Significance
Amyloid fibrils are generally related to neurodegenerative diseases, but they can also be part of normal protein function. Amyloid formation involves numerous steps and intermediate species. In this study, we investigated a fish protein, beta-parvalbumin, which readily forms amyloid on ligand removal. Using biophysical experiments, we provide evidence that the underlying mechanism of amyloid formation includes primary nucleation and elongation processes; we also reveal a key role for a disulfide-bridged dimer in the nucleation step. Little is known about intermolecular disulfides in amyloid formation, but covalent dimers and dimer-induced aggregation may be of clinical relevance, because oxidative stress, which can trigger covalent bond formation, is often a hallmark of human neurodegenerative diseases.
Keywords: amyloid fibrils, dimer formation, disulfide bond, dimer-induced aggregation, kinetic analysis
Abstract
Amyloid formation involves the conversion of soluble protein species to an aggregated state. Amyloid fibrils of β-parvalbumin, a protein abundant in fish, act as an allergen but also inhibit the in vitro assembly of the Parkinson protein α-synuclein. However, the intrinsic aggregation mechanism of β-parvalbumin has not yet been elucidated. We performed biophysical experiments in combination with mathematical modeling of aggregation kinetics and discovered that the aggregation of β-parvalbumin is initiated by the formation of dimers stabilized by disulfide bonds and then proceeds via primary nucleation and fibril elongation processes. Dimer formation is accelerated by H2O2 and hindered by reducing agents, resulting in faster and slower aggregation rates, respectively. Purified β-parvalbumin dimers readily assemble into amyloid fibrils with similar morphology as those formed when starting from monomer solutions. Furthermore, addition of preformed dimers accelerates the aggregation reaction of monomers. Aggregation of purified β-parvalbumin dimers follows the same kinetic mechanism as that of monomers, implying that the rate-limiting primary nucleus is larger than a dimer and/or involves structural conversion. Our findings demonstrate a folded protein system in which spontaneously formed intermolecular disulfide bonds initiate amyloid fibril formation by recruitment of monomers. This dimer-induced aggregation mechanism may be of relevance for human amyloid diseases in which oxidative stress is often an associated hallmark.
Amyloid fibrils are highly ordered protein assemblies characterized by β-strands arranged perpendicularly to the fibril axis forming cross-β sheets of indefinite length (1). They are generally associated with human disorders, such as Alzheimer’s disease, Parkinson’s disease and type 2 diabetes (2–4); however, it is clear that organisms also exploit the cross-β sheet structure for function. For example, amyloid fibrils are part of the human melanosome ultrastructure (5) and are common structural components of bacterial biofilms (6). Amyloid fibrils can form either from intrinsically disordered proteins, such as α-synuclein, which is linked to Parkinson’s disease, or by folded globular proteins, such as β2-microglobulin. Amyloid formation proceeds via at least two reaction steps: primary nucleation and elongation of fibrils (1, 7). The process is also typically catalyzed by secondary processes including secondary nucleation (1, 7) and, in some cases, fibril fragmentation (8). There are also cases of amyloid formation without rate-limiting nucleation; for example, on dissociation of tetrameric transthyretin to monomers, amyloid formation follows a downhill polymerization mechanism (9).
Disulfide bonds, which stabilize the folded structure of proteins, are present in several amyloidogenic proteins, but their role in amyloid formation is complex and remains a subject of some controversy (10–14). It has been demonstrated that the reduction of the single disulfide bond of the C-terminal domain of the human prion protein (PrP) accelerates amyloid formation in denaturing solution at neutral pH compared with the oxidized form (15). In contrast, at acidic pH and denaturing conditions, only the oxidized form of the C-terminal of PrP aggregates into amyloid fibrils (10). The reduction of the conserved disulfide bond in islet amyloid polypeptide, whose aggregation is a pathological hallmark of type 2 diabetes, accelerates amyloid formation in solution and in the presence of membranes, but cytotoxicity is not affected (16). Studies on β2-microglobulin have shown that at neutral pH, reduction of its single disulfide bond leads to aggregation but not amyloid formation (12, 13, 17). At acidic pH, however, reduced β2-microglobulin can either form (at high salt concentrations) or not form (in the presence of seeds) amyloid fibrils depending on the solution conditions (12, 13). In the case of lysozyme, the presence of intact disulfide bonds in the protein was observed to decrease the rate of amyloid fibril formation (11), but in another study, those bonds were found to be required for amyloid formation (18). It has been proposed that the disulfide bonds in lysozyme inhibit amyloid formation by stabilizing the folded state, but under structure-destabilizing conditions, a partially unfolded state can be populated, in which case they are essential for fibril formation (11). Moreover, in the case of insulin with several intramolecular disulfide bonds, these bonds have been shown to be important for aggregation (19).
Our understanding is more limited when considering intermolecular disulfide bonds in amyloid formation. Interprotein disulfide bonds have been found in amyloid fibrils of Cu/Zn superoxide dismutase, whose mutations are linked to familial amyotrophic lateral sclerosis, but interprotein disulfides are not required for fibril formation (20). In addition, a transthyretin mutant, the Tyr114Cys variant, which is implicated in familial amyloidotic polyneuropathy, was found to form intermolecular disulfide bonds that partly impaired amyloid fibril formation (21). However, the Cys10Ala/Tyr114Cys transthyretin variant formed dimers via disulfide bridges that then proceeded to amyloid protofibrils (22). In the case of the human tumor suppressor p16INK4A, with one cysteine residue, the addition of oxidants was found to induce the formation of disulfide-bridged dimers, which then aggregate into amyloid fibrils (23).
Here we investigated the role of intermolecular disulfide bonds in amyloid fibril formation of Gadus morhua β-parvalbumin (β-PV). β-PV is an α-helical protein with a single conserved cysteine residue (Cys19) (24) that was recently suggested to be necessary for amyloid fibril formation (25). β-PV is abundantly expressed in fish muscles (26) and has three EF-hand (helix-loop-helix motif) domains, of which two bind metals (Mg2+ and/or Ca2+), acting as a Ca2+ buffer in muscle cells (27). It has been reported that β-PV forms amyloid fibrils in vitro as a result of Ca2+ removal, which can be induced by protonation of Ca2+ binding residues at low pH or by using chelating agents such as EDTA (28). Antibodies against β-PV epitopes can be detected in the blood after eating fish (29), and studies have shown that antibodies from fish-allergic people bind 1,000-fold more strongly to β-PV in its amyloid conformation than to the monomeric protein (28, 30). Thus, it is speculated that β-PV forms amyloid fibrils in the gut due to the low pH inducing metal ion dissociation. The amyloid structure then protects β-PV against gastrointestinal degradation (28). We have recently reported that β-PV amyloid fibrils may, in addition to their allergenic properties, have beneficial effects as they are capable of inhibiting aggregation of the Parkinson protein α-synuclein in vitro through sequestration of α-synuclein polypeptides to the β-PV amyloid fibril surface (31). In this study, we provide evidence that β-PV, in its apo state, spontaneously forms disulfide-bridged dimers that stimulate the formation of primary nuclei, which are then capable of templating monomer as well as dimer additions to the ends of growing amyloid fibrils.
