Significance
The cerebral cortex of the human brain is a highly complex, heterogeneous tissue that contains many cell types that are exquisitely regulated at the level of gene expression by noncoding regulatory elements, presumably in a cell-type–dependent manner. However, assessing the regulatory elements in individual cell types is technically challenging, and therefore most of the previous studies on gene regulation were performed with bulk brain tissue. Here we analyze two major types of neurons isolated from the cerebral cortex of humans, chimpanzees, and rhesus macaques, and report complex patterns of cell-type–specific evolution of the regulatory elements in numerous genes. Many genes with evolving regulation are implicated in language abilities as well as psychiatric disorders.
Keywords: H3K27ac histone modification, regulatory elements, glutamatergic neurons, GABAergic neurons, primate evolution
Abstract
The human cerebral cortex contains many cell types that likely underwent independent functional changes during evolution. However, cell-type–specific regulatory landscapes in the cortex remain largely unexplored. Here we report epigenomic and transcriptomic analyses of the two main cortical neuronal subtypes, glutamatergic projection neurons and GABAergic interneurons, in human, chimpanzee, and rhesus macaque. Using genome-wide profiling of the H3K27ac histone modification, we identify neuron-subtype–specific regulatory elements that previously went undetected in bulk brain tissue samples. Human-specific regulatory changes are uncovered in multiple genes, including those associated with language, autism spectrum disorder, and drug addiction. We observe preferential evolutionary divergence in neuron subtype-specific regulatory elements and show that a substantial fraction of pan-neuronal regulatory elements undergoes subtype-specific evolutionary changes. This study sheds light on the interplay between regulatory evolution and cell-type–dependent gene-expression programs, and provides a resource for further exploration of human brain evolution and function.
Among the numerous phenotypic differences between humans and other primates, the most striking are specializations of social and cognitive abilities, including language and executive function, such as abstract reasoning, planning, behavioral inhibition, and understanding mental states of others (1). It has been hypothesized that the evolutionary changes associated with the unique features of human cognition reside, primarily, in the neocortex (2). The neocortex contains multiple cell types, including two major classes of neurons, the excitatory glutamatergic (Glu) projection neurons and the inhibitory GABAergic interneurons, which account for about 70 to 80% and 20 to 30% of all cortical neurons, respectively (3–5). The specification and maintenance of these neurons are determined by transcriptional programs that are themselves controlled by the activity of gene regulatory elements (GRE), such as promoters and enhancers (6). GREs recruit transcription factors and chromatin modifiers to control the expression of genes in a cell-type–dependent manner (7). Multiple lines of evidence suggest that phenotypic variation among mammals and susceptibility to brain diseases are largely due to changes in GREs rather than protein-coding sequences (8, 9). Indeed, enhancer changes could cause tissue- or cell-type–dependent adaptations without causing pleiotropic effects that are often associated with changes to genes (10, 11). Therefore, to understand the molecular and cellular differences in brain organization between human and other primates better, it is essential to characterize the mechanisms that drive GRE evolution in individual cell types.
Previous studies used chromatin immunoprecipitation followed by sequencing (ChIP-seq) to assess evolutionary changes of GREs marked by covalent histone modifications in bulk brain tissue (12–14). However, information on key evolutionary changes that could affect the human brain thus far remains limited. One reason is that regulatory changes affecting a particular cell type cannot be reliably inferred from data on bulk brain specimens that conflate signals from all cell types. Mixed signals in bulk specimens likely mask signatures of lower abundance cells (e.g., GABA neurons). In contrast to bulk tissue analysis, single-cell ChIP-seq techniques that are currently under development lack sufficient coverage to reliably detect regulatory elements (15, 16). To provide insight into the cell-type–dependent changes that underlie evolution of the human neocortex, we used fluorescence-activated nuclei sorting (FANS) to isolate cortical Glu and GABA neuronal nuclei obtained from one of our closest extant relatives, chimpanzee, and a commonly studied nonhuman primate, rhesus macaque, followed by ChIP-seq and RNA-sequencing (RNA-seq) analyses of the epigenome and trancriptome (17, 18). We integrated these data with complementary human transcriptomes and ChIP-seq data from sorted Glu and GABA neurons (19).
We identified numerous GREs that have not been detected in bulk brain specimens, many of which have neuron subtype-specific and species-specific enrichment for histone modifications. We found strong evidence of concordant evolutionary changes in expression and epigenetic regulation for ∼200 genes, highlighting the functional importance of regulatory evolution in neuronal subtypes. These include genes involved in opioid signaling and drug abuse (OPRM1, PENK, SLC17A8) (20–22), as well as genes associated with language impairments (ATP2C2, DCDC2) (23). We also found neuron subtype- and human-enriched regulatory elements in two genes that are considered among the strongest candidates for enabling or facilitating language abilities, FOXP2 and CNTNAP2 (24–26), and identified large clusters of human-specific GREs near genes implicated in neuronal function and brain disorders (e.g., CDH8, ASTN2, CNTNAP4) (27–29). Furthermore, we demonstrated that cell-type–specific GREs are more likely to change their activity during primate evolution than GREs shared by different cell types, and found that positional conservation of a GRE in different cell types is not always associated with functional conservation. Our findings provide insight into regulatory evolution that is relevant for brain function and disorders, and present a resource for future studies on comparative epigenomics and neuroscience.
Results
Epigenomic Profiling of Glu and Medial Ganglionic Eminence-GABA Neurons from Primate Brains Reveals Extensive Differences in Regulatory Landscapes between Neuronal Subtypes.
FANS allows separation and acquisition of nuclei from Glu and medial ganglionic (MGE)-derived GABA neurons (MGE-GABA) from autopsied cortical samples (17, 19) (Fig. 1A). MGE-GABA neurons comprise ∼60 to 70% of all neocortical GABA neurons and have been implicated in schizophrenia, major depression, autism spectrum disorder (ASD), and epilepsy (30, 31). We recently reported RNA-seq transcriptome profiling and ChIP-seq analysis of histone 3 lysine 27 acetylation (H3K27ac) in Glu and MGE-GABA nuclei that were obtained from the dorsolateral prefrontal cortex (DLPFC) (19). H3K27ac is a robust marker of active promoters and enhancers (32, 33). The DLPFC is the neocortical region that is important for cognition and executive function (1). To investigate regulatory changes across primate evolution, we performed H3K27ac ChIP-seq in Glu and MGE-GABA nuclei from four male chimpanzee and four rhesus macaque DLPFC, and integrated these datasets with our previously obtained data for humans (19) (Dataset S1).
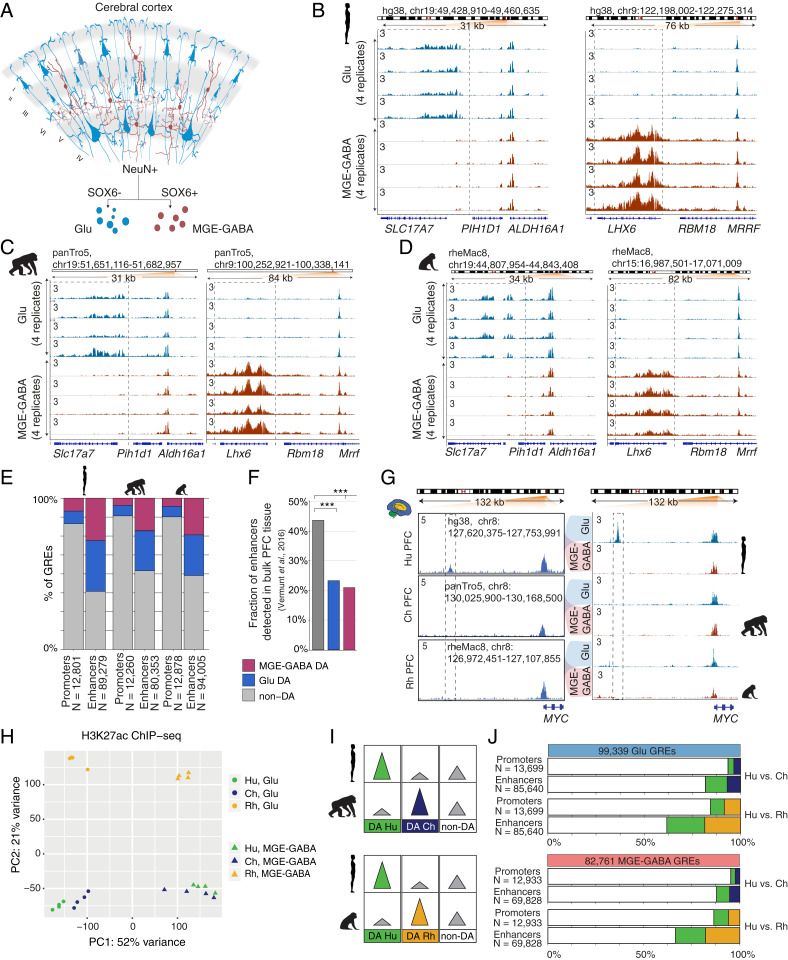
Fig. 1.
