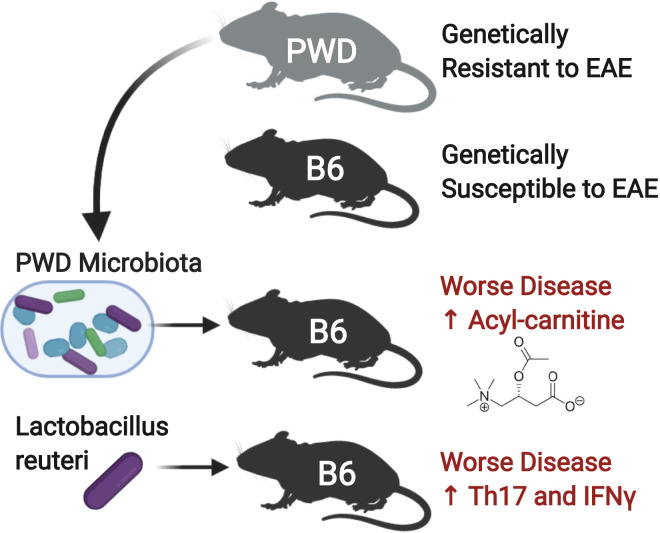
While the risk of developing multiple sclerosis (MS) is recognized to have both genetic and environmental components, little is known about these complex interactions. The microbiome has recently been recognized as an environmental factor that contributes to MS. In Montgomery et al. (1), the authors harnessed the natural genetic diversity between B6 mice, PWD/PhJ (PWD) wild-derived mice, and a panel of 27 B6.ChrPWD consomic mice to investigate gene plus microbiome interactions. They identified chromosomes that affected disease susceptibility and also identified microbes that increased disease severity in disease-susceptible hosts. They found that while the PWD mice are resistant to experimental autoimmune encephalomyelitis (EAE), these mice surprisingly harbor a microbiota that increases central nervous system (CNS) autoimmunity when transferred into the genetically susceptible B6 mice (Fig. 1). This study advances the field by illustrating an important point: that altered microbiota alone may not be sufficient to cause MS, but rather, a perfect storm of host genetic risk plus specific microbes triggers CNS autoimmunity.
Fig. 1.
Genetic susceptibility to EAE shapes interactions with the microbiome. PWD mice are genetically resistant to EAE, whereas B6 mice are susceptible. However, transfer of the microbiota from PWD mice or colonization with PWD-enriched microbe L. reuteri worsens disease in B6 mice and increased inflammatory cytokines interleukin-17 (Th17) and interferon-gamma (IFNγ), highlighting the importance of gene × microbiome interactions.
In the last 5 years, several studies have identified alterations in the microbiome of patients with MS (2–4), with consistent depletions in butyrate producing bacteria and elevations in Akkermansia and Clostridia. Furthermore, the transfer of microbiota from patients with MS can increase disease incidence in spontaneous models of the disease (5, 6), demonstrating that alterations in the microbiome are not just a bystander of disease but may actually contribute to MS. Germfree mice and antibiotic-treated mice are resistant to EAE in C57/BL6 mice, providing further evidence that the microbiome is necessary for disease induction. However, several of the MS-associated bacteria can be found in the microbiome of healthy controls, raising the question of whether alterations in the microbiome are sufficient to cause MS.
