Significance
Sterol regulatory element-binding proteins (SREBPs) are transcription factors that control production of cholesterol and other lipids. In endoplasmic reticulum (ER) membranes, SREBPs form complexes with Scap, a cholesterol-sensing membrane protein. When cholesterol levels are low, Scap escorts SREBPs from the ER to the Golgi where proteolytic cleavage releases the transcription factor domain that turns on genes involved in cholesterol synthesis. The remainder of SREBP still bound to Scap must be eliminated so that Scap can be recycled to bind additional SREBPs. Here, we discover a degradation signal in SREBP that mediates clearance of the cleaved SREBP fragments from Scap. Such clearance is essential for the recycling of Scap and thus it is a central step in regulation of cholesterol homeostasis.
Keywords: Scap, cholesterol, endoplasmic reticulum, Golgi, proteasome
Abstract
Lipid homeostasis in animal cells is maintained by sterol regulatory element-binding proteins (SREBPs), membrane-bound transcription factors whose proteolytic activation requires the cholesterol-sensing membrane protein Scap. In endoplasmic reticulum (ER) membranes, the carboxyl-terminal domain (CTD) of SREBPs binds to the CTD of Scap. When cholesterol levels are low, Scap escorts SREBPs from the ER to the Golgi, where the actions of two proteases release the amino-terminal domains of SREBPs that travel to the nucleus to up-regulate expression of lipogenic genes. The CTD of SREBP remains bound to Scap but must be eliminated so that Scap can be recycled to bind and transport additional SREBPs. Here, we provide insights into how this occurs by performing a detailed molecular dissection of the CTD of SREBP2, one of three SREBP isoforms expressed in mammals. We identify a degradation signal comprised of seven noncontiguous amino acids encoded in exon 19 that mediates SREBP2’s proteasomal degradation in the absence of Scap. When bound to the CTD of Scap, this signal is masked and SREBP2 is stabilized. Binding to Scap requires an arginine residue in exon 18 of SREBP2. After SREBP2 is cleaved in Golgi, its CTD remains bound to Scap and returns to the ER with Scap where it is eliminated by proteasomal degradation. The Scap-binding motif, but not the degradation signal, is conserved in SREBP1. SREBP1’s stability is determined by a degradation signal in a different region of its CTD. These findings highlight a previously unknown role for the CTD of SREBPs in regulating SREBP activity.
Cellular lipid production is controlled by sterol regulatory element-binding proteins (SREBPs), a family of transcription factors that are anchored to the membrane of the endoplasmic reticulum (ER) (1). SREBPs are essential for cellular life and are central players in human diseases ranging from atherosclerosis to cancer to infections (2–6). The mammalian genome encodes three isoforms of SREBPs, designated SREBP1a, SREBP1c, and SREBP2, which combine to activate more than 30 genes involved in the synthesis and uptake of cholesterol, fatty acids, phospholipids, and triglycerides (7). Each SREBP is comprised of an NH2-terminal domain (NTD) that contains a basic helix–loop–helix leucine zipper (bHLH) region for binding DNA, a membrane anchor containing two membrane-spanning helices, and a COOH-terminal domain (CTD) that performs a regulatory function (Fig. 1A). To reach the nucleus and activate genes, the NTD of SREBP is released from the membrane by a proteolytic process that is regulated by cellular cholesterol levels.
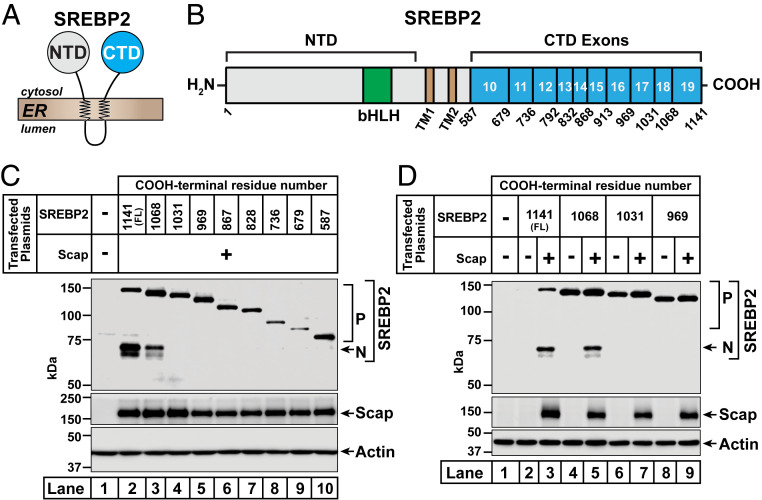
Fig. 1.
Distinct regions of the COOH-terminal domain of SREBP2 are required for its trafficking by Scap and its degradation in the absence of Scap. (A) SREBP2 topology. SREBP2 is comprised of a large NH2-terminal domain (NTD) and a large COOH-terminal domain (CTD), both of which project into the cytosol and are anchored to the ER membrane by two transmembrane helices connected by a short luminal loop. (B) Domain structure of human SREBP2. Starting from the NH2 terminus, SREBP2 contains a bHLH transcription factor domain (amino acids [aa] 330 to 401) (green) in its NTD, the cleavage site for S2P (after L484) in the first transmembrane helix (TM1) (brown), the cleavage site for S1P (after L522) in the short luminal loop connecting TM1 to the second transmembrane helix (TM2) (brown), and a large CTD (aa 588 to 1,141) (cyan). The CTD is comprised of 10 exons (exons 10 to 19), the boundaries of which are indicated. (C and D) Functional analysis of truncated versions of SREBP2. On day 0, SREBP2-deficient TR-4411 cells were set up in medium C at a density of 400,000 cells/well of six-well plates. The compositions of the various media used in this study are listed in SI Appendix, Methods. After 6 h, cells were switched to fresh medium B and transfected as described in SI Appendix, Methods with 0.5 µg of pTK-Scap and 1 µg of either full-length (FL) or truncated versions of pTK-3xFLAG-SREBP2, as indicated. The total amount of DNA in each transfection was adjusted to 1.5 µg per well by addition of control pTK plasmid. After incubation for 40 h, the transfection medium was removed, and cells were switched to cholesterol-depleting medium E. After further incubation for 1 h, cells were harvested and equal fractions of whole cell lysates were subjected to immunoblot analysis of SREBP2 (IgG-22D5), Scap (IgG-4H4), or actin. bHLH, basic helix–loop–helix leucine zipper; P, precursor form of SREBP2; N, cleaved nuclear form of SREBP2.
Cholesterol regulation is mediated by Scap, a cholesterol-sensing protein that contains eight transmembrane helices and a CTD that binds to the CTD of SREBPs (8). The interaction with Scap stabilizes SREBPs and also allows for SREBPs to be escorted to the Golgi (9). When cholesterol in ER membranes is low, Scap serves as the binding site for COPII proteins that cluster the Scap/SREBP complex into COPII vesicles that bud from the ER membrane and fuse with the Golgi (10, 11). In the Golgi, proteolytic cleavage releases the SREBP NTDs to travel to the nucleus and activate lipogenic gene transcription (12). SREBP cleavage is initiated by site-1 protease (S1P) which cleaves the SREBP in the luminal loop that separates the two transmembrane helices. The NTD is then released by a second protease, site-2 protease (S2P). This leaves the CTD still attached to the Golgi membrane and still attached to Scap. It is known that Scap returns to the ER and is recycled to carry out additional rounds of SREBP transport (13–15). In order for this recycling to occur, a mechanism must exist by which the CTD of cleaved SREBP dissociates from Scap and is eliminated. This mechanism is currently unknown.
In the current paper, we begin to solve this mystery by carrying out a systematic dissection of the CTD of SREBP2, one of the three isoforms of SREBP. While the NTD of the three isoforms of SREBP has been the subject of numerous studies (4, 16–19), the CTDs of SREBPs, which comprise almost half of the protein and perform critical regulatory roles, have received far less attention. A pair of early studies showed that the CTDs of SREBPs are required for interacting with the CTD of Scap (20, 21) and this finding was confirmed in two recent structural studies of complexes of the soluble CTDs of the equivalent proteins in fission yeast (22, 23). There have been no further investigations of this domain.
Our studies of the CTD of human SREBP2 reveal a degradation signal that causes the immediate proteasomal degradation of the protein when it is produced in the absence of Scap but is inactive when SREBP2 binds to Scap. After cleavage of SREBP2 by S1P in the Golgi, its CTD returns to the ER bound to Scap. In the ER, the CTD dissociates from Scap, exposing the degradation signal and facilitating its proteasomal degradation. The CTD of human SREBP1 also contains a degradation signal that facilitates its degradation; however, its signal is located in a different region of the protein compared to SREBP2. Such degradation of the CTDs is essential to release Scap so that it can be recycled to bind and activate additional SREBPs. These findings highlight a previously unknown role for the CTD in regulating SREBP activity and lipid homeostasis.
Results
Distinct Regions of SREBP2 Mediate Its Stability and Ability to Interact with Scap.
The CTD of human SREBP2 is comprised of 554 amino acids produced by exons 10 to 19 of the SREBP2 gene (Fig. 1B). To study the functions of this domain without complications from endogenous SREBP2, we first generated a SREBP2-deficient cell line. CRISPR-Cas9 technology was used to delete a portion of exon 2 of SREBP2 in SV-589 cells (a line of transformed human fibroblasts) and the resulting SREBP2-deficient cells are hereafter designated TR-4411 (SI Appendix, Methods and Fig. S1A). Compared to parental SV-589 cells, TR-4411 cells showed no detectable signal for either the precursor or the processed nuclear form of SREBP2, regardless of cholesterol levels (SI Appendix, Fig. S1B). Levels of Scap were unchanged in TR-4411 cells.