Regulatory changes in Glu and MGE-GABA neurons during primate evolution. (A) Schematic of isolation of Glu and MGE-GABA nuclei. (B–D) H3K27ac ChIP-seq profiles for Hu (B), Ch (C), and Rh (D) at the loci of Glu (SLC17A7, Left) and MGE-GABA (LHX6, Right) markers. Read per million (RPM)-normalized reads (axis limit 3 RPM). (E) Fractions of promoters or enhancers that are Glu DA, MGE-GABA DA or non-DA in Glu vs. MGE-GABA. (F) Fractions of Glu DA, MGE-GABA DA, and non-DA Hu enhancers that overlap GREs in bulk prefrontal cortex (PFC) tissue (12). ***P < 0.0005 (Fisher’s exact test). (G) The regulatory landscapes near the MYC locus in bulk PFC (12) (Left) and in Glu and MGE-GABA neurons (Right). (H) PCA of ChIP-seq data for Glu and MGE-GABA neurons from Hu, Ch, and Rh. (I) Schematic of pairwise interspecies comparisons between the ChIP-seq datasets. (J) Fractions of Glu DA, MGE-GABA DA, and non-DA GREs in pairwise species comparisons; Glu (Upper) and MGE-GABA (Lower) neurons.
We produced high-quality H3K27ac ChIP-seq datasets (Dataset S2), with biological replicates showing strong correlation in each cell type and species (all correlation coefficients, r > 0.8) (SI Appendix, Fig. S1A). Well-established Glu markers (e.g., SLC17A7, TBR1) were enriched in H3K27ac specifically in Glu neurons, whereas typical MGE-GABA markers (e.g., LHX6, SOX6) were enriched in MGE-GABA neurons only (Fig. 1 B–D and SI Appendix, Fig. S1 B–D). We defined putative GREs as regions (peaks) of histone acetylation that were significantly enriched over the background in at least three of the four donors, detecting a comparable number of GREs (∼100,000) in at least one neuronal subtype in each species (hereafter, Glu or MGE-GABA GREs) (Methods and Dataset S3). We then identified GREs that were differentially acetylated (DA) between neuronal subtypes in each species separately, focusing on the subset of GREs with the strongest cell type differences (fold change [FC] > 2, false-discovery rate [FDR] < 0.05) (Methods, SI Appendix, Fig. S1E, and Dataset S4). We then subdivided GRE regions into proximal H3K27ac peaks (within 1 kb from annotated transcriptional start sites in the human genome), which are indicative of active promoters, and distal H3K27ac peaks, which are indicative of active enhancers. We confirmed the previously observed greater regulatory diversity of enhancers vs. promoters, detecting significant differential acetylation between cell types in 48 to 61% of enhancers vs. 12 to 17% of promoters (Fig. 1E) (11). A substantial proportion of all active neuronal GREs (promoters and enhancers) was DA between Glu and MGE-GABA neurons (ranging from 43 to 56% in different species), with a comparable ratio of DA GREs between the neuronal subtypes in each species (SI Appendix, Fig. S1E). Gene ontology (GO) analysis using GREAT (Genomic Regions Enrichment of Annotations Tool) (34) validated the cell type-specificity and functional relevance of the regulatory elements we identified (Dataset S5).
To test whether the cell-type–specific approach increases the sensitivity and resolution of GRE analysis, we compared the human Glu and MGE-GABA enhancers in the DLPFC with enhancers that were previously identified in bulk human tissues from several brain regions (cortex, cerebellum, and subcortical regions) using H3K27ac ChIP-seq (12, 35). Over 40% of non-DA enhancers but only ∼20% of either Glu or MGE-GABA DA enhancers had been previously identified in bulk cortex (Fig. 1F). Despite a higher proportion of Glu vs. MGE-GABA neurons in the cortex, a comparable fraction of Glu DA and MGE-GABA DA enhancers was not detected in bulk cortical tissue. This is likely explained by the large number of unique subpopulations of cortical Glu neurons, which were recently uncovered in single-nucleus RNA-seq analyses (36). In contrast, the MGE-derived GABA neurons represent a less diverse subset of cortical neurons (36). Our approach also enables the assignment of neuron-subtype specificity for GREs that were previously detected in bulk brain tissues. For example, the human MYC enhancer that was previously found to be both human-specific and cortex-specific (12), is exclusively active in Glu but not in MGE-GABA neurons (Fig. 1G).
Substantial Regulatory Changes in Glu and MGE-GABA Cortical Neurons during Primate Evolution.
To enable the comparison of the H3K27ac datasets from different species, coordinates of Glu and MGE-GABA H3K27ac-enriched regions from chimpanzee and rhesus macaque were converted to the hg38 human genome assembly using liftOver (12) (Methods). Regions with overlapping coordinates were merged (n = 110,270 for Glu; n = 91,560 for MGE-GABA), and liftOver was used again to convert the coordinates to panTro5 and rheMac8. Only GREs that could be mapped onto all three genomes and did not overlap blacklisted regions were included in the subsequent analyses (99,339 Glu and 82,761 MGE-GABA GREs) (Methods and Dataset S6). We compared the H3K27ac signals within GREs between the species and neuronal subtypes using Pearson correlation, hierarchical clustering, and principal component analysis (PCA) (Methods, Fig. 1H, and SI Appendix, Fig. S1 F and G). Based on correlation and clustering analyses, we detected greater differences between cell types in each species compared with the differences between species in each cell type. Clustering and PCA showed a clear separation of samples into six groups according to cell type and species, with human and chimpanzee clustering closer together vs. rhesus macaque in each cell type.
To investigate the regulatory changes during primate evolution, we analyzed H3K27ac levels within the GREs that were DA between species in each neuronal subtype in pairwise comparisons (i.e., species-enriched GREs) (Methods, Fig. 1 I and J, SI Appendix, Fig. S1H, and Dataset S7). In both Glu and MGE-GABA, ∼14 to 16% of promoters (2,146 in Glu and 1,754 in MGE-GABA) and ∼34 to 38% of enhancers (32,785 in Glu and 23,435 in MGE-GABA) were DA in human vs. rhesus macaque (FDR < 0.05, FC > 2) (Fig. 1J). Consistent with the closer evolutionary relationship between human and chimpanzee, there were fewer DA promoters (896 in Glu and 609 in MGE-GABA) and enhancers (15,625 in Glu and 8,495 in MGE-GABA) between human and chimpanzee. Thus, using a cell-type–specific analysis, we showed that, in each neuronal subtype, ∼30% of the GREs underwent significant changes in primate evolution. Examples of such regulatory changes are shown in SI Appendix, Fig. S1I.
Neuron Subtype–Specific Regulatory Elements Change more Frequently during Evolution than Nonspecific Ones.
Previous work in bulk brain tissues has shown that evolutionary changes preferentially occur in GREs that are mainly active in a single brain region (cortex, midbrain, or cerebellum), but not in multiple structures (12). This result fits a model in which changes in GREs that are not region-specific are more likely to cause pleiotropic effects, which might be detrimental to the fitness of the organism and are, therefore, selected against during evolution. We hypothesized that this trend also extends to evolutionary changes in the activity of cell-type–enriched GREs. Only a limited number of GREs have changed between human and chimpanzee, precluding a reliable statistical analysis. We therefore assembled two sets of GREs that were detected in human and/or rhesus macaque in each neuronal subtype (93,222 for Glu and 79,181 for MGE-GABA), and compared cell-type specificity of DA and non-DA GREs between the two species (Fig. 2A). We found that DA GREs were significantly more often cell type-specific than non-DA ones (Fisher’s exact test, P < 2.2e-16; odds ratios: 2.8 for Glu and 2.2 for MGE-GABA). Conversely, human GREs that were non-DA between neuronal subtypes were significantly less frequently DA between the two species compared to GREs that were DA between Glu and MGE-GABA (Fisher’s exact test, P < 2.2e-16) (SI Appendix, Fig. S2A).
