Abstract
Tumor hypoxia contributes to the development of resistance to chemotherapeutic drugs in several human cancer cell lines. Atovaquone, an anti-malaria drug approved by the US Food and Drug Administration, has recently demonstrated anti-cancer effects in vitro and in vivo in several cancer models. To assess the potential of atovaquone as an anti-cancer agent under hypoxia in colorectal carcinoma, EpCAM+CD44+ colon cancer stem cells were isolated from HCT-116 human colon cancer cells through magnetic-activated cell sorting. The efficacy of atovaquone on cytotoxicity, tumorsphere formation, apoptosis, invasion and cell-cycle progression under hypoxic conditions were evaluated. MTS assays indicated that atovaquone inhibited the proliferation of EpCAM+CD44+ HCT-116 cells with a half-maximal inhibitory concentration of 15 µM. Atovaquone inhibited tumorsphere formation and cell proliferation by causing cell-cycle arrest in S-phase, which induced apoptosis of EpCAM+CD44+ HCT-116 cells, as detected by Annexin V-FITC/PI double staining assays, and caused mitochondrial membrane potential depolarization, as determined by a JC-1 staining assay. Reverse transcription-quantitative PCR demonstrated increased expression of Bax and downregulation of Bcl-2. Transwell invasion assays indicated that atovaquone inhibited the invasiveness of EpCAM+CD44+ HCT-116 cells under hypoxia, which was associated with upregulation of MMP-2 and -9 and increased expression of tissue inhibitor of MMPs (TIMP)-1. Taken together, atovaquone reduced the tumorsphere formation and invasion ability of EpCAM+CD44+ HCT-116 cells, at least in part by increasing the expression of TIMP-1 and downregulating the expression of MMP-2 and -9, as well as the cells' viability by inducing cell-cycle arrest in S-phase and induction of apoptosis via the Bcl-2/Bax pathway under hypoxic conditions. Further studies are warranted to explore the mechanisms of action of atovaquone as a promising anticancer agent in the treatment of colorectal carcinoma.
Keywords: colon cancer, cancer stem cells, atovaquone, hypoxia, in vitro
Introduction
Colorectal carcinoma is the second most common type of cancer. Although conventional treatments such as surgical resection, chemotherapy and radiation therapy have decreased the mortality rate of colon cancer, it has been projected that there will be 148,950 estimated new cases and 53,200 estimated deaths in the US in 2020(1). Accumulating evidence has indicated that the existence of cancer stem cells (CSCs) in human colorectal carcinoma is one of the major causes of treatment failure (2,3). CSCs represent a subpopulation of cells within cancer cells with an inherent capacity for unlimited self-renewal and heterogeneous-lineage differentiation (4). They display distinct immunophenotypes resistant to conventional chemotherapeutics and radiation therapy and are responsible for tumorigenesis, relapse and metastasis (5-9). Hence, the elimination of CSCs in colon cancer may be a novel approach for effective cancer management.
Reduced intratumoral oxygen tension (hypoxia) is a feature that is common in a majority of solid tumors and occurs as a result of the abnormal vasculature's limited capacity to deliver oxygen, which cannot meet the demands of the rapidly proliferating cancer cells (10). Evidence has indicated that up to 60% of locally advanced solid tumors exhibit substantial heterogeneity in the tissue oxygenation levels within the tumor (11). In hypoxia, cancer cells are able to adapt to their new microenvironment via multiple mechanisms: Inhibition of apoptosis via Bcl-2 family proteins (12), upregulation of MMPs (13), and stimulation of angiogenesis by vascular endothelial growth factor (14). Hypoxia also mediates resistance to radiotherapy and chemotherapy, resulting in poor prognosis (15). Several commercially available drugs such as cisplatin, mitomycin C and sorafenib have been associated with hypoxia-induced impairment of chemosensitivity in several human cancer cell lines (16). Therefore, finding a chemotherapeutic agent that is effective in a hypoxic environment is crucial in cancer therapy.
Atovaquone (ATO) is an anti-parasitic drug approved by the US Food and Drug Administration in the treatment of malaria and pneumocystis pneumonia. ATO is structurally similar to coenzyme Q10 and competitively inhibits its binding to the cytochrome bc1 complex, resulting in mitochondrial membrane potential (MMP) collapse and parasite death (17). ATO has attracted attention as a novel anticancer agent for targeting various types of cancer cell (18-22). Ashton et al (23) revealed that ATO reduces the oxygen consumption rate by inhibiting mitochondrial respiration complex III activity, reduces hypoxia in both spheroids and xenografted tumors and causes tumor growth delay in combination with radiation. However, studies on ATO targeting CSCs are limited and the anti-cancer effects of ATO on hypoxic colon CSCs have not been previously investigated. In the present study, epithelial cell adhesion molecule (EpCAM) and CD44, which are robust makers of human colon CSCs (2), were used to isolate EpCAM+CD44+ cells from the HCT-116 colon cancer cell line and the potential of ATO in eradicating colon CSCs under hypoxic conditions was investigated. The present results demonstrated that ATO inhibited cell growth and invasiveness, induced apoptosis and caused S-phase arrest of EpCAM+CD44+ HCT-116 cells under hypoxic conditions.
Materials and methods
Cell lines and culture
The human HCT-116 colon cancer cell line was purchased from the Cell Bank of the Chinese Academy of Sciences and was cultured in high-glucose DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) at 37˚C with 5% CO2. EpCAM+CD44+ HCT-116 cells were cultured in serum-free DMEM/F12 (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 20 ng/ml epidermal growth factor (EGF), 20 ng/ml basic fibroblast growth factor (bFGF; both from PeproTech, Inc.) and 2% B27 (Gibco; Thermo Fisher Scientific, Inc.) at 37˚C with 5% CO2. For hypoxic incubation, cells were cultured in a hypoxic chamber at 37˚C in a humidified atmosphere of 5% CO2, 1% O2 and 94% N2.
