Significance
Circadian rhythms including wake-sleep cycles are driven by molecular time cues generated by a self-sustaining transcriptional negative feedback loop. Among all clock proteins, PERIOD (PER) is considered the pacemaker protein because its rhythm of accumulation and nuclear entry generates the timing and duration of feedback inhibition. Here we provide a new understanding of how robust PER rhythms are generated: the collective action of interacting PER molecules, not a random mass action of individual molecules, allows compensation of spatial and temporal differences (or “noise”) of individual molecules. We also show that the collective PER rhythm requires healthy cytoplasmic trafficking, and that circadian sleep disorders can arise in such conditions as obesity, aging, and neurodegenerative disorders in which the cytoplasm becomes congested.
Keywords: circadian rhythm, PERIOD, negative feedback loop, cytoplasmic trafficking, bistable phospho-switch
Abstract
The circadian clock is based on a transcriptional feedback loop with an essential time delay before feedback inhibition. Previous work has shown that PERIOD (PER) proteins generate circadian time cues through rhythmic nuclear accumulation of the inhibitor complex and subsequent interaction with the activator complex in the feedback loop. Although this temporal manifestation of the feedback inhibition is the direct consequence of PER’s cytoplasmic trafficking before nuclear entry, how this spatial regulation of the pacemaker affects circadian timing has been largely unexplored. Here we show that circadian rhythms, including wake-sleep cycles, are lengthened and severely unstable if the cytoplasmic trafficking of PER is disrupted by any disease condition that leads to increased congestion in the cytoplasm. Furthermore, we found that the time delay and robustness in the circadian clock are seamlessly generated by delayed and collective phosphorylation of PER molecules, followed by synchronous nuclear entry. These results provide clear mechanistic insight into why circadian and sleep disorders arise in such clinical conditions as metabolic and neurodegenerative diseases and aging, in which the cytoplasm is congested.
Biological oscillations, such as the cell cycle and circadian rhythms, rely on negative feedback loops in which time delay and high sensitivity are essential features for their robustness and self-sustainability (1–3). In the mammalian circadian clock, CLOCK:BMAL1 (the activator complex) drives transcription of the pacemaker genes Per1 and Per2 (Per), and PER proteins form an inhibitor complex containing Cryptochrome (CRY) proteins, which inhibits CLOCK:BMAL1 to close the loop (4–7). PER is translated and accumulates for several hours before being allowed to inhibit the activator complex (time delay); PER seems to be a scaffolding protein with a phosphotimer that mediates time-dependent interaction between the activator and the inhibitor complex (8–12). Casein kinases CK1δ/ε are critical for PER phosphorylation-mediated nuclear entry and subsequent interaction with the activator complex: PER is constitutively hypophosphorylated and perinuclear in CK1δ/ε double-knockout cells (11, 13). Thus, a critical time delay in the mammalian clock, which would be followed by rapid feedback inhibition from accumulated inhibitor molecules (high sensitivity), is likely mediated at the phosphorylation step for nuclear entry.
The precision of the time delay and the high sensitivity are challenging to explain due to “noise” in the cellular environment and the stochastic behavior of individual PER molecules. Several thousand PER molecules translated throughout the cytoplasm over several hours would have great heterogeneity in phosphorylation and distance to the nucleus (14), and these molecules must transit through a crowded and noisy cytoplasm, further amplifying the heterogeneity. Because the phosphorylation and nuclear entry of PER molecules must occur in a precise and collective manner at the same time every day, there must be a mechanism that can compensate for the spatiotemporal heterogeneity of the pacemaker molecules to elicit coordinated feedback inhibition from thousands of molecules of the pacemaker. If the cytoplasm is congested with increased macromolecules, such as accumulated waste or energy reserve materials, a state known to increase cytoplasmic viscosity and thus decrease the speed of molecular diffusion (15), these “traffic jams” would interfere with PER trafficking and coordination, creating even more noise in the clock, which can manifest in disrupted output rhythms, such as wake-sleep cycles. If the robustness and preciseness of the circadian clock indeed originate from coordinated precise cytoplasmic trafficking of PER molecules, then disruption in cytoplasmic homeostasis would greatly compromise circadian rhythms, including wake-sleep cycles.
To test this hypothesis, we developed a mathematical model, cell-based assays, and animal models simulating different levels of traffic jams. We initially focused on the autophagy pathway because it is a primary mechanism regulating cytoplasmic flux by trafficking waste, energy materials, and dysfunctional organelles to the lysosomes for recycling. However, we also tested our hypothesis in nonautophagy animal models with increased cytoplasmic crowding, such as P62, ob/ob mutants, and a tauopathy transgenic animal (Tg4510).
In the present study, we show that circadian rhythms are predictably modulated by the level of cytoplasmic crowding. In cell and animal models with congested cytoplasm due to increased cytoplasmic crowding, circadian behavioral rhythms (wake-sleep cycles) and the molecular clock are lengthened and extremely unstable due to disrupted coordination of individual pacemaker molecules before nuclear entry. The key to the coordination is spatially coordinated temporal PER phosphorylation. PER phosphorylation for nuclear entry does not occur until its concentration reaches a certain threshold, which is strongly favored only in the perinucleus, and thus leads to the time delay and high amplitude (robust feedback inhibition) of the clock.
Results
Circadian Rhythms Are Dramatically Modulated by Cytoplasmic Conditions.
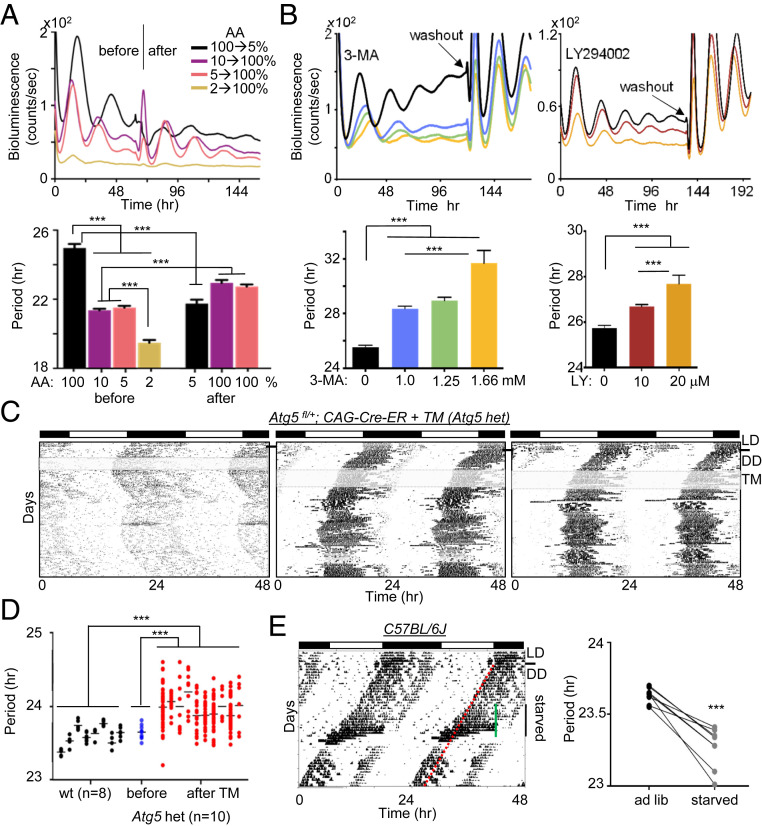
To test whether compromised cytoplasmic traffic affects the circadian clock, we measured circadian rhythms and molecular clock components under conditions in which cytoplasmic flux is either accelerated or slowed. Specifically, since autophagy is a major regulator of cytoplasmic content (16), we first tested how circadian rhythms are affected under autophagy-activating and -inhibiting conditions. We started with two established cell culture models, mouse embryonic fibroblasts (MEFs) and human U2OS cells (17, 18), using circadian bioluminescence rhythms as a reporter. When Per2Luc MEFs were subjected to amino acid (AA) starvation, a strong inducer of autophagy (16, 19), circadian period was dramatically shortened, by up to several hours (Fig. 1A). In contrast, when autophagy was inhibited pharmacologically using 3-MA, LY294002, or wortmannin—all of which inhibit the class III phosphatidylinositol 3-kinase (PI3K) and thereby inhibit autophagosome formation, the cargo-carrying vehicle (20–22)—the cells showed dramatic period lengthening and dampened amplitude in a dose-dependent manner (Fig. 1B and SI Appendix, Fig. S1A). Similar observations for autophagy-regulated circadian rhythms were made in U2OS cells (SI Appendix, Fig. S1 B and C). Circadian rhythms were phase-shifted by the drugs in a phase-dependent manner (SI Appendix, Fig. S2A), consistent with direct action on the clock mechanism (not a “masking” effect). Treatment with 3-MA was confirmed to inhibit autophagy, as it increased levels of LC-II and inhibited degradation of P62 (a known autophagy substrate) under starvation conditions (19) (SI Appendix, Fig. S2 B and C).
