Significance
Fatty liver, which is an initial step in the development of more severe complications such as liver cirrhosis, is prevalent worldwide in our society. This study demonstrates that stimulation of soluble guanylate cyclase (sGC), an enzyme producing the second messenger cGMP, protects against the most common features of fatty liver, namely inflammation and fibrosis, in animal models of the disease. Our study also provides an explanation for this protection and describes how sGC stimulation blocks the inflammasome (a protein complex responsible for the production of the potent proinflammatory cytokine interleukin-1β) in liver macrophages. The results of this study support the investigation of sGC stimulators, which are already approved for treatment in other conditions, in patients with fatty liver disease.
Keywords: liver, inflammation, Kupffer cells, soluble guanylate cyclase
Abstract
Soluble guanylate cyclase (sGC) catalyzes the conversion of guanosine triphosphate into cyclic guanosine-3′,5′-monophosphate, a key second messenger in cell signaling and tissue homeostasis. It was recently demonstrated that sGC stimulation is associated with a marked antiinflammatory effect in the liver of mice with experimental nonalcoholic steatohepatitis (NASH). Here, we investigated the mechanisms underlying the antiinflammatory effect of the sGC stimulator praliciguat (PRL) in the liver. Therapeutic administration of PRL exerted antiinflammatory and antifibrotic actions in mice with choline-deficient l-amino acid-defined high-fat diet-induced NASH. The PRL antiinflammatory effect was associated with lower F4/80- and CX3CR1-positive macrophage infiltration into the liver in parallel with lower Ly6CHigh- and higher Ly6CLow-expressing monocytes in peripheral circulation. The PRL antiinflammatory effect was also associated with suppression of hepatic levels of interleukin (IL)-1β, NLPR3 (NACHT, LRR, and PYD domain-containing protein 3), ASC (apoptosis-associated speck-like protein containing a caspase-recruitment domain), and active cleaved-caspase-1, which are components of the NLRP3 inflammasome. In Kupffer cells challenged with the classical inflammasome model of lipopolysaccharide plus adenosine triphosphate, PRL inhibited the priming (expression of Il1b and Nlrp3) and blocked the release of mature IL-1β. Mechanistically, PRL induced the protein kinase G (PKG)-mediated phosphorylation of the VASP (vasodilator-stimulated phosphoprotein) Ser239 residue which, in turn, reduced nuclear factor-κB (NF-κB) activity and Il1b and Nlrp3 gene transcription. PRL also reduced active cleaved-caspase-1 levels independent of pannexin-1 activity. These data indicate that sGC stimulation with PRL exerts antiinflammatory actions in the liver through mechanisms related to a PKG/VASP/NF-κB/NLRP3 inflammasome circuit.
Nonalcoholic fatty liver disease (NAFLD) is a prevalent condition affecting roughly 25% of the worldwide population (1). NAFLD is characterized by macrovesicular hepatic steatosis and its more aggressive form, nonalcoholic steatohepatitis (NASH), combines steatosis with inflammation and fibrosis (2). Although many molecular pathways contribute to the development of NASH and the mechanisms leading to the disease are highly heterogeneous, inflammation appears to play a crucial role in its progression (1, 2). Moreover, a consistent target and a satisfactory and effective therapy for this clinical entity have so far not been achieved.
Recently, we and others have demonstrated that modulation of cyclic guanosine-3′,5′-monophosphate (cGMP) exerts antiinflammatory and antifibrogenic effects in models of NASH and reduces portal pressure and fibrogenesis in cirrhotic rats (3–5). In these studies, small molecules with the ability to stimulate soluble guanylate cyclase (sGC), an enzyme that catalyzes the conversion of guanosine triphosphate (GTP) to cGMP, proved to be efficacious in the prevention as well as in the treatment of hepatic inflammation and fibrosis (3–5). In particular, using an optimized experimental model of NASH induced by a choline-deficient l-amino acid-defined high-fat diet (CDAHFD) (6), we recently demonstrated that administration of the sGC stimulator praliciguat (PRL) delayed, in a dose-dependent manner, the development of liver inflammation and fibrosis (3). In addition, Schwabl et al., using another sGC stimulator (riociguat), described reductions in portal hypertension and liver fibrosis in cholestatic (bile duct ligation) and toxic (carbon tetrachloride; CCl4) models of cirrhosis in rats (4). More recently, Hall et al. confirmed the antiinflammatory and antifibrotic effects of PRL in different murine models of NASH, including CCl4, streptozotocin plus a high-fat/high-cholesterol diet, and thioacetamide (5). Together, the findings of these studies position the cGMP pathway as a new and promising therapeutic target for the pharmacological modulation of the inflammatory and fibrogenic processes leading to NASH. At present, the sGC stimulator riociguat has approval as treatment for pulmonary arterial hypertension and chronic thromboembolic pulmonary hypertension (7, 8).
There is very limited information on the precise mechanisms that mediate the liver-protective properties of sGC stimulators. Hall et al. recently reported that the antifibrotic effects of PRL in the liver can be ascribed to direct actions of this drug on hepatic stellate cells (HSCs) (5). In fact, these authors described in HSCs in culture that PRL has the ability to antagonize the fibrogenic properties of transforming growth factor-β potentially through interacting with adenosine monophosphate-activated protein kinase. and SMAD7 signaling (5). On the other hand, little is known about the mechanistic aspects linking PRL with its antiinflammatory actions in the liver. The current study was undertaken to comprehensively delineate the mechanisms underlying the antiinflammatory effects of PRL in the liver and included experiments in vivo, in mice with CDAHFD-induced NASH, and in vitro, in circulating monocytes, resident macrophages, Kupffer cells, hepatocytes, and HSCs.
Results
Effects of sGC Stimulation on Whole Body, Organ Weight, and Serum Biochemistry.
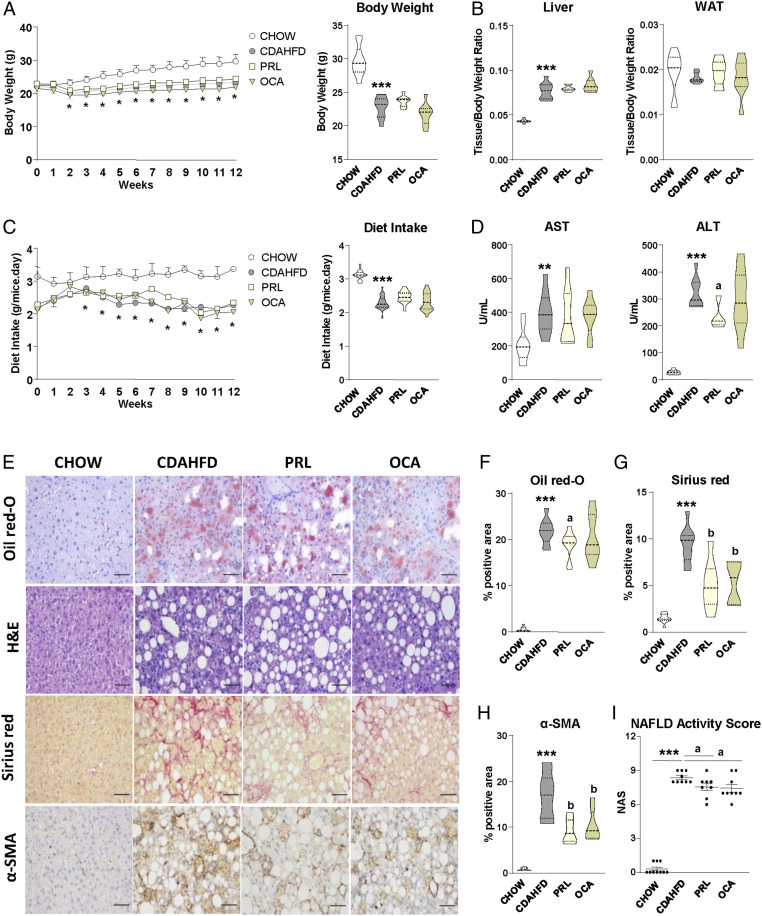
Compared with chow-fed mice, mice with CDAHFD-induced NASH gained less weight across the study and had a higher liver-to-body weight ratio and lower food intake (Fig. 1 A–C). There were no changes in white adipose tissue (WAT)-to-body weight ratio (Fig. 1B). Mice with CDAHFD-induced NASH also had lower brown adipose tissue (BAT)-to-body weight and higher spleen-to-body weight ratios (SI Appendix, Fig. S1A). Mice with CDAHFD-induced NASH exhibited higher serum aspartate aminotransferase (AST) and alanine aminotransferase (ALT) levels (Fig. 1D) together with lower serum glucose, total cholesterol, and triglyceride (TAG) levels (SI Appendix, Fig. S1B). The therapeutic administration of the sGC stimulator PRL to mice with CDAHFD-induced NASH induced minor changes in the above-described parameters, producing only a statistically significant decrease in serum ALT levels (Fig. 1D). This reduction in ALT levels was not seen with the administration of the farnesoid X receptor (FXR) agonist obeticholic acid (OCA), which only produced a significant reduction in serum TAG levels (SI Appendix, Fig. S1B). The sGC stimulator PRL had a greater concentration in the liver (1,324 ± 355 nM) (mean ± SD) as compared with serum (27.3 ± 12.3 nM).