Magnetic-activated cell sorting and FACS
EpCAM+CD44+ HCT-116 cells were obtained by magnetic-activated cell sorting as previously described (24). In brief, dissociated HCT-116 colon cancer cells were labeled with biotin-conjugated EpCAM antibodies (1:50; cat. no. 13-9326-82; eBioscience; Thermo Fisher Scientific, Inc.). The cells were magnetically separated using a CELLection Biotin Binder kit (Invitrogen; Thermo Fisher Scientific, Inc.). The sorted EpCAM+ HCT-116 cells were further labeled with biotin-conjugated CD44 antibody (1:50; cat. no. 13-0441-82; eBioscience; Thermo Fisher Scientific, Inc.) and then fractionated using the CELLection Biotin Binder kit. In the meantime, 1x106 dissociated HCT-116 cells and EpCAM+CD44+ HCT-116 cells in 0.1 ml PBS were incubated with FITC-conjugated anti-EpCAM antibody (1:20; cat. no. 324203) and phycoerythrin-conjugated anti-CD44 antibody (1:20; cat. no. 338807; both from BioLegend, Inc.) in the dark for 10 min at 4˚C. The cells were washed with PBS and then acquired and analyzed using a Beckman Coulter FC500 Flow Cytometer with the CellQuest Pro software (version 6.0; BD Biosciences) to determine the proportion of EpCAM+CD44+ cells.
Tumorsphere-formation assay
In brief, a single-cell suspension of sorted EpCAM+CD44+ HCT-116 cells was cultured in serum-free DMEM/F12 supplemented with 20 ng/ml EGF, 20 ng/ml bFGF and 2% B27. The cells were then seeded on uncoated 6-well culture plates (Corning, Inc.) at a density of 1x104 cells/well. Tumorsphere formation was observed for 4 days and representative images of at least five random fields and were captured using an inverted light microscope (Olympus Corp.) at a magnification of x100.
To evaluate the effect of ATO on tumorsphere formation, a single-cell suspension of EpCAM+CD44+ HCT-116 cells was treated with 15 µM ATO for 3 days under hypoxic conditions, with 50 µM DDP and 0.05% DMSO as a positive and negative control, respectively. The number of tumorspheres was counted under an inverted light microscope (Olympus Corp.) at a magnification of x40.
Serum-induced differentiation
EpCAM+CD44+ HCT-116 cells were resuspended and incubated in DMEM/F12 supplemented with 10% FBS at 37˚C with 5% CO2. Images of cells before and after 48 h of serum induction were acquired using an inverted light microscope (Olympus Corp.) at a magnification of x200.
Reverse transcription-quantitative PCR (RT-qPCR)
To determine the expression of stemness-related genes [OCT-4, SOX-2, Nanog homeobox (NANOG) and C-MYC], total RNA of 1x106 HCT-116 and EpCAM+CD44+ HCT-116 cells was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Complementary DNA was synthesized by RT using a PrimeScript RT kit (Takara Bio, Inc.) according to the manufacturer's instructions and quantified by performing qPCR using the FastStart SYBR Green master mix (Roche Diagnostics, GmbH) on a PikoReal 96 Real-Time PCR system (Thermo Fisher Scientific, Inc.) with the following thermocycling conditions: Initial denaturation for 5 min at 95˚C; and 40 cycles of 95˚C for 5 sec and 60˚C for 30 sec. Likewise, in EpCAM+CD44+ HCT-116 cells treated with 15 µM ATO for 24 h under hypoxic conditions and in the respective controls, the expression levels of apoptosis-associated genes (Bcl-2 and Bax) and invasion-associated genes [MMP-2/-9 and tissue inhibitor of MMPs 1 (TIMP-1)] were evaluated. GAPDH was detected as an internal control. The average Cq values of the target genes were normalized to those of the control genes (∆∆Cq) (25). Primers used for PCR are presented in Table I. All experiments were performed in triplicate.
Table I.
Primer sequences.
| Target gene | Sequence (5'-3') | Product size (bp) |
|---|---|---|
| GAPDH | F: CAGGAGGCATTGCTGATGAT | 138 |
| R: GAAGGCTGGGGCTCATTT | ||
| OCT4 | F: GATGTGGTCCGAGTGTGGTTCTG | 195 |
| R: CGAGGAGTACAGTGCAGTGAAGTG | ||
| NANOG | F: ATGCCTGTGATTTGTGGGCC | 403 |
| R: GCCAGTTGTTTTTCTGCCAC | ||
| c-MYC | F: CACCAGCAGCGACTCTGAGGAG | 239 |
| R: ACTTGACCCTCTTGGCAGCAGG | ||
| SOX2 | F: TCCATGACCAGCTCGCAGA | 152 |
| R: GAGGAAGAGGTAACCACGGG | ||
| MMP-2 | F: GCCTCTCCTGACATTGACCTTGG | 112 |
| R: CACCACGGATCTGAGCGATGC | ||
| MMP-9 | F: GCACCACCACAACATCACCT | 284 |
| R: ACCACAACTCGTCATCGTCG | ||
| TIMP-1 | F: CCTGGCTTCTGGCATCCTGTTG | 162 |
| R: CGCTGGTATAAGGTGGTCTGGTTG | ||
| BCL-2 | F: GGGGAGGATTGTGGCCTTCTTT | 107 |
| R: TAATGTGCAGGTGCCGGTTCAG | ||
| BAX | F: TAACCAAGGTGCCGGAACTGA | 126 |
| R: GGGAGGAGTCTCACCCAACCA |
F, forward; R, reverse; NANOG, Nanog homeobox; TIMP-1, tissue inhibitor of MMPs 1.
MTS assay
EpCAM+CD44+ HCT-116 cells (5x103/well) were seeded in a 96-well plate and cultured under hypoxic conditions. Cells were incubated with ATO at different concentrations (0, 5, 10, 15 and 20 µM) for 24 h. Triplicate wells were set up for each concentration. Cell viability was measured using an MTS kit (Promega Corp.) according to the manufacturer's protocol. The absorbance was measured at 490 nm using a Multiskan GO microplate reader (Thermo Fisher Scientific, Inc.). All experiments were performed at least three times. The cell viability was determined, and the inhibition ratio was calculated using the following formula: Inhibition ratio (%) = (1-optical density of the treatment group/optical density of the solvent control) x100.
