Significance
Although geneticists often emphasize diversity generated during sexual reproduction, genomic alterations that occur during vegetative growth are an important source of genetic diversity. In the current study, we have measured the rate and location of single-base mutations, large deletions/duplications, ploidy alterations, and mitotic recombination events in extensively subcultured diploid isolates of the yeast Saccharomyces cerevisiae. We were able to determine the relative frequency of these events and to predict the frequency of loss of heterozygosity of allelic markers resulting from mitotic recombination. This analysis is relevant to understanding of genomic evolution, as well as the mitotic recombination events in humans that predispose cells to form tumors.
Keywords: chromosome rearrangements, mutations, spontaneous mitotic recombination, loss of heterozygosity
Abstract
Genomic alterations including single-base mutations, deletions and duplications, translocations, mitotic recombination events, and chromosome aneuploidy generate genetic diversity. We examined the rates of all of these genetic changes in a diploid strain of Saccharomyces cerevisiae by whole-genome sequencing of many independent isolates (n = 93) subcloned about 100 times in unstressed growth conditions. The most common alterations were point mutations and small (<100 bp) insertion/deletions (n = 1,337) and mitotic recombination events (n = 1,215). The diploid cells of most eukaryotes are heterozygous for many single-nucleotide polymorphisms (SNPs). During mitotic cell divisions, recombination can produce derivatives of these cells that have become homozygous for the polymorphisms, termed loss-of-heterozygosity (LOH) events. LOH events can change the phenotype of the cells and contribute to tumor formation in humans. We observed two types of LOH events: interstitial events (conversions) resulting in a short LOH tract (usually less than 15 kb) and terminal events (mostly cross-overs) in which the LOH tract extends to the end of the chromosome. These two types of LOH events had different distributions, suggesting that they may have initiated by different mechanisms. Based on our results, we present a method of calculating the probability of an LOH event for individual SNPs located throughout the genome. We also identified several hotspots for chromosomal rearrangements (large deletions and duplications). Our results provide insights into the relative importance of different types of genetic alterations produced during vegetative growth.
As cells divide, the genetic diversity of the population increases as a consequence of multiple processes, including mutations (single-base alterations and small [<100 bp] insertion/deletions [in/dels]), large (>1 kb) deletions and duplications, translocations, and changes in ploidy. In diploid cells, another potential source of genomic diversity is loss of heterozygosity (LOH), an event associated with cross-overs or gene conversions between the two homologs. These processes result in cells with different phenotypes and can contribute to the rapid cellular evolution associated with metastatic cancers (1, 2).
Since the rate of these events is low in most organisms, prior to the advent of whole-genome sequencing, most information about these rates (largely derived from studies in the yeast Saccharomyces cerevisiae) was based on selection of events involving single genes or single chromosomes. More recently, subculturing of yeast strains over many generations, followed by DNA sequencing, has allowed a more global analysis of genomic alterations; this approach is often described as a “mutation accumulation” (MA) study. Below, we summarize the results of single-gene/single-chromosome analysis and MA studies.
Studies of forward mutation rates in the yeast URA3 and CAN1 genes found mutation rates per cell division per base pair of about 4 × 10−10 and 6 × 10−10, respectively (3). In MA experiments, genomic mutation rates were similar, varying between 1 and 3 × 10−10 per base pair per cell division (4–7) for diploids and 3 and 4 × 10−10 for haploids (7, 8).
Another class of genomic alteration is large (>1 kb) interstitial deletions and duplications. In S. cerevisiae, such events usually involve homologous recombination between dispersed repeats such as the Ty class of retrotransposons (9). The locus-specific frequency of this type of event is dependent on a large number of factors: the existence of direct repeats flanking the locus of interest, the distance between the repeats, the sequence similarity between the repeats, and the ability of the cell to tolerate the event. In one early study, the rate of a deletion between Ty elements that removed three adjacent yeast genes was about 10−5 to 10−6 per division (10).
In MA studies, large deletions and duplications also generally involve homologous recombination between nonallelic dispersed repeats (SI Appendix, Fig. S1A) (5, 7, 11–14). The rates of large deletions and duplications are 10−4 to 10−5 per cell division (5, 7, 14).
Translocations in S. cerevisiae also often reflect homologous recombination between dispersed repeats (SI Appendix, Fig. S1 B and C), most frequently Ty elements (9, 15, 16). Umezu et al. (16) found a rate of about 1.2 × 10−5 per division of translocations between Ty elements on the right arm of chromosome III and other Ty elements in the genome. Chan and Kolodner (17) detected translocations formed between a Ty element on chromosome V and other Ty elements in the genome at a rate of about 8 × 10−8 per division.
Translocations can be detected throughout the genome by two methods. First, translocations can result in coupled large terminal deletions and duplications observed by DNA microarrays or DNA sequencing (17, 18). Alternatively, in one MA experiment, translocations were looked for by pulse-field gel electrophoresis, and none were observed in about 5,000 cell divisions of a wild-type diploid (4).
In most MA studies of genomic alterations, ploidy changes in diploid strains are infrequent. In three such studies, whole-chromosome losses or gains in wild-type diploids varied between about 10−5 to 3 × 10−4 per cell division (5, 7, 14), with chromosome gains generally outnumbering chromosome losses. Despite this low rate of aneuploidy, the frequency of aneuploidy in natural isolates is not low. Peter et al. (19) found that about 20% of the 1,000 isolates that were examined were aneuploid. In addition, aneuploidy for specific chromosomes can give a selective growth advantage under specific environmental conditions (20). Last, Hose et al. (21) showed that aneuploidy for chromosome XII slowed the growth rate of a laboratory strain but had little effect on the growth rate of a yeast strain isolated from the wild. Thus, the rate of aneuploidy is likely to be a function of both the genetic background and the specific conditions of subculturing in MA experiments.
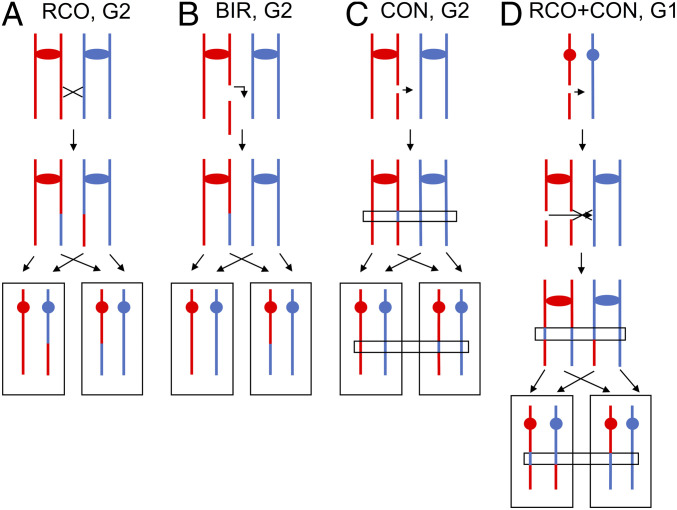
In addition to the genomic alterations that create new DNA sequences or change the copy number of sequences, mitotic recombination events produce genetic diversity. In many diploid organisms, the homologs are heterozygous for many single-nucleotide polymorphisms (SNPs). In two studies in S. cerevisiae, between half and three quarters of natural isolates were heterozygous for thousands of SNPs (19, 22). Mitotic recombination can result in terminal or interstitial loss of heterozygosity (LOH); subsequently, we will refer to these two types of events as T-LOH and I-LOH. Most T-LOH events reflect reciprocal cross-overs or break-induced replication (BIR; Fig. 1 A and B), whereas most I-LOH events are a consequence of gene conversion (Fig. 1C). Although the details of the mechanistic distinction between these two classes of events will be discussed later, T-LOH events result in extensive regions (often >100 kb) of LOH that include all markers located centromere-distal to the breakpoint, whereas I-LOH events generally involve the transfer of relatively small (<10 kb) segments of DNA. However, both types of LOH events can have important phenotypic consequences. For example, loss of a heterozygous tumor-suppressor gene can lead to the production of a malignant cell (23).
Fig. 1.