Results
Kinetic Analysis of Apo β-PV Amyloid Formation.
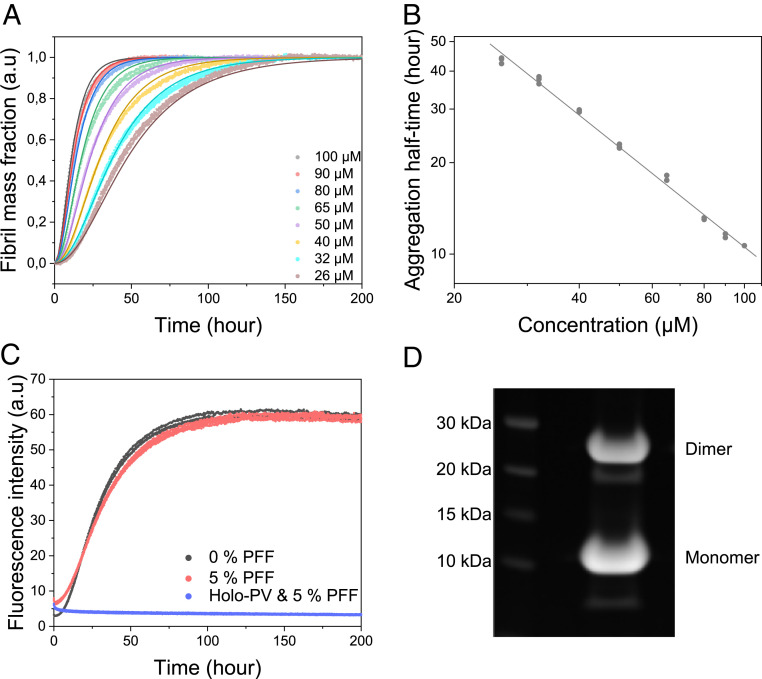
The aggregation of β-PV in the presence of EDTA was monitored at 37 °C using the thioflavin T (ThT) fluorescence assay (32, 33). The presence of only protein monomers at the starting point was ensured by performing size-exclusion chromatography (SEC) of the samples before each aggregation experiment. The EDTA was added at the start of the reaction as apo β-PV begins to aggregate immediately. By monitoring the amyloid formation of apo β-PV in a wide concentration range (26 to 100 µM), we found that the aggregation rate increases in a concentration-dependent manner (Fig. 1A). Notably, the aggregation curves are rather atypical for amyloid formation as they exhibit short lag phases. We plotted the ThT fluorescence half-time (i.e., the time at which 50% final fluorescence has been achieved) against the protein concentration in a double-logarithmic plot to investigate the concentration dependence of apo β-PV aggregation (Fig. 1B). A linear dependence was found with a slope (corresponding to the so-called scaling parameter, γ) of −1.1 ± 0.1 (Fig. 1B and SI Appendix, Table S1). The scaling parameter acts as a rough guide to the involved mechanism (7) and can be related to the reaction order of the nucleation process, n. For a reaction dominated by primary processes, γ corresponds to −nc/2 and a γ value of −1 and means that nc = 2; if secondary processes are also involved, then γ corresponds to −(n2 + 1)/2, and then a γ value of −1 means that n2 = 1 (7). Regardless of the mechanism, the fact that the log-log dependence of the half-time on the protein concentration is linear suggests that a single aggregation mechanism dominates across the entire protein concentration range (7).
Fig. 1.
(A) Aggregation kinetics of apo β-PV as a function of monomer concentration, as measured by ThT fluorescence at 37 °C under quiescent conditions (150 mM NaCl, 1 mM CaCl2, 5 mM EDTA, and 25 mM Tris⋅HCl pH 7.4). Solid lines indicate fits to data based on a primary nucleation model in the AmyloFit web interface (7) (SI Appendix, Fig. S2B, nonnormalized data). (B) Double-logarithmic plot of amyloid formation half-time vs. apo β-PV concentration extracted from the ThT data; the fitted line has a slope (γ) of −1.1. (C) Aggregation kinetics of 50 µM apo β-PV in the presence of 5% monomer equivalent preformed apo β-PV amyloid fibrils (PFFs) at the same conditions as in A. (D) SDS/PAGE of apo β-PV fibrils obtained after incubation of protein monomers at 50 µM for 300 h.
We globally fitted mathematical models of amyloid formation to the kinetic data of apo β-PV aggregation at the different concentrations using the web-based AmyloFit program (7). The analysis revealed that a mechanistic model involving only primary nucleation and elongation steps fitted the data best (Fig. 1A). The convoluted rate constant factor knk+ (7), describing the combined rate of primary nucleation (kn) and elongation (k+), for the global fit shown in Fig. 1A was found to be 2.7 × 106 ± 1.1 × 106 M−2h−2 (SI Appendix, Table S1). Reasonable global fits to the data were also obtained if secondary nucleation was added to the model, besides primary nucleation and elongation (SI Appendix, Fig. S1A), but including a fragmentation step resulted in worse fits (SI Appendix, Fig. S1B).
To assess for the presence of secondary nucleation processes, we performed seeded aggregation experiments in which we added preformed full-length apo β-PV fibrils, up to 5% monomer equivalents, to protein monomers. In an amyloid formation reaction with strong influence from secondary nucleation processes, the addition of small amounts of preformed fibrils (not sonicated) should drastically increase the aggregation rate (34). From our results, however, it is evident that the addition of preformed apo β-PV fibrils (up to 5%) has no effect on apo β-PV amyloid formation kinetics (Fig. 1C). Thus, in accordance with the mathematical modeling, it appears that primary processes (nucleation-elongation) dominate apo β-PV aggregation. However, for reactions with only primary processes, there should also be some acceleration of aggregation on the addition of amyloid seeds, as the presence of sufficient amount of free fibril ends should allow for rapid elongation without nucleation. Therefore, we performed seeding experiments with higher concentrations of amyloid fibril seeds (that had also been sonicated to create more fibril ends). Under these conditions, we observed clear seeding effects such that the lag phase was abolished (SI Appendix, Fig. S2A).