Detection of apoptosis by Annexin V-FITC/PI staining
Flow cytometry was performed to analyze cell apoptosis in hypoxia using an Annexin V/PI-FITC kit (Nanjing KeyGen Biotech Co., Ltd.) according to the manufacturer's protocol. In brief, the cells were seeded in a 6-well plate (1x106 cells/well) and cultured under hypoxic conditions. The cells were then treated with ATO (5, 10 or 15 µM) for 24 h, with 50 µM DDP and DMSO used as a positive and negative control, respectively. After washing with PBS, the cells (5x105) were resuspended in binding buffer (500 µl) and stained with Annexin V-FITC (5 µl) and PI (5 µl) in the dark at room temperature for 5 min. The cells were washed with PBS and analyzed within 1 h using a Beckman Coulter FC500 Flow Cytometer with the CellQuest Pro software (version 6.0; BD Biosciences). The experiment was performed for a total of three times.
Detection of apoptosis by JC-1 assay
The MMP was detected by a JC-1 staining assay using the JC-1 Apoptosis Detection kit (Nanjing KeyGen Biotech Co., Ltd.). In brief, the EpCAM+CD44+ HCT-116 cells were dissociated into single cells using TrypLE reagent and then seeded in a 6-well plate (1x106/well). The cells were incubated under hypoxic conditions and treated with 5, 10 or 15 µM ATO for 24 h, with 50 µM DDP and DMSO as a positive and negative control, respectively. After washing with PBS, cells were resuspended in 500 µl binding buffer, and 5x105 cells were stained with JC-1 (5 µl) and incubated in the dark at room temperature for 15 min at 37˚C with 5% CO2 under hypoxic conditions. The cells were then resuspended in incubation buffer (500 µl), washed with PBS and analyzed using a Beckman Coulter FC500 Flow Cytometer with CellQuest Pro software (version 6.0; BD Biosciences). The experiment was performed three times.
Cell-cycle analysis
In brief, 1x106 EpCAM+CD44+ HCT-116 cells treated with 15 µM ATO were incubated under hypoxic conditions for 24 h. The cells were dissociated into single cells using TrypLE reagent and fixed with ice-cold 70% ethanol for 12 h at 4˚C. The cells were pelleted at 500 x g for 5 min at 4˚C and washed by gently resuspending them in 1 ml PBS. After carefully removing the supernatant, the cells were treated with 50 µl RNaseA (100 µg/ml; Nanjing KeyGen Biotech Co., Ltd.) and stained with 200 µl PI (50 µg/ml) at 37˚C in the dark for 30 min. The stained cells were placed on ice in the dark, washed with PBS and analyzed for PI fluorescence using a Beckman Coulter FC500 flow cytometer with CellQuest Pro software (version 6.0; BD Biosciences). The experiment was performed three times.
Transwell invasion assay
An invasion assay was performed using 6.5-mm Transwell plates with sterile 8.0-µm pore polycarbonate membrane inserts (Corning, Inc.) and covered with a thin layer of BD Matrigel (BD Biosciences). In brief, the EpCAM+CD44+ HCT-116 cells (1x105 cells/well) were seeded in a Transwell plate and treated with 15 µM ATO. As positive and negative controls, 50 µM DDP and DMSO were respectively used. DMEM/F12 medium supplemented with 10% FBS was loaded into the bottom chamber through the insert as a chemostatic factor. After incubation for 24 h under hypoxic conditions, the media in the upper and lower chambers were removed and the cells that had invaded the membranes were fixed with 4% paraformaldehyde (500 µl) for 20 min and stained with 0.01% crystal violet for 20 min at room temperature. The cells were counted on an inverted light microscope (Olympus Corp.) and the mean value was determined from counts of five random fields.
Statistical analyses
Values are expressed as the mean ± standard deviation of data from triplicate experiments. GraphPad Prism (version 6.0; GraphPad Software Inc.) was employed for statistical analysis. Student's t-test or one-way ANOVA followed by Dunnett's test was used in the analysis when appropriate. P<0.05 was considered to indicate statistical significance.
Results
Characteristics of isolated EpCAM+CD44+ HCT-116 cells
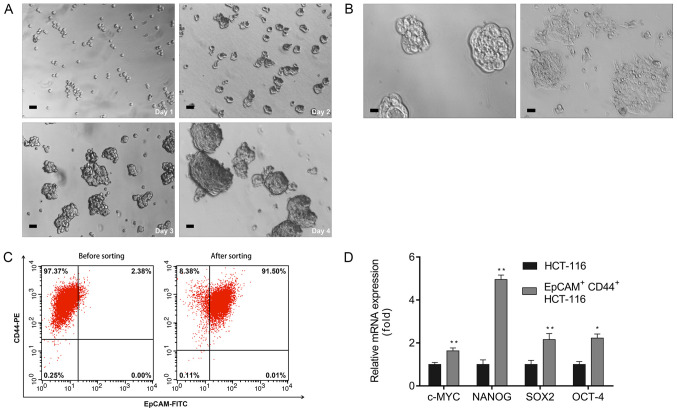
CSCs have a crucial role in tumor initiation, progression and metastasis (26). To study the efficacy of ATO in colon CSCs, the EpCAM+CD44+ cells were isolated from the HCT-116 colon cancer cell line by magnetic-activated cell sorting and the proportion of CSCs and non-CSCs was measured by flow cytometric analysis using EpCAM+ and CD44+ monoclonal antibodies. A single cell suspension of the sorted EpCAM+CD44+ HCT-116 cells cultured in serum-free DMEM/F12 gradually proliferated and formed spheres on day 4 (Fig. 1A) and were considered to be colon CSCs. These spheroid cells switched to adherent confluent cells after adding 10% FBS to the culture medium (Fig. 1B). In addition, flow cytometric analyses revealed that the percentage of EpCAM+CD44+ cells in unsorted and sorted HCT-116 cells was 2.38 and 91.50%, respectively (Fig. 1C). The RT-qPCR results indicated upregulation of pluripotent-associated genes, including C-MYC, NANOG, SOX-2 and OCT-4, in EpCAM+CD44+ HCT-116 cells relative to the HCT-116 cells (Fig. 1D).
Figure 1.