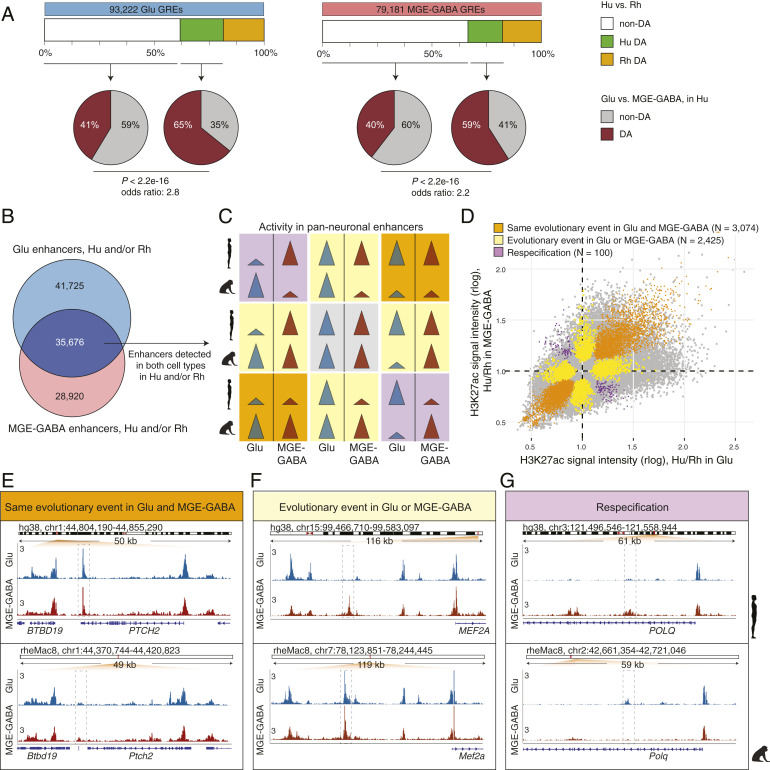
Fig. 2.
Evolutionary changes in neuron subtype-specific and pan-neuronal regulatory elements. (A) Cell type specificity of Hu > Rh DA (green boxes) or non-DA (white boxes) GREs. GREs detected in Glu (Left) or MGE-GABA (Right) neurons in Hu and/or Rh. Pie charts indicate fractions of DA (brown) or non-DA (gray) GREs between neuronal subtypes in Hu. In both neuronal subtypes, Hu > Rh DA GREs were more often neuron-subtype-specific than non-DA GREs (P < 2.2e-16; Fisher’s exact test). (B) Venn diagram of the overlap between Glu and MGE-GABA enhancers in Hu and/or Rh. The overlapping enhancers (dark blue) are positionally shared in Glu and MGE-GABA neurons (pan-neuronal). (C) Schematic of possible evolutionary changes in pan-neuronal enhancers in Hu and/or Rh. Gray, no evolutionary change in any neuronal subtype; orange, same direction of an evolutionary change in both subtypes; yellow, an evolutionary change in only one subtype; purple, different direction of an evolutionary change between subtypes (respecification). (D) Simultaneous cross-species and cross-cell–type analysis of H3K27ac signal intensities for pan-neuronal enhancers in Hu and/or Rh (Methods and SI Appendix, Fig. S2C). Shown are DA GREs confirmed by the analysis to undergo the type of an evolutionary change described in C. (E–G) Examples of enhancers (dashed boxes) with the same direction of evolutionary change in Glu and MGE-GABA (E, the PTCH2 locus), with an evolutionary change in only one neuronal subtype, MGE-GABA (F, near MEF2A), and with evolutionary changes in opposite directions in Glu and MGE-GABA neurons (respecification) (G, the POLQ locus).
To further validate this finding, we used a threshold-free approach by directly comparing the enrichment of enhancers in neuronal subtypes (e.g., difference between Glu and MGE-GABA in human, ΔCellTypehuman = H3K27acGlu,human − H3K27acGABA,human) vs. the evolutionary change (e.g., difference between human and macaque in Glu cells, ΔSpeciesGlu = H3K27acGlu,human − H3K27acGlu,rhesus) (Methods). This analysis used independent biological samples to estimate H3K27acGlu,human at each step of the analysis, to avoid spurious correlations driven by noise in individual datasets. Across all enhancers detected in human Glu cells, we found a significant correlation between the cell type-specificity and the evolutionary change (Spearman r = 0.203, P < 1e-16) (SI Appendix, Fig. S2 B, Upper). No such correlation was observed when the enhancers were randomly shuffled. A similar pattern was observed in human MGE-GABA cells (r = 0.106, P < 1e-16) (SI Appendix, Fig. S2 B, Lower). These analyses confirm that evolutionary divergence of neuronal enhancers preferentially occurs in a cell-type–specific manner.
Pan-Neuronal Enhancers Undergo Neuron Subtype-Specific Evolutionary Changes.
Whereas GREs active in both Glu and MGE-GABA (hereafter, pan-neuronal GREs) are more often evolutionarily conserved between human and rhesus macaque than cell-type–specific ones, a substantial proportion of pan-neuronal GREs change in at least one neuronal subtype (SI Appendix, Fig. S2A). Thus, our data offered a unique opportunity to compare the evolution of enhancers that are active in at least two closely related cell types. We found that, among these shared enhancers (n = 35,676) (dark purple in Fig. 2B), 19,225 (∼54%) showed no evolutionary changes (gray panel in Fig. 2C), whereas the rest (n = 16,451) changed between the species in at least one cell type (DA analyses between human and rhesus macaque in Glu or MGE-GABA, FC > 2; FDR < 0.05) (yellow, orange, and purple panels in Fig. 2C). We then asked whether evolutionary changes in the latter group were similar in Glu vs. MGE-GABA neurons. We categorized the enhancers into 1) enhancers with the same direction of evolutionary change in both Glu and MGE-GABA neurons (orange panels in Fig. 2C), 2) enhancers with an evolutionary change in only one cell type (yellow panels in Fig. 2C), and 3) enhancers with evolutionary changes of opposite directions in Glu and MGE-GABA neurons. The latter group represents a small subset of enhancers that may be functionally respecified (12, 37), with activity switching from one neuronal subtype in one species to a different neuronal subtype in another species (purple panels in Fig. 2C). We used a linear model to further assess the statistical significance of these evolutionary events, assigning events to several categories depending on the proportion of variance explained by the corresponding factor (η2) and the FDR value (Methods and SI Appendix, Fig. S2C). By overlapping the results of this analysis with the DA analysis (Fig. 2C), we identified 3,074 enhancers that changed in the same direction in both cell types, 2,425 enhancers that changed in one but not in the other cell type, and 100 enhancers that underwent a respecification (Fig. 2D and Dataset S8).
Examples of these three types of evolutionary events are highlighted in Fig. 2 E–G. The enhancer in the PTCH2 locus showed increased activity in human vs. rhesus macaque in both neuronal subtypes (Fig. 2E). Another enhancer, located ∼60 kb upstream of the MEF2A promoter, illustrates an evolutionary change that occurred in only one cell type. The pan-neuronal activity of this enhancer in rhesus macaque is contrasted with strictly Glu-specific H3K27ac signal in human (Fig. 2F). Finally, an enhancer in the POLQ locus provides a rare example of respecification, as it is Glu-specific in human and MGE-GABA–specific in rhesus macaque (Fig. 2G). Altogether, these results indicate that enhancers in conserved genomic positions between species in the same tissue can undergo different evolutionary changes in different cell types. Therefore, positional conservation of an enhancer is not always a reliable proxy for its functional conservation, suggesting that variation of enhancer activity between cell types is most likely greater than can be inferred based on positional conservation alone.
Concordant Evolutionary Changes in GREs and Gene Expression Underscore Functional Relevance of Regulatory Evolution in Neuronal Subtypes.
We next examined how the evolutionary changes in the regulatory landscape of the neuronal subtypes are reflected in changes in gene expression. We performed RNA-seq on FANS-separated Glu and MGE-GABA nuclei purified from the same DLPFC specimens that were used for the H3K27ac profiling (Dataset S1), obtaining high-quality transcriptomes for each species and cell type (Methods, SI Appendix, Fig. S3 A–E, and Dataset S9). In agreement with previous findings in various bulk tissues and species (38), the expression of protein-coding genes (n = 16,846) was much more conserved between species than the expression of long intergenic noncoding RNA genes (n = 518) (SI Appendix, Fig. S3F).
