Significance
Pancreatic ductal adenocarcinoma (PDAC) remains one of the most lethal cancers to treat. Existing drugs rarely work as a single agent. Therefore, identifying novel drug combinations that will synergistically and selectively kill PDAC cells is a significant unmet need. Here, we used CRISPR gene knockout screening to identify combinatorial targets of gemcitabine (Gem) in PDAC. We discovered that PRMT5 depletion makes PDAC cells significantly sensitive to Gem. Our mechanistic findings show that PRMT5 inhibition impairs DNA repair mechanism. Hence the combination results in synergistic accumulation of Gem-induced DNA damage. We show that the combination leads to synergistic cell death in PDAC cells in vitro and in vivo.
Keywords: CRISPR screening, pancreatic cancer, cancer genomics and epigenomics, synthetic lethality, combinatorial drugs targets
Abstract
Pancreatic ductal adenocarcinoma (PDAC) remains one of the most challenging cancers to treat. Due to the asymptomatic nature of the disease and lack of curative treatment modalities, the 5-y survival rate of PDAC patients is one of the lowest of any cancer type. The recurrent genetic alterations in PDAC are yet to be targeted. Therefore, identification of effective drug combinations is desperately needed. Here, we performed an in vivo CRISPR screen in an orthotopic patient-derived xenograft (PDX) model to identify gene targets whose inhibition creates synergistic tumor growth inhibition with gemcitabine (Gem), a first- or second-line chemotherapeutic agent for PDAC treatment. The approach revealed protein arginine methyltransferase gene 5 (PRMT5) as an effective druggable candidate whose inhibition creates synergistic vulnerability of PDAC cells to Gem. Genetic depletion and pharmacological inhibition indicate that loss of PRMT5 activity synergistically enhances Gem cytotoxicity due to the accumulation of excessive DNA damage. At the molecular level, we show that inhibition of PRMT5 results in RPA depletion and impaired homology-directed DNA repair (HDR) activity. The combination (Gem + PRMT5 inhibition) creates conditional lethality and synergistic reduction of PDAC tumors in vivo. The findings demonstrate that unbiased genetic screenings combined with a clinically relevant model system is a practical approach in identifying synthetic lethal drug combinations for cancer treatment.
Pancreatic ductal adenocarcinoma (PDAC) is the most common and aggressive form of pancreatic cancer. It arises due to abnormal growth of exocrine ductal cells, the digestive enzyme-producing cells that compose 98% of the pancreas biomass. PDAC remains one of the deadliest of any cancer type. The main reasons for this high rate of mortality are twofold. Firstly, the disease is mostly asymptomatic until the late stages. Secondly, the current treatment strategies, mainly chemotherapy drug combinations, are relatively ineffective. Therefore, surgery, if possible, remains the only curative therapy. However, only 15 to 20% of PDAC patients are eligible for surgery due to presence of metastases or the extension of the tumor to neighboring major arteries. For these unresectable patients, standard treatment involves radiotherapy and chemotherapy combinations. Unfortunately, current chemotherapy combinations have severe side effects due to ineffective selectivity toward the PDAC tumors. Therefore, the median survival rate is only 6 mo, and more than 93% of patients die within the first 5 y (1). As such, despite the significant increase in the survival rates of most cancers, PDAC survival remains unchanged in the last 50 y (2), and it is projected to be the second leading cause of cancer deaths in the United States by 2030 (3). Novel drug combinations that can result in better therapeutic value are desperately needed for PDAC treatment.
Improved understanding of the significant drivers of pancreatic cancer progression and chemotherapy response can potentially yield novel and more effective treatment strategies. To this end, cancer genome sequencing efforts identified several recurrent genetic alterations. Among these, oncogenic KRAS mutations are observed in 93% of PDAC tumors (4). Additionally, loss-of-function mutations in CDKN2A, TP53, and SMAD4 tumor suppressor genes are the primary, recurrent mutations. Unfortunately, none of these genetic alterations are currently targetable. The major signaling pathways downstream of oncogenic RAS mutations, such as the aberrantly active MEK-ERK pathway, present themselves as promising therapeutic targets (5). However, clinical efforts to inhibit these pathways have not been successful yet for PDAC (6, 7). Therefore, identifying effective chemotherapy combinations is a critical unmet need for PDAC treatment. However, the reliance on suboptimal in vitro cellular models and lack of effective unbiased approaches have hampered the ability to find effective drug combinations.
In this study, we used in vivo CRISPR screening to identify effective and potentially synergistic lethal drug combinations for PDAC. We specifically aimed to identify new targets whose inhibition would create conditional vulnerability with the well-established existing chemotherapy, gemcitabine (Gem). Historically, Gem has been the first-line chemotherapy and forms the backbone of several drug combinations for most PDAC patients. Gem is designated as an “essential medicine” (8) and has been in use since 1983. In addition to being the primary chemotherapy for PDAC, it is a critical component of therapy in multiple other carcinomas (9). Although a new multidrug combination (FOLFIRINOX) slightly improves the survival of PDAC patients, due to high toxicity, only a small fraction of patients tolerate this regimen (10). Therefore, Gem remains the first line or second line for chemotherapy for the majority of PDAC patients. We rationalize that, given the poor 5-y survival rate of PDAC patients, finding novel drug combinations that will synergistically increase Gem's therapeutic effects will have significant therapeutic value.
To this end, we utilized CRISPR/Cas9 genetic knockout (KO) screenings in clinically relevant in vivo model systems to find novel targets that synergize with Gem. We recently performed in vivo and in vitro CRISPR KO screens for 4,000 genes to identify synthetic lethal partners of inhibitors of MEK signaling, an aberrantly active pathway due to oncogenic KRAS mutations in PDAC (11). In this study, we focused on chromatin regulators (CRs) essential for the survival of PDAC cells in response to Gem. We aimed to identify CRs whose inhibition creates synthetic lethality when PDAC cells are treated with Gem. We, therefore, constructed a custom sgRNA library of 8,000 sgRNAs that target ∼700 epigenetic regulators and performed both in vitro and in vivo screening to identify synthetic lethal partners of Gem. Our approach revealed protein arginine methyltransferase gene 5 (PRMT5) as a promising druggable candidate whose inhibition creates synergistic vulnerability of PDAC cells to Gem. At the molecular level, our findings suggest that genetic depletion or pharmacological inhibition of PRMT5 results in synergistic cytotoxicity with Gem due to depleted replication protein A (RPA) levels and an impaired homology-directed DNA repair (HDR) mechanism. Thus, the combinatorial treatment results in excessive DNA damage accumulation and subsequent cell deaths both in vitro and in vivo. These findings highlight the unbiased power of CRISPR screening in relevant clinical models to identify novel and more effective combinatorial drug targets.
Results
In Vivo CRISPR Gene KO Screening.
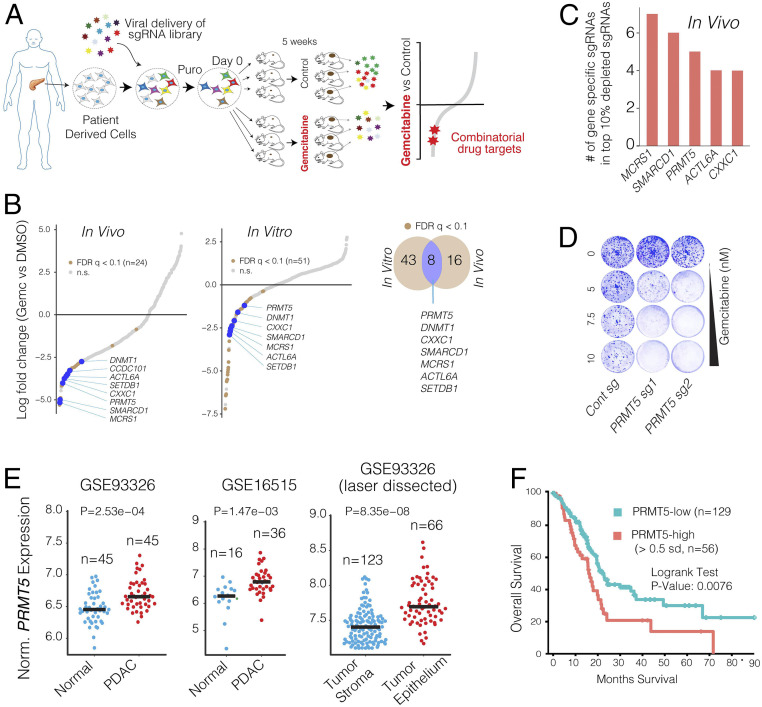
We performed the CRISPR screening using a clinically relevant patient-derived xenograft (PDX) model of PDAC in which a patient’s tumor is propagated in vivo within the pancreas of athymic nude mice (12). The PDX366 line is established from a poorly differentiated metastatic tumor with low stromal content and mutant for KRAS, P53, and SMAD4 but the wild type (WT) for P16 genes (13). In our CRISPR screen (Fig. 1A), we used an 8,031-single-guide RNA (sgRNA) containing library targeting 619 human genes enriched for chromatin modifiers plus 360 control sgRNAs.
Fig. 1.
In vivo CRISPR screening identifies PRMT5 as a combinatorial target of Gem. (A) Schematics for in vivo selection screening to identify novel drug combinations. (B) Dot plots show gene-specific CRISPR viability scores. Significantly depleted genes (FDR < 0.1) are labeled with brown dots, whereas the genes that are depleted both in vitro and in vivo are labeled with blue dots. (C) Bar plots show the number of sgRNAs targeting indicated genes among the top 10% of depleted sgRNAs. (D) Crystal violet colony formation assays show the relative cell proliferation rates of cells expressing control and PRMT5 targeting sgRNAs in response to the indicated Gem concentrations. (E) Dot plots show normalized PRMT5 expression levels in normal and matched PDAC tumors. (F) The Kaplan–Meier plot demonstrates survival rates of PDAC patients whose tumors have high PRMT5 expression (>0.5 SDs) relative to the PRMT5-low patients.