Isolation and characterization of EpCAM+CD44+ HCT-116 cells. (A) Optical micrographs presenting the morphology of EpCAM+CD44+ HCT-116 cells obtained by magnetic-activated cell sorting cultured over 4 days (scale bar, 50 µm). (B) Serum-induced differentiation of EpCAM+CD44+ HCT-116 cells (left) into adherent cells (right). Scale bar, 25 µm. (C) EpCAM+CD44+ cell percentage among HCT-116 cells prior to and after magnetic-activated cell sorting evaluated by flow cytometry. (D) Reverse transcription-quantitative PCR analyses of NANOG, OCT-4, SOX-2 and C-MYC mRNA expression in EpCAM+CD44+ HCT-116 cells. Values are expressed as the mean ± standard deviation of three independent experiments. *P<0.05 and **P<0.01 vs. HCT-116, as determined by Student's t-test for each gene. NANOG, Nanog homeobox; EpCAM, epithelial cell adhesion molecule.
ATO inhibits the viability of EpCAM+CD44+ HCT-116 cells
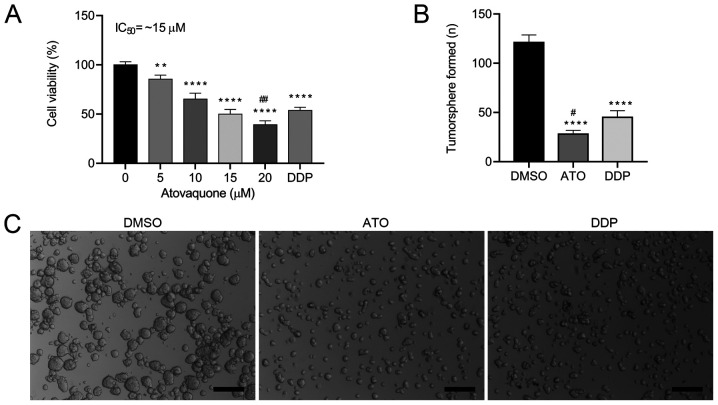
To determine the effects of ATO on cell viability, the EpCAM+CD44+ HCT-116 cells were treated with different concentrations (0-20 µM) of ATO under hypoxic conditions for 24 h. The results indicated that ATO inhibited the proliferation of EpCAM+CD44+ HCT-116 cells in a concentration-dependent manner (Fig. 2A). The proliferation of EpCAM+CD44+ HCT-116 cells was significantly inhibited by 5 and 10 µM ATO and the IC50 values was ~15 µM in hypoxia. When atovaquone was administered with food at the standard regimen of 750 mg twice daily, the average steady-state plasma concentration was ~57 µM (27). Therefore, the obtained IC50 of ATO is pharmacologically achievable and was used for the subsequent experiments. To investigate whether ATO is involved in the tumorigenic activity of the CSCs, a tumorsphere formation assay was performed. ATO treatment induced the formation of a lower number of tumorspheres (Fig. 2B) and smaller tumorspheres (Fig. 2C) in comparison with those of DMSO-treated cells. Likewise, ATO treatment induced the formation of a lower number of tumorspheres, but there were no significant differences in the size of the tumorshperes in comparison with DDP-treated cells. These results suggested that ATO suppressed the tumorsphere-formation capacity of EpCAM+CD44+ HCT-116 cells.
Figure 2.
ATO reduces EpCAM+CD44+ HCT-116 cell viability in hypoxia. (A) EpCAM+CD44+ HCT-116 cells treated with ATO (0-20 µM) for 24 h under hypoxic conditions were assessed for cell viability using an MTS assay. (B and C) ATO (15 µM) inhibited the tumorsphere formation capacity of EpCAM+CD44+ HCT-116 cells under hypoxic conditions. Cells treated with 50 µM DDP used as a positive control. (B) Quantified results and (C) representative microscopy images of the tumorspheres (scale bar, 200 µm). Values are expressed as the mean ± standard deviation of three independent experiments. **P<0.01 and ****P<0.0001 vs. DMSO; #P<0.05 and ##P<0.01 vs. DDP as determined by one-way ANOVA followed by Dunnett's test. DDP, cisplatin; ATO, atovaquone; EpCAM, epithelial cell adhesion molecule.
ATO induced apoptosis of EpCAM+CD44+ HCT-116 cells
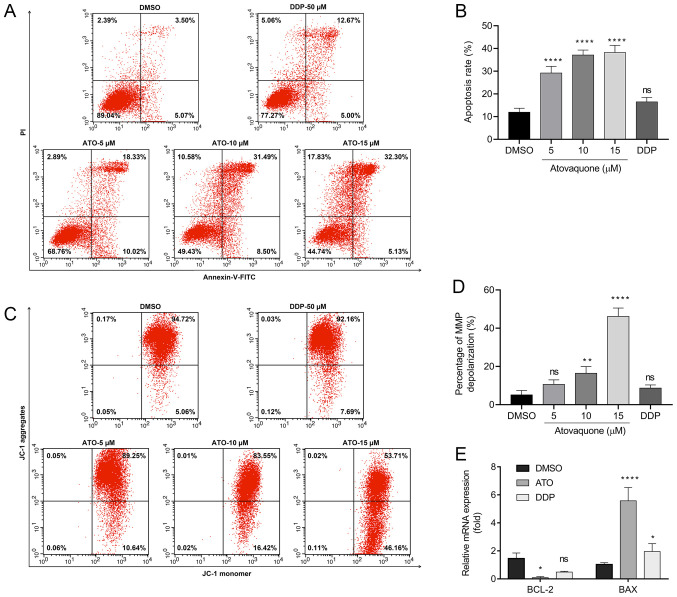
An annexin V-FITC/PI double staining assay and JC-1 assay were performed to determine whether ATO is able to induce apoptosis in EpCAM+CD44+ HCT-116 cells under hypoxic conditions. The results suggested that treatment with 15 µM ATO increased the percentage of Annexin V-positive cells to 38.22±5.18% compared with 8.48±2.75% in the DMSO group and 18.86±2.98% in the DDP group (Fig. 3A and B). Likewise, JC-1 staining revealed that the group treated with 15 µM ATO exhibited loss of the MMP (48.96±4.42%) compared with the DMSO-treated (5.25±1.63%) and DDP-treated groups (8.69±1.71%; Fig. 3C and D). Furthermore, cancer cells are able to adapt to their surrounding hypoxic microenvironment by inhibition of apoptosis via Bcl-2 family proteins. The RT-qPCR results indicated suppression of Bcl-2 expression and upregulation of Bax following treatment of EpCAM+CD44+ HCT-116 cells with ATO under hypoxia (Fig. 3E).
Figure 3.