Fig. 1.
The circadian clock is regulated by integrated spatial and temporal mechanisms. (A) Circadian rhythms were shortened by autophagy-inducing AA starvation. Per2Luc MEFs were placed in different concentrations of AAs by varying the ratio between DMEM (100% AA) and EBSS (0% AA). After approximately 3 d, 2%, 5%, and 10% media were replaced with 100% medium and 100% medium was replaced with 5% medium while bioluminescence rhythms were measured. The low signal in 2% medium after replacement with the normal medium suggests that the starvation condition was not physiologically sustainable. n = 3 each. Data are representative of three experiments. (B) Circadian rhythms were lengthened and dampened by inhibition of autophagy in a dose-dependent manner. Note that robust rhythms were recovered after washout, indicating that the drugs did not affect cell viability at these doses. n = 3 each. Data are representative of three experiments. (C) Actograms of three mice heterozygous for Atg5 deletion. The black line indicates LD (12 h light:12 h dark) to DD (constant dark) transition, and the gray shading indicates TM treatment. No two mice showed a similar stable phase angle after TM treatment. Six more heterozygotes are shown in SI Appendix, Fig. S4. (D) Periods calculated from short intervals of stable phase angle (1 to 2 wk) were compared between WT (C57BL/6J) and Atg5 het mice. The average period of Atg5 het mice (Atg5fl/+; CAG-Cre-ER) was significantly longer than that of WT mice after TM treatment and significantly different before and after TM treatment (P < 0.01, two-tailed t test). The periods of Atg5 het mice before TM treatment were calculated from the actograms in the first 2 wk in DD. (E) Starvation-induced period shortening in WT mice. C57BL/6J mice were entrained in LD followed by DD for 2 wk, which served as baseline activity. A hypocalorie diet was provided at ZT12 of the previous LD for 12 d (indicated by the green line), followed by a return to ad libitum. The red line indicates the predicted continuous activity onset had the mice not been subjected to starvation. n = 8.
We next analyzed the effects of altering autophagy on sleep cycles in mice. Atg5 and Atg7 are critical autophagy genes that encode structural proteins important for the formation of autophagosomes (23, 24). A conventional Atg5 mutant mouse dies due to defective autophagy during the early neonatal starvation period (24). To circumvent this early lethality, we used a floxed Atg5 mutant mice with an estrogen receptor (ER)-inducible Cre driver, CAG-Cre-ER, which allows us to delete the floxed Atg5 in adult mice using tamoxifen (TM)-containing food. Unfortunately, when both alleles of Atg5 were deleted by the CAG-Cre-ER and TM, all the resulting homozygous mutant mice died within 10 d (n = 6) (SI Appendix, Fig. S3A). However, heterozygous Atg5 mutant mice (Atg5fl/+; CAG-Cre-ER) were viable and had no health issues except a circadian phenotype. All of them showed unstable, lengthened circadian rhythms (Fig. 1 C and D and SI Appendix, Fig. S4). The heterozygotes showed abrupt, spontaneous phase shifts and frequent period changes, leading to an unstable phase angle. In contrast, when WT C57BL/6J mice were subjected to starvation to induce autophagy, wake-sleep rhythms were significantly shortened (Fig. 1E). It has been demonstrated that the TM treatment does not affect circadian rhythms (25, 26) (SI Appendix, Fig. S3 B and C).
Temporal Phosphorylation of PER Molecules Is Spatially Coordinated.
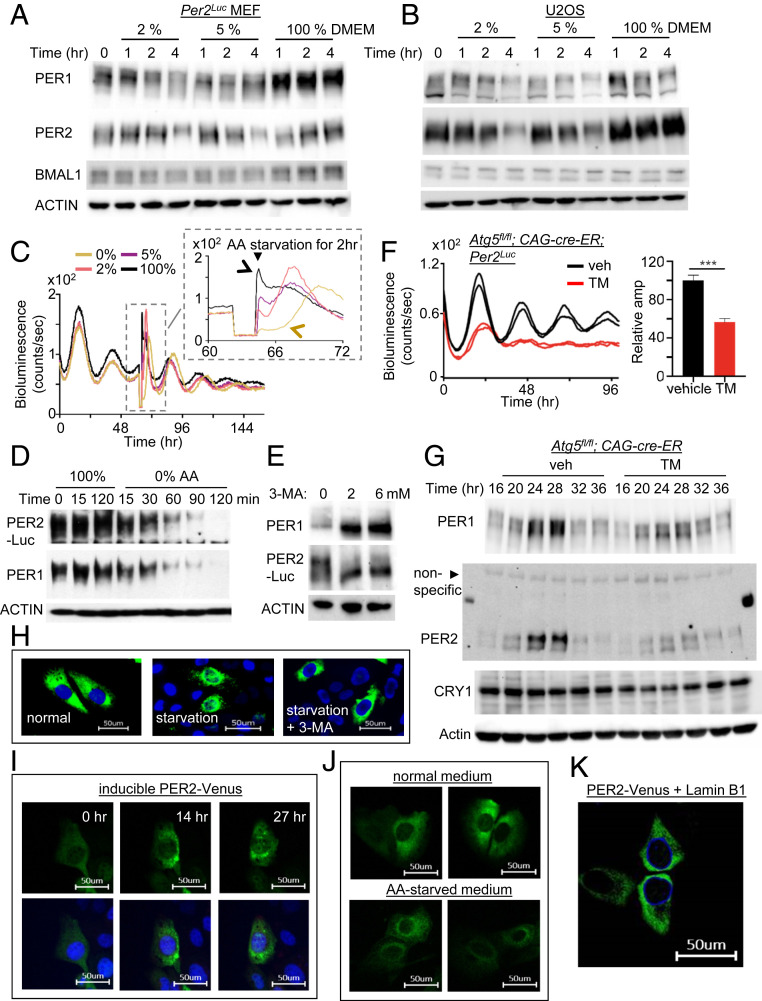
Biochemical PER rhythms are defined by temporal phosphorylation and abundance changes. PER phosphorylation and abundance are temporally regulated in ∼24-h cycles in MEFs and U2OS cells, mirroring the in vivo phosphotimer dynamics (8, 27). Therefore, we next tested how altering autophagy affects the PER rhythms in the cells. In starved MEFs and U2OS cells, PER exhibited accelerated profiles of phosphorylation and breakdown over 4 h compared with control cells, suggesting that nuclear entry of PER was accelerated as well (Fig. 2 A and B) (10). AA starvation given in a 2-h pulse produced a rapid drop in bioluminescence signal in a dose-responsive manner (Fig. 2C, Inset). Accordingly, 0% AA treatment induced greater acceleration in PER phosphorylation and breakdown compared with 2% and 5% AA treatment (Fig. 2 A and D). This acceleration likely is not mediated through the modulation of translation or transcription of Per genes, because PER phosphorylation and degradation seem to occur normally even when translation is blocked by cycloheximide (28). PER degradation has previously been shown to be dependent on phosphorylation (11, 13, 25), and thus the AA starvation treatment likely accelerated PER phosphorylation first, which then led to accelerated PER degradation. Treatment with 3-MA induced the opposite effect: accumulation of hypophosphorylated PER (Fig. 2E). These data suggest that temporal PER phosphorylation is regulated by the speed of PER trafficking (see below for more supporting data). Consistent with this idea and with the noisy rhythms in Atg5 heterozygote mice, circadian rhythms—especially amplitude—were severely disrupted in homozygous Atg5 knockout MEFs (Fig. 2 F and G). Circadian phenotypes were mild in Atg5 mutant mice and cells compared with PI3K-inhibited cells, probably because autophagosomes can be generated by Atg5/7-independent noncanonical autophagy pathways (29).