To maintain sgRNA coverage, we infected ∼50 to 100 million cells at ∼0.25 multiplicity of infection (MOI). After a week of drug selection, the surviving cells were randomly divided into nine batches, each containing ∼2 million cells (∼200× sgRNA coverage). One sample was harvested as “day 0,” and others were maintained in culture for in vitro screening or for xenograft injection into the pancreas of athymic nude mice (∼2 million cells/mouse, six mice total). One week after injection, animals were randomized to receive either vehicle control (n = 3) or gemcitabine treatment (n = 3) for 5 wk as described in Materials and Methods and depicted in Fig. 1A. The relative abundance of each sgRNA was assessed by targeted amplification and deep sequencing of tumor genomic DNA. Data analysis was performed using MAGeCK (14) and R. In parallel, we also performed in vitro screening, in which cultured cells were exposed to control dimethyl sulfoxide (DMSO) or 20% inhibitory concentration (IC20) doses of Gem every 3 d for 5 wk.
The sgRNA read count distribution analyses of the day 0 sample (>99.9% coverage) demonstrated that the sgRNAs in our library were evenly represented with a Gini index of 0.07 [∼0.1 is suggested for initial-state samples (15) and SI Appendix, Fig. S1]. Contrary to the day 0 samples, 86% and 81% of the sgRNAs were detectable in control in vitro and in vivo samples after a month of selection, respectively. Assuming that the ∼15% depletion was due to the functional roles of the target genes, the analyses suggested that ∼95% of cells containing sgRNAs contributed to in vivo tumor formation (SI Appendix, Fig. S2). Notably, only 20 and 12 genes had two or fewer sgRNAs in the in vitro and in vivo control samples, and 7 genes were overlapped (SI Appendix, Fig. S3). The 7 genes include essential DNA repair genes like CHEK1, MSH2, and RAD21 (SI Appendix, Fig. S3). One of the in vivo Gem-treated tumors had substantially more sgRNAs depleted than the other two replicates. Reasoning that this tumor responded to gemcitabine at a higher than expected rate, we excluded it from the downstream analysis (SI Appendix, Fig. S2). Since the nongenomic targeting control sgRNAs were well represented in all of the samples, they were used to profile the null distribution of robust rank aggregation (RRA) scores when calculating the P values (14) (SI Appendix, Fig. S4). Negative selection RRA scores identified genes that were consistently depleted compared to day 0 samples, indicating that these genes are critical for the survival of the PDX cell line (SI Appendix, Fig. S5). To check this, we compared these sets of genes with known essential fitness functions. Critically, more than half of the 104 critical survival genes that we identified from the in vitro and in vivo samples overlapped with the previously identified essential gene list from five independent cell lines (16). It is also notable that nearly one-third of essential fitness genes we identified are in vitro or in vivo specific, indicating their differential essentiality for different growth conditions. Reasoning the limited therapeutic index of targeting these essential genes, we excluded them from the candidate genes that showed synthetic lethality with Gem.
Identifying Genes Whose Depletion Results in Synthetic Lethality with Gemcitabine.
We aimed to identify genes that could be therapeutically targeted to synergistically boost the therapeutic effect of Gem. We, therefore, prioritized our CRISPR screening hits based on three criteria. Firstly, the gene must have been significantly depleted both in vitro and in vivo. We also included the in vitro screening data so that we could robustly validate the screening hits using in vitro assays. Secondly, the potential hit must have been “druggable,” i.e., have an existing inhibitor. And finally, targeting the CRISPR hit should have a high potential for strong therapeutic value. Among these three criteria, the latter one is more ambiguous. To this end, we focused on genes whose high expression have a substantial negative prognostic value for PDAC patients.
This primary screening (Fig. 1A) identified MCRS1, SMARCD1, PRMT5, CXXC1, SETDB1, ACTL6A, and DNMT1 as significant hits whose depletion was potentially lethal with Gem (Fig. 1 B and C). We then performed a validation screening with additional sgRNAs for each of these genes using the PDX366 cell line. PRMT5 scored as the top hit whose depletion synergistically increased Gem cytotoxicity (Fig. 1D). PRMT5 is the primary type II PRMT that is responsible for the majority of symmetric dimethylation (SDMA) on the arginine residues of its targets, which include various histone proteins as well as transcription factors (17). PRMT5 is implicated in diverse functions, including genome organization, transcription, cell cycle, and spliceosome assembly (18). The role of PRMT5 in the proliferation of cancer cells is increasingly appreciated (19–22). Importantly, PRMT5 is a druggable protein with several selective inhibitors available and many of which are currently being tested in clinical trials (e.g., NCT03573310 and NCT03854227). However, its significance in PDAC progression or its potential as a combinatorial therapeutic target in PDAC cells has not been explored.
We, therefore, investigated the potential role of PRMT5 in PDAC progression and aimed to assess whether PRMT5 inhibition has a potential therapeutic value for PDAC. To this end, we initially analyzed whether PRMT5 expression is differentially regulated in PDAC tumors and has a prognostic value for the survival of patients. The gene expression analysis of multiple independently generated datasets such as normal-matched PDAC tumor (GSE28735) (23), tumor-adjacent normal vs. PDAC tumor (GSE16515) (24), and laser microdissected PDAC tumor cells vs. adjacent stromal cells (GSE93326) (25) shows that PRMT5 mRNA expression is significantly up-regulated in PDAC cancer cells compared to normal stromal cells (Fig. 1E). Most critically, the analysis of The Cancer Genome Atlas (TCGA) PDAC patient data shows that tumors with high PRMT5 expression result in significantly shorter overall patient survival (Fig. 1F), indicating that PRMT5 is a critical player in PDAC progression or therapy response, and thus a promising therapeutic target.
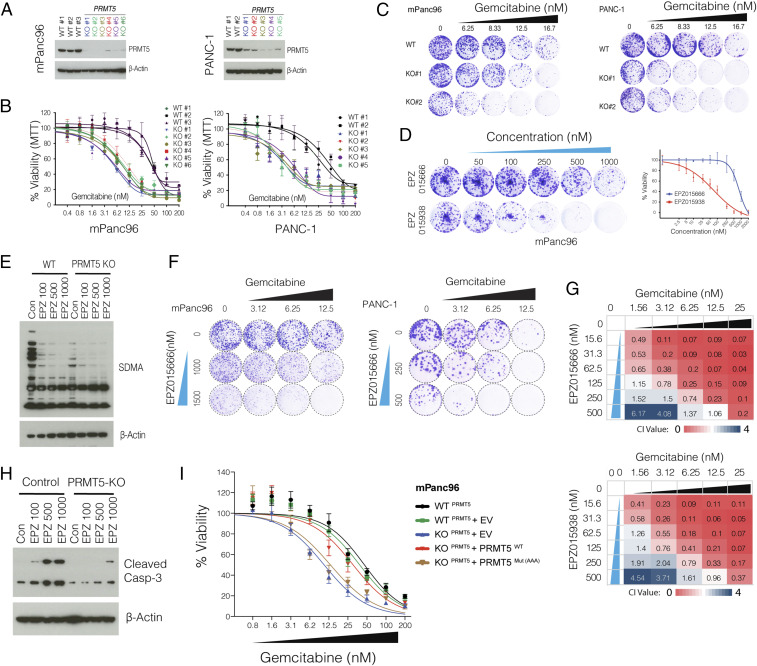
To better study the role of PRMT5 in the cellular response to Gem, we generated multiple single KO clones in two additional pancreatic cancer cell lines (mPanc96 and PANC-1) (Fig. 2A). Notably, despite screening for ∼100 single clones, we seldom observed full depletion of PRMT5 at the protein level, especially in PANC-1 cells. However, the clones with even partial PRMT5 depletion were nearly an order of magnitude more sensitive to Gem compared to WT clones (4 to 5 μM vs. ∼50 μM IC50) as measured by cell viability assay (Fig. 2B). In line with these, longer-term crystal-violet colony formation assays also demonstrated that the PRMT5 KO clones were significantly more sensitive to Gem compared to WT cells (Fig. 2C).
Fig. 2.
PRMT5-depleted cells are hypersensitive to Gem. (A) Western blot shows PRMT5 protein levels in WT cells, as well as single-cell, expanded clones expressing PRMT5 targeting sgRNAs. The beta-actin level is shown as a loading control. (B) The line graph shows % viability of WT and PRMT5 KO PDAC cells in response to increasing doses of Gem. (C) Crystal violet colony formation assay shows the overall survival of indicated WT and PRMT5 KO cells. (D) Crystal violet colony formation assay shows relative growth inhibition activity of two separate PRMT5 inhibitors. (E) Western blot result shows relative levels of symmetric dimethylation of arginine (SDMA) in WT cells and PRMT5 KO cells treated with increasing concentration (nM) of the indicated PRMT5 inhibitor. (F) Crystal violet colony formation assay shows relative survival and proliferation rates of mPanc96 (Left) and PANC-1 cells (Right) treated with various combinatorial doses of Gem and a PRMT5 inhibitor in two separate cell lines. (G) Heatmaps show the combination index (CI) values across multiple combinatorial doses in PDX 366T cells. CI < 1 indicates synergism for two independent PRMT5 inhibitors. (H) Western blot result shows a relative rate of Caspase-3 cleavage in WT and PRMT5 KO cells treated with increasing doses (nM) of PRMT5 inhibitor. (I) Cell viability results are shown for WT cells as well as PRMT5 KO cells that express WT PRMT5 and an enzymatically inactive mutant form of PRMT5.
