Significance
We demonstrate that combining optical superresolution imaging with deep learning classification methods increases the speed and accuracy of assessing the biological affinities of fossil pollen taxa. We show that it is possible to taxonomically separate pollen grains that appear morphologically similar under standard light microscopy based on nanoscale variation in pollen shape, texture, and wall structure. Using a single pollen morphospecies, Striatopollis catatumbus, we show that nanoscale morphological variation within the fossil taxon coincides with paleobiogeographic distributions. This new approach improves the taxonomic resolution of fossil pollen identifications and greatly enhances the use of pollen data in ecological and evolutionary research.
Keywords: Airyscan microscopy, automated classification, Detarioideae, machine learning, palynology
Abstract
Taxonomic resolution is a major challenge in palynology, largely limiting the ecological and evolutionary interpretations possible with deep-time fossil pollen data. We present an approach for fossil pollen analysis that uses optical superresolution microscopy and machine learning to create a quantitative and higher throughput workflow for producing palynological identifications and hypotheses of biological affinity. We developed three convolutional neural network (CNN) classification models: maximum projection (MPM), multislice (MSM), and fused (FM). We trained the models on the pollen of 16 genera of the legume tribe Amherstieae, and then used these models to constrain the biological classifications of 48 fossil Striatopollis specimens from the Paleocene, Eocene, and Miocene of western Africa and northern South America. All models achieved average accuracies of 83 to 90% in the classification of the extant genera, and the majority of fossil identifications (86%) showed consensus among at least two of the three models. Our fossil identifications support the paleobiogeographic hypothesis that Amherstieae originated in Paleocene Africa and dispersed to South America during the Paleocene-Eocene Thermal Maximum (56 Ma). They also raise the possibility that at least three Amherstieae genera (Crudia, Berlinia, and Anthonotha) may have diverged earlier in the Cenozoic than predicted by molecular phylogenies.
Fossil pollen is an incomparable source of data on plant diversity history and the evolution of terrestrial ecosystems (1, 2). Few terrestrial paleontological records are as abundant, geographically widespread, or temporally continuous. However, the majority of pre-Quaternary palynomorphs (older than 2 million years) have no clear biological affinities, limiting ecological and evolutionary interpretations that can be derived from fossil pollen (3–5). Better assessment of fossil taxa taxonomy remains one of the most critical and elusive goals of palynology, requiring pollen morphology to be both efficiently visualized and effectively compared (4–7).
The quality and quantity of morphological features used to discriminate among pollen taxa can be substantially improved using high-resolution microscopy (5, 8–10). Electron microscopy (EM) methods like scanning electron microscopy (SEM, <5-nm resolution) and transmitted electron microscopy (TEM, 0.2-nm resolution) have traditionally been used to increase visual resolution and morphological detail (2, 10). However, EM requires destructive preparation methods that are often too time intensive to scale (9, 11, 12). An alternative high-resolution optical imaging technique—Airyscan confocal superresolution microscopy—produces images comparable to EM, but is nondestructive and uses material mounted on slides (9). Airyscan captures the three-dimensional (3D) morphology of pollen surfaces and internal structures, including features up to 140 nm, smaller than the diffraction limit of light (8, 9). Because Airyscan is an optical microscopy method, high-resolution images of both extant reference and fossil specimens can be amassed rapidly (8, 9).
However, discriminating between morphologically similar pollen taxa can be challenging even with the highest resolution images. Variation among species and genera can be subtle and not easily categorized. Quantitative morphological analyses are more reproducible than visual assessments, and can often distinguish between morphologically similar or closely related species (6, 7, 13, 14). Of these approaches, deep learning methods, such as convolutional neural networks (CNNs), hold the most potential to improve the efficiency and accuracy of fossil pollen identifications (15, 16). CNNs efficiently process and analyze visual data, extracting features from images hierarchically through layer-wise architecture. The features encode morphological characteristics, like shape and texture, which can be used for biological classification (15, 17).
Striatopollis catatumbus (18) is an example of a fossil pollen taxon for which we have limited taxonomic information. It has been associated with several genera within the monophyletic Amherstieae tribe of the Detarioideae subfamily of the legumes, including Crudia, Isoberlinia, Anthonotha, and Macrolobium (19–21). It is found in low abundances (<5% of counted grains per sample) throughout the Cenozoic and is a regular component of lowland tropical forest pollen assemblages (22) (Datasets S1 and S2). The broad geographic and temporal extent of S. catatumbus (SI Appendix, Fig. S1) suggests this fossil morphospecies corresponds to an entire clade rather than a single species. However, reliance on transmitted light microscopy has limited our ability to systematically compare and differentiate morphological variation within the morphospecies. At least 28 extant Amherstieae genera have similar pollen morphology when observed under standard light microscopy (23, 24).
We developed and trained three CNN models to differentiate among the morphological variability inherent in the extant genera of Amherstieae. We then used these models to hypothesize the likely biological affinity of individual S. catatumbus fossil specimens. Our models emulate the human analyst by transferring training from extant reference material to the classification of taphonomically altered fossil material. This is an application of deep learning in deep time. Our findings improve the taxonomic resolution of S. catatumbus identifications. They also provide information and support existing hypotheses about the paleobiogeographic history of the Amherstieae tribe. Our approach represents a significant step forward in automated fossil pollen identification and shows the potential for pollen to provide greater taxonomic insights into the evolutionary history of plants.
Results
Extant Classifications (Training-Validation Stage).
We produced images of 459 extant Amherstieae specimens (45 species from 16 genera with morphology similar to S. catatumbus under light microscopy) using Airyscan (25) (Dataset S3). Images were a series of axial focal planes through the depth of each pollen grain, generating a 3D representation of the external and internal structure (SI Appendix, Fig. S2).
These images were used to train three CNN models: maximum projection (MPM), multislice (MSM), and fused (FM) (see Materials and Methods for details). All three models employed a simple code composition approach for summarizing feature representations and a multiway softmax classifier for pollen grain identification (Fig. 1). We used the maximum softmax values to select the class label (the genus name) for a given pollen grain. In the training-validation stage, we used the results to calculate an average classification accuracy for each genus (a percentage from 0 to 100%) (Fig. 2 and SI Appendix, Figs. S3 and S4). In our fossil specimen analysis, we used the max-softmax score as a measure of the classification confidence (a probability ranging from 0.0 to 1.0) (SI Appendix, Table S1). We explain the confidence scores (cs) of neural network models and our justification for their use in our discussion.
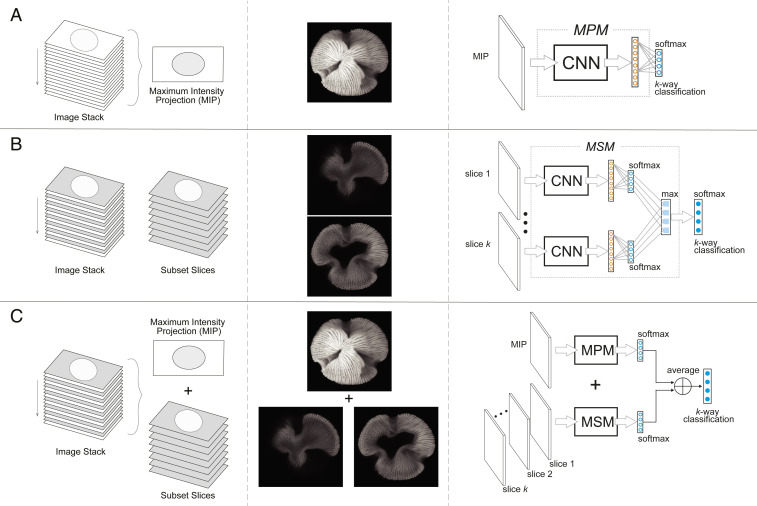
Fig. 1.
