Significance
Surface features of a virus are very important in determining its virility. For example, the spike protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) binds to the ACE2 receptor site of the host cell with a much stronger affinity than did the original SARS virus. Thus, it is clearly important to understand the virion surface structure. To that end, the present paper combines the spatial resolution of atomic force microscopy and the spectral resolution of coherent Raman spectroscopy. This combination of tip-enhanced microscopy using femtosecond adaptive spectroscopic techniques for coherent anti-Stokes Raman scattering (FAST CARS) with enhanced resolution (FASTER CARS) allows us to map a single virus particle with nanometer resolution and chemical specificity.
Keywords: virus detection, coherent anti-Stokes Raman scattering (CARS), tip-enhanced Raman scattering (TERS)
Abstract
From the famous 1918 H1N1 influenza to the present COVID-19 pandemic, the need for improved viral detection techniques is all too apparent. The aim of the present paper is to show that identification of individual virus particles in clinical sample materials quickly and reliably is near at hand. First of all, our team has developed techniques for identification of virions based on a modular atomic force microscopy (AFM). Furthermore, femtosecond adaptive spectroscopic techniques with enhanced resolution via coherent anti-Stokes Raman scattering (FASTER CARS) using tip-enhanced techniques markedly improves the sensitivity [M. O. Scully, et al., Proc. Natl. Acad. Sci. U.S.A. 99, 10994–11001 (2002)].
Scanning probe microscopy, especially in combination with plasmon-enhanced near-field spectroscopy, is used to specifically analyze and study objects below Abbe’s diffraction limit. The scientific goal is, in addition to virus diagnostics, to identify structural changes of the virus surface at an early stage, using so-called tip-enhanced Raman scattering (TERS) together with coherent anti-Stokes Raman spectroscopy (CARS) (Fig. 1). This combined technique will improve the sensitivity and consequently speed up acquisition times considerably. In contrast to known methods, this is unique. Due to its surface specificity our technique allows early detection of changes that alter the antigenic properties of viruses and thus the effectiveness of vaccines, with the smallest sample quantities. Furthermore, identification without specific antibodies is possible. This goes far beyond the current state of the art.
Fig. 1.
(A) Schematics of the transmission TERS setup used in the experiments with the virus mixture spread on a slide and the laser illuminating the sample from below ( = 532 nm, P = 500 W). In the model of an H1N1 virus protein spikes (cyan) protrude from the lipid bilayer (blue). In the model of a coxsackievirus B3 the protein lattice (cyan) is on the surface and the RNA strands (red) are inside the virus. (B) Selected TERS spectra recorded on a single H1N1 virus (measurement point distance 20 nm, acquisition time = 10 s). (C) Selected TERS spectra recorded on a single coxsackievirus B3 (measurement point distance 4 nm, = 10 s). In B and C, lipid marker bands (blue), protein marker bands (cyan), and RNA marker bands (red) are highlighted. See the text in Nanospectroscopic System for Differentiating between Influenza and Picornavirus for further explanation.
In Nanospectroscopic System for Differentiating between Influenza and Picornavirus, we demonstrate how the tip-enhanced Raman technique can be used to characterize and differentiate between influenza and picornaviruses. In Instrumentation and Techniques we explain the instrumentation and show how it provides exciting tools for viral research and detection systems, e.g., femtosecond adaptive spectroscopic techniques with enhanced resolution via coherent anti-Stokes Raman scattering (FASTER CARS). The present paper builds on and extends our earlier femtosecond Raman work in which the focus was on detection of anthrax (1). Discussion presents a conclusion and an outlook.
Nanospectroscopic System for Differentiating between Influenza and Picornavirus
Viruses are infectious agents that spread as nanosized particles, so-called virions outside of cells. Viruses can multiply only within a suitable host cell and as the infection propagates, the virion attacks a cell of a living organism by injecting it with genetic material and inducing the cell to make multiple replicas of the virion. The building blocks of a virion are either DNA or RNA molecules surrounded by a protective scaffold of proteins, the so-called (nucleo)capsid (e.g., coxsasckievirus B3 [CVB3] virus in Fig. 1). Moreover, there can be a lipid bilayer serving as an envelope. Protein spikes can protrude from the bilayer, forming a sea urchin-like structure (e.g., H1N1 virus in Fig. 1).