To further corroborate these genetic depletion results, we tested two separate small molecule pharmacological inhibitors (EPZ015666 and EPZ015938) that specifically target PRMT5. When tested as a single agent, EPZ015938 had substantially more growth inhibition activity on colony formation (Fig. 2D). As anticipated, EPZ015666 treatment significantly inhibited global SDMA (Fig. 2E). These inhibitors significantly potentiated Gem growth inhibition activity at multiple dose combinations as measured by long-term colony formation assay (Fig. 2F). To better assess whether PRMT5 inhibitors are synergistic with Gem, we calculated the combination index (CI) values for each dose combination (26). The CI < 1 indicates synergy between two drugs, whereas CI ∼ 1 is additive, and CI ≥ 1.2 suggests an antagonistic effect. Importantly, of the 24 dose combinations for two separate inhibitors, we observed robust synergistic activity for ∼80% of EPZ015666 + Gem and ∼78% of EPZ015938 + Gem dose combinations (Fig. 2G). Critically, the inhibitor is specific toward PRMT5 as it results in selective apoptosis (caspase-3 cleavage) in WT control cells but not in PRMT5 KO cells (Fig. 2H).
To further understand whether PRMT5 enzymatic activity is required for survival of PDAC cells under the Gem treatment pressure, we performed rescue experiments in the KO cells. Significantly, exogenous expression of WT PRMT5 fully rescues the viability of PRMT5 KO cells, making them as resistant as WT cells. (Fig. 2I). On the other hand, the expression of an enzymatically mutant form of PRMT5 (Glu392A, Asp419A, and Glu435A) did not yield any significant increase in the viability of PRMT5 KO mPanc96 cells. These findings further highlight that the enzymatic activity of PRMT5 is a critical determinant of cellular survival in response to Gem treatment.
Understanding Pathways Underlying PRMT5 Depletion-Mediated Vulnerability to Gem.
Encouraged by the genetic depletion and pharmacological inhibition studies, we then aimed to understand the molecular mechanism of conditional sensitivity of PRMT5-depleted cells to Gem and assess the therapeutic value of this combination in vivo. At the chemical level, Gem is composed of di-fluoro-deoxycytidine (dFdC). Mechanistically, it exerts its biological effects by inducing replication stress in fast-dividing cancer cells. Once taken up by the cells, dFdC is converted into dFdC-diphosphate (dFdCDP) and dFdC-triphosphate (dFdCTP). dFdCTP incorporates into DNA as a cytosine analog and blocks DNA synthesis due to strand termination. Additionally, dFdCDP also inhibits the ribonucleotide reductase enzyme, thereby resulting in depletion of the dNTP pool necessary for DNA synthesis.
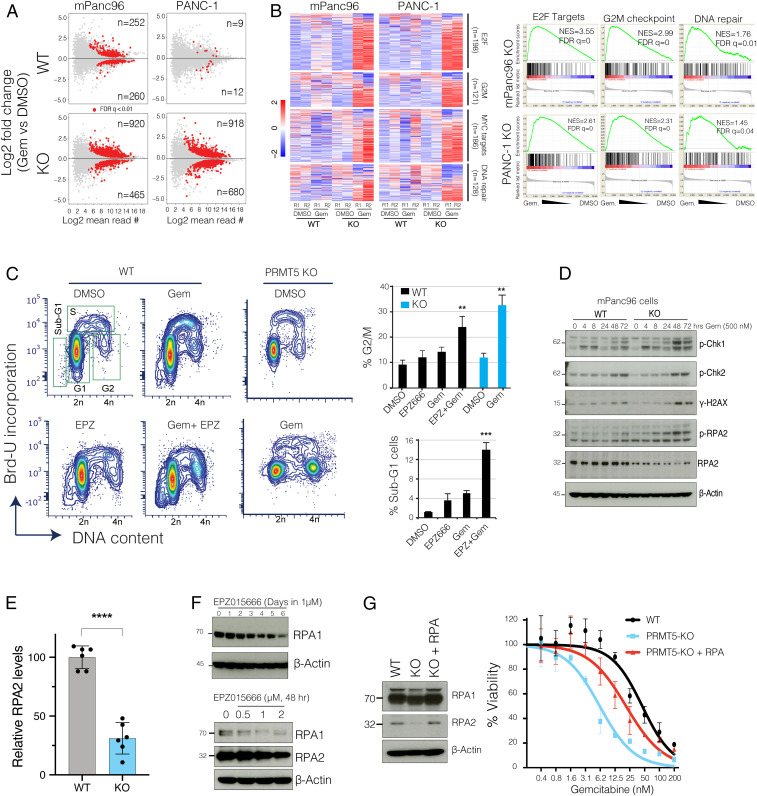
Since PRMT5 is a major transcriptional regulator, we initially investigated whether PRMT5-depleted cells had a differential transcriptional response to Gem. We, therefore, comparatively analyzed the transcriptional responses of PRMT5 WT and KO cells to Gem in two independent PDAC cancer cell lines. Critically, the KO cells responded to Gem by differentially regulating a much larger number of genes. For example, while only 21 genes (9 up, 12 down) in WT PANC-1 cells and 512 genes (252 up, 260 down) in mPanc96 WT cells were significantly altered, 1,589 genes (918 up, 680 down) in PANC-1 KO cells and 1,385 genes (920 up, 465 down) in the mPanc96 KO cells were significantly altered in response to treatment with IC30 Gem for 24 h (Fig. 3A). These results suggested that physiological levels of PRMT5 are required to buffer global transcriptional response to Gem. Comparative gene set enrichment analysis (GSEA) demonstrated that genes implicated in cell cycle and DNA repair pathways were aberrantly active in the KO cells when treated with Gem (Fig. 3B). Gene sets identifying cell cycle-related genes such as G2/M checkpoints, and E2F and MYC targets were all more strongly up-regulated in Gem-treated KO cells compared to WT cells. Furthermore, genes involved in DNA repair were among the most highly differentially regulated genes when the KO cells were treated with Gem (Fig. 3B).
Fig. 3.
PRMT5 depletion results in the aberrant transcriptional program of cell cycle and DNA repair genes in response to Gem. (A) The MA plots (log fold change vs. log mean expression of each gene) show the number of differentially regulated genes in WT and PRMT5 KO cells due to Gem treatment. (B) Heatmaps and GSEA show relative levels of expression changes in genes involved in indicated cellular processes. (C) Flow cytometry cell cycle analysis (DNA content vs. BrdU incorporation) of control vs. Gem (250 nM)-treated WT, PRMT5 KO, or PRMT5 inhibitor EPZ015666 (500 nM)-treated cells. The bar plot shows the percent of cells at the indicated cell cycle stage. ** and *** indicate P values less than 0.01 and 0.001, respectively. (D) Western blots show relative levels of phosphorylated or total levels of indicated proteins. (E) Bar plots of RPA2 protein levels quantified from Western blots. **** indicates P value less than 0.0001. (F) Western blots show levels of RPA1 and RPA2 proteins in PDAC cells treated with various times and doses of the indicated PRMT5 inhibitor. (G) Western blots show relative levels of RPA1 and RPA2 protein levels in WT, PRMT5 KO, and PRMT5 KO cells expressing RPA cDNA. The line plots show the relative viability of indicated cells in response to increasing doses of Gem.
These results support the hypothesis that PRMT5 depletion-mediated conditional vulnerability to Gem was partially due to the aberrant regulation of cell cycle and DNA repair pathways. To test this hypothesis, we set out several molecular assays to study the mechanism of PRMT5 depletion-mediated aberrant cell cycle and DNA repair programs. The analysis of cell cycle position through bromodeoxyuridine (BrdU) incorporation showed that Gem treatment of WT cells resulted in a partial delay in cell cycle with a substantial accumulation of cells in S phase and partial increases in G2/M cells (Fig. 3C). On the other hand, the combination of Gem and PRMT5 inhibitor resulted in a significant accumulation of G2/M cells and sub-G1 dead cells (Fig. 3C). In line with the pharmacological inhibition of PRMT5, Gem treatment resulted in a significantly higher number of G2/M cells in the PRMT5 KO cells compared to WT cells.
PRMT5 Depletion Results in RPA Exhaustion.
Coordinated activation of cell cycle checkpoints and cell cycle arrest is one of the primary mechanisms that enable cells sufficient time to repair DNA against external cues (27). The robust arrest of cells at the G2/M cell cycle led us to study the activation of checkpoints further. Excessive DNA damage in S and G2/M activates ATR-Chk1-Cdc25C (28–30). Therefore, we performed time-course experiments to study whether Gem treatment resulted in differential activation of DNA damage and cell cycle checkpoints in the KO cells. Notably, we detected sustained and stronger phospho-Chk1 (a marker of DNA damage, S and/or G2/M arrest), gamma-H2AX (a marker of DNA damage) as well as phospho-RPA2 (a marker of DNA damage and replication stress) in the KO cells compared to WT cells with Gem treatment (Fig. 3D).
This analysis also revealed something unexpected to us. Although we observed a strong induction of phospho-RPA2 in the KO cells, the total levels of RPA2 were substantially lower in the KO cells (Fig. 3D). Further quantitative analyses suggested that the depletion of PRMT5 resulted in a significant reduction in RPA2 protein levels (P < 0.0001). We observed a ∼60 to 70% reduction in RPA2 levels in the PRMT5 KO cells compared to WT cells (Fig. 3E). These findings led us to investigate whether the depletion of RPA was due to enzymatic activity of PRMT5. Critically, both time-course, as well as dose-escalation experiments, showed that depletion of PRMT5 activity through small molecule inhibitors resulted in RPA depletion (Fig. 3F).