LOH patterns resulting from gene conversion and cross-overs. Red and blue lines represent the homolog pairs. (A) Terminal LOH resulting from a cross-over in G2 of the cell cycle. Both daughter cells have terminal LOH, although one cell is homozygous for red SNPs and the other is homozygous for blue SNPs. (B) Terminal LOH resulting from break-induced replication in G2. Terminal LOH is observed in only one of the two daughter cells. (C) Interstitial LOH resulting from repair of a DSB in G2. DSB repair usually produces a region of gene conversion, but only a fraction of these conversions is associated with cross-overs (28). This type of conversion event results in an interstitial LOH event (right daughter cell). (D) Terminal LOH and gene conversions resulting from repair of a G1-associated DSB. Spontaneous cross-overs are often initiated by a DSB in G1 that is replicated to produce two sister chromatids broken at the same place. If one DSB is repaired by conversion unassociated with a cross-over and the second by conversion associated with a cross-over, the left cell will have an interstitial blue region of LOH and a terminal red LOH region. The right cell will have only a terminal blue region of LOH.
The rate of mitotic cross-overs is usually measured by selecting loss of heterozygous CAN1 or URA3 markers, although nonselective loss of a visible colony color marker has also been used. Extrapolating the rates of these chromosome-specific methods (24–26) over the entire genome, one calculates a rate of about 5 × 10−4 cross-overs per genome per cell division. Since only half of the segregation events following a cross-over result in detectable LOH (27), the likely rate is about 10−3 per cell division. The rate of spontaneous mitotic conversions per gene is usually estimated for a single site within a gene; this rate is about 10−6 per cell division per bp (28, 29); based on an average conversion tract of 6 kb, the expected genomic rate of conversion is about 5 × 10−3 per genome per cell division.
To measure accurate LOH rates in MA experiments and to locate the genomic regions impacted, one must use diploids that are heterozygous for closely spaced SNPs. The rate of T-LOH events per cell division in four studies (6, 14, 30, 31) varied from 4 × 10−4 to 1.3 × 10−3 per division. The average rate of I-LOH events (gene conversions) in the same studies varied from 4 × 10−3 to 1.2 × 10−2 per division.
Several further points concerning conversions and cross-overs should be mentioned. First, most mitotic cross-overs are associated with an adjacent gene conversion tract (26). Second, most meiotic conversions unassociated with cross-overs involve a different mechanism (synthesis-dependent strand annealing) than cross-overs (32). Third, in studies in which recombination events in both daughter cells are examined, it is clear that many spontaneous mitotic events are initiated by double-stranded DNA breaks (DSBs) that occur in unreplicated chromosomes (26). The broken chromosome is replicated, and the resulting two broken chromatids are subsequently repaired (Fig. 1D).
In this study, we emphasize LOH events rather than other classes of genome alterations for several reasons. First, in most previous MA studies of genome instability in diploid cells, relatively few events were observed, and the diploids had the potential to undergo meiotic levels of recombination. In the current study, we identified more than 1,000 LOH events in spo11 diploids that lack the ability to perform meiotic recombination. Second, in our study, we mapped a sufficient number of I- and T-LOH events to determine that their distributions were not the same, suggesting that these two classes of events were initiated by different types of DNA lesions or that the recombinogenic lesions were processed differently. Third, LOH events are an important contributor to one pathway of tumor formation. Knudson (33) first suggested that retinoblastomas were formed as a consequence of a “two-hit” process: generation of a mutation in one allele followed by loss of the wild-type allele by a somatic event. Although there are a number of types of somatic events that eliminate the wild-type allele (mutation or deletion of the wild-type copy, for example), LOH events resulting from mitotic recombination is one important pathway (23, 34). Thus, our analysis of the mechanism of mitotic recombination should contribute to our understanding of the formation of cancers that are promoted by mitotic recombination.
Results
Experimental System and Rationale.
To monitor the spontaneous genomic alterations of a diploid genome, we constructed a diploid that was heterozygous for about 55,000 single-nucleotide polymorphisms (SNPs) by crossing haploid strains derived from a RAD5 derivative of W303-1A (35, 36) and the clinical isolate YJM789 (37) (SI Appendix, Table S1-1). Both haploids are wild-type for known mutations affecting genome stability, although W303-1A has a mutation in the SSD1 gene that affects the growth rate of aneuploid derivatives (21); this mutation would be heterozygous in the diploid used in our experiments. In addition, the experimental diploid (WYspo11) was constructed to be homozygous for mutations in SPO11. Strains with this mutation cannot undergo meiosis (38), ensuring that any genomic alterations observed after passaging diploid isolates are a consequence of mitotic recombination rather than meiotic recombination.
We subcultured 93 isolates of WYspo11 from single cells to colonies on rich growth medium. To avoid selecting fast-growing variants, we picked the colony on each plate that was closest to a mark placed on the plate before streaking. Ten isolates were subcultured 60 times, and 83 were subcultured 120 times. Since a colony contains about 2 × 107 cells, these passages correspond to 1,500 (60 × 25) and 3,000 (120 × 25) cell divisions per isolate, or a total of 264,000 cell divisions for all isolates. Following subculturing, the diploids were sequenced using Illumina technology; coverage was ∼140-fold. After removing the SNPs shared between W303-1A and YJM789 relative to S288c and the SNPs in repeated genes, 45,174 SNPs were used to analyze genomic alterations (13, 39).
Rate and Spectra of Single-Base Mutations and Small Insertion/Deletions (In/Dels).
We identified 1,265 single-base mutations among the 93 sequenced isolates. Because of the ambiguities in comparing sequences within repeated genes, we restricted our analysis to single-copy yeast genes as described in Sui et al. (40). Based on the size of the yeast genome (excluding the ribosomal RNA genes) of about 23 Mb and the total number of cell divisions for the isolates (264,000, assuming 25 cell divisions per subcloning), the rate of mutations per bp per cell division is about 2.1 × 10−10; this rate is similar to that previously reported in MA experiments in yeast diploids as described in the Introduction. All mutations are shown in Dataset S1.
As expected for diploid strains, most of the mutations were heterozygous, but about 2.3% were homozygous. Of the 29 homozygous mutations detected, 28 were in regions of LOH and likely reflect mutations that occurred prior to the LOH event. Homozygous mutations induced in a wild-type diploid were previously shown to involve this mechanism (mutation in one homolog followed by a mitotic cross-over) (41). The single homozygous mutation that was not associated with a detectable LOH event may reflect a gene conversion event that did not include a heterozygous SNP and that was therefore undetectable. It should be pointed out that our explanation for the homozygous mutations is very similar to the two-hit mechanism for retinoblastoma hypothesized by Knudson (33).
The distribution of point mutations on the chromosomes is shown in SI Appendix, Fig. S2. The number of mutations per chromosome is strongly correlated with chromosome length (r2 = 0.96). Although some degree of clustering is observed, no very strong mutational hotspots were observed. The types of base substitutions are shown in SI Appendix, Fig. S3. Although we have not investigated the sources of these mutations, about half to two thirds of spontaneous mutations in wild-type yeast reflect the activity of the error-prone DNA polymerase zeta (42, 43).
In addition to the single-base mutations, we observed 72 short in/dels (<100 bp; Datasets S1–S3 and S1-4) and 12 complex mutations (Dataset S1-5). The short in/dels can be separated into two classes, those that are less than 10 bp (59 mutations), most of which occur in mononucleotide tracts (Dataset S1-3), and those that are ≥10 bp and are flanked by short direct repeats (13 mutations; Dataset S1-4). Both of these classes are likely to be a consequence of DNA polymerase slippage, with the second class reflecting a more extensive dissociation of primer and template strands (44, 45).
The complex events are defined as those in which more than one sequence change occurs within 10 bp. Most of these clustered changes are consistent with the mutagenic effects of DNA polymerase epsilon that can induce multiple closely linked mutations (43). In three of the complex mutations, however, the alterations reflect a short inversion that occurs between short palindromic sequences (Dataset S1-5). Such mutations may represent template switching during DNA replication (SI Appendix, Fig. S4).
Most Large Deletions/Duplications and Translocations Are Mediated by Homologous Recombination between Ectopic Repeats.
