Abstract
Iron is essential to life, but surprisingly little is known about how iron is managed in non-vertebrate animals. In mammals, the well-characterized transferrins bind iron and are involved in iron transport or immunity, whereas other members of the transferrin family do not have a role in iron homeostasis. In insects, the functions of transferrins are still poorly understood. The goals of this project were to identify the transferrin genes in a diverse set of insect species, resolve the evolutionary relationships among these genes, and predict which of the transferrins are likely to have a role in iron homeostasis. Our phylogenetic analysis of transferrins from 16 orders of insects and two orders of non-insect hexapods demonstrated that there are four orthologous groups of insect transferrins. Our analysis suggests that transferrin 2 arose prior to the origin of insects, and transferrins 1, 3 and 4 arose early in insect evolution. Primary sequence analysis of each of the insect transferrins was used to predict signal peptides, carboxyl-terminal transmembrane regions, GPI-anchors, and iron binding. Based on this analysis, we suggest that transferrins 2, 3 and 4 are unlikely to play a major role in iron homeostasis. In contrast, the transferrin 1 orthologs are predicted to be secreted, soluble, iron-binding proteins. We conclude that transferrin 1 orthologs are the most likely to play an important role in iron homeostasis. Interestingly, it appears that the louse, aphid, and thrips lineages have lost the transferrin 1 gene and, thus, have evolved to manage iron without transferrins.
Keywords: hemolymph, insect, iron homeostasis, melanotransferrin, phylogeny, transferrin
Introduction
Iron is an essential micronutrient, but it is also potentially toxic; therefore, the amount, location and form of iron within an animal must be tightly regulated (Kosman 2010; Frazer & Anderson 2014). Different animal lineages have evolved distinct mechanisms of iron homeostasis that provide an adequate amount of iron to cells while limiting iron toxicity (Lambert 2012; Tang & Zhou 2013; Anderson & Leibold 2014; Galay et al. 2015). We are particularly interested in iron homeostasis in insects. Iron is essential for many aspects of an insect’s life, including energy metabolism, DNA synthesis, and detoxification of xenobiotics (Locke & Nichol 1992; Mandilaras et al. 2013). In addition, iron homeostatic mechanisms prevent iron overload in blood-feeding insects and are likely to have an important role in immuntiy (Graça-Souza et al. 2006; Ong et al. 2006).
In vertebrates, some well-characterized members of the transferrin family participate in iron homeostasis by sequestering and transporting iron (Lambert 2012). Those transferrins are extracellular, soluble proteins that are composed of two homologous lobes, each of which binds one ferric ion (Mizutani et al. 2012). They include lactoferrin, which functions in immunity, serum transferrin, which transports iron, and ovotransferrin, which has both immune and transport functions (Farnaud and Evans 2003; Anderson and Vulpe 2009; Giansanti et al. 2012). In contrast, two transferrin family members, melanotransferrin and inhibitor of carbonic anhydrase, have functions unrelated to iron homeostasis (Wang et al. 2007; Suryo Rahmanto et al. 2012). Membrane-bound melanotransferrin binds just one ferric ion whereas inhibitor of carbonic anhydrase is not an iron-binding protein (Lambert 2012).
Much less is known about invertebrate transferrins, including those from insects. A previous study has indicated that there are four orthologous groups of insect transferrins (Bai et al. 2016). These groups are designated transferrin 1 (Tsf1 or hemolymph transferrin), transferrin 2 (Tsf2 or melanotransferrin-like), transferrin 3 (Tsf3) and transferrin 4 (Tsf4). Note that for our study, we have used nomenclature that has been in place for many years (for example, Dunkov & Georgieva 2006; Geiser & Winzerling 2012); this nomenclature is different from that used in the study by Bai et al. (2016), in which Tsf1, Tsf2, Tsf3 and Tsf4 are referred to as Tsf4, Tsf1, Tsf2 and Tsf3, respectively. Three Tsf1 orthologs are known to bind iron. Tsf1 from the cockroach Blaberus discoidalis binds two ferric ions, whereas Tsf1 from the fly Drosophila melanogaster and the moth Manduca sexta each bind one (Huebers et al. 1988; Bartfeld & Law 1990; Gasdaska et al. 1996; Weber et al. 2018). Tsf1 has been detected in hemolymph and other extracellular fluids (Geiser & Winzerling 2012; Simmons et al. 2013; Qu et al. 2014; Zhang et al. 2014; Bonilla et al. 2015; Hattori et al. 2015; Brummett et al. 2017). Direct evidence suggests that Tsf1 orthologs can function in iron transport, immune-related iron sequestration, and protection against oxidative stress (Huebers et al. 1988; Kurama et al. 1995; Hirai et al. 2000; Lee et al. 2006; Kim et al. 2008; Brummett et al. 2017; Xiao et al. 2019). Tsf2 from D. melanogaster binds a single ferric iron (Tiklová et al. 2010). It is a membrane-bound epithelial protein that functions in septate junction formation and does not appear to be involved in iron homeostasis (Tiklová et al. 2010; Hall et al. 2014). Little is known about Tsf3 and Tsf4, including iron-binding status and function.
The mammalian transferrins that have a high affinity for iron have six conserved amino acid residues in each lobe that facilitate iron binding. Four of these residues directly bind iron, and the other two bind a carbonate anion that binds iron (Lambert et al. 2005; Mizutani et al. 2012). For example, in the amino-lobe of human serum transferrin, the iron-binding residues are Asp63, Tyr95, Tyr188 and His249, and the carbonate-binding residues are Thr120 and Arg124 (Mizutani et al. 2012). The iron-binding and carbonate-binding residues of insect transferrins have not yet been biochemically identified; however, sequence alignments suggest that they may be similar to the mammalian signature residues (Lambert et al. 2005; Tiklová et al. 2010; Geiser & Winzerling 2012).
The goals of this project were to identify the transferrin genes in a diverse set of insect species, perform a phylogenetic analysis to resolve the evolutionary relationships among these genes, and predict which of the insect transferrins are likely to have a role in iron homeostasis. We found that almost all insect transferrins cluster into one of four orthologous groups, that Tsf2 must have arisen prior to the evolution of insects, and that the other three orthologous groups probably arose early in insect evolution. In addition, the Tsf1 orthologs are the only insect transferrins likely to have a significant role in iron homeostasis, and, surprisingly, some insect lineages lack a Tsf1 ortholog.
Materials and Methods
Identification of transferrin sequences
To identify transferrin sequences from insect and non-insect hexapod species, we performed BLAST searches of species-specific databases at the National Center for Biotechnology Information (NCBI Resource Coordiators, 2016) and the i5k Workspace at i5k.nal.usda.gov (Poelchau et al. 2015) using the D. melanogaster Tsf1, Tsf2 and Tsf3 sequences as queries. Our objective was to identify all of the transferrin family members in each of the 24 selected species. When possible, predicted protein databases (NCBI reference proteins or i5k Workspace proteins) were used. NCBI transcriptome shotgun assembly databases were used when adequate genome information was unavailable. Gene predictions for transferrins from Rhodnius prolixus were previously reported (Walter-Nuno et al. 2018), and we edited these based on recent transcriptomic data. Transferrin sequences that were much shorter than a typical transferrin were omitted from further analysis because we assumed that they were probably incomplete. Several gene predictions contained apparent gaps or insertions. For those genes, when possible, we used additional information (mainly from transcript data) to edit the sequences. In four cases, it was obvious that open reading frames were encoded by two incomplete gene predictions (each with their own accession number); therefore, we combined partial sequences to create complete open reading frames. We also added to our dataset the sequence of the well-studied hemolymph transferrin from B. discoidalis (Gasdaska et al. 1996). Table 1 summarizes information about the identification of transferrin sequences, including query accession numbers, the species analyzed, the type and source of the databases for each species, the accession numbers of the transferrins identified, and an indication of which sequences were edited.
Table 1.
