Significance
Little is known regarding the mechanisms involved in loss of androgen receptor (AR) function in the development of castration resistance. Recently, the clinical use of new potent AR therapies has led to the emergence of a new lethal subtype of castration-resistant prostate cancer (CRPC) lacking AR protein and signaling, termed ARL/negative CRPC. We discovered that DDX3 (DEAD-box helicase 3 X-linked) is a key repressor of AR protein translation—a mechanism that leads to development of castration resistance. Inhibition of DDX3 in ARL/negative CRPC promotes the expression of AR protein/signaling and hence sensitivity to hormone therapy. These data provide a molecular mechanism for hormone action and describe a synthetic lethality approach to the clinical treatment of therapy-resistant diseases such as CRPC.
Keywords: androgen independence, castration-resistant prostate cancer, double-negative prostate cancer, posttranscriptional regulation, prostate cancer
Abstract
Prostate cancer (CaP) driven by androgen receptor (AR) is treated with androgen deprivation; however, therapy failure results in lethal castration-resistant prostate cancer (CRPC). AR-low/negative (ARL/−) CRPC subtypes have recently been characterized and cannot be targeted by hormonal therapies, resulting in poor prognosis. RNA-binding protein (RBP)/helicase DDX3 (DEAD-box helicase 3 X-linked) is a key component of stress granules (SG) and is postulated to affect protein translation. Here, we investigated DDX3-mediated posttranscriptional regulation of AR mRNA (messenger RNA) in CRPC. Using patient samples and preclinical models, we objectively quantified DDX3 and AR expression in ARL/− CRPC. We utilized CRPC models to identify DDX3:AR mRNA complexes by RNA immunoprecipitation, assess the effects of DDX3 gain/loss-of-function on AR expression and signaling, and address clinical implications of targeting DDX3 by assessing sensitivity to AR-signaling inhibitors (ARSI) in CRPC xenografts in vivo. ARL/− CRPC expressed abundant AR mRNA despite diminished levels of AR protein. DDX3 protein was highly expressed in ARL/− CRPC, where it bound to AR mRNA. Consistent with a repressive regulatory role, DDX3 localized to cytoplasmic puncta with SG marker PABP1 in CRPC. While induction of DDX3-nucleated SGs resulted in decreased AR protein expression, inhibiting DDX3 was sufficient to restore 1) AR protein expression, 2) AR signaling, and 3) sensitivity to ARSI in vitro and in vivo. Our findings implicate the RBP protein DDX3 as a mechanism of posttranscriptional regulation for AR in CRPC. Clinically, DDX3 may be targetable for sensitizing ARL/− CRPC to AR-directed therapies.
Background
Prostate cancer (CaP) is an androgen-driven disease with signaling mediated through androgen receptor (AR) (1). This hormone-induced signaling cascade is targeted with androgen deprivation therapy, initially resulting in tumor regression; however, therapy failure occurs in 10–20% of cases, and subsequent recurrence is known as castration-resistant prostate cancer (CRPC) (2). Mechanisms driving CRPC can be generally classified as AR-dependent or -independent (2, 3). Recent introduction of more effective AR-signaling inhibitors (ARSI), enzalutamide (ENZ) and abiraterone, has led to increased incidences of AR-low/negative (ARL/−) CRPC (4, 5). While most AR-dependent CRPCs can be treated with ARSI, there are few options for treatment of ARL/− CRPC. Furthermore, patients who have developed resistance to a first-line ARSI rarely respond to treatment with a different ARSI (6). These accumulated clinical data highlight the need to identify novel mechanisms driving resistance that can be therapeutically targeted.
Several types of CRPC exhibit an ARL/− expression pattern including neuroendocrine prostate cancer (NEPC), double-negative prostate cancer (DNPC), and AR-low prostate cancer (ARLPC) (4, 5); however, mechanisms of AR repression [i.e., methylation (7, 8), AR protein degradation (9, 10), RNA-binding-proteins (RBPs) (11–13), and microRNAs (14, 15)] are not fully understood in this context. Recently, studies in Drosophila melanogaster have revealed the RBP, Belle, mediates translational regulation of target mRNAs (messenger RNAs) in response to steroid hormone pulses during fruit fly development (16). DDX3 (DEAD-box helicase 3 X-linked; gene symbol DDX3X), the ortholog of Belle in mammalian cells, has been implicated in multiple cancers including CaP (17, 18), but its role as a translational regulator in prostatic disease remains unstudied. As an RNA helicase, DDX3 aids or prevents translation of target mRNA depending on the cellular conditions (19, 20). Under normal conditions, DDX3 facilitates translation of mRNA by resolving the secondary structure of the 5′ untranslated region (21). Alternatively, under stress conditions, DDX3, along with other translation initiation factors (i.e., PABP1) form ribonucleoprotein congregates known as stress granules (SGs) that prevent or delay translation of the sequestered mRNA (19, 20). In this role, DDX3 has been identified as an SG-nucleating factor and is necessary for SG formation (19, 22).
In the present study, we explored RBP-mediated translational regulation of AR mRNA in CRPC. Our findings identify AR mRNA present in ARL/− CRPC subtypes, provide insight into mechanisms of posttranscriptional regulation of AR, and suggest a potential targetable pathway for treatment of ARL/− CRPC.
Methods
Patient Samples and Immunostaining.
Patient samples were acquired from the Prostate Cancer Biorepository Network. The hormone-resistance tissue microarray (TMA) (Institutional Review Board #8723) contains 56 hormone-naive and -resistant cores, with four cores per patient (n = 14) (SI Appendix, Table S4). The patient-derived xenograft (PDX) TMA contains 41 PDXs with multiple cores per xenograft. These PDXs include CRPC subtypes AR+ (n = 5), ARL/− (DNPC, ARLPC) (n = 3), and NEPC (n = 4) (SI Appendix, Table S3). All antibodies are detailed in SI Appendix, Table S1, and multiplexed immunohistochemistry (IHC) was performed as described (23, 24) (SI Appendix, Fig. S1). Western immunoblotting and immunofluorescence (IF) were performed according to Abcam protocol (17) and are described in greater detail in SI Appendix.
Cell Culture and Induced Cell Stress.
The BPH1 to cancer progression (BCaP) cell lines were from our own stocks (17, 25), lymph node carcinoma of the prostate (LNCaP) cell line and derivatives were obtained from American Type Culture Collection (ATCC), and Los Angeles Prostate Cancer 4 (LAPC4) cell line and derivatives were gifted from Donald Vander Griend (University of Illinois at Chicago, Chicago, IL) (26). All cell lines were grown as described (17, 26) and authenticated by ATCC or University of Wisconsin (UW)–Madison core services between 6 mo and 1 y prior to experimental use. To induce hypoxic stress, 0.25% sodium azide in full media was incubated on cells for 3 h (22). Additional details regarding cell lines used in this study can be found in SI Appendix.
RNA Immunoprecipitation, In Situ Hybridization, and qPCR.
For RNA immunoprecipitation (RIP) experiments, Protein G dynabeads (Thermo, 10004D) were bound to DDX3 or IgG antibodies and incubated with lysates for 4 h at 4 °C. RNA was isolated using Qiagen RNAeasy Mini kit (Cat. No. 74104). RNA samples were analyzed using 1) Agilent 2100 bioanalyzer or 2) quantitative PCR (qPCR) according to Biorad protocol (SI Appendix, Table S2). RIP qPCR was quantified using Sigma Imprint Analysis Calculations, and in situ hybridization (ISH) was performed as described (27). RNAscope was performed according to Advanced Cell Diagnostics recommendations and quantified using CellProfiler (28). Details on RIP protocols and analysis can be found in SI Appendix.
DDX3 Overexpression and Inhibition.