Fig. 2.
PER molecular rhythms are also modulated by autophagy-enhancing and inhibiting conditions. (A and B) PER rhythms were accelerated after MEFs and U2OS were subjected to starvation. Unsynchronized cells were subjected to starvation and harvested at indicated times. PER phosphorylation is reflected by slower electrophoretic mobility, as established previously (10). Note that PER phosphorylation and breakdown were accelerated by the starvation. (C) A pulse of starvation induced a rapid drop in bioluminescence in a dose-dependent manner. Note that the new phases after 100% and 0% AA pulses are indicated by black and brown arrows, respectively. All four samples show baseline signals during the 2-h pulse in the Inset because the samples were removed from the LumiCycle for the pulse. Note that the phase of 0% (brown) seems delayed relative to that of the control (black) in the Inset, but the 0% (brown) peak is from the next cycle relative to that of the control (black) peak (see D). However, it is less clear for the 2% and 5% cases, because the current cycle during the pulse was partially affected and could be recovering after the pulse, which will result in delays. In any case, our data show that pulses of starvation can induce stable phase shifts through modulating the pacemaker PER. (D) PER phosphorylation and breakdown were dramatically accelerated by starvation with 0% AA. Note that the current cycle of PER ended within 2 h for the 0% sample. (E) Treatment with autophagy inhibitor 3-MA induced accumulation of hypophosphorylated PER. (F and G) Homozygous Atg5 null mutant MEFs combined with the endogenous Per2Luc reporter (18) showed dramatically dampened bioluminescence and molecular rhythms. Period could not be determined from the Atg5 mutant cells due to the low amplitude. n = 3 each. Data are representative of two experiments. (H) Nuclear accumulation of PER2-Venus was accelerated by starvation (5% AA) in U2OS, but this was abolished by 3-MA treatment. (I) Nuclear entry of PER2-Venus was gated. Note that PER2-Venus is highly enriched in the perinuclear area. The same cell was monitored for 27 h after discontinuation of PER2-Venus induction (Movie S1). Nuclear entry was not complete even after 27 h, because endogenous CK1δ/ε were limiting to the transgenic PER2-Venus (SI Appendix, Fig. S5) (Scale bar: 50 μm.) (J) Perinuclear enrichment of PER2-Venus was more pronounced by starvation. U2OS cells were incubated with normal or 5% AA medium for 12 h after discontinuation of PER2-Venus induction (Scale bar: 50 μm.) (K) Perinuclear localization of PER2-Venus was confirmed by coexpressing a nuclear membrane protein, Lamin B1. Lamin B1 was expressed as an mCerulean3 fluorescent protein.
To determine whether cytoplasmic trafficking of PER was indeed affected by altered autophagy, we analyzed PER subcellular localization under different conditions by labeling it with the fluorescent protein Venus. Venus fusion to the C terminus of PER2 did not affect the clock properties of PER2, as demonstrated by Smyllie et al. (14) and described in SI Appendix, Fig. S5. In accordance with the data on bioluminescence and phospho-timer rhythms in different autophagy conditions, nuclear accumulation of PER2-Venus protein was accelerated and delayed by starvation and 3-MA, respectively (Fig. 2H). During the course of these imaging studies, we noticed that PER strongly accumulated in the perinuclear region before nuclear accumulation, which was accelerated by starvation (Fig. 2 I–K and Movie S1). Because PER phosphorylation by CK1δ/ε is required for nuclear entry (11, 13) (SI Appendix, Fig. S5), our data suggest that the PER phosphorylation needed for nuclear entry most likely occurs in the perinucleus. If this phosphorylation occurred independent of location and was only temporally regulated, then PER would not accumulate strongly in the perinucleus.
Cooperative PER Phosphorylation Underlies the Robustness and Time Delay.
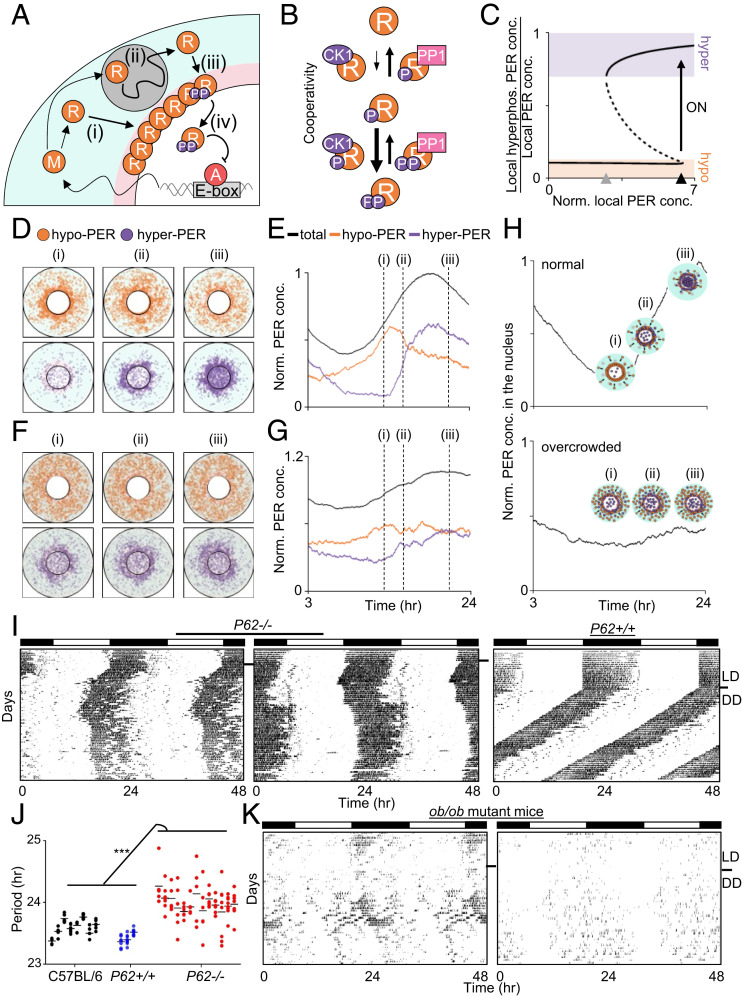
Because our data suggest that phosphorylation of PER molecules occurs in a collective manner in the perinucleus for coordinated nuclear entry of PER molecules, we evaluated whether a computer simulation based on our experimental data could generate such a spatially regulated collective phosphorylation of PER molecules (Fig. 3A). In this model, hyperphosphorylation of PER for nuclear entry occurs faster than its hypophosphorylation in a cooperative manner (Fig. 3B), which is supported by experimental data and several analogous models of the cell cycle (9, 30, 31). This leads to a sharp, switch-like collective hyperphosphorylation of PER molecules; as the local level of PER exceeds a threshold in the perinucleus, the majority of hypophosphorylated PER is synchronously hyperphosphorylated, generating a bistable transition (Fig. 3C and SI Appendix, Fig. S6). When cytoplasmic flux is normal, PER accumulates to threshold levels only in the perinuclear region, which can trigger synchronous hyperphosphorylation and nuclear entry within a narrow time window (Fig. 3 D and E). Although not included in our current model, this switch-like phosphorylation in the perinucleus would be even more robust if the model were to reflect the fact that the affinity of CK1δ/ε to PER is stronger than that of phosphatase PP1 (11). Because CK1δ/ε are stoichiometric binding partners of PER through dedicated protein-binding domains, the local concentration of the kinases would resemble that of PER, leading to more local enrichment of CK1δ/ε over PP1 in the perinucleus (13). When the cytoplasm is overcrowded, PER can accumulate near the threshold level beyond the narrow perinucleus area, which will generate hyperphosphorylated PER in a random spatiotemporal manner, disabling the circadian perinuclear bistable transition (Fig. 3 F and G and SI Appendix, Fig. S7). This model explains all the foregoing data (Figs. 1 and 2) and predicts noisy rhythms in any mouse models with increased cytoplasmic congestion (Fig. 3H).