Workflows illustrating the three CNN models developed for the identification of fossil pollen: (A) maximum projection model; (B) multislice model; (C) fused model. The Left column shows a graphic representation of the image data used by the models. The image stack can contain between 60 and 120 images per specimen at 0.19-μm increments in the z-axis. The Central column shows examples of this image data (maximum intensity projections and image stack slices). The Right column illustrates the analysis pipeline of each model.
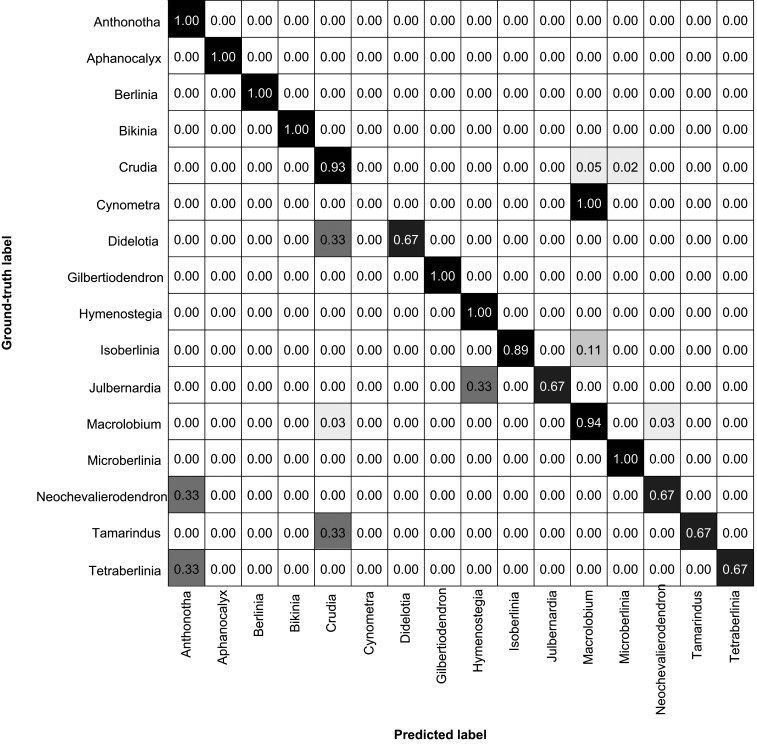
Fig. 2.
Confusion matrix of classification accuracy for the fused model (FM). Rows represent the true taxonomy of the pollen images, and columns represent the machine identifications. Average accuracy was 90.30%.
The first model, MPM, took as input the maximum intensity projection of the specimen image stack and carried out a multiway classification (Fig. 1A and SI Appendix, Fig. S2, see Materials and Methods). This model considered only the external morphology of the pollen grains (Fig. 1A) and achieved 83.59% classification accuracy (SI Appendix, Fig. S3). Five genera (Aphanocalyx, Bikinia, Microberlinia, Macrolobium, and Crudia) were classified at accuracies >90%. Gilbertiodendron, Neochevalierodendron, and Tetraberlinia had accuracies <50%.
The second model, MSM, used a subset of the Airyscan image stack that the model determined was the most diagnostic (Fig. 1B, see Materials and Methods). As a result, this model captured differences in the internal structure of the pollen walls (Fig. 1B). MSM performed better than MPM with 89.5% average accuracy (SI Appendix, Fig. S4). Eight genera (Aphanocalyx, Berlinia, Bikinia, Crudia, Gilbertiodendron, Hymenostegia, Macrolobium, and Microberlinia) were classified at accuracies >90%. Neochevalierodendron increased to 67% accuracy while Tetraberlinia had <50% accuracy.
The third model, FM, used as input both the MPM maximum intensity projection and the MSM image stack (Fig. 1C), allowing the model to work with the external and internal morphology of the pollen wall (Fig. 1C). FM had the best performance with 90.3% accuracy (Fig. 2). Ten genera (Anthonotha, Aphanocalyx, Berlinia, Bikinia, Crudia, Gilbertiodendron, Hymenostegia, Isoberlinia, Macrolobium, and Microberlinia) were identified with accuracies >89%. Didelotia, Julbernardia, Neochevalierodendron, and Tamarindus remained at 67% accuracy while Tetraberlinia identifications increased to 67% accuracy (Fig. 2).
All three models had difficulty in differentiating the polyphyletic Cynometra from the more species-rich South American genus, Macrolobium (Fig. 2 and SI Appendix, Figs. S3 and S4). The Cynometra species in our study, C. marginata, is South American (Dataset S3). After visual comparison, we concluded that the external and internal wall morphology of the two taxa are indistinguishable (SI Appendix, Figs. S5 and S6, see Morphological Discussion in SI Appendix). Another challenging genus, Neochevalierodendron, was misclassified as Crudia (33%, MPM, SI Appendix, Fig. S3), Isoberlinia (33%, MPM, SI Appendix, Fig. S3), Macrolobium (33%, MSM, SI Appendix, Fig. S4), and Anthonotha (33%, FM, Fig. 2). Neochevalierodendron has several features of the external and internal structure that resemble the other four genera, such as similarities in striation pattern and shape of the striae in cross-section (SI Appendix, Fig. S7, see Morphological Discussion in SI Appendix).
Fossil Classifications (Testing Stage).
We next used the three trained CNN models to classify 48 fossil S. catatumbus specimens from Paleocene, Eocene, and Miocene deposits of Africa and South America (25) (SI Appendix, Fig. S1 and Dataset S4). S. catatumbus is typically found in low abundances (Datasets S1 and S2) and those 48 specimens represent the majority of occurrences of the species in the 25 samples analyzed. Each model assigned the 48 specimens a classification label and a classification score (SI Appendix, Table S1). Most identifications (86.7%) were consistent among at least two models, and only six specimens had no consistent identifications among the three models (SI Appendix, Table S1). The majority of specimens without consistent identifications showed equal likelihood of classification between multiple genera in the FM model (SI Appendix, Table S2), suggesting possible intermediate morphologies or morphologies associated with genera not included in this study.
We found that the classification confidence scores (cs) for Macrolobium identifications were negatively correlated with age (SI Appendix, Fig. S8, see Classification Discussion in SI Appendix). For all other identifications, we did not find a significant relationship between age and classification scores. South American specimens identified as extant African genera were identified with lower classification scores than African fossils identified as extant African genera (SI Appendix, Fig. S8).
Comparison to Traditional Morphometric Analysis.
We manually measured morphological features from a small subset of extant specimens, which included 60 grains from 10 genera (Isoberlinia, Crudia, Anthonotha, Macrolobium, Berlinia, Julbernardia, Didelotia, Neochevalierodendron, Microberlinia, and Cynometra), all 48 fossil grains of S. catatumbus, and one fossil specimen of Ephedripites. This last specimen was included to test the ability of traditional morphometric and CNN analyses to identify outliers. We measured 14 morphological features from the Airyscan images (Dataset S5, see Materials and Methods for details). We then used nonmetric multidimensional scaling (NMDS) to compare the extant genera and fossil specimens (SI Appendix, Fig. S9 and Dataset S6) and to calculate a similarity index (sim).