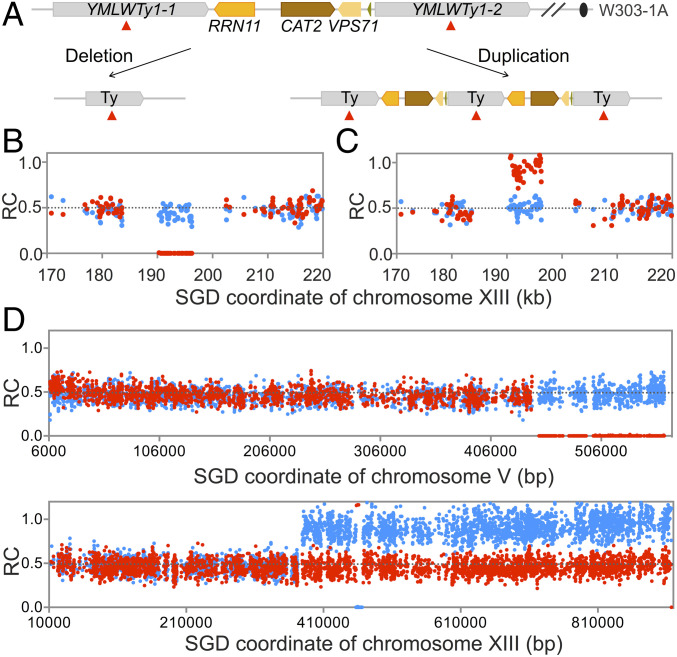
We observed 47 chromosomal rearrangements in which a chromosome segment (>1 kb) of one homolog was deleted (n = 35) or duplicated (n = 12) while the other homolog remained unchanged. The rate of these events is 1.8 × 10−4 per isolate per cell division, considerably less than rates of gene conversions or cross-overs (to be discussed below). Examples of internal deletions and internal duplications are shown in Fig. 2 A–C. The deletions and duplications had repeated genes at their breakpoints (Dataset S2-1), with Ty elements or delta elements (the long terminal repeats [LTRs] that flank Ty elements or are solo elements) being the most common type of repeats at the breakpoints; such events likely reflect unequal sister-strand recombination between nonallelic repeats (SI Appendix, Fig. S1A).
Fig. 2.
Patterns of deletions and duplications resulting from recombination between nontandem repeats. (A) Chromosomal region that is a hotspot for deletions and duplications on chromosome XIII. Both deletions and duplications are a consequence of homologous recombination between YMLWTy1-1 and YMLWTy1-2. (B) Heterozygous deletion between YMLWTy1-1 and YMLWTy1-2. Red and blue dots reflect the number of “reads” of W303-specific and YJM789-specific SNPs, respectively. The number of reads of W303-specific and YJM789-specific SNPs are divided by the average number of W303-specific plus the YJM789-specific reads for all SNPs in the genome, resulting in the RC (ratio of coverage) of W303-specific and YJM789-specific SNPs. Thus, SNPs represented in zero, one, or two copies in the genome have RC values of about 0, about 0.5, and about 1, respectively. The deleted region corresponds to the region containing the RRN11, CAT2, and VSP71 genes on the W303-derived chromosome. (C) Heterozygous duplication between YMLWTy1-1 and YMLWTy1-2. (D) Terminal deletion on chromosome V and terminal duplication on chromosome XIII in isolate Spo11-188. The breakpoint of the deletion is at YERCTy1-1, and the duplication breakpoint is at YMRCTy1-4. As shown in SI Appendix, Fig. S1 B and C, pairs of terminal deletions and duplications likely reflect translocations formed by recombination between repeats on nonhomologous chromosomes. This translocation was confirmed by other methods as described in the text.
Several regions were found to be duplicated or deleted in multiple independent isolates (SI Appendix, Fig. S5). For example, a region of 6 kb on chromosome XIII containing the RRN11, CAT2, and VPS71 genes located between two flanking Ty1 elements was deleted in four isolates and duplicated in two isolates (Fig. 2 A–C). On the YJM789-derived XV homolog, there is a 2-kb duplication of the sequences encoding RDL1 and RDL2. This region underwent frequent deletion (eight isolates) and duplication (one isolate) events, a rate of about 3.4 × 10−5 per division. In addition, one duplication was a consequence of formation of a circular derivative of chromosome III, as a consequence of recombination between two directly oriented Ty elements flanking the centromere (SI Appendix, Fig. S6A).
In addition to interstitial duplications and deletions, we observed three terminal duplications and five terminal deletions (Datasets S2-2 and S2-3). Most of these terminal events (six of eight) are located within 30 kb of a telomere. One event, involving breakpoints displaced from the telomeres, was an isolate that had both a terminal deletion with a breakpoint near YERCTy1-1 on chromosome V and a terminal duplication with a breakpoint at YMRCTy1-4 on chromosome XIII (Fig. 2D). Such paired terminal deletions and duplications with breakpoints in repetitive elements reflect a translocation formed by an ectopic cross-over or BIR event (SI Appendix, Fig. S1 B and C); translocations formed by this mechanism are common in strains under replication stress (13). This translocation was confirmed by analysis of chromosomal DNA using contour-clamped homogeneous electric field (CHEF) gels (SI Appendix, Fig. S6B). It is noteworthy that we found only a single translocation by detecting coupled deletions and duplications. Although we would not be able to detect balanced translocations by this method, our results indicate that the rate of unselected translocations is very low, 3.8 × 10−6 per isolate per cell division. This rate is about 370-fold less than the rate of allelic cross-overs (discussed below).
Deletions and Duplications from the Ribosomal RNA Gene Cluster and the Cluster of CUP1 Genes.
In addition to dispersed repeats, S. cerevisiae has two large tandem arrays of genes, the rRNA gene cluster (about 100 9.1 kb repeats per array located on chromosome XII) (46) and the CUP1 cluster (variable numbers of repeats in different strains) (47). Since both the rRNA genes and the CUP1 genes have polymorphisms that distinguish those on the two homologs (13), we could monitor the numbers of repeats on each homolog separately by determining their sequence coverage relative to single-copy genes.
In the parental diploid WYspo11, before subculturing, the W303-1A- and the YJM789-derived homologs had about 123 and 72 ribosomal DNA repeats, respectively. As shown in Dataset S3, the numbers of repeats on the W303-1A–derived homolog varied between 24 and 233; values of 0 indicated that a cross-over/BIR event occurred centromere-proximal to the rRNA gene cluster. The number of repeats on the YJM789-derived chromosome varied between 2 and 177. The changes in the numbers of rRNA genes in each isolate is shown in SI Appendix, Fig. S7A.
The total number of repeats in each isolate (relative to the starting strain) varies less than the number of repeats per homolog. In many of the isolates (76 of 93), a reduction in the number of one type of repeat is partially compensated for by an increase in the other type of repeats (Dataset S3). In the 60 of 76 isolates with compensating changes in the numbers of repeats, the W303-1A repeats were preferentially elevated relative to the YJM789 repeats. Two mechanisms that could produce this pattern of rDNA alterations are discussed in the SI Appendix and are depicted in SI Appendix, Fig. S8.
Based on the fraction of the genome occupied by the rRNA gene cluster (about 0.07) and the total number of cross-over/BIR events (n = 356), we expect 25 events within the cluster if the cluster has the same rate of recombination per kb as the rest of the genome. We observed 19 events within the cluster (Dataset S4-1), an insignificant difference by χ2 analysis (P = 0.26), indicating that the rRNA gene cluster is not a preferred target for mitotic recombination between homologs. It is likely that a substantial fraction of the DSBs that occur in the rRNA genes are repaired by sister-chromatid exchange or single-strand annealing rather than interhomolog events; such events cannot be detected by LOH.
Changes in the numbers of the CUP1 repeats were less frequent than observed in the rRNA genes, presumably reflecting target size (Dataset S3 and SI Appendix, Fig. S7B). Most alterations occurred in the W303-associated array (12 repeats) instead of the YJM789-associated array (6 repeats).
Rate of Aneuploidy.
We observed 17 aneuploid events among 15 different isolates, including 12 trisomes, 1 tetrasome, 3 monosomes, and 1 uniparental disome (2 copies of 1 parental homolog and none of the other). The rate of aneuploidy was about 6.4 × 10−5 per cell division, similar to the rate observed in previous studies (5, 7). As pointed out in the Introduction, it is possible that this rate will be different in other genetic backgrounds or in cells subcultured in different environmental conditions. Ten chromosomes (I, II, III, IV, V, VII, VIII, IX, XI, and XIV) were involved in these aneuploidy events (Dataset S5). Consistent with previous observations (5), trisomies outnumber monosomies by a factor of four, and the monosomes involve two of the smallest yeast chromosomes (chromosomes I and III). A simple explanation of this bias is that monosomes are likely to grow more slowly than trisomes.
To determine whether aneuploidy would affect the rate of other genetic events, we compared the numbers of mitotic recombination events and point mutations in euploid and aneuploid isolates that were subcultured 120 times (Dataset S5-3). By the Mann–Whitney U test, no significant differences were found in either the number of recombination events (P = 0.67) or the number of mutations (P = 0.34) in euploid and aneuploid isolates.
Rates of LOH Events Resulted from Mitotic Gene Conversions or Cross-Overs.