ATO induces apoptosis of EpCAM+CD44+ HCT-116 cells in hypoxia. (A and B) Flow cytometric analysis of Annexin V/PI staining of EpCAM+CD44+ HCT-116 cells treated with ATO (5, 10 and 15 µM) under hypoxic conditions. (A) Flow cytometry dot plots of cells with Annexin V/FITC and PI fluorescence staining. Apoptotic cells are contained in the lower-right and upper-right quadrants. (B) Quantification of apoptotic cells. (C) Flow cytometric analysis of JC-1 staining assay of EpCAM+CD44+ HCT-116 cells treated with ATO (5, 10 and 15 µM) under hypoxic conditions. (D) Percentage of MMP depolarization. (E) Reverse transcription-quantitative PCR analysis of Bcl-2 and Bax mRNA expression in EpCAM+CD44+ HCT-116 cells treated with ATO (15 µM) under hypoxia. Cells treated with 50 µM DDP were used as the positive control. The relative mRNA expression is presented relative to the DMSO control. Values are expressed as the mean ± standard deviation of three independent experiments. *P<0.05, and ****P<0.0001 vs. DMSO as determined by one-way ANOVA by following Dunnett's test. ATO, atovaquone; DDP, cisplatin; ns, no significance; EpCAM, epithelial cell adhesion molecule; MMP, mitochondrial membrane potential.
ATO induces cell-cycle arrest of EpCAM+CD44+ HCT-116 cells
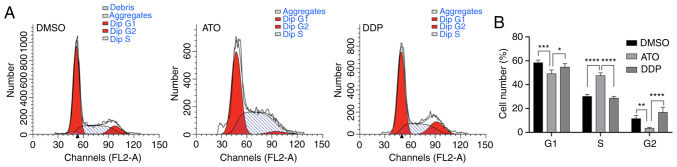
Flow cytometric analysis of the cell-cycle profile of EpCAM+CD44+ HCT-116 cells after treatment with 15 µM ATO revealed that the number of cells in S phase significantly increased compared with that of the untreated cells (P<0.01), while the number of cells in G1 and G2 phase decreased accordingly (P<0.01). These results suggested that ATO treatment of EpCAM+CD44+ HCT-116 cells under hypoxia induced cell-cycle arrest in S-phase (Fig. 4A and B).
Figure 4.
ATO inhibits cell proliferation by causing cell-cycle arrest in S-phase. (A) Flow cytometric analysis of cell-cycle progression in EpCAM+CD44+ HCT-116 cells following treatment with 15 µM ATO for 24 h under hypoxic conditions. (B) Number of EpCAM+CD44+ HCT-116 cells in each cell-cycle phase. Cells treated with 50 µM DDP served as the positive control and DMSO was used as a negative control. Values are expressed as the mean ± standard deviation of three independent experiments. *P<0.05, **P<0.01 and ***P<0.001 and ****P<0.0001 as determined by one-way ANOVA followed by Dunnett's test. ATO, atovaquone; DDP, cisplatin; EpCAM, epithelial cell adhesion molecule.
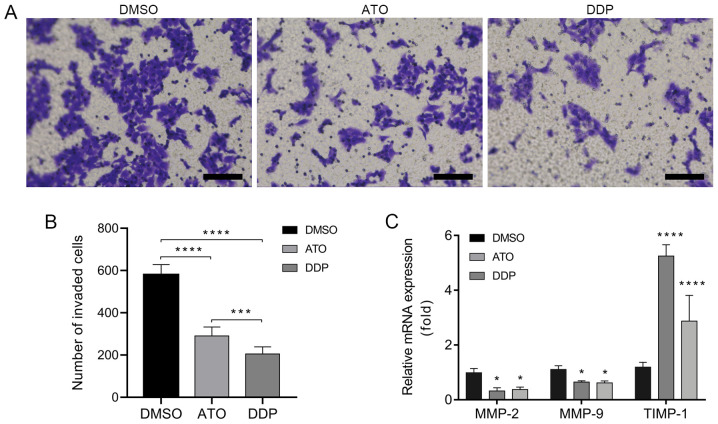
ATO inhibits the invasion of EpCAM+CD44+ HCT-116 cells
A Transwell invasion assay was performed to investigate the invasiveness of EpCAM+CD44+ HCT-116 cells following treatment with ATO under hypoxic conditions for 24 h. The results indicated that treatment with ATO significantly inhibited the invasion of EpCAM+CD44+ HCT-116 cells as compared with that in the DMSO group (Fig. 5A and B). In addition, the RT-qPCR results indicated downregulation of the invasion-associated genes MMP-2 and MMP-9 and upregulation of TIMP-1 in EpCAM+CD44+ HCT-116 cells after treatment with ATO under hypoxic conditions for 24 h (Fig. 5C).
Figure 5.
ATO inhibits invasion of EpCAM+CD44+ HCT-116 cells in hypoxia. (A) Optical micrographs presenting the inhibition of EpCAM+CD44+ HCT-116 cell invasiveness after treatment with 15 µM ATO for 24 h under hypoxic conditions according to Transwell invasion assays. Cells treated with 50 µM DDP were used as a positive control. Scale bar, 100 µm. (B) Number of invaded EpCAM+CD44+ HCT-116 cells. (C) Reverse transcription-quantitative PCR analysis of MMP-2, MMP-9 and TIMP-1 mRNA expression in EpCAM+CD44+ HCT-116 cells treated with 15 µM ATO for 24 h under hypoxia. Cells treated with 50 µM DDP served as the positive control. The mRNA expression is presented relative to the DMSO control. Values are expressed as the mean ± standard deviation of three independent experiments. *P<0.05, ***P<0.001 and ****P<0.0001 vs. DMSO as determined by one-way ANOVA followed by Dunnett's test. ATO, atovaquone; DDP, cisplatin, TIMP, tissue inhibitor of MMPs; EpCAM, epithelial cell adhesion molecule.
Discussion
Hypoxia is a major feature of the tumor microenvironment and results from an imbalance between oxygen supply and oxygen consumption of cancer cells (10). Tumor hypoxia was observed to diminish the efficacy of several chemotherapeutic drugs (28). Recently, ATO has gained attention due to its anticancer effects against various types of cancer, including brain, breast and cervical cancer, as well as hepatocellular carcinoma and retinoblastoma (18-22). To the best of our knowledge, the present study was the first to investigate the anticancer efficacy of ATO against colon CSCs in vitro under hypoxic conditions. The results suggested that ATO treatment inhibited the proliferation and invasion, induced apoptosis and caused S-phase arrest of EpCAM+CD44+ HCT-116 cells in vitro.