Some viruses are pathogenic and can cause severe diseases, which makes accurate diagnostics essential. The goal is to characterize the specific virus based on single viral particles. One common technique is the PCR, which has simplified and accelerated the detection of pathogens over culturing techniques (2–4). In PCR, the nucleic acid—either DNA or RNA—has to be extracted from the specimen first. Next, characteristic viral sequences are amplified employing varying primer sets followed by further molecular analysis (2, 3). In general, multiple DNA target molecules are necessary for sufficient amplification and the examination of a single virus particle is still challenging. Although PCR technologies are quite powerful, several drawbacks limit the application in microbiological diagnostics. On the one hand, high sensitivity of DNA/RNA amplification makes this process susceptible to contamination that might yield false-positive results. Furthermore, false-negative results, e.g., due to failed amplification, have to be considered, too (4). The nature of the PCR process is prone to several disturbing factors that may hamper exponential DNA amplification. The implementation of the quantitative real-time PCR (qRT-PCR) in recent years, where fluorescent molecules are added, enables exact quantitation of template DNA (3, 5, 6).
Another approach to identifying viruses is the immunoassay-based detection of viral proteins, which is in some cases less sensitive. Thus, the existing molecular analysis tools do not permit a combination of multiplexing and quantitation and in every case the viral components have to be extracted. Working on a single viral particle via the PCR technique is not possible.
Ideally, a different technique would combine the abilities of qualitative and quantitative analyses at the single virus particle level, rendering the need for separation of the different components unnecessary. A promising technique for that endeavor is tip-enhanced Raman and potentially other nonlinear optical spectroscopies like FASTER CARS. TERS has been demonstrated to operate in a very specific and sensitive mode down to the single-molecule level, with nanometer resolution and below (7–10). In TERS, scanning probe techniques (atomic force microscopy [AFM] or scanning tunneling microscopy [STM]) are paired with Raman spectroscopy. The former component enables morphological imaging with subnanometer lateral resolution, and the latter provides detailed spectral information on every specifically selected position on the sample surface.
The heart piece in TERS is the probe, which generally is a commercial AFM tip (for AFM-based setups) that is commonly evaporated with silver and exactly positioned in the laser focus. Upon laser irradiation of the metallized tip a so-called evanescent field is generated. Molecules located in some nanometer proximity to the tip experience a Raman signal enhancement (up to ) of their vibrational modes according to the surface-enhanced Raman spectroscopy theory (for instance refs. 11–15).
Once the tip is positioned in the laser spot, the sample is moved under the tip and the region of interest is selected. After setting a grid of profile lines on the sample, the AFM is coupled to the Raman spectrometer and enhanced Raman spectra are recorded. Such a setup does not need special sample pretreatments or tagging and allows a direct chemical characterization. The same setup also provides topographic imaging of the sample surface in a single experiment. The spatial resolution in TERS is limited by the diameter of the silver particle at the tip and lately has been pushed down to around 0.5 to 1 nm for biosamples (9, 16, 17). A not-to-scale setup is sketched in Fig. 1A. For specific instrumental details the reader is referred to ref. 18. Depending on the number of acquired data, subsequent multivariate data analysis might be useful for data assessment. In the context of a general tip-enhanced approach of a virus identification, it is important to point out that not only all major components to be expected in viruses [namely DNA/RNA (19–21), proteins, lipids, and even glycoproteins] have been already detected by TERS. Also, different studies on viruses (22–26) already proved the feasibility of the concept. In contrast to PCR-related techniques, in TERS a separation of the different components or labeling is not required, and ultimately only a single virion is sufficient for an identification.
So far, reported TERS experiments on single virus particles were performed on more or less randomly chosen points, which is apparently not sufficient for a thorough characterization (22–26). It was shown that grid-based TERS mapping of varicella-zoster and porcine teschovirus allowed a linear discriminant supported distinction of the two viruses. Clearly, the identification and discrimination of different virus strains demand a comprehensive characterization in terms of spectral surface imaging which is in line with the abovementioned extreme lateral resolution (25).