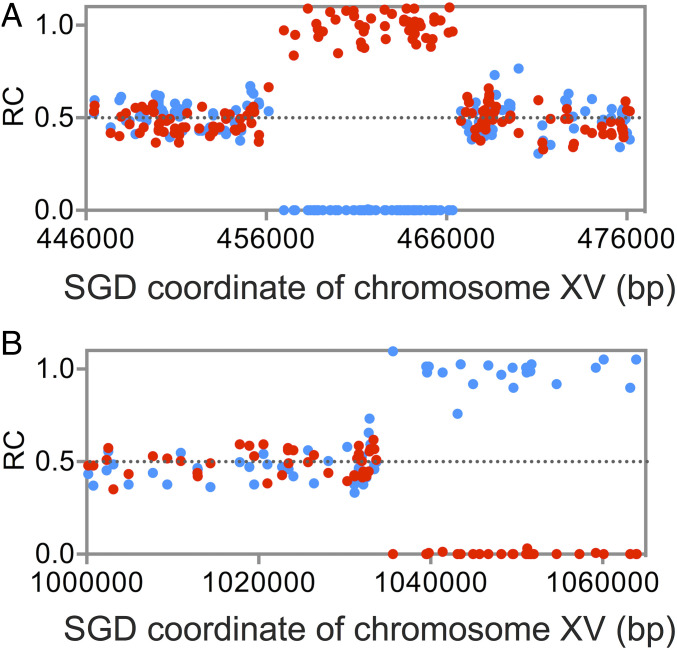
The two most common genetic alterations detected by DNA sequencing were interstitial (I) and terminal (T) LOH events (Fig. 1). Although I-LOH can be a consequence of deletion of sequences from one homolog (SI Appendix, Fig. S1A), such heterozygous deletions were rare (30 events) relative to interstitial events reflecting gene conversion (859 events). The distinction between these events is based on allelic-specific SNP coverage. For heterozygous deletions, the SNPs representing one homolog are present in one copy per cell, while those of the other homolog are reduced to zero (Fig. 2B). For gene conversion events, the SNPs representing one homolog are elevated by a factor of two, whereas those representing the other homolog are reduced to zero (Fig. 3A).
Fig. 3.
Examples of interstitial and terminal LOH events as determined by DNA sequence analysis. As in Fig. 2, we show the RC (ratio of coverage) for each SNP, with W303- and YJM789-derived SNPs depicted as red and blue dots, respectively. (A) Interstitial LOH event. There is a ∼10-kb region in which there are no YJM789-derived SNPs and a double dose of W303-derived SNPs, as expected for the gene conversion event. (B) Terminal LOH event. The RC values of SNPs are about 0.5 until coordinate 1035000. Distal to that coordinate, the isolate has no sequences derived from W303 and a double dose of counts from the YJM789 homolog. This pattern is expected for a reciprocal cross-over or a BIR event.
Of the 859 conversion events, there were 765 simple conversions in which a continuous region of one homolog is transferred to the other and 94 complex conversions in which a conversion tract derived from one homolog is interrupted with sequences derived from the other homolog or by heterozygous regions (Dataset S4-2). Complex events have been observed in our previous studies and likely reflect “patchy” repair of mismatches in heteroduplexes, branch migration of Holliday junctions, and/or template switching, all initiated by DSBs formed in either G1 or G2 (26, 48–50); examples of mechanisms that could give rise to complex conversion events associated with a cross-over are shown in SI Appendix, Fig. S9. The total rate of I-LOH events, summed over the 93 sequenced isolates, is 859 of 264,000 or 3.3 × 10−3 per cell division. It should be pointed out that we cannot distinguish conversion events initiated in G1 from those initiated in G2, since we did not analyze both daughter cells involved in the event in the present study, unlike some previous studies (26, 51).
The locations, sizes, and patterns for I-LOH events are given in Dataset S4. If two LOH events on the same chromosome were separated by more than 10 kb, they were counted as two separate events. The median size of gene conversion tracts was 2.8 kb. In a study of spontaneous conversion at the URA3 locus, the median conversion tract unassociated with cross-overs was 6.4 kb in length (29), and the median tract lengths in other MA studies (6, 14, 31) varied between 1.6 and 3.4 kb. Some of the variation in the tract lengths in the MA studies may reflect the relatively small number of events analyzed in the previous studies (about 20).
There were 356 T-LOH events (example in Fig. 3B). Such events could reflect a reciprocal cross-over (RCO) in G2 (Fig. 1A) or a BIR event (Fig. 1B). The distinction between RCO and BIR requires genetic methods in which all products of the mitotic event can be detected. In such studies, in wild-type strains, reciprocal cross-overs are about 10 times more common than BIR (30, 52). The rate of T-LOH events was 356 of 264,000 or 1.4 × 10−3 per cell division. As noted previously, the rate of mitotic cross-overs is about twice the rate of T-LOH events (27), or 2.8 × 10−3 per cell division.
Most cross-overs are associated with a contiguous region of gene conversion (26, 28). If all mitotic products derived from a single G2-associated event can be analyzed, conversion events associated with a cross-over are detectable as the difference in LOH junctions in the two daughter cells (26). If only one of the daughter cells is examined, as in our experiments, the conversion tract associated with the cross-over cannot be detected for G2-initiated events. However, a fraction of the conversion tracts associated with a G1-initiated recombination event can be detected (Fig. 1D). A total of 79% of the T-LOH events had no observable conversion tracts (simple cross-overs), whereas the remainder had an adjacent conversion tract. The median size of conversion tracts associated with cross-overs was 9.0 kb, considerably longer than the conversion tracts unassociated with cross-overs (2.8 kb). This observation is consistent with studies showing greater conversion tract lengths associated with cross-overs in both spontaneous (53) and DNA damage-induced events (48).
Distribution of I-LOH Events (Gene Conversions) within the Yeast Genome.
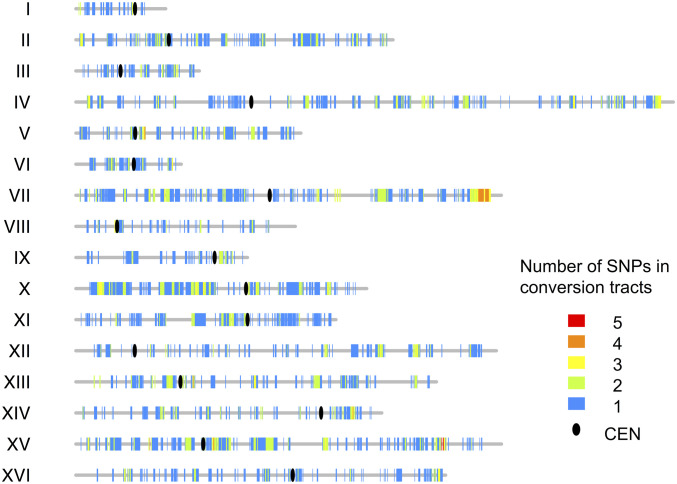
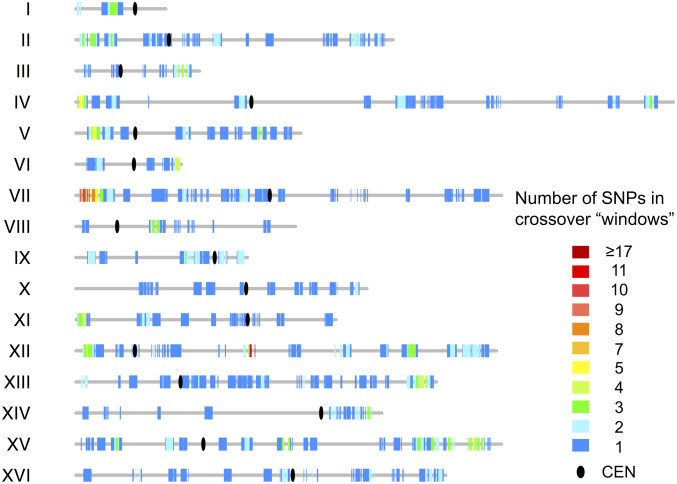
The distribution of I-LOH events along the chromosomes is shown in Fig. 4. For interstitial events, the heterozygous SNPs flanking the region(s) of LOH were used to define the conversion tracts, and all SNPs within this region were counted. The conversion events are widely and relatively uniformly distributed over the yeast chromosomes, in contrast to the more uneven distribution of meiotic recombination events (54, 55). The numbers of conversion events per chromosome were proportional to the chromosome size (Dataset S4-5).
Fig. 4.