The first NMDS axis identified the fossil specimen Ephedripites and the African genus Julbernardia as the most morphologically dissimilar specimens in the dataset. The second axis separated genera by geographic region. The sole South American genus, Macrolobium, was distinct from the pantropical and African genera (SI Appendix, Fig. S9). NMDS categorized 96% of the fossil specimens as similar to two or more extant genera (SI Appendix, Table S3). Specimens were identified as similar to as many as five different genera. Only two South American fossil specimens clustered with a single genus. These specimens were identified as Macrolobium by both NMDS (sim: 0.96, 0.92) and the FM (cs: 0.98, 1.00) (SI Appendix, Table S3). Thirty-two of the remaining fossil specimens were classified by the FM as one of the multiple genera identified as most similar in the NMDS (SI Appendix, Table S3). These included 19 African fossils: four Thanetian identified by the FM as Crudia, and 15 Ypresian identified as Crudia (9 specimens), Anthonotha (3), Berlinia (1), Isoberlinia (1), and Microberlinia (1) (SI Appendix, Table S3). These also include 14 South American specimens: six Paleocene-Eocene Thermal Maximum (PETM) identified as Crudia, one Burdigalian identified as Anthonotha, one Langhian identified as Macrolobium, three Serravallian specimens identified as Crudia (1) and Macrolobium (2), and two Tortonian fossils identified as Crudia (1) and Macrolobium (1) (SI Appendix, Table S3).
For the remaining 13 fossil specimens, the FM identifications disagreed with the NMDS results (SI Appendix, Table S3). None of the four fossils identified as Neochevalierodendron in the FM (cs: 0.493 to 0.501) clustered with the genus in the NMDS (SI Appendix, Table S3). Two fossil specimens, one African and one South American, identified by the FM as Julbernardia/Isoberlinia (cs: 0.496/0.335) and Anthonotha/Julbernardia (cs: 0.497/0.447), clustered with Didelotia and Crudia (sim: 0.85, 0.95) in the NMDS (SI Appendix, Table S3). Two African Ypresian specimens were identified in the FM as Berlinia/Isoberlinia (cs: 0.50/0.491) and Berlinia (cs: 0.998), but clustered with Didelotia and Crudia in the NMDS (sim: 0.90 to 0.94, 0.88 to 0.92). A third African Ypresian specimen was identified as Crudia/Anthonotha by the FM (cs: 0.53/45), but clustered with Macrolobium and Cynometra in the NMDS (sim: 0.86, 0.86) (SI Appendix, Table S3). Four Miocene South American specimens, identified as Berlinia (2 specimens, cs: 0.848, 0.609) and Crudia (2 specimens, cs: 0.841, 0.924) in the FM, clustered with Macrolobium (sim: 0.80 to 0.94) and Cynometra (sim: 0.82 to 0.94) in the NMDS (SI Appendix, Table S3).
Morphological Verification of CNN Results.
We confirmed each CNN FM identification using qualitative morphological comparisons.
Review of African fossils identifications.
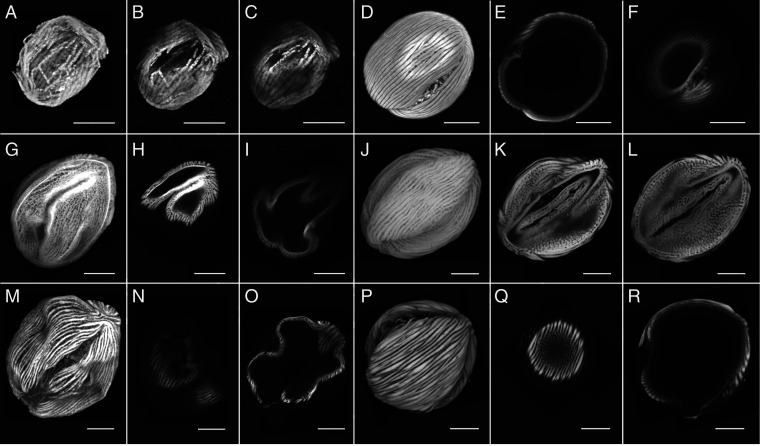
The pantropical genus Crudia is distinguished by mushroom-shaped striae over a reticulate tectum (SI Appendix, Fig. S6). The four Thanetian fossil specimens identified as Crudia (cs: 0.72 to 1.00) present striation similar to Crudia, and, in some, distinct reticulation under the striae (Fig. 3). Poor preservation made it difficult to visualize the internal structure of the pollen wall (Fig. 3). Of the 11 Ypresian specimens identified as Crudia, six had cs >0.56 with ornamentation similar to Crudia (Fig. 3 and SI Appendix, Fig. S6 and Table S3). In contrast, five Ypresian specimens identified as Crudia with cs <0.6 resembled multiple African genera (SI Appendix, Table S2). The tectum of two specimens identified as Crudia/Anthonotha (cs: 0.52/0.47, 0.53/0.45) are reticulate like Crudia, but the striae of one is more similar to Anthonotha (SI Appendix, Table S3). Two specimens identified as Crudia/Didelotia (cs: 0.52/0.45, 0.51/0.34) have Crudia-like striation and tectum reticulation. One is poorly preserved and there are no obvious similarities to Didelotia (SI Appendix, Table S3). The final specimen, identified as Crudia/Isoberlinia (cs: 0.51/0.34), is also not preserved well enough for morphological verification (SI Appendix, Table S3).
Fig. 3.
African fossil specimens of S. catatumbus compared with modern specimens. Fossils: Thanetian: 027.Striatopollis AF (A–C); Ypresian: 016.Striatopollis AF (G–I), 06.Striatopollis AF (M–O). Modern specimens: Crudia (D–F), Berlinia (J–L), and Anthonotha (P–R). (Scale bars: 10 µm.)
We confirmed the identifications of four Ypresian specimens, three identified as Anthonotha (cs: 0.59 to 0.99) and one identified as Berlinia (cs: 0.99) (SI Appendix, Table S1), based on striation, striae shape, and pollen wall arrangement (Fig. 3 and SI Appendix, Table S3). Three additional specimens identified as Berlinia/Isoberlinia (cs: 0.5/0.49, 0.52/0.47) and Berlinia/Cynometra (cs: 0.55/0.45) (SI Appendix, Table S1) have striation similar to Berlinia, but the pollen wall structure appears more similar to other Amherstieae genera, including Isoberlinia (SI Appendix, Table S3).
We also reviewed the morphology of the fossil African specimens identified as Neochevalierodendron/Berlinia and Neochevalierodendron/Cynometra (SI Appendix, Fig. S10 and Table S3). The Thanetian fossil identified as Neochevalierodendron/Berlinia (cs: 0.5/0.49) has striation similar to both genera (SI Appendix, Fig. S10). However, the fossil tectum is more similar to Neochevalierodendron (SI Appendix, Fig. S10). The two Ypresian fossils identified as Neochevalierodendron/Cynometra (cs: 0.5/0.45, 0.49/0.29) also have striation similar to both genera (SI Appendix, Fig. S10). The tectum in both fossils appears more similar to Neochevalierodendron, but the internal shape of the fossils’ striae appears baculate to gemmate, which is different from either Neochevalierodendron or Cynometra (SI Appendix, Figs. S6, S10, see SI Appendix, Discussion).
One specimen identified as Julbernardia/Isoberlinia (cs: 0.5/0.34) has a striate supratectum and tectum ornamentation similar to Crudia (SI Appendix, Fig. S11 and Table S3). The fossil’s striae are mushroom shaped, while those in Isoberlinia are clavate, and in Julbernardia more verruca shaped (SI Appendix, Fig. S11). This fossil was flattened and broken, limiting the extraction of more diagnostic characters.
Review of South American fossils identifications.
All six South American specimens identified as Macrolobium (cs: 0.5 to 1.00) are similar to extant species of this genus (SI Appendix, Fig. S12 and Table S3). Both extant species and fossils are distinguished by thick, rectangular striae and a perforate to psilate exine (SI Appendix, Figs. S6 and S12). In contrast, the morphology of the 15 South American fossil specimens with pantropical and African affinities was more diverse (SI Appendix, Fig. S13).