Fig. 1.
Antisteatotic, antiinflammatory, and antifibrotic effects of sGC stimulation with praliciguat in mice with CDAHFD-induced NASH. (A, Left) Body weight changes during the 12 wk of treatment in mice receiving either chow diet (n = 10), CDAHFD (n = 10), CDAHFD plus PRL at 3 mg/kg (n = 10), or CDAHFD plus the FXR agonist obeticholic acid at 15 mg/kg (n = 10). (A, Right) Body weight at week 12. (B) Liver and white adipose tissue weights, expressed as tissue/body weight ratio. (C, Left) Diet intake changes during the 12 wk of treatment in the different groups of mice. (C, Right) Mean diet intake expressed in grams per mouse per day. (D) Serum AST and ALT levels. (E) Representative photomicrographs (magnification 200×) of liver sections stained with oil red O, H&E, sirius red, and specific α-SMA antibody. (Scale bars, 50 μm.) (F–H) Histomorphometric analysis of the area stained with oil red O, sirius red, and α-SMA. (I) NAFLD activity score calculated from the inflammation, steatosis, and fibrosis values as scored by a registered pathologist. Results are expressed as mean ± SEM. *P < 0.05; **P < 0.005 and ***P < 0.001 vs. chow; aP < 0.05 and bP < 0.001 vs. CDAHFD.
Effects of sGC Stimulation on Hepatic Steatosis, Inflammation, and Fibrosis.
As expected, mice with CDAHFD-induced NASH showed marked hepatic steatosis, inflammation, and fibrosis as compared with chow-fed mice, as revealed by histological (oil red O, hematoxylin and eosin [H&E], and sirius red) and immunohistochemical (α-smooth muscle actin; α-SMA) staining (Fig. 1 E–H). In addition, mice with NASH had a higher NAFLD activity score (NAS), which combines the values of steatosis, inflammation, and fibrosis (Fig. 1I). The administration of PRL to mice with CDAHFD-induced NASH was associated with a significant amelioration of liver injury as demonstrated by significantly lower oil red O, sirius red, and α-SMA staining and NAS, which includes H&E staining in its calculation (Fig. 1 E–I). Mice with CDAHFD-induced NASH receiving OCA showed similar changes in inflammation and fibrosis but not in hepatic steatosis (Fig. 1 E–I).
Effects of sGC Stimulation on Hepatic Gene Expression.
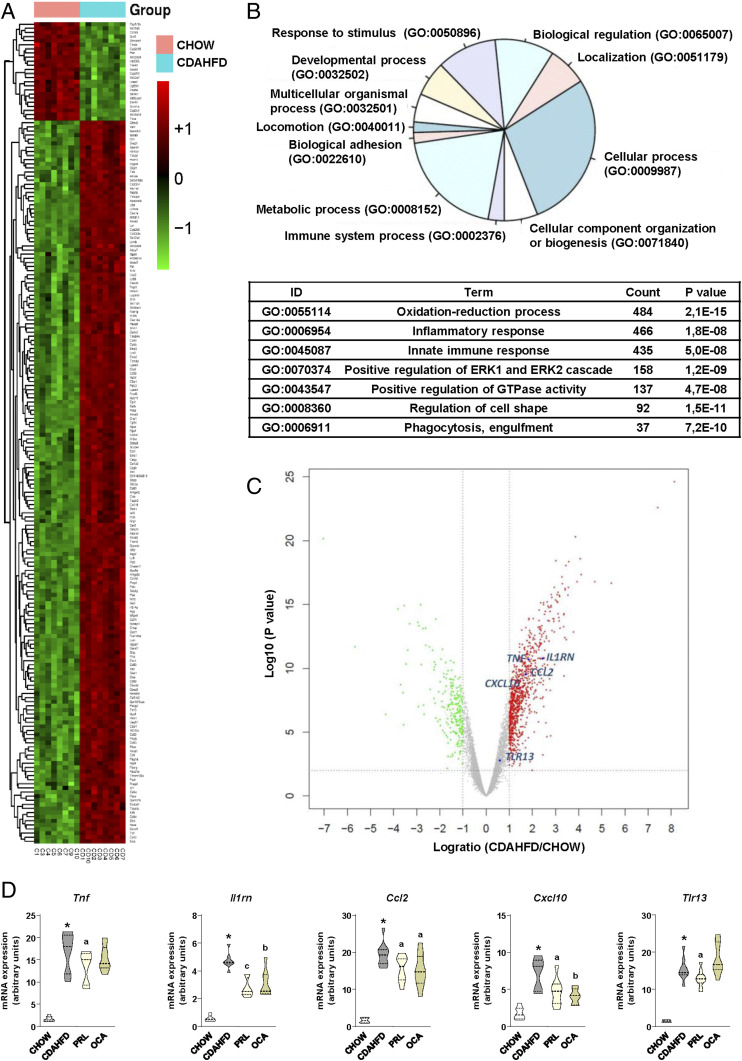
Since the liver transcriptome in the NASH model induced by CDAHFD is poorly characterized, we performed a microarray comparison between these mice and control chow mice. Fig. 2A shows a heatmap of the dataset including the 200 genes most differentially regulated, which reveals profound changes in the liver transcriptome of CDAHFD mice. To reduce the dimension of this dataset, we performed functional Gene Ontology (GO) enrichment analysis and identified that a number of cellular processes were modified during the development of NASH (Fig. 2B). A deeper insight into this analysis identified that, in addition to the oxidation–reduction process, inflammation and innate immune response were two of the top differentially regulated processes in the liver of mice with CDAHFD-induced NASH, with an aggregate count of 901 genes listed with a highly significant P value (Fig. 2B). To further reduce the dimension of this analysis, we focused on genes related to inflammation, and among them we selected a number of representative genes that were up-regulated and evenly distributed across the volcano plot (Fig. 2C). According to these criteria, we assessed the effects of sGC stimulation on the expression of Tnf, Il1rn, Ccl2, Cxcl10, and Tlr13 by real-time PCR. As shown in Fig. 2D, administration of PRL to mice with CDAHFD-induced NASH exerted a generalized inhibition of the expression of genes coding for cytokines (i.e., Tnf and Il1rn), chemokines (i.e., Ccl2 and Cxcl10), and receptors that trigger the innate immune response such as Tlr13. The effects of OCA on the expression of these inflammatory genes were milder and only affected the transcription levels of Il1rn, Ccl2, and Cxcl10 (Fig. 2D), suggesting that sGC stimulation has a broader and more profound impact on the hepatic inflammatory gene transcriptome.
Fig. 2.
Analysis of the liver transcriptome of CDAHFD-fed mice. Effects of PRL on the expression of inflammatory genes. (A) Heatmap showing the top 200 differentially regulated genes ranked by fold change (up-regulated, red; down-regulated, green) in the liver from CDAHFD-fed mice (n = 8) in comparison with chow-fed mice (n = 8). (B, Upper) Gene Ontology database (http://www.geneontology.org) functional classification of GO enrichment in liver of CDAHFD-fed mice in comparison with chow. (B, Lower) List of top GO terms in the database ranked by count and P value. (C) Volcano plot representing a number of selected genes up-regulated in CDAHFD and evenly distributed in the plot. (D) Expression of Tnf, Il1rn, Ccl2, Cxcl10, and Tlr13 in liver samples from the animals included in the different groups of the study as determined by real-time PCR. Results are expressed as mean ± SEM. *P < 0.001 vs. chow; aP < 0.05, bP < 0.005, and cP < 0.001 vs. CDAHFD.
Effects of sGC Stimulation on Liver Innate Immune Cells.