Distribution of interstitial LOH events (gene conversions) along the chromosomes. The vertical lines of different colors indicate the number of times that SNPs were included in gene conversion tracts. The ovals represent centromeres, and the gray lines show chromosomes.
As in previous studies (for example, ref. 26), we determined whether the conversion tracts, which presumably contain the site of the DNA lesion initiating the conversion event, had overrepresentations of a variety of chromosome elements (Dataset S4-4). We found significant associations between the breakpoints of conversion events and replication-termination (Ter) sequences and regions with unusually high or low GC content. In our previous study of cross-over–associated conversion tracts on chromosome IV (26), we found an overrepresentation of Ter sequences, G-rich quadruplex sequences, delta/Ty elements, and Rrm3p pause sites. Most of these elements are associated with slow-moving or stalled DNA replication forks.
Distribution of T-LOH Events (BIR/CO) within the Yeast Genome.
We also examined the distribution of the breakpoints of T-LOH events within the chromosomes (Fig. 5); the breakpoints were defined as a 20-kb window flanking the midpoint of the transition between homozygous and heterozygous SNPs at the border of the LOH event. As for the conversion events, the BIR/CO events were distributed at many positions over all of the chromosomes, although the left end of chromosome VII had an unusually large number of cross-overs. We examined the associations between cross-over breakpoints and various elements of chromosome structure as described for conversion events. There were overrepresentations of G4 quadruplex sequences, regions with high levels of gamma H2AX, regions with high GC content, and noncoding RNA genes (Dataset S4-4). There was also a significant underrepresentation of delta repeats and the promoters of weakly transcribed genes. The only motif associated with both I- and T-LOH events was high-GC base composition.
Fig. 5.
Distribution of terminal LOH breakpoints along the chromosomes. For each terminal LOH event, we defined a “window” of 20 kb that was centered on the middle of the interval between the heterozygous SNP and the homozygous SNP that define the LOH breakpoint. As in Fig. 4, the vertical lines of different colors indicate the number of times that SNPs were included in these windows. The hotspot for cross-overs on chromosome XII is misleading because the SGD (Saccharomyces Genome Database) coordinates show the 100-repeat rRNA gene cluster as a 2-repeat cluster. As discussed in the text, the rRNA gene cluster does not have more cross-overs than expected based on its physical length.
Differences in Breakpoints between I- and T-LOH Events.
Aside from the differences in associations described above, the two types of LOH events had different patterns with respect to the telomeres and centromeres. The T-LOH events, but not the I-LOH events, were significantly enriched in the region within 20 kb of the SNPs closest to the telomeres (P < 0.0001; SI Appendix, Table S2). In contrast, I-LOH events were significantly more common in regions within 15 kb of the centromere (P = 0.02). There were large chromosomal regions with very few terminal LOH events, particularly on chromosomes IV and XIV. In addition, cross-over events were particularly enriched near the left telomere of chromosome VII (P < 0.0001; calculated in SI Appendix), and this enrichment was not observed for I-LOH events. Possible explanations for these differences will be given in the Discussion.
Discussion
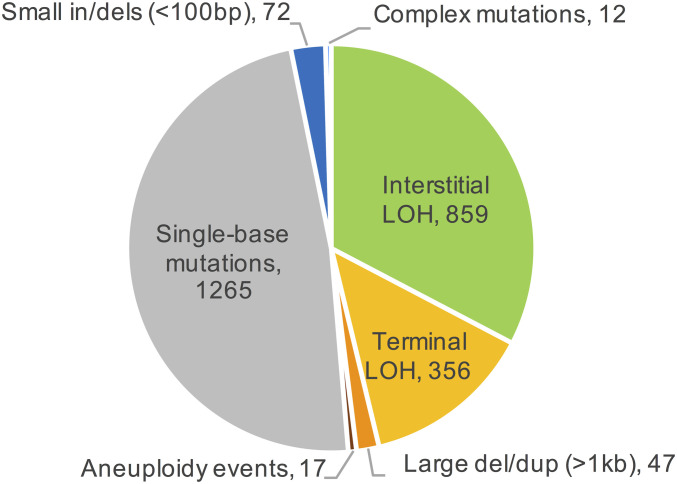
By investigating a large number of diploid isolates subcultured for many generations, we have obtained accurate quantitative and qualitative measurements of the rates of multiple types of mitotic genetic alterations including mutations, deletions and duplications, and LOH events. A summary of the numbers of these alterations is shown in Fig. 6. Our findings are as follows. 1) Rates and spectra of mutations are in general agreement with previous studies. 2) Large (>1 kb) deletions and duplications are primarily the result of homologous recombination between nonallelic, but closely linked, retrotransposons; the tandemly repeated rRNA and CUP1 genes have high rates of instability. 3) Translocations and aneuploidy are rare relative to other types of gross chromosome rearrangements. 4) Gene-conversion events and cross-overs have different genomic distributions, suggesting that these events may be initiated and/or resolved by different mechanisms. 5) Finally, mitotic LOH events, both interstitial and terminal, are common, occurring at a rate of about 4.7 × 10−3 events per genome per cell division. We present a simple method of calculating the probability per cell division of an LOH event for an individual SNP. In S. cerevisiae, these probabilities are largely a function of the distance of the SNP from the centromere.
Fig. 6.
Numbers of different classes of genomic alterations summed over 93 subcultured WYspo11 isolates. A total of 2,628 events were observed.
Frequency and Spectra of Mutations.
The rate of single-bp mutations in our study was about 2 × 10−10 per bp per cell division, similar to the rates of 1 to 3 × 10−10 per bp per cell division observed by others. As in previous studies, the rates of small (<10 bp) in/dels are less than 10% of the rates of single-bp mutations. Most of the in/dels occur within mononucleotide tracts or between short direct repeats, and likely reflect DNA polymerase slippage. These data, based on diploids without genome-destabilizing mutations grown under unstressed conditions, serve as a baseline for examining strains under replication stress or strains with a mutator phenotype.
Large Deletions and Duplications.
In agreement with many previous studies done in S. cerevisiae, most large deletions and duplications are a consequence of homologous recombination between repeated genes. These events usually involve closely linked repeats. Only one translocation was observed, consistent with previous results indicating that recombination occurs more frequently between repeats on the same homolog than between repeats on nonhomologs (56). Translocations were also infrequent in a diverse collection of yeast strains (47); 79 of 100 samples had colinear chromosomes.
As expected from previous mitotic studies of the rRNA and CUP1 gene tandem arrays (57–60), we observed frequent size changes of the arrays. These size changes likely reflect a number of mechanisms, including unequal sister-strand recombination, gene conversion, single-strand annealing, and BIR. The relative importance of these mechanisms has not been completely resolved. We found that loss of repeats from one cluster is often associated with gain of repeats from the other cluster, suggesting that there is selection for an optimal number of repeats under the growth conditions of our experiment.
Differences in the Distribution of Interstitial and Terminal LOH Events.
The distributions of interstitial and cross-over/BIR breakpoints (Figs. 4 and 5) differ in a variety of ways. In general, the distribution of events appears more uniform for conversion events than the terminal LOH events. As discussed previously, the T-LOH events, unlike the I-LOH events, are enriched near the telomeres. Such patterns are unexpected if both types of events are initiated by the same type of DNA lesion and processed to generate conversions and cross-overs with a fixed probability.