Seven fossil specimens, five from the PETM and two from the Serravallian, were identified as Crudia (cs: 0.49 to 0.99). The PETM specimens have striation and wall ornamentation similar to Crudia (SI Appendix, Fig. S12). One Serravallian specimen has striate ornamentation resembling Crudia (SI Appendix, Fig. S12). The other has striation similar to Macrolobium (SI Appendix, Table S3), with a perforate-fossulate tectum, similar to some species of Crudia (SI Appendix, Fig. S12). Two additional PETM specimens were identified as Crudia/Berlinia (cs: 0.52/0.39) and Crudia/Cynometra (cs: 0.46/0.32) (SI Appendix, Table S3). The striation of the first specimen is similar to Berlinia, but the tectum is reticulate as in Crudia. The internal ornamentation is not visible (SI Appendix, Table S3). The striation and tectum of the second specimen resemble Crudia, but other morphological characters were not visible because of poor preservation (SI Appendix, Table S3). The Tortonian specimen identified as Crudia/Anthonotha (cs: 0.49/0.48) has a striation pattern similar to Crudia. However, the striae varies from clavate to mushroom shaped, a characteristic observed in both Crudia and Anthonotha (SI Appendix, Table S3).
The Burdigalian specimen identified as Anthonotha in the FM (cs: 0.67) and as Crudia (sim: 0.90) in the NMDS analysis (SI Appendix, Table S3) has a reticulate exine with striae above the tectum, like herbarium specimens in both Anthonotha and Crudia (SI Appendix, Fig. S13). The striae are thicker in Anthonotha than in the fossil or Crudia. The striae in both Anthonotha and the fossil specimen appear clavate. However, the structure of the fossil’s supratectum is more similar to Crudia (SI Appendix, Table S3). The South American PETM specimen identified as Anthonotha/Julbernardia (cs: 0.5/0.45) has thick striation similar to Anthonotha (SI Appendix, Fig. S13). The exine in the fossil appears reticulate, as in Julbernardia, but this reticulation is caused by pollen wall degradation (SI Appendix, Fig. S11). The internal ornamentation of this fossil is not well preserved (SI Appendix, Fig. S11).
The two South American fossils identified as Berlinia (cs: 0.85 to 0.61) have similar external and internal structure to Macrolobium (SI Appendix, Fig. S13 and Table S3). Striae are rectangular, and the tectum is perforate. In the Serravallian fossil (cs: 0.61) the striae are not continuous from pole to pole (SI Appendix, Figs. S6 and S13). This is different to the older Burdigalian fossil, where the striae appear continuous (SI Appendix, Fig. S13). The internal columellae are visible in all specimens, but are more distinctive in the Burdigalian specimen (SI Appendix, Fig. S14). The Burdigalian specimen identified as Neochevalierodendron/Berlinia (cs: 0.49/0.47), have rectangular striae as Macrolobium (SI Appendix, Table S3 and Fig. S13).
Discussion
Our study demonstrates that the morphological diversity of fossil pollen is far richer than previously recognized and that greater taxonomic resolution is possible through the combination of superresolution imaging and automated classification. Our results confirm that the fossil pollen assigned to the morphotype S. catatumbus were produced by more than one genus and the majority share morphological similarities with extant Amherstieae genera. All African S. catatumbus specimens shared morphological features with extant African and pantropical genera and most South American fossils were similar to extant Neotropical or pantropical genera (SI Appendix, Table S1). These identifications support previous palynological and phylogenetic research which hypothesized that Amherstieae originated in Africa and later dispersed to the Neotropics (26, 27). Our results largely follow this anticipated biogeographic pattern (Movie S1) and provide indirect support for our CNN classifications. These identifications are also supported by standard morphometric measurements and visual expert assessments (SI Appendix, Table S3).
Capturing Morphological Diversity.
Airyscan confocal superresolution microscopy efficiently visualized the external and internal morphology of pollen specimens (8, 9 and SI Appendix, Fig. S2), allowing us to separate morphotypes that appear indistinguishable under the lower resolution of traditional brightfield microscopy (9). The morphological detail visible in Airyscan has been shown to be comparable to EM (8, 9); and as a nondestructive optical technique, Airyscan opens up the possibility of imaging and analyzing far greater numbers of fossil specimens than would be possible with EM (8, 11, 12). We were able to capture the morphological diversity in a low-abundance morphospecies like Striatopollis because Airyscan works with specimens directly on a microscope slide. Taxa like Striatopollis are rarely candidates for detailed morphological investigations because of the difficulty in isolating rare fossil pollen grains for EM.
Comparison to Visual and Morphometric Palynological Analysis.
The results of the CNN analysis agreed with the majority of our visual expert assessments and also largely captured the generic similarities identified by the NMDS morphometric analysis (SI Appendix, Table S3). All fossil specimens identified as the South American genus Macrolobium by the FM CNN were from South America. These specimens were also clustered with the genus in the NMDS analysis (SI Appendix, Table S3). Most South American and African specimens identified as Crudia or an African genus by the FM were also most similar to the pantropical Crudia or African genera in the NMDS analysis (SI Appendix, Table S3). We found that specimens that showed no consistency in the identifications among the three CNN models or had low classification confidence scores (FM cs: <0.55) were either poorly preserved, displayed morphological characteristics of multiple genera, or had morphologies that were not present in the 16 extant genera included in the study (SI Appendix, Fig. S14). Our results also showed that CNN were better at narrowing the possible taxonomic affinities of fossil specimens than the NMDS. Our CNN models gave one or two possible classifications per fossil, while the NMDS identified as many as five candidates.
The effect of sample degradation on the accuracy of the CNN identifications was most evident for specimens identified as the African genus Julbernardia. While Julbernardia has an external ornamentation pattern that is striate, the pattern differs from the fossil S. catatumbus (SI Appendix, Fig. S11). In Striatopollis, as in the extant Amherstieae genera Anthonotha, Crudia, Berlinia, and Macrolobium (SI Appendix, Fig. S14), the striae are clearly defined by furrows from pole to pole (SI Appendix, Fig. S11). In Julbernardia, striation is fused with a reticulate pattern, and the striae are not as defined (SI Appendix, Figs. S11 and S18). The features used in the NMDS analysis easily differentiated modern Julbernardia specimens from other striate Amherstieae genera.
However, degradation resulted in some S. catatumbus specimens displaying Julbernardia-like reticulation, which affected the FM predictions (SI Appendix, Tables S1 and S3). The Ypresian African fossil identified as Julbernardia/Isoberlinia (FM cs: 0.49/0.34) presents a striation pattern that overall is different to Julbernardia (SI Appendix, Fig. S11). Degradation resulted in visible reticulation under the supratectum, superficially resembling the ornamentation of Julbernardia. In the NMDS analysis, the African specimen was most similar to Didelotia and Crudia (sim: 0.96, 0.95). The PETM South American specimen identified as Anthonotha/Julbernardia (FM cs: 0.50/0.45) was similarly damaged and flattened in equatorial view. Its striation is similar to Anthonotha, but some fossil sections appear reticulate due to degradation of the pollen wall (SI Appendix, Fig. S11) and the internal wall structure is not clear. In the NMDS analysis, this specimen was closest to Crudia, Cynometra, and Didelotia (sim: 0.85, 0.87, 0.9) (SI Appendix, Table S3).
Many of the fossil specimens could not be easily classified as a single extant genus (SI Appendix, Table S2). Eighteen fossils (37.5% of the total) could not be classified with cs >0.55 in the FM model (SI Appendix, Tables S1 and S2). Many of these specimens share morphological characteristics with multiple extant Amherstieae genera and may represent extinct morphologies or genera not reviewed in our analysis. Previous studies of fossil angiosperm pollen ultrastructure recognize the possibility of intermediate morphological characteristics in the arrangements of extant and fossil pollen walls (28, 29).