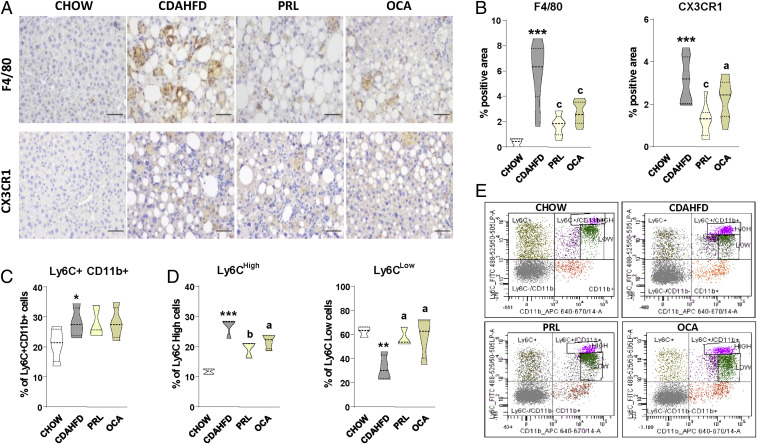
Since cytokines and chemokines as well as other inflammatory mediators are mainly produced by innate immune cells, we next explored the effects of sGC stimulation on liver macrophages, the principal immune cell type contributing to NASH in the CDAHFD model. Livers from mice with CDAHFD-induced NASH had a more intense F4/80 staining, indicating a higher macrophage abundance (Fig. 3 A, Upper). Liver sections from these mice also showed a more extensive staining for C-X3-C motif chemokine receptor 1 (CX3CR1), which is a marker of infiltrating macrophages and monitors the recruitment of monocytes into tissues (Fig. 3 A, Lower). The quantitative morphometric assessment of these parameters is shown in Fig. 3B. Administration of PRL to mice with CDAHFD-induced NASH was associated with significantly lower levels of F4/80 signal and a lower percentage of the area positive for CX3CR1 (Fig. 3 A and B). Changes in CX3CR1 indicated reduced infiltrating monocyte-derived macrophages into the liver and were consistent with the observation that although mice with CDAHFD-induced NASH showed a higher number of circulating monocytes (Ly6C+CD11b+) (Fig. 3C), those receiving PRL had a lower percentage of monocytes with high expression of Ly6C (Ly6CHigh) and a larger proportion of these cells with low expression of this marker (Ly6CLow) (Fig. 3 D and E). This phenotypic switch is characteristic of “proresolutive/patrolling” monocytes with less capacity to infiltrate tissues (9), thus indicating that PRL administration could also limit the infiltration of inflammatory cells into the injured liver. Of interest, in the microarray dataset, Ly6c2 and Cx3cr1 were in the list of genes with strong correlations with the inflammatory process (r = 0.82 and r = 0.74, respectively) in mice with CDAHFD-induced NASH (SI Appendix, Fig. S2A). The effects of PRL on the monocyte/macrophage phenotype appeared not to be exclusive because similar effects were seen with OCA administration (Fig. 3).
Fig. 3.
Effects of PRL on liver and peripheral blood monocytes. (A) Representative photomicrographs (magnification 200×) of liver sections stained with specific F4/80 and CX3CR1 antibodies used for the assessment of total liver macrophages and infiltrated liver macrophages, respectively. (Scale bars, 50 μm.) (B) Histomorphometric analysis of the area stained with F4/80 and CX3CR1 antibodies. (C) Percentage of total monocytes in the peripheral blood of mice determined by flow cytometry (n = 4 for each group). (D) Percentage of Ly6CHigh and Ly6CLow monocytes among the Ly6C+Cd11b+ cells. (E) Representative flow cytometry plots for Ly6C-expressing monocytes. Results are expressed as mean ± SEM. *P < 0.05; **P < 0.005 and ***P < 0.001 vs. chow; aP < 0.05, bP < 0.005, and cP < 0.001 vs. CDAHFD.
The Antiinflammatory Actions of sGC Stimulation Are Associated with Inhibition of the NLR Protein 3 Inflammasome in Liver Macrophages.
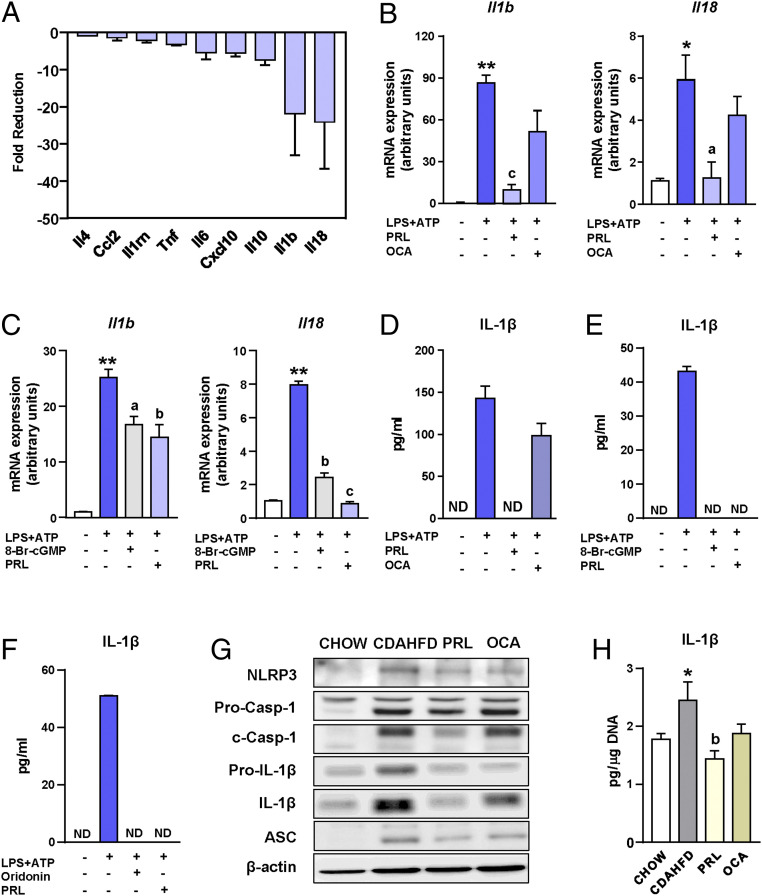
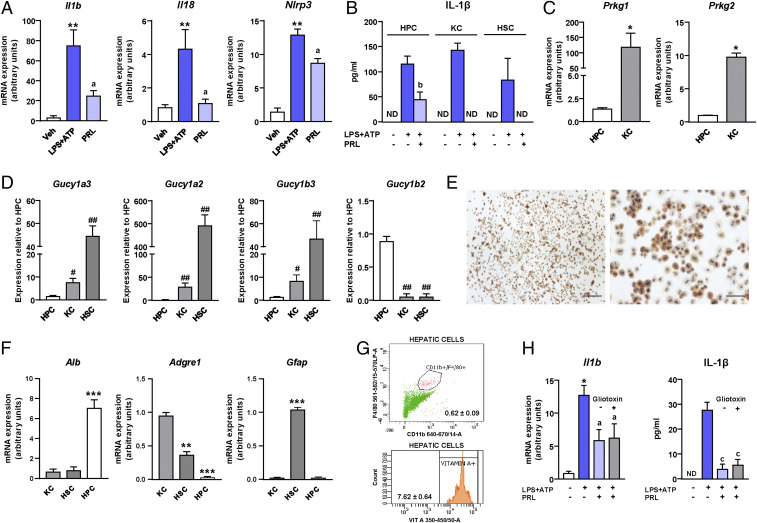
To investigate whether the antiinflammatory effects of sGC stimulation could be ascribed to the inhibition of the release of cytokines by liver macrophages, we next assessed the in vitro effects of PRL on Kupffer cells, the liver resident macrophages. In these experiments, Kupffer cells were challenged with the common proinflammatory stimulus lipopolysaccharide (LPS) in the presence or absence of PRL, and changes in the expression of genes coding for cytokines were assessed by real-time PCR. Il18 and Il1b, coding for interleukin (IL)-18 and IL-1β, respectively, were the two genes with the largest fold reduction induced by PRL in Kupffer cells (Fig. 4A). Given that IL-1β and IL-18 are both processed by the NLRP3 (NACHT, LRR, and PYD domain-containing protein 3) inflammasome (10–13), these findings suggested that PRL might exert regulatory actions on the macrophage inflammasome system. In the microarray dataset, four genes (i.e., Pycard, Casp1, P2rx7, and Aim2) coding for inflammasome components were also in the list of genes with strong correlations with the inflammatory process (between r = 0.79 and r = 0.62) and all of them were up-regulated in mice with CDAHFD-induced NASH (SI Appendix, Fig. S2).
Fig. 4.
Antiinflammatory actions of PRL are associated with inhibition of the NLRP3 inflammasome. (A) Fold reduction in the expression of inflammatory genes induced by the addition of PRL to incubations of Kupffer cells exposed to LPS. (B) Il1b and Il18 mRNA expression in isolated Kupffer cells challenged with LPS + ATP and treated with PRL or OCA. (C) Il1b and Il18 mRNA expression in isolated Kupffer cells challenged with LPS + ATP and treated with PRL or the cGMP analog 8-Br-cGMP. (D) IL-1β levels in supernatants of Kupffer cells from B as determined by ELISA. (E) IL-1β levels in supernatants of Kupffer cells from C. (F) IL-1β levels in supernatants of Kupffer cells challenged with LPS + ATP and treated with PRL or the NLRP3 inflammasome inhibitor oridonin. (G) NLRP3, pro-caspase-1, cleaved-caspase-1, pro-IL-1β, IL-1β, ASC, and β-actin protein levels in liver tissue from the mice included in the four groups of the study as determined by SDS/PAGE Western blot. (H) IL-1β levels in the liver relative to the DNA content. Results are expressed as mean ± SEM of n = 4 separate experiments. *P < 0.05 and **P < 0.001 vs. chow or vehicle; aP < 0.05, bP < 0.005, and cP < 0.001 vs. CDAHFD or LPS + ATP. ND, not detectable.