Next, in each neuronal subtype, we identified DA GREs and differentially expressed (DE) genes that were enriched or depleted in a particular species compared to the two other species (Fig. 3 A, Left and SI Appendix, Fig. S3 G, Upper). We denoted these up-regulated or down-regulated GREs or genes as species-specific up- or down-DA GREs or DE genes. To focus on high-confidence DA GREs and DE genes, we applied conservative FC thresholds in both analyses (FC > 2, FDR < 0.05) (Methods, SI Appendix, Fig. S3 G, Lower, and Datasets S10–S12). We found thousands of DA GREs and hundreds of DE genes for each comparison, with the largest numbers of species-specific GREs and genes detected in rhesus macaque (Fig. 3 B and C). The analysis of DE genes showed that approximately half of the genes that were species-specific in one neuronal subtype were also specific for the same species in another neuronal subtype (Dataset S12). In addition, for both neuronal subtypes, the species-specific up-DE genes significantly overlapped with the up-DE genes that have been recently reported for the same species in sorted neuronal (NeuN+) nuclei from the DLPFC (11 to 31% overlap for Glu DE genes and 12 to 23% overlap for MGE-GABA DE genes, P < 2.5e-6 by hypergeometric test) (Dataset S13) (39). Also, our gene-expression data were in good agreement with the results of human vs. rhesus macaque DE analysis in bulk adult DLPFC reported by the PsychENCODE consortium (17 to 19% overlap, P < 2.8e-14 by hypergeometric test) (Dataset S14) (40).
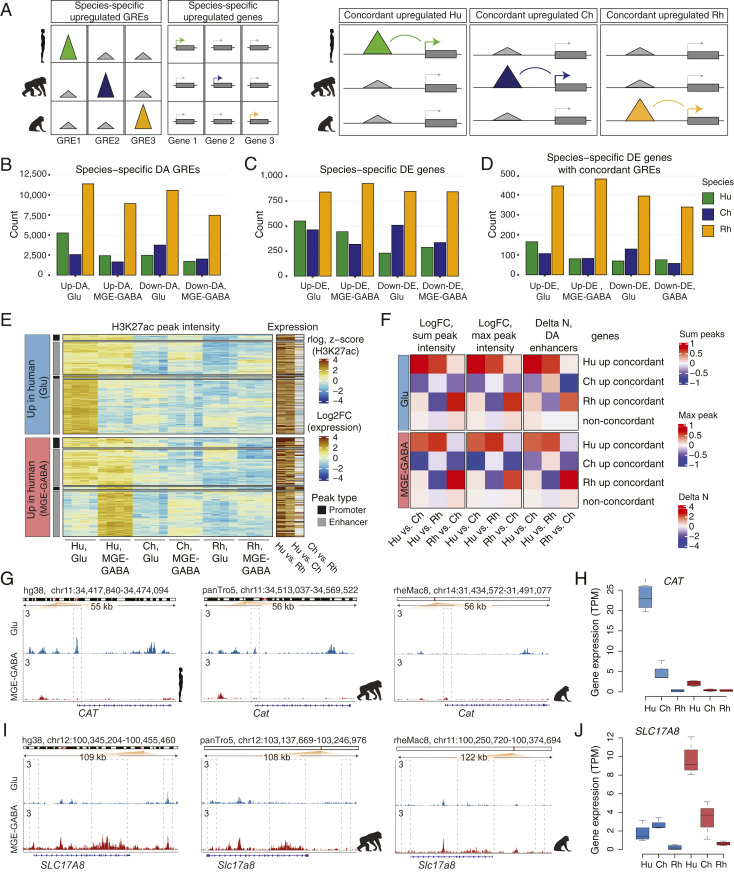
Fig. 3.
Concordant evolutionary changes in GREs and gene expression. (A) Schematic of the analysis of association between species-specific up-regulated GREs and genes. (Left) identification of species-specific DA GREs and DE genes. (Right) identification of concordant pairs of species-specific up-regulated DA GREs and DE genes. (B) Numbers of species-specific up-regulated or down-regulated DA GREs detected in Glu or MGE-GABA neurons. (C) Numbers of species-specific up-regulated or down-regulated DE genes. (D) Numbers of species-specific DE genes with concordant species-specific DA GREs. (E) Heatmap of H3K27ac signal intensities and interspecies changes in gene expression for concordant pairs of Hu-specific up-DA GREs and up-DE genes. Shown are concordant GRE-gene pairs in Glu (Upper) and MGE-GABA (Lower). K-means clustering of the H3K27ac signal in individual Hu, Ch, and Rh samples resulted in two major clusters for each neuronal subtype, corresponding to neuron-subtype-specific or nonspecific GREs. Concordant promoter- and enhancer-gene pairs are shown separately for each cluster. Heatmap colors indicate normalized H3K27ac signal intensities (rlog) for each replicate sample and log2 FC of normalized RNA-seq reads (transcripts per million, TPM). (F) Mean interspecies changes of quantitative features of gene regulatory domains. The features are detailed on the top of the panel. Shown are data for species-specific up-regulated concordant genes and for non-concordant genes. (G and H) Evolutionary changes in regulation (G) and expression (H) for the CAT gene. (I and J) Same as in G and H for the SLC17A8 gene.
We performed GO analysis for species-specific DE genes using Enrichr (41), as well as for genes located near species-specific DA GREs using GREAT. The human-specific DE gene sets in Glu or MGE-GABA neurons were not enriched for genes from any functional GO categories; the chimpanzee-specific Glu DE genes were enriched for a few GO terms, including “axon development,” adjusted P = 0.014. (Dataset S15). Species-specific GREs were enriched for gene sets from several GO terms; however, the majority of these terms were not related to the nervous system (Dataset S15). One exception was the enrichment of the human-specific DA GREs in Glu neurons for the “regulation of glutamate secretion” term (adjusted P value 0.019). Also of interest were the results of GO analysis for the pairwise interspecies comparisons (Dataset S15). In MGE-GABA neurons, the GREs that were up-regulated in human vs. rhesus macaque were significantly enriched for the “L-type voltage gated calcium channels” term as well as for genes involved in “Wnt-activated receptor activity.” The Wnt pathway-related genes (e.g., FZD8) are known to influence cortical size (42, 43).
We then linked species-specific GREs to nearby genes using GREAT (Methods) and overlapped the resulting gene sets with genes showing species-specific expression (Fig. 3 A, Right, SI Appendix, Fig. S3H, and Datasets S16 and S17). This analysis yielded hundreds of genes with evidence of concordant species-specific evolutionary changes in gene expression and in at least one associated GRE (Fig. 3D). The fraction of species-specific DE genes that had a concordant DA GRE varied for each species and was in the range of 18 to 30% in human, 17 to 26% in chimpanzee, and 40 to 53% in rhesus macaque for up- or down-regulated DE genes in Glu or MGE-GABA (Dataset S18). We used random shuffling (Methods) to assess statistical significance of the observed concordant association between GREs and genes. For all categories of concordant changes (species-specific up- or down-regulated, Glu or MGE-GABA neurons, promoters or enhancers), concordant GRE-gene pairs were significantly overrepresented compared with random pairs (all Ps < 1e-7) (SI Appendix, Fig. S3I), indicating that the genes linked to DA regulatory elements are more likely to be DE than expected by chance. Thus, a sizable proportion of the identified concordant GRE-gene pairs likely represents functional associations that underlie evolutionary differences in gene expression between the primate species, rather than resulting from coincidental colocalizations of species-specific DA peaks and DE genes.
Among the concordant human up-DA GREs, ∼50 to 60% were specific for Glu or MGE-GABA, whereas the remaining group displayed a strong H3K27ac signal in both neuronal subtypes (Fig. 3E). Separation of GREs into promoters and enhancers showed that the majority of genes with concordant evolutionary changes in expression and promoter signal also had an evolutionary change in at least one enhancer (70% in Glu and 64% in MGE-GABA) (SI Appendix, Fig. S3J). Finally, in all three species, concordant genes often associated with multiple concordant enhancers (23 to 45% of human- or chimpanzee-specific and ∼60% of rhesus macaque-specific concordant up- or down-regulated DE genes in Glu or MGE-GABA) (SI Appendix, Fig. S3K).