Fig. 3.
Unique spatiotemporal regulation of PER generates a bistable switch in hyperphosphorylation for nuclear entry. (A) Diagram of the spatial stochastic model of the circadian clock. After Per mRNA, M, is translated to protein, R, in the cytoplasm (i), PER transits toward the perinucleus, past obstacles while being hypophosphorylated (ii; gray circle). The accumulated PER in the perinucleus is hyperphosphorylated (iii). Then it enters the nucleus and inhibits the transcriptional activity of the activator, A (iv). More details are provided in SI Appendix. (B) In the model, hyperphosphorylation of PER occurs faster than hypophosphorylation due to cooperativity. For simplicity, one phosphate group and two phosphate groups were modeled as hypophosphorylation and hyperphosphorylation, respectively. (C) This leads to the bistability in PER hyperphosphorylation. When the “local” concentration of cytoplasmic PER reaches the switch-on threshold (black triangle), the fraction of hyperphosphorylated PER sharply increases. Then the high fraction persists for awhile even when the local concentration of PER decreases, until the switch-off threshold is reached (gray triangle). The solid and dashed lines indicate the stable and unstable steady states, respectively. More details are provided in SI Appendix, Fig. S6. (D and E) Snapshots of the simulated spatial distribution of PER (D) and the simulated trajectories of PER concentration (E). The cytoplasmic flux over several hours increases PER abundance in the perinucleus (D, pink region in A) compared with the peripheral cytoplasm (D, cyan region in A). This induces a sharp switch-like hyperphosphorylation in the perinucleus due to the cooperativity (ii), followed by synchronous nuclear entry within a narrow time window (iii). (F and G) When a cell is overcrowded (dark background), the cytoplasmic flux is hindered, and thus PER does not accumulate in the same gradient as in the normal cell (i). This disables the sharp switch-like PER hyperphosphorylation and nuclear entry (ii and iii) (details in SI Appendix, Fig. S7). (H) Simulated temporal nuclear PER in a normal and an overcrowded cell. Note that PER concentration in C, E, and G and nuclear PER concentration in H are normalized by the peak level of total PER and that of nuclear PER in a normal cell, respectively. (I and J) P62 mutant mice showed unstable, lengthened rhythms. Periods were calculated as in Fig. 1D. Data for C57BL/6J were copied from Fig. 1D. (K) ob/ob mutant mice showed lengthened and unstable rhythms (n = 6).
P62 is one of many autophagy adaptor molecules but is not an autophagy gene and is not involved in autophagosome formation (32). It has been shown that P62 mutant mice exhibit adult-onset obesity and increased fat deposition, which can cause cytoplasmic congestion (33). When P62 mutant mice were subjected to behavioral analysis, all showed lengthened and unstable rhythms indistinguishable from those in Atg5 heterozygotes (Fig. 3 I and J). Furthermore, instability of their wake-sleep rhythms worsened with age, which also correlated with weight gain (SI Appendix, Fig. S8). Another line of nonautophagy mutant mice, ob/ob (34), which exhibit cytoplasmic congestion due to increased fat deposition, also showed similarly lengthened and unstable rhythms (Fig. 3K).
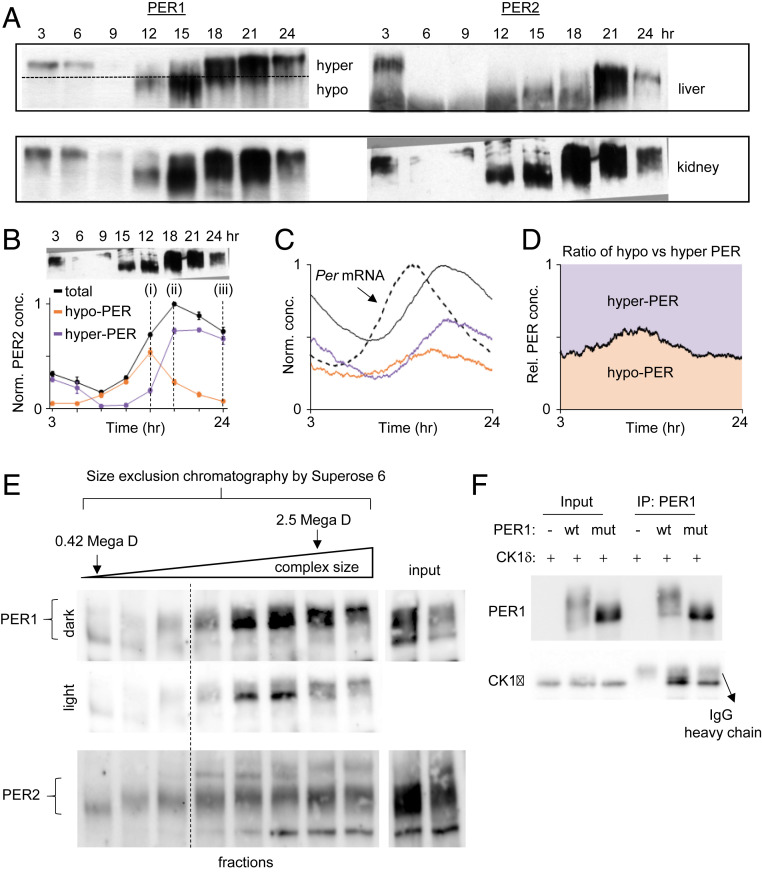
Our model of the spatiotemporally regulated bistable PER phosphorylation is further supported by several lines of in vivo biochemical data. When PER protein rhythms were analyzed in in vivo tissues by immunoblotting, all exhibited a switch-like pattern of phosphorylation: a sudden jump between hypophosphorylated and hyperphosphorylated states (Fig. 4 A and B), consistent with previous publications (e.g., ref. 10). This pattern would not be observed if PER molecules produced several hours apart were progressively phosphorylated; according to our simulation, this would lead to significant amounts of hypophosphorylated PER at all times, even in the presence of robust mRNA oscillations (Fig. 4 C and D). Notably, size fractionation of cell extracts revealed sharp hyperphosphorylation of PER when the size of PER complexes reached a certain threshold, likely via multimerization among PER molecules, supporting our model of cooperativity (Fig. 4E). Furthermore, hypophosphorylated species were minimally detectable once the PER complexes reached a certain size, after which they jumped to the hyperphosphorylation state (Fig. 4E). While not explicitly incorporated in our model for simplicity (Fig. 3 A and B), previous studies have suggested that a switch-like phosphorylation can occur through cooperative multimerization of substrates (1, 35). As PER-containing complexes can contain multiple copies of PER molecules (10, 36), we hypothesized that the switch-like phosphorylation (Fig. 3C) can occur via multimerization of PER molecules, which would be greatly facilitated by local enrichment. Because the PAS domain is the only known motif required for PER multimerization—PER homodimers, heterodimers, or possibly multimers—we tested how PER phosphorylation is affected when PAS-PAS dimerization is disrupted by mutating a key residue in the direct interface.
Fig. 4.
The bistable phosphoswitch model is further supported by other lines of experimental evidence. (A and B) PER phosphorylation in vivo exhibits two different levels of phosphorylation and jumps from hypophosphorylated to hyperphosphorylated states around the nuclear entry time (ZT15 to 18) (10). Hypophosphorylated PER species are not detectable after approximately CT21 because Per mRNA levels would be at trough levels once the majority of PER molecules enter the nucleus and inhibit CLOCK:BMAL1 (10). (C and D) If there were no bistability, both hypophosphorylated and hyperphosphorylated PER species would be observed at all times despite robust mRNA rhythms. This result was obtained by removing bistability from our mathematical model (Fig. 3A). (E) Size exclusion chromatography reveals that phosphorylation states are associated with complex size. U2OS cell extracts were fractionated in a Superose 6 column (GE Healthcare) and immunoblotted for PER1 and PER2. Fractions after the void volume of the column are shown. Note that the first fraction and the second to last fractions were where proteasome lid-L20 (0.42 MDa) and whole particles (2.5 MDa) were eluted (63, 64). We believe that hyperphosphorylation can occur only when PER complexes grow to a certain size through PER-PER multimerization, which is facilitated by local enrichment. (F) The PAS dimerization domain is critical for hyperphosphorylation of PER. WT and mutant (W448E) PER1 were transiently expressed along with CK1δ in 293A cells. Cell extracts were immunoprecipitated with anti-PER1 antibody, and the immune complexes were immunoblotted for CK1δ and PER1.