EpCAM and CD44 were used as markers for the isolation of colon CSCs from the colon cancer cell line HCT-116 by magnetic-activated cell sorting; in the native cell population, EpCAM+CD44+ HCT-116 cells accounted for 2.38% of all HCT-116 cells, which is consistent with the result of previous studies (29,30). Highly purified (>91%) EpCAM+CD44+ HCT-116 cells were subsequently obtained and the identity of CSCs was confirmed by performing a tumorsphere formation assay and serum-differentiation assay, and by evaluating the expression of stemness-related transcription factors, including C-MYC, OCT-4, SOX-2 and NANOG. The expression of these genes is associated with poor prognosis and malignant progression (31). In the present study, ATO inhibited the proliferation of EpCAM+CD44+ HCT-116 cells in a concentration-dependent manner with an IC50 of ~15 µM under hypoxic conditions. However, cell inhibition showed a linear trend with increasing ATO concentrations, while normally, a sigmoidal dose-response relation would be expected. Several factors may affect the accuracy of IC50 estimation (32), such as the total number of assay concentrations, the number of concentrations beyond the lower and upper bend point, dilution factor and the viability of the responses at the same concentration. A linear trend obtained in the present study may be partly due to the dilution factor and the total number of drug concentrations. Five drug concentrations (0, 5, 10, 15 and 20 µM) were used and only one concentration was below the IC50. Moreover, a difference of 5 µM between each concentration point instead of various concentrations with a wide range, for example, covering six logs of magnitude (0.001, 0.01 0.1, 1, 10, 100 and 1,000 µM) was used in the assay. A linear trend was also observed in other studies, possibly due to similar reasons (19,33). In the present study, the IC50 (~15 µM) was lower than the average plasma concentration (~57 µM) achieved in patients when ATO was administered with food at a standard dose of 750 mg twice daily to treat Pneumocystis carinii pneumonia (27) or when four 250/100 mg tablets of ATO-proguanil were administered daily to treat malaria (34,35). In an in vitro study, ATO treatment inhibited the viability of the breast cancer MCF-7 cell line with an IC50 of ~10 µM and MCF-7-derived CSCs with an IC50 of ~1 µM, examined by sulforhodamine B assay and mammosphere assay, respectively (19). Similarly, ATO at 10 µM markedly inhibited the proliferation in retinoblastoma cells and renal cell carcinoma cell lines (22,33). These results suggested that the effective concentration against EpCAM+CD44+ HCT-116 cells and certain other cancer cells is pharmacologically achievable (27).
ATO is a cytotoxic and apoptosis-inducing agent that is potent against various cancer cell lines (36). In the present study, ATO treatment induced apoptosis as detected by annexin V-FITC/PI double staining assays and caused mitochondrial membrane potential depolarization, as determined by a JC-1 staining assay. However, the trend of different ATO concentrations to induce apoptosis in the former assay appears to not be in accordance with the trend observed on JC-1 staining. This may be caused by several factors, such as the variation in trypsin digestion time and incubation time. In apoptotic cells, phosphatidylserine translocates from the inner to outer leaflet of the plasma membrane. Prolonged cell digestion time or incubation time may increase the translocation of phosphatidylserine, which can be identified and bound by Annexin V and ultimately cause an increase in the number of apoptotic cells. Cells may also undergo autophagy, which is another factor that should be considered in the two assays. Autophagy can lead to nonapoptotic cell death. Yu et al demonstrated that the degradation of the reactive oxygen species scavenger, catalase, results in the accumulation of reactive oxygen species in the cell, leading to loss of membrane integrity and ultimately cell death (37). The loss of membrane integrity may cause an increase in the apoptotic rate in the annexin V/PI double staining assays. In the JC-1 staining assay, 15 µM ATO treatment markedly increased the percentages of mitochondrial membrane depolarization compared with the DMSO control group and 5 and 10 µM ATO treatment groups. ATO-induced mitochondrial membrane permeabilization is limited to only a subset of mitochondria, autophagosomal elimination of the depolarized mitochondria (known as mitophagy) will take place and promote cell survival (38). On the other hand, 15 µM ATO treatment can lead to extensive mitochondrial membrane permeabilization and finally results in apoptotic cell death, as shown by increased percentages of mitochondrial membrane potential depolarization. ATO treatment altered the expression of apoptosis-related genes, as evidenced by downregulation of the mRNA expression of Bcl-2 and upregulation of Bax. While the induction of Bcl-2 leads to inhibition of apoptotic mechanisms and the development of drug resistance (39), downregulation of Bcl-2 may be one of the underlying mechanisms of apoptosis induced by ATO. The present results suggested that cell proliferation was inhibited by inducing cell-cycle arrest in S-phase. Similar results were also obtained in a study on hepatocellular carcinoma, demonstrating that ATO significantly inhibited the cell proliferation via inducing cell-cycle arrest in S phase and the apoptotic pathway associated with downregulation of cyclin A2 and cyclin-dependent kinase 2, and upregulation of the expression of p53 and p21(21). mTOR is a central regulator of cell growth, proliferation and differentiation. It also participates in the regulation of tumor cell migration, invasion and cancer metastasis (40). ATO induces proapoptotic signaling and inhibits the mTOR pathway via upregulation of activating transcription factor 4(41). In the present study, ATO suppressed the invasiveness of EpCAM+CD44+ HCT-116 cells and inhibited the activity of the zinc-dependent endopeptidases MMP-2/-9 with an increased expression of TIMP-1. These proteins are crucial in cell migration and invasion by controlling the degradation of extracellular matrix. Previous studies suggested that MMP-2/-9 expression is associated with the Akt/mTOR and JAK2/STAT3 signaling pathways (42). Of note, ATO inhibits Akt/AMPK/mTOR signaling and is a potent STAT3 inhibitor (43).