To establish these cells as a system for studying SREBP2, we transfected TR-4411 cells with SREBP2, depleted the cells of cholesterol, and then subjected detergent-solubilized whole cell lysates to immunoblot analysis (SI Appendix, Fig. S1C). We detected the precursor form of SREBP2; however, very little of the SREBP2 precursor was processed to the mature, nuclear form in the cholesterol-depleted cells (lane 3). We reasoned that the amount of Scap in TR-4411 cells was insufficient for trafficking the transfected SREBP2. Indeed, when we cotransfected Scap, much higher levels of both forms of SREBP2 were detected (lane 4), consistent with Scap’s well-known functions in stabilizing full-length SREBP2 as well as in trafficking this protein to the Golgi for proteolytic activation (9). In this initial analysis, we used two antibodies to detect Scap—the previously described monoclonal IgG-4H4 (24), which detects hamster (transfected), but not human (endogenous) Scap, and the newly developed monoclonal IgG-2G10, which detects Scap from both species. In most of the remaining experiments in this study, we used IgG-4H4 to detect transfected hamster Scap.
With these reagents in hand, we generated plasmids encoding truncated fragments of SREBP2 that terminate at or near the boundaries of 8 of the 10 exons that encode the CTD (Fig. 1B). Truncations after exon 12 or exon 15 were not examined in this study. We transfected TR-4411 cells with full-length or truncated SREBP2 together with Scap, depleted the cells of cholesterol, and analyzed whole cell lysates by immunoblot (Fig. 1C). As expected, full-length SREBP2 was trafficked to the Golgi by Scap and processed to its nuclear form in the cholesterol-depleted cells (lane 2). Similar results were obtained with SREBP2 truncated after exon 18 (lane 3). In contrast, SREBP2 truncated after exon 17 showed no Scap-dependent processing (lane 4). Mutants of SREBP2 harboring the remaining truncations (after exons 9, 10, 11, 13, 14, and 16) also failed to be processed (lanes 5 to 10).
Experiments analyzing the processing of the truncated versions of SREBP2 in the absence or presence of Scap revealed an unexpected role for exon 19 (Fig. 1D). In the absence of Scap, the precursor form of full-length SREBP2 was barely detectable, whereas the precursor form of SREBP2 lacking exon 19 was readily detected (compare lane 2 to lane 4). This Scap-independent enhancement of expression was also observed for precursor forms of SREBP2s truncated after exon 17 (lane 6) or exon 16 (lane 8). Upon cotransfection of Scap, levels of the precursor form of full-length SREBP2 were increased and a fraction of this SREBP2 was processed to its nuclear form, as expected (lane 3). However, a similar Scap-dependent enhancement was not observed for SREBP2 lacking exon 19, which was also processed to its nuclear form (lane 5). Levels of the precursor forms of SREBP2s truncated after exon 17 or exon 16 were also not enhanced by Scap; however, these mutants failed to be processed to their respective nuclear forms (lanes 7 and 9). Taken together, these results suggest that residues in exon 19 of SREBP2 mediate its degradation in the absence of Scap, while residues in exon 18 are required for its interaction with Scap and subsequent processing.
Seven Amino Acids in Exon 19 of SREBP2 Define a Motif that Mediates Its Degradation.
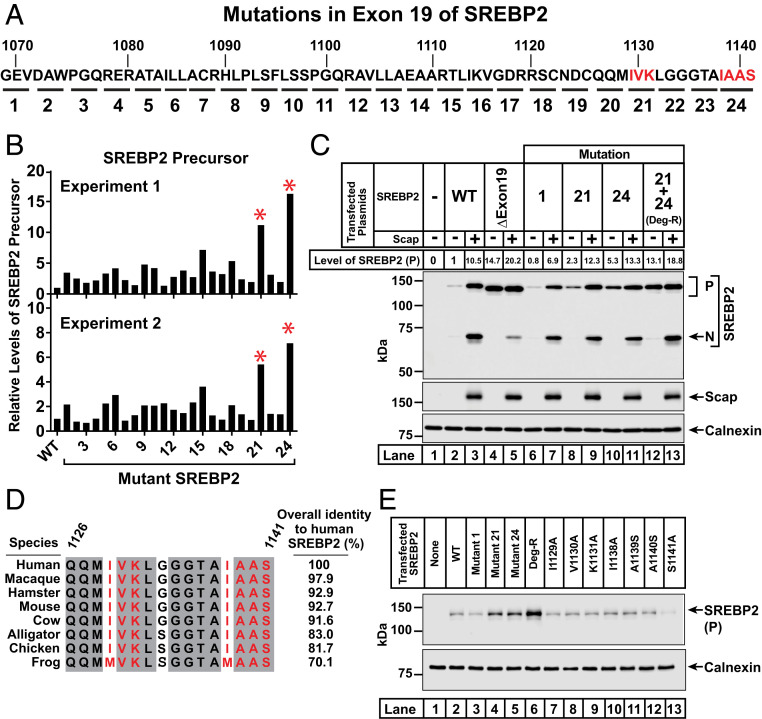
To identify residues that mediate SREBP2’s degradation in the absence of Scap, we began by conducting an alanine-scanning mutagenesis study of the entire exon 19 by systematic replacement of every trio or quartet of contiguous residues with alanine (Fig. 2A). In cases where the wild-type (WT) residue was alanine, it was mutated to serine. TR-4411 cells were transfected with these plasmids and whole cell lysates were subjected to immunoblot analysis. We then used densitometry to quantify levels of the precursor form of each SREBP2 mutant. In two separate experiments, mutant 21 (I1129A, V1130A, and K1131A) and mutant 24 (I1138A, A1139S, A1140S, and S1141A) showed a more than fivefold increase in the levels of the precursor form of SREBP2 relative to the WT control (Fig. 2B).
Fig. 2.
Identification of 7 amino acids in exon 19 of SREBP2 that mediate its degradation in the absence of Scap. (A) Strategy for alanine-scanning mutagenesis of exon 19. A panel of 24 mutant plasmids was generated in which 3 or 4 amino acids in exon 19 (denoted by each underline and numbered 1 to 24) were individually mutated to alanine. In cases where the wild-type residue was alanine, it was mutated to serine. Mutants 21 and 24 are in red to highlight their role in mediating the degradation of SREBP2 as described below. (B) Analysis of levels of the precursor form of WT and mutant versions of SREBP2. On day 0, SREBP2-deficient TR-4411 cells were set up in medium C at a density of 400,000 cells/well of six-well plates. After 6 h, cells were switched to fresh medium B and transfected with 1 µg of either WT or indicated mutant versions of pTK-3xFLAG-SREBP2 as described in SI Appendix, Methods. After incubation for 40 h, the transfection medium was removed and cells were switched to cholesterol-depleting medium E for 1 h, after which cells were harvested. Equal fractions of whole cell lysates were subjected to immunoblot analysis of SREBP2 (IgG-22D5). The intensity value obtained by LI-Cor imaging for the band representing the precursor form of SREBP2(WT) was set to 1 and the intensities of the precursor form of all mutant versions of SREBP2 were normalized relative to this set point. Results from two independent experiments are shown. Red asterisks denote mutant versions of SREBP2 whose precursor levels were increased in both experiments (>10-fold in experiment 1, >5-fold in experiment 2). (C) Mutation of a 7-amino acid motif stabilizes SREBP2 in the absence of Scap. On day 0, TR-4411 cells were set up in medium C at a density of 400,000 cells/well of six-well plates. Cells were then transfected as described in SI Appendix, Methods with 0.5 µg of pTK-Scap and 1 µg of either WT or mutant versions of pTK-3xFLAG-SREBP2, as indicated. The total amount of DNA in each transfection was adjusted to 1.5 µg per well by addition of control pTK plasmid. After treatment and cholesterol depletion as in B, cells were harvested and subjected to immunoblot analysis of SREBP2 (IgG-22D5), Scap (IgG-4H4), and calnexin. The intensity value obtained by LI-Cor imaging for the band representing the precursor form of SREBP2(WT) in the absence of Scap was set to 1 and the intensities of all other precursor forms of SREBP2 were normalized relative to this set point. (D) Amino acid sequence of the COOH-terminal 16 amino acids of SREBP2. Residues that are identical in eight animal species are shaded (gray). Residues that mediate the degradation of SREBP2 in the absence of Scap are red. GenBank accession numbers for SREBP2 sequences are listed in SI Appendix, Methods. Sequences were aligned and % identity was calculated using Clustal Omega. (E) Analysis of individual mutations of each of the 7 amino acids of the degradation motif. TR-4411 cells were set up and transfected as described in SI Appendix, Methods with 1 µg of either pTK control plasmid (lane 1) or indicated versions of pTK-3xFLAG-SREBP2 (lanes 2 to 13). After treatment and cholesterol depletion as in B, cells were harvested and subjected to immunoblot analysis of SREBP2 (IgG-22D5) and calnexin. P, precursor form of SREBP2; N, cleaved nuclear form of SREBP2.
Based on the results of this initial scan, we generated a plasmid encoding a version of SREBP2 containing the seven mutations corresponding to both mutant 21 and mutant 24. We then conducted transfection studies to compare the behavior of this hepta-mutant version of SREBP2 to that of WT and other mutant forms of SREBP2 (Fig. 2C). As expected, we observed low levels of SREBP2(WT) precursor in the absence of Scap and its levels increased by 10.5-fold after cotransfection of Scap (lanes 2 and 3). Consistent with our observation in Fig. 1D, expression of the precursor form of SREBP2 lacking exon 19 was greatly enhanced compared to WT even in the absence of Scap (14.7-fold, lane 4). Expression in the presence of Scap increased slightly (lane 5). SREBP2 harboring mutation 1 behaved similarly to SREBP2(WT) (Fig. 2C, lanes 6 and 7), in line with its minimal effect in the scan of Fig. 2B. Compared to SREBP2(WT), the precursor forms of SREBP2s harboring either mutation 21 or mutation 24 were enhanced by 2.3-fold and 5.3-fold, respectively, in the absence of Scap, and stabilized to levels similar to that observed for SREBP2(WT) in the presence of Scap (Fig. 2C, lanes 8 to 11). The precursor form of SREBP2 harboring both mutations 21 and 24 was enhanced by 13.1-fold in the absence of Scap and increased even further by Scap (18.8-fold), levels that are similar to that observed for SREBP2 lacking the entire exon 19 (Fig. 2C, lanes 12 and 13). Thus, the seven amino acids in SREBP2 that were mutated in mutations 21 and 24 constitute a degradation motif. The combined hepta-mutant that is stable in the absence of Scap is hereafter referred to as SREBP2(Deg-R) (degradation resistant).