As described previously, Ca2+-bound β-PV (holo β-PV) does not form amyloid fibrils (28, 31). To test whether holo β-PV could be forced to form amyloid via seeding, we added preformed apo β-PV fibrils to holo β-PV, but found that they did not trigger amyloid formation of holo β-PV (Fig. 1C). In addition, as β-PV can bind both Ca2+ and Mg2+, we also tested whether Mg2+-bound β-PV would aggregate into amyloid fibrils, but the data showed that the Mg2+-bound protein also does not aggregate (SI Appendix, Fig. S3). Thus, only the apo-state of the protein forms amyloid fibrils. Surprisingly, when the apo β-PV amyloid fibrils were analyzed by nonreducing SDS/PAGE, we detected ∼40% covalent apo β-PV dimers (Fig. 1D).
Oxidizing and Reducing Conditions Influence Apo β-PV Amyloid Formation.
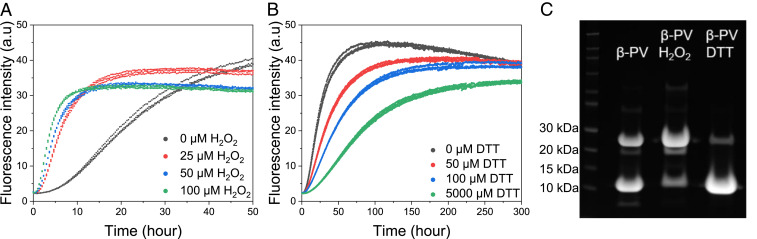
The dimers detected in samples of apo β-PV amyloid fibrils likely result from disulfide-bond formation involving β-PV’s cysteine, Cys19. Therefore, the influence of redox agents on the aggregation kinetics of apo β-PV was studied by adding H2O2 (Fig. 2A) or DTT (Fig. 2B). We found that oxidizing conditions strongly accelerate apo β-PV amyloid formation in a H2O2-concentration dependent manner, whereas reducing conditions (using DTT) delay apo β-PV amyloid formation. Densitometric analysis of SDS/PAGE data for amyloid samples show that the fraction of dimers increased from 40% under normal conditions to 77% of total protein in the amyloid fibrils under oxidizing conditions, whereas dimers corresponded to only 1% of total amyloid protein under reducing conditions (Fig. 2C). Thus, the β-PV dimers appear to be linked by disulfide bonds, and the presence of disulfide-bridged dimers clearly accelerate amyloid formation.
Fig. 2.
(A and B) ThT fluorescence curves of 50 µM apo β-PV at 37 °C under quiescent conditions in 150 mM NaCl, 1 mM CaCl2, 5 mM EDTA, 25 mM Tris⋅HCl pH 7.4, and 0 to 100 µM H2O2 (A) or 0 to 5 mM DTT (B). (C) SDS/PAGE analysis of apo β-PV amyloid fibrils from ThT kinetic experiments with or without either 100 µM H2O2 or 100 µM DTT.
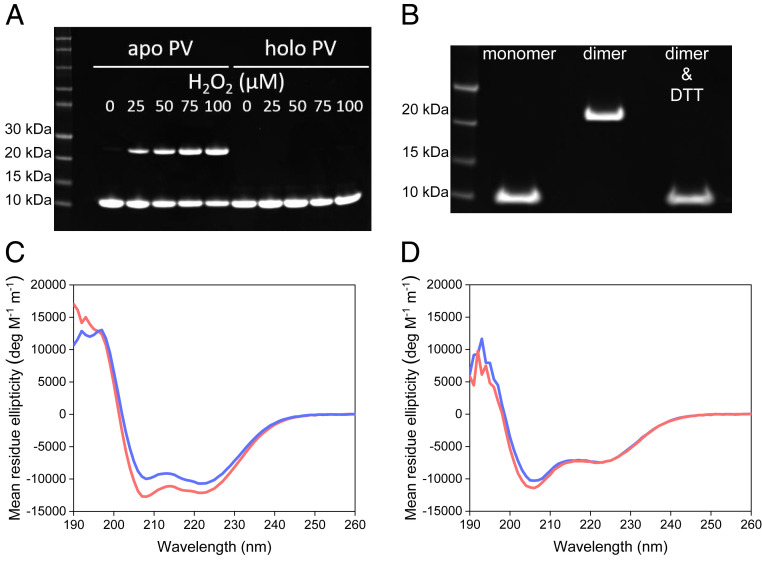
To obtain pure dimers for further biophysical analysis, monomeric apo β-PV was incubated with increasing amounts of H2O2 at 4 °C. Increasing amounts of dimers were found in an H2O2-concentration dependent manner according to SDS/PAGE (Fig. 3A). Notably, H2O2 did not induce dimerization of Ca2+- or Mg2+-bound β-PV (Fig. 3A, and SI Appendix, Fig. S4A). Dimers were purified by SEC (using Ca2+ in the running buffer to avoid aggregation). When such purified dimers were treated with DTT, dimers resolved to monomers according to SDS/PAGE (Fig. 3B), indicating that the dimerization process is reversible and that the covalent bonds involved are disulfides.
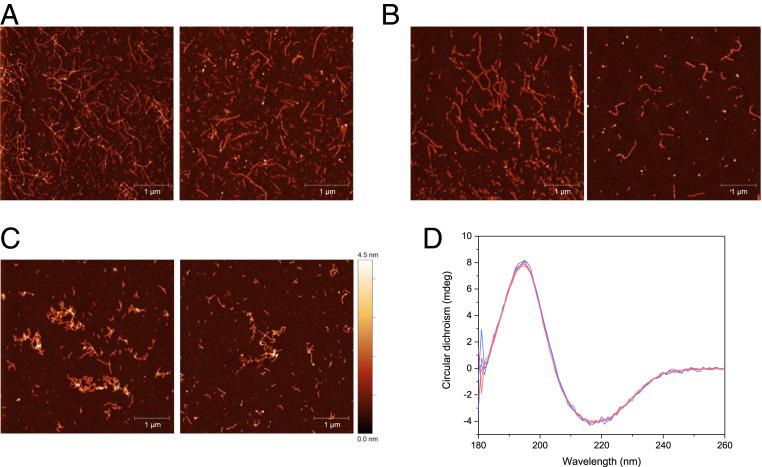
Fig. 3.