To test whether PRL can modulate the macrophage inflammasome, we then exposed Kupffer cells to LPS plus adenosine triphosphate (ATP), which is the classical NLRP3 inflammasome activation model. Messenger RNA (mRNA) expression of Il1b and Il18 was robustly stimulated by incubation of Kupffer cells with LPS and ATP, a response that was significantly inhibited in the presence of PRL (Fig. 4B). Under these conditions, the cGMP analog 8-Br-cGMP exerted similar inhibitory actions to those of PRL (Fig. 4C). Consistent with this, the release of mature IL-1β protein was increased by LPS and ATP and completely blocked by PRL, as the levels of this cytokine were undetectable in the supernatants of PRL-treated Kupffer cells (Fig. 4D). In contrast, OCA was ineffective in inhibiting Il1b and Il18 expression (Fig. 4B) and in blocking the secretion of mature IL-1β (Fig. 4D). PRL inhibition of LPS+ATP-induced mature IL-1β secretion was similar to that of 8-Br-cGMP (Fig. 4E). The effects of PRL on the expression of the inflammasome components and the release of mature IL-1β were also seen in peritoneal macrophages exposed to LPS plus ATP (SI Appendix, Fig. S3A), indicating that the inhibitory actions of PRL on the inflammasome are not exclusive to liver macrophages. Of note, PRL appeared to also affect inflammasome assembly, since this sGC stimulator exerted a comparable inhibitory effect on IL-1β secretion to that of oridonin, a specific blocker of NLRP3 inflammasome assembly (Fig. 4F).
To translate these findings to in vivo conditions, we next assessed the expression at the protein level of the inflammasome components NLRP3, pro-caspase-1 (Pro-Casp-1), cleaved-caspase-1 (c-Casp-1), pro-IL-1β, IL-1β (mature), and ASC (apoptosis-associated speck-like protein containing a caspase-recruitment domain) in livers of mice with CDAHFD-induced NASH treated with PRL. As shown in Fig. 4G, mice with CDAHFD-induced NASH had increased protein levels of the inflammasome components in the liver, while mice treated with PRL did not. Densitometric analysis of the bands is shown in SI Appendix, Fig. S3B. Importantly, PRL decreased the protein levels of the active form of caspase-1 (c-Casp-1), which is cleaved after the assembly of the inflammasome (Fig. 4G), supporting the finding with oridonin and the view that PRL not only inhibits priming but also might interfere with inflammasome assembly. Reduction of the hepatic levels of IL-1β in mice with CDAHFD-induced NASH receiving PRL further solidified these findings (Fig. 4H). Similar to that observed in vitro, OCA treatment in mice with CDAHFD-induced NASH did not modify the protein levels of the inflammasome components, especially those that are critical for its functioning such as active c-Casp-1 and mature IL-1β (Fig. 4 G and H and SI Appendix, Fig. S3B). The lack of effects of OCA on the NLRP3 inflammasome was not due to improper activation of FXR since OCA induced the expression of Abcb11, a bona fide FXR target gene that codes for the bile salt export pump (SI Appendix, Fig. S4).
Modulation of the NLRP3 Inflammasome by PRL in Kupffer Cells Is Mediated by VASP Phosphorylation and NF-κB Inhibition.
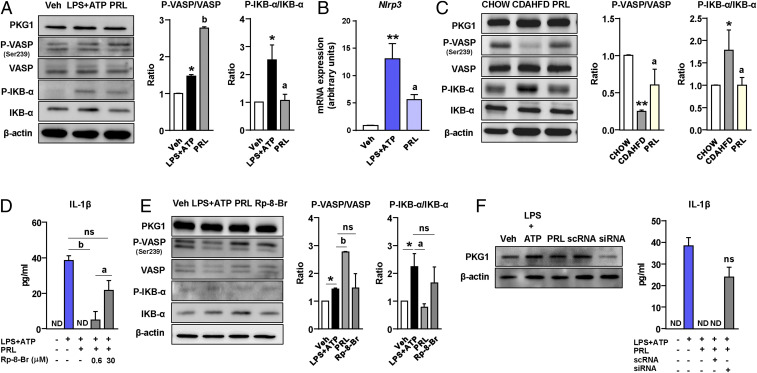
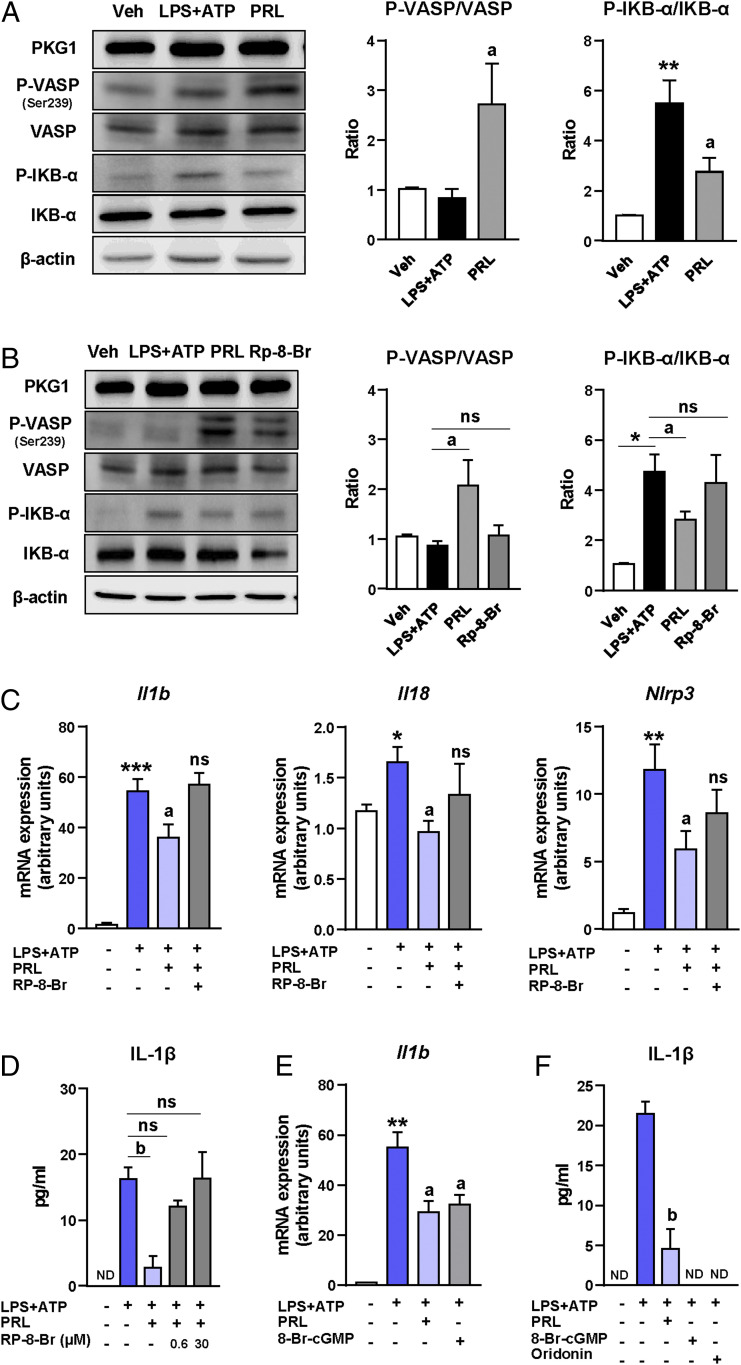
To investigate the mechanisms by which PRL modulates the macrophage NLRP3 inflammasome, we dissected the potential pathways linking cGMP signaling with the priming (first hit) and assembly or activation (second hit) of the inflammasome complex in Kupffer cells. We first explored the canonical cGMP signal transduction pathway implicating protein kinase G1 (PKG1) and the phosphorylation of its downstream effector vasodilator-stimulated phosphoprotein (VASP). As shown in Fig. 5A, incubation of Kupffer cells with PRL during the activation of the inflammasome with LPS + ATP was associated with no changes in PKG1 protein levels but increased VASP Ser239 phosphorylation and decreased LPS+ATP-induced NF-κB inhibitor alpha (IKB-α) phosphorylation, indicative of reduced nuclear factor-κB (NF-κB) activity. Since NF-κB is a major driver of inflammatory gene expression, inhibition of NF-κB activity by PRL was consistent with reductions in the expression of downstream target genes such as Nlrp3 (Fig. 5B) and Il1b and Il18 (Fig. 4B). To further support our findings on VASP and IKB-α phosphorylation, experiments were repeated and reproduced in Kupffer cells stimulated with LPS for a shorter time (30 min) (SI Appendix, Fig. S5A). The phosphorylation of VASP and inhibition of NF-κB in response to PRL were also clearly confirmed in vivo as VASP phosphorylation was higher and IKB-α phosphorylation was lower in livers from mice with CDAHFD-induced NASH receiving PRL than in livers from CDAHFD controls (Fig. 5C). Although we did not observe changes in the protein levels of PKG1, the involvement of this kinase in PRL inhibition of the NLRP3 inflammasome-mediated IL-1β secretion was confirmed by performing concentration–response experiments with the PKG-blocking agent Rp-8-Br-PET-cGMPS (Rp-8-Br) in which efficacy was observed at 30 μM concentration (Fig. 5D). Furthermore, blockage of PKG with Rp-8-Br prevented PRL induction of VASP phosphorylation and reduction of IKB-α phosphorylation, confirming that PRL actions were mediated by this kinase (Fig. 5E and SI Appendix, Fig. S5A). To further confirm the involvement of PKG in PRL actions, we performed knockdown experiments using a small interfering RNA (siRNA) directed against Prkg1, the gene encoding for PKG1. As shown in Fig. 5F, protein expression for PKG1 was significantly reduced 72 h after siRNA transfection, an effect that was associated with the loss of the ability of PRL to inhibit LPS+ATP-induced IL-1β secretion.