Overexpression was achieved by transient transfection of DDX3 in a mammalian expression vector driven by the cytomegalovirus promoter (pcDNA3.1-DDX3+YFP) plasmid using TransIT-X2 (Mirus, MIR6000) according to MirusBio protocol. DDX3 was inhibited genetically by transfection of six pooled small interfering RNAs (siRNA) targeting DDX3 (Dharmacon, L-006874-02-0005; nontargeting, D-001810-10-05) and pharmacologically with 2 µM RK33 (Selleck Chem, S8246) for 48 h. Cell viability was assessed by MTT (29). Briefly, cells were treated with dimethyl sulfoxide (DMSO), 10 μM bicalutamide (BICA; Sigma-Aldrich B9061) or ENZ (Selleck Chem, S1250), 2 uM RK33, or cotreatment, followed by colorimetric quantification of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) precipitate.
Renal Capsule Xenografts.
All animal experiments were conducted under protocols approved by the UW Animal Care and Use Committee and detailed in SI Appendix. Briefly, C42 xenografts (350,000 cells per graft in rat-tail collagen) and BCaPMT10 (∼5 mg tissue) were engrafted under the renal capsule of adult male athymic Nu/Nu mice as described (30). Treatment groups included untreated control (UNT), 25 mg BICA, 20 mg/kg RK33, and cotreatment, n = 4–6 per treatment. RK33 injections were administered as previously described (18, 31).
Statistics.
Student’s t tests and one-way ANOVAs with Tukey’s test were performed using GraphPad/Prism version 7.05 (GraphPad Software, Inc.). Bar graphs in figures 1–5 (1 B–C, G; 2 C–D; 3E, G; 4 D–F; 5 A–B, F, H) represent sample mean ± SEM. Significance is represented by *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
Results
ARL/− CRPC Expresses AR mRNA and High Levels of RBP DDX3.
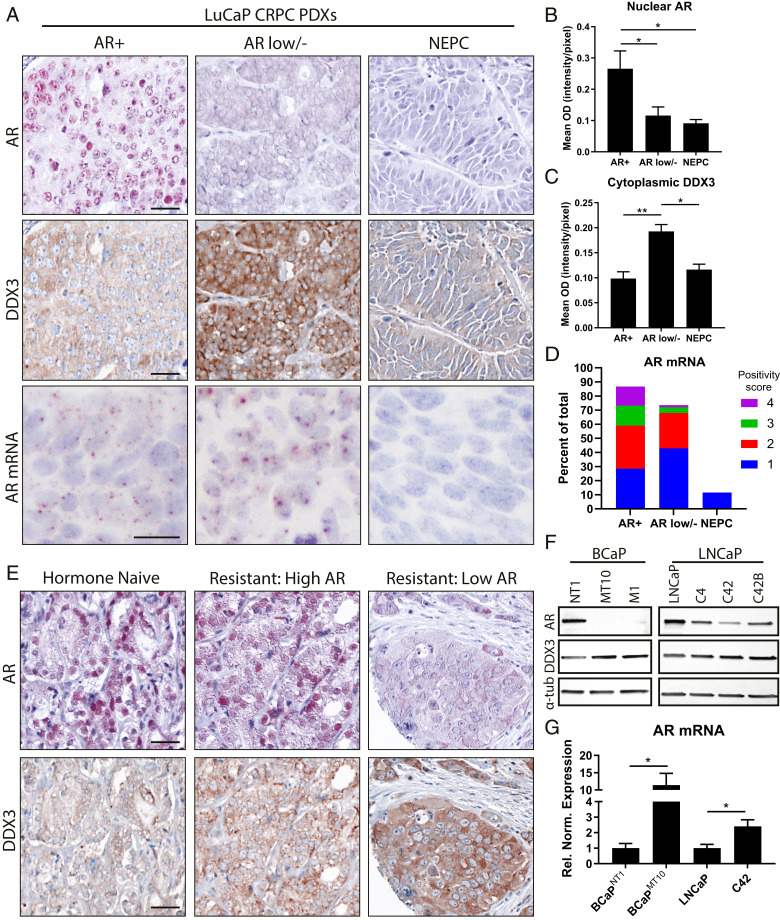
To determine AR and DDX3 expression in CRPC, we costained patient samples and objectively quantified protein expression. In PDXs, tissues were stratified based on CRPC subtype: AR+, ARL/− (DNPC, ARLPC), and NEPC (Fig. 1A). Nuclear AR was highly expressed in AR+ samples with significantly lower expression in ARL/− (P = 0.046) and NEPC (P = 0.017), supporting previous results (4, 5) (Fig. 1B and SI Appendix, Fig. S1). Cytoplasmic DDX3 expression was significantly higher in ARL/− CRPC compared to AR+ (P = 0.002) and NEPC (P = 0.011) (Fig. 1C). AR mRNA was detected in 87.42% of cells in AR+ samples, 73.7% of ARL/− (DNPC, ARLPC), and 11.52% of NEPC (Fig. 1 A and D). In hormone-naive/resistant patient samples, we observed high nuclear AR protein expression in hormone-naive cases, while hormone-resistant samples exhibited a range of AR expression from high to low (Fig. 1E). Cytoplasmic DDX3 expression was significantly (P = 0.044) higher in resistant vs. naive (SI Appendix, Fig. S1C), and within CRPC, DDX3 was highest in ARL/− samples, supporting the inverse association between DDX3 and AR protein (Fig. 1E). Taken together, these data indicate that AR expression is variable in CRPC and that in some cases, AR negativity occurs at the protein level despite the presence of mRNA. Additionally, DDX3 is highly expressed in CRPCs with low AR protein expression, potentially implicating it as a negative regulator of AR protein expression in CRPC.
Fig. 1.
ARL/− CRPC retains AR mRNA and expresses high levels of DDX3. AR and DDX3 expression were assessed in human CRPC samples. (A) Representative images of AR protein (red, first row), DDX3 protein (brown, second row), and AR mRNA (brown, third row) in PDXs from three subtypes of CRPC: AR+, ARL/−, and NEPC. Nuclei were counterstained with hematoxylin (blue). (Scale bar, 25 µm.) (B) Mean optical density of nuclear AR expression from PDXs showed nuclear AR was significantly higher in the AR+ compared to ARL/− (P = 0.046) and NEPC (P = 0.017). AR+, n = 5; ARL/− (DNPC, ARLPC), n = 3; NEPC, n = 4. (C) Mean optical density of cytoplasmic DDX3 expression from prostate cancer PDX model (LuCaP) showed DDX3 was significantly higher in DNPC compared to AR+ and NEPC (P = 0.002 and 0.011, respectively). AR+, n = 5; ARL/− (DNPC, ARLPC), n = 3; NEPC, n = 4. (D) AR mRNA positivity scoring in CRPC where “1” indicates 1–3 dots per cell, “2” indicates 4–9 dots per cell, “3” indicates 10–15 dots per cell, and “4” indicates >15 dots/cell. (E) Representative images of IHC colocalization for AR protein (red) and DDX3 protein (brown) in hormone-naive and hormone-resistant (CRPC) patient specimens. Nuclei were counterstained with hematoxylin (blue). (Scale bar, 10 µm.) (F) Representative Western blot analysis in two models of CaP progression (BCaP, LNCaP-C4) showed decreased amounts of AR protein coincident with high amounts of DDX3 protein in CRPC; α-tubulin (α-tub) was used as a loading control. (G) Analysis of AR mRNA expression via qPCR in cell line models of CRPC (BCaPMT10 and C42) showed significantly higher amounts compared to parental cell lines (BCaPNT1, P = 0.011; LNCaP, P = 0.045, n = 3) after normalization to reference genes TBP and YWHAZ. Bar graphs represent mean ± SEM. Significance is represented by *P ≤ 0.05, **P ≤ 0.01.