To test how the CNN models responded to outliers, we analyzed a specimen of the fossil morphospecies Ephedripites, a taxon associated with Ephedraceae (SI Appendix, Fig. S15). Ephedripites has parallel ridges that superficially appear striate. We found that although this taxon can be confused with Striatopollis by the nonexpert, the FM classification score was low (Neochevalierodendron/Anthonotha, cs: 0.48/0.42) (SI Appendix, Fig. S15). The NMDS also identified Ephedripites as distinct from Amherstieae (SI Appendix, Fig. S9). However, the NMDS similarity score was relatively high. The fossil clustered with Julbernardia (sim: 0.89), the Amherstieae genus with pollen morphology distinct from other striate Amherstieae genera (SI Appendix, Fig. S9 and Table S3).
There were only 13 mismatches between the CNN and the NMDS results. Among these mismatches, the expert assessments for four specimens agreed with the FM classifications (SI Appendix, Table S3). The expert assessment for the remaining specimens agreed with at least one of the genera identified by the NMDS (SI Appendix, Table S3). The pollen walls of these specimens were poorly preserved (SI Appendix, Table S3). Three were from South America and identified by the FM as Berlinia (cs: 0.61, 0.85) and Neochevalierodendron/Berlinia (cs: 0.49/0.465), but they were more similar to Macrolobium (sim: 0.89 to 0.93) in the NMDS and expert assessments (SI Appendix, Table S3).
The majority of identifications were consistent between the FM CNN, NMDS morphometric analysis, and visual assessments. What distinguished the FM was the speed of the analysis. The CNN models analyzed more than 400 images in minutes. A human analyst takes days to analyze the same number of images. Both the FM and the NMDS quantified the relative uncertainty of each classification, the FM with a max-softmax classification score, and the NMDS with a numerical measurement of similarity. This uncertainty is often difficult to measure and not always recorded for visual identifications (5, 16). However, the FM identifications were more definitive than those from the NMDS (SI Appendix, Table S3), with one or two potential affinities, whereas the NMDS identified up to five potential generic labels. The morphometric analysis was able to identify clear outliers, like Ephedripites and Julbernardia, but was unable to discriminate among more morphologically similar taxa. Additionally, although NMDS is quantitative, it is still somewhat subjective. Experts chose the morphological characters to include or exclude, and error is introduced through manual measurements. This choice of characters and precision of measurements can alter the analysis results.
Fossil Classifications and Biogeographic Context.
S. catatumbus first appears in Africa during the Paleocene (30) (Dataset S7). The oldest specimens in our analysis are from the late Paleocene (Thanetian, 59.2 to 56 Ma) of Nigeria (Dataset S4 and SI Appendix, Fig. S1). We identified four of these specimens as the pantropical Crudia (cs: 0.7 to 1.00) (SI Appendix, Table S1), supporting an African Paleocene origin for the genus (27, 31, 32). If our fossil grains are Crudia, this would suggest an earlier origination date than previously inferred from middle Eocene age macrofossils (fruits and leaflets), which have placed the origin of Crudia in the Eocene (∼45 Ma) (26, 33–36).
Notably, Crudia was not easily confused with other Amherstieae genera in our models (Fig. 2 and SI Appendix, Figs. S3 and S4). We captured the range of morphological variability by including pollen from 14 extant species from different tropical regions to train our CNN models (Dataset S3). However, although the classification scores of the Thanetian fossil identifications were high and consistent among the three CNN models (SI Appendix, Table S1), visually, we were only able to identify the specimens as cf Crudia. The poor preservation made it difficult to extract diagnostic characters (Fig. 3). However, these fossils are more similar to Crudia than other Amherstieae genera, and the CNN results support the hypothesis that Crudia-type pollen is the oldest pollen morphology in Amherstieae (20 and SI Appendix, Table S1).
Among the Eocene African fossils, seven Ypresian (56 to 47.8 Ma) specimens were identified as Berlinia (cs: 0.5 to 0.9) and Anthonotha (cs: 0.5 to 0.9) (SI Appendix, Table S1). We were able to visually corroborate most of these identifications, suggesting that both genera were present in Africa before the Oligocene and early Miocene originations proposed by current molecular phylogenies (26, 35, 37).
S. catatumbus spread across the tropics during the Paleocene-Eocene (Movie S1) and is present in South America from the PETM, 56.8 Ma (38) (Dataset S7). The South American specimens in this study are from deposits from the PETM/early Eocene (56.8 to 56.2 Ma), Burdigalian (20.4 to 16 Ma), Langhian (16 to 13.8 Ma), Serravallian (13.8 to 11.6 Ma), and Tortonian (11.6 to 7.2 Ma) (Dataset S4 and SI Appendix, Fig. S1). Six specimens from the PETM/early Eocene were identified as Crudia (cs: 0.50 to 0.98, SI Appendix, Table S1). Of these specimens, we visually confirmed four as Crudia and one as Crudia/Berlinia. One specimen was too poorly preserved to assess (SI Appendix, Table S3). Visual inspection revealed that a seventh specimen, identified as Anthonotha/Julbernardia (cs: 0.49/0.44) (SI Appendix, Tables S1 and S2), shared features of both Anthonotha and Crudia (SI Appendix, Table S3).
South American Burdigalian fossils were all identified as African genera: Anthonotha (one specimen, cs: 0.67), Berlinia (one specimen, cs: 0.85), and Neochevalierodendron/Berlinia (one specimen, cs: 0.49/0.46) (SI Appendix, Tables S1 and S2). Following the morphometric analysis and visual inspection, we determined that the fossil identified as Anthonotha is morphologically similar to Crudia and Anthonotha (SI Appendix, Table S3). The fossils identified as Berlinia and Neochevalierodendron/Berlinia have a perforate exine and baculate striae more similar to Macrolobium (SI Appendix, Fig. S13 and Table S3). A single South American Serravallian specimen was identified as Berlinia (cs: 0.60, SI Appendix, Table S1), but the structure of the pollen wall was poorly preserved.
If accurate, the Burdigalian classifications suggest that Anthonotha, a genus which is restricted to Africa today (39–41), was present in the Neotropics from the early Eocene through at least the Miocene. Alternatively, these specimens may be of extinct South American lineages that share plesiomorphic traits with these extant African genera (SI Appendix, Fig. S13). In contrast, all but one of the younger South American fossils from the Langhian, Serravallian, and Tortonian were classified and visually verified as the pantropical Crudia (three specimens, cs: 0.49 to 0.99) and the Neotropical Macrolobium (six specimens, cs: 0.50 to 1.00) (SI Appendix, Table S3).
Interpreting CNN Classification Confidence Scores.
The softmax classification scores output by the CNN give a discrete probability distribution over the class labels conditioned on the input data. During training, the network parameters are optimized using maximum likelihood (often referred to as cross-entropy loss). While the max-softmax classification score does not reflect confidence intervals or predictive uncertainty in the Bayesian sense (42, 43), the relative ranking of softmax scores do reflect the likelihood of the prediction being correct (44). In practice, CNN models tend to be overconfident (predicting scores that are higher than the empirical probability with which they are correct in their classification), particularly on examples which are far from the training data distribution. When additional validation data are available, this overconfidence can be addressed by calibrating the model scores using scaling functions (e.g., histogram binning or isotonic regression) that monotonically adjust scores such that the calibrated scores indicate real world classification accuracies (43). In this way, calibrated scores can indicate a true classification accuracy of the model’s predictions. Such a calibration, while retaining the original ranking of predictions, requires a set of held-out examples with ground-truth labels. In our work, we do not have labels for our testing (fossil) examples, as their true identity is not known. However, the relative ranking is sufficient for generating a hypothesis of taxonomy, so we use the max-softmax as a raw indicator of the prediction confidence (43).
