pdr12 was also examined and it revealed that both transporters can confer tolerance to some fusel acids but have different modes of activation (Fig. 4B). While ESBP6 is activated by Aro80 when AAAs are used as nitrogen sources, Pdr12 is activated through War1 possibly by direct activation by fusel acids.
Besides providing insight into the metabolism of aromatic compounds in S. cerevisiae, the work described here can also have implications in the design of cell factories for natural product synthesis. By up-regulating the transport of aromatic acids, cell factories that produce this type of compounds can be improved by increasing the flux of product from the cytosol to the medium and by allowing cells to tolerate higher levels of product in the medium (33). Fig. 4A shows that increased levels of ESBP6 can lead to a significant improvement in the production titer and higher cell density. Since diffusion of extracellular organic acids into the cells is increased at lower pH (10), by performing these production tests at pH 3.5 we replicated the harsh conditions of industrial fermentations where the product is present in severely inhibiting amounts. The improvement in product titer and OD600 showed that ESBP6 overexpression can promote the export of acid anions from the cytosol and improve the biomass yield probably by reducing the stress burden imposed by excess aromatic acids inside the cell. We also would like to point out that previous studies achieved much higher coumaric acid titers in fed-batch fermentations but using a considerably higher pH. Although using a higher pH can reduce the toxicity of organic acids (10), in industrial conditions it is desirable to keep pH low to reduce the costs related to product purification (34).
Furthermore, cell factories that use aromatic acid as intermediates can potentially benefit from the down-regulation of the transporters for these classes of compounds if these intermediates are not toxic. By forcing the aromatic acid precursors to accumulate in the cytosol, the flux to natural products (such as flavonoids) can possibly be increased.
Materials and Methods
A more detailed description of the materials and methods used in this work is provided in SI Appendix, Supplementary Information Text, Materials and Methods.
Yeast Strains and Medium.
The strain GL01 (31) was used for all ALE experiments and the strain IMX585 (35) was used for the reverse engineering work. Both strains were derived from the S. cerevisiae CEN.PK113-7D background. The full list of strains used and constructed in this work is provided in SI Appendix, Table S6.
ALE experiments and growth tests were performed in the minimal medium described in ref. 36 and buffered at pH 3.5 by adding 140 mL/L of 0.5 M citrate solution and 60 mL/L of 1 M Na2HPO4. For testing strains in alternative nitrogen sources, (NH4)2SO4 was replaced with 6.6 g/L of K2SO4 and the required amino acid was added in the appropriate concentration (5 g/L of l-phenylalanine, 5 g/L of l-tryptophan, or 0.5 g/L of l-tyrosine).
Strain Construction.
The reverse engineered strains containing deletions, point mutations, and insertions were constructed using the single gRNA method described elsewhere (35). The list of primers used to engineer each mutation is available in SI Appendix, Table S7 and the list of plasmids used and constructed in this study is provided in SI Appendix, Table S8.
Adaptive Laboratory Evolution.
Adaptive laboratory evolution experiments were conducted on an automated platform using a liquid handling robot as described previously (17, 37).
Growth Screening.
The growth characterization of the postevolution mutants and reverse engineered strains was performed in a Growth Profiler 960 (Enzyscreen) using 96-half-deep-well microplates (with transparent bottom) with a total culture volume of 250 μL, agitation at 250 rpm, temperature controlled at 30 °C, and initial OD600 of 0.05.
DNA Extraction, Sequencing, and Data Analysis.
The Blood & Cell Culture DNA Mini Kit (Qiagen) was used to extract genomic DNA from 3 mL of the overnight yeast culture (∼5 × 108 cells) using the protocol recommended by the manufacturer. Sequencing was performed with the NextSeq (High Output Kit) with 2 × 150 paired-end reads targeting a genome coverage of 100× per sample.
Mutations in evolved clones were identified using bresEq. 0.30.2 (38) with Bowtie 2.2.8 as aligner (39) and using the reference genome of CEN.PK113-7D (40) as described before (31).
Phenylacetic Acid and Phenylethanol Quantification.
Extracellular concentration of phenylacetic acid and phenylethanol in cultures of a reference strain (IMX585) and a double deletion mutant (ΔPDR12+ΔESBP6) were quantified by HPLC using a method previously described by Li et al. (32).