It should be noted that RPA2 is one of the three subunits of the RPA complex, which is viewed as the guardian of the genome (31, 32) because it binds and protects any single-stranded DNA (ssDNA) that forms during DNA replication, transcription, and repair. The cytotoxicity of Gem in fast-dividing cancer cells is mostly due to the creation of replication stress (RS) by blocking DNA synthesis and diminishing the dNTP pool by inhibiting ribonucleotide reductase enzyme. During RS, RPA becomes essential to protect ssDNA at the stalled replication forks. Critically, the global RPA level is a crucial determinant as to whether cells can resolve the stalled forks. In low RPA conditions, RS leads to “replication catastrophe,” where chromosomes shatter with thousands of double-strand breaks (DSBs) (33). These findings point to the “RPA exhaustion” hypothesis, which states that when RPA is not sufficient, cells can’t survive RS, and the stalled replication forks collapse, which results in the breakage of forks, and ultimately replication catastrophe (34).
Our results so far support the hypothesis that PRMT5 depletion results in RPA exhaustion, and therefore, cells are not able to cope with Gem-mediated DNA damage. To test this hypothesis, we aimed to replenish the RPA complex to see if it could rescue the PRMT5 depletion phenotype. Since RPA works as a tripartite complex where each subunit is needed at an equimolar ratio, we exogenously provided cells with a vector that expresses all three subunits (33). Importantly, replenishing the RPA complex in PRMT5 KO cells to near equal levels to WT cells (Fig. 3G) results in significant resistance to Gem in PRMT5 KO cells. These results suggest that, at the molecular level, the PRMT5 depletion-mediated Gem-sensitivity phenotype is, in part, due to exhaustion of the RPA complex.
PRMT5 Depletion Results in Excessive DNA Damage Accumulation.
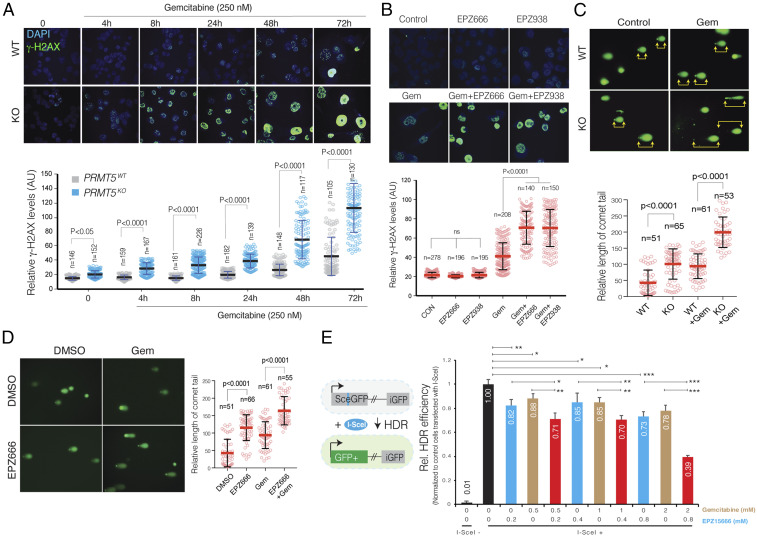
Our expression analysis also highlighted that genes involved in DNA repair pathways were aberrantly regulated when PRMT5 KO cells were treated with Gem. Furthermore, the above results suggested that due to RPA exhaustion, PRMT5 depleted cells are not able to resolve a stalled replication fork, which may result in the collapse of the fork and accumulation of DNA DSB. We, therefore, utilized two independent molecular assays to detect and quantify Gem-induced DNA damage in control and PRMT5-depleted cells. Initially, we used immunofluorescence (IF) assays to detect the phosphorylated gamma-H2AX (γ-H2AX), which is a modified histone variant deposited into a mega-base chromatin region around the DSB. Critically, strong γ-H2AX foci can be detected as early as 4 h post-Gem treatment in the KO cells. On the other hand, it took 48 to 72 h to detect similar levels of γ-H2AX foci in WT cells using the same concentration of Gem (Fig. 4A). Quantitative analysis of γ-H2AX foci formation levels over a period of 72 h showed that PRMT5 KO cells consistently had significantly higher levels of γ-H2AX, indicating higher levels of DNA DSBs due to Gem treatment (Fig. 4 A, Lower). In line with the genetic depletion of PRMT5, pharmacological inhibition of PRMT5 with two separate small molecule inhibitors also demonstrated that depletion of PRMT5 activity resulted in significant accumulation of DNA DSBs, as detected by levels of γ-H2AX foci formation (Fig. 4B). In addition to γ-H2AX foci formation, we also measured the level of DNA strand breaks through a comet assay, which measures the overall levels of DNA damage, as done through single-cell gel electrophoresis. As the frequency of DNA breaks increases, so does the fraction of the DNA extending toward the anode, forming the comet tail. The length of the tail is an indication of levels of fragmented DNA in individual cells. In line with the γ-H2AX IF results, we observed a significantly longer comet tail when PRMT5 KO cells or PRMT5-inhibited cells were treated with Gem, compared to WT and control-treated cells, respectively (Fig. 4 C and D).
Fig. 4.
PRMT5 depletion results in impaired DNA repair and excessive DNA damage accumulation in the PDAC cells treated with Gem. (A and B) IF images of γ-H2AX relative levels in WT, PRMT5 KO, and PRMT5 inhibitor EPZ015666 or 938 (500 nM)-treated WT mPanc96 cells in response to Gem treatment (250 nM) for indicated times (Upper). The dot plot (Lower) shows quantified IF γ-H2AX levels at the indicated number of single cells. N indicates the number of cells quantified. (C and D) IF images of comet assay indicate levels of overall DNA strand breaks in WT, PRMT5 KO, and EPZ015666 (500 nM)-treated WT mPanc96 cells in response to Gem treatment (250 nM) (Left). The Right show individual cell level quantified length of the comet tail in the indicated number of cells. (E) The bar plot shows results of I-SceI endonuclease-based genetic reporter assays indicating relative repair efficiency of DNA strand breaks through HDR. *, **, and *** indicate P values less than 0.05, 0.01, and 0.001, respectively.
PRMT5 Depletion Results in Impaired DNA Repair Activity.
Depending on the time and kind of DNA damage, the DSB is repaired through either precise HDR or error-prone nonhomologous end joining (NHEJ) (27). NHEJ is active throughout the cell cycle, whereas HDR is restricted to the late S and G2 phases of growing cells (35). On the other hand, DNA double-strand breaks due to stalled and collapsed replication fork in S phase can be precisely repaired by HDR. During this process, RPA levels are a critical determinant of whether the stalled replication fork will resolve and resume (36). Our differential gene expression results and the finding that RPA is significantly depleted in PRMT5 KO cells led us to test the hypothesis that the excessive DNA damage accumulation in Gem-treated PRMT5 KO cells is due to impaired HDR activity. To test this, we used I-SceI endonuclease-based genetic reporters where relative efficiency of DSB repair by the HDR pathway could be robustly quantified through a fluorescent reporter system. In the HDR GFP-reporter system (Fig. 4E, schematics), the construct contains two defective GFP genes; the first one contains an I-SceI site. The engineered cells (HeLa) that stably express this construct are GFP−. However, exogenous expression of I-SceI leads to a DSB, and when this break is repaired through the HDR pathway, it creates intact GFP. The level of GFP+ cells can be quantified as a direct measure for relative HDR repair efficiency (37, 38).
Our results show that both Gem and PRMT5 inhibitor treatment substantially inhibits the HDR activity as single agents (Fig. 4E). Importantly, the combination of these two drugs results in a significant further reduction of HDR activity. Significantly, this additional reduction is especially notable at higher dose combinations. These findings support the overall hypothesis that depletion of PRMT5, using small molecule inhibitors, results in depleted RPA complex, which results in impaired HDR-mediated DNA repair (Fig. 4E). When cells are exposed to additional DNA damaging agents such as Gem, they cannot survive due to increased DNA damage accumulation.
The Combinatorial Treatment Results in Synergistic Tumor Growth Inhibition In Vivo.
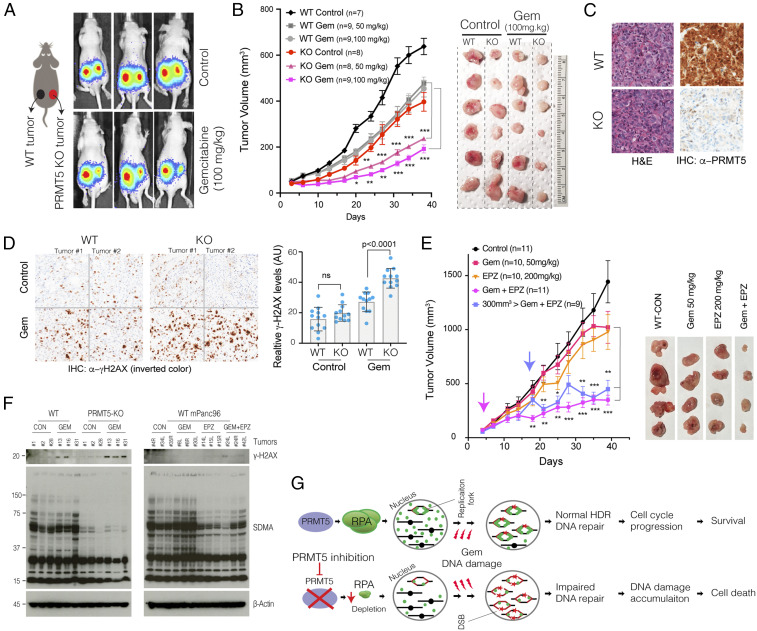
Next, we investigated to see if the combination would result in synergistic growth inhibition of PDAC tumors. To this end, we explored both genetic depletion and pharmacological inhibition of PRMT5 in a xenograft model of PDAC. Initially, we tested whether tumors formed by WT and PRMT5 KO cells were differentially sensitive to Gem treatment. To be able to better compare the tumors from these two genetic backgrounds, we injected 5 × 105 WT mPanc96 cells in the left flank and the same number of the PRMT5 KO cells in the right flank of the same mouse. This strategy enabled us to compare the two tumors grown in the same mouse. After 1 wk of tumor formation, the mice were randomly divided into three groups where one received control, and the other two received two separate Gem doses (50 mg/kg or 100 mg/kg). Notably, the PRMT5 KO cells were able to form tumors. However, these tumors were slightly smaller than the tumors formed by WT cells. The Gem treatments did result in a notable reduction of WT tumors. However, the most significant decrease in tumor volumes was in the PRMT5 KO tumors treated with Gem (Fig. 5 A and B). Starting from the fifth treatment (day 20 of tumor formation), the Gem-treated PRMT5 KO tumors were significantly smaller than their WT counterparts or the untreated KO tumors.