For the present experiments an enveloped influenza A virus H1N1 and a nonenveloped CVB3 were chosen. (To apply the present technique to the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virion, the experiment must be carried out in a biosafe environment and such studies will be reported elsewhere.) The swine H1N1 influenza virus particle is composed of a 2- to 6-nm lipid bilayer decorated with viral proteins and eight ribonucleoprotein (RNP) complexes. The RNPs consist of RNA strands, which interact with numerous nucleoprotein molecules and some viral polymerase complexes. The consistency of the lipid envelope depends on the type of cell membrane of host cell the virus originates. The inner surface is covered completely with viral matrix protein. From the lipid layer flexible protein spikes with a protrusion height of 10 to 14 nm stick out (27). In Fig. 1 the structure of an influenza virus is sketched. From AFM measurements it is known that H1N1 shows pleomorphism that means size (150 to 400 nm) and shape (spherical or rodlike) vary (27, 28). The dimension of the CVB3 virus (Fig. 1) is 10 times smaller (20 to 30 nm), with an icosahedral structure (determined by crystallography), where the RNA is packed in a protein coat (capsid) (29). Clearly, the different surface components of the viruses yield TERS spectra with information on proteins and lipids for H1N1 (Fig. 1B) and protein and RNA for CVB3 (Fig. 1C). Again, it is obvious that spectral variability on the virus surface occurs and a larger virus surface area must be considered. Nevertheless, a discrimination of both viruses solely based on the TERS spectra is quite straightforward. The experiment with the full statistical evaluation (multiple maps, different particles, different tips) will be published elsewhere. Here we want to emphasize that the method is potentially able to identify any single virus based on the surface composition. The main challenge is the time required for an assessment, particularly the spectral acquisition time. Interestingly, the necessary AFM topography which is always detected to locate a sample can also provide substantial information for a virus prescreening, thus considerably limiting the potential candidates for TERS investigations (30).
Clearly, the extreme intrinsic lateral resolution of TERS requires the acquisition of a statistically relevant number of spectra on the virus surface to catch all relevant structural features of a given virus. If a mere virus classification rather than nanometer-scale structural information is the goal, a different experimental approach could be advantageous, particularly by decreasing the overall acquisition time considerably. The approach shown here artificially decreases the spatial resolution by deliberately scanning the tip during the acquisition. This way, the spectral information is based on the entire scanned surface and as the tip is scanned continuously rather than in a step-scan manner, this also ensures a “gap-free” TERS experiment. It is important to note that this approach is fundamentally different from a conventional SERS experiment, where one would bring the plasmonic substrate in contact with a virus sample. In such an experiment no averaging can be achieved on a single virus and consequently a large number of samples must be investigated. Furthermore, specific substrate–sample interactions will lead to preferential binding sites that will also strongly affect the results.
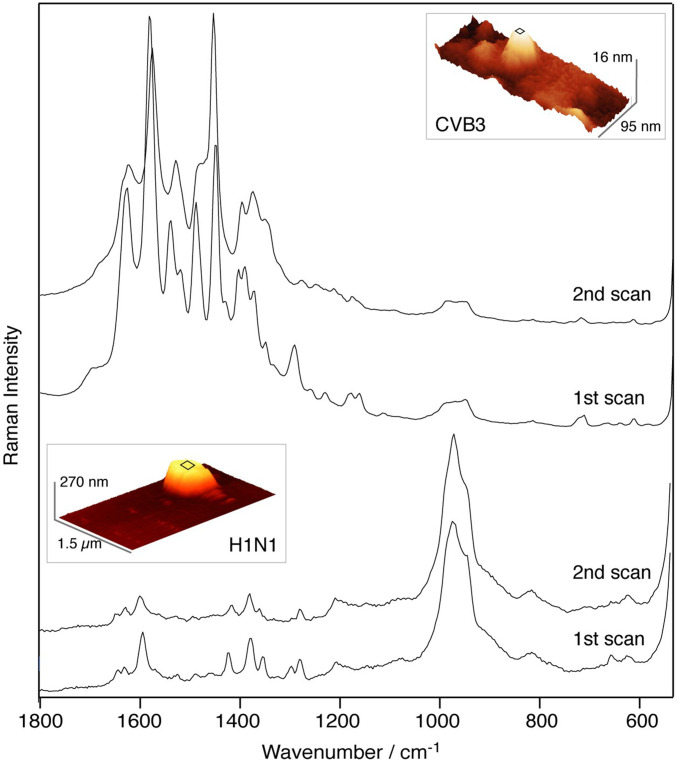
In Fig. 2 we present two straightforward TERS experiments, acquired while scanning a defined area on a single H1N1 and CVB3 virus, respectively. This way, all Raman signals during the scan were detected while the lateral resolution was intentionally decreased. The measurement parameters were adjusted such that exactly one spectrum was recorded during one topography scan.