Coumaric Acid Production Tests.
Coumaric acid production tests were performed in 100-mL shake flasks containing 20 mL of the minimal medium without glucose and pH adjusted to 3.5. Six tablets of FeedBeads (SMFB08001, Kuhner Shaker) were added to the culture as the source of glucose. Cultures were inoculated with an initial OD600 of 0.05 and shaken at 200 rpm in an orbital incubator, at 30 °C during 96 h. For the strains QL38_URA and QL38_ESBP6, 1% of galactose was also added to induce genes controlled by the GAL promoters. p-coumaric acid was quantified using HPLC as described by Li et al. (32).
Supplementary Material
Acknowledgments
This work was funded by the Novo Nordisk Foundation (NNF10CC1016517 and NNF18OC0034844), the Knut and Alice Wallenberg Foundation, and Ångpanneföreningens Forskningsstiftelse. We thank Dr. Quanli Liu for his assistance on the coumaric acid production tests.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2013044117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Paddon C. J., Keasling J. D., Semi-synthetic artemisinin: A model for the use of synthetic biology in pharmaceutical development. Nat. Rev. Microbiol. 12, 355–367 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Cravens A., Payne J., Smolke C. D., Synthetic biology strategies for microbial biosynthesis of plant natural products. Nat. Commun. 10, 2142 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee J. H., Wendisch V. F., Biotechnological production of aromatic compounds of the extended shikimate pathway from renewable biomass. J. Biotechnol. 257, 211–221 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Adeboye P. T., Bettiga M., Olsson L., The chemical nature of phenolic compounds determines their toxicity and induces distinct physiological responses in Saccharomyces cerevisiae in lignocellulose hydrolysates. AMB Express 4, 46 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li M., et al. , Common problems associated with the microbial productions of aromatic compounds and corresponding metabolic engineering strategies. Biotechnol. Adv. 41, 107548 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Gottardi M., Reifenrath M., Boles E., Tripp J., Pathway engineering for the production of heterologous aromatic chemicals and their derivatives in Saccharomyces cerevisiae: Bioconversion from glucose. FEMS Yeast Res. 17, 17 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Rodrigues J. L., Prather K. L. J., Kluskens L. D., Rodrigues L. R., Heterologous production of curcuminoids. Microbiol. Mol. Biol. Rev. 79, 39–60 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chambel A., Viegas C. A., Sá-Correia I., Effect of cinnamic acid on the growth and on plasma membrane H+-ATPase activity of Saccharomyces cerevisiae. Int. J. Food Microbiol. 50, 173–179 (1999). [Google Scholar]
- 9.Sandberg T. E., Salazar M. J., Weng L. L., Palsson B. O., Feist A. M., The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology. Metab. Eng. 56, 1–16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarboe L. R., Royce L. A., Liu P., Understanding biocatalyst inhibition by carboxylic acids. Front. Microbiol. 4, 272 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skoneczny M., Skoneczna A., “Response mechanisms to chemical and physical stresses in yeast and filamentous fungi” in Stress Response Mechanisms in Fungi, Skoneczny M., Ed. (Springer International Publishing, 2018), pp. 35–85. [Google Scholar]
- 12.Mira N. P., Teixeira M. C., Sá-Correia I., Adaptive response and tolerance to weak acids in Saccharomyces cerevisiae: A genome-wide view. Integr. Biol. 14, 525–540 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hazelwood L. A., et al. , A new physiological role for Pdr12p in Saccharomyces cerevisiae: Export of aromatic and branched-chain organic acids produced in amino acid catabolism. FEMS Yeast Res. 6, 937–945 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Piper P., et al. , The pdr12 ABC transporter is required for the development of weak organic acid resistance in yeast. EMBO J. 17, 4257–4265 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandes A. R., Mira N. P., Vargas R. C., Canelhas I., Sá-Correia I., Saccharomyces cerevisiae adaptation to weak acids involves the transcription factor Haa1p and Haa1p-regulated genes. Biochem. Biophys. Res. Commun. 337, 95–103 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Kildegaard K. R., et al. , Evolution reveals a glutathione-dependent mechanism of 3-hydroxypropionic acid tolerance. Metab. Eng. 26, 57–66 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Mohamed E. T., et al. , Generation of a platform strain for ionic liquid tolerance using adaptive laboratory evolution. Microb. Cell Fact. 16, 204 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LaCroix R. A., Palsson B. O., Feist A. M., A model for designing adaptive laboratory evolution experiments. Appl. Environ. Microbiol. 