The sequence of this seven-amino acid degradation motif is highly conserved in eight vertebrate species of SREBP2 that we examined (Fig. 2D). We attempted to deconvolute this motif by preparing plasmids where each of the seven residues were individually mutated to alanine or serine (Fig. 2E). Transfection studies with these plasmids in TR-4411 cells showed low levels of the precursor form of each of the seven point mutants (lanes 7 to 13), comparable to the low level of the precursor form of SREBP2(WT) (lane 2). In contrast, the hepta-mutant SREBP2(Deg-R) once again showed robust enhancement of its precursor form (lane 6).
Full-Length SREBP2 Is Degraded by the Proteasome in the Absence of Scap.
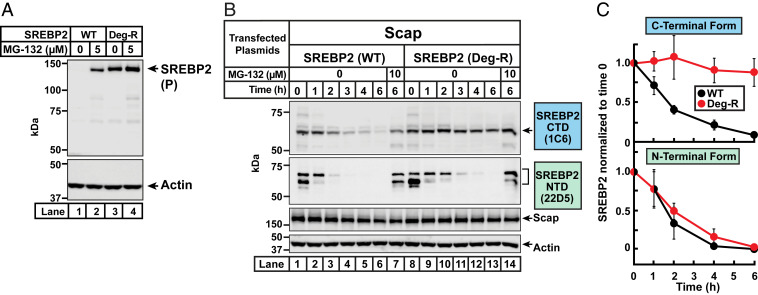
We asked next how the precursor form of SREBP2 was degraded in the absence of Scap. Since it has been shown previously that the processed, soluble nuclear form of SREBP2 is degraded by proteasomes (16, 17, 25), we wondered whether this same pathway was also responsible for degrading the precursor, ER membrane-bound form of SREBP2. To test this idea, Scap-null SRD-13A cells were transfected with either SREBP2(WT) or SREBP2(Deg-R). The transfected cells were then treated without or with MG-132, a proteasome inhibitor. To prevent new protein synthesis during the duration of MG-132 treatment, cells were cotreated with cycloheximide, an inhibitor of protein translation. After 4 h, we harvested the cells and performed immunoblot analysis for SREBP2 (Fig. 3A). Transfected SREBP2(WT) was only detectable when cells were treated with MG-132 (compare lane 1 to lane 2). In contrast, transfected SREBP2(Deg-R) was detected at similar levels irrespective of MG-132 treatment (lanes 3 and 4). This suggests that, in the absence of Scap, the precursor form of SREBP2 is degraded by the proteasome by a process that requires the degradation motif.
Fig. 3.
Proteasome-mediated degradation of both the precursor form and the cleaved COOH-terminal fragment of SREBP2 is blocked by mutation of the degradation motif in exon 19. (A) MG-132 treatment. On day 0, Scap-deficient SRD-13A cells were set up in medium G at a density of 200,000 cells/well of six-well plates. The compositions of the various media used in this study are listed in SI Appendix, Methods. On day 1, cells were transfected as described in SI Appendix, Methods with 1 µg of the indicated version of pTK-3xFLAG-SREBP2. On day 2, cells were switched to 2 mL of fresh medium F containing 50 µM cycloheximide and the indicated concentrations of MG-132 (solubilized in 1 µL of dimethyl sulfoxide [DMSO]). After incubation for 4 h, cells were harvested and equal fractions of whole cell lysates were subjected to immunoblot analysis of SREBP2 (IgG-22D5) and actin. (B) Stability of SREBP2 fragments generated after proteolytic cleavage by S1P and S2P. On day 0, CHO-K1 cells were set up in medium F at a density of 250,000 cells/well of six-well plates. On day 1, cells were switched to fresh medium F and transfected as described in SI Appendix, Methods with 0.5 µg of pTK-Scap and 1 µg of the indicated version of pTK-3xFLAG-SREBP2. On day 2, the transfection medium was removed, and cells were switched to cholesterol-depleting medium H. After incubation for 2 h, the medium was removed and cells were switched to medium F containing 50 µM cycloheximide, 1 µg/mL 25-HC, and the indicated concentrations of MG-132. After further incubation for the indicated times, cells were harvested and equal fractions of cell lysates were subjected to immunoblot analysis of SREBP2 (IgG-1C6 and IgG-22D5), Scap (IgG-4H4), and actin. (C) Quantification of cleaved fragments of SREBP2(WT) and SREBP2(Deg-R). The intensities of bands corresponding to the CTD of SREBP2 (detected by IgG-1C6) and the NTD of SREBP2 (detected by IgG-22D5) in B were quantified by LI-Cor densitometry imaging. The intensity values for the 0 time point [lane 1 for SREBP2(WT); lane 8 for SREBP2(Deg-R)] were set to 1 and the intensities of all other bands were normalized relative to these set points. Plots show the average values obtained from three independent experiments (the experiment in B and two other experiments conducted in the same manner). Error bars represent SEM. P, precursor form of SREBP2.
Proteasome-mediated degradation of proteins is initiated by ubiquitinylation of lysine residues in the substrate protein. The topology of the ubiquitinylation machinery suggests that potential sites of ubiquitinylation should be accessible to the cytosol. The cytosol-facing CTD of SREBP2 contains 29 lysine residues that are distributed throughout this domain (SI Appendix, Fig. S2A). To determine whether degradation is mediated by these CTD lysines, we generated a plasmid encoding a version of SREBP2 where all 29 CTD lysines were mutated to arginines. We then conducted transfection studies in TR-4411 cells to compare the behavior of this mutant version of SREBP2 to that of SREBP2(WT) and SREBP2(Deg-R) (SI Appendix, Fig. S2B). As before, levels of the precursor form of SREBP2(WT) were barely detectable in the absence of Scap and were greatly enhanced by Scap (lanes 1 and 2), while levels of the precursor form of SREBP2(Deg-R) were detected at a high level irrespective of Scap (lanes 3 and 4). Levels of the precursor form of SREBP2 lacking all 29 CTD lysines were enhanced to a similar degree as those of SREBP2(Deg-R) (lanes 3 to 6). It is noteworthy that mutation of all 29 CTD lysines to arginines did not affect SREBP2’s ability to be processed to its nuclear form in a Scap-mediated fashion (compare lane 6 to lane 2).
We next sought to identify the specific CTD lysine(s) that mediate degradation of the precursor form of SREBP2. We began by analyzing the conservation of these lysine residues in SREBP2s from eight vertebrate species. Unfortunately, most of the lysines are highly conserved, with 18 of the 29 being identical in the eight sequences (SI Appendix, Fig. S2C). In the absence of clear candidates, we divided the 29 CTD lysines into two groups and generated two mutant SREBP2 plasmids where we mutated either all 20 lysines in exons 10 through 14 to arginines (group 1) or all 9 lysines in exons 15 through 19 to arginines (group 2). When we transfected these two plasmids in TR-4411 cells, we found that elimination of lysines in either group 1 or in group 2 could each partially enhance levels of the precursor form of SREBP2 (SI Appendix, Fig. S2D, compare lanes 4 and 5 to lane 2). Further enhancement was observed when all 29 CTD lysines were eliminated (lane 6). Combined, these results suggest that multiple lysines throughout the CTD mediate proteasomal degradation of the precursor form of SREBP2 in the ER by a process requiring the degradation motif in exon 19. Inasmuch as the 22 cytoplasmically disposed lysines in the NTD of SREBP2 were left intact during our mutational analysis of CTD lysines, we also conclude that degradation of the precursor form of SREBP2 is not mediated by NTD lysines.
Degradation of the Cleaved COOH-Terminal Fragment of SREBP2 Occurs after It Returns to the ER with Scap.
We next studied the effects of the degradation motif in exon 19 on the stability of the CTD of SREBP2 generated after cleavage in the Golgi by S1P. A simple way to track the CTD would be to append an epitope tag to the carboxy terminus of SREBP2. To this end, we generated plasmids encoding versions of SREBP2 with His6, Myc, or FLAG tags fused to the COOH terminus. Since the tags are in close proximity to the degradation motif at the COOH terminus, we first carried out transfection studies in Scap-null SRD-13A cells to ensure that the tags did not affect SREBP2’s stability (SI Appendix, Fig. S3A). As expected, in the absence of Scap, SREBP2 with no COOH-terminal tag was barely detectable (lane 2). Unfortunately, all three COOH-terminal tags that we tested stabilized the precursor form of SREBP2 in the absence of Scap to a similar degree as mutation of the degradation motif (lanes 3 to 6). Even addition of single amino acids such as proline, arginine, asparagine, glutamine, aspartate, glutamate, tyrosine, or tryptophan to the very end of the COOH terminus (after S1141) stabilized the precursor form of SREBP2 to a similar degree as SREBP2(Deg-R) (SI Appendix, Fig. S3B). Addition of any of the other 12 amino acids did not stabilize SREBP2. These results highlight the importance of the very end of the COOH terminus of SREBP2 in determining its stability.