(A) SDS/PAGE of apo and Ca2+ β-PV at a concentration of 50 µM exposed to 0 to 100 µM H2O2 in 25 mM Tris⋅HCl, 5 mM EDTA (for apo), and 1 mM CaCl2, pH 8.22 at 4 °C. (B) SDS/PAGE of apo β-PV treated with H2O2 for 20 h (after SEC separation of monomer and dimer fractions with a 25 mM Tris⋅HCl and 1 mM CaCl2 running buffer) and after adding 5 mM DTT to the purified dimers. (C and D) Monomer (blue) and dimer (red) CD spectra for 15 µM monomer equivalent β-PV in 25 mM Tris⋅HCl and 1 mM CaCl2, without (C; holo) or with (D; apo) 5 mM EDTA, at 21 °C and pH 7.8.
Using far-UV circular dichroism (CD) spectroscopy, we found that the secondary structure of purified apo β-PV dimers is similar to that of apo β-PV monomers (Fig. 3D). In the presence of Ca2+, the dimers adopt the same folded structure as monomeric holo β-PV with minima at 208 and 222 nm, typical of predominantly α-helical protein conformations (Fig. 3C).
Apo β-PV Dimers Form Amyloid Fibrils and Accelerate the Aggregation of Apo-β-PV Monomers.
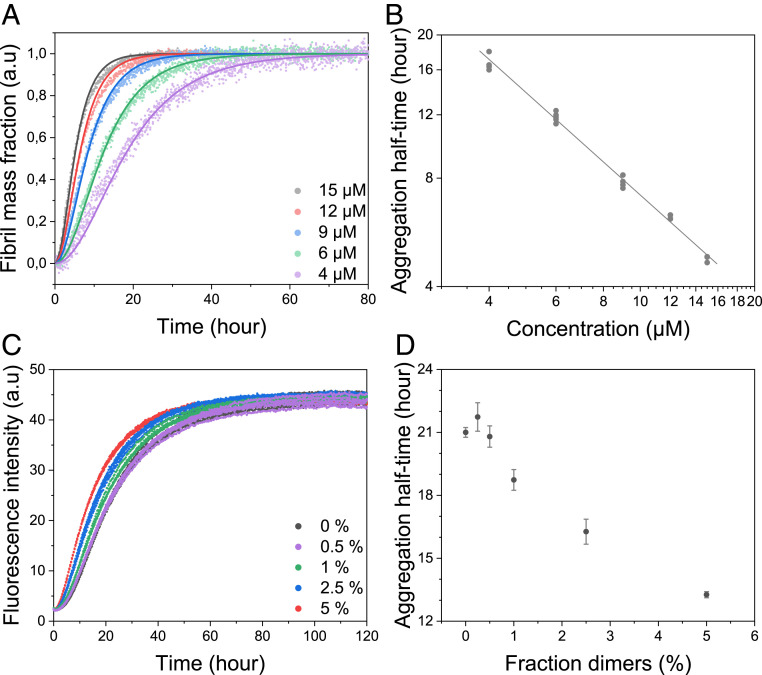
We next tested whether purified apo β-PV dimers could form amyloid fibrils in the absence of monomers by performing ThT-detected kinetic experiments with dimers at a concentration range between 4 and 15 µM (monomer equivalents). As observed for apo β-PV monomers, the ThT data showed faster aggregation reactions for higher concentrations of apo β-PV dimers (Fig. 4A). Plotting fluorescence half-time vs. protein concentration in a double-logarithmic plot again revealed a linear dependence, with a slope of −1.0 ± 0.1 (Fig. 4B and SI Appendix, Table S1). Using AmyloFit, the kinetic profiles for apo β-PV dimer aggregation were well fitted with the same nucleation-elongation model (Fig. 4A) as used to fit the kinetics of apo β-PV monomer amyloid formation; the addition of secondary processes to the model again resulted in worse fits. Concerning the apo β-PV dimer aggregation reaction, the convoluted rate constant factor knk+ was determined to be 5.7 × 107 ± 1.7 × 107 M−2h−2 (monomer equivalents) (SI Appendix, Table S1), which is 20-fold higher than the corresponding value found for the monomer reaction.
Fig. 4.
(A) Aggregation kinetics of dimeric apo β-PV as function of concentration (monomer equivalents given), as measured by ThT fluorescence at 37 °C under quiescent conditions (150 mM NaCl, 1 mM CaCl2, 5 mM EDTA, and 25 mM Tris⋅HCl pH 7.4). The solid lines represent fits to data based on a primary nucleation model in the AmyloFit web interface (7) (SI Appendix, Fig. S2C, nonnormalized data). (B) Double-logarithmic plot of half-time vs. apo β-PV dimer concentration from ThT aggregation data. (C) ThT fluorescence of apo β-PV aggregation in the presence of 0 to 5% (monomer equivalent) apo β-PV dimers (50 µM total apo β-PV) under the same conditions as described in A. (D) Half-time plot of the data reported in C vs. fraction of added dimers.
We also tested whether purified apo β-PV dimers could accelerate the amyloid fibril formation of apo β-PV monomers. A concentration-dependent acceleration of apo β-PV monomer aggregation was observed on the addition of 1 to 5% (monomer equivalents) dimers at the start of the reaction (50 μM total protein; Fig. 4 C and D). The addition of 5% preformed dimers (the highest concentration tested) shortened the half-time from 21.7 ± 1.5 h to 13.6 ± 1.7 h (Fig. 4 C and D). Notably, this dimer-catalyzed reaction is still slower than the aggregation of pure dimers (15 μM protein in monomer-equivalent dimers display an aggregation half-time of only approx. 5 h; Fig. 4B), implying that fibril elongation by addition of dimers is highly efficient.
Amyloid Fibrils Formed from Apo β-PV Monomers and Dimers Are Morphologically Similar.
Apo β-PV amyloid fibrils formed from either monomeric or dimeric starting material, as well as fibrils obtained from dimer-accelerated monomer aggregation experiments, were analyzed by atomic force microscopy (AFM) to characterize amyloid fibril morphology. We observed that all analyzed amyloid fibrils had similar morphology, including fiber heights, clustering preference, and curvilinear appearance (Fig. 5 A–C). Moreover, based on far-UV CD, the different amyloid fibrils exhibited similar content of β-sheet secondary structure (Fig. 5D).
Fig. 5.
(A–C) AFM images of fibrils obtained from apo β-PV monomers (A), monomers accelerated with dimers (B), or dimers only (C) . (D) CD spectra of fibrils obtained from dimers (red) and monomers (blue). The concentration of fibrils is unknown; the CD signal was matched to 222 nm.