Conclusions
The combination of superresolution imaging and automated classification has the potential to transform the way pollen data are used in ecological and evolutionary research. Machine models allow experts to focus their time on the most challenging and ambiguous identifications by narrowing the universe of potential identifications. By drawing on existing palynological knowledge, we were able to conduct very focused morphological comparisons. We circumscribed our classification problem by including only fossil specimens that had been expertly identified as Striatopollis. We compared them to the striate genera of the Amherstieae tribe, recognized as the most likely extant analogs of this fossil.
The extant diversity of Amherstieae is the net result of nearly 65 million years of origination and extinction. Our results demonstrate that pollen can preserve more evidence of this evolutionary history than has been previously recognized. Because S. catatumbus is not abundant (<5% of grains per sample), 48 pollen grains represent a large sample of its morphological variability. Our analysis of this locally rare, but geographically and temporally widespread, taxon supports the hypothesis that Amherstieae first evolved in Africa during the Paleocene and later dispersed to South America. Our model identifications aligned well with expert assessments of fossil morphology. However, 37% of our fossil specimens were not easily categorized as extant genera, suggesting that the evolution of Amherstieae may include extinct diversity that is only visible in the fossil record. Our results also suggest that several genera, including Crudia and Anthonotha, may have diverged earlier than proposed by current molecular phylogenies (34–36). Specimens that morphologically resemble extant African genera in the South American fossil record raise the possibility that several endemic African genera, or shared ancestors, may have been pantropical in the past.
S. catatumbus is just one example of a fossil morphospecies, but there are thousands of fossil pollen morphotypes for which taxonomic affinities remain unexplored. Our methods offer an effective, higher-throughput workflow for identifying fossil morphospecies. Our results demonstrate the degree to which fossil pollen data can be explicitly incorporated into evolutionary and paleoecological research. The widespread adoption of these new microscopy and computation tools would set the stage for a new era of comparative morphological research in palynology.
Materials and Methods
Collection of Specimens.
Extant specimens.
The Amherstieae tribe contains 48 genera (34). We restricted our search to the 28 extant genera with pollen morphology similar to S. catatumbus: semitectate, striate, and triaperturate with long colpi (23, 24, 45), and were able to obtain pollen from 16 of these genera (Dataset S3 and SI Appendix, Fig. S14). We assessed that four genera (Tetraberlinia, Aphanocalyx, Bikinia, and Julbernardia) were morphologically different from S. catatumbus under light microscopy (e.g., different striation and much wider colpi, SI Appendix, Fig. S14). However, we did not exclude these from the CNN analysis.
Pollen was extracted from 45 herbarium specimens (Dataset S3) from three herbaria, including the University of Illinois at Urbana–Champaign, the Missouri Botanical Garden, and the Smithsonian Institution. We included 1 to 14 species per genus. The unbalanced number of species per genus reflects the underlying diversity within Amherstieae (Dataset S3). There is a low morphological variation within the genera of Amherstieae analyzed (24) (Dataset S3).
Fossil specimens.
A total of 48 fossil specimens of S. catatumbus were obtained from 25 geologic samples from historical palynological collections (Dataset S4). The samples were from five northern South American and four western African geologic sections (SI Appendix, Fig. S1 and Dataset S4). We imaged all Striatopollis grains in each sample. Twenty-seven specimens were from the Paleocene and Eocene of Africa and 21 from the Paleocene-Eocene and Miocene of South America (Dataset S4). For South America, we included seven specimens named Striatopollis “crassitectatus” and S. “pseudocrassitectatus.” These taxa have morphological characteristics similar to S. catatumbus, but with enough differences that they have been informally assigned to different morphospecies (Dataset S4).
Chemical Preparation.
Modern Amherstieae specimens were prepared following a modified version of the protocol described in Urban et al. (46). Fossil samples were processed by Paleoflora, Bucaramanga, Colombia. Samples were prepared following standard palynological techniques of digesting sediments in mineral acids (HF and HCl), alkaline treatment in KOH, heavy liquid separation using ZnBr2, and sieving (2) and mounted with a film of polyvinyl alcohol and Canada balsam (30, 47).
Airyscan Imaging.
We imaged both modern and fossil specimens using Airyscan confocal superresolution microscopy (Zeiss LSM 880 Airyscan; 63×/NA 1.4 oil DIC [differential interference contrast]) (25) (Datasets S3 and S4). We imaged 8 to 14 pollen grains from each extant species (Dataset S3). Each pollen grain was imaged as a stack of axial cross-sectional planes (0.19-µm increments), capturing its full 3D morphology across >60 planes per grain (SI Appendix, Fig. S2). We used the 488-nm (blue) and 405-nm (violet) excitation frequencies. Resulting images are of fluorescence intensities (25).
Morphological Analysis Using CNNs.
We used both the full cross-sectional image stack and its maximum intensity projection (MIP). MIP is a two-dimensional (2D) projection of an image stack (SI Appendix, Fig. S2). Due to attenuation and scattering, the MIP captures primarily the surface features of our pollen grains (8) while the individual cross-sectional images capture the internal structure.
Three CNN models were developed with the MatConvNet toolbox (48) (Fig. 1): maximum projection model (MPM), multislice model (MSM), and fused model (FM). For our CNN architecture, we built upon the state-of-the-art residual neural networks (ResNet) (49). We chose the 50-layer ResNet model pretrained on the ImageNet dataset (50). We tested three different fine-tuning strategies: 1) fine tuning the whole model; 2) training only the top new layer; and 3) fine tuning from a specific layer to the top. We found that fine tuning the whole model consistently led to the best performance. During fine tuning, we also fixed batch normalization (51) by using the global moments instead of local batch statistics. By doing this, we maintained the statistics of features at intermediate layers, and made the training more stable. We also used cross-entropy classification loss to fine tune the model. When feeding our data into the model, we stacked the same grayscale slice into a three-channel image, as the ResNet50 model is trained on RGB (red, green, and blue) color images (see additional details in SI Appendix).
The first model, MPM, used as input the MIP of the specimen and carried out a standard multiway classification task of external morphology (Fig. 1A). The second model, MSM, used as input subsets of the image stack—a series of banded axial projections (Fig. 1B), and then used a bag of instances (the series of banded projections) to classify the sample (52). The final classification was the banded projection with the highest classification confidence. In effect, the model selected the best representation of internal morphology for taxonomic classification. The third model, FM, combined representations of both external and internal morphological characteristics of the pollen wall. This model used the MIP of MPM and the stack of images of MSM as its input (Fig. 1C). In the FM, we tested three different fusion methods: the sum of the classification scores of two classifiers for the two views; the sum of the features from the two views as the final representation into a single classifier; and concatenating the two features, which is fed into the model as a single classifier. The first approach performed the best, as it did not overfit the small training data and generalized better to unseen test data. This was the final approach used in our fossil classifications. For all three models, we cropped images to a standard 640 × 640 pixels (45 × 45 µm, Fig. 1). The number of image slices per specimen varied with the depth of each grain, but this variation does not affect the analysis.
We first trained and tested the models using the extant genera. The sample dataset was randomly divided into a training set (70% of images per genus, 325 samples) and a validation set (30% of images per genus, 134 samples). We augmented the training samples by randomly mirroring the images during training. In our evaluation, we report performance accuracy without augmentation of the validation set. Classification accuracies are represented as confusion matrices (i,j) indicating the proportion of samples from a given genus (i) that are classified into each genus (j) (Fig. 2 and SI Appendix, Figs. S3 and S4).
We next applied the three trained models to the classification of our fossil specimens. Each specimen received predicted classifications with their associated class scores from each model (SI Appendix, TableS1). We emphasize the FM results because of the model’s higher classification accuracy. We visually reviewed the morphology of all fossil pollen identifications given by the CNN models to identify morphological similarities and differences.
Traditional Morphometric Analysis.