Fig. 2.
TERS spectra acquired during two repetitive topography scans on a single CVB3 (Top) and a single H1N1 virus (Bottom), respectively. Measurement conditions for CVB3 scan are nm, pixels (px), scan rate 2 Hz, = 5 s, 14 accumulations; those for H1N1 scan are nm, px, scan rate 1 Hz, = 10 s, 14 accumulations. The shown three-dimensional topography images were generated with Gwyddion (31). The gray rectangles indicate the investigated areas on the particles.
In both samples the respective spectra acquired during subsequent topography scans indicate that the apparent spectral reproducibility is much higher compared to the single acquisitions in Fig. 1, thus demonstrating the expected location averaging. This leads to a much faster overall data acquisition and much more representative spectral data. This comes at a cost that some information is consequently buried in an averaged dataset; i.e., the clearly visible lipid marker bands of the H1N1 virus of the “high-resolution” data acquisition are not evident in Fig. 2, hence indicating that lipids patches that are accessible for the TERS tip are comparatively rare. While it is theoretically possible to achieve the same result by combining all high-resolution spectra of a sample, e.g., combining all spectra from a virus scan in Fig. 1 B and C, for a comparison it must be warranted that the specimen is sampled correctly; otherwise an imbalance of certain chemical distributions will affect the resulting postexperimental averaged data. This is particularly important since recent experimental and theoretical high-resolution TERS studies indicate a resolution down to the ångström regime. For a TERS experiment this leads to very small step sizes, i.e., to a prohibitively long acquisition time. For an analytical/diagnostic application the average data are most likely much more useful as they can be accessed much faster and due to the intrinsic spectral averaging provide a typical fingerprint of the virus. It is of course still important to determine the minimum representative scan area for a virus, which allows the clear identification of the virus strand. This is essential for comparison with other experiments, e.g., to avoid geometrical influences of the tip.
Another possibility to increase the sensitivity will be introduced in the following section where we propose a combined tip/surface enhanced Raman technique with femtosecond adaptive spectroscopic techniques for coherent anti-Stokes Raman scattering (FAST CARS) (1).
Instrumentation and Techniques
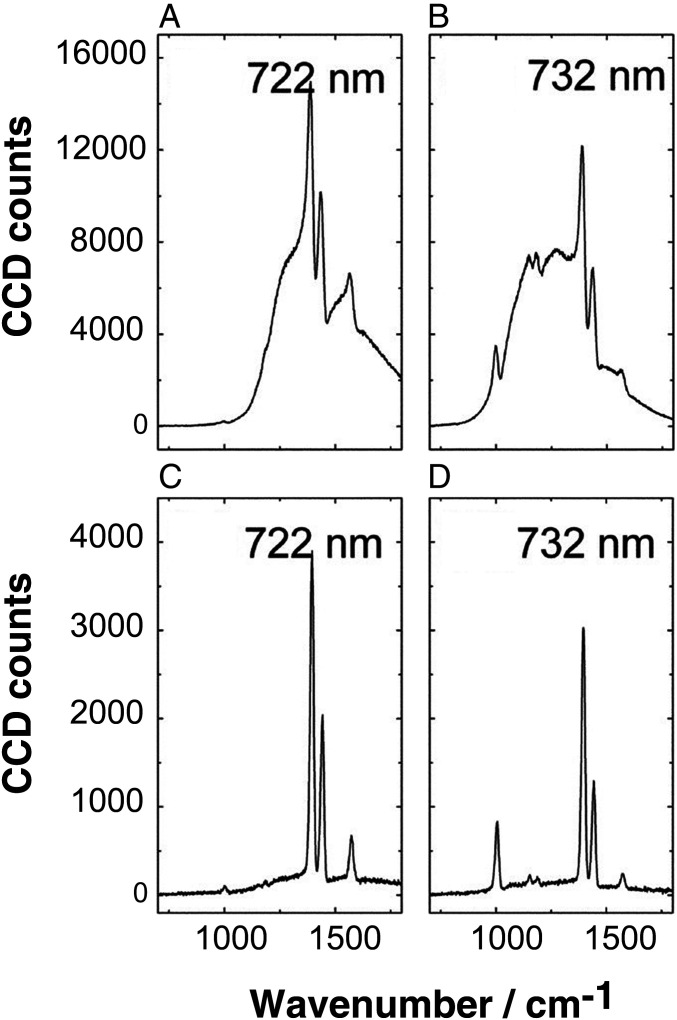
To our best knowledge, tip-enhanced CARS has been demonstrated in the early 2000s (32). Stimulated by our success in detecting anthrax using FAST CARS (33) (Fig. 3), we developed additional techniques for enhancing the sensitivity and resolution of coherent Raman spectroscopy. Specifically, we have developed additional vibrational spectroscopic techniques for nanoscale real-time molecular sensing having large signal enhancement, small background, short detection time, and high spectral resolution (34, 35). On the other hand, other groups have pushed the surface-enhanced CARS to single-molecule sensitivity (36, 37). Our time-resolved tip enhanced coherent anti-Stokes Raman spectroscopy is an exciting additional tool for virus research. Our early “proof of principle” research was used to detect hydrogen-bonded molecular complexes of pyridine with water in the near field of gold nanoparticles with large signal enhancement (34). This yields an improved FAST CARS (1) approach which is aptly called FASTER CARS.