To address whether the disulfide bonds within the amyloid fibrils are exposed or hidden in the core, we added DTT to preformed amyloids made from either monomeric or dimeric apo β-PV. After incubation with DTT, SDS/PAGE revealed no change in the amount of apo β-PV dimer amount compared with no DTT incubation (SI Appendix, Fig. S5A), suggesting that the disulfide bonds are not accessible to the solvent in the amyloid fibrils. Furthermore, the addition of DTT to preformed amyloid fibrils did not affect their morphology according to AFM (SI Appendix, Fig. S5B; data for amyloid fibrils formed from dimers).
Discussion
Although β-PV amyloid formation on Ca2+ removal remains poorly understood, a recent study suggested that the conserved cysteine residue (Cys19) is critical for amyloid fibril formation (25). On exchanging Cys19 for a serine residue, the resulting variant aggregated more slowly than wild-type apo β-PV and formed rod-like aggregates instead of the protofibrillar structures that are typical for wild-type β-PV (25). Therefore, we carried out a comprehensive study of apo β-PV aggregation at different concentrations and assessed the effect of reducing and oxidizing conditions, as well as the influence of seed fibrils on amyloid formation. Our results show that aggregation of apo β-PV into amyloid fibrils at 37 °C under nonreducing conditions involves the formation of disulfide-bridged dimers. The process is accelerated in the presence of oxidants and delayed by reducing agents in a concentration-dependent manner. Moreover, the addition of dimers to monomeric apo β-PV increases the protein aggregation rate, suggesting that the dimers induce nuclei formation. The dimers (when isolated and purified; before aggregation) were found to be folded and to bind Ca2+ like the monomers. Pure dimers, in the absence of Ca2+, aggregated faster than the monomers but formed amyloid fibrils with similar morphology and β-sheet secondary structure as those formed from monomers.
Global data analysis of the concentration dependence of the apo β-PV aggregation kinetic curves revealed that the data were best fitted to a simple nucleation-elongation model. The failure of attempts to seed β-PV aggregation with preformed full-length fibrils (up to 5%) supports an aggregation mechanism without secondary processes. The aggregation data for both monomer and dimer β-PV starting material fitted to the same aggregation mechanism, demonstrating that the dimers are not competent nuclei. Instead, the dimers need to assemble, partially unfold, or structurally convert in the rate-limiting nucleation step before rapid fibril elongation occurs. Notably, the kinetic models in the AmyloFit software do not include reaction steps preceding the formation of an elongation-competent primary nucleus; thus, we cannot assess the role of dimers in the early steps via global kinetic fitting.
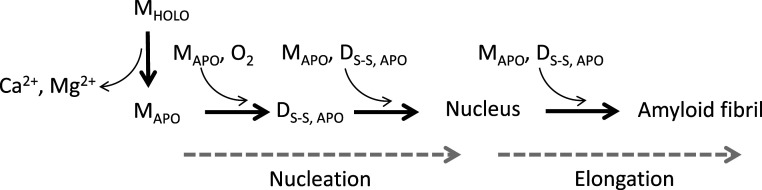
The observed γ value of −1, indicating a linear dependence between the logarithms of protein concentration and half-times, supports a simple nucleation-elongation mechanism but is also expected for a downhill polymerization mechanism. A downhill polymerization mechanism was demonstrated for amyloid formation of transthyretin (9) which, like β-PV, is a folded protein. However, several observations exclude this mechanism as a likely scenario for apo β-PV aggregation. First, apo β-PV amyloid formation exhibits a lag phase, which should not be the case for downhill polymerization. Second, in contrast to what is expected for downhill polymerization, the apo β-PV aggregation reaction can be seeded by the addition of preformed disulfide-bridged dimers and the addition of high concentrations of amyloid fibril seeds. Third, the apo β-PV kinetics experimental data are well fitted to the nucleation-elongation model when analyzed by the AmyloFit software. Fig. 6 shows a proposed mechanism, based on our data, for apo β-PV amyloid formation that involves nucleation and elongation steps, in which the first step is catalyzed by disulfide-bridged dimers. Importantly, the involvement of disulfide-bridged dimers accelerates amyloid formation, but it remains unclear whether these species are compulsory intermediates.
Fig. 6.
Proposed dominant mechanism of fish apo β-PV amyloid fibril formation in the absence of reducing agents and in oxidizing conditions. Aggregation is triggered by metal ion (e.g., Ca2+) release from the holo protein (MHOLO). The resulting apo monomers (MAPO) may associate to form dimers (DAPO), via disulfide bond formation between exposed cysteines (through O2 mediated oxidation; DS-S). These dimers promote nuclei formation that then proceed to amyloid fibrils via elongation by either monomeric (MAPO) or dimeric (DS-S, APO) apo β-PV additions. Here disulfide bridged dimers are shown as obligatory intermediates because our data point toward this conclusion; however, we cannot exclude that amyloid fibrils may also form via pathways that do not include disulfide-bridged dimers.
On metal ion release, β-PV can spontaneously form intermolecular disulfide bonds, resulting in apo β-PV dimers, which then catalyze the nucleation step in amyloid fibril formation. The role of intermolecular disulfide bonds in amyloid fibril formation has been investigated to some degree for both natural and engineered cysteines in proteins (17, 18, 22, 23, 35). In close similarity to the results presented here for β-PV, the human tumor suppressor p16INK4A was shown to form amyloid fibrils on disulfide-bridged dimerization when exposed to excess oxidizers under agitation (23). In contrast to the β-PV reaction, which does not require oxidizing agents nor agitation and involves a mixture of dimers and monomers, only dimeric p16INK4A could form fibrils. In addition, disulfide-bridged dimers of the transthyretin Cys10Ala/Tyr114Cys mutant were found to be compatible with aggregation toward amyloid protofibrils. However, the formation of disulfide-bridged dimers in the case of the transthyretin Tyr114Cys variant prevented fibril formation and instead resulted in amorphous precipitates, indicating that the latter dimers could not adopt amyloidogenic conformations (21).
Synthetic disulfide-linked dimers of amyloid-β (the amyloidogenic peptide linked to Alzheimer’s disease), generated by introducing a cysteine in place of serine 26, were observed to aggregate more rapidly than wild-type and mutated monomers, but the dimers formed toxic protofibrils that did not convert into typical amyloid fibrils (36). Synthetic disulfide-bridged dimers of ⍺-synuclein were also engineered by introducing a cysteine at different positions in the sequence. Depending on whether the cysteine residue was in the C terminus or the N terminus, dimer formation resulted in either increased or reduced aggregation rates compared with the monomer (37), suggesting that the orientation of monomers in the dimers is important in the amyloid formation process of α-synuclein.