We manually collected morphological measurements from a small subset of modern specimens and all fossil specimens directly from the Airyscan images (Dataset S5). We measured 48 Striatopollis grains from Africa and South America and an African fossil specimen of Ephedripites. We also measured a total of 60 pollen grains from 12 extant species (5 grains per species), corresponding to 10 genera: Isoberlinia, Anthonotha, Crudia, Macrolobium, Berlinia, Julbernardia, Didelotia, Neochevalierodendron, Microberlinia, and Cynometra (Dataset S5). We defined four categorical variables and 18 quantitative measurements (Dataset S5). For each quantitative variable, we used the mean of three measurements (Dataset S5). We excluded 8 variables that could not be measured consistently. The final 14 variables were: external ornamentation, internal ornamentation, thickness of the striae, shape of the stria in transversal view, thickness of the endexine, diameter of the head of the columellae, thickness of the columellae, equatorial length, thickness of the tectum, columellae length, and the ratios of thickness of the tectum to thickness of the striae, thickness of the stria to polar length, thickness of the striae to equatorial length, and diameter of the columellae to stria thickness.
We used NMDS to compare the extant genera and the fossil specimens (53), using the R software packages “cluster” (54) and “vegan” (55). We used Gower distance, developed for morphometrics, in our NMDS analysis (56), using the complement of the “daisy” function in the R package cluster. This distance allowed us to combine categorical and quantitative variables and include missing values. We also calculated all of the pairwise similarities of our NMDS using an Euclidean distance. These similarities were used to compare with our CNN analysis (Dataset S6 and SI Appendix, Table S3).
We additionally analyzed and compared the overall morphology of grains visually, relying on palynological experience. The analyst develops an intuition for the diverse morphology of pollen taxa that is more than the collection of individual measurements (SI Appendix, Table S3). This ability is developed with decades of experience and is difficult to describe with words. This approach captures characters that cannot be measured or scored in a standard morphometric analysis and is in some ways akin to the CNN approach to classification.
Paleobiogeography.
We compiled geographic localities and age estimates (∼56 to 0 Ma) for all published records of S. catatumbus included in the Palynodata database (57) (Dataset S7). Because Palynodata (57) does not include the geographic coordinates of the localities reported, we used the original publications to find this information when possible. When exact coordinates could not be found, we estimated a broad geographical location based on the locality name found in the database or the literature. We used GPlates 2.0.0 (58) to plot the occurrence data (58).
Access to Airyscan Images and Code.
The images used in this study were submitted to Illinois Databank (https://doi.org/10.13012/B2IDB-9133967_V1). A readme file guides how to access data and meta information.
The codebase is available in GitHub (https://github.com/aimerykong/deepPollen). A readme file demonstrates how to run the models, which can be downloaded through a link therein.
Supplementary Material
Acknowledgments
This project was funded by the NSF, grants NSF-DBI–Advances in Bioinformatics (NSF-DBI-1262561 to S.W.P. and NSF-DBI-1262547 to C.C.F.), and NSF Information and Intelligent Systems (NSF-IIS-1618806 and NSF-IIS-1253538 to C.C.F.). We thank Mayandi Sivaguru, Glenn Fried, and Austin Cyphersmith at the Carl R. Woese Institute for Genomic Biology, University of Illinois at Urbana–Champaign for microscopy support. We also thank David Seigler at the University of Illinois at Urbana–Champaign, James Solomon at the Missouri Botanical Garden, and Meghann Toner at the US National Herbarium for facilitating the loan of the herbarium specimens for pollen extraction. We also want to thank Michael Peterson, the four reviewers, and the editor for feedback that significantly improved this manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2007324117/-/DCSupplemental.
Data Availability.
Airyscan images data have been deposited in Illinois Databank (https://doi.org/10.13012/B2IDB-9133967_V1). All study data are included in the article and supporting information.
References
- 1.Doyle J. A., Palynological patterns: Pollen and spores. Science 238, 557–558 (1987). [DOI] [PubMed] [Google Scholar]
- 2.Traverse A., Paleopalynology, Traverse A., Ed. (Springer, ed. 2, 2007), pp. 45–54. [Google Scholar]
- 3.Kutzbach J. E., et al. , “Epilogue” in Global Climates Since the Last Glacial Maximum, Wright J. E., et al., Eds. (Minnesota Press, 1993), pp. 536–642. [Google Scholar]
- 4.Ritchie J. C., Tansley review No. 83. Current trends in studies of long-term plant community dynamics. New Phytol. 130, 469–494 (1995). [DOI] [PubMed] [Google Scholar]
- 5.Mander L., Punyasena S. W., On the taxonomic resolution of pollen and spore records of Earth’s vegetation. Int. J. Plant Sci. 175, 931–945 (2014). [Google Scholar]
- 6.Punyasena S. W., Tcheng D. K., Wesseln C., Mueller P. G., Classifying black and white spruce pollen using layered machine learning. New Phytol. 196, 937–944 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Mander L., Li M., Mio W., Fowlkes C. C., Punyasena S. W., Classification of grass pollen through the quantitative analysis of surface ornamentation and texture. Proc. Biol. Sci. 280, 20131905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sivaguru M., et al. , Comparative performance of airyscan and structured illumination superresolution microscopy in the study of the surface texture and 3D shape of pollen. Microsc. Res. Tech. 81, 101–114 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Romero I. C., Urban M. A., Punyasena S. W., Airyscan superresolution microscopy: A high-throughput alternative to electron microscopy for the visualization and analysis of fossil pollen. Rev. Palaeobot. Palynol. 276, 104192 (2020). [Google Scholar]
- 10.Zavialova N., Tekleva M., Polevova S., Bogdanov A., Electron Microscopy for Morphology of Pollen and Spores (RIPOL Classic Press, 2018). [Google Scholar]
- 11.Gavrilova O., Zavialova N., Tekleva M., Karasev E., Potential of CLSM in studying some modern and fossil palynological objects. J. Microsc. 269, 291–309 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Hoorn C., et al. , Going north and south: The biogeographic history of two Malvaceae in the wake of Neogene Andean uplift and connectivity between the Americas. Rev. Palaeobot. Palynol. 264, 90–109 (2019). [Google Scholar]
- 13.Zhang Y., Fountain D. W., Hodgson R. M., Flenley J. R., Gunetileke S., Towards automation of palynology: Pollen recognition using Gabor transforms and digital moments. J. Quat. Sci. 19, 763–768 (2004). [Google Scholar]
- 14.Zhang W. X., et al. , Study on relationship between pollen exine ornamentation pattern and germplasm evolution in flowering crabapple. Sci. Rep. 7, 39759 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong S., Fowlkes C. C., Low-rank bilinear pooling for fine-grained classification. arXiv:1611.05109 (30 November, 2017). [Google Scholar]
- 16.Kong S., Punyasena S., Fowlkes C., Spatially aware dictionary learning and coding for fossil pollen identification. arXiv:1605.00775 (3 May 2016).