Fortunately, we were able to find an alternative method to track the CTD using the previously described IgG-1C6 monoclonal antibody (26) that was raised against the CTD of SREBP2 (SI Appendix, Fig. S4A). To overcome the detection limitations of IgG-1C6, we used CHO-K1 cells instead of TR-4411 cells for this experiment because of the much higher transfection efficiency obtained in CHO-K1 cells. As shown in SI Appendix, Fig. S4B, IgG-1C6 detected the precursor and CTD forms of transfected, but not endogenous, SREBP2 in CHO-K1 cells. To simultaneously monitor the NTD fragment of SREBP2, we used IgG-22D5, a robust monoclonal antibody that has been used in all of the experiments described above (SI Appendix, Fig. S4A). We transfected CHO-K1 cells with plasmids encoding Scap and either SREBP2(WT) or SREBP2(Deg-R), depleted the cells of sterols, and subjected whole cell lysates to immunoblot analysis (Fig. 3B). As expected, both SREBP2s were trafficked to the Golgi and cleaved, generating the CTD fragment (lane 1 and lane 8, first panel) and the NTD fragment (lane 1 and lane 8, second panel). We then treated cells with cycloheximide to stop synthesis of additional SREBP2 and also with 25-hydroxycholesterol (25HC) to trap any residual SREBP2 in the ER and prevent generation of new cleaved fragments. These treatments allowed us to follow the fates of only the SREBP2 cleavage fragments generated during the cholesterol depletion step described above. The intensity of the band corresponding to the CTD of SREBP2(WT) declined over time, whereas that of SREBP2(Deg-R) was unchanged even after the longest treatment time of 6 h (compare lanes 1 through 6 to lanes 8 through 13 in the first panel). In contrast, the NTD fragments of both SREBP2(WT) and SREBP2(Deg-R) were rapidly eliminated (compare lanes 1 through 6 to lanes 8 through 13 in the second panel). The disappearance of both the CTD and NTD were rescued by treatment with MG-132, a proteasome inhibitor (lanes 7 and 14). Unlike the cleaved fragments of SREBP2, Scap levels were unchanged even after the longest time point of 6 h. We then used densitometry to quantify levels of the CTD and NTD fragments of SREBP2 in Fig. 3B and two other independent experiments that were conducted in the same manner. This analysis (shown in Fig. 3C) confirmed the result in Fig. 3B that the CTD, but not the NTD, of SREBP2 was stabilized by mutation of the degradation motif in exon 19.
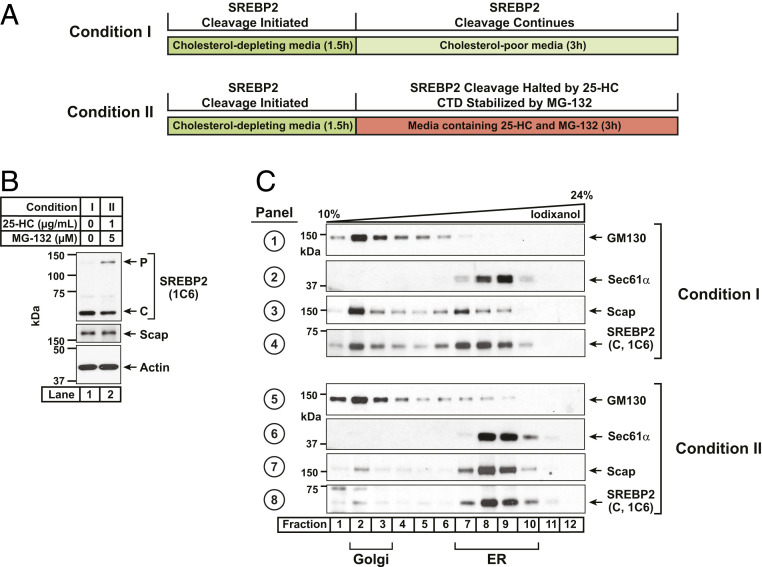
While ER-associated degradation (ERAD) is the main pathway for proteasomal degradation of membrane proteins, recent work has identified an additional pathway termed endosome and Golgi-associated degradation (EGAD) that extracts membrane proteins from the Golgi membrane for proteasomal degradation (27). Is the residual CTD-containing fragment of SREBP2 degraded by EGAD in the Golgi immediately after cleavage by S1P or does it remain bound to Scap and accompany it on its retrograde journey back to the ER to undergo ERAD? To distinguish between these possibilities, we performed organelle fractionation of SV-589 cells to pinpoint the location of endogenous SREBP2’s CTD. We first depleted cells of sterols to trigger transport of Scap/SREBP2 from the ER to the Golgi and generate the CTD fragment of SREBP2 (see experimental scheme in Fig. 4A). The sterol-depleted cells were then switched to fresh media containing lipoprotein-deficient serum either without sterols to maintain SREBP2 processing or with 25HC to trap SREBP2 in the ER and prevent production of additional CTDs. The 25HC-treated cells were also treated with MG-132 to prevent degradation of the already-generated CTD so that we could track its itinerary. Both sets of cells were then harvested, and postnuclear supernatants were collected. Immunoblot analysis using IgG-1C6 showed that upon sterol depletion, most of the SREBP2 had been processed to its CTD form (Fig. 4B, lane 1). The combined 25HC and MG-132 treatment resulted in a buildup of precursor SREBP2 while still preserving the CTD generated during the sterol depletion step (Fig. 4B, lane 2). We then subjected these postnuclear supernatants to centrifugation through an iodixanol density gradient (28) and performed immunoblot analysis of the gradient fractions for Scap, CTD of SREBP2, as well as Golgi and ER-specific markers (Fig. 4C). ER and Golgi membranes were well separated after either treatment condition, with a Golgi-resident protein (GM130) concentrating in lighter fractions (panels 1 and 5, lanes 1 to 4) and an ER-resident protein (Sec61α) concentrating in denser fractions (panels 2 and 6, lanes 7 to 10). At steady state in sterol-depleted cells, Scap was evenly distributed between the ER and Golgi membranes (panel 3). This finding is consistent with previous observations that sterol depletion allows a transient translocation of Scap from ER to Golgi, whereupon Scap quickly returns to the ER (13, 15). The CTD of SREBP2 showed the exact same distribution as Scap (panel 4), suggesting that this residual fragment of SREBP2 remains bound to Scap and also returns to the ER. Upon addition of 25HC to prevent translocation of Scap, any remaining Golgi-resident Scap quickly returned to the ER (panel 7). The CTD of SREBP2 generated during the sterol depletion step also returned to the ER with Scap where it accumulated due to MG-132 inhibition of ERAD (panel 8). These results suggest that the CTD of SREBP2 generated in the Golgi after S1P cleavage remains bound to Scap and returns to the ER with Scap to undergo ERAD.
Fig. 4.
Itinerary of the cleaved COOH-terminal domain of SREBP2. (A) Experimental scheme. On day 0, six 10-cm dishes of SV-589 cells were set up in medium B at a density of 1 × 106 cells per dish. The compositions of the various media used in this study are listed in SI Appendix, Methods. On day 1, cells were switched to medium D supplemented with 1.5% (wt/vol) hydroxypropyl-β-cyclodextrin (HPCD) to deplete cellular cholesterol. After incubation for 90 min, cells were washed once with prewarmed phosphate buffered saline (PBS), after which one group of cells (three dishes) were switched to fresh medium D (condition I) while the remaining cells (three dishes) were switched to medium D supplemented with 1 µg/mL of 25-HC and 5 µM MG-132 (condition II). After further incubation for 3 h, both groups of cells were washed twice with ice-cold PBS, harvested, and subjected to organelle fractionation by density gradient centrifugation as described in SI Appendix, Methods. (B and C) Immunoblot analysis. After processing, aliquots representing equal volumes of postnuclear supernatants (B) or each fraction from an iodixanol gradient (C) were subjected to immunoblot analysis of the CTD of SREBP2 (IgG-1C6), Scap (IgG-2G10), actin, Golgi marker GM130, and ER marker Sec 61α. P, precursor form of SREBP2; C, cleaved COOH-terminal form of SREBP2.
Mutation of a Single Arginine Residue in Exon 18 of SREBP2 Disrupts Its Interaction with Scap.
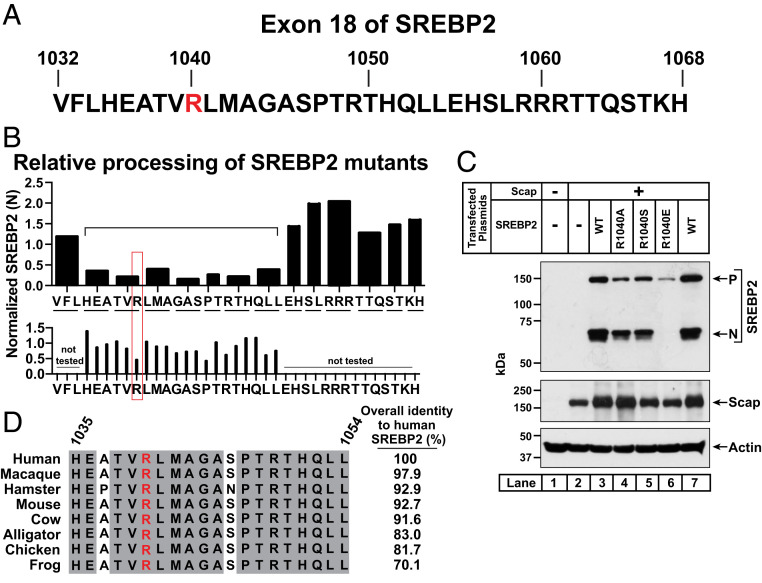
We next returned to our initial analysis of SREBP2’s CTD exons which showed that while exon 19 imparted stability to SREBP2 in the absence of Scap, a different region of SREBP2 encoded by exon 18 was required for interaction with Scap and subsequent processing (Fig. 1). To pinpoint the exact residues in this 37-amino acid exon (Fig. 5A) that are responsible for interacting with Scap, we followed a strategy similar to that used to identify the degradation motif in exon 19 (Fig. 2). We began by generating expression plasmids where each pair or trio of contiguous residues in exon 18 was replaced by alanine. In cases where the WT residue was alanine, it was mutated to serine. For this initial scan, we used SREBP2 lacking exon 19 as template due to the high levels of expression of this version of SREBP2 even in the absence of Scap (Fig. 1D). TR-4411 cells were transfected with plasmids encoding the mutant SREBP2s along with a plasmid encoding Scap. After depletion of cholesterol, whole cell lysates were subjected to immunoblot analysis for the cleaved NH2-terminal fragment of SREBP2, generation of which is indicative of proper interaction with Scap. Densitometry was used to quantify levels of the NH2-terminal fragment of each SREBP2 mutant. This initial scan revealed seven contiguous mutants spanning 20 residues that showed a >60% decrease in levels of the NH2-terminal fragment of SREBP2 when compared to that observed with SREBP2(WT) (Fig. 5 B, Top).
Fig. 5.