Disulfide bonds between two cysteine sulfurs can form spontaneously in vitro in the presence of dissolved O2 (38, 39). Our results show that dimer formation of β-PV readily occurs for the apo protein, but not when the protein is loaded with Ca2+ or Mg2+. The differential behavior of apo and holo β-PV correlates with high-resolution structural data of the mammalian homolog (also known as oncomodulin), showing that the cysteine is buried in the holo form of β-PV but solvent-exposed in the apo form of β-PV (SI Appendix, Fig. S6) (24, 40, 41). In accordance with this, we found using Ellman’s assay that the cysteine in monomeric apo β-PV readily reacts with Ellman’s reagent, indicating high solvent accessibility, whereas the cysteine residue is more protected in monomeric Ca2+- and Mg2+-loaded β-PV proteins (SI Appendix, Fig. S4B).
As β-PV cycles between Ca2+- and Mg2+-bound forms during fish muscle contractions (27), there should never be true “apo” β-PV, and thus no amyloid formation, under normal conditions in vivo. Only when fish β-PV is placed in strongly destabilizing environments, such as in the human gut, can dissociation of metal ions result in disulfide bond formation and assembly into amyloid fibrils. In our experiments, EDTA addition was used to trigger the reaction by removing all metal ions from β-PV. To link to biology, we tested whether apo β-PV amyloid fibrils formed under gut pH conditions (i.e., low pH) contained disulfide-bridged dimers. Although disulfide bonds are disfavored at low pH, we detected a fraction of dimers in amyloid formed at pH 2.3 (SI Appendix, Fig. S7). The proteins responsible for redox regulation of muscle adaptation and oxidant-mediated muscle fatigue are unknown, but high levels of reactive oxygen species (ROS) have been shown to result in contractile dysfunction and muscle fatigue (42). Therefore, in fish muscles, the normal function of the conserved cysteine residue in β-PV may be that of a redox sensor, reacting with ROS to form sulfenic or sulfinic acid (43). Although less reactive in Ellman’s assay than the apo protein, there is slow reactivity (and thus exposure to solvent) of the cysteine in Mg2+- and Ca2+-loaded β-PV forms (SI Appendix, Fig. S4B).
Taken together, our findings demonstrate a protein system (folded, globular) in which intermolecular disulfide-bond formation primes amyloid formation. In amyloid formation of the apo form of β-PV, disulfide-bridged (folded) dimers accelerate the formation of primary nuclei, which can then be readily elongated with monomers and dimers into amyloid fibrils in a process lacking secondary processes. Although we studied a fish protein, our findings have direct implications for possible scenarios in human neurodegenerative diseases in which oxidative stress is often a signature element (44). Under such conditions, various oxidation reactions (e.g., disulfide, dityrosine, nitration) may create covalent dimers of human amyloidogenic proteins that in turn affect their aggregation potential.
Materials and Methods
Protein Expression and Purification.
The G. morhua β-parvalbumin (A51874, Gad m1) gene in a pET15b vector (with a His-tag) was transformed into BL21 (DE3) competent cells and grown in LB medium containing 100 mg/L carbenicillin at 37 °C until an OD600 of ∼0.5. Protein expression was induced by 1 mM isopropyl β-d-1-thiogalactopyranoside and overnight incubation at 26 °C. The cells were centrifuged at 4,100 × g for 20 min, after which the supernatant was decanted and the pellet resuspended in 10 mM Tris⋅HCl, 1 mM CaCl2, and protease inhibitor mixture (05892791001; Roche), pH 7.8. The resuspended pellet was then sonicated on ice for 5 min (30 s on, 30 s off) at 20% amplitude, followed by centrifugation at 15,000 × g for 30 min. The supernatant was recovered, mixed 1:1 with 25 mM Tris⋅HCl, 1 mM CaCl2, 0.2 M NaCl, and 80 mM imidazole, and cleared by 30 min of centrifugation at 15,000 × g.
The supernatant was then filtered (Nalgene rapid-flow filter, 0.2-µm aPES membrane; Thermo Fisher Scientific) and loaded on an equilibrated (20 mM Tris⋅HCl, 0.1 mM NaCl, and 40 mM imidazole) HisPrep FF 16/10 column (GE Healthcare), followed by a gradient elution in an Äkta purifier 10 (GE Healthcare) with increasing concentrations of 20 mM Tris⋅HCl, 0.1 M NaCl, and 0.5 M imidazole. The protein was then collected and concentrated, followed by loading onto a HiLoad 16/600 Superdex 75 pg (GE Healthcare) equilibrated with 25 mM Tris⋅HCl and 1 mM CaCl2. The eluate was flash-frozen in aliquots and stored at −80 °C. Just before the experiments, the protein His-tag was cleaved with thrombin (35.5 NIH units/µM protein, 1 mM β-PV) for 4 to 5 h at room temperature and 800 rpm shaking, and then separated by an SEC column (Superdex 75 10/300; GE Healthcare) in 25 mM Tris⋅HCl with 1 mM CaCl2 to create Ca2+-loaded β-PV. For Mg2+-loaded β-PV experiments, 0.1 M MgCl2 and 20 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′ tetraacetic acid (EGTA) was added to the thrombin/PV solution just before SEC, equilibrated in 25 mM Tris⋅HCl and 2 mM MgCl2.
To create apo β-PV, 5 mM EDTA was added just before the aggregation experiments. The concentration of β-PV was determined by absorption using an extinction coefficient of 1,950 cm−1 M−1 at 257 nm. Dimers were prepared by incubating 100 µM β-PV in 25 mM Tris⋅HCl pH 8.2, 1 mM CaCl2, 5 mM EDTA, 7.7 mM NaN3, and 50 µM H2O2 for 20 h at 4 °C, followed by 0.2-µm filtration and the addition of 0.1 M CaCl2 before injection into a Superdex 75 10/300 column, equilibrated with 25 mM Tris⋅HCl and 1 mM CaCl2. Preformed amyloid fiber seeds for experiments were prepared by incubating 500 µM apo β-PV under the same conditions as for ThT aggregation assays but in an Eppendorf tube without ThT for 20 h. The resulting amyloid fibers were then used for experiments directly (full-length fibers), or sonicated on ice for 10 s (5 s on, 10 s off) at 20% amplitude (to create shorter fibrils and thus more amyloid fibril ends), followed by flash-freezing of aliquots in liquid nitrogen and storage at −80 °C when specified.
ThT Assay.