Fig. 5.
Genetic depletion or pharmacological inhibition of PRMT5 results in synergistic tumor growth inhibition with Gem. (A) Schematics show the experimental strategy where WT and PRMT5 KO mPanc96 cells are xenografted in the left and right side of the mice, respectively. The bioluminescence imaging results show relative levels of WT and PRMT5-depleted tumors in control and Gem-treated mice. (B) The line plot shows caliper-measured relative tumor volumes over time in WT and PRMT5-depleted tumors treated with control and two separate Gem doses. The images show extracted tumors at the end of the experiments. (C) H&E and IHC staining, respectively, show tumor architecture and relative levels of PRMT5 protein in tumors originated from WT and PRMT5 KO cells. (D) IHC images and bar plots show relative levels of DNA damage (γ-H2AX staining) in WT and PRMT5-depleted tumors treated with control vehicle or Gem. (E) The line plots show caliper measured relative tumor volumes in vehicle control, single agent, or combinatorial Gem, and PRMT5 inhibitor (EPZ015666)-treated mice. Pink and blue arrows indicate treatment start times for respective modalities. The images show extracted tumors at the end of the experiment. (F) Western blots indicate relative levels of DNA damage (γ-H2AX) and SDMA in multiple different tumor tissues receiving indicated treatments. (G) Schematic describing the model on the mechanism of increased cytotoxicity due to combined Gem and PRMT5 depletion. *, **, and *** indicate P values less than 0.05, 0.01, and 0.001, respectively. ns stands for not significant.
We also extracted tumors to analyze their morphology and molecular structure through hematoxylin and eosin (H&E) and immunohistochemistry (IHC) for selected markers. The H&E staining of WT and PRMT5 KO tumors demonstrated comparable cellular architecture (Fig. 5C). The IHC staining confirmed that the tumors from PRMT5 KO cells did not express PRMT5 protein (Fig. 5C). We then performed IHC to investigate whether the Gem treatment resulted in more significant DNA damage in PRMT5 KO cells in vivo as was seen in in vitro experiments. Consistent with the in vitro experiments, we observed significantly more γ-H2AX staining in the PRMT5 KO tumors when treated with Gem (Fig. 5D), indicating that Gem resulted in considerably higher amount of DNA damage in the PRMT5 KO tumors than in the WT tumors.
We next performed in vivo xenograft experiments to assess whether pharmacological inhibition of PRMT5 would result in a synergistic reduction of PDAC tumors when combined with Gem. To this end, we designed two separate strategies: early treatment and delayed treatment. One set of tumors was treated with control, single-agent drugs or combination of Gem and PRMT5 inhibitor as soon as the tumors reached ∼100 mm3. Importantly, as soon as a week after treatment, we observed a statistically significant reduction in tumor volume only in the tumors receiving the combination treatment. It is notable that the therapeutic effect of PRMT5 inhibitor plus Gem is much stronger than the Gem or PRMT5 inhibitor treatment alone (Fig. 5E). Importantly, we also allowed a set of tumors to grow to significant sizes (∼300 mm3) before starting the combination treatment. Interestingly, we observed a notable decrease in tumor volume after just one cycle of combined treatment (Fig. 5E). These tumors started to grow for the next couple of treatment cycles but then stopped growing and remained significantly smaller in volume compared to the control or single-agent-treated tumors. In the end, these tumors were almost indistinguishable from the tumors that received combination treatment from the beginning.
To assess whether the combination treatment abolished PRMT5 function in vivo, we evaluated the overall levels of the γ-H2AX and the symmetric arginine dimethylation (SDMA) in WT tumors, PRMT5 KO tumors, as well as single-agent and combination-treated WT tumors. As anticipated, the PRMT5 KO tumors had significantly lower SDMA levels. In line with genetically depleted tumors, the EPZ015666-treated tumors had lower overall levels of SDMA, indicating that the inhibitor doses that we used resulted in substantial inhibition of PRMT5 function in the tumors in vivo (Fig. 5F). Furthermore, the γ-H2AX was significantly activated in both the GEM-treated PRMT5 KO tumors and the combination-treated tumors (Fig. 5F), suggesting that activation of the γ-H2AX may be due to the low level of SDMA.
Discussion
Our main objective in this study was to identify novel drug targets that could synergistically increase the therapeutic efficacy of existing chemotherapy in PDAC, which remains one of the deadliest cancers despite extensive efforts. PDAC comprises 95% of pancreatic cancers, and the 5-y survival rate is less than 8%. Rationalizing that the nucleoside analog Gem is the most widely used chemotherapy agent and the backbone of other combinatorial therapies (10), we specifically focused on identifying novel combinatorial targets that may result in synthetic lethality with Gem. To achieve this, we employed multiplex CRISPR gene KO screening technology with a library of sgRNAs to screen chromatin regulators whose depletion may create conditional lethality with Gem. The screening was performed in an orthotopic PDX model where a tumor is growing in the pancreas of the mice. In this screening, we used a custom sgRNA library targeting ∼700 genes implicated in epigenetic control of chromatin regulation. The initial screening and subsequent validation experiments demonstrated that PRMT5 inhibition resulted in the synergistic vulnerability of PDAC cells to Gem.
The role of PRMT5 as a critical driver of cancer progression is an emerging topic. More importantly, PRMT5 is a druggable protein. There are several ongoing clinical trials of PRMT5 inhibitors for multiple advanced-stage cancers (NCT03573310 and NCT03854227). Although PDAC patients are currently not recruited for these trials, our findings presented here show that PRMT5 is a significant therapeutic target in PDAC patients who mostly receive Gem as a chemotherapeutic agent. We show that PRMT5 expression is significantly induced in PDAC tumors and more critically, higher PRMT5 expression is associated with significantly poorer survival in patients with PDAC. These findings indicate that inhibiting PRMT5 will positively impact patient survival. Notably, our screening results indicate that PRMT5 inhibition is synthetically lethal with Gem. Since PDAC cells express higher levels of PRMT5, these findings suggest that combinatorial PRMT5 inhibition with Gem will have stronger and potentially selective efficacy toward PDAC cells.
Recently, several complementary reports demonstrated that combining PRMT1 inhibitors with PRMT5 inhibitors resulted in synergistic antitumor activity (39, 40). Although PRMT1 did not score as one of the top hits in our screening, it is tempting to speculate that the triple combination of inhibitors targeting PRMT1 and PRMT5 with Gem may result in far greater synergistic cytotoxicity toward PDAC cancer cells. In such a combinatorial strategy, achieving a high therapeutic index is the ultimate goal in cancer treatment. Notably, the PRMT5 inhibitor dose that achieved significant and synergistic cytotoxicity with Gem in our study is an order of magnitude less than what has been shown to have a cytotoxic effect in hematopoietic cancers (250 nM vs. 10 μM) (19). These results indicate that combinatorial strategies may have a strong therapeutic index.
How PRMT5 inhibition results in synergistic cytotoxicity with Gem is yet to be understood entirely. The accumulated DNA damage in the combination treatment sheds some mechanistic insights. Our results support the model that PRMT5 depleted cells cannot tolerate the replication stress elicited by the Gem treatment. PRMT5 inhibition leads to significant depletion of RPA, which acts as the guardian of the genome by binding and protecting ssDNA that forms during DNA replication and increases during replication stress and repair (31, 32). Binding of RPA to ssDNA constitutes a key physiological signal which activates the master ATR kinase to protect and repair stalled or collapsed replication forks during replication stress (41). We observe significant DNA damage accumulation in the combination of Gem plus PRMT5 depletion. Although the molecular mechanism is not entirely clear, replication fork stalling due to Gem treatment is expected to make RPA the limiting factor in preventing replication catastrophe (33, 34). We hypothesize that PRMT5 inhibition is a tipping point by depleting RPA and decreasing HDR. These two activities could be linked to each other as RPA is also needed for efficient DSB repair through HDR (42, 43). Critically, we see significant reduction in HDR when cells are treated with either Gem or PRMT5 inhibitors in a reporter assay that uses a single DSB. However, the combination of these two drugs results in further significant reduction in HDR activity. This finding supports and at least in part explains the synergistic increase in overall DNA damage as measured by γ-H2AX staining and alkaline-condition comet assays that measure both single and DSBs.