Identification of an arginine residue in exon 18 of SREBP2 that is required for its interaction with Scap. (A) Amino acid sequence of exon 18 of human SREBP2. Amino acid residue R1040 is in red to highlight its role in mediating the interaction between SREBP2 and Scap as described below. (B) Analysis of levels of the cleaved nuclear forms of WT and mutant versions of SREBP2. On day 0, SREBP2-deficient TR-4411 cells were set up in medium C at a density of 400,000 cells/well of six-well plates. After 6 h, cells were switched to fresh medium B and transfected as described in SI Appendix, Methods with 0.5 µg of pTK-Scap together with 1 µg of either WT or indicated mutant versions of pTK-3xFLAG-SREBP2 lacking exon 19. After incubation for 40 h, the transfection medium was removed and cells were switched to cholesterol-depleting medium E for 1 h, after which cells were harvested. Equal fractions of whole cell lysates were subjected to immunoblot analysis of SREBP2 (IgG-22D5). The intensity value obtained by LI-Cor imaging for the band representing the cleaved nuclear form of SREBP2(WT) was set to 1 and the intensities of the cleaved nuclear forms of all mutant versions of SREBP2 were normalized relative to this set point. The Top shows results for SREBP2 mutants where two or three contiguous residues (indicated by underline) were mutated to alanine (or serine in cases where the WT residue was alanine). The bracketed region denotes mutant versions of SREBP2 whose cleaved nuclear forms were reduced by more than 60%. The Bottom shows results for SREBP2 mutants where each residue in the bracketed region was mutated to alanine (or serine when the WT residue was alanine). Mutation of residue R1040 (red box) resulted in reduction of the cleaved nuclear form by more than 50% and was chosen for further study. (C) Effect of amino acid substitutions at residue 1040 of SREBP2. TR-4411 cells were set up as in B and transfected with 1 µg of pTK-Scap together with 1 µg of either WT or the indicated mutant versions of full-length pTK-3xFLAG-SREBP2. The total amount of DNA in each transfection was adjusted to 2 µg per well by addition of control pTK plasmid. After incubation for 40 h, cells were treated as in B, harvested, and equal fractions of whole cell lysates were subjected to immunoblot analysis of SREBP2 (IgG-22D5), Scap (IgG-4H4), and actin. (D) Conservation analysis of residues 1,035 to 1,054 of human SREBP2 (bracketed region in B). Residues that are identical in eight animal species are shaded (gray). Shown in red is the arginine residue that eliminates the ability of SREBP2 to be processed to its cleaved nuclear form in the presence of Scap. Uniprot accession numbers for SREBP2 sequences are listed in SI Appendix, Methods. Sequences were aligned and % identity was calculated using Clustal Omega. P, precursor form of SREBP2; N, cleaved nuclear form of SREBP2.
We then generated plasmids encoding SREBP2 lacking exon 19 where each of the 20 residues identified in the initial scan were individually mutated to alanine (or serine). Transfection studies as above showed that only two single mutations—R1040A and P1047A—decreased levels of the NH2-terminal fragment of SREBP2 by more than 50% when compared to SREBP2(WT) (Fig. 5 B, Bottom). We selected the R1040 residue for further investigation and generated a series of plasmids encoding full-length SREBP2 with various substitutions at this position. We again conducted transfection studies as above and found that substitution of arginine with glutamic acid dramatically reduced both the precursor and the cleaved NH2-terminal fragment of SREBP2 (Fig. 5C, lane 6). In contrast, substitutions with alanine or serine had little effect (Fig. 5C, lanes 4 and 5). These results suggest that SREBP2 requires a positive charge at residue 1040 for its interaction with Scap. Comparison of sequences from eight different vertebrate species of SREBP2 focusing on the 20 amino acids identified in the initial scan results of Fig. 5B shows that this entire region is very highly conserved with R1040 being invariant (Fig. 5D).
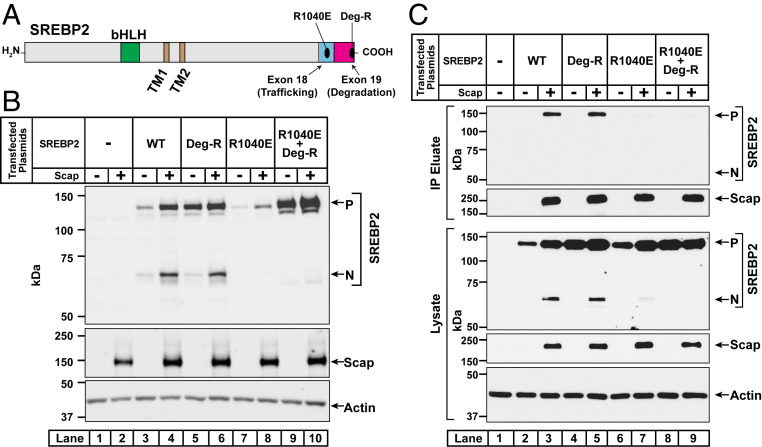
To determine if the reduced levels of precursor and cleaved NH2-terminal fragments of SREBP2(R1040E) were due to its reduced interaction with Scap, we conducted three additional sets of experiments. For the first experiment, we generated a plasmid encoding a version of full-length SREBP2 harboring the R1040E mutation along with the previously described mutation of the seven amino acids comprising the degradation motif (Fig. 6A). We then conducted transfection studies in TR-4411 cells to compare the behavior of this octa-mutant of SREBP2, designated as SREBP2(R1040E; Deg-R), to that of WT and other mutant versions of SREBP2 (Fig. 6B). As before, we observed low levels of the precursor form of SREBP2(WT) in the absence of Scap and this level was enhanced by Scap (lanes 3 and 4). In contrast, the precursor form of SREBP2(Deg-R) was detected at a high level even in the absence of Scap and was not dramatically enhanced by Scap (lanes 5 and 6). The precursor form of SREBP2(R1040E) was barely detectable in the absence of Scap and its level was only weakly enhanced by Scap (lanes 7 and 8). However, when we combined the two mutations in SREBP2(R1040E; Deg-R), the precursor form of this mutant SREBP2 was now detected at a level similar to that observed for SREBP2(Deg-R), regardless of Scap (lanes 9 and 10). This enhancement occurred even though SREBP2(R1040E; Deg-R), just like SREBP2(R1040E), was unable to interact with Scap as judged by the lack of detection of their cleaved nuclear forms (lanes 8 and 10). The cleaved forms were readily detected for SREBP2(WT) and SREBP2(Deg-R), since these versions interact with Scap (lanes 4 and 6). These data indicate that the instability of the precursor form of SREBP2(R1040E) is not likely due to gross protein misfolding since it can be rescued by mutation of the degradation motif.
Fig. 6.
Mutation of the degradation motif in exon 19 stabilizes SREBP2 even when it is rendered incapable of interacting with Scap. (A) Domain architecture of SREBP2. Starting from its NH2 terminus, SREBP2 contains a bHLH transcription factor domain (green), two transmembrane helices (brown), and a large COOH-terminal domain. Indicated are mutations in exons 18 and 19 that disrupt distinct properties of SREBP2. (B) SREBP2 analysis. On day 0, SREBP2-deficient TR-4411 cells were set up in medium C at a density of 400,000 cells/well of six-well plates. After 6 h, cells were switched to fresh medium B and transfected as described in SI Appendix, Methods with 1 µg of pTK-Scap together with 1 µg of either WT or the indicated mutant versions of pTK-3xFLAG-SREBP2. The total amount of DNA in each transfection was adjusted to 2 µg per well by addition of control pTK plasmid. After incubation for 40 h, the transfection medium was removed and cells were switched to cholesterol-depleting medium E for 1 h, after which cells were harvested. Equal fractions of whole cell lysates were subjected to immunoblot analysis of SREBP2 (IgG-22D5), Scap (IgG-4H4), and actin. (C) Coimmunoprecipitation of full-length SREBP2 with full-length Scap. On day 0, SREBP2-deficient TR-4411 cells were set up in medium C at a density of 106 cells per 10-cm dish. On day 1, cells were switched to fresh medium B and transfected as described in SI Appendix, Methods with 3 µg of pEZT-3xFLAG-Scap together with 3 µg of either WT or mutant versions of pEZT-2xMyc-TEV-SREBP2, as indicated. The total amount of DNA in each transfection was adjusted to 6 µg per well by addition of control pcDNA 3.0 plasmid. On day 2, the transfection medium was removed and cells were switched to cholesterol-depleting medium E. After incubation for 1 h, cells were harvested, and membranes were solubilized as described in SI Appendix, Methods. A portion of the solubilized membranes (8% of total) was saved and the remainder (92% of total) was used for immunoprecipitation of FLAG-tagged Scap with anti-FLAG M2 resin as described in SI Appendix, Methods. Equivalent volumes of the solubilized membranes (lysate) and the immunoprecipitated material (IP eluate) were subjected to immunoblot analysis of SREBP2 (IgG-22D5), Scap (IgG-2G10), and actin. P, precursor form of SREBP2; N, cleaved nuclear form of SREBP2; Deg-R, degradation-resistant form of SREBP2.
In a second experiment, we conducted coimmunoprecipitation studies to directly examine the interaction between full-length Scap and full-length SREBP2 without or with the R1040E mutation (Fig. 6C). TR-4411 cells were cotransfected with FLAG-tagged Scap along with various versions of Myc-tagged SREBP2, including the octa-mutant described above. In this experiment, we used plasmids under control of the strong cytomegalovirus (CMV) promoter to ensure adequate expression of SREBP2(R1040E). Cells were then depleted of sterols, membranes were solubilized with digitonin, and the FLAG-tagged Scap was precipitated on anti-FLAG beads. Bound proteins were eluted by incubation with FLAG peptides and subjected to immunoblot analysis for Scap and SREBP2. In the absence of Scap, none of the versions of SREBP2 were precipitated by the anti-FLAG beads (lanes 2, 4, 6, and 8). When Scap was coexpressed, the precursor form, but not the cleaved NH2-terminal fragment, of SREBP2(WT) as well as SREBP2(Deg-R) were coimmunoprecipitated (lanes 3 and 5). However, neither SREBP2(R1040E) nor SREBP2(R1040E; Deg-R) were coprecipitated by Scap, even though the precursor forms of these SREBP2s were detected at levels similar to that of SREBP2(WT) or SREBP2(Deg-R) (lanes 7 and 9). These data indicate that SREBP2(R1040E) loses the ability to bind to Scap in detergent solutions.