Aggregation assays of SEC-purified β-PV was performed in quiescent conditions at 37 °C in 25 mM Tris⋅HCl pH 7.4, 150 mM NaCl, 1 mM CaCl2 (or 2 mM MgCl2 when specified), 5 mM EDTA (for apo experiments), 7.7 mM NaN3, and 20 µM recrystallized ThT in 96-well plates, and fluorescence was measured with a plate reader (FLUOstar Optima or FLUOstar Omega; BMG Labtech). Excitation was at 440 nm, and fluorescence was recorded at 480 nm every 20 min.
Analysis of ThT Kinetic Curves.
The half-times of PV amyloid fibril formation were computed from ThT fluorescence kinetic data in the AmyloFit web interface (7). The microscopic rate constants for PV amyloid formation were determined using a global fitting strategy based on previously described integrated rate laws for amyloid fibril growth (45). Theoretical curves describing the generation of amyloid aggregates were globally fitted to experimental ThT data to provide estimates of the involved rate constants using the AmyloFit web interface. All fits were conducted using a 10-basin hop algorithm with errors and using normalized ThT kinetics data, as described previously (7).
Ellman’s Assay for Exposed Cysteines.
Here 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB; Merck) was dissolved in 99.9% ethanol at a concentration of 8 mg/mL. The reaction was performed at a concentration of 35 µM β-PV monomer samples in 25 mM Tris⋅HCl pH 7.8 and 180 µM DTNB, with either 2 mM CaCl2 with or without 5 mM EDTA or 2 mM MgCl2 with 100 µM EGTA. The samples were incubated at room temperature for 200 min and 412-nm absorption was measured at timed intervals. The spectrophotometer (Lambda Bio+, PerkinElmer) was blanked against the same working solution at each time point but without the protein. Reacted thiols were calculated using the extinction coefficient of 14,150 M−1 cm−1 at 412 nm.
Nonreducing SDS/PAGE.
SDS/PAGE was performed with precast gels (NuPAGE 4 to 12% bis-Tris gel; Invitrogen) in MES buffer (NuPAGE MES SDS Running Buffer; Invitrogen) at a constant 200 V for 40 min. Before β-PV fibrils were run on SDS/PAGE, they were first partially sedimented by centrifugation at 100,000 × g for 2 h, followed by resuspension into 15 µL of 25 mM Tris⋅HCl with 0.1 M CaCl2. In the case of fibrils exposed to DTT, the pellet was resuspended with 25 mM Tris⋅HCl to remove trace DTT and then sedimented again by centrifugation at 100,000 × g for 2 h before proceeding. The resuspended pellet was then further diluted with 200 µL of 1,1,1,3,3,3-hexafluoro-2-propanol (Merck), followed by complete evaporation of the liquid by N2 gas. Finally, 15 µL of Milli-Q water was used to solubilize the dried material, after which 4x LDS buffer (NuPAGE LDS Sample Buffer 4x, Invitrogen) was added before running the gel. The gel was then stained with a Coomassie solution and imaged with 715/30 far-red epi settings (Chemidoc MP; Bio-Rad). Analysis of the gels was performed with Image Lab Software 6.1 (Bio-Rad).
AFM.
PV fibril samples were diluted with Milli-Q water (10–20 times) and deposited on freshly cleaved mica. After 15 min, the mica was rinsed with Milli-Q water and dried under a gentle nitrogen stream. AFM images were recorded in intermittent contact mode in air using an NTEGRA Prima setup (NT-MDT) and single crystal silicon cantilever (NSG01; TipsNano) with a force constant of ∼5.1 N/m at a resonance frequency of ∼180 kHz. Images were analyzed using Gwyddion software (46).
CD Spectroscopy.
Far-UV CD spectra (190 to 260 nm) of PV monomers, dimers, and fibrils were collected at room temperature (21 ± 1 °C) using a Chirascan CD spectropolarimeter (Applied Photophysics) and a quartz cuvette with a path length of 1 mm. Five scans were collected and averaged for each sample at a time-per-point of 0.8 s, with a bandwidth of 1 nm and a step size of 1 nm. The signal of the buffer was subtracted, and the resulting intensities in millidegrees of the PV monomer and dimer samples were converted to mean residue molar ellipticity (degrees M−1 m−1).
Supplementary Material
Acknowledgments
This work was supported by grants to P.W.-S. from the Knut and Alice Wallenberg foundation and the Swedish Research Council. E.K.E acknowledges support from Chalmers Excellence Initiative Nano and the Swedish Research Council.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015503117/-/DCSupplemental.
Data Availability.
All study data are included in the main text and SI Appendix.
References
- 1.Chiti F., Dobson C. M., “Protein misfolding, amyloid formation, and human disease: A summary of progress over the last decade” in Annual Review of Biochemistry, Kornberg R. D., Ed. (Annual Reviews, 2017), vol. 86, pp. 27–68. [DOI] [PubMed] [Google Scholar]
- 2.Jarrett J. T., Berger E. P., Lansbury P. T. Jr, The carboxy terminus of the beta amyloid protein is critical for the seeding of amyloid formation: Implications for the pathogenesis of Alzheimer’s disease. Biochemistry 32, 4693–4697 (1993). [DOI] [PubMed] [Google Scholar]
- 3.Wakabayashi K., Matsumoto K., Takayama K., Yoshimoto M., Takahashi H., NACP, a presynaptic protein, immunoreactivity in Lewy bodies in Parkinson’s disease. Neurosci. Lett. 239, 45–48 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Cooper G. J. S., et al. , Purification and characterization of a peptide from amyloid-rich pancreases of type 2 diabetic patients. Proc. Natl. Acad. Sci. U.S.A. 84, 8628–8632 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fowler D. M., et al. , Functional amyloid formation within mammalian tissue. PLoS Biol. 4, e6 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanco L. P., Evans M. L., Smith D. R., Badtke M. P., Chapman M. R., Diversity, biogenesis and function of microbial amyloids. Trends Microbiol. 20, 66–73 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meisl G., et al. , Molecular mechanisms of protein aggregation from global fitting of kinetic models. Nat. Protoc. 11, 252–272 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Bolognesi B., et al. , Single point mutations induce a switch in the molecular mechanism of the aggregation of the Alzheimer’s disease associated Aβ42 peptide. ACS Chem. Biol. 9, 378–382 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Hurshman A. R., White J. T., Powers E. T., Kelly J. W., Transthyretin aggregation under partially denaturing conditions is a downhill polymerization. Biochemistry 43, 7365–7381 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Gerum C., Silvers R., Wirmer-Bartoschek J., Schwalbe H., Unfolded-state structure and dynamics influence the fibril formation of human prion protein. Angew. Chem. Int. Ed. Engl. 48, 9452–9456 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Mossuto M. F., et al. , Disulfide bonds reduce the toxicity of the amyloid fibrils formed by an extracellular protein. Angew. Chem. Int. Ed. Engl. 50, 7048–7051 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohhashi Y., et al. , The intrachain disulfide bond of beta(2)-microglobulin is not essential for the immunoglobulin fold at neutral pH, but is essential for amyloid fibril formation at acidic pH. J. Biochem. 