DDX3 Binds AR mRNA in ARL/− CRPC.
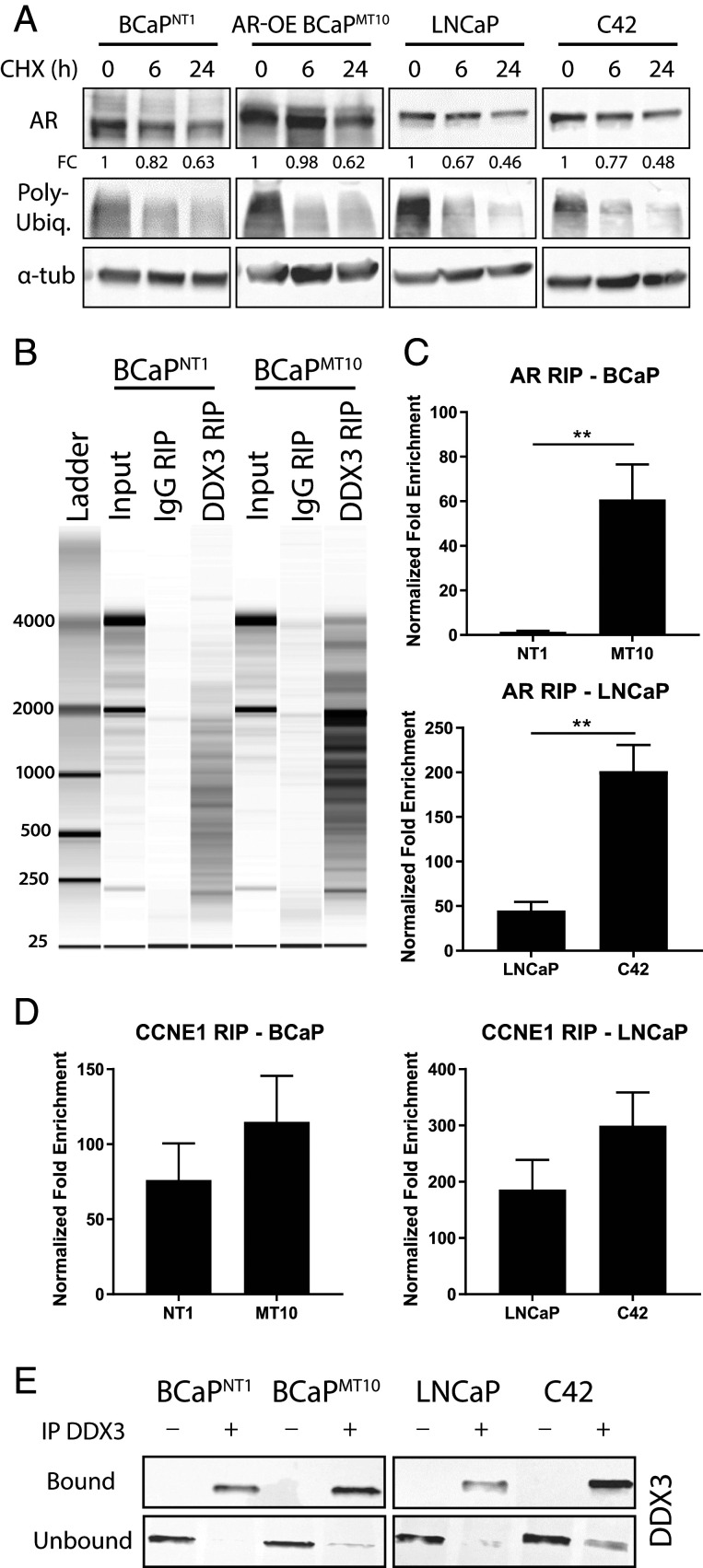
We hypothesized that DDX3 could bind AR mRNA to modulate AR protein expression. AR and DDX3 expression were assessed in two CRPC models: BCaP and LNCaP-C4 series (17). AR protein was decreased in CRPC (BCaPMT10 and C42) compared to their parental lines (BCaPNT1 and LNCaP, respectively), whereas DDX3 protein increased (Fig. 1F and SI Appendix, Fig. S2A). Similar to patient data, the lack of AR was not seen at the RNA level; rather, AR mRNA was significantly higher in CRPC (BCaPMT10, C42) compared to parental (BCaPNT1, LNCaP, respectively) (BCaP, P = 0.011; LNCaP, P = 0.045) (Fig. 1G); this mRNA expression pattern was supported by patient-derived metadata (SI Appendix, Fig. S1 E and F). To determine if other protein-regulating processes regulated AR protein, we performed further experiments including 1) translation inhibition by cycloheximide (CHX), 2) 26S proteasome inhibition by bortezomib, and 3) qPCR for microRNA, which demonstrated decreased AR expression was not due to increased protein degradation or microRNA-mediated repression (Fig. 2A and SI Appendix, Fig. S2 B–D). To test the potential RNA-binding capacity of DDX3, we performed RIP, by pulling down DDX3. A number of RNA species were present in input controls and DDX3 RIP samples, with little RNA in IgG RIP controls (Fig. 2B). To test if DDX3 bound AR mRNA, we performed RIP-qPCR. We observed a significant increase in AR mRNA bound to DDX3 in CRPC compared to parental (BCaP, P = 0.009; LNCaP, P = 0.002) (Fig. 2C). Positive control CCNE1 mRNA was bound to DDX3 in all cell lines (21) (Fig. 2D), whereas negative control mRNA (TBP) was not detected in any of the samples (SI Appendix, Fig. S3E). Western blot analysis of IP DDX3+ lysates confirmed robust DDX3 pulldown in “bound” samples, while “unbound” controls showed depletion of DDX3 (Fig. 2E and SI Appendix, Fig. S3). Additionally, although visualization of an AR mRNA/DDX3 complex is technically challenging, ISH for AR mRNA followed by staining for DDX3 revealed AR mRNA can colocalize with punctate DDX3 protein in some instances, but not others (SI Appendix, Fig. S3 B and F). Taken together, these results suggest DDX3 binds AR mRNA in CRPC.
Fig. 2.
DDX3 binds AR mRNA in ARL/− CRPC. Posttranscriptional regulation of AR was assessed via degradation and DDX3-dependent mechanisms, where AR was identified as a target mRNA of RBP DDX3. (A) Degradation rates of AR protein were determined using CHX pulse–chase assays. Expression of AR was determined at 0, 6, and 24 h post-CHX treatment. Poly-ubiquitin expression decreased when translation was inhibited, as expected, and α-tub as a loading control. AR protein expression fold change (FC) was determined relative to the 0-h time point using densitometry (n = 3). (B) Analysis of RNA binding using DDX3 RIP followed by bioanalyzer visualization of RNA in BCaPNT1 and BCaPMT10 showed robust RNA yields for the RNA input control and DDX3 RIP, but lower RNA yields for the IgG RIP controls, suggesting significant RNA binding to DDX3 and little nonspecific binding to IgG control. (C) RIP followed by qPCR for AR mRNA showed significantly increased binding of DDX3 to AR mRNA in CRPC cell lines BCaPMT10 and C42 compared to parental cell lines BCaPNT1 and LNCaP (BCaP, P = 0.009; LNCaP, P = 0.002). These results were normalized to input RNA, and represented as the FC for specific antibody pulldown over the IgG control pulldown. (D) Validation of RIP-DDX3 RNA binding using qPCR for CCNE1 showed mRNA was bound to DDX3 in all cell lines. (E) Western analysis showed “bound” samples, taken directly after the IP before RNA isolation, exhibited robust DDX3 pulldown in samples given DDX3 antibody (IP DDX3 +), while IgG controls (IP DDX3 -) did not pull down DDX3. Conversely, the “unbound”, i.e., the remainder of the lysate after the magnetic beads were removed, showed a decrease of DDX3 protein in the DDX3 IP pulldowns, while the IgG controls retained high DDX3 protein content, as expected. Bar graphs represent mean ± SEM. Significance is represented by **P ≤ 0.01.