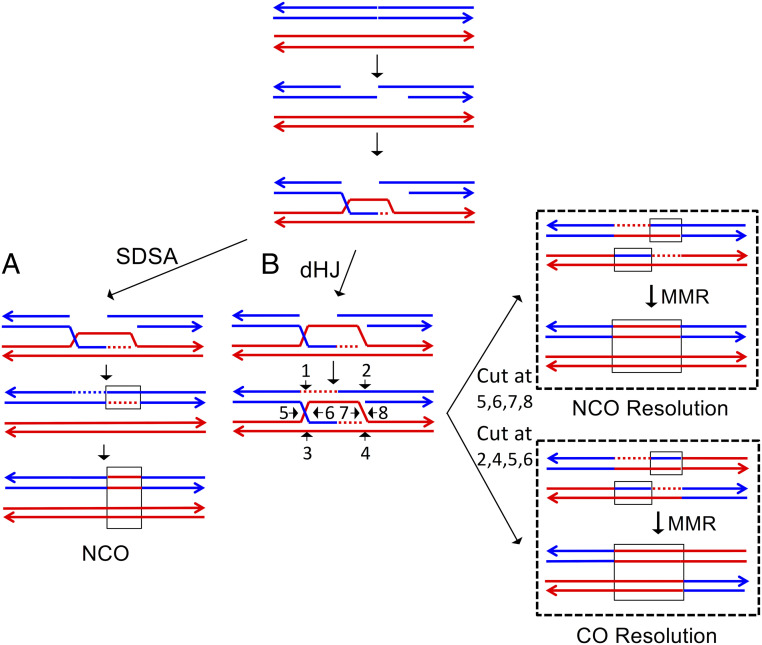
In meiosis in S. cerevisiae, Allers and Lichten (32) showed that intermediates that lead to non–cross-over conversions appear earlier than those that result in cross-overs, and that the two types of exchange are under different genetic regulation. They also showed physical evidence that conversions unassociated with cross-overs are primarily a consequence of the synthesis-dependent strand annealing pathway (SDSA; Fig. 7A), whereas cross-overs with associated conversions were produced by formation and resolution of double Holliday junctions (Fig. 7B). In addition, Mancera et al. (61) found that meiotic hotspots for cross-overs and conversions were not always coincident. Our results suggest the same separation may exist in mitosis. Based on the relatively few relevant studies, it is difficult to generalize about the relative frequencies of I- and T-LOH events in human cancers. In retinoblastomas, T-LOH events appear more common (34), whereas, in other types of solid tumors (gastric, glioma, and lung), I-LOH events are the predominant class (62).
Fig. 7.
Simplified form of the double-strand break-repair model. In this figure, recombination is initiated on the blue chromatid, and the broken ends are resected 5′ to 3′. The broken end invades the homologous chromatid, forming a D-loop. Two possible outcomes of the strand invasion are shown in A and B. It should be emphasized that recent studies of patterns of heteroduplex formation during meiotic and mitotic recombination indicate that additional steps (branch migration, “patchy” mismatch repair, and strand switching) are required to explain some recombination events. (A) Synthesis-dependent strand annealing (SDSA). Following strand invasion, the end of the invading strand is used as a primer for DNA synthesis, resulting in a longer D-loop. The invading strand is then extruded, pairing with the other broken end. The resulting heteroduplex may contain mismatches that can be repaired to produce a conversion event (enclosed in a rectangle) unassociated with a cross-over. (B) Formation of a double Holliday junction. Following strand invasion and DNA synthesis primed from the invading strand, the D-loop pairs with the second broken end. The resulting junctions can be cleaved in a variety of ways as indicated by the numbered arrows. Cleavage at positions 5, 6, 7, and 8 results in a region of conversion without an associated cross-over. Cleavage at positions 2, 4, 5, and 6 results in a conversion tract associated with a cross-over.
The difference in the location of breakpoints for I- and T-LOH events can be explained by a number of mechanisms. It is possible that the DNA lesions responsible for spontaneous I- and T-LOH events are different. For example, spontaneous conversion events could be primarily a consequence of repair of nicked DNA (28) and T-LOH events could reflect repair of DSBs. In addition, recombinogenic DSBs could be generated in two ways: by an interstitial break on the chromosome or by terminal degradation of the end as a consequence of telomere disfunction. One interpretation of the observation that T-LOH events are nonrandomly near the telomeres is that a subset of terminal LOH events result from repair of a terminally degraded chromosome.
Alternatively, the different distribution of I- and T-LOH events could result from different modes of processing the same initiating lesion. In the current models of recombination (28), both the SDSA and DSBR pathways are initiated by a DSB (Fig. 7). In both pathways, the initial step involves 5′-3′ excision of the broken end, followed by invasion of the broken end into the unbroken homolog. In the SDSA model, after limited DNA synthesis, the invading end dissociates and anneals with the other broken end (Fig. 7A). In the DSBR pathway, after more extensive DNA synthesis, the noninvading broken end pairs with the resulting D-loop, forming a double Holliday junction (Fig. 7B). Consistent with these models, the lengths of both meiotic and mitotic conversion tracts are longer for cross-over–associated events than for non–cross-over conversions (53, 61). Based on these observations, one explanation for the different distribution of I-LOH and T-LOH events is that DNA synthesis primed by the invading strand is more processive in some chromosome regions than others, and this processive synthesis is more likely to result in a T-LOH event than an I-LOH event.
Another interesting conclusion from our data is that the numbers of I- and T-LOH events are different by only a factor of 2.4. Assuming that the cross-overs are associated with conversions (26), we conclude that about 30% of the conversions are associated with cross-overs. Although about two thirds of meiotic conversion events are associated with cross-overs (61), in previous mitotic studies, the percentage of conversion events associated with cross-overs varied from 10% to 50% [reviewed by Yim et al., 2014 (29)]. Many of these previous studies, however, were done by selecting recombination between heteroalleles at ectopic chromosome locations, which may bias the results. In studies of allelic gene conversion, Yim et al. (29) found that about 40% of mitotic gene conversions were associated with cross-overs. Our current results are consistent with this estimate.
Calculation of Expected SNP-Specific Rates of LOH.
The expected rates of LOH (RLOH) for individual SNPs are the sum of two rates: the rate expected from I-LOH events (RI-LOH) and the rate expected from T-LOH events (RT-LOH). Since mitotic gene conversions are distributed reasonably uniformly in the genome in our analysis, we will assume that RI-LOH is the same for all SNPs. There are two related methods for calculating RI-LOH: 1) multiply the genomic rate of interstitial LOH times the ratio of the average conversion tract size divided by the total genome size or 2), using Dataset S6, calculate the average number of SNPs that undergo LOH in each isolate and divide by the number of cell divisions per isolate. The details of these calculations are in the SI Appendix. Both methods lead to an RI-LOH value of about 10−6 per SNP per division.
The second rate (RT-LOH) is a function of the distance of the SNP from the centromere of the chromosome, since a cross-over occurring anywhere in this interval will result in LOH for centromere-distal SNPs. Therefore, we can estimate the rate of LOH caused by cross-overs/BIR events for individual SNPs by multiplying the rate of terminal LOH events in the genome (1.4 × 10−3 per cell division) by the ratio of the distance of the SNP from the centromere (in kb) divided by the total genome length (12.5 Mb). Thus, RT-LOH will vary from 0 (for SNPs very close to the centromere) to about 2 × 10−4 per SNP per cell division for a marker located at the end of the right arm of chromosome XII, the longest chromosome arm in the genome.
The RT-LOH values can be used to calculate the expected number of isolates undergoing LOH by multiplying these values by the aggregate number of cell divisions (N = 264,000). The details of these calculations are in the SI Appendix, and a summary of the expected numbers of T-LOH events for SNPs located near the ends of the chromosome arms is in Dataset S7. In general, there is good agreement between the expected and observed numbers of T-LOH events. For example, for an SNP located near the end of chromosome XII, the predicted number of events is about 52; the observed number is 47. One exception to this generalization is the right arm of chromosome IV, where we expected 32 events and observed only 11. As discussed in the SI Appendix, the likely explanation of this discrepancy is that cross-overs on the right arm of IV result in loss of the ade2-1–suppressing SUP4 insertion. Cells homozygous for the unsuppressed ade2-1 allele accumulate a red pigment that results in slow growth and, therefore, a reduced level of such cells in an MA experiment.
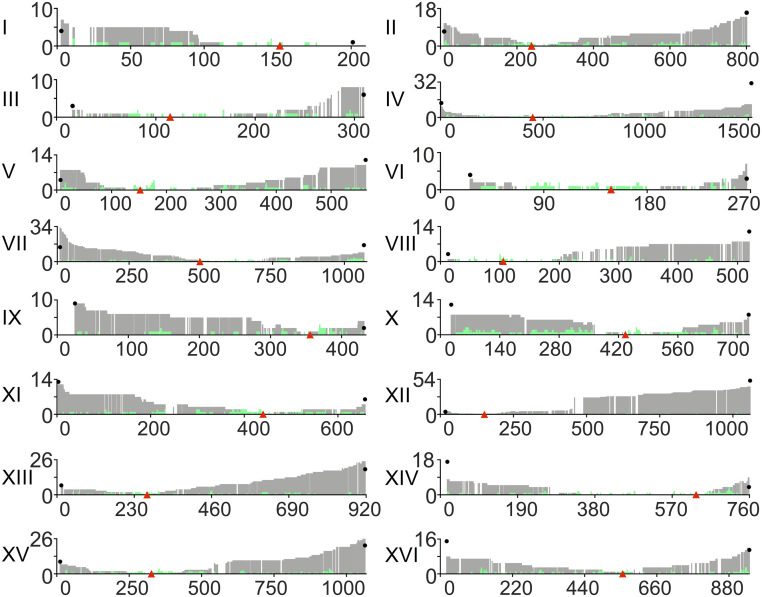
In summary, the expected rate of LOH per cell division for SNPs can be estimated by the following equation: rate of LOH per SNP per cell division = 10−6 + [(CEN–SNP distance in kb/12.5 Mb) × 1.4 × 10−3]. For SNPs located more than 100 kb from the centromere, >90% of the LOH rate will be a consequence of T-LOH events rather than gene conversion. This conclusion assumes that most terminal LOH events are a consequence of repair of random DSBs. The observed numbers of LOH events for each SNP are shown in Fig. 8. A similar pattern of increasing LOH as SNPs get further from the centromere has also been observed in other studies with smaller datasets (14, 22, 31).