131, 45–52 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Smith D. P., Radford S. E., Role of the single disulphide bond of beta(2)-microglobulin in amyloidosis in vitro. Protein Sci. 10, 1775–1784 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mossuto M. F., Disulfide bonding in neurodegenerative misfolding diseases. Int. J. Cell Biol. 2013, 318319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honda R., Role of the disulfide bond in prion protein amyloid formation: A thermodynamic and kinetic analysis. Biophys. J. 114, 885–892 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ridgway Z., et al. , Analysis of the role of the conserved disulfide in amyloid formation by human islet amyloid polypeptide in homogeneous and heterogeneous environments. Biochemistry 57, 3065–3074 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamamoto K., et al. , Thiol compounds inhibit the formation of amyloid fibrils by beta 2-microglobulin at neutral pH. J. Mol. Biol. 376, 258–268 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Wang S. S. S., Liu K. N., Lu Y. C., Amyloid fibrillation of hen egg-white lysozyme is inhibited by TCEP. Biochem. Biophys. Res. Commun. 381, 639–642 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Ivanova M. I., Sievers S. A., Sawaya M. R., Wall J. S., Eisenberg D., Molecular basis for insulin fibril assembly. Proc. Natl. Acad. Sci. U.S.A. 106, 18990–18995 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chattopadhyay M., et al. , Initiation and elongation in fibrillation of ALS-linked superoxide dismutase. Proc. Natl. Acad. Sci. U.S.A. 105, 18663–18668 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eneqvist T., et al. , Disulfide-bond formation in the transthyretin mutant Y114C prevents amyloid fibril formation in vivo and in vitro. Biochemistry 41, 13143–13151 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Karlsson A., Olofsson A., Eneqvist T., Sauer-Eriksson A. E., Cys114-linked dimers of transthyretin are compatible with amyloid formation. Biochemistry 44, 13063–13070 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Göbl C., et al. , Cysteine oxidation triggers amyloid fibril formation of the tumor suppressor p16INK4A. Redox Biol. 28, 101316 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moraes A. H., et al. , Solution and high-pressure NMR studies of the structure, dynamics, and stability of the cross-reactive allergenic cod parvalbumin Gad m 1. Proteins 82, 3032–3042 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Castellanos M., Torres-Pardo A., Rodríguez-Pérez R., Gasset M., Amyloid assembly endows Gad m 1 with biomineralization properties. Biomolecules 8, 13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuehn A., Scheuermann T., Hilger C., Hentges F., Important variations in parvalbumin content in common fish species: A factor possibly contributing to variable allergenicity. Int. Arch. Allergy Immunol. 153, 359–366 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Rall J. A., Role of parvalbumin in skeletal muscle relaxation. News Physiol. Sci. 11, 249–255 (1996). [Google Scholar]
- 28.Martínez J., et al. , Fish β-parvalbumin acquires allergenic properties by amyloid assembly. Swiss Med. Wkly. 145, w14128 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Scheers N., Lindqvist H., Langkilde A. M., Undeland I., Sandberg A. S., Vitamin B12 as a potential compliance marker for fish intake. Eur. J. Nutr. 53, 1327–1333 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Sánchez R., et al. , The amyloid fold of Gad m 1 epitopes governs IgE binding. Sci. Rep. 6, 32801 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Werner T., Kumar R., Horvath I., Scheers N., Wittung-Stafshede P., Abundant fish protein inhibits α-synuclein amyloid formation. Sci. Rep. 8, 5465 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naiki H., Higuchi K., Hosokawa M., Takeda T., Fluorometric determination of amyloid fibrils in vitro using the fluorescent dye, thioflavin T1. Anal. Biochem. 177, 244–249 (1989). [DOI] [PubMed] [Google Scholar]
- 33.Xue C., Lin T. Y. W., Chang D., Guo Z., Thioflavin T as an amyloid dye: Fibril quantification, optimal concentration and effect on aggregation. R. Soc. Open Sci. 4, 160696 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Linse S., Monomer-dependent secondary nucleation in amyloid formation. Biophys. Rev. 9, 329–338 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y., Yan J., Zhang X., Huang K., Disulfide bonds in amyloidogenesis diseases related proteins. Proteins 81, 1862–1873 (2013). [DOI] [PubMed] [Google Scholar]
- 36.O’Nuallain B., et al. , Amyloid beta-protein dimers rapidly form stable synaptotoxic protofibrils. J. Neurosci. 30, 14411–14419 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pivato M., et al. , Covalent α-synuclein dimers: Chemico-physical and aggregation properties. PLoS One 7, e50027 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cavallini D., De Marco C., Dupré S., Luminol chemiluminescence studies of the oxidation of cysteine and other thiols to disulfides. Arch. Biochem. Biophys. 124, 18–26 (1968). [DOI] [PubMed] [Google Scholar]
- 39.Rehder D. S., Borges C. R., Cysteine sulfenic acid as an intermediate in disulfide bond formation and nonenzymatic protein folding. Biochemistry 49, 7748–7755 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Babini E., et al. , Solution structure of human beta-parvalbumin and structural comparison with its paralog alpha-parvalbumin and with their rat orthologs. Biochemistry 43, 16076–16085 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Henzl M. T., Tanner J. J., Solution structure of Ca2+-free rat beta-parvalbumin (oncomodulin). Protein Sci. 16, 1914–1926 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Powers S. K., Ji L. L., Kavazis A. N., Jackson M. J., Reactive oxygen species: Impact on skeletal muscle. Compr. Physiol. 1, 941–969 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paulsen C. E., Carroll K. S., Cysteine-mediated redox signaling: Chemistry, biology, and tools for discovery. Chem. Rev. 113, 4633–4679 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim G. H., Kim J. E., Rhie S. J., Yoon S., The role of oxidative stress in neurodegenerative diseases. Exp. Neurobiol. 24, 325–340 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knowles T. P. J., et al. , An analytical solution to the kinetics of breakable filament assembly. Science 326, 1533–1537 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Necas D., Klapetek P., Gwyddion: An open-source software for SPM data analysis. Cent. Eur. J. Phys. 10, 181–188 (2012). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and SI Appendix.