Sequences used for phylogenetic analysis of insect transferrins.
| Order | Species | Genome, Transcriptome or cDNA | Source | Accession number | Orthologous Group (based on this study) |
|---|---|---|---|---|---|
| Diptera | Drosophila melanogaster (fly) | Genome | NCBI | AAC67389 | Tsf1 |
| Genome | NCBI | NP_524044 | Tsf2 | ||
| Genome | NCBI | NP_523759 | Tsf3 | ||
| Aedes aegypti (mosquito) | Genome | NCBI | XP_001647719 | Tsf1 | |
| Genome | NCBI | XP_021699709 | Tsf2 | ||
| Genome | NCBI | EAT34844 edited | Tsf3 | ||
| Genome | NCBI | XP_001661801 | Tsf4 | ||
| Siphonaptera | Ctenocephalides felis (flea) | Genome | NCBI | XP_026466165 | Tsf1 |
| Genome | NCBI | XP_026479930 | Tsf2 | ||
| Genome | NCBI | XP_026471608 and 026471609 | Tsf3 | ||
| Lepidoptera | Manduca sexta (moth) | cDNA | NCBI | P22297 | Tsf1 |
| Genome | i5k | Msex2.11184 edited | Tsf2 | ||
| Genome | i5k | Msex2.10792 and 10793 | Tsf3 | ||
| Genome | i5k | Msex2.12754-RB | Tsf4 | ||
| Papilio xuthus (butterfly) | Genome | NCBI | XP_013173464 | Tsf1 | |
| Genome | NCBI | KPJ03186 | Tsf2 | ||
| Genome | NCBI | KPI99226 | Tsf3 | ||
| Genome | NCBI | KPI99233 edited | Tsf4 | ||
| Trichoptera | Annulipalpia species (caddisfly) | Transcriptome | NCBI | GATX01086443 | Tsf1 |
| Transcriptome | NCBI | GATX01000541 | Tsf2 | ||
| Transcriptome | NCBI | GATX01016449 edited | Tsf3 | ||
| NCBI | GATX01086805 | Tsf4 | |||
| Micropterna lateralis (caddisfly) | Transcriptome | NCBI | GELV01013828 | Tsf2 | |
| Transcriptome | NCBI | GELV01010679 | Tsf3 | ||
| Transcriptome | NCBI | GELV01015247 | Tsf4 | ||
| Coleoptera | Tribolium castaneum (beetle) | Genome | NCBI | XP_001808066 | Tsf1 |
| Genome | NCBI | XP_015839046 | Tsf2 | ||
| Genome | NCBI | XP_015838610 | Tsf3 | ||
| Genome | NCBI | XP_008199941 edited | Tsf4 | ||
| Hymenoptera | Apis mellifera (bee) | Genome | NCBI | NP_001011572 | Tsf1 |
| Genome | NCBI | XP_396618 | Tsf2 | ||
| Genome | XP_001122328 edited | Tsf3 | |||
| Psocodea | Pediculus humanus (louse) | Genome | NCBI | XP_002422999 | Tsf2 |
| Genome | NCBI | XP_002425773 edited | Tsf3 | ||
| Hemiptera | Acyrthosiphon pisum (aphid) | Genome | NCBI | XP_001947699 | Tsf2 |
| Genome | NCBI | XP_016660805 | Tsf2 | ||
| Genome | NCBI | XP_001946481 | Tsf3 | ||
| Rhodnius prolixus (bug) | Transcriptome | NCBI | GECK01013297 | Tsf1 | |
| Transcriptome | NCBI | GECK01101790 | Tsf2 | ||
| Transcriptome | NCBI | GECK01023379 | Tsf4 | ||
| Thysanoptera | Frankliniella occidentalis (thrips) | Genome | NCBI | XP_026287309 | Tsf2 |
| Genome | NCBI | XP_026287716 | Tsf4 | ||
| Blattodea | Zootermopsis nevadensis (termite) | Genome | NCBI | XP_021919348 | Tsf1 |
| Genome | NCBI | XP_021922858 | Tsf2 | ||
| Genome | NCBI | XP_021919332 edited | Tsf3 | ||
| Genome | NCBI | XP_021919264 | Tsf4 | ||
| Blaberus discoidalis (cockroach) | cDNA | NCBI | Q02942 | Tsf1 | |
| Phasmatodea | Medauroidea extradentata (walking stick) | Transcriptome | NCBI | GAWD01077063 | Tsf1 |
| Transcriptome | NCBI | GAWD01046554 | Tsf2 | ||
| Transcriptome | NCBI | GAWD01030369 | Tsf3 | ||
| Transcriptome | NCBI | GAWD01074570 | Tsf4 | ||
| Orthoptera | Locusta migratoria (locust) | Genome | NCBI | BBE27867 | Tsf1 |
| Genome | i5k | JAMg_model_8133 | Tsf2 | ||
| Genome | i5k | JAMg_model_4881 | Tsf3 | ||
| Ephemeroptera | Ephemera danica (mayfly) | Genome | i5k | Ed_EDAN016810 edited | Tsf2 |
| Genome | i5k | EDAN009681_PA | Tsf3 | ||
| Genome | i5k | EDAN001636_PA | Tsf4 | ||
| Odonata | Ladona fulva (dragonfly) | Genome | i5k | LFUL008155_PA edit | Tsf1 |
| Genome | i5k | LFUL002433 and LFUL002434 (has gap) | Tsf2 | ||
| Genome | i5k | LFUL016472 and LFUL016473 (has gap) | Tsf3 | ||
| Zygentoma | Atelura formicaria (silverfish) | Transcriptome | NCBI | GAYJ02040055 | Tsf1 |
| Transcriptome | NCBI | GAYJ02040158 | Tsf3 | ||
| Transcriptome | NCBI | GAYJ02044273 | Tsf4 | ||
| Thermobia domestica (firebrat) | Transcriptome | NCBI | GASN02065056 | Tsf1 | |
| Transcriptome | NCBI | GASN02058069 | Tsf3 | ||
| Transcriptome | NCBI | GHEH01000467 | Tsf4 | ||
| Archaeognatha | Meinertellus cundinamarcensis (jumping bristletail) | Transcriptome | NCBI | GAUG02039070 | |
| Transcriptome | NCBI | GAUG02047150 | |||
| Diplura | Occasjapyx japonicus (two-pronged bristletail) | Transcriptome | NCBI | GAXJ02018425 | |
| Transcriptome | NCBI | GAXJ02018310 | |||
| Transcriptome | NCBI | GAXJ02017766 | |||
| Transcriptome | NCBI | GAXJ02019413 | |||
| Transcriptome | NCBI | GAXJ02017728 | |||
| Transcriptome | NCBI | GAXJ02018531 | |||
| Transcriptome | NCBI | GAXJ02019650 | Tsf2 | ||
| Collembola | Folsomia candida (springtail) | Genome | NCBI | XP_021946057 | |
| Genome | NCBI | XP_021948718 | |||
| Genome | NCBI | XP_021960906 | Tsf2 | ||
| Orchesella cincta (springtail) | Genome | NCBI | ODM93913 | ||
| Genome | NCBI | ODM92907 | |||
| Genome | NCBI | ODM95217 | |||
| Genome | NCBI | ODM96803 | Tsf2 | ||
| Phylum Placozoa | Trichoplax adhaerens | Genome | NCBI | XP_002108117 | outgroup |
We did not find Tsf1 sequences in the genomes of Pediculus humanus (louse), Acyrthosiphon pisum (aphid) or Frankliniella occidentalis (thrips). To determine whether the genomes of lice, aphids and thrips contain Tsf1, we used the T. castaneum Tsf1 sequence as a query to search the following datasets: the NCBI transcriptome shotgun assembly databases for Psocodea and Phthiraptera (lice), the NCBI non-redundant protein database for Aphidoidea (aphids), and the NCBI transcriptome shotgun assembly database for Thysanoptera (thrips). For Phthiraptera, Aphidoidea and Thysanoptera, all BLAST hits with an E value exponent less than zero were evaluated, and for Psocodea, the top 50 BLAST hits were evaluated. Redundant sequences and those with large gaps were omitted from further analysis. Phylogenetic analysis (described below) was used to determine whether or not the sequences were Tsf1 orthologs.
We did not find Tsf1 sequences in aphid genomes but did identify Tsf1 in true bugs; therefore, we wanted to know whether other types of Hemipteran insects have a Tsf1 ortholog. We used the T. castaneum Tsf1 sequence as a query to search the NCBI non-redundant protein databases for Sternorrhyncha, Heteroptera, and Hemiptera with heteropteran sequences excluded. Putative Tsf1 sequences were evaluated by phylogenetic analysis as described below. Hemipteran phylogeny is described by (Li et al. 2017).