Whereas changes in promoter signals were associated with larger changes in gene expression compared with changes in enhancers (Fig. 3E and SI Appendix, Fig. S3L), the neuronal regulatory landscapes often encompass more than one enhancer per gene (6, 44). Therefore, we asked if additional quantitative features that represent aggregate measures of an entire gene regulatory domain (as defined by GREAT with the basal plus extension setting) (Methods), such as the number of DA regulatory elements per gene (45) or the intensity of an enhancer signal (maximum and sum), could provide independent validation of the functional evolutionary change in a concordant DA enhancer-gene pair. We found that, in both neuronal subtypes, each of these regulatory features significantly correlated with gene expression (SI Appendix, Fig. S3 M and N) and that evolutionary changes in these features were significantly correlated with evolutionary changes in gene expression (Spearman correlation, all Ps < 2.2e-16) (SI Appendix, Fig. S3 O and P). Finally, we confirmed that the majority of the concordant enhancer-gene pairs represent genes with an evolutionary change not only in individual enhancer but also in the entire regulatory domain (Fig. 3F). These gene sets, with their associated epigenetic features, are compiled in Dataset S19, which provides a resource to enable future hypothesis-driven functional and evolutionary studies on primate brains.
Two representative examples of human-specific concordant GRE-gene pairs are CAT and SLC17A8 loci (Fig. 3 G–J). CAT encodes the hydrogen peroxide-degrading enzyme catalase. CAT is more strongly expressed in Glu vs. MGE-GABA neurons and is preferentially expressed in hominids (i.e., humans and chimpanzees) vs. rhesus macaque, with a significantly higher expression in human vs. chimpanzee. In addition to the strong human-specific H3K27ac enrichment in the promoter, we also detected an enrichment in several Glu-specific CAT enhancers in human or chimpanzee vs. rhesus macaque. Accumulation of H2O2 in oxidative metabolism has been linked to oxidative-stress–associated neurodegenerative disorders (46). Elevated expression of CAT in human neurons could, therefore, reflect the evolution of protective mechanisms that limit oxidative damage resulting from the higher metabolic activity of the human brain compared to that of chimpanzee (47–49). SLC17A8 encodes vesicular glutamate transporter 3 and is selectively up-regulated in human MGE-GABA neurons, with corresponding gains in human-specific enhancers. SLC17A8 is implicated in cocaine abuse (22) as well as in anxiety-related behaviors (50).
In line with earlier observations based on gene expression and DNase I hypersensitivity data (51, 52), GO analysis of human-specific concordant genes (enriched or depleted) resulted in no enrichment for groups of genes with similar biological features. The time span for evolutionary divergence between human and chimpanzee is likely too short for groups of functionally related genes to have coevolved (12, 52).
Evolutionary Changes in Regulation and Expression of Genes Associated with Language Ability.
One of the most striking differences between humans and other primate species is language ability. We examined 10 human genes (ATP2C2, CMIP, CNTNAP2, DCDC2, DYX1C1, FOXP2, KIAA0319, NFXL1, ROBO1, ROBO2) that had been associated with language impairment or developmental dyslexia (23, 53). Among them, ATP2C2, which is linked to language impairment (54), is strongly expressed in Glu neurons specifically in human (five- or eightfold change vs. chimpanzee or rhesus macaque, adjusted P < 1.2e-5), which was concordant with changes in multiple regulatory features (Dataset S19). In addition, DCDC2, which is implicated in reading proficiency (55), showed concordant up-regulation of expression and GRE signals in hominids compared with rhesus macaque, specifically in MGE-GABA neurons.
Whereas the other eight genes associated with language impairment did not show any concordant evolutionary changes in their regulation and expression, we detected many human-specific (and often cell type-specific) DA GREs that were linked to several of these genes (Dataset S20). Human-specific enhancers were found in CNTNAP2 (n = 8 in Glu, n = 6 in MGE-GABA) and FOXP2 (n = 1 in Glu) loci (Fig. 4A and SI Appendix, Fig. S4 A and B). These two genes have been suggested to be required for the proper development of speech and language in humans (24–26). The functional importance of these regulatory changes remains unclear, as we did not observe any significant human-specific changes in gene expression for FOXP2 or CNTNAP2. Nevertheless, it appears plausible that the increase in regulatory complexity engenders context-dependent mechanisms of regulation of FOXP2 or CNTNAP2 expression in anatomical (including brain areas and subpopulations of Glu or MGE-GABA neurons), environmental, or developmental contexts, which could, in turn, facilitate the emergence of language skills. Notably, cortical areas other than the DLPFC, such as the inferior frontal cortex and temporal cortex, constitute the core of the brain language network (56, 57). The evolutionary changes in regulation and expression of FOXP2 or CNTNAP2 in these cortical areas remain to be investigated.
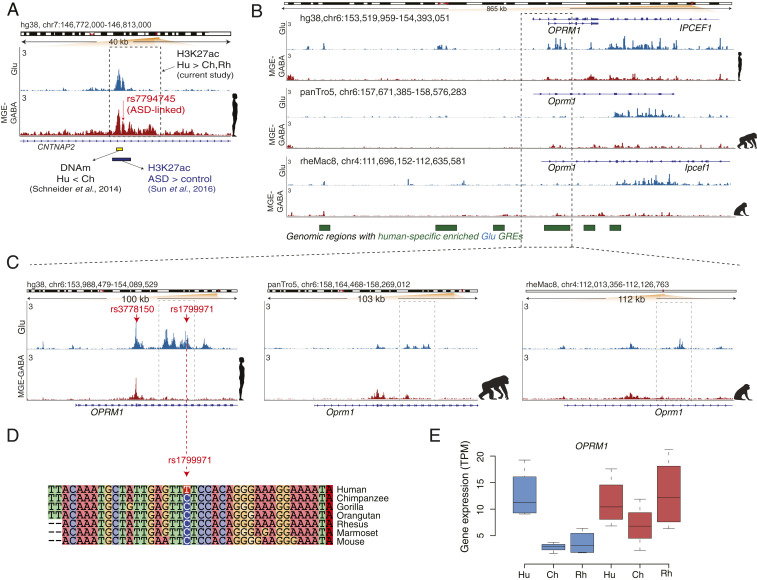
Fig. 4.
Human-specific up-regulated GREs harbor genes implicated in language, ASD, and opioid addiction. (A) Evolutionary regulatory (H3K27ac and DNA methylation) and ASD-associated changes at an enhancer within the CNTNAP2 locus in humans (also see SI Appendix, Fig. S4B). The enhancer (shown in a dashed box) is located within the second intron of CNTNAP2 and shows human-specific up-regulation of the H3K27ac signal in MGE-GABA neurons. Shown are: ASD-associated SNP rs7794745 (red arrow) (59), the region with the largest decrease in DNA methylation (DNAm) in Hu vs. Ch (green bar) (53), and the position of an H3K27ac peak (brown bar) that is up-regulated in ASD vs. control subjects (55). (B) The OPRM1 locus shows a high density of Glu human-specific up-DA GREs (the regions marked as green boxes depict areas with one or several Hu up-DA GREs). The leftmost region exemplifies an evolutionary respecification change from a Rh-enriched enhancer in MGE-GABA to a Hu-enriched enhancer in Glu. (C) H3K27ac profiles within the OPRM1 locus in three species and two neuronal subtypes. Evolutionary regulatory changes were found within the 5′ region, including human up-regulated promoter and enhancer GREs. The opioid abuse-associated SNPs rs3778150 and rs1799971 (red arrows) overlap with a human-specific up-DA promoter and human-specific up-DA enhancer, respectively. (D) Genomic alignment of the 40-bp region centered on the rs1799971 SNP in humans. The human-specific nucleotide substitution at the SNP position (C → T) is highlighted. Notice a high level of sequence conservation in the immediate vicinity of the SNP. The C nucleotide in humans represents the minor allele, which has been associated with opioid abuse (65). (E) Evolutionary changes in gene expression of OPRM1 in Glu neurons. Gene-expression changes were concordant with the regulatory changes, suggesting human-specific and Glu-specific up-regulation of OPRM1.