It is known that genetics plays an important role in MS. The strongest association of genetic risk is linked to the human leukocyte antigen (HLA) system, which was first identified more than 40 years ago (7, 8), with a greater MS risk associated with DRB1*15:01 (9). In addition, large-scale genetic studies have identified hundreds of loci across the genome that predispose to MS. Most of these loci regulate genes in the immune system, both peripherally and centrally (microglia) (10). How these genes relate to the microbiome in MS is yet unknown. In animals, different inbred animal models exhibit different disease susceptibility to EAE, the animal model for MS (11). For example, the commonly used C57/BL6 (hereafter referred to as B6) mice immunized with myelin oligodendrocyte glycoprotein (MOG) and complete Freund’s adjuvant present with acute paralysis and then recover. In contrast, the closely related B10.S mice are resistant to EAE and skew toward a T helper cell type 2 (Th2) immune response (12). There are minor genetic differences between the B6 and B10.S mice, which result in altered susceptibility to gastrointestinal parasitic infection (13), suggesting that in addition to alterations in autoimmune disease, these mice differ in their immune responses in the gut. The nonobese diabetic (NOD) mice immunized with MOG initially present with a relapsing–remitting disease course that eventually transitions into a progressive disease. The SJL mouse immunized with proteolipid protein results in a relapsing remitting disease. Finally, the wild-derived inbred strain PWD is resistant to EAE when immunized with MOG and effect linked to the altered transcription of MS-associated genes (1).
Multiple genomic differences between the C57, SJL, NOD, and PWD mouse strains present a challenge to narrow down genetic elements that confer resistance or susceptibility to CNS autoimmunity. To overcome this, Montgomery et al. (1) dissected the contribution of each chromosome from the PWD mice by utilizing 27 consomic mice, which had the genetic background of EAE-susceptible B6 mice and carried one chromosome from the EAE-resistant PWD mice (notated as B6.ChrPWD). They found that B6.ChrPWD mice carrying PWD chromosomes 2, 10.2, 15, and 17 were protected from EAE, whereas mice carrying B6.ChrPWD chromosomes 1, 11.3, and 16 had markedly worse disease compared with B6 mice, identifying genomic regions that may alter susceptibility to CNS autoimmune disease. Examining the microbiome in these 27 B6.ChrPWD lines, the authors identified one operational taxonomic unit (OTU) in the Bacteroidales order and two OTUs in the Porphyromonadaceae family linked with worse EAE and six OTUs in the Porphyromonadaceae family and one OTU in the Erysipelotrichaceae family linked with decreased EAE scores. They further identified microbial genes involved in propionate and butyrate metabolism linked with worse EAE. However, the overall composition of microbiota from EAE-resistant mice did not cluster separately from the microbiota from EAE-susceptible mice, and the variation in disease susceptibility that could be explained by microbiota difference was relatively small (0.5 to 1.3%), suggesting that altered genes in each of the B6.ChrPWD chromosomes have a stronger effect on disease susceptibility and microbiota composition.
To directly test whether PWD mice resist disease because of a protective microbiome, the authors next transferred gut microbiota from PWD mice or from B6 mice into genetically susceptible B6 germfree mice. Unexpectedly, the PWD microbiome worsened EAE in B6 mice. This was the case when germfree mice were colonized at 4 wk of age or if colonized at birth by transferring the microbiota to germfree breeder pairs. In an independent experiment, the authors also found that cohousing B6 mice with PWD mice, a common practice to transfer microbiota. In an independent study, and consistent with the microbiota transfer, B6 mice cohoused with PWD mice had more severe disease compared with B6 mice housed with their own genotype. Surprisingly, PWD mice cohoused with B6 mice developed EAE, whereas PWD mice housed alone were resistant, as is typical for PWD mice. Taken together, this may indicate that there is an optimal gene + microbiota combination, and altering these relationships may result in aggravation of induced autoimmune disease. These results raise the question of whether B6.ChrPWD consomic mice harbor a microbiota with altered capacity to induce CNS autoimmunity. Additional transfers of microbiota from consomic B6.ChrPWD mice into B6 would provide an elegant system to investigate how genetics may select for a microbiota with the capacity to modulate neuroinflammation.
In order to identify microbial culprits that account for enhanced disease severity in the B6 model, the authors performed metabolomics and 16S ribosomal RNA (rRNA) sequencing. In B6 mice colonized with PWD microbiota, they found decreased propionyl-carnitine, hexanyl-l-carnitine and indoxyl and increased acetyl-carnitine and indole-3-acetaldehyde, suggesting the PWD microbiota produced compounds that could worsen disease in susceptible mice, and provide targets for future therapeutic applications. In the cohoused animals, they found that B6 exposed to PWD had a specific increase in Lactobacillus reuteri, whereas another Lactobacillus species (Lactobacillus murinis) was unaltered by cohousing. Moving beyond observation to causation, the authors then colonized BL6 mice with L. murinis and found that it worsened EAE and skewed immune responses toward a proinflammatory phenotype, with elevated granulocyte macrophage–colony-stimulating factor production by CD4 T cells and interleukin-17 production by CD8 T cells.