Phylogenetic analysis
Phylogenetic analyses were performed with MEGA 10.0.5 software (Kumar et al. 2018). Sequences were aligned with MUSCLE with the following settings: gap open penalty of −2.90, gap extend penalty of 0, hydrophobicity multiplier of 1.20, maximum memory of 2048 MB, maximum iterations of 16, cluster method of UPGMA, and minimum diagonal length of 24. Rooted phylogenetic trees were constructed with the maximum likelihood method using the following settings: the Jones-Taylor Thornton amino acid substitution model, uniform rates among sites, use all sites in cases of gaps and missing data, the Nearest-Neighbor-Interchange maximum likelihood heuristic method of tree inference, the default NJ/BioNJ for making the initial tree, no branch swap filter, and bootstrapping with 500 replications. The outgroup sequence (see Table 1) was the single transferrin sequence identified from the placozoan species Trichoplax adhaerens, a very simple animal near the base of the metazoan tree (Srivastava et al. 2008).
Prediction of signal peptides, transmembrane regions and GPI anchor sites
Signal sequences were predicted with Signal P (Bendtsen et al. 2004), and those sequences that were not predicted by Signal P to have a signal peptide were evaluated with PSORT II (Nakai & Horton 1999). Putative carboxyl-terminal transmembrane regions were predicted with TMPred software (Hofmann & Stoffel 1993). The presence or absence of GPI anchor sites was predicted with GPI-SOM (Fankhauser & Mäser 2005). A protein was categorized as membrane-bound if either a carboxyl-terminal membrane region or GPI anchor was predicted.
Prediction of iron-binding and carbonate-binding residues
To predict iron-binding and carbonate-binding residues of insect transferrins, all members of a particular orthologous group were aligned with bovine serum transferrin (NP_803450), lactoferrin (NP_851341) and melanotransferrin (NP_001179241) with the use of Clustal Omega (Sievers et al. 2011). Amino acid residues that aligned with conserved iron-binding or carbonate-binding residues in the bovine transferrins were analyzed.
Results
Insects have four orthologous groups of transferrins
To accomplish our goal of a comprehensive phylogenetic analysis of insect transferrins, we attempted to identify all the transferrin family members in 21 species of insects and three species of non-insect hexapods. The selected insect species represent 16 orders that range from more recent lineages, such as the dipterans and siphonopterans, to more ancient lineages, such as the archaeognathans and zygentomans (Figure 1). We identified a total of 82 transferrin sequences and performed a phylogenetic analysis to establish the evolutionary relationships among these genes. Our analysis demonstrates that the insect transferrins fall into four clades (Figure 2). The only insect transferrins that did not cluster with one of the four orthologous groups were two sequences from M. cundinamarcensis, a species of jumping bristletail that represents the most ancient insect lineage in our study. Although these two sequences are shown grouped with the Tsf1 orthologs in the phylogenetic tree, statistical support for this grouping is weak. In contrast, all of the orthologous groups have strong statistical support, with bootstrap values of at least 95%.
Figure 1. A diverse group of insect orders was chosen for a phylogenetic analysis of insect transferrins.
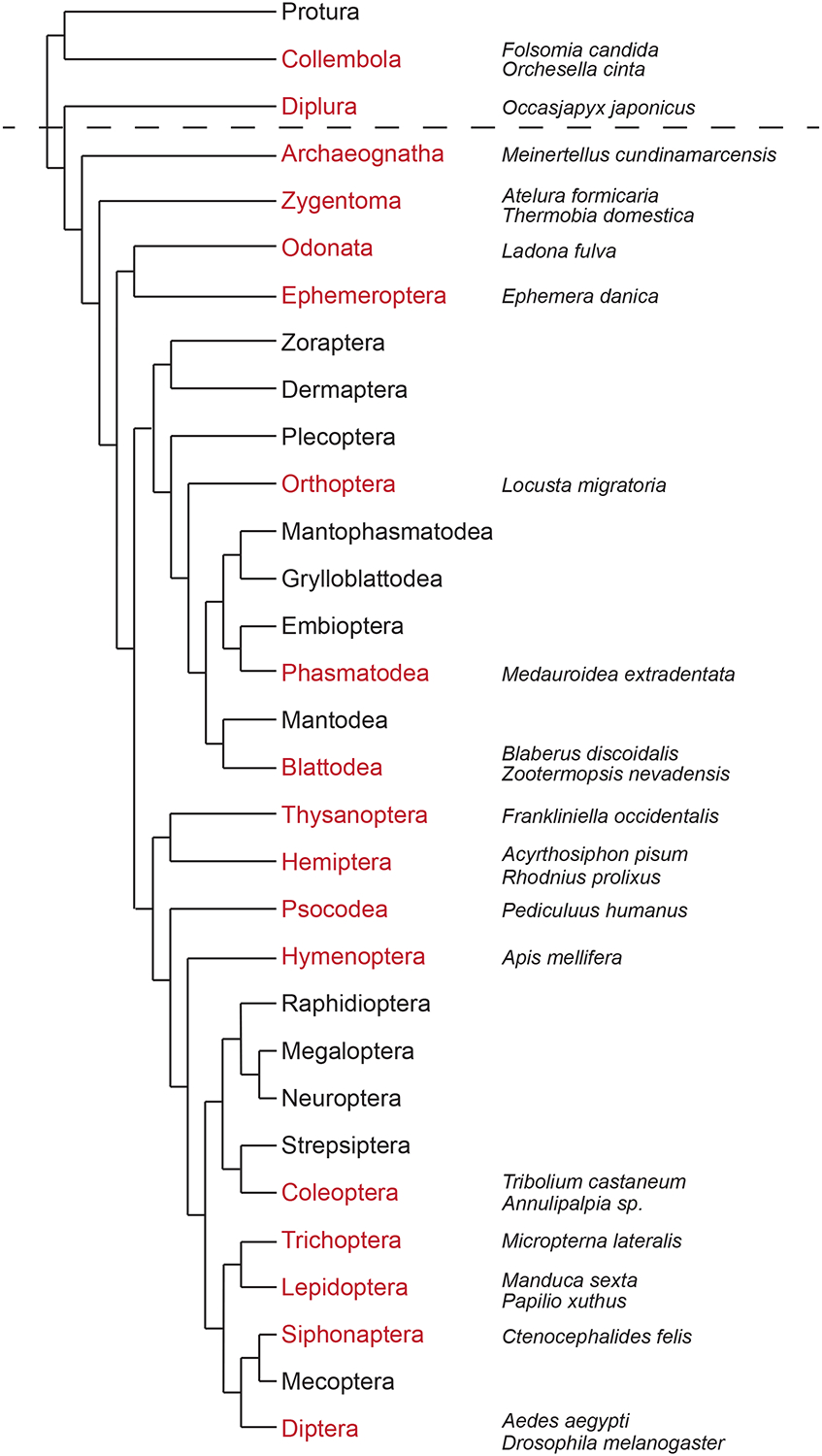
The tree shown is based on the phylogeny described by Misof et al. (2014) with a modification to include termites in the order Blattodea (Inward et al. 2007; Harrison et al. 2018). (Branch lengths do not represent evolutionary time.) The 18 orders represented in our phylogenetic analysis are in red text. The three non-insect hexapod orders are shown above the dashed line. The species included in our analyses are listed on the right.
Figure 2. Insects have four orthologous groups of transferrins.