A recent DNA methylation study in the PFC of human and chimpanzee identified four regions within the CNTNAP2 locus that were differentially methylated between the two hominids (58). Remarkably, the region with the largest decrease in DNA methylation in human vs. chimpanzee (region B in that study) overlapped with one of the MGE-GABA– and human-specific up-regulated enhancers detected in our study (Fig. 4A). In addition, an ASD-linked SNP, rs7794745 (59), which is located ∼280 bp from differentially methylated region B, colocalizes with the same CNTNAP2 enhancer (Fig. 4A). The major allele nucleotide of rs7794745 (A > T) is a human-specific substitution compared to other primate species (SI Appendix, Fig. S4C). This enhancer was also identified and found to be up-regulated in ASD in a recent H3K27ac study in bulk brain tissues, including the PFC (Fig. 4A) (60). These findings link a cell-type–specific regulatory element that underwent an evolutionary change in humans to a risk locus associated with a neuropsychiatric disorder.
Human-Specific Up-Regulated GREs Form Neuron Subtype-Dependent Clusters Located near Genes Associated with Neuronal Function and Neuropsychiatric Disorders.
We found contiguous genomic regions with a high density of human-specific up-regulated GREs, which suggested a nonuniform distribution of these GREs across the genome. This trend is exemplified by an ∼1-Mb-long genomic region upstream of CDH8 on chromosome 16 that contains a cluster of 11 human-specific up-DA GREs in Glu neurons (SI Appendix, Fig. S4D). Applying stringent statistical analysis (Methods), we identified a significant enrichment of human up-DA GREs over the background distribution of all human GREs in 23 clusters, with 18 and 5 nonoverlapping clusters detected in Glu and MGE-GABA neurons, respectively (SI Appendix, Fig. S4E). Four Glu clusters contained a human-specific DE gene (OPRM1, GULP1, NIT2, PKDCC). Several genes with important roles in the nervous system or neuropsychiatric disorders overlapped with the human up-DA GRE clusters. In particular, CDH8 encodes a neurite outgrowth-regulating membrane protein cadherin-8 and has been linked to ASD and learning disability (27). Similarly, ASTN2 is located within a Glu cluster on chromosome 9 and has been associated with ASD (29). Among the 76 genes located at Glu-neuronal DA clusters, 9 are associated with ASD according to the SFARI database (https://www.sfari.org/resource/sfari-gene/) (61), which is a significant enrichment (P = 0.02 by hypergeometric distribution). Notably, recent publications suggest that human-specific evolutionary changes in gene regulation and expression could be associated with risk for ASD (14, 62). A previously described class of human genomic regions that harbor signatures of human-specific evolutionary changes consists of human-accelerated regions (HARs) (63). Three of the 18 Glu clusters overlapped with HARs (HAR2, HAR22, HAR47; Fisher’s exact test, P = 0.005), and each of these 3 HARs overlapped with a human Glu enhancer identified in our study.
We were particularly interested in the cluster containing OPRM1, a gene that encodes a receptor of endogenous opioid peptides and is implicated in opioid addiction (Fig. 4 B–E) (20, 21). OPRM1 is expressed in the PFC, and μ-opioid receptor signaling in the PFC is implicated in altering executive control and motivational functions, which are important in drug addiction (64). The OPRM1 locus overlaps with one of the Glu clusters on chromosome 6 that contains 17 human- and Glu-specific up-DA enhancers, as well as a human- and Glu-specific up-DA promoter (Fig. 4B). Concordant with these regulatory changes, OPRM1 expression is up-regulated in humans compared with the two primate species in Glu but not in MGE-GABA neurons (Fig. 4E). We also found that two human-specific up-DA GREs within OPRM1 harbor risk SNPs associated with opioid addiction (rs1799971 and rs3778150), which are situated within the OPRM1 promoter and one of its enhancers, respectively (65, 66) (Fig. 4C). Strikingly, the major allele nucleotide of rs3778150 (T > C) is a strictly human-specific substitution within a region that otherwise shows high sequence conservation across mammals (Fig. 4D). The OPRM1 regulatory domain also provides an example of a respecification event, as it contains an enhancer that is active in Glu neurons in human but in MGE-GABA neurons in rhesus macaque (Fig. 4B). Thus, the OPRM1 locus shows strong evidence of extensive evolutionary changes of its regulatory landscape that are coupled with concordant changes in gene expression specific to Glu neurons. In addition to OPRM1, two other genes implicated in drug abuse, SLC17A8 (discussed above) (Fig. 3J) and PENK (SI Appendix, Fig. S4 F and G), showed concordant evolutionary changes in gene expression and regulation. In contrast to OPRM1, the changes in these two genes were specific to MGE-GABA neurons. Notably, among the eight genes that comprise the family of opioid ligands and receptors (OPRM1, OPRD1, OPRK1, OPRL1, POMC, PENK, PDYN, PNOC), we detected two concordant human-up-regulated DE genes (OPRM1 and PENK) (enrichment P = 0.01 by hypergeometric test). In summary, our findings suggest human-specific modification of cellular and molecular pathways implicated in drug addiction.
Discussion
Here, we demonstrate the value of analyzing isolated neuronal cell populations to increase the sensitivity and specificity of gene expression and GRE analysis compared with analyses of bulk cortical tissue. Our findings provide insight into the cell-type–specific regulatory landscape of the primate brain and its evolution. Our analyses show that neuron-subtype–specific regulatory elements preferentially changed during primate evolution as compared with elements that are active in multiple neuronal subtypes. This greater evolutionary plasticity of neuron-subtype–specific GRE suggests that an alteration of a regulatory element that is beneficial or neutral when it occurs in a single cell type could be detrimental when it involves multiple cell or tissue types (67). We also detected regulatory elements that, despite being active in both Glu and MGE-GABA neurons of one species, were active in a specific cell type in other species. Thus, the assumption that a GRE that is active in multiple cell types (or tissues) represents a functionally conserved regulatory element is likely to be overly simplistic. Rather, these enhancer regions might employ different sets of transcription factor binding sites in different cell types and species, activating distinct regulatory programs.
Supporting these observations, we identified numerous regulatory elements whose activity showed neuron subtype-specific evolutionary changes in humans (5,259 in Glu and 2,415 in MGE-GABA). We also detected 165 genes in Glu and 80 genes in MGE-GABA in humans with strong evidence of concordant evolutionary changes in their expression and the activity of at least one GRE. For the majority of these genes, the evolutionary change was detected across the entire regulatory domain (quantified as the number of evolved enhancers per gene and the magnitude of the enhancer H3K27ac signal). These findings demonstrate the functional importance of regulatory evolution in different neuronal subtypes. For example, enrichment of H3K27ac in the promoter and enhancers of the catalase gene (CAT) in human Glu neurons is associated with a concordant human-specific up-regulation of gene expression, which could protect these neurons against oxidative stress caused by the high metabolic activity of the human brain (47–49). The limitation of our study is that genes were linked to their putative enhancers using the distance-based assignment, which is complicated by the mostly unknown high-order chromatin structure. The latter can be assessed by genome-wide chromosome conformation capture (e.g., Hi-C) (68, 69). However, currently, Hi-C data are not available for Glu or MGE-GABA subtypes, precluding more accurate linking between promoters and their distal regulatory elements.
Complex spoken language is unique for humans, and the development of language was essential to human evolution, through enabling efficient exchange of information, higher-order social organization as well as specializations of cognition and symbolic thinking. Our study emphasizes the importance of regulatory evolution in the development of language abilities and in the emergence of disorders associated with language. We identified two language-associated genes with strong human-specific (ATP2C2) or hominid-specific (DCDC2) as well as neuron subtype-dependent concordant changes in expression and regulatory landscapes. We also discovered human-specific regulatory changes in Glu and GABA neurons in the FOXP2 and CNTNAP2 genes that are crucial for brain development, neural plasticity, and language abilities (2, 25). FOXP2 encodes Forkhead box protein P2 (FoxP2), a transcription factor that is expressed at high levels in the brain during fetal development (70). Many FoxP2 targets (including CNTNAP2) are implicated in schizophrenia and ASD (71, 72). ASD is characterized by difficulties in social communication (including language), and previous analyses have uncovered a link between ASD and HARs, suggesting that certain aspects of ASD evolved specifically in humans (62). Notably, here we found that human-specific regulatory elements in neurons are not uniformly distributed in the human genome, and that clusters of these GREs in Glu neurons are enriched for HARs and genes associated with ASD.