DDX3 Localization to SGs Represses AR Protein Expression in ARL/− CRPC.
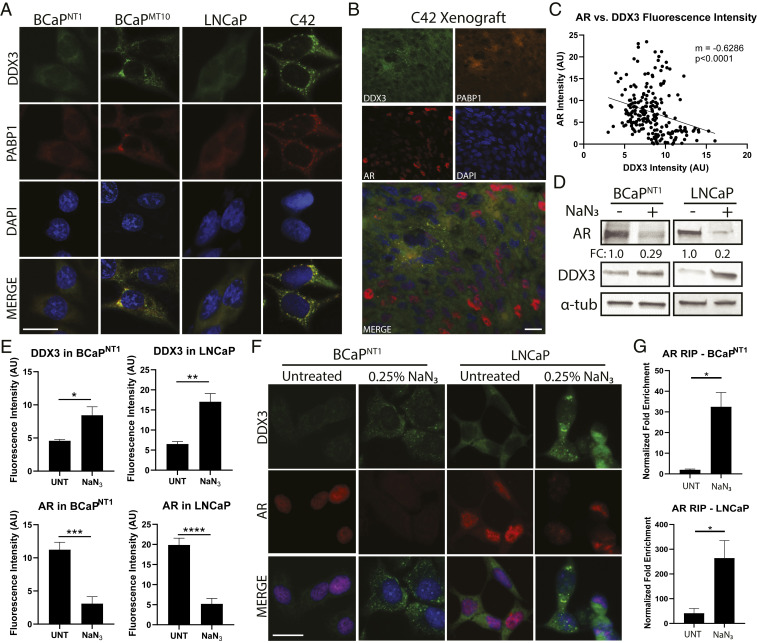
DDX3-mediated protein translational repression is dependent on its localization to SGs; because DDX3 and AR protein expression is inversely associated, we hypothesized that DDX3 localized to SGs in CRPC. To test this, we costained CRPC cell lines for DDX3 and SG-marker PABP1 and, as expected, observed colocalization of DDX3 and PABP1 in cytoplasmic puncta in CRPC (BCaPMT10 and C42), but not in parental (BCaPNT1 and LNCaP) (Fig. 3A). Additionally, DDX3, PABP1, and AR protein localization was assessed in CRPC xenografts, confirming an inverse relationship between AR protein and DDX3/PABP1 localization to SGs (Fig. 3B and SI Appendix, Fig. S4B). When quantified, there was a statistically significant negative correlation between DDX3 and AR expression (P < 0.0001) (Fig. 3C and SI Appendix, Fig. S4A). SGs can form in response to cellular stresses including hypoxia (20, 32). To assess induction of SGs by hypoxic stress, we treated AR-positive cells with sodium azide (NaN3) as previously described (22). NaN3-treated cells showed a decrease of AR protein by Western blot and a localization of DDX3 to putative SGs (i.e., cytoplasmic puncta) with concurrent lack of AR protein expression by IF (Fig. 3 D and F). DDX3 expression was increased with NaN3 treatment (Fig. 3 D and F). When quantified, DDX3 fluorescence intensity was significantly increased with NaN3 treatment (BCaPNT1, P = 0.037; LNCaP, P = 0.002), while AR was significantly decreased (BCaPNT1, P = 0.0001; LNCaP, P < 0.0001) (Fig. 3E). Importantly, RIP analysis showed NaN3 treatment increased AR mRNA bound to DDX3 compared to UNTs (BCaPNT1, P = 0.043; LNCaP, P = 0.039) (Fig. 3G and SI Appendix, Fig. S3D), suggesting the induced localization of DDX3 to SGs is sufficient to cause binding to AR mRNA. Additionally, DDX3 overexpression (DDX3-OE) in parental lines was sufficient to induce SG formation; DDX3-OE cells had significantly decreased AR protein, while adjacent cells without DDX3-OE had high AR protein expression (BCaPNT1, P = 0.0001; LNCaP, P = 0.0004) (SI Appendix, Fig. S4 C–E). Taken together, these data provide a potential mechanism for DDX3-mediated posttranscriptional regulation of AR in CRPC, where DDX3 binds AR mRNA in SGs and prevents the translation of AR mRNA transcripts.
Fig. 3.
DDX3 localizes to SGs in CRPC and represses AR protein expression. Consistent with translational repression, DDX3-mediated regulation of AR occurred when DDX3 was localized to SGs. Induction of SGs resulted in increased AR mRNA bound to DDX3 and decreased AR protein expression. (A) Immunofluorescence for DDX3 (green) showed localization to cytoplasmic puncta (SGs) in CRPC lines BCaPMT10 and C42 grown in vitro, as compared to diffuse cytoplasmic staining in the parental cell lines BCaPNT1 and LNCaP. Merged images show colocalization (yellow) of DDX3 with SG marker PABP1 (red). Nuclei were counterstained with DAPI (blue). (B) IHC of a C42 xenograft grown in vivo showed colocalization of DDX3 (green) and PABP1 (orange) puncta and AR protein expression in red. In these xenografts, DDX3/PABP1 puncta are only present in AR protein negative cells, despite robust AR protein expression in the surrounding area (red nuclei). Nuclei were counterstained with DAPI (blue). (C) Linear regression for DDX3 and AR protein fluorescence intensity in C42 xenografts showed a significant negative correlation between DDX3 and AR expression (n = 3, P < 0.0001). (D) Western blot analysis of AR protein expression in BCaPNT1 and LNCaP bulk populations after induced hypoxic stress by 3 h treatment with 0.25% sodium azide (NaN3) showed a decrease of overall AR protein (FC = 0.29 for BCaPNT1 and 0.2 for LNCaP), with a concurrent increase of DDX3 protein expression. α-tub was used as a loading control. (E) Quantification of DDX3 and AR protein fluorescence intensity after treatment with NaN3 showed a significant increase of DDX3 intensity (P = 0.037 in BCaPNT1 and P = 0.002 in LNCaP) and a significant decrease of AR intensity (P = 0.0001 in BCaPNT1 and P < 0.0001 in LNCaP) compared to UNTs. Fluorescence intensity was averaged between at least three separate experiments. (F) Representative images for hypoxia-induced stress from E showed NaN3 treatment increased DDX3 expression (green) and induced localization to cytoplasmic puncta concurrent with decreased AR protein expression (red) in parental cell lines BCaPNT1 and LNCaP. Nuclei were counterstained with DAPI (blue). (G) RIP analysis following treatment with NaN3 significantly increased AR mRNA bound to DDX3 compared to UNT (BCaPNT1, P = 0.043; LNCaP, P = 0.039). (Scale bars, 10 µm.) Bar graphs represent mean ± SEM. Significance is represented by *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
Genetic and Pharmacological Inhibition of DDX3 Increases AR Protein Expression and Signaling.