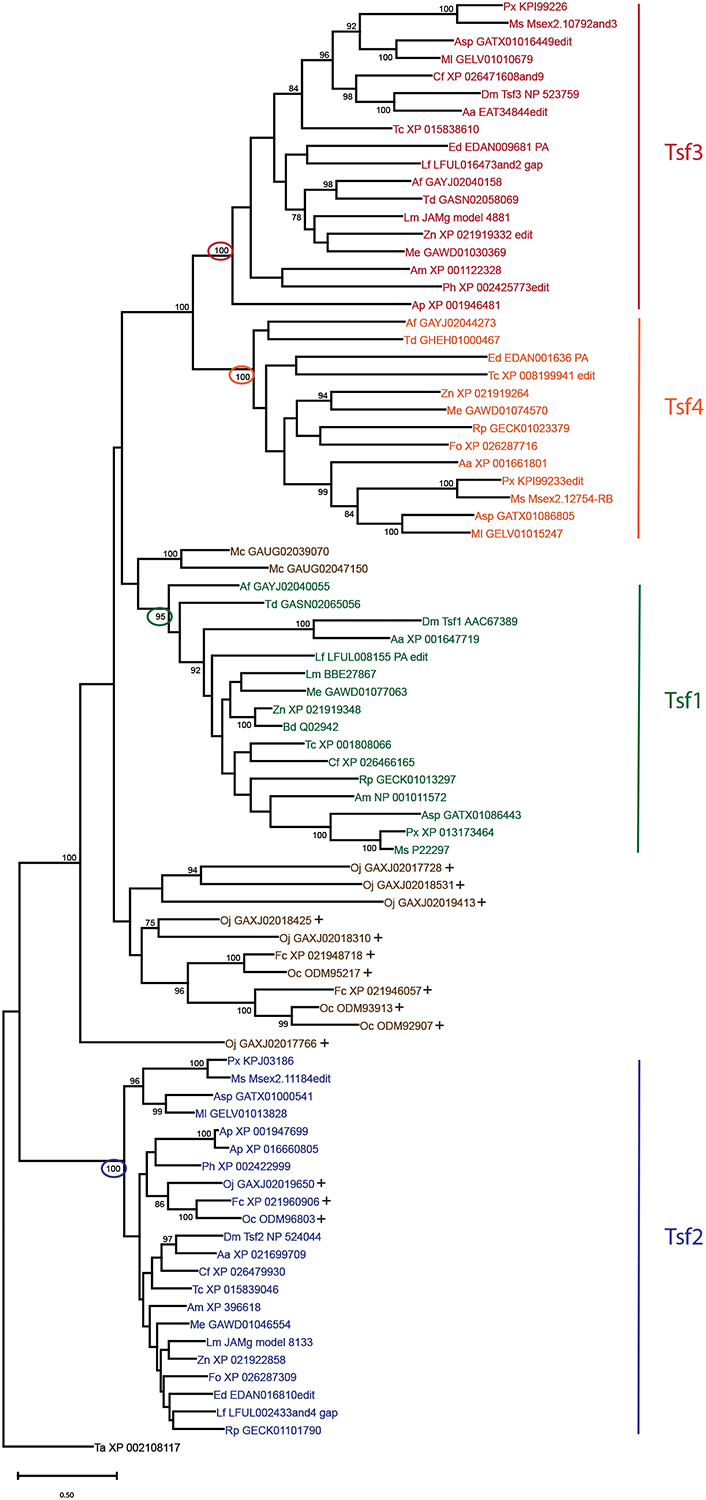
A phylogenetic analysis of transferrin sequences from 21 insect and three non-insect hexapod species was performed with the maximum likelihood method, and the resulting rooted tree is shown. Bootstrap values greater than 70 percent are shown. Tsf1 orthologs are indicated by green text, Tsf2 by blue, Tsf3 by red, and Tsf4 by orange. Non-insect hexapod transferrins are marked with a plus sign. The bootstrap value for each orthologous group is circled. The branch length scale indicates number of substitutions per site. Species abbreviations are as follows: Aa, Aedes aegypti; Af, Atelura formicaria; Am, Apis mellifera; Ap, Acyrthosiphon pisum; Asp, Annulipalpia species; Bd, Blaberus discoidalis; Cf, Ctenocephalides felis; Dm, Drosophila melanogaster; Ed, Ephemera danica; Fc, Folsomia candida; Fo, Frankliniella occidentalis; Lf, Ladona fulva; Lm, Locusta migratoria; Mc, Meinertellus cundinamarcensis; Me, Medauroidea extradentata; Ml, Micropterna lateralis; Ms, Manduca sexta; Oc, Orchesella cincta; Oj, Occasjapyx japonicus; Ph, Pediculus humanus; Px, Papilio xuthus; Rp, Rhodnius prolixus; Ta, Trichoplax adhaerens (outgroup); Tc, Tribolium castaneum; Td, Thermobia domestica; Zn, Zootermopsis nevadensis.
Tsf2 orthologs were identified in the non-insect hexapod species, including a species of two-pronged bristletail (O. japonicus), and two species of springtail (F. candida and O. cincta); therefore, Tsf2 must have arisen prior to the origin of insects (Figure 2). Shorter branch lengths suggest that Tsf2 orthologs are more highly conserved than the other insect transferrins. The Tsf1, Tsf3 and Tsf4 orthologs cluster with the non-Tsf2 sequences from the non-insect hexapods (Figure 2). Within this large cluster, the Tsf3 and Tsf4 groups appear to be more closely related to each other than they are to the Tsf1 group (Figure 2). Tsf1, Tsf3 and Tsf4 orthologs were identified in the silverfish (A. formicaria) and firebrat (T. domestica) but not in the jumping bristletail or the non-insect hexapods; therefore, Tsf1, Tsf3 and Tsf4 must have arisen prior to the zygentoman lineage, probably very early in the process of insect evolution.
Tsf1 orthologs are the only transferrins likely to have a major role in iron homeostasis
The vertebrate transferrins that function in iron homeostasis are extracellular, soluble, iron-binding proteins; therefore, we assumed that insect transferrins that function in iron homeostasis are likely to have similar characteristics (although membrane-bound transferrins could also play a role in iron homeostasis).
Extracellular proteins typically have an amino-terminal signal peptide that targets the protein to the secretory pathway (Nielsen 2017). Because all of the well-studied vertebrate transferrin family members are extracellular proteins (Lambert 2012), we expected that the insect transferrins would be secreted. Consistent with that expectation, we found that all but six of the insect transferrins have a predicted signal peptide (Table 2 and Table S1). Given the possibility of false negatives and missing 5’ sequence information, it seems likely that all of the insect transferrins are secreted.
Table 2.
Predicted signal peptide and membrane association of orthologous groups of transferrins.
| Orthologous Group | Signal Peptide | Membrane Bound |
|---|---|---|
| Tsf1 | yes (15/16) | no (0/16) |
| Tsf2 | yes (18/19) | yes (18/19) |
| Tsf3 | yes (15/18) | yes (17/18) |
| Tsf4 | yes (12/13) | yes (13/13) |
Extracellular proteins can be membrane-bound (attached to the plasma membrane) or soluble (released from the cell). We categorized a transferrin as membrane-bound if it had either a predicted carboxyl-terminal transmembrane region or a GPI anchor, whereas we categorized a transferrin as soluble if it had neither. All of the Tsf1 orthologs were predicted to be soluble proteins (Table 2). In contrast, all but two of the remaining transferrin orthologs were predicted to be membrane-bound, with 42 of the 48 sequences having both a predicted carboxyl-terminal transmembrane region and a GPI anchor (Table 2 and Table S1). It is not obvious whether the two atypical transferrins are actually soluble or if the gene predictions are missing carboxyl-terminal codons.
Next, we used two types of sequence information to predict whether or not each insect transferrin is an iron-binding protein. First, we analyzed sequence alignments of insect and mammalian transferrins to identify conserved iron-binding and carbonate-binding residues. In particular, we looked for the presence of the iron-binding signature (Asp Tyr Tyr His) and carbonate-binding signature (Thr Arg) of the mammalian transferrins (Lambert et al. 2005; Mizutani et al. 2012). Second, we compared each of the insect sequences to the four insect transferrins that are known to bind iron (Huebers et al. 1988; Bartfeld and Law 1990; Gasdaska et al. 1996; Tiklová et al. 2010; Weber et al. 2018). Although the iron-binding and carbonate-binding residues of these three insect transferrins have not been biochemically verified, sequence alignments strongly suggest that they are similar to the mammalian signatures (Lambert et al. 2005; Tiklová et al. 2010; Geiser & Winzerling 2012).
Our analyses indicate that all of the Tsf1 orthologs are likely to be iron-binding proteins. Although the Tsf1 amino-lobe sequences lack a signature iron-binding histidine, and four have an aspartate to glutamate substitution (Figure 3A), biochemical studies have demonstrated that the signature histidine is not essential for iron binding in either insect or mammalian transferrins and that an aspartate to glutamate substitution does not interfere with iron binding (Huebers et al. 1988; Bartfeld & Law 1990; Gasdaska et al. 1996; He et al. 2000; MacGillivray et al. 2000; Weber et al. 2018). Taken together, the data suggest that all the Tsf1 orthologs have amino-lobe iron binding. In contrast, only six of the Tsf1 carboxyl-lobes are likely to bind iron; these six have complete iron-binding and carbonate-binding signatures, whereas the other Tsf1 carboxyl-lobes are missing at least one conserved iron-binding residue and at least one carbonate-binding residue (Figure 3A). Our conclusion is that all of the Tsf1 orthologs bind iron, with some binding one ferric ion and some (mostly from the older insect lineages) binding two.