Fig. 8.
Number of times SNPs underwent LOH at different sites along the yeast chromosomes. The gray and green colors indicate T-LOH and I-LOH, respectively. Red triangles show the positions of centromeres. Numbers on the y axis are the numbers of LOH events at each SNP, and numbers on the x axis are SGD coordinates (in kb). Black dots at the ends of the chromosome indicate the expected level of LOH at the ends of the chromosomes resulting from cross-overs (as calculated in Dataset S7).
Based on Dataset S6, we calculated that the average LOH rate per SNP per division was 2.6 × 10−5. As expected, the rates for different SNPs have a very wide range. SNPs near the end of chromosome XII have a rate of LOH of about 1.6 × 10−4, and those near the centromere of XII have a rate of 3.8 × 10−6. In several other species in the Saccharomycodaceae family, Nguyen et al. (63) calculated average LOH rates per SNP/division varying between 2 and 11 × 10−6. In our view, since the rate of LOH per SNP in S. cerevisiae is largely a function of the distance between the SNPs and the centromere, the average rate has limited utility.
Several other points should be mentioned. First, our conclusions are based on S. cerevisiae. Other organisms could have higher or lower rates of recombination or a different ratio of gene conversions to cross-overs. For example, in Daphnia pulex, the rate of LOH per SNP per generation is about 8 × 10−8, and most of the LOH events are deletions rather than conversions or cross-overs (64). It is likely that, in organisms (such as S. cerevisiae) in which LOH events per SNP increase as a function of distance from the centromere, cross-overs and BIR events are the drivers of LOH. Second, the LOH events in our experiments were distributed relatively evenly over the isolates (Datasets S4–S6). Thus, our conclusions are not derived from a small subset of cells with unusually high levels of recombination.
In summary, our results provide a global view of spontaneous genomic alterations in unstressed diploid yeast cells. Our analysis and those of others show that mitotic recombination events are frequent enough to be an important source of diversity. In organisms such as S. cerevisiae, these events will produce variants that may have selective advantages in certain environments (65–67). In humans, such events can release cells from normal growth regulation, initiating tumor formation.
Materials and Methods
Strain Construction and Subculturing Procedure.
The diploid strain used in our study, WYspo11, was a spo11/spo11 derivative isogenic with diploid strains used in our previous study (26). The haploids used to generate the diploid (Wspo11 and Yspo11) were derived from W303-1A and YJM789 (SI Appendix, Table S1-1); the resulting diploid is heterozygous for about 50,000 SNPs. Wspo11 and Yspo11 were constructed by transforming W1588-4C and JSC20-1 with PCR fragments that allow the replacement of SPO11 with the kanMX gene. These fragments were generated by amplification of the plasmid pUG6 DNA (68) with primers dspo11S and dspo11A (SI Appendix, Table S1-2).
For subculturing, we grew independent isolates of WYspo11 at 30 °C from single cells to colonies (about 25 divisions per passage) on solid YPD media (2% glucose, 2% peptone, 1% yeast extract, and 2% agar). A total of 83 isolates were grown for 120 passages, and 10 were grown for 60 passages.
Whole-Genome Sequencing of Diploid Strains.
Genomic DNA was extracted using the Omega yeast DNA kit (Life Science Products) and sequenced by 150-bp pair-end strategy of the Illumina NextSeq 500 platform. Details of the sequencing are described in the SI Appendix.
Supplementary Material
Acknowledgments
We thank all members of the Petes and Jinks-Robertson labs and H. Klein, K. T. Nishant, P. Magwene, S. Otto, T. James, and L. Argueso for useful suggestions. The research was supported by grants from the National Natural Science Foundation of China (32022004), the Natural Science Foundation of Zhejiang Province (LY18C060002), and the Zhoushan City–Zhejiang University Joint Specific Project (2019C81055) to D.-Q.Z., the National Natural Science Foundation of China (31800055) to K.Z., and the NIH and the US Army (R35GM118020 and W911NF1920082) to T.D.P.
Footnotes
The authors declare no competing interest.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2018633117/-/DCSupplemental.
Data Availability.
All Illumina DNA sequencing data are available in the National Center for Biotechnology Information (NCBI) Bioproject: Sequence Read Archive (accession no. PRJNA314677) (69).
References
- 1.Aguilera A., Gómez-González B., Genome instability: A mechanistic view of its causes and consequences. Nat. Rev. Genet. 9, 204–217 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Jeggo P. A., Pearl L. H., Carr A. M., DNA repair, genome stability and cancer: A historical perspective. Nat. Rev. Cancer 16, 35–42 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Lang G. I., Murray A. W., Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics 178, 67–82 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishant K. T., et al. , The baker’s yeast diploid genome is remarkably stable in vegetative growth and meiosis. PLoS Genet. 6, e1001109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu Y. O., Siegal M. L., Hall D. W., Petrov D. A., Precise estimates of mutation rate and spectrum in yeast. Proc. Natl. Acad. Sci. U.S.A. 111, E2310–E2318 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dutta A., et al. , Genome dynamics of hybrid Saccharomyces cerevisiae during vegetative and meiotic divisions. G3 (Bethesda) 7, 3669–3679 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharp N. P., Sandell L., James C. G., Otto S. P., The genome-wide rate and spectrum of spontaneous mutations differ between haploid and diploid yeast. Proc. Natl. Acad. Sci. U.S.A. 115, E5046–E5055 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynch M., et al. , A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc. Natl. Acad. Sci. U.S.A. 105, 9272–9277 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mieczkowski P. A., Lemoine F. J., Petes T. D., Recombination between retrotransposons as a source of chromosome rearrangements in the yeast Saccharomyces cerevisiae. DNA Repair (Amst.) 5, 1010–1020 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Liebman S. W., Singh A., Sherman F., A mutator affecting the region of the iso-1-cytochrome c gene in yeast. Genetics 92, 783–802 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCulley J. L., Petes T. D., Chromosome rearrangements and aneuploidy in yeast strains lacking both Tel1p and Mec1p reflect deficiencies in two different mechanisms. Proc. Natl. Acad. Sci. U.S.A. 107, 11465–11470 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H., et al. , Gene copy-number variation in haploid and diploid strains of the yeast Saccharomyces cerevisiae. Genetics 193, 785–801 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng D.-Q., Zhang K., Wu X.-C., Mieczkowski P. A., Petes T. D., Global analysis of genomic instability caused by DNA replication stress in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 113, E8114–E8121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pankajam A. V., Dash S., Saifudeen A., Dutta A., Nishant K. T., Loss of heterozygosity and base mutation rates vary among Saccharomyces cerevisiae hybrid strains. G3 (Bethesda) 10, 3309–3319 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaleff D. T., Fink G. R., Genetic events associated with an insertion mutation in yeast. Cell 21, 227–237 (1980). [DOI] [PubMed] [Google Scholar]
- 16.Umezu K., Hiraoka M., Mori M., Maki H., Structural analysis of aberrant chromosomes that occur spontaneously in diploid Saccharomyces cerevisiae: Retrotransposon Ty1 plays a crucial role in chromosomal rearrangements. Genetics 160, 97–110 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan J. E., Kolodner R. D., A genetic and structural study of genome rearrangements mediated by high copy repeat Ty1 elements. PLoS Genet. 7, e1002089 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemoine F. J., Degtyareva N. P., Lobachev K., Petes T. D., Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell 120, 587–598 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Peter J., et al. , Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 556, 339–344 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Selmecki A. M., et al. , Polyploidy can drive rapid adaptation in yeast. Nature 519, 349–352 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hose J., et al. , The genetic basis of aneuploidy tolerance in wild yeast. eLife 9, e52063 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Magwene P. M., et al. , Outcrossing, mitotic recombination, and life-history trade-offs shape genome evolution in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 108, 1987–1992 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lasko D., Cavenee W., Nordenskjöld M., Loss of constitutional heterozygosity in human cancer. Annu. Rev. Genet. 25, 281–314 (1991). [DOI] [PubMed] [Google Scholar]