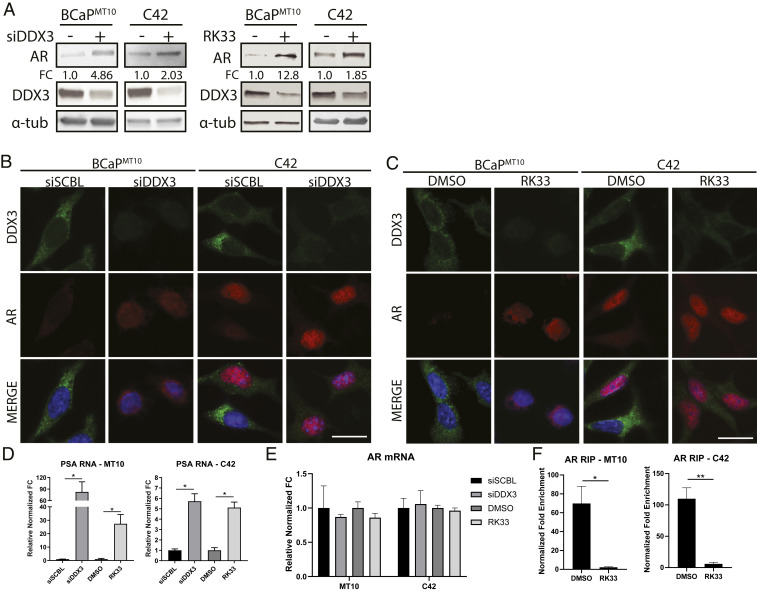
Because DDX3-nucleated SG induction decreased AR protein, we hypothesized that inhibition of DDX3 resolves SGs and restores AR protein in ARL/− CRPC models (BCaPMT10 and C42). Genetic inhibition by siRNAs targeting DDX3 (siDDX3) decreased DDX3 expression (BCaPMT10, P = 0.008; C42, P = 0.032) and increased AR protein expression (BCaPMT10, P = 0.019; C42, P = 0.049) compared to siRNA scramble (siSCBL) (Fig. 4 A and B and SI Appendix, Fig. S5B). Similarly, chemical inhibition using small-molecule RK33 resulted in decreased DDX3 protein levels (BCaPMT10, P = 0.043; C42, P = 0.047), decreased localization to puncta, and increased AR protein expression (BCaPMT10, P = 0.001; C42, P = 0.002) (Fig. 4 A and C and SI Appendix, Fig. S5C). Additionally, RK33 treatment was sufficient to decrease sodium azide-induced SG induction, supporting previous studies showing DDX3 is an SG-nucleating factor (19, 22) (SI Appendix, Fig. S5E). An in vitro dose curve for RK33 in multiple prostate cell lines revealed a dose–response consistent to previous findings (18, 31) (SI Appendix, Fig. S5A). To assess AR activity, we evaluated AR-target gene, prostate-specific antigen (PSA), expression (1). Here, we observed a significant increase of PSA mRNA in both siDDX3 (BCaPMT10, P = 0.036; C42, P = 0.021) and RK33 (BCaPMT10, P = 0.019; C42, P = 0.029) treated groups, compared to controls (Fig. 4D). PSA protein was also increased with DDX3 inhibition (SI Appendix, Fig. S5D). When quantified, AR mRNA was not significantly changed with siDDX3 or RK33, suggesting transcriptional regulation of AR is not involved in this mechanism (Fig. 4E). Finally, RIP analysis confirmed that inhibition of DDX3 by RK33 decreased AR mRNA bound to DDX3 compared to DMSO controls (BCaPMT10, P = 0.02; C42, P = 0.004) (Fig. 4F and SI Appendix, Fig. S3D). Taken together, these data provide additional evidence that DDX3 represses AR protein expression in CRPC and further suggest that inhibition of DDX3 is sufficient to increase AR protein expression and signaling in ARL/− CRPC.
Fig. 4.
Genetic and pharmacological inhibition of DDX3 increases AR protein expression and signaling. Inhibition of DDX3 resulted in increased AR protein expression and AR signaling in CRPC. (A) Western blot analysis of AR protein expression in BCaPMT10 and C42 bulk populations after inhibition of DDX3 by siRNA and small-molecule inhibitor RK33 showed an increase of overall AR protein and decrease of DDX3 protein. With siRNA, AR protein increased 4.86-fold in BCaPMT10 and 2.03-fold in C42 compared to scramble control. With RK33 treatment, AR protein increased 12.8-fold in BCaPMT10 and 1.85-fold in C42 compared to DMSO control. α-tub was used as a loading control. (B) Immunofluorescence analysis of DDX3 expression and localization after inhibition with siRNAs showed decreased cytoplasmic DDX3 expression (green) and increased AR expression (red), compared to scramble control (siSCBL) in two CRPC models (BCaPMT10 and C42). Nuclei were counterstained with DAPI (blue). (C) Immunofluorescence analysis of DDX3 expression and localization after pharmacologic inhibition using 2 µM RK33 showed decreased cytoplasmic DDX3 expression (green) and increased AR expression (red) compared to DMSO control in BCaPMT10 and C42. Nuclei were counterstained with DAPI (blue). (D) Assessment of AR signaling, using PSA, a transcriptional target of AR, was significantly increased with siDDX3 compared to siSCBL (BCaPMT10, P = 0.036; C42, P = 0.021) and with RK33 compared to DMSO (BCaPMT10, P = 0.019; C42, P = 0.029) in both CRPC models. (E) qPCR for AR mRNA showed no significant difference in expression between siSCBL vs. siDDX3 and RK33 vs. DMSO. (F) RIP analysis of AR mRNA bound to DDX3 after RK33 treatment (RK33) showed a significant decrease in DDX3 binding to AR mRNA compared to control (DMSO) in CRPC cell lines BCaPMT10 (P = 0.02) and C42 (P = 0.004). (Scale bars, 10 µm.) Bar graphs represent mean ± SEM. Significance is represented by *P ≤ 0.05, **P ≤ 0.01.
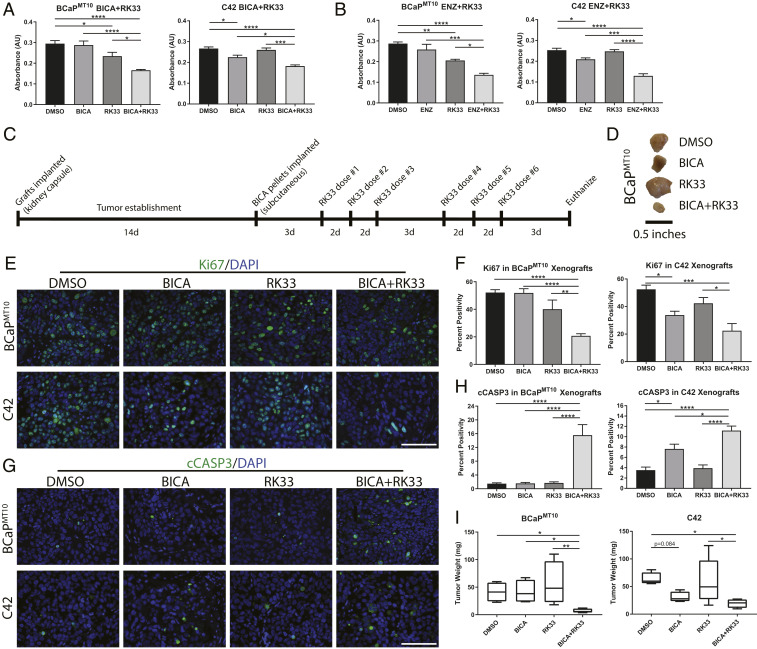
DDX3 Inhibition Resensitizes ARL/− Cells to Anti-Androgen Therapy.