Previous work has demonstrated that PER homodimers are stabilized by the conserved tryptophan residue in the PAS B domain (W448mPER1, W419mPER2, and W359mPER3), and mutation of this residue to glutamate effectively disrupted homodimers in all PERs (37, 38). Therefore, we assessed phosphorylation of transiently expressed WT and W448E PER1 mutant in the presence of CK1δ. While WT PER1 was robustly phosphorylated by CK1δ, phosphorylation of the mutant PER1 was largely abolished, exhibiting only hypophosphorylated species (Fig. 4F). Both WT and mutant PER1 interacted with CK1δ (Fig. 4F), demonstrating that the mutation did not affect CK1δ binding (12, 39). Finally, we found that, as with overexpressed PER2-Venus, a ring shape of perinuclear accumulation was detected with endogenous PER1 and PER2 at a specific phase in U2OS cells (SI Appendix, Fig. S9A). Taken together, our data suggest that the PAS domain, which has not been characterized in terms of clock function, is the key motif for generating a time cue for time-delayed, high-amplitude nuclear entry through mediation of cooperative phosphorylation of PER.
Defective Clocks in Diseased Cytoplasm Can Be Corrected by Restoring Stoichiometry of PER.
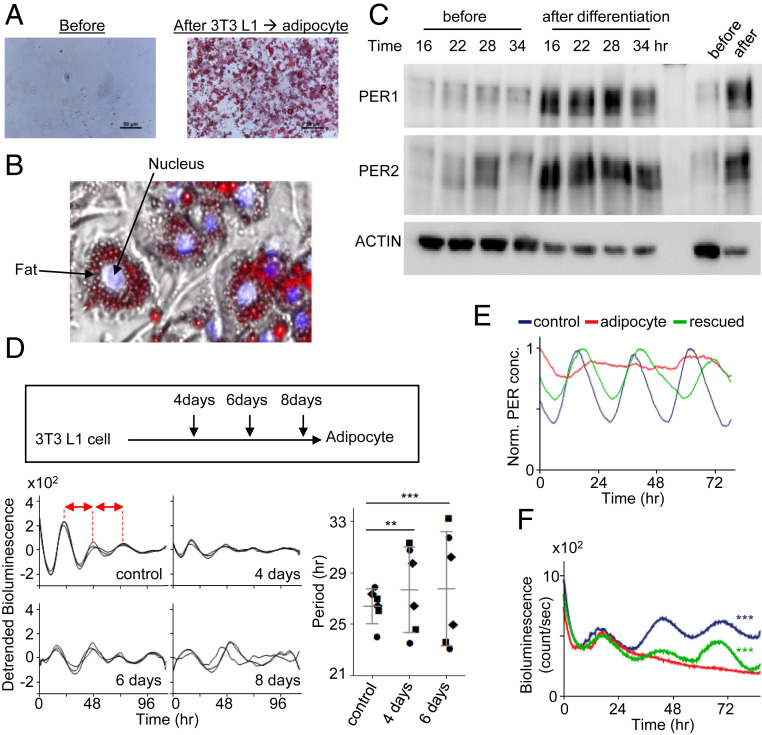
According to our hypothesis, the severity of rhythm disruption and collective phosphorylation should be worsened as the level of crowding increases in clock cells. To test this prediction, we monitored circadian rhythms during the differentiation of 3T3 L1 fibroblasts into adipocytes, which should produce a gradual increase in fat storage vacuoles over the course of differentiation. Indeed, the development of fat vacuoles was dramatic after differentiation, especially in the perinuclear area (Fig. 5 A and B). Consistent with our model (Fig. 3H) and the foregoing data, hypophosphorylated PER species remained pronounced at all times of the day, and the overall abundance of PER increased dramatically, as hypophosphorylated species are more stable than hyperphosphorylated species (25) (Fig. 5C). PER2-Venus was cytoplasmic predominantly in the adipocytes even after starvation (SI Appendix, Fig. S9B). Circadian rhythms became gradually unstable and ultimately were completely lost as L1 cells differentiated into adipocytes (Fig. 5D). As demonstrated by the behavioral data from Atg5 and p62 mutant mice, when the clock is unstable due to mildly increasing crowding, period can go in either direction unpredictably, which is predicted by our modeling.
Fig. 5.
Spatial regulation of PER is directly linked to temporal manifestation of PER and circadian rhythms. (A and B) Fat vacuoles (Oil Red O) were enriched in the cytoplasm of adipocytes differentiated from 3T3 L1 cells. A zoomed-in image from the right image in A is shown in B. (C) Both hypophosphorylated and hyperphosphorylated PER species were visible at all times in the adipocytes. Unsynchronized cells are shown in the last two lanes. (D) Circadian rhythms were gradually compromised as L1 cells were differentiated into adipocytes and completely lost on day 8. Note that circadian period is unstable and generally longer in partially differentiated day 4 and day 6 cells compared with controls. The periods were estimated by peak-to-peak (red arrows) analysis in the samples. Three different symbols represent three independent samples. Data are mean ± SD, representative of two experiments. (E) Our model predicts that arrhythmicity in adipocytes can be rescued by increasing Per promoter activity. (F) Robust bioluminescence rhythms were recovered in adipocytes when PER2 was overexpressed using an adenoviral vector-delivered Per2 transgene (Per2 promoter and Per2 coding sequence). n = 3 each. Data are representative of two experiments.
Our mathematical model predicted that circadian rhythms could be restored in adipocytes if rhythmic Per2 gene expression were augmented to compensate for the loss of the bistable phosphorylation in the perinucleus (Fig. 5E). Indeed, when we overexpressed a Per2 promoter-Per2 cDNA (Per2-cPer2) transgene in adipocytes using an adenoviral vector, as has been done previously (8), circadian rhythms were successfully restored, confirming our prediction (Fig. 5F).
Circadian Rhythms Are Lengthened in a Nonmetabolic Disease Model with Increased Cytoplasmic Crowding.
If the underlying mechanism of the circadian phenotypes in our models is indeed through disruption of the pacemaker trafficking and not by disruption of metabolic pathways, then a similar phenotype would be observed in any animal model with increased cytoplasmic crowding. To test this hypothesis, we measured circadian rhythms in a tauopathy model, Tg4510, which harbors the mutant P301L mutant tau and has been shown to have evident tanglelike cytoplasmic inclusions, brain atrophy, and behavioral phenotypes, including circadian disorders at age 5 to 8 mo (40, 41). Although circadian rhythms, including wake-sleep cycles, are known to be disrupted in neurodegenerative diseases such as Alzheimer’s disease (AD), the underlying mechanistic causes of the circadian disorders have not been fully characterized (40, 42).