The fact that the EAE-resistant PWD mice harbor a microbiome that promotes CNS autoimmunity in a genetically susceptible host at first seems counterintuitive. However, the authors are able to dissect this complexity of gene × environment interaction by performing several experiments designed to test causality. Furthermore, a microbe enriched in absence of disease does not necessarily mean that it is protective, and a microbe enriched in disease does not necessarily mean that it pathogenic. One of the most consistent alterations in the MS microbiome is an elevation in Akkermansia muciniphila (2, 5, 6), and B6 mice at peak disease also have elevated Akkermansia. However, the transfer microbiota from B6 mice at peak disease or transfer Akkermansia alone can ameliorate disease (14). This suggests that throughout the disease course of MS or EAE, the host may be selecting for a beneficial microbiota. Since the PWD mice are not susceptible to the disease, there is no evolutionary benefit for the PWD mouse to exclude L. reuteri. We are only beginning to learn about the multiple mechanisms of how the host may tune the microbiome, but in addition to selection through the immune responses, we have found that the host may select for a beneficial microbiome through the secretion of microRNAs (15). Whether the PWD mice have altered intestinal production of microRNAs and whether this is a mechanism by which L. reuteri is selected are yet unknown.
L. reuteri is part of the endogenous microbiota in mice and is a lactic acid-producing member of the class Bacilli. Lactobacillus species have been long studied as probiotic microorganisms, and their potential beneficial effects have been shown to be strain and disease specific. However, several studies find conflicting associations between L. reuteri and autoimmune disease. In a study from He et al. (16), the authors found that L. reuteri ameliorated EAE. Since He et al. (16) and Montgomery et al. (1) used different strains of L. reuteri, comparative genomics could be employed to identify bacterial strain-specific pathogenicity factors for EAE. Differing properties of the same species are common in well-studied commensals. For example, commensal Escherichia coli can prevent infection with other gastrointestinal pathogens, whereas E. coli 0157:57 can result in gastroenteritis and septicemia. Commensal strains of Bacteroides fragilis that have polysaccharide in their cell wall can induce T regulatory cells and ameliorate EAE, whereas B. fragilis strains that encode the matrix metalloprotease fragilysin toxin can contribute to inflammatory bowel disease (IBD) and colon cancer (17). Another possibility is that combination of L. reuteri with other inflammatory bacteria is responsible for triggering diseases, as was recently shown in Miyauchi et al. (18). Given the complexity of strain-specific and compounded microbiome interactions, we should not be surprised that L. reuteri strains can have divergent effects on EAE.
Understanding disease risk is critical because it could lead to preventative measures. Individuals with a family history of MS or other autoimmune diseases could theoretically take a probiotic to help prevent the development of disease. However, it is critical to understand disease-specific probiotics. This paper, as well as well as another recently published paper, has identified L. reuteri as a detrimental organism that can contribute to CNS autoimmunity (1, 18). L. reuteri can be found in several over-the-counter probiotics, raising the question of whether probiotics with L. reuteri are safe for MS patients. Our group found that a probiotic with five types of Lactobacillus, two types of Bifidobacterium, and one strain of Streptococcus was safe to administer to patients with MS and lowered inflammatory markers on circulating monocytes both during and after probiotic administration (19). Critically, this formulation did not have L. reuteri. There is currently little guidance on which probiotics could be beneficial or detrimental to patients with MS; however, these recent findings in EAE (1, 18) suggest that patients and physicians should exercise caution when using modifying the microbiota to treat MS.