In ARL/− CRPC, anti-androgens are ineffective due to the lack of targetable AR (3, 4). We hypothesized that treatment with DDX3 inhibitor, RK33, resensitizes ARL/− cells to ARSIs. Cotreatment of RK33 and BICA (or ENZ) in CRPC models in vitro significantly reduced cell viability and proliferation and increased apoptosis, compared to either treatment alone (Fig. 5 A and B and Table 1). To investigate the effectiveness of RK33 and anti-androgen cotreatment in vivo, CRPC xenografts (BCaPMT10 and C42) were implanted into BICA and/or RK33-treated mice, as described (18, 30, 31) (Fig. 5C). Xenografts were harvested, weighed, and stained for Ki67 and cCASP3 (Fig. 5 E and G). In BCaPMT10, proliferation was significantly decreased with cotreatment compared to single treatment with BICA (P < 0.0001) and RK33 (P = 0.007) (Fig. 5F). In C42 xenografts, proliferation was significantly decreased with BICA and cotreatment compared to control (P = 0.048 and P = 0.0006, respectively) and with cotreatment compared to RK33 alone (P = 0.015) (Fig. 5F). Apoptosis was significantly increased in cotreated BCaPMT10 (vs. BICA, P < 0.0001; vs. RK33, P < 0.0001) and C42 (vs. BICA, P = 0.034; vs. RK33, P < 0.0001) xenografts compared to either treatment alone (Fig. 5H). Tumor mass was significantly decreased in cotreatment of BCaPMT10 xenografts compared to control (P = 0.049), BICA (P = 0.046), and RK33 (P = 0.008), while C42 tumor size was significantly decreased compared to control (P = 0.0255) and RK33 (P = 0.0263) (Fig. 5 D and I). These findings suggest cotreatment with RK33 sensitizes ARL/− CRPC xenografts to AR-targeted therapy.
Fig. 5.
DDX3 inhibition resensitizes ARL/− cells to anti-androgen therapy. By increasing AR protein expression with DDX3 inhibition, ARL/− CRPC was sensitized to AR-targeted therapy. Cotreatment with a DDX3 inhibitor and BICA resulted in decreased cell viability, decreased proliferation, and increased apoptosis. (A) MTT assay in BCaPMT10 and C42 showed cotreatment with BICA, and RK33 significantly decreased cell viability compared to BICA alone (BCaPMT10, P < 0.0001; C42, P = 0.014) and RK33 alone (BCaPMT10, P = 0.011; C42, P = 0.0001). RK33 treatment alone was sufficient to decrease cell viability in BCaPMT10 compared to DMSO control (P = 0.041). (B) Cotreatment with ENZ and RK33 significantly decreased cell viability in BCaPMT10 and C42 compared to ENZ alone (BCaPMT10, P = 0.0003; C42, P = 0.0002) and RK33 alone (BCaPMT10, P = 0.021; C42, P < 0.0001). RK33 treatment alone was sufficient to decrease cell viability in BCaPMT10 (P = 0.007s). (C) Schematic for experimental design using RK33 and BICA cotreatment in vivo. d = days. (D) Representative images of tumors from each treatment group harvested from mice at necropsy. (E) Representative images of proliferation (Ki67, green) in each treatment group in BCaPMT10 and C42 xenografts. Nuclei were counterstained with DAPI (blue). (F) Labeling index of Ki67-positivity showed a significant decrease in cotreatment compared to DMSO control (BCaPMT10, P < 0.0001; C42, P = 0.0006). In BCaPMT10, cotreatment significantly decreased Ki67 compared to BICA alone and RK33 alone (P < 0.0001 and P = 0.007, respectively). In C42, cotreatment significantly decreased Ki67 compared to RK33 alone (P = 0.015) but was not significantly different from BICA alone (P = 0.512). (G) Representative images of apoptosis (cCASP3, green) from each treatment group in BCaPMT10 and C42 xenografts. Nuclei were counterstained with DAPI (blue). (H) Calculation of percent positivity of cCASP3 showed a significant increase in cotreatment compared to DMSO in both BCaPMT10 and C42 (P < 0.0001 for both). In BCaPMT10, cotreatment significantly increased cCASP3 compared to BICA alone (P < 0.0001) and RK33 alone (P < 0.0001). In C42, cotreatment significantly increased cCASP3 compared to BICA alone (P = 0.034) and RK33 alone (P < 0.0001). (I) Quantification of tumor weight at necropsy showed mice receiving cotreatment (BICA+RK33) had significantly smaller tumors than the control mice for both BCaPMT10 (P = 0.049) and C42 (P = 0.0255). In BCaPMT10, tumor weight after cotreatment was significantly smaller than BICA alone (P = 0.046) and RK33 alone (P = 0.008). In C42, tumor weight after cotreatment was significantly smaller than RK33 alone (P = 0.0263), but not BICA alone (P = 0.534). (Scale bars in E and G, 100 µm.) Bar graphs represent mean ± SEM. Significance is represented by *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
Table 1.
Proliferation and apoptosis with cotreatment in vitro
| DMSO | BICA | RK33 | BICA+RK33 | ||
| Ki67 | Cell Line | ||||
| Percent | BCaPMT10 | 51.4 (43.2–59.5) | 53.2 (48.3–58.2) | 43.4 (40.4–46.5) | 26.9 (22.4–31.5) |
| Positivity | C42 | 85.7 (79–92.5) | 75.5 (72.2–78.8) | 73.4 (71.2–75.5) | 54.1 (48.6–59.6) |
| cCASP3 | |||||
| Percent | BCaPMT10 | 4.76 (4.29–5.24) | 5.26 (4.13–6.37) | 6.76 (6.6–6.92) | 9.45 (7.84–11.1) |
| Positivity | C42 | 1.09 (0.91–1.28) | 1.73 (0.85–2.61) | 1.96 (1.69–2.21) | 4.3 (2.92–5.68) |
*Values show mean percent positivity for n = 3 with the 95% confidence intervals in parentheses. Bolded text indicates significance compared to DMSO (P < 0.05), and italicized text indicates significance compared to BICA alone (P < 0.05).
Discussion
The advent of near-complete AR suppression therapies has given rise to new, more difficult to treat, CRPC. Recent evidence shows some CRPC subtypes lack AR protein expression, and we report here that this AR negativity occurs at the protein level despite the presence of AR mRNA. Our results indicate that the RBP DDX3 is highly expressed in ARL/− CRPC, where it binds AR mRNA in SGs. Strikingly, when AR mRNA is bound to DDX3 in SGs, AR protein expression and downstream signaling is greatly diminished or absent. As an SG-nucleating factor, inhibition of DDX3 results in inhibition of SG formation. Moreover, targeting DDX3 can resensitize ARL/− CRPC to anti-androgen therapy. There are several immediate implications for these findings.
First, we have proposed a regulatory pathway for AR at the posttranscriptional level. Here, the repressive regulatory role for DDX3 in CRPC is dependent on its localization to SGs; however, the conditions underlying this localization have not been identified. Mutations in the DDX3 ortholog Belle result in a persistent SG phenotype in D. melanogaster salivary glands, and DDX3 is commonly mutated in medulloblastoma (16, 33). However, interrogation of cBioPortal showed DDX3 is not commonly mutated in CaP or CRPC (SI Appendix, Fig. S6 A and B) (34, 35). SGs can also form due to hypoxic/metabolic stress (32, 36–39). We assessed metabolic stress-induced SG formation by growing androgen-dependent cells in low-glucose media, which induced localization of DDX3 to SGs concurrent with a loss of AR protein (SI Appendix, Fig. S6C). Finally, several studies have identified increased reactive oxygen species after chemical castration (40, 41). When androgen-dependent cells were treated with anti-androgens, we observed DDX3 localization to SGs and decreased AR protein expression (SI Appendix, Fig. S6 C and D). Taken together, hypoxic, metabolic, and/or therapeutic stress may underlie DDX3-mediated regulation of AR in CRPC and should be addressed in future studies.