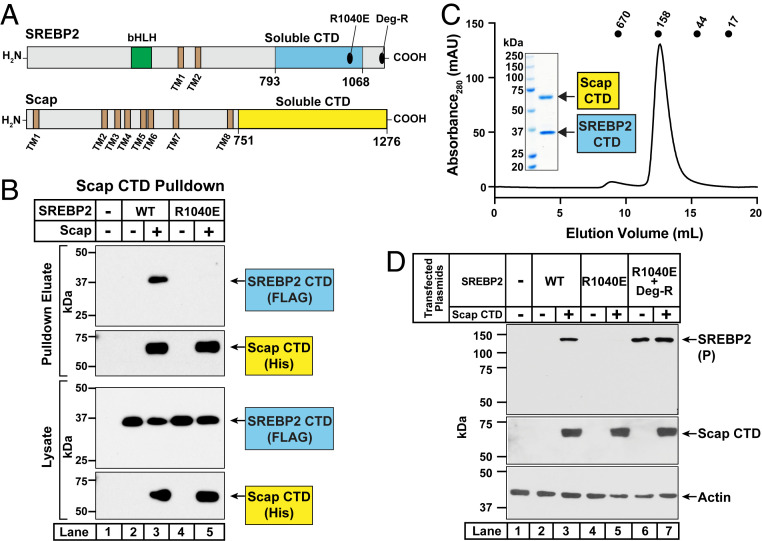
In a third approach, we examined the interaction between the soluble CTD of Scap and the soluble CTD of either SREBP2(WT) or SREBP2(R1040E) in the absence of membrane anchors. For this study, we shifted to the baculovirus expression system in insect cells. We generated baculoviruses to produce the soluble CTDs of His10-tagged Scap and FLAG-tagged SREBP2 (see Fig. 7A for domain boundaries that yielded stable, soluble proteins). After infecting insect cells with these baculoviruses, cell lysates were subjected to 20,000 × g centrifugation and the soluble supernatants were incubated with nickel beads to precipitate the His10-tagged Scap. The eluted proteins were then subjected to immunoblot analysis for Scap and SREBP2 (Fig. 7B). In the absence of Scap, neither the CTD of SREBP2(WT) nor the CTD of SREBP2(R1040E) were precipitated by the nickel beads (lanes 2 and 4). When the CTD of Scap was coexpressed, the CTD of SREBP2(WT), but not that of SREBP2(R1040E), was coprecipitated (compare lane 3 to lane 5). Combined, the results of these three experiments show that arginine 1040 in exon 18 of SREBP2 is crucial for the interaction of the CTD of SREBP2 with the CTD of Scap. To further characterize their interaction, we purified the soluble complex of CTDs using a three-step process as outlined in SI Appendix, Methods. Analytical gel filtration of the final product revealed a single peak corresponding to ∼150 kDa (Fig. 7C), and Coomassie staining showed that this peak contained highly pure forms of the CTDs of both Scap and SREBP2 (Fig. 7 C, Inset). Based on the calculated molecular weights of the tagged CTDs of Scap (58 kDa) and SREBP2 (35 kDa), our data are consistent with a Scap:SREBP2 complex of a 1:1 or 2:2 stoichiometry.
Fig. 7.
Purification of complexes of the CTDs of SREBP2 and Scap and demonstration that the soluble CTD of Scap is sufficient to mask the degradation motif on full-length SREBP2. (A) Domain architecture of SREBP2 and Scap. Starting from its NH2 terminus, SREBP2 contains a bHLH transcription factor domain (green), two transmembrane helices (TM1 and TM2, brown), and a large CTD. Indicated are mutations in exons 18 and 19 of the CTD that disrupt distinct properties of SREBP2. A portion of this CTD is soluble and sufficient to interact with Scap (aa 793 to 1,068, cyan). Starting from its NH2 terminus, Scap contains eight transmembrane helices (TM1 to TM8, brown) and a large CTD. A portion of this region is soluble and sufficient to interact with SREBP2 (aa 751 to 1,276, yellow). (B) Coprecipitation of the soluble CTD of SREBP2 with the soluble CTD of Scap. On day 0, 20-mL cultures of Sf9 insect cells were set up at a density of 1 × 106 cells/mL. After 4 h, cultures were infected with 200 µL each of baculoviruses encoding the soluble CTD of Scap and either WT or mutant versions of the soluble CTD of SREBP2, as indicated. After 48 h, cells were harvested, lysed, and subjected to centrifugation at 20,000 × g for 30 min. A portion of the soluble supernatant (1% of total) was saved and the remainder was used for precipitation of His10-tagged Scap on nickel beads, as described in SI Appendix, Methods. Equal fractions of the soluble supernatants (lysate) and the precipitated material (pulldown eluate) were subjected to immunoblot analysis of SREBP2 (anti-FLAG) and Scap (anti-His). (C) Purification of complexes of the CTD of Scap and the CTD of SREBP2. On day 0, 1-L cultures of Sf9 insect cells were set up at a density of 1 × 106 cells/mL. After 4 h, cultures were infected with 10 mL each of baculoviruses encoding the soluble CTD of Scap and the soluble CTD of SREBP2. After 72 h, cells were harvested, lysed, and subjected to centrifugation at 20,000 × g for 30 min. The soluble supernatant was subjected to a three-step purification process as described in SI Appendix, Methods. An aliquot of the final purified product (650 µg) was subjected to gel filtration chromatography on a Superdex 200 Increase column. Absorbance at 280 nm was monitored continuously to identify the purified complex. Standard molecular weight markers (thyroglobulin, Mr 670,000; γ-globulin, Mr 158,000; ovalbumin, Mr 44,000; and myoglobin, Mr 17,000) were chromatographed on the same column using identical conditions (black circles). (Inset) An aliquot of the peak fraction (10 µg) was subjected to 10% SDS/PAGE followed by Coomassie staining. (D) Soluble CTD of Scap masks the degradation motif on membrane-bound full-length SREBP2. On day 0, Scap-deficient SRD-13A cells were set up in medium G at a density of 700,000 cells per 10-cm dish. On day 1, cells were switched to fresh medium F and transfected as described in SI Appendix, Methods with 2 µg of either WT or the indicated mutant versions of pTK-3xFLAG-SREBP2 along with the indicated amounts of pTK-2xMyc-Scap-CTD. The total amount of DNA in each transfection was adjusted to 6 µg per dish by addition of control pcDNA 3.0 plasmid. On day 2, cells were harvested and equal fractions of whole cell lysates were subjected to immunoblot analysis of SREBP2 (IgG-22D5), Scap CTD (anti-Myc), and actin. P, precursor form of SREBP2; Deg-R, degradation-resistant form of SREBP2.
The results of Fig. 7B confirm the role of R1040 in mediating the binding of the CTD of SREBP2 to the CTD of Scap; however, they do not shed light on whether this binding masks the degradation motif in SREBP2. To answer this question, we conducted cotransfection studies in Scap-null SRD-13A cells to test whether the soluble CTD of Scap would neutralize full-length SREBP2’s COOH-terminal degradation signal (Fig. 7D). Indeed, we detected the precursor form of SREBP2(WT), but not that of SREBP2(R1040E), upon cotransfection of the CTD of Scap (lanes 2 to 5). The CTD of Scap had no effect on SREBP2(R1040E; Deg-R), levels of which were enhanced even in the absence of Scap due to disruption of the degradation signal (lanes 6 and 7).
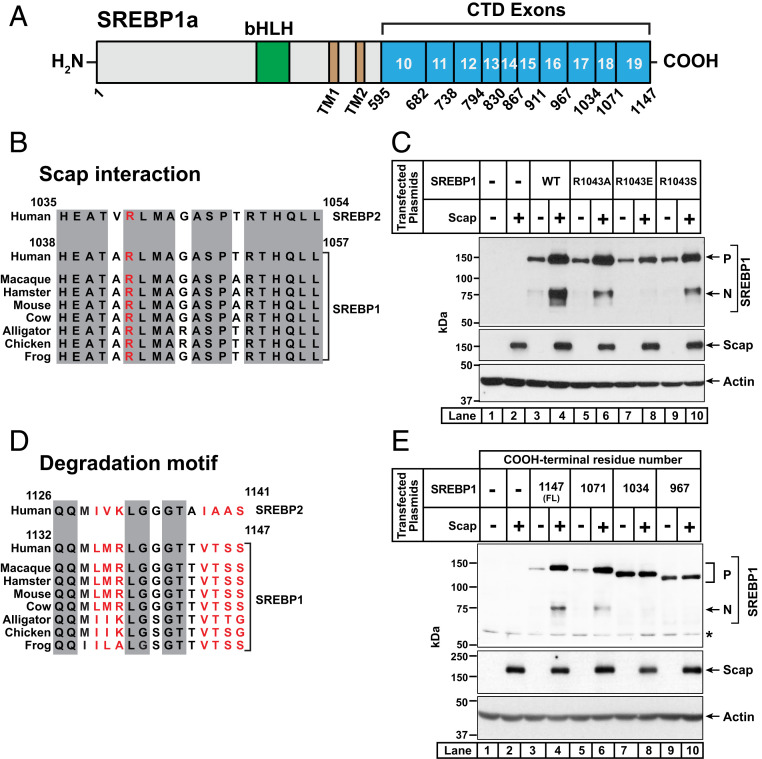
Finally, we asked whether the residues in SREBP2 that mediate interaction with Scap and constitute the degradation motif are conserved in SREBP1. Like SREBP2, SREBP1 also interacts with Scap and is unstable in the absence of Scap (9). The mammalian genome encodes two SREBP1 isoforms, SREBP1a and SREBP1c. The CTDs of SREBP1a and SREBP1c are identical, since these two isoforms are derived from a single gene through use of alternative transcription start sites that produce different forms of exon 1 in the NTD (1). In this study, we used expression plasmids encoding full-length SREBP1a to analyze SREBP1’s CTD. The CTD of human SREBP1 is comprised of 553 amino acids produced by exons 10 through 19 of the SREBP1 gene (Fig. 8A).