- 24.Hartwell L. H., Smith D., Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics 110, 381–395 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein H. L., Spontaneous chromosome loss in Saccharomyces cerevisiae is suppressed by DNA damage checkpoint functions. Genetics 159, 1501–1509 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.St Charles J., Petes T. D., High-resolution mapping of spontaneous mitotic recombination hotspots on the 1.1 Mb arm of yeast chromosome IV. PLoS Genet. 9, e1003434 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chua P., Jinks-Robertson S., Segregation of recombinant chromatids following mitotic crossing over in yeast. Genetics 129, 359–369 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Symington L. S., Rothstein R., Lisby M., Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics 198, 795–835 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yim E., O’Connell K. E., St Charles J., Petes T. D., High-resolution mapping of two types of spontaneous mitotic gene conversion events in Saccharomyces cerevisiae. Genetics 198, 181–192 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Connell K., Jinks-Robertson S., Petes T. D., Elevated genome-wide instability in yeast mutants lacking RNase H activity. Genetics 201, 963–975 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.James T. Y., et al. , Adaptation by loss of heterozygosity in Saccharomyces cerevisiae clones under divergent selection. Genetics 213, 665–683 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allers T., Lichten M., Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106, 47–57 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Knudson A. G., Jr, Mutation and cancer: Statistical study of retinoblastoma. Proc. Natl. Acad. Sci. U.S.A. 68, 820–823 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hagstrom S. A., Dryja T. P., Mitotic recombination map of 13cen-13q14 derived from an investigation of loss of heterozygosity in retinoblastomas. Proc. Natl. Acad. Sci. U.S.A. 96, 2952–2957 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas B. J., Rothstein R., Elevated recombination rates in transcriptionally active DNA. Cell 56, 619–630 (1989). [DOI] [PubMed] [Google Scholar]
- 36.Zhao X., Muller E. G., Rothstein R., A suppressor of two essential checkpoint genes negatively affects dNTP pools. Mol. Cell 2, 329–340 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Wei W., et al. , Genome sequencing and comparative analysis of Saccharomyces cerevisiae strain YJM789. Proc. Natl. Acad. Sci. U.S.A. 104, 12825–12830 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Esposito R. E., Frink N., Bernstein P., Esposito M. S., The genetic control of sporulation in Saccharomyces. II. Dominance and complementation of mutants of meiosis and spore formation. Mol. Gen. Genet. 114, 241–248 (1972). [DOI] [PubMed] [Google Scholar]
- 39.Zheng D.-Q., Petes T. D., Genome instability induced by low levels of replicative DNA polymerases in yeast. Genes (Basel) 9, 539 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sui Y., et al. , Analysis of APOBEC-induced mutations in yeast strains with low levels of replicative DNA polymerases. Proc. Natl. Acad. Sci. U.S.A. 117, 9440–9450 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gordenin D. A., Inge-Vechtomov S. G., [Mechanism of mutant induction in the ade2 gene of diploid Saccharomyces cerevisiae yeasts by ultraviolet rays]. Genetika 17, 822–831 (1981). [PubMed] [Google Scholar]
- 42.Quah S.-K., von Borstel R. C., Hastings P. J., The origin of spontaneous mutation in Saccharomyces cerevisiae. Genetics 96, 819–839 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Makarova A. V., Burgers P. M., Eukaryotic DNA polymerase ζ. DNA Repair (Amst.) 29, 47–55 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strand M., Prolla T. A., Liskay R. M., Petes T. D., Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature 365, 274–276 (1993). [DOI] [PubMed] [Google Scholar]
- 45.Kiktev D. A., Sheng Z., Lobachev K. S., Petes T. D., GC content elevates mutation and recombination rates in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 115, E7109–E7118 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petes T. D., Yeast ribosomal DNA genes are located on chromosome XII. Proc. Natl. Acad. Sci. U.S.A. 76, 410–414 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strope P. K., et al. , The 100-genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome Res. 25, 762–774 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yin Y., Petes T. D., Genome-wide high-resolution mapping of UV-induced mitotic recombination events in Saccharomyces cerevisiae. PLoS Genet. 9, e1003894 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hum Y. F., Jinks-Robertson S., DNA strand-exchange patterns associated with double-strand break-induced and spontaneous mitotic crossovers in Saccharomyces cerevisiae. PLoS Genet. 14, e1007302 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marsolier-Kergoat M.-C., Khan M. M., Schott J., Zhu X., Llorente B., Mechanistic view and genetic control of DNA recombination during meiosis. Mol. Cell 70, 9–20.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Lee P. S., Petes T. D., From the cover: Mitotic gene conversion events induced in G1-synchronized yeast cells by gamma rays are similar to spontaneous conversion events. Proc. Natl. Acad. Sci. U.S.A. 107, 7383–7388 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ho C. K., Mazón G., Lam A. F., Symington L. S., Mus81 and Yen1 promote reciprocal exchange during mitotic recombination to maintain genome integrity in budding yeast. Mol. Cell 40, 988–1000 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aguilera A., Klein H. L., Yeast intrachromosomal recombination: Long gene conversion tracts are preferentially associated with reciprocal exchange and require the RAD1 and RAD3 gene products. Genetics 123, 683–694 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gerton J. L., et al. , Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 97, 11383–11390 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pan J., et al. , A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144, 719–731 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lichten M., Haber J. E., Position effects in ectopic and allelic mitotic recombination in Saccharomyces cerevisiae. Genetics 123, 261–268 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Szostak J. W., Wu R., Unequal crossing over in the ribosomal DNA of Saccharomyces cerevisiae. Nature 284, 426–430 (1980). [DOI] [PubMed] [Google Scholar]
- 58.Gangloff S., Zou H., Rothstein R., Gene conversion plays the major role in controlling the stability of large tandem repeats in yeast. EMBO J. 15, 1715–1725 (1996). [PMC free article] [PubMed] [Google Scholar]
- 59.Casper A. M., Mieczkowski P. A., Gawel M., Petes T. D., Low levels of DNA polymerase alpha induce mitotic and meiotic instability in the ribosomal DNA gene cluster of Saccharomyces cerevisiae. PLoS Genet. 4, e1000105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao Y., et al. , Properties of mitotic and meiotic recombination in the tandemly-repeated CUP1 gene cluster in the yeast Saccharomyces cerevisiae. Genetics 206, 785–800 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mancera E., Bourgon R., Brozzi A., Huber W., Steinmetz L. M., High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454, 479–485 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kumar Y., et al. , Massive interstitial copy-neutral loss-of-heterozygosity as evidence for cancer being a disease of the DNA-damage response. BMC Med. Genomics 8, 42–62 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nguyen D. T., et al. , Variable spontaneous mutation and loss of heterozygosity among heterozygous genomes in yeast. Mol. Biol. Evol., 10.1093/molbev/msaa150 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Flynn J. M., Chain F. J., Schoen D. J., Cristescu M. E., Spontaneous mutation accumulation in Daphnia pulex in selection-free vs. competitive environments. Mol. Biol. Evol. 34, 160–173 (2017). [DOI] [PubMed] [Google Scholar]
- 65.Magwene P. M., “Revisiting Mortimer’s genome renewal hypothesis: heterozygosity, homothallism, and the potential for adaptation in yeast” in Ecological Genomics, Landry C., Aubin-Horth N., Eds (Springer, 2014), pp. 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smukowski Heil C. S., et al. , Loss of heterozygosity drives adaptation in hybrid yeast. Mol. Biol. Evol. 34, 1596–1612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sampaio N. M. V., Watson R. A., Argueso J. L., Controlled reduction of genomic heterozygosity in an industrial yeast strain reveals wide 2 cryptic phenotypic variation. Front. Genet. 10, 782 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gueldener U., Heinisch J., Koehler G. J., Voss D., Hegemann J. H., A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 30, e23 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sui Y., et al. , Bioproject: Saccharomyces cerevisiae Raw Sequence Reads. National Center for Biotechnology Information Sequence Read Archive. https://www.ncbi.nlm.nih.gov/sra/PRJNA314677. Accessed 12 August 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All Illumina DNA sequencing data are available in the National Center for Biotechnology Information (NCBI) Bioproject: Sequence Read Archive (accession no. PRJNA314677) (69).