Fig. 5.
PRL blocks the NLRP3 inflammasome in Kupffer cells through mechanisms based on VASP phosphorylation and NF-κB inhibition. (A, Left) PKG1, phospho-VASP (Ser239) (P-VASP), VASP, phospho-IKB-α (P-IKB-α), IKB-α, and β-actin protein levels in Kupffer cells challenged with LPS + ATP alone or in the presence of PRL. (A, Right) Densitometric analysis of the P-VASP/VASP and P-IKB-α/IKB-α ratios. (B) Nlrp3 mRNA expression in Kupffer cells incubated in the conditions described in A. (C, Left) PKG1, P-VASP (Ser239), VASP, P-IKB-α, IKB-α, and β-actin protein levels in liver tissue from the mice included in the four groups of the study. (C, Right) Densitometric analysis of the P-VASP/VASP and P-IKB-α/IKB-α ratios. (D) Levels of mature IL-1β in supernatants from Kupffer cells challenged with LPS + ATP and treated with PRL in the presence or absence of the PKG-blocking agent Rp-8-Br at 0.6 and 30 μM concentrations. (E, Left) PKG1, P-VASP (Ser239), VASP, P-IKB-α, IKB-α ,and β-actin protein levels in Kupffer cells challenged with LPS + ATP and treated with PRL in the presence or absence of Rp-8-Br. (E, Right) Densitometric analysis of the P-VASP/VASP and P-IKB-α/IKB-α ratios. (F, Left) PKG1 and β-actin protein levels in Kupffer cells incubated with vehicle (Veh) and LPS + ATP alone or in combination with PRL, PRL plus scrambled nontargeting RNA (scRNA), and PRL plus siRNA targeting the Prkg1 gene. (F, Right) IL-1β levels in supernatants of Kupffer cells incubated under these conditions. Results are expressed as mean ± SEM of n = 3 or 4 separate experiments. *P < 0.05 and **P < 0.001 vs. chow or vehicle; aP < 0.05 and bP < 0.005 vs. CDAHFD or LPS + ATP. ns, not statistically significant vs. LPS + ATP.
We next explored mechanisms by which PRL might interfere with the second hit leading to the oligomerization of the inactive inflammasome complex (NLRP3, ASC, and Casp-1) and the processing of mature IL-1β. A potential mechanism that could link the cGMP pathway to inhibition of the NLRP3 inflammasome is pannexin-1, a hemichannel protein that interacts with the ATP receptor P2X7 to allow inflammasome assembly during LPS + ATP challenge (14–16). To explore this possibility, we incubated Kupffer cells with LPS + ATP and the selective pannexin-1 mimetic inhibitory peptide 10Panx1 or its control scrambled peptide. The presence of 10Panx1 in the Kupffer cell media did not modify the inhibitory actions of PRL on the expression of Il1b, Il18, and Nlrp3 (SI Appendix, Fig. S5 B–D) as well as on the secretion of mature IL-1β (SI Appendix, Fig. S5E), excluding the involvement of pannexin-1 in PRL inhibition of the NLRP3 inflammasome.
Participation of Other Liver Cell Types in Inflammasome-Mediated Release of IL-1β.
Hepatocytes are also able to produce mature IL-1β through the NLRP3 inflammasome complex (17), especially when these cells are exposed to inflammatory stimuli such as LPS (18). Indeed, we were able to induce the expression of Il1b, Il18, and Nlrp3 (Fig. 6A) and to trigger the secretion of mature IL-1β (Fig. 6B) by incubating murine hepatocytes with LPS plus ATP. On a per-cell basis, the release of mature IL-1β by hepatocytes was comparable with that of HSCs and lower than its release by Kupffer cells (Fig. 6B). In hepatocytes, PRL also down-regulated the expression of Il1b, Il18, and Nlrp3 (Fig. 6A) and reduced IL-1β secretion (Fig. 6B), although its inhibitory effect was more modest than in Kupffer cells, in which PRL completely suppressed LPS+ATP-induced IL-1β secretion. Of interest, the expression of Prkg1 and Prkg2, the genes encoding for PKG1 and PKG2, was marginal in hepatocytes as compared with Kupffer cells (Fig. 6C).
Fig. 6.
Participation of other liver cell types in inflammasome-mediated release of IL-1β. (A) Il1b, Il18, and Nlrp3 mRNA expression in isolated hepatocytes exposed to LPS + ATP and treated with PRL. (B) Comparison of the IL-1β levels in the supernatants of hepatocytes (HPCs), Kupffer cells (KCs), and hepatic stellate cells challenged with LPS + ATP and treated with PRL. (C) Expression of Prkg1 and Prkg2 in resting HPCs and KCs. (D) Expression of Gucy1a3, Gucy1a2, Gucy1b3, and Gucy1b2 in resting HPCs, KCs, and HSCs. (E) Representative photomicrographs of isolated HPCs stained with a specific GUCY1B2 antibody. (E, Left) Magnification 40×. (Scale bar, 200 μm.) (E, Right) Magnification 200×. (Scale bar, 50 μm.) (F) Expression of Alb, Adgre1, and Gfap in KCs, HSCs, and HPCs. (G) Representative flow cytometry plots for KCs (F4/80+Cd11b+; 0.62 ± 0.09% of total cells) (Upper) and HSCs (vitamin A+; 7.62 ± 0.64% of total cells) (Lower) present in HPC isolations. (H) Il1b mRNA expression and levels of mature IL-1β in the supernatants of HPCs challenged with LPS + ATP and treated with PRL in the presence or absence of gliotoxin. Results are expressed as mean ± SEM of n = 3 or 4 separate experiments. *P < 0.05; **P < 0.005 and ***P < 0.001 vs. vehicle; aP < 0.05 and bP < 0.005 vs. LPS + ATP; #P < 0.005 and ##P < 0.001 vs. HPCs.
We next assessed the expression in hepatocytes of Gucy1a3, Gucy1a2, Gucy1b3, and Gucy1b2, which encode for the soluble guanylate cyclase α1, α2, β1, and β2 subunits, respectively. As shown in Fig. 6D, as compared with Kupffer cells and HSCs, expression of Gucy1a3, Gucy1a2, and Gucy1b3 was marginal in hepatocytes, whereas the expression of Gucy1b2, encoding for the β2-subunit of sGC, which has been reported to produce cGMP in its homomeric configuration (19), was detected in hepatocytes but not in Kupffer cells or HSCs (Fig. 6D). Interestingly, PRL stimulation of Kupffer cells and HSCs resulted in the up-regulation of the α1-, α2-, and β1-subunits (i.e., Gucy1a3, Gucy1a2, and Gucy1b3) whereas the β2-subunit (i.e., Gucy1b2) was the only isoform up-regulated during the incubation of hepatocytes with PRL (SI Appendix, Fig. S6A). The expression of Gucy1b2 in hepatocytes was confirmed at the protein level by immunocytochemistry (see the positive signal for binucleated cells in Fig. 6E). To confirm that the effect of PRL on IL-1β secretion was not reflecting the presence of contaminating Kupffer cells and/or HSCs in the hepatocyte cultures, we tested the expression of specific markers for the different liver cell types, as previously described by others (17). As shown in Fig. 6F, cultures of hepatocytes predominantly expressed the albumin gene (Alb) and were virtually negative for the expression of Adgre1 (coding for F4/80, a specific macrophage marker) and Gfap (a specific marker for HSCs). The percentage of contaminating Kupffer cells and HSCs in hepatocyte cultures was quantitated by flow cytometry, resulting in less than 0.7% of Kupffer cells and 7.6% of vitamin A-positive cells (mostly HSCs) (Fig. 6G). To further minimize the involvement of contaminating HSCs in our findings, we incubated hepatocytes with gliotoxin, which specifically depletes HSCs without affecting hepatocyte viability (20). As shown in Fig. 6H, the inhibitory effect of PRL on hepatocyte LPS+ATP-induced Il1b expression and IL-1β secretion was not affected by gliotoxin, despite this toxin significantly depleting the HSC population (SI Appendix, Fig. S6B).
NLRP3 Inflammasome Blockade by sGC Stimulation in Hepatocytes Is Also Mediated by VASP Phosphorylation and NF-κB Inhibition.