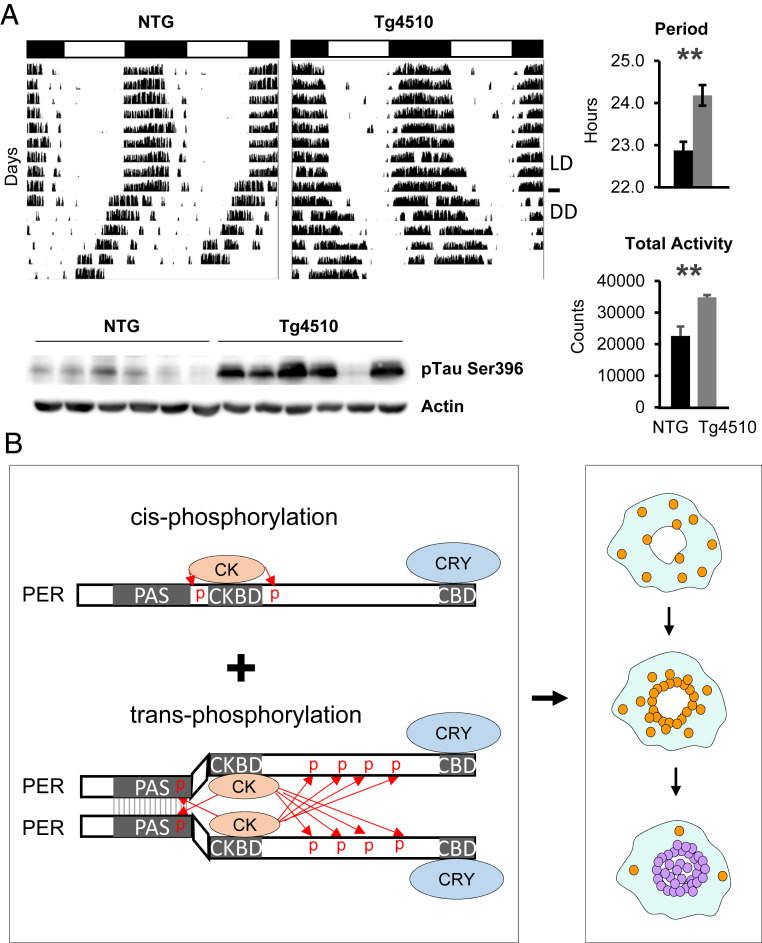
Tauopathy is a hallmark feature of several neurodegenerative diseases, including AD, progressive supranuclear palsy, and frontotemporal dementias, all of which develop intracellular tau tangles in the cytoplasm of neurons, generating a significant amount of cytoplasmic congestion (43). Hyperphosphorylation of tau, in particular pSer396, is strongly associated with abnormal cleavage and folding of tau, leading to tauopathy (44). Because most behavioral assays involving circadian locomotor activity in Tg4510 have been done at middle age to old age, when brain atrophy is evident, the phenotypes could be related to loss of relevant neurons or disruption in the neuronal network rather than to disruption in cell-intrinsic mechanisms. To avoid this complication, we measured behavioral rhythms at 3 mo of age, before severe tauopathy and atrophy are evident. As shown in Fig. 6A, the free-running period was lengthened in 3-mo-old Tg4510 mice compared with control mice, consistent with our prediction. The presence of tau tangles was confirmed in the hypothalamus, where the SCN (suprachiasmatic nuclei) resides, as measured by dramatically increased pSer396 levels (Fig. 6 A, Bottom). These data suggest that the clock impairment may precede brain atrophy and canonical tauopathy, leading to cell death.
Fig. 6.
Increased cytoplasmic crowding disrupts circadian rhythms. (A) Circadian rhythms are lengthened in a tauopathy model, Tg4510. Period was significantly longer in Tg4510 mice compared with littermate nontransgenic mice (n = 8 each; P < 0.01, two-tailed t test). Data are mean ± SEM. Tg mice showed increased activity levels (n = 8 each; P < 0.01, two-tailed t test), suggesting that lengthened rhythms are not due to degenerative neuronal activity at this stage. Data are mean ± SEM. Increased tau tangles were confirmed by immunoblotting for pSer396. (B) Cooperative phosphorylation of PER is driven by transacting phosphorylation of CK1δ/ε. PER multimerization, at least dimerization, is required for hyperphosphorylation of PER. PER molecules are trafficked toward the nucleus while being hypophosphorylated (brown) throughout the cytoplasm. PER multimerization is highly favorable only in the perinucleus, where they are hyperphosphorylated (blue) for nuclear entry by transacting CK1δ/ε.
Discussion
Our model can explain how a long and precise time delay before rapid feedback inhibition can be generated by a delayed and switch-like collective phosphorylation of PER molecules for nuclear entry. Although PER can be hypophosphorylated or prime-phosphorylated by the kinases in a cis-acting manner (30), our data suggest that dimerization or multimerization is required for nuclear entry-mediating hyperphosphorylation, because CK1δ/ε phosphorylate only trans-PER in the complex (Fig. 6B). The key to this collective hyperphosphorylation is transacting phosphorylation by CK1δ/ε in PER multimers, which is facilitated only when PER levels reach a certain threshold and is further facilitated by increasing the local CK1/PP1 activity ratio in the perinucleus. We believe that perinuclear accumulation of PER molecules is actively managed by the actin and/or microtubule transport networks, rather than via passive diffusion of PER molecules. We also believe that active cytoskeletal transport of PER toward the nucleus is likely, given that many other transcription factors with nuclear localization signals are transported in this way (e.g., ref. 45). PER accumulates in the perinucleus because this active transport drives PER to the perinucleus, but without hyperphosphorylation, PER cannot enter the nucleus. Only after accumulating to a certain level can PER sufficiently dimerize and then be phosphorylated for nuclear entry. We speculate that efficient PER phosphorylation and nuclear entry cannot occur until the majority of PER-interacting importin molecules are occupied by PER monomers, which then start to become dimerized in the perinucleus. This idea is supported by the previous studies showing that nuclear import of the PER:CRY inhibitory complex is mediated by specific importin molecules (46–48). This spatiotemporally regulated collective behavior of interacting pacemaker PER molecules can compensate for the stochastic noise of individual molecules and differences in cell size at the system level. According to our model, variances in trafficking speed and distance of individual molecules are highly tolerable at the group level. Spatiotemporally regulated bistable phosphorylation is also a key feature of several critical steps in the cell cycle; for example, phosphorylation of Aurora B is similarly regulated during mitosis (31).
The presence of a time delay mechanism before nuclear entry was first discovered in Drosophila and has been proposed to be conserved in mammals (49). However, Smyllie et al. (14), using a Per2-Venus knock-in mouse model, showed that endogenous PER2-Venus is not detectable exclusively in the cytoplasm at any circadian phase throughout the whole circadian cycle, arguing against a Drosophila-like time delay mechanism. We believe that our data are not incompatible with those reported by Smyllie et al. According to our model (SI Appendix, Fig. S10), cytoplasmic PER forms a strong gradient throughout the cytoplasm, with the perinucleus having the highest levels, not the diffuse cytoplasmic staining. As soon as perinuclear PER levels reach the threshold, those PER species are collectively phosphorylated and enter the nucleus. This occurs continuously within a narrow time window (high sensitivity), over ∼4 h because of the collective, cooperative phosphorylation (Figs. 3H and 4B). Thus, our model does not predict exclusive and diffusive cytoplasmic accumulation of PER at any circadian phase. Indeed, we observed a faint perinuclear ring and both cytoplasmic and nuclear staining at a peak phase of endogenous PER (SI Appendix, Fig. S9A).
Interestingly, the temporal profile of PER2-Venus nuclear accumulation measured by Smyllie et al. (14) also occurred on a similar time scale, ∼3.7 h. This fast nuclear accumulation rate is highly unlikely without the collective phosphorylation followed by synchronous nuclear entry, considering the presence of PER over 20 h each day (Fig. 4A). A similarly fast nuclear entry of PER through distinctive cytoplasmic foci has been observed in Drosophila, suggesting that the cooperative dPER (Drosophila PERIOD protein) phosphorylation mechanism by DBT (Doubletime) may be conserved in Drosophila (50, 51).
Similar phenotypes as in our mutant/Tg models have also been reported in many other disease conditions, including high-fat diet-induced obese mice and mice of advanced age (40, 52, 53). Although it has been demonstrated that all these models have increased cytoplasmic congestion (e.g., refs. 33, 41, 54, 55), it is possible that the clock could also be modulated by disruption in specific metabolic pathways, such as mTOR, AMPK, and oxidative stress pathways, in these animals (56–58).
Based on our data and model, we propose that the essential time delay before feedback inhibition and the high amplitude of circadian rhythms are generated at the nuclear gating step, and this can be severely disrupted by many disease conditions with compromised cytoplasmic homeostasis.
Materials and Methods
Animals.