Figure 3. Tsf1 orthologs are predicted to bind iron.
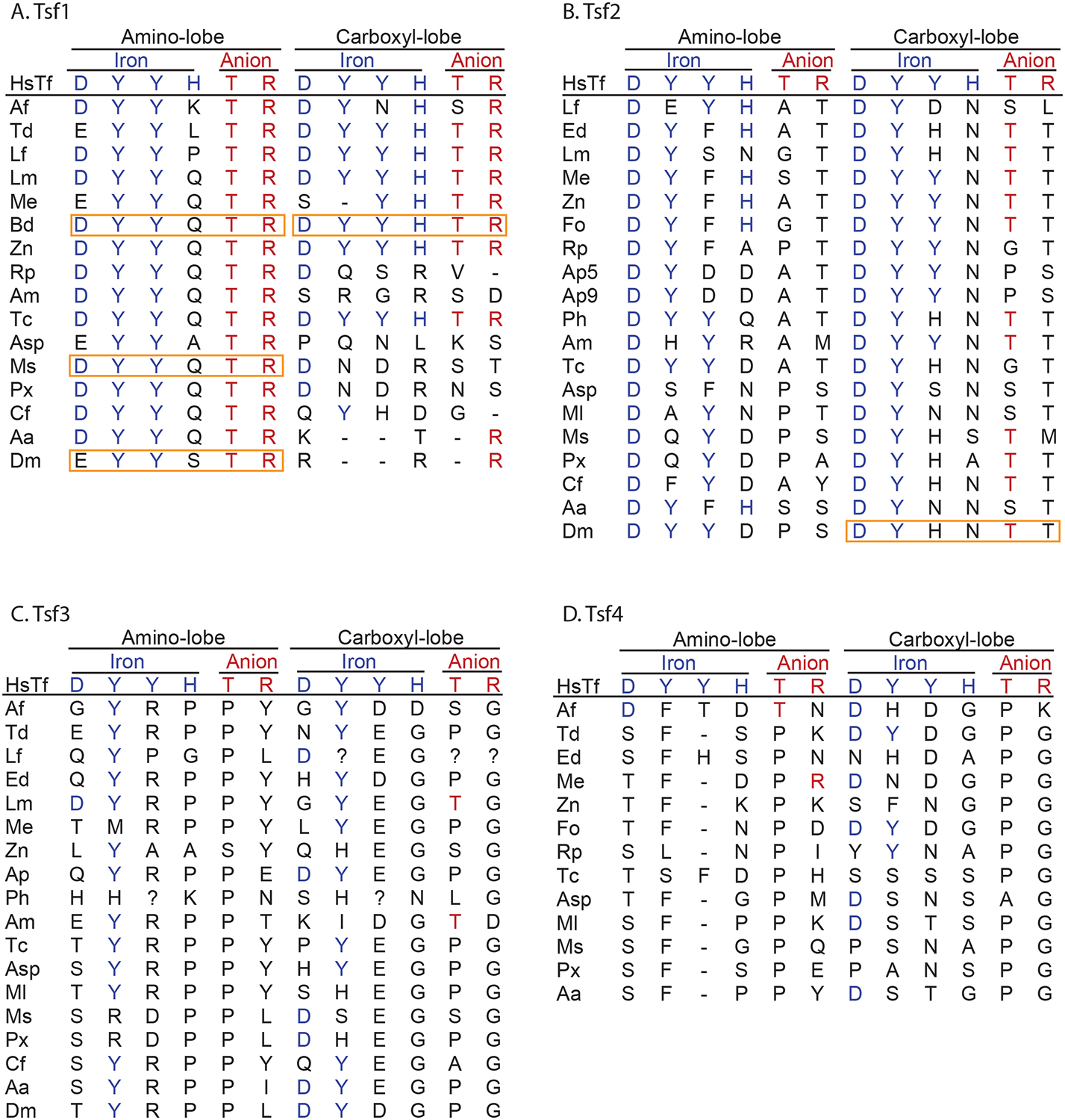
Sequences from each insect transferrin group were aligned with mammalian transferrin sequences to identify positions of putative iron-binding and putative carbonate-binding residues. The residues in these positions are shown. Insect transferrin lobes that are known to bind iron are boxed in orange. Species abbreviations are the same as those used for Figure 2. Human serum transferrin residues are labeled HsTf. Acyrthosiphon pisum has two Tsf2 orthologs, indicated by Ap5 and Ap9 (accession numbers XP_016660805 and XP_001947699). All of the Tsf1 orthologs are predicted to bind to one or two ferric ions, whereas only some of the Tsf2 orthologs and none of the Tsf3 and Tsf4 orthologs are predicted to bind iron.
Predicting whether or not the Tsf2 orthologs bind iron was challenging. The carboxyl-lobe of D. melanogaster Tsf2 is known to bind iron, even though it is lacking two signature iron-binding residues and one signature carbonate-binding residue (Tiklová et al. 2010). The D. melanogaster Tsf2 signature DYHNTT is present in four of the other Tsf2 orthologs and the similar DYYNTT is present in four additional sequences (Figure 3B); therefore, we predict that these eight Tsf2 orthologs also bind iron. Predicting iron-binding in the other Tsf2 orthologs was difficult due to an absence of relevant biochemical information, but their amino acid substitions suggest that they may not have a high affinity for iron. Unlike the carboxyl-lobe sequences, all of the Tsf2 amino-lobe sequences appear to be non-iron-binding (Figure 3B). We suggest that many but not all Tsf2 orthologs are iron-binding proteins.
The Tsf3 and Tsf4 orthologs have almost none of the iron-binding or carbonate-binding signature residues (Figure 3C and 3D); therefore, we predict that these transferrin family members do not bind iron.
Some insect lineages lack a Tsf1 ortholog
Our sequence analyses suggested that Tsf1 is the only orthologous group to have an important role in insect iron homeostasis; therefore, we were surprised to find that four of the species in our analysis had no identified Tsf1 ortholog. These four species included P. humanus (louse), A. pisum (aphid), F. occidentalis (thrips), and Ephemera danica (mayfly). A trivial explanation for this result is that the datasets used for our analyses were incomplete (i.e., that Tsf1 sequences were mistakenly absent in the datasets). A more interesting explanation is that the louse, aphid, thrips, and mayfly lineages may lack a Tsf1 gene. We reasoned that if the explanation was an incomplete dataset, we should find Tsf1 sequences in related species, but if the four lineages are missing the Tsf1 gene, we would not identify Tsf1 in other louse, aphid, thrips, or mayfly datasets. There were not enough data from mayfly species to pursue this line of reasoning, but we were able to search datasets for the other three lineages.
We identified transferrin sequences from 23 species of lice (Order Psocodea), including parasitic lice, booklice and barklice, and then did a phylogenetic analysis to evaluate whether any of the louse transferrins were Tsf1 orthologs. We found that all of the louse transferrins clustered with either the Tsf2, Tsf3 or Tsf4 groups (Figure S1); therefore, we conclude that lice are missing a Tsf1 ortholog. We also did a phylogenetic analysis of transferrins from seven species of aphid (order Hemiptera) and eight species of thrips (order Thysanoptera). We found that the aphid sequences clustered with the Tsf2 and Tsf3 groups, and the thrips sequences clustered with the Tsf2 and Tsf4 groups (Figures S2 and S3). These results suggest that aphids lack Tsf1 and Tsf4 and that thrips lack Tsf1 and Tsf3.
Within the order Hemiptera, the bug R. prolixus (suborder Heteroptera) has a Tsf1 ortholog whereas aphids (suborder Sternorrhyncha) apparently do not. To evaluate which types of hemipteran insects have a Tsf1 ortholog, we searched hemipteran datasets to identify putative Tsf1 sequences and then performed a phylogenetic analysis to verify Tsf1 orthology. We identified Tsf1 in ten hemipteran species: six bugs (suborder Heteroptera), two planthoppers (suborder Fulgoromorpha), one leafhopper (suborder Cicadamorpha), and one psyllid (suborder Sternorrhyncha) (Figure S4). We conclude that while aphids lack a Tsf1 gene, other hemipteran species, including the closely related psyllids, have a Tsf1 ortholog.