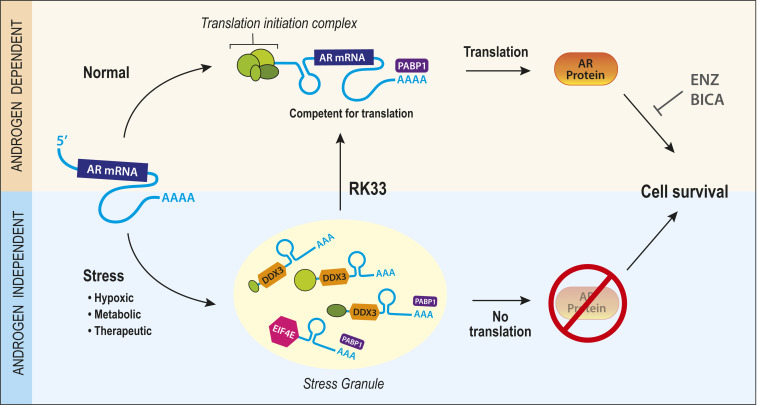
Second, we have identified a potential therapeutic target for ARL/− CRPC. Currently, there are no hormonal-based therapies for ARL/− CRPC due to the lack of targetable AR protein. This study proposes DDX3 inhibition by RK33 may be a valuable cotreatment to restore androgen sensitivity in ARL/− CRPCs. In this paradigm, normal protein translation is disrupted in CRPC due to exogenous stressors that precipitate the formation of SGs (Fig. 6). Because AR mRNA is bound by DDX3 in SGs, this complex represses AR protein expression and renders the stressed cell resistant to ARSI. By inhibiting DDX3 pharmacologically, SGs are resolved, AR expression and signaling are restored, and cells are resensitized to ARSI (Fig. 6). This concept of cotreatment producing synergistic deleterious effects is known as synthetic lethality and has been implicated in overcoming therapeutic resistance in several cancer types (42, 43). However, because the characterization of ARL/− CRPCs that are not neuroendocrine is relatively recent, clinical samples representing this subset are not abundant and should be further explored in future studies. Importantly, targeting DDX3 to induce therapeutic sensitivity has been tested in multiple cancer types including lung and medulloblastoma (31, 44). In early stages of CaP, targeting nuclear DDX3 was an effective strategy to induce sensitivity to radiation therapy (18). Targeting DDX3 could be effective in other diseases in which AR is implicated (e.g., breast, liver, salivary gland cancers, benign prostatic hyperplasia, and others) and could be of interest in future studies.
Fig. 6.
Model for DDX3 regulation of AR in CRPC. Schematic for DDX3-mediated regulation of AR in CRPC. Under normal conditions, AR mRNA is translated into AR protein, resulting in AR protein and androgen-mediated cell survival, which can be inhibited using androgen/AR targeting therapies BICA or ENZ. Under stress conditions (hypoxic, metabolic, therapeutic), DDX3 is up-regulated and localizes to SGs. Here, within SGs, AR mRNA is bound to DDX3 causing repression of AR protein translation. This process is associated with low sensitivity to anti-androgens due to the absence of targetable AR protein, resulting in AR-independent cell survival. Therefore, by pharmacologically inhibiting the formation of SGs with chemical DDX3 inhibitors, one can resensitize castration-resistant (stressed) cells to hormonal therapy through the restoration of AR protein translation.
Finally, we have highlighted the significance of AR negativity in bulk tumors that show diminished AR protein expression (DNPC, ARLPC). Because these tumors lack AR protein, they are insensitive to ARSI; however, we have shown here that reexpression of AR in ARL/− CRPCs is sufficient to induce sensitivity to ARSI. In previous publications, reintroduction of AR expression in CRPC models reduced proliferation (45). This does not appear to be the case in models for DNPC, as we observed an increase of cell growth with AR overexpression (SI Appendix, Fig. S2C), suggesting a continued reliance on AR. Examples of increased sensitivity upon reexpression of steroid hormone receptors have previously been described in breast cancer (46, 47). Importantly, these CRPC subtypes are not the only case of AR negativity in CaP. It has previously been reported that within a bulk AR+ population, some luminal/carcinoma cells lack AR protein (23, 48, 49). This AR heterogeneity is associated with poor clinical outcomes and disease progression (3, 23, 48, 50–53). In theory, a subpopulation of cells lacking AR would survive ARSI therapy despite the reduction of overall tumor volume; this population may then give rise to a recurrent tumor. It is possible that targeting DDX3 in these cells would allow for resensitization to ARSI if cotreated with DDX3 inhibitors. Therefore, another potential implication for targeting DDX3 in CaP could be in prevention of recurrence.
Conclusions
We have shown that RBP DDX3 can regulate AR at the posttranscriptional level by sequestering mRNA in SGs. Resolving SGs by inhibiting DDX3 was sufficient to induce AR protein expression and signaling, resensitizing these cells to ARSI. These data provide insight into several longstanding questions in CaP research including 1) mechanisms of AR regulation, 2) therapeutic interventions to target refractory CRPC, and 3) significance of AR negativity. Identification of DDX3 as a mediator of AR posttranscriptional repression is significant from a scientific and clinical perspective, and continued investigation of the role of DDX3 may unveil mechanisms of disease recurrence and strategies for therapeutic intervention.
Supplementary Material
Acknowledgments
We thank the University of Wisconsin Translation Research Initiatives in Pathology (TRIP) laboratory and the UW–Madison Carbone Cancer Center (UWCCC) for core services provided. We thank Drs. Colm Morrissey and Johnathan Melamed for creating and sharing the TMAs used in this study. We also acknowledge Dalton McLean for assistance with xenografting experiments, Dr. Jordan Becker for sharing the DDX3-OE vector, Glen Leverson for biostatistical assistance, Dr. Donald Vander Griend for providing LAPC4 cells, and Drs. Chad Vezina and Diya Joseph for assistance with ISH assays. Finally, we thank Drs. Arash Bashirullah, Teresa Liu, Yunsik Kang, Sarah Neuman, and Taryn James for experimental advice and assistance, and Dr. Joshua Lang for his insights on clinical significance and translation. This work was supported by NIH Grant U54 DK104310 (to W.A.R.). Carbone Cancer Center core services were funded by P30 CA014520 (UWCCC), and funding for the TMA construction was provided by the Department of Defense Prostate Cancer Research Program, Award No. W81XWH-18-2-0013,15-19 (Prostate Cancer Biorepository Network - PCBN). J.E.V. is a trainee in the Cancer Biology Graduate Program at the University of Wisconsin–Madison and was funded by T32 CA009135.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008479117/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information.