All mice were maintained in a climate-controlled room and used according to Florida State University or University of South Florida Animal Care and Use Committee guidelines. All experiments involving animals were performed according to approved protocols. We used approximately equal numbers of male and female mice for behavioral studies. Sex differences in behavioral rhythms and cellular clock rhythms were very subtle to insignificant (59).
The floxed Atg5 mutant mice in the C57BL/6 background were obtained from the Mizushima laboratory under a material transfer agreement (MTA). We combined it with an inducible cre driver, CAG-cre-ER (The Jackson Laboratory; 004682), which allowed us to delete Atg5 throughout the whole body in adults (Atg5fl/fl; CAG-cre-ER) using TM-containing food. The cre driver has been used successfully in our laboratory to delete several genes, including CK1δ (11), Dicer (26), and βTrcp2 (25). According to Mizushima et al. (60), Atg5 mutant mice generated by low-level recombination with a CAG-cre driver were viable. However, in our study, when Atg5 was deleted by the CAG-cre-ER driver and TM, all the resulting homozygous mutant mice died within 10 d (n > 6). Our recombination rates were higher than those reported by Hara et al. (60), which could explain the discrepancy. Heterozygous Atg5 mutant mice (Atg5fl/+; CAG-cre-ER) were viable and did not have any health issues.
The P62 mutant mice in C57BL/6 background were obtained from the Yanagawa laboratory under an MTA (61). The behavioral data presented in Fig. 3I were obtained when the P62 mutant mice started to show unstable phase angles. The majority of the mice showed such unstable rhythms at age 8 mo or older.
The leptin knockout ob/ob mice were obtained from The Jackson Laboratory (B6.Cg-Lepob/J: 000632). The behavioral rhythms shown in Fig. 3K were obtained when these mice were ∼3 mo old, while they were still active and showed discernible circadian rhythms.
C57BL/6J (000664) mice were purchased from The Jackson Laboratory. The behavioral rhythms shown in SI Appendix, Fig. S3C were obtained twice: once at age ∼3 mo and then after they were reentrained by LD (light:dark cycles) at age ∼5 mo before they were placed in DD (constant darkness) again.
Tg4510 transgenic mice harboring tetO-P301L tau (The Jackson Laboratory; 015815) were crossed with Camk2a-tTA transgenic mice (The Jackson Laboratory; 007004) to express the mutant tau (40). Then 3-mo-old double transgenic and nontransgenic littermates were used to assess the circadian period.
MEFs.
Per2Luc MEFs have been described previously. MEFs from Atg5fl/fl; CAG-cre-ER; Per2Luc/Luc were prepared from embryos isolated from pregnant female mice at 13 d postcoitum. Embryos were removed, finely minced, treated with 0.25% trypsin, and then incubated at 37 °C for 30 min. The mixture was passed through a fine 100-µm membrane to remove debris, and the resulting cells were maintained at 37 °C in 5% CO2 in Dulbecco’s Modified Eagle Medium (DMEM), supplemented with 10% fetal bovine serum (FBS). Atg5 deletion was induced by 4-hydroxytamoxifen as described previously (11). Both male and female embryos were used.
Other Cell Lines.
3T3 L1 (ATCC CL-173) and U2OS (ATCC HTB-96) cells were obtained from the commercial vendor American Type Culture Collection. The U2OS-Bmal1-luc cell line has been described previously (27). All cells were maintained at 37 °C and 5% CO2 in DMEM supplemented with 10% FBS.
Cell Culture and Bioluminescence Recording.
For bioluminescence recordings, cells were plated into 24-well plates to be ∼90% confluent at 24 h before the start of the experiment. Immediately before the start of the experiment, cells were given a 2-h serum shock with 50% horse serum in DMEM, washed with PBS and fresh DMEM supplemented with 1% FBS, 7.5 mM sodium bicarbonate, 10 mM Hepes, 25 U/mL penicillin, and 25 μg/mL streptomycin, after which 0.1 mM luciferin was added. The plates were sealed with cellophane tape and placed into a LumiCycle (Actimetrics). For bioluminescence experiments, the results were reproduced in at least two independent experiments. AA starvation media were generated by mixing a complete medium DMEM (Corning; 10-017-CV) with 0% AA medium EBSS (Sigma; E6267). In SI Appendix, Fig. S1C, one sample per wortmannin concentration is shown. The results are representative of three experiments. 3-MA (M9281), LY294002 (L9908), wortmannin (W1628), and MG-132 (474790) were from Sigma-Aldrich.
For live cell snapshots and movies in Fig. 2 and Movie S1, an Andor Revolution spinning-disk laser confocal microscope with 20× objective was used.
For Fig. 2 F and G, Atg5 mutant cells were treated with 4-hydroxytamoxifen or ethanol 3 d before the cells were serum-shocked, placed in the LumiCycle, and harvested at indicated times.
Differentiation of 3T3 L1 into adipocytes was done following the protocol described by Zebsch et al. (62). In brief, 3T3 L1 cells were grown to 70% confluency. To initiate differentiation, cells were treated with MDI medium (0.5 mM IBMX, 1 mM dexamethasone, and 10 μg/μL insulin in DMEM) for 3 d, followed by insulin-only medium for 3 d. Fat vacuoles were visible after day 4, and differentiation was complete in 10 d. To visualize fat vacuoles, the cells were stained with 10% Oil Red O (Sigma-Aldrich; O0625).
More detailed information about the materials and methods used in this study is available in SI Appendix.
Data and Materials Availability.
All data presented in this study are available in the main text or SI Appendix. The noncommercial antibodies are available under an MTA with Florida State University. The computer codes used in this study are available at https://github.com/daewookkim/The-spatial-stochastic-model-of-the-circadian-clock.
Supplementary Material
Acknowledgments
We thank Dennis Chang for assistance with manuscript preparation and Life Science Editors for editorial assistance. This work was supported by NIH Grants NS099813 and GM131283 (to C.L.), a bridge grant from the Florida State University Department of Biomedical Sciences (to C.L.), Human Frontiers Science Program Organization Grant RGY0063/2017 (to J.K.K.), a National Research Foundation of Korea Grants NRF-2016 RICIB 3008468 (to J.K.K.) and NRF-2017-Fostering Core Leaders of the Future Basic Science Program/Global PhD Fellowship Program (to D.W.K.), and an award from the Florida Department of Health’s Ed and Ethel Moore Alzheimer’s Disease Program (9AZ31, to J.G.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2003524117/-/DCSupplemental.