Fig. 3.
CARS (A and B) and FAST CARS (C and D) spectra for probe delay of 0 ps (A and B) and 1.5 ps (C and D). The pump wavelengths are 722 and 732 nm, as indicated. From ref. 33. Reprinted with permission from The American Association for the Advancement of Science.
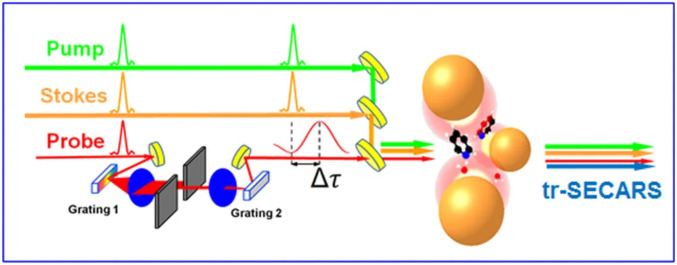
Combining this technique with modern quantum molecular calculation is natural for studying complex biological systems. Fig. 4 shows a simplified experimental arrangement used in ref. 34 and Fig. 5 shows typical data.
Fig. 4.
A schematic for the time-resolved surface-enhanced CARS spectroscopy. Femtosecond pump (green) and Stokes (orange) laser pulses excite coherent vibration of molecules, which scatter properly delayed sinc-shaped probe pulse coherently to generate the CARS signal. All these optical fields are enhanced in the near field of gold nanoparticles. Adapted from ref. 34, which is licensed under CC BY-NC-ND 3.0.
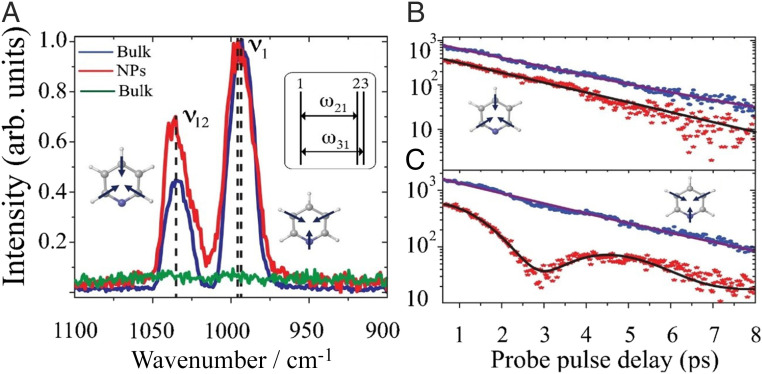
Fig. 5.
(A) Normalized spectra of pyridine with probe pulse delay of 1 ps and (B and C) temporal traces of peak intensity count with (red) and without (blue, green) gold nanoparticles (NPs). The 12-m thick pyridine with NPs (red) generates the same order of magnitude of CARS signals as a 2-mm bulk (blue) under the same laser pulse conditions, while 12-m thick pure pyridine (green) is not detectible. After a deconvolution from the probe pulse sinc shape, we can extract the ring breathing mode of the pyridine–water complex from the red trace in C. Adapted from ref. 34, which is licensed under CC BY-NC-ND 3.0.