In light of these findings, we propose a model (Fig. 5G) which suggests that enzymatically active PRMT5 is needed for cells to survive replication stress caused by DNA damaging agents. The model postulates that inhibition of PRMT5 leads to reduced HDR through RPA exhaustion and hence enhanced accumulation of DNA DSB and cell death. Importantly, our model predicts that other DNA damaging chemotherapeutic agents, such as cisplatin, could also result in enhanced cytotoxicity when PRMT5 is depleted. Furthermore, the synergistic cytotoxicity induced by PRMT5 inhibition and DNA damaging agents may not be specific to PDAC cells. Therefore, it is plausible to speculate that such combinations may have a strong therapeutic index in other cancers where PRMT5 is significantly expressed.
In conclusion, our findings indicate the power of unbiased in vivo CRISPR screening in a clinically relevant model system. One limitation of in vivo screening is the limited number of target genes that can be screened. This limitation emerges due to the maximum number of cells that can be injected into the relevant anatomical site in the in vivo model. Since the complexity of the sgRNA library needs to be preserved at ∼200× during all experimental steps, screening with a library targeting the whole genome is currently not feasible with the CRISPR/Cas9 system. However, alternative CRISPR systems, such as the CRISPR/AsCpf1 system, can be exploited to construct significantly smaller libraries (44), which may potentially enable genome level screening in vivo.
Materials and Methods
In Vitro Cell Culture.
The PDX366 cell line had been established from deidentified pancreatic patients’ tumors prior to this study and has been previously described (12). PDX366, mPanc96, and PANC-1 pancreatic carcinoma cells were cultured in RPMI1640 medium supplemented with 10% fetal bovine serum (FBS) and 1% streptomycin/penicillin. Cells were treated with gemcitabine (GEMZAR; Eli Lilly) and/or either EPZ015666 (GSK3235025; SelleckChem) or EPZ015938 (GSK3326595; ChemieTek).
Generation of CRISPR sgRNA Library Pool and Viral Infection.
The PDX366 cell line was produced from pancreatic patients, as described earlier (12). WT Cas9 expressing lentivirus was generated in HEK293T cell line by cotransfection of WT Cas9 (modified from GeCKO plasmid by removing gRNA), psPAX, and pMD2.6 plasmid with a 5:4:1 ratio. A total of 10 µg total DNA was used in the presence of 30 µL of Fugene6 reagent in a 10-cm plate dish that had 70% confluency. The PDX366 cell line was infected with this lentivirus for 1 d and then treated with 0.5 µg/mL puromycin for 4 d. The nuclear sgRNA libraries were kind gifts from D. M. Sabatini’s laboratory, Massachusetts Institute of Technology, Cambridge, MA (45); (Addgene catalog no. 51047). The libraries were amplified using the published protocol at Addgene. (www.addgene.org/static/data/08/61/acb3ad96-8db6-11e3-8f62-000c298a5150.pdf). The library pool targets 619 epigenetic regulators with ∼10 sgRNA/gene. A total of 360 nongenomic targeting control sgRNAs are included in the library. The sgRNA library expressing viruses was generated in 2 × 15 cm plates by using a total of 20 µg DNA and the condition mentioned above. Serial dilutions of a virus were used to find the MOI of ∼0.25 after selection with 5 µg/mL blasticidin for 4 d. Cells were harvested from 12 × 15 cm plates to get at least 200× fold coverage (∼2 million cells per sample) for the in vitro and in vivo (orthotopic injection into mouse pancreas) screening.
In Vivo CRISPR Screening in an Orthotopic PDX Model of PDAC.
Six- to seven-wk-old athymic nude mice (Envigo) were used for in vivo screening and selection. The sgRNA library, WT Cas9-expressing PDX366 cells, were resuspended in 150 µL Matrigel Growth Factor Reduced Basement Membrane Matrix (Corning). After the anesthesia, the left flank of the mouse was opened to exteriorize the pancreas, and 8 × 106 PDX366 cells were injected directly into the pancreas. At this stage, one batch of cells was harvested as day 0 control sample. For in vitro screening, cells were passaged every 3 to 4 d by a 1:3 split with fresh media in 15-cm plates. At least 12 million cells were passaged each time using 3 × 15 cm plates.
Tumor volumes were monitored by MRI. MRI measurement (University of Virginia Molecular Imaging Core, Charlottesville, VA) was performed after 4 wk, at the conclusion of the experiment. Tumors were harvested and weighed and samples collected for further analysis. Formalin-fixed tumor samples were submitted to the University of Virginia Research Histology Core Lab for processing and H&E staining. Tumor sections were scored by a board-certified pathologist who specializes in gastrointestinal cancers. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the NIH. The animal protocol was approved by the Animal Care and Use Committee of the University of Virginia (PHS Assurance A3245-01).
Targeted Amplification of CRISPR/sgRNA Library and Sequencing.
Tumors from mice and in vitro cultured cells were harvested after 4 wk. Entire tumors and all cell pellets were used to obtain genomic DNA. Briefly, tumor samples were minced into small pieces and lysed with 8 mL SDS lysis buffer (100 mM NaCl, 50 mM Tris-Cl pH 8.1, 5 mM EDTA, and 1% wt/vol SDS). Cell pellets were processed in a similar way. Minced tumor samples or cell pellets were treated with 100 µL proteinase K (20 mg/mL) at 55 °C for overnight incubation. The next day, entire lysis solutions were used in EtOH precipitation, and genomic DNA pellets washed with 70% EtOH twice. Pellets were resuspended in RNase-containing water and quantified by Nanodrop. For each DNA sample, 100 µg genomic DNA was used for the first PCR. We ran 10 separate PCR reactions with 10 µg DNA in a single PCR tube. We used the same outer forward primer and outer reverse primer from Sabatini sgRNA library-specific primers for all of the samples (these primers are different from GeCKO Array Forward and Reverse). Q5-High Fidelity 2× master mix was used as polymerase from NEB (M0429L). PCR conditions for the first PCR were 98 °C for 30 s, 18× (98 °C for 10 s, 63 °C for 10 s, and 72 °C for 25 s) and 72 °C for 2 min. After the first PCR, all reactions were combined (10 × 100 µL) in one single Eppendorf tube and vortexed well. For the second PCR, 5 µL PCR mix from the first PCR step was used in 100 µL total PCR. PCR conditions for the second PCR were 98 °C for 30 s, 24× (98 °C for 10 s, 63 °C for 10 s, and 72 °C for 25 s) and 72 °C for 2 min. In the second PCR, each sample was amplified with specific forward primers that had a 6-bp barcode sequence for demultiplexing of our reads during next-generation sequencing and common reverse primer. In this setting, custom sequencing and custom indexing primers for Illumina Sequencing were used. The entire solution from the second PCR was loaded on a 2% gel, and the bands around 270 bp were cut and cleaned with the Qiagen gel extraction kit (a faint band above 270 bp was noticed, likely due to carrying over of primers from the first PCR). Purified PCR products were quantified by using Qubit (Invitrogen), and equimolar amounts of each PCR fragment were mixed and used for subsequent high-throughput sequencing (40 nM DNA in 20 µL). The library was sequenced using the Illumina Miseq platform to get an average of 10 million reads for each sample.
Data Analysis for CRISPR/Cas9 Screening.
Sequencing reads from CRISPR/Cas9 screenings were first demultiplexed with cutadapt (v. 1.8.3). Sequences of a total length of 56 nt (sequencing barcode and sample barcode) were supplied to the program with the requirements that at least 36 nt of this barcode had to be present in the read, so that it could be assigned to an individual tumor isolated from the PDX model. More than 99% of reads were assigned to one of the three in vitro and six in vivo samples: cells from the day of injection (further referred to as day 0), control and gem treated in vitro samples (one each), and control and gem treated in vivo samples (three each). After demultiplexing and removing sequencing and sample barcodes, the abundance of each sgRNA was assessed and normalized among samples with the use of MAGeCK v. 0.5.2. About 87% of the reads contained correct sgRNA sequences.
Downstream data analysis was performed in RStudio v. 0.99.484 with R v. 3.3.0 following the previous publications (45) with slight modifications. We performed the following analysis to identify potential combinatorial targets of gemcitabine: The first step of this analysis was to calculate the relative abundance of sgRNAs targeting each gene between day 0 and one of the other eight samples by comparing normalized average counts of all of the sgRNAs targeting the particular gene. Since the nongenomic targeting control sgRNAs were well represented in all of the samples, they were used to profile the null distribution of RRA scores when calculating the P values. Based on the negative selection RRA scores, one of the in vivo Gem-treated samples had a substantially higher sgRNA depletion rate compared to the other two replicates, and thus was excluded from the downstream analysis. Genes consistently depleted in all of the samples compared to day 0 were likely to be essential genes for the PDX cell line and were removed from the downstream analysis. The second step of the analysis was to calculate log fold change (LFC) of mean read counts between Gem-treated and control samples for all of the retained genes in in vitro and in vivo settings, respectively. In the third step, we ranked all of the retained genes based on LFC, and genes significantly depleted (false discovery rate [FDR] q < 0.1) in both in vitro and in vivo screenings were selected as candidate combinatorial targets of gemcitabine. The sgRNA-specific normalized read counts are provided in Dataset S1.
Validation of PRMT5 as a Viable CRISPR Screening Hit.
For validation of PRMT5 after the initial screening, the following sgRNA guiding sequences (sgRNAs) were designed and cloned to generate PRMT5 knockout cells:
sgRNA1: GGTACCCTTGGTGGCACCAG,
sgRNA2: GGTGATGGCCAGTGTGGATG,
sgRNA3: GTAAGGGGCAGCAGGAAAGC.