Discussion
A previous phylogenetic study that included transferrin sequences from five insect orders found that there are four orthologous groups of insect transferrins (Bai et al. 2016). We have extended these previous findings by performing a phylogenetic analysis of transferrins from 16 orders of insects. Our analysis supports the results of the previous study. We identified only two insect transferrins that did not cluster with one of the four groups, and they were both from the oldest insect lineage in our analysis. The previous study noted that one insect sequence, from the termite Z. nevadensis, was not orthologous to the others (Bai et al. 2016). To explain this outlying sequence, the authors suggested that Z. nevadensis had acquired an ortholog of a vertebrate serum transferrin by horizontal transfer; however, this termite sequence (accession #KDR19735.1) is actually the carboxyl-terminal part of Z. nevadensis Tsf4 and thus is not a unique sequence. Based on the results of our study and the one by (Bai et al. 2016), we conclude that almost all insect transferrins fall into one of four orthologous groups.
The Tsf2 orthologs appear to be more highly conserved than the other insect transferrins, given their shorter branch lengths in the phylogenetic tree. In addition, the Tsf2 group appears to be the most ancient of the insect transferrin groups. Our study demonstrates that Tsf2 must have arisen prior to the origin of insects, and this conclusion is consistent with the previous finding that insect Tsf2 sequences and flatworm melanotransferrin-like sequences cluster together in a phylogenetic tree that includes insects and flatworms as representative invertebrates (Bai et al. 2016). Many Tsf2 sequences are annotated as melanotransferrin-like. Although insect Tsf2 orthologs and vertebrate melanotransferrins are not orthologous proteins (Geiser & Winzerling 2012; Bai et al. 2016), overexpression of mouse melanotransferrin partially rescued a Tsf2 null phenotype in D. melanogaster, suggesting that Tsf2 and mammalian melanotransferrin have at least one shared function (Tiklová et al. 2010). D. melanogaster Tsf2 is a membrane-associated protein that functions in septate junction formation (Tiklová et al. 2010; Hall et al. 2014). It is unknown whether the other insect Tsf2 orthologs have a similar function, but their high sequence conservation and their predicted membrane association suggest that they do. The role of iron in Tsf2 function is unclear. D. melanogaster Tsf2 binds iron in its carboxyl-lobe, and a predicted iron-binding tyrosine residue is required for Tsf2 function (Tiklová et al. 2010). On the other hand, the putative iron-binding residues of D. melanogaster Tsf2 are not highly conserved in other Tsf2 orthologs, and we predict that some Tsf2 orthologs do not actually bind iron. In addition, while mouse melanotransferrin and D. melanogaster Tsf2 appear to have a similar function, mouse melanotransferrin binds iron in its amino-lobe, whereas D. melanogaster Tsf2 binds iron in its carboxyl-lobe (Tiklová et al. 2010; Suryo Rahmanto et al. 2012). Clearly, more information is needed about the role of iron binding by Tsf2 orthologs; however, the results from this and previous studies suggest that the Tsf2 orthologs are more ancient and more highly conserved than the Tsf1, Tsf3 and Tsf4 orthologs, and that they are likely to be membrane-associated proteins with a role in septate junction formation.
Little previous information was available about the Tsf3 and Tsf4 orthologs. The presence of Tsf3 and Tsf4 in A. formicaria and T. domestica suggest that these genes evolved early in insect evolution, prior to the emergence of the zygentoman lineage. We identified a Tsf3 ortholog in all insect species analyzed except R. prolixus (true bug), F. occidentalis (thrips) and M. cundinamarcensis (jumping bristletail); therefore, Tsf3 is present in a diverse set of insect species. Tsf4 was identified in only 13 of 20 species; therefore, it is likely that Tsf4 was lost in multiple insect lineages. Because Tsf3 and Tsf4 are predicted to be membrane-bound, and neither is expected to bind iron, we conclude that Tsf3 and Tsf4 are unlikely to be involved in iron homeostasis.
Most studies of insect transferrins have focused on the Tsf1 orthologs, which are present in hemolymph and other extracellular fluids (Geiser & Winzerling 2012; Simmons et al. 2013; Qu et al. 2014; Zhang et al. 2014; Bonilla et al. 2015; Hattori et al. 2015; Brummett et al. 2017). The Tsf1 orthologs that have been biochemically characterized were found to bind iron, and studies suggest that they participate in iron transport, immunity, and protection from oxidative stress (Huebers et al. 1988; Huebers et al. 1988; Bartfeld & Law 1990; Kurama et al. 1995; Gasdaska et al. 1996; Hirai et al. 2000; Lee et al. 2006; Kim et al. 2008; Brummett et al. 2017; Xiao et al. 2019). The results of our phylogenetic and primary sequence analyses, which predict that the Tsf1 orthologs are soluble, iron-binding proteins, are compatible with the conclusions of these previous studies. Because the Tsf1 orthologs appear to be the only transferrins that function in iron homeostasis, we were surprised to find that lice, aphids and thrips lack Tsf1. Given their different predicted biochemical characteristics, it seems improbable that Tsf2, Tsf3 or Tsf4 could compensate for the loss of Tsf1; therefore, our study suggests the interesting possibility that some insect lineages have evolved iron homeostatic mechanisms that do not involve transferrins. In those insects, the iron-binding protein ferritin is likely to play a major role in iron transport (Tang & Zhou, 2013), but, because binding of iron to ferritin involves ferrous rather than ferric ions, it is not likely that ferritin could directly substitute for Tsf1 in immune-related iron sequestration or in protection against iron-induced oxidative stress. Future research will be needed to learn how lice, aphids and thrips have have evolved to accomplish these functions without transferrins.
Supplementary Material
Figure S1. Lice lack a Tsf1 ortholog. A phylogenetic tree of insect transferrins, including transferrin sequences from 23 louse species, is shown. Louse accession numbers are preceded by the word LOUSE and are in red text. Species abbreviations for non-louse sequences are the same as those used for Figure 2. Louse species abbreviations are as follows: Asp, Aaroniella sp.; Aj, Amphipsocus japonicus; Bt, Badonnelia titei; Bsp, Bertkauia sp.; Cc, Campanulotes compar; Cg, Cerobasis guestfalica; Cc, Columbicola columbae; Csp, Craspedorrhynchus sp., Eh, Echmepteryx hageni; Ek, Elipsocus kuriliensis; Ge, Geomydoecus ewingi; Gc, Graphopsocus cruciatus; Hc, Hemipsocus chloroticus; Hs, Heterocaecilius solocipennis; La, Lachesilla abiesicola; Mr, Matsumuraiella radiopicta; Mu, Mesopsocus unipunctatus; Np, Neoblaste papillosa; Ph, Pediculus humanus; Pp, Peripsocus phaeopterus; Pj, Ptycta johnsoni; Sj, Stimulopalpus japonicus; Vb, Valenzuela badiostigma.
Figure S2. Aphids lack a Tsf1 and Tsf4 ortholog. A phylogenetic tree of insect transferrins, including transferrin sequences from seven aphid species, is shown. Aphid accession numbers are preceded by the word APHID and are in red text. Species abbreviations for non-aphid sequences are the same as those used for Figure 2. Aphid species abbreviations are as follows: Ap, Acyrthosiphon pisum; Ag, Aphis gossypii; Dn, Diuraphis noxia; Ms, Melanaphis sacchari; Mp, Myzus persicae; Rm, Rhopalosiphum maidis; Sf, Sipha flava.
Figure S3. Thrips lack a Tsf1 and Tsf3 ortholog. A phylogenetic tree of insect transferrins, including transferrin sequences from eight thrips species, is shown. Thrips accession numbers are preceded by the designation THRIP and are in red text. Species abbreviations for non-thrips sequences are the same as those used for Figure 2. Thrips species abbreviations are as follows: Fc, Frankliniella cephalica; Fo, Frankliniella occidentalis; Fv, Franklinothrips vespiformis; Gf, Gynaikothrips ficorum; Ok, Orothrips kelloggi; Psp, Phlaeothripidae sp; Tp, Thrips palmi, Tt, Thrips tabaci.