In summary, the present in vitro study indicated that ATO inhibited the proliferation of EpCAM+CD44+ HCT-116 cells via induction of S-phase arrest, triggered apoptosis and inhibited cell invasion under hypoxic conditions. However, the present study has several limitations: Several factors, such as the differences in the incubation time may affect the apoptotic rate in the Annexin V/PI double staining assays and JC-1 staining assays; autophagy and mitophagy may influence apoptosis and the interplay between autophagy and apoptosis was not examined and the study focused on the antitumor effects of ATO on colon cancer stem cells in vitro. In future studies a xenograft model will be used to determine the role of ATO in vivo. Nevertheless, the present study laid a foundation for further investigation of the in-depth mechanisms of the antitumor activity of ATO on colon CSCs and provide a novel therapeutic regimen for colorectal carcinoma.
Acknowledgements
Not applicable.
Funding
The current study was supported by the Jilin Province Science and Technology Support Program (grant no. 20200204036YY), the Education Department of Jilin Province (grant no. JJKH20201122KJ) and the Jilin Province Health Technology Innovation Project (grant no. 2017J062).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
CF, WL and YW conceived and designed the experiments. CF and XX performed the experiments. CF drafted the manuscript and supervised the experiments. CF, XX and HX performed data analysis and interpretation. WL and YW verified the results of the experiments, helped with the statistical analysis and revised the manuscript critically for intellectual content. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 4.Nguyen LV, Vanner R, Dirks P, Eaves CJ. Cancer stem cells: An evolving concept. Nat Rev Cancer. 2012;12:133–143. doi: 10.1038/nrc3184. [DOI] [PubMed] [Google Scholar]
- 5.Nandy SB, Lakshmanaswamy R. Cancer stem cells and metastasis. Prog Mol Biol Transl Sci. 2017;151:137–176. doi: 10.1016/bs.pmbts.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 6.Todaro M, Alea MP, Di Stefano AB, Cammareri P, Vermeulen L, Iovino F, Tripodo C, Russo A, Gulotta G, Medema JP, Stassi G. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell. 2007;1:389–402. doi: 10.1016/j.stem.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 7.Was H, Czarnecka J, Kominek A, Barszcz K, Bernas T, Piwocka K, Kaminska B. Some chemotherapeutics-treated colon cancer cells display a specific phenotype being a combination of stem-like and senescent cell features. Cancer Biol Ther. 2018;19:63–75. doi: 10.1080/15384047.2017.1385675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szarynska M, Olejniczak A, Kmieć Z. The role of cancer stem cells in pathogenesis of colorectal cancer. Postepy Hig Med Dosw (Online) 2016;70:1469–1482. doi: 10.5604/17322693.1227897. [DOI] [PubMed] [Google Scholar]
- 9.Dawood S, Austin L, Cristofanilli M. Cancer stem cells: Implications for cancer therapy. Oncology (Williston Park) 2014;28:1101–1107. 1110. [PubMed] [Google Scholar]
- 10.Muz B, de la Puente P, Azab F, Azab AK. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia (Auckl) 2015;3:83–92. doi: 10.2147/HP.S93413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaupel P, Mayer A, Höckel M. Tumor hypoxia and malignant progression. Methods Enzymol. 2004;381:335–354. doi: 10.1016/S0076-6879(04)81023-1. [DOI] [PubMed] [Google Scholar]
- 12.Zhou J, Schmid T, Schnitzer S, Brune B. Tumor hypoxia and cancer progression. Cancer Lett. 2006;237:10–21. doi: 10.1016/j.canlet.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 13.Stamenkovic I. Matrix metalloproteinases in tumor invasion and metastasis. Semin Cancer Biol. 2000;10:415–433. doi: 10.1007/s000180050495. [DOI] [PubMed] [Google Scholar]
- 14.Hicklin DJ, Ellis LM. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. J Clin Oncol. 2005;23:1011–1027. doi: 10.1200/JCO.2005.06.081. [DOI] [PubMed] [Google Scholar]
- 15.Graham K, Unger E. Overcoming tumor hypoxia as a barrier to radiotherapy, chemotherapy and immunotherapy in cancer treatment. Int J Nanomedicine. 2018;13:6049–6058. doi: 10.2147/IJN.S140462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strese S, Fryknas M, Larsson R, Gullbo J. Effects of hypoxia on human cancer cell line chemosensitivity. BMC Cancer. 2013;13(331) doi: 10.1186/1471-2407-13-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srivastava IK, Rottenberg H, Vaidya AB. Atovaquone, a broad spectrum antiparasitic drug, collapses mitochondrial membrane potential in a malarial parasite. J Biol Chem. 1997;272:3961–3966. doi: 10.1074/jbc.272.7.3961. [DOI] [PubMed] [Google Scholar]
- 18.Takabe H, Warnken ZN, Zhang Y, Davis DA, Smyth HDC, Kuhn JG, Weitman S, Williams Iii RO. A Repurposed drug for brain cancer: Enhanced atovaquone amorphous solid dispersion by combining a spontaneously emulsifying component with a polymer carrier. Pharmaceutics. 2018;10(60) doi: 10.3390/pharmaceutics10020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fiorillo M, Lamb R, Tanowitz HB, Mutti L, Krstic-Demonacos M, Cappello AR, Martinez-Outschoorn UE, Sotgia F, Lisanti MP. Repurposing atovaquone: Targeting mitochondrial complex III and OXPHOS to eradicate cancer stem cells. Oncotarget. 2016;7:34084–34099. doi: 10.18632/oncotarget.9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian S, Chen H, Tan W. Targeting mitochondrial respiration as a therapeutic strategy for cervical cancer. Biochem Biophys Res Commun. 2018;499:1019–1024. doi: 10.1016/j.bbrc.2018.04.042. [DOI] [PubMed] [Google Scholar]
- 21.Gao X, Liu X, Shan W, Liu Q, Wang C, Zheng J, Yao H, Tang R, Zheng J. Anti-malarial atovaquone exhibits anti-tumor effects by inducing DNA damage in hepatocellular carcinoma. Am J Cancer Res. 2018;8:1697–1711. [PMC free article] [PubMed] [Google Scholar]