References
- 1.Heinlein C. A., Chang C., Androgen receptor in prostate cancer. Endocr. Rev. 25, 276–308 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Kirby M., Hirst C., Crawford E. D., Characterising the castration-resistant prostate cancer population: A systematic review. Int. J. Clin. Pract. 65, 1180–1192 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Deng Q., Tang D. G., Androgen receptor and prostate cancer stem cells: Biological mechanisms and clinical implications. Endocr. Relat. Cancer 22, T209–T220 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bluemn E. G., Coleman I. M., Lucas J. M. et al. , Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell. 32, 474–489.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Labrecque M. P., et al. , Molecular profiling stratifies diverse phenotypes of treatment-refractory metastatic castration-resistant prostate cancer. J. Clin. Invest. 129, 4492–4505 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khalaf D. J., et al. , Optimal sequencing of enzalutamide and abiraterone acetate plus prednisone in metastatic castration-resistant prostate cancer: A multicentre, randomised, open-label, phase 2, crossover trial. Lancet Oncol. 20, 1730–1739 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Jarrard D. F., et al. , Methylation of the androgen receptor promoter CpG island is associated with loss of androgen receptor expression in prostate cancer cells. Cancer Res. 58, 5310–5314 (1998). [PubMed] [Google Scholar]
- 8.Kinoshita H., et al. , Methylation of the androgen receptor minimal promoter silences transcription in human prostate cancer. Cancer Res. 60, 3623–3630 (2000). [PubMed] [Google Scholar]
- 9.Litvinov I. V., et al. , Androgen receptor as a licensing factor for DNA replication in androgen-sensitive prostate cancer cells. Proc. Natl. Acad. Sci. U.S.A. 103, 15085–15090 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vummidi Giridhar P., Williams K., VonHandorf A. P., Deford P. L., Kasper S., Constant degradation of the androgen receptor by MDM2 conserves prostate cancer stem cell integrity. Cancer Res. 79, 1124–1137 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeap B. B., et al. , Novel binding of HuR and poly(C)-binding protein to a conserved UC-rich motif within the 3′-untranslated region of the androgen receptor messenger RNA. J. Biol. Chem. 277, 27183–27192 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Zhou H., Zhang Y., Hamburger A. W., EBP1 inhibits translation of androgen receptor mRNA in castration resistant prostate cancer cells. Anticancer Res. 31, 3129–3135 (2011). [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou H., et al. , Post-transcriptional regulation of androgen receptor mRNA by an ErbB3 binding protein 1 in prostate cancer. Nucleic Acids Res. 38, 3619–3631 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nadiminty N., et al. , MicroRNA let-7c suppresses androgen receptor expression and activity via regulation of Myc expression in prostate cancer cells. J. Biol. Chem. 287, 1527–1537 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandes R. C., Hickey T. E., Tilley W. D., Selth L. A., Interplay between the androgen receptor signaling axis and microRNAs in prostate cancer. Endocr. Relat. Cancer 26, R237–R257 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Ihry R. J., Sapiro A. L., Bashirullah A., Translational control by the DEAD Box RNA helicase Belle regulates ecdysone-triggered transcriptional cascades. PLoS Genet. 8, e1003085 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vellky J. E., Ricke E. A., Huang W., Ricke W. A., Expression and localization of DDX3 in prostate cancer progression and metastasis. Am. J. Pathol. 189, 1256–1267 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie M., et al. , RK-33 radiosensitizes prostate cancer cells by blocking the RNA helicase DDX3. Cancer Res. 76, 6340–6350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shih J.-W., et al. , Critical roles of RNA helicase DDX3 and its interactions with eIF4E/PABP1 in stress granule assembly and stress response. Biochem. J. 441, 119–129 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Anderson P., Kedersha N., Stress granules: The Tao of RNA triage. Trends Biochem. Sci. 33, 141–150 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Lai M.-C., Chang W.-C., Shieh S.-Y., Tarn W.-Y., DDX3 regulates cell growth through translational control of cyclin E1. Mol. Cell. Biol. 30, 5444–5453 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hondele M., et al. , DEAD-box ATPases are global regulators of phase-separated organelles. Nature 573, 144–148 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Vellky J. E., Bauman T. M., Ricke E. A., Huang W., Ricke W. A., Incidence of androgen receptor and androgen receptor variant 7 coexpression in prostate cancer. Prostate 79, 1811–1822 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicholson T. M., Sehgal P. D., Drew S. A., Huang W., Ricke W. A., Sex steroid receptor expression and localization in benign prostatic hyperplasia varies with tissue compartment. Differentiation 85, 140–149 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu T., et al. , Modeling human prostate cancer progression in vitro. Carcinogenesis 40, 893–902 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kregel S., et al. , Acquired resistance to the second-generation androgen receptor antagonist enzalutamide in castration-resistant prostate cancer. Oncotarget 7, 26259–26274 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keil K. P., et al. , Visualization and quantification of mouse prostate development by in situ hybridization. Differentiation 84, 232–239 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barriga F. M., Montagni E., Mana M. et al. , Mex3a marks a slowly dividing subpopulation of Lgr5+ intestinal stem cells. Cell Stem Cell 20, 801–816.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar P., Nagarajan A., Uchil P. D., Analysis of cell viability by the MTT assay. Cold Spring Harb. Protoc. pdb.prot095505 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Nicholson T. M., et al. , Renal capsule xenografting and subcutaneous pellet implantation for the evaluation of prostate carcinogenesis and benign prostatic hyperplasia. J. Vis. Exp., e50574 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bol G. M., et al. , Targeting DDX3 with a small molecule inhibitor for lung cancer therapy. EMBO Mol. Med. 7, 648–669 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buchan J. R., Yoon J.-H., Parker R., Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J. Cell Sci. 124, 228–239 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oh S., et al. , Medulloblastoma-associated DDX3 variant selectively alters the translational response to stress. Oncotarget 7, 28169–28182 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao J., et al. , Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cerami E., et al. , The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data (AACR, 2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milosevic M., et al. , Tumor hypoxia predicts biochemical failure following radiotherapy for clinically localized prostate cancer. Clin. Cancer Res. 18, 2108–2114 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Cheng H. H., et al. , Circulating microRNA profiling identifies a subset of metastatic prostate cancer patients with evidence of cancer-associated hypoxia. PLoS One 8, e69239 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tonry C., Armstrong J., Pennington S. R., Probing the prostate tumour microenvironment I: Impact of glucose deprivation on a cell model of prostate cancer progression. Oncotarget 8, 14374–14394 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomida A., Tsuruo T., Drug resistance mediated by cellular stress response to the microenvironment of solid tumors. Anticancer Drug Des. 14, 169–177 (1999). [PubMed] [Google Scholar]
- 40.Ming L., et al. , Androgen deprivation results in time-dependent hypoxia in LNCaP prostate tumours: Informed scheduling of the bioreductive drug AQ4N improves treatment response. Int. J. Cancer 132, 1323–1332 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Shiota M., et al. , Antioxidant therapy alleviates oxidative stress by androgen deprivation and prevents conversion from androgen dependent to castration resistant prostate cancer. J. Urol. 187, 707–714 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Heinzel A., et al. , Synthetic lethality guiding selection of drug combinations in ovarian cancer. PLoS One 14, e0210859 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fechete R., et al. , Synthetic lethal hubs associated with vincristine resistant neuroblastoma. Mol. Biosyst. 7, 200–214 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Tantravedi S., et al. , Targeting DDX3 in medulloblastoma using the small molecule inhibitor RK-33. Transl. Oncol. 12, 96–105 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niu Y., et al. , Androgen receptor is a tumor suppressor and proliferator in prostate cancer. Proc. Natl. Acad. Sci. USA 105, 12182–12187 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fan J., et al. , ER alpha negative breast cancer cells restore response to endocrine therapy by combination treatment with both HDAC inhibitor and DNMT inhibitor. J. Cancer Res. Clin. Oncol. 134, 883–890 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Keen J. C., et al. , A novel histone deacetylase inhibitor, scriptaid, enhances expression of functional estrogen receptor alpha (ER) in ER negative human breast cancer cells in combination with 5-aza 2′-deoxycytidine. Breast Cancer Res. Treat. 81, 177–186 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Sehgal P. D., et al. , Tissue-specific quantification and localization of androgen and estrogen receptors in prostate cancer. Hum. Pathol. 89, 99–108 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Masai M., et al. , Immunohistochemical study of androgen receptor in benign hyperplastic and cancerous human prostates. Prostate 17, 293–300 (1990). [DOI] [PubMed] [Google Scholar]
- 50.Wan X., et al. , Activation of β-catenin signaling in androgen receptor-negative prostate cancer cells. Clin. Cancer Res. 18, 726–736 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Winter J. A. R., et al. , Androgen receptor heterogeneity in human prostatic carcinomas visualized by immunohistochemistry. J. Pathol. 160, 329–332 (1990). [DOI] [PubMed] [Google Scholar]
- 52.Liu X., et al. , Systematic dissection of phenotypic, functional, and tumorigenic heterogeneity of human prostate cancer cells. Oncotarget 6, 23959–23986 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Q., et al. , Linking prostate cancer cell AR heterogeneity to distinct castration and enzalutamide responses. Nat. Commun. 9, 3600 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.