Fig. 8.
Identification of regions in the COOH-terminal domain of SREBP1a that are required for its trafficking by Scap and its degradation in the absence of Scap. (A) Domain structure of human SREBP1a. Starting from the NH2 terminus, SREBP1a contains a bHLH transcription factor domain (aa 323 to 394) (green) in its NH2-terminal domain, two transmembrane helices (TM1 and TM2, brown), and a large CTD (aa 595 to 1,147) (cyan). The CTD is comprised of 10 exons (exons 10 to 19), the boundaries of which are indicated. (B and D) Comparison of sequences of SREBP2 and SREBP1. Conservation analyses of residues 1,035 to 1,054 in exon 18 (B) or residues 1,126 to 1,141 in exon 19 (D) of human SREBP2 with the corresponding residues in the indicated species of SREBP1 are shown. Residues that are identical in all species are shaded (gray). Shown in red is the arginine residue, which when mutated to glutamic acid disrupts the interaction of SREBP2 with Scap (B) and the seven residues that mediate degradation of SREBP2 in the absence of Scap (D). Uniprot accession numbers for SREBP1 and SREBP2 sequences are listed in SI Appendix, Methods. Sequences were aligned and % identity was calculated using Clustal Omega. (C and E) Functional analysis of mutant versions of SREBP1a. On day 0, SV-589 cells were set up in medium B at a density of either 300,000 cells/well of 6-well plates (C) or 200,000 cells/well of 12-well plates (E). After 6 h, cells were switched to fresh medium B and transfected as described in SI Appendix, Methods. In C, transfection was carried out with 0.5 µg of pTK-Scap and 1 µg of either WT or mutant versions of pTK-3xFLAG-SREBP1a, as indicated. In E, transfection was carried out with 0.25 µg of pTK-Scap and 0.5 µg of either full-length (FL) or truncated versions of pTK-3xFLAG-SREBP1a, as indicated. The total amount of DNA in each transfection was adjusted to 2 µg per well (C) or 0.75 µg per well (E) by addition of control pTK plasmid. After incubation for 40 h, the transfection medium was removed, and cells were switched to cholesterol-depleting medium E. After further incubation for 1 h, cells were harvested and equal fractions of whole cell lysates were subjected to immunoblot analysis of SREBP1a (anti-FLAG), Scap (IgG-4H4), or actin. bHLH, basic helix–loop–helix leucine zipper; P, precursor form of SREBP1a; N, cleaved nuclear form of SREBP1a; *, non-specific band.
The arginine residue in exon 18 of SREBP2 (R1040) that we identified as being crucial for interacting with Scap (Fig. 5) corresponds to R1043 in exon 18 of SREBP1 and is conserved in all SREBP1 sequences that we examined (Fig. 8B). To test whether R1043 in SREBP1 was important for its interaction with Scap, we generated a series of plasmids encoding full-length SREBP1a with various substitutions of R1043. SV-589 cells were transfected with plasmids encoding the mutant SREBP1s along with either a control plasmid or a plasmid encoding Scap. After depletion of cholesterol, whole cell lysates were subjected to immunoblot analysis for the cleaved NH2-terminal fragment of SREBP1a, generation of which is indicative of proper interaction with Scap. As shown in Fig. 8C, substitution of arginine with glutamic acid dramatically reduced the levels of the cleaved NH2-terminal fragment of SREBP1a (lane 8), whereas substitutions with alanine or serine had little effect (lanes 6 and 10). These results suggest that SREBP1 requires a positive charge at residue 1043 to interact with Scap, similar to SREBP2’s requirement of a positive charge at its residue 1040 (Fig. 5C).
Unlike the conservation of residues crucial for interacting with Scap, the degradation motif in exon 19 of SREBP2 that mediates its stability is not conserved in any of the SREBP1 sequences that we examined (Fig. 8D). To identify regions of SREBP1 required for its degradation in the absence of Scap, we generated plasmids encoding truncated fragments of SREBP1a that terminate after exons 16, 17, and 18. We transfected SV-589 cells with full-length or truncated SREBP1a plasmids along with either a control plasmid or a plasmid encoding Scap. We then depleted the cells of cholesterol and analyzed whole cell lysates by immunoblot (Fig. 8E). As expected, in the absence of Scap, the precursor form of full-length SREBP1a was barely detectable and its levels increased upon cotransfection of Scap (lanes 3 and 4). Scap also mediated the processing of SREBP1 to its nuclear form (lane 4). However, in contrast to our observations with SREBP2, the precursor form of SREBP1a lacking exon 19 was not stable in the absence of Scap (lane 5). Upon cotransfection of Scap, levels of the precursor form of this truncated SREBP1a were increased and processing to its cleaved nuclear form occurred to a similar degree as with full-length SREBP1a (lane 6). Stabilization of the precursor form of SREBP1a in the absence of Scap was only observed when SREBP1a was truncated after exon 17 (lane 7). Cotransfection of Scap did not further increase the level of the precursor form of this truncated form of SREBP1a (lane 8). Moreover, this truncated SREBP1a lacking exons 18 and 19 was not processed to its nuclear form by Scap, consistent with our earlier result in Fig. 8C showing that R1043 in exon 18 of SREBP1a is crucial for its interaction with Scap. Truncation of SREBP1a after exon 16 led to similar results as that observed for truncation after exon 17 (lanes 9 and 10).
This analysis reveals that although both SREBP1 and SREBP2 bind Scap through a common motif, they contain distinct signals that mediate their destruction when not bound to Scap. The degradation signal in SREBP2 is located in exon 19, whereas the degradation signal in SREBP1 is located in another portion of the protein encoded by exon 18.
Discussion
The current study identifies two distinct functional motifs in the CTD of SREBP2 that control cholesterol homeostasis. The first motif is located in the final 13 amino acids at the very end of SREBP2’s COOH-terminal exon 19 and acts as a degradation signal (Fig. 2). This signal triggers the proteasomal degradation of full-length SREBP2 in the ER when it is not bound to Scap (Fig. 3). The second motif, located in the adjoining exon 18, contains a conserved arginine residue (R1040) and acts as a protective signal (Fig. 5). This motif allows the CTD of SREBP2 to bind to the CTD of Scap, an interaction which counteracts the degradation signal and prevents the proteasomal degradation of full-length SREBP2 in the ER (Figs. 6 and 7). Mutation of the degradation signal stabilizes SREBP2 even in the absence of Scap (Fig. 3). Mutation of the arginine residue in exon 18 to oppositely charged glutamic acid disrupts the binding interaction between the CTDs of SREBP2 and Scap and leads to the degradation of SREBP2 even in the presence of Scap (Figs. 6 and 7). These findings give insights into how Scap protects full-length SREBP2 from degradation and how two exons in SREBP2 act in opposing fashions to control this process.
In addition to protecting SREBP2 from degradation in the ER, the Scap-binding motif in exon 18 is also crucial for transporting SREBP2 from the ER to the Golgi when cholesterol levels are low (Figs. 5 and 6). In the Golgi, SREBP2 is sequentially cleaved by S1P and S2P to release its NTD for translocation to the nucleus to activate lipogenic gene transcription. This cleavage leaves the CTD of SREBP2 still bound to Scap in the Golgi. It has long been appreciated that Scap returns to the ER where it is recycled to carry out additional rounds of SREBP2 transport (13–15), but how and where the CTD of the cleaved SREBP2 is removed to liberate Scap remains unknown. Our discovery of the degradation signal in SREBP2’s CTD has allowed us to begin to answer this question. Organelle fractionation studies reveal that the CTD of SREBP2 returns to the ER bound to Scap where it undergoes ERAD (Fig. 4). Mutation of the degradation signal stabilizes the CTD of SREBP2, similar to its effects in stabilizing full-length SREBP2 (Fig. 3). These findings on the cellular itinerary of SREBP2 are summarized in the model shown in Fig. 9. We hope that future structural studies will yield insights into the nature of the conformational changes that must occur in the Scap/SREBP2 complex after cleavage to expose the degradation signal and degrade the CTD. Such clearance of SREBP2’s CTD ensures the recycling of Scap to maintain proper functioning of the SREBP pathway (Fig. 9).
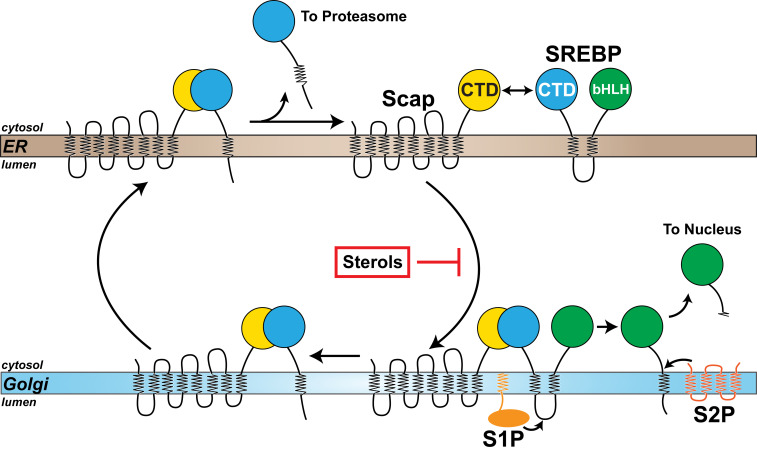
Fig. 9.
A model of the SREBP pathway. SREBP is comprised of a large NTD (green) and a large CTD (cyan), both of which project into the cytosol and are anchored to the ER membrane by two transmembrane helices connected by a short luminal loop. The NTD contains a basic helix–loop–helix leucine zipper transcription factor domain (bHLH) and the CTD binds to the cytosol-facing CTD of Scap (yellow), a cholesterol-sensing membrane protein containing eight transmembrane helices. When cellular cholesterol levels fall below a threshold, Scap escorts SREBP from the ER to the Golgi. Transport is blocked when sterols accumulate. Upon arrival at the Golgi, SREBP is sequentially cleaved by two Golgi-resident proteases, site-1 protease (S1P) and site-2 protease (S2P), releasing its NTD from the membrane for translocation to the nucleus to up-regulate expression of lipogenic genes. Following cleavage by S1P, the CTD of SREBP remains bound to Scap and returns to the ER with Scap. Once in the ER, the CTD of SREBP dissociates from Scap and is degraded by the proteasome, liberating Scap to bind to additional SREBPs and undergo transport to the Golgi if cellular cholesterol levels are still below a threshold set point.