- 22.Ke F, Yu J, Chen W, Si X, Li X, Yang F, Liao Y, Zuo Z. The anti-malarial atovaquone selectively increases chemosensitivity in retinoblastoma via mitochondrial dysfunction-dependent oxidative damage and Akt/AMPK/mTOR inhibition. Biochem Biophys Res Commun. 2018;504:374–379. doi: 10.1016/j.bbrc.2018.06.049. [DOI] [PubMed] [Google Scholar]
- 23.Ashton TM, Fokas E, Kunz-Schughart LA, Folkes LK, Anbalagan S, Huether M, Kelly CJ, Pirovano G, Buffa FM, Hammond EM, et al. The anti-malarial atovaquone increases radiosensitivity by alleviating tumour hypoxia. Nat Commun. 2016;7(12308) doi: 10.1038/ncomms12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu C, Zhou N, Zhao Y, Duan J, Xu H, Wang Y. Dendritic cells loaded with CD44 CT-26 colon cell lysate evoke potent antitumor immune responses. Oncol Lett. 2019;18:5897–5904. doi: 10.3892/ol.2019.10952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Phi LTH, Sari IN, Yang YG, Lee SH, Jun N, Kim KS, Lee YK, Kwon HY. Cancer stem cells (CSCs) in drug resistance and their therapeutic implications in cancer treatment. Stem Cells Int. 2018;2018(5416923) doi: 10.1155/2018/5416923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Falloon J, Sargent S, Piscitelli SC, Bechtel C, LaFon SW, Sadler B, Walker RE, Kovacs JA, Polis MA, Davey RT Jr, et al. Atovaquone suspension in HIV-infected volunteers: Pharmacokinetics, pharmacodynamics, and TMP-SMX interaction study. Pharmacotherapy. 1999;19:1050–1056. doi: 10.1592/phco.19.13.1050.31598. [DOI] [PubMed] [Google Scholar]
- 28.Cosse JP, Michiels C. Tumour hypoxia affects the responsiveness of cancer cells to chemotherapy and promotes cancer progression. Anticancer Agents Med Chem. 2008;8:790–797. doi: 10.2174/187152008785914798. [DOI] [PubMed] [Google Scholar]
- 29.Zhang C, Tian Y, Song F, Fu C, Han B, Wang Y. Salinomycin inhibits the growth of colorectal carcinoma by targeting tumor stem cells. Oncol Rep. 2015;34:2469–2476. doi: 10.3892/or.2015.4253. [DOI] [PubMed] [Google Scholar]
- 30.Xiong B, Ma L, Hu X, Zhang C, Cheng Y. Characterization of side population cells isolated from the colon cancer cell line SW480. Int J Oncol. 2014;45:1175–1183. doi: 10.3892/ijo.2014.2498. [DOI] [PubMed] [Google Scholar]
- 31.Müller M, Hermann PC, Liebau S, Weidgang C, Seufferlein T, Kleger A, Perkhofer L. The role of pluripotency factors to drive stemness in gastrointestinal cancer. Stem Cell Res. 2016;16:349–357. doi: 10.1016/j.scr.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 32.Larsson P, Engqvist H, Biermann J, Werner Rönnerman E, Forssell-Aronsson E, Kovács A, Karlsson P, Helou K, Parris TZ. Optimization of cell viability assays to improve replicability and reproducibility of cancer drug sensitivity screens. Sci Rep. 2020;10(5798) doi: 10.1038/s41598-020-62848-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen D, Sun X, Zhang X, Cao J. Targeting mitochondria by anthelmintic drug atovaquone sensitizes renal cell carcinoma to chemotherapy and immunotherapy. J Bioch Mol Toxicol. 2018;32(e22195) doi: 10.1002/jbt.22195. [DOI] [PubMed] [Google Scholar]
- 34.Baggish AL, Hill DR. Antiparasitic agent atovaquone. Antimicrob Agents Chemother. 2002;46:1163–1173. doi: 10.1128/aac.46.5.1163-1173.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nixon GL, Moss DM, Shone AE, Lalloo DG, Fisher N, O'Neill PM, Ward SA, Biagini GA. Antimalarial pharmacology and therapeutics of atovaquone. J Antimicrob Chemother. 2013;68:977–985. doi: 10.1093/jac/dks504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou J, Duan L, Chen H, Ren X, Zhang Z, Zhou F, Liu J, Pei D, Ding K. Atovaquone derivatives as potent cytotoxic and apoptosis inducing agents. Bioorg Med Chem Lett. 2009;19:5091–5094. doi: 10.1016/j.bmcl.2009.07.044. [DOI] [PubMed] [Google Scholar]
- 37.Yu L, Wan F, Dutta S, Welsh S, Liu Z, Freundt E, Baehrecke EH, Lenardo M. Autophagic programmed cell death by selective catalase degradation. Proc Natl Acad Sci USA. 2006;103:4952–4957. doi: 10.1073/pnas.0511288103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rambold AS, Lippincott-Schwartz J. Mechanisms of mitochondria and autophagy crosstalk. Cell Cycle. 2011;10:4032–4038. doi: 10.4161/cc.10.23.18384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kinoshita M, Johnson DL, Shatney CH, Lee YL, Mochizuki H. Cancer cells surviving hypoxia obtain hypoxia resistance and maintain anti-apoptotic potential under reoxygenation. Int J Cancer. 2001;91:322–326. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1064>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 40.Zhou H, Huang S. Role of mTOR signaling in tumor cell motility, invasion and metastasis. Curr Protein Pept Sci. 2011;12:30–42. doi: 10.2174/138920311795659407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stevens AM, Xiang M, Heppler LN, Tošić I, Jiang K, Munoz JO, Gaikwad AS, Horton TM, Long X, Narayanan P, et al. Atovaquone is active against AML by upregulating the integrated stress pathway and suppressing oxidative phosphorylation. Blood Adv. 2019;3:4215–4227. doi: 10.1182/bloodadvances.2019000499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fang Z, Tang Y, Fang J, Zhou Z, Xing Z, Guo Z, Guo X, Wang W, Jiao W, Xu Z, Liu Z. Simvastatin inhibits renal cancer cell growth and metastasis via AKT/mTOR, ERK and JAK2/STAT3 pathway. PLoS One. 2013;8(e62823) doi: 10.1371/journal.pone.0062823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiang M, Kim H, Ho VT, Walker SR, Bar-Natan M, Anahtar M, Liu S, Toniolo PA, Kroll Y, Jones N, et al. Gene expression-based discovery of atovaquone as a STAT3 inhibitor and anticancer agent. Blood. 2016;128:1845–1853. doi: 10.1182/blood-2015-07-660506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.