The review article of Lis and Cecchet (38) characterizes this technique nicely; they say
“Investigations demonstrated that the electronic background that derives from the water and the metal could also be reduced on such kind of solid substrates. Voronine et al. used time-resolved surface-enhanced CARS, which combines delayed laser pulses of different widths, to obtain a high spectral resolution with the suppression of the non-resonant back ground.”
They go on to say that this technique yields “astonishing sensitivity.” Indeed, this improvement over our earlier FAST CARS anthrax detection scheme holds real promise for detection and identification of single virus particles such as SARS-CoV-2 virus which causes COVID-19.
Discussion
We have shown that on one hand, the tip-enhanced Raman technique can be used to characterize and differentiate between influenza and picornavirus. On the other hand, surface-enhanced CARS results in “astonishing” enhancement in the sensitivity (38). The FASTER CARS system, which combines nanometer spatial resolution and enhanced sensitivity, forged by both the tip and coherent Raman enhancement, provides exciting tools for viral research and detection, e.g., the SARS-CoV-2 virus.
Acknowledgments
We express our deep appreciation to The Texas A&M Univeristy System chancellor John Sharp without whom this project would not have happened. We thank the Office of Naval Research, Air Force Office of Scientific Research, NSF, Chancellor’s Research Initiative/Governor’s University Research Initiative, the Robert A. Welch Foundation (Grants A-1261 and A-1547), King Abdulaziz City for Science and Technology, and the Deutsche Forschungsgemeinschaft (DFG) (German Research Foundation, Grant DFG-CRC1375NOA) for support of this work. We also thank G. Agarwal, B. Brick, K. Chamakura, J. Mogford, J. Sharp, R. Young, and A. Zheltikov for stimulating and helpful discussions.
Footnotes
The authors declare no competing interest.
Data Availability.
All study data are included in this article.
References
- 1.Scully M. O., Sokolov A. V., Zubairy M. S. et al. , FAST CARS: Engineering a laser spectroscopic technique for rapid identification of bacterial spores. Proc. Natl. Acad. Sci. U.S.A. 99, 10994–11001 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang Y. W., Procop G. W., Persing D. H., Molecular diagnostics of infectious diseases. Clin. Chem. 43, 2021–2038 (1997). [PubMed] [Google Scholar]
- 3.Ellis J. S., Zambon M. C., Molecular diagnosis of influenza. Rev. Med. Virol. 12, 375–389 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Vaneechoutte M., Van Eldere J., The possibilities and limitations of nucleic acid amplification technology in diagnostic microbiology. J. Med. Microbiol. 46, 188–194 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Valones M. A. A., et al. , Principles and applications of polymerase chain reaction in medical diagnostic fields: A review. Braz. J. Microbiol. 40, 1–11 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mackay I. M., Arden K. E., Nitsche A., Real-time PCR in virology. Nucleic Acids Res. 30, 1292–1305 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang R., et al. , Chemical mapping of a single molecule by plasmon-enhanced Raman scattering. Nature 498, 82–86 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Deckert-Gaudig T., Kämmer E., Deckert V., Tracking of nanoscale structural variations on a single amyloid fibril with tip-enhanced Raman scattering. J. Biophot. 5, 215–219 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Deckert-Gaudig T., et al. , Spatially resolved spectroscopic differentiation of hydrophilic and hydrophobic domains on individual insulin amyloid fibrils. Sci. Rep. 6, 33575 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee J., Crampton K. T., Tallarida N., Apkarian V. A., Visualizing vibrational normal modes of a single molecule with atomically confined light. Nature 568, 78–82 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Deckert-Gaudig T., Taguchi A., Kawata S., Deckert V., Tip-enhanced Raman spectroscopy–From early developments to recent advances. Chem. Soc. Rev. 46, 4077–4110 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Kawata S., Ichimura T., Taguchi A., Kumamoto Y., Nano-Raman scattering microscopy: Resolution and enhancement. Chem. Rev. 117, 4983–5001 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Richard-Lacroix M., Zhang Y., Dong Z., Deckert V., Mastering high resolution tip-enhanced Raman spectroscopy: Towards a shift of perception. Chem. Soc. Rev. 46, 3922–3944 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Verma P., Tip-enhanced Raman spectroscopy: Technique and recent advances. Chem. Rev. 117, 6447–6466 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Langer J., et al. , Present and future of surface-enhanced Raman scattering. ACS Nano 14, 28–117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He Z., et al. , Tip-enhanced Raman imaging of single-stranded DNA with single base resolution. J. Am. Chem. Soc. 141, 753–757 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Lin X. M., et al. , Direct base-to-base transitions in ssDNA revealed by tip-enhanced Raman scattering. arXiv:1604.06598 (22 April 2016).