Similar to that observed in Kupffer cells, PRL induced VASP phosphorylation and markedly reduced IKB-α phosphorylation in hepatocytes challenged with the classical inflammasome activation model of LPS plus ATP (Fig. 7A). These effects were blocked by the PKG blocker Rp-8-Br (Fig. 7B) and were reproduced in hepatocytes stimulated with LPS for a shorter time (30 min) (SI Appendix, Fig. S6C). Consistent with this, Rp-8-Br blocked PRL-induced down-regulation of Il1b, Il18, and Nlrp3 expression (Fig. 7C) and suppressed the PRL-induced reduction of IL-1β secretion by hepatocytes (Fig. 7D). Moreover, the reduction in inflammasome priming was similar in PRL-treated hepatocytes to those treated with the cGMP analog 8-Br-cGMP (Fig. 7E). Finally, the reduction in IL-1β secretion in response to PRL was similar to the reduction exerted by 8-Br-cGMP and oridonin (Fig. 7F).
Fig. 7.
Inhibition of the NLRP3 inflammasome by PRL in hepatocytes is also mediated by VASP phosphorylation and NF-κB inhibition. (A, Left) PKG1, P-VASP (Ser239), VASP, P-IKB-α, IKB-α, and β-actin protein levels in hepatocytes challenged with LPS + ATP and treated with PRL. (A, Right) Densitometric analysis of the P-VASP/VASP and P-IKB-α/IKB-α ratios. (B, Left) PKG1, P-VASP (Ser239), VASP, P-IKB-α, IKB-α, and β-actin protein levels in hepatocytes challenged with LPS + ATP in the presence or absence of the PKG antagonist Rp-8-Br and treated with PRL. (B, Right) Densitometric analysis of the P-VASP/VASP and P-IKB-α/IKB-α ratios. (C) Expression of Il1b, Il18, and Nlrp3 in isolated hepatocytes challenged with LPS + ATP in the presence or absence of Rp-8-Br and treated with PRL. (D) IL-1β levels in the supernatants from hepatocytes incubated with LPS + ATP and treated with PRL in the presence or absence of Rp-8-Br at 0.6 and 30 μM concentrations. (E) Il1b mRNA expression in isolated Kupffer cells challenged with LPS + ATP and treated with PRL or the cGMP analog 8-Br-cGMP. (F) IL-1β levels in the supernatants from Kupffer cells challenged with LPS + ATP and treated with PRL, 8-Br-cGMP, or the NLRP3 inflammasome inhibitor oridonin. Results are expressed as mean ± SEM of n = 3 or 4 separate experiments. *P < 0.05; **P < 0.005 and ***P < 0.001 vs. vehicle; aP < 0.05 and bP < 0.005 vs. LPS + ATP.
Discussion
This study describes a mechanism by which sGC stimulators modulate liver inflammation in an experimental model of NASH: inhibition of NLRP3 inflammasome-mediated IL-1β production. The involvement of IL-1β in the progression of liver injury and evidence that IL-1 receptor-deficient mice are protected from liver damage have previously been demonstrated (21, 22). Evidence that NLRP3 inflammasome activation is required for development of liver inflammation and fibrosis in NASH has also been provided (23). Therefore, our study contributes to the understanding of the antiinflammatory actions associated with the stimulation of sGC with PRL in experimental NASH, which, together with its antisteatotic and antifibrogenic effects, supports further investigation of PRL in the prevention and treatment of chronic liver diseases. Other important aspects of our study are the following: 1) We used a therapeutic approach in which the administration of PRL was initiated after appearance of the first signs of NASH development in mice, extending results from a previous investigation administering PRL in a preventive mode (3); 2) our study directly compared the therapeutic effects of PRL with those of the FXR agonist OCA, the only drug with a good clinical profile in patients with NAFLD (24); and 3) our study comprises a systematic analysis of the expression of the different sGC subunits in individual liver cell types, including liver resident macrophages, hepatocytes, and HSCs.
IL-1β is one of the most potent proinflammatory cytokines and plays a critical role in the so-called cytokine storm, a descriptor of the dramatic consequences of the massive release of cytokines during uncontrolled inflammation (10). Unlike many other cytokines, IL-1β is produced via nonconventional multiprotein complexes called inflammasomes, which are required for the posttranslational processing of an inactive 31-kDa precursor, termed pro-IL-1β, into mature IL-1β (11). Inflammasomes also process the maturation of pro-IL-18 into mature IL-18 (11, 12). The most well characterized inflammasome is the NLRP3 inflammasome, which is assembled when NLRP3 homotypically engages ASC to recruit the inactive zymogen pro-Casp-1 (11–13). Oligomerization of pro-Casp-1 proteins induces their autoproteolytic cleavage into active cleaved-Casp-1, a cysteine-dependent protease that cleaves pro-IL-1β and pro-IL-18 to generate the biologically active inflammatory cytokines IL-1β and IL-18, respectively (11–13). Commonly, the activation of the NLRP3 inflammasome is the result of a two-step process referred to as priming and assembly. The first step is the priming signal triggered by Toll-like receptor (TLR) ligands such as LPS, which leads to the transcriptional up-regulation of pro-IL-1β and NLRP3, whereas the second step implicates the assembly of the inflammasome components (i.e., NLRP3, ASC, and pro-Casp-1), a process that can be triggered by different stimuli such as ATP (11–13). Our data provide evidence that PRL inhibits IL-1β secretion by regulating both processes, thus providing an approach to reversing the adverse effects of IL-1β that is complementary to the use of monoclonal antibodies such as canakinumab, which blocks the binding of IL-1β to its receptor [see results from the CANTOS Trial (25)].
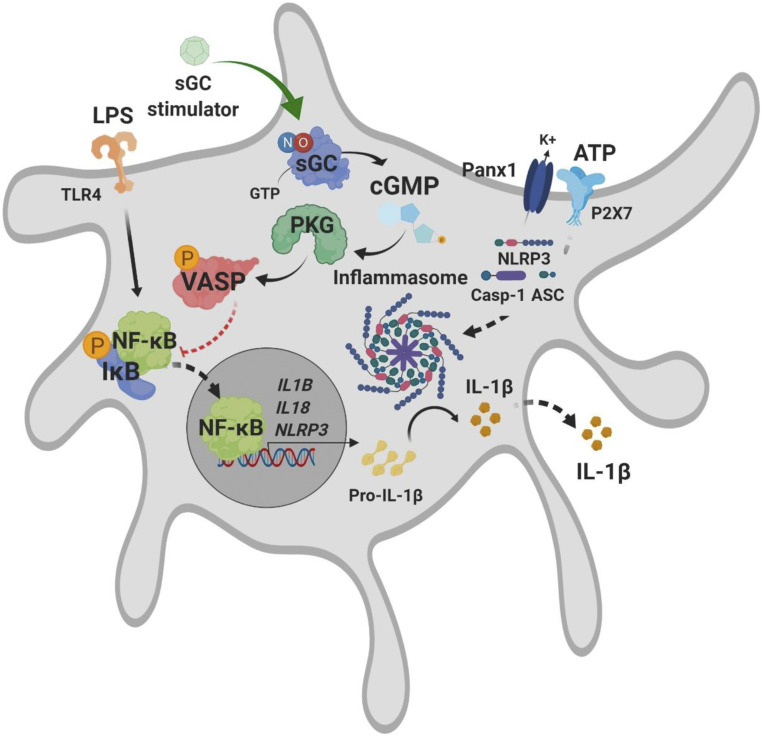
Our study also provides data toward elucidating the mechanism underlying the inhibition of the priming of the hepatic NLRP3 inflammasome by PRL. These findings are schematically illustrated in Fig. 8 and support the view that the down-regulation of the expression of pro-IL-1β and NLRP3 in Kupffer cells by PRL is associated with reduced phospho-IKB-α, and thus with NF-κB inhibition. These results are in agreement with a recent study by Hall et al. reporting a reduction of CCl4-induced hepatic inflammation in rats receiving PRL, an effect that was accompanied by a decrease in the protein levels of p65/NF-κB (5). Of interest, in our study, inhibition of NF-κB by PRL was associated with increased VASP Ser239 phosphorylation, which is a well-stablished downstream target of cGMP-dependent kinases and plays a crucial role in the antiinflammatory signaling of sGC (26, 27). These findings are consistent with those reported by Tateya et al., who demonstrated that Kupffer cells from VASP-deficient mice had increased NF-κB activation and that the cGMP analog 8-Br-cGMP was not able to exhibit antiinflammatory effects in cells lacking VASP expression (27). At present, the connection between VASP phosphorylation and NF-κB inhibition awaits further investigation.
Fig. 8.
Schematic illustration summarizing the potential mechanisms of action of sGC stimulation in Kupffer cells.
PRL produced a marked reduction of the active form of caspase-1 (cleaved-caspase-1), the formation of which requires the assembly of the inflammasome. Of interest, PRL exerted a comparable inhibitory effect on IL-1β secretion with that of oridonin, a specific blocker of the NLRP3 inflammasome assembly. These findings are in agreement with previous work by Kim and collaborators demonstrating that nitric oxide (NO) can suppress caspase-1 cleavage and IL-1β and IL-18 release in macrophages (28). Investigation of the mechanisms underlying the reduction of inflammasome assembly induced by PRL revealed that it was not mediated by pannexin-1, since inhibition of this hemichannel did not modify PRL actions. This finding does not support previous data in human and murine cell lines showing that the blockage of pannexin-1 was associated with inhibition of caspase-1 cleavage and IL-1β processing (15), but are in agreement with data in primary macrophages from pannexin-1 knockout mice, which still have the ability to release IL-1β (29).