83, e03115-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adeboye P. T., Bettiga M., Aldaeus F., Larsson P. T., Olsson L., Catabolism of coniferyl aldehyde, ferulic acid and p-coumaric acid by Saccharomyces cerevisiae yields less toxic products. Microb. Cell Fact. 14, 149 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iraqui I., Vissers S., André B., Urrestarazu A., Transcriptional induction by aromatic amino acids in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 3360–3371 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Godard P., et al. , Effect of 21 different nitrogen sources on global gene expression in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 27, 3065–3086 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hazelwood L. A., Daran J. M., van Maris A. J. A., Pronk J. T., Dickinson J. R., The Ehrlich pathway for fusel alcohol production: A century of research on Saccharomyces cerevisiae metabolism. Appl. Environ. Microbiol. 74, 2259–2266 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee S. B., Kang H. S., Kim T., Nrg1 functions as a global transcriptional repressor of glucose-repressed genes through its direct binding to the specific promoter regions. Biochem. Biophys. Res. Commun. 439, 501–505 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Lamb T. M., Mitchell A. P., The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 23, 677–686 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teixeira M. C., et al. , YEASTRACT: An upgraded database for the analysis of transcription regulatory networks in Saccharomyces cerevisiae. Nucleic Acids Res. 46, D348–D353 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dubreuil B., et al. , YeastRGB: Comparing the abundance and localization of yeast proteins across cells and libraries. Nucleic Acids Res. 47, D1245–D1249 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yofe I., et al. , One library to make them all: Streamlining the creation of yeast libraries via a SWAp-tag strategy. Nat. Methods 13, 371–378 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sugiyama M., Akase S. P., Nakanishi R., Kaneko Y., Harashima S., Overexpression of ESBP6 improves lactic acid resistance and production in Saccharomyces cerevisiae. J. Biosci. Bioeng. 122, 415–420 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Oud B., et al. , Genome duplication and mutations in ACE2 cause multicellular, fast-sedimenting phenotypes in evolved Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 110, E4223–E4231 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dohrmann P. R., et al. , Parallel pathways of gene regulation: Homologous regulators SWI5 and ACE2 differentially control transcription of HO and chitinase. Genes Dev. 6, 93–104 (1992). [DOI] [PubMed] [Google Scholar]
- 31.Pereira R., et al. , Adaptive laboratory evolution of tolerance to dicarboxylic acids in Saccharomyces cerevisiae. Metab. Eng. 56, 130–141 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Liu Q., et al. , Rewiring carbon metabolism in yeast for high level production of aromatic chemicals. Nat. Commun. 10, 4976 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mundhada H., et al. , Increased production of L-serine in Escherichia coli through adaptive laboratory evolution. Metab. Eng. 39, 141–150 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Chen Y., Nielsen J., Biobased organic acids production by metabolically engineered microorganisms. Curr. Opin. Biotechnol. 37, 165–172 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Mans R., et al. , CRISPR/Cas9: A molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae. FEMS Yeast Res. 15, fov004 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verduyn C., Postma E., Scheffers W. A., Van Dijken J. P., Effect of benzoic acid on metabolic fluxes in yeasts: A continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 8, 501–517 (1992). [DOI] [PubMed] [Google Scholar]
- 37.Sandberg T. E., et al. , Evolution of Escherichia coli to 42 °C and subsequent genetic engineering reveals adaptive mechanisms and novel mutations. Mol. Biol. Evol. 31, 2647–2662 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deatherage D. E., Barrick J. E., Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 1151, 165–188 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jenjaroenpun P., et al. , Complete genomic and transcriptional landscape analysis using third-generation sequencing: A case study of Saccharomyces cerevisiae CEN.PK113-7D. Nucleic Acids Res. 46, e38 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.