Briefly, the oligos that have 5′-CACC and 5′-AAAC overhangs of the sgRNA guiding sequence were ordered from Eurofins and hybridized to get sticky end double-strand DNA for ligation. The plasmid containing the sgRNA backbone was digested with Bbs.i restriction enzyme at 55 °C for 2 h, followed by calf intestinal treatment at 37 °C for a half hour. Purified vector backbone from a 2% gel (60 ng) and hybridized oligos (1 µL from 1 to 10 nM) was used for the ligation reaction in the presence of T4 ligase.
WT Cas9 and gRNA-expressing lentivirus were generated using the HEK293T cell line. mPanc96 and PANC-1 cells were virally infected to express Cas9 and sgRNA to produce stable cell lines. After 4 d of puromycin selection (2 µg/mL), serial dilution was performed to generate single clones. Once the desired number of clones was obtained, lysates were prepared in RIPA buffer, and Western blot was performed to determine PRMT5 knockout efficiency.
MTT Cell Viability.
PDX366, mPanc96, and PANC-1 cells were seeded in a flat-bottom 96-well plate (Corning) in triplicate at a density of 1 to 2 × 103 cells per well. The following day, cells were treated with gemcitabine (GEMZAR; Eli Lilly) and EPZ015666 (GSK3235025; SelleckChem) or EPZ015938 (GSK3326595; ChemieTek) for 4 to 5 d prior to MTT [3-(4,5-dimethylthiazolyl)-2,5-diphenyltetrazolium bromide] to determine effects of drugs on cell viability. Culture media were replaced with fresh RPMI, which had 10% FBS and 10% MTT (5 mg/mL) and incubated for 4 h in a humidified (37 °C, 5% CO2) incubator. A total of 100 µL MTT solvent (10% SDS in 0.01M HCL) was added to each well, and cells were incubated overnight. The absorbance was read at 595 nm.
Crystal Violet Assay.
Pancreatic cancer cells were seeded in a flat-bottom 12-well plate (Corning) at a density of 1 to 2 × 103 cells per well. The following day, cells were treated with gemcitabine and EPZ015666 or EPZ015938 for 2 wk. Culture media were replaced every week with fresh medium in the presence of drugs. Wells were washed with phosphate-buffered saline (PBS), then stained for 30 min with crystal violet solution (0.4% crystal violet, 10% formaldehyde, 80% methanol). After staining, wells were washed once with PBS and water. The plate was dried out overnight and imaged using a scanner. Colonies were measured and analyzed with ImageJ (NIH).
Annexin V Staining.
Annexin V staining was performed to determine the percentage of apoptotic cells. After treatment with gemcitabine and EPZ015666 or EPZ015938, the pancreatic cancer cells were washed with cold PBS, resuspended in Annexin V binding buffer (10 mM Hepes, 140 mM NaCl, and 2.5 mM CaCl2, pH 7.4) with an appropriate amount of FITC-conjugated Annexin V antibody (Life Technologies A13199), and incubated at room temperature (RT) for 15 min. After washing with binding buffer, the cells were resuspended in 2 μg/mL propidium iodide (PI) (Sigma) in PBS plus RNase, incubated at RT for 15 min in the dark, and then analyzed using a FACSCalibur flow cytometer (Becton-Dickinson).
BrdU Staining.
Pancreatic cancer cells were treated with gemcitabine and EPZ015666 or EPZ015938, incorporated with BrdU (Sigma) for 1 h, and then fixed by 70% ethanol. BrdU staining was performed according to the manufacturer’s instructions (BD Biosciences). Briefly, the fixed cells were washed with PBS and then resuspended in 2 N HCl for 20 min to denature the DNA. After washing with 0.1 M Na2B4O7, pH 8.5, to stop acid denaturation, the cells were resuspended and washed with 180 μL 0.5% Tween 20 (Sigma) with 1% normal goat serum (NGS) (Dako) in PBS. Then, the cells were incubated with Alexa Fluor 647-conjugated anti-BrdU (mAb) (Invitrogen) for 1 h at room temperature in the dark. After washing with PBS, the cells were resuspended in 2 μg/mL PI (Sigma) in PBS plus RNase, incubated at 37 °C for 30 min in the dark, and then analyzed by FACSCalibur flow cytometer (Becton-Dickinson).
Western Blot.
Cells were washed with cold PBS, and then lysed in RIPA buffer (Cell Signaling Technology). After centrifugation at 14,000 rpm for 15 min at 4 °C, the supernatants were collected, and the protein concentrations were measured using bicinchoninic acid protein assay reagent (Bio-Rad). Subsequently, equal amounts of proteins were separated in NuPAGE 4 to 12% Bis-Tris gradient gel (Invitrogen NP0335), and transferred onto nitrocellulose membranes (Invitrogen B301002). After blocking with 5% milk, the membranes were then probed at 4 °C overnight with various primary antibodies: anti-γ-H2AX (Cell Signaling), anti-phospho-Chk1 (Ser345) (Cell Signaling), anti-phospho-Chk2 (Thr68) (Cell Signaling), anti-PRMT5 (Abcam), anti-RPA1 (Abcam), anti-phospho-RPA2 (S4/S8) (Bethyl Laboratories), anti-RPA2 (Ab-2) (Calbiochem), cleaved caspase-3 (Cell Signaling), and anti-β-actin (Sigma), washed with TBST (20 mM Tris, 150 mM NaCl, 0.1% Tween 20; pH 7.6), and incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (Promega) at room temperature for 1 h. Finally, after washing with TBST, the antibody-bound membranes were treated with enhanced chemiluminescent Western blot detection reagents (GE Healthcare) and visualized with an X-ray film (GE Healthcare).
Immunofluorescence Staining.
Cells grown on glass coverslips (VWR) were rinsed with PBS and then fixed in 4% formaldehyde for 15 min. The cells were subsequently treated with 0.2% Triton X-100 in PBS for 10 min. After blocking with 2% BSA in PBS containing 5% FBS at RT for 30 min, cells were incubated with an appropriate primary antibody γ-H2AX (Cell Signaling) for 2 h. Then the cells were washed with PBS and incubated for 1 h with secondary antibody (Alexa Fluor-488 goat anti-mouse IgG [heavy chain (H) + light chain (L)] conjugate or anti-rabbit IgG [H+L] conjugate [Invitrogen]). After washing with PBS, the coverslips were dried and then reversely covered onto slides (Fisher Scientific) by adding mounting medium with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (Vector Laboratories). A LSM-710 confocal microscope (Zeiss) was used to obtain fluorescence images.
Comet Assay.
The comet assay measures DNA damage in individual cells. It was performed according to the instructions of the OxiSelect Comet Assay Kit (Cell Biolabs). Briefly, microscope slides were first covered with a normal melting point agarose to create a base layer. Then, 1 to 2 × 105 of cells were embedded into 75 μL of low-melting-point agarose at 37 °C, and the gel was cast over the first agarose layer. Then slides were immersed into a lysis buffer and kept for 1 h at 4 °C. After cell lysis, the slides were electrophoresed in alkaline electrophoresis buffer (300 mM NaOH, 1 mM EDTA, pH 13). The slides were then stained with Vista Green DNA dye. Comet tails were measured using ImageJ.
Homologous Repair Reporter Assay.
Homologous repair (HR) reporter assay to examine the repair efficiency of I-SceI inducible DSBs was performed using the HeLa DR13-9 cell line as previously described with slight modifications (46, 47). Briefly, 3 × 105 cells were plated on six-well dishes, and 2 μg I-SceI expression vector pCβASce was transfected using Lipofectamine 2000 (Invitrogen). After 24 h posttransfection, the indicated amount of gemcitabine and/or EPZ015666 was incubated with cells for 24 h. The GFP-expressing cells were counted in flow cytometric analysis (BD FACS Calibur and CellQuest Pro) by the FL2 and FL1 channels for HR repair efficiency. The % of fluorescent positive cells in treatment of gemcitabine and/or EPZ015666 was normalized to that of the nontreatment cells (Ctrl) transfected with pCβASce to calculate the relative repair efficiency.
RPA Overexpression.
WT RPA1/2/3 and GFP were overexpressed in the PRMT5 KO cell line by cotransfection of WT RPA1/2/3 (Addgene) and pCMV-GFP plasmid with a 5:1 ratio. The wild-type cell line was transfected with pCMV-GFP plasmid alone as a control. A total of 10 µg total DNA was used in the presence of 30 µL of Fugene6 reagent in 10-cm plate dishes that had 70% confluency. After transfection for 24 h, GFP+ cells were sorted by a FACS Aria cell sorter.
In Vivo Xenograft Experiments.
All animal care and experimental procedures were carried out in accordance with protocols approved by the University of Virginia School of Medicine Animal Care and Use Committee (PHS Assurance A3245-01). To develop xenograft tumors, control sgRNA-infected WT cells and PRMT5 KO cells were s.c. injected into the dorsal flanks of 8-wk-old nude mice, which were obtained from The Jackson Laboratory. When the tumors were visible (∼30 mm3 in volume), the mice received respective gemcitabine treatments via i.p. injection. After weekly monitoring, time to appearance of the tumor was recorded, and the tumor volume was measured by caliper. The tumor volume was calculated as follows: volume = longest tumor diameter × (shortest tumor diameter)2/2. After 35 d of treatment, the mice were killed by CO2 inhalation, and the tumor tissues were collected for further analyses.
RNA-Seq and Library Preparation.
Control sgRNA (CgRNA) and PRMT5 KO cells were treated with gemcitabine (200 nM) or EPZ015666 (500 nM) for 48 h. Total RNA was purified using the RNeasy mini kit (Qiagen 74104) by following the kit instructions. mRNA was isolated by using the NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs 7490S). RNA-Seq libraries were prepared using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs E7420S) by following the company’s protocol. A Qubit measurement and bioanalyzer were used to determine the library quality.