Our data indicate that the primary cell target of PRL antiinflammatory effects in the liver is resident (i.e., Kupffer cells) and/or infiltrated macrophages, although hepatocytes might also contribute to PRL actions. This view is supported by the observation that PRL completely suppressed IL-1β secretion in Kupffer cells, whereas its inhibitory effect was more modest in hepatocytes. This difference could be due to the fact that Kupffer cells showed considerably higher expression of Prkg1 (∼100 times) and Prkg2 (∼10 times) than hepatocytes. Since Prkg1 and Prkg2 encode for the two PKG isoforms that effectively mediate cGMP signaling, increased content of PKG likely translates into higher inhibition of the NLRP3 inflammasome by PRL. In any event, hepatocytes are sensitive to inflammatory stimuli such as LPS and the saturated fatty acid palmitic acid, and release IL-1β secondary to NLRP3 inflammasome activation (17, 18). Moreover, these cells have the ability to respond to NO with increased cGMP production as reported by Wood and Ignarro (30) in liver slices and by Billiar et al. (31) in isolated hepatocytes. However, the expression of sGC in hepatocytes remains controversial. Hall et al. previously reported that these cells are negative for sGCα1 and sGCβ1 (5). On the other hand, Gobeil and collaborators demonstrated constitutive localization of sGC in cytoplasmic and nuclear spaces in rat hepatocytes by in situ cryoimmunogold electron microscopy (32). In our study, we detected marginal expression of the genes encoding for the sGCα1-, sGCα2-, and sGCβ1-subunits in hepatocytes but positive expression at both mRNA and protein levels of the sGCβ2-subunit, which according to Koglin et al. is able to produce cGMP in its homomeric configuration (19). One might argue that inhibition of IL-1β secretion by PRL in hepatocytes could reflect the presence of contaminating Kupffer cells or HSCs. However, our hepatocyte cultures contained less than 0.7% of Kupffer cells and about 7.6% of HSCs. Furthermore, depletion of HSCs with gliotoxin in our hepatocyte incubations did not significantly modify the inhibitory actions of PRL on IL-1β secretion. Therefore, we propose that inhibitory effects of PRL on IL-1β secretion by hepatocyte cultures are mediated by direct actions of PRL on these parenchymal liver cells.
In summary, our study provides evidence that the administration of the sGC stimulator PRL to mice with experimentally induced NASH exerts antiinflammatory actions in the liver, mainly in Kupffer cells, through inhibition of NLRP3 inflammasome priming and activation. In liver cells, the inhibitory actions of PRL on NLRP3 inflammasome priming appeared to be mediated by the interaction of cGMP with the PKG/VASP/NF-κB pathway, whereas the effects on NLRP3 inflammasome activation appeared to be mediated through cleaved-caspase-1 inhibition. This study provides insights into the mechanisms of action of sGC stimulators at the experimental level in NASH. Future investigations in patients with NASH to further explore the clinical relevance of our findings are warranted.
Materials and Methods
Animals and Experimental Design.
Male C57BL/6J mice were randomly assigned to four independent groups, which received the following treatments: 1) control chow diet (n = 10) for 12 wk; 2) CDAHFD (60% kcal from fat) (n = 10) for 12 wk; 3) CDAHFD diet for 3 wk and then CDAHFD supplemented with the sGC stimulator PRL (3 mg·kg−1·d−1) (n = 10) for 9 more weeks; and 4) CDAHFD for 3 wk followed by CDAHFD plus the FXR agonist OCA (15 mg·kg−1·d−1) (n = 10) for 9 more weeks. See SI Appendix for more details. All studies were conducted in accordance with the criteria of the Investigation and Ethics Committee of the Universitat de Barcelona and the European Community laws governing the use of experimental animals.
Isolation of Primary Liver Cells.
Hepatocytes, Kupffer cells, and HSCs were isolated from C57BL/6J mice (n = 56) by a three-step in situ collagenase perfusion through the portal vein. After pelleting the hepatocytes, Kupffer cells and HSCs were isolated by centrifugation using 16 and 14% density Nycodenz gradients, respectively (SI Appendix).
Isolation of Peritoneal Macrophages.
Peritoneal exudates from C57BL/6J mice (n = 8) were collected by peritoneal lavage with 7 mL of ice-cold Dulbecco’s phosphate-buffered saline−/− and peritoneal macrophages were isolated by adhesion in a humidified 5% CO2 incubator at 37 °C for 2 h (SI Appendix).
Cell Incubations.
Hepatocytes, Kupffer cells, HSCs, and peritoneal macrophages were seeded in 12-well plates and exposed to LPS (100 ng/mL) for 30 min followed by treatment with either vehicle, PRL (10 µM), or OCA (2 µM) (Selleckchem) for 3 h, 30 min at 37 °C before the addition of ATP (5 mM) for 1 h. For mechanistic experiments, cells were pretreated with the PKG blocker Rp-8-Br (0.6 to 30 µM) (Tocris). In additional experiments, Kupffer cells were incubated with Accell mouse Prkg1 siRNA or scrambled nontargeting RNA (1 µM) (Horizon Discovery) for 72 h followed by LPS + ATP treatment in the presence or absence of PRL. In some experiments, cells were treated with the NLRP3 inflammasome blocker oridonin (2 µM) (Tocris) or the cGMP analog 8-Br-cGMP (50 µM) (Sigma-Aldrich) instead of PRL. In another set of experiments, cells were pretreated with the pannexin-1 inhibitory peptide 10Panx1 trifluoroacetate salt (100 µM) or the control scrambled peptide 10Panx1 trifluoroacetate salt (100 µM), both from Sigma-Aldrich. For the depletion of HSCs in hepatocyte cultures, we pretreated the hepatocytes with gliotoxin (1.5 µM) (Sigma-Aldrich) followed by the LPS + ATP treatment in the presence or absence of PRL.
Biochemical Analyses and Measurement of Cytokine Levels.
Serum concentrations of glucose, total cholesterol, TAG, AST, and ALT were determined by standard laboratory procedures. Levels of IL-1β in liver tissue were assessed in a Luminex 100 System using a custom-made MILLIPLEX MAP Mouse High Sensitivity T Cell Magnetic Bead Panel (Merck Millipore). IL-1β levels in cell supernatants were assessed by enzyme-linked immunosorbent assay (ELISA) (Beckton Dickinson) (SI Appendix).
Liver and Serum Levels of PRL and OCA.
Drug levels in liver tissue and serum samples were analyzed by liquid chromatography-tandem mass spectrometry in positive-ion–mode electrospray ionization (SI Appendix).
Histological Analysis of Liver Steatosis, Inflammation, and Fibrosis.
Steatosis was assessed by oil red O staining of OCT-embedded cryosections, and inflammatory injury was scored by analyzing the amount of inflammatory foci/fields in H&E-stained sections and fibrosis by sirius red staining (SI Appendix).
Immunohistochemistry and Immunocytochemistry.
Immunostainings for F4/80, α-SMA, and CX3CR1 were performed using primary species-specific antibodies developed with diaminobenzidine, visualized at 200×, and quantified by histomorphometry. Hepatocytes seeded on collagen I-coated eight-well Lab-Tek slides (5 × 104 cells per well) were immunostained with primary rabbit anti-mouse GUCY1B2 antibody (LS-C383668; 1:200; LSBio), developed with diaminobenzidine, and counterstained with hematoxylin (SI Appendix).
Flow Cytometry Analysis.
For the analysis of peripheral blood monocytes and Cd45+Cd11b+Ly6C+ cells, Ly6CHigh and Ly6CLow subpopulation blood (100 µL) was extracted from mouse tail vein and a total of 6 × 105 white blood cells were incubated with APC anti-mouse CD11b (Thermo Fisher Scientific), fluorescein isothiocyanate anti-mouse Ly6C (BioLegend), and V450 anti-mouse CD45.2 (Beckton Dickinson) for 20 min at 4 °C in the dark. For the analysis of Kupffer cells in hepatocyte cultures, a total of 106 hepatic cells were incubated with APC anti-mouse CD11b and PE anti-mouse F4/80 (eBioscience) for 20 min at 4 °C in the dark. HSCs were discriminated by vitamin A autofluorescence. Analysis was performed on a BD LSRFortessa cytometer using BD FACSDiva software (SI Appendix).
DNA Extraction and Quantification.
Genomic DNA from liver tissue was isolated using the Omni-Pure Tissue Genomic DNA Purification System (Gene Link) and its concentration was assessed on a NanoDrop 1000 spectrophotometer (NanoDrop Technologies).
Gene Expression Analysis by Real-Time PCR.
Isolation of total RNA was performed using TRIzol reagent (MRC), complementary DNA (cDNA) synthesis was performed using the High-Capacity cDNA Archive Kit (Applied Biosystems), and real-time PCR quantification was performed using validated and predesigned TaqMan Gene Expression Assays on a 7900HT Fast System (Applied Biosystems) (SI Appendix).