Our study may also explain an interesting observation made in cell lines deficient in S1P (9, 14). When S1P-deficient cells were depleted of cholesterol, Scap escorted SREBP2 to the Golgi as expected, but the absence of cleavage of SREBP2 by S1P led to degradation of Scap in the lysosome. Interestingly, even though Scap was eliminated, SREBP2 was stable (9). Our current findings provide a solution to this paradox. Usually, Scap carries SREBP2 from the ER to the Golgi and then carries the cleaved CTD of SREBP2 from the Golgi back to the ER for elimination by ERAD (Fig. 9). However, in S1P-deficient cells, since Scap is degraded after escorting SREBP2 from the ER to the Golgi, there is no mechanism to return SREBP2 to the ER. Consequently, SREBP2 levels are stable not because of protection by Scap, but because SREBP2 is marooned in the Golgi and kept apart from the ERAD machinery.
While our study focused on SREBP2, our results likely extend to the SREBP1a and SREBP1c isoforms, which are also stabilized and transported from the ER to the Golgi by Scap to ensure homeostasis of fatty acids, phospholipids, and triglycerides (1, 29–31). The CTDs of SREBP1a/c and SREBP2 are not highly conserved (∼50% identity). Our initial analysis revealed that SREBP2 and SREBP1a use the same arginine residue in their respective exon 18 regions for binding to Scap (Fig. 8C). The amino acids adjacent to the arginine residue in both SREBP1a and SREBP2 are also highly conserved, suggesting a common interface for binding to Scap (Figs. 5D and 8B). This interface could be targeted for small molecule binders. A high-affinity binder could disrupt the Scap/SREBP interaction and lead to degradation of SREBPs when they are not bound to Scap. Such compounds could serve as leads into eliminating SREBPs in liver, an outcome that could prevent fatty liver progression (32). In contrast to the common Scap-binding motif, the degradation signal in SREBP1a is located in a different part of the protein (exon 18) compared to SREBP2’s degradation signal which is located in exon 19 (Fig. 8E). In future studies we will study whether this difference plays a role in the regulation of SREBP1, but not SREBP2, by unsaturated fatty acids (33).
Methods
Reagents and materials used in this study, buffers and media, antibodies, plasmids, sequence analysis, cell culture, generation of SREBP2-deficient cells, transient transfection of cells, coimmunoprecipitation assays, organelle fractionation, immunoblot analysis, protein overexpression in Sf9 cells, purification of SREBP2-CTD/Scap-CTD complexes, assay to measure interaction between SREBP2-CTD and Scap-CTD, and reproducibility of data are described in detail in SI Appendix, Methods.
Supplementary Material
Acknowledgments
We thank Mike Brown and Joe Goldstein for their continued encouragement, advice, and critical reading of the manuscript. We also thank Jay Horton, Russell Debose-Boyd, and Jin Ye for many helpful discussions; Yulian Zhou and Gustavo Torres-Ramirez for their assistance during an early phase of this work; Linda Donnelly and Angela Carroll for producing the 2G10 antibody; Karen Chapman, Danya Vazquez, and Daphne Rye for expert technical assistance; and Lisa Beatty, Ijeoma Dukes, Camille Harry, Briana Carter, Elise Morgan, Leticia Esparza, and Shomanike Head for assistance with cell culture. This work was supported by the Welch Foundation (I-1793 to A.R. and I-1770 to D.M.R.), NIH (HL20948 to A.R. and GM116387 to D.M.R.), Fondation Leducq (19CVD04 to A.R.), and the Mallinckrodt Foundation Scholar Award (to D.M.R.). D.L.K. is a recipient of a postdoctoral fellowship from the American Heart Association (18POST34080141).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2018578117/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information.
References
- 1.Brown M. S., Goldstein J. L., The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89, 331–340 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Goldstein J. L., Brown M. S., A century of cholesterol and coronaries: From plaques to genes to statins. Cell 161, 161–172 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shimano H., Sato R., SREBP-regulated lipid metabolism: Convergent physiology–Divergent pathophysiology. Nat. Rev. Endocrinol. 13, 710–730 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Luo J., Yang H., Song B. L., Mechanisms and regulation of cholesterol homeostasis. Nat. Rev. Mol. Cell Biol. 21, 225–245 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Dang E. V., McDonald J. G., Russell D. W., Cyster J. G., Oxysterol restraint of cholesterol synthesis prevents AIM2 inflammasome activation. Cell 171, 1057–1071.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abrams M. E., et al. , Oxysterols provide innate immunity to bacterial infection by mobilizing cell surface accessible cholesterol. Nat. Microbiol. 5, 929–942 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horton J. D., et al. , Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. U.S.A. 100, 12027–12032 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown M. S., Radhakrishnan A., Goldstein J. L., Retrospective on cholesterol homeostasis: The central role of Scap. Annu. Rev. Biochem. 87, 783–807 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rawson R. B., DeBose-Boyd R., Goldstein J. L., Brown M. S., Failure to cleave sterol regulatory element-binding proteins (SREBPs) causes cholesterol auxotrophy in Chinese hamster ovary cells with genetic absence of SREBP cleavage-activating protein. J. Biol. Chem. 274, 28549–28556 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Nohturfft A., Yabe D., Goldstein J. L., Brown M. S., Espenshade P. J., Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes. Cell 102, 315–323 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Sun L. P., Li L., Goldstein J. L., Brown M. S., Insig required for sterol-mediated inhibition of Scap/SREBP binding to COPII proteins in vitro. J. Biol. Chem. 280, 26483–26490 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Sakai J., et al. , Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell 85, 1037–1046 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Nohturfft A., DeBose-Boyd R. A., Scheek S., Goldstein J. L., Brown M. S., Sterols regulate cycling of SREBP cleavage-activating protein (SCAP) between endoplasmic reticulum and Golgi. Proc. Natl. Acad. Sci. U.S.A. 96, 11235–11240 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shao W., Espenshade P. J., Sterol regulatory element-binding protein (SREBP) cleavage regulates Golgi-to-endoplasmic reticulum recycling of SREBP cleavage-activating protein (SCAP). J. Biol. Chem. 289, 7547–7557 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takashima K., et al. , COPI-mediated retrieval of SCAP is crucial for regulating lipogenesis under basal and sterol-deficient conditions. J. Cell Sci. 128, 2805–2815 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Hirano Y., Yoshida M., Shimizu M., Sato R., Direct demonstration of rapid degradation of nuclear sterol regulatory element-binding proteins by the ubiquitin-proteasome pathway. J. Biol. Chem. 276, 36431–36437 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Sundqvist A., et al. , Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF(Fbw7). Cell Metab. 1, 379–391 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Nagoshi E., Yoneda Y., Dimerization of sterol regulatory element-binding protein 2 via the helix-loop-helix-leucine zipper domain is a prerequisite for its nuclear localization mediated by importin beta. Mol. Cell. Biol. 21, 2779–2789 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S. J., et al. , The structure of importin-beta bound to SREBP-2: Nuclear import of a transcription factor. Science 302, 1571–1575 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Sakai J., et al. , Identification of complexes between the COOH-terminal domains of sterol regulatory element-binding proteins (SREBPs) and SREBP cleavage-activating protein. J. Biol. Chem. 272, 20213–20221 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Sakai J., Nohturfft A., Goldstein J. L., Brown M. S., Cleavage of sterol regulatory element-binding proteins (SREBPs) at site-1 requires interaction with SREBP cleavage-activating protein. Evidence from in vivo competition studies. J. Biol. Chem. 273, 5785–5793 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Gong X., et al. , Structure of the WD40 domain of SCAP from fission yeast reveals the molecular basis for SREBP recognition. Cell Res. 25, 401–411 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong X., et al. , Complex structure of the fission yeast SREBP-SCAP binding domains reveals an oligomeric organization. Cell Res. 26, 1197–1211 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ikeda Y., et al. , Regulated endoplasmic reticulum-associated degradation of a polytopic protein: p97 recruits proteasomes to insig-1 before extraction from membranes. J. Biol. Chem. 284, 34889–34900 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X., Sato R., Brown M. S., Hua X., Goldstein J. L., SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell 77, 53–62 (1994). [DOI] [PubMed] [Google Scholar]
- 26.Hua X., Sakai J., Ho Y. K., Goldstein J. L., Brown M. S., Hairpin orientation of sterol regulatory element-binding protein-2 in cell membranes as determined by protease protection. J. Biol. Chem. 270, 29422–29427 (1995). [DOI] [PubMed] [Google Scholar]
- 27.Schmidt O., et al. , Endosome and Golgi-associated degradation (EGAD) of membrane proteins regulates sphingolipid metabolism. EMBO J. 38, e101433 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu D., et al. , PAQR3 modulates cholesterol homeostasis by anchoring Scap/SREBP complex to the Golgi apparatus. Nat. Commun. 6, 8100 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tontonoz P., Kim J. B., Graves R. A., Spiegelman B. M., ADD1: A novel helix-loop-helix transcription factor associated with adipocyte determination and differentiation. Mol. Cell. Biol. 13, 4753–4759 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horton J. D., Goldstein J. L., Brown M. S., SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109, 1125–1131 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rong S., et al. , Expression of SREBP-1c requires SREBP-2-mediated generation of a sterol ligand for LXR in livers of mice. eLife 6, e25015 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moon Y. A., et al. , The Scap/SREBP pathway is essential for developing diabetic fatty liver and carbohydrate-induced hypertriglyceridemia in animals. Cell Metab. 15, 240–246 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hannah V. C., Ou J., Luong A., Goldstein J. L., Brown M. S., Unsaturated fatty acids down-regulate srebp isoforms 1a and 1c by two mechanisms in HEK-293 cells. J. Biol. Chem. 276, 4365–4372 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.