- 17.Krizhevsky A., Sutskever I., Hinton G., ImageNet classification with deep convolutional neural networks, (NIPS Proc. B, 2012). [Google Scholar]
- 18.Gonzales-Guzman E., A Palynological Study on the Upper Los Cuervos and Mirador Formations, (Brill Archive, Leiden, 1967). [Google Scholar]
- 19.Germeraad J. H., Hopping C. A., Muller J., Palynology of tertiary sediments from tropical areas. Rev. Palaeobot. Palynol. 6, 189–348 (1968). [Google Scholar]
- 20.Muller J., Fossil pollen records of extant Angiosperms. Bot. Rev. 47, 1–142 (1981). [Google Scholar]
- 21.Pan A. D., Jacobs B. F., Herendeen P. S., Detarieae sensu lato (Fabaceae) from the Late Oligocene (27.23 Ma) Guang River flora of north-western Ethiopia. Bot. J. Linn. Soc. 163, 44–54 (2010). [Google Scholar]
- 22.Jaramillo C., et al. , Effects of rapid global warming at the Paleocene-Eocene boundary on neotropical vegetation. Science 330, 957–961 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Ferguson I. K., Skvarla J. J., “The pollen morphology of the subfamily Papilionoideae (Leguminosae)” in Advances in Legume Systematics, Polhill R. M., Raven P. H., Eds. (Royal Botanic Gardens, Kew, 1981), pp. 859–896. [Google Scholar]
- 24.Banks H., Klitgaard B. B., “Palynological contribution to the systematics of detarioid legumes (Leguminosae: Caesalpinioideae)” in Advances in Legume Systematics, Herendeen P. S., Bruneau A., Eds. (Royal Botanic Gardens, Kew, ed. 9, 2000), pp. 79–106. [Google Scholar]
- 25.Romero I., Urban M. A., Punyasena S. W., Airyscan confocal superresolution images of fossil and modern pollen of Amherstieae (Fabaceae). Illinois Data Bank; 10.13012/B2IDB-9133967_V1 (2020). Deposited 24 June 2019. [DOI] [Google Scholar]
- 26.de la Estrella M., Forest F., Wieringa J. J., Fougère-Danezan M., Bruneau A., Insights on the evolutionary origin of Detarioideae, a clade of ecologically dominant tropical African trees. New Phytol. 214, 1722–1735 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Mackinder B., “Detariae sensu lato” in Legumes of the World, Lewis G., Schrire B., Mackinder B., Lock M., Eds. (Kew Publishing, UK, 2005), pp. 69–109. [Google Scholar]
- 28.Denk T., Tekleva M. V., Comparative pollen morphology and ultrastructure of Platanus: Implications for phylogeny and evaluation of the fossil record. Grana 45, 195–221 (2007). [Google Scholar]
- 29.Kriebel R., Khabbazian M., Sytsma K. J., A continuous morphological approach to study the evolution of pollen in a phylogenetic context: An example with the order Myrtales. PLoS One 12, e0187228 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oboh-Ikuenobe F. E., Obi C. G., Jaramillo C. A., Lithofacies, palynofacies, and sequence stratigraphy of Palaeogene strata in Southeastern Nigeria. J. Afr. Earth Sci. 41, 79–101 (2005). [Google Scholar]
- 31.Doyle J. J., Luckow M. A., The rest of the iceberg. Legume diversity and evolution in a phylogenetic context. Plant Physiol. 131, 900–910 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Domenech B., Systématique, biogéographie et divertification du genre Crudia (Leguminosae, Detarioideae), (Université de Montréal Biodiversity Centre, 2018). [Google Scholar]
- 33.Herendeen P. S., Dilcher D. L., Reproductive and vegetative evidence for the occurrence of Crudia (Leguminosae, Caesalpinioideae) in the Eocene of Southeastern North America. Bot. Gaz. 151, 402–413 (1990). [Google Scholar]
- 34.de la Estrella M., et al. , A new phylogeny-based tribal classification of subfamily Detarioideae, an early branching clade of florally diverse tropical arborescent legumes. Sci. Rep. 8, 6884 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bruneau A., Mercure M., Lewis G. P., Herendeen P. S., Phylogenetic patterns and diversification in the Caesalpinioid legumes. Botany 86, 697–718 (2008). [Google Scholar]
- 36.LPWG , A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny. Taxon 66, 44–77 (2017). [Google Scholar]
- 37.Bruneau A., Klitgaard B. B., Prenner G., Fougère-Danezan M., Tucker S. C., Floral evolution in the detarieae (Leguminosae): Phylogenetic evidence for labile floral development in an early-diverging legume lineage. Int. J. Plant Sci. 175, 392–417 (2014). [Google Scholar]
- 38.Jaramillo C. A., Dilcher D. L., Microfloral diversity patterns of the Late Paleocene-Eocene interval in Colombia, northern South America. Geology 28, 815–818 (2000). [Google Scholar]
- 39.Ojeda D. I., et al. , Phylogenomics within the Anthonotha clade (Detarioideae, Leguminosae) reveals a high diversity in floral trait shifts and a general trend towards organ number reduction. bioRxiv:511949 (4 January 2019).
- 40.Cowan R. S., A taxonomic revision of the genus Macrolobium (Leguminosae-Caesalpinioideae). Mem. N. Y. Bot. Gard. 8, 22557–342 (1953). [Google Scholar]
- 41.Murphy B., de la Estrella M., Schley R., Forest F., Klitgaard B., On the monophyly of Macrolobium Schreb., an ecologically diverse neotropical tree genus (Fabaceae-Detarioideae). Int. J. Plant Sci. 179, 75–86 (2018). [Google Scholar]
- 42.Gal Y., Ghahramani Z., Dropout as a bayesian approximation: Representing model uncertainty in deep learning. arXiv:1506.02142 (4 October 2016).
- 43.Guo C., Pleiss G., Sun Y., Weinberger K. Q., On calibration of modern neural networks. arXiv:1706.04599 (3 August 2017).
- 44.Hendrycks D., Gimpel K., A baseline for detecting misclassified and out-of-distribution examples in neural networks. arXiv:1610.02136 (3 October 2018).
- 45.Banks H., Structure of pollen apertures in the Detarieae sensu stricto (Leguminosae: Caesalpinioideae), with particular reference to underlying structures (Zwischenkörper). Ann. Bot. 92, 425–435 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Urban M. A., Romero I. C., Sivaguru M., Punyasena S. W., Nested cell strainers: An alternative method of preparing palynomorphs and charcoal. Rev. Palaeobot. Palynol. 253, 101–109 (2018). [Google Scholar]
- 47.Jaramillo C., et al. , Miocene flooding events of western Amazonia. Sci. Adv. 3, e1601693 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vedaldi A., Lenc K., Matconvnet: Convolutional neural networks for matlab. CoRR, abs/1412.4564 (2014).
- 49.He K., Zhang X., Ren S., Sun J., Deep residual learning for image recognition. arXiv:1512.03385 (10 December 2015).
- 50.Deng J., et al. , “ImageNet: A large-scale hierarchical image database” in IEEE Conf. Computer Vision Pattern Recognition, (CVPR, 2009), pp. 248–255. [Google Scholar]
- 51.Ioffe S., Szegedy C., Batch Normalization: Accelerating deep network training by reducing internal covariate shift. arXiv:1502.03167 (2 March 2015).
- 52.Andrews S., Tsochantaridis I., Hofmann T., “Support vector machines for multiple-instance learning”” in Proceedings of the 15th International Conference on Neural Information Processing Systems, (NIPS, 2003), pp. 577–584. [Google Scholar]
- 53.Legendre P., Legendre L., Numerical Ecology (Elsevier, 2012), pp. 425–523. [Google Scholar]
- 54.Maechler M., Rousseeuw P., Struyf A., Hubert M., Hornik K., Cluster: Cluster analysis basics and extensions, version 2.0.7-1 (2018), pp. 1–79.
- 55.Oksanen J., et al. , Vegan: Community Ecology Package (R Foundation, Vienna, 2019). Version 2.5-6.
- 56.Gower J. C., A general coefficient of similarity and some of its properties. Biometrics 27, 857–871 (1971). [Google Scholar]
- 57.Palynodata Inc , Palynodata Datafile, (Natural Resources of Canada, Canada, 2007). [Google Scholar]
- 58.Matthews K. J., et al. , Global plate boundary evolution and kinematics since the Late Paleozoic. Global Planet. Change 146, 226–250 (2016). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Airyscan images data have been deposited in Illinois Databank (https://doi.org/10.13012/B2IDB-9133967_V1). All study data are included in the article and supporting information.