- 18.Langelüddecke L., Singh P., Deckert V., Exploring the nanoscale: Fifteen years of tip-enhanced Raman spectroscopy. Appl. Spectrosc. 69, 1357–1371 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Domke K., Zhang D., Pettinger B., Tip-enhanced Raman spectra of picomole quantities of DNA nucleobases at Au(111). J. Am. Chem. Soc. 129, 6708–6709 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Hennemann L. E., Meixner A. J., Zhang D., Surface- and tip-enhanced Raman spectroscopy of DNA. Spectroscopy 24, 119–124 (2010). [Google Scholar]
- 21.Bailo E., Deckert V., Tip-enhanced Raman scattering. Chem. Soc. Rev. 37, 921–930 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Hermann P., et al. , Evaluation of tip-enhanced Raman spectroscopy for characterizing different virus strains. Analyst 136, 1148–1152 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Hermann P., Fabian H., Naumann D., Hermelink A., Comparative study of far-field and near-field Raman spectra from silicon-based samples and biological nanostructures. J. Phys. Chem. C 115, 24512–24520 (2011). [Google Scholar]
- 24.Cialla D., et al. , Raman to the limit: Tip-enhanced Raman spectroscopic investigations of a single tobacco mosaic virus. J. Raman Spectrosc. 40, 240–243 (2009). [Google Scholar]
- 25.Olschewski K., et al. , A manual and an automatic TERS based virus discrimination. Nanoscale 7, 4545–4552 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Hermelink A., et al. , Towards a correlative approach for characterising single virus particles by transmission electron microscopy and nanoscale Raman spectroscopy. Analyst 142, 1342–1349 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Giocondi M. C., et al. , Organization of influenza A virus envelope at neutral and low pH. J. Gen. Virol. 91, 329–338 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Liu Y., et al. , Study on the morphology of influenza virus A by atomic force microscopy. Chin. J. Virol. 24, 106–110 (2008). [PubMed] [Google Scholar]
- 29.Muckelbauer J. K., et al. , The structure of coxsackie B3 at 3.5Å resolution. Structure 3, 653–667 (1995). [DOI] [PubMed] [Google Scholar]
- 30.Bocklitz T., et al. , Single virus detection by means of atomic force microscopy in combination with advanced image analysis. J. Struct. Biol. 188, 30–38 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Nečas D., Klapetek P., Gwyddion: An open-source software for SPM data analysis. Cent. Eur. J. Phys. 10, 181–188 (2012). [Google Scholar]
- 32.Ichimura T., Hayazawa N., Hashimoto M., Inouye Y., Kawata S., Application of tip-enhanced microscopy for nonlinear Raman spectroscopy. Appl. Phys. Lett. 84, 1768–1770 (2004). [Google Scholar]
- 33.Pestov D., et al. , Optimizing the laser-pulse configuration for coherent Raman spectroscopy. Science 316, 265–268 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Voronine D. V., et al. , Time-resolved surface-enhanced coherent sensing of nanoscale molecular complexes. Sci. Rep. 2, 891 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shutov A. D., et al. , Giant chemical surface enhancement of coherent Raman scattering on MoS2. ACS Photonics 5, 4960–4968 (2018). [Google Scholar]
- 36.Zhang Y., et al. , Coherent anti-Stokes Raman scattering with single-molecule sensitivity using a plasmonic Fano resonance. Nat. Commun. 5, 4424 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Yampolsky S., et al. , Seeing a single molecule vibrate through time-resolved coherent anti-Stokes Raman scattering. Nat. Photon. 8, 650–656 (2014). [Google Scholar]
- 38.Lis D., Cecchet F., Localized surface plasmon resonances in nanostructures to enhance nonlinear vibrational spectroscopies: Towards an astonishing molecular sensitivity. Beilstein J. Nanotechnol. 5, 2275–2292 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All study data are included in this article.