In summary, the authors demonstrated the utility of comparing mice with different genetic backgrounds to investigate the microbiome as an environmental modifier of risk in neuroinflammatory disease. The connection between PWD mouse genetics that contribute to EAE and human MS loci is unknown. However, by studying divergent gene × environmental interactions in PWD mice, they identified L. reuteri as a bacterial strain with enhanced capacity to drive disease in a genetically susceptible host, and link disease susceptibility to altered microbial metabolites and immune responses. This work has broad implications for the challenges of investigating the role of the microbiota in diverse patient populations, as well as for the development of microbiome-based therapeutics to prevent and treat MS.
Acknowledgments
This work was supported by an award from the Race to Erase MS Foundation.
Footnotes
The authors declare no competing interest.
See companion article, “Interactions between host genetics and gut microbiota determine susceptibility to CNS autoimmunity,” 10.1073/pnas.2002817117.
References
- 1.Montgomery T. L., et al. , Interactions between host genetics and gut microbiota determine susceptibility to CNS autoimmunity. Proc. Natl. Acad. Sci. U.S.A. 117, 27516–27527 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jangi S., et al. , Alterations of the human gut microbiome in multiple sclerosis. Nat. Commun. 7, 12015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miyake S., et al. , Dysbiosis in the gut microbiota of patients with multiple sclerosis, with a striking depletion of species belonging to Clostridia XIVa and IV clusters. PLoS One 10, e0137429 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J., et al. , Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci. Rep. 6, 28484 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berer K., et al. , Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc. Natl. Acad. Sci. U.S.A. 114, 10719–10724 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cekanaviciute E., et al. , Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc. Natl. Acad. Sci. U.S.A. 114, 10713–10718 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertrams J., Kuwert E., HL-A antigen frequencies in multiple sclerosis. Significant increase of HL-A3, HL-A10 and W5, and decrease of HL-A12. Eur. Neurol. 7, 74–78 (1972). [DOI] [PubMed] [Google Scholar]
- 8.Naito S., Namerow N., Mickey M. R., Terasaki P. I., Multiple sclerosis: Association with HL-A3. Tissue Antigens 2, 1–4 (1972). [DOI] [PubMed] [Google Scholar]
- 9.Sawcer S., Franklin R. J., Ban M., Multiple sclerosis genetics. Lancet Neurol. 13, 700–709 (2014). [DOI] [PubMed] [Google Scholar]
- 10.International Multiple Sclerosis Genetics Consortium , Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science 365, eavv7188 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato F., Omura S., Martinez N. E., Tsunoda I., “Animal models of multiple sclerosis” in Neuroinflammation, Minagar A., Ed. (Academic Press, ed. 2, 2018), chap. 3, pp. 37–72. [Google Scholar]
- 12.Maron R., et al. , Genetic susceptibility or resistance to autoimmune encephalomyelitis in MHC congenic mice is associated with differential production of pro- and anti-inflammatory cytokines. Int. Immunol. 11, 1573–1580 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Smith P. M., Sproule T. J., Philip V. M., Roopenian D. C., Stadecker M. J., Minor genomic differences between related B6 and B10 mice affect severity of schistosome infection by governing the mode of dendritic cell activation. Eur. J. Immunol. 45, 2312–2323 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu S., et al. , Oral administration of miR-30d from feces of MS patients suppresses MS-like symptoms in mice by expanding Akkermansia muciniphila. Cell Host Microbe 26, 779–794.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu S., et al. , The host shapes the gut microbiota via fecal microRNA. Cell Host Microbe 19, 32–43 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He B., et al. , Lactobacillus reuteri reduces the severity of experimental autoimmune encephalomyelitis in mice by modulating gut microbiota. Front. Immunol. 10, 385 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wexler H. M., Bacteroides: The good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 20, 593–621 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyauchi E., et al. , Gut microorganisms act together to exacerbate inflammation in spinal cords. Nature 585, 102–106 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Tankou S. K., et al. , A probiotic modulates the microbiome and immunity in multiple sclerosis. Ann. Neurol. 83, 1147–1161 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]