Figure S4. Tsf1 orthologs were identified in non-aphid hemipterans. A phylogenetic tree of insect transferrins, including Tsf1 sequences from 10 hemipteran species is shown. Hemipteran Tsf1 accession numbers are preceded by “HEM” and are in green text. Non-aphid hemipteran species abbreviations are as follows: Dc, Diaphorina citri (psyllid), Hh, Halyomorpha halys (bug), Ls, Laodelphax striatellus (planthopper), Ld, Lethocerus distinctifermur (bug), Nc, Nephotettix cincticeps (leafhopper), Nl, Nilaparvata lugens (planthopper), Pp, Pristhesancus plagipennis (bug), Pa, Pyrrhocoris apterus (bug), Rp, Rhodnius prolixus (bug), Rc, Riptortus clavatus (bug), Rpe, Riptortus pedestris (bug). The other species abbreviations are the same as those used for Figure 2.
Acknowledgements
We thank Yoonseong Park for helpful suggestions regarding this work and Averi Baker for her help with identifying some insect transferrin sequences. This work was supported by National Science Foundation Grant 1656388 and National Institute of General Medical Sciences grant R37 GM041247. This is contribution 20-121-J from the Kansas Agricultural Experiment Station.
Footnotes
Disclosure
The authors disclose that they have no conflicts of interest.
References
- Anderson CP, and Leibold EA (2014) Mechanisms of iron metabolism in Caenorhabditis elegans. Frontiers in Pharmacology, 5, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GJ, and Vulpe CD (2009) Mammalian iron transport. Cellular and Molecular Life Sciences, 66, 3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Qiao M, Zheng R, Deng C, Mei S, and Chen W (2016) Phylogenomic analysis of transferrin family from animals and plants. Comparative Biochemistry and Physiology - Part D: Genomics & Proteomics, 17, 1–8. [DOI] [PubMed] [Google Scholar]
- Bartfeld NS, and Law JH (1990) Isolation and molecular cloning of transferrin from the tobacco hornworm, Manduca sexta: sequence similarity to the vertebrate transferrins. Journal of Biological Chemistry, 265, 21684–21691. [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, and Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. Journal of Molecular Biology, 340, 783–795. [DOI] [PubMed] [Google Scholar]
- Bonilla ML, Todd C, Erlandson M, and Andres J (2015) Combining RNA-seq and proteomic profiling to identify seminal fluid proteins in the migratory grasshopper Melanoplus sanguinipes (F). BMC Genomics, 16, 1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummett LM, Kanost MR, and Gorman MJ (2017) The immune properties of Manduca sexta transferrin. Insect Biochemistry and Molecular Biology, 81, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkov B, and Georgieva T (2006) Insect iron binding proteins: insights from the genomes. Insect Biochemistry and Molecular Biology, 36, 300–309. [DOI] [PubMed] [Google Scholar]
- Fankhauser N, and Mäser P (2005) Identification of GPI anchor attachment signals by a Kohonen self-organizing map. Bioinformatics, 21, 1846–1852. [DOI] [PubMed] [Google Scholar]
- Farnaud S, and Evans RW (2003) Lactoferrin—a multifunctional protein with antimicrobial properties. Molecular Immunology, 40, 395–405. [DOI] [PubMed] [Google Scholar]
- Frazer DM, and Anderson GJ (2014) The regulation of iron transport. BioFactors, 40, 206–214. [DOI] [PubMed] [Google Scholar]
- Galay RL, Umemiya-Shirafuji R, Mochizuki M, Fujisaki K, and Tanaka T (2015) Iron metabolism in hard ticks (Acari: Ixodidae): the antidote to their toxic diet. Parasitology International, 64, 182–189. [DOI] [PubMed] [Google Scholar]
- Gasdaska JR, Law JH, Bender CJ, and Aisen P (1996) Cockroach transferrin closely resembles vertebrate transferrins in its metal ion-binding properties: a spectroscopic study. Journal of Inorganic Biochemistry, 64, 247–258. [DOI] [PubMed] [Google Scholar]
- Geiser DL, and Winzerling JJ (2012) Insect transferrins: multifunctional proteins. Biochimica et Biophysica Acta - General Subjects, 1820, 437–451. [DOI] [PubMed] [Google Scholar]
- Giansanti F, Leboffe L, Pitari G, Ippoliti R, and Antonini G (2012) Physiological roles of ovotransferrin. Biochimica et Biophysica Acta – General Subjects, 1820, 218–225. [DOI] [PubMed] [Google Scholar]
- Graça-Souza AV, Maya-Monteiro C, Paiva-Silva GO, Braz GRC, Paes MC, Sorgine MHF, et al. (2006) Adaptations against heme toxicity in blood-feeding arthropods. Insect Biochemistry and Molecular Biology, 36, 322–335. [DOI] [PubMed] [Google Scholar]
- Hall S, Bone C, Oshima K, Zhang L, McGraw M, Lucas B, et al. (2014) Macroglobulin complement-related encodes a protein required for septate junction organization and paracellular barrier function in Drosophila. Development, 141, 889–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MC, Jongepier E, Robertson HM, Arning N, Bitard-Feildel T, Chao H, et al. (2018) Hemimetabolous genomes reveal molecular basis of termite eusociality. Nature Ecology & Evolution, 2, 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M, Komatsu S, Noda H, and Matsumoto Y (2015) Proteome analysis of watery saliva secreted by green rice leafhopper, Nephotettix cincticeps. PloS One, 10, e0123671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He QY, Mason AB, Pakdaman R, Chasteen ND, Dixon BK, Tam BM, et al. (2000) Mutations at the histidine 249 ligand profoundly alter the spectral and iron-binding properties of human serum transferrin N-lobe. Biochemistry, 39, 1205–1210. [DOI] [PubMed] [Google Scholar]
- Hirai M, Watanabe D, and Chinzei Y (2000) A juvenile hormone-repressible transferrin-like protein from the bean bug, Riptortus clavatus: cDNA sequence analysis and protein identification during diapause and vitellogenesis. Archives of Insect Biochemistry and Physiology, 44, 17–26. [DOI] [PubMed] [Google Scholar]
- Hofmann K, and Stoffel W (1993) TMbase - A database of membrane spanning proteins segments. Nucleic Acids Research, 31, 406–409. [Google Scholar]
- Huebers HA, Huebers E, Finch CA, Webb BA, Truman JW, Riddiford LM, et al. (1988) Iron binding proteins and their roles in the tobacco hornworm, Manduca sexta (L.). Journal of Comparative Physiology B: Biochemical, Systemic, and Environmental Physiology, 158, 291–300. [DOI] [PubMed] [Google Scholar]
- Inward D, Beccaloni G, and Eggleton P (2007) Death of an order: a comprehensive molecular phylogenetic study confirms that termites are eusocial cockroaches. Biology Letters, 3, 331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BY, Lee KS, Choo YM, Kim I, Je YH, Woo SD, et al. (2008) Insect transferrin functions as an antioxidant protein in a beetle larva. Comparative Biochemistry and Physiology - Part B: Biochemistry and Molecular Biology, 150, 161–169. [DOI] [PubMed] [Google Scholar]
- Kosman DJ (2010) Redox cycling in iron uptake, efflux, and trafficking. The Journal of Biological Chemistry, 285, 26729–26735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, and Tamura K (2018) MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Molecular Biology and Evolution, 35, 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurama T, Kurata S, and Natori S (1995) Molecular characterization of an insect transferrin and its selective incorporation into eggs during oogenesis. European Journal of Biochemistry, 228, 229–235. [PubMed] [Google Scholar]
- Lambert LA (2012) Molecular evolution of the transferrin family and associated receptors. Biochimica et Biophysica Acta - General Subjects, 1820, 244–255. [DOI] [PubMed] [Google Scholar]
- Lambert LA, Perri H, Halbrooks PJ, and Mason AB (2005) Evolution of the transferrin family: conservation of residues associated with iron and anion binding. Comparative Biochemistry and Physiology - Part B, Biochemistry & Molecular Biology, 142, 129–141. [DOI] [PubMed] [Google Scholar]