Data Analysis for RNA-Seq.
General sequencing data quality was examined using FastQC (v. 0.11.5). RNA-Seq data were aligned to the human reference genome (hg19) using HISAT2 (48) (v. 2.1.0) with the default paired-end mode settings. The resulting sam files were sorted by reading names and converted to bam files using samtools (49) (v. 1.9) sort command. Then the bam files were sorted by mapping position and indexed using corresponding samtools commands. The sorted and indexed bam files were first converted to bigwig files for visualization in the University of California Santa Cruz Genome Browser (https://genome.ucsc.edu/) to avoid technical alignment errors. Next, the bam files were quantified against gencode (v27lift37) annotation using Stringtie (50) (v. 1.3.4d) with the default settings.
After obtaining the gene count matrix from Stringtie, we imported it into R and normalized the data following the pipeline of DESeq2 (51). Specifically, to ensure a roughly equal distribution for all of the genes across samples, we used rlog transformation to stabilize expression variance for genes with different expression levels. Then samples were clustered according to Euclidean/Poisson distances to make sure replicates are clustered together. By calling the DESeq function, we determined genes with significant expression changes between the PRMT5 WT and KO samples thresholding at an adjusted P value of 0.01. Heatmaps were produced using the pheatmap R package. All other plots were generated using ggplot2. Gene set enrichment analysis (52) was performed using the GSEA website (https://www.gsea-msigdb.org/gsea/index.jsp) and the stand-alone GSEA program referencing the Molecular Signatures Database (MSigDB). The gene-specific normalized read counts are provided in Dataset S2.
Data Analysis for Three Publicly Available PDAC Studies from GEO and TCGA.
Data were downloaded from Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) for studies GSE28735, GSE16515, and GSE93326. Normalized PRMT5 expression was compared between relevant sample groups using an appropriate Student’s t test. Analyses were performed and plots were generated in RStudio v. 0.99.484 with R v. 3.3.0. The survival analysis for PDAC patients with high (>0.5 SD) and low expression (<0.5 SD) of PRMT5 was carried out through cBioPortal (www.cbioportal.org).
Supplementary Material
Acknowledgments
The study was initially funded by a pilot project award (to M.A. and T.W.B.) from the University of Virginia Cancer Center and through the National Cancer Center (NCI) Support Grant 5P30CA044579. Additional resources from Pinn Scholar Award (to M.A.) and from NIH/NCI 1R01 CA211648-01 award were mobilized to complete the work. K.Y.L. and A.D. were supported by R01 CA060499. The University of Virginia Biorepository and Tissue Research Facility as well as the Research Histology Core were utilized for tissue staining. We are thankful to all the members of the M.A. and T.W.B. laboratories for critical discussions during this study.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. S.H.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009899117/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information.
References
- 1.Siegel R. L., Miller K. D., Jemal A., Cancer statistics, 2016. CA Cancer J. Clin. 66, 7–30 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Kamisawa T., Wood L. D., Itoi T., Takaori K., Pancreatic cancer. Lancet 388, 73–85 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Rahib L., et al. , Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 74, 2913–2921 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Biankin A. V. et al.; Australian Pancreatic Cancer Genome Initiative , Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491, 399–405 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones S., et al. , Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321, 1801–1806 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Misale S., et al. , Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 486, 532–536 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Little A. S., et al. , Amplification of the driving oncogene, KRAS or BRAF, underpins acquired resistance to MEK1/2 inhibitors in colorectal cancer cells. Sci. Signal. 4, ra17 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Robertson J., Barr R., Shulman L. N., Forte G. B., Magrini N., Essential medicines for cancer: WHO recommendations and national priorities. Bull. World Health Organ. 94, 735–742 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mini E., Nobili S., Caciagli B., Landini I., Mazzei T., Cellular pharmacology of gemcitabine. Ann. Oncol. 17 (suppl. 5), v7–v12 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Spadi R., et al. , Current therapeutic strategies for advanced pancreatic cancer: A review for clinicians. World J. Clin. Oncol. 7, 27–43 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szlachta K., et al. , CRISPR knockout screening identifies combinatorial drug targets in pancreatic cancer and models cellular drug response. Nat. Commun. 9, 4275 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walters D. M., et al. , Clinical, molecular and genetic validation of a murine orthotopic xenograft model of pancreatic adenocarcinoma using fresh human specimens. PLoS One 8, e77065 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindberg J. M., et al. , Co-treatment with panitumumab and trastuzumab augments response to the MEK inhibitor trametinib in a patient-derived xenograft model of pancreatic cancer. Neoplasia 16, 562–571 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W., et al. , MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koike-Yusa H., Li Y., Tan E. P., Velasco-Herrera Mdel. C., Yusa K., Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat. Biotechnol. 32, 267–273 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Hart T., et al. , High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163, 1515–1526 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Blanc R. S., Richard S., Arginine methylation: The coming of age. Mol. Cell 65, 8–24 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Stopa N., Krebs J. E., Shechter D., The PRMT5 arginine methyltransferase: Many roles in development, cancer and beyond. Cell. Mol. Life Sci. 72, 2041–2059 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamard P. J., et al. , PRMT5 regulates DNA repair by controlling the alternative splicing of histone-modifying enzymes. Cell Rep. 24, 2643–2657 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clarke T. L., et al. , PRMT5-dependent methylation of the TIP60 coactivator RUVBL1 is a key regulator of homologous recombination. Mol. Cell 65, 900–916.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng S., et al. , Arginine methylation-dependent reader-writer interplay governs growth control by E2F-1. Mol. Cell 52, 37–51 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jansson M., et al. , Arginine methylation regulates the p53 response. Nat. Cell Biol. 10, 1431–1439 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Zhang G., et al. , Integration of metabolomics and transcriptomics revealed a fatty acid network exerting growth inhibitory effects in human pancreatic cancer. Clin. Cancer Res. 19, 4983–4993 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pei H., et al. , FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell 16, 259–266 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Renz B. W., et al. , β2 adrenergic-neurotrophin feedforward loop promotes pancreatic cancer. Cancer Cell 33, 75–90.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chou T. C., Talaly P., A simple generalized equation for the analysis of multiple inhibitions of Michaelis-Menten kinetic systems. J. Biol. Chem. 252, 6438–6442 (1977). [PubMed] [Google Scholar]
- 27.Curtin N. J., DNA repair dysregulation from cancer driver to therapeutic target. Nat. Rev. Cancer 12, 801–817 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Blackford A. N., Jackson S. P., ATM, ATR, and DNA-PK: The trinity at the heart of the DNA damage response. Mol. Cell 66, 801–817 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Maréchal A., Zou L., DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 5, a012716 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartek J., Lukas C., Lukas J., Checking on DNA damage in S phase. Nat. Rev. Mol. Cell Biol. 5, 792–804 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Wu Y., Lu J., Kang T., Human single-stranded DNA binding proteins: Guardians of genome stability. Acta Biochim. Biophys. Sin. (Shanghai) 48, 671–677 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Liu T., Huang J., Replication protein A and more: Single-stranded DNA-binding proteins in eukaryotic cells. Acta Biochim. Biophys. Sin. (Shanghai) 48, 665–670 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Toledo L. I., et al. , ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 155, 1088–1103 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Toledo L., Neelsen K. J., Lukas J., Replication catastrophe: When a checkpoint fails because of exhaustion. Mol. Cell 66, 735–749 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Panier S., Boulton S. J., Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 15, 7–18 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Alexander J. L., Orr-Weaver T. L., Replication fork instability and the consequences of fork collisions from rereplication. Genes Dev. 30, 2241–2252 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gunn A., Stark J. M., I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods Mol. Biol. 920, 379–391 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Golding S. E., et al. , Pro-survival AKT and ERK signaling from EGFR and mutant EGFRvIII enhances DNA double-strand break repair in human glioma cells. Cancer Biol. Ther. 8, 730–738 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fedoriw A., et al. , Anti-tumor activity of the type I PRMT inhibitor, GSK3368715, synergizes with PRMT5 inhibition through MTAP loss. Cancer Cell 36, 100–114.e25 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Fong J. Y., et al. , Therapeutic targeting of RNA splicing catalysis through inhibition of protein arginine methylation. Cancer Cell 36, 194–209.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zou Y., Liu Y., Wu X., Shell S. M., Functions of human replication protein A (RPA): From DNA replication to DNA damage and stress responses. J. Cell. Physiol. 208, 267–273 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plate I., et al. , Interaction with RPA is necessary for Rad52 repair center formation and for its mediator activity. J. Biol. Chem. 283, 29077–29085 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruff P., Donnianni R. A., Glancy E., Oh J., Symington L. S., RPA stabilization of single-stranded DNA is critical for break-induced replication. Cell Rep. 17, 3359–3368 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J., et al. , Pooled library screening with multiplexed Cpf1 library. Nat. Commun. 10, 3144 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang T., Wei J. J., Sabatini D. M., Lander E. S., Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ransburgh D. J., Chiba N., Ishioka C., Toland A. E., Parvin J. D., Identification of breast tumor mutations in BRCA1 that abolish its function in homologous DNA recombination. Cancer Res. 70, 988–995 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee K. Y., Im J. S., Shibata E., Dutta A., ASF1a promotes non-homologous end joining repair by facilitating phosphorylation of MDC1 by ATM at double-strand breaks. Mol. Cell 68, 61–75.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim D., Langmead B., Salzberg S. L., HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H. et al.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pertea M., Kim D., Pertea G. M., Leek J. T., Salzberg S. L., Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.