Microarray Analysis.
Processing of RNA samples, fragmentation, and labeling of single-stranded cDNA were performed according to the Affymetrix WT PLUS Reagent Kit user guide on an automated system (Beckman FX robot). Following fragmentation and terminal labeling, single-stranded cDNAs were hybridized for 17 h at 45 °C on Clariom S Array Plate Mouse 24, using the automated GeneTitan System. See SI Appendix for details on normalization and analysis of microarray data.
Analysis of Protein Expression by Western Blot.
Extracted total proteins were separated for 90 min at 120 V at 4 °C by 10% sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS/PAGE). Transfer onto polyvinylidene difluoride membranes was performed by the iBlot Dry Blotting System (Invitrogen) and membranes were incubated overnight at 4 °C with primary species-specific antibodies. Bands were visualized using the EZ-ECL Chemiluminescence Detection Kit (Biological Industries) on an LAS 4000 Imaging System (GE Healthcare Life Sciences) and quantified using Image GE ImageQuant TL analysis software. See SI Appendix for more details.
Statistical Analysis.
Statistical analysis was performed with Prism version 8.0 (GraphPad Software). For multiple comparisons, an ANOVA of the data in which a significant (P < 0.05) main effect followed by Dunnett’s post hoc test was performed. For single comparisons, Student’s unpaired t test was used. Results are expressed as mean ± SEM. All measurements were undertaken in at least two technical replicates.
Supplementary Material
Acknowledgments
We thank Albert Salvatella, Ana Isabel Martínez-Puchol, Silvia Sanz, and Monica Romo for their technical help in the completion of the experiments. We are indebted to the Biobanc-Banc de Tumors facility and Flow Cytometry platforms of the IDIBAPS for technical help. This work was supported by grants from the Spanish Ministerio de Ciencia e Innovación (SAF15-63674-R and PID2019-105240RB-I00) under European Regional Development Funds. Our laboratory is a Consolidated Research Group recognized by the Generalitat de Catalunya (2017SGR1449). The CIBERehd is funded by the Instituto de Salud Carlos III. This study was carried out at the Center Esther Koplowitz (IDIBAPS), which is a member of the Centres de Recerca de Catalunya Programme/Generalitat de Catalunya. Our laboratory receives funding from the European Union’s Horizon 2020 research and innovation programme (Grant Agreement 825694).
Footnotes
Competing interest statement: R.S. is an employee of Cyclerion Therapeutics. K.H. and J.L.M. were employees of Ironwood Pharmaceuticals.
This article is a PNAS Direct Submission. T.M. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2000466117/-/DCSupplemental.
Data Availability.
The microarray gene expression data reported in this article have been deposited in the Gene Expression Omnibus (accession no. GSE154892). All study data are included in the article and SI Appendix.
References
- 1.Friedman S. L., Neuschwander-Tetri B. A., Rinella M., Sanyal A. J., Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 24, 908–922 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanyal A. J., Mechanisms of disease: Pathogenesis of nonalcoholic fatty liver disease. Nat. Clin. Pract. Gastroenterol. Hepatol. 2, 46–53 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Flores-Costa R., et al. , The soluble guanylate cyclase stimulator IW-1973 prevents inflammation and fibrosis in experimental non-alcoholic steatohepatitis. Br. J. Pharmacol. 175, 953–967 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwabl P., et al. , The soluble guanylate cyclase stimulator riociguat reduces fibrogenesis and portal pressure in cirrhotic rats. Sci. Rep. 8, 9372 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall K. C., et al. , sGC stimulator praliciguat suppresses stellate cell fibrotic transformation and inhibits fibrosis and inflammation in models of NASH. Proc. Natl. Acad. Sci. U.S.A. 116, 11057–11062 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsumoto M., et al. , An improved mouse model that rapidly develops fibrosis in non-alcoholic steatohepatitis. Int. J. Exp. Pathol. 94, 93–103 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sandner P., Becker-Pelster E. M., Stasch J. P., Discovery and development of sGC stimulators for the treatment of pulmonary hypertension and rare diseases. Biol. Chem. 77, 88–95 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Montani D., et al. , Targeted therapies in pulmonary arterial hypertension. Pharmacol. Ther. 141, 172–191 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Ramachandran P., et al. , Differential Ly-6C expression identifies the recruited macrophage phenotype, which orchestrates the regression of murine liver fibrosis. Proc. Natl. Acad. Sci. U.S.A. 109, E3186–E3195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dinarello C. A., Interleukin-1β and the autoinflammatory diseases. N. Engl. J. Med. 360, 2467–2470 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Lamkanfi M., Dixit V. M., Mechanisms and functions of inflammasomes. Cell 157, 1013–1022 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Guo H., Callaway J. B., Ting J. P. Y., Inflammasomes: Mechanism of action, role in disease, and therapeutics. Nat. Med. 21, 677–687 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schroder K., Tschopp J., The inflammasomes. Cell 140, 821–832 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Poornima V., Vallabhaneni S., Mukhopadhyay M., Bera A. K., Nitric oxide inhibits the pannexin 1 channel through a cGMP-PKG dependent pathway. Biol. Chem. 47, 77–84 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Pelegrin P., Surprenant A., Dynamics of macrophage polarization reveal new mechanism to inhibit IL-1beta release through pyrophosphates. EMBO J. 28, 2114–2127 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanneganti T. D., et al. , Pannexin-1-mediated recognition of bacterial molecules activates the cryopyrin inflammasome independent of Toll-like receptor signaling. Immunity 26, 433–443 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Csak T., et al. , Fatty acid and endotoxin activate inflammasomes in mouse hepatocytes that release danger signals to stimulate immune cells. Hepatology 54, 133–144 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boaru S. G., Borkham-Kamphorst E., Tihaa L., Haas U., Weiskirchen R., Expression analysis of inflammasomes in experimental models of inflammatory and fibrotic liver disease. J. Inflamm. (Lond.) 9, 49 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koglin M., Vehse K., Budaeus L., Scholz H., Behrends S., Nitric oxide activates the β2 subunit of soluble guanylyl cyclase in the absence of a second subunit. J. Biol. Chem. 276, 30737–30743 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Orr J. G., et al. , Mechanism of action of the antifibrogenic compound gliotoxin in rat liver cells. Hepatology 40, 232–242 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Gieling R. G., Wallace K., Han Y. P., Interleukin-1 participates in the progression from liver injury to fibrosis. Am. J. Physiol. Gastrointest. Liver Physiol. 296, G1324–G1331 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gehrke N., et al. , Hepatocyte-specific deletion of IL1-RI attenuates liver injury by blocking IL-1 driven autoinflammation. J. Hepatol. 68, 986–995 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Mridha A. R., et al. , NLRP3 inflammasome blockade reduces liver inflammation and fibrosis in experimental NASH in mice. J. Hepatol. 66, 1037–1046 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neuschwander-Tetri B. A. et al.; NASH Clinical Research Network , Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): A multicentre, randomised, placebo-controlled trial. Lancet 385, 956–965 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ridker P. M. et al.; CANTOS Trial Group , Antiinflammatory therapy with canakinumab for atherosclerotic disease. N. Engl. J. Med. 377, 1119–1131 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Butt E., et al. , cAMP- and cGMP-dependent protein kinase phosphorylation sites of the focal adhesion vasodilator-stimulated phosphoprotein (VASP) in vitro and in intact human platelets. J. Biol. Chem. 269, 14509–14517 (1994). [PubMed] [Google Scholar]
- 27.Tateya S., et al. , Endothelial NO/cGMP/VASP signaling attenuates Kupffer cell activation and hepatic insulin resistance induced by high-fat feeding. Diabetes 60, 2792–2801 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim Y. M., Talanian R. V., Li J., Billiar T. R., Nitric oxide prevents IL-1beta and IFN-gamma-inducing factor (IL-18) release from macrophages by inhibiting caspase-1 (IL-1beta-converting enzyme). J. Immunol. 161, 4122–4128 (1998). [PubMed] [Google Scholar]
- 29.Qu Y., et al. , Pannexin-1 is required for ATP release during apoptosis but not for inflammasome activation. J. Immunol. 186, 6553–6561 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Wood K. S., Ignarro L. J., Hepatic cyclic GMP formation is regulated by similar factors that modulate activation of purified hepatic soluble guanylate cyclase. J. Biol. Chem. 262, 5020–5027 (1987). [PubMed] [Google Scholar]
- 31.Billiar T. R., et al. , Association between synthesis and release of cGMP and nitric oxide biosynthesis by hepatocytes. Am. J. Physiol. 262, C1077–C1082 (1992). [DOI] [PubMed] [Google Scholar]
- 32.Gobeil F., Jr, et al. , Nitric oxide signaling via nuclearized endothelial nitric-oxide synthase modulates expression of the immediate early genes iNOS and mPGES-1. J. Biol. Chem. 281, 16058–16067 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray gene expression data reported in this article have been deposited in the Gene Expression Omnibus (accession no. GSE154892). All study data are included in the article and SI Appendix.