Unexpectedly, we also identified concordant evolutionary changes in GREs and expression for several genes that have been implicated in drug addiction, with OPRM1 showing an extensive reorganization of its regulatory domain and a pronounced up-regulation of expression, specifically, in human Glu neurons compared with chimpanzee and rhesus macaque. Because OPRM1 is one of the strongest candidates for affecting risk for opioid-use disorder (65, 66, 73), our findings suggest that vulnerability to opioid addiction might have a unique human component. However, to the best of our knowledge, there is currently no support for the involvement of regulatory evolution in propensity to opioid addiction.
The findings presented in this work highlight the importance of differential regulatory changes in major neuronal subtypes in brain evolution and brain disorders. These results call for further analyses that will connect evolutionary changes in regulation with those in gene expression in multiple subpopulations of neuronal and glial cells, in different brain areas, and across developmental trajectories, perhaps using single-cell based approaches currently under development (16). These future studies will help to uncover the complex regulatory interaction networks that underlie the evolution of human brain function and human-specific traits, and hence will further advance the understanding of neuropsychiatric disorders.
Methods
Specimens.
Human brain specimens (DLPFC tissue samples from four clinically unremarkable male subjects) were described in Kozlenkov et al. (19) (Dataset S1). Tissue samples were dissected from the lateral part of Brodmann area 9 (BA9). The rhesus macaque brain tissues were obtained from the Texas Biomedical Research Institute and California National Primate Research Center from their established biospecimen distribution programs (Dataset S1). The chimpanzee brains were obtained from the National Chimpanzee Brain Resource. Brains of both rhesus macaques and chimpanzees were collected and snap frozen at the time of necropsy within 5 h or less postmortem. The DLPFC was sampled from frozen brains in a region corresponding to BA9 as described in macaques and chimpanzees (74).
Glu and MGE-GABA Nuclei Isolation by FANS.
The isolation of nuclei was performed by FANS as described in Kozlenkov et al. (19). In short, to distinguish between the two neuronal populations, we employed antibodies against RNA-Binding Protein RBFOX3 (also known as NeuN; mouse anti-NeuN, Alexa488-conjugated, MAB377X, Millipore) that is expressed in all neuronal nuclei, and antibodies against SOX6 (guinea pig anti-SOX6) (75). SOX6 is a transcription factor that regulates the ontogeny of the MGE-derived GABA neurons and is robustly expressed in these cells in the adult human PFC. This experimental approach has been previously validated using immunocytochemistry, immunohistochemistry, and RNA-seq. (17). As we previously reported (17), in addition to glutamatergic neurons, the FANS-isolated Glu population contained a small fraction of non–MGE-GABA neurons (∼8% of the all sorted Glu neurons) (17). We used ∼800 mg and 150 mg of primate tissue to isolate nuclei for ChIP-seq and RNA-seq, respectively.
ChIP-Seq.
H3K27ac ChIP was performed for each subject and neuronal subtype as described in refs. 18 and 19, using 100,000 to 150,000 FANS-separated nuclei and anti-H3K27ac antibody (rabbit polyclonal, Active Motif cat# 39133). We employed the “native” ChIP protocol that uses enzymatic chromatin fragmentation with micrococcal nuclease (MNase) without cross-linking proteins to DNA (18, 76). This approach yields ChIP-seq data with a high signal-to-noise ratio and is, therefore, advantageous for profiling various histone modifications. ChIP-seq libraries were constructed using the NEBNext Ultra DNA Library Prep kit (New England Biolabs) as described in Kozlenkov et al. (19). For each sample, both ChIP and input (MNase-digested chromatin) libraries were generated and sequenced. Sequencing was performed on an Illumina HiSeq 2500 instrument, using a paired-end 50 (PE50) protocol to an average of ∼40 million read pairs per sample.
RNA-Seq.
RNA was isolated from each subject and neuronal subtype as previously described (19), using 40,000 FANS-separated nuclei. To preserve RNA integrity, the RNase inhibitor (Clontech) was added during each step of the nuclear preparation. Nuclei were sorted directly into tubes containing 3:1 by volume of the Extraction Buffer from PicoPure RNA Isolation kit (ThermoFisher Scientific). RNA was then extracted from the sorted nuclei using the PicoPure kit, and the RNA-seq libraries were constructed using SMARTer Stranded Total Pico-Input RNA-seq kit (Clontech) and 10 ng RNA from each sample, as previously described (19). Libraries were sequenced on HiSeq 2500, using PE50 protocol to an average of ∼50 million read pairs per sample.
Supplementary Material
Acknowledgments
We gratefully acknowledge the help of Cheryl Stimpson for technical assistance with chimpanzee brain dissections and the help of Dr. Rob Patro with the use of Salmon software package. This work was supported by PsychEncode consortium NIH/National Institute of Mental Health MH103877 and MH122590 (to S.D.); NIH/National Institute on Drug Abuse DA043247 (to S.D.); VA Merit Awards BX001829 and BX002876 (to S.D.); Intramural funds of the US Department of Health and Human Services to National Library of Medicine (E.V.K.); The Dutch Royal Academy of Sciences (M.P.C.); the Parkinson’s Foundation (Stichting ParkinsonFonds, M.P.C.); James S. McDonnell Foundation 220020293 (to C.C.S.); and National Science Foundation INSPIRE SMA-1542848 (to C.C.S.). The chimpanzee brains were obtained from the National Chimpanzee Brain Resource (supported by NIH/National Institute of Neurological Disorders and Stroke NS092988).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2011884117/-/DCSupplemental.
Data Availability.
The data reported in this paper have been deposited in the Gene Expression Omnibus database (accession no. GSE158934). The data for human samples are from Kozlenkov et al. (19), and are available in the PsychENCODE Knowledge Portal at Synapse (https://doi.org/10.7303/syn12034263). All other study data and methods are included in SI Appendix and Datasets S1–S21.
References
- 1.Bonelli R. M., Cummings J. L., Frontal-subcortical circuitry and behavior. Dialogues Clin. Neurosci. 9, 141–151 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sousa A. M. M., Meyer K. A., Santpere G., Gulden F. O., Sestan N., Evolution of the human nervous system function, structure, and development. Cell 170, 226–247 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma T. et al., Subcortical origins of human and monkey neocortical interneurons. Nat. Neurosci. 16, 1588–1597 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Hendry S. H., Schwark H. D., Jones E. G., Yan J., Numbers and proportions of GABA-immunoreactive neurons in different areas of monkey cerebral cortex. J. Neurosci. 7, 1503–1519 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torres-Gomez S. et al., Changes in the proportion of inhibitory interneuron types from sensory to executive areas of the primate neocortex: Implications for the origins of working memory representations. Cereb. Cortex 30, 4544–4562 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long H. K., Prescott S. L., Wysocka J., Ever-changing landscapes: Transcriptional enhancers in development and evolution. Cell 167, 1170–1187 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ren B., Yue F., Transcriptional enhancers: Bridging the genome and phenome. Cold Spring Harb. Symp. Quant. Biol. 80, 17–26 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Carroll S. B., Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell 134, 25–36 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Wray G. A., The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 8, 206–216 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Cotney J. et al., The evolution of lineage-specific regulatory activities in the human embryonic limb. Cell 154, 185–196 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Villar D. et al., Enhancer evolution across 20 mammalian species. Cell 160, 554–566 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vermunt M. W. et al.; Netherlands Brain Bank , Epigenomic annotation of gene regulatory alterations during evolution of the primate brain. Nat. Neurosci. 19, 494–503 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Reilly S. K. et al., Evolutionary genomics. Evolutionary changes in promoter and enhancer activity during human corticogenesis. Science 347, 1155–1159 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castelijns B. et al., Hominin-specific regulatory elements selectively emerged in oligodendrocytes and are disrupted in autism patients. Nat. Commun. 11, 301 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rotem A. et al., Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat. Biotechnol. 33, 1165–1172 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grosselin K. et al., High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer. Nat. Genet. 51, 1060–1066 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Kozlenkov A. et al., Substantial DNA methylation differences between two major neuronal subtypes in human brain. Nucleic Acids Res. 44, 2593–2612 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brind’Amour J. et al., An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations. Nat. Commun. 6, 6033 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Kozlenkov A. et al., A unique role for DNA (hydroxy)methylation in epigenetic regulation of human inhibitory neurons. Sci. Adv. 4, eaau6190 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hurd Y. L., O’Brien C. P., Molecular genetics and new medication strategies for opioid addiction. Am. J. Psychiatry 175, 935–942 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berrettini W., A brief review of the genetics and pharmacogenetics of opioid use disorders. Dialogues Clin. Neurosci. 