References
- 1.Novák B., Tyson J. J., Design principles of biochemical oscillators. Nat. Rev. Mol. Cell Biol. 9, 981–991 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim J. K., Forger D. B., A mechanism for robust circadian timekeeping via stoichiometric balance. Mol. Syst. Biol. 8, 630 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferrell J. E. Jr, Tsai T. Y., Yang Q., Modeling the cell cycle: Why do certain circuits oscillate? Cell 144, 874–885 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Hardin P. E., Hall J. C., Rosbash M., Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 343, 536–540 (1990). [DOI] [PubMed] [Google Scholar]
- 5.Harmer S. L., Panda S., Kay S. A., Molecular bases of circadian rhythms. Annu. Rev. Cell Dev. Biol. 17, 215–253 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Lowrey P. L., Takahashi J. S., Genetics of circadian rhythms in mammalian model organisms. Adv. Genet. 74, 175–230 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reppert S. M., Weaver D. R., Coordination of circadian timing in mammals. Nature 418, 935–941 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Chen R., et al. , Rhythmic PER abundance defines a critical nodal point for negative feedback within the circadian clock mechanism. Mol. Cell 36, 417–430 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiu J. C., Ko H. W., Edery I., NEMO/NLK phosphorylates PERIOD to initiate a time-delay phosphorylation circuit that sets circadian clock speed. Cell 145, 357–370 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee C., Etchegaray J. P., Cagampang F. R., Loudon A. S., Reppert S. M., Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107, 855–867 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Lee H. M., et al. , The period of the circadian oscillator is primarily determined by the balance between casein kinase 1 and protein phosphatase 1. Proc. Natl. Acad. Sci. U.S.A. 108, 16451–16456 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee C., Weaver D. R., Reppert S. M., Direct association between mouse PERIOD and CKIepsilon is critical for a functioning circadian clock. Mol. Cell. Biol. 24, 584–594 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee H., Chen R., Lee Y., Yoo S., Lee C., Essential roles of CKIdelta and CKIepsilon in the mammalian circadian clock. Proc. Natl. Acad. Sci. U.S.A. 106, 21359–21364 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smyllie N. J., et al. , Visualizing and quantifying intracellular behavior and abundance of the core circadian clock protein PERIOD2. Curr. Biol. 26, 1880–1886 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Theillet F. X., et al. , Physicochemical properties of cells and their effects on intrinsically disordered proteins (IDPs). Chem. Rev. 114, 6661–6714 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizushima N., Komatsu M., Autophagy: Renovation of cells and tissues. Cell 147, 728–741 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Liu A. C., Lewis W. G., Kay S. A., Mammalian circadian signaling networks and therapeutic targets. Nat. Chem. Biol. 3, 630–639 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Yoo S. H., et al. , PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. U.S.A. 101, 5339–5346 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klionsky D. J., et al. , Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8, 445–544 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez J., et al. , Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc. Natl. Acad. Sci. U.S.A. 108, 17396–17401 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Y. T., et al. , Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J. Biol. Chem. 285, 10850–10861 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petiot A., Ogier-Denis E., Blommaart E. F., Meijer A. J., Codogno P., Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem. 275, 992–998 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Komatsu M., et al. , Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169, 425–434 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuma A., et al. , The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036 (2004). [DOI] [PubMed] [Google Scholar]
- 25.D’Alessandro M., et al. , Stability of wake-sleep cycles requires robust degradation of the PERIOD protein. Curr. Biol. 27, 3454–3467.e8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen R., D’Alessandro M., Lee C., miRNAs are required for generating a time delay critical for the circadian oscillator. Curr. Biol. 23, 1959–1968 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin Y. H., et al. , Streamlined procedure for gene knockouts using all-in-one adenoviral CRISPR-Cas9. Sci. Rep. 9, 277 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Alessandro M., et al. , A tunable artificial circadian clock in clock-defective mice. Nat. Commun. 6, 8587 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishida Y., et al. , Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 461, 654–658 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Narasimamurthy R., et al. , CK1δ/ε protein kinase primes the PER2 circadian phosphoswitch. Proc. Natl. Acad. Sci. U.S.A. 115, 5986–5991 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gelens L., Qian J., Bollen M., Saurin A. T., The importance of kinase-phosphatase integration: Lessons from mitosis. Trends Cell Biol. 28, 6–21 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Pankiv S., et al. , p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez A., et al. , Mature-onset obesity and insulin resistance in mice deficient in the signaling adapter p62. Cell Metab. 3, 211–222 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Pelleymounter M. A., et al. , Effects of the obese gene product on body weight regulation in ob/ob mice. Science 269, 540–543 (1995). [DOI] [PubMed] [Google Scholar]
- 35.Chung J. K., et al. , Switch-like activation of Bruton’s tyrosine kinase by membrane-mediated dimerization. Proc. Natl. Acad. Sci. U.S.A. 116, 10798–10803 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aryal R. P., et al. , Macromolecular assemblies of the mammalian circadian clock. Mol. Cell 67, 770–782.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hennig S., et al. , Structural and functional analyses of PAS domain interactions of the clock proteins Drosophila PERIOD and mouse PERIOD2. PLoS Biol. 7, e94 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kucera N., et al. , Unwinding the differences of the mammalian PERIOD clock proteins from crystal structure to cellular function. Proc. Natl. Acad. Sci. U.S.A. 109, 3311–3316 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vielhaber E., Eide E., Rivers A., Gao Z. H., Virshup D. M., Nuclear entry of the circadian regulator mPER1 is controlled by mammalian casein kinase I epsilon. Mol. Cell. Biol. 20, 4888–4899 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stevanovic K., et al. , Disruption of normal circadian clock function in a mouse model of tauopathy. Exp. Neurol. 294, 58–67 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Santacruz K., et al. , Tau suppression in a neurodegenerative mouse model improves memory function. Science 309, 476–481 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scarmeas N., et al. , Disruptive behavior as a predictor in Alzheimer disease. Arch. Neurol. 64, 1755–1761 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rizzo G., et al. , Diffusion-weighted brain imaging study of patients with clinical diagnosis of corticobasal degeneration, progressive supranuclear palsy and Parkinson’s disease. Brain 131, 2690–2700 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Mondragón-Rodríguez S., Perry G., Luna-Muñoz J., Acevedo-Aquino M. C., Williams S., Phosphorylation of tau protein at sites Ser(396-404) is one of the earliest events in Alzheimer’s disease and Down syndrome. Neuropathol. Appl. Neurobiol. 40, 121–135 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Salman H., et al. , Nuclear localization signal peptides induce molecular delivery along microtubules. Biophys. J. 89, 2134–2145 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakakida Y., et al. , Importin alpha/beta mediates nuclear transport of a mammalian circadian clock component, mCRY2, together with mPER2, through a bipartite nuclear localization signal. J. Biol. Chem. 280, 13272–13278 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Lee Y., Jang A. R., Francey L. J., Sehgal A., Hogenesch J. B., KPNB1 mediates PER/CRY nuclear translocation and circadian clock function. eLife 4, e08647 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim Y. H., Han M. E., Oh S. O., The molecular mechanism for nuclear transport and its application. Anat. Cell Biol. 50, 77–85 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Curtin K. D., Huang Z. J., Rosbash M., Temporally regulated nuclear entry of the Drosophila period protein contributes to the circadian clock. Neuron 14, 365–372 (1995). [DOI] [PubMed] [Google Scholar]
- 50.Meyer P., Saez L., Young M. W., PER-TIM interactions in living Drosophila cells: An interval timer for the circadian clock. Science 311, 226–229 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Top D., et al. , CK1/Doubletime activity delays transcription activation in the circadian clock. eLife 7, e32679 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kohsaka A., et al. , High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 6, 414–421 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Valentinuzzi V. S., Scarbrough K., Takahashi J. S., Turek F. W., Effects of aging on the circadian rhythm of wheel-running activity in C57BL/6 mice. Am. J. Physiol. 273, R1957–R1964 (1997). [DOI] [PubMed] [Google Scholar]
- 54.Kuma A., Komatsu M., Mizushima N., Autophagy-monitoring and autophagy-deficient mice. Autophagy 13, 1619–1628 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trak-Smayra V., et al. , Pathology of the liver in obese and diabetic ob/ob and db/db mice fed a standard or high-calorie diet. Int. J. Exp. Pathol. 92, 413–421 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lamia K. A., et al. , AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 326, 437–440 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramanathan C., et al. , mTOR signaling regulates central and peripheral circadian clock function. PLoS Genet. 14, e1007369 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilking M., Ndiaye M., Mukhtar H., Ahmad N., Circadian rhythm connections to oxidative stress: Implications for human health. Antioxid. Redox Signal. 19, 192–208 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuljis D. A., et al. , Gonadal- and sex chromosome-dependent sex differences in the circadian system. Endocrinology 154, 1501–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hara T., et al. , Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Komatsu M., et al. , Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131, 1149–1163 (2007). [DOI] [PubMed] [Google Scholar]
- 62.Zebisch K., Voigt V., Wabitsch M., Brandsch M., Protocol for effective differentiation of 3T3-L1 cells to adipocytes. Anal. Biochem. 425, 88–90 (2012). [DOI] [PubMed] [Google Scholar]
- 63.Howell L. A., Peterson A. K., Tomko R. J. Jr, Proteasome subunit α1 overexpression preferentially drives canonical proteasome biogenesis and enhances stress tolerance in yeast. Sci. Rep. 9, 12418 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nemec A. A., Peterson A. K., Warnock J. L., Reed R. G., Tomko R. J. Jr, An allosteric interaction network promotes conformation state-dependent eviction of the Nas6 assembly chaperone from nascent 26S proteasomes. Cell Rep. 26, 483–495.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.