- Lee KS, Kim BY, Kim HJ, Seo SJ, Yoon HJ, Choi YS, et al. (2006) Transferrin inhibits stress-induced apoptosis in a beetle. Free Radical Biology & Medicine, 41, 1151–1161. [DOI] [PubMed] [Google Scholar]
- Li H, Leavengood JM, Chapman EG, Burkhardt D, Song F, Jiang P, et al. (2017) Mitochondrial phylogenomics of Hemiptera reveals adaptive innovations driving the diversification of true bugs. Proceedings of the Royal Society B: Biological Sciences, 284, 20171223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke M, and Nichol H (1992) Iron economy in insects: transport, metabolism, and storage. Annual Review of Entomology, 37, 195–215. [Google Scholar]
- MacGillivray RT, Bewley MC, Smith CA, He QY, Mason AB, Woodworth RC, et al. (2000) Mutation of the iron ligand His 249 to Glu in the N-lobe of human transferrin abolishes the dilysine ‘trigger’ but does not significantly affect iron release. Biochemistry, 39, 1211–1216. [DOI] [PubMed] [Google Scholar]
- Mandilaras K, Pathmanathan T, and Missirlis F (2013) Iron absorption in Drosophila melanogaster. Nutrients, 5, 1622–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misof B, Liu S, Meusemann K, Peters RS, Donath A, Mayer C, et al. (2014). Phylogenomics resolves the timing and pattern of insect evolution. Science, 346, 763–767. [DOI] [PubMed] [Google Scholar]
- Mizutani K, Toyoda M, and Mikami B (2012) X-ray structures of transferrins and related proteins. Biochimica et Biophysica Acta - General Subjects, 1820, 203–211. [DOI] [PubMed] [Google Scholar]
- Nakai K, and Horton P (1999) PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends in Biochemical Sciences, 24, 34–36. [DOI] [PubMed] [Google Scholar]
- NCBI Resource Coordiators (2016) Database resources of the National Center for Biotechnology Information. Nucleic Acids Research, 44, D7–D19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H (2017) Predicting secretory proteins with SignalP. Methods in Molecular Biology, 1611, 59–73. [DOI] [PubMed] [Google Scholar]
- Ong ST, Ho JZS, Ho B, and Ding JL (2006) Iron-withholding strategy in innate immunity. Immunobiology, 211, 295–314. [DOI] [PubMed] [Google Scholar]
- Poelchau M, Childers C, Moore G, Tsavatapalli V, Evans J, Lee C-Y, et al. (2015). The i5k Workspace@NAL—enabling genomic data access, visualization and curation of arthropod genomes. Nucleic Acids Research, 43, D714–D719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu M, Ma L, Chen P, and Yang Q (2014) Proteomic analysis of insect molting fluid with a focus on enzymes involved in chitin degradation. Journal of Proteome Research, 13, 2931–2940. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology, 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons LW, Tan Y-F, and Millar AH (2013) Sperm and seminal fluid proteomes of the field cricket Teleogryllus oceanicus: identification of novel proteins transferred to females at mating. Insect Molecular Biology, 22, 115–130. [DOI] [PubMed] [Google Scholar]
- Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, et al. (2008) The Trichoplax genome and the nature of placozoans. Nature, 454, 955–960. [DOI] [PubMed] [Google Scholar]
- Suryo Rahmanto Y, Bal S, Loh KH, Yu Y, and Richardson DR (2012) Melanotransferrin: search for a function. Biochimica et Biophysica Acta – General Subjects, 1820, 237–243. [DOI] [PubMed] [Google Scholar]
- Tang X, and Zhou B (2013) Iron homeostasis in insects: Insights from Drosophila studies. IUBMB Life, 65, 863–872. [DOI] [PubMed] [Google Scholar]
- Tiklová K, Senti K-A, Wang S, Gräslund A, and Samakovlis C (2010) Epithelial septate junction assembly relies on melanotransferrin iron binding and endocytosis in Drosophila. Nature Cell Biology, 12, 1071–1077. [DOI] [PubMed] [Google Scholar]
- Walter-Nuno AB, Taracena ML, Mesquita RD, Oliveira PL, and Paiva-Silva GO (2018) Silencing of iron and heme-related genes revealed a paramount role of iron in the physiology of the hematophagous vector Rhodnius prolixus. Frontiers in Genetics, 9, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Lothrop AP, James NG, Griffiths TAM, Lambert LA, Leverence R, et al. (2007) A novel murine protein with no effect on iron homoeostasis is homologous with transferrin and is the putative inhibitor of carbonic anhydrase. Biochemical Journal, 406, 85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J, Brummett L, Geisbrecht B, Kanost M, and Gorman M (2018) A biochemical and structural look into the functional role of transferrin in D. melanogaster. Faseb Journal, 32, 1_supplement: 652.39. [Google Scholar]
- Xiao G, Liu Z-H, Zhao M, Wang H-L, and Zhou B (2019) Transferrin 1 functions in iron trafficking and genetically interacts with ferritin in Drosophila melanogaster. Cell Reports, 26, 748–758. [DOI] [PubMed] [Google Scholar]
- Zhang J, Lu A, Kong L, Zhang Q, and Ling E (2014) Functional analysis of insect molting fluid proteins on the protection and regulation of ecdysis. Journal of Biological Chemistry, 289, 35891–35906. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Lice lack a Tsf1 ortholog. A phylogenetic tree of insect transferrins, including transferrin sequences from 23 louse species, is shown. Louse accession numbers are preceded by the word LOUSE and are in red text. Species abbreviations for non-louse sequences are the same as those used for Figure 2. Louse species abbreviations are as follows: Asp, Aaroniella sp.; Aj, Amphipsocus japonicus; Bt, Badonnelia titei; Bsp, Bertkauia sp.; Cc, Campanulotes compar; Cg, Cerobasis guestfalica; Cc, Columbicola columbae; Csp, Craspedorrhynchus sp., Eh, Echmepteryx hageni; Ek, Elipsocus kuriliensis; Ge, Geomydoecus ewingi; Gc, Graphopsocus cruciatus; Hc, Hemipsocus chloroticus; Hs, Heterocaecilius solocipennis; La, Lachesilla abiesicola; Mr, Matsumuraiella radiopicta; Mu, Mesopsocus unipunctatus; Np, Neoblaste papillosa; Ph, Pediculus humanus; Pp, Peripsocus phaeopterus; Pj, Ptycta johnsoni; Sj, Stimulopalpus japonicus; Vb, Valenzuela badiostigma.
Figure S2. Aphids lack a Tsf1 and Tsf4 ortholog. A phylogenetic tree of insect transferrins, including transferrin sequences from seven aphid species, is shown. Aphid accession numbers are preceded by the word APHID and are in red text. Species abbreviations for non-aphid sequences are the same as those used for Figure 2. Aphid species abbreviations are as follows: Ap, Acyrthosiphon pisum; Ag, Aphis gossypii; Dn, Diuraphis noxia; Ms, Melanaphis sacchari; Mp, Myzus persicae; Rm, Rhopalosiphum maidis; Sf, Sipha flava.
Figure S3. Thrips lack a Tsf1 and Tsf3 ortholog. A phylogenetic tree of insect transferrins, including transferrin sequences from eight thrips species, is shown. Thrips accession numbers are preceded by the designation THRIP and are in red text. Species abbreviations for non-thrips sequences are the same as those used for Figure 2. Thrips species abbreviations are as follows: Fc, Frankliniella cephalica; Fo, Frankliniella occidentalis; Fv, Franklinothrips vespiformis; Gf, Gynaikothrips ficorum; Ok, Orothrips kelloggi; Psp, Phlaeothripidae sp; Tp, Thrips palmi, Tt, Thrips tabaci.
Figure S4. Tsf1 orthologs were identified in non-aphid hemipterans. A phylogenetic tree of insect transferrins, including Tsf1 sequences from 10 hemipteran species is shown. Hemipteran Tsf1 accession numbers are preceded by “HEM” and are in green text. Non-aphid hemipteran species abbreviations are as follows: Dc, Diaphorina citri (psyllid), Hh, Halyomorpha halys (bug), Ls, Laodelphax striatellus (planthopper), Ld, Lethocerus distinctifermur (bug), Nc, Nephotettix cincticeps (leafhopper), Nl, Nilaparvata lugens (planthopper), Pp, Pristhesancus plagipennis (bug), Pa, Pyrrhocoris apterus (bug), Rp, Rhodnius prolixus (bug), Rc, Riptortus clavatus (bug), Rpe, Riptortus pedestris (bug). The other species abbreviations are the same as those used for Figure 2.