19, 229–236 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakae D. Y. et al., The absence of VGLUT3 predisposes to cocaine abuse by increasing dopamine and glutamate signaling in the nucleus accumbens. Mol. Psychiatry 20, 1448–1459 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Mozzi A. et al., The evolutionary history of genes involved in spoken and written language: Beyond FOXP2. Sci. Rep. 6, 22157 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Enard W. et al., Molecular evolution of FOXP2, a gene involved in speech and language. Nature 418, 869–872 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Konopka G., Roberts T. F., Insights into the neural and genetic basis of vocal communication. Cell 164, 1269–1276 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graham S. A., Fisher S. E., Decoding the genetics of speech and language. Curr. Opin. Neurobiol. 23, 43–51 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Pagnamenta A. T. et al., Rare familial 16q21 microdeletions under a linkage peak implicate cadherin 8 (CDH8) in susceptibility to autism and learning disability. J. Med. Genet. 48, 48–54 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shangguan Y. et al., CNTNAP4 impacts epilepsy through GABAA receptors regulation: Evidence from temporal lobe epilepsy patients and mouse models. Cereb. Cortex 28, 3491–3504 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Glessner J. T. et al., Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 459, 569–573 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Magueresse C., Monyer H., GABAergic interneurons shape the functional maturation of the cortex. Neuron 77, 388–405 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Kepecs A., Fishell G., Interneuron cell types are fit to function. Nature 505, 318–326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Creyghton M. P. et al., Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 107, 21931–21936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nord A. S. et al., Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell 155, 1521–1531 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McLean C. Y. et al., GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vermunt M. W. et al.; Netherlands Brain Bank , Large-scale identification of coregulated enhancer networks in the adult human brain. Cell Rep. 9, 767–779 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Lake B. B. et al., Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science 352, 1586–1590 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vierstra J. et al., Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science 346, 1007–1012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Necsulea A. et al., The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505, 635–640 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Berto S. et al., Accelerated evolution of oligodendrocytes in the human brain. Proc. Natl. Acad. Sci. U.S.A. 116, 24334–24342 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu Y. et al., Spatiotemporal transcriptomic divergence across human and macaque brain development. Science 362, eaat8077 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuleshov M. V. et al., Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chenn A., Walsh C. A., Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science 297, 365–369 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Boyd J. L. et al., Human-chimpanzee differences in a FZD8 enhancer alter cell-cycle dynamics in the developing neocortex. Curr. Biol. 25, 772–779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Torbey P. et al., Cooperation, cis-interactions, versatility and evolutionary plasticity of multiple cis-acting elements underlie krox20 hindbrain regulation. PLoS Genet. 14, e1007581 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berthelot C., Villar D., Horvath J. E., Odom D. T., Flicek P., Complexity and conservation of regulatory landscapes underlie evolutionary resilience of mammalian gene expression. Nat. Ecol. Evol. 2, 152–163 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andersen J. K., Oxidative stress in neurodegeneration: Cause or consequence? Nat. Med. 10 (suppl.), S18–S25 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Bauernfeind A. L. et al., High spatial resolution proteomic comparison of the brain in humans and chimpanzees. J. Comp. Neurol. 523, 2043–2061 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bozek K. et al., Exceptional evolutionary divergence of human muscle and brain metabolomes parallels human cognitive and physical uniqueness. PLoS Biol. 12, e1001871 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Preuss T. M., The human brain: Rewired and running hot. Ann. N. Y. Acad. Sci. 1225 (suppl. 1), E182–E191 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakae D. Y. et al., Differential expression of VGLUT3 in laboratory mouse strains: Impact on drug-induced hyperlocomotion and anxiety-related behaviors. Genes Brain Behav. 18, e12528 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Shibata Y. et al., Extensive evolutionary changes in regulatory element activity during human origins are associated with altered gene expression and positive selection. PLoS Genet. 8, e1002789 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Somel M., Liu X., Khaitovich P., Human brain evolution: Transcripts, metabolites and their regulators. Nat. Rev. Neurosci. 14, 112–127 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Peterson R. L., Pennington B. F., Developmental dyslexia. Lancet 379, 1997–2007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Newbury D. F. et al., CMIP and ATP2C2 modulate phonological short-term memory in language impairment. Am. J. Hum. Genet. 85, 264–272 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scerri T. S. et al., DCDC2, KIAA0319 and CMIP are associated with reading-related traits. Biol. Psychiatry 70, 237–245 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Michon M., López V., Aboitiz F., Origin and evolution of human speech: Emergence from a trimodal auditory, visual and vocal network. Prog. Brain Res. 250, 345–371 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Friederici A. D., Hierarchy processing in human neurobiology: How specific is it? Philos. Trans. R. Soc. B 375, 20180391 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schneider E. et al., Widespread differences in cortex DNA methylation of the “language gene” CNTNAP2 between humans and chimpanzees. Epigenetics 9, 533–545 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arking D. E. et al., A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am. J. Hum. Genet. 82, 160–164 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun W. et al., Histone acetylome-wide association study of autism spectrum disorder. Cell 167, 1385–1397.e11 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Abrahams B. S. et al., SFARI Gene 2.0: A community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol. Autism 4, 36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Doan R. N. et al., Mutations in human accelerated regions disrupt cognition and social behavior. Cell 167, 341–354.e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pollard K. S. et al., An RNA gene expressed during cortical development evolved rapidly in humans. Nature 443, 167–172 (2006). [DOI] [PubMed] [Google Scholar]
- 64.van Steenbergen H., Eikemo M., Leknes S., The role of the opioid system in decision making and cognitive control: A review. Cogn. Affect. Behav. Neurosci. 19, 435–458 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hancock D. B. et al., Cis-expression quantitative trait loci mapping reveals replicable associations with heroin addiction in OPRM1. Biol. Psychiatry 78, 474–484 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou H. et al.; Veterans Affairs Million Veteran Program , Association of OPRM1 Functional Coding Variant With Opioid Use Disorder: A Genome-Wide Association Study. JAMA Psychiatry, e201206, 10.1001/jamapsychiatry.2020.1206 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wittkopp P. J., Kalay G., Cis-regulatory elements: Molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 13, 59–69 (2011). [DOI] [PubMed] [Google Scholar]
- 68.Won H. et al., Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 538, 523–527 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Won H., Huang J., Opland C. K., Hartl C. L., Geschwind D. H., Human evolved regulatory elements modulate genes involved in cortical expansion and neurodevelopmental disease susceptibility. Nat. Commun. 10, 2396 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lai C. S., Fisher S. E., Hurst J. A., Vargha-Khadem F., Monaco A. P., A forkhead-domain gene is mutated in a severe speech and language disorder. Nature 413, 519–523 (2001). [DOI] [PubMed] [Google Scholar]
- 71.Adam I., Mendoza E., Kobalz U., Wohlgemuth S., Scharff C., CNTNAP2 is a direct FoxP2 target in vitro and in vivo in zebra finches: Complex regulation by age and activity. Genes Brain Behav. 16, 635–642 (2017). [DOI] [PubMed] [Google Scholar]
- 72.Chen Y. C. et al., Foxp2 controls synaptic wiring of corticostriatal circuits and vocal communication by opposing Mef2c. Nat. Neurosci. 19, 1513–1522 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Egervari G., Kozlenkov A., Dracheva S., Hurd Y. L., Molecular windows into the human brain for psychiatric disorders. Mol. Psychiatry 24, 653–673 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Petrides M., Pandya D. N., Dorsolateral prefrontal cortex: Comparative cytoarchitectonic analysis in the human and the macaque brain and corticocortical connection patterns. Eur. J. Neurosci. 11, 1011–1036 (1999). [DOI] [PubMed] [Google Scholar]
- 75.Stolt C. C. et al., SoxD proteins influence multiple stages of oligodendrocyte development and modulate SoxE protein function. Dev. Cell 11, 697–709 (2006). [DOI] [PubMed] [Google Scholar]
- 76.Kundakovic M. et al., Practical guidelines for high-resolution epigenomic profiling of nucleosomal histones in postmortem human brain tissue. Biol. Psychiatry 81, 162–170 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this paper have been deposited in the Gene Expression Omnibus database (accession no. GSE158934). The data for human samples are from Kozlenkov et al. (19), and are available in the PsychENCODE Knowledge Portal at Synapse (https://doi.org/10.7303/syn12034263). All other study data and methods are included in SI Appendix and Datasets S1–S21.